Abstract
Through a computer search of DNA databases, we have identified the homologs of the mgU6-47 snoRNA gene from the yeast Schizosaccharomyces pombe, the fly Drosophila melanogaster and human. The three box C/D-containing snoRNA genes showed no significant similarity in their sequences except for an 11 nt long complementarity to U6 snRNA, suggesting that the mechanism of snoRNA guided snRNA methylation is conserved from mammals to yeast. The corresponding snoRNAs have been positively detected by reverse transcription and northern blotting. Taking advantage of the fission yeast system, we have disrupted the yeast mgU6-47 gene and demonstrated that it is absolutely required for site-specific 2′-O-methylation of U6 at position A41. No growth differences between mgU6-47 gene-disrupted and wild-type cells were observed, suggesting that the mgU6-47 gene, as for most rRNA methylation guides, is dispensable in yeast. Nevertheless, it was revealed by temperature shift assay that abolition of A41 methylaion in yeast U6 snRNA might cause a small decrease in mRNA splicing efficiency. The timing of S.pombe U6 pre-RNA transport in the nucleus for splicing and methylation was also analyzed and is described.
INTRODUCTION
The spliceosomal small nuclear RNAs (snRNAs) contain a number of modified nucleotides (1), most of which occur in or around phylogenetically conserved regions (2). This implies radical roles in common required in various organisms. Post-transcriptional modification is an essential process for snRNA maturation and a large cellular machinery is devoted to introduce different kinds of modified nucleotides into snRNAs. The biosynthetic pathway of the different snRNAs is complicated and may differ from one snRNA to another. Transit to the cytoplasm is necessary for N2, N2, 7-trimethylguanosine (TMG) capping of U1, U2, U4 and U5 snRNAs (3,4). However, it is likely that internal modifications are introduced into some snRNAs only after return to the nucleus (5,6). Conversely, U6 snRNA, the only snRNA transcribed by RNA polymerase III, is not transported to the cytoplasm (7), but transits the nucleoli for post-transcriptional modification (8–10).
It has been shown that the highly modified regions in snRNAs correspond to functionally important regions, which participate in hydrogen bonding with pre-mRNA or interact with other snRNAs (2,11). This is also reflected in the conservation of some modified nucleotides in phylogenetically diverse organisms. Nevertheless, apart from the conserved modified nucleotides, a greater number of modified nucleotides are observed in mammals than in yeast (2,12). It is possible that modified nucleotides may be more important in higher organisms than single cell organisms such as yeast. The requirement for modified nucleotides for snRNA function in small nuclear ribonucleoprotein particle (snRNP) biogenesis has been assayed in several reconstitution systems. Clearly the modifications of mammalian U2 snRNA are required for snRNP assembly and pre-mRNA splicing in HeLa splicing extract and Xenopus oocytes (5,13,14). However, in the case of yeast, in vitro synthesized U2 snRNA can restore splicing (15,16). This is probably because mammalian U2 snRNA is more extensively modified than its counterpart in the yeast Saccharomyces cerevisiae, and yeast is well known to differ from other eukaryotes in some aspects of RNA splicing (17). Moreover, although various in vitro synthesized snRNAs have been shown to be functional in reconstitution systems, the influence of modified nucleotides on overall splicing efficiency in vivo remains to be evaluated. Information about modified nucleotides in snRNAs are well documented from mammalian cells, plants and yeast (12), however, no specific function has been assigned to particular site-specific modifications in snRNAs except for the TMG cap in nuclear import of Sm-antigen binding snRNP (3,4).
Recent progress on 2′-O-methylation of animal U6 snRNA (10) has led to a breakthrough in the understanding of the mechanism of snRNA modification. 2′-O-Methylation of U6 snRNA is guided by snoRNAs that were initially thought to be specific for rRNA modification (18,19). Recently, characterization of brain-specific and imprinted snoRNAs that display the hallmarks of the two guide RNA families point to a potential role in mRNA processing (20). The nucleolus has been shown to play an essential role in post-transcriptional modification and processing of nuclear components and other cellular RNAs (21). The finding of snoRNA guides for internal modification of U6 snRNA and for other cellular RNAs also shows a new way to investigate in vivo the function of a particular site-specific modification in RNA molecules using snoRNA gene disruption. Thus, the fission yeast is an attractive model system for molecular genetics studies.
In this work we report the identification of three homologs of mouse mgU6-47 RNA from human, Drosophila melanogaster and the fission yeast Schizosaccharomyces pombe. Using the fission yeast system, we have demonstrated that the mgU6-47 gene is required for site-specific 2′-O-methylation of S.pombe U6 snRNA. We also report the effects on cell growth and mRNA splicing of disruption of the snoRNA gene. Our results also provide a glimpse into the timing of S.pombe U6 pre-RNA transport in the nucleus for splicing and methylation.
MATERIALS AND METHODS
All techniques used for manipulation of Escherichia coli, DNA, RNA and oligonucleotides were performed essentially as described by Sambrook et al. (22). Molecular genetics analysis of the fission yeast S.pombe was carried out according to a standard protocol (23).
Computer search of the nucleic acids databases
The nucleic acids databases GenBank and EMBL were screened using the BLAST (24) and Fasta (25) programs. Searches for perfect 10 nt complementarity to snRNA ribose-methylated sequences immediately followed by the sequence NCUGA were carried out as previously described (26). Sequences exhibiting snoRNA gene features were selected and further analyzed using the Pcgene 6.0 package.
Strains and media
The S.pombe wild-type haploid strain sp972 was used for transformations and all RNA and DNA analyses. This strain was grown in rich (YPD) medium (1% yeast extract, 1% peptone, 2% glucose) at 30°C or the temperature specified below for the temperature shift assays. Yeast were transformed by the lithium acetate method. Transformants were screened on selective plates with 200 mg/l G418 and the chromosomal allele was checked by PCR. Escherichia coli strains TG1 [F′/supE, hsdΔ5, thiΔ(lac-proAB)] and DH5α [F′, endA1, hsdr17 (rk-mk+), supE44, thi-1, recA1, gyrA (Nalr), relA1, Δ(lacIZYA-argF)U169, deoR, (f80dlacΔ(lacZ)M15)] grown on 2YT (1.6% Bacto tryptone, 1% Bacto yeast extract, 0.5% NaCl) liquid or solid medium were used for all cloning procedures.
