Abstract
The G-quadruplexes (G4s) in the genome are important drug targets because they regulate gene expression and the genome structure. Several small molecules that bind the G4 have been developed, but few artificial G4 binding proteins have been reported. We previously reported a novel DNA G4 binding protein (RGGF) engineered using the Arg-Gly-Gly repeat (RGG) domain of TLS (translocated in liposarcoma), also known as FUS (fused in sarcoma) protein (TLS/FUS). Here, we show that RGGF recognizes DNA loops in the G4 and preferentially binds DNA G4 with long loops in vitro. Furthermore, RGGF binds to the DNA G4 of the bcl-2 promoter in vitro. RGGF overexpression in HeLa cells represses bcl-2 transcription. On the basis of these findings, G4 binding protein engineered from the RGG domain will be useful for investigating G4 transcriptional function in the genome.
Introduction
G-quadruplexes (G4s) are noncanonical secondary structures formed in guanine-rich sequences that are involved in important biologic roles in the genome, such as replication, gene expression, genome stability, telomere maintenance, and histone modification.1,2 The G4 conformation provides selective DNA and RNA structures targeted by small molecules, and several such molecules have been developed. Functional studies of the G4 in DNA and RNA using G4-binding small molecules revealed that these molecules stabilize G4s and repress the transcriptional and translational activities of promoters and mRNA with G4s, respectively.1,2 Artificial G4-binding proteins constructed from the G4-binding domain of helicase RHAU (also named DHX36 or G4R1) facilitate the detection of the G4 in DNA and RNA.3,4 We reported the assembly of DNA- and RNA-G4-binding proteins from the Arg-Gly-Gly repeat (RGG) domain of TLS (translocated in liposarcoma) protein, also known as FUS (fused in sarcoma) protein (TLS/FUS).5,6 These molecules revealed that telomere DNA and telomeric repeat-containing RNA (TERRA), DNA, and RNA G4, respectively, promote histone methylation at different amino acid sites in the telomere region. The RGG domain is conserved in several G4 binding proteins, such as TLS/FUS, Ewing’s sarcoma protein (EWS), heterogeneous ribonucleoprotein A1, nucleolin, cold-inducible RNA-binding protein, Dbp2, Ded1, fragile X mental retardation protein, and METTL14, but the RGG domain as a molecule to regulate G4 function has not been well investigated.7−17 Here, we found that the engineered RGG3 domain of TLS/FUS containing Phe (RGGF) recognizes the DNA loop in the G4 and inhibits the transcription of bcl-2. These data confirm that the RGG domain is a potentially useful tool for regulating and exploring the function of G4s.
Materials and Methods
Plasmid Constructs
For the polymerase chain reaction (PCR) template, we used pGEX6P-1-GST-RGG3, which was previously cloned as RGG3 of TLS/FUS into the pGEX6P-1 vector (Cytiva, Tokyo, JP).7 pGEX6P-1-GST-RGGF was obtained by deletion in pGEX6P-1-GST-RGG3 using a KOD-Plus-Mutagenesis Kit (Toyobo, Osaka, JP). pGEX6P-1-GST-RGGF was constructed by PCR using pGEX6P-1-RGG3 as the template and the following RGGF primers: forward, d(CGG GGC CGC GGC GGG GAC CG), and reverse, d(GTT ACC CCC CAT GTG AGA GCC ACC). The nucleolin RBDs-RGG plasmid, which contains globular RNA binding domains (RBDs) and an RGG domain (267–710), was constructed as described previously.18 The FLAG- and GFP-tagged RGGF plasmid (pLPC-FLAG-GFP-RGGF) was constructed by PCR using pGEX6P-1-GST-RGGF as the template. pLPC-FLAG-GFP-RGGF was constructed by PCR using pGEX6P-1-GST-RGGF as the template with the RGGF forward and reverse primers. All constructs underwent automated DNA sequencing for verification. All oligomers used for plasmid constructs, the EMSA, and circular dichroism spectroscopies were obtained from Operon Biotechnologies (JP).
Expression and Purification of Glutathione S-Transferase (GST) Fusion Proteins
For the in vitro experiments, the recombinant proteins were fused to the N-terminus of GST and overexpressed in Escherichia coli. For the protein expression, E. coli strain BL21 (DE3) pLysS-competent cells were transformed with the plasmids of pGEX6p-1-GST-RGGF and pGEX6p-1-GST-nucleolin RBDs-RGG, and the transformants were grown at 37 °C in a Luria Bertani medium containing ampicillin (0.1 mg/mL). Protein expression was induced at A600 = 0.6 with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. The cells were then grown for an additional 16 h at 25 °C for nucleolin RBDs-RGG and 19 °C for RGGF. The harvests were centrifugated by centrifugation (3000g for 15 min). The protein purification used the GST-column as previously reported.19 The E. coli pellets were resuspended in W buffer [100 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA·2Na (pH 8.0), and 1 mM dithiothreitol (DTT)] with 30 mg/mL phenylmethylsulfonyl fluoride (PMSF) and RNase G.S (Nippon Gene, JP). The equilibrium buffer for column is 10 mL of W buffer. The supernatants containing the expressed proteins were lysed by sonication (model UR-20P, Tobcl-2my Seiko, JP) at 4 °C and centrifuged at 16,200g for 15 min at 4 °C. The resulting supernatants were applied to a 1 mL GSTrap FF column (GE Healthcare, USA) and washed with 20 mL of WT buffer [100 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA·2Na (pH 8.0), 10% of Triton X-100]. PreScission protease (8 units/mL, Cytiva) in buffer was used to remove the GST tags from the RGGF, which was then loaded on a column for 16 h at 4 °C and eluted with a potassium-Tris buffer [50 mM Tris–HCl (pH 7.5), 50 mM KCl]. The nucleolin RBDs-RGG GST tags were eluted by adding reduced glutathione and changing the buffer to a potassium-Tris buffer by dialysis. The protein concentration was measured using a BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, US). The proteins were stored at 4 °C and used within 12 h of purification.
