Abstract
Tumour cells have exquisite flexibility in reprogramming their metabolism in order to support tumour initiation, progression, metastasis and resistance to therapies. These reprogrammed activities include a complete rewiring of the bioenergetic, biosynthetic and redox status to sustain the increased energetic demand of the cells. Over the last decades, the cancer metabolism field has seen an explosion of new biochemical technologies giving more tools than ever before to navigate this complexity. Within a cell or a tissue, the metabolites constitute the direct signature of the molecular phenotype and thus their profiling has concrete clinical applications in oncology. Metabolomics and fluxomics, are key technological approaches that mainly revolutionized the field enabling researchers to have both a qualitative and mechanistic model of the biochemical activities in cancer. Furthermore, the upgrade from bulk to single-cell analysis technologies provided unprecedented opportunity to investigate cancer biology at cellular resolution allowing an in depth quantitative analysis of complex and heterogenous diseases. More recently, the advent of functional genomic screening allowed the identification of molecular pathways, cellular processes, biomarkers and novel therapeutic targets that in concert with other technologies allow patient stratification and identification of new treatment regimens. This review is intended to be a guide for researchers to cancer metabolism, highlighting current and emerging technologies, emphasizing advantages, disadvantages and applications with the potential of leading the development of innovative anti-cancer therapies.
Subject terms: Biochemistry, Cancer metabolism
Introduction
Tumour cells rewire their metabolism in order to drive and promote the growth and spreading of cancer cells.1 This metabolic rewiring causes a unique metabolic phenotype, whose features can be exploited for biomarkers discovery, disease prediction, cancer diagnosis, patient stratifications and personalized therapies.2–4 Metabolic alterations of malignant cells have long been a central focus in the field of cancer research and currently the deregulation of cellular metabolism is a recognized core hallmark of cancer in modern medicine5,6 (Fig. 1). The field of cancer metabolism put down roots in the 1920s with the pioneered studies of Otto H. Warburg who hypothesized, for the first time, that cancer, compared to a normal tissue, utilizes enormous amounts of glucose to generate lactic acid, even in normoxia or in the presence of non-hypoxic conditions. This process was named the “Warburg effect” and its molecular explanation completely prioritized the following years of cancer research.7–10 During the 1980s, the search for abnormal metabolic pathways that can be therapeutically exploited drastically accelerated, leading to the discovery of oncogenes and tumour suppressor genes that are the basis for alternative metabolic pathways in cancer. The uncontrolled cell proliferation, increased cell survival, cell differentiation, immune surveillance, aberrant angiogenesis, and many other metabolic events linked to cancer are strictly associated with the oncogenic activities of mutant KRAS, mutant p53, activation of mTOR signalling and of the transcription factor c-MYC, among the others.11–19 Human cancers are characterized by an extreme context-dependent plasticity and diversity; indeed, they develop multiple metabolic alterations, as part of a multistep process, to fulfil the bioenergetic, biosynthetic, and reduction–oxidation (redox) demands of malignant cells.20 The most common metabolic alterations include: (i) cell survival and anabolic growth during nutrient deprivation, which is sustained via the acquisition of nutrients from the extracellular space and their conversion into macromolecules through the core metabolic pathways, such as glycolysis, Pentose phosphate pathway (PPP) and Tricarboxylic acid cycle (TCA).21–24 The increased uptake of amino acids and glucose, with consequent release of lactate and protons to the tumour microenvironment, is further enhanced via more mechanical processes, such as phagocytosis, macro-pinocytosis and entosis;25–31 (ii) increased acquisition and utilization of nitrogen, building block for the de novo synthesis of a variety of biological compounds including nucleotides, amino acids, polyamines, hexosamines, Glutathione (GSH), porphyrins, ammonia, creatine, nitric oxide;32,33 (iii) extensive metabolic enzyme-mediated and metabolite-mediated modulation of gene expression including methylation, acetylation, phosphorylation, ubiquitination of histones and succinylation; (iv) balanced Reactive oxygen species (ROS) by fine-tuning redox systems: ROS cause DNA damage, contributing to the appearance of oncogenic mutations, and acting as pro-growth signals thus sustaining tumour initiation and progression as well as angiogenesis and metastasis. Therefore, a tumour generally depends more than a normal tissue upon the antioxidants, including PPP-derived Nicotinamide adenine dinucleotide phosphate (NADPH), GSH, thioredoxin (TXN), antioxidant enzymes [i.e., mitochondrial and cytosolic peroxiredoxins (PRXs), glutathione peroxidase (GPXs), catalase and Superoxide dismutase (SOD)] and their transcriptional master regulators, as the Nuclear factor erythroid 2-related factor 2 (NRF2)34–43; (v) accumulation of oncometabolites, such as succinate, fumarate, and 2-hydroxyglutarate (2HG), which have higher concentration in malignant cells as a consequence of respectively loss-of-function mutations within the genes encoding succinate dehydrogenase (SDH), Fumarate hydratase (FH) or gain-of-function mutations in the Isocitrate dehydrogenase 1 or 2 (IDH1 or IDH2) genes. The increased level of any of these oncometabolites has been associated with a pro-tumorigenic effect in certain tissues.44–48
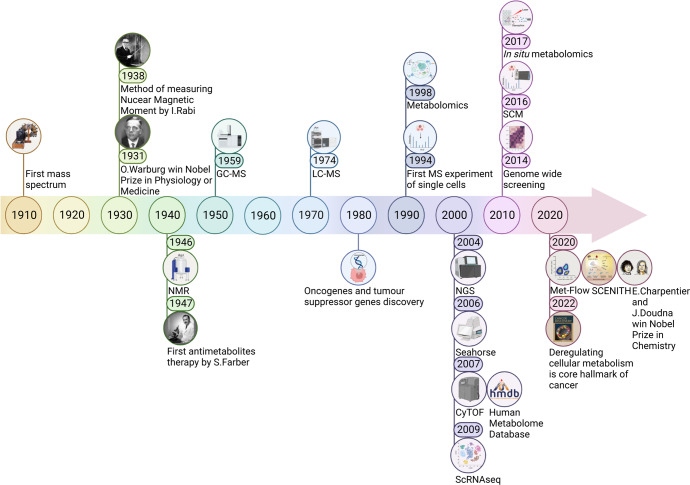
Fig. 1.
Timeline of the milestone events for cancer metabolism. The first mass spectrum of a molecule was measured by Joseph J. Thomson in 1910. In 1931 Otto H. Warburg won the Nobel Prize in Physiology or Medicine for characterizing the respiratory enzyme. In 1938 Isidor I. Rabi detected the Nuclear magnetic resonance (NMR) for the first time in a beam of lithium chloride thus developing the methodology and further expanded in 1946 by Felix Bloch and Edward M. Purcell for use on liquids and solids. Gas chromatography (GC)-MS was described in 1959 and Liquid chromatography (LC)-MS was introduced in 1974. The discovery of oncogenes and tumour suppressor genes dates back to the 1980s. In 1994 Tsutomu Nomizu and colleagues realized the first single cells MS experiment, while in 1998 Steven Oliver firstly introduced the concept of metabolomics. 2004 is the year of Next-generation sequencing (NGS), 2006 of the Seahorse real-time cell metabolic analyser, 2007 of the first prototype of Cytometry by Time-of-flight (CyTOF) and of the first version of The Human metabolome database (HMDB). In 2009 there was the development of Single-cell RNA sequencing (scRNAseq), in 2016 of Single-cell metabolomics (SCM) and in 2017 of the In situ metabolomics. The first genome-wide functional screening was performed in 2014 and in 2020 Emmanuelle Charpentier and Jennifer Doudna were awarded the Nobel Prize in Chemistry for discovering the CRISPR/Cas9 system. In 2020 the flow-cytometry-based technologies Met-flow and SCENITH have been proposed. In 2022 the deregulation of cellular metabolism was eventually recognized core hallmark of cancer by Douglas Hanahan. This figure was created with Biorender.com
The idea of targeting metabolism is not a new concept; the first antimetabolites therapy dates back to 1947 when Dr. Sidney Farber, the father of the modern chemotherapy, first discovered that aminopterin administration to children affected by Acute lymphoblastic leukaemia (ALL) may arrest tumour progression (Fig. 1). Aminopterin is a folate analogue that inhibits the de novo nucleotide biosynthesis by blocking the one-carbon transfer reaction and its clinical success led to the development of an entirely new class of drugs.49–55 The introduction of antimetabolites probably constitutes one of the biggest achievements in the field of cancer metabolism: Methotrexate (MTX, amethopterin) a synthetic and less toxic derivative of aminopterin, the uracil analogue 5-fluorouracil (5-FU) and the nucleoside analogues gemcitabine and cytarabine are all notable examples of widely used antimetabolites employed in a variety of cancer settings over the years.56–58 Despite these successes, until now only a few metabolic liabilities have been translated into effective therapies. Toxicity versus normal cells and the intrinsic plasticity of cancer, which allow cells to activate alternative pathways in response to metabolic drugs, make particularly challenging targeting metabolism in these cells thus highlighting the urgent need to further investigate cancer biology with the final goals of the identification of successful screening and personalized therapeutic strategies to improve patient care.
Despite the massive effort, the metabolic reprogramming of cancer cells remains far from being fully characterized because of its complexity and due to technical limitations. Currently, we recognize the multidisciplinary of cancer metabolism that sits at the interface between numerous technical-scientific disciplines and approaches, in particular biochemistry, immunology, genetics and microscopy. This knowledge brought tremendous technological advancement in metabolites characterization and quantification, functional analysis of metabolic activities, innovative single-cell and genetic approaches that have given researchers specific and potent tools to untangle the complexity of cancer metabolism. Selecting the best method or the combination of methods that is more appropriate for a given study depends on equipment availability, budget, and scientific hypothesis. The purpose of this review is to provide an overview of the available innovative methods to study cancer metabolism, discussing experimental limitations, future directions and clinical applications.
Metabolomics: a powerful tool in cancer research and treatment
Genomic and transcriptomic studies often fail to explain the complexity of a biological system, thus, leaving us with a huge gap in the map from genotype to phenotype. The approach that is closest to fill this gap is metabolomics, first introduced by Steven Oliver in a review article on yeast functional genomics published in 1998, and now widely recognized as a cornerstone to all of the systems biology4,59,60 (Fig. 1). Metabolomics is a high-throughput study of the metabolome which includes all the small molecules (50–1500 Da) with diverse physiochemical characteristics and dynamic range of abundance, commonly known as metabolites, within cells, biofluids, tissues or organisms.61 The identification of metabolites and their concentrations, unlike other omics measurements, directly represents the molecular phenotype. Metabolomics, more than any other methods, significantly revolutionized the field of cancer research; it is one of the most powerful omics techniques, mainly employed in cancer research to effectively detect metabolites whose level is affected by the neoplastic progression in a biological sample with a wide range of applications such as biomarker identification, drug discovery or development, clinical toxicology, nutritional studies, and quantitative phenotyping.62,63 As with any omics techniques, metabolomics is highly dependent on the availability and quality of public databases. In this regard, the biggest achievement in the field was reached in 2007 with the completion of the first draft of the human metabolome, the chemical equivalent of the human genome (Fig. 1). The Human metabolome database (HMDB) is a freely accessible electronic database (current version HMDB 5.0, https://hmdb.ca/) designed with the possibility to link data to other databases (KEGG, PubChem, MetaCyc, UniProt, and GenBank) thus combining chemical data, clinical information and molecular biology/biochemistry models.64–68 The database contains more than 220,000 metabolite entries including both water-soluble and lipid-soluble metabolites.
However, despite increasing technological progress, none of the current analytical platforms can completely measure the whole metabolome. Reaching an adequate metabolome coverage remains a major challenge, which can be achieved only through a combination of approaches.47,69–73 The most critical step in a metabolomics workflow is the fast quenching of all metabolic pathways and the isolation of metabolites in order to completely block all enzymes and chemical activities and generate a stable extract with metabolite ratios and concentrations reflecting the levels of endogenous metabolites from the original living cell at a chosen time.47,74 In addition, the collection and extraction of the samples have to maintain the original analyte concentration, increase the instrument productivity, and lower the matrix effect in the analysis. Sample collection and storage can depend on target metabolites and are the source of the greatest variation; therefore, harmonization of these procedures is mandatory for a correct analysis.75 High-resolution spectroscopic techniques include Mass Spectrometry (MS) and Nuclear Magnetic Resonance (NMR) spectroscopy with specific and complementary applications in the field that may require a prior separation step and different ion sources75 (Fig. 2a).
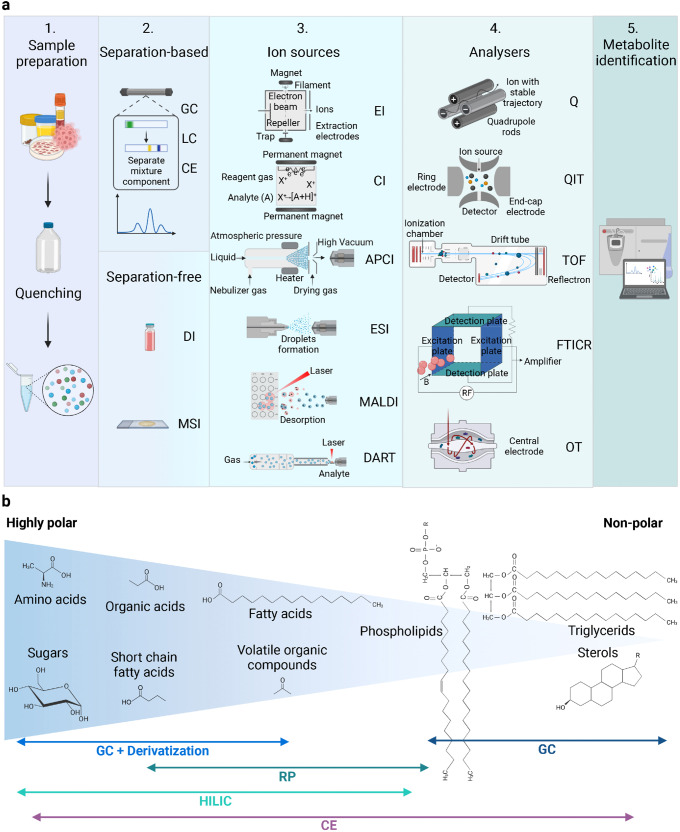
Fig. 2.
a Mass spectrometry (MS)-metabolomic workflow. 1. Samples preparation consists of metabolism quenching and metabolites extraction. 2. Metabolites may need a separation step with Gas chromatography (GC), Liquid chromatography (LC), Capillary electrophoresis (CE) or can be directly ionized in the Direct infusion (DI) and in the Mass spectrometry imaging (MSI). 3. Different ionization techniques can be employed: Electron impact ionization (EI), Chemical ionization (CI), Atmospheric pressure chemical ionization (APCI), Electrospray ionization (ESI), Matrix-assisted laser desorption ionization (MALDI) and Direct real-time analysis (DART). 4. Single (MS) or tandem (MS/MS) mass analysers can be alternatively employed to separate ions according to their m/z: Quadrupole (Q), Quadrupole ion trap (QIT), Time-of-flight analyser (TOF), Fourier transform ion cyclotron resonance (FTICR), Orbitrap (OT). 5. Data processing includes conversion of m/z values, detection, filtering, normalization and identification. b Schematic depicting the most suitable techniques to separate metabolites with distinct polarity. This figure was created with Biorender.com
Metabolomics approaches: untargeted and targeted strategies
Metabolome profiling is typically performed through two different strategies: targeted or untargeted analysis (Table 1). The goal of targeted (or validation-based) metabolomic approaches is the identification and quantification of a relatively low number (generally less than one hundred) of known analytes, while the untargeted (or discovery-based) metabolomics is used for a more comprehensive analysis and relative quantification of metabolites; in this case, the analysis must be further validated with an orthogonal approach.76 In targeted metabolomics, by using native and isotopically labeled internal standards (IS) it is possible to reduce the false positives, thus more accurately identifying and quantifying the metabolites. Furthermore, the use of IS improves the matrix-induced ionization effects, thus enhancing the sensitivity. However, a limitation of targeted approaches is the partial metabolome coverage, which increases the chance of a wrong interpretation of the metabolic process of interest.77 Oppositely, the analysis with an untargeted metabolomic approach does not need any pre-existing knowledge or hypothesis even though, sample preparation and analytical methods affect the subset of metabolites detected, thus they must be chosen wisely. In untargeted metabolomics studies, two different approaches for data acquisition can be used: Data-dependent acquisition (DDA) and Data independent acquisition (DIA) mode. In DDA the metabolites with the highest signal intensity in a full-scan MS1 spectrum are selected to generate fragmentation patterns. The DIA approach integrates full MS1 with MS/MS fragmentation for all precursor ions, thus producing an extremely complex spectrum of fragmentation where decoding the connection between precursor and end-products is very hard. To perform metabolite identification, it is necessary to search the experimental MS1 or MS/MS data into the metabolome database. To increase the metabolome coverage, several libraries must be simultaneously interrogated therefore requiring a massive bioinformatic effort to reduce redundant matching. However, this process is further challenged by the fact that the nomenclature of metabolites is not fully standardized and varies widely across databases.76
Table 1.