RNA extraction and analyses
RNA from HeLa cells, mouse liver, D.melanogaster and S.pombe cells were isolated by guanidinium thiocyanate/phenol-chloroform extraction as described by Chomoczynski and Sacchi (27). Temperature shift assays were done by heat shock treatment of yeast cells grown at 23°C for 2 days, then 37°C for 30 min, 2 h and 7 h, before isolating the RNA. For northern analysis, 50 µg total RNA was fractionated on 10% acrylamide–7 M urea gels, electroblotted onto a hybond-N nylon membrane (Amersham), hybridized with 5′-end-labeled probes and washed as described (28). Reverse transcription was carried out in a 20 µl reaction mixture containing 25 µg total RNA, 20 ng 5′-end-labeled primer and appropriate concentrations of dNTPs as required. After denaturion at 65°C for 5 min and cooling to 42°C, 200 U MLV reverse transcriptase (Promega) was added and extension carried out at 42°C for 30 min. The cDNAs were then analyzed on 10% acrylamide–7 M urea gels.
Detection of ribose-methylated nucleotides
Ribose-methylated nucleotides in U6 snRNA were detected by reverse transcription at low dNTP concentrations with oligonucleotide U6sp. In brief, primer extension was carried out in parallel on two aliquots in the presence of either 0.1 or 1.5 mmol/l dNTPs (26). To remove the intron sequence, a U6 snRNA sequence ladder was prepared from the full-length U6 cDNA. After adding a 3′ poly(G) tail using terminal transferase, U6 cDNA was amplified by PCR with the primers poly(C) and U6sp. The PCR product was purified and cloned into the SmaI site of plasmid pTZ18. The recombinant plasmid was sequenced with 5′-end-labeled primer U6sp and run in parallel with reverse transcription of U6 snRNA as a molecular weight marker. The U6 gene sequence ladder used in U6 snRNA precursor analysis was prepared by cloning a PCR fragment of the U6 snRNA gene with primers U6sp-Fw and U6sp.
Construction of plasmids for the snoRNA gene disruption
For S.pombe mgU6-47 gene disruption an ∼1.2 kb fragment of S.pombe genomic DNA encompassing the mgU6-47 snoRNA gene and flanking sequences was PCR amplified with the Fu6sp and Ru6sp primer pair. After digestion with EcoRI and KpnI, the amplified fragment was cloned into the corresponding restriction site of pTZ19. The EcoRV–SalI region in the amplified fragment was replaced by a 1.4 kb selectable marker module from pFA6-kanmx4 (29), which permits efficient selection of transformants resistant to geneticin (G418), and the SalI–BglII fragment encompassing the 5′-region of the mgU6-47 snoRNA gene was substituted by a 18 bp linker produced from sal-link and bgl-link, giving rise to plasmid pU6sp3. pU6sp3 was linearized with KpnI and used to transform the wild-type haploid strain sp972 of S.pombe by the lithium acetate procedure. Transformants were screened on selective plates with 200 mg/l G418. Disruption of the chromosomal alleles of the mgU6-47 snoRNA gene was verified by PCR with the Fu6sp and Ru6sp primer pair. Depletion of expression of mgU6-47 snoRNA was analyzed by reverse transcription and northern analysis using oligonucleotide Pz30sp.
Oligonucleotides
Oligonucleotides were synthesized and purified by Sangon Co. (Shanhai, China). The sequences of oligonucleotides used for northern and reverse transcription analyses of mgU6-47 snoRNAs were as follows: Pz30hs, 5′-AGCTCAGAGAGAAGATTAAGRG-3′; Pz30dm, 5′-GCGTCAGCGAGAAGATTAGTTG-3′; Pz30sp, 5′-GGATCAGAGAGAAGATTAGCTT-3′. These oligonucleotides are complementary to the 3′-end of the mgU6-47 snoRNAs from human, D.melanogaster and S.pombe, respectively. The following oligonucleotides were used to disrupt the S.pombe mgU6-47 gene: Fu6sp, 5′-CAGTGAGCAAATGAAGTCACTC-3′; Ru6sp, 5′-CACAAAGATTTAAAGTCTCACGC-3′; sal-link, 5′-TCGACCCAAGGATCCCCA-3′; bgl-link, 5′-GATCTGGGGATCCTTGGG-3′. The probe and primer used for northern and reverse transcription analyses of S.pombe U6 snRNA and its precursor were as follows: U6sp, 5′-AATGGGTTTTCTCTCAATGTCGCAG-3′; U6sp-In, 5′-CTAAACAACGAGTTAGTATGACTCG-3′. The poly(C) primer used for PCR amplification of oligo(G)-tailed U6 cDNAs was 5′-GGAATTCGGATC16-3′. The sequence of U6sp-Fw, used for PCR amplification of the U6 snRNA gene, was 5′-TCCTGCATACTGTAAATCTCAGAGC-3′. The oligonucleotides were 5′-32P-end-labeled as described previously (28) and used directly as probes for northern hybridization or submitted to purification by electrophoresis on a 10% acrylamide–7 M urea gel before utilization as reverse transcription primers.
Nucleotide sequence accession numbers
The S.pombe, D.melanogaster and human mgU6-47 sequences have been deposited in the EMBL database under accession nos AJ007736, AJ007735 and AJ007733, respectively.
RESULTS
mgU6-47 is a conserved snoRNA between S.pombe and human
It has been shown that most, if not all, sites of rRNA ribose-methylation are specified through a process that involves many snoRNAs (30,31). This mechanism of methylation also seems applicable for some snRNAs, such as U6 snRNA (10). Knowing the modification pattern of snRNAs from various eukaryotes (12), we performed a search of the DNA databases to find novel snoRNA guides for snRNA modification with complementarity to the sequence flanking the methylated nucleotide together with the conserved elements of snoRNAs, i.e. box C/D snRNAs. A number of DNA sequences coding for potential snoRNAs were identified. Here we focus our attention to three candidate mgU6-47 snoRNA homologs (initially termed Z30), which correspond to the methylation guides for U6 snRNA from S.pombe, D.melanogaster and human (Fig. 1).
