Abstract
A novel cDNA, insulinoma-associated antigen-1 (IA-1), containing five zinc-finger DNA-binding motifs, was isolated from a human insulinoma subtraction library. IA-1 expression is restricted to fetal but not adult pancreatic and brain tissues as well as tumors of neuroendocrine origin. Using various GAL4 DNA binding domain (DBD)/IA-1 fusion protein constructs, we demonstrated that IA-1 functions as a transcriptional repressor and that the region between amino acids 168 and 263 contains the majority of the repressor activity. Using a selected and amplified random oligonucleotide binding assay and bacterially expressed GST–IA-1DBD fusion protein (257–510 a.a.), we identified the consensus IA-1 binding sequence, TG/TC/TC/TT/AGGGGG/TCG/A. Further experiments showed that zinc-fingers 2 and 3 of IA-1 are sufficient to demonstrate transcriptional activity using an IA-1 consensus site containing a reporter construct. A database search with the consensus IA-1 binding sequence revealed target sites in a number of pancreas- and brain-specific genes consistent with its restricted expression pattern. The most significant matches were for the 5′-flanking regions of IA-1 and NeuroD/β2 genes. Co-transfection of cells with either the full-length IA-1 or hEgr-1AD/IA-1DBD construct and IA-1 or NeuroD/β2 promoter/CAT construct modulated CAT activity. These findings suggest that the IA-1 protein may be auto-regulated and play a role in pancreas and neuronal development, specifically in the regulation of the NeuroD/β2 gene.
INTRODUCTION
Insulinoma-associated antigen-1 (IA-1) encodes a zinc-finger DNA-binding protein that was originally isolated from a human insulinoma subtraction library (1). The IA-1 gene is unique because it contains a high percentage of alanine and proline residues (∼40%, 250 a.a.) at its N-terminus and five zinc-finger motifs (250 a.a.) symmetrically arranged at the C-terminus. IA-1 expression was detected in a large number of neuroendocrine tumors including insulinoma, retinoblastoma, medullablastoma, pheochromocytoma, medullary thyroid carcinoma, pituitary tumor (1; M.S.Lan, M.Zhu and M.B.Breslin, unpublished observation), and small cell lung carcinoma (2). IA-1 expression, detected in early human fetal brain and pancreas development, is dramatically reduced in later stages of development, and absent in normal adult tissues (3; M.S.Lan, M.Zhu and M.B.Breslin, manuscript in preparation) suggesting that the IA-1 gene might function as a developmentally regulated transcription factor in neuroendocrine cells.
Using a rat pancreatic amphicrine cell line, AR42J, induction of IA-1 expression was found to coincide with AR42J cells converting to insulin-producing endocrine cells. Additionally, an increase in islet-specific transcription factors Pdx1, NeuroD/β2 and Nkx6.1 was seen in parallel with IA-1 induction (3). Therefore, the expression of IA-1 is closely associated with other pancreas-specific transcription factors whose role in endocrine pancreas development is well established. Based on the expression pattern of IA-1, it is reasonable to suggest that it may function as an upstream regulator of key islet differentiation factors.
Pancreas development proceeds by the interaction of numerous transcription factors to orchestrate a complex cascade of events resulting in the development of both endocrine and exocrine tissues (4–6). The endocrine pancreas is composed of clusters of cells, the islets of Langerhans, which include α, β, δ and pancreatic polypeptide cells embedded in the exocrine pancreas. Extensive genetic studies have revealed a complex network of transcription factors including Pdx1/IPF-1 (7,8), Isl1 (9), Pax4 (10), Pax6 (11,12), NeuroD/β2 (13), Nkx2.2 (14), Nkx6.1 (15), Hes-1 (16), Hlxb9 (17,18) and neurogenin 3 [ngn3 (19)], which regulate the development of islet cells at different stages.
To understand the functions of IA-1 as a transcription factor, it is crucial to identify the downstream target genes it may regulate. In this study, we used a selected and amplified binding site selection (SAAB) strategy to determine the consensus sequence that is recognized by the IA-1 DNA-binding domain (DBD). We report here the consensus IA-1 binding sequence and the potential target genes containing the IA-1 binding site in their 5′-flanking regions. A database sequence search and electrophoretic mobility shift assay (EMSA) competition analyses revealed that both the IA-1 gene itself and a neuroendocrine transcription factor, NeuroD/β2, are the potential downstream target genes for IA-1. Further functional studies using fusion constructs between the GAL4 DBD and IA-1 protein or the full-length IA-1 protein revealed that IA-1 possesses transcriptional repressor activity suggesting a primary role in IA-1 gene auto-regulation and NeuroD/β2 gene expression in pancreas and nervous system development.
MATERIALS AND METHODS
Cell culture and transient transfection for chloramphenicol acetyltransferase (CAT) reporter assay
HeLa (human cervical carcinoma) and βTC-1 (mouse insulinoma) cells were maintained in low glucose Dulbecco’s Minimal Essential Medium supplemented with 10% heat inactivated fetal bovine serum (Gibco), 100 U/ml penicillin and 100 µg/ml streptomycin. For transient transfection studies, the cells were seeded to a density of 1 × 105 cells per well in a six-well culture dish the day before transfection. On the day of transfection, the cells were washed three times in serum-free media. A 1:1 ratio of 5 µl lipofectamine 2000 reagent (1 mg/ml, Life Technologies) to 5 µg of test DNA [2 µg GAL4 fusion construct, 2 µg GAL4(5)tkCAT reporter construct, and 1 µg cytomegalovirus (CMV)-βgal or 2 µg of either IA-1, Pax6 or NeuroD/β2 promoter/CAT construct, 2 µg expression vector and 1 µg CMV-βgal] were used according to the manufacturer’s instructions. The cells were assayed 48 h post-transfection for β-galactosidase (Promega) and CAT (Promega) activities.
