Abstract
Dlm-1 is a recently described gene which is upregulated in murine stromal cells lining tumors. The function of the 40 kDa DLM-1 protein is poorly understood. DLM-1 contains an N-terminal Z-DNA binding domain homologous to the Zα domain in the RNA editing enzyme ADAR1. We report the cloning of human and rat DLM-1. In addition to the Zα domain, three further conserved regions were identified. One of these is homologous to the second Z-DNA binding domain, Zβ, of ADAR1. We find that human DLM-1 is predominantly expressed in lymphatic tissues. The gene spans 17 kb and consists of 10 exons. DNA transcripts are extremely heterogeneous as a result of alternative splicing and the usage of exon variants combined with at least two transcriptional start sites and 3′-terminal exons. The exon coding for the Zα domain was present in approximately one-third of the analyzed mRNAs. Nearly half of the transcripts contained exon variants that had premature stop codons incorporated. Based on our analysis, over 2000 different mRNAs may be produced due to alternative splicing and usage of different 5′ and 3′ ends. The cellular function of DLM-1 appears to call for a high degree of adaptation by this complex regulation.
INTRODUCTION
Left-handed Z-DNA can be formed by sequences containing alternating purine–pyrimidine nucleotides (1). Under physiological conditions, Z-DNA formation is a dynamic process, largely controlled by the amount of supercoiling (2,3). A tight correlation between Z-DNA formation and transcriptional activity has been observed (4–6). Biological roles for left-handed Z-DNA have not yet been established. Several functions for Z-DNA formation have been proposed (7). It has been shown that sequences which contain potential Z-DNA forming sequences have the potential to modulate promoter activity in reporter gene assays (reviewed in 8,9). The Z-DNA forming sequences may either enhance (10,11) or repress (12,13) promoter activity.
In an attempt to identify proteins with a high affinity for Z-DNA, the RNA editing enzyme ADAR1 has been found to contain two Z-DNA binding domains (14). Crystallization of the Z-DNA binding domain Zα revealed that it belongs to the family of winged-helix proteins and that it recognizes Z-DNA in a structure-specific manner (15). A function of this domain may be to direct ADAR1 to a location where RNA editing is supposed to take place. Since Z-DNA formation is favored by negative supercoiling as generated behind moving RNA polymerases, ADAR1 would be directed to loci where transcription takes place (7). It has been further demonstrated that Zα can also bind left-handed, double-stranded RNA (16).
It was recently shown that the murine protein DLM-1 contains a region homologous to ZαADAR (17). Co-crystallization of the Zα domain of mDLM-1 with Z-DNA revealed that Zα of DLM-1 binds left-handed Z-DNA in a structure-specific manner, similarly to the Zα domain of ADAR1 (15,17). Both domains adopt a winged-helix conformation. Murine DLM-1 was identified because it was upregulated in tumor stromal cells. It is induced upon activation in macrophages (18). It has been proposed that DLM-1 plays an important role in the host response to tumor formation (18). Besides the Z-DNA binding domain, no further significant similarities have been found that may explain the function of DLM-1, especially in the context of nucleic acid binding.
In order to characterize conserved regions in DLM-1, we cloned the human and rat homologs and analyzed the tissue expression pattern as well as the gene structure of human DLM-1.
MATERIALS AND METHODS
Rapid amplification of cDNA ends (RACE) PCR
RACE PCRs were performed with human spleen Marathon-Ready cDNA (Clontech). A first amplification round was performed for 5′-RACE with primers hDLMcap1R (5′-CCG TTG TTG GCT GAA CTG AGG GCC AG-3′) and adaptor primer AP1 (Clontech) and for 3′-RACE with primers hDLM1.5f (5′-AGT CCA AAG CAT GGA CGA TTT ACC GCC C-3′) and AP1 for 30 cycles of 30 s at 95°C and 2 min at 68°C. Nested PCR was performed using 1 µl of first amplification rounds with primers hDLMcap2R (5′-GGC CAG GGG TCT CTG GAA TTG TAG CTG C-3′) and nested adaptor primer AP2 (for 5′-RACE) or AP2 combined with hDLM2f (5′-GGA CCC AAC AGC TGG ATT TCC-3′) or hDLM3f (5′-AAT CGT GCT TTC TCG AGG ACG C-3′) (for 3′-RACE) for 30 cycles of 30 s at 95°C, 30 s at 64°C and 90 s at 68°C.