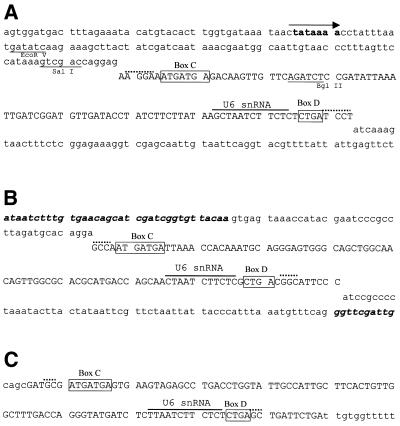
Figure 1.
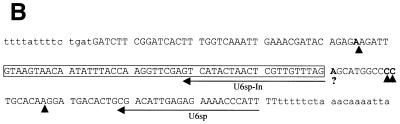
The sequences of mgU6-47 homolog genes from S.pombe (A), D.melanogaster (B) and human (C). SnoRNA genes are in capital letters; the C/D boxes are denoted. A bar is drawn over sequences complementary to U6 snRNA and dashed lines indicate nucleotides involved in the terminal stem. A putative promoter element in the S.pombe mgU6-47 gene is denoted by an arrow and restriction sites used for disruption of the snoRNA gene are also indicated. The exon sequences of the D.melanogaster RNA helicase gene are presented in italic. (D) Alignment of mgU6-47 sequences. Only the nucleotides that differ from the top line sequence (human) are shown. Identity is denoted by a hyphen and deletion by a star. Hs, human; Mm, mouse (10); Ce, C.elegans (10); Dm, D.melanogaster; Sp, S.pombe.
Schizosaccharomyces pombe mgU6-47 was found as an independent transcript gene located on a 1.4 kb genomic DNA spacer fragment of chromosome II (accession no. AL022299), which is between two protein coding genes. A promoter element (TATAAA box) was found 95 bp upstream of the snoRNA coding region. Drosophila melanogaster mgU6-47 was identified as an intronic RNA encoded in the first intron of a RNA helicase gene (accession no. AF017777). Human mgU6-47 was from an unidentified EST sequence (accession no. AA552905). All three mgU6-47 sequences are box C/D-containing snoRNAs with 11–13 nt long complementarity to U6 snRNA. According to the relationship of antisense snoRNA structure and function (32–34), this sequence complementarity precisely targets a conserved methylated nucleotide of U6 snRNA, determined to be Am47 for mammals (1) and D.melanogaster (35), and Am41 for S.pombe (2). A short terminal stem, derived from the inverted sequences at the 5′- and 3′-ends of the mgU6-47 genes, has been revealed to be important for snoRNA biogenesis (36,37). Comparison of the novel mgU6-47 snoRNAs with their homologs in mouse and Caenorhabditis elegans (10) showed extensive sequence divergence (Fig. 1). Besides the box C/D element, the sequence complementary to U6 snRNA in the snoRNAs is only strictly maintained among distant organisms, although two mammal mgU6-47 RNAs have ∼85% sequence similarity.
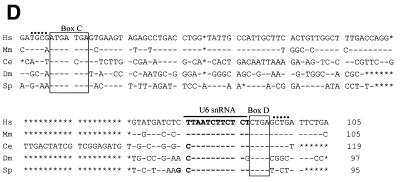
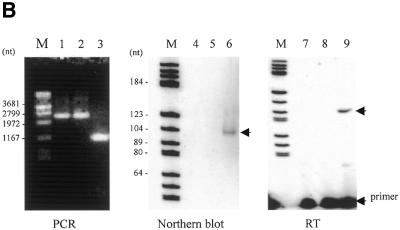
With oligonucleotide probes specific for the 3′-region of each putative snoRNA the three RNAs were positively determined from human, fly and S.pombe by northern blotting (Fig. 2A). The lengths of the RNAs were determined to be 105, 97 and 95 nt for human, D.melanogaster and S.pombe, respectively. Mouse mgU6-47 RNA was also analyzed as a reference. In addition, northern analysis of RNA isolated from purified HeLa cell and S.pombe nuclei showed that mgU6-47 was enriched in the nucleus (data not shown). Reverse transcription was carried out to map the 5′-end of the snoRNAs (Fig. 2B), followed by cloning and sequencing of the cDNAs. As expected, all cDNA sequences perfectly matched the sequences of the snoRNA coding regions, as shown in Figure 1.
Figure 2.
Characterization of snoRNAs. (A) Northern analyses. Aliquots of 50 µg total cellular RNA were separated on 10% acrylamide–7 M urea gels, electroblotted onto a hybond-N nylon membrane and hybridized with 5′-32P-end-labeled oligonucleotide probes as described in Materials and Methods. (B) Reverse transcription analysis. Primer extension was carried out with the same oligonucleotides as used in the northern analyses. Hu, human; Mm, mouse; Dm, D.melanogaster; Sp, S.pombe. M, molecular weight marker (pBR322 digested with HaeIII).
Although mgU6-47 appears to be a well-conserved snoRNA, our attempt to search for a mgU6-47 gene in the DNA database of the yeast S.cerevisiae failed to give any significant result. 2′-O-ribose methylation analysis of U6 snRNA by reverse transcription at low dNTP concentrations confirmed that the four methylated sites in S.pombe were not methylated in S.cerevisiae (L.-H.Qu and H.Zhou, unpublished results).
Disruption of the yeast mgU6-47 gene abolishes 2′-O-methylation of A41 in U6 snRNA
Although an analysis of mouse mgU6-47 sequence complementarity to U6 snRNA has suggested that it could direct 2′-O-methylation of A47 in U6 snRNA (10), identification of the S.pombe mgU6-47 gene provides us with a useful genetic system to prove this function and to evaluate the effects on cell growth of disruption of the mgU6-47 gene in S.pombe.
