Abstract
Bacillus subtilis is a soil‐dwelling bacterium that can interact with a plethora of other microorganisms in its natural habitat. Due to the versatile interactions and its ability to form nanotubes, i.e., recently described membrane structures that trade cytoplasmic content between neighbouring cells, we investigated the potential of HGT from B. subtilis to industrially‐relevant members of lactic acid bacteria (LAB). To explore the interspecies HGT events, we developed a co‐culturing protocol and provided proof of transfer of a small high copy non‐conjugative plasmid from B. subtilis to LABs. Interestingly, the plasmid transfer did not involve conjugation nor activation of the competent state by B. subtilis. Moreover, our study shows for the first time non‐conjugative cell‐to‐cell intraspecies plasmid transfer for non‐competent Lactococcus lactis sp. cremoris strains. Our study indicates that cell‐to‐cell transformation is a ubiquitous form of HGT and can be potentially utilized as an alternative tool for natural (non‐GMO) strain improvement.
In this work, we investigated the interspecies HGT events in a mix co‐cultures of Bacillus subtilis and members of lactic acid bacteria. We developed a co‐culturing protocol and provided proof of transfer of a small high copy non‐conjugative plasmid from B. subtilis to LABs. Interestingly, the plasmid transfer did not involve conjugation, nor activation of the competent state by B. subtilis. Moreover, our study shows for the first time non‐conjugative cell‐to‐cell intraspecies plasmid transfer for non‐competent Lactococcus lactis sp. cremoris strains. Our study indicates that cell‐to‐cell transformation is a ubiquitous form of HGT and can be potentially utilized as an alternative tool for natural (non‐GMO) strain improvement.
INTRODUCTION
Horizontal gene transfer (HGT) plays a crucial role in bacterial evolution and ecology. In adaptation to changing environments, bacteria acquire foreign DNA that may offer beneficial properties under various selection pressures (Cohan & Koeppel, 2008; Wiedenbeck & Cohan, 2011). Prevalent evidence of the ecological importance of HGT is the acquisition and spread of antibiotic resistance and virulence determinants in closely related species, which can have significant consequences for the emergence of antibiotic‐resistant pathogens (Deng et al., 2019; Schmidt & Hensel, 2004; Siddaramappa et al., 2011).
The generally recognized and well‐known mechanisms of HGT in bacteria are conjugation, transduction, and natural transformation. In the transformation process, bacteria enter a transient state of competence and activate the assembly of the DNA‐uptake machinery to acquire naked DNA directly from the environment (Chen et al., 2005; Chen & Dubnau, 2004; Lorenz & Wackernagel, 1994). This process is genetically encoded in the recipient's cell, and it is regulated by many physiological and environmental factors such as growth phase, nutrient availability, starvation, cell density, and even antibiotic stress (Claverys et al., 2006; Dubnau, 1991a; Hamoen et al., 2003; Håvarstein et al., 1996; Slager et al., 2014). While the DNA‐uptake machinery is generally conserved across species, the induction conditions and type of inducers vary between species. For instance, Bacillus subtilis induces competence at the onset of the stationary phase or in response to high cell density (Anagnostopoulos & Spizizen, 1961; Dubnau, 1991b). In contrast, in Streptococcus pneumoniae, competence activation was observed at a specific time during the exponential phase and was inhibited at the stationary phase (Håvarstein et al., 1995). Notably, many bacteria display a complete set of competence‐induced genes; however, the transformation‐promoting conditions are still not always elucidated (David et al., 2017; Mulder et al., 2017; Wydau et al., 2006).
Unlike natural transformation, conjugation and transduction require the presence of a donor cell, or a donor phage, respectively, and engage specific conduits for DNA exchange. In the case of conjugation, after the mating pair formation between a compatible donor and recipient, the genetic material is transferred through a mating channel or conjugative pilus, typically encoded on the conjugative elements (Auchtung et al., 2016; Cabezón et al., 2015). During transduction, genetic information is carried through phage virions that acquired random fragments of the host's DNA during capsid assembly and were able to infect both donor and recipient (Chiang et al., 2019; Clokie et al., 2011; Marcelli et al., 2020). While conjugation and transduction certainly contribute to HGT in bacteria, their transfer capacity is limited by many factors, including surface exclusion, type and size of genetic material, host‐range specificity, restriction‐modification systems present in the recipient, and sexual isolation (Dahmane et al., 2017; Mahony et al., 2017; Majewski, 2001; Thomas & Nielsen, 2005).
Besides these classical HGT mechanisms, recent evidence support the idea of alternative means of DNA transfer in bacteria, including membrane vesicles (Domingues & Nielsen, 2017), cell‐to‐cell natural transformation (Blesa et al., 2015; Etchuuya et al., 2011; Matsumoto et al., 2016; Zhang et al., 2018), and nanotubes (Dubey & Ben‐Yehuda, 2011). Interestingly, B. subtilis has been shown to transfer genetic material not only via conjugation (Grohmann, 2010a, 2010b; Lee et al., 2012; Rösch et al., 2014; Singh et al., 2013) or phage transduction (Deichelbohrer et al., 1985; Tzipilevich et al., 2017; Yasbin & Young, 1974) but also through cell‐to‐cell non‐conjugative plasmid transfer (Dubey & Ben‐Yehuda, 2011; Zhang et al., 2018), which makes it an excellent candidate to study intra‐ and interspecies HGT events. B. subtilis and members of lactic acid bacteria (LAB) are of great importance in food and feed production and the probiotic market. However, the current strain improvement methods for the production of new starter cultures are limited due to public resistance and reinforced by the European Union directives on the use of genetically modified organisms. Thus, in recent years, the interest in naturally occurring HGT processes for the mobilization of genetic traits has been renewed (Bron et al., 2019; Kuipers, 2015; Marcelli et al., 2020). Here, we explore the potential of HGT within co‐cultures of B. subtilis and industrially‐relevant LAB, including Lactococcus lactis and Streptococcus thermophilus. For the first time, we demonstrate the inter‐ and intraspecies transfer of a high copy, non‐conjugative plasmid in non‐competent and conjugation‐negative B. subtilis and L. lactis strains with co‐culturing techniques.
EXPERIMENTAL PROCEDURES
Strains and growth conditions
B. subtilis strains were grown at 37°C in Lysogeny Broth (LB), either aerated by shaking at 220 rpm or on solidified LB with 1.5% (w/v) agar; L. lactis strains were cultured at 30°C in Difco M17 medium supplemented with glucose at the final concentration 0.5% (w/v) (GM17), as standing cultures or on solidified GM17 with 1.5% (w/v) agar. S. thermophilus strains were grown anaerobically as standing cultures at 37°C in Difco M17 medium supplemented with glucose (0.5% w/v) and lactose (1% w/v) (GLM17) or on solidified GLM17 with 1.5% (w/v) agar. When needed, growth media were supplemented with antibiotics at the following final concentrations: chloramphenicol (cm) 5 μg/ml, tetracycline (tet) 5 μg/ml, spectinomycin (spec) 100 μg/ml, and erythromycin (ery) 5 μg/ml.
All strains used in this study are listed in Table 1. All plasmids and oligonucleotides used are listed in Tables 2 and 3, respectively.
TABLE 1.
Strains used in this study.
| Strain | Species | Genotype | Abr | Reference |
|---|---|---|---|---|
| PY79 | B. subtilis | Prototroph SPβ, ICEBs1 − | – | BGSC |
| PY79 ΔcomK | B. subtilis | PY79; comK::spec | spec | This work |
| PY79 pNZ8048 | B. subtilis | PY79; pNZ8048 | cm | This work |
| 168 | B. subtilis | trpC2 | – | BGSC |
| 168 pNZ8048 | B. subtilis | 168 trpC2, pNZ8048 | – | BGSC |
| 168 ΔcomK | B. subtilis | 168 trpC2, comK::spec | spec | Laboratory stock |
| 168 ΔamyE | B. subtilis | 168 trpC2, amyE::tet | tet | Laboratory stock |
| MG1363 | L. lactis | Plasmid‐free derivative of NCDO712 (Prt‐, Lac‐) | – | Gasson (1983) |
| MG1363 GFP+ | L. lactis | MG1363; pseudo10::Pusp45‐sfGFP(Bs) | ery | Overkamp et al. (2013) |
| MG1363 ΔcluA | L. lactis | MG1363; ΔcluA | – | This work |
| MG1363 GFP+ ΔcluA | L. lactis | MG1363_sfgfp(Bs); ΔcluA | ery | This work |
| MG1363 pNZ8048 | L. lactis | MG1363; pNZ8048 | cm | This work |
| MG1363 ΔcluA pNZ8048 | L. lactis | MG1363; ΔcluA, pNZ8048 | cm | This work |
| MG1363 pNZ521 | L. lactis | MG1363; pNZ521 | cm | This work |
| MG1363 ΔcluA pNZ521 | L. lactis | MG1363; ΔcluA, pNZ521 | cm | This work |
| CNRZ302 pNZ8048 | S.thermophilus | pNZ8048 | cm | This work |
| ST11 pNZ8048 | S.thermophilus | pNZ8048 | cm | This work |
TABLE 2.
