Highlights
-
•
A model to classify multiple sclerosis disability with a single MRI is proposed.
-
•
The analyses of attention maps help to understand deep learning model decisions.
-
•
Frontotemporal cortex and cerebellum were highlighted as relevant anatomical areas.
Keywords: Multiple sclerosis, Structural MRI, Deep learning, Attention maps, Disability
Abstract
The application of convolutional neural networks (CNNs) to MRI data has emerged as a promising approach to achieving unprecedented levels of accuracy when predicting the course of neurological conditions, including multiple sclerosis, by means of extracting image features not detectable through conventional methods. Additionally, the study of CNN-derived attention maps, which indicate the most relevant anatomical features for CNN-based decisions, has the potential to uncover key disease mechanisms leading to disability accumulation.
From a cohort of patients prospectively followed up after a first demyelinating attack, we selected those with T1-weighted and T2-FLAIR brain MRI sequences available for image analysis and a clinical assessment performed within the following six months (N = 319). Patients were divided into two groups according to expanded disability status scale (EDSS) score: ≥3.0 and < 3.0. A 3D-CNN model predicted the class using whole-brain MRI scans as input. A comparison with a logistic regression (LR) model using volumetric measurements as explanatory variables and a validation of the CNN model on an independent dataset with similar characteristics (N = 440) were also performed. The layer-wise relevance propagation method was used to obtain individual attention maps.
The CNN model achieved a mean accuracy of 79% and proved to be superior to the equivalent LR-model (77%). Additionally, the model was successfully validated in the independent external cohort without any re-training (accuracy = 71%). Attention-map analyses revealed the predominant role of frontotemporal cortex and cerebellum for CNN decisions, suggesting that the mechanisms leading to disability accrual exceed the mere presence of brain lesions or atrophy and probably involve how damage is distributed in the central nervous system.
1. Introduction
Multiple sclerosis (MS) is a chronic disease of the central nervous system characterised by inflammation, demyelination and neurodegeneration, being one of the main non-traumatic causes of irreversible disability in young adults. The exact cause of MS and the pathological mechanisms ultimately leading to an irreversible accumulation of disability are still unknown. Furthermore, its disease course can be highly variable among individuals (Thompson et al., 2018).
Magnetic Resonance Imaging (MRI) is an essential tool to diagnose and predict prognosis of MS and is routinely used to assess disease activity and treatment effectiveness through serial MRI analyses over time. MRI biomarkers such as the number of brain lesions and their evolution over time (Thompson et al., 2018, Tintore et al., 2015), and the quantification of brain volume (i.e., grey matter [GM] and white matter [WM] volume) (Calabrese and Castellaro, 2017, de Stefano et al., 2003), are known to be good measurements to establish patients’ prognosis. The accumulation of patient disability has been associated with progression on some of these biomarkers (Bonacchi et al., 2022, Sastre-Garriga et al., 2020).
In recent years, the combination of MRI and deep learning (DL)-based models, especially convolutional neural networks (CNN), has gained popularity thanks to their ability to solve complex classification tasks. DL techniques are not dependent on pre-defined features, instead are capable of automatically extracting relevant information from raw or minimally processed data.
In neuroimaging, the use of CNN approaches has been useful not only to extract biomarkers from MRI images, but also to address the classification and the prediction of different diseases (Lian et al., 2021, van der Burgh et al., 2017, Venugopalan et al., 2021). However, the use of DL techniques to predict disease progression is still largely unexplored. Some recent studies have presented promising results for future prediction of disability (Roca et al., 2020, Storelli et al., 2022, Tousignant et al., 2019) and cross-sectional patient stratification (Cruciani et al., 2021, Marzullo et al., 2019).
New tendencies in the field have also put emphasis on producing explainable DL-based techniques, with the goal of disentangling the reason behind DL decisions, therefore providing additional information that could be extremely useful to the end users enhancing data insights (Bach et al., 2015, Simonyan et al., 2013, Springenberg et al., 2014). This task of deciphering the “black box” of DL-based models in MS has recently been studied for disease diagnosis (Eitel et al., 2019, Lopatina et al., 2020) and MS phenotype signature decrypting (Cruciani et al., 2021). All these studies concluded that the use of the layer-wise relevance propagation (LRP)(Bach et al., 2015), among other (similar) methods, is the most promising tool for these analyses providing individual heatmaps for each subject, called attention maps, reflecting the voxel-specific relevance to the classification output, according to the DL model, in an easy and intuitive way.
The main goal of this work was to build a DL-model able to accurately classify MS patients based on their disability level, while analysing the CNN-derived attention maps in order to understand the reasons behind the decisions taken by the DL-model. We believe these analyses may get us closer to deciphering the physiopathological mechanisms responsible for clinical progression in MS.
2. Materials and methods
2.1. Datasets
2.1.1. In-house dataset
The data used for our experiments is part of a larger in-house cohort of patients followed over time after their first demyelinating attack (or clinically isolated syndrome, CIS), the Barcelona CIS cohort, from the Multiple Sclerosis Centre of Catalonia (Cemcat), Vall d'Hebron University Hospital (VHUH) (Tintore et al., 2015). This is a prospective cohort started in 1995 and is still in course. The selected subjects' acquisition dates run from 2010 to 2020. The inclusion criteria was not dependent on the clinical phenotype at the scan acquisition time point. Our only requirement was that patients had to have experienced a CIS. Approval was received from the Vall d’Hebron Institute of Research – Research and Ethics Committee (XMG-INT-2014-01 and PR(AG)389/2021) and informed consent was obtained from each patient conforming the cohort.
For this cross-sectional study, we included all patients after their first demyelinating attack who had at least one brain MRI scan available for image analysis and a clinical examination within six months after the scan. Clinical examinations included the assessment on the Expanded Disability Status Scale (EDSS) score (Kurtzke, 1983). EDSS ranges from 0 to 10. An EDSS of 3.0 is commonly recognised as the boundary between mild or no-disability vs moderate disability status (Amato et al., 2008, Tintoré et al., 2006, Tintore et al., 2015).
We included 319 unique patients in the study, making a total dataset of 382 scans i.e., for 33 patients we used one or more MRI scans, performed at different time-points after the first attack. The experiments were carried out with a fold cross validation strategy. Not to bias the results of the models during the cross validation strategy, we ensured that all MRI scans from a given patient were included in the same fold, preventing their use for training and testing at the same time. Each scan was matched with the first EDSS score obtained within the following six months after the MRI acquisition.
The whole in-house dataset was acquired in the same centre with five different Siemens scanner models at two different magnetic fields (1.5 T – 3.0 T). For all scanners, the MRI protocol included sagittal 3D T1 magnetization prepared rapid gradient-echo (MPRAGE) and transverse T2 fluid-attenuated inversion recovery (T2-FLAIR). The acquisition protocols of each scanner are summarised in Table 1.
Table 1.
MRI acquisition parameters for each scanner used in the in-house database.
| Avanto | Avanto Fit | Symphony | Symphony Tim | Tim Trioa | |
|---|---|---|---|---|---|
| Field Strength (T) | 1.5 | 1.5 | 1.5 | 1.5 | 3.0 |
| MPRAGE | |||||
| TR (ms) | 1980 | 2300 | 2700 | 1980 | 2300 |
| TE (ms) | 3.1 | 3.05 | 4.8 | 3.08 | 2.98 |
| TI (ms) | 1100 | 900 | 850 | 1100 | 900 |
| Voxel size (mm) | 1x1x1 | 1x1x1 | 1x1x1.2 | 1x1x1 | 1x1x1 |
| Plane | Sagittal | Sagittal | Sagittal | Sagittal | Sagittal |
| T2-FLAIR | |||||
| TR (ms) | 8500 | 8500 | 9000 | 8500 | 9000//2300 |
| TE (ms) | 92 | 99 | 114 | 95 | 87//392 |
| TI (ms) | 2439 | 2440 | 2500 | 2440 | 2500//1800 |
| Voxel size (mm) | 0.97x0.97x3 | 0.97x0.97x3 | 0.49x0.49x3 | 0.97x0.97x3 | 0.49x0.49x3//1x1x1 |
| Plane | Axial | Axial | Axial | Axial | Axial//Sagittal |
| In dataset, n (%) | 64 (17) | 64 (17) | 10 (3) | 51 (13) | 193 (50) |
TR: repetition time, TE: echo time, TI: inversion time.
With Tim Trio there are T2-FLAIR scans acquired with 2D and 3D. In table are specified by 2D parameters//3D parameters. The 3D T2-FLAIR acquisition parameters are the same used in the external validation dataset.
2.1.2. External validation (MS PATHS) dataset
MS PATHS (Multiple Sclerosis Partners Advancing Technology and Health Solutions)(Mowry et al., 2020) is a learning health system in MS, started in 2016, comprising a collaborative network of 10 healthcare centres, providing standardised routinely-acquired clinical and MRI data. From this large database, we randomly selected a subset, with representation of all grades of disability, to be used as an independent test set from the models trained with the in-house dataset. The resultant set was composed of 440 patients with 3D T1 MPRAGE and 3D T2-FLAIR sequences from four different Siemens 3 T scanner models from six different sites (excluding the provider of our in-house dataset). All images acquired using standardised image acquisition protocols, which correspond to the 3D acquisition protocol of the Tim Trio in Table 1. All patients completed the (patient-reported outcome) Patient Determined Disease Steps (PDDS) score (Rizzo et al., 2004). The PDDS score ranges between 0 and 8 and has been proven to have a strong correlation with the EDSS score (Learmonth et al., 2013).