Electrophoretic Mobility Shift Assay (EMSA)
The 32P-labeled G4s were formed by heating the samples with a thermal heating block to 95 °C and then cooling them in 50 mM Tris–HCl (pH 7.5) and 100 mM KCl in 2 °C/min steps to 4 °C. The binding reactions were conducted in 20 μL (final volume) with 1 nM labeled oligonucleotides with 50 nM protein and 0.1 mg/mL bovine serum albumin in 50 mM Tris–HCl (pH 7.5) and 100 mM KCl.6,8,18 The competition assay was performed with 1 nM labeled BCL-2 with 50 nM nucleolin and 50, 250, or 500 nM RGGF and 0.1 mg/mL bovine serum albumin in 50 mM Tris–HCl (pH 7.5) and 100 mM KCl. The binding reactions were performed with 1 nM labeled BCL-2 with varying concentrations (0–500 nM) of purified proteins and 0.1 mg/mL bovine serum albumin in 50 mM Tris–HCl (pH 7.5) and 100 mM KCl. The samples were then incubated for 30 min at 4 °C and then electrophoresed on a 6% polyacrylamide (acrylamide/bisacrylamide = 19:1) nondenaturing gel at 10 V/cm for 100 min at 4 °C. The gel and electrophoresis buffer both contained 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, and 0.5 mM EDTA) with or without 20 mM KCl. Following electrophoresis, the gels were placed in a phosphor imager cassette and imaged using the Personal Molecular Imager FX (Bio-Rad Laboratories, Hercules, CA, USA). The equilibrium dissociation constants (Kd) were determined by plotting the data from four replicate experiments as ϕ (1 fraction of free DNA) versus the protein concentration, which is equivalent to the amount of protein that binds half of the free DNA. The binding reactions were carried out in a final volume of 20 μL with 1 nM labeled oligonucleotide and various concentrations of purified protein dissolved in a solution of 0.1 mg/mL bovine serum albumin in potassium buffer. The Kd was calculated by nonlinear regression using Microsoft Excel for Microsoft 365 MSO according to the following equation: ϕ = [P]/{Kd + [P]}.19
Cell Culture and Transfection
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 1% penicillin–streptomycin. For the assays, the HeLa cells were cultured in six-well plates. The plasmids for protein expression were transfected into the HeLa cells using the Xfect transfection reagent (Takara Bio, Shiga, JP) incubated at 37 °C for 16 h according to the manufacturer’s instruction. We used the transiently transfection for the experiment after 48 h of incubation.
Western Blot Analysis
Expression of RGGF was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12% polyacrylamide gel. For visualization of FLAG-tagged proteins, they were transferred to polyvinylidene difluoride (PVDF) membranes and probed using a mouse monoclonal anti-FLAG M2 antibody (MilliporeSigma, St. Louis, MO). Anti-mouse horseradish peroxidase was used as the secondary antibody (Cell Signaling Technology, Danvers, MA, USA). In addition, the translational level of BCL-2 effected by expression of RGGF was confirmed by SDS-PAGE using a 10% polyacrylamide gel. For visualization of BCL-2 proteins, they were transferred to PVDF membranes and probed using a mouse monoclonal anti-BCL-2 antibody (Abcam, Cambridge, UK). Anti-mouse horseradish peroxidase was used as the secondary antibody (Cell Signaling Technology, Danvers, MA, USA). Protein bands were visualized using the ECL Western Blotting Analysis System from GE Healthcare (UK).
Reverse Transcription (RT)-qPCR Analysis
The total RNA was isolated from HeLa cells with an RNeasy Mini Kit (Qiagen, Hilden, DE). RNA (0.5 μg) was reverse-transcribed in 20 μL of 1× buffer and 1.25 μM random primer using a ReverTra Ace Kit (Toyobo, Osaka, JP) for 10 min at 30 °C, 30 min at 42 °C, and then 5 min at 99 °C according to the manufacturer’s instructions. The levels of gene expression were quantified by real-time PCR using a Thunderbird SYBR qPCR Mix (Toyobo, Osaka, JP) and specific primer set (each 200 nM) with a Dice Thermal Cycler (Takara, Shiga, JP) using the following of protocol: 1 cycle of 50 °C for 2 min and 95 °C for 10 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min.20 Sequences of specific primers used are: bcl-2 cDNA, 5′-GGGATGCCTTTGT GGAACTGTA-3′ and 5′-AGAGACAGCCAGGAGAAATCAAAC-3′ (size: 67 bp); β-actin cDNA, 5′-GACAGGATGCAGAAGGAGATCACT-3′ and 5′-CGCTCAG GAGGAGCAATGA-3′ (size: 74 bp). The quantification was determined by applying the 2–Cq formula and calculating the average of the values obtained for each sample. Eligibility of this formula was verified by qPCR using the RT-qPCR product of total RNA as a template at different concentrations that covered five orders of magnitude. We used the β-actin as the reference gene.
