Abstract
In this study, new hybrids of quinazolinone-1,2,3-triazole-acetamide were designed, synthesized, and screened for their α-glucosidase inhibitory activity. The results obtained from the in vitro screening indicated that all analogs exhibited significant inhibitory activity against α-glucosidase (IC50 values ranging from 4.8–140.2 μM) in comparison to acarbose (IC50 = 750.0 μM). The limited structure–activity relationships suggested the variation in the inhibitory activities of the compounds affected by different substitutions on the aryl moiety. The enzyme kinetic studies of the most potent compound 9c, revealed that it inhibited α-glucosidase in a competitive mode with a Ki value of 4.8 μM. In addition, molecular docking studies investigated the structural perturbation and behavior of all derivatives inside the α-glucosidase active site. Next, molecular dynamic simulations of the most potent compound 9c, were performed to study the behavior of the 9c-complex during the time. The results showed that these compounds can be considered as potential antidiabetic agents.
In this study, new hybrids of quinazolinone-1,2,3-triazole-acetamide were designed, synthesized, and screened for their α-glucosidase inhibitory activity.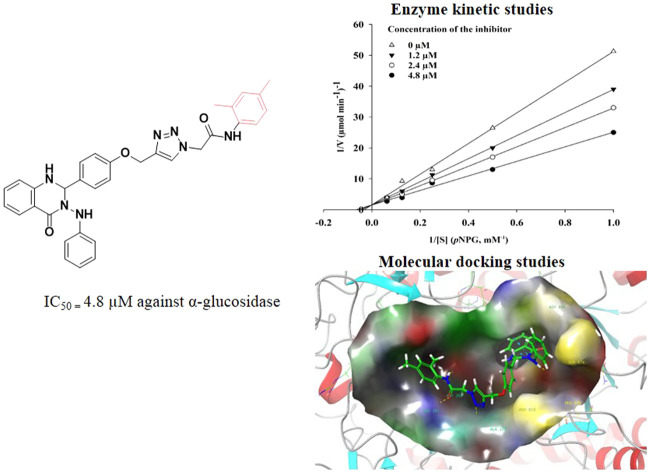
1. Introduction
Diabetes mellitus (DM) is a chronic metabolic disorder described by hyperglycemia, which can lead to serious health issues such as cardiovascular disease, nephropathy, retinopathy, and thrombosis.1 The prevalence of DM is estimated to be around 415 million people and this number will increase to about 642 million by 2040. Type 2 DM (T2DM) for 85–90% of all diabetic patients around the world, results from pancreatic β-cell dysfunction and resistance to insulin action in the muscle and adipose tissues.2,3
Inhibiting the digestion of dietary carbohydrates has emerged a strong tool to control hyperglycemia in diabetic patients.4,5 The α-glucosidase (EC.3.2.1.20) enzyme located in the small intestine, is responsible for the hydrolysis of dietary carbohydrates such as starch and disaccharides to glucose that enters the bloodstream resulting hyperglycemia.6 Inhibition of the enzyme reduces the digestion and absorption of simple carbohydrates in the intestine. α-Glucosidase inhibitors such as miglitol, acarbose, and voglibose reduce the postprandial blood glucose level. These drugs are also effective to decrease the risk of T2DM, obesity, hyperlipoproteinemia, and cancer.7,8 However, regarding the side effects recorded for α-glucosidase inhibitors such as diarrhea and flatulence, it is essential to find new active molecules to target T2DM with minimal risks of side effects.
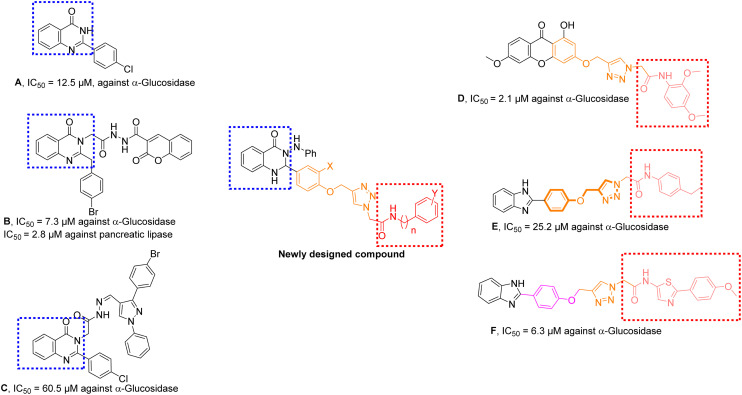
Quinazolin-4(1H)-one is undoubtedly one of the important nuclei in medicinal chemistry. Taking the advantage of its versatile reactivity, a huge library of quinazolin-4(1H)-one derivatives is developed with diverse applications like antibacterial,9 antifungal,10 antiviral,11 anticancer,12 anti-Alzheimer's,13 and anti-inflammatory,14 and anti-α-glucosidase15 effects. Very recently, anti-diabetic properties of quinazolin-4(1H)-one as α-glucosidase inhibitors were introduced. In this context, compound A with an IC50 value of 12.5 μM as a non-competitive inhibitor16 and compound B bearing quinoxaline ring with IC50 value of 7.3 μM in comparison with the standard acarbose 26.1 μM (ref. 17) are good examples (Fig. 1). Recently quinazolinone–pyrazole derivatives were synthesized and screened as anti-α-glucosidase agents. All derivatives were more potent (IC50 = 60.5–186.6 μM) than the standard inhibitor and among them, compound C was the most potent derivative (Fig. 1). Kinetic studies revealed that C inhibited α-glucosidase in a competitive mode with a Ki of 56 μM.18
Fig. 1. Chemical structures of some reported α-glucosidase inhibitors and newly designed compounds.
Aromatic heterocyclic compounds are the focus of research in pharmaceutical sciences.19,20 1,2,3-Triazole as an electron-rich five-membered heterocyclic ring can participate in several non-bounded interactions in the chemical and biological systems. 1,2,3-Triazole-based pharmacophore according to its chemical and physiological properties got much attention in medicinal chemistry due to its easy synthetic procedure and versatile functions as a key entity in design and synthesis of pharmacologically active agents. 1,2,3-Triazole derivatives are known to possess various biological activities such as antimicrobial,21 anticancer,22 anti-inflammatory,23 and anti-Alzheimer's24 properties. Phenoxy-1,2,3-triazole moiety was also introduced as promising anti-α-glucosidase inhibitors with provided optimum conformation to penetrate the deep gorge of the binding site and participate in several interactions with the binding site to get the suppressed conformer of the enzyme.25–27 Apart from these, aryl-acetamide bearing skeleton also finds applications as α-glucosidase inhibitor.28 The binding and proper structure of inhibitors with targeted molecules are crucial to improve the therapeutic properties of designed compounds in drug development. There is great evidence that substituted acetohydrazide derivatives as the key skeleton of anchoring agents, can be effective in interacting and stabilizing α-helices, β-sheets, and other secondary structures of biological macromolecules via hydrogen bonding, nucleophile–carbonyl, carbonyl–carbonyl (CO/CO) interactions.29 This pharmacophore also provides a suitable site for derivatization to evaluate the structure–activity relationships (SARs). Take the example of the recent research, compound D (Fig. 1) containing 1,2,3-triazole acetamide exerted an IC50 value of 2.1 μM in comparison with the standard acarbose with low toxicity to the human normal hepatocyte cell line.30 In another research, compound E proved the potent effect of 1,2,3-triazole and acyl acetamide moieties as the α-glucosidase inhibitor vs. standard acarbose (IC50 = 750.0 μM).31 All tested phenoxymethybenzimidazoles bearing 1,2,3-triazole acetamide (such as compound F) displayed excellent α-glucosidase inhibitory potential with IC50 values in the range of 6.31 to 49.9 μM compared to standard drug acarbose (IC50 = 750.0 μM) and the critical role of phenoxy-1,2,3-triazole-acetamide were confirmed via in silico assessment.32 Therefore, the quinazolin-4(1H)-one as well as 1,2,3-triazole were united according to fragments-based drug design strategies to design a novel set of compounds. Derivatives were then synthesized (Scheme 1) and characterized by IR, 1H-NMR, and 13C-NMR spectroscopy as well as CHN elemental analysis. Next, the inhibitory potency of all derivatives were invested against α-glucosidase. The interaction of the top-ranked compound with the enzyme was determined using enzymatic assay methods and molecular docking analysis.
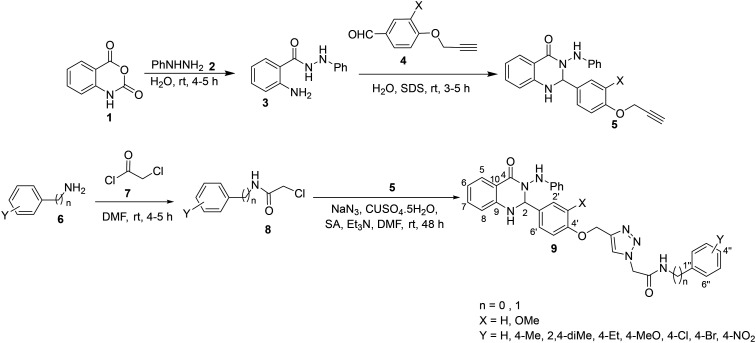
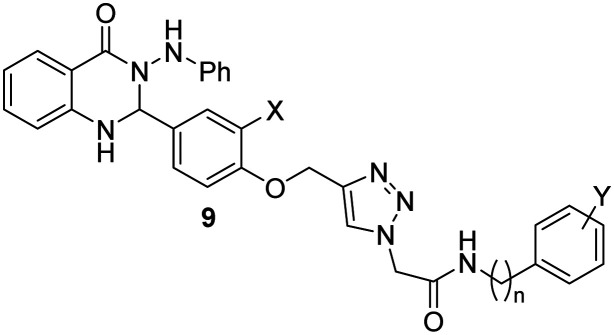
Scheme 1. Synthesis of compounds 9a–t.
2. Experimental section
2.1. General
All chemicals and reagents were purchased from Merck and Aldrich. Melting points were determined using Kofler hot stage apparatus and are uncorrected. The IR spectra were obtained using a Nicolet Magna FTIR 550 spectrometer (potassium bromide disks). NMR spectra were recorded on a Varian-INOVA 500 MHz.
2.2. Synthesis of compound 9a–t
A mixture of isatoic anhydride 1 (10 mmol) and phenylhydrazine 2 (10 mmol) in water (50 mL) was stirred at room temperature for 4–5 h. After completion of the reaction (checked by TLC), the precipitate was filtered off to obtain 2-amino-N′-phenylbenzohydrazide 3 in cream color and 80% yield. Then, compound 3 (10 mmol) reacted with 4-(prop-2-yn-1-yloxy)benzaldehyde or 3-methoxy-4-(prop-2-yn-1-yloxy)benzaldehyde 4 (10 mmol) in water (25 mL) in the presence of sodium dodecyl sulfate (SDS, 30 mol%) at room temperature for 3–5 h. After completion of the reaction (checked by TLC), the precipitate was filtered off and compound 5 was afforded in cream color and 80–85% yield. It should be noted that compound 4 was prepared by the reaction of 4-hydroxy benzaldehyde or vanillin (5 mmol) and propargyl bromide (6 mmol) in the presence of potassium carbonate (K2CO3, 5 mmol) in DMF at 80 °C for 4–5 h. After the expected time, the mixture was poured into the ice-water and the product was collected by filtration.33
On the other, chloroacetyl chloride 7 (12 mmol) was added to a solution of desired amine 6 (10 mmol) in DMF (20 mL) which was kept in an ice bath for 15 min and then, the mixture was stirred at room temperature for 4–5 h. After completion of the reaction (checked by TLC), the mixture was poured into the crushed ice and the precipitate was filtered off to give compound 8 in cream color. Finally, click reaction of compounds 5 and 8 was conducted. For this purpose, a mixture of 8 (0.8 mmol), sodium azide (1.6 mmol), and trimethylamine (Et3N, 1.3 mmol) in DMF (4 mL) was stitted for 1 h at room temperature. Then, compound 7 (0.6 mmol), CuSO4·5H2O (7 mol%), and sodium ascorbate (SA, 1.3 mmol) were added to the reaction mixture and it was continued for 48 h at the same temperature. Upon completion of the reaction (monitored by TLC), the mixture was poured in water, extracted with ethyl acetate, and dried over anhydrous Na2SO4. Evaporation of solvent afforded compound 9 as an off-white powder which was recrystallized from ethyl acetate and petroleum (60–70% yield).