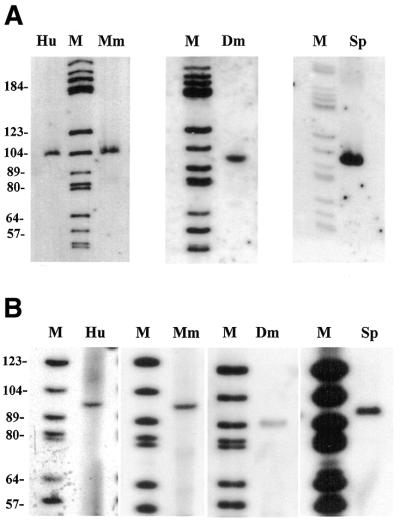
Disruption of the mgU6-47 gene was carried out in a wild-type haploid strain of S.pombe. We first disrupted the mgU6-47 gene by inserting a 1.4 kb long Kan selectable marker module between the promoter TATA box element and the mgU6-47 RNA coding region, resulting in the construct Pu6sp2 (Fig. 3A). Unexpectedly, this disruption was insufficient to suppress expression of the mgU6-47 gene after transformation of the haploid wild-type strain by the disrupted allele (data not shown). Making use of the endonuclease sites at the 5′-end of the mgU6-47 gene, direct modification in the gene was carried out by replacing the 36 bp fragment of Pu6sp1 with a synthetic 18 bp SalI–BglII linker (Fig. 3A). This manipulation deleted the 5′-end TGATGA (box C), a key sequence for snoRNA stability and nucleolar localization (38,39), of the mgU6-47 gene (plasmid pU6sp3). After transformation of S.pombe with delineated pU6sp3, suppression of expression of the mgU6-47 gene by the disrupted allele was observed in Kan+ colonies. PCR analysis of genomic DNA with a specific primer demonstrated that the mgU6-47 locus was disrupted and modified in the Kan+ cells. An expected 2.5 kb PCR product, instead of the 1.1 kb band, was amplified with the Fu6sp and Ru6sp primer pair (Fig. 3B). Disruption of the mgU6-47 gene in the Kan+ cells was further confirmed by primer extension and nothern blot analysis (Fig. 3B). In both cases no signal was detected with the specific primer for mgU6-47 RNA when probing total RNA from the Kan+ cells, in contrast to what was observed for the parental wild-type strain RNA.
Figure 3.
snoRNA gene disruption. (A) Construction of plasmids. The top diagram shows PCR amplification of the ∼1.2 kb genomic DNA fragment encompassing the mgU6-47 gene of S.pombe with primers Fu6sp and Ru6sp and cloning of the amplified fragment, after digestion with KpnI and EcoRI, into the corresponding restriction sites of E.coli plasmid Bluescript M13– (MCS, multiple cloning site). The second diagram shows how the EcoRV–SalI region in the amplified fragment was replaced by the selectable marker module from pFA6-kanmx4. The bottom diagram shows how the SalI–BglII region at the 5′-end of the mgU6-47 gene was substituted by the SalI–BglII linker, giving rise to plasmid pU6sp3, which was used to transform a haploid S.pombe after linearization with KpnI. (B) Analyses of transformants. PCR amplification of genomic DNA from two Kan+ transformants, sp5m and sp14m (lanes 1 and 2), and the wild-type strain (lane 3) with primers Fu6sp and Ru6sp. PCR products were analyzed by electrophoresis on a 1.0% agarose gel followed by ethidium bromide staining. Nothern analysis of total RNAs from the two transformants (lanes 4 and 5) and the wild-type strain (lane 6) with the Pz30sp oligonucleotide probe, and reverse transcription of total RNAs from the two Kan+ transformants (lanes 7 and 8) and the wild-type strain (lane 9) with the Pz30sp oligonucleotide primer.
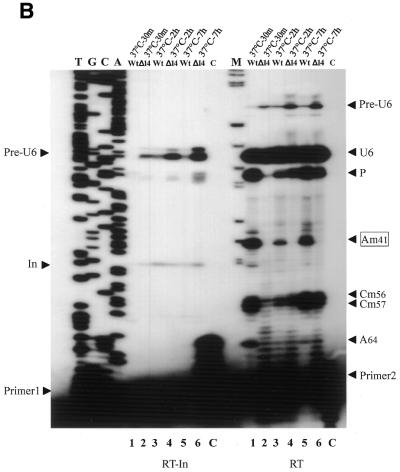
The impact on U6 snRNA methylation of disruption of the mgU6-47 gene was analyzed using primer extension at different dNTP concentrations. 2′-O-Methylated nucleotides in the RNA template present severe conformational blocks to the passage of reverse transcriptase at low dNTP concentrations, but this effect can be dramatically attenuated at higher concentrations of dNTPs (40). As expected, disruption of the mgU6-47 gene indeed resulted in abolition of U6 snRNA methylation at A41. The reverse transcription pause disappeared completely at 0.1 mmol/l dNTPs when RNA from the mgU6-47 gene-disrupted strain was analyzed (Fig. 4). In contrast, a clear pause of reverse transcription could be observed at 0.1 mmol/l dNTPs which became less evident at 1.5 mmol/l dNTPs when the wild-type strain RNA was used (Fig. 4). Note that in the reverse transcription analyses of 2′-O-methylated nucleotides in rRNA, dNTP concentrations as low as 0.004 mmol/l were used (26). However, much higher dNTP concentrations (0.01–0.15 mmol/l) can be used for U6 snRNA analysis, probably because of the lower abundance of U6 snRNA in total RNA.
Figure 4.