Results and Discussion
Preferential Binding of RGGF to G4s with DNA Loops
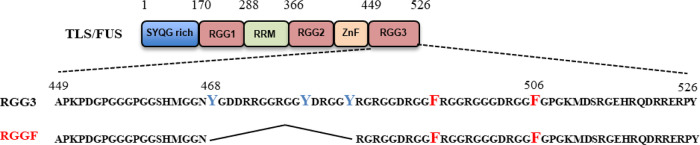
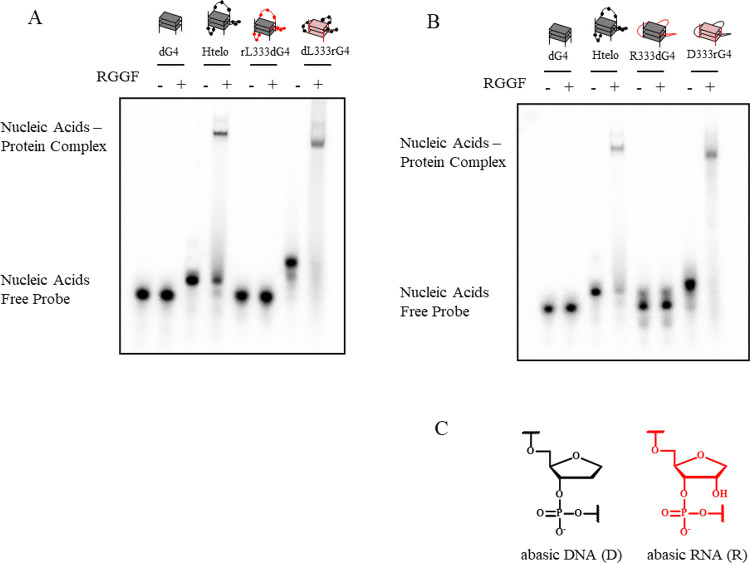
Previously, we identified that the RGG domain of the C-terminal region (RGG3) of TLS/FUS containing two Phe and three Tyr binds to DNA and RNA G4, respectively (Figure 1).7 Nuclear magnetic resonance analysis and EMSA revealed that RGG3 binds the loops and G-tetrads in the G4.5,7,13 Moreover, we reported that the engineered RGG3 domain of TLS/FUS containing Phe (RGGF) specifically binds and stabilizes the folded DNA G4 (Figure 1).6 Moreover, RGGF inhibits specific histone modifications in the telomere region, although the binding mechanism is unclear.6 To investigate whether RGGF mainly recognizes the loops and/or G-tetrad of DNA G4, we examined the binding of RGGF to 32P-labeled human telomere DNA (Htelo), four d(GGG) repeat without loops (dG4), four d(GGG) repeat with r(UUA) loops (rL333dG4), and four r(GGG) repeat with d(TTA) loops (dL333rG4) by EMSA (Figure 2A and Table 1). The purification of all proteins reported herein was confirmed by SDS-PAGE (Supplementary Figure S1), and circular dichroism (CD) spectra of Htelo, dG4, rL333dG4, and dL333rG4 had been confirmed and already reported.5,8 We found that dG4 and dL333rG4 are typical of the parallel strand G4s, and Htelo and rL333dG4 are typical of the hybrid (3 + 1) G4s in 100 mM KCl. The EMSA revealed that the G4 of Htelo is favorable for binding, whereas the G4 of dG4 is unfavorable for binding. This result suggests that RGGF prefers G4s with loops. Moreover, the EMSA revealed that Htelo and dL333rG4 are favorable for binding, whereas the G4 of rL333dG4 is unfavorable for binding. However, the RGGF cannot distinguish the G4 topology of the parallel and the hybrid (3 + 1) forms. The EMSA in Figure 2A revealed that RGGF binds to the hybrid (3 + 1)-formed Htelo and parallel-formed dL333rG4, of which both the loops consist of DNA. The finding suggests the preferential binding of RGGF to DNA G4s with loops, regardless of different G4 topologies. We next evaluated how the base and ribose in the nucleotide on the loop in the G4 are singled out by RGGF for binding (Figure 2B and Table 1), we examined the binding of RGGF to 32P-labeled Htelo, dG4, four d(GGG) repeat with three RNA abasic loops (R333dG4), and four r(GGG) repeat with three DNA abasic loops (D333rG4) by EMSA (Figure 2B,C and Table 1). The CD spectra of R333dG4 had been confirmed and already reported.5 We found that R333dG4 is typical of the parallel strand G4 in 100 mM KCl. Moreover, the G4 structure of D333rG4 was confirmed by CD spectra and melting curves (Supplementary Figure S2). We found that D333rG4 is typical of the parallel strand G4 in 100 mM KCl. The EMSA revealed that the G4 of D333rG4 is favorable for binding, whereas the G4 of R333dG4 is unfavorable for binding. These observations indicate that RGGF is able to discriminate between DNA and RNA loops in the G4.