DNA construct
The pSG424 GAL4 (1–147) fusion vector (20) was used for mapping the functional domains of the IA-1 protein. Briefly, all of the GAL4–IA-1 fusion constructs, with the exception of the 257–510 a.a. IA-1 construct, were made by restriction endonuclease digestion of the full-length IA-1 library clone IA-1-18 (1) and subcloned in-frame with the GAL4 1–147 coding sequence. The 257–510 a.a. IA-1 fragment was generated by PCR amplification and subcloned into both the pSG424 GAL4 vector and pGEX4T2 [glutathione S-transferase (GST)] vector (Pharmacia). The sequences were verified to be correct and were in-frame with the GAL4 1–147 fusion protein or GST fusion protein except for the 1–263fs construct which contained an additional nucleotide resulting in a frameshift in the IA-1 reading frame. The various human early growth response factor-1/IA-1 fusion constructs (hEgr-1/IA-1) were made by PCR amplification of the various regions of interest. Human Egr-1 (1–147 a.a.) region was amplified from human heart cDNA using PCR primers, 5′-GCCAAGCTTTCCAGGATGGCCGCGGCCAAG-3′ and 5′-GCTGAATTCGGAAAAGCGGCCAGTATAGGT-3′, and subcloned into the HindIII/EcoRI site of the pcDNA3 (Invitrogen) expression vector. The IA-1 DBD 257–510 a.a. was PCR amplified using a plasmid DNA template with PCR primers 5′-AGCGAATTCGCGGGGGGCGCGGCGCGGCCG-3′ and 5′-GTCCTCGAGCTAGCAGGCCGGGCGCACGGG-3′ and subcloned into the EcoRI/XhoI sites of the pcDNA3 vector. Other IA-1 zinc-finger region clones were generated by PCR using primers 262 a.a. 5′-ACGGAATTCCGGCCGCTGGGCGAGTTCATCTGC-3′, 290 a.a. 5′-AGTGAATTCATCGTGCGTGTGGAGTACCGCTGT-3′, 312 a.a. 5′-ACGGAATTCTCGCACCGCCGCTGGCACAAACCGCGG-3′, 320 a.a. 5′-CACCTCGAGCTACGCGGTTTGTGCCAGCGGCGGTGCGA-3′, 439 a.a. 5′-CACCTCGAGCTACTCGGCGGACGCACTCAGGCCCAGCAC-3′, 440 a.a. 5′-ACGGAATTCTGCCACCTGTGCCCAGTGTGCGGAGAG-3′, 469 a.a. 5′-TATCTCGAGGAACACCTGGGCGGCGTGCAGCAG-3′, 510 a.a. 5′-CACCTCGAGCTAGCAGGCCGGGCGCACGGGCACCTGCAG-3′. The clones were sequenced to confirm that they were correct. The full-length IA-1 expression construct was prepared by digestion of the pBS34-18 clone with EcoRI/PvuI. A 2.8 kb IA-1 cDNA fragment was subcloned into the EcoRI site of the pcDNA3 vector (Invitrogen).
SAAB
To identify the consensus IA-1 target binding sequence, a SAAB approach was performed as described (21) with the following modifications. Briefly, oligonucleotides were synthesized which contained 25 randomized nucleotides in between fixed 5′ and 3′ PCR primer sequences. Labeled, double-stranded templates were generated by using a 5′-primer, [α-32P]dCTP (3000 Ci/mmol; NEN) and the Klenow fill-in reaction. After the initial round of selection, the PCR products were gel purified on a 2% agarose gel and labeled using [γ-32P]ATP (NEN; 3000 Ci/mmol), and T4 polynucleotide kinase (New England Biolabs). The labeled oligonucleotides were subjected to EMSA analysis. The reaction mixture for EMSA contained the following components: ∼40 000 c.p.m. labeled oligonucleotide, 3 µg purified GST–IA-1DBD (also named as Cpep, amino acids 257–510) protein (pGEX4T2 vector, Pharmacia) in the binding reaction buffer [13 mM HEPES pH 7.9, 67 mM KCl, 13% glycerol, 0.3 mM DTT, 0.1 mM ZnSO4, 0.3 mg/ml BSA, 0.13 mg/ml poly(dIdC)]. The protein–DNA complexes were resolved on a 4% polyacrylamide gel (40:1) gel in 0.25× TBE buffer. The protein–DNA complexes were eluted from the gel using 500 µl elution buffer (0.5 M NH4OAc, 10 mM MgCl2, 1% SDS, 1 mM EDTA) at 37°C for 4 h. Four rounds of selection were performed and the final PCR products were TA cloned into a pCR2.1 TOPO cloning vector (Invitrogen). Individual clones were selected and sequenced using an Amersham Thermosequenase Kit. Following the initial selection of individual clones, three additional rounds of selection were performed on a pool of 72 unique, individual clones. The resulting products from the final round of the selection were TA cloned into the pCR2.1 TOPO cloning vector. Alignment of various clones revealed a consensus binding site as shown in Table 1.
Table 1. Alignment of consensus sequences selected from SAAB protocola.
| Consensus | T | G/T | C/T | C/T | T/A | G | G | G | G | G/T | C | G/A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9/27b | T | G | T | C | A | G | G | G | G | G | C | A |
| 5/27 | T | G | C | C | T | G | G | G | G | T | T | G |
| 3/27 | G | T | T | A | A | G | G | G | G | G | C | G |
| 2/27 | T | G | C | T | T | G | G | G | G | C | A | T |
| 1/27 | T | T | G | T | T | G | G | G | G | T | C | G |
aSeven rounds of EMSA and PCR selection as detailed in Materials and Methods.
bDominant C5 appears 9 out of 27 times. Additional seven individual clones do not shown any sequence homology with the consensus sequence.
