ABSTRACT
Mycoplasma genitalium is an important sexually transmitted pathogen affecting both men and women. Its extremely slow growth in vitro and very demanding culture requirements necessitate the use of molecular-based diagnostic tests for its detection in clinical specimens. The recent availability of U.S. Food and Drug Administration (FDA)-cleared commercial molecular-based assays has enabled diagnostic testing to become more widely available in the United States and no longer limited to specialized reference laboratories. Advances in the knowledge of the epidemiology and clinical significance of M. genitalium as a human pathogen made possible by the availability of molecular-based testing have led to updated guidelines for diagnostic testing and treatment that have been published in various countries. This review summarizes the importance of M. genitalium as an agent of human disease, explains the necessity of obtaining a microbiological diagnosis, describes currently available diagnostic methods, and discusses how the emergence of antimicrobial resistance has complicated treatment alternatives and influenced the development of diagnostic tests for resistance detection, with an emphasis on developments over the past few years.
KEYWORDS: HIV, Mollicutes, Mycoplasma genitalium, antimicrobial treatment, cervicitis, lab detection, macrolide, pelvic inflammatory disease, quinolone, urethritis
MYCOPLASMA GENITALIUM AS AN AGENT OF HUMAN DISEASE
The 2019 Centers for Disease Control and Prevention (CDC) report of the top antibiotic resistance threats in the United States named Mycoplasma genitalium as one of three organisms included in the watch list (1). The heightened concern over M. genitalium is due to the rapidity with which it is moving toward becoming an untreatable infection. Since the most recent review on M. genitalium diagnostics was published in the Journal of Clinical Microbiology 5 years ago (2), three molecular-based assays that detect M. genitalium have received U.S. Food and Drug Administration (FDA) clearance. The more widespread availability of diagnostic testing and the introduction of assays for the detection of antimicrobial resistance in some countries have increased the knowledge of M. genitalium epidemiology and disease associations as well as the extent of antimicrobial resistance worldwide.
Data on M. genitalium infections are limited in comparison to those for other Mycoplasma species largely due to its more recent discovery, extremely slow growth (generation time of about 16 h), and demanding cultivation requirements. By as early as the 1970s, there was suspicion that a noncultivable microorganism could be responsible for nongonococcal urethritis (NGU) that was not due to chlamydia or ureaplasma because of the clinical improvement in men treated with tetracyclines. One possibility that was considered was that such infections could be due to spiroplasma-like organisms. This consideration prompted Taylor-Robinson and colleagues to collect urethral swabs from 13 men with urethritis and send them to the National Institutes of Health Mycoplasma Laboratory in 1980, where the samples were incubated for several weeks in SP4 medium, originally developed for the cultivation of spiroplasmas. Ultimately, two strains of a glucose-fermenting mycoplasma, designated G37 and M30, were grown in vitro. These strains were shown by serological methods to be distinct from all other known human Mycoplasma species, and the organism came to be known as M. genitalium (3, 4).
Experimental urogenital inoculation of male and female nonhuman primates in the mid-1980s demonstrated that M. genitalium could induce an inflammatory response, result in bloodstream dissemination, and elicit a humoral immune response (5), further supporting the potential role of M. genitalium as an important agent of human disease. More recent studies using female pig-tailed macaques inoculated with M. genitalium intracervically demonstrated that infection can persist for 16 to 18 weeks in the midst of a mild to moderate inflammatory response, ascend into the upper reproductive tract, and induce a humoral immune response directed against the MgpB and MgpC adhesins (6).
PATHOGENESIS AND EPIDEMIOLOGY OF M. GENITALIUM INFECTIONS
Pathogenesis.
While it is beyond the scope of this review to discuss the pathogenesis and epidemiology of M. genitalium infections in depth, reviews of these topics provide further information (7–10). M. genitalium has a terminal organelle comprised of numerous cytoskeletal proteins, heat shock proteins, metabolic enzymes, and cytadherence-related proteins, one of which is adhesin P140, also known as the MgPa adhesin. This adhesin is similar to the P1 adhesin of Mycoplasma pneumoniae, which facilitates its attachment to and invasion of host epithelial cells and stimulation of the immune response (8). M. genitalium also attaches to spermatozoa and erythrocytes; invades epithelial cells, with evidence of nuclear localization; and exhibits gliding motility (7, 11). A family of repetitive DNA elements with homology to the MgPa adhesin gene provides a reservoir for sequence variation. Antigenic variation helps the organism avoid the host immune response, optimize adhesion, and adapt to diverse host microenvironments, thus facilitating persistent infection (12). Other important pathogenic mechanisms that may elicit a host inflammatory response, cause cell death, and/or help the organism avoid host immunity include the secretion of nucleases, membrane lipoproteins, and various other enzymes and biofilm formation (8, 13).
Epidemiology.
Only after the introduction of nucleic acid amplification tests (NAATs) in the early 1990s did the epidemiology of M. genitalium infections and disease associations become apparent. M. genitalium is often present as a coinfection, especially with chlamydia and trichomoniasis (14). Due to the likelihood of coinfections and the asymptomatic nature of many M. genitalium infections (15–17), these factors must be considered when evaluating data for an association or causality between M. genitalium and a particular clinical condition. M. genitalium is typically detected in the lower urogenital tract of about 1% to 6.4% of men and women in the general population (16, 18–22). However, it has a much higher prevalence (sometimes exceeding 20%) in persons with urogenital symptoms and in those who attend clinics providing sexual health services (22–24). The prevalence of M. genitalium is similar to that of chlamydia and exceeds that of Neisseria gonorrhoeae (14).
M. GENITALIUM DISEASE ASSOCIATIONS
Infections in men.
M. genitalium causes 20 to 35% of NGU cases (7, 25) and has been detected in up to 40% of men with recurrent/persisting NGU after treatment with doxycycline or azithromycin (7). Sparse evidence suggests that M. genitalium may cause acute epididymitis, but there is insufficient evidence that it causes prostatitis or male infertility (7, 10, 25). Data regarding the association of M. genitalium with proctitis are inconsistent (26–28). One recent study and a meta-analysis concluded that there was an association with rectal symptoms in men who have sex with men (MSM) (29, 30). M. genitalium is also significantly more common in MSM with rectal symptoms who are HIV positive than in those who are HIV negative (31). Balanoposthitis has been associated with M. genitalium in one study (32).
Infections in women.
Bacterial vaginosis (BV) has been linked to an increased risk of sexually transmitted infections (STIs) (33). The association of M. genitalium with BV is controversial, as in some studies, an association has been shown (33, 34), while in others, a relationship has not been demonstrated (35–37). However, BV has been associated with cervicitis, endometritis, salpingitis, and tubal factor infertility (38), and M. genitalium may play a role in each of these conditions (7, 16, 25). M. genitalium can be detected in 10% to 30% of women with mucopurulent cervicitis (15). Many women with M. genitalium infection have vaginal discharge and may also manifest with dysuria; however, it is unclear to what extent M. genitalium is involved in the acute urethral syndrome (7, 10, 16, 39). M. genitalium may cause pelvic inflammatory disease (PID) based on (i) its ability to adhere to Fallopian tube epithelial cells and cause ciliary damage in organ culture, (iii) the production of endometritis and salpingitis after experimental inoculation of nonhuman primates, (iii) its presence in endometrial biopsy specimens in most women undergoing this procedure who harbor the organism in the cervix, (iv) its detection in the Fallopian tube of a woman with laparoscopically diagnosed acute salpingitis, (v) the association of tubal factor infertility with a previous infection with M. genitalium, and (vi) the demonstration of M. genitalium antibody responses in women with acute PID (7, 40). The association of M. genitalium with PID is also indicated by its significant associations with cervicitis (7, 25, 36, 41, 42) and endometritis (43–45). The prevalence of M. genitalium among women with PID ranges from 4% to 22% (15, 46) but may be as high as 60% in women with postabortal PID (47). Significant associations were also found between M. genitalium and cervicitis and PID in a recent meta-analysis (48). However, some studies were unable to prove a significant association between M. genitalium infection and progression to PID (49–52). Serological and direct evidence showed an association of M. genitalium infection with tubal infertility (48, 53–57), but some investigations have been unable to demonstrate this association (58). Data on M. genitalium and the risk of ectopic pregnancy are scant, and the results are mixed (59, 60).