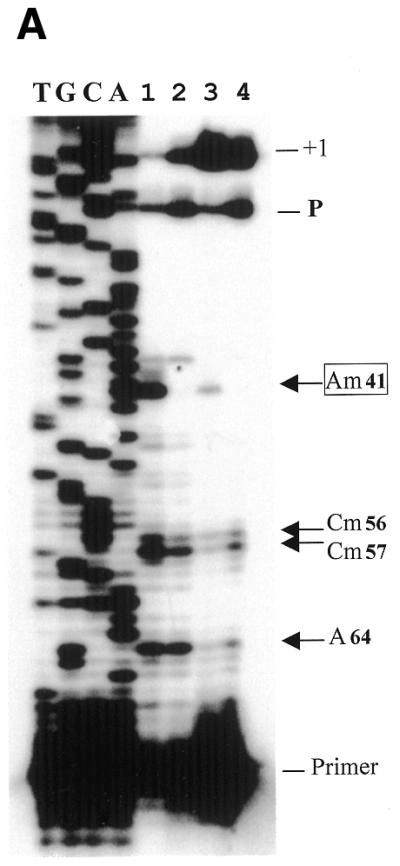
Abolition of 2′-O-methylation at A41 in U6 snRNA after disruption of the mgU6-47 gene. (A) Determination of U6 snRNA ribose methylation sites by reverse transcription at different dNTP concentrations with 5′-32P-end-labeled primer U6sp. Lane1, control reaction at 0.1 mmol/l dNTPs on wild-type strain RNA; lane 2, primer extension at 0.1 mmol/l dNTPs on disrupted strain RNA; lane 3, control reaction at 1.5 mmol/l dNTPs on wild-type strain RNA; lane 4, primer extension at 1.5 mmol/l dNTPs on disrupted strain RNA; T, G, C and A, the U6 cDNA sequence ladder generated with the same primer as reverse transcription. Arrows indicate the sites of 2′-O-methylation revealed by reverse transcriptase pauses at low dNTP concentrations. P, a reverse transcriptase pause at a secondary structure of U6 snRNA; +1, full-length U6 cDNA. (B) 2′-O-methylated nucleotides in S.pombe U6 snRNA. The coding sequence of the U6 snRNA gene is represented in capital letters. The intron in the U6 snRNA gene is boxed. The four 2′-O-methylated nucleotides in U6 snRNA (2) are in black, while arrowheads denote the 2′-O-methylated sites detected by reverse transcription at low dNTP concentrations. Arrows indicate the locations of two oligonucleotides used for U6 snRNA analyses.
Compared to nine 2′-O-methylated nucleotides in U6 snRNA of mammals, only four 2′-O-methylated nucleotides were identified in S.pombe U6 snRNA using RNase hydrolysis and DEAE–cellulose separation (2). As shown in Figure 4, our analysis determined three 2′-O-methylated nucleotides, Am41, Cm56 and Cm57, as previously reported by Gu et al. (2). However, A47, although previously reported as a 2′-O-methylated nucleotide, has no obvious effect on reverse transcription at low concentrations of dNTPs when RNA from either the wild-type or a disrupted strain was used. The unusual behavior of A47, which was very different from the other three 2′-O-methylated nucleotides, made the methylation at this site suspect. Conversely, our experiment indicated another possible 2′-O-methylation site, A64, which had the same repressive effect on reverse transcription as the three 2′-O-methylated nucleotides, and this effect largely depended on dNTP concentration (Fig. 4). Note that A64 lies in the region interacting with U4 snRNA and is also a very conserved methylated site among D.melanogaster, human and mouse.
Disruption of the mgU6-47 gene by U6sp3 did not completely suppress transcription of truncated mgU6-47 RNA in Kan+ cells; truncated mgU6-47 RNA could still be detected by RT–PCR (data not shown) in spite of a negative result on northern blots. Therefore, abolition of A41 methylation resulted directly from instability of the truncated mgU6-47 RNA, whose 5′-end box C sequence was completely deleted.
No phenotypic differences between the Kan+ and parental cells were observed when grown at different temperatures such as 25, 30 and 37°C (data not shown), suggesting that the mgU6-47 gene is dispensable in yeast, as are most rRNA methylation guides.
Abolition of methylation of U6 snRNA slightly affects the efficiency of splicing
Although no phenotypic differences were observed under various growth conditions, an investigation of the impact at the mRNA precursor level of abolition of A41 methylation of U6 snRNA was carried out. The S.pombe U6 snRNA gene is split by a mRNA-type intron (41,42), which could be spliced by the same machinery as carries out mRNA processing in the cell. U6 pre-RNA was easily detected in the wild-type strain by temperature shift assay (43). Therefore, the experiment analyzed U6 snRNA and its precursor from mgU6-47 gene-disrupted and wild-type cells grown under normal and restrictive conditions.
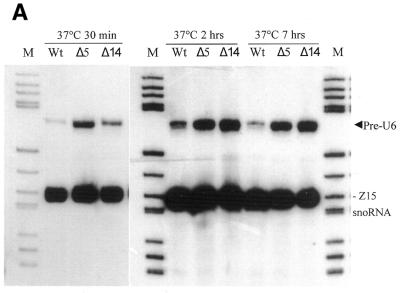
Total RNA was extracted from two mgU6-47 gene-disrupted strains, sp5m and sp14m, and the parental wild-type strain. Primer extension was performed with the exon-specific oligonucleotide U6sp, complementary to the 3′-end of S.pombe U6 snRNA, and intron-specific oligonucleotide U6sp-In, complementary to the intron sequence of S.pombe U6 snRNA (Fig. 4). The latter was also used to detect the U6 precursor by northern hybridization. There were no evident differences in the levels of mature U6 snRNA and its precursor among disrupted and wild-type cells grown at 30°C (data not shown). However, when the cells were shifted from 23 to 37°C, slightly more U6 pre-RNA accumulated in the disrupted cells as compared to the wild-type cells, as detected by northern hybridization (Fig. 5A). The same tendency was confirmed by primer extension analyses of total RNA. In the primer extension with U6sp-In, increasing accumulation of U6 snRNA precursor over time after cells were shifted from 23 to 37°C was clearly observed in disrupted strain sp14m as compared to the wild-type strain (Fig. 5B). By primer extension with U6sp we could detect intact U6 precursor and mature U6 snRNA in the same assay; it reproducibly indicated slightly more accumulation of U6 pre-RNA in disrupted sp14m cells than wild-type cells (Fig. 5B). It is worth noting that slightly increased accumulation of U6 pre-RNA did not significantly diminish mature U6 snRNA levels in the disrupted cells because the amount of U6 pre-RNA was less than 1/20 that of mature U6 snRNA under the above growth conditions (Fig. 5B). These results suggest that abolition of A41 methylation in U6 snRNA caused a small, but perceptible, defect in mRNA splicing level and that this impairment in mRNA splicing was temperature sensitive.
Figure 5.