2-(4-((4-(4-Oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-phenylacetamide (9a)
Mp = 131–133 °C; IR (KBr): 3350, 3058, 2957, 2850, 1660, 1614, 1480 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.48 (s, 1H, NH), 8.30 (s, 1H, NH), 8.27 (s, 1H, triazole), 7.65 (d, J = 7.8 Hz, 1H, H5), 7.59 (d, J = 8.0 Hz, 2H, H2′′, H6′′), 7.54 (s, 1H, NH), 7.37 (d, J = 8.0 Hz, 2H, Ph), 7.33 (t, J = 8.0 Hz, 2H, H3′′, H5′′), 7.28 (t, J = 7.8 Hz, 1H, H7), 7.17 (t, J = 8.0 Hz, 2H, Ph), 7.09 (t, J = 8.0 Hz, 1H, H4′′), 7.01 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.85 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.75 (d, J = 7.8 Hz, 1H, H8), 6.75 (t, J = 8.0 Hz, 1H, Ph), 6.70 (t, J = 7.8 Hz, 1H, H6), 5.86 (s, 1H, CH), 5.35 (s, 2H, CH2), 5.16 (s, 2H, CH2) ppm. 13CNMR (DMSO-d6, 125 MHz): δ = 164.6 (C O), 163.2 (C O), 158.6 (C4′), 152.1 (C1 of Ph), 148.4 (C9), 147.4 (C of triazole), 138.8 (C1′′), 134.2 (C1′), 133.6 (C7), 129.5 (C10), 129.4 (C3,5 of Ph), 129.3 (C3′′,5′′), 128.2 (C5), 128.0 (C4′′), 127.0 (C2′,6′), 126.9 (C4 of Ph), 125.1 (C of triazole), 124.3 (C2′′,6′′), 119.7 (C6), 117.8 (C8), 114.8 (C3′,5′), 112.9 (C2,6 of Ph), 73.8 (CH), 66.1 (CH2), 61.5 (CH2) ppm. Anal. calcd for C31H27N7O3: C, 68.24; H, 4.99; N, 17.97. Found: C, 68.41; H, 5.21; N, 18.15.
2-(4-((4-(4-Oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(p-tolyl)acetamide (9b)
Mp = 115–117 °C; IR (KBr): 3345, 3060, 2955, 2848, 1660, 1614, 1447 cm−1.1HNMR (DMSO-d6, 500 MHz): 10.39 (s, 1H, NH), 8.30 (s, 1H, NH), 8.29 (s, 1H, triazole), 7.65 (d, J = 7.7 Hz, 1H, H5), 7.54 (s, 1H, NH), 7.48 (d, J = 8.0 Hz, 2H, H2′′, H6′′), 7.35 (d, J = 7.5 Hz, 2H, Ph), 7.28 (t, J = 7.7 Hz, 1H, H7), 7.20 (m, 4H, Ph, H3′′, H5′′), 7.02 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.85 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.77 (d, J = 7.7 Hz, 1H, H8), 6.73 (t, J = 7.5 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.84 (s, 1H, CH), 5.33 (s, 2H, CH2), 5.31 (s, 2H, CH2), 2.25 (s, 3H, CH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.4 (C O), 163.5 (C O), 155.3 (C4′), 150.3 (C1 of Ph), 148.2 (C9), 147.5 (C of triazole), 136.4 (C1′), 133.2 (C1′′), 132.3 (C7), 130.4 (C10), 129.7 (C4′′), 129.3 (C3,5 of Ph), 129.2 (C3′′,5′′), 128.2 (C5), 128.1 (C2′,6′), 127.0 (C4 of Ph), 122.4 (C of triazole), 120.3 (C2′′,6′′), 119.7 (C6), 117.7 (C8), 115.7 (C3′,5′), 112.8 (C2,6 of Ph), 72.2 (CH), 67.1 (CH2), 61.9 (CH2), 20.9 (CH3) ppm. Anal. calcd for C32H29N7O3: C, 68.68; H, 5.22; N, 17.52. Found: C, 68.83; H, 5.50; N, 17.31.
N-(2,4-Dimethylphenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9c)
Mp = 134–136 °C; IR (KBr): 3350, 3057, 2948, 2855, 1660, 1623, 1456 cm−1. 1HNMR (DMSO-d6, 500 MHz): 9.78 (s, 1H, NH), 8.29 (s, 1H, NH), 8.24 (s, 1H, triazole), 7.63 (d, J = 7.7 Hz, 1H, H5), 7.53 (s, 1H, NH), 7.35 (d, J = 8.0 Hz, 2H, Ph), 7.27 (t, J = 7.7 Hz, 1H, H7), 7.16 (t, J = 8.0 Hz, 2H, Ph), 7.09–7.05 (m, 3H, H3′′, H5′′, H6′′), 7.01 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.78 (d, J = 7.7 Hz, 1H, H8), 6.75 (t, J = 8.0 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.84 (s, 1H, CH), 5.38 (s, 2H, CH2), 5.14 (s, 2H, CH2), 2.17–2.15 (m, 6H, 2× CH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.4 (C O), 163.1 (C O), 158.6 (C4′), 150.5 (C1 of Ph), 148.4 (C9), 147.4 (C4′′), 147.3 (C of triazole), 135.5 (C1′), 134.6 (C2′′), 134.2 (C1′′), 133.6 (C7), 129.3 (C3′′), 128.6 (C10), 128.2 (C3,5 of Ph), 128.0 (C5), 127.2 (C2′,6′), 126.7 (C5′′), 123.6 (C4 of Ph), 123.2 (C of triazole), 119.6 (C6), 117.8 (C8), 115.1 (C6′′), 114.8 (C3′,5′), 112.9 (C2,6 of Ph), 73.7 (CH), 65.9 (CH2), 61.4 (CH2), 18.5 (CH3) ppm. Anal. calcd for C33H31N7O3: C, 69.09; H, 5.45; N, 17.09. Found: C, 69.26; H, 5.68; N, 17.25.
N-(4-Ethylphenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9d)
Mp = 126–128 °C; IR (KBr): 3380, 3058, 2955, 2850, 1660, 1620, 1442 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.39 (s, 1H, NH), 8.30 (s, 1H, NH), 8.24 (s, 1H, triazole), 7.63 (d, J = 7.7 Hz, 1H, H5), 7.54 (s, 1H, NH), 7.47 (d, J = 8.0 Hz, 2H, H2′′, H6′′), 7.36 (d, J = 7.5 Hz, 2H, Ph), 7.28 (t, J = 7.7 Hz, 1H, H7), 7.20 (m, 4H, Ph, H3′′, H5′′), 7.02 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.78 (d, J = 7.7 Hz, 1H, H8), 6.74 (t, J = 7.5 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.32 (s, 2H, CH2), 5.14 (s, 2H, CH2), 2.55 (q, J = 7.6 Hz, 2H, CH2), 1.15 (t, J = 7.6 Hz, 3H, CH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.3 (C O), 163.0 (C O), 158.6 (C4′), 150.3 (C1 of Ph), 148.4 (C9), 147.4 (C4′′), 139.7 (C of triazole), 136.5 (C1′), 134.2 (C1′′), 133.0 (C7), 129.3 (C10), 128.5 (C3,5 of Ph), 128.2 (C5), 126.8 (C2′,6′), 126.7 (C3′′,5′′), 126.2 (C4 of Ph), 125.8 (C of triazole), 120.3 (C2′′,6′′), 119.8 (C6), 117.8 (C8), 114.8 (C C3′,5′), 112.9 (C2,6 of Ph), 73.7 (CH2), 65.9 (CH2), 61.4 (CH2), 28.0 (CH2), 16.1 (CH3) ppm. Anal. calcd for C33H31N7O3: C, 69.09; H, 5.45; N, 17.09. Found: C, 68.87; H, 5.35; N, 16.89.
N-(4-Methoxyphenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9e)
Mp = 125–127 °C; IR (KBr): 3375, 3050, 2955, 2842, 1660, 1621, 1445 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.34 (s, 1H, NH), 8.30 (s, 1H, NH), 8.23 (s, 1H, triazole), 7.64 (d, J = 8.0 Hz, 1H, H5), 7.54 (s, 1H, NH), 7.49 (d, J = 7.2 Hz, 2H, H2′′, H6′′), 7.37 (d, J = 7.1 Hz, 2H, Ph), 7.28 (t, J = 8.0 Hz, 1H, H7), 7.17 (t, J = 7.1 Hz, 2H, Ph), 7.02 (d, J = 7.5 Hz, 2H, H2′, H6′), 6.90 (d, J = 7.2 Hz, 2H, H3′′, H5′′), 6.85 (d, J = 7.5 Hz, 2H, H3′, H5′), 6.79 (d, J = 8.0 Hz, 1H, H8), 6.75 (t, J = 7.1 Hz, 1H, Ph), 6.70 (t, J = 8.0 Hz, 1H, H6), 5.86 (s, 1H, CH), 5.31 (s, 2H, CH2), 5.15 (s, 2H, CH2), 3.72 (s, 3H, OCH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.1 (C O), 163.0 (C O), 158.6 (C4′), 156.0 (C1 of Ph), 148.4 (C4′′), 147.4 (C9), 142.9 (C of triazole), 134.2 (C1′), 133.5 (C7), 131.9 (C1′′), 131.5 (C10), 129.3 (C3,5 of Ph), 128.2 (C5), 128.0 (C2′,6′), 126.7 (C4 of Ph), 121.9 (C of triazole), 121.2 (C2′′,6′′), 119.6 (C6), 117.9 (C8), 114.8 (C3′,5′), 114.5 (C3′′,5′′), 112.9 (C2,6 of Ph), 73.8 (CH2), 66.0 (CH2), 61.5 (CH2), 55.6 (OCH3) ppm. Anal. calcd for C32H29N7O4: C, 66.77; H, 5.08; N, 17.03. Found: C, 66.58; H, 4.85; N, 17.32.