Infections in pregnant women and neonates.
Earlier studies on the prevalence of M. genitalium in pregnant women found that it was low, around 1% (61), but more recent studies have reported a higher prevalence of 12% to 18% (62–65). Two meta-analyses reported that M. genitalium infection was associated with preterm birth (48, 66), one of which also found an association with stillbirth (48). Other studies did not find an association of M. genitalium with preterm birth or pregnancy loss (67–70). Infants born to women with M. genitalium infection had lower birth weights according to one study, but otherwise, there were no significant differences in pregnancy outcomes among infected women and those who had no other STIs (64).
Infections in persons living with HIV.
A meta-analysis reported a consistent association of M. genitalium with HIV infection (71). Women who had a high load of M. genitalium were more likely to shed HIV than were M. genitalium-negative women and, thus, could facilitate viral transmission (10, 72). In a study of HIV-positive pregnant women, 21.4% had M. genitalium detected by PCR, and the infection was associated with a significantly higher HIV RNA load in the plasma. However, no association between exposure to M. genitalium and the perinatal transmission of HIV was detected (73).
Extragenital infections.
M. genitalium has been detected in the rectum of 1% to 26% of MSM and 3% of women, mostly asymptomatic, and, less commonly, in the pharynx (15, 29, 74). There have also been reports of its detection in the joint fluid of patients with arthritis, but there is insufficient evidence to conclude that it is a significant cause of septic arthritis or a postinfectious arthritic syndrome (75, 76). An adult with urethritis and conjunctivitis tested positive for M. genitalium in the urine and conjunctiva, but no formal studies have been performed to determine how often this occurs (77).
WHEN TO PERFORM DIAGNOSTIC TESTING FOR M. GENITALIUM
Escalating antimicrobial resistance and the associated treatment failures contributed to the CDC adding diagnostic recommendations for M. genitalium in their 2015 treatment guidelines for STIs and updating them in 2021 (15). Men with recurrent NGU and women with recurrent cervicitis should be tested for M. genitalium using an FDA-cleared molecular-based test. However, these guidelines stop short of recommending testing for primary cases of NGU in men or cervicitis in women. Testing should be considered for women with PID. If testing to detect rRNA gene mutations conferring macrolide resistance (macrolide-resistance-associated mutations [MRMs]) is available, this should also be done, and the results should be used to guide therapy. Resistance testing for persons who test positive for M. genitalium was also recommended in current European and British guidelines (17, 78). However, resistance testing in the United States is rarely performed due to the lack of FDA-cleared commercial assays and the very limited availability of laboratory-developed assays in reference facilities such as the University of Alabama at Birmingham (UAB) Diagnostic Mycoplasma Laboratory. Therefore, even if M. genitalium is confirmed by a molecular-based test in a symptomatic person in the United States, treatment will likely still be empirical. A comparison of recommendations for M. genitalium testing and treatment from the United States, Europe, and the United Kingdom is provided in Table 1. CDC guidelines do not recommend M. genitalium testing for primary cases of NGU or cervicitis, which makes it challenging to manage patients when they first return with recurrent NGU or cervicitis since M. genitalium results would not be available. Moreover, we believe that testing for M. genitalium should also be considered for men with primary cases of NGU and women with cervicitis in whom no other microbial etiology is identified during initial testing.
TABLE 1.
Comparison of current recommendations from three published guidelines for the performance of diagnostic tests for M. genitalium
| NAAT testing recommendation | Indication in guideline |
||
|---|---|---|---|
| United Statesc | United Kingdomd | Europee | |
| Asymptomatic persons | No | No | Not specified |
| Test of cure following treatment of persons who test positive | Only if resistance testing is not available and moxifloxacin cannot be used | Yes, no earlier than 3 wks | Consider, no earlier than 3 wks |
| Sex partners of symptomatic persons who test positive | Yes | Yes | Yes |
| Resistance testing on all persons who test positive | Yes, if availablea | Yes (macrolide) | Yes (macrolide)b |
| Men | |||
| NGU | Yes, if persistent or recurrent after treatment | Yes | Yes |
| Epididymo-orchitis | Not specified | Consider | Yes, if aged <50 yrs |
| Sexually acquired proctitis | Consider if symptoms persist after treatment | Consider | Yes, if C. trachomatis and N. gonorrhoeae are excluded |
| Women | |||
| Mucopurulent cervicitis | Yes, if recurrent | Consider | Yes |
| Symptoms of PID | Consider | Yes | Yes |
| Intermenstrual or postcoital bleeding | Not specified | Yes | Yes |
| Dysuria with no other known etiology | Not specified | Not specified | Yes |
| Sexually acquired proctitis | Consider if symptoms persist after treatment | Consider | Yes, if C. trachomatis and N. gonorrhoeae are excluded |
| Termination of pregnancy | Not specified | Not specified | Consider |
In 2023, there are no FDA-cleared NAATs that incorporate resistance testing that are commercially available in the United States. Resistance testing for macrolides and fluoroquinolones is available through the UAB Diagnostic Mycoplasma Laboratory in Birmingham, AL, using validated laboratory-developed PCR assays.
Testing for parC mutations is not indicated on a routine basis but is useful after moxifloxacin treatment failure.
See reference 15.
See reference 78.
See reference 17.
European and British guidelines recommend the testing of all men with urethritis and to consider retesting following antimicrobial treatment to ensure microbiological cure after at least 3 weeks have passed. In patients who clinically respond to treatment, test results could be negative after a week of treatment, but in some instances, a person who eventually fails treatment may appear negative on a transient basis due to a decreased bacterial load (79), hence the need to wait at least 3 weeks before retesting. All of these guidelines support the testing of sexual contacts of persons who are positive for M. genitalium. The testing of pregnant women and neonates is not addressed in any of these guidelines. None of these guidelines recommends the screening of asymptomatic persons for M. genitalium, in contrast to recommendations for Chlamydia trachomatis and N. gonorrhoeae (15). Major arguments against screening for M. genitalium include (i) the lack of knowledge about the consequences associated with asymptomatic M. genitalium infection, (ii) the lack of studies that have addressed whether the screening of women reduces the likelihood of tubal infertility or PID, (iii) whether screening would be cost-effective, and (iv) limited treatment options that can be associated with serious adverse effects (80, 81). Given that persons who test positive for M. genitalium may be likely to acquire other STIs, concomitant testing for Chlamydia trachomatis, Neisseria gonorrhoeae, Trichomonas vaginalis, syphilis, and HIV is indicated (17).
DIAGNOSTIC APPROACHES TO LABORATORY DETECTION OF M. GENITALIUM
Culture.
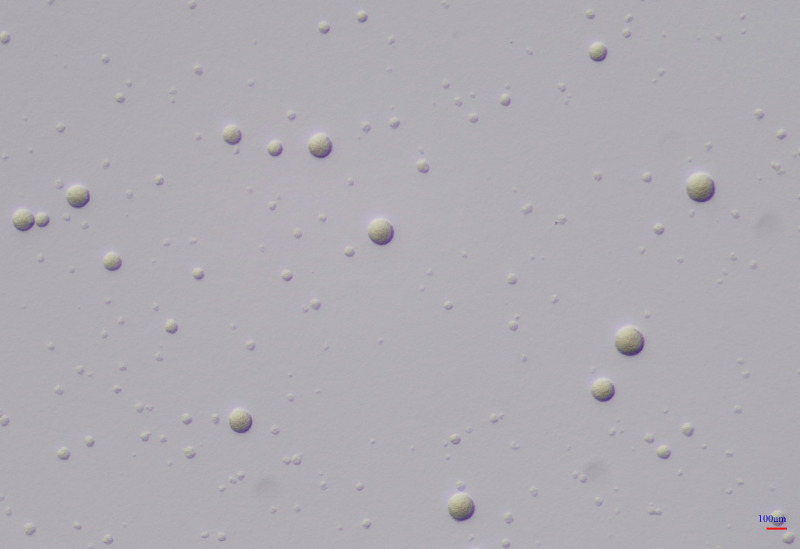
The method of direct specimen inoculation into SP4 medium used by Tully and coworkers to obtain the first clinical isolates of M. genitalium from male urethral swabs (3) proved to be insensitive, leading to the development of alternative methods. Jensen et al. used a strategy in which Vero cell cultures were inoculated with specimens and PCR was used to monitor bacterial multiplication (82, 83). When PCR showed evidence of multiplication, the cell culture was subcultured onto mycoplasma medium. The UAB Diagnostic Mycoplasma Laboratory has been successful in isolating M. genitalium from urine specimens by utilizing a modification of the direct method of Tully et al., inoculating concentrated urine into SP4 broth, incubating the sample for several weeks, and performing periodic blind subcultures in broth and on SP4 agar. Successful cultures have been performed on urine specimens from persons who had a high load of M. genitalium as determined by quantitative PCR (qPCR). However, given the difficulty in direct culture, we adapted a Vero cell-PCR method (84) in order to obtain more clinical isolates. This enhanced culture technique is still very time-consuming, requiring several weeks to obtain colonies on agar, as shown in Fig. 1, and is quite expensive. While culture is not useful for diagnostic purposes, it has a role in obtaining isolates for antimicrobial resistance testing and research. Once clinical isolates have been grown on agar, they can be preserved for prolonged periods by freezing them at −80°C in suitable transport medium or SP4 culture medium. Storage at −20°C is not recommended as it may result in a loss of viability.