Plasmids used in this study.
| Plasmid | Genotype | Abr | Reference |
|---|---|---|---|
| pNZ8048 | NICE inducible vector | cm | de Ruyter et al. (1996) |
| pseudo10::Pusp45‐sfGFP(Bs) | Integrative vector pSEUDO10 with Pusp45‐sfGFP(Bs) sequence | ery | Overkamp et al. (2013) |
| pGh9::cluA | pGh9 thermosensitive plasmid (a replication thermosensitive derivative of pWV01; Otto et al., 1982) with L. lactis cluA construct for deletion, cloned by Gibbson assembly to pGh9 through SmaI, unpublished | ery | Strain collection of Dr. Kulakauskas (unpublished) |
| pNZ521 | 10.7 kb pNZ122 derivative carrying complete prtP and prtM gene of pSK111 | cm | Marugg et al. (1995) |
TABLE 3.
List of primers used in this study.
| Name | Sequence 5′ → 3′ | Amplified fragment | Product size (bp) |
|---|---|---|---|
| cmR_FW | GCAGACAAGTAAGCCTCCTA | 1 | 767 |
| cmR_RV | GGGGCAGGTTAGTGACATTAGA | 1 | |
| repA_FW | TGCGGCGTTAGCTATAGAAG | 2 | 1225 |
| repA_RV | CTGCTTTCTTCATTAGAATCAATC | 2 | |
| Pnis_FW | CCAAGATCTAGTCTTATAACTATAC | 3 | 803 |
| Pnis_RV | CGGCTTTCATAATCTAACAGAC | 3 | |
| repC_FW | TATGAAAGCCGATGACTGAATG | 4 | 539 |
| repC_RV | AACCGCAGATTTTGAAAAAC | 4 |
Construction of Lactococcus lactis strains lacking the sex factor
For the knock‐out of cluA in L. lactis MG1363, the pGh9 thermosensitive plasmid system, a gift from Dr. Saulius Kulakauskas, was used. Transformation of electrocompetent L. lactis MG1363 with the pGh9 plasmid containing the cluA sequence was performed as previously described (Maguin et al., 1996). In L. lactis, pGh9 replicates at 28°C, but it is lost when cells are cultured at temperatures above 37°C. The pGh9‐cluA plasmid derivative was used to integrate the plasmid in the native cluA locus at the permissive temperature of 28°C. Further excision of the plasmid at 37°C resulted in the L. lactis MG1363 ΔcluA strain.
Mating procedure—cell‐to‐cell plasmid transfer
Monocultures of donor and recipient were inoculated from an overnight culture, diluted to a final OD600 of 0.05, supplemented with an appropriate antibiotic, and incubated at the optimal temperature separately: 30°C for L. lactis and 37°C for B. subtilis. Subsequently, cultures were grown until they reached mid‐log growth phase (OD600 ~ 0.5). After the incubation period, cells were harvested and washed in 1XSMM (Harwood & Cutting, 1990) media to a final density of 107 cells per 1 μl of culture. Donor and recipient cells were mixed in the cell ratio 1:1 (donor: recipient), and 5 μl of mixed culture was spotted on nonselective GM17 medium, air‐dried, and incubated O/N at the optimal temperature: 37°C for B. subtilis matings and 30°C for L. lactis matings only. For S. thermophilus, matings were performed on GM17 media supplemented with 1% lactose (GLM17), and the co‐culture was incubated at 37°C. Cells from the O/N mating plates were harvested using 1 ml of 1XSMM medium and plated on selective rich medium plates with appropriate antibiotics and subsequently incubated at the indicated temperature for O/N. Plates with transformants were replicated on rich selective medium, GM17 for B. subtilis and L. lactis and GLM17 for S. thermophilus, with appropriate antibiotics to exclude the nonheritable transfer of chloramphenicol acetyltransferase in case of chloramphenicol resistance. After the overnight incubation, transformants were propagated in selective liquid medium and plasmids were isolated (NucleoSpin Plasmid, Mini kit for plasmid DNA, Macherey‐Nagel, GmbH & Co. KG, Germany) for gel electrophoresis (horizontal gel electrophoretic system from Bio‐Rad Laboratories, Inc., The Netherlands). Additionally, to confirm the presence of pNZ8048 in L. lactis transformants, a PCR reaction (T100 Thermal Cycler, Bio‐Rad Laboratories, Inc., The Netherlands) with pNZ8048 annealing primers (Table 3) was performed.
DnaseI treatment
DnaseI treatment was performed similarly to previously published work on nanotubes (Dubey & Ben‐Yehuda, 2011). The donor and recipient strains were separately grown to the mid‐exponential phase, subsequently pelleted and washed with DNaseI buffer (50 mM Tris, pH 7.2, 10 mM MgCl2, 5 mM CaCl2), and incubated separately in the presence of 100 μg/ml of DNaseI for 15 min at 37°C. Donor and recipient were mixed in a 1:1 ratio, and the mixtures were supplemented with DNaseI (100 μg/ml) and spotted on LB agar. DNaseI buffer (without DNaseI) was added as a control. Mixed co‐cultures were grown for 18 h at 30°C or 37°C and replica plated on respective antibiotic plates.
Microscopy techniques
TIRF microscopy—membrane visualization with FM5‐95
Cultures were prepared as above and spotted on 1% (w/v) agarose in GM17 supplemented with FM5‐95 membrane dye at a final concentration of 1 μg/ml and incubated at 37°C for 1 h prior to the experiment. For the visualization of nanotubes at high resolution, the TIRF 60× objective was used (Olympus 60X/1.49, APON 60XOTIRF, UIS2, 1‐U2B720). To observe membrane structures with FM5‐95 dye, a filter set Quad‐mCherry was used: excitation with laser at 568 nm and emission at 594 nm.
SEM
In mixed cultures, B. subtilis and L. lactis were grown separately in rich media to mid‐exponential growth phase and mixed in 1:1 ratio to visualize nanotubes in mixed cultures. The mixture was spotted on GM17 agar, and EM copper grids were placed on top of it and incubated for 6 h at 37°C. After the incubation period, cells were fixed with glutaraldehyde (2%) for overnight at 4°C. The next day, cells were washed in 0.1 M sodium cacodylate buffer and incubated with 1% osmium tetroxide (prepared in 0.1 M sodium cacodylate) for 1 h at room temperature. Post‐fixed sample was washed 3 times with water and subsequently dehydrated with 30%, 50%, and 70% ethanol for 15 min and finally with 100% ethanol (3 × 30 min). Last, samples were incubated in 100% ethanol:tetramethylsilane (TMS) (1:1). After 10 min, incubation samples were treated with pure TMS for 15 min and subsequently air‐dried prior imaging.
RESULTS
B. subtilis, L. lactis and S. thermophilus are able to form membranous structures that resemble bacterial nanotubes
Visualization of membranous structures that might facilitate HGT in studied mixed co‐cultures
In the recent decade, the presence of membranous structures bridging neighbouring bacterial cells was extensively studied (Bhattacharya et al., 2019; Dubey et al., 2016; Dubey & Ben‐Yehuda, 2011; Pal et al., 2019; Pande et al., 2015; Stempler et al., 2017). With the use of microscopic techniques, these membrane connections have been identified and characterized in Gram‐positive (B. subtilis, Staphylococcus aureus) and Gram‐negative (Escherichia coli) bacteria. It has been proposed that structures like nanotubes can conduit the HGT events in an intra‐ and interspecies manner (Dubey & Ben‐Yehuda, 2011). Prompt by these recent findings, we wondered whether these membranous connections are present in L. lactis, S. thermophilus, and more importantly, we explored interspecies interactions of these microorganisms with B. subtilis—a known nanotube producer, in mixed co‐culutres.
Accordingly, we optimized co‐culturing conditions for chosen microorganisms and performed microscopic analysis in tested environment. We observed that B. subtilis and L. lactis can co‐exist when spotted on rich medium GM17 and co‐incubated at 37°C. For B. subtilis and S. thermophilus, the optimal conditions were GM17 with 1% of lactose and incubation at 37°C. Given the environment, we examined co‐cultures after 6 h incubation with scanning electron microscopy (SEM) and visualized inter‐ and intraspecies membrane connections, nanotubes, known to facilitate molecular transfer events, using total internal reflection fluorescence microscopy (TIRF) (Figure 1 and Supplementary Materials).
FIGURE 1.
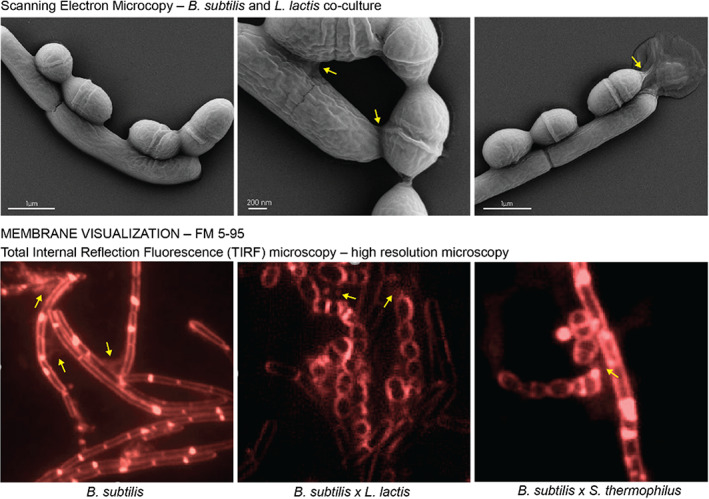
SEM of B. subtilis PY79 and L. lactis MG1363 after 6 h of co‐culture. Top row: Scanning electron microscopy of B. subtilis PY79 and L. lactis mg1363 co‐cultured for 6 h on solid GM17 media show that B. subtilis can co‐exist with L. lactis in close proximity. The close contact places are marked with yellow arrows. Bottom row: micrographs of FM5‐95 membrane staining of studied co‐cultures reveal the presence of membranous connections (yellow arrows) similar to previously reported bacterial nanotubes.