Features of untargeted and targeted metabolomics approaches
| Metabolomics | |
|---|---|
| Untargeted | Targeted |
| Discovery strategy (hypothesis generating) | Validation strategy (hypothesis driven) |
| Global and comprehensive analysis | Subset analysis |
| High number of metabolites studied | Small number of metabolites |
| Correlated to database/libraries | Correlated to reference standards |
| Qualitative identification | Identification of metabolites known a priori |
| Relative quantification | Absolute quantification |
| Challenging data interpretation | Easy data analysis and interpretation |
| Samples preparation requires a global metabolites extraction | Samples preparation is metabolite-specific |
| Biomarker discovery | Biomarker validation |
| Global/untargeted screening | Pathway mapping targeted metabolomic profiling |
Separation-based MS techniques for metabolites investigation
To analyse the myriad of metabolites, which constitutes a biological sample, a separation through chromatographic systems may be necessary.78 The separation is required to avoid ion suppression or over-loading of the MS system, which causes a general loss of sensitivity. The interaction between the metabolites and the chromatographic system depends on the physical-chemical proprieties of the metabolites, the stationary phase and it determines the order of elution in the chromatographic system. Usually, lipids, volatile organic metabolites and derivative compounds are analysed by Gas chromatography (GC)-MS, originally described in 1959, while the analysis of the polar molecules is performed through Liquid chromatography (LC)-MS, introduced for the first time in 1974 (Figs. 1, 2a, b).79
Gas chromatography (GC)
GC coupled with MS is an analytical technique applied in metabolomics optimal for the analysis of small molecular substances (<650 Daltons). GC is a separative tool to quantitively and qualitatively measure volatile and thermally stable compounds. As a mobile carrier it uses an inert gas, such as nitrogen, helium, or hydrogen, while the stationary phase can be a liquid with low vapour pressure that coats the inner walls of a capillary (Gas liquid chromatography, GLC) or a solid that exhibits interactions with analytes by physical adsorption (Gas solid chromatography, GSC). Usually, the first choice is a stationary phase with similar polarity to the analytes while increasing the column temperature over time facilitates the elution of metabolites. Since most primary metabolites with a pivotal role in cancer metabolism such as lactic acid, pyruvic acid, malic acid, glucose, and palmitic acid have high boiling points, a derivatization step is necessary with the aim of making them volatile enough to be analysed by GC-MS. Among the derivatization protocols, the more employed ones use trimethylsilylation or thereof-like variants, which detach acidic protons from hydroxyl-, carboxyl-, amino- or thiol-group.80–82 The derivatization process is fast, with high yield, carried out under mild conditions, and break the molecular proton bridge bond making the boiling points lower and the metabolites more stable for GC-MS.83 Due to the complexity of biological samples, enzyme activity, and often the need to increase the concentration of low abundant molecules, the choice of the right procedure is dictated by the molecular target.84,85 With two-dimensional gas chromatography (GCxGC), a second independent separation is performed by importing the analytes eluted from the first column into the second column increasing both the peak capacity and the sensitivity thus facilitating the characterization of metabolites.86,87
Liquid chromatography (LC)
LC is a potent separative technique ideal for studying complex biological matrices, and it has been further optimized to ensure efficient, easy, and robust analysis. LC coupled to a mass spectrometer (LC-MS) revolutionized the field, allowing the characterization of non-volatile or thermally labile compounds with high molecular weight that were not possible to analyse with GC-MS. By reducing the diameter of the packing particles (3–50 µm) in the column and by employing high pressure to improve the mobile phase speed, it has been possible to make LC operating in a highly efficient mode, the so-called High-performance liquid chromatography (HPLC).88 To gain in selectivity and sensitivity some metabolomic studies have employed the Ultra-performance liquid chromatography (UPLC) system using columns with even smaller particle size (<2 µm) and higher pressure. Nevertheless, the low injection volumes and short column life do not make this technique widely used.75 A further development is the Ultra-high-pressure liquid chromatography (UHPLC), which is a further optimization capable of producing the best resolution and peak capacity for liquid chromatography known and therefore it is mainly applied to analyse complex samples, including biofluids.88 The analysis of metabolites with different physicochemical characteristics cannot be performed with a single stationary phase. Given its high reliability and robustness, Reversed-phase chromatography (RP) is one of the most widely employed technique, thanks also to the possibility of repeatable separations covering a broad-spectrum of chemical compounds. In a RP chromatographic system, the stationary phase consists of an organic species chemically bound to silica particles that form the chromatographic support, and it has a lower polarity compared to the mobile one. Non-polar columns, as C18, are ideal for metabolomic analysis because of their ability to separate semipolar compounds such as phenolic acids, flavonoids, glycosylated steroids, alkaloids, and other glycosylated species. However, other types of stationary phases are reported in RP chromatography like short alkyl chains (C8) and amides. In this type of separation, the mobile phase is polar (water, methanol, acetonitrile), and the elution can be isocratic (constant composition of the mobile phase) or gradient (variable composition); the latter mode is used for heterogenous samples because it improves separation and reduces the elution times of substances.75 Many biofluids analysed in metabolomic studies are aqueous thus containing many polar molecules that are almost not kept by the RP stationary phase. The separation of these polar analytes is mostly carried out by the Hydrophilic interaction liquid chromatography (HILIC) technique, which is a variation of the Normal phase (NP) chromatography, that uses a highly polar stationary phase (Fig. 2b).75 Many metabolites linked to tumorigenesis have hydrophilic chemical proprieties; in these cases, both ion-paring reagent-based RP chromatography and HILIC can be employed.
Reversed-phase ion-pair (RPIP) chromatography has a hydrophobic stationary phase and a mobile phase containing an ion-pair reagent, which consists of both hydrophilic and hydrophobic residues in a single molecular structure, added at low concentration. HILIC-based separation technique uses a hydrophilic stationary phase with which the water-soluble metabolites interact and a hydrophobic organic mobile phase, which has a percentage of solvent increasing over time enabling the elution of the metabolites from the column according to their affinity. The HILIC columns allow the retention of polar molecules and the fast elution of lipophilic compounds, which are usually difficult to elute from other types of columns and can therefore accumulate causing ion suppression due to background bleeding.72,75 Supercritical fluid chromatography (SFC) uses a supercritical fluid as mobile phase with temperature and pressure higher than a critical point. Among the most widely used eluents in the supercritical state is Carbon dioxide (CO2) is non-toxic, easily handled and environmentally friendly since it needs just little amount of organic solvent as auxiliary solvent.89
Capillary electrophoresis (CE)
CE is an analytical separative method applicable to a broad spectrum of chemical substances, primarily polar and charged ones (Fig. 2b). The most widely used separation modes for metabolomic analysis are Capillary zonal electrophoresis (CZE) and Capillary micellar electrokinetic chromatography (MEKC).90,91 CZE separates compounds according to the different electrophoretic mobility of the analytes, which is determined by their charge and size; the separation occurs in a capillary that contains a separation buffer, and it is subjected to an electric field.92 MEKC extends the functionality of CE to uncharged compounds; thus, the separation of the analytes happens through differential splitting between micelles and a surrounding aqueous solution. CE allows for different selectivity and higher efficiency than HPLC as well as shorter analysis times. However, this technique has low concentration sensitivity compared to HPLC, and, for this reason, sample pre-concentration is often necessary. CE-MS has great potentiality for microscale analytical technique, meeting the need of analytical methods to profile metabolism of (sub)microliters of samples.93 However, the CE-MS-based metabolomics is not widely diffused in comparison to other analytical separation approaches described above, as it is perceived as a technically demanding approach that suffers of low reproducibility and sensitivity.92
Separation-free MS techniques for metabolites investigation
For fast and high-throughput analysis the option of a direct MS analysis, without any prior separation, may be particularly useful (Fig. 2a). In Direct infusion mass spectrometry (DI-MS) samples are directly injected or infused into high-resolution and high-accuracy MS, without a separation step. DI-MS enables rapid sample analysis, improving repeatability and accuracy among samples.94–96 However, isomeric compounds cannot be separated and contamination of the ion source by compounds residues is tedious but overcome by using chip-based nano-electrospray ionization (ESI, see “MS-Ion sources” paragraph below).89 Mass spectrometry imaging (MSI) is an interesting emerging approach to analyse metabolites in situ directly in organs, organoids or cells. MSI utilizes several ionization sources: Matrix-assisted laser desorption ionization (MALDI), Secondary ion mass spectrometry (SIMS), Desorption electrospray ionization (DESI), and Laser ablation electrospray ionization (LAESI). In MALDI-MSI, the laser targets the matrix-coated tissue surface to achieve tissue scanning.97 In SIMS, first a high-energy primary ion beam (Ar,+ Ga,+ and In+) hits the sample surface and then the secondary ions are assembled and analysed. DESI derives from electrospray and desorption ionization, and it exploits the conduction of electrosprayed charged droplets and solvent ions into the surface to analyse. LAESI is a mix between ambient ionization source grounded on mid-infrared laser ablation with charged droplets produced by Electrospray ionization (ESI).89
The mass spectrometer: the metabolomics core instrument
The first mass spectrum of a molecule was measured in the 1910s by Joseph J. Thomson, who constructed the first mass spectrometer98,99 (Fig. 1). Initially, mass spectrometers were mainly employed by physicists to study the atomic weights of elements and isotopes and their respective relative abundance.100 This innovative technology was applied in biological settings in the 1940s, when heavy stable isotopes were used as tracers to study CO2 production in animals.101 Since that time, technological advances have increased the range of sample types that can be analyzed by MS. In recent years, MS has been widely used to investigate cancer-specific changes in the biomass composition of human tumours, including metabolic alterations since it qualitatively and quantitatively measures the compounds. A mass spectrometer is always composed by three elements: an ion source, a mass analyser, and a detector (Figs. 1, 2a).
MS-Ion sources
The analytes are first introduced into the ionization source, where they acquire either a positive or negative charge. At this point, the ions pass through the mass analyser and, on the basis of their mass-to-charge (m/z) ratio, arrive to distinct parts of the detector, where signals are generated and recorded by a computer system, which in turn graphically displays these signals as a mass spectrum. A hard ionization method like the Electron impact ionization (EI) renders GC-MS the right approach to identify metabolites. In EI a molecule in gaseous phase is ionized by collision with an electron flux of typically 70 eV of energy, giving rise to an excited molecular ion that can dissociate to generate fragment ions related to a structure. Structural information is extrapolated by disrupting the compounds and producing molecule-specific fragments. Then, the pattern of unique fragments is employed to identify metabolites by taking advantage of the available libraries.102 Chemical ionization (CI) is also commonly used in GC-MS analysis. It produces ions of the analyte of interest by ion/molecule reactions from ions of a reactant gas that is present in excess. This type of ionization is not the first choice in metabolomics since the obtained fragments are limited as well as the libraries for the following analysis. On the other hand, CI uses a soft ionization by applying low energy to the molecules and it can be used for unknown metabolite identification.103 Atmospheric pressure chemical ionization (APCI) is another soft ionization source which gives rise to some degree of fragmentation enabling structural characterization. Ions are formed in the gas phase using a corona discharge to ionize metabolites present in the aerosol, then the ions released in the gas phase are analysed with the analyser. APCI is commonly used to analyse small polar and non-polar compounds that are poorly ionized by ESI (see below) and it is commonly coupled with HPLC. APCI is a useful tool for lipidomic; in particular, it generates ions from large neutral molecules such as triacylglycerols that are still a challenge to analyse with other techniques.104–106 Electrospray ionization (ESI) is the more widely used ion source in metabolomic studies; it generates ions from metabolites out of a solution and it is coupled to liquid-based separation techniques like LC, CE or DI. Its soft ionization produces vast number of ions through charge exchange in solution and forms intact molecular ions that aid the initial identification. This ionization technique provides a sensitive, robust, and reliable tool to study at femto-mole quantities non-volatile and thermally labile analytes that are difficult to analyse with other conventional techniques. Clusters of charged droplets consisting of analytes surrounded by many solvent molecules are produced by introducing a fine spray of a liquid solution of molecules into an electric field (2–4 kV) outstanding between the capillary and the counter electrode of the MS inlet at atmospheric pressure. Then, a flow of nitrogen (drying gas) enhances the evaporation and elimination of solvent from the charged analyte. The droplets size decreases due to the evaporation of solvent and the charge density increases till the coulombic explosion of the droplet. At this point, the resulting analyte ions enter the MS via electrostatic lenses.75,107,108 Matrix-assisted laser desorption ionization (MALDI) is a tool for soft ionization and transfer of complex samples, which are co-crystallized with an organic matrix on a metal target, from the solid phase to the gas phase. A pulsed laser excites the matrix and induces a fast heating of the molecules and the desorption of ions to the gas phase; the selection of the matrix is a fundamental aspect that affects the experimental results. The most used MALDI matrices are α-cyano-4-hydroxycinnamic acid (CHCA), 2,5-dihydroxybenzoic acid (DHB) and 3,5-dimethoxyl-4-hydrocinnamic acid (SA).97 This technique is suitable for high molecular weight analytes, it is fast, consumes little amount of sample and has high tolerance towards impurities (e.g., salts).109 By contrast, it must have low vapour pressure and the pulsed nature of the laser limits compatibility with the analyser. In fact, normally, the MALDI is associated to a MS capable of measuring a full mass spectrum without scanning a mass range.89 MALDI-MS is a potent analytical tool and a valuable method in cancer research and tissue imaging, due to its ease of use and high mass resolution.110–114 Direct real-time analysis (DART) is another recent ionization technology that allows metabolites to be analysed without sample preparation and at atmospheric pressure. In DART, there is first the thermo-desorption of condensed-phase analytes by a stream of a hot gas (such as helium, argon or nitrogen), which carries active species derived from a plasma discharge. Then, the ionization enables the acquisition of respective mass spectra. DART is used with small molecular compounds with minor cross-contamination and provides a simple and high-throughput analysis.89,115
MS analysers
Improving resolution and sensitivity is the main goal of metabolomics studies. Nevertheless, often the two parameters are mutually exclusive, and this remains the main limitation of metabolomics. Single (MS) or tandem (MS/MS) mass analysers can be alternatively employed with distinct output and final resolution. The single-configuration mass analysers mostly used in metabolomics are the Quadrupole (Q), Quadrupole ion trap (QIT), Time-of-flight (TOF), Fourier transform ion cyclotron resonance (FTICR), and Orbitrap (OT) (Fig. 1a).116 Quadrupole (Q) has four parallel cylindrical rods, which are responsible for filtering the sample ions based on their m/z.117 Ions are separated according to their trajectory stability in the oscillating electric field applied to the rods. By continuously modulating the applied voltages, it is possible to select a particular m/z or scan for a range of m/z values. Quadrupole ion trap (QIT) is a variant of the quadrupole analyser with three electrodes (an annular electrode placed between two hemispherical inlet and outlet electrodes) to trap and accumulate ions in a small cavity, called ion trap. The mass spectrum is obtained by varying the electric potential so that the ions are sequentially ejected from the trap toward the detector according to an increasing m/z value.118 Time-of-flight analyser (TOF) consists of a field-free drift chamber held under high vacuum through which ions run. The ions are separated according to their velocity, and they reach the detector at separate times since they possess the same kinetic energy but a different velocity depending on the m/z ratio. The same analytes can be desorbed at slightly different times, therefore, a reflectron composed of sequential electrodes is placed at the end of the TOF resulting in an increased mass resolution and accuracy of TOF analysers.119 Fourier transform ion cyclotron resonance (FTICR) is a high resolution and high accuracy but also very expensive analyser, which works by applying a strong magnetic field (9–15 T), thus the ions assume a circular motion in a plane perpendicular to that of the magnetic field (cyclotron). Ions start to rotate with an angular frequency inversely proportional to m/z. By applying a radiofrequency voltage pulse corresponding to the angular frequency of a specific analyte, this ion with a specific m/z value absorbs energy and produces a cycloidal path moving with a wider orbit. The way the excited ions move creates the image current measured on electrodes. Since the ions oscillate as a function of their m/z, by measuring the image current during time and using a Fourier transform (FT), it is possible to extrapolate the m/z values. By applying a scan of voltages, a complete spectrum is obtained.120,121 Orbitrap (OT) traps in an electrostatic field positive ions because of their attraction to the inner electrode (set at about –3200 V). The ions start to rotate around the inner electrode and together oscillate along the z-axis according to their m/z ratios. An image current is detected and converted by a FT to obtain a mass spectrum with the frequency and the intensity of each ion.122,123 Quadrupole and ion trap analysers are extremely sensitive but limited in resolution, whereas TOF, FTICR and OT offer the highest mass resolution. Mass analysers in a tandem configuration include diverse types of analysers.124 Triple quadrupole (TQ) and Triple-quadrupole ion trap (QTrap) analysers, which have high sensitivity and selectivity, are the most common MS-spectrometers coupled to LC and employed in targeted metabolic studies. In contrast, the high mass-resolving capacity of Quadrupole-TOF (Q-TOF), Linear-quadrupole ion trap-Orbitrap (LTQ-Orbitrap), and FTICR analysers is ideal for globally profile and to identify metabolites. GC is mainly coupled with single quadrupoles or TOFs, but novel instruments also exploit QTOF or TQ MSs.116,125,126
Nuclear magnetic resonance (NMR) spectroscopy: an alternative approach in metabolomics
NMR spectroscopy, originally described by Isidor Rabi in 1938 and later used for the analysis of liquids and solids by Felix Bloch and Edward M. Purcell in 1946, is a consolidated approach for the analysis of cancer metabolism69,127 (Fig. 1). This analytical tool measures the chemical shifts of atomic nuclei with non-zero spin (i.e. 1H, 31P and 13C), which are dependent on the atom environment in a chosen analyte, and enable detecting and elucidating the structure thanks to shifts in the magnetic resonances.128 NMR spectroscopy allows the characterization of new compounds and it does not require sample destruction, chromatographic separation, sample treatment, or chemical derivatization.
