Abstract
T4 polynucleotide kinase (Pnk) is a bifunctional 5′-kinase/3′-phosphatase that aids in the repair of broken termini in RNA by converting 3′-PO4/5′-OH ends into 3′-OH/5′-PO4 ends, which are then sealed by RNA ligase. Here we have employed site-directed mutagenesis (introducing 31 mutations at 16 positions) to locate candidate catalytic residues within the 301 amino acid Pnk polypeptide. We found that alanine substitutions for Arg38 and Arg126 inactivated the 5′-kinase, but spared the 3′-phosphatase activity. Conservative substitutions of lysine or glutamine for Arg38 and Arg126 did not restore 5′-kinase activity. These results, together with previous mutational studies, highlight a constellation of five amino acids (Lys15, Ser16, Asp35, Arg38 and Arg126) that likely comprise the 5′-kinase active site. Four of these residues are conserved at the active sites of adenylate kinases (Adk), suggesting that Pnk and Adk are structurally and mechanistically related. We found that alanine substitutions for Asp165, Asp167, Arg176, Arg213, Asp254 and Asp278 inactivated the 3′-phosphatase, but spared the 5′-kinase. Conservative substitutions of asparagine or glutamate for Asp165, Asp167 and Asp254 did not revive the 3′-phosphatase activity, nor did lysine substitutions for Arg176 and Arg213. Glutamate in lieu of Asp278 partially restored activity, whereas asparagine had no salutary effect. Alanine substitutions for Arg246 and Arg279 partially inactivated the 3′-phosphatase; the conservative R246K change restored activity, whereas R279K had no benefit. The essential phosphatase residues Asp165 and Asp167 are located within a 165DxDxT169 motif that defines a superfamily of phosphotransferases. Our data suggest that the 3′-phosphatase active site incorporates multiple additional functional groups.
INTRODUCTION
T4 polynucleotide kinase (Pnk) is the founding member of a family of 5′-kinase/3′-phosphatase enzymes that heal broken termini in RNA or DNA by converting 3′-PO4/5′-OH ends into 3′-OH/5′-PO4 ends, which are then available for sealing by RNA or DNA ligases (1–3). Pnk catalyzes the transfer of the γ-phosphate from ATP or other nucleoside triphosphates to the 5′-OH terminus of DNA, RNA and nucleoside 3′-monophosphates. Pnk also catalyzes the hydrolytic removal of 3′-PO4 termini from DNA, RNA and deoxynucleoside 3′-monophosphates (4–8). The 5′-kinase and 3′-phosphatase reactions both require a divalent cation. The stereochemical course of the 5′-kinase reaction entails inversion of configuration of the transferred phosphate, indicative of an in-line mechanism in which the polynucleotide 5′-OH directly attacks the γ-phosphorus of ATP (9). The mechanism of the 3′-phosphatase reaction is not known.
Recombinant Pnk produced in Escherichia coli is a homotetramer of a 301 amino acid polypeptide (10). We are undertaking a dissection of the domain structure of T4 Pnk, via site-directed mutagenesis and biochemical methods, to localize the essential constituents of the active sites. We initially characterized deletion mutants Pnk(42–301) and Pnk(1–181), which correspond to domains defined by proteolysis with chymotrypsin. Pnk(1–181) is a monomer with no 3′-phosphatase and low residual 5′-kinase activity. Pnk(42–301) is a dimer with no 5′-kinase and low residual 3′-phosphatase activity (10). These results confirmed and extended earlier studies of Pnk purified from T4-infected bacteria (11), which had implicated the N-terminus of Pnk in the 5′-kinase reaction and the C-terminus in the 3′-phosphatase function.
To map the active sites, we introduced alanine and conservative mutations at 17 residues of T4 Pnk that are conserved in its closest homolog, a putative 5′-kinase/3′-phosphatase encoded by the AcNPV baculovirus (Fig. 1B) (10). Alanine substitutions for Lys15, Ser16 and Asp35 inactivated the 5′-kinase, but did not affect the 3′-phosphatase or the tetrameric quaternary structure. 5′-Kinase activity was ablated by the conservative mutations K15R, K15Q and D35N; however, kinase activity was restored by the S16T change. Lys15 and Ser16 are located within an A box motif, GxxGxGK(S/T), found in many NTP phosphohydrolases and phosphotransferases. Mutation D167A inactivated the 3′-phosphatase, without affecting the 5′-kinase or tetramerization. Asp167 resides within a putative phosphatase motif FDLDGTL. Mutation D85A caused a severe decrement in 5′-kinase activity and had only a modest effect on the 3′-phosphatase; the nearby N87A mutation resulted in a significantly reduced 3′-phosphatase activity and slightly reduced 5′-kinase activity. D85A and N87A both affected the quaternary structure, resulting in a mixed population of tetramer and dimer species. Alanine mutations at 11 other conserved positions (indicated by + in Fig. 1B) had no significant effect on either 5′-kinase or 3′-phosphatase activity.