2.2. Image Pre-processing
The same image pre-processing was applied to both datasets, in-house dataset and MS PATHS (external validation dataset from now on). All T1-weighted (T1-w) and T2-FLAIR sequences were pre-processed with (i) bias correction (Tustison et al., 2010), (ii) skull-stripping (Isensee et al., 2019), (iii) registration (Jenkinson & Smith, 2001) to MNI152 space (1x1x1mm3), as well as co-registration of T2-FLAIR sequences to T1-w space, and (iv) min–max voxel intensity normalisation.
2.3. Proposed CNN method
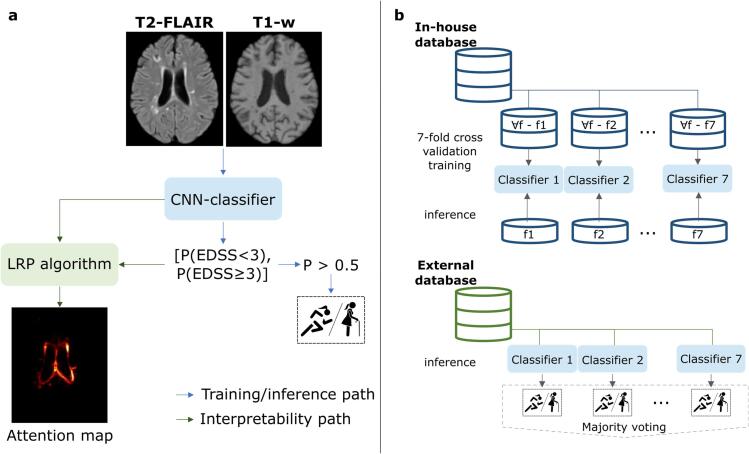
The proposed pipeline to stratify patients based on EDSS < 3.0 and EDSS ≥ 3.0 is summarised in Fig. 1. For the external validation experiment, patients were classified into PDDS < 3.0 (no or mild disability) and PDDS ≥ 3.0 (moderate disability), since PDDS scores ≥ 3.0 indicate moderate-marked disability, equivalent to EDSS ≥ 3.0 (Learmonth et al., 2013).
Fig. 1.
Pipeline followed in this work. a For training and inference, whole brain input is evaluated by the classifier model to predict the probability belonging to each disability status. We set a threshold of 0.5 for this probability. After inference, the probability obtained from the model is fed into the LRP algorithm to backpropagate through the CNN and obtain the attention map. As shown in b, the training procedure is only performed with the in-house database in a cross-validation strategy, hence obtaining different models. Afterwards, these models are used to evaluate the external database, where the final classification decision is obtained with the majority voting of the different models. P: probability, f: fold, LRP: layer-wise relevance propagation.
After fully pre-processing the whole-brain T2-FLAIR and T1-w scans, the volumes were cropped to the brain region obtaining the input patch with a fixed size of 144x184x152mm3. These patches were used as input to train and test the CNN. When testing on new data, the output prediction was propagated to provide the LRP heatmap, complementing the numerical classification decision.
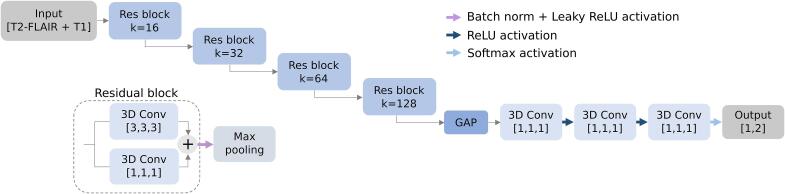
2.3.1. Network architecture
The proposed DL network was inspired by the ResNet CNN architecture (He et al., 2016), built with three-dimensional (3D) layers. Each residual block is based on 3D convolutional layers that produce 3x3x3 and 1x1x1 kernel convolution layers, normalised with batch normalisation and activated with a leaky rectified linear unit (LeakyReLU). As shown in Fig. 2, the architecture was composed of four residual blocks with an increasing number of kernels k (16, 32, 64 and 128), followed by a 2x2x2 downscale max pooling operation. Afterwards, the feature map extracted was projected in a global adaptive max pooling (GAP) layer to reduce feature dimensionality. The produced vector was fed into three successive 1x1x1 3D convolutional layers, with k = 128, 64, 2, where the first two were activated with a ReLU and the last one with a Softmax, providing the probability to belong to one class or another.
Figure 2.
Network architecture. Residual convolutional neural network architecture. Res block: residual block, conv: convolutional layer, k: kernels, GAP: global adaptive max pooling, ReLU: rectified linear unit.
2.3.2. Training procedure
The model was trained using only the in-house dataset. A 7-fold patient cross-validation strategy was used to train and test the model. We sampled the folds to keep the same class distribution in each one, while following the distribution present along the dataset. In each iteration five folds (275 scans) were used for training, one fold for validation (55 scans) and the last fold for inference (55 scans).
To mitigate class imbalance and relatively small cohort data, we augmented the available data for training applying an axial flip to all subjects with an EDSS ≥ 3.0 (i.e., doubling the samples) and to 75% from the other class, EDSS < 3.0, considering the difference in class-size. A random Gaussian noise (σ = 0.02) was also used as data augmentation to create intensity variation on both channels (T1-w and T2-FLAIR).
We trained the model for a maximum of 200 epochs, with a fixed batch size of two, and an early stopping strategy based on the validation loss behaviour to prevent overfitting. Every epoch the whole training set was evaluated. The model was optimised with Adam (Kingma & Ba, 2015) with a learning decay strategy depending on the validation performance and trained by minimising a weighted cross-entropy loss as cost function.
2.3.3. Inference
Following the same sampling procedure described for training, the whole 3D brain was used as input through the trained model providing the output probabilities to belong to one class or another. The final classification was determined by the maximum of both probabilities, with a threshold fixed at 0.5.
2.4. Logistic regression model
To assess whether our CNN method was superior to a more conventional statistical approach, we also built a logistic regression (LR) model. For the LR, the disability class (EDSS < 3.0/EDSS ≥ 3.0) was considered as the dependent variable (output) and the different volumetric measures were considered as the explanatory variables (input). Volumetric measures included: WM, GM, total intracranial, and lateral ventricle volumes, closely related to atrophy measures. These were calculated from the extracted brain parcellation atlas (Henschel et al., 2020) using the T1-w lesion filled (Prados et al., 2016). Brain lesion volume was also computed from the automatically extracted masks obtained using LST (Schmidt et al., 2012) and included as explanatory variable.
2.5. External validation
Inference on the external validation dataset was computed using directly the trained models on the in-house dataset. As seen in Fig. 1b, the reported results were obtained using a majority voting of the seven different models.
Additionally, the LR-models built on the in-house dataset were applied to the external validation dataset in exactly the same way.
2.6. Attention Maps: LRP
To investigate the decisions made by the CNN, attention maps were computed in the input image space to show which were the regions that support the decisions taken by the CNN. The method used for that was the LRP (Bach et al., 2015). This technique decodes the resulting classification output through the network, propagating the relevance layer by layer, obtaining a heatmap on the input space with each voxel contribution.
For this study, we extracted the individual LRP heatmaps for each patient of the in-house dataset. The implementation of the method was inspired in Pytorch-LRP (Böhle et al., 2019), and adapted to our specific CNN, with β = 0, only reflecting the positive contributions that led the classification to the winning class. The decision of choosing only the positive contributions was made based on previous literature findings (Böhle et al., 2019) and corroborated with preliminary analyses on our own data that showed that the negative contributions did not add additional information.
The resulting LRP heatmaps were evaluated (i) individually and (ii) as a class-average prediction.
2.6.1. Individual attention map analyses
Individually, the attention maps were assessed visually and qualitatively showing which voxels contributed the most to the classification given by the model inference. Additionally, semi-quantitative analyses of the individual attention maps were carried out, multiplying such maps by a parcellation map. This allowed us to classify the relevant voxels for the CNN decision into the different anatomical areas. For this purpose, we set a threshold at the 95% percentile of positive relevance, to catch up the most relevant areas in each case, although other thresholds have been studied (see Supplementary Material).
2.6.2. Class-average attention map analyses
Class-average attention maps were also built for visual inspection, through averaging the individual values of the attention maps across the subjects of each one of the prediction results: true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN). In addition, each class-average map was multiplied by the parcellation map, and a mean value per anatomical area was obtained. This allowed us to identify the anatomical areas with greatest relevance.
2.6.3. Voxel-wise regression to explain individual attention maps
We then carried out a quantitative analysis based on the LRP heatmaps aiming at investigating to what extent the variability within each voxel (across independent subjects) of the attention map could be explained by the presence of lesions and atrophy, the best-known contributors to disability in MS, at the voxel level.
For that, we first grouped our subjects by the predicted vs real class (TP, FP, FN, TN). We then computed voxel-wise regression models, one per each prediction group (i.e., TP, FN, FP, TN), where the LRP value at each voxel (considering all individual LRP heatmaps of the same group) was the dependent variable. As explanatory variables of these voxel-wise regression models we included: i) voxel-wise binary indicator of lesion (obtained from individual T2-FLAIR scans), and ii) voxel-wise value describing the deformation suffered by the T1-w scan when moving it to a common space (MNI), calculated with the Jacobian determinant (Lungu et al., 2019). The latter variable (ii) was used as proxy for atrophy, being aware that the common template is based on healthy controls. After this step we obtained four voxel-wise maps of R-squared values, where each voxel indicated the proportion of the variability of the LRP value that could be explained by the presence of lesions and native-to-MNI deformations. Additionally, we estimated the standardised beta coefficient for each one of the explanatory variables, which indicated the relative importance of each one of these for the prediction of the dependent variable, being the two standardised beta maps comparable (since they were in the same scale).
