Abstract
Mangiferin, a natural flavonoid compound with multiple biological activities (e.g., anti-inflammatory, anti-oxidant, anti-diabetic, and anti-tumor), has gained increased research interest in recent years. Nevertheless, the metabolic processing of mangiferin has not been fully investigated. In this study, a rapid and efficient analytical strategy named “Drug Metabolite Clusters” was applied for comprehensive profiling of mangiferin metabolites in rat plasma, urine, and feces samples in vivo following oral administration and liver microsomes in vitro. First, the biological samples were pretreated with methanol, acetonitrile, and solid phase extraction (SPE) for further liquid chromatography–mass spectrometry (LC–MS) analysis. Second, the raw data were acquired using ultra-high performance liquid chromatography quadrupole exactive orbitrap high-resolution mass spectrometry (UHPLC-Q-Exactive Orbitrap HRMS) under the positive and negative full-scan/dd MS2 modes. Third, mangiferin and its basic metabolites (norathyriol, trihydroxyxanthone, and dihydroxyxanthone) were selected as mangiferin metabolite cluster centers by referring to the relevant literature. Subsequently, according to the pyrolysis law of mass spectrometry, literature reports, and reference material comparison, especially the diagnostic product ions (DPIs), the candidate metabolites were accurately preliminarily identified, and mangiferin metabolite clusters based on metabolite cluster center changes were formed. As a result, a total of 67 mangiferin metabolites (mangiferin included) were detected, including 29 in plasma, 48 in urine, 12 in feces, and 6 in liver microsomes. Among them, trihydroxyxanthones were first detected in rat urine samples after oral mangiferin. We found that mangiferin mainly underwent deglucosylation, dehydroxylation, methylation, glucuronidation, sulfation, and other composite reactions in rats. Herein, we have elucidated the metabolites and metabolic pathways of mangiferin in vivo and in vitro, which provided an essential theoretical basis for further pharmacological studies of mangiferin and a comprehensive research method for the identification of drug metabolites.
1. Introduction
Mangiferin (2-β-d-glucopyranosyl-1,3,6,7-tetrahydroxy-9H-xanthen-9-one), one of C-glycosylated flavonoids, was originally isolated from Mangifera indica L and Anemarrhena asphodeloides Bge.1,2 Studies have shown that mangiferin is richly expressed in the pulp, peel, seed, bark, and leaves of mangoes and the dry rhizome of Anemarrhenae as well.3,4 Mangiferin has recently gained increased research interest due to its wide range of pharmacological effects, including anti-inflammatory,5,6 anti-oxidant,7 anti-diabetic,8 anti-tumor,9,10 liver protection,11 and neuroprotection.12,13 Moreover, studies on mangiferin have shown that it plays a crucial role in the prevention and treatment of various types cancer and neurodegenerative diseases.14−17 Mangiferin was primarily implicated in down-regulating inflammation, reducing metastasis in malignant cells, and ameliorating cognitive dysfunction.6,18−20 Taken together, mangiferin could be served as a potential natural therapeutic candidate for cancers and neurodegenerative diseases.
In vivo and in vitro metabolism studies allow for the determination of the main metabolic mode, biotransformation pathways, drug metabolites, etc. On this basis, further comparison of the activity or toxicity of prototype drugs and their metabolites can be carried out, so as to elucidate their mechanisms of pharmacodynamics or toxicity. Due to the low bioavailability and remarkable pharmacological effects, some mangiferin metabolites have been thought to accumulate to exert pharmacological effects.21−23 Therefore, it is important to reveal the metabolic profile of mangiferin for the study of its pharmacology. Liu et al. developed a methodology for the identification of mangiferin, and 33 metabolites were inferred in plasma, urine, feces, and various organs based on electrospray ionization (ESI) tandem hybrid ion trap mass spectrometry.22 However, the metabolism study of mangiferin has been not comprehensive enough owing to the relatively low separation efficiency, detection sensitivity, and resolution to some extent, and difficulty remains hard regarding how to screen out the metabolites from the complex biological samples.
Consequently, our team first proposed the “Drug Metabolite Clusters” analytical strategy to search for drug metabolites.24,25 The prototype drug generated primary metabolites through drug metabolic enzymes according to certain metabolic rules and then formed primary metabolite cluster centers. The primary metabolites were further metabolized according to the corresponding reactions to form secondary metabolite cluster centers, and finally, Drug Metabolite Clusters was formed. In addition, with the rapid development of ultra-high performance liquid chromatography quadrupole exactive orbitrap high resolution mass spectrometry (UHPLC-Q-Exactive Orbitrap HRMS) with high efficiency and high sensitivity, it accelerates the process of efficiently screening and characterizing known and unknown compounds. As a consequence, in the present study, mangiferin metabolites were systematically characterized using UHPLC-Q-Exactive Orbitrap HRMS coupled with the “Drug Metabolite Clusters” analytical strategy, so as to aid our understanding of the biotransformation of mangiferin in vivo and in vitro.
2. Results
2.1. Establishment of the Analytical Strategy
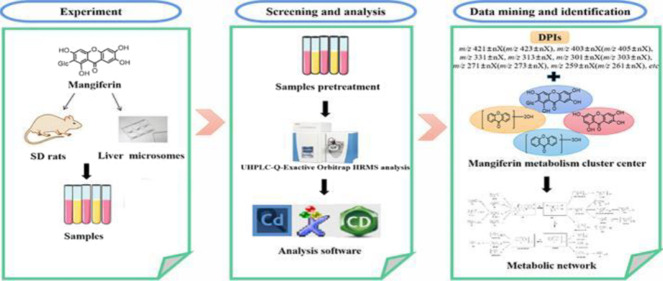
Due to the complexity of the metabolic profile and interference from endogenous substances, it is important to establish an efficient analytical strategy for metabolite identification. To identify as many metabolites as possible, we established an analytical method based on the “Drug Metabolite Clusters” and used numerous data processing techniques to search for mangiferin metabolites in vivo and in vitro in this study (Figure 1). First, UHPLC-Q-Exactive Orbitrap HRMS technology was adopted to preliminarily screen candidates after the acquisition of high-quality accurate raw data in positive as well as negative ion modes. Second, for subsequent background removal, Compound Discoverer Software was utilized to get rid of large amounts of interferential components and further search for potential metabolites. Third, the prototype drug (mangiferin) and its primary metabolites (norathyriol, trihydroxyxanthone, and dihydroxyxanthone) were selected as the primary cores of Drug Metabolite Clusters to predict metabolites. Moreover, the Drug Metabolite Clusters strategy was developed for the screening and prediction of mangiferin metabolites, combined with multiple metabolic pathway templates. Finally, the diagnostic product ion (DPI), a data mining method, was performed to rapidly identify the mangiferin metabolites.
Figure 1.
Summary diagram of the experimental process for the identification of mangiferin metabolites in vivo and in vitro.
2.1.1. Establishment of Cores for Drug Metabolite Clusters
We applied a validated strategy termed “Drug Metabolite Clusters” to the process of metabolite identification. Since mangiferin is a special flavonoid glycoside with the stable C-glycoside bond, it can be hydrolyzed to generate norathyriol by intestinal flora and then absorbed into the blood. Norathyriol can undergo the dehydroxylation reaction to form trihydroxyxanthone and dihydroxyxanthone. We confirmed that mentioned above by preliminary analysis results and literature-related results.22,23,26 Thus, the prototype drug (mangiferin) and its basic metabolites (norathyriol, trihydroxyxanthone, and dihydroxyxanthone) were regarded as the core points of the Drug Metabolite Clusters to further search for secondary metabolites.
2.1.2. Establishment of Analytical Templates
In vivo biotransformation reactions of drugs can be divided into phase I metabolism and phase II metabolism. Phase I metabolism mainly consists of oxidation, reduction, and hydrolysis reaction, and the phase II metabolism is the conjugation reaction, including glycosylation, sulfation, glucuronidation, and so on. Therefore, according to the above common metabolic reactions and the four core points of the Drug Metabolite Clusters, we established individual templates to allow us to scan and search for a large number of compounds so that more mangiferin metabolites could be found. Four primary templates were set in parallel as follows: mangiferin-type template, norathyriol-type template, trihydroxyxanthone-type template, and dihydroxyxanthone-type template. Then, the main metabolites found by the primary templates were used as the centers of the secondary templates to continue the search for more metabolites, eventually forming Drug Metabolite Clusters.