EMSA
EMSA binding reactions were performed as described above. The oligonucleotides were made double stranded by incubating equal molar amounts of the sense and antisense oligonucleotides at 95°C for 5 min and cooled to room temperature by 1°C/min. The oligonucleotides used included: clone 5 (C5) 5′-TACAGGCAAACACATCATACTGTCAGGGGGCAAT-3′ and the complementary strand; clone 5 left (C5L) 5′-TCCGAATTCCTACAGGCAAACACATGA-3′ and the complementary strand; clone 5 right (C5R) 5′-TACTGTCAGGGGGCAATGGATCCGTCT-3′ and the complementary strand; 2X wild-type (wt) C5 5′-(TGTCAGGGGCA)2-3′ and the complementary strand; 2X mutant1 (mut1) C5 5′-(TGTCATTGGGCA)2-3′ and the complementary strand; 2X mutant2 (mut2) C5 5′-(TGTCAGGTTTCA)2-3′ and the complementary strand; 2X mutant3 (mut3) C5 5′-(TGTCAGTTGGCA)2-3′ and complementary strand; IA-1 promoter (–103/–63 bp) 5′-GAGGAGCTGCGGACGCGCTGATTGGCTCCAGGGGAAGCGGG-3′ and the complementary strand; 3X Pax4 site 5′-(CAAACCCTGGAG)3-3′ and the complementary strand; 3X Pdx-1 site 5′-(CTCCAGGGTAAA)3-3′ and the complementary strand; 3X Pax6 site 5′-(CGCCCCCTTGCT)3-3′ and the complementary strand; 3X NeuroD/β2 site 5′-(TGGAAGGGGGCG)3-3′ and the complementary strand. The 3X tandem repeat/E1bTATA/CAT constructs were made by cloning the 3X binding site oligonucleotides (Pax6 and NeuroD/β2) in the pCR2.1 TOPO vector. Constructs were sequenced to determine orientation and subcloned into the HindIII/XbaI cloning site upstream of the E1bTATA/CAT construct or subcloned using XhoI/HindIII sites for the opposite orientation.
IA-1 promoter/CAT constructs
The –189/–16 bp IA-1 promoter fragment was amplified using PCR with the upstream –189 bp primer 5′-TCAGGTACATCTGCCGCACCTA-3′ and the downstream –16 bp primer 5′-GGCAGCCGCTCCCTTTTAAC-3′. The resulting –189/–16 bp IA-1 promoter fragment was TA cloned into the pCR2.1 TOPO cloning vector (Invitrogen) and the orientation as well as sequence was confirmed by sequencing. The –189/–16 bp IA-1 promoter fragment was subcloned into the HindIII/XbaI site upstream of the E1bTATA box driven CAT reporter gene. The –426/+40 bp IA-1 promoter/CAT construct was generated by subcloning a NheI/XhoI –426/+40 bp IA-1 promoter fragment into the NheI/XhoI site of the pCAT3 vector (Promega). The ΔE1bTATACAT vector was prepared by digestion with HindIII/BamHI to remove the E1bTATA box and religated. The –426/+40 bp IA-1 promoter fragment was blunt-end ligated into the SmaI site of the ΔE1bTATACAT vector to test the suppressor activity of the full-length IA-1 protein.
RESULTS
Characterization of IA-1 as a transcriptional repressor
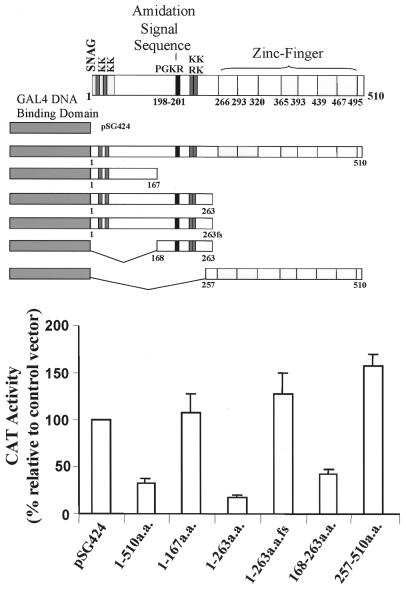
The IA-1 coding sequence shares a significant degree of homology with proteins from the Snail/Slug family of transcriptional repressor proteins in the seven amino acid SNAG domain responsible for transcriptional repression activity (22). Therefore, in order to map the functional domains of the IA-1 protein, regions of the IA-1 protein were fused in frame with the GAL4 DBD (Fig. 1). Transient co-transfection studies in HeLa cells with GAL4DBD–IA-1 fusion constructs and a reporter construct containing five tandem copies of the GAL4 DNA binding element located upstream of the thymidine kinase minimal promoter driven CAT reporter gene demonstrated that the full-length IA-1 sequence, amino acids 1–510, repressed CAT activity by ∼75% (Fig. 1). Fine mapping of the region responsible for the reduction in CAT activity revealed that a region between amino acids 168 and 263 contained the majority of the observed repression activity (Fig. 1). Within this region there is large number of proline and alanine residues, 39 out of 95 amino acids (∼41%). This type of motif has been described in other transcriptional repressor molecules (23).
Figure 1.
IA-1 functions as a transcriptional repressor in HeLa cells. Schematic representation of the various IA-1 fragments fused with the GAL4 DBD (amino acids 1–147) as an effector vector that was co-transfected with a reporter vector containing the GAL4 DNA-binding sequence, the thymidine kinase minimal promoter driven CAT reporter gene. The pSG424 vector contains the GAL4 DBD alone. The full-length IA-1 hybrid construct yielded 25% CAT activity of the pSG424 control vector. Amino acids 168–263 of the IA-1 protein possess repressor activity. A 263fs fragment represents a nucleotide deletion that causes a frame shift of the 1–263 amino acid sequence. Transient transfections were performed as described in the Materials and Methods in HeLa cells. The data are expressed as the percentage change as compared with the GAL4DBD construct alone (control). The CMV–βgal construct was used to normalize the transfection efficiency. The graph shows the average of four separate experiments and the SEM.