N-(4-Chlorophenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9f)
Mp = 139–141 °C; IR (KBr): 3355, 3058, 2950, 2842, 1665, 1622, 1442 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.61 (s, 1H, NH), 8.30 (s, 1H, NH), 8.24 (s, 1H, triazole), 7.64–7.60 (m, 3H, H5, H2′′, H6′′), 7.54 (s, 1H, NH), 7.40–7.35 (m, 4H, Ph, H3′′, H5′′), 7.28 (t, J = 7.7 Hz, 1H, H7), 7.16 (t, J = 7.5 Hz, 2H, Ph), 7.02 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.78 (d, J = 7.7 Hz, 1H, H8), 6.74 (t, J = 7.5 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.35 (s, 2H, CH2), 5.14 (s, 2H, CH2) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.8 (C O), 163.0 (C O), 158.6 (C4′), 151.1 (C1 of Ph), 148.4 (C9), 147.4 (C of triazole), 137.8 (C1′), 134.2 (C1′′), 133.6 (C4′′), 132.3 (C7), 129.4 (C10), 129.3 (C3,5 of Ph), 128.2 (C3′′,5′′), 128.0 (C5), 127.8 (C2′,6′), 126.8 (C4 of Ph), 121.9 (C of triazole), 121.2 (C2′′,6′′), 119.6 (C6), 117.8 (C8), 114.8 (C3′,5′), 112.9 (C2,6 of Ph), 73.8 (CH), 66.0 (CH2), 61.5 (CH2) ppm. Anal. calcd for C31H26ClN7O3: C, 64.19; H, 4.52; N, 16.90. Found: C, 64.48; H, 4.21; N, 16.71.
N-(4-Bromophenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9g)
Mp = 116–118 °C; IR (KBr): 3350, 3050, 2955, 2840, 1660, 1615, 1445 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.61 (s, 1H, NH), 8.30 (s, 1H, NH), 8.24 (s, 1H, triazole), 7.63 (d, J = 7.7 Hz, 1H, H5), 7.54–7.51 (m, 4H, H2′′, H3′′, H5′′, H6′′), 7.46 (d, J = 8.0 Hz, 1H, NH), 7.36 (d, J = 8.0 Hz, 2H, Ph), 7.28 (t, J = 7.7 Hz, 1H, H7), 7.16 (t, J = 8.0 Hz, 2H, Ph), 7.02 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.78 (d, J = 7.7 Hz, 1H, H8), 6.74 (t, J = 8.0 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.35 (s, 2H, CH2), 5.14 (s, 2H, CH2) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.9 (C O), 163.1 (C O), 158.6 (C4′), 151.6 (C1 of Ph), 148.4 (C9), 147.4 (C of triazole), 138.2 (C1′′), 134.2 (C1′), 133.6 (C7), 133.2 (C3′′,5′′), 132.3 (C10), 132.2 (C3,5 of Ph), 129.7 (C5), 129.3 (C2′,6′), 128.2 (C4 of Ph), 128.0 (C of triazole), 122.2 (C4′′), 121.6 (C2′′,6′′), 119.7 (C6), 117.8 (C8), 114.8 (C3′,5′), 112.9 (C2,6 of Ph), 73.7 (CH), 66.0 (CH2), 61.5 (CH2) ppm. Anal. calcd for C31H26BrN7O3: C, 59.62; H, 4.20; N, 15.70. Found: C, 59.38; H, 4.37; N, 15.92.
N-(4-Nitrophenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9h)
Mp = 129–131 °C; IR (KBr): 3350, 3058, 2955, 28 455, 1663, 1620, 1470, 1375 cm−1. 1HNMR (DMSO-d6, 500 MHz): 11.11 (s, 1H, NH), 8.29–8.24 (m, 4H, NH, triazole, H3′′, H5′′), 7.82 (d, J = 8.6 Hz, 2H, H2′′, H6′′), 7.63 (d, J = 8.0 Hz, 1H, H5), 7.54 (s, 1H, NH), 7.36 (d, J = 8.0 Hz, 2H, Ph), 7.28 (t, J = 8.0 Hz, 1H, H7), 7.16 (t, J = 8.0 Hz, 2H, Ph), 7.02 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.77 (d, J = 8.0 Hz, 1H, H8), 6.75 (t, J = 8.0 Hz, 1H, Ph), 6.70 (t, J = 8.0 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.43 (s, 2H, CH2), 5.15 (s, 2H, CH2) ppm. 13CNMR (DMSO-d6, 125 MHz): 165.8 (C O), 163.1 (C O), 158.6 (C4′), 148.4 (C1 of Ph), 147.4 (C9), 144.9 (C1′′), 143.1 (C4′′), 143.0 (C of triazole), 134.2 (C1′), 133.6 (C7), 132.3 (C10), 129.3 (C3,5 of Ph), 128.2 (C5), 128.0 (C2′,6′), 126.7 (C4 of Ph), 125.6 (C2′′,6′′), 125.5 (C of triazole), 119.6 (C2′′,6′′), 119.5 (C6), 117.8 (C8), 114.8 (C3′,5′), 112.9 (C2,6 of Ph), 73.7 (CH), 66.0 (CH2), 61.5 (CH2) ppm. Anal. calcd for C31H26N8O5: C, 63.04; H, 4.44; N, 18.97. Found: C, 62.87; H, 4.21; N, 19.25.
N-Benzyl-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9i)
Mp = 128–130 °C; IR (KBr): 3370, 3055, 2955, 2850, 1661, 1620, 1450 cm−1. 1HNMR (DMSO-d6, 500 MHz): 8.84 (bs, 1H, NH), 8.29 (bs, 1H, NH), 8.21 (s, 1H, triazole), 7.63 (d, J = 7.7 Hz, 1H, H5), 7.53 (s, 1H, NH), 7.37–7.26 (m, 8H, H7, H2′′, H3′′, H4′′, H5′′, H6′′, Ph), 7.16 (t, J = 7.5 Hz, 2H, Ph), 7.01 (d, J = 8.0 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.0 Hz, 2H, H3′, H5′), 6.78 (d, J = 7.7 Hz, 1H, H8), 6.75 (t, J = 7.5 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.84 (s, 1H, CH), 5.18 (s, 2H, CH2), 5.12 (s, 2H, CH2), 4.32 (d, J = 5.6 Hz, 2H, CH2). 13CNMR (DMSO-d6, 125 MHz): 165.8 (C O), 163.1 (C O), 158.6 (C4′), 150.6 (C1 of Ph), 148.4 (C9), 147.7 (C of triazole), 139.1 (C1′′), 134.2 (C1′), 133.6 (C7), 132.3 (C10), 129.4 (C3,5 of Ph), 129.3 (C2′,6′), 128.8 (C3′′,5′′), 128.2 (C5), 128.0 (C4′′), 127.8 (C2′′,6′′), 127.5 (C4 of Ph), 126.8 (C of triazole), 119.6 (C6), 117.8 (C8), 114.8 (C3′,5′), 112.9 9 (C2,6 of Ph), 73.7 (CH), 65.9 (CH2), 61.4 (CH2), 42.8 (CH2) ppm. Anal. calcd for C32H29N7O3: C, 68.68; H, 5.22; N, 17.52. Found: C, 68.44; H, 5.48; N, 17.21.
N-(4-Methylbenzyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9j)
Mp = 112–114 °C; IR (KBr): 3375, 3055, 2950, 2850, 1662, 1615, 1450 cm−1. 1HNMR (DMSO-d6, 500 MHz): 8.79 (t, J = 5.9 Hz, 1H, NH), 8.29 (bs, 1H, NH), 8.19 (s, 1H, triazole), 7.63 (d, J = 7.8 Hz, 1H, H5), 7.53 (s, 1H, NH), 7.35 (d, J = 8.1 Hz, 2H, H2′′, H6′′), 7.16 (t, J = 7.8 Hz, 1H, H7), 7.17–7.13 (m, 6H, H3′′, H5′′, Ph), 7.01 (d, J = 8.2 Hz, 2H, H2′, H6′), 6.84 (d, J = 8.2 Hz, 2H, H3′, H5′), 6.78 (d, J = 7.8 Hz, 1H, H8), 6.75 (t, J = 7.2 Hz, 1H, Ph), 6.70 (t, J = 7.8 Hz, 1H, H6), 5.84 (s, 1H, CH), 5.16 (s, 2H, CH2), 5.12 (s, 2H, CH2), 4.27 (d, J = 5.9 Hz, 2H, CH2), 2.27 (s, 3H, CH3). 13CNMR (DMSO-d6, 125 MHz): 165.7 (C O), 163.1 (C O), 158.6 (C4′), 150.7 (C1 of Ph), 148.4 (C9), 147.4 (C of triazole), 136.6 (C1′), 136.1 (C4′′), 134.2 (C1′′), 133.5 (C7), 129.5 (C10), 129.4 (C3,5 of Ph), 129.3 (C5), 129.0 (C3′′,5′′), 128.2 (C2′′,6′′), 128.0 (C4 of Ph), 127.9 (C2′,6′), 126.7 (C of triazole), 119.6 (C6), 117.8 (C8), 114.8 (C3′,5′), 112.8 (C2,6 of Ph), 73.6 (CH), 65.9 (CH2), 61.4 (CH2), 42.6 (CH2), 21.1 (CH3) ppm. Anal. calcd for C33H31N7O3: C, 69.09; H, 5.45; N, 17.09. Found: C, 68.85; H, 5.22; N, 16.85.
2-(4-((2-Methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-phenylacetamide (9k)
Mp = 119–121 °C; IR (KBr): 3380, 3058, 2955, 2845, 1660, 1614, 1445 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.47 (s, 1H, NH), 8.32 (s, 1H, NH), 8.31 (s, 1H, triazole), 7.64 (d, J = 7.7 Hz, 1H, H5), 7.58 (d, J = 8.0 Hz, 2H, H2′′, H6′′), 7.52 (s, 1H, NH), 7.33 (t, J = 8.0 Hz, 1H, H4′′), 7.29 (t, J = 7.7 Hz, 1H, H7), 7.16 (t, J = 7.5 Hz, 2H, Ph), 7.12–7.07 (m, 4H, Ph, H3′′, H4′′), 6.91 (d, J = 8.3 Hz, 1H, H6′), 6.86–6.84 (m, 2H, H2′, H5′), 6.80 (d, J = 7.7 Hz, 1H, H8), 6.75 (t, J = 7.5 Hz, 1H, Ph), 6.71 (t, J = 7.7 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.35 (s, 2H, CH2), 5.11 (s, 2H, CH2), 3.69 (s, 3H, OCH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.6 (C O), 163.1 (C O), 149.2 (C1 of Ph), 148.3 (C3′), 148.1 (C4′), 147.5 (C9), 142.9 (C of triazole), 138.9 (C1′′), 134.2 (C1′), 133.4 (C7), 129.5 (C10), 129.4 (C3,5 of Ph), 129.3 (C3′′,5′′), 128.0 (C5), 126.8 (C4′′), 126.3 (C4 of Ph), 125.1 (C of triazole), 124.2 (C2′′,6′′), 120.3 (C6′), 119.7 (C6), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 112.8 (C2,6 of Ph), 78.7 (C2), 62.0 (CH2), 60.2 (CH2), 55.8 (OCH3) ppm. Anal. calcd for C33H29N7O4: C, 67.77; H, 5.08; N, 17.03. Found: C, 67.50; H, 5.32; N, 16.85.
