Summary
We tested the hypothesis that conserved placental mammal-specific microRNAs and their targets facilitate endometrial receptivity to implantation. Expression of miR-340-5p, -542-3p, and -671-5p was regulated by exposure of endometrial epithelial cells to progesterone (10 μg/ml) for 24 h coordinate with 1,713 of their predicted targets. Proteomic analysis of cells transfected with miRNA mimic/inhibitor (48 h: n = 3) revealed 1,745 proteins altered by miR-340-5p (mimic; 1,369, inhibitor; 376) of which 171 were predicted targets and P4-regulated. MiR-542-3p altered 2,353 (mimic; 1,378, inhibitor; 975) 100 which were mirDB predicted, including 46 P4-regulated. MiR-671-5p altered 1,744 proteins (mimic; 1,252, inhibitor; 492) 95 of which were predicted targets and 46 P4-regulated. All miRNAs were detected in luteal phase endometrial biopsies, irrespective of pregnancy outcomes. miR-340-5p expression increased in biopsies from individuals suffering previous and subsequent miscarriage compared to those with subsequent live birth. Dysfunction of these miRNAs and their targets contribute to endometrial-derived recurrent pregnancy loss.
Subject areas: Biological sciences, Molecular biology, Developmental biology, Embryology
Graphical abstract
Highlights
-
•
Progesterone upregulates miR-340-5p, -542-3p and -671-5p in endometrial epithelium
-
•
Progesterone alters transcripts involved in proliferation and epithelial development
-
•
Over- or under-expressing these microRNAs results in altered abundance of proteins
-
•
Functions of altered proteins are involved in implantation and uterine receptivity
Biological sciences; Molecular biology; Developmental biology; Embryology
Introduction
Successful pregnancy in all placental mammals, including humans, is contingent on a carefully orchestrated series of events, including fertilisation between competent gametes, appropriate embryo development, successful implantation, and the establishment of a functional placenta. Perturbations at any of these key developmental points, such as chromosomal abnormalities in the embryo, are known to result in pregnancy loss. However, the contribution of an inadequately regulated endometrium is considerably underexplored. In humans, the uterine endometrium is a heterogeneous tissue composed of luminal and glandular epithelial cells, underlying fibroblast-like stromal cells, and populations of immune cells, vasculature and stem cells.1 Spatial and temporal responsiveness of these cells to hormonal cues and other signaling molecules throughout the menstrual cycle facilitates rapid proliferation and differentiation of the endometrial lining to prepare for successful implantation. The appropriate physiological response of the endometrium depends not only on the cellular response to systemic cues, such as circulating estradiol and progesterone (P4) levels, but also on the inter- and intra-cellular communication between different cell types.
Inappropriate remodeling of the endometrium, both before and at the time of implantation, can lead to pregnancy failure such as miscarriage even when a developmentally competent embryo implants.2,3,4 Indeed, miscarriage can affect up to 1 in 6 pregnancies.5 The dynamic transcriptional responses of the endometrium during the menstrual cycle and implantation (reviewed by6) have been investigated in detail in relation to how they contribute to miscarriage.6,7,8 However, the regulatory signaling networks responsible for establishing a receptive endometrium to facilitate implantation, and their dysregulation in recurrent pregnancy loss (RPL), have yet to be determined.
Given their flexible and dynamic regulatory affects, miRNAs may be key in these processes. Once processed from pri- and pre-miRNA forms, these small RNA molecules are usually 22 nucleotides long and bind primarily to the 3′UTR of mRNAs and either suppress the initiation of translation or promote the degradation of the mRNA itself.9 They play key roles in inter-cellular communication as components of extracellular vesicles (EVs) or modify direct cargo loading to EVs.9 Some candidate microRNAs (miRNAs) that are altered in the endometrium have been reported to relate to early miscarriage.6,7,8 The expression of miRNAs varies dramatically and dynamically during pregnancy,10 with dysregulation of specific miRNAs also having been associated with other pregnancy disorders including preeclampsia and intrauterine growth restriction.11,12 However, conflicting data between studies as well as cell-specific patterns of expression and regulation of miRNA mean we do not fully understand their role in normal and dysfunctional endometria.
The origin of placental mammals is synonymous with the emergence of placentation, lactation and also of hair. This burst of morphological innovation was mediated by a combination of novel and repurposed genomic elements (both protein coding and non-protein coding), regulatory mechanisms, signaling pathways, cells and tissues.13,14,15 Indeed, bilateral signaling between fetal and maternal tissues is key to establishment of the mammalian placenta. In humans in particular, invasion of the maternal endometrium by the fetal trophoblast required coordinate specialization of these distinct tissues with the co-evolution of the underlying molecular signaling networks facilitating these interactions and communications.13 Recent data using an evolutionary approach has identified a key role for the transcription factor, HAND2, and its target genes as important in the pathogenesis of pre-term birth and gestation length.16,17 We have adopted a similar strategy, leading to the identification of 13 miRNA families (17 microRNA genes) that arose coordinate with placental mammals and have been retained in all modern placental mammals.18 These miRNAs are detectable in the endometrial epithelium and their expression is altered in a species-specific manner by proteins known to facilitate signaling between the embryo and endometrium.19,20
Given that (1) the endometrium and the trophoblast had to co-evolve regulatory networks to support implantation and early pregnancy success, (2) miRNAs can be important regulators in establishing these types of networks and (3) miRNA (and their targets) are regulated by molecules important to establish receptivity to implantation, we tested the hypothesis that these mammal-specific miRNA genes and their targets are regulated in the endometrium to facilitate receptivity to implantation. We specifically focus on the endometrial epithelium as this is where uterine receptivity is established and also the site of the first biophysical interaction between the embryo and endometrium. Furthermore, we propose that dysregulated expression of these miRNAs in the endometrium may be a feature in women with early pregnancy loss.
Results
Progesterone alters selected miRNAs and predicted targets in vitro
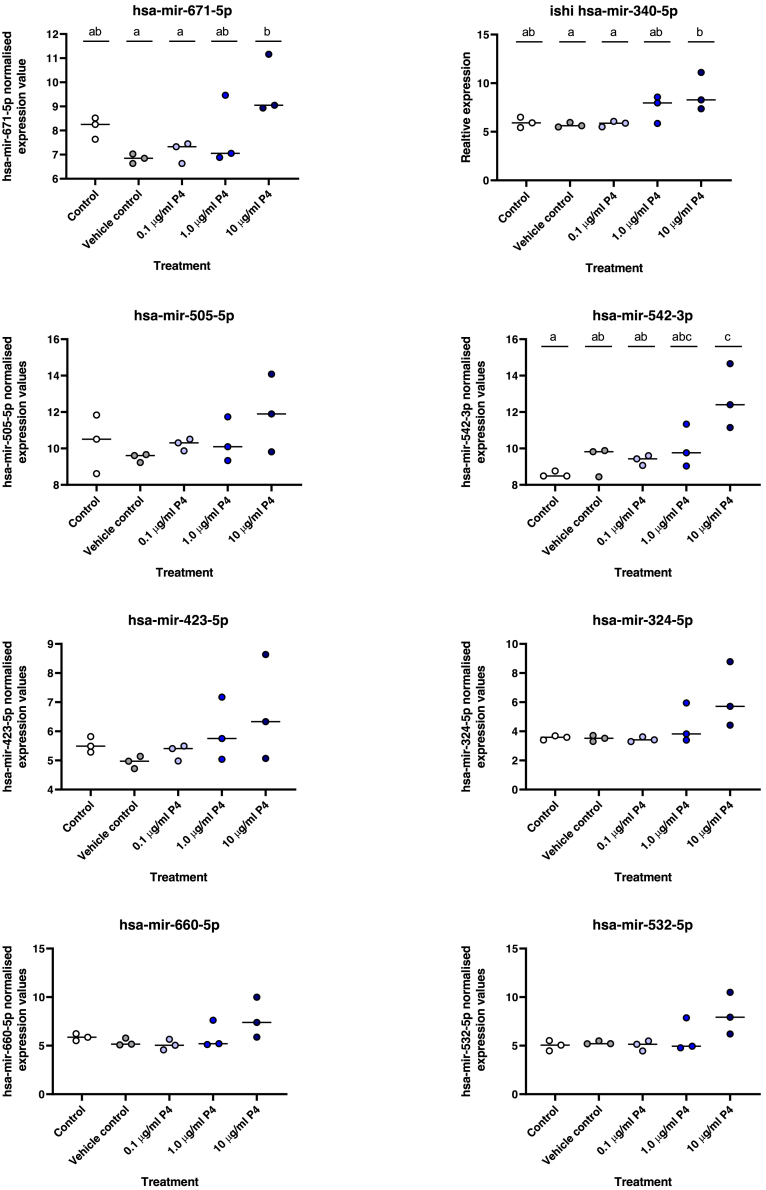
Relative expression levels of miR-340-5p, -542-3p, and -671-5p all increased following treatment with progesterone (10 μg/mL) for 24 h (Figure 1) compared to control(s). No treatment effect was observed for the other miRNAs (p>0.05). TargetScan predicted miR-340-5p has binding sites in 6,558 human transcripts. Of these predicted targets 68% (4,434) have functional assignments to ontologies/pathways in the interactome database, i.e., have an assigned function. For miR-542-3p, 4,748 targets were predicted using TargetScan and 2041 have assigned functions in the reactome database. In the case of miR-671-5p 2820 of 4,224 predicted targets have a function in the reactome database.
Figure 1.
Expression of selected miRNAs in human endometrial epithelial (Ishikawa) cells
Cells were cultured for 24 h with: (1) Control (open circle), (2) Vehicle Control (10% EtOH: gray circle), (3) 0.1ug/ml P4 (light blue circle), (4) 1.0 μg/ml P4 (royal blue circle), or (5) 10 μg/ml P4 (dark blue circle: n = 3 biological replicates). Expression values of miRNAs were determined using LNA Sybr Green method and the relative expression values following normalization. Differences in expression were determined relative to vehicle control using an ANOVA and considered statistically different when p< 0.05. Differences between groups are denoted by difference in letters (a, b, c, d).
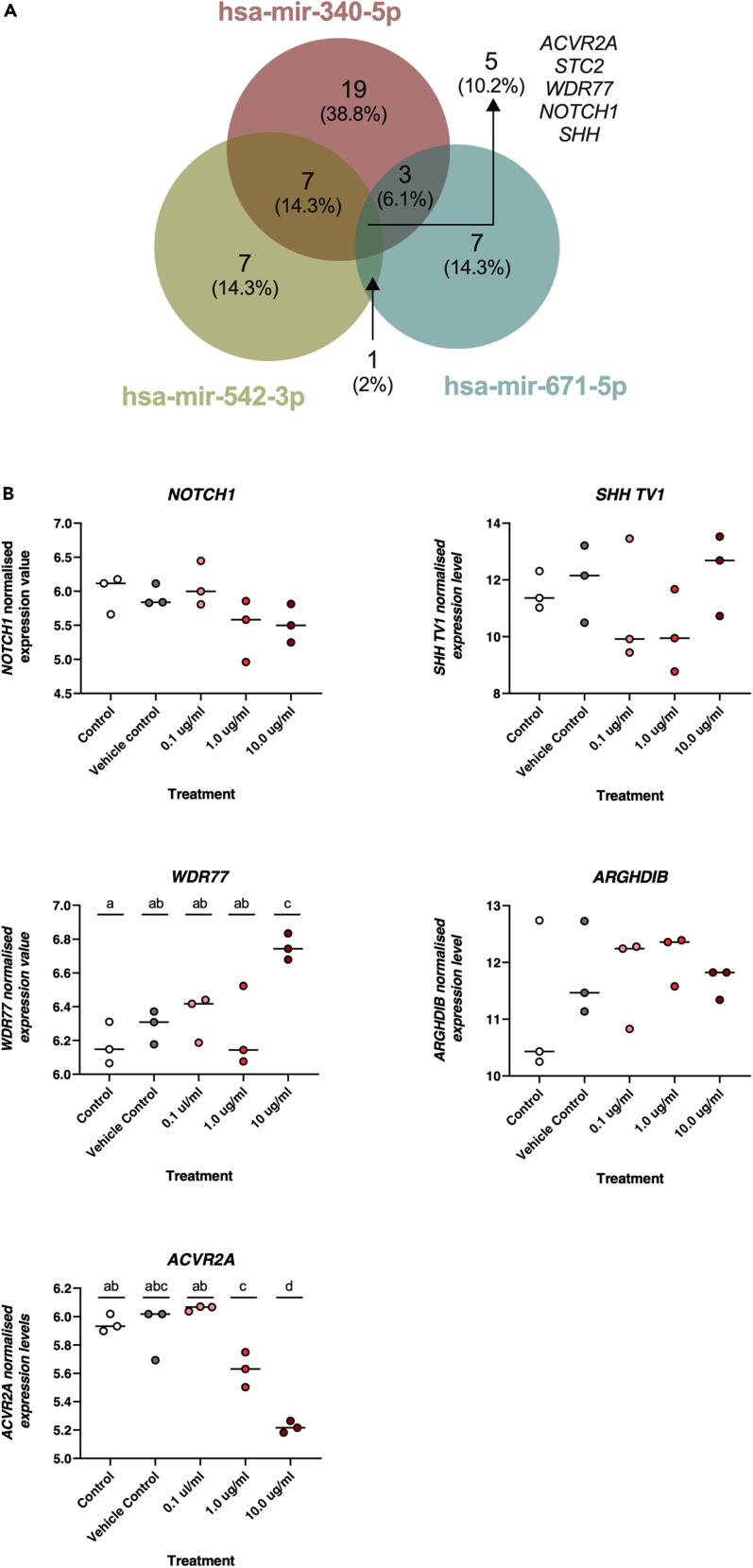
In total, 34 predicted targets of mir-340-5p, 20 predicted targets of mir-542-3p, and 16 predicted targets of mir-671-5p were associated with the GO term ‘reproductive function’ and were thus selected for further investigation. There was considerable overlap in the predicted ‘reproductive function’ targets across these three microRNA mature sequences where mir-542-3p and mir-671-5p had seven unique target predictions each (Figure 2A), conversely 19 were predicted to be targets of miR-340-5p alone. Common to all three P4-dependent miRNAs (i.e., mir-340-5p, mir-542-3p and mir-671-5p) were five ‘reproductive function’ predicted targets (Activin A Receptor, Type IIA [ACVR2A], Stanniocalcin-Related Protein [STC2], WD Repeat-Containing Protein 77 [WDR77], Notch Receptor 1 [NOTCH1], and Sonic Hedgehog Signaling Molecule [SHH]. On examining these five common mRNA targets (Table 1) we found there was no effect of P4 on expression of ARGHDIB, NOTCH1, or SHH between any groups (Figure 2B:(p > 0.05)). Expression of WDR77 increased in cells treated with the highest dose of P4 compared to controls (p < 0.05), whereas expression of ACVR2A expression decreased in cells treated with 1.0 and 10.0 μg/ml of P4 compared to control(s) (p < 0.05).
Figure 2.
