Abstract
In this research, the LAB and yeast isolations and identifications of homemade traditional sourdoughs were investigated. Both LAB and yeasts were identified by the PCR method and used in the production of sourdough bread (SDB). Twelve types of SDB were produced from combinations of isolated LAB and yeasts. Eleven LAB and yests were identified from 36 sourdoughs. The most frequently isolated LAB species were Lactobacillus brevis (45.0%), Pediococcus acidilactici (20.0%) and Lactobacillus plantarum (18.3%) while other LAB species were isolated by only 1.7–3.5%. On the other hand, 27.5% of the isolates were Saccharomyces cerevisiae, which was followed by Pichia kudriavzevii (25.0%) and Kluyveromyces marxianus (12.5%). The total titratable acidity and pH of the sourdough ranged from 3.10 to 34.45% and from 4.05 to 4.80 respectively. High acceptable loaf height (7.2 cm), moisture (30.05%), dry matter (69.95%), volume (1370 cm3), specific volume (4.35 cm3 g−1) and sensory value (62 scores) were determined for SDB12. Isolated LAB is mainly composed of heterofermentative species (75%). Among the bread produced with the combination of LAB and yeasts, the SDB12 was the most preferred SDB in terms of sensory analysis and other quality characteristics. The use of homofermentative and heterofermentative LAB and yeast combinations provided high-quality SDB. This would be due to the symbiotic growth of these microorganisms. The use of L. plantarum by L. brevis and by other species resulted in high-quality SDB.
Keywords: Polymerase chain reaction, Sourdough bread, Lactic acid bacteria, Yeasts
Graphical abstract
Highlights
-
•
LAB and yeasts were identified from traditional homemade sourdough.
-
•
LAB and yeasts were identified by the 23S rRNA and ITS sequences.
-
•
Some properties of sourdough breads were detected.
-
•
Better acceptable was determined in SD12 sourdough bread.
1. Introduction
Sourdough is a fermented product that is a mixture of flour and water. Yeast and lactic acid bacteria (LAB) are responsible for sourdough fermentation and it is an intermediate product in the production of sourdough bread (SDB). The use of LAB in sourdough fermentation has significantly increased globally because of their nutraceutical role (Erkmen and Bozoglu, 2016). LAB are Gram-positive rods, catalase-negative, nonmotile, non-spore-forming, obligate fermentative, microaerophilic, acidophilic, producing lactic acid as the main product during fermentation (Erkmen and Bozoglu, 2016; Perez-Alvarado et al., 2022). Yeasts have a rapid fermentative metabolism and are resistant to many stress factors in breadmaking. Isolation and identification of LAB and yeasts from traditional sourdough allow safe use as starter cultures in sourdough fermentation (Ozulku, 2019; Chiv et al., 2021; Landis et al., 2021; Bazalova et al., 2022). LAB and yeast identification rely heavily on polymerase chain reaction (PCR) methods based on barcode sequences.
Traditional homemade sourdough is mostly type I sourdough. It is a hard dough that contains a spontaneous selection of microorganisms and is produced by conventional methods; it is characterized by self-initiated fermentation at room temperature (20–30 °C) by the microbial species present in the first sourdough (Gorkem, 2019; Ozulku, 2019). The first sourdough can often be prepared using just flour and water or by adding another raw material that is naturally rich in microorganisms (such as fruits, yogurt, tripe cuts and manure). Selection of the sourdough microbiota occurs spontaneously during the daily refreshing. Daily reverse screeding is repeated three to ten times, depending on the microorganisms present at the beginning of the process and the desired sensory properties of the final product (Siepmann et al., 2018).
Different studies have been carried out on the sourdough flora in different regions of Turkey and different types of LAB and yeasts have been isolated (Dertli et al., 2016; Yagmur et al., 2016; Ispirli and Dertli, 2022; Boyaci-Gunduz and Erten, 2020). Sourdoughs with different microbiota enabled the production of SDB with varying quality characteristics. However, no studies have been conducted to identify the sourdough LAB and yeast flora from Konya, Gaziantep and Mardin cities. These cities have an ancient history dating back to people first started to live there, as well as a long history of the production of different types of SDB. This manuscript provides an important opportunity to advance the understanding of homemade traditional sourdough LAB and yeast microflora and SDB characterization by physicochemical, texture and sensory analysis. Another aim of the research is to enable the sourdough production with least refreshing (3 times) by combining appropriate LAB and yeast species.
2. Materials and methods
2.1. Sourdough sampling
Samples (n = 36) of homemade traditional sourdough were obtained from three cities in Turkey: Konya (in the central region of Turkey), Mardin and Gaziantep (in the southeast region of Turkey). Sourdoughs (n = 12) were obtained from Konya Özkan bakery and Alsancak patisserie in center, Derbent, Ilgın and Kadınhanı districts. Sourdoughs (n = 12) were obtained from the Mardin Araplar district in the center, Kızıltepe and Midyat districts. Sourdoughs (n = 12) were obtained from Gaziantep Metro Inc. in the center, Cevizler, Morcalı village and Nizip districts. All of them were type I sourdoughs. Samples were transferred into sterile containers and stored in a refrigerator for further use.
2.2. Isolation of yeasts and LAB
LAB and yeasts were isolated from sourdoughsby the methods indicated by Erkmen (2022a, 2022b). Each isolated LAB and yeast colony was transferred into 8 mL of de Man Rogosa-Sharp broth (MRS broth; Difco, Detroit) and 8 ml of potato dextrose broth (PDB; Difco, Detroit), respectively, and incubated anaerobically for LAB and aerobically for yeast at 28 °C for 48 and 72 h. Then, the isolates were purified by streak plating on the respective Petri plates. The single colony of the purified isolates was added into respective broths containing 20% (v v−1) glycerol and stored at −20 °C until use in molecular identification and SDB production.