Analysis of DLM-1 expression and splice variants
cDNAs from human multiple tissue cDNA panels (Clontech) were used as templates for PCR amplification. DLM-1 transcripts were amplified using primers hDLM1f (5′-CCG ACT CCT TGC AGC TGC TGT C-3′) and hDLM3r (5′-CCA GCC ACC CCT GGG CTG ATA G-3′) in 40 µl reaction volumes containing 0.8 µl AdvanTaq DNA Polymerase (Clontech) for 32 cycles of 30 s at 95°C, 30 s at 67°C and 2 min at 68°C. In control reactions, cyclophilin-specific primers cphilin H1 (5′-ATG GTC AAC CCC ACC GTG T-3′) and cphilin H2 (5′-TCT CCT GAG CTA CAG AAG G-3′) were used and PCR was carried out for 25 cycles of 30 s at 95°C, 30 s at 56°C and 1 min at 68°C. Additional DLM-1-specific PCR products were amplified with primers hDLM1f × hDLM4R (5′-TCA CTT CTT ATT CTT CAA TAA ATA TAT TTT ATG AGG GC-3′) or hDLM1f × hDLM6r (5′-ACT CCC TGT CAT CTA CTC CTG GCC-3′).
Cloning of Rattus norvegicus DLM-1
Reverse transcription (RT) of 2 µg of total spleen RNA from a LEW rat was primed with random hexamers (for RT–PCR) or oligo(dT) containing oligonucleotide AXT (5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′) (for 3′-RACE). The reaction was carried out with M-MLV reverse transcriptase (Life Technologies) in a 20 µl reaction volume for 10 min at 24°C, 30 min at 42°C and 2 min at 94°C. One microliter of the RT reaction was used for PCR amplification.
PCR reactions for 3′-RACE were performed with primers rDLM2F (5′-CCA AGG CTC TGG GAA TGA CGA C-3′) or rDLM3F (5′-TGA CAG CTA TGG CTC TGG GAG A-3′) and AXT. PCR was carried out using primers mDLM1f (5′-CCC CAA GGC TGC TGT GAG CAT GGC-3′) and r/mDLM3r (5′-CAY CCA GGA GTG GTA SAA CTG GCT C-3′) and for 3′-RACE experiments using primers rDLM2F (5′-CCA AGG CTC TGG GAA TGA CGA C-3′) or rDLM3F (5′TGA CAG CTA TGG CTC TGG GAG A-3′) and AXT, using AmpliTaq Gold (PE Applied Biosystems) at 94°C for 8 min, followed by 34 cycles of 30 s at 95°C, 1 min at 59°C and 2 min at 72°C.
Cloning of PCR
Products. PCR products were cloned into the pCR2.1 vector using the TA-cloning kit (Invitrogen), and sequenced using the dye terminator protocol (PE Applied Biosystems) according to the manufacturer’s instructions. Sequence analysis was performed with DNASTAR software. Accession numbers of sequences: human DLM-1, AJ300575; rat DLM-1, AJ302054.
RESULTS
Organization and sequence of human DLM-1
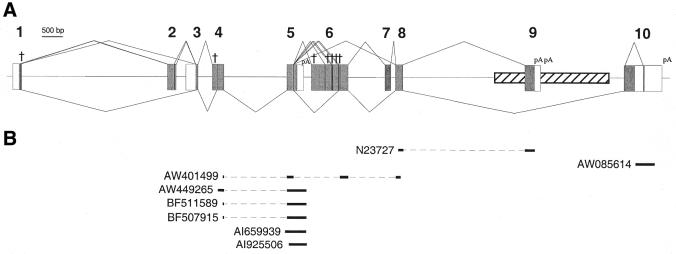
The human genome database was searched with the cDNA sequence of the murine Z-DNA binding protein gene Dlm-1 to identify its human homolog. Homologous regions were identified on chromosome 20q13.31. Primers specific for human DLM-1 were derived from predicted exonic regions and RT–PCRs were performed. The genomic organization of the 10 identified exons of human DLM-1 is shown (Fig. 1). It spans over 17 kb.
Figure 1.
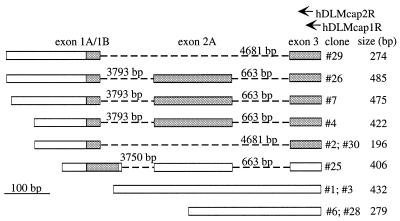
Organization of the human DLM-1 gene. (A) The exon–intron structure of the human DLM-1 gene is shown. The 5′-untranslated regions of exons 1 and 3 and the 3′-untranslated regions of exons 5, 9 and 10 are depicted as white boxes. Other exonic parts are depicted as gray boxes. Polyadenylation sites (pA) and premature stop codons (†) in exon variants are indicated. The splicing events observed most frequently are indicated by lines below, alternative splicing events are shown above the gene. The location of a Tigger transposable element, which includes exon 9 is indicated (striped box). (B) The composition and accession nos of EST clones representing human DLM-1 found in the database are shown.