2-(4-((2-Methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(p-tolyl)acetamide (9l)
Mp = 114–116 °C; IR (KBr): 3350, 3050, 2950, 2850, 1665, 1625, 1445 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.39 (s, 1H, NH), 8.33 (s, 1H, NH), 8.23 (s, 1H, triazole), 7.65 (d, J = 7.5 Hz, 1H, H5), 7.53 (s, 1H, NH), 7.47 (d, J = 8.3 Hz, 2H, H2′′, H6′′), 7.28 (t, J = 7.5 Hz, 1H, H7), 7.18–7.10 (m, 6H, Ph, H2′′, H6′′), 6.91 (d, J = 7.0 Hz, 1H, H6′), 6.86–6.84 (m, 2H, H2′, H5′), 6.80 (d, J = 7.5 Hz, 1H, H8), 6.75 (t, J = 7.5 Hz, 1H, Ph), 6.71 (t, J = 7.5 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.33 (s, 2H, CH2), 5.12 (s, 2H, CH2), 3.69 (s, 3H, OCH3), 2.26 (s, 3H, CH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.4 (C O), 163.2 (C O), 149.4 (C1 of Ph), 149.2 (C3′), 148.4 (C4′), 148.3 (C9), 148.1 (C of triazole), 142.9 (C1′), 136.3 (C4′′), 135.5 (C1′′), 134.2 (C7), 133.2 (C10), 129.8 (C3′′,5′′), 129.7 (C3,5 of Ph), 129.6 (C5), 128.0 (C4 of Ph), 127.2 (C of triazole), 126.8 (C2′′,6′′), 126.3 (C6′), 119.7 (C6), 117.8 (C8), 115.1 (C5′), 114.5 (C2′), 112.9 (C2,6 of Ph), 78.7 (C2), 62.0 (CH2), 60.2 (CH2), 55.8 (OCH3), 20.9 (CH3) ppm. Anal. calcd for C33H31N7O4: C, 67.22; H, 5.30; N, 16.63. Found: C, 67.50; H, 5.52; N, 16.45.
N-(2,4-Dimethylphenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9m)
Mp = 134–136 °C; IR (KBr): 3365, 3055, 2950, 2850, 1670, 1625, 1440 cm−1. 1HNMR (DMSO-d6, 500 MHz): 9.78 (s, 1H, NH), 8.35 (s, 1H, NH), 8.32 (s, 1H, triazole), 7.64 (d, J = 7.7 Hz, 1H, H5), 7.52 (s, 1H, NH), 7.28 (t, J = 7.7 Hz, 1H, H7), 7.17 (t, J = 7.7 Hz, 2H, Ph), 7.11–7.08 (m, 5H, Ph, H3′′, H5′′, H6′′), 6.91 (d, J = 8.35 Hz, 1H, H6′), 6.86–6.84 (m, 2H, H2′, H5′), 6.78 (d, J = 7.7 Hz, 1H, H8), 6.75 (t, J = 7.7 Hz, 1H, Ph), 6.70 (t, J = 7.7 Hz, 1H, H6), 5.84 (s, 1H, CH), 5.38 (s, 2H, CH2), 5.11 (s, 2H, CH2), 3.68 (s, 3H, OCH3), 2.19–2.12 (m, 6H, 2× CH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.5 (C O), 163.4 (C O), 149.2 (C1 of Ph), 148.4 (C3′), 148.0 (C4′), 147.5 (C9), 146.2 (C4′′), 144.6 (C of triazole), 141.6 (C1′), 135.5 (C2′′), 134.6 (C1′′), 134.2 (C7), 133.9 (C3′′), 129.3 (C10), 128.2 (C3,5 of Ph), 128.0 (C5), 127.2 (C5′′), 126.8 (C4 of phenyl), 126.7 (C of triazole), 123.2 (C6′), 119.6 (C6), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 113.2 (C6′′), 112.8 (C2,6 of Ph), 74.0 (CH), 62.0 (CH2), 55.8 (OCH3), 52.1 (CH2), 18.5 (CH3) ppm. Anal. calcd for C34H33N7O4: C, 67.65; H, 5.51; N, 16.24. Found: C, 67.42; H, 5.30; N, 16.41.
N-(4-Ethylphenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9n)
Mp = 127–129 °C; IR (KBr): 3345, 3050, 2950, 2840, 1665, 1620, 1445 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.40 (s, 1H, NH), 8.32 (s, 1H, NH), 8.23 (s, 1H, triazole), 7.65 (d, J = 8.0 Hz, 1H, H5), 7.52 (s, 1H, NH), 7.48 (d, J = 7.0 Hz, 2H, H2′′, H6′′), 7.29 (t, J = 8.0 Hz, 1H, H7), 7.21–7.09 (m, 6H, Ph, H3′′, H5′′), 6.92 (d, J = 8.3 Hz, 1H, H6′), 6.86–6.84 (m, 2H, H2′, H5′), 6.80 (d, J = 8.0 Hz, 1H, H8), 6.75 (t, J = 7.1 Hz, 1H, Ph), 6.71 (t, J = 8.0 Hz, 1H, H6), 5.82 (s, 1H, CH), 5.33 (s, 2H, CH2), 5.11 (s, 2H, CH2), 3.69 (s, 3H, OCH3), 2.56 (q, J = 7.5 Hz, 2H, CH2), 1.15 (t, J = 7.5 Hz, 3H, CH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.4 (C O), 163.2 (C O), 149.7 (C1 of Ph), 149.2 (C3′), 148.4 (C4′), 148.1 (C9), 147.5 (C4′′), 140.6 (C of triazole), 139.7 (C1′), 136.5 (C1′′), 134.2 (C7), 133.8 (C10), 129.3 (C3′′,5′′), 128.6 (C5), 128.5 (C3,5 of phenyl), 128.0 (C2′′,6′′), 127.4 (C4 of phenyl), 126.9 (C of triazole), 126.8 (C6′), 119.8 (C6), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 112.9 (C2,6 of Ph), 74.0 (C2), 66.0 (CH2), 62.0 (CH2), 55.8 (CH2), 28.0 (CH2), 16.1 (CH3) ppm. Anal. calcd for C34H33N7O4: C, 67.65; H, 5.51; N, 16.24. Found: C, 67.80; H, 5.81; N, 16.38.
2-(4-((2-Methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(4-methoxyphenyl)acetamide (9o)
Mp = 127–129 °C; IR (KBr): 3365, 3055, 2950, 2850, 1665, 1620, 1447 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.33 (s, 1H, NH), 8.32 (s, 1H, NH), 8.22 (s, 1H, triazole), 7.64 (d, J = 7.5 Hz, 1H, H5), 7.52–7.49 (s, 3H, NH, H2′′, H6′′), 7.28 (t, J = 7.5 Hz, 1H, H7), 7.18–7.09 (m, 4H, Ph), 6.92–6.90 (m, 3H, H6′, H3′′, H5′′), 6.86–6.84 (m, 2H, H2′, H5′), 6.80 (d, J = 7.5 Hz, 1H, H8), 6.76–6.70 (m, 2H, Ph, H6), 5.85 (s, 1H, CH), 5.31 (s, 2H, CH2), 5.11 (s, 2H, CH2), 3.72 (s, 3H, OCH3), 3.69 (s, 3H, OCH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.1 (C O), 161.7 (C O), 156.2 (C4′′), 149.2 (C1 of phenyl), 148.4 (C3′), 148.1 (C4′), 147.5 (C9), 143.0 (C of triazole), 134.2 (C1′), 133.9 (C7), 131.9 (C1′′), 131.4 (C10), 129.3 (C3,5 of phenyl), 128.0 (C5), 126.8 (C4 of phenyl), 121.9 (C of triazole), 121.2 (C2′′,6′′), 120.8 (C6′), 119.6 (C6), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 114.5 (C3′′,5′′), 112.8 (C2,6 of Ph), 74.0 (C2), 62.0. (CH2), 60.3 (CH2), 55.8 (OCH3), 55.6 (OCH3) ppm. Anal. calcd for C33H31N7O5: C, 65.44; H, 5.16; N, 16.19. Found: C, 65.62; H, 5.38; N, 15.91.
N-(4-Chlorophenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9p)
Mp = 125–127 °C; IR (KBr): 3350, 3057, 2950, 2850, 1667, 1621, 1442 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.62 (s, 1H, NH), 8.32 (s, 1H, NH), 8.23 (s, 1H, triazole), 7.65 (d, J = 8.0 Hz, 1H, H5), 7.61 (d, J = 8.0 Hz, 2H, H2′′, H6′′), 7.53 (s, 1H, NH), 7.39 (d, J = 7.5 Hz, 2H, Ph), 7.29 (t, J = 8.0 Hz, 1H, H7), 7.17 (t, J = 7.5 Hz, 2H, Ph), 7.10 (d, J = 8.0 Hz, 2H, H3′′, H5′′), 6.92 (d, J = 8.3 Hz, 1H, H6′), 6.86–6.85 (m, 2H, H2′, H5′), 6.80 (d, J = 8.0 Hz, 1H, H8), 6.74 (t, J = 7.5 Hz, 1H, Ph), 6.71 (t, J = 8.0 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.36 (s, 2H, CH2), 5.11 (s, 2H, CH2), 3.69 (s, 3H, OCH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 167.0 (C O), 165.4 (C O), 151.4 (C1 of Ph), 150.5 (C3′), 150.2 (C4′), 149.7 (C9), 140.0 (C of triazole), 136.3 (C1′), 136.1 (C1′′), 131.6 (C4′′), 131.5 (C3′′,5′′), 130.2 (C7), 130.0 (C10), 129.0 (C3,5 of Ph), 124.1 (C5), 123.4 (C4 of Ph), 121.7 (C of triazole), 121.2 (C6′), 120.0 (C2′′,6′′), 117.3 (C6), 117.0 (C8), 115.4 (C5′), 115.0 (C2′), 113.3 (C2,6 of Ph), 76.2 (C2), 68.2 (CH2), 64.2 (CH2), 58.0 (OCH3) ppm. Anal. calcd for C32H28ClN7O4: C, 63.00; H, 4.63; N, 16.07. Found: C, 62.81; H, 4.48; N, 16.33.
N-(4-Bromophenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9q)
Mp = 121–123 °C; IR (KBr): 3355, 3060, 2950, 2850, 1668, 1623, 1447 cm−1. 1HNMR (DMSO-d6, 500 MHz): 10.65 (s, 1H, NH), 8.33 (s, 1H, NH), 8.23 (s, 1H, triazole), 7.65 (d, J = 7.7 Hz, 1H, H5), 7.58–7.51 (m, 5H, NH, H2′′, H3′′, H5′′, H6′′), 7.28(t, J = 7.7 Hz, 1H, H7), 7.17 (t, J = 8.0 Hz, 2H, Ph), 7.11 (d, J = 8.0 Hz, 2H, Ph), 6.92 (d, J = 8.3 Hz, 1H, H6′), 6.87–6.85 (m, 2H, H2′, H5′), 6.80 (d, J = 7.7 Hz, 1H, H8), 6.75 (t, J = 8.0 Hz, 1H, Ph) 6.71 (d, J = 7.7 Hz, 1H, H6), 5.86 (s, 1H, CH), 5.36 (s, 2H, CH2), 5.11 (s, 2H, CH2), 3.70 (s, 3H, OCH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 164.9 (C O), 163.2 (C O), 149.2 (C1 of Ph), 148.4 (C3′), 148.1 (C4′), 147.5 (C9), 142.9 (C of triazole), 138.2 (C1′), 137.6 (C1′′), 134.2 (C7), 132.2 (C3′′,5′′), 130.4 (C10), 129.3 (C3,5 of phenyl), 128.0 (C5), 127.2 (C4′′), 126.8 (C4 of phenyl), 126.3 (C of triazole), 122.2 (C6′), 121.6 (C2′′,6′′), 119.6 (C6), 117.8 (C8), 115.9 (C5′), 115.1 (C2′), 112.9 (C2,6 of Ph), 74.0 (CH), 62.0 (CH2), 60.2 (CH2), 55.8 (OCH3) ppm. Anal. calcd for C32H28BrN7O4: C, 58.72; H, 4.31; N, 14.98. Found: C, 58.48; H, 4.50; N, 15.21.