2.3. Molecular identification of LAB and yeasts
Each of the LAB and yeast DNA was extracted using PureLinkTM genomic DNA kits (Thermo Fisher, Bleiswijk, Netherlands), according to the manufacturer’s protocols. Polymerase chain reaction (PCR) was used to amplify the LAB 23S rRNA region using ISR forward (5′ GCTGGATCACCTCCTTTC-3′) and ISR reverse (5′ CCTTTCCCTCACGGTACTG-3′) primer pairs. Yeasts were identified from the internal transcribed spacer (ITS) region by ITS1 forward (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 reverse (5′-GGAAGTAAAAGTCGTAACAAGG-3′) primers. The PCR reactions were conducted (Saitou and Nei, 1987). Both strands of DNA sequences were FASTA formatted and homology was determined using BLAST within the Biotechnology Information National Center database. The isolated LAB and yeast species were identified using the neighbor-joining method (Saitou and Nei, 1987; Nei and Kumar, 2000), and analyses were conducted in MEGA 11 version (Tamura et al., 2021). Identified LAB and yeast species were given in Table 1.
Table 1.
Types of LAB and yeast species identified from sourdoughs by PCR analysis.
| LAB | % (n = 60) | Yeasts | % (n = 40) |
|---|---|---|---|
| Lactobacillus brevis | 45.0 | Saccharomyces cerevisiae | 27.50 |
| Pediococcus acidilactici | 20.0 | Pichia kudriavzevii | 25.00 |
| Lactobacillus plantarum | 18.3 | Kluyveromyces marxianus | 12.50 |
| Lactobacillus pentosus | 3.3 | Geotrichum candidum | 7.50 |
| Lactobacillus paraplantarum | 1.7 | Kazachstania humilis | 5.00 |
| Lactobacillus paralimentarius | 1.7 | Kazachstania unispora | 5.00 |
| Weissella confusa | 1.7 | Wickerhamomyces anomalus | 5.00 |
| Enterococcus hirae | 3.3 | Candida kefyr | 2.50 |
| Enterococcus faecalis | 1.7 | Candida glabrata | 5.00 |
| L. mesenteroides subsp. mesenteroides | 1.7 | Galactomyces candidum | 2.50 |
| L. mesenteroides subsp. cremoris | 1.7 | Candida tropicalis | 2.50 |
2.4. Sourdough bread production
Sourdough preparation. Isolated LAB and yeast species from sourdoughs were grown separately in 100 ml of MRS broth and PDB respectively. After amplification, the cultures were centrifuged (6000×g, 15 min). The pellets of cultures were washed twice with physiological saline solution (PSS) and then centrifuged again. Stock cultures were prepared by resuspending pellet in PSS. Experimental culture (EC) was prepared by mixing equal amounts of each stock microbial culture in different combinations (Table 2). Isolated Lactobacillus paralimentarius and Weissella confusa were not used in the production of SDB.
Table 2.
Sourdough starter culture combinations in the production of sourdough breads.
| City | Sourdough | Combination of sourdough starter cultures |
|---|---|---|
| Gaziantep |
SD1 | L.paraplantarum + L.brevis + S.cerevisiae |
| SD2 | L.paraplantarum + L.pentosus + P.kudriavzevii | |
| SD3 | L.plantarum + L.brevis + E.hirae + W anomalus | |
| SD4 | P.acidilactici + L.plantarum + E.faecalis + C.glabrata + C.tropicalis | |
| SD5 |
L.pentosus + L.brevis + P.kudriavzevii |
|
| Konya |
SD6 | L.pentosus + L.plantarum + G.candidum |
| SD7 | P.acidilactici + E. hirae + L. brevis + C. keyfr + K. unispora | |
| SD8 |
L.plantarum + L.brevis + E.hirae + L.pentosus + P.kudriavzevii + K.marxianus + K.unispora |
|
| Mardin | SD9 | L.mesenteroides subsp. cremoris + L.brevis + S. cerevisiae |
| SD10 | L.brevis + L. plantarum + K. marxianus | |
| SD11 | L.mesenteroides subsp. cremoris + L.plantarum + P.acidilactici + K.marxianus | |
| SD12 | L.brevis + L.mesenteroides subsp. mesenteroides + P.acidilactici + L.plantarum + S.cerevisiae + K. marxianus |
Wheat (Golia type wheat) flour (WF) was supplied from Özmen Flour Industry and Trade Inc. (Gaziantep, Turkey). The following ingredients, based on their percent composition, were used in the preparation of mother sourdough culture (MSC): 100 g WF, 1.5 g NaCl, 200 mL topwater, 1.5 mL LAB EC, and 1.5 mL yeast EC. Twelve types of sourdough (named from SD1 to SD12) in triplicate were produced from combinations of LAB and yeasts (Table 2) and they used in the production of sourdough bread. Ingredients except culture were added to the orbital mixer (HY-10M, Mateka, Tekirda) except culture, mixed for a while, and kneaded at 400 rpm for 5 min. Then the microbial cultures were added to the mixture and mixed for 2 min. The kneading process was continued for 10–15 min. After the kneading, it was left to ferment at 28 °C for 24 h. With the following steps, three enrichment (refreshing) step through intermediate culture (IMC) were performed.