FIG 1.
Spherical M. genitalium colonies (G37) growing on SP4 agar after 10 days of incubation at 37°C in 5% CO2 in air. Magnification, ×100.
Serology.
Prior to the availability of NAAT technology, attempts were made to develop serological tests as an indirect means of identifying M. genitalium infections, but problems occurred because of cross-reactions of the reagents with M. pneumoniae. In addition to enzyme immunoassays (85), a microimmunofluorescence assay (86) was developed to detect antibody responses in men with NGU (87) and women with salpingitis (88). This method was proven to be rapid, reproducible, sensitive, and specific, with less cross-reactivity with M. pneumoniae than other methods. A serological assay for M. genitalium using lipid-associated membrane proteins (LAMPs) as antigens was used in combination with Western immunoblotting to assess the immunoreactivity of women who were culture positive for M. genitalium (89). Immunoblotting using a cloned fragment of the C-terminal part of the MgPa adhesin as the antigen has been used to assess the serological response to M. genitalium in infertile women (90, 91). However, with the development of NAATs, serological testing for M. genitalium has never gained widespread use, and no FDA-cleared assays are available.
MOLECULAR-BASED DETECTION
Early molecular-based assays.
Nonamplified DNA probes for M. genitalium were developed in the 1980s but were abandoned once the more sensitive NAAT technology was introduced in the early 1990s. NAATs are now considered the recommended methods for the detection of M. genitalium in clinical specimens (15).
Early NAATs amplified various fragments of the MgPa adhesin gene and enabled the detection of as few as 10 organisms (92, 93). A variety of PCR assays using the 16S rRNA gene target followed (94–96). Other molecular-based formats included RNA-based transcription-mediated amplification (TMA) (97–99). Real-time PCR assays that allowed the measurement of bacterial loads in clinical specimens using targets such as the MgPa gene, 16S rRNA, 23S rRNA, the 115-kDa-protein-encoding gene, and gap, encoding glyceraldehyde-3-phosphate dehydrogenase (96, 99–105), were also developed. The MgPa gene was initially believed to be an ideal target since the operon contains repetitive elements. However, there are valid concerns about the use of the MgPa target relating to the variations in the repetitive elements among M. genitalium strains and the 98% identity of the 16S rRNA genes of M. pneumoniae and M. genitalium.
Commercial molecular-based assays.
As the use of laboratory-developed NAATs began to verify that M. genitalium is an important cause of STIs, commercial development soon followed. NAATs for the detection of M. genitalium alone or multiplexed with C. trachomatis, N. gonorrhoeae, T. vaginalis, Mycoplasma hominis, Ureaplasma parvum, and Ureaplasma urealyticum in various combinations have been developed using several amplification and detection techniques, gene targets, and platforms. Some products are designed to work only on instrumentation provided by the manufacturer (e.g., Hologic Panther), while others can be used on various thermocycler platforms from different manufacturers. Some are fully automated and incorporate nucleic acid extraction with amplification, while others require a separate extraction step.
Table 2 summarizes several commercial NAATs for M. genitalium detection that have received the CE-IVD mark (approved Communauté Européenne marking on in vitro diagnostic medical devices) and the three assays that are FDA cleared. The CE-IVD mark indicates the manufacturer’s declaration that the product complies with relevant European protective legislation and that it has been assessed to meet high safety, health, and environmental protection requirements, and it ensures free movement within the European Union. However, the CE-IVD mark does not require the validation of clinical performance against a reference standard as is required by the FDA (106). Several independent evaluations of commercial NAATs have been published, but many NAATs have not been extensively or rigorously compared with laboratory-developed reference methods using large sample sizes or with one another, so their true performance characteristics are not well documented.
TABLE 2.
Examples of commercial molecular tests for detection of M. genitalium that have been CE marked and/or FDA cleareda
| Product | Manufacturer(s); platform(s) | Method(s) of detection | Pathogen(s) detected | M. genitalium target(s) | Detection of macrolide resistance mutationsb | Regulatory status | Selected reference(s) |
|---|---|---|---|---|---|---|---|
| Aptima Mycoplasma genitalium | Hologic, Marlborough, MA; Panther | TMA, HPA, PCR | M. genitalium | 16S rRNA | No | FDA, CE | 107–110, 113, 174, 200–207 |
| cobas TV/MG detection | Roche Molecular Systems, Pleasanton, CA; cobas 6800/8800 | PCR | M. genitalium, T. vaginalis | 16S rRNA | No | FDA, CE | 112 – 114 |
| Alinity m STI | Abbott Molecular Diagnostics, Des Plaines, IL; Alinity m | PCR | N. gonorrhoeae, C. trachomatis, T. vaginalis, M. genitalium | rRNA | No | FDA, CE | 208 |
| STI Plus ELITe MGB | ELITechGroup, Bothell, WA; ELITe InGenius | PCR | N. gonorrhoeae, C. trachomatis, T. vaginalis, M. genitalium | 23S rRNA | No | CE | 114 |
| Allplex STI Essential | Seegene, Seoul, South Korea; Bio-Rad Cfx96 | PCR | N. gonorrhoeae, C. trachomatis, M. genitalium, T. vaginalis, M. hominis, U. urealyticum, U. parvum | mgpA | No | CE | 108, 109, 114, 178, 207 |
| Bio-Rad Dx CT/NG/MG | Bio-Rad, Marnes-la-Coquette, France; Bio-Rad Dx real-time system | PCR | N. gonorrhoeae, C. trachomatis, M. genitalium | mgpA | No | CE | 2, 116, 117 |
| Hyplex STD mycoplasma test system | Amplex Biosystems, Lich, Germany | PCR | M. genitalium, M. hominis, U. urealyticum, U. parvum | mgpA | No | CE | 2, 118 |
| N. gonorrhoeae/C. trachomatis/M. genitalium/T. vaginalis Real-TM | Sacace Biotechnologies, Como, Italy | PCR | N. gonorrhoeae, C. trachomatis, T. vaginalis, M. genitalium | Undisclosed | No | CE | 114 |
| FTD Urethritis Plus | Fast-Track Diagnostics, Luxembourg | PCR | N. gonorrhoeae, C. trachomatis, T. vaginalis, M. genitalium, U. urealyticum, U. parvum, M. hominis | Undisclosed | No | CE | 2, 114 |
| AmpliSens N. gonorrhoeae/C. trachomatis/M. genitalium/T. vaginalis-Multiprime FRT assay | InterLabService, Moscow, Russia | PCR | N. gonorrhoeae, C. trachomatis, T. vaginalis, M. genitalium | gyrB | No | CE | 2, 111 |
| S-DiaMGTV | Diagenode, Liege, Belgium | PCR | M. genitalium, T. vaginalis | mgpA, MG219 | No | CE | 2, 115, 209, 210 |
| ResistancePlus MG FleXible | SpeeDx/Cepheid, Sydney, Australia, Cepheid GeneXpert, Sunnyvale, CA | PlexZyme, PlexPrime technology, PCR | M. genitalium | mgpA | Yes, 4 mutations, A2058G/C/T and A2059C | CE | 167, 177–180 |
| ResistancePlus MG | SpeeDx, Sydney, Australia | PlexZyme, PlexPrime technology, PCR | M. genitalium | mgpA | Yes, 5 mutations, A2058G/C/T and A2059C/G | CE | 109, 110, 123, 133, 136, 154, 170–175, 203 |
| Macrolide-R/MG ELITe MGB | ELITechGroup, Bothell, WA; ELITe InGenius | PCR | M. genitalium | 23S rRNA | Yes, 5 mutations, A2058G/C/T and A2059G/C, and WT | CE | 160, 180 |
| Allplex MG & AziR | Seegene, Seoul, South Korea; Bio-Rad Cfx 96 | PCR | M. genitalium | mgpA | Yes, 6 mutations, A2058G/C/T and A2059G/C/T | CE | 109, 180 |
| S-DiaMGRes | Diagenode, Liege, Belgium | PCR | M. genitalium | MG219 | Yes, 5 mutations, A2058G/C/T and A2059G/C | CE | 173, 176, 208 |
| Real-Accurate TVMGres | Pathofinder, Maastricht, Netherlands | PCR | M. genitalium, T. vaginalis | mgpA | Yes, 4 mutations, A2058G/C/T and A2059G | CE | 173 |
Adapted from Table 4 in reference 211. TMA, transcription-mediated amplification; HPA, hybridization protection assay; CE-IVD, approved Communauté Européenne marking on in vitro diagnostic medical devices; FDA, U.S. Food and Drug Administration; WT, wild type.