2.1.3. Establishment of DPIs
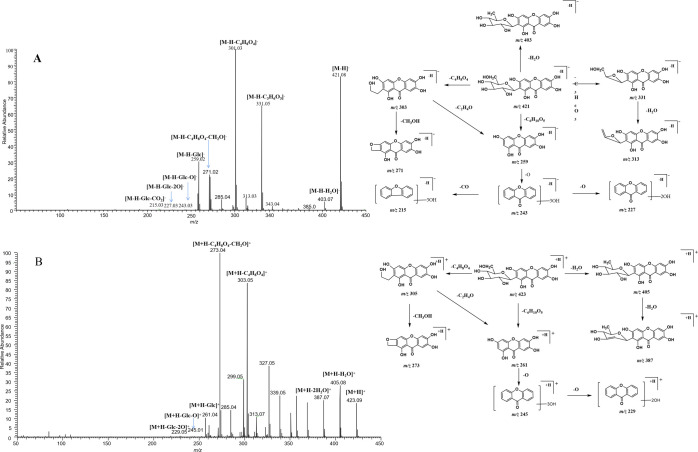
Mangiferin was eluted at 9.06 and 9.07 min in negative and positive ion modes, respectively. The accurate mass measurement gave [M-H]− ions at m/z 421.07620 (mass error of −0.802 ppm) and [M + H]+ ions at m/z 423.09116 (mass error of −2.429 ppm) with a theoretical elemental composition of C11H18O11. ESI-MS/MS spectra revealed the fragment ions of mangiferin at m/z 421, m/z 403, m/z 331, m/z 313, m/z 301, m/z 285, m/z 271, m/z 259, m/z 243, m/z 227, and m/z 215 in negative ion mode. Furthermore, the fragment ions at m/z 423, m/z 405, m/z 387, m/z 303, m/z 273, m/z 273, m/z 245, and m/z 229 were all observed in positive ion mode. It was inferred that the fragment ion at m/z 403 [M-H-18]− (m/z 405 [M + H-18]+) was formed by the neutral loss of H2O from mangiferin. The representative ions at m/z 331 [M-H-90]− and m/z 301 [M-H-120]− (m/z 303 [M + H-120]+) might be formed by the loss of C3H6O3 and C4H8O4 groups, which were adopted as the characteristic fragmentation pathways of C-glycosyl flavonoids.27 The loss of C3H6O3 + H2O and CO in turn from mangiferin generated the product ions m/z 313 and m/z 285. The fragment ion at m/z 271 (m/z 273) was formed by the loss of the CH2O group of the ion at m/z 301 (m/z 303). Additionally, the fragment ion at m/z 259 (m/z 261) was derived from the ion at m/z 421 (m/z 423) of mangiferin by the loss of a glucose unit (C6H10O5, 162 Da), which also corresponds to the molecular ion of norathyriol. The fragment ions at m/z 243 (m/z 245) and m/z 227 (m/z 229) were produced by further cleavage of norathyriol, corresponding to trihydroxyxanthone and dihydroxyxanthone. During metabolism, the basic skeletal structure of mangiferin tends to remain unchanged. Therefore, the product ions at m/z 421 ± nX, m/z 403 ± nX, m/z 331 ± nX, m/z 313 ± nX, m/z 301 ± nX, m/z 271 ± nX, m/z 259 ± nX m/z 243 ± nX, m/z 227 ± nX, m/z 215 ± nX in negative ion mode and m/z 423 ± nX, m/z 405 ± nX, m/z 303 ± nX, m/z 273 ± nX, m/z 261 ± nX, m/z 245 ± nX, m/z 229 ± nX in positive ion mode (X = substituent groups, such as CH2 (14 Da), Glc (162 Da), GluA (176 Da), SO3 (80 Da), O (16 Da), etc.; n = the number of substituents) were selected as the DPIs, which could be used to identify the mangiferin metabolites. The ESI-MS/MS spectra and the predominant fragmentation patterns of mangiferin in negative and positive ion modes are shown in Figure 2.
Figure 2.
ESI-MS/MS spectra and mass fragmentation behavior of mangiferin in negative (A) and positive (B) ion modes.
2.2. Identification of Mangiferin Metabolites In Vivo and In Vitro
In all, a total of 67 metabolites were discovered and identified in biological samples by using UHPLC-Q-Exactive Orbitrap HRMS combined with the “Drug Metabolite Clusters” analytical strategy, including 29 in plasma (SPE treatment: 24; methanol treatment:16, acetonitrile treatment: 17), 48 in urine, 12 in feces, and 6 in liver microsomes. Information regarding these metabolites (retention times, formula proposed, product ions, etc.) is shown in Table 1.
Table 1. Metabolites and Distribution of Mangiferin In Vivo and In Vitroa.
| no. | molecular formula | retention time (min) | mode | theoretical value [M-H]−/[M + H]+ | measured value [M-H]−/[M + H]+ | RDB | error | MS2 | U | PS | PM | PA | F | LM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M0 | C19H18O11 | 9.08 | N | 421.07654 | 421.07620 | 11.5 | –0.802 | 301.03(100), 421.08(88), 331.04(65), 259.02(31), 271.02(23), 285.04(7), 403.07(6) | + | + | + | + | + | |
| 9.07 | P | 423.09219 | 423.09116 | 10.5 | –2.429 | 273.04(100),303.05(83),405.08(28),423.09(19),261.04(7) | + | + | + | |||||
| M1 | C19H18O14S | 3.05 | N | 501.03443 | 501.03326 | 11.5 | –2.373 | 501.03(100), 411.00(29), 301.03(21), 271.02(14), 331.04(11), 259.02(4) | + | |||||
| M2 | C25H28O16 | 8.47 | N | 583.12936 | 583.12927 | 12.5 | –2.037 | 301.03(100), 583.13(89), 331.05(55), 259.02(23), 273.04(7) | + | |||||
| M3 | C26H28O17 | 8.74 | N | 611.12428 | 611.12421 | 13.5 | –0.107 | 435.09(100), 315.05(70), 345.06(47), 330.04(21), 611.12(21), 272.03(13), 273.04(12) | + | + | ||||
| M4 | C25H28O16 | 8.85 | N | 583.12936 | 583.12982 | 12.5 | –1.094 | 583.13(74), 301.03(50), 258.12(17), 315.05(16) | + | |||||
| M5 | C25H26O17 | 9.36 | N | 597.10862 | 597.10876 | 13.5 | –1.612 | 259.02(100), 435.06(23), 597.11(9) | + | + | ||||
| M6 | C20H20O11 | 9.52 | N | 435.09219 | 435.09256 | 11.5 | –1.665 | 435.09(49), 287.02(17), 335.20(15), 272.03(13), 315.05(11) | + | |||||
| 9.51 | P | 437.10784 | 437.10733 | 10.5 | –1.162 | 287.05(100), 317.06(39), 313.07(32), 401.09(20), 437.11(3) | + | |||||||
| M7 | C20H20O14S | 9.55 | N | 515.04900 | 515.04889 | 11.5 | –0.218 | 435.09(100), 315.05(79), 345.06(67), 515.05(32), 300.03(28), 330.04(26), 259.02(10) | + | + | ||||
| M8 | C20H20O11 | 10.10 | N | 435.09219 | 435.09262 | 11.5 | –1.527 | 272.03(100), 435.09(82), 315.05(58), 345.06(22), 259.02(9) | + | |||||
| 10.11 | P | 437.10784 | 437.10757 | 10.5 | –0.613 | 287.05(100), 317.07(80), 419.10(30), 313.07(30), 401.08(21), 383.07(16), 261.04(11) | + | |||||||
| M9 | C20H20O10 | 10.32 | N | 419.09727 | 419.09967 | 11.5 | 3.102 | 243.03(100), 419.14(4) | + | |||||
| M10 | C21H22O11 | 10.32 | N | 449.10784 | 449.10815 | 11.5 | 0.695 | 273.04(100), 258.02(89), 449.07(23), 259.02(12), 315.05(4) | + | |||||
| M11 | C20H20O10 | 10.61 | N | 419.09727 | 419.09616 | 11.5 | –2.656 | 243.03(100), 419.06(25), 259.02(5) | + | |||||
| M12 | C21H22O11 | 10.62 | N | 449.10784 | 449.10913 | 11.5 | 2.877 | 258.02(100), 273.04(91), 449.07(18), 259.02(15) | + | |||||
| M13 | C20H20O11 | 10.71 | N | 435.09219 | 435.09177 | 11.5 | –0.96 | 259.02(100), 260.03(13), 435.09(3), 272.03(2), 287.02(2) | + | + | ||||
| M14 | C20H20O10 | 11.98 | N | 419.09727 | 419.09753 | 11.5 | 0.613 | 419.13(100), 243.10(48) | + | |||||
| M15 | C13H8O6 | 11.94 | N | 259.02371 | 259.02417 | 10.5 | 1.759 | 259.02(100), 215.11(6), 187.04(2), 231.03(2), 241.14(1) | + | + | + | + | ||
| 11.95 | P | 261.03936 | 261.03897 | 9.5 | –1.511 | 261.04(100), 243.03(14) | + | + | + | |||||
| M16 | C13H8O9S | 3.99 | N | 338.98053 | 338.98306 | 10.5 | 4.231 | 292.98(100), 203.08(64), 271.07(53), 338.98(15), 310.99(41), 225.06(39) | + | + | ||||
| M17 | C13H8O9S | 4.12 | N | 338.98053 | 338.98309 | 10.5 | 4.32 | 292.98(100), 203.08(66), 225.06(45), 271.07(43), 310.99(41), 338.98(19) | + | |||||
| M18 | C13H8O9S | 4.82 | N | 338.98053 | 338.98041 | 10.5 | –0.351 | 339.06(100), 259.11(98), 260.11(17) | + | |||||
| M19 | C25H24O18 | 7.98 | N | 611.08789 | 611.08765 | 14.5 | –0.393 | 259.02(100), 435.06(42), 187.01(2), 611.09(2) | + | |||||
| M20 | C25H24O18 | 9.00 | N | 611.08789 | 611.08807 | 14.5 | 0.294 | 259.02(100), 435.06(39), 611.09(14), 242.01(4) | + | |||||
| M21 | C25H24O18 | 9.39 | N | 611.08789 | 611.08789 | 14.5 | –1.795 | 259.02(100), 611.09(45), 435.05(11) | + | |||||
| M22 | C25H24O18 | 9.44 | N | 611.08789 | 611.08838 | 14.5 | 0.802 | 259.02(100), 435.06(43), 611.09(2) | + | |||||