2-(4-((2-Methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(4-nitrophenyl)acetamide (9r)
Mp = 129–131 °C; IR (KBr): 3365, 3050, 2955, 2851, 1660, 1616, 1448 cm−1. 1HNMR (DMSO-d6, 500 MHz): 11.10 (s, 1H, NH), 8.33 (s, 1H, NH), 8.29–8.24 (m, 3H, triazole, H3′′, H5′′), 7.82 (d, J = 9.1 Hz, 2H, H2′′, H6′′), 7.64 (d, J = 8.0 Hz, 1H, H5), 7.52 (s, 1H, NH), 7.28 (t, J = 8.0 Hz, 1H, H7), 7.16 (t, J = 8.0 Hz, 2H, Ph), 7.10 (d, J = 8.0 Hz, 2H, Ph), 6.92 (d, J = 8.3 Hz, 1H, H6′), 6.86–6.84 (m, 2H, H2′, H5′), 6.79 (d, J = 8.0 Hz, 1H, H8), 6.75 (t, J = 8.0 Hz, 1H, Ph) 6.71 (d, J = 8.0 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.44 (s, 2H, CH2), 5.12 (s, 2H, CH2), 3.69 (s, 3H, OCH3) ppm. 13CNMR (DMSO-d6, 125 MHz): 165.8 (C O), 163.4 (C O), 149.2 (C1 of Ph), 148.4 (C3′), 148.0 (C4′), 147.5 (C9), 145.0 (C4′′), 143.1 (C3′′), 143.0 (C of triazole), 134.2 (C1′), 129.3 (C7), 128.0 (C10), 126.8 (C3,5 of phenyl), 126.3 (C5), 126.2 (C3′′,5′′), 125.6 (C4 of phenyl), 125.5 (C of triazole), 120.2 (C6′), 119.6 (C6), 119.5 (C2′′,6′′), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 112.9 (C2,6 of Ph), 74.0 (C2), 66.0 (CH2), 62.0 (CH2), 55.8 (OCH3) ppm. Anal. calcd for C32H28N8O6: C, 61.93; H, 4.55; N, 18.06. Found: C, 62.26; H, 4.31; N, 17.80.
N-Benzyl-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9s)
Mp = 111–113 °C; IR (KBr): 3365, 3055, 2955, 2849, 1658, 1614, 1451 cm−1. 1HNMR (DMSO-d6, 500 MHz): 8.84 (t, J = 5.8 Hz, 1H, NH), 8.32 (bs, 1H, NH), 8.17 (s, 1H, triazole), 7.64 (d, J = 7.7 Hz, 1H, H5), 7.52 (s, 1H, NH), 7.36–7.09 (m, 10H, H7, H2′′, H3′′, H4′′, H5′′, H6′′, Ph), 6.91 (d, J = 8.3 Hz, 1H, H6′), 6.85–6.84 (m, 2H, H2′, H5′), 6.79 (d, J = 7.7 Hz, 1H, H8), 6.74 (t, J = 7.3 Hz, 1H, Ph), 6.71 (t, J = 7.7 Hz, 1H, H6), 5.84 (s, 1H, CH), 5.18 (s, 2H, CH2), 5.10 (s, 2H, CH2), 4.32 (d, J = 5.8 Hz, 2H, CH2), 3.66 (s, 3H, OCH3). 13CNMR (DMSO-d6, 125 MHz): 165.9 (C O), 163.3 (C O), 149.2 (C1 of Ph), 148.4 (C3′), 148.0 (C4), 147.5 (C9), 142.9 (C of triazole), 134.2 (C1′), 133.8 (C1′′), 129.7 (C7), 129.6 (C10), 129.3 (C5), 128.8 (C3,5 of phenyl), 128.0 (C3′′,5′′), 127.9 (C2′′,6′′), 127.5 (C4′′), 126.9 (C4 of phenyl), 126.8 (C of triazole), 126.7 (C6′), 119.6 (C6), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 112.8 (C2,6 of Ph), 74.0 (CH), 65.9 (CH2), 62.0 (CH2), 55.8 (OCH3), 42.8 (CH2) ppm. Anal. calcd for C33H31N7O4: C, 67.22; H, 5.30; N, 16.63. Found: C, 67.00; H, 5.12; N, 16.81.
2-(4-((2-Methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(4-methylbenzyl)acetamide (9t)
Mp = 118–120 °C; IR (KBr): 3350, 3052, 2944, 2845, 1660, 1614, 1461 cm−1. 1HNMR (DMSO-d6, 500 MHz): 8.79 (t, J = 5.9 Hz, 1H, NH), 8.32 (bs, 1H, NH), 8.18 (s, 1H, triazole), 7.63 (d, J = 7.9 Hz, 1H, H5), 7.52 (s, 1H, NH), 7.29 (t, J = 7.9 Hz, 1H, H7), 7.18–7.09 (m, 8H, H2′′, H3′′, H5′′, H6′′, Ph), 6.92 (d, J = 8.3 Hz, 1H, H6′), 6.86–6.84 (m, 2H, H2′, H5′), 6.80 (d, J = 7.9 Hz, 1H, H8), 6.75 (t, J = 7.3 Hz, 1H, Ph), 6.71 (t, J = 7.9 Hz, 1H, H6), 5.85 (s, 1H, CH), 5.17 (s, 2H, CH2), 5.10 (s, 2H, CH2), 4.27 (d, J = 5.9 Hz, 2H, CH2), 3.69 (s, 3H, OCH3), 2.28 (s, 3H, CH3). 13CNMR (DMSO-d6, 125 MHz): 165.7 (C O), 163.2 (C O), 149.2 (C1 of Ph), 148.4 (C3′), 148.0 (C4′), 147.5 (C9), 142.2 (C of triazole), 136.6 (C1′), 136.1 (C4′′), 134.2 (1′′), 133.9 (C7), 129.5 (C10), 129.4 (C3,5 of phenyl), 129.3 (C3′′,5′′), 128.5 (C5), 128.0 (C2′′,6′′), 127.9 (C4 of phenyl), 126.8 (C of triazole), 126.7 (C6′), 119.6 (C6), 117.8 (C8), 115.1 (C5′), 114.8 (C2′), 112.8 (C2,6 of Ph), 74.0 (C2), 66.0 (CH2), 62.0 (CH2), 55.8 (OCH3), 42.6 (CH2), 21.1 (CH3) ppm. Anal. calcd for C34H33N7O4: C, 67.65; H, 5.51; N, 16.24. Found: C, 67.41; H, 5.73; N, 16.38.
2.3. α-Glucosidase inhibition assay
The anti-α-glucosidase activity of synthesized compounds 9a–t, was evaluated according to the previously reported method. Briefly, to 135 μL of potassium phosphate buffer in the 96-well plate, 20 μL of α-glucosidase solution and 20 μL of the target entries at different concentrations were added and incubated at 37 °C for 10 min. Then, p-nitrophenyl glucopyranoside as the substrate (25 μL, 4 mM) was added and further incubated for an extra 20 min at 37 °C. Finally, the absorbance was measured at 405 nm by a spectrophotometer.25
2.4. Enzyme kinetic study
The mode of inhibition of the most active compound 9c, was investigated against α-glucosidase with different concentrations of p-nitrophenyl α-d-glucopyranoside (1–16 mM) as the substrate in the absence and presence of 9c at different concentrations of 0, 1.2, 2.4, and 4.8 μM. A Lineweaver–Burk plot was generated to identify the type of inhibition and the Michaelis–Menten constant (Km) value was determined from the plot between the reciprocal of the substrate concentration (1/[S]) and reciprocal of enzyme rate (1/V) over various inhibitor concentrations.25
2.5. Molecular docking
To perform the molecular docking studies, the Maestro Molecular Modeling platform (version 10.5) by Schrödinger, LLC was used. The X-ray crystal structure of the receptor was downloaded from the PDB database (PDB ID: 5NN8). The protein was then prepared using a protein preparation wizard so that all water molecules and co-crystallized ligands were removed, the missing side chains and loops were filled using the prime tool, and PROPKA assigned H-bonds at pH: 7.4. To prepare the ligands, the 2D structures of the ligands were drawn in ChemDraw (ver. 16) and converted into SDF files, which were used further by the ligprep module. Ligands were prepared by OPLS_2005 force field using EPIK. The grid box was generated for each binding site using entries with a box size of 25 A, all derivatives were docked on binding sites using induced-fit docking, reporting 10 poses per ligand to form the final complex.26
2.6. Molecular dynamic simulation
The molecular simulation was performed using the Desmond v5.3 (Schrödinger 2018-4 suite). To build the system for MD simulation, the protein–ligand complex was solvated with SPC explicit water molecules and placed in the center of an orthorhombic box of appropriate size in the periodic boundary condition. Sufficient counter-ions and a 0.15 M solution of NaCl were also utilized to neutralize the system and to simulate the real cellular ionic concentrations, respectively. The MD protocol involved minimization, pre-production, and finally production MD simulation steps. In the minimization procedure, the entire system was allowed to relax for 2500 steps by the steepest descent approach. Then, the temperature of the system was raised from 0 to 300 K with a small force constant on the enzyme in order to restrict any drastic changes. MD simulations were performed via NPT (constant number of atoms, constant pressure i.e. 1.01325 bar, and constant temperature i.e. 300 K) ensemble. The Nose–Hoover chain method was used as the default thermostat with 1.0 ps interval and Martyna–Tobias–Klein as the default barostat with 2.0 ps interval by applying isotropic coupling style. Long-range electrostatic forces were calculated based on the particle-mesh-based Ewald approach with the cut-off radius for Columbia forces set to 9.0 Å. Finally, the system was subjected to produce MD simulations for 30 ns for each protein–ligand complex. The dynamic behavior and structural changes of the systems were analyzed by the calculation of the RMSD and RMSF.34
3. Results and discussion
3.1. Chemistry
The synthetic route was described in Scheme 1. Initially, 3-(phenylamino)-2-(4-(prop-2-yn-1-yloxy)phenyl)-2,3-dihydroquinazolin-4(1H)-one 5 was prepared starting from isatoic anhydride 1. The reaction of 1 and phenylhydrazine 2 in water at room temperature gave 2-amino-N′-phenylbenzohydrazide 3. The reaction of compound 3 and aldehyde 4 (ref. 33) in water in the presence of sodium dodecyl sulfate (SDS) gave compound 5. On the other, different aromatic and aliphatic amine derivatives 6 reacted with chloroacetyl chloride 7 in DMF at room temperature to prepare desired chloride derivative 8 which are prone to participate in the click reaction with 5 in the presence of Et3N, CuSO4·5H2O, and sodium ascorbate (SA) in DMF at room temperature, affording the title compounds 9a–t.