NMR is highly automatable and, if the same instruments and analytical strategies are employed, the reproducibility between different laboratories is guaranteed.129 Unlike other metabolomic platforms, NMR is not limited to the analysis of biofluids or tissue extracts, but it can be employed for the study of any biological sample, including a solid or semi-solid tissue or organ130–133 via either the solid-state NMR (ssNMR) or the magic-angle sample spinning (MAS-NMR).134 NMR allows live analysis coupling the imaging and the metabolic profiling with the Magnetic resonance spectroscopy (MRS) and the Magnetic resonance imaging (MRI). Moreover, it is widely employed to profile metabolic fluxes in real time.129 The primary limitation of this technique is the low sensitivity which is 10 to 100 times lower in comparison to LC-MS and GC-MS. A significant signal enhancement has been achieved by increasing the magnetic field, using a cryo-probe and the processing of digital signals; however, most of the low-concentrated small molecules are still undetectable with NMR. In order to make NMR analysis high-throughput, innovative fast and ultrafast multidimensional NMR approaches have been introduced while NMR sensitivity has been enhanced by altering the nuclear polarization with the Dynamic nuclear polarization (DNP).
However, the NMR applications in metabolomics are limited mainly because NMR spectrometers are more expensive than many other analytical instruments including mass spectrometers and they need experienced users and vast laboratory infrastructure with ad hoc non-vibrational floors and specific area isolated from magnetic and radio frequency interference.135
Metabolic flux analysis (MFA)
Global metabolomics analyses are robust techniques that allow the identification of tumour-associated metabolic liabilities; however, often they fail to reveal the plasticity and dynamism of metabolic pathways, providing only a static snapshot of cancer cell metabolism. Metabolic flux analysis (MFA), using stable-isotope-labelled substrates, can determine flux rates by tracing the isotope enrichment of particular atoms on downstream metabolites, thus really connecting omics analysis and phenotypes (Fig. 3a).72,136 As depicted in the workflow of Fig. 3b, the first step is the design of the experimental setting that includes the in silico identification of the optimal tracer for the highest flux resolution and the substrate labelling. Following administration of the stable-isotope tracer, samples are analysed both for isotopic labelling and external rates that takes into account substrate uptake and product secretion. The metabolite labelling can be measured either by MS or NMR and integrated into the metabolic network model to which the labelling measurements are fit and must include all relevant reactions and their respective carbon atom transitions. The network is constructed based on information from metabolome databases.
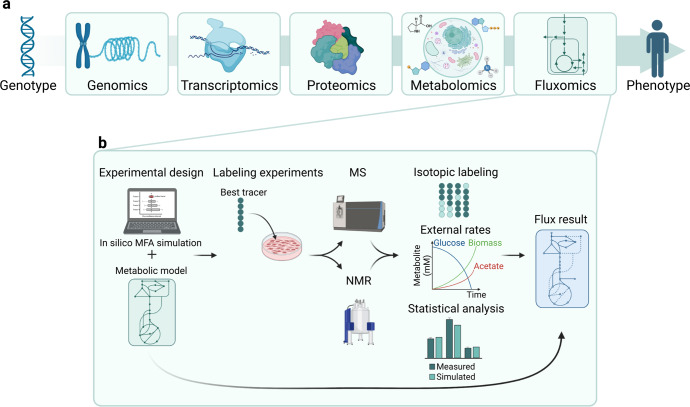
Fig. 3.
a Fluxomics is the omics approach that gets closer to the phenotype. b Experimental workflow of a standard Metabolic flux analysis (MFA) experiment. The first step is the experimental design and the definition of the best tracer and metabolic model by in silico analysis. Cells or tissues are incubated with the tracer and samples are analysed by either MS or NMR. The isotope tracing is analysed considering the metabolites external rates and the statistical value. The results are thus integrated into the original metabolic model. This figure was created with Biorender.com
Two different experimental settings are mainly used in MFA: stationary or non-stationary. The stationary design deals with metabolites at their basal state: the labelling is not linked to the analyte’s concentration thus simplifying the flux measurement and allowing the study of the involvement of different metabolic routes to the final amount of a given metabolite. By contrast, to analyse the flux to a downstream pathway a non-stationary design should be employed. In this setting, a labelled substrate is applied for a brief time, in order to detect the label incorporation into downstream molecules.137 According to the pathway of interest, it is possible to choose among substrates that can be uniformly or positionally labelled. Generally, the first type of substrates is employed to analyse complex fluxes, while positionally labelled substrates allows the detection of small changes into key node of metabolic pathways. 1,2–13C2 glucose is used to accurately quantify glycolysis, PPP and gets a general picture of the energy metabolism of a tumour cell, while U-13C5 glutamine is the best substrate for the measurement of the TCA cycle.128 In cancer metabolism research, the most common labelling is with 13C for substrates as glucose, glutamine, pyruvic acid, lactic acid, palmitic acid, amino acids, and acetic acid.
Carbon flux distribution into downstream and interconnected metabolic pathways of 13C labelled substrates are determined by MS or NMR techniques; it is thus possible to understand how dependent the biosynthesis of a metabolite is to a specific source of carbon and what is the flux into downstream metabolic routes. To measure specific metabolic activities there are many tracers. 1–2H and 3–2H-glucose can be employed to quantify the biosynthesis of NADPH from the Oxidative pentose phosphate pathway (OxPPP)138,139 and Nicotinamide adenine dinucleotide (NAD) synthesis-breakdown fluxes.140 MFA is an extremely useful method to identify metabolic shifts, but both the measures and the bioinformatics and statistical analysis are extremely complex requiring skilled workforce and cutting-edge infrastructures.128
Translational metabolomics: from bench to bedside
The substantial technological, methodological and computational advancements in metabolomics platforms and the growing accessibility of the technologies over the last 40 years have led to the development of personalized metabolic profiling that in concert with personalized genomics, represent the foundation of personalized medicine. Metabolomics can be employed to analyse a wide range of samples (i.e. tissue, biofluids, cells), thus spacing outside the academic institutions and finding applications in many different disciplines including human health, wellness and food chemistry.141 The new metabolomic platforms have provided opportunities for cancer screening, diagnosis and treatment.63,142–145 The most successful example of application is the employment of metabolomics to discover and validate biomarkers for tumour progression and metastasis in different sample types including plasma or serum,146–156 urine,157–160 saliva161–164 and cerebrospinal fluid.165–167 Metabolomics can be used for non-invasive diagnosis and prognostic evaluation of cancer147,168–171 and for tumour subtyping.168,172–175 Moreover, metabolomics constitutes a cost-effective and productive platform for monitoring anti-cancer treatment effects, both as a predictive measure of efficacy and as a pharmacodynamic marker as well as for evaluation of drug resistance.176–181 One of the most interesting and recent application is the use of metabolomics as an alternative method to the most traditional immune and biochemistry assays to investigate the metabolic alterations in response to immunotherapy.182 Employing metabolic profiling by metabolomics, to evaluate the immune responses in cancer patients, is an innovative area of cancer research with the potential for identifying host immune factors that would prevent the effective anti-tumour immunity thus compromising the clinical output.183–192
Extracellular flux analysis (EFA)
Extracellular flux analysis (EFA), with the Agilent Seahorse XF analyser as leading technology counting more than 5000 peer-reviewed publications, is the most common and feasible method to broadly quantify the bioenergetic activity of live cells, organoids or tissues193 (Fig. 4a). Sketchily, EFA quantifies Oxygen consumption rate (OCR) as a quantitative measurement of mitochondrial electron transport rate named mitochondrial OXPHOS and the Extracellular acidification rate (ECAR) as a read out of glycolysis. The instrument performs live measures of OCR and ECAR by taking just few μL of medium from monolayer of cells in microplates. Two sensors installed into fibre-optic probes quantify oxygen level and medium pH for several minutes, then the software extrapolates the OCR (pmol/minute) and ECAR (mpH/min) quantifications. The versatility of the instrument is given by a continuous and systematic temperature regulation that controls the extracellular microenvironment and an integrated drug delivery equipment for serial injection of up to four metabolic inhibitors to each well at pre-set time points, thus allowing measurement of metabolic capacity and adaptation.
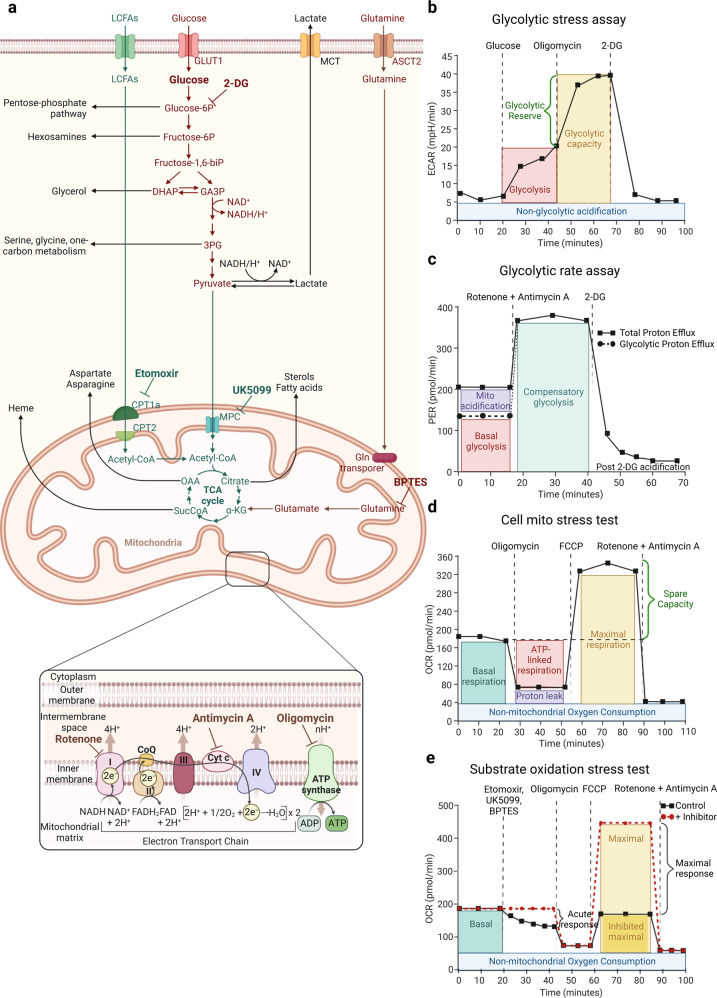
Fig. 4.
a Key metabolic pathways governing cancer cell growth that can be measured by Extracellular flux analysis (EFA). b Glycolysis stress assay is performed by serial injections of Glucose, Oligomycin and 2-deoxyglucose (2-DG) to get as a read out glycolysis, glycolytic capacity, the glycolytic reserve and non-glycolytic acidification measurements. ECAR stands for extracellular acidification rate. c The Glycolytic rate assay reports multiple key parameters, such as basal glycolysis, compensatory glycolysis achieved by shutting down mitochondrial respiration with rotenone and antimycin A. Proton efflux rate (PER) is a quantitative measure of protons extruded into the extracellular medium during glycolysis. d In the Cell mito stress test, mitochondrial respiration is measured by quantifying the Oxygen consumption rate (OCR). Cells are sequentially exposed to oligomycin, Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) and rotenone and antimycin A thus allowing the measurement of the basal and maximal respiration and spare respiratory capacity. e The Substrate oxidation stress test measures the contribution of Long-chain fatty acids (LCFAs), glucose/pyruvate and glutamine as primary substrates that fuel mitochondrial metabolism by using specific inhibitors in combination with a standard Cell mito stress assay. Etomoxir inhibits the Carnitine palmitoyl transferase 1a (CPT1a), UK5099 blocks glucose and/or pyruvate through inhibition of the Mitochondrial pyruvate carrier (MPC) and BPTES inhibits glutamine through Glutaminase-1 (GLS-1). This figure was created with Biorender.com
Glycolysis is a hallmark of cancer cells and its analysis together with the associated metabolic rewiring are considered reliable diagnostic means for detection and prognosis of malignant phenotypes and to lead to the identification of new biomarkers and therapeutic strategies.194,195 The Glycolytic stress assay is the easiest method to gain a qualitative overview of the bioenergetic phenotype (Fig. 4a, b). In this assay, first ECAR is measured right after incubation of the cells in a medium in which glucose was depleted. This is followed by a first injection of glucose until saturation causing a rapid extrusion of protons into the medium quantified by a rise in the ECAR. This glucose-induced response is considered as basal glycolysis rate under normal conditions. Injection of oligomycin, a Complex V (ATPase) inhibitor, interferes with mitochondrial respiration by blocking the proton channel of the ATP synthase thus shifting the cellular bioenergetics towards glycolysis, which is detected by an increased in ECAR: the so called, cellular maximum glycolytic capacity. The last injection is with 2-deoxy-glucose (2-DG), a synthetic glucose analog that blocks glycolysis through competitive binding to glucose Hexokinase (HK), which causes a drastic decrease in ECAR and serves as positive control of ECAR shifts as read out of glycolysis. The measurement of ECAR, before the injection of glucose, is the non-glycolytic acidification (due to any intracellular metabolic activity other than glycolysis) while the difference between the glycolytic capacity and the glycolysis rate allows the calculation of the glycolytic reserve. At pH of 7 the physiological conversion of glucose into lactate causes protons release and acidifies the medium, thus providing a quantitative assessment of the glycolytic rate. However, this quantification does not consider that the final ECAR is the result of two processes: glycolytic and respiratory acidification. More in detail, CO2 derived from the TCA can be spontaneously or enzymatically hydrated to carbonic acid, H2CO3, that dissociates to HCO3− and H+ in a medium with physiological pH. The conversion of one molecule of glucose into lactate yields 2 lactate− and 2 H,+ whereas the complete oxidation of one glucose to CO2 generates molecules of HCO3− and 6 H+ molecules; as a consequence, the extracellular acidification when a glucose molecule is oxidized to CO2 is three times greater than when it is converted to lactic acid. The ratio of glycolytic and respiratory acidification is significantly affected by the experimental conditions, such as cell type and media composition, and it can fluctuate from nearly 100% glycolytic acidification to nearly 100% respiratory acidification. Given the linear correlation between O2 consumption and CO2 production during OXPHOS, the Glycolytic rate assay implements the previous analysis by calculating the contribution of mitochondria/CO2 to extracellular acidification and subtract the CO2-dependent acidification from the total Proton efflux rate (PER). The resulting value, named “glycoPER”, is a quantitative and reliable measure of protons extruded in the culture medium during glycolysis (Fig. 4c).
Complementary, glycolytic flux can be measured by quantifying glucose uptake and lactic acid excretion: several commercially available methods based on colorimetric and fluorimetric read-outs are optimized to quantify glucose and lactate levels within the extracellular media. In addition, measurement of the activity of the three rate-limiting glycolytic enzymes [HK, Phosphofructokinase-1 (PFK-1), and Pyruvate kinase (PK)] is crucial for understanding the energetic metabolic status of the cells.196
The Cell mito stress test is a common EFA to fully characterize mitochondrial respiration based on the quantification of the OCR after interference with the complexes of the Electron transport chain (ETC) (Fig. 4a, d). After the initial basal OCR measurement, in the stress test, cells are exposed to oligomycin, which affects ATP synthase, causing a drastic decrease in mitochondrial respiration or OCR. Subsequently, electron flow through the ETC is simulated by the injection of the uncoupling agent Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) so that oxygen consumption by complex IV reaches its maximum. This increase in OCR is used to extrapolate the cellular spare respiratory capacity, by definition the difference between maximal respiration and basal respiration, which indicates the capability of the cells to cope with stress and respond to changes in energetic demand thus indicating the cellular fitness. The third injection is a mixture of rotenone and antimycin A, respectively complex I and complex III inhibitors. The two inhibitors completely shut down mitochondrial respiration and allow the quantification of non-mitochondrial respiration caused by processes independent from mitochondria. This assay is particularly useful in pharmaceuticals pipelines; indeed, damages to mitochondrial function are important indicators of drug-mediated liver, cardiac and neurological disfunctions. Therefore, the integration of this test into the toxicology studies would allow a better evaluation of the drug side effects and improve the chemical substance synthesis upstream the later development steps.197–201
To generate ATP via OXPHOS, the nutrients taken up from the extracellular environment are ultimately oxidized via the TCA cycle and the ETC. The Substrate oxidation stress test is a useful method to evaluate the contribution of the three primary substrates in generating energy: Long-chain fatty acids (LCFAs), glucose/pyruvate and glutamine. The assays combine the Cell mito stress test for the interrogation of mitochondrial function (described above in this paragraph) and anaplerotic pathway specific inhibitors such as Etomoxir for LCFAs by inhibiting the Carnitine palmitoyl transferase 1a (CPT1a),202–204 UK5099 to block glucose and/or pyruvate by interfering with the Mitochondrial pyruvate carrier (MPC)205–207 and BPTES that blocks the Glutamine through Glutaminase-1 (GLS-1).208,209 When we are in the situation of substrate oxidation, the basal and especially the maximal respiration rates are significantly affected by the capacity of the cells to transport and utilize the available substrates revealing dependence on a specific metabolic pathway (Fig. 4a, e).