Non‐conjugative plasmids can be transferred between B. subtilis and members of LAB
The microscopy analysis of mono and co‐cultures of each studied microorganism indicates that cells grown on solid rich media produce membranous structures that resemble bacterial nanotubes (Figure 1 ). Since the occurrence of membrane connections might facilitate HGT and so‐called cell‐to‐cell transformation, we examined interspecies plasmid transfer between B. subtilis and LAB members and designed a collection of donor strains harbouring high‐copy, nonconjugative shuttle vector pNZ8048 (3.4 kb, cmR) replicating in both, donor and recipient cells (de Ruyter et al., 1996). To facilitate further selection of newly engineered strains, recipient strains contained a chromosomally integrated antibiotic cassette (Experimental Procedures; Table 1).
Plasmid transfer between B. subtilis strains
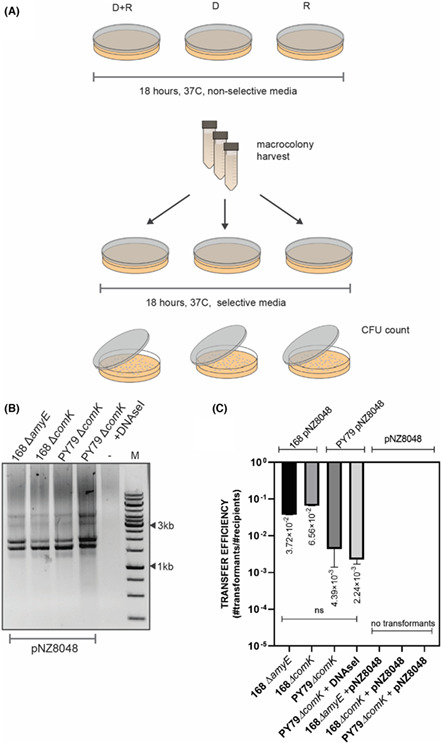
First, we investigated cell‐to‐cell transformation between two B. subtilis strains. For the B. subtilis matings, we started with a well‐characterized B. subtilis 168 laboratory strain. Since B. subtilis is able to internalize free DNA from its environment upon induction of the competence state, we decided to use a recipient strain with a knocked‐out comK gene, encoding the competence master regulator ComK (Hahn et al., 1996; van Sinderen et al., 1994), to ensure that during the mating procedure the transferred plasmid was not taken up by the recipient cell from adjacent lysed donor cells. Additionally, the comK − strain carried a spectinomycin antibiotic cassette for later selection on LB agar plates. For the donor strain, we used B. subtilis 168 harbouring the pNZ8048 (cmR) non‐conjugative shuttle plasmid. To examine the plasmid transfer, we spotted a mix of donor and recipient cells on a rich non‐selective agar plate and incubated overnight without antibiotic selection pressure. As a control, we spotted donor and recipient cells separately on the same type of non‐selective agar plates. To additionally exclude DNA uptake via activation of the natural competence and confirm the cell‐to‐cell DNA transfer, we separately spotted recipient cells on the mating plates and added 1 μg of exogenous pNZ8048 DNA. After the incubation time, the formed mixed macrocolonies were collected, and the recipient strain's antibiotic resistance was tested. The analysis of selectable plates showed that all colonies resistant to both antibiotics, specR and cmR, were derived only from the mixed population of donor and recipient (Figure 2A). We did not observe any resistant colonies of monocultures of donor or recipient on selectable media, indicating that recipient cells should be in the same environment as the donor cells in order to form colonies on spectinomycin and chloramphenicol agar plates. Moreover, the recipient strain could not take up externally provided plasmid DNA from the media, which excluded the possibility of natural plasmid transformation. After replica plating, the colonies obtained were additionally examined for plasmid presence (Figure 2B). The transfer efficiency was estimated as a fraction of recipients that received the genetic material from donor cells and was estimated on 10−2 transformants per CFU recipient (Figure 2C).
FIGURE 2.
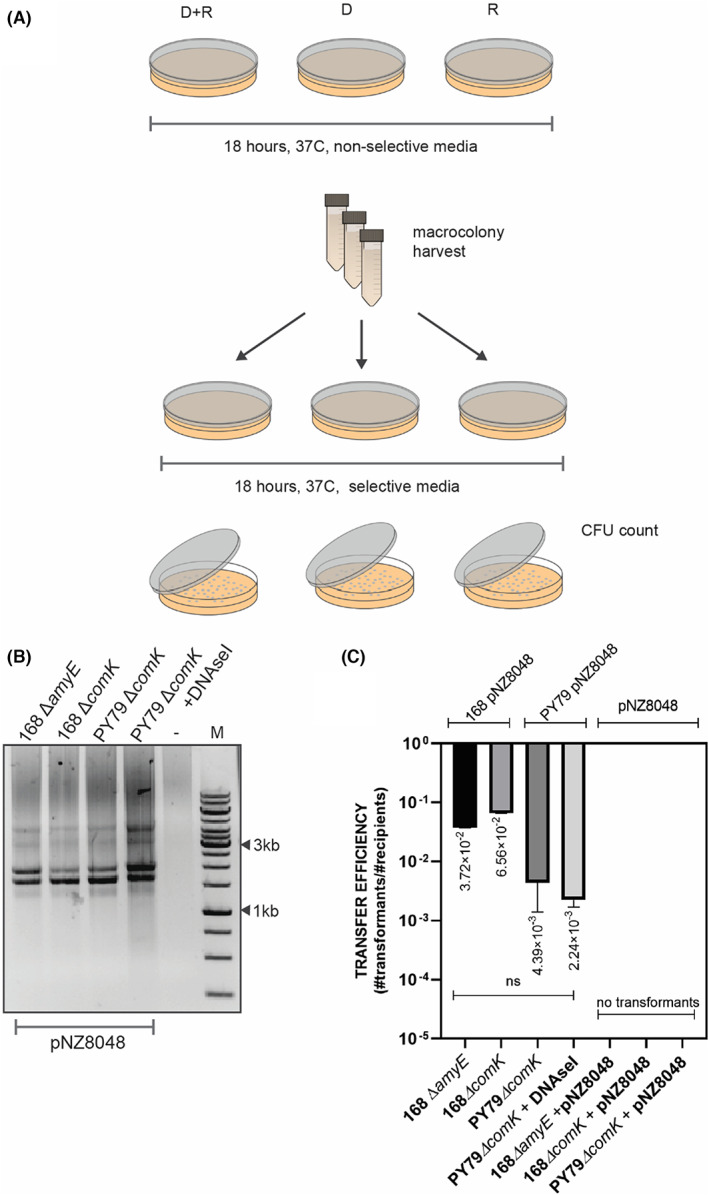
Cell‐to‐cell transformation of nonconjugative plasmid between B. subtilis strains. (A) A graphic representation of mating procedure. (B) Gel electrophoresis of isolated pNZ8048 from the recipient strain after the replica plating. (C) The average values of plasmid transfer efficiency for B. subtilis to B. subtilis matings. Donors of the genetic material are indicated on top of the graph. Mating without donor strain but with exogenously added pNZ8048 did not yield in colonies (indicated with 0). The statistical analysis was performed in Prism (Kruskal–Wallis test, ns, not significant).
To further investigate the relevance of functional natural competence in the cell‐to‐cell transformation, we compared plasmid transfer efficiencies when used 168 ∆comK (specR) strain 168 ΔamyE (tetR), comK+ strain as recipient strains. It is worth mentioning that lack of a functional amyE gene, encoding an alpha‐amylase used by B. subtilis to degrade starch, should not affect the transfer efficiency or diminish the efficiency of natural transformation on a solid medium. Following the mating procedure, the same donor strain of B. subtilis 168 pNZ8048 was co‐cultured with either 168 ΔcomK (specR) or 168 ΔamyE (tetR) on solid media and the plasmid transfer was examined accordingly (Figure 2A). On average, the efficiency of plasmid transfer to 168 ΔamyE (tetR) was not significantly different when compared with the 168 ΔcomK (cmR) (Figure 2C), demonstrating that under the mating conditions, the putative activation of natural competence in B. subtilis cells does not facilitate plasmid acquisition.
Genome analysis of the B. subtilis 168 strain has revealed the presence of the temperate phage SPß (Warner et al., 1977) and the integrative and conjugative element ICEBs1 (Auchtung et al., 2005, 2007). Therefore, to ensure that the plasmid transfer is cell‐to‐cell contact‐dependent and not a result of conjugative events or induced prophage transduction, we tested the B. subtilis PY79 strain, cured from ICEBs1 and SPβ. Subsequently, we designed a competence negative strain of PY79 by inserting spectinomycin antibiotic cassette in the comK gene, resulting in B. subtilis PY79 ΔcomK (specR) recipient strain. For the donor strain, we used B. subtilis PY79 pNZ8048 (cmR) strain. Estimated transfer efficiencies revealed that PY79 performed similarly to the 168 strain, indicating that pNZ8048 was transferred in a non‐conjugative manner, and it did not involve phage transduction (Figure 2C). Additionally, we found that plasmid transfer was resistant to DNaseI, showing that pNZ8048 was likely transferred via protective conduit rather then taken up directly from the environment. Transfer efficiency for B. subtilis matings was estimated to yield 10−2 transformants per CFU recipient (Figure 2C).