Analyses of U6 snRNA and its precursor by temperature shift assay. Total RNA was extracted from two disrupted strains (sp5m and sp14m) and the wild-type strain which were cultured at 23°C for 48 h before shifting to 37°C for 30 min, 2 and 7 h, respectively. (A) Northern hybridization was performed with probe U6sp-In. Another probe, Pz15, which hybridizes to a rRNA methylation guide, Z15 snoRNA, was added to the hybridization mix as an internal control for total RNA quantity in each lane. Lane Wt, wild-type strain RNA; lanes Δ5 and Δ14, sp5m and sp14m strain RNA; lane M, molecular weight marker (pBR322 digested with HaeIII). The arrowhead indicates the band corresponding to the U6 snRNA precursor. (B) Reverse transcription at 0.2 mmol/l dNTP with 5′-end-labeled U6sp-In (primer 1) and U6sp (primer 2). Lane Wt, wild-type strain RNA; lane Δ14, sp14m strain RNA; lane Co, control reaction without added RNA; lanes T, G, C and A, U6 snRNA gene sequence ladder generated with primer U6sp-In.
The timing of S.pombe U6 snRNA splicing and methylation
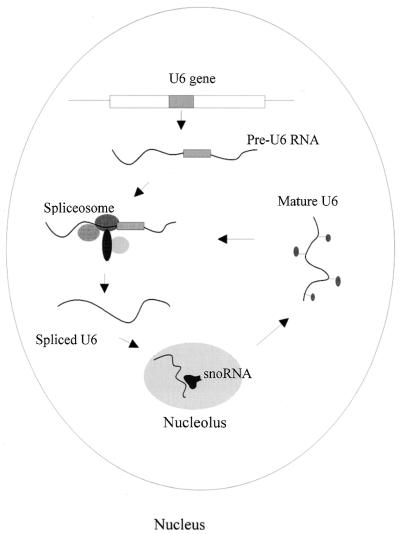
All U6 snRNA genes are transcribed by polymerase III (44), but intronic sequences in U6 snRNA were found in only three yeasts: S.pombe, Rhodotorula hasegawae and Rhodosporidium dacryoidum (41,42). This structure is characteristic and provides us with a unique system to study the timing of U6 pre-RNA transportation in the nucleus for processing. An intron-possessing U6 pre-RNA needs both splicing and chemical modification during maturation. It is evident that the two events should take place in different loci in the nucleus and occur in a temporal order. This processing would involve complex trafficking of U6 pre-RNA from the spliceosome to the nucleolus or vice versa. Direct examination of 2′-O-methylated nucleotides in U6 pre-RNA by intron-specific primer extension provides an easy means to follow this because the methylated nucleotide, Am41, of U6 snRNA in the wild-type strain is just 31 nt upstream of the primer U6sp-In (see Fig. 4). It has been demonstrated by the primer extension experiment reported above that methylated nucleotides cause reverse transcriptase pausing in mature U6 snRNA even at high concentration of dNTPs, while no methylation pause effect was apparent when inton-specific primer U6sp-In was used (Fig. 5B). In the primer extension with U6sp-In, only a slight band, probably corresponding to the reverse transcription product of a lariat intronic sequence released from exon 1 of U6 pre-RNA, appeared in the lanes for both the disrupted and wild-type strains. It is likely that A41 is not methylated in U6 pre-RNA before the intron is eliminated. To further investigate this hypothesis, primer extension with U6sp-In was carried out at low dNTP concentrations to determine the methylation site in U6 pre-RNA. As expected, no reverse transcription pause was observed at position A41 of U6 pre-RNA from the wild-type strain at as low as 4 µmol/l dNTPs as compared to the disrupted strain (Fig. 6). In conclusion, after transcription S.pombe U6 pre-RNA is first transported to the spliceosome for intron processing and then the spliced U6 snRNA travels through the nucleolus, in which chemical modifications guided by snoRNAs at specific sites of U6 snRNA are carried out. A schematic representation of this process is described in Figure 7.
Figure 6.
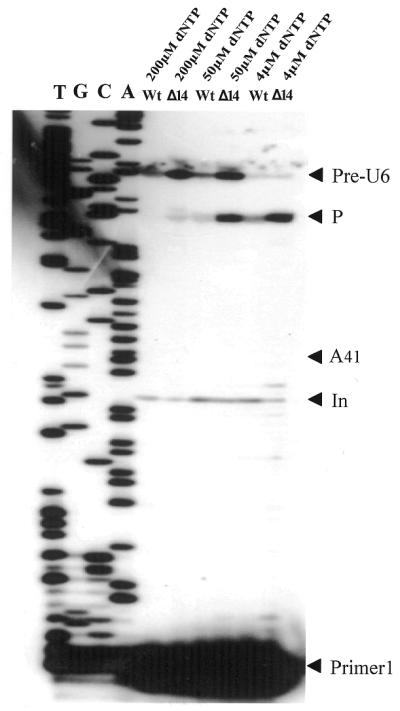
Analysis of the U6 RNA precursor by reverse transcription at low dNTP concentrations. Total RNA was extracted from the disrupted and wild-type strains cultured at 23°C for 48 h before shifting to 37°C for 2 h. Primer extension was carried out with primer U6sp-In at different dNTP concentrations. An arrowhead indicates the position of nucleotide A41. Pre-U6, full-length U6 RNA precursor; P, the reverse transcriptase pause at the secondary structure at the 5′-end of U6 snRNA; In, lariat product from intron splicing. The sequence ladder of the U6 snRNA gene was generated with the same primer as for reverse transcription.
Figure 7.
A schematic representation of the pathway of S.pombe U6 snRNA biogenesis in the nucleus.