23S rRNA mutations that are known to confer macrolide resistance in M. genitalium (E. coli numbering).
The FDA-cleared Aptima MG (Mycoplasma genitalium) (Hologic, Inc.), cobas TV (T. vaginalis)/MG (Roche Molecular Systems), and Alinity m STI (Abbott Molecular Diagnostics) assays are fully automated, complete sample-to-result assays that provide qualitative results without the need for a separate DNA or RNA isolation procedure. All three assays are classified as high-complexity assays by the Clinical Laboratory Improvement Amendments (CLIA). The Aptima MG assay, which targets the 16S rRNA gene, has been more extensively evaluated than any other commercial NAAT. This assay involves target capture, a TMA step, and target detection by hybridization with complementary probes linked to chemiluminescent labels. The TMA step involves a target nucleic acid amplification method using RNA polymerase and reverse transcription (RT) to produce an amplicon from the specific target. Clinical specimens are run on the Panther instrument. According to the manufacturer’s specifications, the analytical sensitivity is 0.01 CFU/mL. The Aptima MG assay was FDA cleared in 2019 following the performance of a multicenter clinical study involving 11,556 specimens obtained from 1,778 women and 1,583 men across the United States (107). As the Aptima MG assay is based on the detection of 16S rRNA genes, for which there are many more copies than targets of DNA-based assays, it has been proven to have higher clinical sensitivity than other laboratory-developed PCRs or CE-marked commercially sold DNA-based tests in numerous studies (Table 2). In one study, the Allplex STI Essential assay had a 41.7% clinical sensitivity compared with the Aptima MG assay (108). No additional reference method was included to resolve discrepancies. Another comparison of Aptima MG with the Allplex STI Essential, ResistancePlus MG (RPMG) (SpeeDx), and Allplex MG & AziR (Seegene) assays reported that the Aptima MG assay detected the most positive specimens, all of which were confirmed by a national reference laboratory. Allplex STI Essential had a sensitivity of 85.7%, while SpeeDx and Allplex MG & AziR had sensitivities of 71.4%. The number of positive samples (7) was very small (109). Another study (110) reported 100% sensitivity for Aptima MG versus 83.3% for RPMG. The AmpliSens N. gonorrhoeae/C. trachomatis/M. genitalium/T. vaginalis-Multiprime FRT assay (InterLabService) had an overall sensitivity of 81.9% compared to the Aptima MG assay (111). Very few false-positive results have been reported with any of the commercial NAATs, so specificities are not a problem.
cobas TV/MG is a real-time PCR assay that detects M. genitalium and T. vaginalis. The M. genitalium target is the 16S rRNA gene. This assay was FDA cleared in 2020. Specimens are run on the cobas 6800 or 8800 instrument. The cobas TV/MG assay was evaluated in comparison to the UAB laboratory-developed PCR targeting the 23S rRNA gene (105), a second laboratory-developed assay targeting mgpA, and the Aptima MG assay in a multicenter study of 2,150 asymptomatic and symptomatic men and women (112). Patients were considered to be infected with M. genitalium if two or more tests were positive. There were 59 women and 60 men who were positive for M. genitalium. The sensitivity of cobas TV/MG for the detection of M. genitalium in vaginal swabs was 96.6%, whereas for female urine and endocervical specimens, the sensitivity ranged from 83.1% to 86.4%. For male urine and meatal swab specimens, the sensitivities were 100% and 85%, respectively. The specificities for all specimen types from men and women ranged from 96% to 99.8%, respectively.
Another study that compared these two FDA-cleared assays directly with one another used the Allplex STI Essential assay and/or Sanger sequencing to resolve discrepancies. Both assays detected M. genitalium in 6 specimens (113). An evaluation of the clinical performances of 4 multiplex PCR kits, STI Plus ELITe MGB (ELITech Group), Allplex STI Essential, FTD Urethritis Plus (Fast-Track Diagnostics), and N. gonorrhoeae/C. trachomatis/M. genitalium/T. vaginalis Real-TM (Sarace Biotechnologies), compared the results of the kits to those obtained with cobas TV/MG (114). All 4 PCRs had low percent positive agreement (PPA) values for the detection of M. genitalium, ranging from 63.3% to 74.1%, in comparison to cobas TV/MG. A low negative percent agreement (NPA) value was found for the Fast-Track kit, whereas the other PCRs had NPAs exceeding 98.1%.
The Abbott Alinity m STI assay was FDA cleared in May 2022. It is a random-access multiplex reverse transcription-PCR (RT-PCR) assay that detects N. gonorrhoeae, C. trachomatis, M. genitalium, and T. vaginalis. The gene target for M. genitalium is rRNA. Specimens are processed on the Alinity m instrument. The M. genitalium limit of detection according to the product specifications is 33 genome equivalents/assay. This assay was compared to a laboratory-developed PCR assay with extraction on the Abbott 2000sp system. Alinity m detected 22 positive samples, versus 18 with the laboratory-developed PCR assay. An additional 167 Alinity-positive samples were tested using the S-DiaMGRes kit (Diagenode), and 159 were positive by both assays, with an additional 8 positive samples being detected only by Alinity m. This study showed the general comparability of the Alinity m, S-DiaMGRes, and laboratory-developed PCR assays (115).
Several other CE-IVD-marked commercial PCRs have been compared to one another or to laboratory-developed PCRs in mostly small studies (Table 2). A French study (116) compared the Bio-Rad Dx CT/NG (N. gonorrhoeae)/MG assay to a laboratory-developed PCR assay. The clinical sensitivity was 100%, and the specificity ranged from 99.5% to 100%, based on the specimen type. However, there were only 11 specimens that had M. genitalium DNA detected, 1 of which was deemed a false-positive specimen. A comparison of Bio-Rad Dx CT/NG/MG PCR with a laboratory-developed PCR assay found concordance for 7 positive samples (117). The clinical sensitivity of the S-DiaMGTV kit (Diagenode) was 91.3%, and the specificity was 95.3%, compared to a laboratory-developed PCR assay (115). In an evaluation of the Hyplex ST mycoplasma test (Amplex Biosystems) using laboratory-developed PCR as a reference, 11 urine specimens were confirmed to be positive for M. genitalium by both tests (118).
SPECIMEN TYPES FOR MOLECULAR-BASED ASSAYS
Universal recommendations regarding the optimum specimen types and collection and processing procedures for NAATs for M. genitalium are not possible because of variations in nucleic acid extraction methods, some of which are incorporated into automated sample-to-result systems that do not require a separate extraction procedure. Additionally, the use of specific transport medium required for the chosen test method, how specimens are stored and transported, performance characteristics, types of specimens validated for individual assays, and manufacturer instructions of individual commercial kits can vary (7). Treatment guidelines for M. genitalium infections (15, 17, 78) provide practical guidance for specimen choice based on clinical studies comparing various specimen types and the ones that have been validated and approved for use with commercial FDA-cleared assays.
