Ovarian cancer is the leading cause of deaths from gynecologic cancer, and although there are nomograms that use pathological variables to predict outcome for advanced-stage disease,1 few if any molecular mechanisms that accurately predict outcome and can potentially guide therapy have been identified. The article by Merritt et al.2 in this issue of the Journal provides evidence for a simple mechanism, based on the biologic characteristics of microRNAs (miRNAs), for formulating a prognosis and potentially guiding therapy in ovarian cancer.
The past decade has heralded in a new era in the understanding of gene regulation in diseases such as cancer. We now appreciate that normal human cells express thousands of noncoding RNAs and that cancer cells misexpress these RNAs. Many of the noncoding RNAs, epitomized by the miRNA, have regulatory functions in normal cells. The aberrant expression of miRNA promotes tumorigenesis, metastasis, and other features of cancer.3
The miRNAs are a large class of small, regulatory noncoding RNAs in plants and animals that inhibit gene expression by binding to imperfect complementary sites within the 3′ untranslated regions of their target — messenger RNA (mRNA) transcripts.4 The miRNAs arise from large RNA precursors (termed pri-miRNAs), which are generally transcribed from genomes by RNA polymerase II and processed in the nucleus — by the RNase III enzyme Drosha and the protein Pasha (also known as DiGeorge syndrome critical region gene 8 [DGCR8]) that binds to double-stranded RNA — into shorter sequences of approximately 70 nucleotides, called pre-miRNAs (Fig. 1). These molecules fold into imperfect stem–loop structures.
Figure 1. Drosha and Dicer and the Processing and Function of MicroRNA (miRNA).
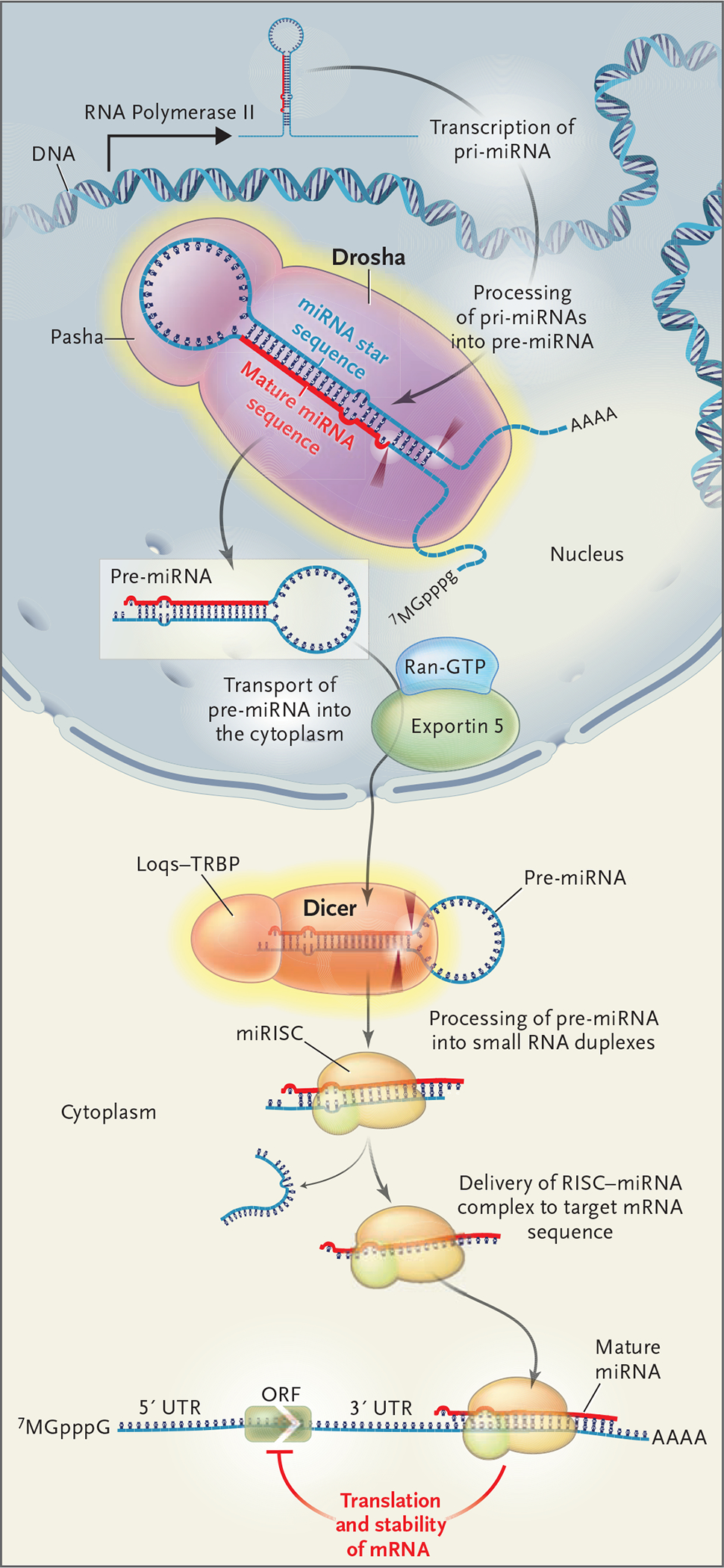
Pri-miRNAs, large miRNA precursors, are transcribed from the genome by RNA polymerase II and fold into a hairpin structure that is the substrate for RNA processing by Drosha (cleavage sites shown as dark red arrows) to a pre-miRNA. Pre-miRNAs enter the cytoplasm, where they are further processed by Dicer (and its partner, Loqs–TRBP), resulting in an RNA duplex containing the mature miRNA and the miRNA star sequences. The mature miRNA preferentially enters the miRNA-associated, multiprotein RNA–induced silencing complex (miRISC) and guides it to partially complementary sequences in target messenger RNA (mRNA), leading to repression of their translation. AAAA represents the poly-A tail, and 7MGpppG denotes the 5′ cap. ORF denotes open reading frame, Ran-GTP Ran guanosine triphosphate, and UTR untranslated region.