Progesterone regulated microRNA targets
(A)Venn diagram representing overlap in predicted mRNA targets of the three miRNAs altered following 24 h treatment of endometrial epithelial cells with P4 in vitro. These were associated with the Gene Ontology Biological Process of Reproductive Biology.
(B) RT-qPCR analysis of predicted targets of all three P4-regulated miRNAs that are associated with the Gene Ontology Biological Process of Reproductive Function. Expression was determined in Ishikawa cells treated with control (open circles), vehicle control (gray circles), 0.1 μg/mL (pale red circle), 1.0 μg/mL (medium red circle), or 10.0 μg/mL (dark red circle) of P4 for 24 hin vitro (n = 3 independent cultures). Differences in expression compared were determined using ANOVA with differences depicted by different subscripts (a, b, c, d) when p < 0.05.
Table 1.
Primers used for q-RT-PCR analysis of P4 modified transcripts related to reproductive function
| Gene abbreviation | Accession No | Forward primer sequence | Primer length | Amplicon length |
|---|---|---|---|---|
| ACVR2A | NM_001278579.1 | Forward: TCCGAGGAAGACCCAGGGAA | 20 | 133 |
| Reverse: GTATAGCACCTGAAGAACAGGAGA | 24 | |||
| STC2 | NM_003714.2 | Forward: CAGTTGCAGCGGGAATGCTA | 20 | 123 |
| Reverse: GAGGTCCACGTAGGGTTCGT | 20 | |||
| WDR77 | NM_001317062.1 | Forward: CCCAACGAAGGCTTCTGCTC | 20 | 111 |
| Reverse: TTCAACAGCACCTGAATCGGA | 21 | |||
| NOTCH1 | NM_017617.5 | Forward: GGCCACCCCTCCTAGTTTGG | 20 | 148 |
| Reverse: CCTCACTGGCATGACACACAA | 21 | |||
| SHH | NM_000193.4 | Forward: CACCCCCAATTACAACCCCG | 20 | 80 |
| Reverse: TACACCTCTGAGTCATCAGCCT | 22 |
Gene name, accession number, forward primer sequence, length, reverse primer sequence, length, and amplicon length used for q-RT-PCR analysis of P4-regualted transcripts associated with reproduction function that are also predicted targets of P4-regualted miRNAs.
Coordinate changes in mRNAs in endometrial epithelial cells treated with progesterone
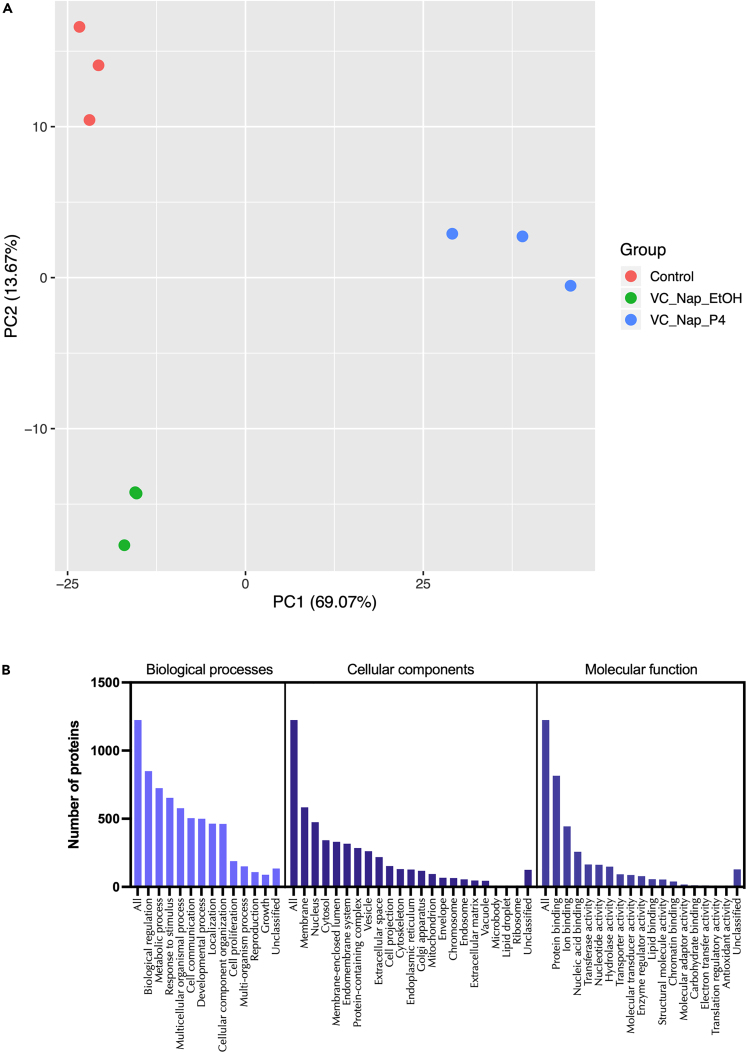
Next, we sought to identify coordinate changes in mRNA expression occurring with the changes in miRNA expression at the highest concentration of P4. Principal Component Analysis (PCA) of RNA sequencing data revealed clear separation between controls and P4 treated cells (Figure 3A). Following data quality control step and a Benjamini–Hochberg FDR correction, differentially expressed protein coding genes (DEGs) were determined. In total, 6,367 DEGs were identified, 1,238 of which had a log2 fold change of greater than 1 (Table S1).
Figure 3.
RNA-Seq analysis of Ishikawa cells treated with P4 in vitro
(A) Principal component analysis depicting the overall transcriptional differences between cells via RNA sequencing of Ishikawa cells treated with (1) Control (orange circle), (2) Vehicle control (green circle), and (3) 10 μg/ml P4 (blue circle) for 24 h. Controls clearly separate on the left-hand side whereas P4 treatment clearly cluster together on the right-hand side of the graph (n = 3 biological replicates per group).
(B) Bar chart of Biological Processes (red), Cellular Component (blue), and Molecular Function (green) associated with P4 modified transcripts.
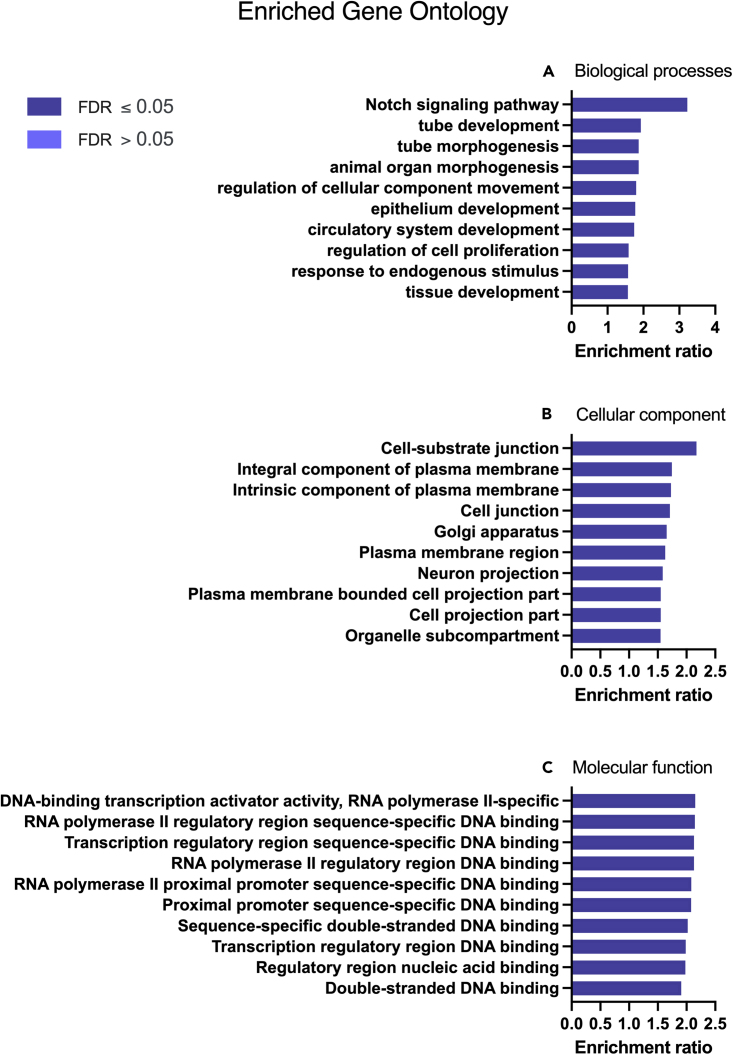
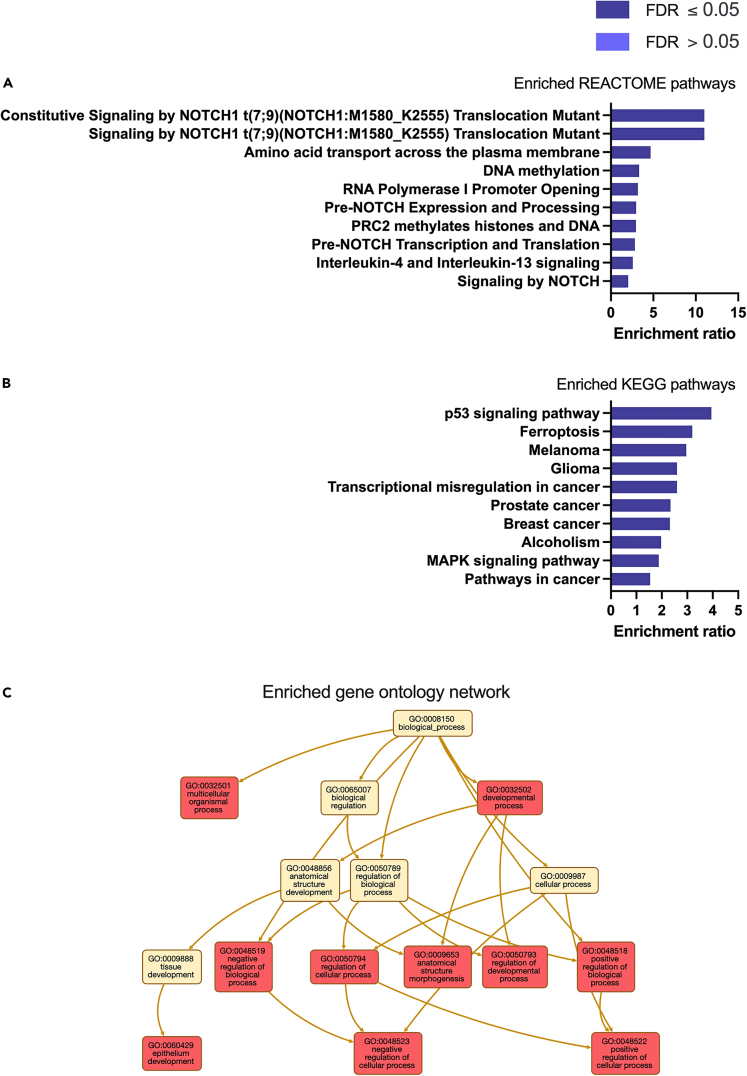
P4 altered the expression of transcripts in the categories of: biological regulation, metabolic processes, localized to the membrane and to the nucleus, and molecular functions of protein, ion, and nucleic acid binding more than was expected by chance (Figure 3B). Biological processes enriched in these P4 treated cells included Notch signaling pathway, tube development, tube morphogenesis, and epithelium development (Figure 4A: Table S2). Cellular components were enriched in ‘cell-substrate junction’, and ‘integral component of plasma membrane’ (Figure 4B: Table S3), whereas molecular functions influenced by P4 treatment included DNA-binding transcription activator activity, RNA polymerase II regulatory region sequence-specific interactions (Figure 4C: Table S4). The signaling pathways that were enriched involved signaling by NOTCH1, amino acid transport and DNA methylation (Figure 5A: Table S5) whereas enriched KEGG pathways were involved in p53 signaling, ferroptosis, melanoma, and glioma (Figure 5B: Table S6). The gene ontology enrichment network used a total of 22,194 genes with 1,089 seeds in the selected network. A total of 844 seeds were identified in the expanded sub-network and they produced an enriched biological process network including those involved in developmental processes, epithelial development, and both positive and negative regulation of cellular processes (Figure 5C).
Figure 4.
Enriched Gene Ontology associated with P4 modified transcripts
(A) Biological processes, (B) Cellular component, and (C) Molecular functions associated with P4 modified transcripts determined following RNA sequencing of human endometrial epithelial cells (Ishikawa cells) treated with 10 μg/ml P4 for 24 h. These are significantly enriched i.e. more transcripts associated with these ontologies than one would expect by chance (FDR <0.05).
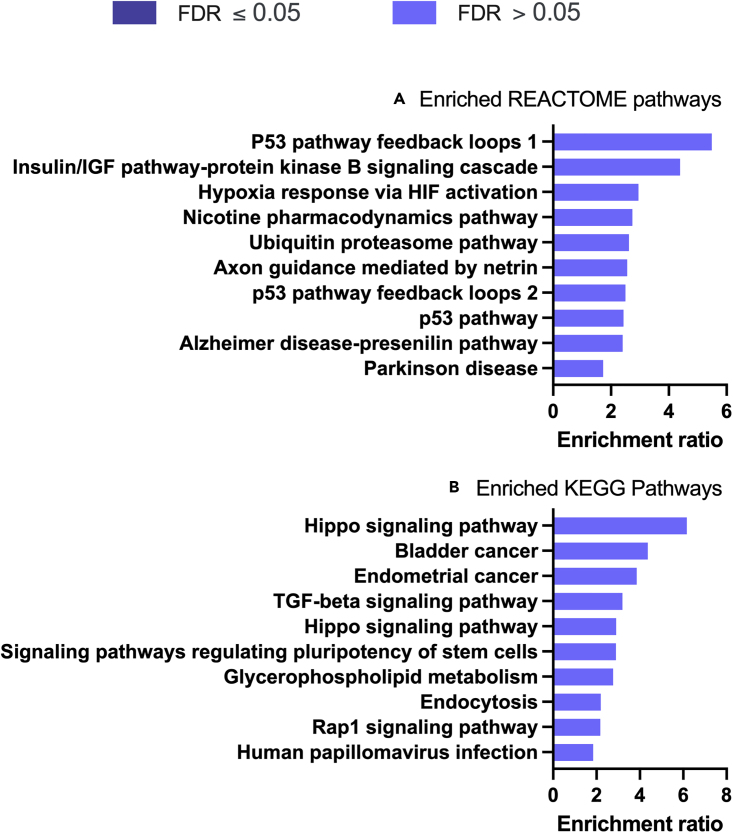
Figure 5.
Enriched pathways and networks associated with P4-modified transcripts
(A) REACTOME pathways, (B) KEGG pathways, and (C) ontology network, associated with P4 modified transcripts determined following RNA sequencing of human endometrial epithelial cells (Ishikawa cells) treated with 10 μg/ml P4 for 24 h. These are significantly enriched i.e. more transcripts associated with these pathways than one would expect by chance (FDR <0.05).