Expression pattern of DLM-1
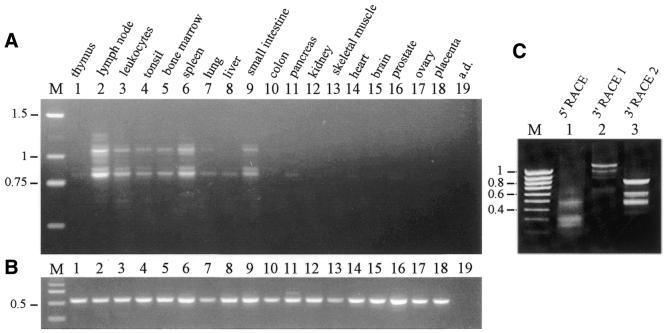
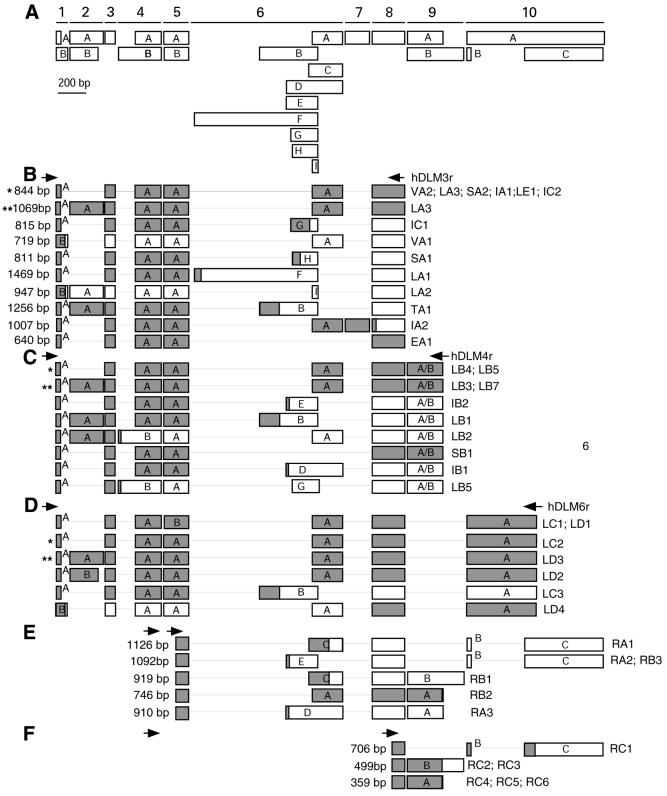
RT–PCRs with primers derived from exon 1 and exon 8 were performed from human cDNAs from various human tissues. Expression of hDLM-1 is tissue specific. It was most prominently expressed in lymph node and splenic cells (Fig. 2A). Significant amounts were also detected in peripheral leukocytes, tonsil, bone marrow and small intestine. Low levels were seen in thymus, lung, liver and pancreas. Alternative splicing led to the detection of reaction products of multiple sizes. Control RT–PCR with cyclophilin-specific primers demonstrated that comparable amounts of cDNA were present in all cDNA preparations (Fig. 2B). DLM-1 amplification products were cloned and sequenced. The exon composition of individually analyzed cDNA isoforms is shown schematically in Figure 3. The most prominent band seen in the PCR analysis evidently represents transcripts comprising exons 1A, 3A, 4A, 5A, 6A and 8 (844 bp; marked by an asterisk in Fig. 3), which was cloned in 10 out of 32 cases. The second most abundant band includes exons 1A, 2A, 3A, 4A, 5A, 6A and 8 (1069 bp; marked by two asterisks in Fig. 3), which was cloned in four cases. Sequences derived from reactions with forward primer DLM1f in combination with a reverse primer from one of the two different 3′-terminal exons are shown in Figure 3C and D. In 56% (18 out of 32) of all analyzed clones the open reading frame (ORF) was continuous, whereas it was interrupted in the remainder. In 9 out of 14 cases the interruption was due to premature stop codons that originated from exon 6 variants. All nine exon 6 variants share a stretch of 39 bp, which encode the complete variant I of exon 6. In three cases the ORF was interrupted by a premature stop codon in the exon 1 variant B, and in two cases by a stop in exon 4, variant B. Exon 2, coding for the Z-DNA binding domain ZαDLM, was subject to alternative splicing and was included in 28% of analyzed clones. A shortened form of exon 2, exon 2B, was identified in one clone, where the last 36 nt in the 3′ region of this exon were spliced out. For the relative locations of exons and the frequency of their occurrence see Supplementary Material Table S1.
Figure 2.
Expression pattern of DLM-1. (A) PCR with DLM-1 specific primers was performed with cDNA from the indicated tissues for 32 cycles. (B) Control PCR was performed with primers specific for cyclophilin for 25 cycles. PCR products were visualized on 1.6% agarose gels, containing ethidium bromide. (C) Amplification products of 5′- (lane 1; see also Fig. 5) and of two 3′-RACE PCRs with different forward primers (lanes 2 and 3; see also Fig. 3E and F) are shown. Sizes of molecular weight markers are shown in kilobases.