Identification of IA-1 target binding site
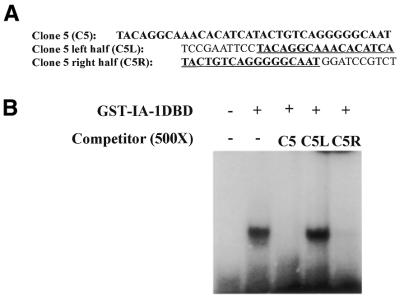
The IA-1 protein, containing five Cys2-His2 type zinc-finger motifs in the C-terminus, belongs to a conserved family of transcriptional repressor proteins, and demonstrates transcriptional repressor activity in a GAL4 DBD reporter gene assay. This suggests that IA-1 can function as a sequence-specific DNA-binding protein. Therefore, we wanted to determine its target-binding site. The consensus IA-1 binding site was identified using a SAAB method (21). Oligonucleotides with 25 random nucleotides in between a pair of fixed PCR primer sequences [5′-TCCGAATTCCTACAG(N)25GCAATGGATCCGTCT-3′] were used in multiple rounds of binding and selection with purified GST fused IA-1 DBD (a.a. 257–510) protein. Following four rounds of binding site selection, the bound pool of oligonucleotides were subcloned into pCR2.1 TOPO TA vector (Invitrogen). Seventy-two individual clones were isolated, sequenced, and established to be unique from one another. The pool of the 72 unique clones was then subjected to three additional rounds of selection to identify high-affinity IA-1 binding sequences. The resulting selected pool was subcloned into pCR2.1 TOPO TA vector and individual clones were sequenced. Alignment of the most frequently observed sequences revealed the putative consensus IA-1 binding site, TG/TC/TC/TT/AGGGGG/TCG/A (Table 1). The most abundant sequence (one-third of the sequences observed), C5, was shown to bind to the purified GST–IA-1DBD protein in an EMSA (Fig. 2). Using 500-fold excess unlabeled cold competitor oligonucleotides, which represented the left (C5L) or right (C5R) half of the full-length C5 oligonucleotide sequence, only the full-length or C5R oligonucleotide competed away the specific protein–DNA complex (Fig. 2). Inspection of the consensus IA-1 binding site reveals a highly conserved nucleotide ‘GGGG’ core. Mutation analysis of any two G nucleotides in the GGGG core abolished IA-1 binding, demonstrating that the core GGGG nucleotides are critical for IA-1 binding activity (Fig. 3).
Figure 2.
Competitive EMSA. IA-1 protein binds to a dominant C5 sequence. A competitive EMSA study revealed that the sequence located at the right-hand half of clone 5 accounts for the binding activity. The underlined sequence represents either half of the full-length C5 oligonucleotide (A). 500-fold molar excess cold competitor full-length C5 or the right hand half of the C5 sequence (C5R) competed away the specific protein/DNA complex. The left hand half sequence (C5L) had no effect on the protein/DNA complex (B). EMSA were performed as described in Materials and Methods.
Figure 3.
Mutation analysis of the IA-1 consensus binding site. Two tandem repeats of the 12 bp C5 binding site (wild-type) as well as mutated sequences (Mut1, Mut2 and Mut3) were synthesized for the EMSA binding study. EMSA was performed as described in Materials and Methods. Mutated nucleotides are underlined. A stretch of G nucleotides is essential for IA-1 protein binding.
A region of the IA-1 promoter binds IA-1 protein
Previous studies on the human IA-1 promoter revealed that deletion of a region between –111 and –66 bp resulted in a 3-fold reduction in promoter activity and the remaining promoter activity was almost undetectable (24). Comparison of this region with the consensus IA-1 binding site shows that it shares a high degree of homology with the consensus IA-1 binding site. To test whether the IA-1 protein binds to this region, a –103 to –63 bp double-stranded probe was labeled and used in an EMSA. Bacterially expressed GST–IA-1DBD protein bound to this site (Fig. 4A). Increasing amounts from 10, 50, 100 and 500 times molar excess cold competitor –103/–63 bp IA-1 oligonucleotide shows that 50 times excess competitor is sufficient to completely block the DNA/IA-1DBD protein complex (Fig. 4B). Competition between the –103/–63 bp IA-1 promoter probe, the full-length C5, C5L and C5R oligonucleotides revealed that the full-length or C5R oligonucleotide sequence competed, but less efficiently than the cold IA-1 promoter probe (Fig. 4A). The ability of IA-1 protein to bind to its own promoter region strongly suggests that the IA-1 gene is active in auto-regulating its own level of expression in target cells.
Figure 4.
IA-1 promoter contains a consensus target sequence. The IA-1 promoter sequence (–103/–63 bp) binds to the IA-1 protein in EMSA analysis. The 500-fold cold IA-1p, C5 and C5R oligonucleotides compete efficiently but not the C5L probe showing that the interaction is specific (A). Different concentrations (10-, 50-, 100- and 500-fold) of cold excess IA-1 promoter sequence reveals dosage-dependent competition for GST–IA-1DBD protein binding (B).
Auto-regulation of IA-1 promoter activity
In vitro binding assays showed that IA-1 can bind to the –103/–63 bp region in its own promoter. In order to assess the ability of IA-1 to affect its own gene transcription, a portion of the IA-1 promoter (–189/–16 bp), containing the IA-1 binding site was subcloned upstream of an E1bTATA box driven CAT reporter gene. To better assess IA-1 binding and transcriptional activity, we switched the IA-1 protein N-terminal repression domain (a.a. 1–256) with the hEgr-1 activation domain (AD) and fused it in frame with the IA-1 DBD (Fig. 5A). In this experiment, instead of assessing IA-1 transcriptional repression activity we observed transcriptional activation activity. Co-transfection of the –189/–16 bp IA-1 promoter/E1bTATA/CAT construct with the chimeric hEgr-1AD/IA-1DBD construct increased CAT activity ∼3-fold in βTC-1 cells (Fig. 5A). The hEgr-1AD or IA-1DBD construct alone had no effect on the CAT activity (Fig. 5A) demonstrating that the observed induction of IA-1 5′-upstream sequence activity requires the specific DNA binding of the IA-1 protein. We also examined the transcriptional repressor activity of the full-length IA-1 using IA-1p(–426/+40 bp)CAT reporter vector, which has a higher basal CAT activity. The full-length CMV–IA-1 cDNA expression vector resulted in an 80% repression in IA-1 promoter driven CAT activity in HeLa cells (Fig. 5B). This result is consistent with the GAL4 reporter assay and demonstrates that IA-1 functions as a transcriptional repressor in regulating its own promoter.