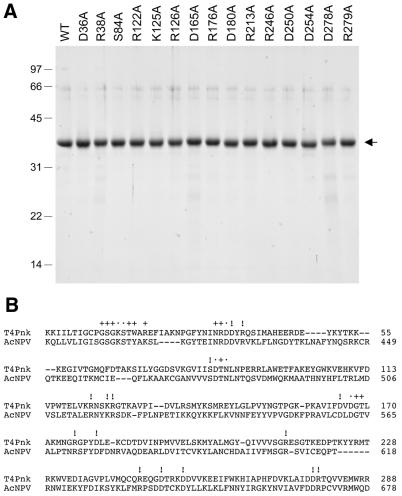
Figure 1.
Purification of wild-type Pnk and Pnk-Ala mutants. (A) Pnk purification. Aliquots (5 µg) of the indicated nickel–agarose Pnk preparations were analyzed by SDS–PAGE. Polypeptides were visualized by staining the gel with Coomassie brilliant blue dye. A scan of the stained gel is shown. The positions and sizes (in kDa) of marker proteins are indicated on the left. (B) The amino acid sequence of T4 Pnk from amino acids 2 to 288 is aligned with that of the related ORF86 polypeptide encoded by Autographa californica nuclear polyhedrosis virus (AcNPV). The amino acids of Pnk that were previously mutated to alanine are denoted by + (non-essential for activity) or • (important for either 5′-kinase or 3′-phosphatase activity). The amino acids in Pnk that were mutated to alanine in the present study are indicated by !.
These findings suggested a complex functional organization of T4 Pnk in which the 5′-kinase and 3′-phosphatase sites are distinct. Here we performed a further round of mutational analysis of 15 new residues of T4 Pnk. We found by alanine scanning that Arg38 and Arg126 are essential for the 5′-kinase and Asp165, Arg176, Arg213, Asp254 and Asp278 are essential for 3′-phosphatase activity. Structure–activity relationships at the essential positions were gauged by introducing conservative substitutions.
MATERIALS AND METHODS
T4 Pnk mutants
Missense mutations were introduced into the Pnk gene using the two-stage PCR-based overlap extension method (12). pET-PNK was used as the template for the first stage PCR reaction. NdeI–BamHI restriction fragments of the mutated second stage PCR products were inserted into pET16b. The inserts of the resulting plasmids were sequenced to confirm the presence of the desired mutations and the absence of any unwanted coding changes.
The wild-type and mutant pET-PNK plasmids were transformed into E.coli BL21(DE3). Recombinant protein production was induced by adjusting exponentially growing cultures to 0.3 mM isopropyl-β-d-thiogalactopyranoside and then incubating them at 17°C for 15 h with continuous shaking. The His-tagged Pnk proteins were purified from soluble bacterial extracts by Ni–NTA–agarose affinity chromatography as described previously (10). Protein concentrations were determined using the Bio-Rad dye reagent with bovine serum albumin as the standard.
5′-Kinase assay
Reactions mixtures (10 µl) containing 70 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 5 mM DTT, 25 µM [γ-32P]ATP, ∼50 pmol of a synthetic 5′-OH oligodeoxyribonucleotide d(ATTCCGATAGTGACTACA), and Pnk as specified were incubated for 20 min at 37°C. The products were analyzed by electrophoresis through a 15 cm 15% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris–borate, 2.5 mM EDTA). The radiolabeled oligonucleotide products were visualized and quantitated by scanning the gel with a Fujix BAS2500 phosphorimager.
3′-Phosphatase assay
Reaction mixtures (25 µl) containing 100 mM imidazole, pH 6.0, 10 mM MgCl2, 10 mM β-mercaptoethanol, 0.1 mg/ml BSA, 1.6 mM 3′-dTMP (Sigma) and Pnk as specified were incubated for 20 min at 37°C. The reactions were quenched by adding 75 µl of cold water and 1 ml of malachite green reagent (Biomol Green; Biomol Research Laboratories, Plymouth Meeting, PA). Phosphate release was determined by measuring A620 and extrapolating the value to a phosphate standard curve.
Velocity sedimentation
Aliquots (40 µg) of the wild-type and mutant Pnk preparations were mixed with catalase (30 µg), BSA (30 µg) and cytochrome c (30 µg) and the mixtures were applied to 4.8 ml 15–30% glycerol gradients containing 50 mM Tris–HCl, pH 8.0, 0.2 M NaCl, 1 mM EDTA, 2.5 mM DTT and 0.1% Triton X-100. The gradients were centrifuged in a SW50 rotor at 50 000 r.p.m. for 15.5 h at 4°C. Fractions were collected from the bottoms of the tubes. Aliquots (20 µl) of odd numbered gradient fractions were analyzed by SDS–PAGE.