These EFA assays have significantly facilitated the identification of novel metabolic liabilities to genetic manipulation, pharmaceutical interventions and, thanks to the screening assays that allow the simultaneous analysis of up to 80 compounds in one plate, the systematic analysis of metabolic dependence of cancer-derived cells and organoids.210 However, cell media composition, cell density, inhibitor concentration and post-run normalization strategy (i.e., cell count or protein/DNA quantification) can dramatically impact the interpretations of the analysis as well as the interlaboratory standardization. OCR and ECAR analyses do not reflect individual pathway activity, but several variables can influence their proportionality to the metabolic processes they are meant to represent. Therefore, the interpretation of these analyses cannot negate the activity of secondary bioenergetic pathways; however, these assays can be usefully employed to compare samples and categorize the cells whether they are broadly more glycolytic or oxidative. Moreover, OCR and ECAR analyses can be complemented with additional techniques such as single-cell imaging using commercially available fluorescent dyes (see “Fluorescent metabolic probes” paragraph below) and ultrastructural morphological and morphometric analysis. For example, Transmission electron microscopy (TEM) evaluation of mitochondria (i.e., mitochondrial length, inner/outer membrane ratio, cristae extension, width, and junction diameter) is a valuable and complementary tool to investigate mitochondrial morphology and functionality.211
Single-cell metabolic analysis
Up to date, most of the findings regarding cancer metabolism have been achieved using cell culture models and in vivo measurements obtained from bulk tumours. Indeed, one of the biggest limitations of standard metabolomics and EFA is that they do not allow the multiplexing of metabolic state and phenotyping. The result is always shaped by genetic and environmental factors for each cell and thus largely incompatible with heterogeneous cellular population. Moreover, standard cell culture media do not resemble the human physiological nutrient milieu and nutrient availability in a tumour microenvironment significantly modulates metabolic dependencies.212,213 The emergence of RNA-sequencing (scRNAseq) provided the unprecedented opportunity to interrogate genomic profiles of tumours with high resolution.214–222 Detection of changes in the expression of metabolic genes is a valuable tool to extend our comprehension of the metabolic rewiring of cancer and to identify metabolic vulnerabilities.223,224 For this analysis, individual cells collected in sub-microliter droplets are sorted into multiwell plates using microfluidic devices; after lysis, the cells are barcoded in order to assign sequencing reads to each cell (Fig. 5a). This technological advance was achieved thanks to the possibility of capturing and sequencing exceptionally small quantities of RNA. scRNAseq studies showed that malignant cells globally up-regulate genes involved in almost all functional categories of metabolic pathways suggesting a high metabolic plasticity that confers great adaptability to different genetic and environmental factors. This global transcription rewiring of metabolic genes suggests that cancer cells reserve more transcriptional resources for the expression of such those genes with consequently increased fluxes for most metabolic reactions.225 Single-cell, high-dimensional profiling allowed the identification of most effective immunotherapeutic approach, the discovery of biomarkers for early-stage of the disease and drug-resistance mechanisms in a variety of cancer settings. Indeed, scRNAseq analysis revealed a high-resolution snapshot of drug-resistant cells to Immune checkpoint inhibitors (ICIs), thus providing the rational for further investigating cell-cell communications and drug effect.226–230
Fig. 5.
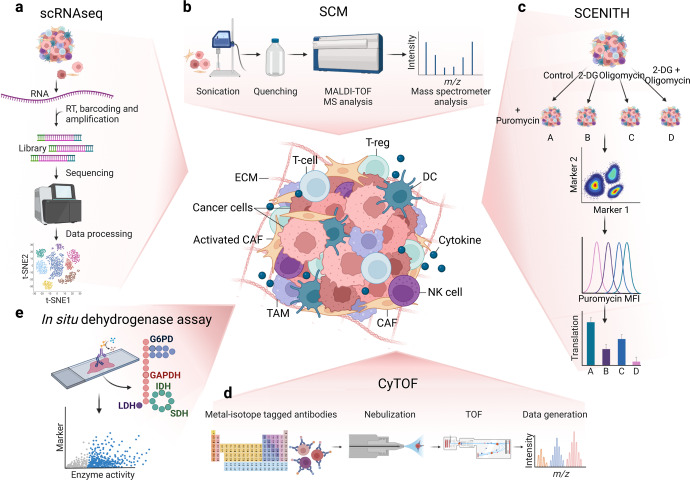
Global overview of the cutting-edge techniques available to investigate the Tumour microenvironment (TME). a Single-cell RNA-sequencing (scRNAseq) representative workflow: individual cells are isolated, lysed and barcoded before retro-transcription, library amplification and sequencing. b Single-cell metabolomics (SCM) includes single cells isolation, sample processing, quenching of metabolism and metabolomics. c In the single-cell energetic metabolism by profiling translation inhibition (SCENITH) the sample is divided into 4 and either left untreated or treated with 2-deoxyglucose (2-DG), oligomycin and both inhibitors. The addition of puromycin allows protein synthesis quantification coupled with flow-cytometry phenotyping. d In Cytometry by Time-of-flight (CyTOF), the cells are labelled using stable heavy metals, nebulized, and vaporized to form ion clouds and analysed by a Time-of-flight (TOF)-MS. e With the In situ dehydrogenase activity assay, the activity of G6PD, GAPDH, LDH, IDH and SDH is coupled with immune-staining to distinguish single cells directly on a tissue slide. This figure was created with Biorender.com
The recent technological advances offered the opportunity to analyse the complexity of biological systems at the single-cell level also in metabolomics.231 Single-cell metabolomics (SCM) provides a snapshot of all metabolites, intermediates, and end-products of cellular metabolism within a biological system decoding the biochemical heterogeneity within cells.232,233 As previously discussed for bulk metabolomics, the biggest challenge in preparing single-cell samples is avoiding, or at least reducing, the impact on cellular metabolism due to the sample processing. The standard methods for single-cell sample preparation are the Fluorescence-activated cell sorting (FACS) and the microfluidic arrays that maintain intact cell morphology or extraction via an Atomic force microscopy (AFM) probe that keeps only the metabolites from the cell in the probe.234,235 Cellular metabolism should be quenched and can be analysed by MS-based technologies (Fig. 5b). Previous genomic analysis targeting metabolic genes at the single-cell level has revealed that individual malignant cells have elevated metabolic activity and variations that were not observed in bulk tumour studies. As for scRNAseq, SCM helps in dissecting cell heterogeneity236; SCM is particularly useful in oncology for disease and metastasis detection, precision drug design and drug assessment and toxicity.237 Moreover, SCM could be employed for profiling rare or Circulating tumour cells (CTCs)238–240 as well as for cancer subtypes discrimination and new therapies development.225,241–244
In addition, several cytometry-based methods that combine metabolic analysis with single-cell phenotypes have been developed. Fluorescent metabolic probes and analogues of metabolites can be analysed by flow cytometry or microscopy enabling single-cell resolution and they are often used for metabolic pre-screening assays since they are fast, relatively cheap, and easily adaptable to different experimental settings. Some examples are: 2-NBDG (2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)−2-Deoxyglucose), a fluorescent indicator to monitor glucose uptake into living cells; BODIPY (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) employed as a synthetic precursor to a wide variety of fluorescent phospholipids; CM-H2DCFDA, a chloromethyl derivative of 2,7-Dichlorodihydrofluorescein diacetate (H2DCFDA), useful indicator for ROS in cells; cystine–Fluorescein isothiocyanate (FITC) conjugate to monitor the uptake and accumulation of the amino acid; Monodansylcadaverine (MDC) to track autophagic vacuoles; JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimi-dazolylcarbocyanine iodide), a carbocyanine dye that can be used as a ratiometric indicator of mitochondrial potential and MitoTracker dyes that are cell permeable probes containing a mildly thiol-reactive chloromethyl moiety for mitochondrial labelling.245–249
In 2020, an innovative flow-cytometry-based method to profile the energetic metabolism with single-cell resolution has been reported (Fig. 1). Single-cell energetic metabolism by profiling translation inhibition (SCENITH) measures metabolic profiles based on metabolism-dependent translation rates and puromycin incorporation into nascent proteins.250 As for EFA, incubation of given samples with on-target inhibitors enables to functionally estimate the glucose and mitochondrial dependence, glycolytic, fatty acid and amino acid oxidation capacity. It is possible to employ puromycinylation detection in combination with multiparametric flow-cytometry analysis thus analysing at single-cell complex and heterogenous samples251–253 So far, SCENITH has been applied to the analysis of ex vivo whole blood and human tumour biopsies. Interestingly, the combination of SCENITH and scRNAseq analysis of renal carcinomas and juxta-tumoral tissues succeeded in the correlation of the metabolic profile and the metabolic gene expression.250 Moreover, SCENITH can be employed in many other tumour settings and physio-pathological conditions including for example the comprehension of cell death pathways that have important and multiple connections with metabolism and redox imbalance254–259 (Fig. 5c).
Other powerful methods are based on single-cell proteomic analysis. In 1994 Tsutomu Nomizu and his collaborators discovered that it was possible to nebulize, dry and ignite individual cells in a hot plasma in order to create ion clouds that could be detected by emission spectrometry. This is the first real MS experiment of single cells.260,261 Over the years the technology had a big evolution until 2007, when Scott D. Tanner, inspired by the flow-cytometry technology, invented the mass cytometry, also termed Cytometry by Time-of-flight (CyTOF), which is the most promising technology for high-dimensional and high-throughput protein (and metabolic) single-cell analysis (Fig. 1). CyTOF uses non-biologically available metal isotopes, with concise mass spectrometry parameters, in replacement to standard fluorescent labels, normally employed in flow-cytometry. For the staining, the cells are incubated with a mixture of probes/antibodies tagged with unique non-radioactive heavy metal isotopes. Afterwards the single-cell suspension passes through an argon (Ar) plasma, by which the sample is atomized and ionized thus converting each cell into a cloud containing ions of the elements present inside or on that cell. Low-mass ions derived from each cloud are extruded by a high-pass optic thus generating a cloud containing only ions associated to the isotope-conjugated probes. Ions are then separated by m/z in the TOF chamber, with subsequent amplification and conversion into electrical signals. Up to 50 parameters can be studied simultaneously, overcoming all pitfalls associated with overlapping emission spectra that are normal for fluorescent-based analysis. Mass cytometry has tremendous potential for the analysis of highly heterogenous clinical samples as well as for the diagnosis and for studying the evolution of malignant disorders.262,263 The applications include the study of cell phenotype and function, signalling networks, apoptosis, cell cycle analysis, and many other complex biological processes.264–269 In addition, CyTOF analysis have been employed in several clinical research trials around the world to investigate multiple areas of human disease to understand and improve prevention and therapeutics. The biggest limitation that remains is inherent to the complexity of the data analysis which requires advanced biostatistical and bioinformatic skills and often makes its application in a clinical setting very complicated (Fig. 5d).270,271
Another single-cell proteomic method is Met-flow, a high-parameter flow-cytometry technique that uses specific antibodies against key metabolic proteins that are crucial in their representative pathway such as enzymes and transporters of the PPP, TCA, fatty-acid synthesis and oxidation, OXPHOS, and amino acid metabolism. Using 10 metabolic proteins, Met-flow can define cell subsets comparably to the resolution obtained by 500 genes by scRNAseq.272
Cellular metabolism in the Tumour microenvironment (TME) is affected by both genetic and environmental variables including intrinsic features of the tissue of origin, somatic mutations that appear during the tumour progression, the local nutrient environment, and the complex network of interactions between tumour, stromal and immune cells. Therefore, defining the metabolic signatures of the cells within their native microenvironment is mandatory to identify metabolic intercellular patterns. To this aim, new methods that employ metabolic imaging to quantify enzyme activity of single cells within tissue slices have been developed.273 With the in situ dehydrogenase activity assay, the quantification of the activity of five enzymes catalyzing key steps in the main metabolic routes [Glucose-6-phosphate dehydrogenase (G6PD) in the PPP, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in glycolysis, Lactate dehydrogenase (LDH) in lactate fermentation, and IDH and SDH in the TCA cycle] is combined with staining to distinguish and characterize cell populations. Enzyme activities are quantified on different tumour tissue cryosections and the amount of formed product is measured at optimal and constant assay conditions, including saturating substrate and co-factor levels. The resulted values depict the amount of active enzyme in the analysed samples, associated with a phenotype and to a localization. The interpretation of the results must consider that the activity of these distinct dehydrogenases is quantified at saturated substrate concentrations; therefore, they do not reflect a precise in vivo setting but can then be used to compare different samples (Fig. 5e).
Functional genetic screening
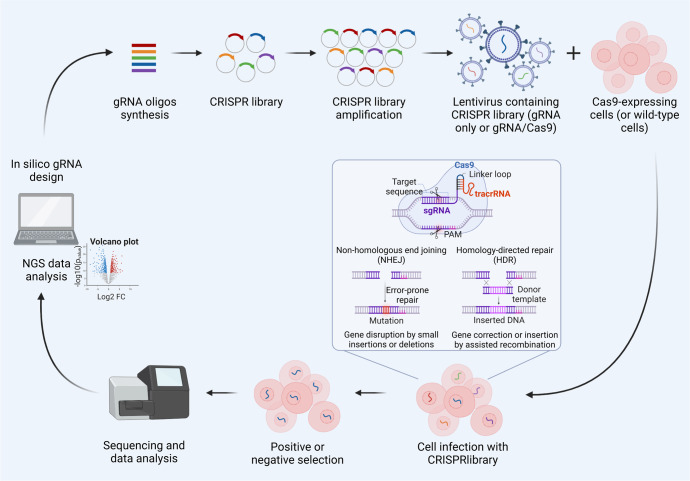
Combining functional screening with metabolic profiling gives the incomparable opportunity to identify context-dependent vulnerabilities that may be therapeutically targeted. Recent innovations in genome editing technology and the advent of the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system, have hugely accelerated the functional genomic research in cancer. The first genome-wide genetic screen in human cells was performed in 2014 by lentiviral delivery of a genome-scale CRISPR-Cas9 knockout (GeCKO) library targeting more than 18,000 genes274 (Fig. 1). Genetic screens, by introducing perturbations into genes at a large-scale, represent an important tool to systematically classify the human genetic elements into functional groups and biological processes, in particular for metabolic pathways (Fig. 6). Compared to traditional short hairpin RNA (shRNA)-based system for performing lethality screens, CRISPR/Cas9 loss-of-function libraries provide a much greater screening sensitivity, since incomplete knockdown by shRNA sometimes does not produce loss-of-function phenotypes. Moreover, CRISPR/Cas9 library screen displays less variation in the data, less off-target effects thus resulting in a low False-discovery rate (FDR) and better consistency across cell lines.275,276 CRISPR/Cas9 screens have been able to identify genes essential for OXPHOS,277,278 for redox homeostasis,279 ferroptosis280 and cell fitness.281–283 CRISPR/Cas9 screening and MFA, allow the study of dispensability and interactions between set of genes encoding enzymes leading to the identification of key nodes within glycolysis and PPP.284 By performing CRISPR/Cas9 screen in Human plasma-like medium (HPLM) compared to traditional media, a massive effect of medium composition on gene essentiality in human cells has been described. Entire sets of essential genes vary with natural cell-intrinsic heterogeneity, suggesting that future genetic screens in HPLM will define new targetable liabilities across diverse human cancers.285 The advent of CRISPR screening technology now provides a rapid, high-throughput means to perform functional characterization of large gene sets. The functional relationships between key enzymes, transcription factors and transporters allowed the identification of multiple interactions within and across metabolic pathways. More importantly, its wide application in concert with the metabolome analysis allowed the annotation of many essential metabolic genes in human tumours and the identification of context-dependent dispensability that can be therapeutically targeted.
Fig. 6.