Figure 5.
Auto-regulation of the IA-1 gene expression. The –189/–16 bp IA-1 5′-upstream region, which contains the IA-1 target binding site, was cloned upstream of the E1bTATA/CAT reporter gene. Co-transfection of hEgr-1AD/IA-1DBD and the IA-1 –189/–16 bp-E1bTATA/CAT vector were performed as described in Materials and Methods. A stronger CAT activity was observed in insulinoma (β-TC-1) cells (A). The empty expression vector, the hEgr-1AD alone or the IA-1DBD construct had no effect on the CAT activity. Alternatively, we tested the IA-1 full-length suppressive activity using –426/+40 bp IA-1 pCAT and CMV–IA-1 cDNA expression vectors (B). Co-transfection of HeLa cells demonstrated 80% repression in IA-1 promoter activity. Transfections were performed in β-TC-1 or HeLa cells on three separate occasions. The data are expressed as fold induction or suppression over the control pcDNA3 (empty) expression vector. A CMV–βgal vector is used to normalize transfection efficiency. The graph represents the average of three separate experiments and SEM.
IA-1 zinc-fingers 2 and 3 are sufficient for transcriptional activity
Using SAAB, we demonstrated that the IA-1 protein could bind DNA in a sequence-specific manner. Therefore, we further characterized the IA-1 zinc-fingers critical for transcriptional activity. This analysis was performed using the hEgr-1 AD fused in frame with different IA-1 zinc-finger regions (Fig. 6). The various hEgr-1AD/IA-1 zinc-finger constructs were transiently transfected along with a –426/+40 bp IA-1 promoter/CAT construct into βTC-1 cells. The IA-1 zinc-finger constructs were assessed for their ability to activate the IA-1 promoter/CAT activity. The intact zinc-finger region (fingers 1–5) induced ∼15-fold increase in CAT activity (Fig. 6). Zinc-finger 3 alone could activate the IA-1 promoter to approximately half the level (∼6.5-fold) of the intact zinc-finger 1–5 construct (Fig. 6). The constructs that contain zinc-fingers 2 and 3 can completely mimic the same activation level as the intact 1–5 zinc-finger domain (Fig. 6). This experiment shows that zinc-fingers 2 and 3 contain all the information necessary to specifically modulate target gene transcription.
Figure 6.
IA-1 zinc-fingers 2 and 3 are essential for transcriptional activity. Various zinc-finger constructs were fused in frame with hEgr-1 AD (a.a. 1–147). Co-transfection of zinc-finger mutants with the IA-1 –426/+40 bp promoter/pCAT3 reporter gene into β-TC-1 cells revealed that zinc-fingers 2 and 3 are the key motifs that contribute to the transcriptional activity. Zinc-finger 3 alone exhibits 50% of the control activity, whereas the combination of zinc-finger 3 and 4 only exhibits 25% of the control activity. The data are expressed as fold increase over the empty pcDNA3 expression vector. A CMV–βgal vector is used to normalize transfection efficiency. The graph represents the average of three separate experiments and SEM.
Database search for potential IA-1 target genes
An 83% or better sequence homology search using the IA-1 consensus sequence of the eukaryotic promoter and GenBank database revealed a selected number of genes from brain and pancreas that may be regulated by IA-1. Most notable in the list are the transcription factors from mouse Pax6, and NeuroD/β2. These factors are expressed in pancreatic endocrine tissues and have been shown to be critical in pancreas development. Further sequence search with the IA-1 promoter binding site revealed two additional potential target genes, Pax4 and Pdx-1. Comparison of the consensus IA-1 binding site with the IA-1, Pax4, Pdx-1, Pax6 and NeuroD/β2 promoters revealed that these sites share a significant degree of homology even outside the consensus binding site with one another (Table 2). However, Pax4 and Pdx-1 sites differ from the consensus binding site within the GGGG region. Double-stranded oligonucleotides containing three tandem repeats of the mouse Pax4, Pdx-1, Pax6 and NeuroD/β2 IA-1 binding sites were used to perform an EMSA (Fig. 7A). As seen in Figure 7A, only the Pax6 and NeuroD/β2 binding sites were able to bind the GST–IA-1DBD protein. This is consistent with the mutation analysis that we performed on the C5 sequence that showed a mutation of any two G nucleotides abolishes IA-1 protein binding (Fig. 3).
Table 2. Alignment of consensus sequence with 5′-flanking region of potential target genesa.
| Consensus | T | G/T | C/T | C/T | T/A | G | G | G | G | G/T | C | G/A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IA-1 (–80/–67 bp) | c | T | C | C | A | G | G | G | G | a | a | G |
| Pax-4 (+116/+103 bp)b | c | T | C | C | A | G | G | G | t | T | t | G |
| Pdx-1 (–116/–103 bp) | c | T | C | C | A | G | G | G | t | a | a | A |
| Pax-6 (P0 promoter)b | a | G | C | a | A | G | G | G | G | G | C | G |
| NeuroD/β2 (–177/–166 bp) | T | G | g | a | A | G | G | G | G | G | C | G |
aUpper case letters represent nucleotides identical to the consensus sequence whereas lower case letters indicate those that do not match.
bPax4 and Pax6 target sequences are in the opposite orientation.