Figure 3.
Composition of individual DLM-1 splice variants. (A) The identified exons and their sizes are shown. The exons are numbered. Variants of individual exons are marked by letters. (B–F) Schematic composition of cloned transcripts. Clones shown in (E) and (F) were derived from 3′-RACE reactions. Arrows indicate the positions of primers used for PCR amplification. Predicted ORFs are shaded gray, presumably untranslated regions are in white. The exon combination identified most often is marked by an asterisk, the second most abundant by two asterisks. The clones are annotated as follows: The first letter defines the tissue origin of the cDNA used as PCR templates. L, lymph node; S, spleen; I, small intestine; V, liver; T, thymus; E, leukocyte; R, spleen (marathon cDNA). The second letter denotes the PCR reaction followed by the number of the clone.
Analyses of 5′- and 3′-terminal exons
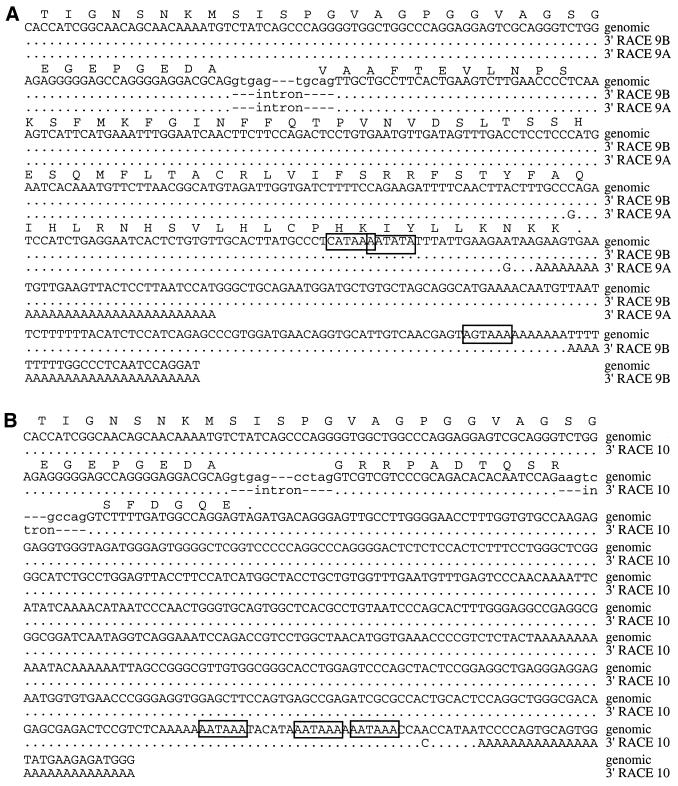
3′-RACE PCRs were performed with gene-specific primers indicated in Figure 3E and F. Three major amplification products were detected (Fig. 2C, lanes 2 and 3). Cloning and sequencing of these revealed that the different products, which were all polyadenylated, resulted from different 3′-terminal exons. In the largest and most prominent product, variants of exon 10 were present, whereas the others contained exon 9. Two variants of exon 9 with different polyadenylation sites were found. The sequences of 3′-RACE PCR products and the genomic sequences, as well as the deduced C-terminal amino acid sequences are shown (Fig. 4). Three consensus polyadenylation signals precede the polyadenylation sites in exon 10. In exon 9 the two polyA tails are preceded by atypical polyadenylation signals. The complete exon 9 is part of a 2.5 kb Tigger repetitive element (Fig. 1), which belongs to the family of DNA transposons (19). Sequence similarity compared with the Tigger1 consensus sequence is 86%. The Tigger element integrated in the opposite direction relative to the orientation of DLM-1. The deduced 82 C-terminal amino acids of DLM-1 ending with exon 9 are encoded by the antisense strand of the ORF1 of the Tigger1 element.
Figure 4.
Nucleotide and deduced amino acid sequences of the two alternative 3′-terminal exons. Three different polyadenylated 3′ ends were identified. Sequences shown are from PCR products using nested primers in exon 8 (Fig. 3F). cDNA derived clones are compared with the genomic sequences. The first and last 5 bp of introns are shown in lower case letters. The potential polyadenylation signals are boxed. The sequences shown here are identical to the sequences obtained from the 3′-RACE where the second primer was located in exon 5 (Fig. 4E). (A) Two isoforms ended with exon 9, designated as 9A and 9B. (B) The third isoform ended with exon 10. Due to an internal splice site in exon 10, parts of exon 10 are skipped.
Sixty-four percent of clones obtained from 5′-RACE PCR (Fig. 2C, lane 1) started with exon 1 (Fig. 5). Four different 5′ ends were identified (see Supplementary Material, Fig. S1). In 50% (three out of six) of these clones exon 2 was included. Four clones started 5′ to exon 3. Two of them started 397 bp and two 231 bp 5′ to exon 3. The potential start codon of transcripts beginning with exon 3 lies in exon 4.