3.2. In vitro inhibition of α-glucosidase and the structure–activity relationships
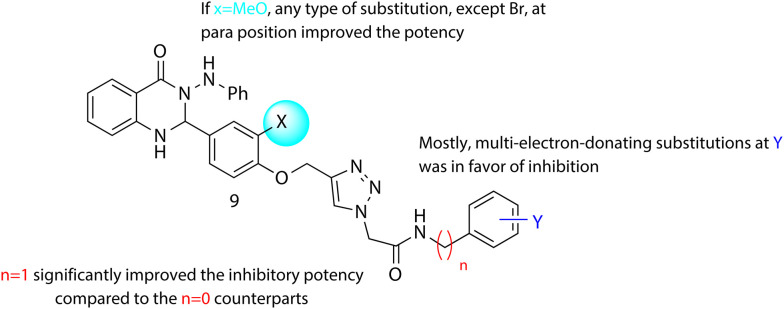
Quinazolin-4(1H)-one derivatives, 9a–t were divided into two categories based on the phenoxy ring substitution at the X position (Table 1). Category “A” has ten compounds 9a–h with X = H and variable aryl group (Y) and different lengths of chain at n position (IC50 values in the range of 4.8 ± 0 to 95.6 ± 1.3 μM). Category “B” has ten analogs 9k–t with methoxy at X position and variable aryl group (Y) and the different number of carbons at n position (IC50 values in the range of 5.0 ± 0.1 to 140.2 ± 1.7 μM). Overall, it can be understood that good to excellent inhibitory activities was observed for all the synthetic derivatives against α-glucosidase when compared to the standard acarbose (IC50 = 750.0 ± 2.1 μM).
α-Glucosidase inhibitory activity of compounds 9a–t.
| |||||
|---|---|---|---|---|---|
| Entry | Compound 9 | X | n | Y | IC50a (μM) |
| 1 | 9a | H | 0 | H | 16.6 ± 0.6 |
| 2 | 9b | H | 0 | 4-Me | 95.6 ± 1.3 |
| 3 | 9c | H | 0 | 2,4-diMe | 4.8 ± 0 |
| 4 | 9d | H | 0 | 4-Et | 14.7 ± 0.1 |
| 5 | 9e | H | 0 | 4-MeO | 42.6 ± 0.5 |
| 6 | 9f | H | 0 | 4-Cl | 15.3 ± 0.7 |
| 7 | 9g | H | 0 | 4-Br | 8.0 ± 0.1 |
| 8 | 9h | H | 0 | 4-NO2 | 12.0 ± 0.5 |
| 9 | 9i | H | 1 | H | 12.0 ± 0.4 |
| 10 | 9j | H | 1 | 4-Me | 8.3 ± 0 |
| 11 | 9k | OCH3 | 0 | H | 11.7 ± 0.4 |
| 12 | 9l | OCH3 | 0 | 4-Me | 13.0 ± 0.7 |
| 13 | 9m | OCH3 | 0 | 2,4-diMe | 10.6 ± 0.1 |
| 14 | 9n | OCH3 | 0 | 4-Et | 7.2 ± 0 |
| 15 | 9o | OCH3 | 0 | 4-MeO | 5.0 ± 0.1 |
| 16 | 9p | OCH3 | 0 | 4-Cl | 6.7 ± 0.1 |
| 17 | 9q | OCH3 | 0 | 4-Br | 140.2 ± 1.7 |
| 18 | 9r | OCH3 | 0 | 4-NO2 | 6.0 ± 0.1 |
| 19 | 9s | OCH3 | 1 | H | 11.6 ± 0.2 |
| 20 | 9t | OCH3 | 1 | 4-Me | 5.4 ± 0 |
| 21 | Acarbose | 750.0 ± 2.1 | |||
Data are expressed as mean ± SE (three independent experiments).
Category “A”
The limited SARs suggested that 2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-phenylacetamide (9a, IC50 = 16.6 ± 0.6 μM) bearing unsubstituted aryl ring demonstrated good inhibition against α-glucosidase. The introduction of electron-donating groups35 into compounds 9b, 9c, and 9d, displayed variable inhibitory activity. In detail, the insertion of a methyl group into the para position of aryl ring resulted in reduced activity in 2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(p-tolyl)acetamide (9b, IC50 = 95.6 ± 1.3 μM), however, N-(2,4-dimethylphenyl)-2-(4-((4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide 9c bearing 2,4-diMe exhibited IC50 value of 4.8 μM with many folds better than the standard acarbose and is the most potent scaffold among the synthesized derivatives. The improvement in the activity was also observed in 9d bearing 4-Et with IC50 = 14.7 ± 0.1 μM compared with 9a. Compound 9e with para-MeO substitution, exhibited less inhibitory activity (IC50 = 45.04 ± 0.52 μM) than 9a. It seems that multi-electron donating substitution is in favor of inhibition.
The electron-withdrawing bearing compounds, 9f, 9g, and 9h, substituted at the para position showed better inhibition compared to the unsubstituted one, 9a. From IC50 values of enzyme inhibition pattern, it was confirmed that 4-Br substituted > 4-NO2 substituted > 4-Cl substituted scaffold which showed the positive role of inductive effect as well as increased bulkiness.
Interesting results were obtained by increasing the length at n position in which a significant increase in the inhibitory potencies was seen in 9a (X = H, n = 0, Y = H, IC50 = 16.6 μM) vs.9i (X = H, n = 1, Y = H, IC50 = 12.0 μM) and 9b (X = H, n = 0, Y = H, IC50 = 95.6 μM) vs.9j (X = H, n = 1, Y = 4-Me, IC50 = 8.3 ± 0).
Category “B”
Geometric mean analysis of compounds 9k–t having methoxy substitution at the X position of phenoxy moiety displayed no significant differences in inhibitory activities as compared with compounds 9a–h categorized as unsubstituted derivatives at the X position.
2-(4-((2-Methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-phenylacetamide 9k with an unsubstituted derivative displayed good inhibition with an IC50 value of 11.7 ± 0.4 μM. Among electron-donating-substituted derivatives, 2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(4-methoxyphenyl)acetamide (9o), Y = 4-MeO was the most active compound of this series, which may be due to the electron-donating power by resonance. N-(2,4-Dimethylphenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9m) and N-(4-ethylphenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9n) also improved the inhibitory activities compared to 2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-phenylacetamide (9k), and there were no significant differences between 9l (Y = 4-Me) and 9k (Y = H). Like the previous set, multi-electron donating substitutions improved the inhibition.
N-(4-Chlorophenyl)-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9p) and 2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(4-nitrophenyl)acetamide (9r) with 4-chloro and 4-nitro electron withdrawing substitutions revealed an improvement in the potency compared to 9k. However, bromo substituted compound, 9q, was the least active compound among all derivatives which might be due to the bulkiness of the bromine group at Y as well as methoxy group at X resulting in poor interaction with the enzyme.
As can be seen in N-benzyl-2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)acetamide (9s) and 2-(4-((2-methoxy-4-(4-oxo-3-(phenylamino)-1,2,3,4-tetrahydroquinazolin-2-yl)phenoxy)methyl)-1H-1,2,3-triazol-1-yl)-N-(4-methylbenzyl)acetamide (9t), elongation of linker at n position was also in favor of potency with IC50 values of 11.6 ± 0.2 and 5.4 ± 0 μM, respectively.
The overall straightforward trend cannot be seen in the comparison of category A vs. category B; however, increased inhibition potential in derivatives with longer chain at n might be due to better rotation of substituted group and occupation of the active site. In category A, SARs indicated that the number of substitutions affects inhibitory activities. While in category B any type of position at the para position except, para bromo is in favor of inhibition. Assessment of the electron-donating group of category B compared to category A showed that the electron donating group bearing OCH3 at X showed better activity compared to their counterparts in group A, the exception in this trend came back to 9c (Fig. 2).
Fig. 2. Summary of SARs of quinazolinone-bearing 1,2,3-triazole acetamide as α-glucosidase inhibitor.
3.3. Enzyme kinetic studies
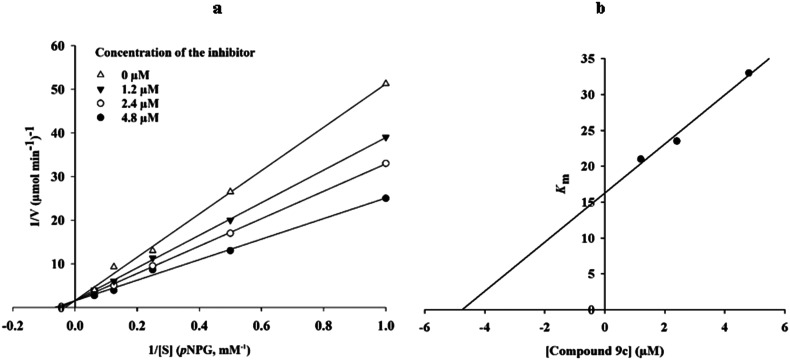
According to Fig. 3a, the Lineweaver–Burk plot showed that the Km gradually increased, and Vmax remained unchanged with increasing concentration of inhibitor, indicating a competitive inhibition. The results showed that 9c bound to the active site in competition with the substrate. Furthermore, the plot of the Kmversus different concentrations of inhibitor gave an estimate of the inhibition constant, Ki = 4.8 μM (Fig. 3b).
Fig. 3. Kinetics of α-glucosidase inhibition by compound 9c. (a) The Lineweaver–Burk plot in the absence and presence of different concentrations of the 9c. (b) The secondary plot between Km and various concentrations of the 9c.
3.4. Molecular docking studies
Docking studies were performed by using the Schrodinger software package. First, to validate the molecular docking procedures, acarbose as a native ligand was docked inside the α-glucosidase.36 Alignment of the best pose of acarbose in the active site of the enzyme and crystallographic ligand recorded an RMSD value of 1.71 Å (RMSD should be less than 2 Å), which confirms the accuracy of docking. Then, the docking procedures were applied to all synthesized derivatives. The detailed interaction of all derivatives is presented in Table 2. The in silico studies showed the score values for compounds 9a–t ranged from −4.350 to −8.973. The top GlideScore value belonged to 9c with a value of −8.973, followed by 9n (−7.741) and 9r (7.69). Overall, developed pharmacophores presented in the designed structure, including quinazolin-4(1H)-one and arylacetamide moieties, effectively interacted with the α-glucosidase active site.