Figure 7.
Target gene regulation by IA-1. EMSA analysis of potential target gene binding sites were performed using three consecutive repeats (12 bp)3 of the putative IA-1 binding sites from the mouse Pax4, Pdx-1, Pax6 and NeuroD/β2 genes and incubated with GST–IA-1DBD protein. Both Pax4 and Pax6 sequences are in the reverse orientation with respect to the NeuroD/β2 site. The Pax6 and NeuroD/β2 sequences demonstrated binding activity to the GST–IA-1DBD protein (A). The 3X repeat sequences of the mouse Pax6 and NeuroD/β2 IA-1 binding sites were cloned upstream of the E1bTATA/CAT construct to assess the ability of IA-1 to modulate their activity (B). The hEgr-1AD/IA-1DBD fusion construct was used in the co-transfection experiments. A CMV–β-gal construct was used to normalize the transfection efficiency. The NeuroD/β2 binding site shows strong activity suggesting it is the target gene for the IA-1 transcription factor. Transient transfections were performed in β-TC-1 cells. The graph represents the average of three separate experiments and SEM.
Regulation of Pax6 and NeuroD/β2 promoters by IA-1
In order to study the potential role IA-1 plays in the regulation of its target genes, three tandem copy repeats of the Pax6 or NeuroD/β2 IA-1 binding sites were subcloned upstream of an E1bTATA/CAT reporter gene. Co-transfection of the 3X Pax6 or 3X NeuroD/β2 E1bTATA/CAT constructs with hEgr-1AD/IA-1DBD constructs into βTC-1 cells revealed that the NeuroD/β2 construct was induced ∼6-fold over the E1bTATA/CAT construct (Fig. 7B). No effect was seen on the 3X Pax6 binding site construct (Fig. 7B). Most notable about this observation is that although the Pax6 site can bind IA-1DBD protein in vitro, the Pax6 IA-1 binding site is in the opposite orientation (on the minus strand) with respect to the IA-1 or NeuroD/β2 binding sites. This suggests that the IA-1 binding site may function in an orientation-dependent manner.
DISCUSSION
IA-1 belongs to a class of transcription factors that contain a SNAG repressor domain in the N-terminus and Cys2-His2 type zinc-finger motifs in the C-terminus (22). Currently, there are 20 proteins that belong to this class and they can be subdivided into three groups: (i) Snail/Slug proteins (25), (ii) growth factor independence 1 (Gfi-1) proteins (26), and (iii) IA-1 proteins (27). The IA-1 subclass contains the IA-1 and methylated in liver tumor-1 (mlt-1) genes. In this study, we have shown that either the IA-1 full-length or fused with the GAL4DBD construct demonstrated transcriptional repressor activity. The repressor domain was localized to a region between amino acids 168 and 263 (Fig. 1). Within this portion of the IA-1 protein, a high number of proline and alanine residues, 39 out of 95 (41%), are present. Comparison of this type of motif with another transcriptional repressor molecule, the Drosophila Even-skipped protein, shows that this is a similar motif required for transcriptional repressor activity (28). Surprisingly, the seven amino acid SNAG domain, MPRGFLV, located in the N-terminus of IA-1, does not function as a repression domain in the cell type tested. The seven amino acid SNAG domain, MPRSFLV, present in the Gfi-1/Gfi-1b proteins (22,29) and the Snail/Slug proteins (30) were shown to be critical for maximum repression activity of these molecules. The IA-1 SNAG domain differs from the Snail/Slug, Gfi-1/Gfi-1b SNAG domains by a single amino acid substitution (G for S); however, this residue was not shown to be important for the repression activity for Gfi-1 (22), but may render the IA-1 SNAG domain inactive. Alternatively, the IA-1 SNAG domain may function in a cell-type-specific manner since IA-1 expression is restricted to a limited number of cell types. Further analysis of the functional activity of IA-1 in other cell types not tested here is required.
Although we showed that the IA-1 protein is a transcriptional repressor, the downstream target genes regulated by the IA-1 protein remained unknown. Therefore, we designed an in vitro binding assay using randomized oligonucleotides and bacterial expressed recombinant protein. SAAB analysis revealed that the purified GST–IA-1DBD protein bound to a specific 12 bp consensus sequence (Table 1). EMSA analysis with the most frequently observed clone, C5, bound the GST–IA-1DBD protein (Fig. 2). Further studies showed that zinc-fingers 2 and 3 were sufficient for transcriptional activity. A zinc-finger motif consists of ∼27–30 amino acid residues. It exists as an independently folded unit that binds a zinc ion and is responsible for sequence-specific DNA interaction (32). The C-terminus of the IA-1 protein contains four intact Cys2-His2 zinc-finger motifs and a truncated first zinc-finger motif (missing the second histidine residue). Therefore, the number of bases recognized by the IA-1DBD should not be more than 12 bp given that each zinc-finger is expected to bind three bases (31,32). Mutation analysis of the consensus IA-1 sequence (C5 sequence) showed that the GGGG core is critical for binding because mutation of any two of these G nucleotides abolishes IA-1 protein binding activity (Fig. 3). The IA-1 protein belongs to a subclass of proteins within the SNAG domain family and shares a significant degree of homology with another protein, mlt-1 (27). The highest degree of homology between these two proteins is in the zinc-finger domain. Comparing each of the 27–28 amino acids in the zinc-fingers, 25 (89%), 26 (96%), 16 (57%), 10 (37%) and 24 (89%) amino acids are identical in the first, second, third, fourth and fifth zinc-finger motifs between human IA-1 and mouse mlt-1 proteins. The high degree of homology within the zinc-finger regions of IA-1 and mlt-1 suggests that they can bind similar DNA binding elements. Analysis of the zinc-finger regions revealed that zinc-fingers 2 and 3 were required for transcriptional activity (Fig. 6). Whereas zinc-finger 2 of mlt-1 is ∼90% identical to IA-1, zinc-finger 3 is only 57% identical suggesting that these two proteins would have different DNA binding sequences and targets. Mlt-1 is expressed in normal adult brain, spleen, stomach and liver and its expression is silenced by methylation of the promoter region in liver and neuroblastoma tumors (27). In contrast, IA-1 is expressed in fetal brain and pancreas as well as tumors of neuroendocrine origin (3). Due to their difference in tissue distributions, IA-1 and mlt-1 probably regulate completely different sets of genes.