DISCUSSION
The fission yeast is an attractive organism for studying post-transcriptional modification of snRNA
We have identified the homologs of mgU6-47 RNA from S.pombe, D.melanogaster and human. The three C/D box snoRNAs show no significant similarity in their sequences except for the 11 nt long complementarity to U6 snRNA at the 3′-end, which is strictly conserved among human, fly and yeast. The conservation of mgU6-47 RNA, which guides a conserved site-specific methylated nucleotide in U6 snRNA, implies a biological significance in common throughout evolution. However, failure to determine the counterpart of mgU6-47 RNA and its related methylated nucleotides in S.cerevisiae indicates that mgU6-47 RNA is not ubiquitous in eukaryotes. It would be interesting to know why the conservation of mgU6-47 RNA is so strictly maintained between mammals and S.pombe, but not between the two yeasts, which both belong to the Ascomycetes. The fission and budding yeasts probably diverged from a common ancestor around one billion years ago (2). One distinct difference between the two yeasts, with respect to mRNA processing, is the scarcity of introns in the S.cerevisiae genome. There are only 235 introns within 6275 hypothetical protein coding ORFs (45). It is reasonable to suppose that a higher efficiency of mRNA splicing is required in mammals and fission yeast, which possess many more introns as compared to budding yeast. Taking into account the fact that there are nine 2′-O-methylated nucleotides in mammalian U6, seven in a plant and only four in S.pombe, with possibly none in S.cerevisiae (12; L.-H.Qu and H.Zhou, unpublished results), the role played by 2′-O-methylated nucleotides in mRNA splicing can only be speculated at. Being more closely related to mammalian cells with respect to snRNA methylation pattern as compared to budding yeast, S.pombe is an attractive organism in which to study the occurrence and function of post-transcriptional modifications of snRNA. Characterization of the homolog of mgU6-47 RNA from the fission yeast S.pombe provides the first example in the field.
The mgU6-47 gene is required for S.pombe U6 snRNA methylation
The internal modification of U6 snRNA specified by the snoRNA guide process was first revealed by identification of mgU6-47 and mgU6-77 RNA (10). mgU6-77 RNA, which possesses an unusual sequence complementarity to both U6 snRNA and 28S rRNA, was suggested to function in the methylation of two different kinds of RNA. This suggestion was proved by microinjection of antisense RNA into Xenopus laevis oocytes. In contrast to mgU6-77 RNA, preliminary data suggested that mgU6-47 RNA functions as a RNA guide for site-specific methylation of U6 snRNA. However, there is no direct evidence for this yet. In our experiment, disruption of the S.pombe mgU6-47 gene resulted in complete abolition of methylation at A41 of U6 snRNA. This result is consistent with the sequence comparison between U6 snRNA and mgU6-47 RNA (10). The absolute effect of the gene disruption on methylation of A47 in U6 snRNA indicates that the S.pombe mgU6-47 gene is a single copy gene and that its function in site-specific methylation of U6 snRNA cannot be fulfilled in an alternative way in S.pombe. However, the other methylated nucleotides in U6 snRNA were unaffected by this operation. Therefore, formation of the 2′-O-methylated nucleotides in U6 snRNA proceeds independently. In addition to the previous analysis (10), our results provide genetic evidence for a clear relationship between site-specific methylation of U6 snRNA and a snoRNA gene from S.pombe, supporting the function of the nucleolus in the modification of U6 snRNA.
Regulation of splicing at the post-transcriptional level
Although many snRNAs require 2′-O-methylation as a post-transcriptional modification, the function of this modification remains largely unknown. 2′-O-methylation introduces a positive charge and increases the hydrophobicity. This effect may prevent the 2′-hydroxyl group from forming hydrogen bonds with phosphodiester bridges in RNA and/or with peptide bonds in proteins (46). Recently, a conserved pseudouridine modification in U2 snRNA has been reported to induce a change in the structure and stability of the branch site sequence (47). It is likely that conserved modifications in snRNAs confer a slight advantage to the organism by strengthening or weakening the interactions through hydrogen bonding between RNA and RNA or protein. Among all snRNAs, U6 snRNA is the most highly conserved. U6 snRNA is very sensitive to mutation and chemical modification, probably because it is directly involved in the catalysis of splicing (48,49). In this work we have demonstrated that the abolition of U6 pre-RNA methylation at A41 by disrupting the mgU6-47 gene does not necessarily result in the inhibition of U6 pre-RNA maturation and snRNP assembly in yeast. Normal growth of the disrupted strain was observed at different temperatures, revealing that mgU6-47 RNA is dispensable in fission yeast. However, a slight accumulation of RNA precursors was observed when cell cultures were shifted from 23 to 37°C, suggesting that fine tuning by site-specific 2′-O-methylation is involved in the regulation of splicing. This tuning is sensitive to the temperature shift. Our results imply that the efficiency of splicing may be regulated at the post-transcriptional level, although a low importance of U snRNA post-transcriptional modification for the yeast splicing machinery has been suggested (15). It is worth noting that the effect was produced by deletion of only one of four methylations normally present in S.pombe U6 snRNA. A more significant effect would possibly be evident if all four methylations in U6 snRNA were abolished.
How U6 snRNA trafficks through the nucleolus
The existence of an intron sequence in S.pombe U6 pre-RNA makes its biogenesis complicated. It appears that the spliced U6 RNA does not immediately integrate into the spliceosome, although it must first pass into the spliceosome for intron processing. A further trafficking through the nucleolus is necessary for the modification of U6 snRNA. It would be of interest to know how the spliced U6 snRNA trafficks through the nucleolus for modification. In addition to the individual U6 and U4 snRNPs, the U4 and U6 snRNAs can interact by base paring and organize as a U4/U6 snRNP in the nucleus. It has been reported that pseudouridine formation in U6 snRNA is dependent on its interaction with U4 snRNA (50) and that some internal methylations occur in mammalian U4 snRNA and maximal pseudouridine formation in mammalian U4 snRNA requires nuclear factors (6). It is very likely that unmodified U6 snRNA is accompanied by U4 snRNA in trafficking through the nucleolus and that U4 snRNA also requires a snoRNA for internal modification. The recent identification of a C/D box snoRNA predicted to direct U4 methylation (51) provides further evidence for this speculation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jean-Pierre Bachellerie and Yves Henry for helpful suggestions on our experiments. We are particularly grateful to Xin-Jie Chen and Marlene Faubladier for their experimental help and helpful discussions. We thank Wei-Xin Zhou and Jin Zhao for their assistance in the analysis of D.melanogaster RNA. We also thank Dr Henri Grosjean for revising the text of the manuscript. This research was supported by the National Natural Science Foundation of China (key projects 39730300, 39970171 and 30170216), by a Fund for Distinguished Young Scholar from the Education Ministry of China and the Natural Science Foundation of Guangdong province (990242).