General workflow for screening using CRISPR/Cas9 libraries. The guide RNA (gRNA) library is synthetized after in silico optimization and cloned into plasmids for the amplification. A library of lentiviruses is then produced and used to infect the cells harbouring or not the Cas9 enzyme. A positive or a negative selection can be applied to identify specific phenotypes and next-generation sequencing is used to determine which genes are disrupted and which are not. This figure was created with Biorender.com
Metabolomics and bioinformatic approaches in precision medicine
High-throughput approaches such as metabolomics involve, even for the simplest assay, the processing of a huge quantity of data, which must be manipulated and mined to get to the underlying biological information. As for the other omics, this need is addressed by the heavy use of computational algorithms. After metabolite detection through the appropriate analytical approach (discussed above), pre-processing of the raw signals is performed to generate the correct data format for subsequent data processing, in which data are normalized to minimize technical and system bias, and in untargeted approaches metabolites are identified thanks to the availability of spectral databases. Over the last two decades, even more advanced signal processing techniques for MS- and NMR-based metabolomics have been proposed, including software for peak detection and alignment (e.g. XCMS, MZmine2, Open-MS, MS-DIAL, eRah, ADAP-GC, BinBase), which enable spectral annotations and identifications from big and complex data.286–295 Statistical analysis, including univariate and multivariate analysis, is used to identify significantly expressed metabolites, which are then linked to a biological process by functional analysis performed with specialized tools that map metabolites to known biochemical pathways according to the information collected in public databases such as KEGG.296 Finally, metabolomics data may be integrated with other omics data (transcriptomics, proteomics, or the microbiome) to fully picture the complexity of a biological system.297,298 Thus, computational methods are involved in metabolomics at least at three levels: primary data production and conversion into standardized formats, bioinformatic analysis of data collections for prediction or classification, and integrative analysis of metabolomics with other omics data. While the first level is heavily linked to the specific technology used for data production, the second and the latter approaches are applicable within a more general analysis framework and have been increasingly employed to the field of precision medicine. Bioinformatics is the perfect tool for precision medicine since it enables the integration of the omics biomarkers recognized in individuals or cohorts of patients. According to the guidelines established by the US National Research Council, multilayered molecular or omics data should be deposited in the Information Commons, which is an easily accessible repository containing also all the clinical and epidemiological information, when available, enabling a complete analysis of the linkages between the data layers, thus generating a network of knowledge. This network permits a categorization of diseases in taxonomic ranks, thus allowing precise diagnosis and therapy design.299 Data processing and statistical analysis for metabolomics have been extensively reviewed300–302 and are not the scope of this manuscript. Like for several other high-throughput disciplines, these tools are quite mature, while the field of integrative and artificial intelligence-based analysis of multiple datasets still poses significant challenges,303 including the harmonization of data from different metabolomic platforms and their combination with data from other high throughput technologies.
Historically, genomics was the first high-throughput discipline in which bioinformatic analysis was used to stratify patients.304 Over the years, using data generated with different omics platforms, there was the development of molecular classifiers capable of discriminating between samples derived from patients and normal individuals thus allowing patient stratification. Nonetheless, all the studies tackling the development of classifiers face the problem of multiple comparisons, large number of parameters determined on a (relatively) small number of cases, and the need for extensive validation to avoid overfitting issues. Despite these challenges, studies producing molecular classifiers are appealing to researcher in the oncology field due to the need to subset patients’ cohorts in potential responders and non-responders to specific treatments.305 To overcome the technical issues related to molecular classifiers, integrative analysis of multiomics data and spatially and temporally dynamic sampling of tumours have been suggested and attempted.306,307
The vast majority of approaches to integrative multiomics data analysis relies heavily on machine learning. In the recent years, there has been a lot of hype on the use of Artificial intelligence (AI) for multilayered data analysis, which has somehow obscured the notion that machine learning, an AI implementation, has been around for several decades and have been at the core of omics data analysis from the beginning. Certainly, the huge amount of information produced by the “big data era” has pushed the application of these computational techniques even further. Machine learning has been classified primarily into supervised and unsupervised; the first one requires labelled data (training data) to allow the model to learn the “known” patterns underlying the labelled data and then discriminate these patterns in data not yet seen (test data); conversely, the second type of learning does not require labelled data and its goal is to distinguish the underlying unknown patterns from large datasets in an unbiased manner. The most employed supervised learning tools are neural networks, support vector machines, decision trees, random forests, and hidden Markov models. Classic examples of unsupervised methods are clustering algorithms, such as hierarchical agglomerative clustering, k-means clustering, Principal component analysis (PCA), self-organizing map, and non-negative matrix factorization. Machine learning has been largely used in precision medicine, and it is now the main tool employed at the core of “deep data integration” (i.e. integration of datasets from exploration of interdependent systems, like gene expression and transcription factor binding data)308 and “broad data integration” (i.e. integration of several omics and clinical data to derive high-level properties or subclassifications of a disease, like cancer).309 While computational algorithms are already known, one of the major challenges in their application is the generation of a multidisciplinary attitude between physicians, biologists and bioinformaticians. As outlined by Duerr-Specht et al., in order to improve precision medicine, it is now mandatory not only to improve the organization and the technologies in informatics and data management, but it is also necessary (in a high sense) to focus on training of researchers.303
Indeed, the issue of combining multiomics data is more linked to devising a proper strategy than to developing novel analytical tools. Association analysis and classifier development, in fact, have already been successfully employed to metabolomics data like it was previously done on genomic, transcriptomic, or proteomic data to discriminate tumour subtypes.310,311 Similar to genome-wide association studies, metabolomic analysis have been performed on biofluids or tissue with the aim of identifying markers of susceptibility to different diseases, including breast, prostate, lung, colorectal, ovarian, and pancreatic cancer.151,156,312–315 These studies, however, are restricted to a single dimension of the sample’s biology, require validation on independent cohorts and can only find associations between patterns and a particular physiopathological state, which is not proof of a cause-effect nexus. This is usually provided by searching for specific biomarkers previously identified by the association analysis in disease models, as exemplified by the work of Sreekumar et al. on human and mouse prostate cancer.146 As genomics, transcriptomics and proteomics, metabolomics can be targeted or untargeted, and applied on biofluids, tissues, cell lines or model organisms. This gives the chance to integrate different layers of data from each omics platform, and to move from association studies to the identification of causal relationship and complex interrelations between the different levels of regulation of gene expression and metabolism – and their dysregulation—in cancer. As a simple example, data from genetic profiling and subtyping derived from gene expression analysis can be employed to perform supervised machine learning and to cross-validate metabolic data. Deep data integration is used to combine datasets underlying a common regulatory network (e.g. gene expression, metabolic, and transcription factor data), whilst broad data integration can be performed by parallel integration of multiomics and clinical data.306,308,309 These approaches allow to step forward from single level biology to systems biology, revealing interactions between the different layers of data that can be further exploited in disease models and validated in prospective cohorts. To avoid the generation of misleading data, it is critical to deeply characterize the disease models; as an example of this caveat, a recent meta-analysis demonstrated that most of the studies using JCA-1 cell line referred to it as a prostate cancer cell line, while it was actually derived from a bladder carcinoma.316 The same caution must be applied to avoid strain variability in both cell lines and animal models,317 and in the harmonization of data/sample collection from patients cohorts and tissue and biofluids biobanks.
Conclusion and prospect
The intrinsically dynamic nature of cancer metabolism requires an equally dynamic research attitude.318 The recent technological and conceptual advances discussed in this review, from metabolomics to single-cell approaches, have significantly broaden the research scenario making cancer metabolism one of the most vibrant and prolific area of cancer biology research.3 Despite being the youngest and less employed omics technique, metabolomics is the technology that has mostly driven this change and it has demonstrated an enormous potential to influence the future of cancer research with concrete clinical applications in oncology. In particular, the analysis of biological fluids, which have an easy access and require minimal sample processing, is what is closest to be translated into a clinical setting and it is already widely employed for biomarker discovery, diagnosis, identification of new drug targets and for clinical trials monitoring.319,320 However, these analyses still require a deeper comprehension of how the read outs and the quantifications depict a realistic picture of the human physiology and to what extent a metabolite profile in biofluids reflects the metabolic milieu of the tumour. Moreover, metabolomic analyses and the underlying data processing present several challenges in the standardization and often there is not a unique approach, but each study is context-specific thus researchers should have deep multidisciplinary and computational education.321 Besides metabolomics, over the last 40 years, the development, the increased accessibility, power and resolution of new technologies have pushed cancer research as never before. In particular, the advent of high-throughput and high-content single-cell technologies provided unprecedented tools to investigate cancer biology at cellular resolution (Fig. 1). The upgrade from bulk to single-cell analysis enabled the researchers to face the pressing challenge of elucidating the complexity of heterogeneous diseases, like cancer, and the underlying molecular pathways driving them thus describing the cancer framework in depth with both a quantitative and qualitative analysis. Caution must be still taken because none of the above-mentioned technologies, if employed alone, can completely analyse the metabolic status of a tissue. The choice of the most appropriate technique to extricate oneself from a complex biological system, such as TME, must always balance between sensitivity and resolution, cost and feasibility (Table 2). These metabolic tools should be employed in a complementary way, merging descriptive research to a functional study, thus paving the way to innovative approaches to research, diagnosis, and therapies in cancer. By combining these techniques—and thus exploiting several levels of orthogonality—we will be able to expand our understanding of the tumour and pave the way for new anti-cancer strategies. The biggest challenge, which everyone working in field is currently facing, is the lack of interlaboratory harmonization strategies for analytical procedures, which must be one of the next goals in the field in order to implement the computational tools for the development of open-source databases and ultimately advance metabolic studies in cancer research.
Table 2.
Pros, cons and sample types of the main metabolomic technologies for the analysis of cancer metabolism
| Method | Pros | Cons | Sample types |
|---|---|---|---|
| Gas chromatography-Mass spectrometry (GC-MS) |
• High sensitivity for volatile metabolites • High-resolution separation • Analysis of different groups of metabolites simultaneously • Large linear range |
• Long sample preparation (derivatization step for non-volatile metabolites) • Thermolabile compounds cannot be analysed • Slow dynamic range speed • Slow analysis |
• Cultured cells • Supernatant • Biofluids • Tissues • Organoids |
| Liquid chromatography-Mass spectrometry (LC-MS) |
• Simple and fast sample preparation (derivatization not usually required) • Wide coverage of metabolites • Thermolabile compounds can be analysed • High sensitivity • Soft ionization |
• Ion suppression • Expensive • Slow analysis |
• Cultured cells • Supernatant • Biofluids • Tissues • Organoids |
| Capillary electrophoresis-Mass spectrometry (CE-MS) |
• Low sample volume • High resolution • Rapid analysis • No derivatization required |
• Affected by salt • Low stability compared to GM- and LC-MS • Poor reproducibility and sensitivity |
• Cultured cells • Supernatant • Biofluids • Tissues • Organoids |
| Direct infusion-Mass spectrometry (DI-MS) |
• High-throughput • Simple data processing |
• Do not distinguish the isomers |
• Supernatant • Biofluids |
| Matrix-assisted laser desorption ionization-Mass spectrometry (MALDI-MS) |
• Low sample volume • Fast analysis • High tolerance towards salts • Suitable for high MW metabolites • Non-destructive |
• Low reproducibility • Hard identification due to complex matrix |
• Cultured cells • Supernatant • Biofluids • Tissues • Organoids |
| Mass spectrometry imaging (MSI) |
• In situ detection • Preserve histological integrity |
• High resolution is time-consuming • No functional profile |
• Cultured cells • Tissues • Organoids |
| Direct real-time analysis (DART) |
• No sample processing • Direct analysis |
• Not suitable for polar compounds |
• Supernatant • Biofluids |
| Nuclear magnetic resonance (NMR) |
• No separation • Structural information • High reproducibility • Fast sample preparation • Non-destructive |
• Low sensitivity • Expensive instrument • Some chemical classes are not detected |
• Cultured cells • Supernatant • Biofluids • Tissues • Organoids |
| Metabolic flux analysis (MFA) | • Quantitative analysis and information on metabolites fate |
• Isotope tracing is expensive • Compartment specific flux |
• Cultured cells • Tissues • Organoids |
| Extracellular flux analysis (EFA) |
• Real time measurement • High feasibility • Relatively cheap |
• Bulk analysis • Only relative and indirect measurement • Cell purification is required |
• Cultured cells • Organoids |
| Single-cell RNA-sequencing (scRNAseq) |
• Low cell number • High-resolution • Unbiased gene expression analysis • Metabolic phenotype at mRNA level |
• Expensive • Temporal discordance between mRNA and protein/functional effect • Do not consider post-transcriptional and post-translational mechanisms |
• Cultured cells • Tissues • Organoids |
| Single-cell metabolomics (SCM) |
• Low cell number • High resolution • High-throughput |
• Challenge of combining single cells sorting and metabolism quenching • Need of high sensitivity and throughput analytical platform |
• Cultured cells • Tissues • Organoids |
| Single-cell energetic metabolism by profiling translation inhibition (SCENITH) |
• Functional analysis coupled to large phenotype • Fast and simple sample preparation and analysis |
• Only relative and indirect measurement • Not suitable for cells with undetectable level of protein synthesis |
• Cultured cells • Tissues • Organoids |
| Cytometry by Time-of-flight (CyTOF) |
• High-dimensional • High-throughput • Metabolic phenotype at protein level |
• Not suitable for weakly expressed markers • Requires advanced biostatistics and bioinformatics |
• Cultured cells • Tissues • Organoids |
| Met-flow |
• Fast and single-cell analysis • Metabolic phenotype at protein level |
• Measurements are indirect • No functional profile |
• Cultured cells • Tissues • Organoids |
| In situ dehydrogenase activity assay |
• Single-cell analysis in native microenvironment • Functional profile |
• Measurement at saturated substrate concentrations |
• Cultured cells • Tissues • Organoids |
| Genetic screening |
• Precise gene targeting (few off-targets) • Robust signal derived by permanent gene disruption |
• Complicated to perform • For some types of studies, it is not good having a permanent gene disruption |
• Cultured cells • Organoids |
Acknowledgements
All figures were created with Biorender.com. This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC, Project: 27080), by NextGenerationEU (PNRR “HEAL ITALIA - Health Extended Alliance for Innovative Therapies, Advanced Lab-research, and Integrated Approaches of Precision Medicine”, project: PE00000019, CUP: B33C22001030006), Directorial Decree No. 1559 of 11 October 2022 and by Italian Ministry of Health, HUB Diagnostica Avanzata PNC-E3-2022-23683266 PNCHLS-DA.