Computer analysis of potential IA-1 regulated genes identified several pancreas-specific transcription factors. EMSA analysis and the transient transfection studies of promoter/CAT constructs showed that IA-1 and NeuroD/β2 are two downstream target genes modulated by IA-1. A high-affinity IA-1 binding site was identified in the IA-1 promoter. The –103/–63 bp IA-1 promoter site was effectively competed away by itself but not completely competed away with full-length C5 or C5R oligonucleotides indicating that the IA-1 promoter binds IA-1 protein better than the C5 sequence (Fig. 4A). This suggests that the IA-1 gene can auto-regulate its own expression as a negative feedback mechanism and/or modulate downstream genes in cooperation with other transcription factors. IA-1 gene expression is transient occurring during embryogenesis, whereas NeuroD/β2 gene expression is maintained at a relatively constant level during endocrine and neuronal development, which suggest that the effect of IA-1 on the NeuroD/β2 gene is transient. Within a narrow window of differentiation, certain genes are being turned on or off as required. Similarly, another upstream regulator of the NeuroD/β2 gene, ngn3, performs its critical role in NeuroD/β2 gene activation as well as in the control of endocrine cell precursor differentiation transiently (33–35). NeuroD/β2 is a helix–loop–helix transcription factor. The null mutation of the NeuroD/β2 gene arrests islet cell development at a late stage and results in mice suffering severe postnatal diabetes (13). Furthermore, the NeuroD/β2 expression is restricted to primitive neuroectodermal tumors including medulloblastoma, retinoblastoma and small cell lung carcinoma, which correlate with the IA-1 gene expression patterns (36). Recently, ngn3 was shown to be required for the specification of a common precursor for the four pancreatic endocrine cell types thus placing it upstream of all the other previously identified transcription factors in the cascade. Deletion and mutation analyses of the NeuroD/β2 promoter revealed that the two proximal E box sequences, E1 and E3, can bind to ngn3/E47 heterodimer and mediate ngn3-induced activation of the NeuroD/β2 promoter (33). Since the IA-1 binding site is located in between the TATA box and the E1, E2 and E3 elements of the NeuroD/β2 promoter, it is tempting to speculate that IA-1 may counter-regulate ngn3 to modulate NeuroD/β2 gene expression. Further analysis of the NeuroD/β2 promoter is necessary to determine what role, if any, IA-1 may play in its regulation during pancreatic endocrine cell development. Identification of IA-1 as a regulator of NeuroD/β2 during pancreatic development would place a zinc-finger transcriptional repressor molecule in the complex cascade of factors important for endocrine cell development.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Nancy Zeleznik-Le (Loyola University at Chicago, IL) for providing GAL4 effector and reporter vectors and the E1bTATA/CAT plasmid. This work was supported by a grant from the Juvenile Diabetes Research Foundation International (to M.S.L.).
REFERENCES
- 1.Goto Y., DeSilva,M.G., Toscani,A., Prabhakar,B.S., Notkins,A.L. and Lan,M.S. (1992) A novel human insulinoma-associated cDNA, IA-1, encodes a protein with zinc-finger DNA-binding motifs. J. Biol. Chem., 267, 15252–15257. [PubMed] [Google Scholar]
- 2.Lan M.S., Russell,E.K., Lu,J., Johnson,B.E. and Notkins,A.L. (1993) IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res., 53, 4169–4171. [PubMed] [Google Scholar]
- 3.Zhu M., Breslin,M.B. and Lan,M.S. (2001) A novel zinc-finger cDNA, IA-1, expression is associated with rat AR42J cell differentiation into insulin-positive cells. Pancreas, in press. [DOI] [PubMed] [Google Scholar]
- 4.Huang H.P. and Tsai,M.J. (2000) Transcription factors involved in pancreatic islet development. J. Biomed. Sci., 7, 27–34. [DOI] [PubMed] [Google Scholar]
- 5.Edlund H. (2001) Developmental biology of the pancreas. Diabetes, 50, S5–S9. [DOI] [PubMed] [Google Scholar]
- 6.Sander M. and German,M.S. (1997) The beta cell transcription factors and development of the pancreas. J. Mol. Med., 75, 327–340. [DOI] [PubMed] [Google Scholar]
- 7.Ohlsson H., Karlsson,K. and Edlund,T. (1993) IPF-1, a homeodomain-containing transactivator of the insulin gene. EMBO J., 12, 4251–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonsson J., Carlsson,L., Edlund,T. and Edlund,H. (1994) Insulin promoter-factor 1 is required for pancreas development in mice. Nature, 371, 606–609. [DOI] [PubMed] [Google Scholar]
- 9.Ahlgren U., Pfaff,S.L., Jessell,T.M., Edlund,T. and Edlund,H. (1997) Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature, 385, 257–260. [DOI] [PubMed] [Google Scholar]
- 10.Sosa-Pineda B., Chowdhury,K., Torres,M., Oliver,G. and Gruss,P. (1997) The Pax-4 gene is essential for differentiation of insulin-producing b cells in the mammalian pancreas. Nature, 386, 399–402. [DOI] [PubMed] [Google Scholar]
- 11.Sander M., Neubuser,A., Kalamaras,J., Ee,H.C. Martin,G.R. and German,M.S. (1997) Genetic analysis reveals that Pax6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev., 11, 1662–1673. [DOI] [PubMed] [Google Scholar]