| M23 | C19H16O12 | 9.46 | N | 435.05580 | 435.05734 | 12.5 | 1.014 | 435.09(100), 287.02(45), 272.03(27), 315.05(23), 259.02(6) | + | |||||
| M24 | C19H16O12 | 9.72 | N | 435.05580 | 435.05594 | 12.5 | –2.204 | 259.02(100), 435.05(31), 187.01(6), 108.04(5) | + | + | ||||
| M25 | C26H26O18 | 9.89 | N | 625.10354 | 625.10327 | 14.5 | –0.432 | 273.04(100), 449.07(33), 625.10(27), 259.02(7) | + | + | ||||
| M26 | C19H16O15S | 9.99 | N | 515.01262 | 515.01160 | 12.5 | –4.104 | 283.08(100), 259.02(29), 435.06(9), 515.01(7) | + | |||||
| M27 | C14H10O9S | 10.27 | N | 352.99728 | 352.99597 | 10.5 | –0.592 | 353.13(89), 273.17(19), 227.16(3) | + | |||||
| M28 | C20H18O12 | 10.45 | N | 449.07145 | 449.07196 | 12.5 | 1.131 | 273.04(100), 258.02(89), 449.07(23), 259.02(12), 315.05(4) | + | |||||
| M29 | C19H16O12 | 10.58 | N | 435.05580 | 435.05606 | 12.5 | –1.929 | 259.02(100), 435.06(28) | + | + | + | + | ||
| 10.57 | P | 437.07145 | 437.07108 | 11.5 | –0.852 | 261.04(100), 437.07(27), 262.04(14) | + | |||||||
| M30 | C19H16O12 | 10.73 | N | 435.05580 | 435.05624 | 12.5 | –1.515 | 259.02(100), 435.05(3) | + | + | + | + | ||
| 10.74 | P | 437.07145 | 437.0708 | 11.5 | –1.492 | 261.04(100), 262.04(15), 437.07(7), 243.03(4) | + | |||||||
| M31 | C20H18O12 | 10.75 | N | 449.07145 | 449.07193 | 12.5 | 1.064 | 258.02(100), 273.04(62), 449.07(28), 259.02(20) | + | |||||
| M32 | C20H18O12 | 11.08 | N | 449.07145 | 449.07156 | 12.5 | 0.24 | 273.04(100), 258.02(90), 449.07(13), 259.02(12) | + | + | + | + | ||
| M33 | C20H18O12 | 11.38 | N | 449.07145 | 449.07138 | 12.5 | –0.161 | 273.04(100), 258.02(77), 259.02(10), 230.02(5), 449.07(3) | + | + | + | + | ||
| 11.37 | P | 451.08710 | 451.08649 | 11.5 | –1.358 | no secondary product ions | + | + | ||||||
| M34 | C21H20O12 | 11.45 | N | 463.08710 | 463.08704 | 12.5 | –2.503 | 287.06(100), 273.03(75), 463.08(10), 257.01(10) | + | |||||
| M35 | C13H8O9S | 11.94 | N | 338.98053 | 338.98068 | 10.5 | 0.446 | 259.02(100), 338.98(12), 201.02(6), 321.12(5), 79.96(7) | + | |||||
| M36 | C20H18O12 | 12.48 | N | 449.07145 | 449.07172 | 12.5 | 0.596 | 273.04(100), 258.02(36), 259.02(4), 449.07(4) | + | + | + | + | ||
| 12.50 | P | 451.08710 | 451.08682 | 11.5 | –0.626 | 275.05(100), 276.06(15), 451.09(7), 257.04(3), 229.05(1) | + | + | ||||||
| M37 | C14H10O6 | 12.51 | N | 273.03936 | 273.03903 | 10.5 | –3.887 | 273.04(100), 258.02(4) | + | + | ||||
| M38 | C20H18O12 | 12.87 | N | 449.07145 | 449.07181 | 12.5 | 0.797 | 273.04(100), 449.07(39), 258.02(33), 259.02(4) | + | + | + | + | ||
| 12.88 | P | 451.08710 | 451.08670 | 11.5 | –0.892 | 275.05(100), 451.09(26), 276.06(13), 257.04(2) | + | + | ||||||
| M39 | C15H12O6 | 13.00 | N | 287.05501 | 287.05548 | 10.5 | –2.199 | 272.03(100), 287.06(90), 257.01(58), 258.01(8), 243.12(3) | + | |||||
| M40 | C21H20O12 | 13.00 | N | 463.08710 | 463.08783 | 12.5 | –0.797 | 287.06(100), 273.03(87), 257.01(20), 463.08(10) | + | |||||
| M41 | C14H10O6 | 13.44 | N | 273.03936 | 273.04001 | 10.5 | 2.364 | 258.02(100), 273.04(63), 259.02(15), 228.17(6) | + | |||||
| M42 | C14H10O6 | 14.70 | N | 273.03936 | 273.03976 | 10.5 | 1.449 | 273.04(100), 258.02(94), 259.02(13), 227.16(5), 211.17(4) | + | + | ||||
| M43 | C15H12O6 | 16.48 | N | 287.05501 | 287.05502 | 10.5 | –3.802 | 287.06(100), 272.03(96), 257.01(73), 258.01(10) | + | |||||
| M44 | C13H8O5 | 10.39 | N | 243.02993 | 243.02904 | 10.5 | 0.988 | 243.03(100), 199.04(7), 181.12(7) | + | + | ||||
| M45 | C13H8O5 | 10.83 | N | 243.02993 | 243.02904 | 10.5 | 0.988 | 243.03(41), 199.04(8), 181.12(7) | + | |||||
| M46 | C25H24O17 | 7.62 | N | 595.09297 | 595.09351 | 14.5 | –0.945 | 243.03(100), 419.06(52), 595.09(20), 212.00(25) | + | |||||
| M47 | C13H8O8S | 9.27 | N | 322.98561 | 322.98721 | 10.5 | 1.545 | 223.11(100), 323.12(24), 205.10(74), 243.16(3) | + | |||||
| M48 | C13H8O8S | 10.38 | N | 322.98561 | 322.98468 | 10.5 | –2.893 | 323.12(33), 279.17(24), 243.10(9), 235.18(21) | + | |||||
| M49 | C19H16O11 | 10.40 | N | 419.06089 | 419.06119 | 12.5 | –1.896 | 243.03(100), 175.02(12), 419.06(4) | + | + | + | |||
| 10.41 | P | 421.07654 | 421.07629 | 11.5 | –0.588 | 245.04(100), 246.05(13), 421.08(7) | + | + | + | |||||
| M50 | C19H16O11 | 10.67 | N | 419.06089 | 419.06116 | 12.5 | –1.967 | 243.03(100), 419.06(25), 59.01(11), 259.02(5) | + | + | + | |||
| 10.68 | P | 421.07654 | 421.07602 | 11.5 | –1.229 | 245.04(100), 246.05(13), 421.08(5) | + | |||||||
| M51 | C19H16O11 | 10.83 | N | 419.06089 | 419.06131 | 12.5 | –1.609 | 243.03(100), 419.06(24), 59.01(4) | + | + | + | |||
| M52 | C13H8O8S | 11.78 | N | 322.98561 | 322.98578 | 10.5 | –2.883 | 243.03(100), 322.99(38), 244.03(13) | + | |||||
| M53 | C13H8O8S | 13.73 | N | 322.98561 | 322.98563 | 10.5 | –3.347 | 243.03(100), 219.00(40), 323.01(18) | + | |||||
| M54 | C13H8O4 | 11.44 | N | 227.03388 | 227.03358 | 10.5 | –1.344 | 227.03(100), 199.04(5), 209.12(1), 107.09(1) | + | |||||
| 11.44 | P | 229.04953 | 229.04948 | 9.5 | –0.241 | 229.05(59), 212.16(2), 201.05(1) | + | + | ||||||
| M55 | C13H8O4 | 12.50 | N | 227.03388 | 227.03349 | 10.5 | –1.741 | 227.03(100), 199.04(5), 183.04(4) | + | |||||
| 12.51 | P | 229.04953 | 229.04935 | 9.5 | –0.809 | 229.05(100), 184.10(9), 201.05(2) | + | |||||||
| M56 | C13H8O4 | 13.67 | N | 227.03388 | 227.03380 | 10.5 | –0.375 | 227.13(100), 183.14(28), 209.12(24), 210.12(3), 109.06(2) | + | |||||
| M57 | C13H8O4 | 15.87 | N | 227.03388 | 227.03418 | 10.5 | 1.298 | 227.03(100), 199.04(6), 107.05(3) | + | |||||
| M58 | C25H24O16 | 9.92 | N | 579.09806 | 579.09802 | 14.5 | –1.965 | 403.07(100), 227.03(90), 404.07(20), 228.04(11), 579.10(2) | + | + | + | + | ||
| M59 | C19H18O9 | 10.45 | N | 389.08671 | 389.08737 | 11.5 | –1.119 | 315.09(42), 389.13(31), 341.10(30), 256.07(13), 345.10(8), 201.11(7) | + | |||||
| M60 | C19H18O9 | 11.18 | N | 389.08671 | 389.08966 | 11.5 | 4.767 | 389.09(21), 371.24(5), 307.19(12), 289.18(11), 201.11(6) | + | |||||
| M61 | C19H16O10 | 11.45 | N | 403.06597 | 403.06598 | 12.5 | –2.704 | 227.03(100), 228.04(14), 403.07(4) | + | + | + | + | ||
| 11.44 | P | 405.08162 | 405.08096 | 11.5 | –1.637 | 229.05(100), 230.05(15), 331.08(3), 252.66(2) | + | |||||||
| M62 | C19H18O9 | 11.62 | N | 389.08671 | 389.09030 | 11.5 | 4.844 | 389.09(100), 235.10(13), 211.13(7), 307.12(4) | + | + | + | + | + | |
| M63 | C19H16O10 | 12.50 | N | 403.06597 | 403.06604 | 12.5 | –2.555 | 227.03(100), 403.06(25), 228.04(15), 359.19(5) | + | + | + | + | ||
| 12.50 | P | 405.08162 | 405.08105 | 11.5 | –0.946 | 229.05(100), 230.05(13), 141.02(7), 405.08(3) | + | |||||||
| M64 | C13H8O7S | 13.67 | N | 306.99070 | 306.99124 | 10.5 | –1.813 | 227.03(100), 306.99(37), 228.04(14) | + | + | ||||
| M65 | C13H8O7S | 13.77 | N | 306.99070 | 306.99115 | 10.5 | –2.106 | 227.03(100), 306.99(40), 228.04(13) | + | + | ||||
| M66 | C13H8O7S | 13.80 | N | 306.99070 | 306.99097 | 10.5 | –2.692 | 227.03(100), 306.99(39), 204.99(8) | + |
Notes: U, urine samples; PS, plasma SPE extraction; PA, plasma acetonitrile precipitation; PM, plasma methanol precipitation; F, feces samples; LM, liver microsomes samples.