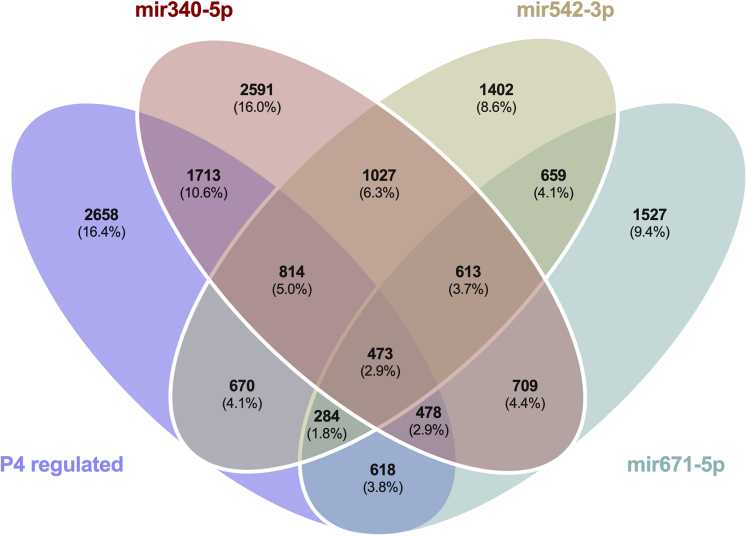
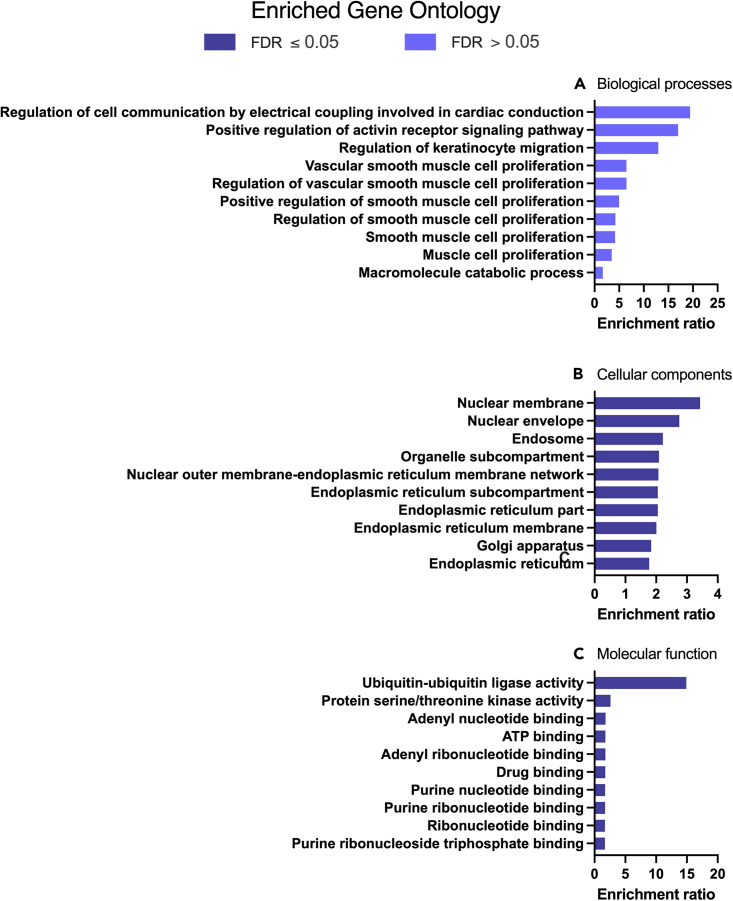
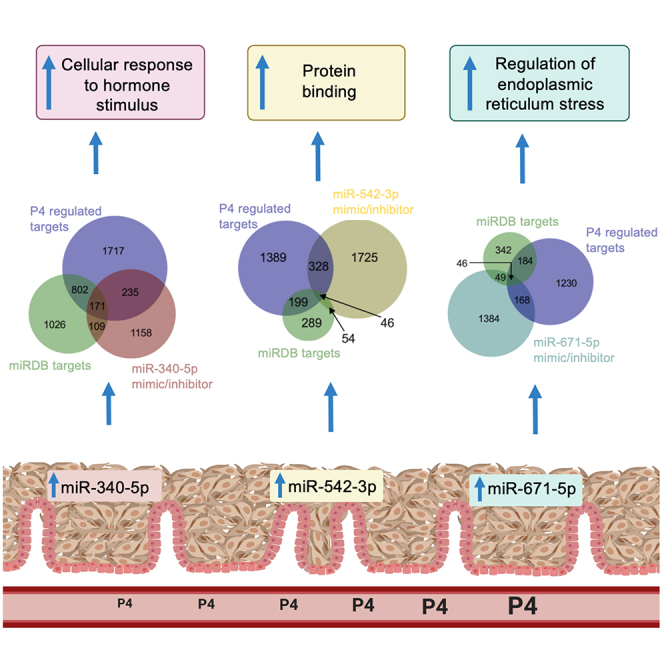
We then investigated the overlap in the P4-dependent expression profiles and the predicted targets from TargetScan of the three P4-regulated miRNAs (Figure 6). Of the predicted targets of miR-340-5p: 1713 transcripts were also changed in P4 treated cells; miR-542-3p: 670 transcripts, and miR-671-5p: 618 transcripts indicating distinct P4-miRNA transcriptomic signatures. Complete details of transcripts are provided in Table S7. A cohort of 473 transcripts that are predicted targets of all three P4-regulated miRNAs exhibit P4-responsive expression in the same cells. No significantly enriched biological processes or KEGG/panther pathways were identified (Figure 7A; Table S8 and Figure 8; Tables S11 and S12). However, significant enrichment for cellular components including those involved in the nuclear membrane, nuclear envelope, and endosome (Figure 7B: Table S9) were observed. Molecular functions of ubiquitin-ubiquitin ligase activity, protein serine/threonine kinase activity, and adenyl nucleotide activity were also significantly enriched (Figure 7C: Table S10).
Figure 6.
Overlap of predicted miRNA targets and P4 altered transcripts
Venn diagram analysis demonstrating the overlap in predicted targets (from miRBase) of the 3 miRNAs modified by treatment of Ishikawa cells with P4 for 24 hin vitro as well as those mRNAs that were identified as altered in the same cells following P4 treatment for 24 h compared to vehicle controls.
Figure 7.
Enriched Gene Ontologies for P4-modified transcripts and miRNA targets
(A) Biological processes, (B) Cellular component, and (C) Molecular functions associated with 473 P4 modified transcripts that are also predicted targets of the 3 miRNAs modified in human endometrial epithelial cells (Ishikawa cells) treated with 10 μg/ml P4 for 24 h. These are significantly enriched i.e. more transcripts associated with these ontologies than one would expect by chance (FDR <0.05).
Figure 8.
Enriched pathways for P4-modified transcripts and miRNA targets
(A) REACTOME pathways, and (B) KEGG pathways associated with 473 P4 modified transcripts that are also predicted targets of the 3 miRNAs modified in human endometrial epithelial cells (Ishikawa cells) treated with 10 μg/ml P4 for 24 h. These are significantly enriched, i.e., more transcripts associated with these ontologies than one would expect by chance when FDR <0.05.
In vitro targets of miR-340-5p in endometrial epithelial cells
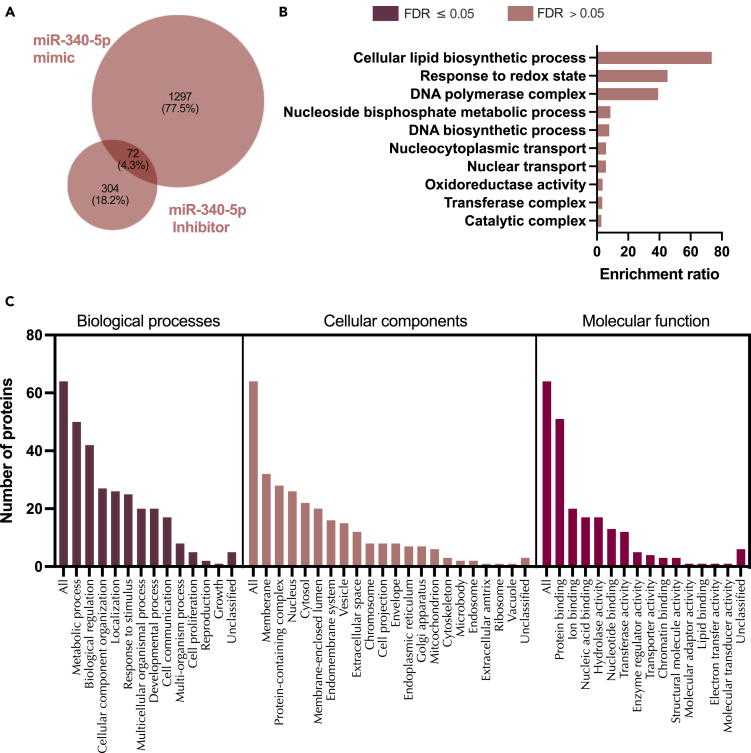
Excluding off target effects, treatment of endometrial epithelial cells with mir-340-5p mimic altered 1,369 proteins as compared to controls (p < 0.05; Figure S1; Table S22). These were significantly enriched in pathways involved in oxidation-reduction function and metabolism pathways (Figure S1). In contrast, inhibition of mir-340-5p changed expression of 376 proteins (p < 0.05; Figure S2; Table S23) including those involved in tRNA and ncRNA processing as well as nuclear transport (Figure S2). A cohort of 72 proteins was altered in endometrial epithelial cells following treatment with both mimic and inhibitor of mir-340-5p (Figure 9; Table S13).
Figure 9.
Proteins altered in Ishikawa cells following treatment with miR-340-5p mimic and inhibitor
(A) Venn diagram depicting total number of significantly differentially expressed proteins (p<0.05) following transfection of Ishikawa cells (n = 3 biological replicates) with miR-340-5p mimic (RHS) and inhibitor (LHS).
(B) Enriched KEGG pathways associated with miR-340-5p mimic and inhibition regulated proteins.
(C) WebGestalt overrepresentation analysis of biological process, cellular component and molecular function categories for identified significantly differentially expressed proteins (p<0.05) in response to miR-340-5p mimic and inhibition (total of 72).
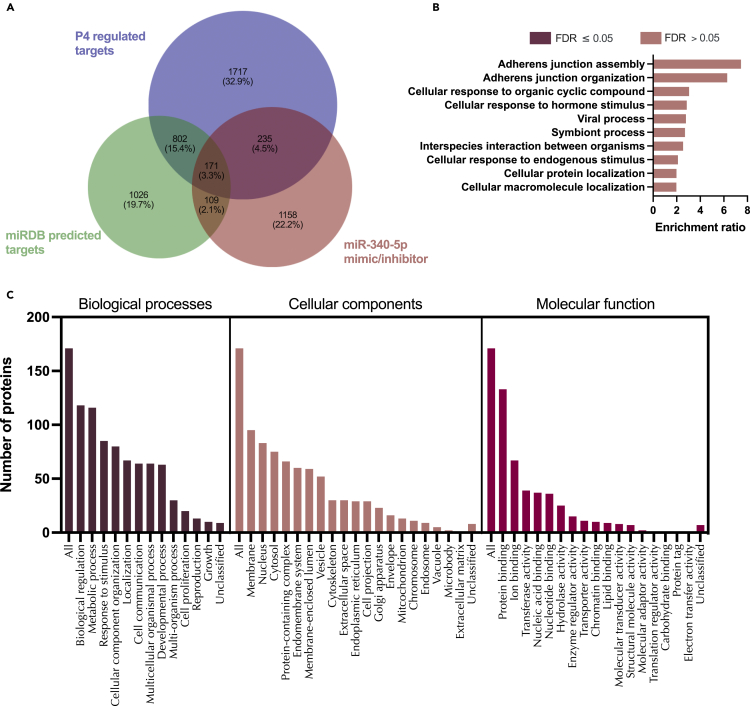
We then sought to determine how many of these in vitro identified targets of mir-340-5p were predicted in mirDB. A total of 280 proteins were identified between predicted (mirDB) and confirmed (in vitro) targets (Figure S3). Only 6.9% of total predicted targets were changed in endometrial cells treated with mir-340-5p mimic whereas 1.3% were altered following inhibition of mir-340-5p (Figure S3: Table S24). These proteins were overrepresented in pathways involving the nuclear pore, mRNA-3′-UTR binding, and cadherin binding (Figure S3). Finally, we compared our in vitro mimic/inhibitor list of proteins, mirDB predicted targets, and those mRNAs altered by P4 in endometrial epithelial cells. In total, 171 proteins predicted to be targets by mirDB were altered in vitro by treatment with miR-340-5p mimic or inhibitor and were also altered by treatment of endometrial epithelial cells with P4. These proteins were involved in biological processes involving developmental processes, cell proliferation, and reproduction amongst others and are likely key components of P4 regulation of the endometrial epithelium (Figure 10, Table S14).
Figure 10.
Proteins altered in Ishikawa cells following treatment with miR-340-5p mimic and inhibitor compared to mirDB predicted targets and P4 regulated mRNAs
(A) Venn diagram depicting total number of significantly differentially expressed proteins (p<0.05) following transfection of Ishikawa cells (n = 3 biological replicates) with miR-340-5p mimic or inhibitor vs non-targeting controls and mirDB predicted targets, or P4-regulated transcripts.
(B) Enriched KEGG pathways associated with miR-340-5p mimic and inhibition regulated proteins, P4-regulated mRNAs and predicted target overlap.
(C) WebGestalt overrepresentation analysis of biological process, cellular component and molecular function categories for identified significantly differentially expressed proteins (p<0.05) in response to miR-340-5p mimic and inhibition, predicted targets, and P4-regulated mRNAs overlap.
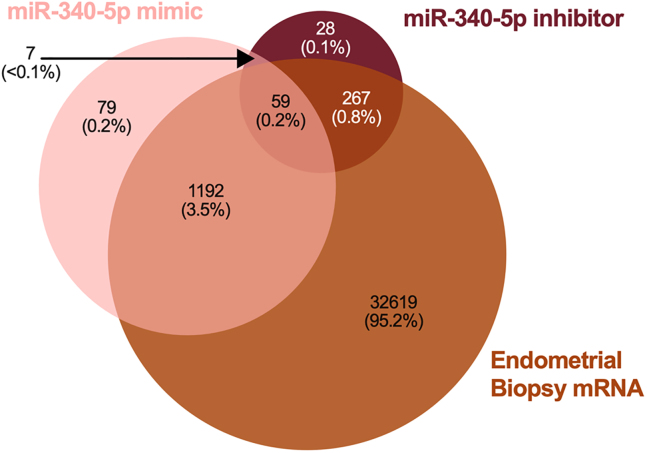
We wished to investigate whether the mRNA counterparts of proteins changed in response to miRNA mimics and inhibitors were present in RNA-Seq data from human endometrial biopsy samples.21 Of the proteins altered by miR-340-5p mimic, 94% of their corresponding mRNAs were detected in patient samples. Furthermore, only 13 of 1251 mRNAs were absent from 1 or more of n = 36 samples. Ninety percent of proteins affected by inhibition of miR-340-5p were identified in RNA-Seq data, with only 5 of 326 mRNAs not present in all of the 36 patient samples. There were 59 of each of these sets of proteins which were changed by both mimic and inhibitor whose mRNA was found in patient samples (Figure 11, Tables S15A–S15D).
