Abstract
Expression of many aminoacyl-tRNA synthetase, amino acid biosynthesis and transport genes in Bacillus subtilis is controlled at the level of transcription termination using the T box system and requires the formation of specific secondary structures in the mRNA leader region. One structure functions as a transcriptional terminator, while an alternate form, the antiterminator, is necessary for transcription of the downstream coding regions. We have investigated the interaction of antiterminator model RNAs, based on the B.subtilis tyrS antiterminator with tRNATyr and tRNA acceptor stem models, using a gel shift assay. Binding of the antiterminator RNA to tRNATyr was dependent on complimentarity with the acceptor end of the tRNA or microhelix; affinity for the microhelix RNA was reduced relative to the tRNA. Alteration of a conserved position in the non-base pairing region of the bulge greatly reduced tRNA binding, consistent with in vivo studies. Therefore, it appears that some of the antiterminator–tRNA binding specificity is dependent on the structure of the antiterminator bulge alone and the complex it forms with tRNA in the absence of additional trans-acting factors. During the course of these studies we also discovered that the antiterminator can form a ‘kissing’ bulge complex, a unique RNA motif. The ease of formation of this RNA homodimer illustrates the propensity for the bulge of the antiterminator to bind RNA.
INTRODUCTION
The T box family of genes is a set of aminoacyl-tRNA synthetase, amino acid biosynthesis and amino acid transport genes that are regulated by a common transcription antitermination mechanism found primarily in Gram-positive species of bacteria (1–3). The mRNA leader regions of T box genes contain a large number of conserved primary sequence and secondary structural elements (1,3). Transcription of these genes is controlled by the formation of specific, secondary structural elements in the untranslated leader region of the mRNA.
Expression of each gene in this family is induced by a reduction in the charging ratio of charged to uncharged cognate tRNA (4). Interaction of the uncharged tRNA with the leader RNA results in the formation and stabilization of an antiterminator structure (Fig. 1); formation of the antiterminator precludes formation of a competing transcriptional terminator stem–loop, allowing synthesis of the full-length mRNA. The specificity of the interaction between the leader RNA and the cognate tRNA depends on base pairing between the anticodon of the tRNA with the complimentary codon sequence, designated the ‘specifier sequence’, which is presented at a defined position within the leader RNA. Changing the specifier sequence to the codon of another amino acid can, in some cases, alter the specificity of the amino acid response (3,5). Base pairing of the acceptor stem of uncharged tRNA with 4 nt in the conserved bulge of the antiterminator is also required for antitermination (Fig. 1) (4). This interaction is postulated to stabilize the thermodynamically less favored antiterminator structure, disfavoring formation of the terminator stem–loop (1).
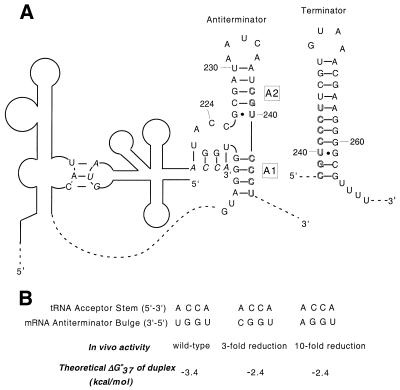
Figure 1.
(A) Schematic of the interaction between B.subtilis tyrS leader RNA and uncharged tRNATyr. The anticodon and the acceptor stem of tRNA (bases in italics) base pair with the specifier sequence in Stem I of the leader region and four of the bases in the antiterminator bulge, respectively. The remainder of the leader region is omitted for clarity (dashed lines). Antiterminator helices A1 and A2 are labeled. The terminator helix, which competes with the antiterminator, is shown in the top right corner. Bases involved in both structures are shown in outline. (B) Theoretical duplex stabilities for select tRNA acceptor stem–antiterminator bulge 4 bp complexes. In vivo activity is from Grundy et al (4). Stabilities were calculated using the nearest-neighbor model (10).
The antiterminator structure consists of two short helical regions (helices A1 and A2) separated by a 7 nt bulge. The first 4 bases of the bulge (5′-UGGN-3′) are involved in base pairing with the conserved 5′-NCCA-3′ acceptor end of tRNA (where the variable base N of the bulge covaries with the discriminator base N of the tRNA) (4). The remaining 3 bases (ACC) of the bulge, as well as the flanking helical regions immediately adjacent to the bulge, are also highly conserved (F.J.Grundy, T.R.Moir, M.T.Haldeman and T.M.Henkin, unpublished results). A single base mutation of C224U (AUC) results in a dramatic negative effect on antitermination in tyrS (6). Phylogenetic analysis of T box antiterminators reveals that C224 is not 100% conserved, but variation at this position may be better tolerated for certain leader–tRNA complexes (F.J.Grundy, T.R.Moir, M.T.Haldeman and T.M.Henkin, unpublished results). Other sequence variations in the ACC element eliminate readthrough (F.J.Grundy, T.R.Moir, M.T.Haldeman and T.M.Henkin, unpublished results). The role of regions of the antiterminator that are not directly involved in base pairing with tRNA is not currently understood. It is possible that these bases interact with an unknown factor involved in the antitermination mechanism. Alternatively, the positions may be a part of a unique RNA tertiary structural element that is required for proper interaction between the bulge and uncharged tRNA. Structural studies to date on the antiterminator have been limited to chemical modification and enzymatic cleavage studies, which have shown that the predicted secondary structure of the antiterminator forms in a truncated leader RNA (7). No detailed tertiary structural information is currently available for the complete antiterminator system, nor has a direct interaction between the antiterminator and uncharged tRNA been demonstrated biochemically.