Interspecies plasmid transfer between B. subtilis and L. lactis
L. lactis is a Gram‐positive bacterium widely used as a starter culture for milk fermentation, a host for heterologous protein production and a platform for delivery of therapeutics. Therefore, the ability to acquire new genetic traits by omitting the genetic engineering steps would greatly contribute to the design and production of novel strains with industrially relevant features. Because of several mutations in competence‐related genes, L. lactis MG1363 is not able to enter the competence state (Mulder et al., 2017). Therefore, it was possible to study cell‐to‐cell transformation, excluding DNA take‐up from the environment.
B. subtilis to L. lactis plasmid transfer
Given that B. subtilis can efficiently deliver non‐conjugative plasmids to its neighbours, we designed our first mating experiment exploiting interspecies plasmid transfer between B. subtilis and L. lactis. As a donor strain, we used B. subtilis 168 pNZ8048 (cmR) and PY79 pNZ8048 (cmR), and as a recipient strain, we used L. lactis MG1363 transformed with pSEUDO10_sfgfp(Bs) (eryR) (MG1363 GFP+). Accordingly, donor and recipient strains were co‐cultured on solid rich media supporting growth of both strains (GM17) at 37°C, and the antibiotic resistance of the recipient strain after mating was analysed. Since the reciepient strain harboured an erythromycin cassette integrated into its genome, selection of L. lactis transformants was performed on erythromycin (5 μg/ml) and chloramphenicol (5 μg/ml) GM17 plates. The cell‐to‐cell plasmid transfer yielded many L. lactis MG1363 colonies resistant to 5 μg/ml of erythromycin and 5 μg/ml of chloramphenicol after the first selection round (Figure 3A), and the plasmid transfer efficiency was estimated on 10−4 to 10−5 transformants per CFU recipient. However, we could not propagate the transformants obtained in the liquid media or solid selectable rich media (GM17, erythromycin 5 μg/ml, chloramphenicol 5 μg/ml). Interestingly, the replica plating of the transformants on rich media with reduced chloramphenicol concentrations to 1 μg/ml and 2.5 μg/ml resulted in L. lactis single colonies as well as a few colonies with B. subtilis morphology (Figure 3B). To additionally confirm the identity of the obtained colonies classified as L. lactis transformants, we performed a microscopy analysis and confirmed that these colonies are MG1363 GFP+ cells (Figure 3C).
FIGURE 3.
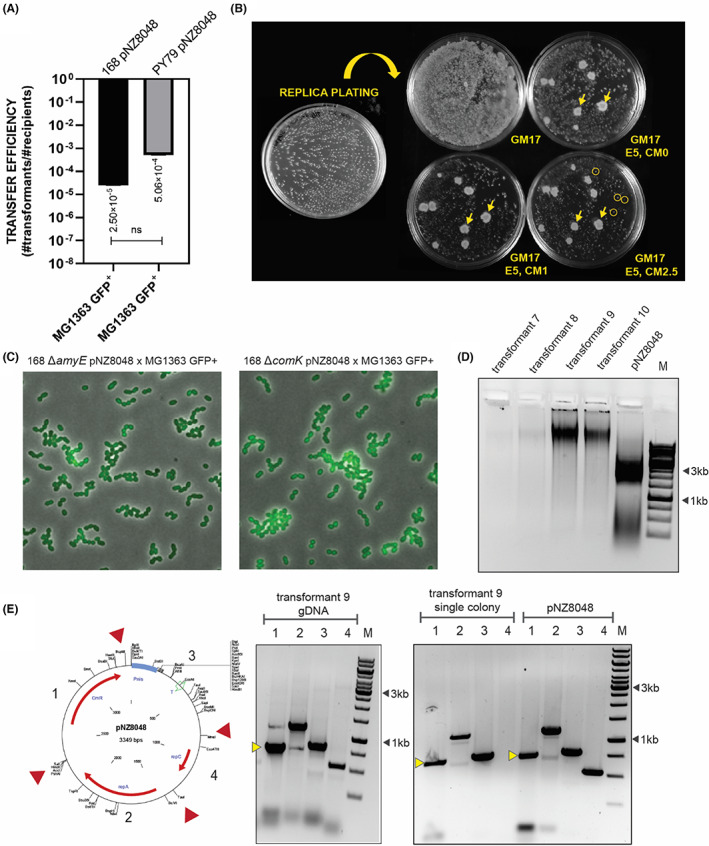
pNZ8048 delivery to L. lactis MG1363 GFP+ from B. subtilis. (A) The average values of pNZ8048 transfer efficiency for B. subtilis to L. lactis matings. B. subtilis 168 and PY79 harbouring pNZ8048 were designed as donor strains and MG1363 with chromosomally integrated and constitutively expressed sfGFP as a recipient strain. The statistical analysis was performed in Prism (t‐test, ns: p = 0.2961), the error bars show the standard deviation (SD), n = 4. (B) Colony formation after replica plating on GM17 selective medium with reduced concentrations of chloramphenicol. The yellow arrows indicate B. subtilis erythromycin resistant colonies. The yellow circles indicate colonies of L. lactis transformants picked for further analysis. (C) Micrographs of L. lactis MG1363 GFP+ transformants. (D) Gel electrophoresis of isolated plasmids from L. lactis transformants, the pNZ8048 lane corresponds to the plasmid isolated from L. lactis MG 1363 pNZ8048 strain. (E) PCR analysis of transformant 9. The yellow markers indicate the band representing chloramphenicol cassette.
Next, we selected single colonies of MG1363 GFP+ pNZ8048, inoculated in GM17 with 5 μg/ml of erythromycin and 2.5 μg/ml of chloramphenicol and verified the presence of pNZ8048 in the L. lactis culture by plasmid extraction and by direct colony PCR. The gel electrophoresis of isolated pNZ8048 from transformed cells did not show pNZ8048‐specific bands (Figure 3D). Conversely, PCR analysis of the MG1363 GFP+ pNZ8048 colony and genomic DNA isolated from the same colony revealed the presence of pNZ8048 fragments, including chloramphenicol cassette (Figure 3E), suggesting that colonies from replica plating likely harboured copies of pNZ8048, however in a reduced amount.
L. lactis to B. subtilis plasmid transfer
To further investigate the non‐conjugative interspecies plasmid transfer, we decided to test the possibility of L. lactis to deliver the genetic material to B. subtilis. This time, as a donor strain, we used L. lactis MG1363 transformed with pNZ8048 (cmR) and as a recipient B. subtilis 168 ΔcomK (specR). Same co‐culturing conditions were applied as in the case of the B. subtilis to L. lactis matings, and the plasmid transfer efficiency was estimated (Figure 4A). Obtained B. subtilis colonies were replated on selective media with appropriate antibiotics (spectinomycin 100 μg/ml and chloramphenicol 5 μg/ml) and inspected for plasmid presence (Figure 4B,C). Additionally, the integrity of pNZ8048 in donor and recipient strains was confirmed by restriction analysis with SalI and BglII restriction enzymes Figure 4D. We did not observe any changes in the plasmid size that might have occurred upon the transfer from recipient to donor cells. The results demonstrated that competence‐ negative B. subtilis can acquire plasmid DNA from neighbouring L. lactis MG1363 cells, and the cell‐to‐cell plasmid transformation efficiency was estimated on 10−6 transformants per CFU recipient.
FIGURE 4.
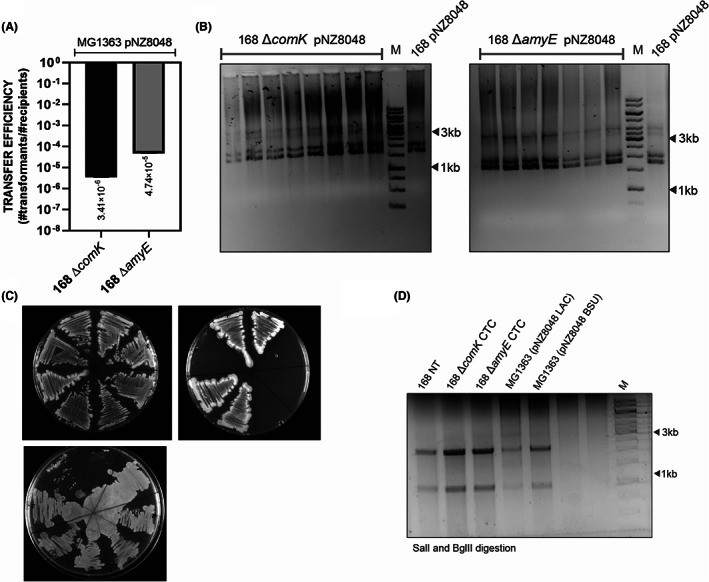
B. subtilis 168 can acquire pNZ8048 from L. lactis MG1363 GFP+. (A) The average values of plasmid transfer efficiency for L. lactis to B. subtilis matings. The statistical analysis was performed in Prism (t‐test, ns: p = 0.3558), and the error bars show the standard deviation (SD), n = 5. (B) Gel electrophoresis of isolated plasmids from B. subtilis transformants. (C) Colony formation after replica plating on LB selective medium with chloramphenicol and kanamycin. (D) Restriction analysis of pNZ8048 isolated from different hosts with SalI and BglII restriction enzymes. The analysis of unmodified pNZ8048 should result in two distinct bands 2313 bp and 1036 bp. 168 NT—pNZ8048 was isolated from B. subtilis 168 donor strain where it was previously introduced via natural transformation; 168 ∆comK CTC and 168 ∆amyE CTC—pNZ8048 isolated from the recipient cells that took up the plasmid DNA from L. lactis MG1363 pNZ8048 strain; MG1363 (pNZ8048 LAC)—plasmid isolated directly from L. lactis MG1363; MG1363 (pNZ8048 BSU)—pNZ8048 was isolated from B. subtilis 168 pNZ8048 strain and further introduced to L. lactis MG1363 via electroporation.