2.2.1. Metabolites Based on the Prototype (M1–M14)
M0 showed [M-H]− ions at m/z 421.07654, and the molecular formula was calculated to be C19H17O11 (mass error of −0.802 ppm). Its ESI-MS/MS spectrum showed the characteristic fragment ions at m/z 403, m/z 331, m/z 301, m/z 259, etc. Compared with the reference standard, M0 was further identified as mangiferin and detected in plasma, urine, feces, and liver microsomes.
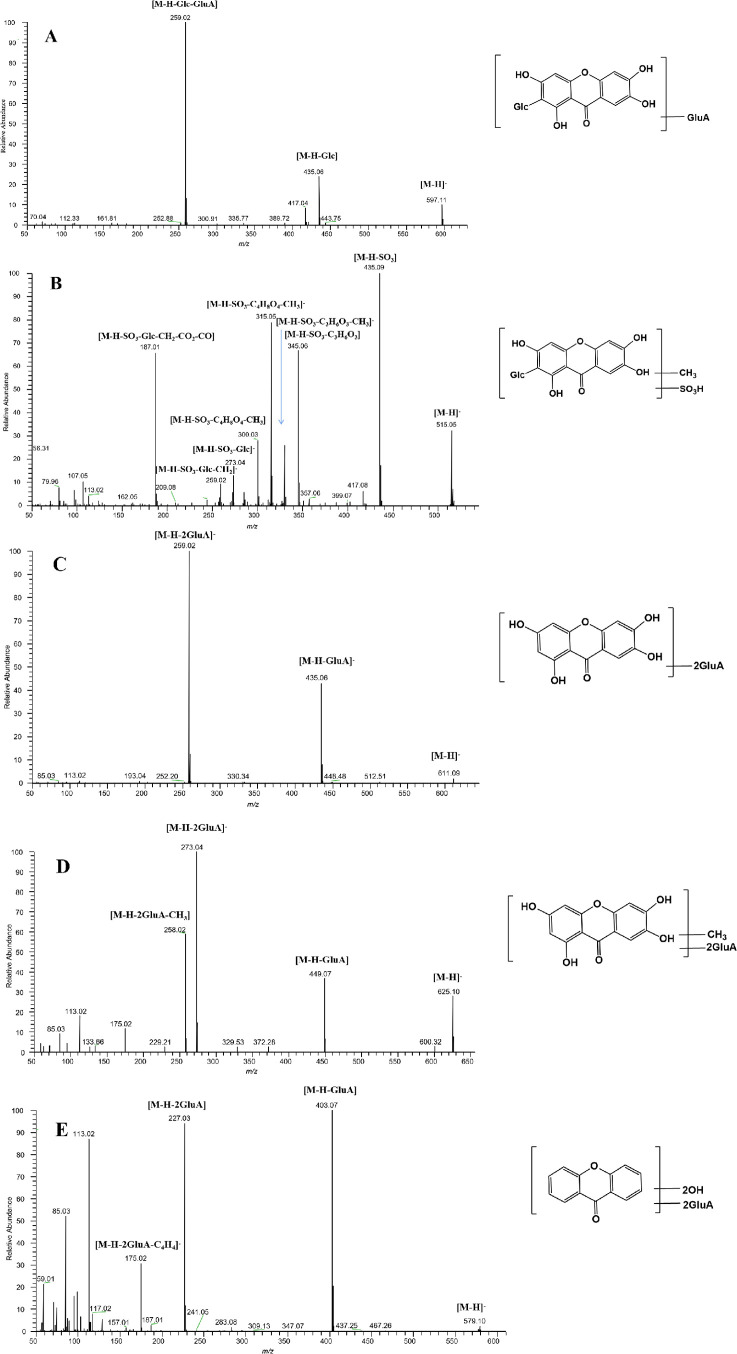
The HRMS spectrum of M5 confirmed an [M-H]− ion at m/z 597.10876. Its molecular formula was established as C25H25O17 with a mass error of −1.612 ppm, which was 176 Da (GluA) more massive than M0. The generation of the DPIs at m/z 435 [M-H-Glc]− and m/z 259 [M-H-Glc-GluA]− resulted from the loss of glucose and glucuronic acid in turn from M5 (Figure 3A). Thus, M5 was identified as the glucuronidation metabolite of mangiferin.
Figure 3.
Mass spectrometry information of M5 (A), M7 (B), M19 (C), M25 (D), and M58 (E) in urine samples in negative ion mode.
M6, M8, and M13 were attributed to three isomeric metabolites with respective [M-H]− ions at m/z 435.09256, m/z 435.09262, and m/z 435.09177 (C20H19O11, mass errors of −1.665, −1.527, and −0.960 ppm). These three metabolites were detected in negative ion mode at 9.52, 10.10, and 10.71 min, respectively. The DPIs at m/z 345 (m/z 331 + CH2) and m/z 315 (m/z 301 + CH2) were correspondingly 14 Da more massive than the fragment ions at m/z 331 and m/z 301, suggesting that mangiferin might occur in the methylation reaction. At the same time, the DPI at m/z 273 (m/z 259 + 14 Da) also confirmed our deduction. Ultimately, M6, M8, and M13 were presumed as methylation metabolites of mangiferin, and the methylation reaction might occur at positions 1, 3, 6, or 7.
M10 and M12 were observed at 10.32 and 10.62 min, with the deprotonated molecular ions at m/z 449.10815 and m/z 449.10913 (C21H21O11, mass errors of 0.695 and 2.877 ppm) in negative ion mode. They showed the same fragment ions but 28 Da higher than that of mangiferin, which was proposed to be dimethylated mangiferin. According to the previous analysis, the DPI at m/z 315 (m/z 301 + CH2) was proved as the occurrence of methylation. The DPI at m/z 273 (m/z 259 + 14 Da) was generated from the fragment ion at m/z 449 by the loss of C6H10O5 and CH2. Finally, both M10 and M12 turned out to be isomeric metabolites, and they could be deduced as the dimethylation products of mangiferin.
M3 gave rise to [M-H]− ions at m/z 611.12421 (C26H27O17, mass error of −1.07 ppm) with a retention time of 8.74 min, which was 190 Da (176 + 14 Da) more massive than mangiferin. In addition, the DPIs at m/z 435 [M-H-GluA]−, m/z 345 [M-H-C3H6O3]−, and m/z 273 [M-H-Glc-GluA]− were all observed in its ESI-MS/MS spectrum, which were 14 Da (CH2) more massive than the corresponding fragment ions at m/z 421, m/z 331, and m/z 259, suggesting that mangiferin underwent the methylation reaction. At the same time, glucuronic acid could be removed from [M-H]− ions and formed the DPI at m/z 435 (m/z 259 + GluA). Therefore, M3 was tentatively identified as methylation and glucuronidation products of mangiferin.
The HRMS spectrum ascertained the molecular formula of M7 as C20H19O14S (m/z 515.04900, mass error of −0.218 ppm). In its ESI-MS/MS spectrum, the fragment ions at m/z 515 [M-H]−, m/z 435 [M-H-SO3]−, m/z 345 [M-H-SO3-C3H6O3]−, m/z 315 [M-H-SO3-C4H8O4]−, m/z 300 [M-H-C4H8O4-CH3]−, m/z 259 [M-H-SO3-Glc-CH2]−, and m/z 187 [M-H-SO3-Glc-CH2-CO2-CO]− were all observed (Figure 3B). The fragment ion at m/z 435 was 80 Da less massive than M7, suggesting the loss of SO3. Simultaneously, the DPIs at m/z 435 (m/z 421 + CH2), m/z 345 (m/z 331 + CH2), and m/z 315 (m/z 301 + CH2) suggested the occurrence of the methylation reaction of mangiferin. Thus, M7 was preliminarily characterized as methylation and sulfation metabolites of mangiferin.
M1 was eluted at 3.05 min, yielding [M-H]− ions at m/z 501.03326 (C19H17O14S) with a mass error of −2.373 ppm, which was 80 Da (SO3) more massive than mangiferin. The DPIs at m/z 331 [M-H-SO3-C3H6O3]−, m/z 301 [M-H-SO3-C4H8O4]−, and m/z 259 [M-H-SO3-Glc]− were observed in its ESI-MS/MS spectra. The fragment ion at m/z 271 [M-H-SO3-C4H8O4-CH2O]− was 30 Da less massive than the fragment ion at m/z 301, which could be attributed to the elimination of OCH2. Meanwhile, the DPI at m/z 501 (m/z 421 + 80 Da) confirmed the existence of the sulfate group. Thus, M1 was thought to be the sulfation metabolite of mangiferin.