Figure 11.
Comparison of miR-340-5p-regulated proteins to endometrial biopsy transcript expression
Venn diagram showing overlap in proteins altered in abundance following transfection of Ishikawa cells (n = 3 biological replicates) with miR-340-5p mimic or inhibitor compared to RNA-Seq data from human endometrial biopsies (n = 36 biological replicates).
In vitro targets of miR-542-3p in endometrial epithelial cells
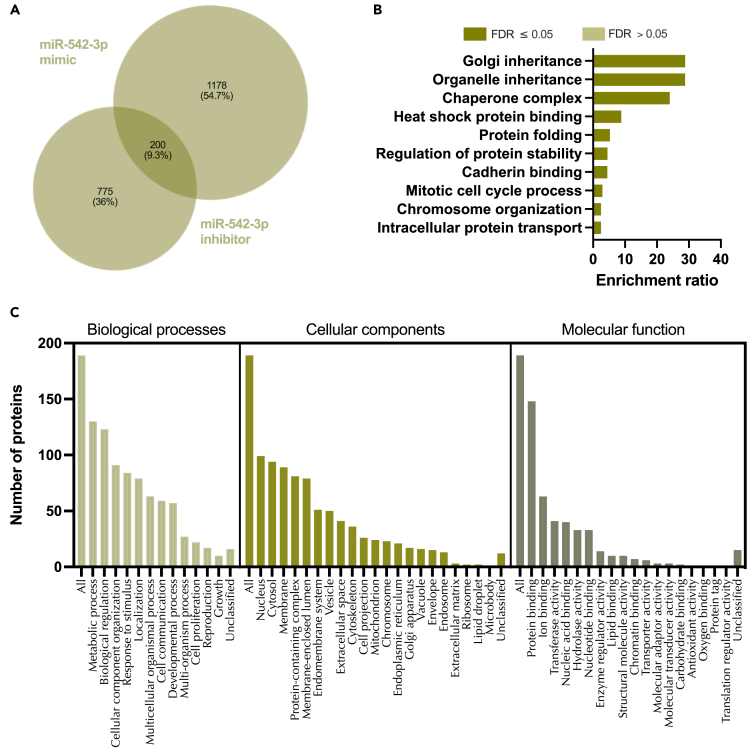
Exclusion of off target effects identified 1,378 proteins altered by treatment of endometrial epithelial cells with miR-542-3p mimic (Figure S4; Table S25). These targets were overrepresented in pathways involved in ribonucleoprotein complex biogenesis and various metabolic processes (Figure S4). Inhibition of miR-542-3p altered 975 proteins including those implicated in pathways related to RNA splicing, mRNA processing, and cellular protein localization (Figure S5, Table S26). A core of 200 proteins was differentially abundant in response to treatment with both miR-542-3p mimic and inhibitor treatment (Figure 12, Table S16). These proteins were enriched pathways involving organelle and Golgi inheritance as well as protein folding (Figure 12).
Figure 12.
Proteins altered in Ishikawa cells following treatment with miR-542-3p mimic and inhibitor
(A) Venn diagram depicting total number of significantly differentially expressed proteins (p<0.05) following transfection of Ishikawa cells (n = 3 biological replicates) with miR-542-3p mimic (RHS) and inhibitor (LHS).
(B) Enriched KEGG pathways associated with miR-542-3p mimic and inhibition regulated proteins.
(C) WebGestalt overrepresentation analysis of biological process, cellular component and molecular function categories for identified significantly differentially expressed proteins (p<0.05) in response to miR-542-3p mimic and inhibition (total of 200).
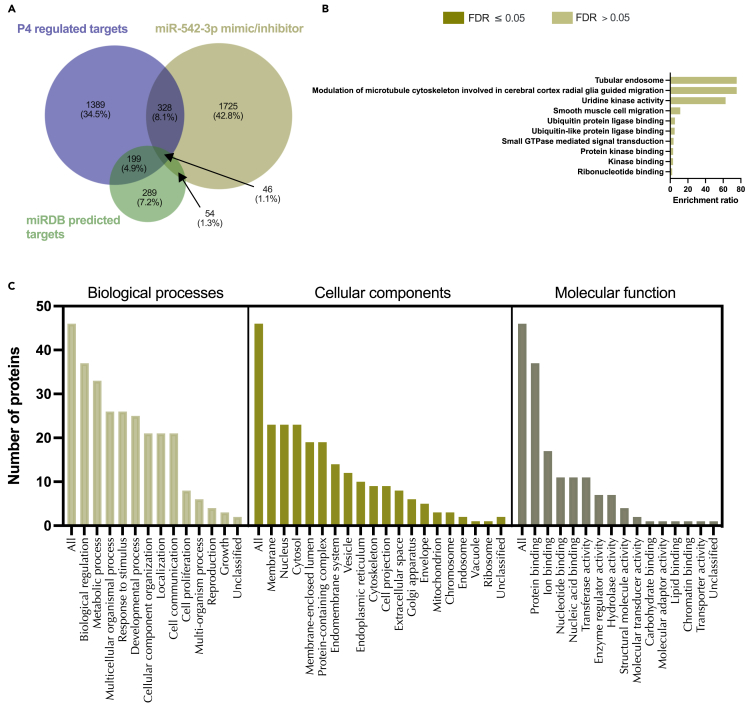
On comparing these in vitro identified targets of miR-542-3p to those predicted by mirDB we identified 100 protein targets in common (Figure S6, Table S27). These were then compared to P4-regulated mRNAs and were assessed for the following criteria: (1) Identified significantly differentially expressed proteins (p<0.05) in response to miR-542-3p mimic and/or inhibitor, (2) mirDB predicted targets of miR-542-3p, and (3) progesterone regulated miRbase targets of miR-542-3p (Figure 13, Table S17). In total, only 46 proteins satisfied all three of our criteria and there are no significantly enriched pathways associated with these 46 proteins; however, they appear to be enriched in thermogenesis (Figure 13).
Figure 13.
Proteins altered in Ishikawa cells following treatment with miR-542-3p mimic and inhibitor compared to mirDB predicted targets and P4 regulated mRNAs
(A) Venn diagram depicting total number of significantly differentially expressed proteins (p<0.05) following transfection of Ishikawa cells (n = 3 biological replicates) with miR-542-3p mimic or inhibitor vs non-targeting controls and mirDB predicted targets, or P4-regulated transcripts.
(B) Enriched KEGG pathways associated with miR-542-3p mimic and inhibition regulated proteins, P4-regulated mRNAs and predicted target overlap.
(C) WebGestalt overrepresentation analysis of biological process, cellular component and molecular function categories for identified significantly differentially expressed proteins (p<0.05) in response to miR-542-3p mimic and inhibition, predicted targets, and P4-regulated mRNAs overlap.
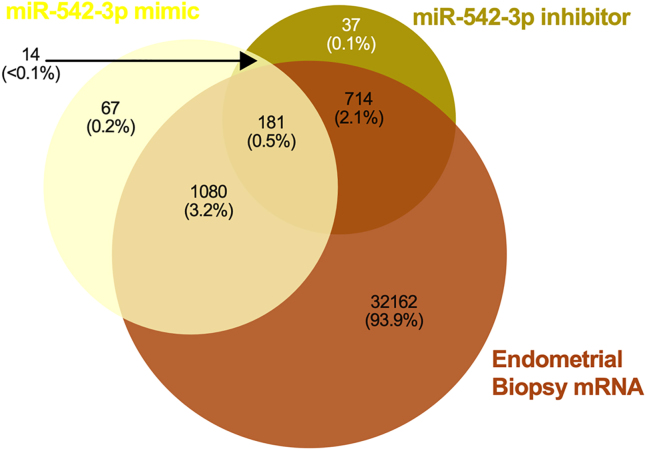
We carried out a comparison of proteomic data to human endometrial biopsies to determine whether proteins altered by miR-542-3p mimic and/or inhibitor were present in clinical samples.21 It was found that 94% of proteins changed in response to miR-542-3p mimic mRNA could be identified in RNA-Seq data. Of these 1,261 proteins, only 11 were absent from 1 or more of n = 36 samples. RNA-Seq data showed presence of mRNA corresponding to 95% of proteins altered in abundance by miR-542-3p inhibitor, with 8 of these 895 mRNAs not recognized in all 36 patients’ endometrial biopsies. A total of 181 proteins common to both mimic and inhibitor altered lists were found in RNA-Seq data (Figure 14, TablesS18A–S18D)
Figure 14.
Comparison of miR-542-3p-regulated proteins to endometrial biopsy transcript expression
Venn diagram showing overlap in proteins altered in abundance following transfection of Ishikawa cells (n = 3 biological replicates) with miR-542-3p mimic or inhibitor compared to RNA-Seq data from human endometrial biopsies (n = 36 biological replicates).
In vitro targets of miR-671-5p in endometrial epithelial cells
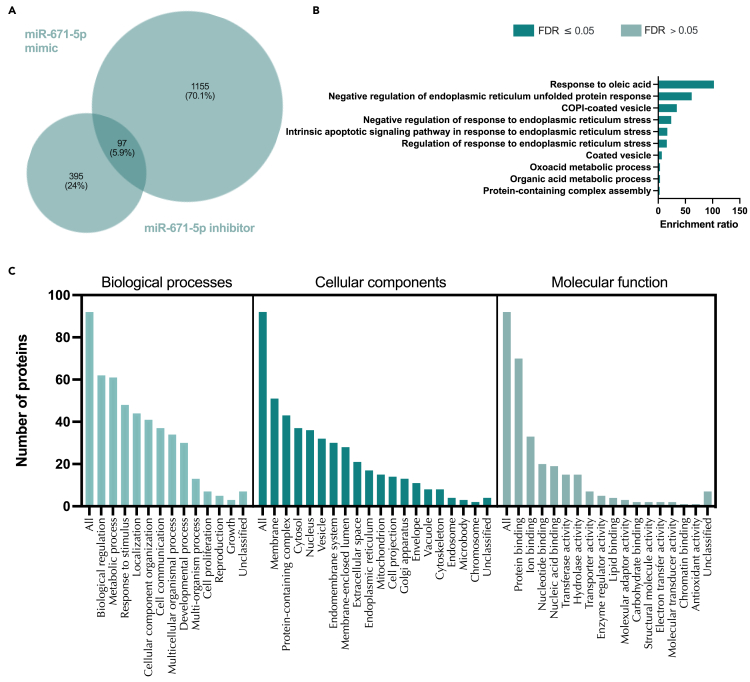
Excluding off target effects, treatment of endometrial epithelial cells with a miR-671-5p mimic, 1,252 proteins were significantly changed (p<0.05) following exclusion of off target effects (Figure S7; Table S13). These proteins were involved in processes involving protein transport and localization (Figure S7). Inhibition of miR-671-5p altered 492 proteins enriched in functions such as rRNA metabolic process, and generation of precursor metabolites and energy (Figure S8; Table S13). The number of proteins that were changed in response to both miR-671-5p mimic and inhibitor was 97, with overrepresented of functions related to response to ER stress (Figure 15; Table S28).
Figure 15.
Proteins altered in Ishikawa cells following treatment with miR-671-5p mimic and inhibitor
(A) Venn diagram depicting total number of significantly differentially expressed proteins (p<0.05) following transfection of Ishikawa cells (n = 3 biological replicates) with miR-671-5p mimic (RHS) and inhibitor (LHS).
(B) Enriched KEGG pathways associated with miR-671-5p mimic and inhibition regulated proteins.
(C) WebGestalt overrepresentation analysis of biological process, cellular component and molecular function categories for identified significantly differentially expressed proteins (p<0.05) in response to miR-671-5p mimic and inhibition.
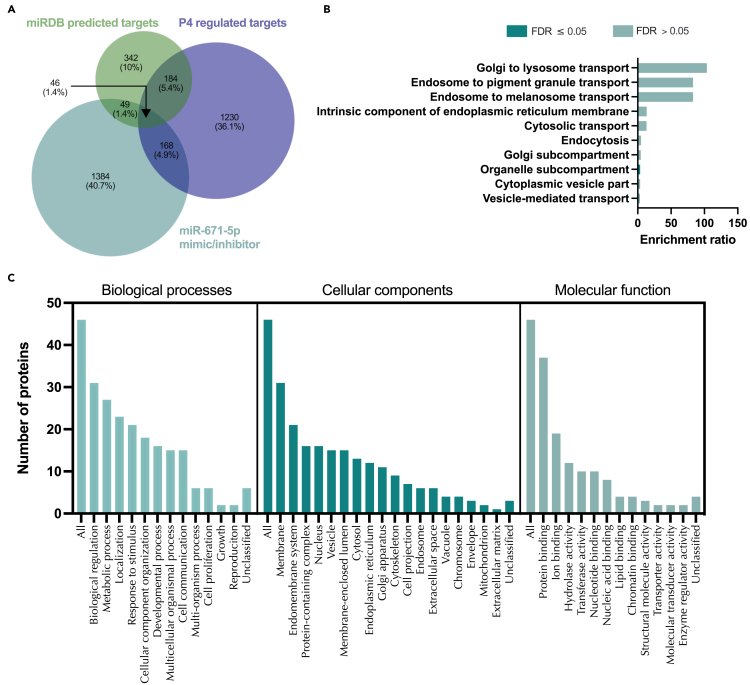
Comparing the set of proteins that were significantly changed following treatment with miR-671-5p mimic or inhibitor to those predicted to be targets of miR-671-5p, identified 95 proteins in common (Figure S9; Table S30). These proteins were significantly overrepresented (FDR <0.05) for processes involving the Golgi apparatus, cytosolic, intra-cellular, and vesicle-mediated transport (Figure S9). Finally, by comparing proteins that fit each of the following 3 criteria: 1) identified significantly differentially expressed proteins (p < 0.05) in response to miR-671-5p mimic and/or inhibitor, 2) mirDB predicted targets of miR-671-5p, and 3) progesterone regulated miRBase targets of miR-671-5p discovers 46 that are common to all (Figure 16, Table S20). The only significantly enriched category is in their cellular component organelle subcompartment (FDR <0.05). Highly represented processes comprise various transport functions including Golgi to lysosome (Figure 16).
Figure 16.
Proteins altered in Ishikawa cells following treatment with miR-671-5p mimic and inhibitor compared to mirDB predicted targets and P4 regulated mRNAs
(A) Venn diagram depicting total number of significantly differentially expressed proteins (p<0.05) following transfection of Ishikawa cells (n = 3 biological replicates) with miR-671-5p mimic or inhibitor vs non-targeting controls and mirDB predicted targets, or P4-regulated transcripts.
(B) Enriched KEGG pathways associated with miR-671-5p mimic and inhibition regulated proteins, P4-regulated mRNAs and predicted target overlap.