RESULTS
Alanine scanning mutagenesis of T4 Pnk
A goal of the present study was to extend the maps of the active sites of T4 Pnk by identifying individual amino acid functional groups required for the kinase and phosphatase reactions. To do this, we introduced single alanine changes at 15 amino acids of the Pnk polypeptide that are conserved in its baculovirus homolog (13). The mutated positions are indicated by ! in Figure 1B. We focused in particular on: (i) conserved basic and acid residues in the N-terminal kinase domain (Asp36, Arg38, Arg122, Lys125 and Arg126) as potential ligands for the ATP substrate or the divalent cation cofactor; (ii) Ser84, a conserved position adjacent to Asp85 and Asn87, which are implicated in enzymatic activity and stabilization of the tetrameric quaternary structure; (iii) Asp165 within the putative phosphatase motif 164FDVDGTL170; (iv) conserved basic and acidic residues in the C-terminal phosphatase domain (Arg176, Asp180, Arg213, Arg246, Aps250, Asp254, Asp278 and Arg279) as potential ligands for the 3′-phosphate or the divalent cation cofactor.
Wild-type Pnk and the 15 Pnk-Ala mutants were produced in bacteria as His10-tagged fusions and purified from soluble bacterial extracts by Ni–agarose chromatography. SDS–PAGE analysis of the imidazole eluate fractions showed that the preparations were highly enriched with respect to the His-Pnk polypeptide (Fig. 1A).
Mutational effects on 5′-OH polynucleotide kinase activity
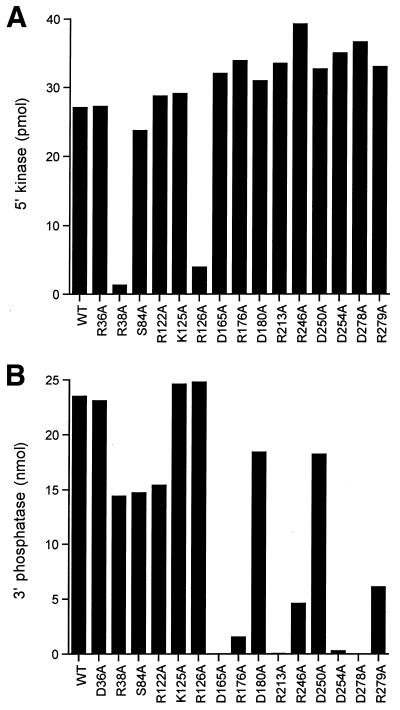
The 5′-kinase activity was measured by label transfer from [γ--32P]ATP to a 5′-OH DNA oligonucleotide. All of the enzyme preparations were assayed in parallel; the kinase reaction mixtures contained 80 ng input Pnk, an amount sufficient for saturating levels of DNA labeling by the wild-type enzyme (see Fig. 3). Mutants R38A and R126A were defective (Fig. 2A). (Our operational criterion for a significant mutational effect is one that elicits at least a 4-fold decrement in activity compared to wild-type Pnk.) The other mutant proteins retained 5′-kinase activity. We surmise that the Asp36, Ser84, Arg122, Lys125, Asp165, Arg176, Asp180, Arg213, Arg246, Asp250, Asp254, Asp278 and Arg279 side chains do not contribute significantly to catalysis of the kinase reaction.
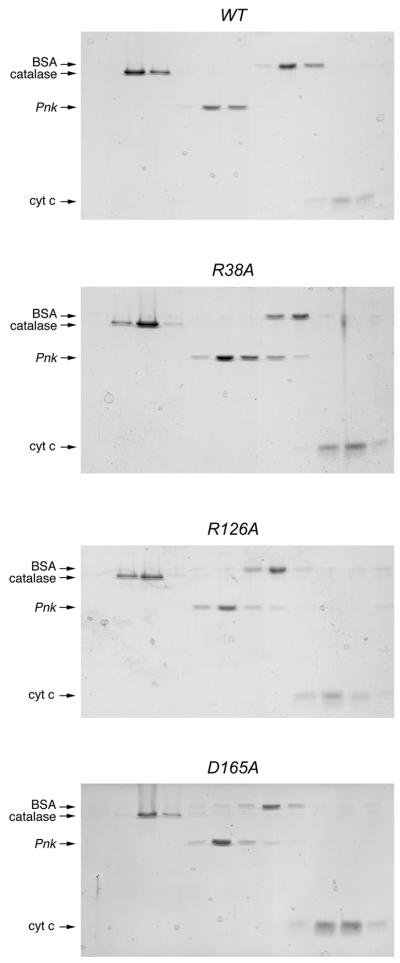
Figure 3.
Glycerol gradient sedimentation of Pnk and Pnk-Ala mutants. Sedimentation analysis was performed as described in Materials and Methods. The distributions of Pnk (either wild-type, R38A, R126A or D165A as indicated) and the marker proteins catalase, BSA and cytochrome c in each gradient were analyzed by SDS–PAGE. Scans of the Coomassie blue stained gels are shown.
Figure 2.
5′-Kinase and 3′-phosphatase activity of wild-type Pnk and Pnk-Ala mutants. (A) 5′-Kinase activity. Aliquots (80 ng) of the indicated Pnk preparations were assayed for 5′-kinase activity as described in Materials and Methods. (B) Aliquots (50 ng) of the indicated Pnk preparations were assayed for 3′-phosphatase activity as described in Materials and Methods.