The predicted binding energy of synthesized compounds 9 with the desired enzyme.
| Compound | GlideScore | Moiety | Residue | Type of interaction |
|---|---|---|---|---|
| 9a | −6.402 | Phenylacetamide | Trp376 | pi–pi stacking |
| CO of phenylacetamide | Leu677 | H-Bonding | ||
| Aminophenyl | Phe525 | pi–pi stacking | ||
| 9b | −5.445 | C O of phenylacetamide | Phe525 | H-Bonding |
| 1,2,3-Triazole | Arg281 | pi–cation | ||
| Quinazolin-4(1H)-one | Trp481 (2)a | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Phe649 | pi–pi stacking | ||
| 9c | −8.973 | C O of quinazolin-4(1H)-one | Leu768 | H-Bonding |
| C O of phenylacetamide | Arg600 | H-Bonding | ||
| C O of phenylacetamide | Ash616 | H-Bonding | ||
| 2,4-Dimethylphenyl | Trp481 (2) | pi–pi stacking | ||
| 9d | −7.252 | C O of quinazolin-4(1H)-one | Ash616 | H-Bonding |
| Quinazolin-4(1H)-one | Trp481 | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Phe649 | pi–pi stacking | ||
| Methoxy | Trp376 | pi–pi stacking | ||
| 9e | −6.316 | C O of quinazolin-4(1H)-one | Ash616 | H-Bonding |
| 4-Methoxyphenyl | Gly651 | H-Bonding | ||
| 9f | −6.510 | C O of phenylacetamide | Phe525 | H-Bonding |
| Aminophenyl | Trp481 | pi–pi stacking | ||
| 9g | −6.077 | C O of quinazolin-4(1H)-one | Arg600 | H-Bonding |
| C O of quinazolin-4(1H)-one | Ash616 | H-Bonding | ||
| Methoxy | Trp481 | pi–pi stacking | ||
| 1,2,3-Triazole | Phe525 | pi–pi stacking | ||
| 9h | 5.827 | NH of aminophenyl | Asp281 | H-Bonding |
| Phenyl | Arg281 | pi–cation | ||
| 9i | −6.357 | C O of quinazolin-4(1H)-one | Ash616 | H-Bonding |
| Quinazolin-4(1H)-one | Phe525 | pi–pi stacking | ||
| Aminophenyl | Trp376 | pi–pi stacking | ||
| 9j | −5.350 | NH of quinazolin-4(1H)-one | Asp282 | H-Bonding |
| Quinazolin-4(1H)-one | Arg281 | pi–cation | ||
| 1,2,3-Triazole | Ash616 | H-Bonding | ||
| 1,2,3-Triazole | Trp481 | pi–pi stacking | ||
| 9k | −6.118 | C O of quinazolin-4(1H)-one | Ash616 | H-Bonding |
| Quinazolin-4(1H)-one | Trp481 | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Trp376 | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Hip674 | pi–cation | ||
| 9l | −6.349 | C O of phenylacetamide | Arg411 | H-Bonding |
| C O of phenylacetamide | Phe525 | H-Bonding | ||
| Quinazolin-4(1H)-one | Phe649 | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Trp481 | pi–pi stacking | ||
| Aminophenyl | Trp481 | pi–pi stacking | ||
| 1,2,3-Triazole | Arg281 | pi–cation | ||
| 9m | −6.580 | C O of quinazolin-4(1H)-one | Ash616 | H-Bonding |
| Quinazolin-4(1H)-one | Phe649 (2) | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Phe649 | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Trp376 (2) | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Phe674 | pi–cation | ||
| 9n | −7.741 | C O of phenylacetamide | Arg411 | H-Bonding |
| C O of quinazolin-4(1H)-one | Ash616 | H-Bonding | ||
| C O of quinazolin-4(1H)-one | Arg600 | H-Bonding | ||
| C O of quinazolin-4(1H)-one | Ash281 | H-Bonding | ||
| Phenoxy | Phe525 | pi–pi stacking | ||
| 9o | −6.326 | Quinazolin-4(1H)-one | Phe281 (2) | pi–cation |
| Quinazolin-4(1H)-one | Asp28 | H-Bonding | ||
| Phenoxy | Arg600 | H-Bonding | ||
| 1,2,3-Triazole | Trp481 | pi–pi stacking | ||
| 1,2,3-Triazole | Phe649 | pi–pi stacking | ||
| 4-Methoxyphenyl | Trp376 | pi–pi stacking | ||
| 9p | −7.607 | C O of quinazolin-4(1H)-one | Ash616 | H-Bonding |
| Amide | Arg281 | H-Bonding | ||
| 9q | −7.441 | Quinazolin-4(1H)-one | Phe525 (2) | pi–pi stacking |
| NH of quinazolin-4(1H)-one | Asp282 | H-Bonding | ||
| Phenoxy | Ash616 | H-Bonding | ||
| C O of phenylacetamide | Leu677 | H-Bonding | ||
| 9r | 7.690 | Quinazolin-4(1H)-one | Trp481 (2) | pi–pi stacking |
| Quinazolin-4(1H)-one | Phe649 (2) | pi–pi stacking | ||
| C O of quinazolin-4(1H)-one | Arg600 | H-Bonding | ||
| C O of quinazolin-4(1H)-one | Arg616 | H-Bonding | ||
| 1,2,3-Triazole | Gly651 | H-Bonding | ||
| C O of acetamide | Leu678 | H-Bonding | ||
| 4-Nitrophenyl | Arg411 | H-Bonding | ||
| 9s | −6.393 | Quinazolin-4(1H)-one | Phe649 (2) | pi–pi stacking |
| Quinazolin-4(1H)-one | Trp481 | pi–pi stacking | ||
| Quinazolin-4(1H)-one | Trp4376 | pi–pi stacking | ||
| Aminophenyl | Ash616 | H-Bonding | ||
| 9t | −6.118 | NH of quinazolin-4(1H)-one | Phe525 | H-Bonding |
| NH of (4-methylbenzyl)acetamide | Asp282 | H-Bonding | ||
| 4-Methylbenzyl | Phe649 | pi–pi stacking | ||
| 4-Methylbenzyl | Trp481 | pi–pi stacking |
(2): two interactions.
3.5. Molecular dynamic simulations
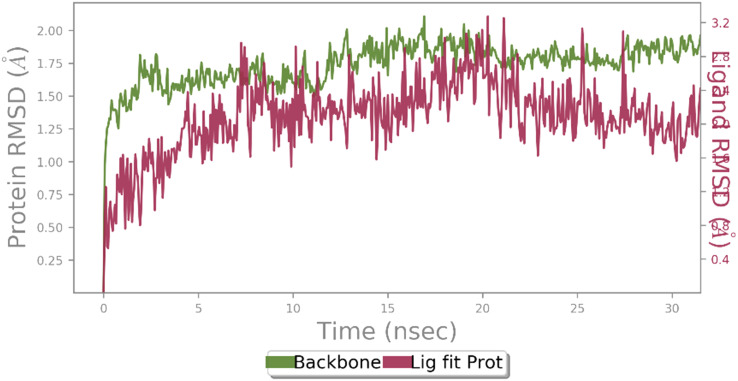
In order to investigate the binding interaction and the stability of the most potent compound 9c in the α-glucosidase active site, molecular dynamics simulations were performed. The root mean square deviation (RMSD) of the protein's backbone compared with initial conformation was used to study the stability of the protein-ligand complex. As can be seen in Fig. 4, the RMSD of 9c-α-glucosidase complex reached to stability after seven ns of MD simulation time with an average value of 1.25 Å. These results indicated that the simulation time was adequate to obtain an equilibrium structure to investigate the structural specificity of the ligand–protein complexes properly.
Fig. 4. RMSD plot of the α-glucosidase backbone (green) and α-glucosidase in complex with 9c (red).
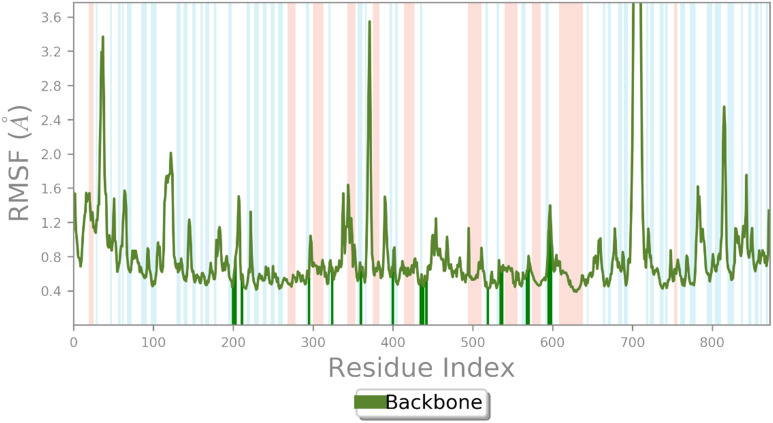
The root mean square fluctuation (RMSF) value displayed the fluctuation and flexibility of the protein's residues from its average position throughout the simulation. The α-helical and β-strand regions as secondary structures are highlighted in red and blue backgrounds (Fig. 5). Comparing the RMSF values showed that the nonstructural region exhibited more fluctuations and the residues of α-helical and β-strand regions are almost fixed during the simulation time.
Fig. 5. RMSF plot of the α-glucosidase residue in complexed with 9c.
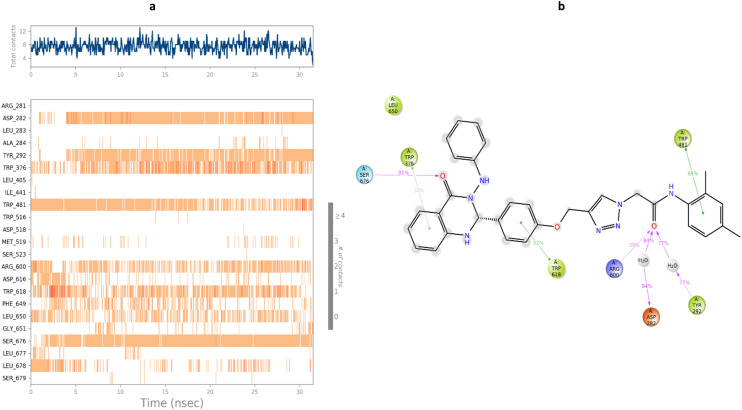
The timeline interaction of compound 9c with the residue of the enzyme was demonstrated in Fig. 6. The interaction pattern of 9c against α-glucosidase showed that quinazolinone participated in H-bonding interaction with Ser676 and pi–pi stacking with Trp376. On the other side of the molecule, 2,4-dimethylphenyl moiety disclosed pi-pi stacking with Trp481, and the amide linker recorded three H-bonding interactions with Asp282, Tyr292, and Arg600. The methoxy group also recorded pi–pi stacking interaction with Trp618.
Fig. 6. a) Timeline rendering of interacting residues with 9c during the simulation time, b) 2D representation of ligand–residue interactions that occur at least 30% of simulation time at the equilibrated phase of MD simulation, which includes α-glucosidase bound-state with compound 9c.
3.6. Druglikeness properties
The in silico drug-likeness properties of all derivatives were predicted from the SwissADME website (Table 3). To improve permeation, at least three of the following rules should be fulfilled, the molecular weight should be less than 500, no more than 5 hydrogen bond donors, no more than 10 H-bond acceptor groups, and log P value less than 5. According to obtained results, it can be understood that compounds 9a, 9b, 9c, 9f, 9g and 9i might exhibit better permeation compared to the rest of the derivatives.
Drug-likeness properties of synthesized compounds 9.
| Compound | MW | Num. rotatable bonds | Num. H-bond acceptors | Num. H-bond donors | Log P |
|---|---|---|---|---|---|
| 9a | 545.59 | 10 | 5 | 3 | 4.91 |
| 9b | 559.62 | 10 | 5 | 3 | 5.28 |
| 9c | 573.64 | 10 | 5 | 3 | 5.64 |
| 9d | 573.64 | 11 | 5 | 3 | 5.71 |
| 9e | 575.62 | 11 | 6 | 3 | 4.88 |
| 9f | 580.04 | 10 | 5 | 3 | 5.54 |
| 9g | 624.49 | 10 | 5 | 3 | 5.60 |
| 9h | 591.60 | 11 | 7 | 4 | 4.28 |
| 9i | 559.62 | 11 | 5 | 3 | 4.85 |
| 9j | 573.64 | 11 | 5 | 3 | 5.31 |
| 9k | 575.62 | 11 | 6 | 3 | 4.88 |
| 9l | 589.64 | 11 | 6 | 3 | 5.25 |
| 9m | 603.67 | 11 | 6 | 3 | 5.61 |
| 9n | 603.67 | 12 | 6 | 3 | 5.68 |
| 9o | 605.64 | 12 | 7 | 3 | 4.85 |
| 9p | 610.06 | 11 | 6 | 3 | 5.51 |
| 9q | 654.51 | 11 | 6 | 3 | 5.57 |
| 9r | 621.62 | 12 | 8 | 4 | 4.25 |
| 9s | 589.64 | 12 | 6 | 3 | 4.82 |
| 9t | 603.67 | 12 | 6 | 3 | 5.18 |
4. Conclusion
A series of quinazolinone-1,2,3-triazole-acetamides incorporating the quinazolin-4-one scaffold was designed, synthesized, and fully characterized. All analogs showed good inhibitory potential with IC50 values ranging from 4.8 ± 0 to 140.2 ± 1.7 μM when compared with acarbose (IC50 = 750.0 μM). SAR reveals that increased length of chain at n increased inhibition potential. Also, multi-substitutions at Y position like 2,4-diMe made the molecule polar and showed high activity. The kinetics study of compound 9c was further studied and found that it was a competitive inhibitor. Through molecular docking, the binding interaction of the molecule with the active site of enzyme was confirmed. Based on these results, quinazolinone-1,2,3-triazole-acetamide could be considered as an attractive candidate for antidiabetic drug discovery.