Figure 5.
Results of 5′-RACE PCR. Individual clones obtained from 5′-RACE PCRs are schematically shown. Presumptive coding regions of exons are depicted as gray boxes, putative untranslated regions as white boxes. Introns are indicated by lines and their sizes are shown. The locations of gene-specific primers used for PCR amplification, as well as the sizes of clones, are indicated.
Identification of a third polyadenylation site by the analysis of expressed sequence tags (ESTs)
The dbEST data base was searched with different fragments of the DLM-1 gene for human ESTs. Eight ESTs were identified that showed sequence identities ranging between 99 and 100% compared with hDLM-1 (Fig. 1B). One clone (AW401499; origin: squamous cell carcinoma of the esophagus) contained the terminal part of exon 10. Another (N23727; origin: olfactory epithelium) contained part of exon 8 spliced to exon 9. Clone AW401499 (origin: lymph node germinal B-cells) contained part of exon 4, the complete exons 5 and 6A as well as part of exon 8.
Five clones (AW449265, BF507915, BF511589, AI659939 and AI925506, derived from germ cells, chronic lymphocytic leukemia, uncharacterized tissue, colonic mucosa of patients with severe Crohn’s disease, and adenocarinoma of the stomach, respectively) contained poly(A) tails originating in intron 5, 272 bp 3′ of the site where exon 5 was usually spliced to exon 6 variants. Inspection of the sequence of this region revealed the presence of a consensus polyadenylation signal AATAAA 19 bp 5′ of the polyadenylated sequences. Three of these clones contained a part of exon 4 spliced to exon 5, whereas two were not spliced to other exons.
Comparison between mouse, rat and human DLM-1
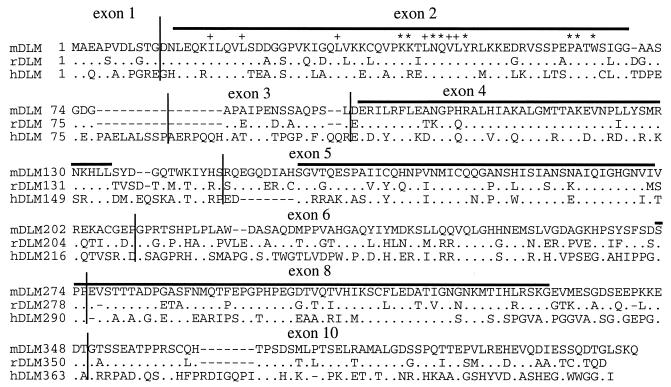
In order to determine conserved regions in DLM-1, we also cloned the rat DLM-1 gene. Primers used for RT–PCR from rat spleen RNA were derived from the 5′- and 3′-untranslated regions of the mouse Dlm-1 gene. These regions have a high degree of sequence homology with a rat EST (AI763632). The obtained sequence was used for primer construction for 3′-RACE PCR. The deduced amino acid identity of rat and mouse DLM-1 is 74%. Protein sequence identity of human to mouse and rat DLM-1 is 47 and 46%, respectively. Four regions have been identified that show a higher degree of conservation between the three species (Fig. 6, lines). The first two conserved domains (CDs) are homologous to the Z-DNA binding domains Zα and Zβ, respectively, of the RNA editing enzyme ADAR1. In particular, those amino acids of Zα that contact DNA or form the hydrophobic protein core are highly conserved. No other homologies were identified for the other two conserved regions. The CDs fit well with the exon/intron structure of human DLM-1 (Fig. 6). The first, Zα homologous, CD is encoded by exon 2, whereas the second, homologous to Zβ, is encoded by exon 4. The third and fourth CDs are encoded by exons 5 and 8, respectively. Sequence identity of the region encoded by exon 6 is relatively low.
Figure 6.
Comparison of mouse, rat and human DLM-1 protein sequences. The deduced amino acid sequences of mouse, rat and human DLM-1 are compared. Dots indicate amino acid identities to mDLM-1. Dashes indicate insertions or deletions. Horizontal lines indicate regions with a high degree of conservation between the species. Vertical lines indicate exon boundaries of hDLM-1. Amino acids important for the recognition of Z-DNA (asterisks) and those forming the hydrophobic core of ZαDLM (+) are marked (17).
Identification of two DLM-1 alleles
Sequence analysis of the cloned PCR products revealed a single nucleotide polymorphism at the position of the third nucleotide of exon 3 (Supplementary Material, Fig. S1). This A440G substitution was present in 32% of the analyzed clones, and would result in a deduced amino acid change from lysine to glutamic acid (K88E). Since the clones were obtained from the amplification of Marathon-Ready cDNA, which is pooled from a large number of human Caucasian individuals, it is likely that this distribution reflects the frequency of the polymorphism in the Caucasian population. This polymorphism affects a position in the predicted linker between the Zα and Zβ domains of DLM-1.