The native sizes of wild-type Pnk and the two catalytically defective mutants were investigated by sedimentation through 15–30% glycerol gradients. The marker proteins catalase, BSA and cytochrome c were included as internal standards. After centrifugation, the polypeptide compositions of the odd numbered gradient fractions were analyzed by SDS–PAGE (Fig. 3). The 37 kDa wild-type Pnk polypeptide sedimented between catalase (248 kDa) and BSA (66 kDa), consistent with a globular tetrameric quaternary structure for Pnk. The R38A and R126A proteins sedimented similarly, although we did note a shoulder on the light side of the R38A tetramer peak, sedimenting just ahead of BSA, which suggested that the R38A preparation contained a minority component of dimeric Pnk.
Structure–activity relationships at Arg38 and Arg126
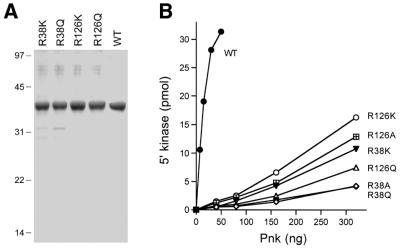
Arg38 and Arg126 were substituted conservatively by lysine and glutamine. The R38K, R38Q, R126K and R126Q proteins were purified from soluble bacterial extracts by Ni–agarose chromatography (Fig. 4A). Kinase activity was measured as a function of enzyme concentration for wild-type Pnk, the two Arg→Ala mutants and the four conservative mutants (Fig. 4B). The specific activity of the R38A mutant was 1% of the wild-type enzyme. Whereas introduction of lysine elicited a slight gain of function (to 3% of wild-type activity), glutamine had no salutary effect. R126A was 3% as active as wild-type Pnk and substitution by lysine (4% of wild-type) or glutamine (2% of wild-type) did not revive activity. These data establish a clear requirement for a positively charged residue, and arginine specifically, at positions 38 and 126 of T4 Pnk.
Figure 4.
Effects of conservative substitutions on 5′-kinase activity. (A) Pnk purification. Aliquots (5 µg) of the indicated nickel–agarose Pnk preparations were analyzed by SDS–PAGE. A scan of the Coomassie blue stained gel is shown. The positions and sizes (in kDa) of marker proteins are indicated on the left. (B) 5′-Kinase activity. The indicated Pnk preparations were assayed for 5′-kinase activity as described in Materials and Methods. Activity is plotted as a function of input Pnk.
Effects of alanine mutations on 3′-phosphatase activity
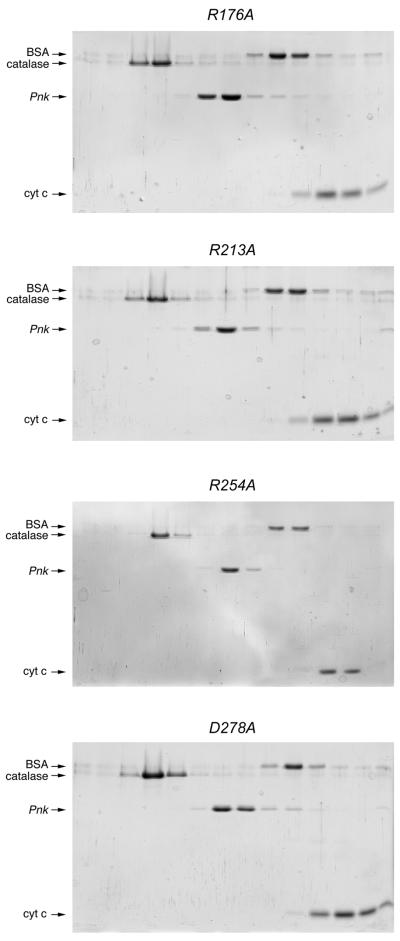
Phosphatase activity was measured by the release of inorganic phosphate from deoxythymidine 3′-monophosphate (3′-dTMP). The wild-type Pnk and 15 Pnk-Ala mutants were assayed in parallel; the 3′-phosphatase reaction mixtures contained 50 ng Pnk, an amount sufficient for dephosphorylation of ∼60% of the input 3′-dTMP substrate. The D165A, R176A, R213A, D254A and D278A mutations abrogated or severely reduced 3′-phosphatase function (Fig. 2B). Glycerol gradient analysis showed that the D165A, R176A, R213A, D254A and D278A proteins sedimented as tetramers (Figs 3 and 5). Because the D165A, R176A, R213A, D254A and D278A mutations did not affect 5′-kinase function or protein quaternary structure, we construe that Asp165, Arg176, Arg213, Asp254 and Asp278 are likely constituents of the 3′-phosphatase active site.
Figure 5.
Glycerol gradient sedimentation of phosphatase-defective Pnk-Ala mutants. Sedimentation analysis was performed as described in Materials and Methods. The distributions of Pnk (either R176A, R213A, R254A or D278A as indicated) and the marker proteins catalase, BSA and cytochrome c in each gradient were analyzed by SDS–PAGE. Scans of the Coomassie blue stained gels are shown.