Presence of the lactococcal sex factor does not facilitate the plasmid transfer between L. lactis cells
So far, our data suggest that L. lactis can deliver plasmid DNA via cell‐to‐cell mediated exchange to B. subtilis. Next, we proceeded to investigate cell‐to‐cell transformation between L. lactis strains.
L. lactis MG1363 contains a chromosomally located cluA gene encoding for sex factor aggregation protein, CluA, involved in conjugative DNA transfer (Godon et al., 1994). To exclude the possibility of plasmid transfer through sex factor mobilization, we transformed L. lactis MG1363 with the pGH9‐cluA thermosensitive plasmid to markerlessly knock‐out cluA, resulting in L. lactis MG1363 ΔcluA. CluA mutant strain was further transformed with pseudo10::Pusp45‐sfGFP(Bs) plasmid to obtain L. lactis MG1363 GFP+ ΔcluA (eryR) recipient strain. To examine the role of the sex factor in non‐conjugative plasmid transfer, we compared the transfer efficiencies for wild type and the cluA mutant used separately as a donor and a recipient of pNZ8048. Additionally, we tested transfer of pNZ521 plasmid encoding industrially desired feature—PrtP protease (Gasson, 1983), the presence of which could be tested on casein agar plates.
After the mating procedure on GM17 agar at 30°C and further antibiotic selection with 5 μg/ml erythromycin and 5 μg/ml chloramphenicol, we observed that L. lactis can transfer both high copy plasmids, pNZ8048 and pNZ521, with an efficiency of 10−3 to 10−4 transformants per CFU recipient, respectively (Figure 5A,B). The analysis of the estimated transfer efficiencies also revealed that the presence of the sex factor in MG1363 strain does not facilitate the plasmid transfer under the studied mating conditions, on a rich, growth‐promoting media (Figure 5A,B).
FIGURE 5.
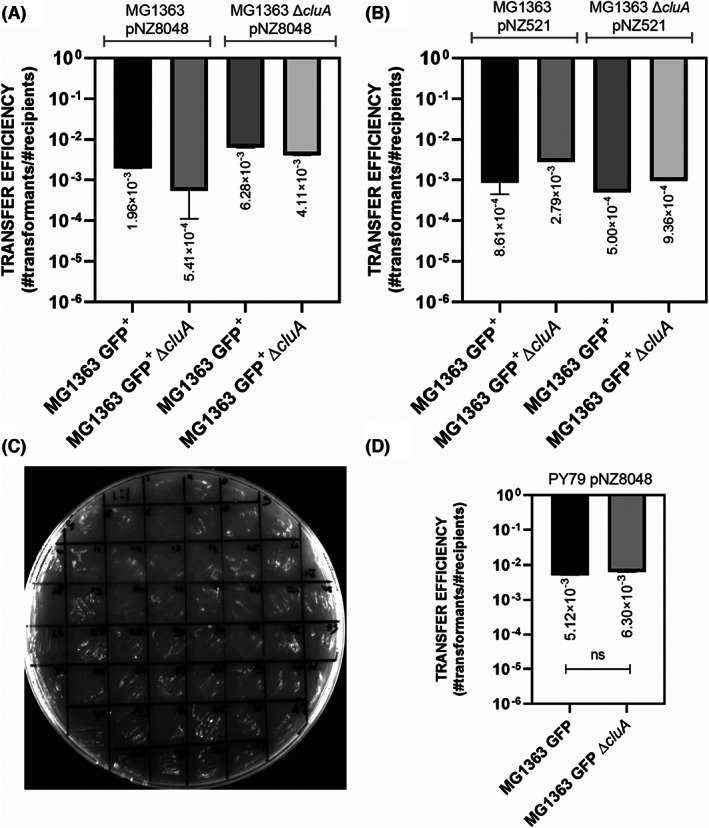
Plasmid transfer in conjugation negative strain of L. lactis MG1363. (A) The average values of plasmid transfer efficiency for L. lactis to L. lactis plasmid‐transfer matings. The statistical analysis was performed in Prism (Kruskal–Wallis test, ns, not significant), the error bars show the standard deviation (SD), n = 5. (B) Transfer efficiency for pNZ521. The statistical analysis was performed in Graph Pad Prism (one‐way ANOVA, ns: p = 0.2770), and the error bars show the standard deviation (SD), n = 2. For both graphs (A) and (B), the names of the donor strains were indicated above the graph, whereas the recipient strains are listed on the x‐axis. (C) Colony‐forming ability of L. lactis MG1363 pNZ521 transformants on casein‐based medium. (D) Transfer efficiencies for B. subtilis to L. lactis matings. The error bars show the standard deviation (SD), n = 5, t‐test, ns p = 0.8718.
Moreover, we compared the plasmid transfer efficiencies to wild type and the cluA mutant strain of L. lactis MG1363 from the B. subtilis PY79 strain and confirmed that a functional cluA is not required for the cell‐to‐cell plasmid transfer under the conditions studied (Figure 5D).
Cell‐to‐cell plasmid transfer from S. thermophilus to B. subtilis
Another industrially relevant bacterium, used for milk fermentation is S. thermophilus. This Gram‐positive bacterium has been reported to be able to induce natural competence. The activation of competence and assembly of the competence machinery is dictated by the ComX pheromone abundance in the environment, whose production and secretion are typically initiated in response to specific environmental factors (Fontaine et al., 2013; Haustenne et al., 2015). Since earlier we observed that B. subtilis and S. thermophilus could be co‐cultured on a lactose supplemented GM17 and form membranous connections, we wondered whether S. thermophilus can deliver plasmid DNA to B. subtilis in a cell‐to‐cell contact manner. Therefore, we tested cell‐to‐cell transfer of pNZ8048 plasmid from S. thermophilus strains, ST‐11 and CNRZ302, available in our strain collection, to B. subtilis 168 ΔcomK (specR). ST11 and CNRZ302 strains were transformed with pNZ8048 plasmid via electrotransformation and used as donor strains. Additionally, we tested B. subtilis 168 ΔamyE (tetR) as a recipient strain. The mating procedure was conducted at 37°C in 1:1 donor to recipient cell ratio on a 1% lactose supplemented GM17 media.
The analysis of the plasmid transfer efficiency showed that B. subtilis can efficiently acquire plasmid DNA from S. thermophilus strains, similarly to what was observed for L. lactis to B. subtilis matings. The transfer efficiency for ST‐11 strain was estimated on 10−6 transformants per CFU recipient, whereas for CNRZ302 10−5 transformants per CFU recipient were found. (Figure 6).
FIGURE 6.
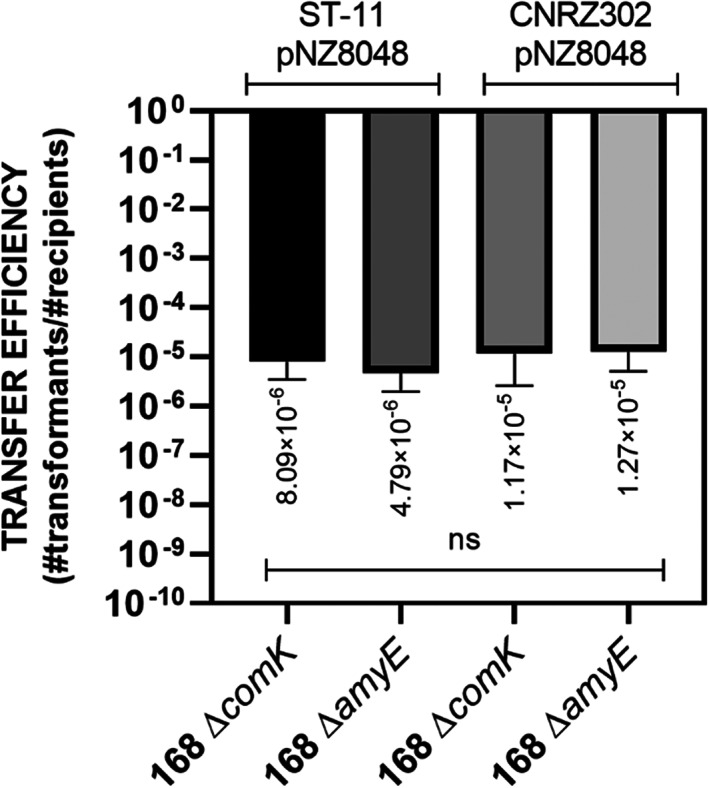
Cell‐to‐cell plasmid transfer from S. thermophilus to B. subtilis. The graph shows the average of the plasmid transfer efficiencies from ST‐11 and CNRZ S. thermophilus donor strains (indicated above the graph) to non‐competent and competent B. subtilis 168 strains, 168 ΔcomK and 168 ΔamyE, respectively (indicated on the x‐axis). The error bars indicate the SD, n = 3. The statistical analysis was performed in Prism (Kruskal–Wallis test, ns, not significant).