DISCUSSION
We describe here the cloning of the human and rat genes coding for the Z-DNA binding protein DLM-1, which is the homolog of murine DLM-1. Comparison between mouse, rat and human DLM-1 amino acid sequences revealed the existence of four domains with a high degree of conservation which show sequence identities of 67, 65, 71 and 61%, respectively, between mouse and human sequences. The first two CDs correspond to the Z-DNA binding domains of ADAR1, Zα and Zβ. No significant similarities to other proteins were identified for the C-terminal part of DLM-1. However, the existence of two CDs in this region indicates that these have been subjected to positive selective pressure and may play important roles.
Human DLM-1 is most strongly expressed in the small intestine as well as in lymphatic tissues, like lymph node, leukocytes and spleen. In lung and liver, DLM-1 was expressed to a lesser degree. This is partly in contrast to the expression pattern of mDlm-1, which was described to be expressed strongest in lung, liver and spleen (18). In both species, no expression was observed in brain and skeletal muscle. Extensive alternative splicing of the 10 exons of human DLM-1 was found. In combination with different 5′ and 3′ termini, the splice variants we observed may lead to the production of over 2000 different mRNAs. However, on the basis of their putative encoded protein products these may be put into a small number of groups. In the majority of analyzed mRNAs (cDNA clones) the ORF was not interrupted through exon 8 (18 out of 32 clones). In the remainder this ORF was interrupted by premature stop codons. Four of these did not contain any significant ORF (>300 bp). The rest contained ORFs of variable lengths.
Approximately one-third of clones included exon 2, which encodes the Z-DNA binding domain Zα. In ADAR1, Z-DNA binding is exclusively dependent on the presence of the Zα domain. Zβ, the second domain, enhances Z-DNA binding when tethered to Zα, but does not bind Z-DNA by itself (20,21). In analogy to ADAR1, we predict that messages lacking exon 2 do not bind Z-DNA efficiently. However, further studies of the tethered ZαβDLM domain and ZβDLM are necessary to prove this statement.
In most cases the ORF was interrupted as a result of alternatively spliced exon 6 variants. Of the nine variants of exon 6, seven (B, C, D, E, F, G and H) terminate the ORF when spliced to exon 5, which was found in all transcripts (Fig. 3B–D). mRNAs containing these exon variants are predicted to encode for truncated proteins with different C-termini. Five out of these seven variants (B, E, F, G and H) use a splice donor site that lies within the most frequent, the exon 6A variant. Only one mRNA was found that contained a variant with the same 5′ exonic sequence as exon 6A combined with the 3′ exonic sequence of exon 6B–H (exon 6I). This exon 6 variant encodes 13 amino acids, and splicing to exon 8 leaves the ORF intact.
The existence of so many different exon 6 variants is remarkable. The alternative incorporation of exons containing premature stop codons into the hDLM-1 mRNA isoforms is a frequent phenomenon and appears to be of functional relevance. Proteins containing ZαDLM, but truncated by premature stop codons, would preserve the potential to bind Z-DNA. However, they would lack the C-terminal domains encoded by exons 6, 8 and 9 or 10. A similar situation applies also for transcripts terminating with exon 5. Although the function of the C-terminus is not known, it is conceivable that these proteins may function as naturally occurring dominant negative variants.
Alternatively, the mRNAs containing premature stop codons may be degraded by nonsense-mediated decay (NMD) (22,23). The mRNA surveillance complex can scan for aberrant mRNAs. Inclusion of different exon variants containing premature translation termination codons occurs frequently and appears to be a regulated process. Degradation of these mRNAs by NMD may thus represent a post-transcriptional mechanism in the control of hDLM-1 expression.
Most of the identified 5′ and 3′ sequences flanking exons are in accord with the consensus sequences for RNA splice junctions obtained by Shapiro and Senapathy (24) (Supplementary Material, Table S1). Only two exceptions regarding the 5′-splice site were identified. The first is an internal splice site in exon 2. This splicing event, which does not interrupt the ORF, was only found in one clone, and occurred 3′ to the Zα domain, in a region linking Zα with Zβ. This linker is of variable length between rodent and human DLM-1 proteins as well as in ADAR1 of different species and its importance for Z-DNA binding is unclear. The splice donor site (AAG/gcgagg) is very similar to the atypical splice junctions found in the human genes for the acetylcholine receptor (25) and superoxide dismutase (26) where (AAG/gcaagg) is found. The second atypical splice donor site (CAG/aagtca) is found in exon 10 and leads to the splicing out of the region between variants B and C of this exon. This variant was only found in 3′-RACE analysis from spleen cDNA, but not in RT–PCR analyses where flanking primers were used for PCR amplification.