The R246A and R279A changes elicited partial loss of 3′-phosphatase activity (Fig. 2B), whereas the D180A and D250A mutations within the C-terminal domain did not have significant effects. All six of the alanine mutants in the N-terminal segment of Pnk generated in the present study retained 3′-phosphatase activity, including R38A and R126A, which were defective in 5′-kinase activity (Fig. 2). These data provide additional evidence for distinct (and perhaps completely separate) 5′-kinase and 3′-phosphatase active sites.
Structure–activity relationships at Asp165 and Asp167
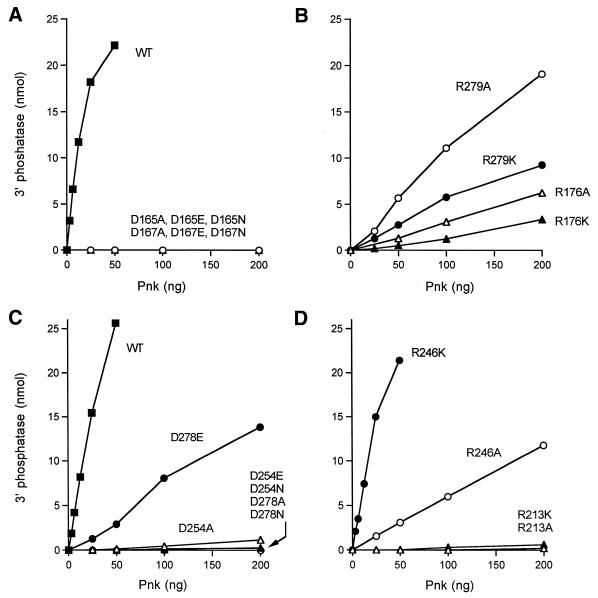
The magnitude of the Asp165A mutational effect was determined by enzyme titration (Fig. 6A). Wild-type Pnk released 0.8 nmol Pi from 3′-dTMP per ng protein in the linear range of enzyme dependence, which translates into a turnover number of ∼24 s–1. In contrast, the D165A mutant released no detectable Pi in the range 25–200 ng input protein. Thus, the specific activity of D165A was <0.1% of the wild-type value. We previously showed that mutant D167A was defective in 3′-phosphatase function, but active as a 5′-phosphatase (10). Here we have quantitated the magnitude of the mutational effect by protein titration. The specific activity of D167A was <0.1% of the wild-type value (Fig. 6A).
Figure 6.
Effects of alanine and conservative substitutions on 3′-phosphatase activity. 3′-Phosphatase activity was determined as a function of input protein for wild-type Pnk, D165A, D65N, D65E, D167A, D167N and D167E (A), R176A, R176K, R279A and R279K (B), wild-type Pnk, D254A, D254N, D254E, D278A, D278N and D278E (C) and R213A, R213K, R246A and R246K (D).
To further evaluate the contributions of Asp165 and Asp167 to the 3′-phosphatase reaction, we tested the effects of conservative asparagine and glutamate substitutions. The D165N, D165E, D167N and D167E proteins were purified from soluble bacterial extracts by Ni–agarose chromatography (Fig. 7). The 3′-phosphatase specific activities of the conservative mutants were <0.1% of the activity of wild-type Pnk (Fig. 6A). These data establish a clear requirement for a carboxylate residue at positions 165 and 167 of T4 Pnk and they indicate a tight steric constraint on the distance from the main chain to the carboxylates, such that the active site is perturbed by the additional methylene group of glutamate.
Figure 7.
Purification of Pnk mutants with conservative substitutions in the phosphatase domain. Aliquots (5 µg) of the indicated nickel–agarose Pnk preparations were analyzed by SDS–PAGE. Polypeptides were visualized by staining the gel with Coomassie brilliant blue dye. A scan of the stained gel is shown. The positions and sizes (in kDa) of marker proteins are indicated on the left.
Structure–function relationships for the phosphatase activity of Pnk
In classifying mutational effects on Pnk function, we adopted the following criteria of significance: (i) functional groups were deemed essential when their replacement by alanine reduced specific activity to ≤5% of wild-type Pnk; (ii) residues were deemed important when alanine substitution lowered activity to between 6 and 25% of wild-type. We found that the specific activity of the R176A mutant was 4% of wild-type and the R176K mutant was 2% as active as wild-type (Fig. 6B). The R213A and R213K mutants had 0.1 and 0.4% of wild-type activity, respectively (Fig. 6D). Thus, Arg176 and Arg213 are essential for the 3′-phosphatase reaction and lysine, though positively charged, cannot substitute for arginine in either case. The R246A and R279A proteins were 7 and 12% as active as the wild-type, respectively (Fig. 6B and D). Hence, Arg246 and Arg279 are important for activity. Whereas introduction of lysine at position 246 restored 3′-phosphatase activity to near wild-type level (73%), the lysine change at position 279 was of no benefit (7% activity).
The 3′-phosphatase activities of the D254A and D278A mutants were 0.7 and <0.1% of wild-type, respectively (Fig. 6C). The D254N (0.1%) and D254E (0.1%) changes were at least as deleterious as the alanine mutation, indicating a requirement for a carboxylate at position 254 and a constraint on the main chain to carboxylate distance. A carboxylate was also essential at position 278, insofar as the D278N change (<0.1% activity) did not restore function. However, the conservative D278E mutation elicited a significant gain of function to 9% of the wild-type level. We surmise that there is less steric constraint at position 278 than at the other essential acidic residues of the phosphatase component of Pnk.