The pre-miRNAs undergo an additional processing step within the cytoplasm (Fig. 1): double-stranded RNA approximately 22 nucleotides in length (the duplex of the miRNA sequence and the miRNA star sequence) is excised from the premiRNA hairpin by another RNase III enzyme, Dicer. The miRNA–miRNA star-sequence duplex is then loaded into the miRNA-associated, multiprotein RNA-induced silencing complex (miRISC). The mature miRNA strand is preferentially retained in the miRISC, where it guides the RISC to target sequences. In most animals, miRNAs direct gene regulation at the level of translation.
Over 1000 miRNAs have been identified in animal genomes through cloning and bioinformatic approaches.5–8 Although the biologic roles of only a small fraction of identified miRNAs have been elucidated, these miRNAs regulate processes essential to cell growth, differentiation, apoptosis, and adhesion, and for this reason they have provided insight into the mechanisms of human cancer. Bioinformatic and microarray studies have revealed that a single miRNA could bind to as many as 200 gene targets and that these targets, which can be diverse in their function, include transcription factors, secreted factors, receptors, and transporters. Hence, miRNAs potentially control the expression of approximately one third of all human mRNAs.9 Complementary sites for multiple types of miRNA have also been identified within the 3′ untranslated regions of a single gene target, indicating the existence of intricate patterns of combinatorial regulation by miRNAs.10–13 The regulatory network controlled by this class of small RNAs is extremely complex, and since miRNAs potentially have a broad influence over a number of diverse genetic pathways, the deletion or misexpression of these tiny RNAs is likely to be pleiotropic and account for their important roles in human cancer.
The miRNAs are invariably found to be misexpressed in every type of cancer examined so far, and miRNA genes can act as both tumor-suppressor genes and oncogenes.14 A study by the Croce group15 revealed that approximately 50% of all annotated human miRNAs are located in areas of the genome associated with cancer or “fragile sites,” and thus miRNAs might have a crucial function in cancer progression. Furthermore, studies indicate that levels of Dicer and let-7 miRNA could be used in cancer prognosis, since patients with non–small-cell lung carcinomas whose tumors expressed lower levels of these RNAs had a poorer prognosis and shortened postoperative survival.16–18 In addition, loss of the enzyme Drosha enhanced tumorigenesis in a mouse model of lung cancer.19 These studies suggest that proteins involved in miRNA biogenesis are likely to play important roles in cancer.