Systematic attempts to force interaction of the tyrS leader with non-cognate tRNAs by alterations in known leader RNA specificity determinants indicated that while certain tRNAs can interact fairly efficiently with the tyrS leader to promote antitermination, others do not (5). This suggests that the leader RNA–tRNA interaction is complex and requires multiple contacts. Mutation of conserved sequence and structural elements of the tyrS leader demonstrated that most of the conserved elements are essential for antitermination (6,8). In contrast, most of the tRNATyr sequence can be varied provided that the tertiary structure and determinants for pairing with the leader RNA remain intact (9). The basis for leader RNA–tRNA specificity beyond the pairing determinants is not yet understood.
Changing the variable base in the antiterminator bulge of Bacillus subtilis tyrS from a U to an A results in a 10-fold drop in expression for tRNAs containing a 5′-ACCA-3′ acceptor stem. Changing this position from a U to a C results in only a 3-fold drop (4). Both the A and C mutants result in a mismatch at the tRNA discriminator position. Based on the predicted stability (10,11) of the short duplexes involving the tRNA acceptor stem/antiterminator bulge sequences (Fig. 1B), it appears that the difference in gene expression for the A versus the C variable base bulge mutants is due to more than just the resulting stabilities of a simple duplex association between the tRNA and the antiterminator bulge. In addition, the added stability of the proposed 4 bp interaction between tRNA and the antiterminator does not appear to be sufficient to overcome the difference in predicted stability (12,13) between the tyrS terminator (ΔG°37 = –18.8 kcal/mol) and the antiterminator (ΔG°37 = –6.4 kcal/mol) in order to promote formation of the antiterminator structure.
While it is currently difficult to accurately predict thermodynamic values for RNA bulges (11), the large difference in ΔG°37 between the two mutually exclusive structures most likely indicates that further tRNA interactions and/or trans-acting factors are involved. For example, oligomers binding to a 4 nt overhang at the 5′ end of a hairpin stem bind up to 1000-fold more tightly than predicted for binding to a free tetramer due to coaxial stacking of the helixes (14). This enhancement in binding is sequence dependent. A similar interaction may be occurring between the helix formed by the tRNA/antiterminator bulge interaction and the lower stem of the antiterminator (helix A1, Fig. 1).
This work addresses the question of whether the tRNA–antiterminator interaction can be demonstrated in the absence of additional factors and to begin to determine whether variations in the bulge region cause RNA structural changes that affect the function of the antiterminator. We have constructed a 29 nt model of the antiterminator region and a variant corresponding to a mutant with reduced function in vivo, to study the structural characteristics of the bulge and surrounding regions as they relate to the function of the antiterminator. Gel shift analysis was used to compare the relative binding affinities of the two antiterminator sequences with several tRNA models.
MATERIALS AND METHODS
Thermodynamic stability predictions
The most stable secondary structure (see Figs 2 and 3B) and thermodynamic stability of the model RNA sequences were predicted using mfold 3.1 (12,13).
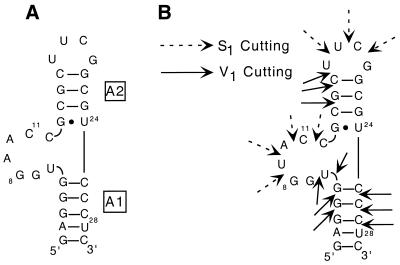
Figure 2.
(A) The 29 nt model system (AM1A) of the antiterminator used in the binding and structural studies. (B) Original antiterminator model RNA (AM1) and the corresponding S1/V1 enzymatic cleavage results. RNase S1 cutting in single-stranded regions is designated with a solid arrow, while RNase V1 cutting in double-stranded or stacked regions is designated by a dashed arrow.
Figure 3.
tRNA and microhelix models used in the binding studies. (A) Bacillus subtilis tRNATyr(A73U); (B) E.coli tRNATyr; (C) mh-UCCA, complimentary acceptor stem to AM1A; (D) mh-AGGU, 4 base mismatch acceptor stem; (E) mhACCA, discriminator mismatch to AM1A. Although there are primary sequence variations between the two tRNA models, previous studies have shown that these changes do not dramatically affect the interaction of the tRNA with the tyrS leader (9).
Design of antiterminator RNA and tRNA models
Antiterminator models. The 29 nt model sequence AM1 (Fig. 2B) is based on the antiterminator sequence found in the B.subtilis tyrS leader region with the following modifications. The A·U and U·A base pairs at the top of the antiterminator helix A2 were replaced by a C·G pair, and the closing loop was replaced with the well-characterized, stable UUCG tetraloop (15,16). The loop region of the antiterminator is not conserved in size or sequence, and a variant of the tyrS leader with these alterations exhibited normal function in vivo (F.J.Grundy, T.R.Moir, M.T.Haldeman and T.M.Henkin, unpublished results). A G·C base pair was also added to the base of the antiterminator helix A1 to facilitate transcription by T7 RNA polymerase (17,18). In AM1A (Fig. 2A), the variable base of the bulge region was changed from a U to an A to avoid dimerization problems resulting from the palindromic sequence present in the wild-type bulge sequence in AM1 (see below). In vivo studies indicate that an A in the variable base position of the bulge is functional, provided that a compensatory change is made in the discriminator base of tRNATyr to maintain base pairing (5).