Both M2 and M4 possessed the same theoretical [M-H]− ion at m/z 583.12936, corresponding to the molecular formula of C25H28O16, eluting at 8.47 and 8.85 min, respectively. They were only detected in feces samples in negative ion mode. The mass weight was 162 Da (C6H10O5) more than that of M0, thereby suggesting that the pairs of isomers were produced by the occurrence of the glucosylation reaction from mangiferin. The DPIs at m/z 331 [M-H-Glc-C3H6O3]− and m/z 301 [M-H-Glc-C4H8O4]− also confirmed our speculation. Therefore, M2 and M4 were considered to be the glucosylation metabolites of mangiferin.
2.2.2. Metabolites Based on the Intermediate Product Norathyriol (M15–M43)
Norathyriol, an aglycone of xanthonoid C-glycoside mangiferin, is also an active metabolite of mangiferin. Mangiferin improves hepatic lipid metabolism mainly through its metabolite, norathyriol, by modulating SIRT-1/AMPK/SREBP-1c signaling. Furthermore, the regulatory effect of norathyriol on liver lipid metabolism is stronger than mangiferin.28
M15 (C13H8O6, mass error of 1.759 ppm) detected at 10.59 min exhibited [M-H]− ions at m/z 259.02417, which was 162 Da (C6H10O5) less massive than mangiferin, indicating that M15 might be the deglycosylation metabolite of mangiferin. In its ESI-MS/MS spectrum, the fragment ions at m/z 241(m/z 259–18 Da) and m/z 231(m/z 259–28 Da) were formed by the respective neutral loss of H2O and CO. The fragment ion at m/z 215 was generated from the [M-H]− ion by the loss of O and CO. Furthermore, the dominant fragment ion at m/z 187 also appeared in its ESI-MS/MS spectrum. Combined with the literature,22,25M15 was identified as norathyriol, which was thought to possess various pharmacological activities.
M37, M41, and M42 produced their [M-H]− ions at m/z 273.03903, m/z 273.04001, and m/z 273.03976 (C14H9O6, mass errors of −3.887, 2.364, and 1.449 ppm), which were eluted at 12.51, 13.44, and 14.70 min, respectively. The DPI at m/z 259 [M-H-CH2]− was 14 Da (CH2) more massive than norathyriol. The DPIs at m/z 227 (m/z 259-2O) and m/z 211 (m/z 259-3O) were formed by the loss of CH2 + 2O and CH2 + 3O from the [M-H]− ion at m/z 273, respectively. M37, M41, and M42 might be isomers, and all of them were deduced as the methylation products of norathyriol. The methylation reaction occurred on hydroxyl groups at positions 1, 3, 6, and 7.
M39 and M43 gave rise to deprotonated molecular ions at m/z 287.05548 and m/z 287.05502, together with the retention time of 13.00 and 16.48 min, respectively. Their [M-H]− ions were 28 Da more massive than norathyriol, thereby suggesting the addition of two CH2 groups to norathyriol. Importantly, the DPI at m/z 273 was formed by the loss of CH2 from the fragment ion at m/z 287. Moreover, the DPIs at m/z 243 (m/z 259-O) and m/z 257 (m/z 273-O) were formed by the stripping of C2H4O and the loss of a CH2O group from the product ion at m/z 287, respectively. Thus, M39 and M43 were preliminarily identified as dimethylation metabolites of norathyriol.
M27 showed [M-H]− ions at m/z 352.99597 (C14H9O9S, mass error of −0.592 ppm) at 10.27 min, which was 80 Da more massive than M37, M41, and M42. It produced the DPIs at m/z 273 (m/z 273 + CH2) and m/z 227 corresponding to fragment ions [M-H-SO3]− and m/z 227 [M-H-SO3-CH2]−. Finally, M27 was deduced as the methylation and sulfation product of norathyriol.
M28, M31, M32, M33, M36, and M38 showed their [M-H]− ions at m/z 449.07196, m/z 449.07193, m/z 449.07156, m/z 449.07138, m/z 449.07172, and m/z 449.07181 with their retention time of 10.45, 10.75, 11.08, 11.38, 11.48, and 12.87 min, respectively. The DPIs at m/z 273 and m/z 259 might be generated due to the sequential loss of C6H8O6 and CH2. Thus, it was inferred that M28, M31, M32, M33, M36, and M38 were isomeric metabolites, and all of them were methylation and glucuronidation products of norathyriol.
M19, M20, M21, and M22 (C25H23O18, mass errors of −0.393, 0.294, −1.975, and 0.802 ppm) displayed [M-H]− ions at m/z 611.09765, m/z 611.08807, m/z 611.08789, and m/z 611.08838 at 7.98, 9.00, 9.39, and 9.44 min, respectively. They were 352 Da (2GluA) more massive than norathyriol. Their ESI-MS/MS spectra gave the characteristic fragment ions at m/z 435 and m/z 259, which yielded correspondingly by the loss of glucuronide (176 Da) and two molecules of glucuronide (352 Da) from the precursor ions. They showed strong responses at m/z 259 and weak responses at m/z 435, indicating that the metabolites were bis-glucuronylation products of norathyriol (Figure 3C).
M25 at 9.89 min showed its [M-H]− ion at m/z 625.10327 (mass error of −0.432 ppm), and its elemental formula was assigned as C26H25O18. On the account of the successive loss of GluA, GluA, and CH2, the DPIs at m/z 449 (m/z 273 + 176 Da), m/z 273 (m/z 259 + 14 Da), and m/z 259 were generated in the ESI-MS/MS spectra (Figure 3D). The appearance of three fragment ions demonstrated that M25 was estimated as the methylation and bis-glucuronylation product of norathyriol.
M34 and M40 were detected at 11.45 and 13.00 min with the deprotonated molecular ions at m/z 463.08704 and m/z 463.08783 (C21H19O12, mass errors of 2.503 and −0.797 ppm). The DPIs at m/z 287 [M-H-GluA]− (m/z 259 + CH2), m/z 273 [M-H-GluA-CH2]−, and m/z 257 [M-H-GluA-CH2O]− (m/z 273-O) were generated in negative ion mode. Finally, M34 and M40 were presumed as bis-methylation and glucuronidation metabolites of norathyriol, respectively.
M16, M17, M18, and M35 showed deprotonated molecular ions at m/z 338.98306, m/z 338.98309, m/z 338.98041, and m/z 338.98069 (C13H7O9S, mass errors of 4.231, 4.320, −0.351, and 0.446 ppm), respectively. The DPI at m/z 339 was 80 Da (SO3) more massive than norathyriol. A sequence of crucial fragment ions at m/z 311 [M-H-H2O]−, m/z 293 [M-H-H2O-CO]−, and m/z 225 [M-H-H2O-O-SO3]− was produced in the ESI-MS/MS spectra. Based on the above information, we inferred that M16, M17, M18, and M35 were isomers, and all of them were initially identified as sulfation products of norathyriol.
M26 (C19H15O15S, mass error of −4.104 ppm) yielded [M-H]− at m/z 515.01160 at 9.99 min in negative ion mode. The typical DPIs at m/z 435[M-H-SO3]− (m/z 259 + 176 Da) and 259 [M-H-SO3-GluA]− indicated that sulfation and glucuronylation reactions took place. Thus, M26 was tentatively deduced to be sulfation and glucuronylation metabolites of norathyriol.
2.2.3. Metabolites Based on the Intermediate Product Trihydroxyxanthone (M44–M53)
M44 and M45 produced the same [M-H]− ions at m/z 243.02904 with the retention time of 10.39 and 10.83 min, meeting the loss of O from norathyriol. The DPIs at m/z 199 and m/z 181 were observed in their ESI-MS/MS spectra, with the successive loss of CO + O and H2O. The presence of M44 and M45 meant that norathyriol went through dehydroxylation to become trihydroxyxanthones.
M46 (C25H23O17, mass error of −0.945 ppm) was eluted at 7.62 min along with [M-H]− ions at m/z 595.09351. The DPIs at m/z 419 and m/z 243 (m/z 259-O) were formed with the successive loss of two glucuronate residues. The DPI at m/z 243 was 16 Da less massive than the fragment ion at m/z 259, suggesting the presence of trihydroxyanthrone. Taken together, M48 was considered as the bis-glucuronidation metabolite of trihydroxyxanthone.
M49, M50, and M51 were eluted at 10.32, 10.61, and 11.98 min in negative ion mode. The same molecular formula was established as C20H19O10, which was 176 Da more massive than trihydroxyxanthone. The DPI at m/z 243 (m/z 259-O) was generated in ESI-MS/MS spectra, which was 16 Da less massive than the ion at m/z 259 and 176 Da less massive than the DPI at m/z 419 (m/z 243 + GluA). Therefore, M49, M50, and M51 could be deduced as glucuronylation metabolites of trihydroxyxanthone.
2.2.4. Metabolites Based on the Intermediate Product Dihydroxyanthrone (M54–M66)
M54, M55, M56, and M57 were observed in negative ion mode and determined to be C13H7O4 (mass errors of −1.334, −1.741, −0.375, and 1.298 ppm), based on their HRMS data. The DPI at m/z 389 was 32 Da (2O) less massive than norathyriol, indicating that they were characterized as dihydroxyanthrones, the debihydroxylation products of norathyriol.