Merritt et al. demonstrate that levels of the miRNA processing enzymes Drosha and Dicer are prognostic indicators in patients with ovarian cancer. They found that low levels of Dicer are associated with advanced tumor stage and that low levels of Drosha are associated with suboptimal surgical cytoreduction,20,21 both known to be poor prognostic factors. High levels of Dicer and Drosha identified a subgroup of patients with ovarian cancer with significantly better survival. The potential of low Dicer levels as a biomarker of poor outcome was validated through the analysis of previously published microarray data in cohorts of patients with ovarian, breast, and lung cancer, confirming for each type of cancer that low Dicer levels indicate a poor prognosis. Although the authors looked for DNA mutations that could account for abnormal levels of Dicer and Drosha in patients, they were unable to find meaningful mutations, and the mechanism leading to their misexpression remains unknown.
However, like most cancers, ovarian cancer is known to have lower miRNA levels than normal tissue. Therefore, it is possible that low Dicer and Drosha levels result in additional abnormalities of miRNA expression, which leads tumors to be associated with a poor prognosis. As evidence supporting the misprocessing of miRNAs in tumors with low Dicer levels, the authors showed that targeting the galectin-3 gene for silencing in ovarian-cancer cells with low Dicer was successful only when small interfering RNA was used, not short hairpin RNA. These findings confirm that low Dicer levels result in decreased processing of short hairpin RNAs (such as miRNAs). This finding may have important implications for future treatment of ovarian cancer based on approaches that involve short hairpin RNAs, since such approaches would not be successful in tumors with low Dicer and Drosha levels. Not only does this work provide useful prognostic markers, but also, perhaps these molecular mechanisms predicting outcome are the first steps toward designing strategies to target miRNA-processing mechanisms with the goal of improving outcomes.
Footnotes
Dr. Slack reports holding two pending patents for the use of microRNAs as potential therapeutic and diagnostic markers in cancer, and Dr. Weidhaas, two pending patents for the use of microRNAs in predicting the risk of ovarian cancer and response to chemotherapy. No other potential conflict of interest relevant to this article was reported.
Contributor Information
Frank J. Slack, Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT.
Joanne B. Weidhaas, Department of Therapeutic Radiology, Yale University, New Haven, CT.
References
- 1.Chi DS, Palayekar MJ, Sonoda Y, et al. Nomogram for survival after primary surgery for bulky stage IIIC ovarian carcinoma. Gynecol Oncol 2008;108:191–4. [DOI] [PubMed] [Google Scholar]
- 2.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 2008;359: 2641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–69. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853–8. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA 2003;9:175–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001;294:858–62. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001;294:862–4. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003;115:787–98. [DOI] [PubMed] [Google Scholar]
- 10.Reinhart BJ, Slack FJ, Basson M, et al. The 21 nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901–6. [DOI] [PubMed] [Google Scholar]
- 11.Lin SY, Johnson SM, Abraham M, et al. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell 2003;4:639–50. [DOI] [PubMed] [Google Scholar]
- 12.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495–500. [DOI] [PubMed] [Google Scholar]
- 13.Hobert O Common logic of transcription factor and microRNA action. Trends Biochem Sci 2004;29:462–8. [DOI] [PubMed] [Google Scholar]
- 14.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 2005;96:111–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753–6. [DOI] [PubMed] [Google Scholar]
- 18.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189–98. [DOI] [PubMed] [Google Scholar]
- 19.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673–7. [DOI] [PubMed] [Google Scholar]
- 20.Gadducci A, Cosio S, Tana R, Genazzani AC. Serum and tissue biomarkers as predictive and prognostic variables in epithelial ovarian cancer. Crit Rev Oncol Hematol 2008. June 30 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 21.Winter WE III, Maxwell GL, Tian C, et al. Tumor residual after surgical cytoreduction in prediction of clinical outcome in stage IV epithelial ovarian cancer: a Gynecologic Oncology Group Study. J Clin Oncol 2008;26:83–9. [DOI] [PubMed] [Google Scholar]