Three different 3′-terminal exons were identified in the 3′-RACE experiments and by the analysis of ESTs. The major product from 3′-RACE experiments ended with polyadenylated exon 10. Three consensus polyadenylation signals are found 35–10 bp 5′ of the polyadenylation site. The two minor bands correspond to products ending with exon 9, which were polyadenylated at different sites. Three atypical polyadenylation signals are present 5′ of the polyadenylation sites, which have been shown to mediate in vitro polyadenylation efficiencies of 18% (CAUAAA), 10% (AAUAUA) or 29% (AGUAAA) as compared with the consensus signal (AAUAAA) (27).
The entire exon 9, including the polyadenylation sites, is part of a Tigger1 DNA transposable element (19). The C-termini of mouse and rat DLM-1 show significant homology only to the amino acids encoded by human exon 10, but not to exon 9. The incorporation of a part of the Tigger1 element into the mRNA with coding capacity provides an example of how transposable elements may increase the genomic flexibility.
Two major transcriptional start sites were identified by 5′-RACE analysis (Fig. 5). The mRNAs starting with exon 1 may include the Z-DNA binding domains encoded by exons 2 and 4. Transcripts beginning with exon 3 most likely use the first in-frame start codon, located in exon 4. It is of interest that this start site in exon 4 would not allow for a properly folded ZβDLM domain, since important parts of the hydrophobic core are encoded 5′ to this start site. Proteins encoded by these mRNAs would be identical to other DLM-1 isoforms in their C-terminal parts, but would be unable to bind Z-DNA.
Two major DLM-1 RNA isoforms were detected by northern blotting in the mouse. The 1.2 kb fragment was sensitive to cyclophosphamide, whereas the larger (2.2 kb) fragment was not. It has been speculated that the two RNA isoforms were most likely generated from alternative splicing (18). However, they may also be a result of alternative transcriptional start or termination sites, analogous to the situation observed for hDLM-1. We have extensively searched various databases in order to identify regions with significant homologies to other sequences. The identified homologous regions were the Zα and Zβ domains of the RNA editing enzyme ADAR1, as well as a domain related to ZαADAR, in the vaccinia virus interferon resistance protein E3L. The Zα homologous domain from E3L can also bind to Z-DNA in vitro (T.Schwartz, unpublished data) and is conserved among orthopoxviridae. It has been shown to be essential for complete viral pathogenesis in a mouse model (28). Since the only two known eukaryotic proteins with Z-DNA binding domains, ADAR1 (29) and DLM-1 (18), are both γ interferon-inducible, it seems possible that these proteins are important in the host-response to pathogens. Proteins of the E3L family could compete with ADAR1 and DLM-1 in nucleic acid binding and may thus prevent these proteins from being located properly.
The potential generation of multiple mRNAs from a single gene due to alternative splicing, the usage of different promoters or polyadenylation sites and RNA editing greatly increases the number of encoded protein isoforms (30). For instance, alternative splicing of RNA transcribed from the Drosophila melanogaster gene encoding the axon guidance receptor Dscam can theoretically result in over 38 000 different mRNAs, by far exceeding the number of genes encoded by the entire D.melanogaster genome (31). The functional consequences of this alternative splicing are unknown.
The human genome is estimated to contain between 30 000 and 40 000 genes (32). However, genomic flexibiltity may be greatly increased by alternative splicing and usage of alternative transcription start and termination sites as shown here for hDLM-1. In addition to the identification of alternative transcripts, it will be a major challenge for the future to study the exact functional biochemical and physiological consequences of such alternative events.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Silvia Schmoock and Julia Nottelmann for excellent technical assistance.