-
1
Enrichment: 100 g of WF was mixed in 205 mL of water in the mixer, and then 60 g of MSC was added and mixed. The mixture was allowed to ferment for 24 h at 28 °C. It was used as IMC-1.
-
2
Enrichment: 135 g of WF was mixed in 200 mL of water in the mixer, and then 65 g of IMC-1 was added to it and mixed. The mixture was allowed to ferment for 24 h at 28 °C. It was used as IMC-2.
-
3
Enrichment: 135 g of WF was mixed in 200 mL of water in the mixer, and then 65 g of IMC-2 was added and mixed. The mixture was allowed to ferment for 24 h at 28 °C. It was used as IMC-3.
2.4.1. Sourdough bread
The SD was prepared in the production of SDB using 900 g WF, 1.5 g salt (w w−1), 20% (w/w) IMC-3 and water. The total amount of water in the SD will be 58%, taking into account the amount of water coming from the ingredients and the remaining water added. Ingredients were added to the mixer and mixed. The SD was left to ferment for 17 h in a bamboo fermentation basket. The fermentation was carried out in two stages: aeration (at room temperature) and cold holding. The SD was aerated by folding it six times. The first folding was allowed to ferment for 30 min, and the next five foldings were allowed to ferment for 1 h. After the sixth folding, the SD in a bamboo basket was placed in a cold room (at 4 °C) for 11.5 h of fermentation. The fermented SD is divided into 350 g pieces and rolled. The rolled SD was left to rest for 10 min. After resting, the SD was shaped. The SD was baked in a baking oven at 240 °C for 25 min. One of the SD from each type was used as sample. Twelve types of SDB (named frm SDB1 to SDB12) in duplicate were produced from 12 types of the SD (Table 2). In the preparation of control bread, commercial yeast was used instead of SDB. After baking, it was cooled to 25 °C (within 1 h), wrapped in a bread cloth, and stored in the refrigerator (4 °C) in the bread basket. Samples were taken from the baked SDB after 1 and 4 h and 1, 3, and 7 days and samples were analyzed within 1 min and remaining SDB was placed back in the storage room.
2.4.1.1. Analysis
2.4.1.1.1. Physicochemical analysis
pH and titratable acidity (TTA) for SD were detected as indicated by Gul et al. (2005). Moisture and dry matter properties were determined on 1 h after production of SDB by keeping them in an oven at 105 °C until a constant weight was reached and the dry matter was calculated (Gul et al., 2005). Each loaf maximum height point was measured in cm on SDB after baking (Park et al., 2017). Bread volume and weight were measured on 1 h SDB after baking (Elgun and Ertugay, 2002).
2.4.1.1.2. Sensory analysis of sourdough bread
The sensory characteristics of SDB were determined by using a group of 10 trained panelists in the Gaziantep University Department of Food Engineering sensory analysis laboratory. Panelist analyses were conducted after 4 and 24 h and after 7 days of stored SDB. The panelists determined the SDB external properties (shape, volume, crust properties, and crust color) and internal properties (crumb dryness, taste, color, wetness, swallowability, acidity odor, tissue softness, pore size, chewing properties, and general acceptability). Sensory analysis of SD has been performed by the method indicated by Gul et al. (2005). Intensities evaluated by panelists were scored on a 15-point numerical scale divided into half-point increments, with 0 meaning none and 15 meaning extremely strong. Intensity scores were discussed to reach a consensus of all panelists. Two replicates of each sample, each from a different batch or loaf, were presented to panelists simultaneously, and panelists were instructed to develop one consensus profile that best represented the replicate samples. Panelists were instructed to note if samples were so different that a representative profile of the replicates was not possible, but that situation did not ocur.
2.5. Statistical analysis
SDB production was repeated three times; each repetition was run in parallel, and a parallel sample was used in each analysis. The chemical, physical, microbiological, and sensory results were evaluated by the IBM SPSS Statistic v.22 program (IBM SPSS Corporation, Chicago, IL, USA). In statistical analysis, one-way ANOVA and ANOVA tests were used. p < 0.05 was considered significant between differences.
