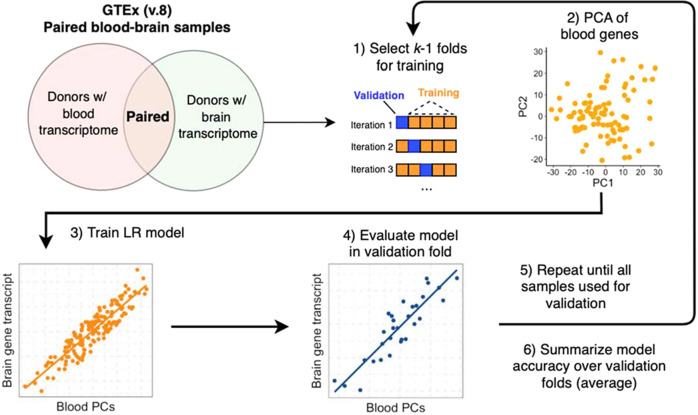
Fig. 1. Schematic illustrating the process for training BrainGENIE using paired blood–brain transcriptome data from the GTEx dataset.
BrainGENIE is trained using paired blood and brain transcriptome profiles collected by GTEx (v8) from adult donors. A single 5-fold cross-validation is performed to estimate the predictive performance of BrainGENIE separately for each brain region. BrainGENIE uses top principal components of transcriptome-wide blood-based gene expression as features to predict brain-regional gene expression levels. The metric used for prediction performance was the coefficinent of determination (R2) to measure how well predicted per-gene expression levels captured observed gene expression levels in the validation folds. Model performance was summarized over the 5 validation folds to obtain an estimate of prediction performance for BrainGENIE.