Figure 1.
Schematic illustration of TLS/FUS, RGG3, and RGGF. SYQG-rich; RGG 1, Arg-Gly-Gly-rich motif 1; RRM, RNA recognition motif; RGG 2, Arg-Gly-Gly-rich motif 2; ZnF, zinc finger; RGG 3, Arg-Gly-Gly-rich motif 3. RGG3 containing two Phe and three Tyr; RGGF containing two Phe.
Figure 2.
RGGF selectively binds the DNA or RNA loops on the G4. EMSA was performed with RGGF and 32P-labeled (A) Htelo, dG4, rL333dG4, or dL333rG4 and (B) Htelo, dG4, R333dG4, or D333rG4. Gray and red in the cartoon show DNA and RNA, respectively. (C) Nucleic acid structures of abasic DNA and abasic RNA. Black and red in the cartoon shows, respectively, DNA and RNA.
Table 1. Oligonucleotide Sequences Used in EMSAa.
The loop sequences are underlined. D = abasic DNA, R = abasic RNA.
Preferential Binding of RGGF to G4 with Longer Loops
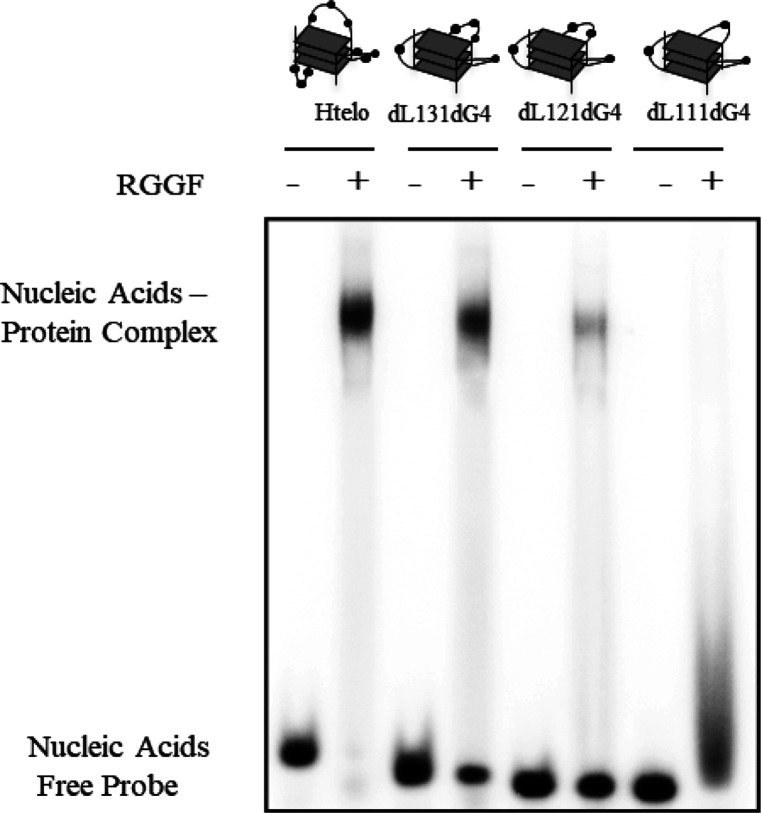
We previously reported preferential binding of the RGG3 of EWS, which is functionally related to TLS/FUS as a subgroup of a ribonucleoprotein family, and nucleolin, comprising four globular RNA binding domains (RBDs) and an RGG domain, to G4s with longer loops.18,19,21 Therefore, we evaluated the influence of loop length in G4 structures on RGGF binding by EMSA (Table 1 and Figure 3). Previously, we already reported that the CD spectra of DNA G4 with four d(GGG) repeats containing d(T)n loops (n = 1–3) in the middle loop (dL111dG4, dL121dG4, and dL131dG4) with two d(T) segments in the loops are typical of the parallel strand G4s.19 An EMSA of RGGF with dL111dG4, dL121dG4, and dL131dG4 and Htelo indicated that the G4 containing Htelo and dL131dG4 was the most favorable for binding, and the G4 containing dL111dG4 was the most unfavorable for binding. We analyzed the binding activities of RGGF to G4s containing d(T)n loops (n = 1–3) in the two lateral loops (dL111dG4, dL212dG4, and dL313dG4) with d(T) in the central loops (Supplementary Figure S3). Previously, we already reported that dL212dG4 and dL313dG4 form parallel strand G4s.19 An EMSA showed that dL313dG4 had the best binding to RGGF. These observations suggest that RGGF binds preferentially to DNA G4 with longer loops. Previously, we reported that the RGG domain recognizes the phosphate and the ribose of the loops in G4.5,19 This binding mode might cause the preferential binding of RGGF to G4 with longer loops. To investigate the RGGF binding to antiparallel-formed DNA G4, we performed an EMSA of RGGF with the Htelo fold in the presence of 100 mM NaCl (Supplementary Figure S4). Previously, we already reported that the CD spectra of Htelo in NaCl showed antiparallel G4.19 An EMSA showed that RGGF bound to antiparallel formed Htelo. Based on Figure 2 and Supplementary Figures S4, these findings suggest that RGGF binds to hybrid (3 + 1), parallel, and antiparallel DNA G4 without preference of different topologies.