(C) WebGestalt overrepresentation analysis of biological process, cellular component and molecular function categories for identified significantly differentially expressed proteins (p<0.05) in response to miR-671-5p mimic and inhibition, predicted targets, and P4-regulated mRNAs overlap.
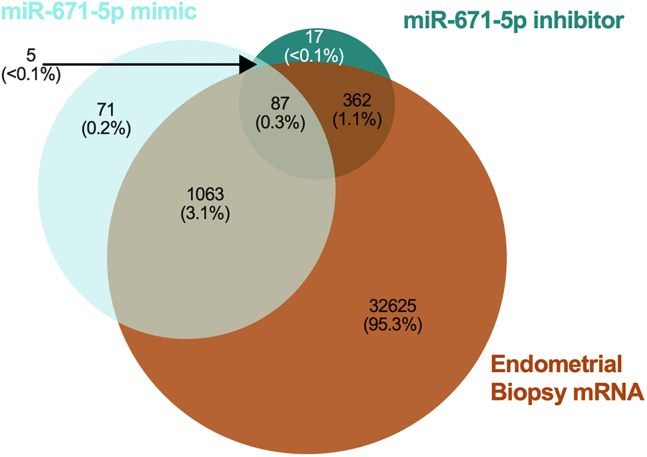
We examined presence of mRNAs in endometrial biopsies21 to find those that correspond to proteins changed in abundance by miR-671-5p mimic and/or inhibitor. This resulted in identification of presence of mRNA for 94% of miR-671-5p mimic affected proteins. Of these 1,150 mRNAs, only 8 were not found in all 36 endometrial samples. Ninety-five percent of proteins changed by the miR-671-5p inhibitor had corresponding mRNAs in patient samples and of those 449, only 8 were absent from 1 or more biopsy. Eighty-seven proteins were altered by both mimic and inhibitor as well as corresponding mRNA detected in endometrial biopsies (Figure 17; Tables S21A–S21D).
Figure 17.
Comparison of miR-671-5p-regulated proteins to endometrial biopsy transcript expression
Venn diagram showing overlap in proteins altered in abundance following transfection of Ishikawa cells (n = 3 biological replicates) with miR-671-5p mimic or inhibitor compared to RNA-Seq data from human endometrial biopsies (n = 36 biological replicates).
Changes to miRNA profiles in patient samples with different clinical outcomes
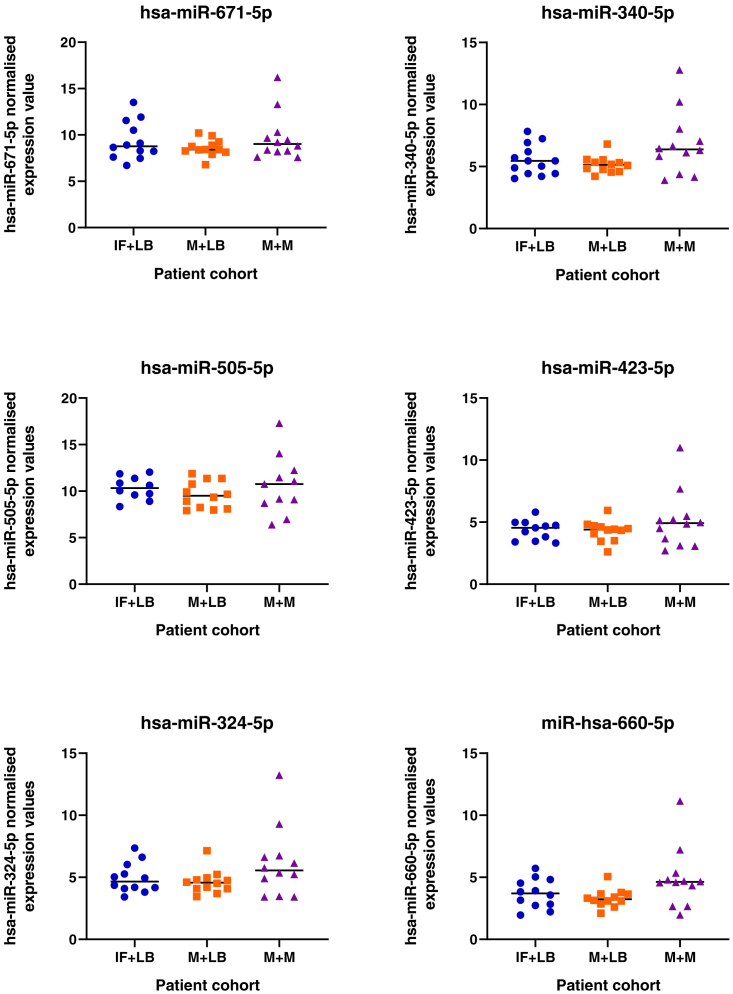
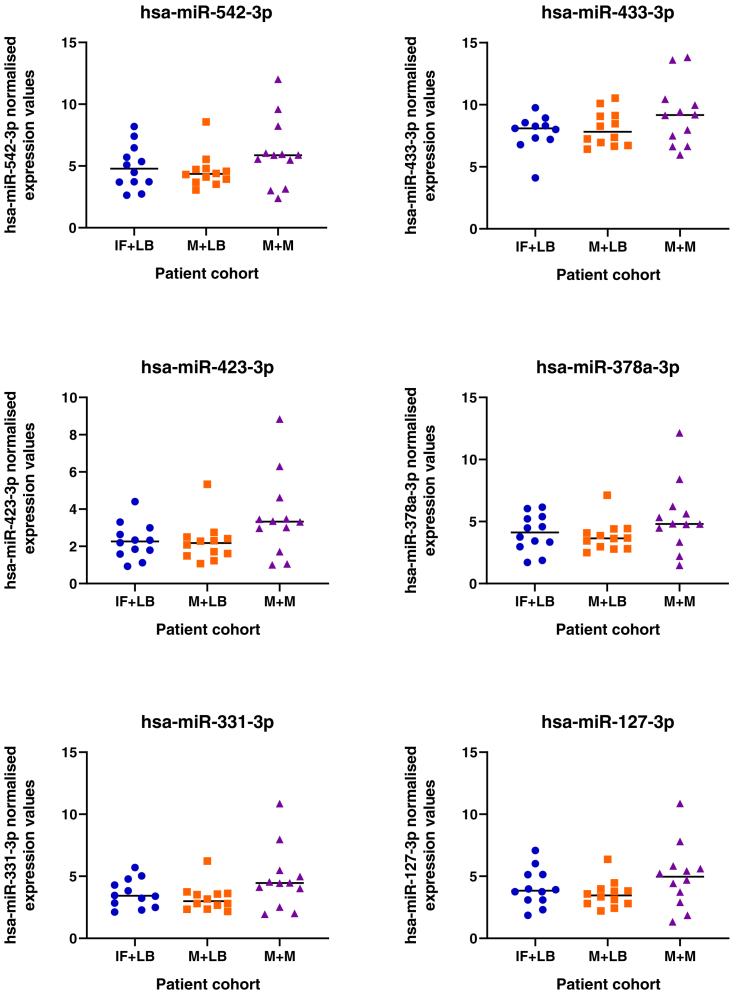
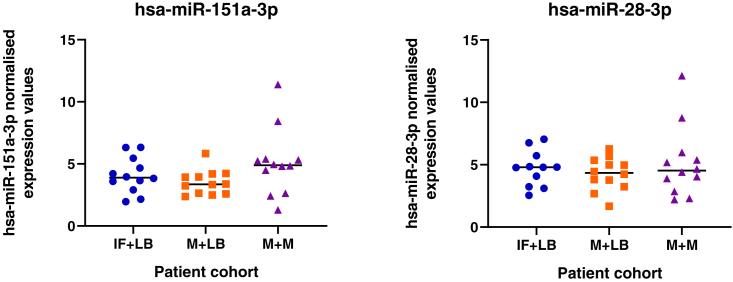
Given the pathways these miRNAs regulate in vitro we sought to determine if their expression is dysregulated in endometrial biopsies from women who experienced pregnancy loss during the luteal phase of the cycle. We detected expression of all miRNAs in endometrial biopsies taken from patients during the luteal phase of their cycle, irrespective of prior or future pregnancy outcomes (Figure 18). No significant difference between patient cohorts was determined for 19 of the 20 mature seeds from the 13 microRNA families. However, expression of mir-340-5p showed an overall increase in expression in patients who had previously suffered a miscarriage and had a subsequent miscarriage, as compared to those who had infertility or previous miscarriage and subsequently went on to have a life birth outcome.
Figure 18.
Expression of miRNAs in endometrial biopsy samples
Samples were collected from patients during the mid-luteal phase of the menstrual cycle (LH+6-LH+9), from patients (n = 12 per group) who have infertility and a subsequent live birth (IF + LB: blue dots), miscarriage and a subsequent live birth (M + LB: orange square), or miscarriage and a subsequent miscarriage (M + M: purple triangles). Following statistical analysis using an ANOVA, there was an overall effect on mir-340-5p expression (p <0.05) between groups.
Discussion
We tested the hypothesis that a set of miRNAs whose evolutionary appearance coincides with the emergence of placental mammals and were retained in all placental mammal lineages thereafter may play a role in uterine function for successful pregnancy in humans.18 The rationale for this is the fact that the endometrium and trophoblast have had to co-evolve regulatory signaling networks and the dysregulation of these networks may contribute to pregnancy loss. P4 signaling during the menstrual cycle is critical to establish receptivity of the endometrium to implantation.22 In contrast with the follicular phase during which the expression of estrogen receptors are high, during the P4-dominant luteal phase, P4 suppresses ESR expression and increases and then modifies PGR expression in a spatial and temporal manner.23 Rising concentrations of P4 results in halted epithelial proliferation and increased stromal cell proliferation, priming the endometrium for implantation and subsequent decidualization.24 Epithelial cells begin to differentiate, and tight junctions and adherens in the luminal epithelium are disrupted.25 An important P4 target, is Indian hedgehog (Ihh), predominantly expressed in the epithelium and is required for endometrial receptivity. It facilitates communication between epithelial and stromal cells, and knockout of its expression in the endometrium results in implantation failure26 as does knockout of its downstream target COUP-TFII. Heart And Neural Crest Derivatives Expressed 2 (Hand2, another P4 regulated transcription factor) is also vital for endometrial receptivity, suppressing the E2-regulated epithelial cell proliferation. Without Hand2-mediated suppression of growth factors, epithelial cells continue to proliferate, leading to implantation failure.27 An immunophilin FKBP52 functions as a co-chaperone for the P4 receptor and absence of this causes the endometrium to be non-receptive.28 Collectively, these data demonstrated the importance of P4-regualted changes to the endometrium to establish receptivity to implantation.
We have determined that miR-340-5p, miR-542-3p, and miR-671-5p were modified by exposure of endometrial epithelial cells to progesterone in vitro. These progesterone-regulated miRNAs changed coordinate with alterations to selected mRNAs, a specific cohort of which they are predicted to target. Following treatment of endometrial epithelial cells with mimics or inhibitors for these three miRNAs we have determined the likely pathways that are P4 regulated by these miRNAs. We have also determined that the overwhelming majority of the proteins altered by these miRNAs are expressed in human endometrial biopsy samples.21 Moreover, we have demonstrated that there is a specific miRNA signature in the endometrium of a subset of women that is associated with miscarriage compared to those with a previous pregnancy loss but a subsequent successful pregnancy outcome. Collectively, these data suggest these P4 modified and mammal-specific miRNAs, and the pathways in which their predicted targets function, are important for successful pregnancy and their dysregulation is associated with pregnancy outcome in a subset of women.
Recent studies have highlighted the transcriptional changes that occur in the endometrium during different stages of the menstrual cycle, but data on miRNA regulation of these processes – specifically in the endometrial epithelium - are more limited. miRNA profiling of whole endometrial biopsies (a heterogeneous mix of cell types) from the proliferative and mid-luteal phase of the menstrual cycle has revealed menstrual stage-specific miRNA signatures.29 This suggests the regulation of some of these 49 miRNAs is under the control of the steroid hormone environment at sampling. Similarly, our study has shown that P4 regulates expression of 3 of the 13 stem lineage miRNA gene families in endometrial epithelial cells suggesting these may contribute to regulation of the transcriptional response of the endometrium to changes in steroid hormones during the menstrual cycle.
The overall effect of P4 exposure to these epithelial cells altered transcripts involved in the processes of epithelium development and cellular proliferation, as well as functions involved in transcriptional regulation. Previous studies29 found that predicted targets of miRNAs upregulated in mid-secretory phase endometrium were found to target cell-cycle regulators, possibly playing a role in cell turnover during the luteal phase. Analysis of the endometrium from women during the receptive stage of the menstrual cycle found ∼21% of differentially expressed genes (653 out of 3,052) directly bind to or are regulated by the progesterone receptor.30 The functions related to these transcripts involve regulation of inflammatory response, estrogen response, cell death and interleukin/STAT signaling – some of which we identified as altered in expression following miRNA mimic/inhibitor treatment in our in vitro study. For example, addition of mimics for our three miRNAs of interest to Ishikawa cells changes expression of proteins involved in cellular proliferation (Figure 14).
Of the stem lineage miRNAs that were investigated we identified 3 that are modified by P4 in vitro in human endometrial epithelial cells, i.e., miR-340-5p, miR-542-3p, and miR-671-5p. We also showed via RNA sequencing that there were coordinate changes to some of the predicted targets of these miRNAs. P4 plays a critical role in endometrial function in a variety of placental mammals. Indeed, exposure of the endometrial epithelial and glandular cells to the sustained action of P4 is required to downregulate its own receptor to facilitate receptivity to implantation – a mechanism that is conserved in all extant placental mammals studied to date.31 Analysis of the 473 mRNAs that were predicted targets of all three P4 modified miRNAs and were altered by P4 themselves (Figure 6) were involved in biological processes including cellular proliferation. In our in vitro treatment of endometrial epithelial cells with mimics/inhibitors of these P4 modified miRNAs, proteins involved in the biological processes of cell proliferation were also regulated by these miRNAs (Figures 9, 12, and 15 panel C). Collectively, we propose that P4 acting on endometrial epithelial cells modifies expression of miR-340-5p, miR-542-3p, and miR-671-5p. This regulates expression of their targets that are involved in cellular proliferation to modify the endometrial epithelium to establish receptivity to implantation.