Author contributions
F.D., M.D. and A.F. conceptualization; F.D., A.F and A.M. writing original draft, table and figures preparation; R.P., M.T.S, A.S. and M.D revised the manuscript; all authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Massimo Donadelli, Alessandra Fiore
References
- 1.Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science368, eaaw5473 (2020). [DOI] [PMC free article] [PubMed]
- 2.Fendt SM, Frezza C, Erez A. Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 2020;10:1797–1807. doi: 10.1158/2159-8290.CD-20-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stine ZE, Schug ZT, Salvino JM, Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022;21:141–162. doi: 10.1038/s41573-021-00339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat. Rev. Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 8.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascale, R. M., Calvisi, D. F., Simile, M. M., Feo, C. F. & Feo, F. The Warburg Effect 97 Years after Its Discovery. Cancers (Basel)12, 2819 (2020). [DOI] [PMC free article] [PubMed]
- 10.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 11.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donati G, Amati B. MYC and therapy resistance in cancer: risks and opportunities. Mol. Oncol. 2022;16:3828–3854. doi: 10.1002/1878-0261.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marei HE, et al. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021;21:703. doi: 10.1186/s12935-021-02396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Leary K. Tracing the origins of KRAS oncogene addiction. Nat. Rev. Cancer. 2021;21:69. doi: 10.1038/s41568-020-00325-x. [DOI] [PubMed] [Google Scholar]
- 15.Felsher DW. Cancer revoked: oncogenes as therapeutic targets. Nat. Rev. Cancer. 2003;3:375–380. doi: 10.1038/nrc1070. [DOI] [PubMed] [Google Scholar]
- 16.Groelly, F. J., Fawkes, M., Dagg, R. A., Blackford, A. N. & Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer23, 78–94 (2022). [DOI] [PubMed]
- 17.Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez A, Hall MN, Lin SC, Hardie DG. AMPK and TOR: The Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 2020;31:472–492. doi: 10.1016/j.cmet.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, DeBerardinis RJ. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 2019;30:434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seyfried TN, Arismendi-Morillo G, Mukherjee P, Chinopoulos C. On the origin of ATP synthesis in cancer. iScience. 2020;23:101761. doi: 10.1016/j.isci.2020.101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer. Oncogene. 2021;40:3351–3363. doi: 10.1038/s41388-020-01639-8. [DOI] [PubMed] [Google Scholar]
- 23.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang MS, et al. Hypoxia-induced macropinocytosis represents a metabolic route for liver cancer. Nat. Commun. 2022;13:954. doi: 10.1038/s41467-022-28618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song S, Zhang Y, Ding T, Ji N, Zhao H. The dual role of macropinocytosis in cancers: promoting growth and inducing methuosis to participate in anticancer therapies as targets. Front. Oncol. 2020;10:570108. doi: 10.3389/fonc.2020.570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F, et al. Macropinocytosis: mechanism and targeted therapy in cancers. Am. J. Cancer Res. 2021;11:14–30. [PMC free article] [PubMed] [Google Scholar]
- 28.Jayashankar V, Edinger AL. Macropinocytosis confers resistance to therapies targeting cancer anabolism. Nat. Commun. 2020;11:1121. doi: 10.1038/s41467-020-14928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H, et al. Cancer cells escape autophagy inhibition via NRF2-induced macropinocytosis. Cancer Cell. 2021;39:678–693.e611. doi: 10.1016/j.ccell.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mlynarczuk-Bialy, I. et al. Entosis: from cell biology to clinical cancer pathology. Cancers (Basel)12, 2481 (2020). [DOI] [PMC free article] [PubMed]
- 31.Koren E, Fuchs Y. Modes of regulated cell death in cancer. Cancer Discov. 2021;11:245–265. doi: 10.1158/2159-8290.CD-20-0789. [DOI] [PubMed] [Google Scholar]
- 32.Kodama M, et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat. Commun. 2020;11:1320. doi: 10.1038/s41467-020-15136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurmi K, Haigis MC. Nitrogen metabolism in cancer and immunity. Trends Cell Biol. 2020;30:408–424. doi: 10.1016/j.tcb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandel NS, Tuveson DA. The promise and perils of antioxidants for cancer patients. N. Engl. J. Med. 2014;371:177–178. doi: 10.1056/NEJMcibr1405701. [DOI] [PubMed] [Google Scholar]
- 35.Xia C, et al. Reactive oxygen species regulate angiogenesis and tumor growth through vascular endothelial growth factor. Cancer Res. 2007;67:10823–10830. doi: 10.1158/0008-5472.CAN-07-0783. [DOI] [PubMed] [Google Scholar]
- 36.Tasdogan A, Ubellacker JM, Morrison SJ. Redox regulation in cancer cells during metastasis. Cancer Discov. 2021;11:2682–2692. doi: 10.1158/2159-8290.CD-21-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis. 2016;7:e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the Hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perillo B, et al. ROS in cancer therapy: the bright side of the moon. Exp. Mol. Med. 2020;52:192–203. doi: 10.1038/s12276-020-0384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forman HJ, Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021;20:689–709. doi: 10.1038/s41573-021-00233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavallini C, et al. Low catalase expression confers redox hypersensitivity and identifies an indolent clinical behavior in CLL. Blood. 2018;131:1942–1954. doi: 10.1182/blood-2017-08-800466. [DOI] [PubMed] [Google Scholar]
- 42.Fiore A, et al. Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling. Mol. Cell. 2022;82:920–932.e927. doi: 10.1016/j.molcel.2022.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Zeitler, L. et al. Anti-ferroptotic mechanism of IL4i1-mediated amino acid metabolism. Elife10, e64806 (2021). [DOI] [PMC free article] [PubMed]
- 44.Bernstock JD, et al. Targeting oncometabolism to maximize immunotherapy in malignant brain tumors. Oncogene. 2022;41:2663–2671. doi: 10.1038/s41388-022-02312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alseekh S, et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat. Methods. 2021;18:747–756. doi: 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dando I, et al. Oncometabolites in cancer aggressiveness and tumour repopulation. Biol. Rev. Camb. Philos. Soc. 2019;94:1530–1546. doi: 10.1111/brv.12513. [DOI] [PubMed] [Google Scholar]
- 49.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 50.Farber S. Some observations on the effect of folic acid antagonists on acute leukemia and other forms of incurable cancer. Blood. 1949;4:160–167. doi: 10.1182/blood.V4.2.160.160. [DOI] [PubMed] [Google Scholar]
- 51.Farber S. Chemotherapeutic studies of tumors, including leukemia, in children. Am. J. Dis. Child. 1950;79:961–962. [PubMed] [Google Scholar]
- 52.Djerassi I, Farber S, Abir E, Neikirk W. Continuous infusion of methotrexate in children with acute leukemia. Cancer. 1967;20:233–242. doi: 10.1002/1097-0142(1967)20:2<233::AID-CNCR2820200209>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 53.Frei E, 3rd, Jaffe N, Farber S. Treatment of acute leukemia. N. Engl. J. Med. 1972;287:1357. doi: 10.1056/NEJM197212282872618. [DOI] [PubMed] [Google Scholar]
- 54.Miller DR. A tribute to Sidney Farber—the father of modern chemotherapy. Br. J. Haematol. 2006;134:20–26. doi: 10.1111/j.1365-2141.2006.06119.x. [DOI] [PubMed] [Google Scholar]
- 55.Willson J. Structural study could aid design of antifolates. Nat. Rev. Cancer. 2022;22:608. doi: 10.1038/s41568-022-00523-9. [DOI] [PubMed] [Google Scholar]
- 56.Lambie DG, Johnson RH. Drugs and folate metabolism. Drugs. 1985;30:145–155. doi: 10.2165/00003495-198530020-00003. [DOI] [PubMed] [Google Scholar]
- 57.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 58.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 59.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/S0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 60.Kell DB, Oliver SG. The metabolome 18 years on: a concept comes of age. Metabolomics. 2016;12:148. doi: 10.1007/s11306-016-1108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.German JB, Hammock BD, Watkins SM. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics. 2005;1:3–9. doi: 10.1007/s11306-005-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han J, Li Q, Chen Y, Yang Y. Recent metabolomics analysis in tumor metabolism reprogramming. Front Mol. Biosci. 2021;8:763902. doi: 10.3389/fmolb.2021.763902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt DR, et al. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021;71:333–358. doi: 10.3322/caac.21670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wishart DS, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wishart DS, et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wishart DS, et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wishart, D. S. et al. NMR and metabolomics—a roadmap for the future. Metabolites12, 678 (2022). [DOI] [PMC free article] [PubMed]
- 70.Alarcon-Barrera JC, Kostidis S, Ondo-Mendez A, Giera M. Recent advances in metabolomics analysis for early drug development. Drug Discov. Today. 2022;27:1763–1773. doi: 10.1016/j.drudis.2022.02.018. [DOI] [PubMed] [Google Scholar]
- 71.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 72.Kang YP, Ward NP, DeNicola GM. Recent advances in cancer metabolism: a technological perspective. Exp. Mol. Med. 2018;50:1–16. doi: 10.1038/s12276-018-0027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Psychogios N, et al. The human serum metabolome. PLoS ONE. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu W, et al. Metabolite measurement: pitfalls to avoid and practices to follow. Annu. Rev. Biochem. 2017;86:277–304. doi: 10.1146/annurev-biochem-061516-044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeki OC, Eylem CC, Recber T, Kir S, Nemutlu E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020;190:113509. doi: 10.1016/j.jpba.2020.113509. [DOI] [PubMed] [Google Scholar]
- 76.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016;27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ribbenstedt A, Ziarrusta H, Benskin JP. Development, characterization and comparisons of targeted and non-targeted metabolomics methods. PLoS ONE. 2018;13:e0207082. doi: 10.1371/journal.pone.0207082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Theodoridis G, Gika HG, Wilson ID. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011;30:884–906. doi: 10.1002/mas.20306. [DOI] [PubMed] [Google Scholar]
- 79.Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/C1AN15605E. [DOI] [PubMed] [Google Scholar]
- 80.Kumari S, Stevens D, Kind T, Denkert C, Fiehn O. Applying in-silico retention index and mass spectra matching for identification of unknown metabolites in accurate mass GC-TOF mass spectrometry. Anal. Chem. 2011;83:5895–5902. doi: 10.1021/ac2006137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fiehn O, et al. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS ONE. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fiehn O, Kopka J, Trethewey RN, Willmitzer L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 2000;72:3573–3580. doi: 10.1021/ac991142i. [DOI] [PubMed] [Google Scholar]
- 83.Laine RA, Sweeley CC. Analysis of trimethylsilyl O-methyloximes of carbohydrates by combined gas-liquid chromatography-mass spectrometry. Anal. Biochem. 1971;43:533–538. doi: 10.1016/0003-2697(71)90284-3. [DOI] [PubMed] [Google Scholar]
- 84.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016;114:30 34 31–30 34 32. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beale DJ, et al. Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics. 2018;14:152. doi: 10.1007/s11306-018-1449-2. [DOI] [PubMed] [Google Scholar]
- 86.Wong YF, Hartmann C, P JM. Multidimensional gas chromatography methods for bioanalytical research. Bioanalysis. 2014;6:2461–2479. doi: 10.4155/bio.14.186. [DOI] [PubMed] [Google Scholar]
- 87.Mostafa A, Edwards M, Gorecki T. Optimization aspects of comprehensive two-dimensional gas chromatography. J. Chromatogr. A. 2012;1255:38–55. doi: 10.1016/j.chroma.2012.02.064. [DOI] [PubMed] [Google Scholar]
- 88.Forcisi S, et al. Liquid chromatography-mass spectrometry in metabolomics research: mass analyzers in ultra high pressure liquid chromatography coupling. J. Chromatogr. A. 2013;1292:51–65. doi: 10.1016/j.chroma.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 89.Ren JL, Zhang AH, Kong L, Wang XJ. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018;8:22335–22350. doi: 10.1039/C8RA01574K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maier TV, Schmitt-Kopplin P. Capillary electrophoresis in metabolomics. Methods Mol. Biol. 2016;1483:437–470. doi: 10.1007/978-1-4939-6403-1_21. [DOI] [PubMed] [Google Scholar]
- 91.Stolz A, et al. Recent advances in capillary electrophoresis-mass spectrometry: instrumentation, methodology and applications. Electrophoresis. 2019;40:79–112. doi: 10.1002/elps.201800331. [DOI] [PubMed] [Google Scholar]
- 92.Zhang W, Hankemeier T, Ramautar R. Next-generation capillary electrophoresis-mass spectrometry approaches in metabolomics. Curr. Opin. Biotechnol. 2017;43:1–7. doi: 10.1016/j.copbio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 93.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Metabolic differentiation of neuronal phenotypes by single-cell capillary electrophoresis-electrospray ionization-mass spectrometry. Anal. Chem. 2011;83:6810–6817. doi: 10.1021/ac2015855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ubhi BK. Direct infusion-tandem mass spectrometry (DI-MS/MS) analysis of complex lipids in human plasma and serum using the Lipidyzer platform. Methods Mol. Biol. 2018;1730:227–236. doi: 10.1007/978-1-4939-7592-1_15. [DOI] [PubMed] [Google Scholar]
- 95.Helmeczi E, et al. A high-throughput platform for the rapid screening of vitamin D status by direct infusion-MS/MS. J. Lipid Res. 2022;63:100204. doi: 10.1016/j.jlr.2022.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lisa M, Cifkova E, Khalikova M, Ovcacikova M, Holcapek M. Lipidomic analysis of biological samples: Comparison of liquid chromatography, supercritical fluid chromatography and direct infusion mass spectrometry methods. J. Chromatogr. A. 2017;1525:96–108. doi: 10.1016/j.chroma.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 97.Karas M, Kruger R. Ion formation in MALDI: the cluster ionization mechanism. Chem. Rev. 2003;103:427–440. doi: 10.1021/cr010376a. [DOI] [PubMed] [Google Scholar]
- 98.Finehout EJ, Lee KH. An introduction to mass spectrometry applications in biological research. Biochem. Mol. Biol. Educ. 2004;32:93–100. doi: 10.1002/bmb.2004.494032020331. [DOI] [PubMed] [Google Scholar]
- 99.McLafferty FW. A century of progress in molecular mass spectrometry. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2011;4:1–22. doi: 10.1146/annurev-anchem-061010-114018. [DOI] [PubMed] [Google Scholar]
- 100.Willard, H. H. Instrumental Methods Of Analysis 7th edn (Wadsworth Pub. Co., 1988).
- 101.Laeter, J. R. d. Applications of Inorganic Mass Spectrometry (Wiley, 2001).