Figure 3.
Effect of G4 loop length on the binding affinity of RGGF. The EMSA was performed using RGGF (lanes 2, 4, 6, and 8) with 32P-labeled Htelo (lanes 1 and 2), dL131dG4 (lanes 3 and 4), dL121dG4 (lanes 5 and 6), or dL111dG4 (lanes 7 and 8). The DNA–protein complexes were resolved by 6% polyacrylamide gel electrophoresis and visualized by autoradiography.
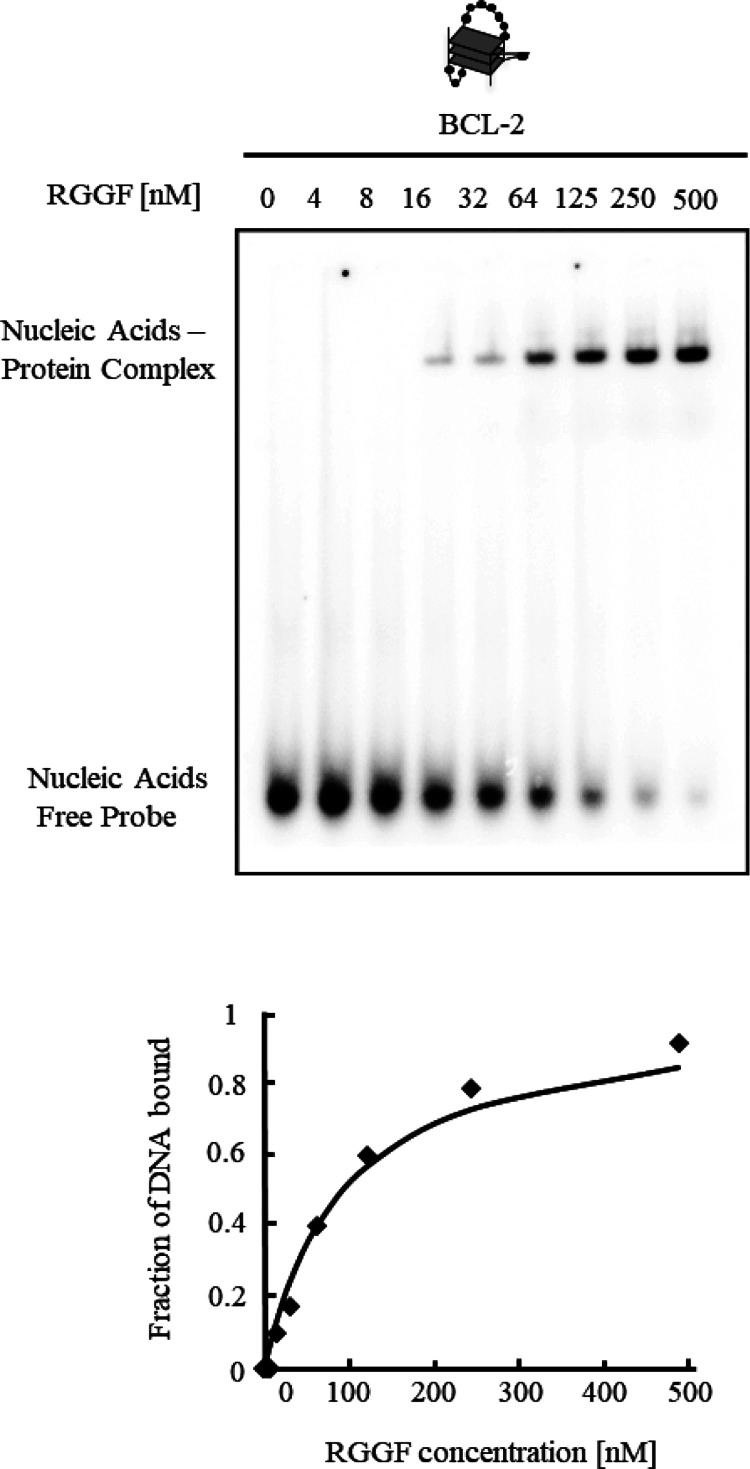
Many G4-binding small molecules that inhibit bcl-2 transcription have been reported.22 Selective stabilization of G4 in the promoter leads to suppression of bcl-2 transcription. The DNA oligomer derived from the human bcl-2 promoter (BCL-2) forms an intramolecular hybrid (3 + 1) G4 in K+-containing solution.23 The BCL-2 structure contains three loops, with the one, three, and seven nucleotides in the loops. Before investigating the effect of RGGF on bcl-2 transcription, an EMSA of RGGF was conducted with various concentrations of BCL-2 to analyze the ability of RGGF to bind BCL-2 (Table 1 and Figure 4). With an increase in the RGGF concentration, there was a decrease in the amount of free DNA as well as an increase in the amount of the higher-molecular weight complex. Fitting the mobility shift data to a hyperbolic equation gave a dissociation constant (Kd) of 96 ± 0.2 nM. This suggests that RGGF binds to G4 BCL-2 with the long loops.