For men, first-catch urine is the diagnostic specimen of choice (15, 17, 78). It is easy to collect and store and is more sensitive than urethral swabs, which can be quite uncomfortable for the patient being swabbed (119). One study reported that self-collected meatal swabs performed better than first-void urine (120), yet another study reported that meatal swabs were inferior to urethral swabs (121). Rectal samples may be obtained, especially in MSM, if there are symptoms of proctitis and no other microbial etiology has been identified. Omitting rectal swabs in MSM may result in missing as many as 70% of M. genitalium infections (27, 122). There are no recommendations for obtaining and testing oropharyngeal samples since carriage is unusual and not associated with symptoms (17).
For women, vaginal swabs have performed better than first-catch urine and are the recommended specimens of choice (15, 17, 78). This recommendation reflects the relative median bacterial loads (genomes per milliliter) that have been reported for these body sites (vaginal swabs, 6,950 genomes/mL; endocervical swabs, 3,970 genomes/mL; urine, 826 genomes/mL) (123). Vaginal swabs can be self-obtained, making specimen collection even easier. Endocervical swabs are an alternate specimen type (98, 124, 125). Some studies suggested obtaining both vaginal and endocervical swabs for maximum sensitivity for detection (121). First-catch urine has been shown to have a lower sensitivity in some but not all studies (111, 116, 126) and is not a preferred specimen for women (15, 17, 78). Rectal swabs may also be obtained for M. genitalium testing in women with proctitis if no other etiology is apparent.
The three FDA-cleared NAATs are each approved for the testing of male or female urine specimens and female vaginal and endocervical swabs. The Aptima MG package insert also states that it can be used to test male urethral and self-collected penile meatal swabs. The cobas assay can also be used to test self- or clinician-collected penile meatal swabs. None of these NAATs are approved for the testing of rectal swabs for M. genitalium. All three NAATs provide a swab and a urine collection kit with appropriate transport media.
M. genitalium has been detected in epididymal fluid, prostatic biopsy specimens, and Fallopian tube specimens (7). If clinical conditions warrant and appropriate specimens are available, they can be processed, but there have been no systematic studies performed to evaluate the performance of FDA-cleared NAATs on them. The processing of specimens before freezing, particularly those from women, appears to be important for maintaining a high sensitivity of detection (7).
REPORTING RESULTS, VALIDATION, VERIFICATION, AND PROFICIENCY TESTING
The UAB Diagnostic Mycoplasma Laboratory provides PCR results for the detection of M. genitalium in clinical specimens with the simultaneous reporting of the presence of MRMs using a laboratory-developed PCR assay. An example of a laboratory report for a specimen in which M. genitalium and MRMs were detected is as follows:
DNA was extracted from the sample and amplified by real-time PCR. A 266-bp fragment was defined by primers and probes targeting the 23S rRNA gene of M. genitalium. Melting curve analysis of the amplicon revealed a macrolide-resistance-associated mutation in this gene. This is a qualitative test for the presence or absence of M. genitalium. A negative result does not rule out the presence of M. genitalium DNA in concentrations below the level of detection by the assay.
Federal regulations also require laboratories to append a disclaimer to laboratory-developed test reports indicating that the test was developed and its performance characteristics were determined by [laboratory name] and that is has not been cleared or approved by the FDA (127). The UAB Diagnostic Mycoplasma Laboratory adds an additional statement that our laboratory-developed M. genitalium PCR is used for clinical purposes and should not be regarded as investigational or for research.
Laboratories in the United States using laboratory-developed assays or any commercially developed assay components designated research use only (RUO), analyte-specific reagents (ASRs), or other non-FDA-cleared products to detect M. genitalium alone or in conjunction with other microorganisms and/or antimicrobial resistance markers must perform and document their internal validations to determine accuracy and precision for all specimen types, transport media, nucleic acid isolation procedures, instrumentation, and reportable and normal reference ranges (if applicable) and independently assess the analytical sensitivity (limit of detection) and specificity of their gene target and methods. This documentation and maintaining copies of the data collected are CLIA requirements (127), and the results of this performance evaluation must be compared to those of another assay for the analyte or to results obtained by a different laboratory.
If using unmodified nonwaived (high-complexity) NAATs that are FDA cleared for in vitro diagnostic use, the process is less complex. It is necessary to verify that the test performs as expected using characteristics described in the manufacturer’s package insert. These characteristics include accuracy, precision, and reportable and normal reference ranges (if applicable). For qualitative FDA-cleared NAATs to detect M. genitalium, the reportable range is not applicable, and the normal reference range would be “negative” or “not detected.” In order to utilize the test and report results to guide patient management, the laboratory must also follow the manufacturer’s recommendations for quality control, interpretation of results, and quality assessment as required by the CLIA. Laboratories that subscribe to the College of American Pathologists (CAP) proficiency testing program can procure three 1-mL liquid specimens (MGEN analyte) to be tested by NAATs for the presence of M. genitalium twice yearly. There is also an independent international quality assessment program that is focused primarily on molecular testing for infectious diseases (QCMD [Quality Control for Molecular Diagnostics]) that provides analytes for M. genitalium proficiency testing.
EMERGING ANTIMICROBIAL RESISTANCE AND THE NEED FOR IN VITRO TESTING
The first clinical isolates of M. genitalium were susceptible to tetracyclines, macrolides, and quinolones. Other agents such as the streptogramin pristinamycin, which is available in France and Australia, also show in vitro activity. Antimicrobial resistance, accompanied by treatment failures, has become widespread in M. genitalium after the first report of macrolide resistance in 2006 (128). Macrolide-resistance-associated mutations (MRMs) (A2058G, A2059G, A2058C, A2059C, and A2058T [Escherichia coli numbering]) in domain V of the 23S rRNA gene that constitute the peptidyl transferase loop region of the ribosome result in a substantial elevation of the MIC and treatment failure with azithromycin due to interference with binding in the 50S bacterial ribosome (81, 129). The selection of MRMs in vivo following azithromycin treatment at 1 g may occur approximately 10 to 12% of the time (130, 131).
MRMs can now be detected in 30 to 100% of M. genitalium specimens, increasing from 10% before 2010 to 51% by 2016 to 2017 (17, 23, 24, 132, 133). Resistance rates may vary geographically and for different patient populations. Macrolide resistance rates are generally higher in the Western Pacific (68%) and the Americas (67%) than in Europe (134). A prevalence of MRMs of 48% was reported for M. genitalium specimens in a multicenter clinical study in the United States (14). Studies in Alabama found MRMs in 74.1% of M. genitalium-positive specimens obtained from HIV-positive MSM attending an STI clinic (24) and in 60.7% of heterosexual African American couples (23). A 67.6% prevalence of M. genitalium macrolide resistance was reported in HIV-positive MSM in France (135). An increased frequency of MRMs in MSM compared to heterosexual men has also been described in Australia (89.7% versus 50%) (136). A recent study (137) reported multiple phenotypic combinations of M. genitalium infecting women at different anatomic sites, with vaginal swab sampling yielding the highest sensitivity for the identification of women with infections by isolates harboring MRMs. A 2015 meta-analysis found an azithromycin cure rate of only 67% after 2009 (138). Treatment failure with pristinamycin, likely caused by a 23S rRNA mutation at position 2062 (E. coli numbering), was recently described in a French patient (139).