DISCUSSION
We employed site-directed mutagenesis of 15 residues to identify seven new essential constituents of the separate active sites for the 5′-kinase and 3′-phosphatase activities of T4 Pnk. In conjunction with an earlier study, we have now pinpointed five amino acids that are essential for the kinase (Lys15, Ser16, Asp35, Arg38 and Arg126) and six amino acids that are essential for the 3′-phosphatase (Asp165, Asp167, Arg176, Arg213, Asp254 and Asp278). None of the alanine mutations at the essential amino acids appear to perturb the quaternary structure of Pnk. In addition, we have identified Arg246 and Arg279 as being important for 3′-phosphatase activity, albeit not essential by our criteria.
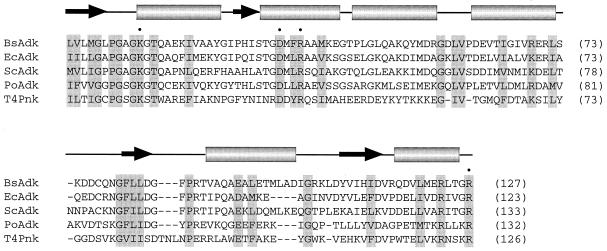
Our results suggest structural and mechanistic similarities between the kinase component of Pnk and adenylate kinase (Adk). Adk catalyzes phosphoryl transfer from ATP to AMP to form two molecules of ADP. The Adk reaction is similar to that of Pnk insofar as it entails nucleophilic attack on the γ-phosphorus of ATP via an in-line mechanism and is dependent on a divalent cation cofactor (14). In the Adk reaction, the attacking nucleophile is a 5′-phosphate oxygen of AMP, whereas in the Pnk reaction it is the 5′-OH of the nucleoside sugar. Figure 8 shows a primary and secondary structure alignment of the N-terminal ∼130 amino acid segments of Bacillus stearothermophilus, E.coli, yeast and porcine Adk enzymes. High-resolution crystal structures are available for these four Adk proteins, including structures of Adk bound to the bisubstrate analog Ap5A (15–18). This segment of the Adk proteins folds into a central four-stranded parallel β-sheet surrounded by α-helices. Blast searches followed by manual alignment of the N-terminal kinase domain of T4 Pnk to the Adk proteins revealed 32 of 123 positions of identity or functional group similarity between Pnk and the four Adk enzymes (shaded in Fig. 8). A single comparison of Pnk to the porcine Adk showed 44 of 123 positions of identity plus similarity. The instructive point was that four of the residues we have identified as essential for the 5′-kinase activity of Pnk (Lys15, Asp35, Arg38 and Arg126) are conserved in all of the Adk enzymes (Fig. 8).
Figure 8.
Structural similarities between T4 Pnk and Adk. The amino acid sequences of B.stearothermophilus (PDB 1ZIN), E.coli (1AKE), Saccharomyces cerevisiae (1AKY) and porcine (3ADK) Adk are aligned with each other and with the N-terminal kinase domain of T4 Pnk. The amino acids of Pnk that are essential for 5′-kinase activity and conserved in Adk are denoted by •. The secondary structure elements of Adk are shown above the aligned sequences. Positions of identity/similarity in all five of the proteins are highlighted in shaded boxes.
The N-terminal kinase domain of T4 Pnk contains the A box motif GxxGxGKS. In Adk the A box comprises a loop connecting a β-strand to an α-helix, the said loop forming a binding pocket for the β-phosphate of ATP via hydrogen bonds with the backbone amides of the loop residues (14–19). The invariant A box lysine (equivalent to Lys15 in Pnk) makes direct contact with the β- and γ-phosphates of the bound nucleotide.
The essential side chains Asp35 and Arg38 of Pnk are located 20 and 23 amino acids downstream of the A box lysine. An aspartate and arginine are located at the identical positions in all four of the Adk proteins (Fig. 8). In the Adk crystal structure the AspOδ1 engages in a hydrogen bond with one of the terminal guanidinium nitrogens of the arginine, while the other terminal nitrogen of arginine donates a hydrogen bond to the 5′-phosphate of AMP. A crystal structure of B.stearothermophilus Adk bound to Ap5A plus magnesium or manganese showed that the Asp and Arg side chains both form hydrogen bonds to a water molecule within the octahedral coordination sphere of the metal ion (18). Our mutational findings are consistent with similar multivalent contacts for Asp35 and Arg38 in the 5′ -kinase reaction of T4 Pnk, i.e. the conservative D35N and R38K mutations did not restore 5′-kinase activity. Asp35 and Arg38 may be involved in metal binding via water, as in Adk. However, because there is no interaction with an attacking phosphate in the Pnk reaction, as there is in Adk, we speculate that Arg38 of Pnk might make other essential contacts, either with the γ-phosphate of ATP or the 5′-OH of the phosphate acceptor. The DxxR motif is conserved in the baculovirus Pnk homolog, but not in the human 5′-OH DNA kinase (2,3). The sequence of the human protein downstream of the A box is NRDTLGSWQR. The NRD triplet (embracing the equivalent of T4 residue Asp35) is conserved in the T4 and baculovirus Pnk proteins, but there is no basic residue in human DNA kinase at the position of T4 Arg38. Conceivably, the Arg located seven amino acids downstream of the Asp in DNA kinase could play a catalytic role, but there has been no mutational analysis of human DNA kinase to bear on this issue. In any event, it would appear that human DNA kinase does not adhere to the arrangement of catalytic side chains in the primary structure that is conserved in T4 Pnk and the Adk family.