Figure 4.
Binding activity of RGGF to G4 BCL-2. The equilibrium binding curve was obtained by calculating the fraction of 32P-labeled BCL-2 at varying RGGF concentrations. The dissociation constant (Kd) was ascertained by fitting the data to the appropriate equation. The DNA–protein complexes were resolved by 6% polyacrylamide gel electrophoresis and visualized by autoradiography.
Nucleolin Binding to BCL-2 Inhibited by Excess RGGF
Nucleolin is a G4-binding protein that activates the bcl-2 promoter.24 The N- and C-terminal ends of the protein contain an acidic region and an RGG domain, respectively, with RBDs located in the central region (Figure 5A). The RBDs of nucleolin binds to guanine in 5′-terminal and 3′-terminal single strands of the G4, while the RGG domain recognizes G4 structures.18 It indicates that the RBDs and RGG domain of nucleolin (nucelolin RBDs-RGG) mainly bind to G4. Moreover, the filter binding assay data of nucelolin RBDs-RGG and BCL-2 were fitted to a hyperbolic equation, giving a Kd of 455 nM.10 To investigate whether RGGF affects nucleolin binding to BCL-2, RGGF and nucleolin RBDs-RGG (Figure 5A) were used in competition assays with 32P-labeled BCL-2 (Table 1 and Figure 5B). Lanes 2 and 3 in Figure 5B show that each position of the RGGF-BCL-2 and nucleolin RBDs-RGG-BCL-2 complex in the gel was different due to the different molecular weights of RGGF and nucleolin RBDs-RGG. Adding excess RGGF inhibited the binding of nucleolin RBDs-RGG to BCL-2 in vitro (slane 4–6, Figure 5B). Adding excess RGGF competitors inhibited nucleolin RBDs-RGG binding to BCL-2.
Figure 5.
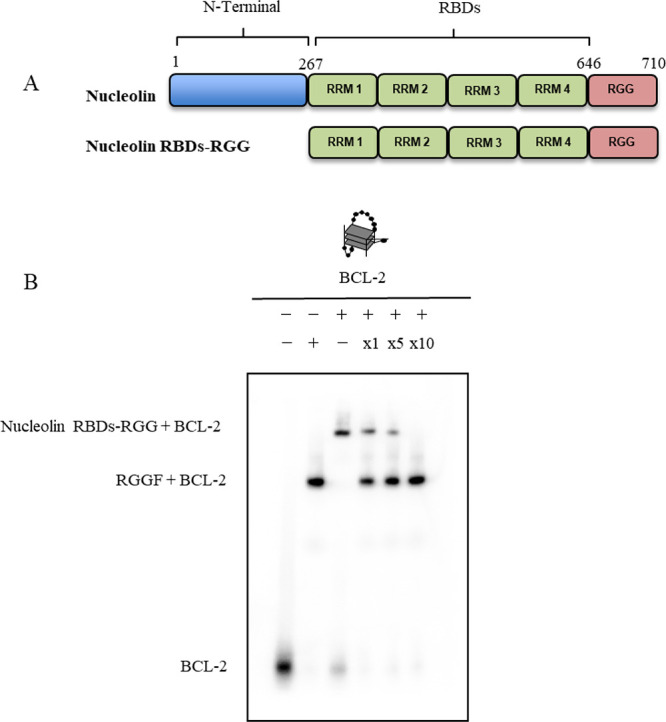
Competitive binding of BCL-2 to nucleolin and RGGF. (A) Schematic illustration of nucleolin and nucleolin RBDs-RGG. (B) EMSA of 32P-labeled BCL-2 with nucleolin and RGGF was performed by 6% polyacrylamide gel electrophoresis and visualized by autoradiography. Labeled BCL-2 and RGGF (lane 2) or nucleolin (lane 3) was incubated and analyzed as a control. A competitive binding assay of 32P-labeled BCL-2 to nucleolin was performed in the presence of RGGF at the indicated molar ratios (lanes 4–6).