Resistance to the second-line treatment moxifloxacin has also increased substantially over the past several years, often in association with treatment failures. While moxifloxacin was able to achieve 100% cure in studies performed prior to 2010, this rate dropped to 89% from 2010 onward (140). There are several possible mutations in parC, parE, gyrA, and gyrB of the quinolone-resistance-determining regions (QRDRs) of the bacterial chromosome that can confer resistance (141). Moxifloxacin treatment failures for M. genitalium infections are typically associated with mutations in the parC gene, which encodes subunit 1 of topoisomerase IV (142). In particular, parC mutations at position 83 (nucleotide G248) and position 87 (nucleotide G259) are involved. The S83I amino acid substitution caused by the single nucleotide polymorphism (SNP) G248T leads to moxifloxacin treatment failure in approximately two-thirds of cases (143–145) and is also associated with phenotypic elevations of MICs (146). Other mutations in parC are less common and appear to be less likely to cause moxifloxacin treatment failure (17, 23, 145). Mutations in gyrA, which encodes subunit 1 of DNA gyrase, are not predictive of treatment failure unless they occur concomitantly with mutations in parC (17). In a meta-analysis (134), the overall prevalence of mutations associated with moxifloxacin resistance was 8% in patients from the Asia-Pacific region (17). Data from Alabama also demonstrated the presence of parC mutations known to be associated with treatment failure in 29.6% of HIV-positive MSM who were PCR positive for M. genitalium (24) but in only 11.1% of heterosexuals (23). Samples from 24% of MSM and 11.1% of heterosexual men and women had evidence of multidrug resistance involving both macrolides and fluoroquinolones according to these two studies. According to a Japanese study of 982 MSM, 89.6% of M. genitalium isolates had MRMs, and 68.3% were fluoroquinolone resistant (147). The high rates of fluoroquinolone resistance mutations in asymptomatic MSM in Australia (148) and Belgium (149) are particularly concerning. In a Chinese study of more than 1,800 men with urethritis, almost 90% of M. genitalium isolates were resistant to both macrolides and fluoroquinolones (150).
The clinical response to doxycycline in patients infected with M. genitalium can be poor, about 30 to 40%, despite fairly low MICs (≤2 μg/mL), and the microbiological response may be even lower (29.6%) (17, 129). Tetracycline resistance (MIC = 32 μg/mL) has been described in a clinical isolate of M. genitalium obtained from an immunocompromised patient who had failed treatment for urethritis with multiple different regimens, including doxycycline (151). The tetM sequence was not detected in this isolate, and there were no polymorphisms in the 16S rRNA gene in the region known to be responsible for tetracycline resistance (152). Preliminary findings indicate that the mechanism may be related to active efflux (L. Xiao, unpublished data). The extent of tetracycline resistance in M. genitalium is unknown since culture is rarely performed, there are few opportunities to perform MIC testing on clinical isolates, and molecular-based tests are rarely available.
The increasing occurrence of mutations that confer resistance to macrolides and/or quinolones in M. genitalium underscores the need for accurate laboratory methods to determine susceptibility and/or resistance in vitro for epidemiological purposes as well as to guide antimicrobial selection aimed at improving patient outcomes as well as curbing the spread of resistance (81). Such resistance-guided therapeutic approaches, often utilizing a combination of and/or sequential antimicrobial agents, have been implemented in Australia, where resistance testing is more readily available than in some other countries (153–155). In addition to MIC determinations using broth microdilution techniques, there are a number of molecular-based methods to measure the inhibitory activity of antimicrobial agents as well as indirect methods aimed at identifying whether mutations conferring resistance are present or absent.
PHENOTYPIC MIC METHODS
Broth microdilution.
The extremely slow growth of M. genitalium makes it difficult to perform traditional phenotypic MIC testing by broth microdilution on clinical isolates according to guidelines for human mycoplasmas from the Clinical and Laboratory Standards Institute (CLSI) (156). Therefore, methods for MIC determination have not been validated and standardized for M. genitalium. However, once a strain has been passaged to become laboratory adapted and can be grown on agar and in SP4 broth to a suitable concentration of 104 to 105 color change units (CCU)/mL, MIC measurement can be undertaken in a 96-well microtiter plate with some modifications. For recent clinical isolates that are not fully laboratory adapted, it is advisable to thaw a frozen stock culture and allow it to preincubate in SP4 broth until it just begins to turn from red to orange before inoculation into a microtiter plate containing 2-fold dilutions of antimicrobial agents. Plates are incubated with 5% CO2 in air at 37°C. Once the growth control changes from red to yellow, indicative of bacterial growth and glucose utilization, the MICs can be read as the lowest concentration of the antimicrobial that prevents the color change from occurring. The time until the MIC can be read in approximately 7 to 14 days. There are no designated quality control strains with defined MIC reference ranges for M. genitalium to use for MIC testing. The UAB Diagnostic Mycoplasma Laboratory uses M. hominis ATCC 23114, for which MIC reference ranges for several drugs have been determined, as a surrogate. M. genitalium strain G37 (ATCC 33530) is also tested with each MIC assay because it has predictable MICs for tetracycline, azithromycin, and moxifloxacin. As there are no published MIC breakpoints, the actual MICs can be reported. For drugs that have published breakpoints for other Mycoplasma species, the results can be reported in comparison to those values. The availability of clinical isolates with known resistance to macrolides and quinolones enables more critical evaluations of new agents with different mechanisms of action or altered chemical structures that can overcome known resistance mechanisms that may not exhibit cross-resistance (151). Broth microdilution can also be used to determine the minimum bactericidal concentrations (MBCs) of antimicrobial agents for M. genitalium (157). MIC testing for M. genitalium is performed primarily for the evaluation of new antimicrobial agents, but this method has also been shown to be useful in some cases of multidrug-resistant M. genitalium infections where there was treatment failure to search for potential alternative treatments.
Vero cell-PCR technique.
Hamasuna and coworkers described in vitro MIC determinations for M. genitalium that could not be grown axenically using a Vero cell culture monitored by quantitative PCR (qPCR) (84). This technique was modified by Jensen et al. and was used to determine the MICs of a new investigational agent, gepotidacin, and comparator agents (158). An inoculum of M. genitalium at 5,000 genome equivalents, as determined by qPCR, is placed into a Vero cell culture containing serial 2-fold dilutions of the antimicrobial agent and incubated for 3 weeks. At this time, the cells and supernatant are harvested for each antimicrobial concentration, and the growth of M. genitalium is determined by qPCR. The MIC is defined as the minimum concentration of the antimicrobial agent causing 99% inhibition of growth compared to the growth control without the antimicrobial. Jensen et al. also adapted the technique for the determination of MBCs (158). The Vero cell culture-based MIC assay is not internationally validated or approved for diagnostic use and may produce MICs that are high for some antimicrobials compared to the broth microdilution assay (84).
GENOTYPIC RESISTANCE TESTING
An alternative approach that can be used to identify antimicrobial resistance quickly in M. genitalium is to test directly for mutations in 23S rRNA that confer macrolide resistance and mutations in the QRDRs that are associated with moxifloxacin resistance. This approach has gained the most widespread use in testing for genetic markers of resistance to macrolides and quinolones since the mutations involved are well known (105, 159–163). Sanger sequencing, which is expensive, time-consuming, and laborious, can be used to identify the presence of the various mutations involved, but other techniques can also be used, such as real-time PCR and coupled techniques, including pyrosequencing, high-resolution melting analysis (HRMA), different primers targeting the wild type (WT) and mutants, labeled hydrolysis probes, and fluorescence resonance energy transfer (FRET) probes coupled with melting curve analysis (14, 133, 161–166). The development of assays that can detect M. genitalium in clinical specimens and simultaneously test for antimicrobial resistance mutations is the most logical approach from both cost- and time-saving perspectives (167).
The UAB Diagnostic Mycoplasma Laboratory currently utilizes a real-time PCR assay (MGMR) that simultaneously detects M. genitalium and the MRMs A2059G, A2058C, A2059C, and A2058T (E. coli numbering) based on a modification of the PCR assay described by Touati et al. (161). This test has been internally validated and is available for diagnostic testing, thus enabling the simultaneous reporting of the presence of M. genitalium and MRMs. The procedure involves the amplification of a fragment from the M. genitalium 23S rRNA gene and the hybridization of two FRET probes covering MRMs coupled with melting curve analysis. This assay was shown to be superior to PCR targeting gap (103) and enables the simultaneous detection of M. genitalium and MRMs in urine and vaginal specimens in a single procedure (105). Considering the high rRNA copy numbers (~1,000-fold higher than the genome copy numbers) in a living cell, the adaptation of this method to a one-step reverse transcription-quantitative PCR (RT-qPCR) method that uses RNA as the input template further improves the sensitivity of this assay, which is comparable to those of the current commercial assays (Xiao, unpublished).