2.7. Evaluation measures of the classification models
To evaluate the performance of the DL-based and LR-models, we used the following metrics:
-
•
Balanced accuracy in correctly classifying each patient by means of their EDSS,
| (1) |
-
•
Sensitivity of correctly classified subjects with EDSS ≥ 3.0,
| (2) |
-
•
Specificity of correctly classified subjects with EDSS < 3.0,
| (3) |
where TP are the number of correctly classified patients with EDSS ≥ 3.0, TN are the number of correctly classified patients with EDSS < 3.0, FP are the number of patients with EDSS < 3.0 classified as they have EDSS ≥ 3.0, and FN are the number of patients with EDSS ≥ 3.0 classified as they have EDSS < 3.0. The results are reported in terms of mean and standard deviation for the 7-fold cross validation computed on the in-house dataset and as majority voting of the 7 models for the external validation dataset.
2.8. Descriptive statistics and other general aspects
Descriptive statistics included comparison between the two groups (EDSS < 3.0/EDSS ≥ 3.0). Chi-square or mixed-effects linear regression models accounting for repeated measures were used, as appropriate.
The proposed method and analyses were entirely implemented in Python (https://www.python.org/), using the Pytorch library (Paszke et al., 2019). We ran all the experiments on a GNU/Linux machine box running Ubuntu 20.04, with 128 GB RAM. For training the model, we used a single Quadro RTX 5000 GPU (NVIDIA Corp, USA) with 16 GB VRAM memory.
3. Results
3.1. Evaluation of DL-models: in-house dataset
Out of the 319 patients, 104 were assigned with EDSS ≥ 3.0 and 215 with EDSS < 3.0. This meant a volume of 215 MRI scans with EDSS < 3.0, and 167 MRI scans with EDSS ≥ 3.0 (non-confirmed). The two stratification groups showed comparable demographic, clinical and MRI characteristics, as summarised in Table 2. The full cohort EDSS median was 2.0, with a median of 1.5 (in range 0.0–2.5) for patients with EDSS < 3.0 and 4.0 (in range 3.0–9.0) for patients with EDSS ≥ 3.0. Patients with EDSS ≥ 3.0 had a longer disease duration than patients with EDSS < 3.0, with a similar age at CIS with a mean age of approximately 30 years old (SD = 8). Compared with patients with EDSS < 3.0, patients with EDSS ≥ 3.0 had lower tissue volumes, GM, WM and total intracranial volume, and higher ventricle volume and lesion load.
Table 2.
Demographic, clinical and brain MRI characteristics of MS patients from the in-house database.
| Full cohort NPAC/SCAN = 319/382 |
EDSS < 3.0 N = 215/215 |
EDSS ≥ 3.0 N = 104/167 |
p-value | |
|---|---|---|---|---|
| Female, n (%) | 207 (65) | 147 (64) | 60 (58) | 0.08 |
| Age at CIS, years, mean [range] | 32.3 [14–50] | 32.4 [14–49] | 32.2 [14–50] | 0.78 |
| Confirmed diagnosisa, n (%) | 260 (82) | 160 (74) | 100 (96) | <0.001 |
| Age at diagnose, years, mean [range] | 33.2 [14–59] | 33.5 [16–59] | 32.7 [14–55] | 0.43 |
| CIS topography, n (%) | <0.001 | |||
| Brain stem | 83 (26) | 59 (27) | 24 (23) | |
| Optic nerve | 98 (31) | 74 (35) | 24 (23) | |
| Spinal cord | 98 (31) | 60 (28) | 38 (37) | |
| Other | 40 (12) | 22 (10) | 18 (17) | |
| MS topography, n (%) | <0.001 | |||
| CIS | 123 (39) | 106 (49.5) | 17 (16) | |
| SP | 41 (13) | 1 (0.5) | 40 (39) | |
| RR | 155 (48) | 108 (50) | 47 (45) | |
| Presence of OB, n (%) | <0.001 | |||
| Positive | 194 (61) | 115 (53) | 79 (75) | |
| Negative | 75 (23) | 64 (30) | 11 (12) | |
| Unknown | 50 (16) | 36 (17) | 14 (13) | |
| DD, years, mean (SD) | 10.4 (7.0) | 7.6 (6.6) | 14.0 (5.6) | <0.001 |
| EDSS, median [range] | 2.0 [0.0–9.0] | 1.5 [0.0–2.5] | 5.0 [3.0–9.0] | <0.001 |
| Lesion load, ml, mean (SD) | 27.5 (39.9) | 10.4 (13.1) | 49.6 (50.7) | <0.001 |
| Ventricles vol, ml, mean (SD) | 29.7 (19.8) | 21.5 (9.6) | 40.2 (25.3) | <0.001 |
| GM vol, ml, mean (SD) | 787.9 (64.0) | 813.1 (48.3) | 755.4 (67.2) | <0.001 |
| WM vol, ml, mean (SD) | 694.8 (68.0) | 707.5 (54.3) | 678.3 (79.6) | <0.001 |
| Brain vol, ml, mean (SD) | 1542.7 (103.7) | 1572.0 (83.7) | 1504.8 (114.4) | <0.001 |
| Scanner model | <0.001 | |||
| Avanto, n (%) | 64 (17) | 19 (9) | 45 (27) | |
| Avanto Fit, n (%) | 64 (17) | 43 (20) | 21 (13) | |
| Symphony, n (%) | 10 (3) | 7 (3) | 3 (2) | |
| Symphony Tim, n (%) | 51 (13) | 13 (6) | 38 (23) | |
| Tim Trio, n (%) | 193 (50) | 133 (62) | 60 (36) |
PAC: patients; EDSS: expanded disability status scale; CIS: clinically isolated syndrome; SP: secondary progressive; RR: relapsing remitting; OB: oligoclonal bands; DD: disease duration; GM: grey matter; WM: white matter.
MS confirmed diagnosis by McDonald 2017 criteria. Two patients were confirmed before their first demyelinating attack.
The average accuracy of the in-house dataset across the seven folds with the whole brain input patch was 79% (SD = 4%), with a sensitivity of 77% (SD = 5%) identifying patients with EDSS ≥ 3.0, and a specificity of 81% (SD = 9%) identifying patients with EDSS < 3.0.
3.2. Comparison with a LR-model
The LR-model built with the six brain volumes corresponding to WM, GM, ventricles, brainstem and cerebellum, intracranial volume and lesion load, achieved an accuracy of 77% (SD = 7%), with a sensitivity of 68% (SD = 10%) when classifying patients with EDSS ≥ 3.0 and a specificity of 86% (SD = 6%). Therefore, the LR-model trained with MRI pre-extracted features showed a 10% lower sensitivity than DL-based models. When considering other input combinations with fewer features, the LR-model obtained always inferior results than the CNN model, and when the LR-models only had a single feature, the accuracies dropped to 50–55%.
3.3. External validation dataset
Table 3 summarises the demographic, clinical and MRI data characteristics of the patients included in the external validation dataset. In this external validation dataset, 220 patients were categorised to the group with PDDS ≥ 3.0 and 220 with PDDS < 3.0. The full median cohort PDDS was 2.5, with a median of 0.5 (in range 0.0–2.0) for the group of no or mild disability patients (PDDS < 3.0) and 5.0 (in range 3.0–7.0) for the group of more disabled patients (PDDS ≥ 3.0). As for the in-house dataset, the disease duration was longer in patients with more disability, with a mean age at diagnosis of 36 years old, and MRI characteristics kept the same relation between groups.
Table 3.
Demographic, clinical and brain MRI characteristics of MS patients from the external database.
| Full cohort N = 440 |
PDDS < 3.0 N = 220 |
PDDS ≥ 3.0 N = 220 |
p-value | |
|---|---|---|---|---|
| Female, n (%) | 310 (70) | 170 (77) | 140 (64) | 0.007 |
| Age at diagnosis, years, mean [range] | 36.8 [19–69] | 36.1 [19–62] | 37.6 [19–69] | 0.24 |
| DD, years, mean (SD) | 11.5 (9.1) | 8.5 (7.6) | 14.9 (9.5) | <0.001 |
| PDDS, median [range] | 2.5 [0.0–7.0] | 0.5 [0.0–2.0] | 5.0 [3.0–7.0] | <0.001 |
| Lesion load, ml, mean (SD) | 13.2 (24.7) | 7.9 (11.7) | 18.5 (32.0) | <0.001 |
| Ventricles vol, ml, mean (SD) | 35.9 (24.0) | 28.7 (17.4) | 43.1 (27.4) | <0.001 |
| GM vol, ml, mean (SD) | 804.6 (73.5) | 828.4 (64.6) | 779.7 (74.0) | <0.001 |
| WM vol, ml, mean (SD) | 706.1 (53.4) | 718.6 (47.3) | 693.6 (56.3) | <0.001 |
| Brain vol, ml, mean (SD) | 1496.4 (92.8) | 1526.2 (79.7) | 1466.5 (95.7) | <0.001 |
PDDS: patient determined disease steps; DD: disease duration; GM: grey matter; WM: white matter.
In this validation dataset, the majority voting system after applying the seven DL models trained on the in-house cohort showed an accuracy of 71%, a sensitivity of 68% and a specificity of 75%. In contrast, when we applied the different LR-models to the validation dataset, we achieved accuracies close to 50% only.