- 12.St-Onge L., Sosa-Pineda,B., Chowdhury,K., Mansouri,A. and Gruss,P. (1997) Pax6 is required for differentiation of glucagon-porducing a-cells in mouse pancreas. Nature, 387, 406–409. [DOI] [PubMed] [Google Scholar]
- 13.Naya F.J., Huang,H.P., Qiu,Y., Mutoh,H., DeMayo,F.J., Leiter,A.B. and Tsai,M.J. (1997) Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev., 11, 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sussel L., Kalamaras,J., Hartigan-O’Connor,D.J., Meneses,J.J., Pedersen,R.A., Rubenstein,J.L.R. and German,M.S. (1998) Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development, 125, 2213–2221. [DOI] [PubMed] [Google Scholar]
- 15.Sander M., Sussel,L., Conners,J., Scheel,D., Kalamaras,J., Cruz,F.D., Schwitzgebel,V., Hayes-Jordan,A. and German,M. (2000) Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of b-cell formation in the pancreas. Development, 127, 5533–5540. [DOI] [PubMed] [Google Scholar]
- 16.Jensen J., Pedersen,E.E., Galante,P., Hald,J., Heller,R.S., Ishibashi,M., Kageyama,R., Guillemot,F., Serup,P. and Madsen,O.D. (2000) Control of endodermal endocrine development by Hes-1. Nature Genet., 24, 36–44. [DOI] [PubMed] [Google Scholar]
- 17.Li H., Arber,S., Jessell,T.M. and Edlund,H. (1999) Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nature Genet., 23, 67–70. [DOI] [PubMed] [Google Scholar]
- 18.Harrison K.A., Thaler,J., Pfaff,S.L., Gu,H. and Kehrl,J.H. (1999) Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nature Genet., 23, 71–75. [DOI] [PubMed] [Google Scholar]
- 19.Gradwohl G., Dierich,A., LeMeur,M. and Guillemot,F. (2000) Neurogenin 3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl Acad. Sci. USA, 97, 1607–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadowski I. and Ptashne,M. (1989) A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res., 17, 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwell T.K., Kretzner,L., Blackwood,E.M., Eisenman,R.N. and Weintraub,H. (1990) Sequence-specific DNA binding by the c-myc protein. Science, 250, 1149–1151. [DOI] [PubMed] [Google Scholar]
- 22.Grimes H.L., Chan,T.O., Zweidler-Mckay,P.A., Tong,B., Tsichlis,P.N. (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol., 16, 6263–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanna-Rose W. and Hansen,U. (1996) Active repression mechanisms of eukaryotic transcription repressors. Trends Genet., 12, 229–234. [DOI] [PubMed] [Google Scholar]
- 24.Li Q., Notkins,A.L. and Lan,M.S. (1997) Molecular characterization of the promoter region of a neuroendocrine tumor marker, IA-1. Biochem. Biophys. Res. Commun., 236, 776–781. [DOI] [PubMed] [Google Scholar]
- 25.Sefton M., Sanchez,S. and Neito,M.O. (1998) Conserved and divergent roles for members of the snail family of transcription factors in the chick and mouse embryo. Development, 125, 3111–3121. [DOI] [PubMed] [Google Scholar]
- 26.Zweidler-Mckay P.A., Grimes,H.L., Flubacher,M.M. and Tsichlis,P.N. (1996) Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol., 16, 4024–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tateno M., Fukunishi,Y., Komatsu,S., Okazaki,Y., Kawai,J., Shibata,K., Itoh,M., Muramatsu,M., Held,W.A. and Hayashizaki,Y. (2001) Identification of a novel member of the Snail/Gfi-1 repressor family, mlt1, which is methylated and silenced in liver tumors of SV40 antigen transgenic mice. Cancer Res., 61, 1144–1153. [PubMed] [Google Scholar]
- 28.Han K. and Manley,J.L. (1993) Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev., 7, 491–503. [DOI] [PubMed] [Google Scholar]
- 29.Tong B., Grimes,H.L., Yang,T., Bear,S.E., Qin,Z., Du,K., El-Deiry,W.S. and Tsichlis,P.N. (1998) The Gfi-1b proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol. Biol. Cell, 18, 2462–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama H., Scott,I.C. and Cross,J.C. (1998) The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev. Biol., 199, 150–163. [DOI] [PubMed] [Google Scholar]
- 31.Desjarlais J.R. and Berg,J.M. (1992) Towards rules relating zinc finger protein sequences and DNA binding site preferences. Proc. Natl Acad. Sci. USA, 89, 7345–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs G.H. (1992) Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J., 11, 4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H.P., Liu,M., El-hodiri,H.M., Chu,K., Jamrich,M. and Tsai,M.J. (2000) Regulation of the pancreatic islet-specific gene beta2 (neuroD) by neurogenin 3. Mol. Biol. Cell, 20, 3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen J., Heller,R.S., Funder-Nielsen,T., Pedersen,E.E., Lindsell,C., Weinmaster,G., Madsen,O.D. and Serup,P. (2000) Independent development of pancreatic a- and b-cells from neurogenin3-expressing precursors. Diabetes, 49, 163–176. [DOI] [PubMed] [Google Scholar]
- 35.Schwitzgebel V.M., Scheel,D.W., Conners,J.R., Kalamaras,J., Lee,J.E., Anderson,D.J., Sussel,L., Johnson,J.D. and German,M.S. (2000) Expression of neurogenin3 reveals an islet cell precursor popolation in the pancreas. Development, 127, 3533–3542. [DOI] [PubMed] [Google Scholar]
- 36.Rostomily R.C., Bermingham-McDonogh,O., Berger,M.S., Tapscott,S.J., Reh,T.A. and Olson,J.M. (1997) Expression of neurogenic basic helix–loop–helix genes in primitive neuroectodermal tumors. Cancer Res., 57, 3526–3531. [PubMed] [Google Scholar]