In addition to roles in cellular proliferation, these P4-regulated targets were also overrepresented in the molecular functions of ubiquitin-ubiquitin ligase activity and protein serine/threonine kinase activity. These are cellular processes (cell communication, proliferation, differentiation:32) that are also regulated by hCG treatment of human endometrial epithelial cells, as well as adhesion of these cells to molecules that make up the extracellular matrix.33 We have demonstrated these three miRNAs target proteins that function in adherens, and their assembly (miR-340), inter- and extracellular transport ((miR-340, miR-542, miR-671). This may indicate that P4 primes the endometrium for the further actions of hCG on the epithelium to facilitate secretion and transport of components of uterine luminal fluid and/or implantation – two key functions of the endometrial epithelium.
miRNAs have been shown previously to play a role in both successful pregnancy and pathophysiology. For example, miR-675 expression increases during placental development and inhibition of the expression of miR-675 negatively regulates cell proliferation in a mouse model of overgrowth.34 Several miRNAs play a role in trophoblast invasion (mir-16, -29b,−34a, −155, −210:35), are serum biomarkers of preeclampsia,36 and have been associated with recurrent pregnancy loss (mir-133a:37). Of the 13 stem lineage miRNA families investigated in this study, 5 (mir-154, mir-185, mir-188, mir-423, mir-542) have previously been implicated as differentially expressed in the placenta from preeclamptic pregnancies.38,39,40 Loss of mir-542 has also been implicated in endometrial stromal cell function by enhancing IGFBP1 expression in decidualisation,41 but this is the first report to examine expression in an epithelial model. We provide evidence of cell compartment-specific regulation of miRNA expression and their predicted targets – in keeping with the known physiological regulation of the endometrium [1]. To further substantiate this hypothesis, we used mimics and inhibitors to determine in vitro endometrial epithelial cell-specific targets of these P4-regulated miRNAs. The in vitro targets of these three miRNAs involved processes of transport and cellular localization – key components of endometrial epithelial cell function. P4 stimulates the secretion of components of uterine luminal fluid (ULF) that supports growth and development of the conceptus before formation of the placenta. Although there are limitations to the use of Ishikawa cells they do display some attributes of a receptive endometrial epithelial layer. As this study investigates the stage during which initial attachment takes place, endometrial epithelial cells were used as opposed to endometrial stromal cells as the innermost layer (and site of attachment) of the endometrium is composed of epithelial cells.42 More recently there have been developments in organoid models of endometrial glands and assembloids.43,44,45 Our approach, though a cellular monolayer of epithelium has allowed us to look at the potential mechanism by which these miRNAs may contribute to endometrial function in vitro and for the first time allow the comparison of computationally predicted targets with confirmed miRNA targets in this model. We propose that P4-regulated modification of these miRNAs and their targets function to provide components of the ULF that supports early pregnancy. Dysregulation of this process could contribute to RPL. In addition to identifying in vitro targets of these mimics, we compared these to computationally predicted targets of these miRNAs and found very limited overlap (Figures 10, 13, and 16). This is not surprising given the high false discovery rate known for target prediction software, of which mirDB is one of the better options.46
We identified a difference in one of these P4-regulated stem-lineage miRNAs in endometrial biopsies from women with different pregnancy outcomes. These biopsies were selected at luteal phase as this study aims to investigate endometrial receptivity, luteal phase being the stage at which endometrium is receptive to implantation of an embryo. This is the only physiologically relevant stage for implantation and is when progesterone is at its peak Luteal phase biopsies were selected as we hypothesized that these miRNAs and their targets play a role in receptivity to implantation. Endometrial receptivity is established in the luteal phase and is the physiologically relevant stage to test expression of these candidate miRNAs s.47 These samples are a heterogeneous combination of cells from endometrial biopsies. miRNAs have been associated with stromal cell decidualization in the human endometrium7 and miRNA profiling of decidua from week 8 (elective termination) compared to those menstrual endometria - found 9 miRNAs to be differentially expressed including one stem lineage miRNA; mir-423.48 This link between miRNAs and uterine receptivity was also established in a study of IVF patients who experienced recurrent implantation failure (RIF)49 with dysregulation in the expression of a set of 13 miRNAs (not investigated in our study) in endometria of RIF compared to fertile women. Moreover, the protein targets of the 3 P4-modified miRNAs (miR-340, miR-542, and miR-671) were enriched for cell adhesion, cell cycle/proliferation and inter- and-extracellular transport, all functions known to be implicated in embryo development/implantation.50 This further supports the important link between miRNAs and endometrial function. miRNA levels in blood plasma from women suffering from recurrent miscarriage to women who were not pregnant also differ including one of the stem linage miRNAs (miR-127-3p).51 Although miR-127-3p is not altered by P4 in our study, there are many factors that could contribute to its modification in circulation, and it may be indicative of dysregulation of the endometrium at a different stage of pregnancy.51 In our study we have identified a subpopulation of endometrial samples that had aberrant miRNA expression profiles and this may identify a distinct cellular phenotype of endometrial from women who may be at risk of RM in the future. These could potentially provide targets for intervention in future.
In conclusion, we propose that 3 mammal-specific and evolutionarily conserved microRNA genes miR-340-5p, miR-542-3p, and miR-671-5p, with some of their protein targets are regulated by progesterone – the key hormone of pregnancy in the endometrial epithelium in humans. Moreover, the regulation of these miRNAs and their protein targets regulate the function of transport and secretion, and adhesion of the endometrial epithelia required for successful implantation in humans. Dysfunction of these miRNAs (and therefore the targets they regulate) may contribute to endometrial-derived recurrent pregnancy loss in women.
Limitations of the study
This study uses Ishikawa cells for endometrial epithelial cell treatment with progesterone to be subsequently analyzed by RNA-Seq and Proteomics. Ishikawa cells are a cancer cell line and so may not completely recapitulate the healthy endometrium. Notwithstanding, Ishikawa cells express functional P4 receptors and respond to signaling cues. Comparison of proteomic results to endometrial biopsy RNA-Seq data to confirm that miRNA targets were present in endometrial tissue sought to increase confidence in the results obtained using the cell line.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Fetal Bovine Serum | Thermo Fisher Scientific (Life Technologies) | 10500064 |
| GSP 100X | GIBCO | 10378016 |
| DMEM low glucose +pyruvate +phenol red | Thermo Fisher Scientific (Life Technologies) | 31885049 |
| Ham’s F12 | Thermo Fisher Scientific (Life Technologies) | 21765037 |
| PBS | Sigma Aldrich | D8537-500ML |
| Lipofectamine 2000 Transfection Reagent | Invitrogen | 12313563 |
| Progesterone | Sigma Aldrich | P8783-5G |
| miRCURY Custom PCR Panel | Qiagen | 339332 |
| miRIDIAN microRNA Hairpin Inhibitor Negative Control #1, 5 nmol | Horizon Discovery | IN-001005-01-05 |
| miRIDIAN microRNA Mimic Negative Control #1, 5 nmol | Horizon Discovery | CN-001000-01- 05 |
| miRIDIAN microRNA hsa-miR340-5p mimic, 2 nmol | Horizon Discovery | C-301081-01- 0002 |
| miRIDIAN microRNA hsa-miR671-5p mimic, 2 nmol | Horizon Discovery | C-301000-03- 0002 |
| miRIDIAN microRNA hsa-miR542-3p mimic, 2 nmol | Horizon Discovery | C-300866-03- 0002 |
| miRIDIAN microRNA hsa-miR340-5p hairpin inhibitor, 2 nmol | Horizon Discovery | IH-301081-02- 0002 |
| miRIDIAN microRNA hsa-miR542-3p hairpin inhibitor, 2 nmol | Horizon Discovery | IH-300866-05- 0002 |
| miRIDIAN microRNA hsa-miR671-5p hairpin inhibitor, 2 nmol | Horizon Discovery | IH-301000-04- 0002 |
| miRCURY LNA™ RT Kit | Qiagen | 339340 |
| miRCURY LNA™ SYBR Green PCR Kit (4000) | Qiagen | 339347 |
| Qiazol® Lysis Reagent | Qiagen | 79306 |
| miRNeasy Mini Kit (50) | Qiagen | 217004 |
| RNase-Free DNase Set (50) | Qiagen | 79254 |
| High Capacity c DNA Rt Kit | Applied Biosystems | 4368814 |
| Deposited data | ||
| Proteomic data | ProteomeXchange | PXD036109 |
| RNASeq data | GEO | GSE211151 |
| Experimental models: Cell lines | ||
| Ishikawa Cells | Cell culture collections | ECACC # 99040201 |
| Oligonucleotides | ||
| Designed primers | IDT | |
| Software and algorithms | ||
| Graphpad Prism | ||
| Webgestalt | www.webgestalt.org | |
Resource availability
Lead contact
Lead contact and further information requests should be directed to and will be fulfilled by the lead contact, Niamh Forde (n.forde@leeds.ac.uk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
All human biopsies were from female patients. The ages of individuals were in the Infertile + live birth group ranged between 25-39 (specifically: 31, 38, 25, 38, 32, 31, 34, 37, 36, 27, 39, 27), for the miscarriage + live birth group ranging between 28-39 (specifically: 34, 28, 30, 33, 36, 31, 27, 34, 38, 32, 39, 36), and in the miscarriage + miscarriage ranging between (specifically: 35, 33, 25, 31, 36, 35, 31, 31, 30, 34, 28, 36). The study was approved by the NHS Research Ethics Committee, Hammersmith and Queen Charlotte’s & Chelsea Research Ethics Committee (1997/5065), and Tommy’s National Reproductive Health Biobank (REC reference: 18/WA/0356). Human endometrial samples were obtained with written informed consent and in accordance with The Declaration of Helsinki (2000) guidelines. Biopsies were obtained from women attending the Implantation Clinic, a dedicated research clinic at University Hospitals Coventry and Warwickshire (UHCW) National Health Service Trust.
Cell lines
Ishikawa cells (adenocarcinoma endometrial epithelial cells) (ECACC # 99040201).
Method details
Unless otherwise stated all consumables were sourced from Sigma Aldrich (UK).
Cell culture
Human immortalised endometrial epithelial cells (Ishikawa cells - ECACC: #99040201) were cultured (50:50 DMEM/F12, 0.5% GSP, 10% charcoal-stripped FBS) at 37 °C and in 5% CO2. Cells were cultured to 70% confluence and seeded into 6-well plates. The cultures were treated with either 1) basal media (as above), 2) vehicle control (70% ethanol, EtOH) or 3), 0.1 mg/ml P4, 4) 1.0 mg/ml P4, or 5) 1.0 mg/ml P4, for 24 hours. Following treatment, cells were harvested by trypsinization and lysed in QIAzol Lysis Reagent. RNA was extracted using the miRNeasy Mini RNA Extraction kit (Qiagen, UK) as per manufacturer’s instructions.
miRNA expression analysis and target prediction
Reverse transcription was carried out using the miRCURY LNA™1 RT Kit (Qiagen) using 10 ng of RNA according to manufacturer’s instructions, alongside reverse transcriptase negative controls. Expression analysis was performed using the miRCURY LNA™ SYBR Green PCR Kit 4000 (Qiagen) and a custom designed plate including miRNAs of interest and controls for normalisation (5S rRNA, U6 snRNA, UniSP3, and UniSP6). Thermal cycling and detection were performed on a ROCHE Light Cycler® 480 II using the following parameters: 2minat 90 °C, followed by 45 cycles of 10secsat 95°C & 60secsat 56°C, and melting curve analysis for 60minat 95 °C. Cq values were determined and normalised to the geometric mean of normalisers. Data were analysed using ANOVA in Graphpad Prism to determine differences (determined as significant when p <0.05).
Predicted targets of miRNAs with P4-dependent expression were identified using TargetScan 7.2,52 with output filtered based on the level of complementary binding between the miRNA seed region and target site: targets with 7mer or 8mer complementary binding to the seed region were selected, since the high number of consecutive matches is indicative of an efficient binding site.53 Ensembl transcript IDs provided by the TargetScan output were then mapped to gene names using Ensembl BioMart (release 98).54
q-RT-PCR of predicted miRNA targets
Reverse transcription from 10 μg of RNA was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) as per the manufacturer’s protocol. Primers were designed for selected transcripts using Primer-BLAST (Table 1) and 20 μM stock dilutions were prepared. PCR reactions were prepared in 20 μl volumes, containing 5 μl Roche SYBR Green Master Mix, 0.5 μl of each forward and reverse primer and 2 μl cDNA. Each sample was analysed in duplicate using the Lightcycler 480 II (Roche) as follows: 95 °C for 5 mins, 45 cycles of 95 °C for 10 secs, then 60 °C for 10 secs and finally 72 °C for secs, followed by melt curve analysis. Data were analysed using the comparative CT method (2-ΔΔCt)55 normalised to the geometric mean of ACTB, GAPDH and PPIA. Significant differences were determined using ANOVA analysis in Graphpad Prism software, including Dunnett’s multiple comparisons test, to identify effects of treatment between each treatment concentration and the vehicle control. Statistical significance was assumed when p < 0.05 after multiple testing correction.
RNA sequencing analysis
Total RNA (100 ng) was used for library preparation using the Illumina TruSeq® Stranded Total kit, according to the manufacturer’s guidelines. The library quality, size range, and sequencing adaptor dimer contamination was assessed with an Agilent TapeStation 2200 using the DNA broad range kit. Excess sequencing adaptor dimer was removed by AxyPrep Mag PCR clean up Kit if present. Final libraries were quantified by the Qubit dsDNA assay kit and Qubit fluorometer (Life Technologies) before creating an equimolar pool of the libraries. The libraries were sequenced by the Next Generation Sequencing Facility at the University of Leeds, using the NextSeq 500 (Illumina, California, USA) to generate single end 75 bp reads. Reads were trimmed by Cutadapt56 and filtered using fastq_quality_filter as part of FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) with parameters including “-q 20” and “-p 90”. The reference genome and gene annotation files of Homo sapiens were retrieved from GENCODE (release 31) (https://www.gencodegenes.org/).57 Read mapping was performed by means of Subread aligners in Rsubread package58 with only uniquely mapped reads reported in the final alignment and read summarisation. Principal Component Analysis (PCA) plot was carried out using all protein-coding genes and long noncoding RNAs with RPKM value ≥1 in at least one sample. Subsequently, log2(RPKM+1) transformation and quantile normalization were applied.
Quantification was carried out by featureCounts function in R. Significant differences in gene expression were identified using DESeq259 with the following cut-offs: log2FoldChange >1 (or <−1) and an FDR-adjusted p value of <0.05. After this analysis, only protein-coding genes were retained based on the gene biotype labels. Overrepresentation enrichment analysis of DEGs were identified using WebGestalt (http://www.webgestalt.org/)60 for gene ontology (GO) terms and pathways, with a significance enrichment level set at an FDR <0.05.
miRNA mimic/inhibitor experiments
Ishikawa cells were plated (∼200,000 cells/well) 24 hours prior to treatment. At the time of treatment, cells were washed 3 times in PBS and 1.6 ml antibiotic & serum-free media added per well. The following treatments (n = 3 biological replicates) were added 1) control medium, 2) Transfection reagent (Lipofectamine 2000), 3) miRNA mimic of interest (80 pmol/well), 4) miRNA inhibitor of interest, 5) miRNA non-targeting mimic, 6) miRNA non-targeting inhibitor. Cells were incubated for 48 hours before media was removed, washed with PBS, and protein extracted using a RIPA buffer with protease inhibitors by passing the cells through a 21′G needle and syringe.