A similar one-step RT-qPCR assay for detecting quinolone-resistance-associated mutations (MGQR) was also developed and internally validated in the UAB Diagnostic Mycoplasma Laboratory, targeting the parC gene and using two hybridization FRET probes covering the mutations reported to be associated with quinolone resistance (Xiao, unpublished). This PCR assay detects any point mutations occurring in the probe region but does not differentiate them. Multiplexed with the MGMR assay, the combined MGMRQR assay enables the triple tasks of the simultaneous detection of M. genitalium and the identification of macrolide and quinolone resistance in one test and is available for clinical diagnostic testing through our reference laboratory.
COMMERCIAL METHODS FOR MOLECULAR DETECTION OF MACROLIDE RESISTANCE
Since the presence of 23S rRNA mutations is an excellent proxy for macrolide resistance and azithromycin treatment failure (166, 168), the detection of the presence of these mutations at positions 2058 and 2059 (E. coli numbering) is sufficient for epidemiological and diagnostic purposes. Several commercial NAATs that can detect M. genitalium and MRMs have been developed, CE marked, and sold in several countries (Table 2). However, no assays capable of resistance detection in M. genitalium are FDA cleared, and none of them are available in the United States. Commercial kits for the detection of MRMs have been compared to other commercial assays as well as laboratory-developed assays, including real-time quantitative PCR, real-time PCR with HRMA, FRET PCR, and 23S rRNA sequencing, to detect macrolide resistance-associated mutations. Commercial products will generally produce results within 4 h, depending on the assay and platform used (133, 160, 167, 169–174). Discussion is limited to those commercial products that have obtained regulatory approval through CE-IVD.
The most extensively studied commercial assay for macrolide resistance is RPMG. It is CE-IVD marked and also cleared for use in Australia and Canada. This PCR utilizes PlexZyme/PlexPrime technology for the detection of M. genitalium (mgpA) and the five predominant 23S rRNA macrolide-resistance-associated mutations. It is validated for the testing of male and female urine, anal, rectal, cervical, endocervical, vaginal, and urethral specimens as well as preextracted samples. RPMG PCR can be run on several different platforms, including the Roche LightCycler 480 II and the Applied Biosystems ABI 7500 platforms. Its performance was generally satisfactory in published studies (Table 2), but the clinical sensitivities for M. genitalium detection are usually lower than those of other commercial assays such as Aptima MG (110). RPMG performed in a satisfactory manner on specimens collected using Hologic Aptima MG collection kits compared to standard specimen collection in phosphate-buffered saline, so it can be used for reflexive testing to allow resistance-guided therapy (175). In a comparison of RPMG with S-DiaMRes, the Real-Accurate TVMGres assay (Pathofinder), and amplification and sequencing of the 23S rRNA gene in a collection of M. genitalium-positive specimens, the PPA value for the detection of the organism for each of the three commercial kits was between 94.8% and 96.4%. The sensitivities and specificities for the detection of MRMs were also similar, ranging from 95% to 100% and 94.6% to 97.3%, respectively (173). Additional evaluations of RPMG (133, 138, 169, 172, 176) reported generally similar results regarding assay sensitivity and specificity for the detection of MRMs compared to reference methods, as did another study involving the S-DiaMRes assay (176). A Canadian study found superior rates of detection of M. genitalium by RPMG in self-collected vaginal swabs versus first-void urine samples (110).
RPMG has been adapted for use on the GeneXpert Dx platform (Cepheid), CE marked, and sold as the RPMG FleXible assay. Incorporation into the GeneXpert platform integrates sample purification, nucleic acid amplification, real-time PCR detection of M. genitalium and MRMs, and reporting of results in a random-access format so that single specimens can be analyzed as needed in a point-of-care setting. This is a significant advantage over other assays to detect MRMs, which are restricted to batch testing of various numbers of specimens. The widespread availability of GeneXpert for the detection of other pathogens, including N. gonorrhoeae and C. trachomatis, can greatly facilitate testing for M. genitalium and MRMs on a widespread basis. Testing is quite simple as the user only has to add the MG FleXible reaction mix, the internal control, and the patient sample to the cartridge and then insert it into the GeneXpert system where extraction, amplification, reporting, and interpretation of the results occur automatically based on measurements of fluorescence signals and embedded calculation algorithms. Multiple independent evaluations of the RPMG FleXible cartridge have been reported (167, 177, 178), in which the assay’s performance characteristics were assessed and collections of positive and negative clinical specimens were analyzed. Compared to a reference PCR assay, RPMG FleXible had an overall sensitivity of 96.1% for M. genitalium detection. The specificities were 100% for urine and 98.5% for swab samples, with analytical sensitivities of detection of 157 genomes/mL for the WT and 387 genomes/mL for strains with MRMs. RPMG FleXible had 93% PPA with sequencing for the detection of specimens with MRMs (167). In another study (177), RPMG FleXible had a sensitivity of 96. 8% and a specificity of 100% compared to a laboratory-developed PCR assay. There was 98.9% agreement between RPMG FleXible and RPMG for the detection of MRMs. A comparison of RPMG FleXible to the Allplex STI Essential assay and Sanger sequencing found a PPA of 94.7% and an NPA of 100% for the detection of M. genitalium. For the detection of MRMs in 84 specimens, there was 94.1% sensitivity and 96% specificity for RPMG FleXible (178). A prospective evaluation of RPMG FleXible in comparison to RPMG using the LightCycler 480 II platform found comparable results, with a PPA of 94.1% and an NPA of 95.2% for the detection of M. genitalium. For the detection of MRMs, the PPA was 97.1%, and the NPA was 78.6%, with 5 of 6 discordant samples determined to be WT specimens by RPMG being shown to be positive for MRMs by RPMG FleXible and Sanger sequencing, indicating its improved performance for mutation detection (179). An evaluation of RPMG FleXible was performed in comparison to Allplex MG & AziR (Seegene) and Macrolide-R/MG ELITe MGB (ELITech Group), using a laboratory-developed real-time PCR assay and Sanger sequencing as references (180). The 23S PCR Macrolide-R/MG ELITe MGB kit (ELITech) is a FRET-probe-based CE-IVD-marked kit that can be run on the ELITe InGenius platform using fully automated extraction, amplification, and result analysis processes. This qPCR assay consists of a hydrolysis probe for the detection of M. genitalium and a melt probe for the discrimination of mutants from the WT based on melting curve analysis. Allplex MG & AziR is a CE-IVD-marked kit that detects M. genitalium and 6 MRMs in the 23S rRNA gene. These investigators reported very similar performances, with an overall agreement of 94.6% to 97.6% for the three assays for the detection of M. genitalium. The sensitivities for MRM detection were 74.5% for Allplex MG & AziR, 96.2% for Macrolide-R/MG ELITe MGB, and 92.8% for RPMG FleXible. The specificities for the three kits were very similar, 97.4% to 97.6%. For the detection of MRMs, the overall agreements with Sanger sequencing were 84.5% to 96.7%. The clinical sensitivities for MRM detection were 76.5% for Allplex MG & AziR, 96.2% for Macrolide-R/MG ELITe MGB, and 92.8% for RPMG FleXible. Allplex MG & AziR missed the detection of MRMs in 14.4% of M. genitalium-positive specimens, which is a major concern. The RPMG FleXible assay had a high percentage (12.1%) of invalid, internal control, or system errors, consistent with other reports (167, 179), indicating that additional modifications and improvements to the analysis software are needed, although this has not been a universal problem with this assay (177, 178).
Another study evaluated the performance characteristics of 23S PCR Macrolide-R/MG ELITe MGB PCR by testing a collection of specimens for M. genitalium by a laboratory-developed PCR assay and Sanger sequencing to detect MRMs (160). 23S PCR Macrolide-R/MG ELITe MGB detected 6 more positive specimens. The detection of MRMs was achieved in 99.5% of specimens in comparison to sequencing.
RPMG, S-DiaMGRes, Real-Accurate TVMGres, and Allplex MG & AziR do not include a 23S rRNA amplification control, which could lead to false-susceptible results in the event of a failure to amplify the 23S rRNA gene, suggesting that the addition of a 23S rRNA amplification control would be beneficial (173). Alternatively, it has been recommended that in cases where a specimen is positive for M. genitalium but no MRMs are detected and the assay quantification cycle (Cq) values for mgpA are high, laboratories should consider reporting the MRM result as indeterminate (167). One explanation for the discrepancies that sometimes occur between organism and MRM detection when the organism load is low is differences in amplification efficacies between the mgpA target and the 23S rRNA gene where only probes for MRMs are present (167). Moreover, none of the current versions of commercial NAATs that detect MRMs include the A2062G mutation, which has been associated with treatment failures with both pristinamycin and josamycin and has been detected in between 2.3% and 7.7% of M. genitalium strains (180). Thus, sequence analysis of specimens from persons with macrolide treatment failure would be required to identify this mutation.