Transcriptional Activity of Bcl-2 Inhibited by Overexpressed RGGF in HeLa Cells
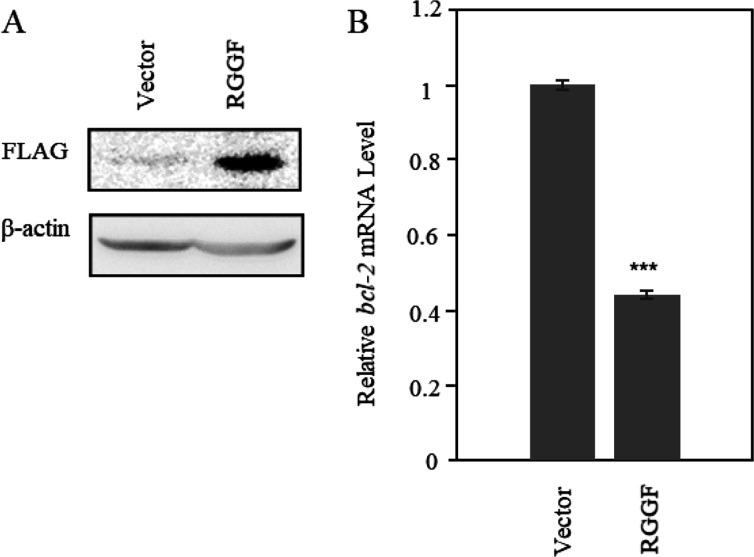
To investigate the bcl-2 transcription in HeLa cells with overexpressed RGGF, we performed RT-qPCR analysis with FLAG-tagged and green fluorescent protein (GFP)-tagged RGGF in HeLa cells (Figure 6). The RGGF was expressed by a vector, and its expression level was detected by Western blot analysis (Figure 6A). The level of bcl-2 transcripts was decreased in RGGF-overexpressing cells (44.2 ± 1.1%), as was determined by RT-qPCR, using β-actin as the internal control (Figure 6B and Supplementary Figure S5). In addition, the relative translation level of Bcl-2 in RGGF-overexpressing cells was decreased (57.6 ± 0.4%), as was determined by Western Blot, using β-actin as the internal control (Supplementary Figure S6). RGGF decreased the transcription level of Bcl-2 in HeLa cells, and it resulted in decreasing the translation level of it. However, RGGF-overexpressing cells’ viability was estimated to be about 96.3% after 48 h of incubation (Supplementary Figure S7). It indicates that RGGF did not have a high cytotoxic to HeLa cells with the transient transfection for this experiment. These findings suggest that RGGF binds to the G4 of the bcl-2 promoters, thereby repressing its transcription.
Figure 6.
Transcription level changes of bcl-2 in RGGF-overexpressing HeLa cells. (A) Overexpressed RGGF was analyzed by Western blot with a FLAG antibody. (B) Relative mRNA expression of bcl-2 in RGGF-overexpressing HeLa cells measured by RT-qPCR and normalized β-actin expression. Student’s test; ***p < 0.001 compared with vector (n = 3). Bars represent mean values (± errors) obtained from three independent experiments.
Conclusions
Here, we demonstrated that RGGF constructed from the RGG domain of TLS/FUS binds to G4s having longer DNA loops (Figures 2 and 3). We previously reported that other engineered RGG domains from TLS/FUS and the RGG domain of EWS recognize loops in the G4.5,19 A recent paper reported that nucleolin, which consists of four RNA recognition motifs and an RGG domain, preferentially binds to G4s with longer loops.21 Loops in G4s are an important common structure recognized by G4-binding proteins with an RGG domain. Furthermore, the dissociation constant of RGGF and BCL-2 was 96 ± 0.2 nM, and excess RGGF inhibited the binding of nucleolin to BCL-2 in vitro (Figures 4 and 5). Moreover, overexpressed RGGF in HeLa cells inhibited the transcriptional activity of bcl-2 (Figure 6). Based on the result of the competition assay in vitro, excess RGGF might compete with nucleolin to bind the bcl-2 promoter and mainly RGGF might inhibit the transcriptional activity of bcl-2. RGGF might be a useful tool for regulating transcription and investigating the role of DNA G4 in the genome.
Glossary
Abbreviations
- TLS/FUS
translocated in liposarcoma/fused in sarcoma
- RRM
RNA recognition motif
- RGG
arginine-glycine-glycine repeat
- RBD
RNA binding domain
- GFP
green fluorescent protein
- EWS
Ewing’s sarcoma
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00050.
SDS-PAGE of RGGF and nucleolin on 12 and 8% polyacrylamide gel (Figure S1), CD spectrum and melting profile of Oligo 5 (Figure S2), effect of G4 loop length on the binding affinity of RGGF (Figure S3), RGGF binding to the Htelo fold in the presence of NaCl (Figure S4), RT-qPCR amplification plots of β-actin and bcl-2 from the vector or RGGF-overexpressing cells (Figure S5), translation level changes of bcl-2 in RGGF-overexpressing HeLa cells (Figure S6), and cell viability assay (Figure S7) (PDF)
Author Contributions
T.O. conceived the study, L.L.U. and R.Y. prepared the protein and performed the EMSA assay, L.L.U. and T.K. performed Western blot, and Y.K. and A.I. performed qPCR. L.L.U., T.O., T.K., R.Y., Y.K., and A.I. analyzed the data and wrote the manuscript.
This work was supported by the JGC-S Scholarship Foundation, a Grant-in-Aid for Scientific Research (C) (no. 20K05704 to T.O.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The authors declare no competing financial interest.