DDBJ/EMBL/GenBank accession no. NM030776
REFERENCES
- 1.Rich A., Nordheim,A. and Wang,A.H. (1984) The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem., 53, 791–846. [DOI] [PubMed] [Google Scholar]
- 2.Wittig B., Dorbic,T. and Rich,A. (1989) The level of Z-DNA in metabolically active, permeabilized mammalian cell nuclei is regulated by torsional strain. J. Cell Biol., 108, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wittig B., Dorbic,T. and Rich,A. (1991) Transcription is associated with Z-DNA formation in metabolically active permeabilized mammalian cell nuclei. Proc. Natl Acad. Sci. USA, 88, 2259–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittig B., Wölfl,S., Dorbic,T., Vahrson,W. and Rich,A. (1992) Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J., 11, 4653–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wölfl S., Martinez,C., Rich,A. and Majzoub,J.A. (1996) Transcription of the human corticotropin-releasing hormone gene in NPLC cells is correlated with Z-DNA formation. Proc. Natl Acad. Sci. USA, 93, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wölfl S., Wittig,B., Dorbic,T. and Rich,A. (1997) Identification of processes that influence negative supercoiling in the human c-myc gene. Biochim. Biophys. Acta, 1352, 213–221. [DOI] [PubMed] [Google Scholar]
- 7.Herbert A. and Rich,A. (1996) The biology of left-handed Z-DNA. J. Biol. Chem., 271, 11595–11598. [DOI] [PubMed] [Google Scholar]
- 8.Kashi Y., King,D. and Soller,M. (1997) Simple sequence repeats as a source of quantitative genetic variation. Trends Genet., 13, 74–78. [DOI] [PubMed] [Google Scholar]
- 9.Rothenburg S., Koch-Nolte,F. and Haag,F. (2001) DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles. Immunol. Rev., 184, 286–298. [DOI] [PubMed] [Google Scholar]
- 10.Liu R., Liu,H., Chen,X., Kirby,M., Brown,P.O. and Zhao,K. (2001) Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell, 106, 309–318. [DOI] [PubMed] [Google Scholar]
- 11.Nordheim A. and Rich,A. (1983) Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature, 303, 674–679. [DOI] [PubMed] [Google Scholar]
- 12.Mori Y., Folco,E. and Koren,G. (1995) GH3 cell-specific expression of Kv1.5 gene. Regulation by a silencer containing a dinucleotide repetitive element. J. Biol. Chem., 270, 27788–27796. [DOI] [PubMed] [Google Scholar]
- 13.Rothenburg S., Koch-Nolte,F., Rich,A. and Haag,F. (2001) A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA, 98, 8985–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert A., Alfken,J., Kim,Y.G., Mian,I.S., Nishikura,K. and Rich,A. (1997) A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl Acad. Sci. USA, 94, 8421–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz T., Rould,M.A., Lowenhaupt,K., Herbert,A. and Rich,A. (1999) Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science, 284, 1841–1845. [DOI] [PubMed] [Google Scholar]
- 16.Brown B.A. II, Lowenhaupt,K., Wilbert,C.M., Hanlon,E.B. and Rich,A. (2000) The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl Acad. Sci. USA, 97, 13532–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz T., Behlke,J., Lowenhaupt,K., Heinemann,U. and Rich,A. (2001) Structure of the DLM-1–Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nature Struct. Biol., 8, 761–765. [DOI] [PubMed] [Google Scholar]
- 18.Fu Y., Comella,N., Tognazzi,K., Brown,L.F., Dvorak,H.F. and Kocher,O. (1999) Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene, 240, 157–163. [DOI] [PubMed] [Google Scholar]
- 19.Smit A.F. and Riggs,A.D. (1996) Tiggers and DNA transposon fossils in the human genome. Proc. Natl Acad. Sci. USA, 93, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y.G., Lowenhaupt,K., Schwartz,T. and Rich,A. (1999) The interaction between Z-DNA and the Zab domain of double-stranded RNA adenosine deaminase characterized using fusion nucleases. J. Biol. Chem., 274, 19081–19086. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz T., Lowenhaupt,K., Kim,Y.G., Li,L., Brown,B.A., Herbert,A. and Rich,A. (1999) Proteolytic dissection of Zab, the Z-DNA-binding domain of human ADAR1. J. Biol. Chem., 274, 2899–2906. [DOI] [PubMed] [Google Scholar]
- 22.Czaplinski K., Ruiz-Echevarria,M.J., Gonzalez,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- 23.Hilleren P. and Parker,R. (1999) Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33, 229–260. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro M.B. and Senapathy,P. (1987) RNA splice junctions of different classes of eukaryotes, sequence statistics and functional implications in gene expression. Nucleic Acids Res., 15, 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibahara S., Kubo,T., Perski,H.J., Takahashi,H., Noda,M. and Numa,S. (1985) Cloning and sequence analysis of human genomic DNA encoding γ subunit precursor of muscle acetylcholine receptor. Eur. J. Biochem., 146, 15–22. [DOI] [PubMed] [Google Scholar]
- 26.Levanon D., Lieman-Hurwitz,J., Dafni,N., Wigderson,M., Sherman,L., Bernstein,Y., Laver-Rudich,Z., Danciger,E., Stein,O. and Groner,Y. (1985) Architecture and anatomy of the chromosomal locus in human chromosome 21 encoding the Cu/Zn superoxide dismutase. EMBO J., 4, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheets M.D., Ogg,S.C. and Wickens,M.P. (1990) Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res., 18, 5799–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandt T.A. and Jacobs,B.L. (2001) Both C- and N-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol., 75, 850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson J.B., Thomis,D.C., Hans,S.L. and Samuel,C.E. (1995) Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by α and γ interferons. Virology, 210, 508–511. [DOI] [PubMed] [Google Scholar]
- 30.Graveley B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- 31.Schmucker D., Clemens,J.C., Shu,H., Worby,C.A., Xiao,J., Muda,M., Dixon,J.E. and Zipursky,S.L. (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell, 101, 671–684. [DOI] [PubMed] [Google Scholar]
- 32.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.