As discussed above, a single base change in the antiterminator bulge from C224 to U224 leads to a significant decrease in antiterminator function in tyrS (6). The reason for this marked decrease upon exchanging one pyrimidine nucleotide for another is not yet known. Consequently, we designed the AM1A(C11U) variant to further investigate tRNA–bulge specificity .
tRNA models. Bacillus subtilis tRNATyr(A73U) (Fig. 3A) is a variant of tRNATyr designed to ensure that the discriminator base would base pair with the appropriate variable base in the bulge of AM1A. Expression of this tRNA in vivo results in efficient antitermination of a matching tyrS leader construct (4). This tRNA was prepared by in vitro transcription using T7 RNA polymerase, and is therefore unmodified. Escherichia coli tRNATyr (Fig. 3B) was purchased in a modified form from Sigma. Previous studies have shown that the E.coli tRNATyr sequence is fully functional when expressed in B.subtilis (9). In this case, the discriminator base is an A and would be expected to base pair with the variable base (U9) of the bulge in AM1, but form a mismatch with the variable base (A9) of AM1A.
Microhelix models of tRNATyr were designed to begin to investigate the extent of tRNA structure needed for binding specificity. tRNA micro and minihelices have been shown to be suitable model substrates for certain tRNA synthetases (19,20) and provided valuable insight into structural studies with synthetases (21). The mh-UCCA (Fig. 3C) and mh-ACCA (Fig. 3E) RNAs were designed to investigate the effect of the discriminator base on specificity, while the mh-AGGU (Fig. 3D) was designed as a control to test for association independent of acceptor stem pairing.
RNA synthesis
Antiterminator RNAs, B.subtilis tRNATyr(A73U) and microhelices were synthesized enzymatically from DNA templates using His-6-tagged T7 RNA polymerase according to the method of Milligan et al. (17,18). Escherichia coli tRNATyr (Type II) was purchased from Sigma. All template DNA for T7 transcription was purchased (Gibco or Oligos Etc.) and purified before use except the template for B.subtilis tRNATyr(A73U). Bacillus subtilis tRNATyr(A73U) double-stranded template DNA was prepared by PCR amplification of a T7 promoter tRNATyr construct using the following primers: 5′-TAATACGACTCACTATAGG-3′ for the T7 promoter region and 5′-TGGAGGAGGGGGGCAGATTCG-3′ where the bold A introduced the A73U mutation at the 3′ end of the tRNA. The non-A-tailing Pfu turbo polymerase (Stratagene) was used for PCR amplification and the PCR product was subsequently transcribed using Ampliscribe (Epicentre) to obtain B.subtilis tRNATyr(A73U). All synthesized RNAs were purified using 20% denaturing acrylamide (19:1 acrylamide:bisacrylamide) gels followed by electroelution and ethanol precipitation. RNAs were dialyzed against 10 mM NaH2PO4 pH 6.5 and 0.01 mM EDTA prior to use.
Enzymatic probing of secondary structure
The structure of 5′-32P-labeled AM1 was probed using nuclease S1 and ribonuclease V1 as described (22). RNase V1 digestions were carried out for 5 min on ice in 15 µl of 25 mM NaOAc, 50 mM KCl and 5 mM MgCl using 0.14 U of RNase V1 (Boehringer Mannheim). RNase S1 digestions were carried out for 5 min at room temperature in 15 µl of 50 mM MES pH 6.3, 100 mM NaCl and 5 mM MgCl using 13 U of RNase S1 (Pharmacia Biotech). The reactions were stopped by the addition of 15 µl of 9 M urea, 0.05% (w/v) xylene cyanol in 50 mM Tris–HCl pH 8.1, 50 mM boric acid and 1 mM EDTA, and freezing at –70°C before fractionating on a 20% polyacrylamide gel (19:1 acrylamide:bisacrylamide).
Gel shift studies
Binding reactions were prepared as follows. Stock solutions of tRNAs, microhelices and 5′-32P-end-labeled antiterminator model RNAs were prepared and heated at 80°C for 1 min and allowed to cool slowly to room temperature. To each reaction the following was added: 2 µl of 5′-end-labeled antiterminator RNA (∼0.2 pmol/µl); an appropriate amount of tRNA or minihelix; 1 µl of 10× loading buffer (5× TBE, 50 mM MgCl2, 500 mM NaCl and 10% glycerol, where 1× TBE is 50 mM Tris–borate pH 8.3 and 1 mM EDTA); and purified water to a final volume of 10 µl. The reactions were gently mixed and incubated at 4°C for 30–40 min prior to loading onto the gel. Gel composition for all reactions was 0.5× TBE, 5 mM MgCl2 and 50 mM NaCl. Binding reactions with tRNA were run on 10% acrylamide gels, while microhelix binding reactions were run on 15% acrylamide gels; all gels were 29:1 acrylamide:bisacrylamide. Running buffer was also 0.5× TBE, 5 mM MgCl2 and 50 mM NaCl. Gels were run between 6–10 W for 4–5 h at room temperature. During this time minimal heating of the gel occurred. Visualization and quantitation of the gels was achieved using a PhosphoImager (Molecular Dynamics). Kd values were determined using the following equation (23): Kd = ([L] – r[L])/r, where [L] is the concentration of ligand and r is the fraction of substrate bound. This equation is valid when the Kd for the ligand (tRNA or tRNA model) is much greater than the concentration of substrate (antiterminator models) as was the case for all the gel shift experiments.