3. Results and discussion
3.1. Genotypic characteristics of the isolated LAB and yeast species
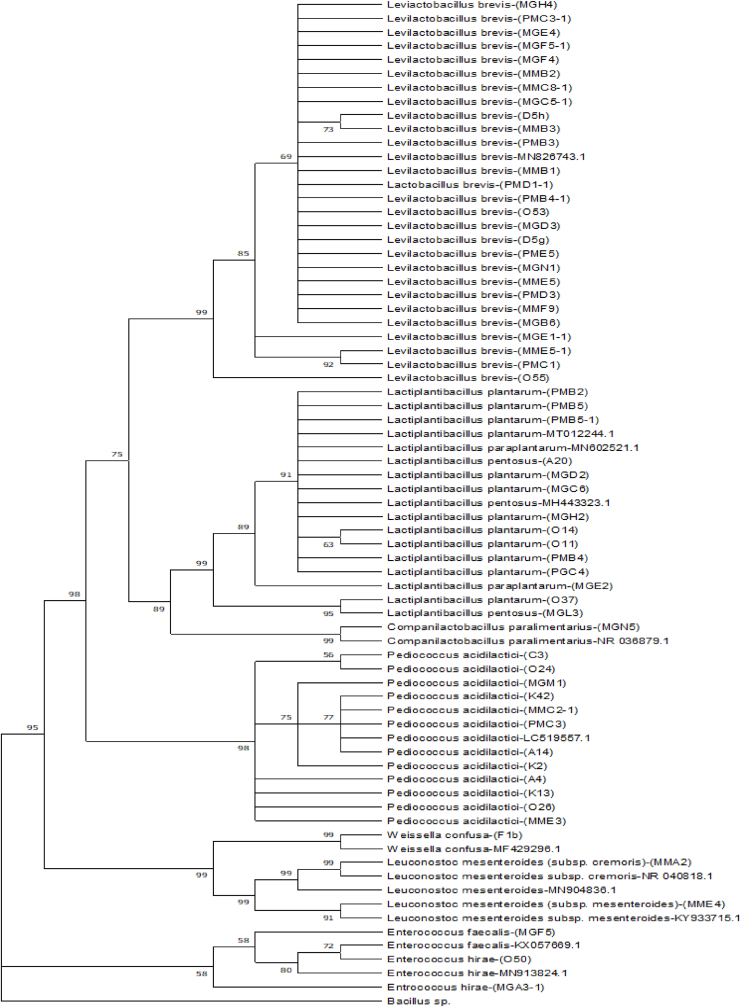
SD has a very complex microbial ecosystem (microbiota), which depends on the ecological factors of the ingredients, equipment and fermentation conditions (Meroth et al., 2003). Fig. 1 shows a sequence analysis of LAB isolates from sourdoughs using 23S rRNA. Five genera of LAB were isolated from 36 sourdoughs (Table 1); Lactobacillus (76.7%), Pediococcus (13.3%), Enterococcus (3.0%), Leuconostoc (3.3) and Weissella (1.7%). Eleven LAB species were isolated from sourdoughs (Table 1). Lactobacillus brevis (43.3%), Pediococcus acidilactici (21.7%) and Lactobacillus plantarum (18.3%) were the dominant LAB species.
Fig. 1.
23S rRNA sequence alignments of LAB strains.
Eight different LAB species were isolated from Gaziantep sourdoughs. L. brevis (47.4%) and L. plantarum (21.1%) were determined as the dominant LAB species. P. acidilactici (41.2%), L. brevis (23.5%) and L. plantarum (17.6%) were determined as the dominant species among six LAB species from Konya sourdoughs. There were 5 different LAB species in the Mardin sourdoughs; L. brevis (58.3%), L. plantarum (16.7%) and P. acidilactici (16.7%) were the dominant LAB species. The most diverse LAB species were determined from Gaziantep sourdoughs, while the least diverse were obtained from Mardin sourdoughs.
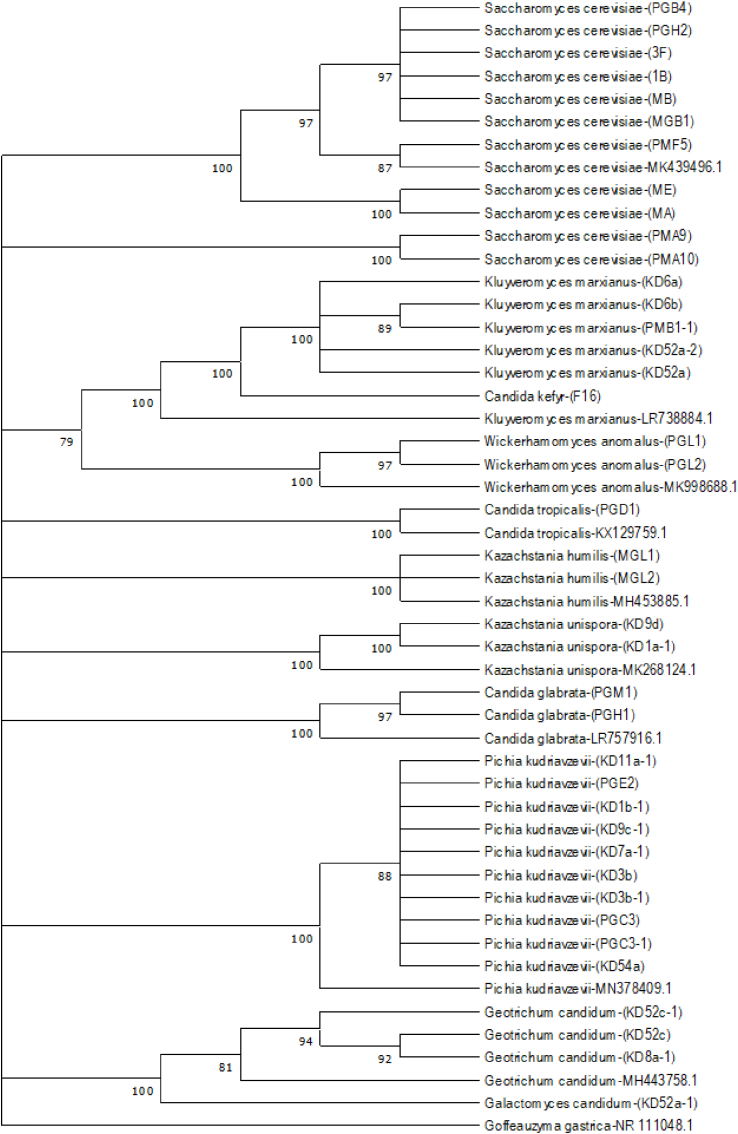
Sequence analysis of yeast isolates from sourdoughs with ITS is given in Fig. 2. Eight genera of yeasts were isolated from 36 sourdoughs (Table 1); Saccharomyces (27.5%), Pichia (25.0%), Kluyveromyces (12.5%), Candida (10.0%), Geotrichium (7.5%), Kazachstania (5.0%), Wickerhamomyces (5.0%) and Galactomyces (2.5%). Eleven yeast species were isolated. The most common yeast species were S. cerevisiae (27.5%), P. kudriavzevii (25.0%), and K. marxianus (12.5%). P. kudriavzevii (38.9%), K. marxianus (22.2%) and G. candidium (16.7%) were determined as dominant yeasts among 6 yeast species from Konya sourdoughs. P. kudriavzevii (23.1%) and S. cerevisiae (23.1%) were determined as dominant yeasts among 6 yeast species from Gaziantep sourdoughs. Only two yeast species were isolated from Mardin sourdoughs; S. cerevisiae (88.9%) and K. marxianus (11.1%).