- 102.Steinhauser D, Kopka J. Methods, applications and concepts of metabolite profiling: primary metabolism. EXS. 2007;97:171–194. doi: 10.1007/978-3-7643-7439-6_8. [DOI] [PubMed] [Google Scholar]
- 103.Capellades J, et al. Exploring the use of gas chromatography coupled to chemical ionization mass spectrometry (GC-CI-MS) for stable isotope labeling in metabolomics. Anal. Chem. 2021;93:1242–1248. doi: 10.1021/acs.analchem.0c02998. [DOI] [PubMed] [Google Scholar]
- 104.Byrdwell WC. Atmospheric pressure chemical ionization mass spectrometry for analysis of lipids. Lipids. 2001;36:327–346. doi: 10.1007/s11745-001-0725-5. [DOI] [PubMed] [Google Scholar]
- 105.Harris KJ, et al. Pressurized liquid extraction followed by liquid chromatography coupled to a fluorescence detector and atmospheric pressure chemical ionization mass spectrometry for the determination of benzo(a)pyrene metabolites in liver tissue of an animal model of colon cancer. J. Chromatogr. A. 2020;1622:461126. doi: 10.1016/j.chroma.2020.461126. [DOI] [PubMed] [Google Scholar]
- 106.Li KM, Rivory LP, Clarke SJ. Rapid quantitation of plasma 2’-deoxyuridine by high-performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry and its application to pharmacodynamic studies in cancer patients. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2005;820:121–130. doi: 10.1016/j.jchromb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 107.Yin L, Zhang Z, Liu Y, Gao Y, Gu J. Recent advances in single-cell analysis by mass spectrometry. Analyst. 2019;144:824–845. doi: 10.1039/C8AN01190G. [DOI] [PubMed] [Google Scholar]
- 108.Chetwynd AJ, David A. A review of nanoscale LC-ESI for metabolomics and its potential to enhance the metabolome coverage. Talanta. 2018;182:380–390. doi: 10.1016/j.talanta.2018.01.084. [DOI] [PubMed] [Google Scholar]
- 109.Rathore R, Corr J, Scott G, Vollmerhaus P, Greis KD. Development of an inhibitor screening platform via mass spectrometry. J. Biomol. Screen. 2008;13:1007–1013. doi: 10.1177/1087057108326143. [DOI] [PubMed] [Google Scholar]
- 110.Ravi VM, et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell. 2022;40:639–655.e613. doi: 10.1016/j.ccell.2022.05.009. [DOI] [PubMed] [Google Scholar]
- 111.Aichler M, Walch A. MALDI Imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Lab Invest. 2015;95:422–431. doi: 10.1038/labinvest.2014.156. [DOI] [PubMed] [Google Scholar]
- 112.Sun, N. et al. Pharmacometabolic response to pirfenidone in pulmonary fibrosis detected by MALDI-FTICR-MSI. Eur. Respir. J.52, 1702314 (2018). [DOI] [PubMed]
- 113.Aichler M, et al. N-acyl taurines and acylcarnitines cause an imbalance in insulin synthesis and secretion provoking beta cell dysfunction in type 2 diabetes. Cell Metab. 2017;25:1334–1347.e1334. doi: 10.1016/j.cmet.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 114.Han Z, et al. Matrix-assisted laser desorption ionization mass spectrometry profiling of plasma exosomes evaluates osteosarcoma metastasis. iScience. 2021;24:102906. doi: 10.1016/j.isci.2021.102906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gu H, et al. Principal component directed partial least squares analysis for combining nuclear magnetic resonance and mass spectrometry data in metabolomics: application to the detection of breast cancer. Anal. Chim. Acta. 2011;686:57–63. doi: 10.1016/j.aca.2010.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gowda GA, Djukovic D. Overview of mass spectrometry-based metabolomics: opportunities and challenges. Methods Mol. Biol. 2014;1198:3–12. doi: 10.1007/978-1-4939-1258-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miller PE, Denton MB. The quadrupole mass filter—basic operating concepts. J. Chem. Educ. 1986;63:617–622. doi: 10.1021/ed063p617. [DOI] [Google Scholar]
- 118.March RE. An introduction to quadrupole ion trap mass spectrometry. J. Mass Spectrom. 1997;32:351–369. doi: 10.1002/(SICI)1096-9888(199704)32:4<351::AID-JMS512>3.0.CO;2-Y. [DOI] [Google Scholar]
- 119.Cotter RJ. Time-of-flight mass spectrometry: an increasing role in the life sciences. Biomed. Environ. Mass Spectrom. 1989;18:513–532. doi: 10.1002/bms.1200180803. [DOI] [PubMed] [Google Scholar]
- 120.Comisarow MB, Marshall AG. The early development of Fourier transform ion cyclotron resonance (FT-ICR) spectroscopy. J. Mass Spectrom. 1996;31:581–585. doi: 10.1002/(SICI)1096-9888(199606)31:6<581::AID-JMS369>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 121.Scigelova M, Hornshaw M, Giannakopulos A, Makarov A. Fourier transform mass spectrometry. Mol. Cell Proteom. 2011;10:M111 009431. doi: 10.1074/mcp.M111.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hu Q, et al. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 123.Makarov A. Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 2000;72:1156–1162. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 124.deHoffmann E. Tandem mass spectrometry: a primer. J. Mass Spectrom. 1996;31:129–137. doi: 10.1002/(SICI)1096-9888(199602)31:2<129::AID-JMS305>3.0.CO;2-T. [DOI] [Google Scholar]
- 125.Yang Q, et al. Metabolomics biotechnology, applications, and future trends: a systematic review. RSC Adv. 2019;9:37245–37257. doi: 10.1039/C9RA06697G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lopes AS, Cruz EC, Sussulini A, Klassen A. Metabolomic strategies involving mass spectrometry combined with liquid and gas chromatography. Adv. Exp. Med. Biol. 2017;965:77–98. doi: 10.1007/978-3-319-47656-8_4. [DOI] [PubMed] [Google Scholar]
- 127.Rabi, I. I., Zacharias, J. R., Millman, S. & Kusch, P. Milestones in magnetic resonance: ‘a new method of measuring nuclear magnetic moment’. 1938. J. Magn. Reson Imaging 2, 131–133 (1992). [DOI] [PubMed]
- 128.Zaimenko I, Lisec J, Stein U, Brenner W. Approaches and techniques to characterize cancer metabolism in vitro and in vivo. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:412–419. doi: 10.1016/j.bbcan.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 129.Emwas, A. H. et al. NMR spectroscopy for metabolomics research. Metabolites9, 123 (2019). [DOI] [PMC free article] [PubMed]
- 130.Tiziani S, Lopes V, Gunther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11:269–276. doi: 10.1593/neo.81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shanaiah N, et al. Class selection of amino acid metabolites in body fluids using chemical derivatization and their enhanced 13C NMR. Proc. Natl Acad. Sci. USA. 2007;104:11540–11544. doi: 10.1073/pnas.0704449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Daly PF, Lyon RC, Faustino PJ, Cohen JS. Phospholipid metabolism in cancer cells monitored by 31P NMR spectroscopy. J. Biol. Chem. 1987;262:14875–14878. doi: 10.1016/S0021-9258(18)48107-0. [DOI] [PubMed] [Google Scholar]
- 133.He X, et al. NMR-based metabolomics analysis predicts response to neoadjuvant chemotherapy for triple-negative breast cancer. Front. Mol. Biosci. 2021;8:708052. doi: 10.3389/fmolb.2021.708052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beckonert O, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Protoc. 2010;5:1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 135.Larive CK, Barding GA, Jr., Dinges MM. NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 2015;87:133–146. doi: 10.1021/ac504075g. [DOI] [PubMed] [Google Scholar]
- 136.Jang C, Chen L, Rabinowitz JD. Metabolomics and isotope tracing. Cell. 2018;173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol. Cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen L, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat. Metab. 2019;1:404–415. doi: 10.1038/s42255-019-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu L, et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 2018;27:1067–1080.e1065. doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 142.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pandey R, Caflisch L, Lodi A, Brenner AJ, Tiziani S. Metabolomic signature of brain cancer. Mol. Carcinog. 2017;56:2355–2371. doi: 10.1002/mc.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Banerjee S. Empowering clinical diagnostics with mass spectrometry. ACS Omega. 2020;5:2041–2048. doi: 10.1021/acsomega.9b03764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fang, C. et al. Metabolic reprogramming and risk stratification of hepatocellular carcinoma studied by using gas chromatography-mass spectrometry-based metabolomics. Cancers (Basel)14, 231 (2022). [DOI] [PMC free article] [PubMed]
- 146.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 147.Luo X, Liu J, Wang H, Lu H. Metabolomics identified new biomarkers for the precise diagnosis of pancreatic cancer and associated tissue metastasis. Pharm. Res. 2020;156:104805. doi: 10.1016/j.phrs.2020.104805. [DOI] [PubMed] [Google Scholar]
- 148.Tao L, et al. Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics. 2019;15:86. doi: 10.1007/s11306-019-1550-1. [DOI] [PubMed] [Google Scholar]
- 149.His M, et al. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019;17:178. doi: 10.1186/s12916-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu, H. et al. Polyamine metabolites profiling for characterization of lung and liver cancer using an LC-tandem MS method with multiple statistical data mining strategies: discovering potential cancer biomarkers in human plasma and urine. Molecules21, 1040 (2016). [DOI] [PMC free article] [PubMed]
- 151.Mayerle J, et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut. 2018;67:128–137. doi: 10.1136/gutjnl-2016-312432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu X, Ao H, Gao H, Zhu Z. Metabolite biomarker discovery for human gastric cancer using dried blood spot mass spectrometry metabolomic approach. Sci. Rep. 2022;12:14632. doi: 10.1038/s41598-022-19061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ostman JR, et al. Identification of prediagnostic metabolites associated with prostate cancer risk by untargeted mass spectrometry-based metabolomics: a case-control study nested in the Northern Sweden Health and Disease Study. Int. J. Cancer. 2022;151:2115–2127. doi: 10.1002/ijc.34223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hang D, et al. Plasma metabolomic profiles for colorectal cancer precursors in women. Eur. J. Epidemiol. 2022;37:413–422. doi: 10.1007/s10654-021-00834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang J, et al. Prospective serum metabolomic profiling of lethal prostate cancer. Int J. Cancer. 2019;145:3231–3243. doi: 10.1002/ijc.32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zeleznik OA, et al. A prospective analysis of circulating plasma metabolites associated with ovarian cancer risk. Cancer Res. 2020;80:1357–1367. doi: 10.1158/0008-5472.CAN-19-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Omran MM, Rashed RE, Darwish H, Belal AA, Mohamed FZ. Development of a gas chromatography-mass spectrometry method for breast cancer diagnosis based on nucleoside metabolomes 1-methyl adenosine, 1-methylguanosine and 8-hydroxy-2’-deoxyguanosine. Biomed. Chromatogr. 2020;34:e4713. doi: 10.1002/bmc.4713. [DOI] [PubMed] [Google Scholar]
- 158.Callejon-Leblic B, Garcia-Barrera T, Pereira-Vega A, Gomez-Ariza JL. Metabolomic study of serum, urine and bronchoalveolar lavage fluid based on gas chromatography mass spectrometry to delve into the pathology of lung cancer. J. Pharm. Biomed. Anal. 2019;163:122–129. doi: 10.1016/j.jpba.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 159.Huang Z, et al. Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol. Cell Proteom. 2011;10:M111 007922. doi: 10.1074/mcp.M111.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang T, et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J. Proteome Res. 2013;12:505–512. doi: 10.1021/pr3009572. [DOI] [PubMed] [Google Scholar]
- 161.Murata T, et al. Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res. Treat. 2019;177:591–601. doi: 10.1007/s10549-019-05330-9. [DOI] [PubMed] [Google Scholar]
- 162.Asai, Y. et al. Elevated polyamines in saliva of pancreatic cancer. Cancers (Basel)10, 43 (2018). [DOI] [PMC free article] [PubMed]
- 163.Ishikawa S, et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 2016;6:31520. doi: 10.1038/srep31520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Soini HA, et al. Analysis of volatile organic compounds in human saliva by a static sorptive extraction method and gas chromatography-mass spectrometry. J. Chem. Ecol. 2010;36:1035–1042. doi: 10.1007/s10886-010-9846-7. [DOI] [PubMed] [Google Scholar]
- 165.Nakamizo S, et al. GC/MS-based metabolomic analysis of cerebrospinal fluid (CSF) from glioma patients. J. Neurooncol. 2013;113:65–74. doi: 10.1007/s11060-013-1090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Reichl, B. et al. Determination of a tumor-promoting microenvironment in recurrent medulloblastoma: a multi-omics study of cerebrospinal fluid. Cancers (Basel)12, 1350 (2020). [DOI] [PMC free article] [PubMed]
- 167.Wang FX, et al. Cerebrospinal fluid-based metabolomics to characterize different types of brain tumors. J. Neurol. 2020;267:984–993. doi: 10.1007/s00415-019-09665-7. [DOI] [PubMed] [Google Scholar]
- 168.Li T, et al. In situ biomarker discovery and label-free molecular histopathological diagnosis of lung cancer by ambient mass spectrometry imaging. Sci. Rep. 2015;5:14089. doi: 10.1038/srep14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Qi SA, et al. High-resolution metabolomic biomarkers for lung cancer diagnosis and prognosis. Sci. Rep. 2021;11:11805. doi: 10.1038/s41598-021-91276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Saorin, A., Di Gregorio, E., Miolo, G., Steffan, A. & Corona, G. Emerging role of metabolomics in ovarian cancer diagnosis. Metabolites10, 419 (2020). [DOI] [PMC free article] [PubMed]
- 171.Grooms AJ, Burris BJ, Badu-Tawiah AK. Mass spectrometry for metabolomics analysis: Applications in neonatal and cancer screening. Mass Spectrom. Rev. 2022;15:e21826. doi: 10.1002/mas.21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang J, et al. Spatial metabolomics identifies distinct tumor-specific subtypes in gastric cancer patients. Clin. Cancer Res. 2022;28:2865–2877. doi: 10.1158/1078-0432.CCR-21-4383. [DOI] [PubMed] [Google Scholar]
- 173.Fan Y, et al. Human plasma metabolomics for identifying differential metabolites and predicting molecular subtypes of breast cancer. Oncotarget. 2016;7:9925–9938. doi: 10.18632/oncotarget.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xiao Y, et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res. 2022;32:477–490. doi: 10.1038/s41422-022-00614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lokshina LA, Solov’eva NI, Orekhovich VN. The role of lysosomal proteinases in tissue destruction. Vopr. Med. Khim. 1987;33:38–43. [PubMed] [Google Scholar]
- 176.Backshall A, Sharma R, Clarke SJ, Keun HC. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin. Cancer Res. 2011;17:3019–3028. doi: 10.1158/1078-0432.CCR-10-2474. [DOI] [PubMed] [Google Scholar]
- 177.Kim KB, et al. Potential metabolomic biomarkers for evaluation of adriamycin efficacy using a urinary 1H-NMR spectroscopy. J. Appl. Toxicol. 2013;33:1251–1259. doi: 10.1002/jat.2778. [DOI] [PubMed] [Google Scholar]
- 178.Zhang Y, et al. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 2019;26:623–633.e629. doi: 10.1016/j.chembiol.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Obrist, F. et al. Metabolic vulnerability of cisplatin-resistant cancers. EMBO J.37 (2018). [DOI] [PMC free article] [PubMed]
- 180.A J, et al. Chronic myeloid leukemia patients sensitive and resistant to imatinib treatment show different metabolic responses. PLoS ONE. 2010;5:e13186. doi: 10.1371/journal.pone.0013186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Poisson LM, et al. A metabolomic approach to identifying platinum resistance in ovarian cancer. J. Ovarian Res. 2015;8:13. doi: 10.1186/s13048-015-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.DePeaux K, Delgoffe GM. Metabolic barriers to cancer immunotherapy. Nat. Rev. Immunol. 2021;21:785–797. doi: 10.1038/s41577-021-00541-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tang Y, Kang Y. Microbial metabolite as icebreaker for immunotherapy. Cell Metab. 2022;34:506–507. doi: 10.1016/j.cmet.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 184.Bird L. Microbial metabolite boosts immunotherapy. Nat. Rev. Immunol. 2020;20:648–649. doi: 10.1038/s41577-020-00465-z. [DOI] [PubMed] [Google Scholar]
- 185.Dastmalchi F, Deleyrolle LP, Karachi A, Mitchell DA, Rahman M. Metabolomics Monitoring of Treatment Response to Brain Tumor Immunotherapy. Front. Oncol. 2021;11:691246. doi: 10.3389/fonc.2021.691246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yoon, S. J., Lee, C. B., Chae, S. U., Jo, S. J. & Bae, S. K. The comprehensive “omics” approach from metabolomics to advanced omics for development of immune checkpoint inhibitors: potential strategies for next generation of cancer immunotherapy. Int. J. Mol. Sci.22, 6281 (2021). [DOI] [PMC free article] [PubMed]
- 187.Li H, et al. Metabolomic adaptations and correlates of survival to immune checkpoint blockade. Nat. Commun. 2019;10:4346. doi: 10.1038/s41467-019-12361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nie X, et al. Serum metabolite biomarkers predictive of response to PD-1 blockade therapy in non-small cell lung cancer. Front Mol. Biosci. 2021;8:678753. doi: 10.3389/fmolb.2021.678753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Botticelli A, et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med. 2020;18:49. doi: 10.1186/s12967-020-02231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bishop SL, et al. Moving beyond descriptive studies: harnessing metabolomics to elucidate the molecular mechanisms underpinning host-microbiome phenotypes. Mucosal Immunol. 2022;15:1071–1084. doi: 10.1038/s41385-022-00553-4. [DOI] [PubMed] [Google Scholar]
- 191.Purohit V, Wagner A, Yosef N, Kuchroo VK. Systems-based approaches to study immunometabolism. Cell Mol. Immunol. 2022;19:409–420. doi: 10.1038/s41423-021-00783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nie M, et al. Evolutionary metabolic landscape from preneoplasia to invasive lung adenocarcinoma. Nat. Commun. 2021;12:6479. doi: 10.1038/s41467-021-26685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 195.Shiratori R, et al. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Sci. Rep. 2019;9:18699. doi: 10.1038/s41598-019-55296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.TeSlaa T, Teitell MA. Techniques to monitor glycolysis. Methods Enzymol. 2014;542:91–114. doi: 10.1016/B978-0-12-416618-9.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Smith RA, Hartley RC, Cocheme HM, Murphy MP. Mitochondrial pharmacology. Trends Pharm. Sci. 2012;33:341–352. doi: 10.1016/j.tips.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 198.O’Malley J, Kumar R, Inigo J, Yadava N, Chandra D. Mitochondrial stress response and cancer. Trends Cancer. 2020;6:688–701. doi: 10.1016/j.trecan.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Giddings EL, et al. Mitochondrial ATP fuels ABC transporter-mediated drug efflux in cancer chemoresistance. Nat. Commun. 2021;12:2804. doi: 10.1038/s41467-021-23071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Acin-Perez R, et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J. 2020;39:e104073. doi: 10.15252/embj.2019104073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Eakins J, et al. A combined in vitro approach to improve the prediction of mitochondrial toxicants. Toxicol. Vitr. 2016;34:161–170. doi: 10.1016/j.tiv.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 202.Ma Y, et al. Functional analysis of molecular and pharmacological modulators of mitochondrial fatty acid oxidation. Sci. Rep. 2020;10:1450. doi: 10.1038/s41598-020-58334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim. Biophys. Acta. 2011;1807:726–734. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 204.Yao CH, et al. Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of beta-oxidation. PLoS Biol. 2018;16:e2003782. doi: 10.1371/journal.pbio.2003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vacanti NM, et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol. Cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Okegawa T, et al. Intratumor heterogeneity in primary kidney cancer revealed by metabolic profiling of multiple spatially separated samples within tumors. EBioMedicine. 2017;19:31–38. doi: 10.1016/j.ebiom.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wiemer EA, Michels PA, Opperdoes FR. The inhibition of pyruvate transport across the plasma membrane of the bloodstream form of Trypanosoma brucei and its metabolic implications. Biochem. J. 1995;312:479–484. doi: 10.1042/bj3120479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Xiang Y, et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 2015;125:2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ludikhuize MC, Meerlo M, Burgering BMT, Rodriguez Colman MJ. Protocol to profile the bioenergetics of organoids using Seahorse. STAR Protoc. 2021;2:100386. doi: 10.1016/j.xpro.2021.100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Arismendi-Morillo G. Electron microscopy morphology of the mitochondrial network in human cancer. Int J. Biochem Cell Biol. 2009;41:2062–2068. doi: 10.1016/j.biocel.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 212.Vande Voorde, J. et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv.5, eaau7314 (2019). [DOI] [PMC free article] [PubMed]