UV melt studies
RNA samples were dialyzed into 10 mM NaH2PO4 pH 6.5 and 0.01 mM EDTA before UV analysis. Samples were prepared at the appropriate concentration (determined by UV absorbance) in the desired buffer and denatured at 85°C for 1 min and then allowed to cool slowly to room temperature before UV analysis. UV melts were measured on a Lambda 10 (Perkin Elmer) UV spectrometer with heating via a peltier device with cuvette pathlengths ranging from 0.01 to 1 cm. Heating rates were either 1 or 0.5°C/min and the absorbance was monitored at 260 nm. The first derivatives of the melt curves were calculated using OD Deriv (24).
RESULTS
Secondary structure of the antiterminator model RNA
The secondary structures shown for all antiterminator and acceptor stem model RNAs were predicted to be the lowest energy structures and at least 2 kcal/mol lower in energy than the next most stable structure (12,13). Nuclease probing of AM1 (Fig. 2B) showed V1 cleavage in both helices A1 and A2 indicative of double-stranded or stacked regions. In addition, V1 cleavage was observed at the first two positions within the bulge indicating possible stacking of U6 and G7 on the lower helix A1. S1 cleavage was observed within the UUCG tetraloop as well as within the bulge, indicating either a lack of stacking in the 3′ portion of the bulge or some degree of conformational flexibility. Preliminary NMR experiments at low concentrations also agreed with the proposed secondary structure (data not shown). The secondary structure of AM1A was confirmed using NMR (M.S.Gerdeman, T.M.Henkin and J.V.Hines, unpublished results).
Palindromic sequence of AM1 forms a ‘kissing’ bulge homodimer
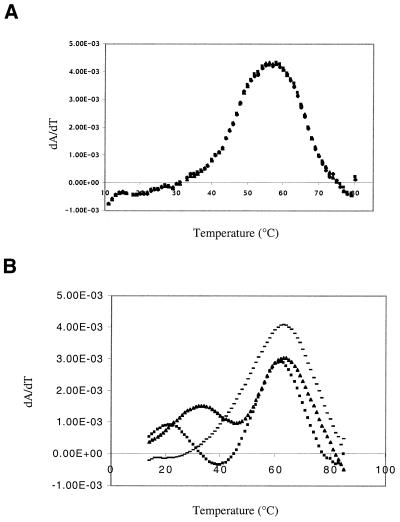
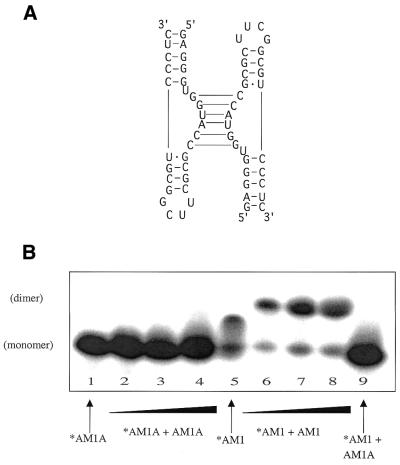
UV melt profiles of AM1 in low salt (Fig. 4A) and high salt (Fig. 4B) indicated that in the presence of salt a concentration-dependent lower melting transition occurs, which we have identified as a ‘kissing’ bulge homodimer (Fig. 5A). This transition was not observed under these conditions in the AM1(C11U) variant model (data not shown) nor in AM1A (M.S.Gerdeman, T.M.Henkin and J.V.Hines, unpublished results) where the kissing bulges would be destabilized by two G·U base pairs or two A·A mismatches, respectively. At higher concentrations (1.8 mM), NMR studies indicated that AM1 dimerized even under low salt conditions while AM1A did not (data not shown). Native gel studies (Fig. 5B) indicated that a shifted species consistent with dimer formation occurs only in the case where the bulge region is fully complimentary. The predicted (11) Tm of 28 and 40°C at 0.4 and 14.3 µM, respectively, for the 6 bp dimerized bulge portion of the AM1 kissing complex agrees well with the observed lower melting transition Tm of 22 and 34°C, respectively (Fig. 4B).
Figure 4.
First derivative plots of UV melts of AM1. (A) At concentrations ranging from 0.15 to 542 µM in 10 mM NaH2PO4 pH 6.5 and 0.1 mM EDTA. (B) At concentrations of 0.4 µM (squares), 14.3 µM (triangles) and 345 µM (dashes) in the same buffer as (A) plus 50 mM NaCl. The lower temperature transition shifts to higher temperature with increasing concentration, while the transition at 63°C remains constant.
Figure 5.