Proteomic analysis
Fifty μg of protein from each sample, described above, was digested with trypsin (1.25 μg trypsin; 37°C, overnight), labelled with Tandem Mass Tag (TMT) eleven plex reagents according to the manufacturer’s protocol (Thermo Fisher Scientific, Loughborough, LE11 5RG, UK) and the labelled samples pooled. One hundred μg of the sample pool was desalted using a SepPak cartridge according to the manufacturer’s instructions (Waters, Milford, Massachusetts, USA). Eluate from the SepPak cartridge was evaporated to dryness and resuspended in buffer A (20 mM ammonium hydroxide, pH 10) prior to fractionation by high pH reversed-phase chromatography using an Ultimate 3000 liquid chromatography system (Thermo Fisher Scientific). In brief, the sample was loaded onto an XBridge BEH C18 Column (130Å, 3.5 μm, 2.1 mm × 150 mm, Waters, UK) in buffer A and peptides eluted with an increasing gradient of buffer B (20 mM Ammonium Hydroxide in acetonitrile, pH 10) from 0–95% over 60 mins. The resulting fractions (15 in total) were evaporated to dryness and resuspended in 1% formic acid prior to analysis by nano-LC MSMS using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). In brief, peptides in 1% (vol/vol) formic acid were injected onto an Acclaim PepMap C18 nano-trap column (Thermo Scientific). After washing with 0.5% (vol/vol) acetonitrile, and 0.1% (vol/vol) formic acid, peptides were resolved on a 250 mm × 75 μm Acclaim PepMap C18 reverse phase analytical column (Thermo Scientific) over a 150 mins organic gradient, using 7 gradient segments (1–6% solvent B over 1 mins, 6–15% B over 58 mins, 15–32% B over 58 mins, 32–40% B over 5 mins, 40–90% B over 1 mins, held at 90% B for 6 mins and then reduced to 1% B over 1 mins) with a flow rate of 300 nl min−1. Solvent A was 0.1% formic acid and Solvent B was aqueous 80% acetonitrile in 0.1% formic acid. Peptides were ionized by nano-electrospray ionization at 2.0 kV using a stainless-steel emitter with an internal diameter of 30 μm (Thermo Scientific) and a capillary temperature of 300 °C. All spectra were acquired using an Orbitrap Fusion Lumos mass spectrometer controlled by Xcalibur 3.0 software (Thermo Scientific) and operated in data-dependent acquisition mode using an SPS-MS3 workflow. FTMS1 spectra were collected at a resolution of 120000, with an automatic gain control (AGC) target of 200000 and a max injection time of 50 ms. Precursors were filtered with an intensity threshold of 5000, according to charge state (to include charge states 2–7) and with monoisotopic peak determination set to Peptide. Previously interrogated precursors were excluded using a dynamic window (60 secs +/−10 ppm). The MS2 precursors were isolated with a quadrupole isolation window of 0.7. m/z. ITMS2 spectra were collected with an AGC target of 10,000, max injection time of 70 ms and CID collision energy of 35%. For FTMS3 analysis, the Orbitrap was operated at 50,000 resolution with an AGC target of 50,000 and a max injection time of 105 ms. Precursors were fragmented by high energy collision dissociation (HCD) at a normalised collision energy of 60% to ensure maximal TMT reporter ion yield. Synchronous Precursor Selection (SPS) was enabled to include up to 10 MS2 fragment ions in the FTMS3 scan.
The raw data files were processed and quantified using Proteome Discoverer software v2.1 (Thermo Scientific) and searched against the UniProt Human database (downloaded January 2021: 169,297 entries) using the SEQUEST HT algorithm. Peptide precursor mass tolerance was set at 10 ppm, and MS/MS tolerance was set at 0.6 Da. Search criteria included oxidation of methionine (+15.995 Da), acetylation of the protein N-terminus (+42.011 Da) and Methionine loss plus acetylation of the protein N-terminus (−89.03 Da) as variable modifications and carbamidomethylation of cysteine (+57.021 Da) and the addition of the TMT mass tag (+229.163 Da) to peptide N-termini and lysine as fixed modifications. Searches were performed with full tryptic digestion and a maximum of 2 missed cleavages were allowed. The reverse database search option was enabled and all data was filtered to satisfy false discovery rate (FDR) of 5%. Protein groupings were determined by PD2.1, with minor modifications to allow inference of biological trends without any loss in the quality of identification or quantification. The MS data were searched against the human Uniprot database retrieved on 2021-01-14 and updated with additional annotation information on 2021-11-15. The protein abundances were normalised within each sample to total peptide amount, then Log2 transformed to bring them closer to a normaldistribution. Statistical significance was then determined using paired t-tests between the conditions of interest with FDR corrected pvalues obtained using the Benjamini-Hochberg method.
Analysis of miRNAs in clinical samples
Human endometrial samples were obtained with written informed consent and in accordance with The Declaration of Helsinki (2000) guidelines. Biopsies were obtained from women attending the Implantation Clinic, a dedicated research clinic at University Hospitals Coventry and Warwickshire (UHCW) National Health Service Trust. Samples were timed relative to the pre-ovulatory LH surge, and obtained using a Wallach Endocell™ endometrial sampler. Following luteal phase endometrial biopsy, pieces of tissue (∼2 mm) were preserved in RNA-later and stored at −80°C prior to analyses. Tissues were homogenised in 1 ml STAT-60 and RNA isolated according to the manufacturer’s instructions (AMS-Bio, UK). RNA was resuspended in 50μl TE buffer (pH 8.0) and assessed by Nanodrop ND-1000 spectrophotometer, before storage at −80°C. Samples underwent reverse transcription and miRNA expression analysis as described above. Expression of miRNAs of interest was measured in patients suffering from infertility and a subsequent live birth (IF+LB; n = 12), patients with miscarriage and a subsequent live birth (M+LB; n = 12), and patients suffering miscarriage and a subsequent miscarriage (M+M; n = 12).
Quantification and statistical analysis
Statistical significance was defined by p <0.05 in all experiments. Statistical analysis of miRNA expression levels (Figures 1 and 18) was determined by an ANOVA. Expression of miRNA in response to P4 (Figure 1) has n =3 biological repeats of Ishikawa cells. Differences between groups are denoted by difference in letters (a, b, c, d). Expression of miRNAs in clinical human endometrial biopsies consists of n = 12 patients per group. Graphpad Prism was used for graphical representation of data. In Figure 2 differences in expression compared were determined using ANOVA with differences depicted by different subscripts (a, b, c, d) when p < 0.05.
RNA Seq data was analysed by quantification carried out by featureCounts function in R. Significant differences in gene expression were identified using DESeq2 (28) with the following cut-offs: log2FoldChange >1 (or <−1) and an FDR-adjusted p value of <0.05. Overrepresentation analyses were performed using WebGestAlt with FDR <0.05.
Proteomic statistical analysis (Figures 9, 10, 11, 12, 13, 14, 15, 16, and 17) was achieved using paired t-tests and a Benjamini Hochberg correction. Proteomic samples were carried out in n = 3 biological replicates of Ishikawa cells. Graphpad Prism was used for graphical representation of data.
Statistical significance of q-RT-PCR of predicted miRNA targets was carried out using the comparative CT method (2-ΔΔCt) (24) normalised to the geometric mean of ACTB, GAPDH and PPIA. Significance differences were determined using ANOVA analysis in Graphpad Prism software, including Dunnett’s multiple comparisons test.
Acknowledgments
We would like to thank all of the women who provided endometrial biopsies for use in this study and the Biomedical Research Unit in Reproductive Health at UHCW for their support. We thank the University of Bristol Proteomics core facility for carrying out the proteomics analysis described. This work was supported by N8 agri-food pump priming, QR GCRF, BBSRCgrant number BB/R017522/1 to NF as well as support from LTHT. JCE is supported by BBSRC DTP studentship: BB/M011151/1.
Author contributions
N.F., M.O’C., K.F., E.S.L., J.J.B., N.A.B.S., and J.S. conceived of the project and designed experiments. L.H., J.C.E., and H.T. performed experiments. D.W., A.S.T., V.O., A.V.G.S., and P.V. performed data analysis. N.F., J.C.E., and L.H. drafted the manuscript. All authors provided components of the manuscript and contributed to editing drafts of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106339.
Supplemental information
Gene ID, gene name, gene type, gene id, baseMean expression value, log2FoldChange, lfcSE, stat, pvalue, and padj for transcripts identified as differentially expressed in Ishikawa cells (n = 3 biological replicates) determined by RNA sequencing. Values reflect changes in P4 treated cells (10 μg/ml) compared to vehicle controls. Raw data are accessible at GEO under accession number GSE211151.
Overrepresented gene ontologies (GO) terms for biological processes (BP), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for cell component (CC), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for molecular function (MF), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched REACTOME pathways description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched KEGG pathways description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Venn diagram analysis of list of transcripts that are common or unique to P4-modified mRNAs determined by RNA sequencing that were altered in Ishikawa cells treated with P4 for 24 h (n = 3 biological replicates) or are predicted targets of three stem lineage. miRNAs that are also altered by P4 treatment of endometrial epithelial cells.
Overrepresented gene ontologies (GO) terms for biological processes (BP), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by TargetScan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for cellular components (CC), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by TargetScan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for molecular function (MF), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by Targetscan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched REACTOME pathways, description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by Targetscan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched KEGG pathways, description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by TargetScan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
15A: List of gene names in segments of Venn diagram related to Figure 11.15B) RNA-Seq data for human endometrial biopsies for mRNAs matching 1,192 proteins changed in response to miR-340-5p mimic, related to Figure 11.15C) RNA-Seq data for human endometrial biopsies for mRNAs matching 267 proteins changed in response to miR-340-5p inhibitor, related to Figure 11.15D) RNA-Seq data for human endometrial biopsies for mRNAs matching 59 proteins changed in response to miR-340-5p mimic AND inhibitor, related to Figure 11.
18A) List of gene names in segments of Venn diagram related to Figure 14.18B) RNA-Seq data for human endometrial biopsies for mRNAs matching 1,080 proteins changed in response to miR-542-3p mimic, related to Figure 14.18C) RNA-Seq data for human endometrial biopsies for mRNAs matching 714 proteins changed in response to miR-542-3p inhibitor, related to Figure 14.18D) RNA-Seq data for human endometrial biopsies for mRNAs matching 181 proteins changed in response to miR-542-3p mimic AND inhibitor, related to Figure 14.
Table 21A: List of gene names in segments of Venn diagram related to Figure 17.
Table S21B: RNA-Seq data for human endometrial biopsies for mRNAs matching 1,063 proteins changed in response to miR-671-5p mimic, related to Figure 17.
Table S21C: RNA-Seq data for human endometrial biopsies for mRNAs matching 362 proteins changed in response to miR-671-5p inhibitor, related to Figure 17.
Table S21D: RNA-Seq data for human endometrial biopsies for mRNAs matching 87 proteins changed in response to miR-671-5p mimic AND inhibitor, related to Figure 17.
Final column lists proteins solely altered by miR-340-5p inhibitor once off target effects are removed (p<0.05), n = 3.
Final column lists genes common to altered proteins in miR-340-5p mimic and/or inhibitor and a miRDB predicted target (p<0.05), n = 3.
Final column lists proteins solely altered by miR-542-3p mimic once off target effects are removed (p<0.05), n = 3.
Final column lists proteins solely altered by miR-542-3p inhibitor once off target effects are removed (p<0.05), n = 3.
Final column lists genes common to altered proteins in miR-542-3p mimic and/or inhibitor and a miRDB predicted target (p<0.05), n = 3.
Final column lists proteins solely altered by miR-671-5p inhibitor once off target effects are removed (p<0.05), n = 3.
Final column lists proteins solely altered by miR-671-5p mimic once off target effects are removed (p<0.05), n = 3
Final column lists genes common to altered proteins in miR-671-5p mimic and/or inhibitor and a miRDB predicted target (p<0.05), n = 3.
Data and code availability
-
•
All RNA sequencing and proteomic data are uploaded in publicly available data repositories - RNA sequencing data is available on GEO under accession number GEO:GSE211151.
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier ProteomeXchange:PXD036109.