Supplementary Material
References
- Varshney D.; Spiegel J.; Zyner K.; Tannahill D.; Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel J.; Adhikari S.; Balasubramanian S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020, 2, 123–136. 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K. W.; Zhang J. Y.; He Y. D.; Gong J. Y.; Wen C. J.; Chen J. N.; Hao Y. H.; Zhao Y.; Tan Z. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 2020, 48, 11706–11720. 10.1093/nar/gkaa841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang D. T.; Phan A. T. Development of a ribonuclease containing a G4-specific binding motif for programmable RNA cleavage. Sci. Rep. 2019, 9, 7432. 10.1038/s41598-019-42143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama K.; Oyoshi T. Specific binding of modified RGG domain in TLS/FUS to G-quadruplex RNA: Tyrosines in RGG domain recognize 2′-OH of the riboses of loops in G-quadruplex. J. Am. Chem. Soc. 2013, 135, 18016–18019. 10.1021/ja4086929. [DOI] [PubMed] [Google Scholar]
- Takahama K.; Miyawaki A.; Shitara T.; Mitsuya K.; Morikawa M.; Hagihara M.; Kino K.; Yamamoto A.; Oyoshi T. G-quadruplex DNA- and RNA-specific-binding proteins engineered from the RGG domain of TLS/FUS. ACS Chem. Biol. 2015, 10, 2564–2569. 10.1021/acschembio.5b00566. [DOI] [PubMed] [Google Scholar]
- Takahama K.; Takada A.; Tada S.; Shimizu M.; Sayama K.; Kurokawa R.; Oyoshi T. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 2013, 20, 341–350. 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Takahama K.; Kino K.; Arai S.; Kurokawa R.; Oyoshi T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 2011, 278, 988–998. 10.1111/j.1742-4658.2011.08020.x. [DOI] [PubMed] [Google Scholar]
- Ghosh M.; Singh M. Structure specific recognition of telomeric repeats containing RNA by the RGG-box of hnRNPA1. Nucleic Acids Res. 2020, 48, 4492–4506. 10.1093/nar/gkaa134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V.; Guo K.; Hurley L.; Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635. 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. L.; Dai J.; Luo W.-H.; Wang X.-G.; Tan J.-H.; Chen S.-B.; Huang Z.-S. Identification of G-quadruplex-binding protein from the exploration of RGG motif/G-quadruplex interactions. J. Am. Chem. Soc. 2018, 140, 17945–17955. 10.1021/jacs.8b09329. [DOI] [PubMed] [Google Scholar]
- Darnell J. C.; Jensen K. B.; Jin P.; Brown V.; Warren S. T.; Darnell R. B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001, 107, 489–499. 10.1016/S0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Kondo K.; Mashima T.; Oyoshi T.; Yagi R.; Kurokawa R.; Kobayashi N.; Nagata T.; Katahira M. Plastic roles of phenylalanine and tyrosine residues of TLS/FUS in complex formation with the G-quadruplexes of telomeric DNA and TERRA. Sci. Rep. 2018, 8, 2864. 10.1038/s41598-018-21142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K. K. P.; Obi I.; Sabouri N. The RGG domain in the C-terminus of the DEAD box helicases Dbp2 and Ded1 is necessary for G-quadruplex destabilization. Nucleic Acids Res. 2021, 49, 8339–8354. 10.1093/nar/gkab620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.; Berroyer A.; Kim M.; Kim N. Yeast Nucleolin Nsr1 Impedes Replication and Elevates Genome Instability at an Actively Transcribed Guanine-Rich G4 DNA-Forming Sequence. Genetics 2020, 216, 1023–1037. 10.1534/genetics.120.303736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyoshi T.; Masuzawa T. Modulation of histone modifications and G-quadruplex structures by G-quadruplex-binding proteins. Biochem. Biophys. Res. Commun. 2020, 531, 39–44. 10.1016/j.bbrc.2020.02.178. [DOI] [PubMed] [Google Scholar]
- Yoshida A.; Oyoshi T.; Suda A.; Futaki S.; Imanishi M. Recognition of G-quadruplex RNA by a crucial RNA methyltransferase component, METTL14. Nucleic Acids Res. 2022, 50, 449–457. 10.1093/nar/gkab1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzawa T.; Oyoshi T. Roles of the RGG domain and RNA recognition motif of nucleolin in G-quadruplex stabilization. ACS Omega 2020, 5, 5202–5208. 10.1021/acsomega.9b04221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama K.; Sugimoto C.; Arai S.; Kurokawa R.; Oyoshi T. Loop lengths of G-quadruplex structures affect the G-quadruplex DNA binding selectivity of the RGG motif in Ewing’s Sarcoma. Biochemistry 2011, 50, 5369–5378. 10.1021/bi2003857. [DOI] [PubMed] [Google Scholar]
- Tamaoki K.; Okada R.; Ishihara A.; Shiojiri N.; Mochizuki K.; Goda T.; Yamauchi K. Morphological, biochemical, transcriptional and epigenetic responses to fasting and refeeding in intestine of Xenopus laevis. Cell Biosci. 2016, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A.; Duchambon P.; Masson V.; Loew D.; Bombard S.; Teulade-Fichou M. P. Nucleolin discriminates drastically between long-loop and short-loop quadruplexes. Biochemistry 2020, 59, 1261–1272. 10.1021/acs.biochem.9b01094. [DOI] [PubMed] [Google Scholar]
- Singh M.; Gupta R.; Comez L.; Paciaroni A.; Rani R.; Kumar V. BCL2 G quadruplex-binding small molecules: Current status and prospects for the development of next-generation anticancer therapeutics. Drug Discovery Today 2022, 9, 2551–2561. [DOI] [PubMed] [Google Scholar]
- Dai J.; Chen D.; Jones R. A.; Hurley L. H.; Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006, 34, 5133–5144. 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstein E.; Du Y.; Santourlidis S.; Christ J.; Uhrberg M.; Wernet P. Nucleolin regulates gene expression in CD34-positive hematopoietic cells. J. Biol. Chem. 2007, 282, 12439–12449. 10.1074/jbc.M608068200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.