(A) Model of ‘kissing’ bulge interaction for AM1. (B) Native gel electrophoresis studies of AM1A and AM1 (15% acrylamide). Lanes 1–4 contain 5′-32P-end-labeled AM1A. Lanes 2–4 have additional unlabeled AM1A added in concentrations of 39, 156 and 390 µM. Lanes 5–8 contain 5′-32P-end-labeled AM1. Lanes 6–8 have additional unlabeled AM1 added in concentrations of 39, 156 and 390 µM. Lane 9 contains 5′-32P-end-labeled AM1 with 390 µM unlabeled AM1A.
Specificity observed in antiterminator–tRNA complexes
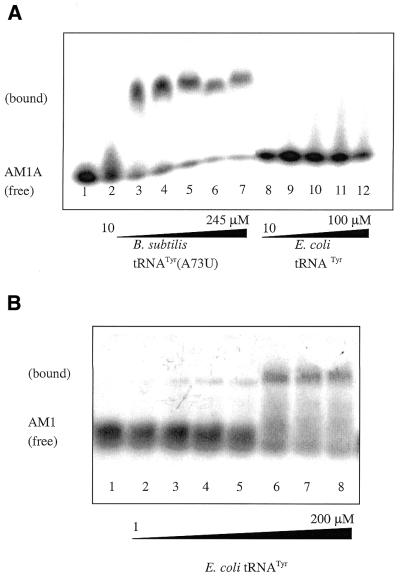
The ability of the model antiterminator RNAs to interact with tRNA and microhelix models was tested by a gel shift assay. Gel shifts are shown for AM1A and AM1 with tRNATyr (Fig. 6); AM1A with microhelices (Fig. 7); and AM1A(C11U) with tRNATyr and microhelices (Fig. 8).
Figure 6.
(A) Native gel studies of AM1A with tRNAs (10% acrylamide). Lanes 1–12 contain 5′-32P-end-labeled AM1A. Lanes 2–7 have increasing concentrations of B.subtilis tRNATyr(A73U) ranging from 10 to 245 µM. Lanes 9–12 contain increasing amounts (10–100 µM) of E.coli tRNATyr in which the discriminator base does not match AM1A. (B) Native gel studies of AM1 with E.coli tRNATyr (10% acrylamide). Lanes 1–8 contain trace amounts 5′-32 P-end-labeled AM1. Lanes 2–8 contain increasing amounts of E.coli tRNATyr (0–200 µM).
Figure 7.
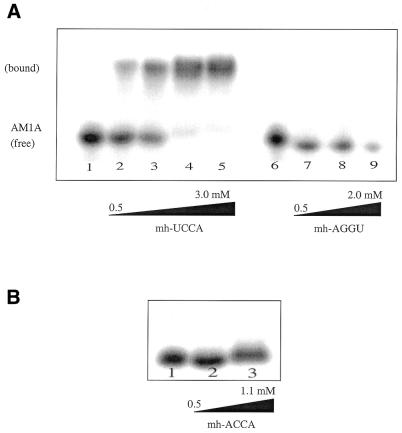
Native gel studies of AM1A with microhelix tRNA models (15% acrylamide). (A) Lanes 1–5 contain 5′-32P-end-labeled AM1A with concentrations of complimentary microhelix (mh-UCCA) of 0, 500, 989, 2967 and 3956 µM, respectively. Lanes 6–9 contain 5′-32P-end-labeled AM1A with concentrations of the complete mismatch microhelix (mh-AGGU) of 0, 500, 989 and 1978 µM, respectively. A Kd of 830 µM was determined for the complimentary minihelix, while the complete mismatched microhelix failed to show an observable complex with AM1A. (B) Same conditions as (A) except using the discriminator mismatch microhelix (mh-ACCA) at concentrations of 0, 277 and 1108 µM in lanes 1–3, respectively. Although no clear shift occurs, there does appear to be a smearing of the band at higher concentrations.
Figure 8.
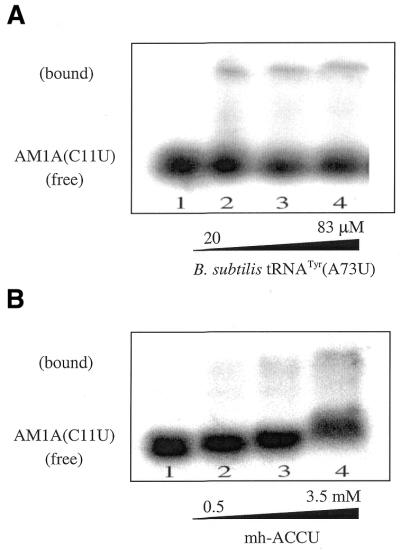
Native gel studies of AM1A(C11U) with tRNA and microhelix tRNA. (A) With the complimentary tRNATyr (A73U) (10% acrylamide). Lanes 1–4 contain 5′-32P-end-labeled AM1A(C11U) variant. The concentration of tRNATyr(A73U) is 0, 20, 50 and 83 µM, respectively. The calculated Kd is 200 µM. (B) With the complimentary microhelix (mh-UCCA) (15% acrylamide). Lanes 1–4 contain a trace amount of 5′-32P-end-labeled AM1A(C11U) variant. The concentration of mh-UCCA is 0, 500, 989 and 3461 µM, respectively. The calculated Kd is 7.5 mM.