3.4. DL-based attention maps analyses
3.4.1. Individual attention maps
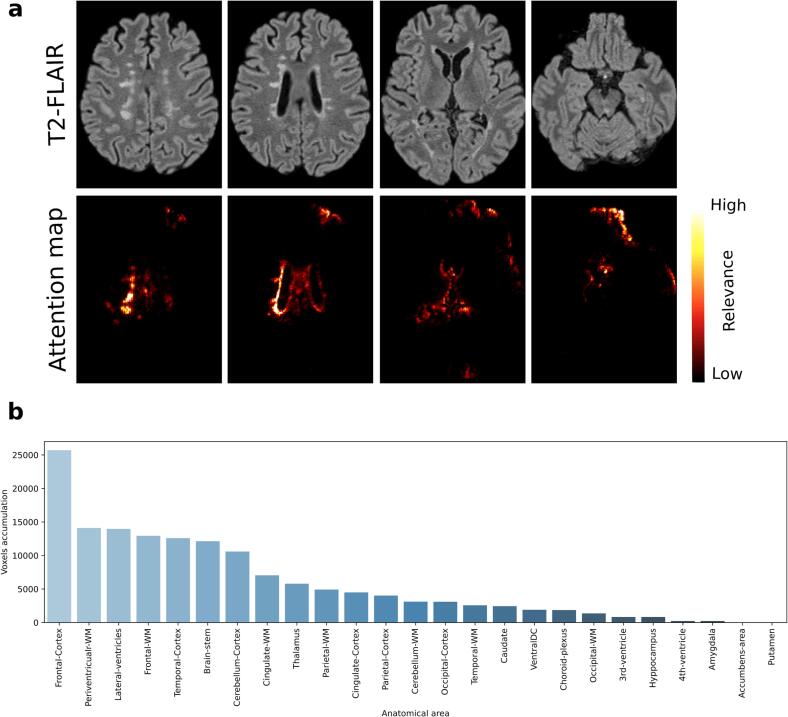
Individual attention maps showed that, in both disability groups, the most relevant voxels that led the classification decisions were mainly located in the periventricular WM regions, which often contained demyelinating lesions, and frontal and temporal cortical areas (see example Fig. 3a). Fig. 3b shows a case-example of the distribution of relevant voxels across the different anatomical regions, considering all voxels with a relevance above a 95% percentile. Other thresholds were also explored (see Supplementary Fig. 1), however insignificant changes in the distribution were observed.
Fig. 3.
Example of an individual attention map analysis. This MS patient was correctly classified as moderate disability with an EDSS = 6.0. a Different T2-FLAIR axial slices with their corresponding computed attention map. b Relevant-voxel accumulation by anatomical area, obtained from the product of the binarised attention map by the brain anatomical parcellation. In this case, the frontal cortex and periventricular WM were the most relevant areas leading the decision.
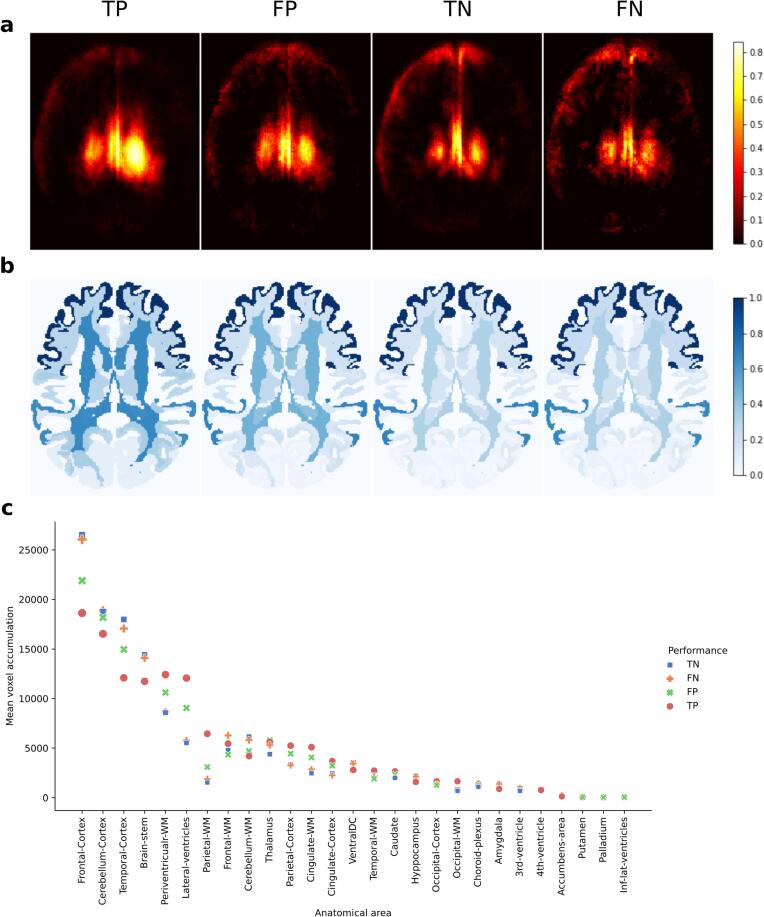
3.4.2. Class-average attention maps
Class-average maps revealed that the most relevant areas were the frontal cortex, cerebellar cortex, periventricular WM, temporal cortex and lateral ventricles, as shown in Fig. 4. The ranking of these regions slightly varied across the different groups (TP, FP, TN, FN). For the patients classified with EDSS ≥ 3.0 (TP, FP), the relevance of periventricular and frontal WM, where most lesions are located, was particularly high. Instead, for no or mild-disability statuses (TN, FN), the relevance of the cortex, especially frontal and temporal cortical areas, and that of the cerebellum were the highest.
Fig. 4.
Class-average attention map analyses. a Class-average attention maps (TP, FP, TN, FN) binarised at 95% percentile. b Brain parcellation with the mean attention value (normalised across groups) attributed to each anatomical region. c Mean voxel accumulation by each anatomical area depending on the class-average group.
3.4.3. Voxel-wise regression analyses
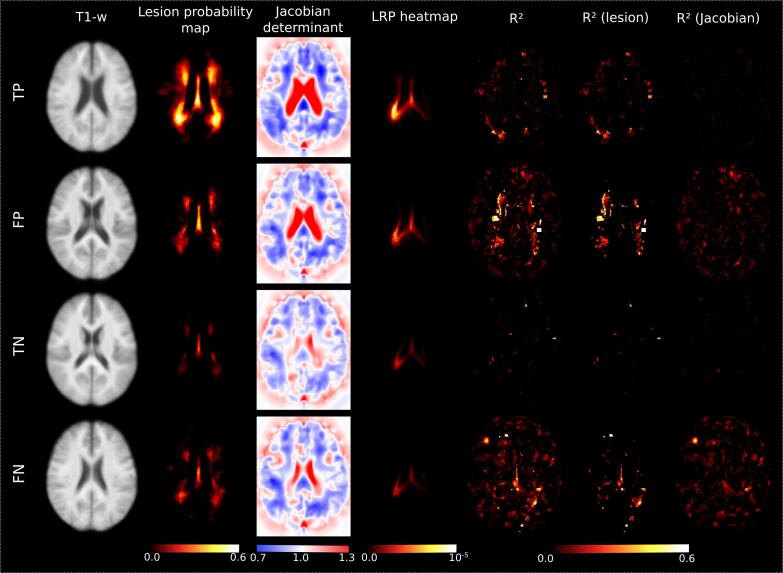
After carrying out the voxel-wise regressions on the in-house dataset, we obtained R-squared maps for the four prediction groups (Fig. 5). In general, the R-squared values were low in all four groups, and mostly ranged between 0 and 0.2. In the correctly classified groups (TP, TN), the R-squared values were even lower (0–0.1). That means that in these groups, at most the 80% of the variability of the attention map could not be explained by the presence of lesions (observed in T2-FLAIR scans) or the native-to-MNI deformation (of the T1-w scans), calculated through the Jacobian determinant.
Fig. 5.
Voxel-wise regression analyses. Average map for the TP, FP, TN and FN of the T1-w scan, lesion probability map, Jacobian determinant, LRP heatmap and the R-squared (R2) map obtained from the voxel-wise regression model built with the individual attention maps as dependent variable, and the individual lesion masks and individual Jacobian determinants as explanatory ones. The partial R2 on each separate variable is also represented. The Jacobian determinant is represented in terms of expansion (values > 1.0) and compression (values < 1.0).
When focused on the maps of partial R-squared, we found that values were always greater for the presence of lesions than for the Jacobian determinant (Fig. 5).
4. Discussion
In this work, we investigated the use of DL-models to classify MS patients according to their disability status while trying to provide an explanation of the decisions taken by the CNN. Our findings showed that a DL-based model using only brain MRI scans, without any guidance, was able to stratify MS patients with high accuracy (79%) when testing data from the same cohort, and 71% when testing images from an unseen database (external validation dataset). Furthermore, the comparison with the LR-model brought to light the superiority of the CNN model. Finally, our attention-map analyses revealed that the most relevant anatomical areas that the CNN model used to decide the level of patients' disability were the frontotemporal cortex and the cerebellum and did not depend on the mere presence of lesions or atrophy in these locations.