DISCUSSION
In this study, we explored the feasibility of intra‐ and interspecies plasmid transfer, mediated by cell‐to‐cell interactions in several members of Gram‐positive bacteria. First, we confirmed a phenomenon of cell‐to‐cell mediated HGT in B. subtilis, previously described for mixed strains of B. subtilis (Dubey & Ben‐Yehuda, 2011; Zhang et al., 2018), and E. coli (Maeda et al., 2006; Matsumoto et al., 2016). Second, for the first time, we demonstrated a transfer of a high copy, non‐conjugative plasmid in mixed co‐cultures of either B. subtilis and L. lactis, or B. subtilis and S. thermophilus, showing that cell‐to‐cell DNA transfer is a ubiquitous form of HGT.
Because B. subtilis is known to cause spoilage of dairy products (Arakawa et al., 2008; Faille et al., 2014; Moschonas et al., 2021), we expected it to co‐exist with LAB in the glucose or lactose supplemented M17 media. To study the HGT between different species, we tested co‐culturing conditions supporting the growth of both donor and recipient strains. We identified the optimal conditions for B. subtilis, L. lactis, and S. thermophilus co‐cultures and visualized the single‐cell interactions with different microscopy techniques (Figure 1 ). The membrane staining with the FM5‐95 fluorescent dye revealed that these Gram‐positive bacteria form membranous structures, previously identified as bacterial nanotubes (Dubey et al., 2016; Dubey & Ben‐Yehuda, 2011). Notably, nanotubes have been recently suggested to contribute to the HGT of non‐conjugative plasmids in B. subtilis on a solid media (Dubey & Ben‐Yehuda, 2011). This prompted us to further investigate the non‐conjugative cell‐to‐cell plasmid transfer and explore its boundaries by executing intraspecies matings.
Our results show that B. subtilis efficiently delivers plasmid DNA to neighbouring B. subtilis strains. Since genetic material can be released to the environment through cell lysis or spontaneous DNA secretion by growing cells (Nielsen et al., 2007), it could be argued that obtained transformants acquired the plasmid DNA from the media. By knocking out the gene encoding for the competence master regulator ComK (Hahn et al., 1996; van Sinderen et al., 1994), we excluded the possibility of donor‐independent DNA uptake through induction of natural competence for transformation (Figure 2C). Additionally, we demonstrated that plasmid transfer is insensitive to DNase treatment, supporting the results of previous works on cell‐to‐cell genetic transfer (Dubey & Ben‐Yehuda, 2011; Zhang et al., 2018). Resistance to DNase treatment indicates that plasmid DNA is enclosed in a protective conduit, similarly to conjugation‐mediated DNA transfer through pili. Yet, here the cell‐to‐cell plasmid transfer could be distinguished from conjugation by differences in the transfer efficiency of B. subtilis 168 strain, harbouring an integrative and conjugative element, ICEBs1, to conjugation‐negative B. subtilis PY79 (Figure 2C). Notably, the cell‐to‐cell non‐conjugative plasmid transfer between B. subtilis strains was more efficient than DNA uptake via transformation (Figure S1A,B). Finally, viable donor cells in the co‐culture are essential for mating. We show that recipient cells with and without functional comK gene could not take up the plasmid when added directly to the plates, confirming the requirement of donor and recipient cells to grow in close proximity (Figure 2C). Additionally, when a donor strain was deliberately killed by heat treatment, the mating was unsuccessful (data not shown).
Significantly, our study adds new insight on the mechanisms of HGT in LAB. We demonstrate that plasmid DNA can be transferred by mixed colonies of B. subtilis and L. lactis (Figures 3 and 4), between L. lactis strains (Figure 5), and finally between B. subtilis and S. thermophilus (Figure 6). Because of several mutations in competence‐related genes, L. lactis MG1363 is not able to enter the competent state (Mulder et al., 2017). Therefore, for L. lactis matings, plasmid transfer did not involve natural transformation. First, we explored the delivery of pNZ8048 (cmR) from B. subtilis to L. lactis and showed that L. lactis displayed resistance to chloramphenicol after the first round of plating on selective media (Figure 3B). However, the transformants obtained could not grow in liquid media with the same chloramphenicol concentration and grow poorly on a solid selective media after replating. Chloramphenicol acetyltransferase confers resistance to chloramphenicol, and it was found to be transferred between bacteria via nanotubes, showing that acquired chloramphenicol resistance can be a nonhereditary feature (Dubey & Ben‐Yehuda, 2011). This would explain the difficulties with the growth of L. lactis transformants after replating or culturing in the liquid media with a killing concentration of chloramphenicol (Figure 3B, Figure S2).
However, PCR analysis of L. lactis colonies and genomic DNA isolated from the liquid cultures of transformants grown in a lower concentration of chloramphenicol revealed pNZ8048 amplicons (Figure 3E), suggesting that pNZ8048 from B. subtilis origin likely resides in the L. lactis transformants, perhaps with a reduced copy number. The reduced copy number of pNZ8048 plasmid could result from the differences in the restriction‐modification (R/M) systems active in B. subtilis and L. lactis. B. subtilis can recognize 5′ CTCGAG 3′ sequences and methylate with BsuM R/M system, which consists of two operons, BsuMM operon (ydiO‐ydiP) for two cytosine DNA methyltransferases, and BsuMR operon (ydiR‐ydiS‐ydjA) for a restriction nuclease and two associated proteins of unknown function (Guha, 1988; Maehara et al., 2011; Matsuoka et al., 2005). Notably, pNZ8048 plasmid contains the recognition sequence for the BsuM R/M system, and it is likely modified in the B. subtilis host. We suggest that during the cell‐to‐cell plasmid transfer from B. subtilis to L. lactis most of the pNZ8048 transferred copies are methylated and could be subsequently recognized by L. lactis R/M as foreign DNA (O'sullivan et al., 2000; Schouler et al., 1998). Moreover, the differences in methylation patterns of incoming DNA via natural transformation and host's DNA greatly influence the transformation efficiency (Beauchamp et al., 2017). Our preliminary experiments revealed that pNZ8048 isolated from B. subtilis was insensitive to the digestion with an isoschisomer of BsuM, XhoI (Figure S3). This points to pNZ8048 methylation of 5′ CTCGAG 3′ sequence in B. subtilis, which protected the plasmid DNA from XhoI activity. Conversely, pNZ8048 isolated from L. lactis host could be cleaved with XhoI. Moreover, we transformed pNZ8048 isolated from different hosts to L. lactis MG1363 and showed 10‐fold reduction in transformation efficiency with pNZ8048 isolated from B. subtilis host. However, to fully understand this phenomenon, future studies are needed. We believe that enumeration of pNZ8048 copy numbers in L. lactis transformants will shed more light on the effect of methylation in B. subtilis donor strain on interspecies transfer of DNA.
Next, we demonstrated that not only B. subtilis could transfer plasmids to adjacent B. subtilis cells, and the intraspecies cell‐to‐cell plasmid transfer also occurred between L. lactis strains. Additionally, with the cluA mutant of L. lactis MG1363, we showed that the transfer was conjugation independent and could be applied to deliver industrially‐relevant plasmid pNZ521 (Figure 5).
The differences in the average transformation efficiencies in studied bacteria revealed that cell‐to‐cell mediated plasmid transfer was the most successful for intraspecies matings of B. subtilis with B. subtilis and L. lactis with L. lactis (Figure S1B). However, in the case of mixed‐species matings with B. subtilis as either donor or recipient strain, the efficiency dropped 1000‐fold on average. It can be speculated that the differences between intra‐ and interspecies transfer of the same non‐conjugative plasmid might be due to different cell surface and colony‐forming properties of B. subtilis, L. lactis, and S. thermophilus, which consequently might affect the formation of stable cell‐to‐cell connections (Figure S1C). Notably, the most efficient transfer was observed for B. subtilis, known for its potent biofilm production abilities (Arnaouteli et al., 2021). Although L. lactis is also able to form biofilms (Habimana et al., 2009; Oxaran et al., 2012), they are less abundant than those found in B. subtilis, which is reflected in the macrocolony morphology (Figure S1C). As biofilms greatly contribute to the HGT in bacteria (Maeda et al., 2006; Molin & Tolker‐Nielsen, 2003), we suggest that the ability to form abundant biofilms by B. subtilis is an important feature for enhanced cell‐to‐cell transfer. Interestingly, the formation of cell‐to‐cell connections via nanotubes and molecular transfer in B. subtilis requires the activity of YmdB (Dubey et al., 2016; Stempler et al., 2017), a phosphodiesterase that affects flagellin expression and biofilm formation (Diethmaier et al., 2014, 2011).
We also showed that within a mixed culture with S. thermophilus, B. subtilis can acquire plasmid DNA without employing natural competence. These promising results will be further explored focusing on DNA transfer to S. thermophilus from B. subtilis and L. lactis donor strains.
To conclude, we showed that co‐residing bacteria employ HGT in a laboratory setup by establishing a physical contact distinct from the classical conjugation mechanism and does not involve natural transformation. This newly described property of non‐conjugative plasmid transfer in LAB is undoubtedly a promising platform for natural strain improvement and should be explored in other industrially‐relevant bacteria.
AUTHOR CONTRIBUTIONS
Luiza P. Morawska: Conceptualization (equal); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Oscar P. Kuipers: Conceptualization (equal); funding acquisition (lead); investigation (supporting); methodology (supporting); resources (lead); supervision (lead); validation (lead); writing – original draft (supporting); writing – review and editing (supporting).
FUNDING INFORMATION
The authors acknowledge funding from BE‐Basic Foundation: TKIBE01001.
CONFLICT OF INTEREST
All authors declare no conflicts of interest.