AM1A binds B.subtilis tRNATyr(A73U) to create a complex that migrates significantly slower than free AM1A (Fig. 6A) with a Kd of 63 µM. We also investigated the binding of tRNATyr from E.coli to AM1A to determine whether the reaction was specific and not simply a non-specific association of two RNA species. Escherichia coli tRNATyr has been shown to effectively induce expression of a tyrS–lacZ fusion in B.subtilis and thus is thought to interact with the antiterminator with similar affinity as B.subtilis tRNATyr (9). The discriminator base in E.coli tRNATyr is an A, which would result in a 1 base mismatch to the bulge of AM1A. No complex formation between AM1A and the E.coli tRNATyr was observed (Fig. 6A). With AM1 the variable base is a U (thus restoring complimentarity with the acceptor end of E.coli tRNATyr), which results in restoration of binding (Fig. 6B).
Due to the tendency of AM1 to homodimerize, an accurate Kd could not be obtained for AM1; however, a Kd range for binding of E.coli tRNATyr was estimated by assuming the Kd to be approximately equal to the concentration of tRNA needed to result in 50% of the AM1 being bound. A similar estimation has been applied to other RNA–RNA complexes (25). To be as conservative as possible with the estimation, a range from ∼25 to 75% bound was chosen resulting in a Kd between 10 and 100 µM.
The microhelix tRNA model mh-UCCA also bound AM1A (Kd = 830 µM, Fig. 7A), but binding was an order of magnitude weaker than observed with B.subtilis tRNATyr(A73U), which has an analogous UCCA acceptor stem. Both B.subtilis tRNATyr(A73U) and mh-UCCA bound AM1A(C11U) with less affinity than AM1A (Fig. 8); however, the full tRNA (Kd = 200 µM) again bound the variant bulge tighter than the corresponding microhelix model (Kd = 7.5 mM). No shift was observed for the completely mismatched acceptor stem mh-AGGU with AM1A (Fig. 7A).
While no shift was directly observed when the discriminator base was mismatched (e.g. E.coli tRNATyr or mh-UCCA with AM1A, Figs 6A and 7B, respectively), extensive streaking of the unbound (or bound) AM1A band was observed (Fig. 6A). This could be due to a dynamic equilibrium between AM1A and acceptor stems with a mismatched A at the discriminator position. A kinetically unstable complex would make it difficult to observe the association by gel shift. Similar explanations have been used for other RNA–RNA (25) and RNA–protein (23) complexes. Further evidence of this possibility is the fact that the free AM1A band exhibited streaking at lower concentrations of matched acceptor stem [e.g. B.subtilis tRNATyr(A73U)]. Consequently, as with other kinetically unstable complexes (26,27), the Kd values observed with these gel shift assays may underestimate the tightness of binding.
DISCUSSION
The gel shift studies were designed to evaluate whether an antiterminator model RNA could bind to tRNA without additional trans-acting factors, and to begin to investigate the specificity determinants for the complex. In the presence of an appropriately matched tRNA acceptor stem, both AM1A and AM1 RNAs formed a complex with tRNA as determined by gel shift studies. The lack of an observed shift with a single mismatch in the acceptor end of the tRNA (when three base pairing interactions are still maintained) suggests that the discriminator position of the tRNA plays an important role in the specificity and affinity of the leader RNA–tRNA interaction, as suggested by in vivo experiments (4). The overall bound structure of the acceptor stem and 5′-NCCA-3′ end of the tRNA may also be important for bulge–tRNA complex stability. This agrees with previous work (9) that examined the ability of a pool of tRNAs with mutations in the acceptor stem to induce expression of the tyrS gene. Only tRNA acceptor stem sequences that retained Watson–Crick base pairing were able to effectively promote tyrS expression. Previous work on recognition of tRNA by cognate aminoacyl tRNA synthetases has shown that the sequence of the acceptor stem is in some cases a determinant for recognition of the tRNA (28).
The structure of the 3′ end of tRNAs and acceptor stem models (21,29), as well as charging efficiencies of tRNA synthetases (20), can be dependent on the identity of the discriminator base. It is possible that discriminator base-induced structural changes in the unbound acceptor end might have an effect on binding to the antiterminator bulge. The binding studies, however, appear to indicate that tRNA–bulge binding is not affected by differences in the tertiary structure of the uncomplexed tRNA acceptor end, since AM1 binds E.coli tRNATyr having a 5′-ACCA-3′ acceptor end with similar efficiency as AM1A binds B.subtilis tRNATyr(A73U) having a 5′-UCCA-3′ acceptor end. In addition, the similar Kd values would appear to indicate that formation of the complex is not dependent on having modified tRNA since B.subtilis tRNATyr(A73U) is an unmodified T7 transcript and E.coli tRNATyr is fully modified.