M58 was detected at 9.92 min, yielding the [M-H]− ion at m/z 579.09802 (mass error of −1.965 ppm), corresponding to the molecular formula of C25H23O16. The DPI at m/z 403 (m/z 259 + GluA) was formed by removing C6H8O6 from its [M-H]− ion. Moreover, the DPI at m/z 227 (m/z 259-2O) was 32 Da less massive than norathyriol and generated by the loss of 2C6H8O6 (Figure 3E). The DPIs at m/z 403 and at m/z 227 were all derived from the important fragment ion at m/z 259. Based on the evidence, M58 was designated to be the bis-glucuronylation metabolite of dihydroxyanthrone.
M64, M65, and M66 gave rise to [M-H]− ions at m/z 306.99124, m/z 306.99115, and m/z 306.99097 (C13H7O7S, mass errors of −1.813, −2.106, and −2.692 ppm) at 13.67, 13.77, and 13.80 min, respectively. These metabolites yielded the same DPI at m/z 227 (m/z 259-2O) by the loss of SO3 (80 Da). Consequently, these data suggested that these three metabolites could be interpreted as sulfation products of dihydroxyxanthone.
M61 and M63 (C19H15O10, mass errors of −2.704 and −2.555 ppm) were presented at 11.45 and 12.50 min, with the deprotonated molecular ions at m/z 403.06598 and m/z 403.06604, respectively. In their ESI-MS/MS spectra, the fragment ions at m/z 359 [M-H-CO-O]− and m/z 227 [M-H-C6H8O6]− were formed by the loss of CO + O and C6H8O6, respectively. Thus, they were all characterized as glucuronidation products of dihydroxyxanthone.
2.3. Metabolic Pathways Analysis of Mangiferin
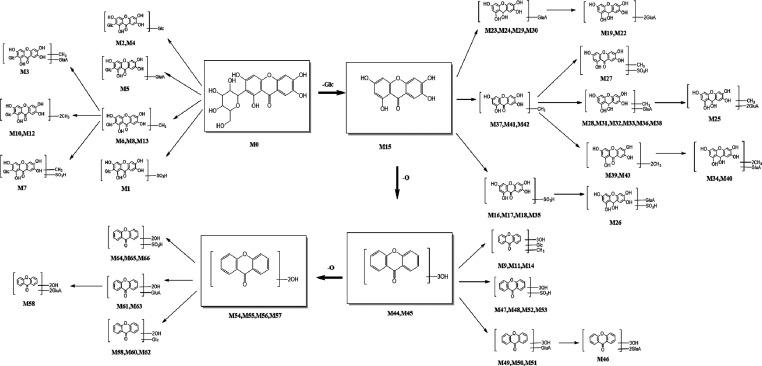
In this study, the metabolism fate of mangiferin in vivo and in vitro was investigated based on the “Drug Metabolite Clusters” analytical strategy using UHPLC-Q-Exactive HRMS. Ultimately, a total of 67 metabolites (mangiferin included) were detected, among which 66 metabolites were identified in vivo and 6 metabolites in vitro. The deduced metabolic pathways of mangiferin in vivo and in vitro are presented in Figure 4. With mangiferin, norathyriol, trihydroxyxanthone, and dihydroxyxanthone as the cores of the Drug Metabolite Clusters, an increasing number of metabolites were produced through further metabolic reactions. The results indicated that the hydroxyl group was the metabolic site of mangiferin. Furthermore, mangiferin mainly underwent phase I metabolism (i.e., deglycosylation, dehydroxylation, methylation, etc.). Deglycosylation and dehydroxylation, that were losses of C6H10O5 and O in mangiferin, were the primary metabolic pathways, followed by further transformation through methylation, glucuronidation, and sulfation. Conjugation reactions with sulfate and glucuronate groups were the predominant phase II metabolite reactions.
Figure 4.
Metabolic pathways of mangiferin in vivo and in vitro.
3. Discussion
Drug metabolism refers to the process in which the chemical structure of the drug changes by various drug-metabolizing enzymes in the body. There are two kinds of results after the biotransformation of drugs in vivo: one is inactivation, becoming no pharmacological activity drugs; the other is activation, from no pharmacological activity to have the pharmacological activity of metabolites or toxic metabolites, or after metabolism still maintains the original pharmacological effects.29 Therefore, the metabolic transformation of drugs plays an important role in the study of drug properties as well as toxicological properties. In this paper, we studied the in vivo and in vitro metabolism of mangiferin by UHPLC-Q-Exactive Orbitrap HRMS based on Drug Metabolite Clusters.
Drug Metabolite Clusters, a rapid and effective method for the identification of metabolites based on primary and secondary metabolites of mangiferin, was applied to this study. This methodology was employed to mine information about drugs and their metabolites on the basis of raw, processed, or interpreted MS2 data. Drug molecules mainly react with functional groups or bind to endogenous polar molecules in metabolic transformation reactions in vivo. To some extent, the basic skeletal structure of a drug does not change. Thus, drug metabolites have characteristic production and neutral loss of the original drug. In negative and positive ion detection modes, the first-order full scanning mass spectrograms of the drug sample were compared to blank samples to identify possible phase I and phase II metabolites of mangiferin. Next, the metabolite structure was predicted by analyzing their product ions and further combining the general metabolic regulation of drugs in vivo. Herein, a total of 67 mangiferin metabolites were identified in vivo and in vitro. In our study, 14, 29, 10, and 13 metabolites were detected in the centers of four metabolic clusters mangiferin, norathyriol, trihydroxyxanthone, and dihydroxyxanthone, respectively. Among them, trihydroxyxanthones (M44 and M45) were first detected in urine after oral mangiferin. In addition, bis-glucuronidation metabolites (M46) and sulfation metabolites (M47, M48, M52, and M53) of trihydroxyxanthone, and the bis-glucuronidation metabolites (M58) and sulfation metabolites (M64, M65, and M66) of dihydroxyxanthone (M46 and M58) were all first discovered in our study as well.
In our study, 29, 48, and 12 mangiferin metabolites were identified in rat plasma, urine, and feces, respectively. We found that more metabolites existed in plasma and urine, while fewer products in feces by comparison, suggesting that mangiferin metabolites might be mainly eliminated through urine. In addition, we found that a total of four products existed simultaneously in the above three biological samples, including the prototype drug M0, the deglycosylation metabolite M15, a glucuronylation metabolite M51, and a glycosylation product M62 (Figure 5A).
Figure 5.
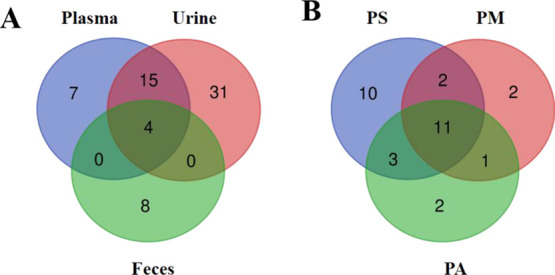
Distribution of mangiferin metabolites in samples. (A) Quantity distribution of mangiferin metabolites in different samples; (B) quantity distribution of mangiferin metabolites in plasma with different preparation methods (PS: plasma - SPE method; PM: plasma - methanol precipitation; PA: plasma - acetonitrile precipitation).
We provided three methods to process plasma samples, which were SPE treatment, methanol treatment, and acetonitrile treatment. In addition, 24, 16, and 17 metabolites were detected in plasma samples with different treatment methods, respectively. Apparently, plasma samples treated with SPE generated significantly more metabolites than plasma samples of precipitated proteins using methanol and acetonitrile. Compared to the protein precipitation method by the organic solvent, the SPE technology has the advantages of a high recovery rate and good compatibility with analytical instruments, allowing for more low-content metabolites to be detected using SPE.27 At the same time, prototypes and 10 glucuronidation metabolites, based on changes with norathyriol, appeared in blood samples treated with three different methods (Figure 5B). The results demonstrated that binding products of glucolycuronic acid might be more stable.
The metabolic pathways of mangiferin in vitro and in vivo have shown that mangiferin underwent extensive phase I metabolism and phase II metabolism, such as deglycosylation, dehydroxylation, methylation, glucuronidation, sulfation, and glycosylation conjugates. We found that mangiferin metabolic pathways communicate with each other, and various metabolic pathways can cross each other through common intermediate metabolites, thus forming a complex metabolic network. The mangiferin metabolites in the phase I reaction can be excreted directly or they can be excreted in the phase II reaction again. Mangiferin can also be excreted directly through phase II reaction. Besides, the study has shown that glucuronidation and sulfation conjugates were found to be the main forms existing in vivo after administration, which is consistent with our experimental results.26 Norathyriol, the deglycosylation metabolite of mangiferin was responsible for the hypouricaemic effect of mangiferin via inhibiting xanthine oxidase activity.30 Moreover, norathyriol could regulate hepatic lipid metabolism, thereby reducing excessive hepatic fat deposition and preventing hepatic steatosis.28 The glucuronidation metabolites of mangiferin led to a loss in free radical scavenging and ferric iron reducing ability, thus exerting antioxidant effects.31 The above studies confirmed the hypothesis that mangiferin exerted some pharmacological effects through its metabolites.