Arg126 of T4 Pnk is essential for phosphoryl transfer. This position is conserved as arginine in all four Adk enzymes and the arginine is essential for Adk activity (14–19). The terminal guanidinium nitrogens of this arginine in Adk donate multiple hydrogen bonds to the α- and γ-phosphates of ATP. Our data are consistent with a similar role for Arg126 in Pnk. Arg126 is situated within a motif, RxxxR, found in all four of the Adk enzymes (Fig. 8). The proximal arginine stacks on the adenine base of ATP in the Adk structure via a cation–π interaction. Although replacing Arg122 of Pnk with alanine did not elicit a gross catalytic defect, our experiments do not rule out a contribution of Arg122 to the affinity of Pnk for ATP at the 5′-kinase active site.
Prior mutational analysis showed that Asp167 was essential for the 3′-phosphatase activity of Pnk. Here we have identified Asp165 as a second essential, and presumably catalytic, constituent of the 3′-phosphatase active site. Both essential aspartates are conserved in the baculovirus Pnk homolog and in the human DNA 5′-kinase/3′-phosphatase and its homologs in Schizosaccharomyces pombe and Caenorhabditis elegans (2,3). The essential aspartates of Pnk reside within a recently defined phosphatase motif (DxDxT) that was identified as essential for phosphoryl transfer by human phosphomannomutase and l-3-phosphoserine phosphatase (20,21). The latter enzymes exemplify a distinct family of phosphotransferases and phosphohydrolases (20,22), many of which have been shown to act via an acyl-phosphoenzyme intermediate in which the phosphate is linked to the first aspartate in the DxDxT motif (23). Although others have speculated that T4 Pnk and its eukaryotic homologs are members of this phosphatase family (2,3,24,25), our demonstration that both aspartates of the DxDxT motif are essential for the 3′-phosphatase activity of T4 Pnk (and not for its 5′-kinase function) provides evidence that polynucleotide 3′-phosphatase may be mechanistically related to the DxDxT family. Specifically, the finding that activity is abolished by introduction of an amide in place of either aspartate is consistent with the idea that one of the Asp side chains (presumably the proximal residue, Asp165) could act as a catalytic nucleophile that forms a phosphoenzyme intermediate (23). Still, we do not exclude an alternative mechanism for the 3′-phosphatase component of Pnk which does not involve an acyl-phosphate intermediate. For example, the DxD motif is also a defining feature of the Toprim domains found in DNA topoisomerases (types IA, IIa and IIB) and DNA primases (26–28). Although the Toprim-containing topoisomerases and primases catalyze phosphoryl transfer reactions, they do not form acyl-phosphate intermediates. Rather, the Toprim domain coordinates a divalent cation. Thus, it is possible that the essential 165DxD167 motif of Pnk serves simply to bind the divalent cation cofactor at the 3′-phosphatase active site. Analysis of the stereochemical course of the 3′-phosphatase reaction, together with crystallography of the enzyme in various functional states, will ultimately distinguish between the two mechanisms.
The mutational data indicate that the 3′-phosphatase active site of Pnk is composed of multiple functional groups outside the DxD motif. The basic residues Arg213 and Arg176 are essential for phosphatase activity and their replacement by lysine does not restore function. These results are consistent with direct catalytic roles for Arg213 and Arg176, perhaps entailing bidentate interactions with the 3′-phosphate that stabilize the transition state. Acidic residues Asp254 and Asp278 are also essential for 3′-phosphatase activity and neither could be replaced by asparagine; these findings are consistent with involvement of the carboxylates in either metal binding, general base catalysis or ionic interactions with one of the essential arginines.