Since binding studies of AM1A with tRNATyr suggested that the sequence of the acceptor end of the tRNA is important, we also wanted to investigate whether any other regions of tRNA are important for binding the antiterminator bulge. Microhelices have been shown to be effective models of the tRNA acceptor stem and are, in some cases, able to be recognized and aminoacylated by the appropriate aminoacyl-tRNA synthetase (19). If only the acceptor end of tRNA is important for recognition and binding of the tRNA to the antiterminator bulge, one would expect similar Kd values for tRNATyr and the corresponding microhelix acceptor stem model. However, the mh-UCCA with a 3′ end identical to that of tRNATyr(A73U) bound AM1A with an order of magnitude weaker Kd than tRNATyr(A73U) bound AM1A. These data suggest that other areas of the tRNA beyond the acceptor stem may be important for binding the antiterminator bulge, either directly or through effects on presentation of the unpaired nucleotides for binding. Studies by Grundy et al. (9) investigated the ability of a tRNA model containing just the anticodon loop and acceptor stem to induce antitermination in vivo. No increase in the expression of a tyrS–lacZ fusion was observed, suggesting that other regions of the tRNA are essential for recognition of the bulge region (although these studies were complicated by the possibility that the tRNA segments failed to fold properly in vivo) (9). As with the full tRNA, if the discriminator base of the microhelix is an A (i.e. mh-ACCA), resulting in a 1 base mismatch with AM1A, no gel shift was observed, supporting the fact that the entire sequence of the acceptor end is important for binding.
We also investigated the binding of the tRNA and microhelices to the variant antiterminator model RNA AM1A(C11U). The corresponding C224U mutation in the leader region of the tyrS gene causes an 18-fold decrease in expression in vivo, which can be partially overcome by increased availability of uncharged tRNATyr resulting from tyrosine-limited growth conditions (6). The Kd for B.subtilis tRNATyr(A73U) was 3.3-fold weaker for the AM1A(C11U) variant than for AM1A. This suggests that at least some of the specificity for binding is dependent on the bulge of the antiterminator itself since no additional factors were present in the binding assay. This may indicate that the C to U substitution causes a structural change in the bulge that decreases the ability of the acceptor stem of tRNATyr(A73U) to bind. This conclusion is supported by preliminary structural information for the two antiterminator model RNAs (M.G.Gerdeman, T.M.Henkin and J.V.Hines, unpublished results). The fact that AM1A(C11U) still binds tRNATyr(A73U) but with less affinity than AM1A may explain why the mutation can be partially overcome in vivo by high levels of uncharged tRNATyr.
The matched microhelix, mh-UCCA, bound AM1A(C11U) with a 9-fold weaker Kd than it bound AM1A. The fact that the decrease in binding is more dramatic when the microhelix models are used suggests that the C11U antiterminator bulge mutation may affect the bulge structure in a manner that primarily alters interaction with the acceptor stem and end structure of the tRNA. Additional contacts between the antiterminator and the full tRNA may partially overcome any decreased acceptor stem recognition, thus moderating the reduction in binding for the C11U variant with the full tRNA versus the microhelices.
The initial antiterminator model (AM1) formed the predicted secondary structure (Fig. 2B). However, the palindromic nature of the bulge resulted in a propensity for homo-dimerization and the formation of a unique ‘kissing’ bulge complex (Fig. 5A). When the Watson–Crick self-complimentarity is disrupted in the bulge [e.g. AM1(C11U), AM1A, AM1A(C11U)] no homo-dimerization was observed by UV (with 50 mM NaCl) nor by gel shift. Yet a concentration-dependent Tm was observed by UV when an appropriate ‘kissing’ bulge heterodimer can form, for example AM1(C11U) in the presence of AM1G8A (data not shown). While this bulge–bulge interaction was detrimental to detailed tRNA–bulge binding studies with AM1, it highlighted the propensity for the wild-type antiterminator bulge to bind RNA. The fact that the bulge region of AM1 readily interacts with another RNA (in this case another bulge RNA) suggests that the bulge structure may be in a conformation that facilitates binding to a complimentary RNA sequence as it must do to bind the 3′ end of tRNA in its normal role as an antiterminator. To the best of our knowledge, this is the first documented observation of a ‘kissing’ bulge motif. This motif is topologically different from the previously observed ‘kissing’ hairpin motif (30) (or loop–loop interaction) such as that seen in the HIV dimerization initiation site (31). To date no biological systems have been identified that utilize a ‘kissing’ bulge motif for biological function. We are currently investigating whether there are any sequence or size dependencies for the formation of kissing bulges.
These in vitro studies have shown that an important component of the sequence–function relationship observed in vivo for the T box system antiterminator is due to specific RNA–RNA interactions between the antiterminator bulge and the tRNA. They have also highlighted the possibility of further antiterminator–tRNA contacts beyond the known base pairing between the bulge and the acceptor end nucleotides. An alternative possibility is that the acceptor end of the tRNA must be presented within the context of the full tRNA to achieve tighter binding. The fact that the in vitro binding data correlate well with antitermination function in vivo indicates that additional trans-acting factors may not be required for specific antiterminator–tRNA recognition. However, whether or not additional cofactors participate in antitermination remains to be determined. Studies are underway to further characterize additional contacts between the tRNA and antiterminator bulge and determine the tertiary structure of the complex.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Dr M. Haldeman, S. Rollins, E. Fingar, K. McQuown and D. Ordaz (OSU fermentation facility) for their assistance with these studies, as well as Dr T. E. Shrader for the plasmid containing the His-6-tagged T7 RNA polymerase. This work was supported by NIH grant R01-GM47823 (T.M.H.), NIH Chemistry at the Biology Interface Training Grant (M.S.G.), and the Department of Chemistry and Biochemistry and the Office of the Vice President for Research, Ohio University.