4. Conclusions
In this work, a rapid and high-throughput UHPLC-Q-Exactive Orbitrap HRMS was applied for systematic research on the metabolic profiling of mangiferin in vivo and in vitro. We also successfully presented an efficient methodology “Drug Metabolite Clusters” to search and identify mangiferin metabolites. As a result, a total of 67 mangiferin metabolites were elucidated and identified, including 29 in plasma, 48 in urine, 12 in feces, as well as 6 in liver microsomes. Moreover, the main metabolic pathways of mangiferin were summarized, involving dehydroxylation, methylation, glucosylation, glucuronylation, and sulfation. This study would be helpful for revealing the in vivo potential effective components of mangiferin and also provide a solid basis for further pharmacology and curative mechanism research. In addition, the systematic analytical strategy can also be used for the discovery and identification of prototypes and metabolites of other drugs.
5. Materials and Methods
5.1. Chemicals and Reagents
Mangiferin (purity ≥98%) was purchased from Chengdu Must Biotechnology Co., Ltd. (Sichuan, China). HPLC-grade acetonitrile, methanol, and formic acid were purchased from Thermo Fisher Scientific (Fair Lawn, NJ, USA). Deionized water used throughout the experiment was purchased from Watsons (Guangzhou, China). Rat liver microsomes were obtained from NEWGAINBIO Co., Ltd. (Wuxi, China) and Grace Pure SPE C18-Low SPE cartridges (200 mg/3 mL, 59 μm, 70 Å) were purchased from Grace Davison Discovery Science (Deerfield, IL, USA). Nicotinamide adenine dinucleotide phosphate (NADPH) and magnesium chloride (MgCl2) were provided by Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China). The 6-well plates were obtained from Corning Incorporated-Life Science (Jiangsu, China).
5.2. In Vitro Liver Microsomes Incubation
In vitro metabolism studies were conducted on the incubation of mangiferin in rat liver microsomes. MgCl2 (3 mM, final concentration) and liver microsomes (1 mg/mL, final protein concentration) were mixed in phosphate buffered solution (pH = 7.4) to form the mixed solvent. Mangiferin reference substance was dissolved in this solution and diluted to 1 mg/mL in methanol ultimately. The administration group and the blank group were set. The above mixture (900 μL) was added to each well of the 6-well plate, while the blank group was given drug negative solution. After preheating at 37 °C for 5 min, NADPH (100 μL, 50 mM) was added to start the reaction, and incubation was continued at 37 °C. At 5, 10, 15, 30, 45, 60, 120, 240, and 480 min, cold acetonitrile (200 μL) was added to the system solution (100 μL) to stop the reaction and was centrifuged at 3500 rpm for 15 min. Finally, the supernatant was dried under nitrogen at room temperature. The residue was then redissolved in methanol (300 μL) and centrifuged for 15 min (14,000 rpm, 4 °C). The obtained supernatant was used for the further LC–MS analysis.
5.3. In Vivo Animal Experiment
5.3.1. Animals and Drug Administration
Six male Sprague–Dawley (SD) rats weighting 200 ± 10 g were obtained from Jinan Pengyue Experimental Animal Breeding Company (Shandong, China, SCXK2019003). The rats were housed in metabolic cages in a controlled room at standard temperature (24 ± 5 °C) and humidity (70 ± 5%) and kept on a 12 h light/12 h dark regime. After 7 days of adaptive feeding, the rats were randomly divided into two groups: drug group (n = 3) and control group (n = 3). The rats fasted for 12 h and drank freely before the experiment. Mangiferin was dissolved in physiological saline solution. The rats in the drug group were administrated mangiferin at a dose of 250 mg/kg body weight once a day for three successive days, while the rats in the control group were given the same volume of physiological saline solution.
5.3.2. Sample Collection and Preparation
Blood samples (0.5 mL) were taken from the suborbital venous plexus of rats at 0.5, 1, 1.5, 2, 4, and 6 h after administration. Each sample was centrifuged at 3500 rpm for 10 min to obtain the supernatant of plasma samples. Urine and feces samples were collected 0–24 h through metabolic cages after administration. All the homogeneous biological samples from the same group were finally merged into a collective sample and stored at −80 °C.
Three methods were used to process plasma samples. The first method (method I) was performed to prepare samples by SPE. Methanol (3 mL) was added to activate the SPE column, and water (3 mL) was added to balance the column. Then, plasma samples (1 mL) were added to the pretreated SPE column and successively washed with water (3 mL) and methanol (3 mL). The methanol eluate was collected into a 5 mL EP tube and evaporated by nitrogen at room temperature. The second method (method II) was to use methanol to precipitate samples. Methanol (3 mL) was added to plasma samples (1 mL) for precipitation, and the supernatant was gained by centrifuging at 14,000 rpm. The third method (method III) used acetonitrile to precipitate plasma samples. Plasma samples (1 mL) were pretreated with acetonitrile (3 mL) for precipitation, and then, they were centrifuged at 14,000 rpm to obtain the supernatant.
In addition, the treatment of urine samples was the same as method I. The feces samples (2 g) were ultrasonically extracted with water (10 mL) for 30 min and then filtered to obtain the supernatant. Meanwhile, normal saline (10 mL) was added to the liver tissue (1 g) to grind, and the supernatant was centrifuged at 3500 rpm. After pretreatment, the above samples were prepared by SPE columns. The supernatant (1 mL) was added to the pretreated SPE column, and then, the same procedure was performed according to method I.
All the treated biological samples were blow-dried with nitrogen at room temperature and stored at −80 °C. Before analysis, they were redissolved in methanol (300 μL), centrifuged at 14,000 rpm at high speed for 15 min, and then, the supernatant was injected into liquid vials with lined tubes for the further LC–MS analysis.
5.4. Instrumentation and Analytical Conditions
5.4.1. Chromatographic Conditions
UHPLC analysis was performed using an Ultimate 3000 system (Thermo Fisher Scientific, MA, USA) equipped with an online vacuum degasser, a quaternary pump, and an automatic sampler. The biological samples were separated by an ACQUITY UPLC BEH-C18 column (2.1 mm × 100 mm, 1.7 μm; Water Company, Milford, Mom, USA). The column temperature was maintained at 30 °C, and the injection volume was 2 μL. The mobile phase was composed of acetonitrile (A) and water with 0.1% formic acid (B). The flowing gradient at the rate of 0.3 mL/min was applied and the gradient elution conditions were set as follows: 0–5 min, 95% A; 5–10 min, 95% → 70% A; 10–15 min, 70% → 50% A; 15–32 min, 50% → 10% A; 32–35 min, 95% A.
5.4.2. Mass Spectrometry Conditions
ESI-MS/MS analyses were performed on Q-Exactive Plus Orbitrap MS (Thermo Fisher, Waltham, MA, USA). Mass spectrometry was analyzed in negative and positive ion modes using a heating ESI source with full MS (resolution 70,000) and dd-MS2 (resolution 17,500), covering m/z 80–1200. The other optimal conditions were set as follows: capillary temperature 320 °C; auxiliary gas heater temperature 350 °C; spray voltage 3.8/3.5 kV (+/−); sheath gas volume flow 45 arbitrary units; and auxiliary gas volume flow 10 arbitrary units.
5.5. Data Processing and Analysis
Thermo Xcalibur Version 3.0 workstation was used for data acquisition and processing. The detected peaks with intensity over 10,000 were selected for identification, with the purpose to obtain as many ESI-MS/MS product ions of mangiferin metabolites as possible. The accurate mass of chemical formulas attributed to the selected peaks was calculated using a formula predictor by setting the parameters as follows: C [0–30], H [0–60], O [0–30], S [0–4] and the ring double bond (RDB) equivalent value [0–20]. Accurate mass measurements were set within the mass error of ±5 ppm.
Acknowledgments
This work was financially supported by the Taishan Young Scholar Program of Shandong (TSQN202103110), the Young and Creative Team for Talent Introduction of Shandong Province (10073004), Major Scientific and Technological Innovation Projects in Shandong Province (2021CXGC010511), and Binzhou Medical University Scientific Research Fund for High-level Talents (2019KYQD06).
Author Contributions
⊥ H.Z. and S.S. contributed equally to this work.
The authors declare no competing financial interest.