-
•
This paper does not report original code and any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Critchley H.O.D., Maybin J.A., Armstrong G.M., Williams A.R.W. Physiology of the endometrium and regulation of menstruation. Physiol. Rev. 2020;100:1149–1179. doi: 10.1152/physrev.00031.2019. [DOI] [PubMed] [Google Scholar]
- 2.Macklon N.S., Brosens J.J. The human endometrium as a sensor of embryo quality. Biol. Reprod. 2014;91:98. doi: 10.1095/biolreprod.114.122846. [DOI] [PubMed] [Google Scholar]
- 3.Weimar C.H.E., Kavelaars A., Brosens J.J., Gellersen B., de Vreeden-Elbertse J.M.T., Heijnen C.J., Macklon N.S. Endometrial stromal cells of women with recurrent miscarriage fail to discriminate between high-and low-quality human embryos. PLoS One. 2012;7:e41424. doi: 10.1371/journal.pone.0041424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosens J.J., Salker M.S., Teklenburg G., Nautiyal J., Salter S., Lucas E.S., Steel J.H., Christian M., Chan Y.-W., Boomsma C.M., et al. Uterine selection of human embryos at implantation. Sci. Rep. 2014;4:3894–3898. doi: 10.1038/srep03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quenby S., Gallos I.D., Dhillon-Smith R.K., Podesek M., Stephenson M.D., Fisher J., Brosens J.J., Brewin J., Ramhorst R., Lucas E.S., et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. 2021;397:1658–1667. doi: 10.1016/S0140-6736(21)00682-6. [DOI] [PubMed] [Google Scholar]
- 6.Salamonsen L.A. Women in reproductive science: my WOMBan’s life: understanding human endometrial function. Reproduction. 2019;158:F55–F67. doi: 10.1530/REP-18-0518. [DOI] [PubMed] [Google Scholar]
- 7.Shah K.M., Webber J., Carzaniga R., Taylor D.M., Fusi L., Clayton A., Brosens J.J., Hartshorne G., Christian M. Induction of microRNA resistance and secretion in differentiating human endometrial stromal cells. J. Mol. Cell Biol. 2013;5:67–70. doi: 10.1093/jmcb/mjs058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas E.S., Vrljicak P., Muter J., Diniz-da-Costa M.M., Brighton P.J., Kong C.-S., Lipecki J., Fishwick K.J., Odendaal J., Ewington L.J., et al. Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun. Biol. 2020;3:1–14. doi: 10.1038/s42003-020-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon A.L., Kiehl M., Fischer E., Proctor J.G., Bush M.R., Givens C., Rabinowitz M., Demko Z.P. Pregnancy outcomes from more than 1,800 in vitro fertilization cycles with the use of 24-chromosome single-nucleotide polymorphism–based preimplantation genetic testing for aneuploidy. Fertil. Steril. 2018;110:113–121. doi: 10.1016/j.fertnstert.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Hayder H., O’Brien J., Nadeem U., Peng C. MicroRNAs: crucial regulators of placental development. Reproduction. 2018;155:R259–R271. doi: 10.1530/REP-17-0603. [DOI] [PubMed] [Google Scholar]
- 12.Morales-Prieto D.M., Ospina-Prieto S., Schmidt A., Chaiwangyen W., Markert U.R. Elsevier Trophoblast Research Award Lecture: origin, evolution and future of placenta miRNAs. Placenta. 2014;35:S39–S45. doi: 10.1016/j.placenta.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Lynch V.J., Nnamani M.C., Kapusta A., Brayer K., Plaza S.L., Mazur E.C., Emera D., Sheikh S.Z., Grützner F., Bauersachs S., et al. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 2015;10:551–561. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner G.P. Evolutionary innovations and novelties: let us get down to business. Zoologischer Anzeiger - A Journal of Comparative Zoology. 2015;256:75–81. [Google Scholar]
- 15.Kin K., Maziarz J., Chavan A.R., Kamat M., Vasudevan S., Birt A., Emera D., Lynch V.J., Ott T.L., Pavlicev M., Wagner G.P. The transcriptomic evolution of mammalian pregnancy: gene expression innovations in endometrial stromal fibroblasts. Genome Biol. Evol. 2016;8:2459–2473. doi: 10.1093/gbe/evw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakabe N.J., Aneas I., Knoblauch N., Sobreira D.R., Clark N., Paz C., Horth C., Ziffra R., Kaur H., Liu X., et al. Transcriptome and regulatory maps of decidua-derived stromal cells inform gene discovery in preterm birth. Sci. Adv. 2020;6:eabc8696. doi: 10.1126/sciadv.abc8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marinić M., Mika K., Chigurupati S., Lynch V.J. Evolutionary transcriptomics implicates HAND2 in the origins of implantation and regulation of gestation length. Elife. 2021;10:e61257. doi: 10.7554/eLife.61257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor A.S., Tinning H., Ovchinnikov V., Smith W., Pullinger A.L., Sutton R.A., Constantinides B., Wang D., Forde N., O'Connell M.J. A burst of regulatory and protein innovation at the origin of placental mammals drove the emergence of placenta and underpins divergent early pregnancy strategies in modern mammals. bioRxiv. 2021 doi: 10.1101/2021.07.22.453388. Preprint at. [DOI] [Google Scholar]
- 19.Forde N., Bazer F.W., Spencer T.E., Lonergan P. ‘Conceptualizing’the endometrium: identification of conceptus-derived proteins during early pregnancy in cattle. Biol. Reprod. 2015;92:156. doi: 10.1095/biolreprod.115.129296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tinning H., Taylor A., Wang D., Constantinides B., Sutton R., Oikonomou G., Velazquez M.A., Thompson P., Treumann A., O’Connell M.J., Forde N. The role of CAPG in molecular communication between the embryo and the uterine endometrium: is its function conserved in species with different implantation strategies? Faseb. J. 2020;34:11015–11029. doi: 10.1096/fj.202000882RR. [DOI] [PubMed] [Google Scholar]
- 21.Lipecki J., Mitchell A.E., Muter J., Lucas E.S., Makwana K., Fishwick K., Odendaal J., Hawkes A., Vrljicak P., Brosens J.J., Ott S. EndoTime: non-categorical timing estimates for luteal endometrium. Hum. Reprod. 2022;37:747–761. doi: 10.1093/humrep/deac006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taraborrelli S. Physiology, production and action of progesterone. Acta Obstet. Gynecol. Scand. 2015;94:8–16. doi: 10.1111/aogs.12771. [DOI] [PubMed] [Google Scholar]
- 23.LESSEY B.A., KILLAM A.P., METZGER D.A., HANEY A.F., GREENE G.L., McCarty K.S., Jr. Immunohistochemical analysis of human uterine estrogen and progesterone receptors throughout the menstrual cycle. J. Clin. Endocrinol. Metab. 1988;67:334–340. doi: 10.1210/jcem-67-2-334. [DOI] [PubMed] [Google Scholar]
- 24.Marquardt R.M., Kim T.H., Shin J.-H., Jeong J.-W. Progesterone and estrogen signaling in the endometrium: what goes wrong in endometriosis? Int. J. Mol. Sci. 2019;20:3822. doi: 10.3390/ijms20153822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhurke A.S., Bagchi I.C., Bagchi M.K. Progesterone-regulated endometrial factors controlling implantation. Am. J. Reprod. Immunol. 2016;75:237–245. doi: 10.1111/aji.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K., Jeong J., Kwak I., Yu C.-T., Lanske B., Soegiarto D.W., Toftgard R., Tsai M.-J., Tsai S., Lydon J.P., DeMayo F.J. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat. Genet. 2006;38:1204–1209. doi: 10.1038/ng1874. [DOI] [PubMed] [Google Scholar]
- 27.Li Q., Kannan A., DeMayo F.J., Lydon J.P., Cooke P.S., Yamagishi H., Srivastava D., Bagchi M.K., Bagchi I.C. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science. 2011;331:912–916. doi: 10.1126/science.1197454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tranguch S., Wang H., Daikoku T., Xie H., Smith D.F., Dey S.K. FKBP52 deficiency-conferred uterine progesterone resistance is genetic background and pregnancy stage specific. J. Clin. Invest. 2007;117:1824–1834. doi: 10.1172/JCI31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuokkanen S., Chen B., Ojalvo L., Benard L., Santoro N., Pollard J.W. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol. Reprod. 2010;82:791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi R.P.A., Wang T., Adams N., Wu S.P., Young S.L., Spencer T.E., DeMayo F. Human endometrial transcriptome and progesterone receptor cistrome reveal important pathways and epithelial regulators. J. Clin. Endocrinol. Metab. 2020;105:e1419–e1439. doi: 10.1210/clinem/dgz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bazer F.W., Spencer T.E., Johnson G.A., Burghardt R.C., Wu G. Comparative aspects of implantation. Reproduction. 2009;138:195–209. doi: 10.1530/REP-09-0158. [DOI] [PubMed] [Google Scholar]
- 32.Greening D.W., Nguyen H.P.T., Evans J., Simpson R.J., Salamonsen L.A. Modulating the endometrial epithelial proteome and secretome in preparation for pregnancy: the role of ovarian steroid and pregnancy hormones. J. Proteonomics. 2016;144:99–112. doi: 10.1016/j.jprot.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 33.Paiva P., Hannan N.J., Hincks C., Meehan K.L., Pruysers E., Dimitriadis E., Salamonsen L.A. Human chorionic gonadotrophin regulates FGF2 and other cytokines produced by human endometrial epithelial cells, providing a mechanism for enhancing endometrial receptivity. Hum. Reprod. 2011;26:1153–1162. doi: 10.1093/humrep/der027. [DOI] [PubMed] [Google Scholar]
- 34.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G., Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bounds K.R., Chiasson V.L., Pan L.J., Gupta S., Chatterjee P. MicroRNAs: new players in the pathobiology of preeclampsia. Front. Cardiovasc. Med. 2017;4:60. doi: 10.3389/fcvm.2017.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munaut C., Tebache L., Blacher S., Noël A., Nisolle M., Chantraine F. Dysregulated circulating miRNAs in preeclampsia. Biomed. Rep. 2016;5:686–692. doi: 10.3892/br.2016.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santamaria X., Taylor H. MicroRNA and gynecological reproductive diseases. Fertil. Steril. 2014;101:1545–1551. doi: 10.1016/j.fertnstert.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 38.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harapan H., Andalas M. The role of microRNAs in the proliferation, differentiation, invasion, and apoptosis of trophoblasts during the occurrence of preeclampsia—a systematic review. Tzu Chi Med. J. 2015;27:54–64. [Google Scholar]
- 40.Hosseini M.K., Gunel T., Gumusoglu E., Benian A., Aydinli K. MicroRNA expression profiling in placenta and maternal plasma in early pregnancy loss. Mol. Med. Rep. 2018;17:4941–4952. doi: 10.3892/mmr.2018.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tochigi H., Kajihara T., Mizuno Y., Mizuno Y., Tamaru S., Kamei Y., Okazaki Y., Brosens J.J., Ishihara O. Loss of miR-542-3p enhances IGFBP-1 expression in decidualizing human endometrial stromal cells. Sci. Rep. 2017;7:40001–40010. doi: 10.1038/srep40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazur E.C., Large M.J., DeMayo F.J. In: T. M. Plant and A. J. B. T.-K. and N. P. of R. Zeleznik F.E., editor. Academic Press; 2015. Chapter 24 - human oviduct and endometrium: changes over the menstrual cycle; pp. 1077–1097. [DOI] [Google Scholar]
- 43.Simintiras C.A., Dhakal P., Ranjit C., Fitzgerald H.C., Balboula A.Z., Spencer T.E. Capture and metabolomic analysis of the human endometrial epithelial organoid secretome. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026804118. e2026804118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawlings T.M., Makwana K., Taylor D.M., Molè M.A., Fishwick K.J., Tryfonos M., Odendaal J., Hawkes A., Zernicka-Goetz M., Hartshorne G.M., et al. Modelling the impact of decidual senescence on embryo implantation in human endometrial assembloids. Elife. 2021;10:e69603. doi: 10.7554/eLife.69603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzgerald H.C., Dhakal P., Behura S.K., Schust D.J., Spencer T.E. Self-renewing endometrial epithelial organoids of the human uterus. Proc. Natl. Acad. Sci. USA. 2019;116:23132–23142. doi: 10.1073/pnas.1915389116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fromm B., Domanska D., Høye E., Ovchinnikov V., Kang W., Aparicio-Puerta E., Johansen M., Flatmark K., Mathelier A., Hovig E., et al. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 2020;48:D132–D141. doi: 10.1093/nar/gkz885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aghajanova L., Hamilton A.E., Giudice L.C. Uterine receptivity to human embryonic implantation: histology, biomarkers, and transcriptomics. Semin. Cell Dev. Biol. 2008;19:204–211. doi: 10.1016/j.semcdb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv Y., Gao S., Zhang Y., Wang L., Chen X., Wang Y. miRNA and target gene expression in menstrual endometria and early pregnancy decidua. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016;197:27–30. doi: 10.1016/j.ejogrb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Revel A., Achache H., Stevens J., Smith Y., Reich R. MicroRNAs are associated with human embryo implantation defects. Hum. Reprod. 2011;26:2830–2840. doi: 10.1093/humrep/der255. [DOI] [PubMed] [Google Scholar]
- 50.Van Mourik M.S.M., Macklon N.S., Heijnen C.J. Embryonic implantation: cytokines, adhesion molecules, and immune cells in establishing an implantation environment. J. Leukoc. Biol. 2009;85:4–19. doi: 10.1189/jlb.0708395. [DOI] [PubMed] [Google Scholar]
- 51.Yang Q., Gu W.-W., Gu Y., Yan N.-N., Mao Y.-Y., Zhen X.-X., Wang J.-M., Yang J., Shi H.-J., Zhang X., Wang J. Association of the peripheral blood levels of circulating microRNAs with both recurrent miscarriage and the outcomes of embryo transfer in an in vitro fertilization process. J. Transl. Med. 2018;16:186–214. doi: 10.1186/s12967-018-1556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S., et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 57.Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene ID, gene name, gene type, gene id, baseMean expression value, log2FoldChange, lfcSE, stat, pvalue, and padj for transcripts identified as differentially expressed in Ishikawa cells (n = 3 biological replicates) determined by RNA sequencing. Values reflect changes in P4 treated cells (10 μg/ml) compared to vehicle controls. Raw data are accessible at GEO under accession number GSE211151.
Overrepresented gene ontologies (GO) terms for biological processes (BP), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for cell component (CC), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for molecular function (MF), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched REACTOME pathways description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched KEGG pathways description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Venn diagram analysis of list of transcripts that are common or unique to P4-modified mRNAs determined by RNA sequencing that were altered in Ishikawa cells treated with P4 for 24 h (n = 3 biological replicates) or are predicted targets of three stem lineage. miRNAs that are also altered by P4 treatment of endometrial epithelial cells.
Overrepresented gene ontologies (GO) terms for biological processes (BP), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by TargetScan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for cellular components (CC), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by TargetScan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Overrepresented gene ontologies (GO) terms for molecular function (MF), description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by Targetscan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched REACTOME pathways, description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by Targetscan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
Enriched KEGG pathways, description, GO term size, expected and degree of enrichment for DEGs identified following RNA sequencing of P4 treated Ishikawa cells (n = 3 biological replicates) compared to vehicle controls that are also predicted targets (as determined by TargetScan) of three miRNA that are P4-modified. Significance of enrichment, p-value, FDR-adjusted p-value and gene IDs are provided. GO overrepresentation analyses were carried out using webgestalt.
15A: List of gene names in segments of Venn diagram related to Figure 11.15B) RNA-Seq data for human endometrial biopsies for mRNAs matching 1,192 proteins changed in response to miR-340-5p mimic, related to Figure 11.15C) RNA-Seq data for human endometrial biopsies for mRNAs matching 267 proteins changed in response to miR-340-5p inhibitor, related to Figure 11.15D) RNA-Seq data for human endometrial biopsies for mRNAs matching 59 proteins changed in response to miR-340-5p mimic AND inhibitor, related to Figure 11.
18A) List of gene names in segments of Venn diagram related to Figure 14.18B) RNA-Seq data for human endometrial biopsies for mRNAs matching 1,080 proteins changed in response to miR-542-3p mimic, related to Figure 14.18C) RNA-Seq data for human endometrial biopsies for mRNAs matching 714 proteins changed in response to miR-542-3p inhibitor, related to Figure 14.18D) RNA-Seq data for human endometrial biopsies for mRNAs matching 181 proteins changed in response to miR-542-3p mimic AND inhibitor, related to Figure 14.
Table 21A: List of gene names in segments of Venn diagram related to Figure 17.
Table S21B: RNA-Seq data for human endometrial biopsies for mRNAs matching 1,063 proteins changed in response to miR-671-5p mimic, related to Figure 17.
Table S21C: RNA-Seq data for human endometrial biopsies for mRNAs matching 362 proteins changed in response to miR-671-5p inhibitor, related to Figure 17.
Table S21D: RNA-Seq data for human endometrial biopsies for mRNAs matching 87 proteins changed in response to miR-671-5p mimic AND inhibitor, related to Figure 17.
Final column lists proteins solely altered by miR-340-5p inhibitor once off target effects are removed (p<0.05), n = 3.
Final column lists genes common to altered proteins in miR-340-5p mimic and/or inhibitor and a miRDB predicted target (p<0.05), n = 3.
Final column lists proteins solely altered by miR-542-3p mimic once off target effects are removed (p<0.05), n = 3.
Final column lists proteins solely altered by miR-542-3p inhibitor once off target effects are removed (p<0.05), n = 3.
Final column lists genes common to altered proteins in miR-542-3p mimic and/or inhibitor and a miRDB predicted target (p<0.05), n = 3.
Final column lists proteins solely altered by miR-671-5p inhibitor once off target effects are removed (p<0.05), n = 3.
Final column lists proteins solely altered by miR-671-5p mimic once off target effects are removed (p<0.05), n = 3
Final column lists genes common to altered proteins in miR-671-5p mimic and/or inhibitor and a miRDB predicted target (p<0.05), n = 3.
Data Availability Statement
-
•
All RNA sequencing and proteomic data are uploaded in publicly available data repositories - RNA sequencing data is available on GEO under accession number GEO:GSE211151.
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier ProteomeXchange:PXD036109.
-
•
This paper does not report original code and any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.