Fig. 2.
ITS regions sequence alignments of yeast strains.
3.2. Sourdough and sourdough bread characteristics
pH and TTA of sourdoughs were given in Table 3. The SD pH varied between 4.08 and 4.80. The pH of SD was significantly (p < 0.05) reduced as the number of LAB increased. The SD with the lowest pH were SD12, SD4 and SD8. TTA of SD ranged from 3.00 to 3.50%.
Table 3.
pH and TTA of sourdough (SD).
| Sample | pH | TTA (%) |
|---|---|---|
| Control | 5.12 ± 0.03a | 2.98 ± 0.08a |
| SD1 | 4.72 ± 0.05h | 3.11 ± 0.03bc |
| SD2 | 4.66 ± 0.02f | 3.17 ± 0.07c |
| SD3 | 4.60 ± 0.03e | 3.21 ± 0.14c |
| SD4 | 4.08 ± 0.01bc | 3.45 ± 0.18de |
| SD5 | 4.70 ± 0.05gh | 3.10 ± 0.02abc |
| SD6 | 4.64 ± 0.02ef | 3.18 ± 0.03c |
| SD7 | 4.65 ± 0.02f | 3.20 ± 0.10c |
| SD8 | 4.10 ± 0.04c | 3.40 ± 0.09de |
| SD9 | 4.80 ± 0.06ı | 3.00 ± 0.03ab |
| SD10 | 4.78 ± 0.05ı | 3.09 ± 0.02abc |
| SD11 | 4.24 ± 0.01d | 3.34 ± 0.12d |
| SD12 | 4.05 ± 0.02b | 3.50 ± 0.07e |
Values mean ± standard deviation. In the columns, different small letters indicate significant difference.
Moisture and dry matter analyses were made on the 1, 3 and 7 days of storage (Table 4). The moisture content of all SDB decreased significantly (p < 0.05) during the storage period. The moisture content of the SDB varied between 21.95 and 30.05% at the end of 7 days. When homofermentative and heterofermentative LAB were used together with three or more species, SDB retained more moisture. The reason for this was the breakdown of more organic compounds (proteins, polysaccharides, etc.) in SDB as a result of the different metabolic activities of microorganisms. This increased the water-holding capacity of the SDB. The dry matter in SDB varied between 69.95 and 78.05%. The dry matter of SDB changed significantly (p < 0.05) during storage depending on the moisture content. The dry matter contents decreased as a result of the increased moisture-holding capacity of SDB.
Table 4.
Moisture and dry matter of sourdough bread (SDB).
| Sourdough bread | Moisture (%) |
Dry matter (%) |
||||
|---|---|---|---|---|---|---|
| 1 day | 3. day | 7. day | 1. day | 3. day | 7. day | |
| Control | 30.15 ± 1.62abA | 25.10 ± 1.25aB | 20.15 ± 1.10aC | 69.85 ± 3.21abA | 74.90 ± 4.12adB | 79.85 ± 4.17acC |
| SDB1 | 32.23 ± 1.84bcdA | 22.98 ± 1.05bB | 21.95 ± 1.22bC | 67.77 ± 3.18abA | 77.02 ± 4.45aB | 78.05 ± 4.15acC |
| SDB2 | 33.16 ± 1.75cdA | 27.84 ± 1.34cdB | 26.32 ± 1.35dC | 66.84 ± 3.05abA | 72.16 ± 4.14abcdB | 73.68 ± 4.05bcC |
| SDB3 | 32.14 ± 1.82bcdA | 30.76 ± 1.28efgB | 26.84 ± 1.37deC | 67.86 ± 3.12abA | 69.24 ± 4.18bcdB | 73.16 ± 4.08bcC |
| SDB4 | 34.18 ± 2.34cdA | 32.55 ± 1.45ghB | 28.63 ± 1.38efC | 65.82 ± 3.14bA | 67.45 ± 4.20bcB | 71.37 ± 4.10bC |
| SDB5 | 29.17 ± 1.26aA | 26.65 ± 1.30acB | 24.32 ± 1.24cC | 70.83 ± 3.34aA | 73.35 ± 4.34acdB | 75.68 ± 4.24abC |
| SDB6 | 33.56 ± 1.67cdA | 31.12 ± 1.32fgB | 26.87 ± 1.36deC | 66.44 ± 3.56abA | 68.88 ± 4.12bcB | 73.13 ± 4.25bcC |
| SDB7 | 31.64 ± 1.01bcA | 29.92 ± 1.26efB | 28.41 ± 1.40efC | 68.36 ± 3.26abA | 70.08 ± 3.98bcdB | 71.59 ± 3.97bC |
| SDB8 | 34.55 ± 2.24dA | 33.17 ± 1.48hB | 29.74 ± 1.42fC | 65.45 ± 3.45bA | 66.83 ± 3.78bB | 70.26 ± 3.87bC |
| SDB9 | 32.56 ± 1.73bcdA | 28.94 ± 1.37deB | 25.32 ± 1.25cdC | 67.44 ± 3.38abA | 71.06 ± 4.05bcdB | 74.68 ± 3.75abcC |
| SDB10 | 32.45 ± 1.56bcdA | 29.97 ± 1.32efB | 27.14 ± 1.36deC | 67.55 ± 3.40abA | 70.03 ± 3.90bcdB | 72.86 ± 3.78bcC |
| SDB11 | 32.87 ± 1.83cdA | 29.92 ± 1.34efB | 27.08 ± 1.36deC | 67.13 ± 3.41abA | 70.08 ± 4.00bcdB | 72.92 ± 4.10bcC |
| SDB12 | 34.78 ± 2.45dA | 32.17 ± 1.50ghB | 30.05 ± 1.46fC | 65.22 ± 3.16bA | 67.83 ± 3.42bcB | 69.95 ± 3.85bC |
Values mean ± standard deviation. In the columns, different small letters indicate significant difference. In the rows, different capitalized letters indicate significant difference.