References
- Loan N. T. T.; Long D. T.; Yen P. N. D.; Hanh T. T. M.; Pham T. N.; Pham D. T. N. Purification Process of Mangiferin from Mangifera indica L. Leaves and Evaluation of Its Bioactivities. Processes 2021, 9, 852. 10.3390/PR9050852. [DOI] [Google Scholar]
- Ji D.; Qiu J. C.; Su X. N.; Qin Y. W.; Hao M.; Li L.; Lu T. L.; Li X. K.; Jiang C. X. Study on pharmacokinetics and tissues distribution of neomangiferin, mangiferin, timosaponin BII, Timosaponin BIII, and timosaponin AIII after oral administration of Anemarrhenae rhizoma extract in rats. Pharmacogn. Mag. 2019, 15, 557–567. 10.4103/pm.pm_65_19. [DOI] [Google Scholar]
- Imran M.; Arshad M. S.; Butt M. S.; Kwon J. H.; Arshad M. U.; Sultan M. T. Mangiferin: a natural miracle bioactive compound against lifestyle related disorders. Lipids Health Dis. 2017, 16, 84. 10.1186/s12944-017-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Huang C.; Fan S. Mangiferin and organ fibrosis: A mini review. BioFactors 2021, 47, 59–68. 10.1002/biof.1693. [DOI] [PubMed] [Google Scholar]
- Qin Z. Z.; Ruan J.; Lee M. R.; Sun K.; Chen P.; Chen Y.; Hong M.; Xia L. H.; Fang J.; Tang H. Mangiferin Promotes Bregs Level, Activates Nrf2 Antioxidant Signaling, and Inhibits Proinflammatory Cytokine Expression in Murine Splenic Mononuclear Cells In Vitro. Curr. Med. Sci. 2021, 41, 454–464. 10.1007/S11596-021-2371-9. [DOI] [PubMed] [Google Scholar]
- Lei L. Y.; Wang R. C.; Pan Y. L.; Yue Z. G.; Zhou R.; Xie P.; Tang Z. S. Mangiferin inhibited neuroinflammation through regulating microglial polarization and suppressing NF-κB, NLRP3 pathway. Chin. J. Nat. Med. 2021, 19, 112–119. 10.1016/S1875-5364(21)60012-2. [DOI] [PubMed] [Google Scholar]
- Kanoi R.; Loachan P.; Das S.; Rao B. S. S. Mangiferin, a naturally occurring polyphenol, mitigates oxidative stress induced premature senescence in human dermal fibroblast cells. Mol. Biol. Rep. 2021, 48, 457–466. 10.1007/S11033-020-06074-2. [DOI] [PubMed] [Google Scholar]
- Wang H.; He X.; Lei T.; Liu Y.; Huai G.; Sun M.; Deng S.; Yang H.; Tong R.; Wang Y. Mangiferin induces islet regeneration in aged mice through regulating p16INK4a. Int. J. Mol. Med. 2018, 41, 3231–3242. 10.3892/ijmm.2018.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Huang J.; Yang B.; Xiang T.; Yin X.; Peng W.; Cheng W.; Wan J.; Luo F.; Li H.; Ren G. Mangiferin exerts antitumor activity in breast cancer cells by regulating matrix metalloproteinases, epithelial to mesenchymal transition, and β-catenin signaling pathway. Toxicol. Appl. Pharmacol. 2013, 272, 180–190. 10.1016/j.taap.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Rajendran P.; Rengarajan T.; Nandakumar N.; Divya H.; Nishigaki I. Mangiferin in cancer chemoprevention and treatment: pharmacokinetics and molecular targets. J. Recept. Signal. Transduct. Res. 2015, 35, 76–84. 10.3109/10799893.2014.931431. [DOI] [PubMed] [Google Scholar]
- Dong M.; Li L.; Li G.; Song J.; Liu B.; Liu X.; Wang M. Mangiferin protects against alcoholic liver injury via suppression of inflammation-induced adipose hyperlipolysis. Food Funct. 2020, 11, 8837–8851. 10.1039/D0FO01436B. [DOI] [PubMed] [Google Scholar]
- Lum P. T.; Sekar M.; Gan S. H.; Pandy V.; Bonam S. R. Protective effect of mangiferin on memory impairment: A systematic review. Saudi J. Biol. Sci. 2021, 28, 917–927. 10.1016/j.sjbs.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S. T.; Wang Z. Z.; Yuan Y. H.; Sun H. M.; Chen N. H.; Zhang Y. Mangiferin: A multipotent natural product preventing neurodegeneration in Alzheimer’s and Parkinson’s disease models. Pharmacol. Res. 2019, 146, 104336 10.1016/j.phrs.2019.104336. [DOI] [PubMed] [Google Scholar]
- Wei Z.; Yan L.; Chen Y.; Bao C.; Deng J.; Deng J. Mangiferin inhibits macrophage classical activation via downregulating interferon regulatory factor expression. Mol. Med. Rep. 2016, 14, 1091–1098. 10.3892/mmr.2016.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S.; Ma H.; Chen X. Anticancer and anti-inflammatory properties of mangiferin: A review of its molecular mechanisms. Food Chem. Toxicol. 2021, 149, 111997 10.1016/J.FCT.2021.111997. [DOI] [PubMed] [Google Scholar]
- Liu T.; Song Y.; Hu A. Neuroprotective mechanisms of mangiferin in neurodegenerative diseases. Drug Dev. Res. 2021, 82, 494–502. 10.1002/ddr.21783. [DOI] [PubMed] [Google Scholar]
- Walia V.; Chaudhary S. K.; Kumar Sethiya N. Therapeutic potential of mangiferin in the treatment of various neuropsychiatric and neurodegenerative disorders. Neurochem. Int. 2021, 143, 104939 10.1016/j.neuint.2020.104939. [DOI] [PubMed] [Google Scholar]
- Zeng Z.; Lin C.; Wang S.; Wang P.; Xu W.; Ma W.; Wang J.; Xiang Q.; Liu Y.; Yang J.; Ye F.; Xie K.; Xu J.; Luo Y.; Liu S. L.; Liu H. Suppressive activities of mangiferin on human epithelial ovarian cancer. Phytomedicine 2020, 76, 153267 10.1016/j.phymed.2020.153267. [DOI] [PubMed] [Google Scholar]
- Deng Q.; Tian Y. X.; Liang J. Mangiferin inhibits cell migration and invasion through Rac1/WAVE2 signalling in breast cancer. Cytotechnology 2018, 70, 593–601. 10.1007/s10616-017-0140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasbe P.; Jangra A.; Lahkar M. Mangiferin ameliorates aluminium chloride-induced cognitive dysfunction via alleviation of hippocampal oxido-nitrosative stress, proinflammatory cytokines and acetylcholinesterase level. J. Trace Elem. Med. Biol. 2015, 31, 107–112. 10.1016/j.jtemb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Mei S.; Perumal M.; Battino M.; Kitts D. D.; Xiao J.; Ma H.; Chen X. Mangiferin: a review of dietary sources, absorption, metabolism, bioavailability, and safety. Crit. Rev. Food Sci. Nutr. 2021, 4, 1–19. 10.1080/10408398.2021. [DOI] [PubMed] [Google Scholar]
- Liu H.; Wang K.; Tang Y.; Sun Z.; Jian L.; Li Z.; Wu B.; Huang C. Structure elucidation of in vivo and in vitro metabolites of mangiferin. J. Pharm. Biomed. Anal. 2011, 55, 1075–1082. 10.1016/j.jpba.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Xie Y. Y.; Wang X. M.; Wang S. H.; Wang Y. M.; Tian H. F.; Yuan Y. S.; Li H. Y.; Liang Q. L.; Luo G. A. Metabolism and pharmacokinetics of major polyphenol components in rat plasma after oral administration of total flavonoid tablet from Anemarrhenae Rhizoma. J. Chromatogr. B: Anal. Technol. Biomed Life Sci. 2016, 1026, 134–144. 10.1016/j.jchromb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Mei X.; Liu Z.; Li J.; Zhang X.; Lang S.; Dai L.; Zhang J. Drug Metabolite Cluster-Based Data-Mining Method for Comprehensive Metabolism Study of 5-hydroxy-6,7,3′,4′-tetramethoxyflavone in Rats. Molecules 2019, 24, 3278. 10.3390/molecules24183278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X.; Li Y.; Li H.; Song S.; Yuan X.; Zhou H.; Chen Q.; Zhang J. Drug Metabolite Cluster Centers-based Strategy for Comprehensive Profiling of Neomangiferin Metabolites in vivo and in vitro and Network Pharmacology Study on Anti-inflammatory Mechanism. Arab. J. Chem. 2022, 104268 10.1016/j.arabjc.2022.104268. [DOI] [Google Scholar]
- Guo X.; Cheng M.; Hu P.; Shi Z.; Chen S.; Liu H.; Shi H.; Xu Z.; Tian X.; Huang C. Absorption, Metabolism, and Pharmacokinetics Profiles of Norathyriol, an Aglycone of Mangiferin, in Rats by HPLC-MS2. J. Agric. Food Chem. 2018, 66, 12227–12235. 10.1021/acs.jafc.8b03763. [DOI] [PubMed] [Google Scholar]
- Dong P.; Shi L.; Wang S.; Jiang S.; Li H.; Dong F.; Xu J.; Dai L.; Zhang J. Rapid Profiling and Identification of Vitexin Metabolites in Rat Urine, Plasma and Faeces after Oral Administration Using a UHPLC-Q-Exactive Orbitrap Mass Spectrometer Coupled with Multiple Data-mining Methods. Curr. Drug Metab. 2021, 22, 185–197. 10.2174/1389200221999210101232841. [DOI] [PubMed] [Google Scholar]
- Li J.; Liu M.; Yu H.; Wang W.; Han L.; Chen Q.; Ruan J.; Wen S.; Zhang Y.; Wang T. Mangiferin Improves Hepatic Lipid Metabolism Mainly Through Its Metabolite-Norathyriol by Modulating SIRT-1/AMPK/SREBP-1c Signaling. Front. Pharmacol. 2018, 9, 201. 10.3389/fphar.2018.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słoczyńska K.; Gunia-Krzyżak A.; Koczurkiewicz P.; Wójcik-Pszczoła K.; Żelaszczyk D.; Popiół J.; Pękala E. Metabolic stability and its role in the discovery of new chemical entities. Acta Pharm. 2019, 69, 345–361. 10.2478/acph-2019-0024. [DOI] [PubMed] [Google Scholar]
- Niu Y.; Liu J.; Liu H. Y.; Gao L. H.; Feng G. H.; Liu X.; Li L. Hypouricaemic action of mangiferin results from metabolite norathyriol via inhibiting xanthine oxidase activity. Pharm. Biol. 2016, 54, 1680–1686. 10.3109/13880209.2015.1120322. [DOI] [PubMed] [Google Scholar]
- Van der Merwe J. D.; Joubert E.; Manley M.; de Beer D.; Malherbe C. J.; Gelderblom W. C. Mangiferin glucuronidation: important hepatic modulation of antioxidant activity. Food Chem. Toxicol. 2012, 50, 808–815. 10.1016/j.fct.2011.11.018. [DOI] [PubMed] [Google Scholar]