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +86 20 84112399; Fax: +86 20 84036551; Email: lsbrc04@zsu.edu.cn AJ007733, AJ007735 and AJ007736
REFERENCES
- 1.Reddy R. and Busch,H. (1988) Small nuclear RNAs: RNA sequences, structure, and modifications. In Birnstiel,M. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer Verlag, Berlin, Germany, pp. 1–37.
- 2.Gu J., Patton,J.R., Shimba,S. and Reddy,R. (1996) Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA, 2, 909–918. [PMC free article] [PubMed] [Google Scholar]
- 3.Mattaj J.W. (1988) UsnRNP assembly and transport. In Birnstiel,M. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer Verlag, Berlin, Germany, pp. 100–114.
- 4.Izaurralde E. and Mattaj,I.W. (1992) Transport of RNA between nucleus and cytoplasm. Semin. Cell Biol., 3, 279–288. [DOI] [PubMed] [Google Scholar]
- 5.Yu Y.Y., Shu,M.D. and Steitz,A. (1998) Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J., 17, 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zerby D.B. and Patton,J.R. (1997) Modification of human U4 RNA requires U6 RNA and multiple pseudouridine synthases. Nucleic Acids Res., 25, 4808–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vankan P., McGuigan,C. and Mattaj,I.W. (1990) Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J., 9, 3397–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganot P., Jady,B.E., Bortolin,M.L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange T.S. and Gerbi,S.A. (2000) Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell, 11, 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- 11.Ares M. Jr and Weiser,B. (1995) Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol., 50, 131–159. [DOI] [PubMed] [Google Scholar]
- 12.Massenet S., Mougin,A. and Branlant,C. (1998) Posttranscriptional modifications in the U small nuclear RNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM press, Washington, DC, pp. 201–227.
- 13.Pan Z.Q. and Prives,C. (1989) U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev., 3, 1887–1898. [DOI] [PubMed] [Google Scholar]
- 14.Segault V., Will,C.L., Sproat,B.S. and Luhrmann,R. (1995) In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J., 14, 4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPheeters D.S., Fabrizio,P. and Abelson,J. (1989) In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev., 3, 2124–2126. [DOI] [PubMed] [Google Scholar]
- 16.McPheeters D.S. and Abelson,J. (1992) Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell, 71, 819–831. [DOI] [PubMed] [Google Scholar]
- 17.Russell P. and Nurse,P. (1986) Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell, 45, 781–782. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 35, 897–934. [DOI] [PubMed] [Google Scholar]
- 19.Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 20.Cavaille J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 25.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu L.H., Henras,A., Lu,Y.J., Zhou,H., Zhou,W.X., Zhu,Y.Q., Zhao,J., Henry,Y., Caizergues-Ferrer,M. and Bachellerie,J.P. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 732–735. [DOI] [PubMed] [Google Scholar]
- 28.Qu L.H., Henry,Y., Nicoloso,M., Michot,B., Azum,M., Renalier,M., Caizergues-Ferrer,M. and Bachellerie,J.B. (1995) U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res., 23, 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 30.Bachellerie J.P. and Cavaille,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. [DOI] [PubMed] [Google Scholar]
- 31.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 32.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 33.Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation rRNA. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- 34.Tollervey D. (1996) Small nucleolar RNAs guide ribosomal RNA methylation. Science, 273, 1056–1057. [DOI] [PubMed] [Google Scholar]
- 35.Saluz H., Choffat,Y. and Kubli,E. (1988) Primary and hypothetical secondary structure of Drosophila melanogaster U6 RNA. Nucleic Acids Res., 16, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavaille J. and Bachellerie,J.B. (1996) Processing of fibrillarin-associated snoRNAs from pre-mRNA intron: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie, 78, 443–456. [DOI] [PubMed] [Google Scholar]
- 37.Darzacq X. and Kiss,T. (2000) Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5′,3′-terminal stem structure. Mol. Cell. Biol., 20, 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caffarelli E., Fatica,A., Prisley,S., De Gregorio,E., Fragapane,P. and Bozzoni,I. (1996) Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing and the stability of the mature snoRNA. EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 39.Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3743–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maden B.E.H. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 41.Tani T. and Ohshima,Y. (1989) The gene for the U6 small nuclear RNA in fission yeast has an intron. Nature, 337, 87–90. [DOI] [PubMed] [Google Scholar]
- 42.Tani T. and Ohshima,Y. (1991) mRNA-type introns in U6 small nuclear RNA genes: implications for the catalysis in pre-mRNA splicing. Genes Dev., 5, 1022–1031. [DOI] [PubMed] [Google Scholar]
- 43.Potashkin J. and Frendewey,D. (1989) Splicing of the U6 RNA precursor is impaired in fission yeast pre-mRNA splicing mutants. Nucleic Acids Res., 17, 7821–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahlberg J.E. and Lund,E. (1988) The genes and transcription of the major small nuclear RNAs. In Birnstiel,M. (ed.), Structure and Functions of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer Verlag, Berlin, Germany, pp. 38–70.
- 45.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M., Louis,E.J., Mewes,H.W., Murakami,Y., Philippsen,P., Tettelin,H. and Oliver,S.G. (1996) Life with 6,000 genes. Science, 274, 546–567. [DOI] [PubMed] [Google Scholar]
- 46.Nichols J.L. and Lane,B.G. (1966) The characteristic alkali-stable dinucleotide sequences in each of the 16S and 23S components of ribonucleates from E. coli. Can. J. Biochem., 44, 1633–1645. [Google Scholar]
- 47.Newby M.I. and Greenbaum,N.L. (2001) A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA, 7, 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsen T.W. (2001) The case for an RNA enzyme. Nature, 408, 782–783. [DOI] [PubMed] [Google Scholar]
- 49.Yean S.L., Wuenschell,G., Termini,J. and Lin,R.J. (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature, 408, 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zerby D.B. and Patton,J.R. (1996) Metabolism of pre-messenger RNA splicing cofactors: modification of U6 RNA is dependent on its interaction with U4 RNA. Nucleic Acids Res., 24, 3583–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]