Loaf height, as an indicator of the increase in volume, was measured based on the highest point of the SDB (Table 4). The loaf height varied between 6.2 and 7.2 cm. Loaf heights of SDB12, SDB4 and SDB8 were significantly (p < 0.05) higher than the others due to the use of heterofermentative LAB together with yeasts. The loaf height of the control bread was significantly (p < 0.05) lower than the SDB. The volume and specific volume were determined from the sourdough bread after 1 h of baking (Table 5). There should be an ideal relationship between bread weight and volume. The highest volume of sourdough bread is 1370 cm3 on SDB12 and 1360 cm3 in SD8 and the lowest volume is obtained on SDB2 (1240 cm3). Significantly (p < 0.05) more volumes of SDB were obtained by using LAB and yeasts together with a higher number of species containing heterofermentative LAB. Volumes did not affect the SDB appearance and texture. The highest and lowest specific volumes were detected on SDB12 (4.35 cm3 g−1) and SDB2 (3.99 cm3 g−1) respectively.
Table 5.
Loaf height, volume, specific volume and sensory score of sourdough bread (SDB).
| Sourdough bread | Loaf height(cm) | Volume(cm3) | Spesific volume(cm3 g−1) | Sensory scorea |
||
|---|---|---|---|---|---|---|
| 4h | 24h | 7d | ||||
| Control | 6.0 ± 0.03a | 1210 ± 24.89a | 3.87 ± 0.1a | 35 ± 2.87aA | 30 ± 1.69aB | 28 ± 2.95abC |
| SDB1 | 7.0 ± 0.08g | 1320 ± 83.76cd | 4.27 ± 0.4c | 61 ± 5.03fA | 51 ± 3.02cB | 42 ± 3.71efC |
| SDB2 | 6.3 ± 0.05d | 1240 ± 33.17b | 3.99 ± 0.6abc | 53 ± 4.47deA | 42 ± 3.25bB | 38 ± 2.10deC |
| SDB3 | 6.5 ± 0.05e | 1250 ± 48.45bc | 4.03 ± 1.0abc | 51 ± 3.62cdA | 42 ± 3.10bB | 36 ± 3.11cdC |
| SDB4 | 6.9 ± 0.06h | 1270 ± 33.15bc | 4.06 ± 1.1abc | 79 ± 6.05gA | 68 ± 3.87dB | 60 ± 3.56hıC |
| SDB5 | 6.5 ± 0.04e | 1250 ± 47.12bc | 4.01 ± 1.9bc | 58 ± 2.59efA | 53 ± 4.37cB | 43 ± 3.57fC |
| SDB6 | 6.8 ± 0.07f | 1260 ± 33.17bc | 4.05 ± 1.0bc | 46 ± 3.65bcA | 35 ± 3.38aB | 31 ± 4.42bC |
| SDB7 | 6.2 ± 0.04g | 1270 ± 64.77b | 4.08 ± 0.6ab | 65 ± 3.52fA | 63 ± 3.02dB | 52 ± 3.42gC |
| SDB8 | 7.1 ± 0.07ı | 1360 ± 56.55d | 4.34 ± 0.1c | 77 ± 4.26gA | 65 ± 7.30dB | 57 ± 3.43hC |
| SDB9 | 6.9 ± 0.07g | 1300 ± 47.29bc | 4.15 ± 0.8bc | 58 ± 3.72efA | 45 ± 3.05bB | 32 ± 2.14bcC |
| SDB10 | 6.8 ± 0.06f | 1265 ± 18.71bcd | 4.04 ± 0.4bc | 60 ± 4.86fA | 52 ± 3.71cB | 48 ± 3.46gC |
| SDB11 | 6.2 ± 0.03b | 1275 ± 56.68bc | 4.10 ± 0.2bc | 63 ± 3.91fA | 55 ± 3.32cB | 50 ± 3.06gC |
| SDB12 | 7.2 ± 0.08j | 1370 ± 78.47d | 4.35 ± 0.1c | 80 ± 4.44gA | 70 ± 4.27dB | 62 ± 3.68ıC |
Evaluation was made on 10 panelists, d = day, 4th = 4 h, 1d = one day, 7d = 7 day, Values mean ± standard deviation. The means within column, different small letters indicate significant difference (p < 0.05) and within rows, different capitalized letters indicate significant difference (p < 0.05).
The sensory characteristics of the SDB were determined by a panel of ten people after 4 h, one day and seven days of storage (Table 5). The SDB was sliced and grading was requested for the external and internal characteristics of the SDB. The bread sensory characteristics significantly (p < 0.05) decreased during the storage period. As a result of the sensory score, SDB12, SDB4 and SDB8 were the most preferred breads in the order. These three breads have significantly different sensory scores (p < 0.05) than others. This would be due to the more acceptable sour taste, high volume, good chewability, swallowability, low hardness and little moisture loss. After 7 days of storage, sensory values of SDB were significantly (p < 0.05) higher than control bread.