Compared with previously reported studies that used DL-based models to perform predictions of MS progression, our work presents some similarities and some major differences. Despite the differences in the target of the classification, i.e., cross-sectional vs future predictions, all published studies (Storelli et al., 2022, Tousignant et al., 2019), like ours, used the whole brain as input to the network. However, in our study, we only used routinely-acquired T1-w and T2-FLAIR MRI data, whereas other studies required the use of several MRI modalities and additional masks (Tousignant et al., 2019) to achieve similar accuracies. Of note, we validated our CNN model on an external, unseen cohort, where the performance of the classification task was more than acceptable, without needing any recalibration or re-training of the model. This utterly necessary step of validating the CNN model (although uncommon in the literature) strengthens our findings, laying the foundation for future longitudinal, comprehensive models of MS disease progression. Furthermore, having at hand an excellent tool to discriminate patients according to their disability level may be extremely useful in contexts where the EDSS score is not obtained as part of routine practice (Bove et al., 2023). Such a discriminating tool may allow the fast identification of patients who already present a certain degree of disability and who, therefore, are at high risk of reaching unfavourable clinical outcomes (Signori et al., 2023). To further assess our model robustness to the actual disability threshold used to classify patients or the distribution of disability scores in the given study population, we carried out a post-hoc analysis on the in-house cohort where we only considered those patients with an EDSS ≤ 2.0 or ≥ 4.0. We observed that the performance of the model only improved by 2%, i.e., we obtained an accuracy, sensitivity, and specificity of 81%, 79%, and 83%, respectively. This slight increase in model performance may highlight the robustness of our CNN model, whose accuracy did not seem too dependent on the threshold used to classify patients into disability groups or how the disability scores were distributed in our cohort. Other data-related aspects that may have affected model performance include the imbalance between classes in terms of scanner models. That is, while 62% of patients in the group with an EDSS < 3.0 were scanned in the Tim Trio scanner, only a 36% of patients with an EDSS ≥ 3.0 did so (Table 2). For this reason, we performed a post-hoc analysis where we assessed, separately, the results obtained with the Trim Trio scans and those obtained with the rest of the scanner models. From this post-hoc analysis we obtained an accuracy of 79% for the Tim Trio scans and an accuracy of 76% for the rest. Thus, considering that the new accuracies were indeed very similar to that obtained with the whole in-house cohort, it might be assumed that the potential impact of such imbalance on model performance was not major. However, we acknowledge that any imbalances between classes are not desirable, since they may introduce biases which are difficult to assess and overcome a posteriori. For that, this may be seen as a limitation of our work. Future studies will evaluate the actual impact that such forms of imbalance, so common in studies which use routinely acquired data, have on deep learning-based models.
Another important remark is that we demonstrated the superiority of our CNN model when compared with a (conventional) LR-model. DL-based models are end-to-end systems which enable the learning of the best feature representation to solve the classification, giving the possibility to extract this representation. Notice that the databases used were conformed of different scanner models and magnetic fields, to which the DL-models were more robust, having<10% drop in performance when testing on the external database. On the other hand, LR-models are based on obtained volumetric measurements that need to be previously computed using different tools which can be affected by changes on the MRI scanners and image protocols used to acquire the data. Thus, considering that volumetric measures have been frequently used as outcome measures in clinical trials (Tur et al., 2018b), our findings suggest that DL-based discriminative tools may also be considered as trial outcomes, given that seem to explain clinical outcome better than conventional volumetric measures feeding into a LR model. Further research in this regard, evaluating the CNN model’s sensitivity to clinical change, is therefore warranted.
Finally, in this study we attempted to understand the reasons behind the decisions made by the CNN through qualitative and quantitative analyses of the individual and class-average attention maps derived from the CNN model. To the best of our knowledge, this is the first large study focusing on giving an explanation to the performance obtained using a CNN when trying to classify patients into different disability categories. Some other studies aiming at solving similar classification problems have also focused on the visualisation of individual attention maps, but without performing further analyses on them (Storelli et al., 2022). Moreover, other studies have mainly investigated the classification between healthy controls and patients (Böhle et al., 2019, Eitel et al., 2019, Lopatina et al., 2020), whereas the task of classifying patients into different disability classes, as in our study, is possibly much more challenging, given the continuous nature of the disease. Thus, we went a step further trying to apply such methods on patients only, i.e., moderately disabled and mildly or non-disabled, with a priori no clear pathological boundaries between them, making it very difficult their identification by providing only brain MRI data to the model. With these analyses we found that the voxels with the highest relevance for the CNN to make the final decisions were in the frontal and temporal cortex, followed by brainstem/cerebellum, and periventricular WM.
The importance of cortical GM for disease progression has been suggested in several studies, although the proposed underlying pathological mechanisms differ across studies. For instance, loss of cortical GM volume (Cordano et al., 2022), microstructural cortical damage, reflected by increased total sodium concentration (Collorone et al., 2021), or grey matter demyelination (Gilmore et al., 2009), including the presence of visible lesions in the cortex (Madsen et al., 2022), may all contribute to the accumulation of disability. Furthermore, the loss of the physiological balance between the cortical thickness of different areas has also been proposed as a process potentially leading to clinical dysfunction in MS (Tur et al., 2018a, Tur et al., 2019). However, no proper head-to-head comparisons across mechanisms have been carried out, which makes it difficult to understand the hierarchy or dynamics of such pernicious pathological events. In any case, it is plausible that our CNN has captured at least some of the imaging features related to them. The consistency across individuals in terms of these areas likely reveal genuine differences between subjects, possibly related to atrophy, although not only: looking at the results of the voxel-wise regression (Fig. 5), it is possible that other pathological aspects, for example changes in image texture denoting underlying non-obvious demyelination, may be playing a role too. On the other hand, it must be acknowledged that some of the highlighted attentions in the cortex might also be caused by non-pathological aspects such as registration inaccuracies. Although the high accuracy of the model might lead us to think that the potential impact of registration inaccuracies on model performance was not major, this would need to be assessed in further studies. That is, we are unable to discern if all the attentions were related or not to pathological aspects, which is a limitation of the current study. It should also be highlighted the involvement of brainstem/cerebellum, whose role in disability progression in MS has been repeatedly seen in the literature (Palesi et al., 2015, Savini et al., 2019, Tintore et al., 2010). Future studies should investigate whether those relevant imaging aspects present in the brainstem/cerebellum have the same pathological translation as those of the relevant cortical areas. An interesting note is that the frontal cortex and the cerebellum have a strong anatomical connection through two distinct fronto-cerebellar pathways, which are known to be damaged in toxic conditions involving the CNS, such as alcohol (Rogers et al., 2012) or heroin abuse (Wang et al., 2013). Future studies will elucidate whether these paths are also altered in MS and, especially, whether such alteration may be a key step in the development of irreversible disability.
The relevance of periventricular WM may be related to the presence of demyelinating lesions, which tend to appear in this location (Thompson et al., 2018), but also to lateral and third ventricle enlargement, which has been shown to play a role too (Brown et al., 2017). Agreeing with that, cases with moderate disability presented a higher mean lesion load in periventricular WM than in mildly or non-disabled cases.
Interestingly, these structures are followed by the thalamus, which also appears to be very relevant for the CNNs, in line with previous studies showing the importance of deep GM structures for disease progression in MS (Eshaghi et al., 2018). All these relevant areas were brought to light through both the individual attention map and the class-average map analyses. To the best of our knowledge, this is the first time that such areas have been identified as relevant for clinical progression in MS in a completely hypothesis-free (unguided) manner, that is, without forcing the model to pay attention to them.
Regarding the voxel-wise regression analysis, the most striking finding was that the attention within a voxel, which indicates the importance of a given voxel to decide the disability class of a given patient, was not (only) explained by the mere presence of lesions or native-to-MNI deformation (used as proxy for volume change or atrophy) in that particular voxel. This may suggest that DL-based methods pay attention to more general aspects of the image, possibly focusing on complex spatial relationships between voxel-wise information, considering that distributional features of brain lesions might impact on disability progression (Tur et al., 2022), or image texture-related information, maybe denoting microscopic processes such as underlying demyelination (Gilmore et al., 2009). Of note, these more general aspects of the image deserve further research and, anyway cannot be summarised as presence/absence of lesions and/or atrophy in a given point.
In any case, our findings strongly support the use of DL-based methods to perform classification/prediction tasks where the input data is derived from images, in line with the superiority observed by our CNN with respect to the LR-model. This confirms the potential of DL-based models to unveil key aspects of the disease which are uncatchable by the human eye. Future studies focusing on unveiling these aspects are therefore warranted.
Potential implications for clinical practice of our findings may include, in the short term, the application of CNN-based models to automatically classify patients according to their disability status using only routinely acquired brain MRI scans. This may be extremely useful in situations where large-scale therapeutic interventions, which may vary depending on the disability status of a given patient population, need to be planned in a relatively rapid manner. Other, mid-term implications include those derived from the development of CNN-based models to predict future disability, which may be extremely powerful to manage patient expectations and design tailored treatment strategies. That is, we should acknowledge that this is a cross-sectional study and, therefore, no strong statements about prediction of disease prognosis based on our specific CNN model can be made. However, we believe that our findings will help build powerful predictive models, possibly focusing on those areas identified as highly relevant through the attention map analyses. Up to now, most of those CNN-based models for future prediction that have been published so far show a relatively limited accuracy, implying that more research is sorely needed (Roca et al., 2020, Storelli et al., 2022, Tousignant et al., 2019).