Supporting information
Figure S1
Video S1
ACKNOWLEDGMENTS
The authors want to thank Dr. Saulius Kulakauskas (INRA) for kindly providing VES6759 strain of E. coli TG1 repA carrying pGh9 thermosensitive Ery100‐r plasmid with L. lactis cluA construct for deletion, and Jelmer Coenradij (RUG) for assistance with protease assay.
Morawska, L.P. & Kuipers, O.P. (2023) Cell‐to‐cell non‐conjugative plasmid transfer between Bacillus subtilis and lactic acid bacteria. Microbial Biotechnology, 16, 784–798. Available from: 10.1111/1751-7915.14195
DATA AVAILABILITY STATEMENT
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.
REFERENCES
- Anagnostopoulos, C. & Spizizen, J. (1961) Requirements for transformation in Bacillus subtilis 1. Journal of Bacteriology, 81, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa, K. , Kawai, Y. , Iioka, H. , Tanioka, M. , Nishimura, J. , Kitazawa, H. et al. (2008) Microbial community analysis of food‐spoilage bacteria in commercial custard creams using culture‐dependent and independent methods. Journal of Dairy Science, 91, 2938–2946. [DOI] [PubMed] [Google Scholar]
- Arnaouteli, S. , Bamford, N.C. , Stanley‐Wall, N.R. & Kovács, Á.T. (2021) Bacillus subtilis biofilm formation and social interactions. Nature Reviews. Microbiology, 19, 600–614. [DOI] [PubMed] [Google Scholar]
- Auchtung, J.M. , Aleksanyan, N. , Bulku, A. & Berkmen, M.B. (2016) Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis . Plasmid, 86, 14–25. [DOI] [PubMed] [Google Scholar]
- Auchtung, J.M. , Lee, C.A. , Garrison, K.L. & Grossman, A.D. (2007) Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis . Molecular Microbiology, 64, 1515–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung, J.M. , Lee, C.A. , Monson, R.E. , Lehman, A.P. & Grossman, A.D. (2005) Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proceedings of the National Academy of Sciences of the United States of America, 102, 12554–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp, J.M. , Leveque, R.M. , Dawid, S. & DiRita, V.J. (2017) Methylation‐dependent DNA discrimination in natural transformation of Campylobacter jejuni . Proceedings of the National Academy of Sciences of the United States of America, 114, E8053–E8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, S. , Baidya, A.K. , Pal, R.R. , Mamou, G. , Gatt, Y.E. , Margalit, H. et al. (2019) A ubiquitous platform for bacterial nanotube biogenesis. Cell Reports, 27, 334–342.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesa, A. , César, C.E. , Averhoff, B. & Berenguer, J. (2015) Noncanonical cell‐to‐cell DNA transfer in Thermus spp. is insensitive to argonaute‐mediated interference. Journal of Bacteriology, 197, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron, P.A. , Marcelli, B. , Mulder, J. , van der Els, S. , Morawska, L.P. , Kuipers, O.P. et al. (2019) Renaissance of traditional DNA transfer strategies for improvement of industrial lactic acid bacteria. Current Opinion in Biotechnology, 56, 61–68. [DOI] [PubMed] [Google Scholar]
- Cabezón, E. , Ripoll‐Rozada, J. , Peña, A. , de la Cruz, F. & Arechaga, I. (2015) Towards an integrated model of bacterial conjugation. FEMS Microbiology Reviews, 39, 81–95. [DOI] [PubMed] [Google Scholar]
- Chen, I. , Christie, P.J. & Dubnau, D. (2005) The ins and outs of DNA transfer in bacteria. Science, 310, 1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I. & Dubnau, D. (2004) DNA uptake during bacterial transformation. Nature Reviews. Microbiology, 2, 241–249. [DOI] [PubMed] [Google Scholar]
- Chiang, Y.N. , Penadés, J.R. & Chen, J. (2019) Genetic transduction by phages and chromosomal islands: the new and noncanonical. PLoS Pathogens, 15, e1007878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys, J.‐P. , Prudhomme, M. & Martin, B. (2006) Induction of competence regulons as a general response to stress in gram‐positive bacteria. Annual Review of Microbiology, 60, 451–475. [DOI] [PubMed] [Google Scholar]
- Clokie, M.R. , Millard, A.D. , Letarov, A.V. & Heaphy, S. (2011) Phages in nature. Bacteriophage, 1, 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan, F.M. & Koeppel, A.F. (2008) The origins of ecological diversity in prokaryotes. Current Biology, 18, R1024–R1034. [DOI] [PubMed] [Google Scholar]
- Dahmane, N. , Libante, V. , Charron‐Bourgoin, F. , Guédon, E. , Guédon, G. , Leblond‐Bourget, N. et al. (2017) Diversity of integrative and conjugative elements of Streptococcus salivarius and their intra‐ and interspecies transfer. Applied and Environmental Microbiology, 83, e00337‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, B. , Radziejwoski, A. , Toussaint, F. , Fontaine, L. , de Frahan, M.H. , Patout, C. et al. (2017) Natural DNA transformation is functional in Lactococcus lactis subsp. cremoris KW2. Applied and Environmental Microbiology, 83, e01074‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichelbohrer, I. , Alonso, J.C. , Lüder, G. & Trautner, T.A. (1985) Plasmid transduction by Bacillus subtilis bacteriophage SPP1: effects of DNA homology between plasmid and bacteriophage. Journal of Bacteriology, 162, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Xu, H. , Su, Y. , Liu, S. , Xu, L. , Guo, Z. et al. (2019) Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis. BMC Genomics, 20, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier, C. , Newman, J.A. , Kovacs, A.T. , Kaever, V. , Herzberg, C. , Rodrigues, C. et al. (2014) The YmdB phosphodiesterase is a global regulator of late adaptive responses in Bacillus subtilis . Journal of Bacteriology, 196, 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier, C. , Pietack, N. , Gunka, K. , Wrede, C. , Lehnik‐Habrink, M. , Herzberg, C. et al. (2011) A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. Journal of Bacteriology, 193, 5997–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues, S. & Nielsen, K.M. (2017) Membrane vesicles and horizontal gene transfer in prokaryotes. Current Opinion in Microbiology, 38, 16–21. [DOI] [PubMed] [Google Scholar]
- Dubey, G.P. & Ben‐Yehuda, S. (2011) Intercellular nanotubes mediate bacterial communication. Cell, 144, 590–600. [DOI] [PubMed] [Google Scholar]
- Dubey, G.P. , Malli Mohan, G.B. , Dubrovsky, A. , Amen, T. , Tsipshtein, S. , Rouvinski, A. et al. (2016) Architecture and characteristics of bacterial nanotubes. Developmental Cell, 36, 453–461. [DOI] [PubMed] [Google Scholar]
- Dubnau, D. (1991a) The regulation of genetic competence in Bacillus subtilis . Molecular Microbiology, 5, 11–18. [DOI] [PubMed] [Google Scholar]
- Dubnau, D. (1991b) Genetic competence in Bacillus subtilis . Microbiological Reviews, 55, 395–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchuuya, R. , Ito, M. , Kitano, S. , Shigi, F. , Sobue, R. & Maeda, S. (2011) Cell‐to‐cell transformation in Escherichia coli: a novel type of natural transformation involving cell‐derived DNA and a putative promoting pheromone. PLoS One, 6, e16355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faille, C. , Bénézech, T. , Midelet‐Bourdin, G. , Lequette, Y. , Clarisse, M. , Ronse, G. et al. (2014) Sporulation of Bacillus spp. within biofilms: a potential source of contamination in food processing environments. Food Microbiology, 40, 64–74. [DOI] [PubMed] [Google Scholar]
- Fontaine, L. , Goffin, P. , Dubout, H. , Delplace, B. , Baulard, A. , Lecat‐Guillet, N. et al. (2013) Mechanism of competence activation by the ComRS signalling system in streptococci. Molecular Microbiology, 87, 1113–1132. [DOI] [PubMed] [Google Scholar]
- Gasson, M.J. (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast‐induced curing. Journal of Bacteriology, 154, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon, J.‐J. , Jury, K. , Shearman, C.A. & Gasson, M.J. (1994) The Lactococcus lactis sex‐factor aggregation gene cluA. Molecular Microbiology, 12, 655–663. [DOI] [PubMed] [Google Scholar]
- Grohmann, E. (2010a) Autonomous plasmid‐like replication of Bacillus ICEBs1: a general feature of integrative conjugative elements? Molecular Microbiology, 75, 261–263. [DOI] [PubMed] [Google Scholar]
- Grohmann, E. (2010b) Conjugative transfer of the integrative and conjugative element ICEBs1 from Bacillus subtilis likely initiates at the donor cell pole. Journal of Bacteriology, 192, 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha, S. (1988) DNA methyltransferase of Bacillus subtilis Marburg: purification, properties and further evidence of specificity. Gene, 74, 77–81. [DOI] [PubMed] [Google Scholar]
- Habimana, O. , Meyrand, M. , Meylheuc, T. , Kulakauskas, S. & Briandet, R. (2009) Genetic features of resident biofilms determine attachment of Listeria monocytogenes . Applied and Environmental Microbiology, 75, 7814–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, J. , Luttinger, A. & Dubnau, D. (1996) Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis . Molecular Microbiology, 21, 763–775. [DOI] [PubMed] [Google Scholar]
- Hamoen, L.W. , Venema, G. & Kuipers, O.P. (2003) Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology, 149, 9–17. [DOI] [PubMed] [Google Scholar]
- Harwood, C.R. & Cutting, S.M. (1990) Molecular biological methods for Bacillus. Chichester: John Wiley, xxxvi + 581 pages. [Google Scholar]