Depending on the differences in carbohydrate fermentation patterns, microorganisms contributes its distinctive characteristics to SDB (Lau et al., 2021). In the production of SDB, homofermentative LAB in combination with yeasts contributed better dough softening than either LAB or yeast alone (Lau et al., 2021). Heterofermentative and homofermentative LAB have different functions in optimizing and maintaining SDB qualities.
In a study, examining the LAB flora of 19 Italian sourdoughs, the most frequent LAB isolates were Lactobacillus sanfranciscensis (28%), L. plantarum (16%) and Lactobacillus paralimentarius (14%) (Minervini et al., 2012). In another study in Italy (Sardinia), Lactobacillus pentosus was dominant in sourdoughs, while L. sanfranciscensis was isolated only from a limited number of sourdoughs (Catzeddu et al., 2006). In central Italy, 85% of the isolates were S. cerevisiae, which is followed by Candida krusei (2.5%), Candida milleri (11%) and Torulaspora delbrueckii (1%) (Valmorri et al., 2010). In our research, the most frequently isolated LAB species were L. brevis (43.33%), P. acidilactici (21.7%), L. plantarum (18.3%) and L. pentosus (3.3%). Low numbers of S. cerevisiae (27.5%) were isolated from sourdoughs than results from Italy. Chinese traditional sourdoughs were dominated by a single yeast species (S. cerevisiae), and L. plantarum and L. brevis were the most commonly isolated LAB species (Landis et al., 2021) as well as our results for LAB species. de Vuyst et al. (2016) indicated that a diversity of yeast genera is isolated worldwide from sourdoughs, with among the most common ones being Saccharomyces, Kazachstania, Pichia, Wickerhamomyces and Candida. Twelve types of LAB have been identified from sourdoughs produced in Italy, and most of these LAB have been identified as heterofermentative species represented between 30 and 60% (Catzeddu et al., 2006). From traditional sourdoughs of the southeast region (Gaziantep and Mardin) and the central region (Konya) of Turkey, most of the LAB species were heterofermentative (75.0%). When comparing the results of sourdoughs from different regions, there may be large differences in the LAB and yeast species. This would be due to the changing microflora of sourdough according to environmental conditions and the ingredients used in the production.
Homofermentative LAB mainly produces lactic acid through glycolysis (homolactic fermentation) while heterofermentative LAB produces lactic acid, CO2, acetic acid and/or ethanol through the 6-phosphogluconate/phosphoketolase pathway (heterofermentative) (Corsetti et al., 2007). LAB acidification characteristics have varied, but in general, heterofermentative LAB produces less acid than homofermentative LAB. In this research, the pH of sourdough produced from more homofermentative and heterofermentative LAB species (as SDB12) had a significantly (p < 0.05) lower pH than those produced from a lower number of homofermentative and heterofermentative LAB species. Variations in the acidification properties of LAB in sourdoughs were mostly due to the characteristics of LAB (such as homofermentative, heterofermentative, etc.) and the number of species used in the fermentation. LAB, in addition to yeasts, causes noticeable swelling and aroma formation in the sourdough.
4. Conclusion
Isolated LAB from homemade traditional sourdough is mainly composed of heterofermentative species. The use of L. plantarum in conjunction with L. brevis and other LAB species and yeasts produced high-quality SDB. This study identifies appropriate combinations of LAB and yeasts in SDB production to obtain high-quality products. The use of homofermentative LAB in conjunction with heterofermentative LAB and yeasts in a higher number of species resulted in sufficient acidification and high-quality products. The major metabolic activities of the sourdough microbiota are acidification (LAB), flavor formation (LAB and yeasts), and leavening (yeasts and heterofermentative LAB). Among the SDB produced with the combination of LAB and yeasts, SD12 was the most preferred in terms of sensory analysis and other quality characteristics. Three cases of refreshing provided high-quality SDB with adequate combinations of LAB and yeast species.
Funding
No funding was received for this work.
We confirm that the manuscript has been read and approved by all named authors.
We confirm that the order of authors listed in the manuscript has been approved by all named authors.
CRediT authorship contribution statement
Ayse Sevgili: Data curation, Investigation, Writing – original draft. Canan Can: Supervision, Conceptualization, Methodology, Writing – original draft. Derya Isler Ceyhan: Methodology, Investigation. Osman Erkmen: Conceptualization, Supervision, Methodology, Software, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The authors have no conflict of interest to declare.
Acknowledgements
This study was funded by the Scientific and Technological Research Council, Turkey (Project number 2180193).
Handling Editor: Yeonhwa Park
Data availability
Data will be made available on request.