Author contributions
Sara Moghadam Farid synthesized compounds. Aida Iraji performed in silico studies and wrote the manuscript. Somayeh Mojtabavi conducted the biological assay. Maliheh Barazandeh Tehrani contributed to characterization of compounds. Mohammad Ali Faramarzi supervised the biological assay. Mehrnaz Ghasemi and Mohammad Mahdavi contributed to the synthesis of compounds. Tahmineh Akbarzadeh contributed to design of compounds. Mina Saeedi designed the project, characterized compounds, wrote the manuscript, and supervised all phases of the project. All authors read and approved the final manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by grants from the Research Council of Tehran University of Medical Sciences with project No. 1400-3-157-55391.
This paper is dedicated to the memory of our unique teacher in Chemistry and Medicinal Chemistry, Professor Abbas Shafiee (1937–2016).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2md00297c
References
- Papachristoforou E. Lambadiari V. Maratou E. Makrilakis K. J. Diabetes Res. 2020;2020:7489795. doi: 10.1155/2020/7489795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumawat V. S. Kaur G. Eur. J. Pharmacol. 2019;862:172628. doi: 10.1016/j.ejphar.2019.172628. [DOI] [PubMed] [Google Scholar]
- Rivera-Mancía S. Lozada-García M. C. Pedraza-Chaverri J. Eur. J. Pharmacol. 2015;756:30–37. doi: 10.1016/j.ejphar.2015.02.045. [DOI] [PubMed] [Google Scholar]
- Mehmood R. Mughal E. U. Elkaeed E. B. Obaid R. J. Nazir Y. Al-Ghulikah H. A. Naeem N. Al-Rooqi M. M. Ahmed S. A. Shah S. W. A. Sadiq A. ACS Omega. 2022;7:30215–30232. doi: 10.1021/acsomega.2c03328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf J. Mughal E. U. Sadiq A. Naeem N. Muhammad S. A. Qousain T. Zafar M. N. Khan B. A. Anees M. J. Mol. Struct. 2020;1218:128458. doi: 10.1016/j.molstruc.2020.128458. [DOI] [Google Scholar]
- Sohrabi M. Binaeizadeh M. R. Iraji A. Larijani B. Saeedi M. Mahdavi M. RSC Adv. 2022;12:12011–12052. doi: 10.1039/D2RA00067A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariya N. Mochizuki K. Inoue S. Saito M. Fuchigami M. Goda T. Osonoi T. Drugs R&D. 2014;14:177–184. doi: 10.1007/s40268-014-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNicolantonio J. J. Bhutani J. O'Keefe J. H. Open Heart. 2015;2:e000327. doi: 10.1136/openhrt-2015-000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehbi O. Riadi Y. Geesi M. H. Anouar E. H. Ibnouf E. O. Azzallou R. Polycyclic Aromat. Compd. 2022:1–9. [Google Scholar]
- Öztürk S. Okay S. Yıldırım A. Russ. Chem. Bull. 2020;69:2205–2214. doi: 10.1007/s11172-020-3023-0. [DOI] [Google Scholar]
- Chen L. Wang X. Tang X. Xia R. Guo T. Zhang C. Li X. Xue W. BMC Chem. 2019;13:1–12. doi: 10.1186/s13065-019-0516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. Zhong T. Yang H. Yang Y. Wang D. Yang X. Xu Y. Fan Y. Eur. J. Med. Chem. 2020;190:112108. doi: 10.1016/j.ejmech.2020.112108. [DOI] [PubMed] [Google Scholar]
- Le-Nhat-Thuy G. Thi N. N. Pham-The H. Thi T. A. D. Thi H. N. Thi T. H. N. Hoang S. N. Van Nguyen T. Bioorg. Med. Chem. Lett. 2020;30:127404. doi: 10.1016/j.bmcl.2020.127404. [DOI] [PubMed] [Google Scholar]
- Ghodge B. Kshirsagar A. Navghare V. Beni-Suef Univ. J. Basic Appl. Sci. 2020;9:1–12. doi: 10.1186/s43088-019-0027-7. [DOI] [Google Scholar]
- Saeedi M. Mohammadi-Khanaposhtani M. Pourrabia P. Razzaghi N. Ghadimi R. Imanparast S. Faramarzi M. A. Bandarian F. Esfahani E. N. Safavi M. Bioorg. Chem. 2019;83:161–169. doi: 10.1016/j.bioorg.2018.10.023. [DOI] [PubMed] [Google Scholar]
- Wei M. Chai W.-M. Wang R. Yang Q. Deng Z. Peng Y. Bioorg. Med. Chem. 2017;25:1303–1308. doi: 10.1016/j.bmc.2016.09.042. [DOI] [PubMed] [Google Scholar]
- Menteşe E. Karaali N. Akyüz G. Yılmaz F. Ülker S. Kahveci B. Chem. Heterocycl. Compd. 2016;52:1017–1024. doi: 10.1007/s10593-017-2002-3. [DOI] [Google Scholar]
- Azimi F. Azizian H. Najafi M. Hassanzadeh F. Sadeghi-aliabadi H. Ghasemi J. B. Ali Faramarzi M. Mojtabavi S. Larijani B. Saghaei L. Mahdavi M. Bioorg. Chem. 2021;114:105127. doi: 10.1016/j.bioorg.2021.105127. [DOI] [PubMed] [Google Scholar]
- Dai R. Li T. Xiao S. Chen Y. Gao J. Su G. Zhao Y. J. Mol. Struct. 2022;1264:133208. doi: 10.1016/j.molstruc.2022.133208. [DOI] [Google Scholar]
- Channar P. A. Saeed A. Larik F. A. Bolte M. Erben M. F. J. Mol. Struct. 2019;1179:11–17. doi: 10.1016/j.molstruc.2018.10.082. [DOI] [Google Scholar]
- Gondru R. Kanugala S. Raj S. Kumar C. G. Pasupuleti M. Banothu J. Bavantula R. Bioorg. Med. Chem. Lett. 2021;33:127746. doi: 10.1016/j.bmcl.2020.127746. [DOI] [PubMed] [Google Scholar]
- Safavi M. Ashtari A. Khalili F. Mirfazli S. S. Saeedi M. Ardestani S. K. Rashidi Ranjbar P. Barazandeh Tehrani M. Larijani B. Mahdavi M. Chem. Biol. Drug Des. 2018;92:1373–1381. doi: 10.1111/cbdd.13203. [DOI] [PubMed] [Google Scholar]
- Nural Y. Acar I. Yetkin D. Efeoglu C. Seferoğlu Z. Ayaz F. Bioorg. Med. Chem. Lett. 2022;69:128800. doi: 10.1016/j.bmcl.2022.128800. [DOI] [PubMed] [Google Scholar]
- Saeedi M. Maleki A. Iraji A. Hariri R. Akbarzadeh T. Edraki N. Firuzi O. Mirfazli S. S. J. Mol. Struct. 2021;1229:129828. doi: 10.1016/j.molstruc.2020.129828. [DOI] [Google Scholar]
- Saeedi M. Mohammadi-Khanaposhtani M. Asgari M. S. Eghbalnejad N. Imanparast S. Faramarzi M. A. Larijani B. Mahdavi M. Akbarzadeh T. Bioorg. Med. Chem. 2019;27:115148. doi: 10.1016/j.bmc.2019.115148. [DOI] [PubMed] [Google Scholar]
- Iraji A. Shareghi-Brojeni D. Mojtabavi S. Faramarzi M. A. Akbarzadeh T. Saeedi M. Sci. Rep. 2022;12:8647. doi: 10.1038/s41598-022-11771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareghi-Boroujeni D. Iraji A. Mojtabavi S. Faramarzi M. A. Akbarzadeh T. Saeedi M. Bioorg. Chem. 2021;111:104869. doi: 10.1016/j.bioorg.2021.104869. [DOI] [PubMed] [Google Scholar]
- Saeedi M. Raeisi-Nafchi M. Sobhani S. Mirfazli S. S. Zardkanlou M. Mojtabavi S. Faramarzi M. A. Akbarzadeh T. Mol. Diversity. 2021;25:2399–2409. doi: 10.1007/s11030-020-10137-8. [DOI] [PubMed] [Google Scholar]
- Mehreen S. Zia M. Khan A. Hussain J. Ullah S. Anwar M. U. Al-Harrasi A. Naseer M. M. RSC Adv. 2022;12:20919–20928. doi: 10.1039/D2RA03307K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G.-J. Lan T. Huang Z.-X. Cheng X.-N. Cai C.-Y. Ding S.-M. Xie M.-L. Wang B. Eur. J. Med. Chem. 2019;177:362–373. doi: 10.1016/j.ejmech.2019.05.045. [DOI] [PubMed] [Google Scholar]
- Asemanipoor N. Mohammadi-Khanaposhtani M. Moradi S. Vahidi M. Asadi M. Faramarzi M. A. Mahdavi M. Biglar M. Larijani B. Hamedifar H. Hajimiri M. H. Bioorg. Chem. 2020;95:103482. doi: 10.1016/j.bioorg.2019.103482. [DOI] [PubMed] [Google Scholar]
- Nasli Esfahani A. Iraji A. Alamir A. Moradi S. Asgari M. S. Hosseini S. Mojtabavi S. Nasli-Esfahani E. Faramarzi M. A. Bandarian F. Larijani B. Hamedifar H. Hajimiri M. H. Mahdavi M. Mol. Diversity. 2022;26:1995–2009. doi: 10.1007/s11030-021-10310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi M. Foroughi N. Saeedi M. Karimi M. Alinezhad H. Foroumadi A. Shafiee A. Akbarzadeh T. Synlett. 2014;25:385–388. [Google Scholar]
- Pedrood K. Rezaei Z. Khavaninzadeh K. Larijani B. Iraji A. Hosseini S. Mojtabavi S. Dianatpour M. Rastegar H. Faramarzi M. A. Hamedifar H. Hajimiri M. H. Mahdavi M. BMC Chem. 2022;16:57. doi: 10.1186/s13065-022-00848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsharif M. A. Naeem N. Mughal E. U. Sadiq A. Jassas R. S. Kausar S. Altaf A. A. Zafar M. N. Mumtaz A. Obaid R. J. Alsantali R. I. Ahmed S. Ahmed I. Altass H. M. Ahmed S. A. J. Mol. Struct. 2021;1244:130965. doi: 10.1016/j.molstruc.2021.130965. [DOI] [Google Scholar]
- Shahzad D. Saeed A. Larik F. A. Channar P. A. Abbas Q. Alajmi M. F. Arshad M. I. Erben M. F. Hassan M. Raza H. Molecules. 2019;24:1511. doi: 10.3390/molecules24081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.