Among the methodological considerations and possible limitations of our study, it is worth mentioning the relatively small sample size, considering the data needs of DL-based models. For this reason and to make the most of the data available, we did not apply any restriction to the disease duration of our patients and considered each individual scan which could be matched to an EDSS score as an independent piece of information. As future work, we would like to analyse the impact of adding clinical data, such as disease duration, age at CIS or sex, on model performance. Additionally, we applied data augmentation strategies when possible. As a result of all these strategies, the final (in-house) database used for training, overall, had enough variability, proving robustness and generalisation of our models to scans from the same vendor. Even though the data from the different MR scanner models used for training were not balanced, they were so in terms of strength field. This was confirmed by the excellent reproducibility of the models when we applied them to the external (i.e., completely unseen) validation cohort, despite the fact the disability score was not the same than the one used for training. Another remark relates to the fact that we did not account for potential disease-modifying treatment effects. Future studies accounting for these are therefore warranted. Concerning the attention maps, they are limited by the lack of a ground truth. Surely, other methodological choices could have been made, providing (probably) slightly different results, which deserve further research. Moreover, we tried to relate the attention maps with well-known biomarkers of the disease, whereas future studies should investigate the association with new, possibly more promising, biomarkers.
5. Conclusions
In conclusion, our CNN model was able to stratify MS patients based on their disability score solely using a single time-point brain MRI (T1-w and T2-FLAIR sequences) providing an excellent performance. Furthermore, our findings bring to light the potential of DL-based models to provide key information about the mechanisms responsible for the accumulation of disability in MS, suggesting the relevant role of frontotemporal cortex and cerebellum for the development of irreversible disability. Importantly, these findings may have immediate and especially long-term implications for clinical practice, laying the foundations for building powerful predictive models of future disability.
CRediT authorship contribution statement
Llucia Coll: Conceptualization, Methodology, Software, Data curation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. Deborah Pareto: Supervision, Conceptualization, Writing – original draft, Writing – review & editing. Pere Carbonell-Mirabent: Writing – original draft. Álvaro Cobo-Calvo: Writing – original draft. Georgina Arrambide: Writing – original draft. Ángela Vidal-Jordana: Writing – original draft. Manuel Comabella: Writing – original draft. Joaquín Castilló: Writing – original draft. Breogán Rodríguez-Acevedo: Writing – original draft. Ana Zabalza: Writing – original draft. Ingrid Galán: Writing – original draft. Luciana Midaglia: Writing – original draft. Carlos Nos: Writing – original draft. Annalaura Salerno: Writing – original draft. Cristina Auger: Writing – original draft. Manel Alberich: Writing – original draft. Jaume Sastre-Garriga: Writing – review & editing. Arnau Oliver: Conceptualization, Writing – review & editing. Xavier Montalban: Writing – review & editing. Àlex Rovira: Writing – review & editing. Mar Tintoré: Writing – review & editing. Xavier Lladó: Supervision, Conceptualization, Writing – original draft, Writing – review & editing. Carmen Tur: Supervision, Conceptualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Ll. Coll: nothing to disclose. D. Pareto: has received a research contract with Biogen Idec, and a grant from Instituto Salud Carlos III (PI18/00823). P. Carbonell-Mirabent: his yearly salary is supported by a grant from Biogen to Fundació privada Cemcat for statistical analysis. A. Cobo-Calvo: has received grant from Instituto de Salud Carlos III, Spain; JR19/00007. G. Arrambide: has received compensation for consulting services, participation in advisory boards or speaking honoraria from Merck, Roche, and Horizon Therapeutics; and travel support for scientific meetings from Novartis, Roche, and ECTRIMS. G. Arrambide is editor for Europe of the Multiple Sclerosis Journal – Experimental, Translational and Clinical; a member of the executive committee of the International Women in Multiple Sclerosis (iWiMS) network, and a member of the European Biomarkers in MS (BioMS-eu) consortium steering committee. She is a recipient of grants PI19/01590 and PI22/01570, awarded by the Instituto de Salud Carlos III (ISCIII), Ministerio de Ciencia e Innovación de España. Á. Vidal Jordana: has engaged in consulting and/or participated as speaker in events organized by Roche, Novartis, Merck, and Sanofi. M. Comabella: has received compensation for consulting services and speaking honoraria from Bayer Schering Pharma, Merk Serono, Biogen-Idec, Teva Pharmaceuticals, Sanofi-Aventis, and Novartis. J. Castilló: nothing to disclose. B. Rodríguez-Acevedo: has received honoraria for consulting services from Wellspect. A. Zabalza: nothing to disclose. I. Galán: nothing to disclose. L. Midaglia: nothing to disclose. C. Nos: has received funding for travel from Biogen Idec and F. Hoffmann-La Roche, Ltd. and speaker honoraria from Novartis. A. Salerno: nothing to disclose. C. Auger: has received speaking honoraria from Novartis, Biogen and Stendhal. M. Alberich: nothing to disclose. J. Río: has received speaking honoraria and personal compensation for participating on Advisory Boards from Biogen-Idec, Genzyme, Merck- Serono, Mylan, Novartis, Roche, Teva, and Sanofi-Aventis. J. Sastre-Garriga: serves as co-Editor for Europe on the editorial board of Multiple Sclerosis Journal and as Editor-in-Chief in Revista de Neurología, receives research support from Fondo de Investigaciones Sanitarias (19/950) and has served as a consultant/speaker for Biogen, Celgene/Bristol Meyers Squibb, Genzyme, Novartis and Merck. A. Oliver: nothing to disclose. X. Montalban: has received speaking honoraria and travel expenses for participation in scientific meetings, has been a steering committee member of clinical trials or participated in advisory boards of clinical trials in the past years with Abbvie, Actelion, Alexion, Biogen, Bristol-Myers Squibb/Celgene, EMD Serono, Genzyme, Hoffmann-La Roche, Immunic, Janssen Pharmaceuticals, Medday, Merck, Mylan, Nervgen, Novartis, Sandoz, Sanofi-Genzyme, Teva Pharmaceutical, TG Therapeutics, Excemed, MSIF and NMSS. A. Rovira: serves on scientific advisory boards for Novartis, Sanofi-Genzyme, Synthetic MR, Roche, Biogen, and OLEA Medical; has received speaker honoraria from Bayer, SanofiGenzyme, Merck-Serono, Teva Pharmaceutical Industries Ltd, Novartis, Roche, and Biogen; and is CMO and co-founder of TensorMedical. M. Tintoré: has received compensation for consulting services and speaking honoraria from Almirall, Bayer Schering Pharma, Biogen-Idec, Genzyme, Merck-Serono, Novartis, Roche, Sanofi-Aventis, and Teva Pharmaceuticals. MT is former co-editor of Multiple Sclerosis Journal. X. Lladó: is currently being supported by the ICREA Academia Program. He has also received support from the DPI2020-114769RBI00 project funded by the Ministerio de Ciencia, Innovación y Universidades. C. Tur: is currently being funded by a Junior Leader La Caixa Fellowship (fellowship code is LCF/BQ/PI20/11760008), awarded by “la Caixa” Foundation (ID 100010434). She has also received the 2021 Merck’s Award for the Investigation in MS, awarded by Fundación Merck Salud (Spain) and a grant awarded by the Instituto de Salud Carlos III (ISCIII), Ministerio de Ciencia e Innovación de España (PI21/01860). In 2015, she received an ECTRIMS Post-doctoral Research Fellowship and has received funding from the UK MS Society. She is a member of the Editorial Board of Neurology. She has also received honoraria from Roche and Novartis and is a steering committee member of the O’HAND trial and of the Consensus group on Follow-on DMTs.