REFERENCES
- 1.Amitsur M., Levitz,R. and Kaufman,G. (1987) Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J., 6, 2499–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jilani A., Ramotar,D., Slack,C., Ong,C., Yang,X.M., Scherer,S.W. and Lasko,D.D. (1999) Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem., 274, 24176–24186. [DOI] [PubMed] [Google Scholar]
- 3.Karimi-Busheri F., Daly,G., Robins,P., Canas,B., Pappin,D.J.C., Sgouros,J., Miller,G.G., Fakhrai,H., Davis,E.M., Le Beau,M.M. and Weinfeld,M. (1999) Molecular characterization of a human DNA kinase. J. Biol. Chem., 274, 24187–24194. [DOI] [PubMed] [Google Scholar]
- 4.Richardson C.C. (1965) Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc. Natl Acad. Sci. USA, 54, 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novogrodsky A. and Hurwitz,J. (1966) The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: phosphorylation at 5′-hydroxyl termini. J. Biol. Chem., 241, 2923–2932. [PubMed] [Google Scholar]
- 6.Novogrodsky A., Tal.,M., Traub,A. and Hurwitz,J. (1966) The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: further properties of the 5′-hydroxyl polynucleotide kinase. J. Biol. Chem., 241, 2933–2943. [PubMed] [Google Scholar]
- 7.Becker A. and Hurwitz,J. (1967) The enzymatic cleavage of phosphate termini from polynucleotides. J. Biol. Chem., 242, 936–950. [PubMed] [Google Scholar]
- 8.Cameron V. and Uhlenbeck,O.C. (1977) 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry, 16, 5120–5126. [DOI] [PubMed] [Google Scholar]
- 9.Jarvest R.L. and Lowe,G. (1981) The stereochemical course of the phosphoryl transfer reaction catalyzed by polynucleotide kinase (bacteriophage-T4-infected Escherichia coli B). Biochem. J., 199, 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L.K. and Shuman,S. (2001) Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem. , 276, 26868–26874. [DOI] [PubMed] [Google Scholar]
- 11.Soltis D.A. and Uhlenbeck,O.C. (1982) Independent location of kinase and 3′-phosphatase activities on T4 polynucleotide kinase. J. Biol. Chem., 257, 11340–11345. [PubMed] [Google Scholar]
- 12.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 13.Durantel D., Croizier,L., Ayres,M.D., Croizier,G., Possee,R.D. and Lopez-Ferber,M. (1998) The pnk/pnl gene (ORF 86) of Autographa californica nucleopolyhedrovirus is a non-essential, immediate early gene. J. Gen. Virol., 79, 629–637. [DOI] [PubMed] [Google Scholar]
- 14.Yan H. and Tsai,M.D. (1999) Nucleoside monophosphate kinases: structure, mechanism and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol., 73, 103–134. [DOI] [PubMed] [Google Scholar]
- 15.Dreusicke D., Karplus,P.A. and Schulz,G.E. (1988) Refined structure of porcine cytosolic adenylate kinase at 2.1 Å resolution. J. Mol. Biol., 199, 359–371. [DOI] [PubMed] [Google Scholar]
- 16.Müller C.W. and Schulz,G.E. (1992) Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 Å resolution: a model for a catalytic transition state. J. Mol. Biol., 224, 159–177. [DOI] [PubMed] [Google Scholar]
- 17.Abele U. and Schulz,G.E. (1995) High-resolution structures of adenylate kinase from yeast liganded with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci., 4, 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berry M.B. and Phillips,G.N. (1998) Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. Proteins, 32, 276–288. [DOI] [PubMed] [Google Scholar]
- 19.Byeon L., Shi,Z. and Tsai,M.D. (1995) Mechanism of adenylate kinase: the essential lysine helps to orient the phosphates and the active site residues to proper conformations. Biochemistry, 34, 3172–3182. [DOI] [PubMed] [Google Scholar]
- 20.Collet J.F., Stroobant,V., Pirard,M., Delpierre,G. and Van Schaftingen,E. (1998) A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J. Biol. Chem., 273, 14107–14112. [DOI] [PubMed] [Google Scholar]
- 21.Collet J.F., Stroobant,V. and Van Schaftingen,E. (1999) Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type ATPases. J. Biol. Chem., 274, 33985–33990. [DOI] [PubMed] [Google Scholar]
- 22.Thaller M.C., Schippa,S. and Rossolini,G.M. (1998) Conserved sequence motifs among bacterial, eukaryotic and archaeal phosphatases that define a new phosphohydrolase family. Protein Sci., 7, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho H., Wang,W., Kim,R., Yokota,H., Damo,S., Kin,S.H., Wemmer,D., Kustu,S. and Yan,D. (2001) BeF3– acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3– complex with phosphoserine phosphatase. Proc. Natl Acad. Sci. USA, 98, 8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vance J.R. and Wilson,T.E. (2001) Uncoupling of 3′-phosphatase and 5′-kinase functions in budding yeast: characterization of Saccharomyces cerevisiae DNA 3′-phosphatase (TPP1). J. Biol. Chem., 276, 15073–15081. [DOI] [PubMed] [Google Scholar]
- 25.Betti M., Petrucco,S., Bolchi,A., Dieci,G. and Ottonello,S. (2001) A plant 3′-phosphoesterase involved in the repair of DNA strand breaks generated by oxidative damage. J. Biol. Chem., 276, 18038–18045. [DOI] [PubMed] [Google Scholar]
- 26.Aravind L., Leipe,D.D. and Koonin,E.V. (1998) Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res., 26, 4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols M.D., DeAngeli,K., Keck,J.L. and Berger,J.M. (1999) Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J., 18, 6177–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keck J.L., Roche,D.D., Lynch,S. and Berger,J.M. (2000) Structure of the RNA polymerase domain of E. coli primase. Science, 287, 2482–2486. [DOI] [PubMed] [Google Scholar]