- Haustenne, L. , Bastin, G. , Hols, P. & Fontaine, L. (2015) Modeling of the ComRS signaling pathway reveals the limiting factors controlling competence in Streptococcus thermophilus . Frontiers in Microbiology, 6. Available from: 10.3389/fmicb.2015.01413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein, L.S. , Coomaraswamy, G. & Morrison, D.A. (1995) An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae . Proceedings of the National Academy of Sciences of the United States of America, 92, 11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein, L.S. , Gaustad, P. , Nes, I.F. & Morrison, D.A. (1996) Identification of the streptococcal competence‐pheromone receptor. Molecular Microbiology, 21, 863–869. [DOI] [PubMed] [Google Scholar]
- Kuipers, O.P. (2015) Back to nature: a revival of natural strain improvement methodologies. Microbial Biotechnology, 8, 17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.A. , Thomas, J. & Grossman, A.D. (2012) The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. Journal of Bacteriology, 194, 3165–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M.G. & Wackernagel, W. (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiological Reviews, 58, 563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S. , Ito, M. , Ando, T. , Ishimoto, Y. , Fujisawa, Y. , Takahashi, H. et al. (2006) Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiology Letters, 255, 115–120. [DOI] [PubMed] [Google Scholar]
- Maehara, T. , Itaya, M. , Ogura, M. & Tanaka, T. (2011) Effect of Bacillus subtilis BsuM restriction–modification on plasmid transfer by polyethylene glycol‐induced protoplast fusion. FEMS Microbiology Letters, 325, 49–55. [DOI] [PubMed] [Google Scholar]
- Maguin, E. , Prévost, H. , Ehrlich, S.D. & Gruss, A. (1996) Efficient insertional mutagenesis in lactococci and other gram‐positive bacteria. Journal of Bacteriology, 178, 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony, J. , Cambillau, C. & van Sinderen, D. (2017) Host recognition by lactic acid bacterial phages. FEMS Microbiology Reviews, 41, S16–S26. [DOI] [PubMed] [Google Scholar]
- Majewski, J. (2001) Sexual isolation in bacteria. FEMS Microbiology Letters, 199, 161–169. [DOI] [PubMed] [Google Scholar]
- Marcelli, B. , Karsens, H. , Nijland, M. , Oudshoorn, R. , Kuipers, O.P. & Kok, J. (2020) Employing lytic phage‐mediated horizontal gene transfer in Lactococcus lactis . PLoS One, 15, e0238988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugg, J. D. , Meijer, W. , van Kranenburg, R. , Laverman, P. , Bruinenberg, P. G. & de Vos, W. M. (1995) Medium‐dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. Journal of Bacteriology 177: 2982–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, A. , Sekoguchi, A. , Imai, J. , Kondo, K. , Shibata, Y. & Maeda, S. (2016) Natural Escherichia coli strains undergo cell‐to‐cell plasmid transformation. Biochemical and Biophysical Research Communications, 481, 59–62. Available from: 10.1016/j.bbrc.2016.11.018 [DOI] [PubMed] [Google Scholar]
- Matsuoka, S. , Asai, K. & Sadaie, Y. (2005) Restriction and modification of SP10 phage by BsuM of Bacillus subtilis Marburg. FEMS Microbiology Letters, 244, 335–339. [DOI] [PubMed] [Google Scholar]
- Molin, S. & Tolker‐Nielsen, T. (2003) Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Current Opinion in Biotechnology, 14, 255–261. [DOI] [PubMed] [Google Scholar]
- Moschonas, G. , Lianou, A. , Nychas, G.‐J.E. & Panagou, E.Z. (2021) Spoilage potential of Bacillus subtilis in a neutral‐pH dairy dessert. Food Microbiology, 95, 103715. [DOI] [PubMed] [Google Scholar]
- Mulder, J. , Wels, M. , Kuipers, O.P. , Kleerebezem, M. & Bron, P.A. (2017) Unleashing natural competence in Lactococcus lactis by induction of the competence regulator ComX. Applied and Environmental Microbiology, 83, e01320‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K.M. , Johnsen, P.J. , Bensasson, D. & Daffonchio, D. (2007) Release and persistence of extracellular DNA in the environment. Environmental Biosafety Research, 6, 37–53. [DOI] [PubMed] [Google Scholar]
- O'sullivan, D. , Twomey, D.P. , Coffey, A. , Hill, C. , Fitzgerald, G.F. & Ross, R.P. (2000) Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis . Molecular Microbiology, 36, 866–875. [DOI] [PubMed] [Google Scholar]
- Otto, R. , de Vos, W.M. & Gavrieli, J. (1982) Plasmid DNA in Streptococcus cremoris Wg2: influence of pH on selection in chemostats of a variant lacking a protease plasmid. Applied and Environmental Microbiology, 43, 1272–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overkamp, W. , Beilharz, K. , Detert Oude Weme, R. , Solopova, A. , Karsens, H. , Kovács, Á. , et al. (2013) Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Applied and Environmental Microbiology 79: 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxaran, V. , Ledue‐Clier, F. , Dieye, Y. , Herry, J.M. , Péchoux, C. , Meylheuc, T. et al. (2012) Pilus biogenesis in Lactococcus lactis: molecular characterization and role in aggregation and biofilm formation. PLoS One, 7, e50989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, R.R. , Baidya, A.K. , Mamou, G. , Bhattacharya, S. , Socol, Y. , Kobi, S. et al. (2019) Pathogenic E. coli extracts nutrients from infected host cells utilizing injectisome components. Cell, 177, 683–696.e18. [DOI] [PubMed] [Google Scholar]
- Pande, S. , Shitut, S. , Freund, L. , Westermann, M. , Bertels, F. , Colesie, C. et al. (2015) Metabolic cross‐feeding via intercellular nanotubes among bacteria. Nature Communications, 6, 6238. [DOI] [PubMed] [Google Scholar]
- Rösch, T.C. , Golman, W. , Hucklesby, L. , Gonzalez‐Pastor, J.E. & Graumann, P.L. (2014) The presence of conjugative plasmid pLS20 affects global transcription of its Bacillus subtilis host and confers beneficial stress resistance to cells. Applied and Environmental Microbiology, 80, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruyter, P.G. , Kuipers, O.P. & de Vos, W.M. (1996) Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Applied and Environmental Microbiology, 62, 3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H. & Hensel, M. (2004) Pathogenicity islands in bacterial pathogenesis. Clinical Microbiology Reviews, 17, 14–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouler, C. , Gautier, M. , Ehrlich, S.D. & Chopin, M.‐C. (1998) Combinational variation of restriction modification specificities in Lactococcus lactis . Molecular Microbiology, 28, 169–178. [DOI] [PubMed] [Google Scholar]
- Siddaramappa, S. , Challacombe, J.F. , Duncan, A.J. , Gillaspy, A.F. , Carson, M. , Gipson, J. et al. (2011) Horizontal gene transfer in Histophilus somni and its role in the evolution of pathogenic strain 2336, as determined by comparative genomic analyses. BMC Genomics, 12, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P.K. , Ramachandran, G. , Ramos‐Ruiz, R. , Peiró‐Pastor, R. , Abia, D. , Wu, L.J. et al. (2013) Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLoS Genetics, 9, e1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager, J. , Kjos, M. , Attaiech, L. & Veening, J.‐W. (2014) Antibiotic‐induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell, 157, 395–406. [DOI] [PubMed] [Google Scholar]
- Stempler, O. , Baidya, A.K. , Bhattacharya, S. , Malli Mohan, G.B. , Tzipilevich, E. , Sinai, L. et al. (2017) Interspecies nutrient extraction and toxin delivery between bacteria. Nature Communications, 8, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M. & Nielsen, K.M. (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Reviews. Microbiology, 3, 711–721. [DOI] [PubMed] [Google Scholar]
- Tzipilevich, E. , Habusha, M. & Ben‐Yehuda, S. (2017) Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell, 168, 186–199.e12. [DOI] [PubMed] [Google Scholar]
- van Sinderen, D. , ten Berge, A. , Hayema, B.J. , Hamoen, L. & Venema, G. (1994) Molecular cloning and sequence of comK, a gene required for genetic competence in Bacillus subtilis . Molecular Microbiology, 11, 695–703. [DOI] [PubMed] [Google Scholar]
- Warner, F.D. , Kitos, G.A. , Romano, M.P. & Hemphill, H.E. (1977) Characterization of SPβ: a temperate bacteriophage from Bacillus subtilis 168M. Canadian Journal of Microbiology, 23, 45–51. [Google Scholar]
- Wiedenbeck, J. & Cohan, F.M. (2011) Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiology Reviews, 35, 957–976. [DOI] [PubMed] [Google Scholar]
- Wydau, S. , Dervyn, R. , Anba, J. , Dusko Ehrlich, S. & Maguin, E. (2006) Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiology Letters, 257, 32–42. [DOI] [PubMed] [Google Scholar]
- Yasbin, R.E. & Young, F.E. (1974) Transduction in Bacillus subtilis by bacteriophage SPP1. Journal of Virology, 14, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Jin, T. , Deng, L. , Wang, C. , Zhang, Y. & Chen, X. (2018) Stress‐induced, highly efficient, donor cell‐dependent cell‐to‐cell natural transformation in Bacillus subtilis . Journal of Bacteriology, 200, e00267‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Video S1
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.