Acknowledgments
Acknowledgments
MS PATHS is funded by Biogen. This study has been possible thanks to a Junior Leader La Caixa Fellowship awarded to C. Tur (fellowship code is LCF/BQ/PI20/11760008) by “la Caixa” Foundation (ID 100010434). The salaries of C. Tur and Ll. Coll are covered by this award.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2023.103376.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Amato M.P., Portaccio E., Stromillo M.L., Goretti B., Zipoli V., Siracusa G., Battaglini M., Giorgio A., Bartolozzi M.L., Guidi L., Sorbi S., Federico A., de Stefano N. Cognitive assessment and quantitative magnetic resonance metrics can help to identify benign multiple sclerosis. Neurology. 2008;71(9):632–638. doi: 10.1212/01.wnl.0000324621.58447.00. [DOI] [PubMed] [Google Scholar]
- Bach S., Binder A., Montavon G., Klauschen F., Müller K.-R., Samek W., Suarez O.D. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS One. 2015;10(7):e0130140. doi: 10.1371/journal.pone.0130140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhle M., Eitel F., Weygandt M., Ritter K. Layer-wise relevance propagation for explaining deep neural network decisions in MRI-based Alzheimer’s disease classification. Front. Aging Neurosci. 2019;10 doi: 10.3389/fnagi.2019.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacchi R., Meani A., Pagani E., Marchesi O., Filippi M., Rocca M.A. The role of cerebellar damage in explaining disability and cognition in multiple sclerosis phenotypes: a multiparametric MRI study. J. Neurol. 2022;269(7):3841–3857. doi: 10.1007/s00415-022-11021-1. [DOI] [PubMed] [Google Scholar]
- Bove R., Poole S., Cuneo R., Gupta S., Sabatino J., Harms M., Cooper T., Rowles W., Miller N., Gomez R., Lincoln R., McPolin K., Powers K., Santaniello A., Renschen A., Bevan C.J., Gelfand J.M., Goodin D.S., Guo C.-Y., Romeo A.R., Hauser S.L., Campbell Cree B.A. Remote observational research for multiple sclerosis: a natural experiment. Neurology(R) Neuroimmunol. Neuroinflamm. 2023;10(2):e200070. doi: 10.1212/NXI.0000000000200070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W.L., Pardini M., Brownlee W.J., Fernando K., Samson R.S., Prados Carrasco F., Ourselin S., Gandini Wheeler-Kingshott C.A.M., Miller D.H., Chard D.T. An abnormal periventricular magnetization transfer ratio gradient occurs early in multiple sclerosis. Brain. 2017;140(2):387–398. doi: 10.1093/brain/aww296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese M., Castellaro M. Cortical Gray Matter MR Imaging in Multiple Sclerosis. Neuroimag. Clin. N. Am. 2017;27(2):301–312. doi: 10.1016/J.NIC.2016.12.009. [DOI] [PubMed] [Google Scholar]
- Collorone S., Prados F., Kanber B., Cawley N.M., Tur C., Grussu F., Solanky B.S., Yiannakas M., Davagnanam I., Gandini Wheeler-Kingshott C.A.M., Barkhof F., Ciccarelli O., Toosy A.T. Brain microstructural and metabolic alterations detected in vivo at onset of the first demyelinating event. Brain. 2021;144:1409–1421. doi: 10.1093/brain/awab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordano C., Nourbakhsh B., Yiu H.H., Papinutto N., Caverzasi E., Abdelhak A., Oertel F.C., Beaudry-Richard A., Santaniello A., Sacco S., Bennett D.J., Gomez A., Sigurdson C.J., Hauser S.L., Magliozzi R., Cree B.A.C., Henry R.G., Green A.J. Differences in age-related retinal and cortical atrophy rates in multiple sclerosis. Neurology. 2022;99(15) doi: 10.1212/WNL.0000000000200977. e1685–e1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F., Brusini L., Zucchelli M., Retuci Pinheiro G., Setti F., Boscolo Galazzo I., Deriche R., Rittner L., Calabrese M., Menegaz G. Interpretable deep learning as a means for decrypting disease signature in multiple sclerosis. J. Neural Eng. 2021;18(4):0460a6. doi: 10.1088/1741-2552/ac0f4b. [DOI] [PubMed] [Google Scholar]
- de Stefano, N., Matthews, P. M., Filippi, M., Agosta, F., de Luca, M., Bartolozzi, M. L., Guidi, L., Ghezzi, A., Montanari, E., Cifelli, A., Federico, A., & Smith, S. M. (2003). Evidence of early cortical atrophy in MS. Neurology, 60(7), 1157 LP – 1162. https://doi.org/10.1212/01.WNL.0000055926.69643.03. [DOI] [PubMed]
- Eitel F., Soehler E., Bellmann-Strobl J., Brandt A.U., Ruprecht K., Giess R.M., Kuchling J., Asseyer S., Weygandt M., Haynes J.D., Scheel M., Paul F., Ritter K. Uncovering convolutional neural network decisions for diagnosing multiple sclerosis on conventional MRI using layer-wise relevance propagation. NeuroImage: Clin. 2019;24 doi: 10.1016/j.nicl.2019.102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A., Prados F., Brownlee W.J., Altmann D.R., Tur C., Cardoso M.J., De Angelis F., van de Pavert S.H., Cawley N., De Stefano N., Stromillo M.L., Battaglini M., Ruggieri S., Gasperini C., Filippi M., Rocca M.A., Rovira A., Sastre-Garriga J., Vrenken H., Leurs C.E., Killestein J., Pirpamer L., Enzinger C., Ourselin S., Wheeler-Kingshott C.A.M.G., Chard D., Thompson A.J., Alexander D.C., Barkhof F., Ciccarelli O. Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann. Neurol. 2018;83(2):210–222. doi: 10.1002/ana.25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore C.P., Donaldson I., Bo L., Owens T., Lowe J., Evangelou N. Regional variations in the extent and pattern of grey matter demyelination in multiple sclerosis: a comparison between the cerebral cortex, cerebellar cortex, deep grey matter nuclei and the spinal cord. J. Neurol. Neurosurg. Psychiatry. 2009;80(2):182–187. doi: 10.1136/jnnp.2008.148767. [DOI] [PubMed] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- Henschel L., Conjeti S., Estrada S., Diers K., Fischl B., Reuter M. FastSurfer – A fast and accurate deep learning based neuroimaging pipeline. Neuroimage. 2020;219 doi: 10.1016/j.neuroimage.2020.117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee F., Schell M., Pflueger I., Brugnara G., Bonekamp D., Neuberger U., Wick A., Schlemmer H.P., Heiland S., Wick W., Bendszus M., Maier-Hein K.H., Kickingereder P. Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp. 2019;40(17):4952–4964. doi: 10.1002/hbm.24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5(2):143–156. doi: 10.1016/S1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Kingma D.P., Ba J.L. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. 2015. Adam: A method for stochastic optimization. [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis. Neurology. 1983;33(11):1444–1452. doi: 10.1212/WNL.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Learmonth Y.C., Motl R.W., Sandroff B.M., Pula J.H., Cadavid D. Validation of patient determined disease steps (PDDS) scale scores in persons with multiple sclerosis. BMC Neurol. 2013;13:37. doi: 10.1186/1471-2377-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C., Liu M., Wang L., Shen D. Multi-task weakly-supervised attention network for dementia status estimation with structural MRI. IEEE Trans. Neural Networks Learn. Syst. 2021;33(8):4056–4068. doi: 10.1109/TNNLS.2021.3055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina A., Ropele S., Sibgatulin R., Reichenbach J.R., Güllmar D. Investigation of deep-learning-driven identification of multiple sclerosis patients based on susceptibility-weighted images using relevance analysis. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.609468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O. Lungu P. Pantano F. Kumfor A. Gallo D. Joel Shaw M.R. Cek K. Czekóová K. Saxunová M. Dufek Marě cek, R., VaníčekVaníˇVaníček, J., & Brázdil, M. Impaired Self-Other Distinction and Subcortical Gray-Matter Alterations Characterize Socio-Cognitive Disturbances in Multiple Sclerosis Front. Neurol 10 2019 525 10.3389/fneur.2019.00525. [DOI] [PMC free article] [PubMed]
- Madsen M.A.J., Wiggermann V., Marques M.F.M., Lundell H., Cerri S., Puonti O., Blinkenberg M., Romme Christensen J., Sellebjerg F., Siebner H.R. Linking lesions in sensorimotor cortex to contralateral hand function in multiple sclerosis: a 7 T MRI study. Brain. 2022;10(145):3522–3535. doi: 10.1093/brain/awac203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzullo A., Kocevar G., Stamile C., Durand-Dubief F., Terracina G., Calimeri F., Sappey-Marinier D. Classification of multiple sclerosis clinical profiles via graph convolutional neural networks. Front. Neurosci. 2019;13(JUN):1–15. doi: 10.3389/fnins.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry E.M., Bermel R.A., Williams J.R., Benzinger T.L.S., de Moor C., Fisher E., Hersh C.M., Hyland M.H., Izbudak I., Jones S.E., Kieseier B.C., Kitzler H.H., Krupp L., Lui Y.W., Montalban X., Naismith R.T., Nicholas J.A., Pellegrini F., Rovira A., Schulze M., Tackenberg B., Tintore M., Tivarus M.E., Ziemssen T., Rudick R.A. Harnessing Real-World Data to Inform Decision-Making: Multiple Sclerosis Partners Advancing Technology and Health Solutions (MS PATHS) Front. Neurol. 2020;11 doi: 10.3389/fneur.2020.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palesi F., Tournier J.-D., Calamante F., Muhlert N., Castellazzi G., Chard D., D’Angelo E., Wheeler-Kingshott C.A.M. Contralateral cerebello-thalamo-cortical pathways with prominent involvement of associative areas in humans in vivo. Brain Struct. Funct. 2015;220(6):3369–3384. doi: 10.1007/s00429-014-0861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., Killeen, T., Lin, Z., Gimelshein, N., Antiga, L., Desmaison, A., Köpf, A., Yang, E., DeVito, Z., Raison, M., Tejani, A., Chilamkurthy, S., Steiner, B., Fang, L., … Chintala, S. (2019). PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the 33rd International Conference on Neural Information Processing Systems (pp. 8026–8037). Curran Associates Inc. https://doi.org/10.5555/3454287.3455008.