- 213.Lagziel S, Gottlieb E, Shlomi T. Mind your media. Nat. Metab. 2020;2:1369–1372. doi: 10.1038/s42255-020-00299-y. [DOI] [PubMed] [Google Scholar]
- 214.Savage P, et al. A targetable EGFR-dependent tumor-initiating program in breast cancer. Cell Rep. 2017;21:1140–1149. doi: 10.1016/j.celrep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 215.Nguyen QH, et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. 2018;9:2028. doi: 10.1038/s41467-018-04334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang P, et al. Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 2019;27:1934–1947 e1935. doi: 10.1016/j.celrep.2019.04.052. [DOI] [PubMed] [Google Scholar]
- 217.Young MD, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zheng H, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–140. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 221.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tang FC, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–U386. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 223.Xu B, et al. Identification of energy metabolism-related gene signatures from scRNA-Seq data to predict the prognosis of liver cancer patients. Front. Cell Dev. Biol. 2022;10:858336. doi: 10.3389/fcell.2022.858336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hrovatin K, Fischer DS, Theis FJ. Toward modeling metabolic state from single-cell transcriptomics. Mol. Metab. 2022;57:101396. doi: 10.1016/j.molmet.2021.101396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xiao Z, Dai Z, Locasale JW. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019;10:3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cazet AS, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018;9:2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rambow F, et al. Toward minimal residual disease-directed therapy in melanoma. Cell. 2018;174:843–855.e819. doi: 10.1016/j.cell.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 228.Ho YJ, et al. Single-cell RNA-seq analysis identifies markers of resistance to targeted BRAF inhibitors in melanoma cell populations. Genome Res. 2018;28:1353–1363. doi: 10.1101/gr.234062.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jain P, et al. Over-expression of PD-1 does not predict leukemic relapse after allogeneic stem cell transplantation. Biol. Blood Marrow Transpl. 2019;25:216–222. doi: 10.1016/j.bbmt.2018.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.De Sanctis, F. et al. Interrupting the nitrosative stress fuels tumor-specific cytotoxic T lymphocytes in pancreatic cancer. J. Immunother. Cancer10, e003549 (2022). [DOI] [PMC free article] [PubMed]
- 231.Guo S, Zhang C, Le A. The limitless applications of single-cell metabolomics. Curr. Opin. Biotechnol. 2021;71:115–122. doi: 10.1016/j.copbio.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wei D, Xu M, Wang Z, Tong J. The development of single-cell metabolism and its role in studying cancer emergent properties. Front Oncol. 2021;11:814085. doi: 10.3389/fonc.2021.814085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liu R, Yang Z. Single cell metabolomics using mass spectrometry: techniques and data analysis. Anal. Chim. Acta. 2021;1143:124–134. doi: 10.1016/j.aca.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Guillaume-Gentil O, et al. Single-cell mass spectrometry of metabolites extracted from live cells by fluidic force microscopy. Anal. Chem. 2017;89:5017–5023. doi: 10.1021/acs.analchem.7b00367. [DOI] [PubMed] [Google Scholar]
- 235.Evers TMJ, et al. Deciphering metabolic heterogeneity by single-cell analysis. Anal. Chem. 2019;91:13314–13323. doi: 10.1021/acs.analchem.9b02410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tellez-Gabriel, M., Ory, B., Lamoureux, F., Heymann, M. F. & Heymann, D. Tumour heterogeneity: the key advantages of single-cell analysis. Int. J. Mol. Sci.17, 2142 (2016). [DOI] [PMC free article] [PubMed]
- 237.Liu R, Sun M, Zhang G, Lan Y, Yang Z. Towards early monitoring of chemotherapy-induced drug resistance based on single cell metabolomics: combining single-probe mass spectrometry with machine learning. Anal. Chim. Acta. 2019;1092:42–48. doi: 10.1016/j.aca.2019.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.DeVilbiss, A. W. et al. Metabolomic profiling of rare cell populations isolated by flow cytometry from tissues. Elife10, e61980 (2021). [DOI] [PMC free article] [PubMed]
- 239.Hiyama E, et al. Direct lipido-metabolomics of single floating cells for analysis of circulating tumor cells by live single-cell mass spectrometry. Anal. Sci. 2015;31:1215–1217. doi: 10.2116/analsci.31.1215. [DOI] [PubMed] [Google Scholar]
- 240.Abouleila Y, et al. Live single cell mass spectrometry reveals cancer-specific metabolic profiles of circulating tumor cells. Cancer Sci. 2019;110:697–706. doi: 10.1111/cas.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wang R, et al. Metabolic discrimination of breast cancer subtypes at the single-cell level by multiple microextraction coupled with mass spectrometry. Anal. Chem. 2019;91:3667–3674. doi: 10.1021/acs.analchem.8b05739. [DOI] [PubMed] [Google Scholar]
- 242.Zuo F, Yu J, He X. Single-cell metabolomics in hematopoiesis and hematological malignancies. Front. Oncol. 2022;12:931393. doi: 10.3389/fonc.2022.931393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rappez L, et al. SpaceM reveals metabolic states of single cells. Nat. Methods. 2021;18:799–805. doi: 10.1038/s41592-021-01198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nascentes Melo LM, Lesner NP, Sabatier M, Ubellacker JM, Tasdogan A. Emerging metabolomic tools to study cancer metastasis. Trends Cancer. 2022;8:988–1001. doi: 10.1016/j.trecan.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 245.O’Neil RG, Wu L, Mullani N. Uptake of a fluorescent deoxyglucose analog (2-NBDG) in tumor cells. Mol. Imaging Biol. 2005;7:388–392. doi: 10.1007/s11307-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 246.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 247.Lin X, et al. A chemical genomics screen highlights the essential role of mitochondria in HIF-1 regulation. Proc. Natl Acad. Sci. USA. 2008;105:174–179. doi: 10.1073/pnas.0706585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Vazquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol. 2009;452:85–95. doi: 10.1016/S0076-6879(08)03606-9. [DOI] [PubMed] [Google Scholar]
- 249.Murphy MP, et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022;4:651–662. doi: 10.1038/s42255-022-00591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Arguello RJ, et al. SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. 2020;32:1063–1075 e1067. doi: 10.1016/j.cmet.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bosc C, et al. Mitochondrial inhibitors circumvent adaptive resistance to venetoclax and cytarabine combination therapy in acute myeloid leukemia. Nat. Cancer. 2021;2:1204–1223. doi: 10.1038/s43018-021-00264-y. [DOI] [PubMed] [Google Scholar]
- 252.Lopes N, et al. Distinct metabolic programs established in the thymus control effector functions of gammadelta T cell subsets in tumor microenvironments. Nat. Immunol. 2021;22:179–192. doi: 10.1038/s41590-020-00848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Verberk SGS, et al. An integrated toolbox to profile macrophage immunometabolism. Cell Rep. Methods. 2022;2:100192. doi: 10.1016/j.crmeth.2022.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Adamik J, et al. Distinct metabolic states guide maturation of inflammatory and tolerogenic dendritic cells. Nat. Commun. 2022;13:5184. doi: 10.1038/s41467-022-32849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hong HS, et al. OXPHOS promotes apoptotic resistance and cellular persistence in T(H)17 cells in the periphery and tumor microenvironment. Sci. Immunol. 2022;7:eabm8182. doi: 10.1126/sciimmunol.abm8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Swatler, J. et al. Dysfunctional subsets of CD39+ T cells, distinct from PD-1+, driven by leukemic extracellular vesicles in myeloid leukemias. Haematologica108 (2022). [DOI] [PMC free article] [PubMed]
- 257.Michaudel, C. et al. Rewiring the altered tryptophan metabolism as a novel therapeutic strategy in inflammatory bowel diseases. Gut 327337 10.1136/gutjnl-2022-327337 (2022). [DOI] [PMC free article] [PubMed]
- 258.Dolfi B, et al. Unravelling the sex-specific diversity and functions of adrenal gland macrophages. Cell Rep. 2022;39:110949. doi: 10.1016/j.celrep.2022.110949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Corral D, et al. ILC precursors differentiate into metabolically distinct ILC1-like cells during Mycobacterium tuberculosis infection. Cell Rep. 2022;39:110715. doi: 10.1016/j.celrep.2022.110715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Nomizu T, et al. Determination of calcium content in individual biological cells by inductively-coupled plasma-atomic emission-spectrometry. Anal. Chem. 1994;66:3000–3004. doi: 10.1021/ac00091a004. [DOI] [Google Scholar]
- 261.Tanner SD, Ornatsky O, Bandura DR, Baranov VI. Multiplex bio-assay with inductively coupled plasma mass spectrometry: towards a massively multivariate single-cell technology. Spectrochim. Acta B. 2007;62:188–195. doi: 10.1016/j.sab.2007.01.008. [DOI] [Google Scholar]
- 262.Behbehani GK. Applications of mass cytometry in clinical medicine: the promise and perils of clinical CyTOF. Clin. Lab Med. 2017;37:945–964. doi: 10.1016/j.cll.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 263.Astle JM, Huang H. Mass cytometry in hematologic malignancies: research highlights and potential clinical applications. Front Oncol. 2021;11:704464. doi: 10.3389/fonc.2021.704464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhu, Y. P. et al. CyTOF mass cytometry reveals phenotypically distinct human blood neutrophil populations differentially correlated with melanoma stage. J. Immunother. Cancer8, e000473 (2020). [DOI] [PMC free article] [PubMed]
- 265.Teh CE, et al. Deep profiling of apoptotic pathways with mass cytometry identifies a synergistic drug combination for killing myeloma cells. Cell Death Differ. 2020;27:2217–2233. doi: 10.1038/s41418-020-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Rein ID, Noto HO, Bostad M, Huse K, Stokke T. Cell cycle analysis and relevance for single-cell gating in mass cytometry. Cytometry A. 2020;97:832–844. doi: 10.1002/cyto.a.23960. [DOI] [PubMed] [Google Scholar]
- 267.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Leelatian N, Diggins KE, Irish JM. Characterizing phenotypes and signaling networks of single human cells by mass cytometry. Methods Mol. Biol. 2015;1346:99–113. doi: 10.1007/978-1-4939-2987-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lun XK, Bodenmiller B. Profiling cell signaling networks at single-cell resolution. Mol. Cell Proteom. 2020;19:744–756. doi: 10.1074/mcp.R119.001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 271.Iyer A, Hamers AAJ, Pillai AB. CyTOF((R)) for the masses. Front. Immunol. 2022;13:815828. doi: 10.3389/fimmu.2022.815828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ahl PJ, et al. Met-Flow, a strategy for single-cell metabolic analysis highlights dynamic changes in immune subpopulations. Commun. Biol. 2020;3:305. doi: 10.1038/s42003-020-1027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Miller A, et al. Exploring metabolic configurations of single cells within complex tissue microenvironments. Cell Metab. 2017;26:788–800.e786. doi: 10.1016/j.cmet.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 274.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Evers B, et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 276.Zhu XG, et al. Functional genomics in vivo reveal metabolic dependencies of pancreatic cancer cells. Cell Metab. 2021;33:211–221.e216. doi: 10.1016/j.cmet.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Arroyo JD, et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 2016;24:875–885. doi: 10.1016/j.cmet.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Thomas LW, et al. Genome-wide CRISPR/Cas9 deletion screen defines mitochondrial gene essentiality and identifies routes for tumour cell viability in hypoxia. Commun. Biol. 2021;4:615. doi: 10.1038/s42003-021-02098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Birsoy K, et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Soula M, et al. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat. Chem. Biol. 2020;16:1351–1360. doi: 10.1038/s41589-020-0613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 282.Rebsamen, M. et al. Gain-of-function genetic screens in human cells identify SLC transporters overcoming environmental nutrient restrictions. Life Sci. Alliance5, e202201404 (2022). [DOI] [PMC free article] [PubMed]
- 283.Li KC, et al. Cell-surface SLC nucleoside transporters and purine levels modulate BRD4-dependent chromatin states. Nat. Metab. 2021;3:651–664. doi: 10.1038/s42255-021-00386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zhao D, et al. Combinatorial CRISPR-Cas9 metabolic screens reveal critical redox control points dependent on the KEAP1-NRF2 regulatory axis. Mol. Cell. 2018;69:699–708.e697. doi: 10.1016/j.molcel.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Rossiter NJ, et al. CRISPR screens in physiologic medium reveal conditionally essential genes in human cells. Cell Metab. 2021;33:1248–1263.e1249. doi: 10.1016/j.cmet.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Giera M, Yanes O, Siuzdak G. Metabolite discovery: biochemistry’s scientific driver. Cell Metab. 2022;34:21–34. doi: 10.1016/j.cmet.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Misra BB. New software tools, databases, and resources in metabolomics: updates from 2020. Metabolomics. 2021;17:49. doi: 10.1007/s11306-021-01796-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 289.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Pfeuffer J, et al. OpenMS—a platform for reproducible analysis of mass spectrometry data. J. Biotechnol. 2017;261:142–148. doi: 10.1016/j.jbiotec.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 291.Tsugawa H, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Tsugawa H, et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020;38:1159–1163. doi: 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- 293.Domingo-Almenara X, et al. eRah: a computational tool integrating spectral deconvolution and alignment with quantification and identification of metabolites in GC/MS-based metabolomics. Anal. Chem. 2016;88:9821–9829. doi: 10.1021/acs.analchem.6b02927. [DOI] [PubMed] [Google Scholar]
- 294.Smirnov A, et al. ADAP-GC 4.0: application of clustering-assisted multivariate curve resolution to spectral deconvolution of gas chromatography-mass spectrometry metabolomics data. Anal. Chem. 2019;91:9069–9077. doi: 10.1021/acs.analchem.9b01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kind T, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ogata H, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Chen, Y., Li, E. M. & Xu, L. Y. Guide to metabolomics analysis: a bioinformatics workflow. Metabolites12, 357 (2022). [DOI] [PMC free article] [PubMed]
- 298.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.National Research Council (US) Committee on a Framework for Developing a New Taxonomy of Disease. in Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease The National Academies Collection: Reports funded by National Institutes of Health (National Academies Press, 2011). [PubMed]
- 300.Ghaste, M., Mistrik, R. & Shulaev, V. Applications of Fourier transform ion cyclotron resonance (FT-ICR) and Orbitrap based high resolution mass spectrometry in metabolomics and lipidomics. Int. J. Mol. Sci.17, 816 (2016). [DOI] [PMC free article] [PubMed]
- 301.Markley, J. L. et al. New bioinformatics resources for metabolomics. Pac. Symp. Biocomput.12, 157–168 (2007). [PubMed]
- 302.Blekherman G, et al. Bioinformatics tools for cancer metabolomics. Metabolomics. 2011;7:329–343. doi: 10.1007/s11306-010-0270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Holzinger, A., Rocker, C. & Ziefle, M. in Information Systems and Applications, incl Internet/Web, and HCI 8700 1 online resource (XIV, 275 pages 268 illustrations) (Springer International Publishing: Imprint: Springer, 2015).
- 304.Sboner A, Elemento O. A primer on precision medicine informatics. Brief. Bioinform. 2016;17:145–153. doi: 10.1093/bib/bbv032. [DOI] [PubMed] [Google Scholar]
- 305.Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51–60. doi: 10.1007/s13167-017-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sun YV, Hu YJ. Integrative analysis of multi-omics data for discovery and functional studies of complex human diseases. Adv. Genet. 2016;93:147–190. doi: 10.1016/bs.adgen.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ferte C, et al. Impact of bioinformatic procedures in the development and translation of high-throughput molecular classifiers in oncology. Clin. Cancer Res. 2013;19:4315–4325. doi: 10.1158/1078-0432.CCR-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Yang A, Troup M, Ho JWK. Scalability and validation of big data bioinformatics software. Comput Struct. Biotechnol. J. 2017;15:379–386. doi: 10.1016/j.csbj.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Greene CS, Tan J, Ung M, Moore JH, Cheng C. Big data bioinformatics. J. Cell Physiol. 2014;229:1896–1900. doi: 10.1002/jcp.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Deja S, et al. Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. J. Pharm. Biomed. Anal. 2014;100:369–380. doi: 10.1016/j.jpba.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 311.Wikoff WR, et al. Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev. Res. (Philos.) 2015;8:410–418. doi: 10.1158/1940-6207.CAPR-14-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kritchevsky SB, Wilcosky TC, Morris DL, Truong KN, Tyroler HA. Changes in plasma lipid and lipoprotein cholesterol and weight prior to the diagnosis of cancer. Cancer Res. 1991;51:3198–3203. [PubMed] [Google Scholar]
- 313.Bamji-Stocke S, van Berkel V, Miller DM, Frieboes HB. A review of metabolism-associated biomarkers in lung cancer diagnosis and treatment. Metabolomics. 2018;14:81. doi: 10.1007/s11306-018-1376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Gunther UL. Metabolomics biomarkers for breast cancer. Pathobiology. 2015;82:153–165. doi: 10.1159/000430844. [DOI] [PubMed] [Google Scholar]
- 315.Costello E. A metabolomics-based biomarker signature discriminates pancreatic cancer from chronic pancreatitis. Gut. 2018;67:2–3. doi: 10.1136/gutjnl-2016-313665. [DOI] [PubMed] [Google Scholar]
- 316.Horbach S, Halffman W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLoS ONE. 2017;12:e0186281. doi: 10.1371/journal.pone.0186281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ben-David U, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Pandit AV, Srinivasan S, Mahadevan R. Redesigning metabolism based on orthogonality principles. Nat. Commun. 2017;8:15188. doi: 10.1038/ncomms15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Zhang A, Sun H, Wang P, Han Y, Wang X. Recent and potential developments of biofluid analyses in metabolomics. J. Proteom. 2012;75:1079–1088. doi: 10.1016/j.jprot.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 320.Evans ED, et al. Predicting human health from biofluid-based metabolomics using machine learning. Sci. Rep. 2020;10:17635. doi: 10.1038/s41598-020-74823-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Winder CL, et al. Providing metabolomics education and training: pedagogy and considerations. Metabolomics. 2022;18:106. doi: 10.1007/s11306-022-01957-w. [DOI] [PMC free article] [PubMed] [Google Scholar]