References
- Bazalová, O., Cihlář, J.Z., Dlouhá, Z., Bár, L., Dráb, V., Kavková, M. 202). Rapid sourdough yeast identification using panfungal PCR combined with high resolution melting analysis. J. Microbiol. Methods 199, 106522.. [DOI] [PubMed]
- Boyaci-Gunduz C.P., Erten H. Predominant yeasts in the sourdoughs collected from some parts of Turkey. Yeast. 2020;37(Special Issue):449–466. doi: 10.1002/yea.3500. [DOI] [PubMed] [Google Scholar]
- Catzeddu P., Mura E., Parente E., Sanna M., Farris G.A. Molecular characterization of lactic acid bacteria from sourdough breads produced in Sardinia (Italy) and multivariate statistical analyses of results. Syst. Appl. Microbiol. 2006;29:138–144. doi: 10.1016/j.syapm.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Chiv R., Celador-Lera L., Uña J.A., Jiménez-López A., Espinosa-Alcantud M., Mateos-Horganero E., Vega S., Santos M.A., Velázquez E., Tamame M. Yeast biodiversity in fermented doughs and raw cereal matrices and the study of technological traits of selected strains isolated in Spain. Microorganisms. 2021;9:47. doi: 10.3390/microorganisms9010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsetti A., Settanni L., Valmorri S., Mastrangelo M., Suzzi G. Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentations. Food Microbiol. 2007;24:592–600. doi: 10.1016/j.fm.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Dertli E., Mercan E., Arici M., Yilmaz M.T., Sagdic O. Characterisation of lactic acid bacteria from Turkish sourdough and determination of their exopolysaccharide (EPS) production characteristics. LWT--Food Sci. Technol. 2016;71:116–124. [Google Scholar]
- Elgun A., Ertugay Z. Atatürk University, Ziraat Faculty Press; Erzurum: 2002. Grain Processing Technology. [Google Scholar]
- Erkmen O. In: Microbiological Analysis of Foods and Food Processing Environments. Erkmen O., editor. Elsevier Inc.; London: 2022. Analysis of fermented foods; pp. 361–380. [Google Scholar]
- Erkmen O. In: Microbiological Analysis of Foods and Food Processing Environments. Erkmen O., editor. Elsevier Inc.; London: 2022. Yeasts and molds counting techniques; pp. 43–52. [Google Scholar]
- Erkmen O., Bozoglu T.F. In: Erkmen O., Bozoglu T.F., editors. vol. 2. Microorganisms in Food Preservation and Processing. John Wiley and Sons, Ltd.; Chichester: 2016. Fermented cereal and grain products; pp. 349–373. (Food Microbiology Principles into Practice). [Google Scholar]
- Gorkem O. Rheological behaviour of type I sourdough during refreshment procedure. Int. J. Food Technol. Nut. 2019;2(3–4):8–14. [Google Scholar]
- Gul H., Ozcelik S., Sagdic O., Certel M. Sourdough bread production with lactobacilli and S. cerevisiae isolated from sourdoughs. Process Biochem. 2005;40:691–697. [Google Scholar]
- Ispirli H., Dertli E. Isolation of distinct Lactobacillaceae spp. with functional characteristics from traditional sourdough samples. Acad. Food. 2022;20(3):211–219. [Google Scholar]
- Landis E.A., Oliverio A.M., McKenney E.A., Nichols L.M., Kfoury N., Biango-Daniels M., Shell L.K., Madden A.A., Shapiro L., Sakunala S., Drake K., Robbat A., Booker M., Dunn R.R., Fierer N., Wolfe B.E. The diversity and function of sourdough starter microbiomes. Elife. 2021;10 doi: 10.7554/eLife.61644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.W., Chong A.Q., Chin N.L., Talib R.A., Basha R.K. Sourdough microbiome comparison and benefits. Microorganisms. 2021;9(7):1355. doi: 10.3390/microorganisms9071355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroth C.B., Walter J., Hertel C., Brandt M.J., Hammes W.P. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2003;69(1):475–482. doi: 10.1128/AEM.69.1.475-482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minervini F., di Cagno R., Lattanzi A., de Angelis M., Antonielli L., Cardinali G., Cappelle S., Gobbetti M. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 2012;78:1251–1264. doi: 10.1128/AEM.07721-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S. Oxford University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- Ozulku G. vol. 2. Taylor and Francis; Oxfordshire: 2019. (Rheological Behaviour of Type I Sourdough during Refreshment Procedure). [Google Scholar]
- Park J., Seo J.S., Kim S.A., Shin S.Y., Park J.H., Han N.S. Microbial diversity of commercial makgeolli and its influence on the organoleptic characteristics of Korean rice sourdough. J. Microbiol. Biotechnol. 2017;27(10):1736–1743. doi: 10.4014/jmb.1708.08003. [DOI] [PubMed] [Google Scholar]
- Perez-Alvarado O., Zepeda-Hernández A., Garcia-Amezquita L.E., Vinderola G., Garcia-Cayuela T. Role of lactic acid bacteria and yeasts in sourdough fermentation during breadmaking: evaluation of postbiotic-like components and health benefits. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.969460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Siepmann F.B., Ripari V., Waszczynskyj N., Spier M.R. Overview of sourdough technology: from production to marketing. Food Bioprocess Technol. 2018;11:242–270. [Google Scholar]
- Tamura K., Stecher G., Kumar S. Mega 11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmorri S., Tofalo R., Settanni L., Corsetti A., Suzzi G. Yeast microbiota associated with spontaneous sourdough fermentations in the production of traditional wheat sourdough breads of the Abruzzo region (Italy) Int. J. Gen. Mol. Microbiol. 2010;97:119–129. doi: 10.1007/s10482-009-9392-x. [DOI] [PubMed] [Google Scholar]
- de Vuyst L., Harth H., van Kerrebroeck S., Leroy F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016;239:26–34. doi: 10.1016/j.ijfoodmicro.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Yagmur G., Tanguler H., Leventdurur S., Elmaci S.B., Turhan E.U., Francesca N., Settanni L., Moschetti G., Erten H. Identification of predominant lactic acid bacteria and yeasts of Turkish sourdoughs and selection of starter cultures for liquid sourdough production using different flours and dough yields, Polish. J. Food Nutr. Sci. 2016;66:99–107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.