- Prados F., Cardoso M.J., Kanber B., Ciccarelli O., Kapoor R., Gandini Wheeler-Kingshott C.A.M., Ourselin S. A multi-time-point modality-agnostic patch-based method for lesion filling in multiple sclerosis. Neuroimage. 2016;139:376–384. doi: 10.1016/j.neuroimage.2016.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M.A., Hadjimichael O.C., Preiningerova J., Vollmer T.L. Prevalence and treatment of spasticity reported by multiple sclerosis patients. Mult. Scler. J. 2004;10(5):589–595. doi: 10.1191/1352458504ms1085oa. [DOI] [PubMed] [Google Scholar]
- Roca P., Attye A., Colas L., Tucholka A., Rubini P., Cackowski S., Ding J., Budzik J.-F., Renard F., Doyle S., Barbier E.L., Bousaid I., Casey R., Vukusic S., Lassau N., Verclytte S., Cotton F., Brochet B., Casey R., Cotton F., De Sèze J., Douek P., Guillemin F., Laplaud D., Lebrun-Frenay C., Mansuy L., Moreau T., Olaiz J., Pelletier J., Rigaud-Bully C., Stankoff B., Vukusic S., Marignier R., Debouverie M., Edan G., Ciron J., Ruet A., Collongues N., Lubetzki C., Vermersch P., Labauge P., Defer G., Cohen M., Fromont A., Wiertlewsky S., Berger E., Clavelou P., Audoin B., Giannesini C., Gout O., Thouvenot E., Heinzlef O., Al-Khedr A., Bourre B., Casez O., Cabre P., Montcuquet A., Créange A., Camdessanché J.-P., Faure J., Maurousset A., Patry I., Hankiewicz K., Pottier C., Maubeuge N., Labeyrie C., Nifle C., Ameli R., Anxionnat R., Attye A., Bannier E., Barillot C., Ben Salem D., Boncoeur-Martel M.-P., Bonneville F., Boutet C., Brisset J.-C., Cervenanski F., Claise B., Commowick O., Constans J.-M., Dardel P., Desal H., Dousset V., Durand-Dubief F., Ferre J.-C., Gerardin E., Glattard T., Grand S., Grenier T., Guillevin R., Guttmann C., Krainik A., Kremer S., Lion S., Menjot de Champfleur N., Mondot L., Outteryck O., Pyatigorskaya N., Pruvo J.-P., Rabaste S., Ranjeva J.-P., Roch J.-A., Sadik J.C., Sappey-Marinier D., Savatovsky J., Tanguy J.-Y., Tourbah A., Tourdias T. Artificial intelligence to predict clinical disability in patients with multiple sclerosis using FLAIR MRI. Diagn. Interv. Imaging. 2020;101(12):795–802. doi: 10.1016/j.diii.2020.05.009. [DOI] [PubMed] [Google Scholar]
- Rogers B.P., Parks M.H., Nickel M.K., Katwal S.B., Martin P.R. Reduced fronto-cerebellar functional connectivity in chronic alcoholic patients. Alcohol Clin Exp Res. 2012;36(2):294–301. doi: 10.1111/j.1530-0277.2011.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre-Garriga J., Pareto D., Battaglini M., Rocca M.A., Ciccarelli O., Enzinger C., Wuerfel J., Sormani M.P., Barkhof F., Yousry T.A., De Stefano N., Tintoré M., Filippi M., Gasperini C., Kappos L., Río J., Frederiksen J., Palace J., Vrenken H., Montalban X., Rovira À. MAGNIMS consensus recommendations on the use of brain and spinal cord atrophy measures in clinical practice. Nat. Rev. Neurol. 2020;16(3):171–182. doi: 10.1038/s41582-020-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G. Savini M. Pardini G. Castellazzi A. Lascialfari D. Chard E. D’Angelo C.A.M. Gandini Wheeler-Kingshott Default Mode Network Structural Integrity and Cerebellar Connectivity Predict Information Processing Speed Deficit in Multiple Sclerosis Front. Cell. Neurosci. 13. [DOI] [PMC free article] [PubMed]
- Schmidt P., Gaser C., Arsic M., Buck D., Förschler A., Berthele A., Hoshi M., Ilg R., Schmid V.J., Zimmer C., Hemmer B., Mühlau M. An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis. Neuroimage. 2012;59(4):3774–3783. doi: 10.1016/J.NEUROIMAGE.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Signori A., Lorscheider J., Vukusic S., Trojano M., Iaffaldano P., Hillert J., Hyde R., Pellegrini F., Magyari M., Koch-Henriksen N., Sørensen P.S., Spelman T., van der Walt A., Horakova D., Havrdova E., Girard M., Eichau S., Grand'Maison F., Gerlach O., Terzi M., Ozakbas S., Skibina O., Van Pesch V., Sa M.J., Prevost J., Alroughani R., McCombe P.A., Gouider R., Mrabet S., Castillo-Trivino T., Zhu C., de Gans K., Sánchez-Menoyo J.L., Yamout B., Khoury S., Sormani M.P., Kalincik T., Butzkueven H. Heterogeneity on long-term disability trajectories in patients with secondary progressive MS: a latent class analysis from Big MS Data network. J Neurol Neurosurg Psychiatry. 2023;94(1):23–30. doi: 10.1136/jnnp-2022-329987. [DOI] [PubMed] [Google Scholar]
- Simonyan, K., Vedaldi, A., & Zisserman, A. (2013). Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps. ICLR 2014. https://doi.org/10.48550/arxiv.1312.6034.
- Springenberg, J. T., Dosovitskiy, A., Brox, T., & Riedmiller, M. (2014). Striving for Simplicity: The All Convolutional Net. ICLR 2015. https://doi.org/10.48550/arxiv.1412.6806.
- Storelli L., Azzimonti M., Gueye M., Vizzino C., Preziosa P., Tedeschi G., de Stefano N., Pantano P., Filippi M., Rocca M.A. A Deep Learning Approach to Predicting Disease Progression in Multiple Sclerosis Using Magnetic Resonance Imaging. Invest. Radiol. 2022;57(7):423–432. doi: 10.1097/RLI.0000000000000854. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Baranzini S.E., Geurts J., Hemmer B., Ciccarelli O. Multiple sclerosis. Lancet. 2018;391(10130):1622–1636. doi: 10.1016/S0140-6736(18)30481-1. [DOI] [PubMed] [Google Scholar]
- Tintoré M., Rovira A., Río J., Nos C., Grivé E., Téllez N., Pelayo R., Comabella M., Sastre-Garriga J., Montalban X. Baseline MRI predicts future attacks and disability in clinically isolated syndromes. Neurology. 2006;67(6):968–972. doi: 10.1212/01.wnl.0000237354.10144.ec. [DOI] [PubMed] [Google Scholar]
- Tintore M., Rovira A., Arrambide G., Mitjana R., Río J., Auger C., Nos C., Edo M.C., Castilló J., Horga A., Perez-Miralles F., Huerga E., Comabella M., Sastre-Garriga J., Montalban X. Brainstem lesions in clinically isolated syndromes. Neurology. 2010;75(21):1933–1938. doi: 10.1212/WNL.0b013e3181feb26f. [DOI] [PubMed] [Google Scholar]
- Tintore M., Rovira L., Río J., Otero-Romero S., Arrambide G., Tur C., Comabella M., Nos C., Jesú Arévalo M., Negrotto L., Galán I., Vidal-Jordana A., Castilló J., Palavra F., Simon E., Mitjana R., Auger C., Sastre-Garriga J., Montalban X. Defining high, medium and low impact prognostic factors for developing multiple sclerosis. Brain. 2015;138(Pt 7):1863–1874. doi: 10.1093/brain/awv105. [DOI] [PubMed] [Google Scholar]
- Tousignant A., Lemaître P., Doina P., Arnold D.L., Arbel T. Prediction of Disease Progression in Multiple Sclerosis Patients using Deep Learning Analysis of MRI Data. Proc. Mach. Learn. Res. 2019;102:483–492. http://proceedings.mlr.press/v102/tousignant19a.html [Google Scholar]
- Tur C., Eshaghi A., Altmann D.R., Jenkins T.M., Prados F., Grussu F., Charalambous T., Schmidt A., Ourselin S., Clayden J.D., Wheeler-Kingshott C.A.M.G., Thompson A.J., Ciccarelli O., Toosy A.T. Structural cortical network reorganization associated with early conversion to multiple sclerosis. Sci. Rep. 2018;8(1):10715. doi: 10.1038/s41598-018-29017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tur C., Moccia M., Barkhof F., Chataway J., Sastre-Garriga J., Thompson A.J., Ciccarelli O. Assessing treatment outcomes in multiple sclerosis trials and in the clinical setting. Nat. Rev. Neurol. 2018;14(2):75–93. doi: 10.1038/nrneurol.2017.171. [DOI] [PubMed] [Google Scholar]
- Tur C., Kanber B., Eshaghi A., Altmann D.R., Khaleeli Z., Prados F., Ourselin S., Thompson A.J., Gandini Wheeler-Kingshott C.A.M., Toosy A.T., Ciccarelli O. Clinical relevance of cortical network dynamics in early primary progressive MS. Mult. Scler. J. 2019;26(4):442–456. doi: 10.1177/1352458519831400. [DOI] [PubMed] [Google Scholar]
- Tur C., Grussu F., de Angelis F., Prados F., Kanber B., Calvi A., Eshaghi A., Charalambous T., Cortese R., Chard D.T., Chataway J., Thompson A.J., Ciccarelli O., Gandini Wheeler-Kingshott C.A.M. Spatial patterns of brain lesions assessed through covariance estimations of lesional voxels in multiple Sclerosis: the SPACE-MS technique. NeuroImage: Clinical. 2022;33 doi: 10.1016/J.NICL.2021.102904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgh H.K., Schmidt R., Westeneng H.J., de Reus M.A., van den Berg L.H., van den Heuvel M.P. Deep learning predictions of survival based on MRI in amyotrophic lateral sclerosis. NeuroImage: Clinical. 2017;13:361–369. doi: 10.1016/j.nicl.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopalan J., Tong L., Hassanzadeh H.R., Wang M.D. Multimodal deep learning models for early detection of Alzheimer’s disease stage. Sci. Rep. 2021;11(1):1–13. doi: 10.1038/s41598-020-74399-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu J., Li Q., Li W., Wu N., Zheng Y., Chang H., Chen J., Wang W., Soriano-Mas C. Altered fronto-striatal and fronto-cerebellar circuits in heroin-dependent individuals: a resting-state fMRI study. PLoS One. 2013;8(3):e58098. doi: 10.1371/journal.pone.0058098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.