REFERENCES
- 1.Henkin T.M. (1994) tRNA-directed transcription antitermination. Mol. Microbiol., 13, 381–387. [DOI] [PubMed] [Google Scholar]
- 2.Grundy F.J. and Henkin,T.M. (1994) Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in Gram-positive bacteria. J. Mol. Biol., 235, 798–804. [DOI] [PubMed] [Google Scholar]
- 3.Grundy F.J. and Henkin,T.M. (1993) tRNA as a positive regulator of transcription antitermination in B.subtilis.Cell, 74, 475–482. [DOI] [PubMed] [Google Scholar]
- 4.Grundy F.J., Rollins,S.M. and Henkin,T.M. (1994) Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol., 176, 4518–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grundy F.J., Hodil,S.E., Rollins,S.M. and Henkin,T.M. (1997) Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination. J. Bacteriol., 179, 2587–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollins S.M., Grundy,F.J. and Henkin,T.M. (1997) Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol., 25, 411–421. [DOI] [PubMed] [Google Scholar]
- 7.Luo D., Condon,C., Grunberg-Manago,M. and Putzer,H. (1998) In vitro and in vivo secondary structure probing the thrS leader in Bacillus subtilis.Nucleic Acids Res., 26, 5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler W.C., Grundy,F.J., Murphy,B.A. and Henkin,T.M. (2001) The GA motif: an RNA element common to bacterial antitermination systems, rRNA and eukaryotic RNAs. RNA, 7, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundy F.J., Collins,J.A., Rollins,S.M. and Henkin,T.M. (2000) tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA, 6, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra M.J. and Turner,D.H. (1995) Predicting thermodynamic properties of RNA. Methods Enzymol., 259, 243–361. [DOI] [PubMed] [Google Scholar]
- 12.Zuker M., Mathew,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43.
- 13.Mathews D.H., Sabina,J., Zuker,M.and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 14.Walter A.E., Turner,D.H., Kim,J., Lyttle,M.H., Muller,P., Mathews,D.H. and Zuker,M. (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA, 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varani G., Cheong,C. and Tinoco,I.,Jr (1991) Structure of an unusually stable RNA hairpin. Biochemistry, 30, 3280–3289. [DOI] [PubMed] [Google Scholar]
- 16.Allain F.H.-T. and Varani,G. (1995) Structure of the P1 Helix from Group 1 self-splicing introns. J. Mol. Biol., 250, 333–353. [DOI] [PubMed] [Google Scholar]
- 17.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 19.Francklyn C. and Schimmel,P. (1989) Aminoacylation of RNA minihelices with alanine. Nature, 337, 478–481. [DOI] [PubMed] [Google Scholar]
- 20.Fischer A., Beuning,P. and Musier-Forsyth,K. (1999) Identification of discriminator base atomic groups that modulate the alanine aminoacylation reaction. J. Biol. Chem., 274, 37093–37096. [DOI] [PubMed] [Google Scholar]
- 21.Ramos A. and Varani,G. (1997) Structure of the acceptor stem of Escherichia coli tRNAAla: role of the G3:U70 base pair in synthetase recognition. Nucleic Acids Res., 25, 2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyatt J., Puglisi,J. and Tinoco,I.J. (1990) RNA pseudoknots. Stability and loop size requirements. J. Mol. Biol., 214, 455–470. [DOI] [PubMed] [Google Scholar]
- 23.Weeks K.M. and Crothers,D.M. (1992) RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry, 31, 10281–10287. [DOI] [PubMed] [Google Scholar]
- 24.Draper D.E. and Gluick,T.C. (1995) Melting studies of RNA unfolding and RNA-ligand interactions. Methods Enzymol., 259, 281–305. [DOI] [PubMed] [Google Scholar]
- 25.Jaeger L., Westhof,E. and Leontis,N.B. (2001) TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res., 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall K.B. and Stump,W.T. (1992) Interaction of N-terminal domain of U1A protein with an RNA stem/loop. Nucleic Acids Res., 20, 4283–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long K. and Crothers,D. (1995) Interaction of human immunodeficiency virus type 1 Tat-derived peptides with TAR RNA. Biochemistry, 34, 8885–8895. [DOI] [PubMed] [Google Scholar]
- 28.McClain W.H., Gabriel,K., Subhra,B., Jou,Y.-Y. and Schnieder,J. (1999) Functional compensation by particular nucleotide substitutions of a critical GU base-pair during aminoacylation of tRNA. J. Mol. Biol., 286, 1025–1032. [DOI] [PubMed] [Google Scholar]
- 29.Puglisi E.V., Puglisi,J.D., Williamson,J.R. and RajBhandary,U.L. (1994) NMR analysis of tRNA acceptor stem microhelices: discriminator base change affects tRNA conformation at the 3′ end. Proc. Natl Acad. Sci. USA, 91, 11467–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang K.-Y. and Tinoco,I.,Jr (1994) Characterization of a ‘kissing’ hairpin complex derived from the human immunodeficiency virus genome. Proc. Natl Acad. Sci. USA, 91, 8705–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lodmell J.S., Ehresmann,C., Ehresmann,B. and Marquet,R. (2000) Convergence of natural and artificial evolution on an RNA loop-loop interaction: The HIV-1 dimerization initiation site. RNA, 6, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]