COMMERCIAL METHODS FOR MOLECULAR DETECTION OF QUINOLONE RESISTANCE
Over the past decade, there have been several descriptions of PCR assays used to detect mutations that may confer quinolone resistance in order to determine the prevalence of this resistance in various geographic regions (24, 141, 181–183). Data from some of these assays were validated by comparison to results obtained by Sanger sequencing. The performance of these assays has varied. Some detected mutations in both parC and gyrA, while others detected only parC mutations. In an Australian study (141), investigators developed two PCRs using molecular beacon and dual-hybridization assays that detected SNPs responsible for amino acid substitution S83I at nucleotide G248 using differences in melting peak temperatures from codons with WT sequences and compared their results with those of Sanger sequencing. They reported 100% agreement with sequencing and readily distinguished specimens containing parC mutations of potential clinical significance, but the sensitivity was reduced in specimens with low organism loads, resulting in only 70% of specimens yielding useful data.
The SpeeDx MG parC (beta) PCR assay detects M. genitalium and 6 common mutations associated with quinolone resistance: SNPs coding for S83R/I (combined), S83N, and D87 (D87Y, D87N, and D87H [combined]) (184). This assay was evaluated using 171 M. genitalium-positive specimens, and the results were compared to those of parC Sanger sequencing (185). There was 94.7% agreement with the results of Sanger sequencing, and 21 M. genitalium isolates with S83 or D87 mutations were detected. However, 9 were shown to have S83 or D87 mutations by the SpeeDx assay but were negative by sequencing. This yielded an overall PPA of 100% and an overall NPA of 93.6%. Because the SpeeDx MG parC (beta) assay detects only groups of parC mutations, it is not possible to distinguish among several of them, and not all mutations may play a role in treatment failure. It also does not assess mutations in gyrA.
A study from Russia (186) evaluated the AmpliSens Mycoplasma genitalium ML/FQ-Resist-FL assay (Central Research Institute of Epidemiology), using Sanger sequencing for validation. This product includes one PCR that detects M. genitalium using the gyrB gene target and the WT 23S rRNA gene. Another PCR detects an internal control and WT parC. This PCR detected potential mutations for macrolide and fluoroquinolone resistance with 100% and 92.3% sensitivities and 99.2% and 100% specificities, respectively. They found macrolide resistance in 13.8%, quinolone resistance in 9%, and dual resistance in 5.5% of the specimens. However, this assay does not identify any specific mutations associated with macrolide or quinolone resistance, which may be problematic since not all mutations, particularly with regard to fluoroquinolones, may be associated with treatment failures. Organism loads of <1,000 genome equivalents/mL may not be sufficient to yield a positive signal, and a quarter of specimens may have such lower loads. Another Russian study reported similar findings with M. genitalium-ML/FQ-Resist-FL, with a specificity of 100% and a sensitivity of 90.7% (176). Other companies have also developed assays to detect M. genitalium and quinolone resistance. These include the Allplex MG and MoxiR assay (Seegene), which detects 6 common mutations associated with quinolone resistance; MGMO qPCR, which detects 4 parC mutations using a TaqMan-based PCR kit (NYtor Research Institute, Nijmegen, Netherlands); and the LightMix modular parC kit (TibMolBiol, Berlin, Germany).
Currently, there are no specific recommendations for attempting to determine the presence of quinolone resistance in urogenital specimens that test positive for M. genitalium to guide therapy except in cases of moxifloxacin treatment failures (17). This position is logical because of the relatively low frequencies of quinolone resistance and treatment failures in many populations and the fact that no commercially produced assays have been thoroughly validated externally or received the CE-IVD mark or FDA clearance, and they are of very limited availability. There is also uncertainty that remains regarding which specific QRDRs are associated most closely with treatment failures and the fact that when amino acid substitutions such as S83I that are most strongly associated with moxifloxacin treatment failures are identified, there are no other treatment options available in some countries. A final concern is the cost, which can be considerable when sequencing multiple gene regions (187). Despite these hindrances, some investigators have advocated that NAATs capable of detecting the S83I mutation and the absence of mutations (WT sequences) should be used to guide therapy, help maintain antimicrobial stewardship, and individualize the need for a test of cure, depending on the results of such testing and patient outcomes (188). The absence of parC mutations at positions 83 and 87 in the QRDRs is known to be highly predictive of moxifloxacin cure (145). All of the above-described assays for detecting macrolide and fluoroquinolone resistance-associated mutations use DNA as the template. During treatment, DNA from dead organisms could be collected, which may lead to false-positive results. RNA templates isolated from live bacteria would help to solve this problem.
STRAIN TYPING FOR EPIDEMIOLOGICAL PURPOSES
Strain typing of human mycoplasmas is generally available only in specialized research or reference laboratories. Even though it is not used for routine clinical purposes, strain typing can be valuable to understand M. genitalium transmission within populations and between individuals, to characterize antimicrobial-resistant strains, and to distinguish relapsing versus persistent infections in persons who experience treatment failures (189, 190). A high rate of concordance of M. genitalium genotypes has been noted for sexual partners, and strain typing data support the concordance of M. genitalium genotypes in infected couples (17).
The first description of M. genitalium strain typing was reported in 1999 based on whole-genome fingerprinting involving the selective amplification of restriction fragments obtained from purified DNA of cultured strains (191). However, the dearth of clinical isolates necessitated the development of molecular-based typing methods that can be performed directly on a clinical specimen without the need for culture. Molecular typing methods used for M. genitalium are based on PCR amplification of a specific genomic locus followed by sequencing or restriction fragment length polymorphism (RFLP) analysis. Specific methods have included short-tandem-repeat (STR) analysis of a surface-localized lipoprotein gene, MG309 (192); analysis of single nucleotide polymorphisms (SNPs) in the rRNA operon (192); analysis of the mgpB gene (101, 193); and RFLP analysis of the mgpC gene (193). A PCR assay based on the mgpB gene identified 29 different sequences from 52 unrelated patients and also demonstrated that 79 M. genitalium-positive specimens from 19 couples had concordant sequence types, indicative of sexual transmission. Those researchers were also able to demonstrate the acquisition of new strains concordantly in sexually active couples. The occurrence of a few large clusters indicated either the spread of certain strains or the presence of particularly common sequence types that were detected in specimens from different countries. These findings confirmed the heterogeneity of M. genitalium isolates from clinical specimens.
Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) based on the variation in the copy number of tandemly repeated sequences (VNTRs) located in different genetic loci was used to identify 18 loci in the M. genitalium G37 reference strain containing STRs and determined that mgpB SNPs and MG309 STRs complement one another, thereby providing greater typing efficiency, and may be more accurate for the determination of defined genetic relationships. The addition of MG307 STRs and MG338 STRs may be potentially useful for studies of the sexual transmission of M. genitalium (194). However, the use of the mgpC gene for RFLP analysis may be unsuitable for genotyping since this region of the genome is undergoing rapid sequence shifts due to homologous recombination with MgPar repeats (194). A French study (195) compared MLVA with mgpB SNP analysis and the combination analysis of VNTR 309 and mgpB. This study also confirmed the heterogeneity of M. genitalium strains and concluded that MLVA was too discriminatory to be used in studies of sexual networks, recommending that mgpB analysis should be used for general epidemiological studies and that the combination of MG309 STR and mgpB SNP methods should be used for sexual-network studies of M. genitalium infection. A study from Australia used mgpB and MG309 PCR and Sanger sequencing to determine that antimicrobial resistance mutations occurred both within and across genotypes from samples that were geographically and temporally diverse, suggesting that while there may be some ongoing transmission of some strains, the spread of resistance is more likely occurring de novo rather than by clonal spread (189). Similar findings were reported in Spain (190). A study from Germany concluded that while there is considerable diversity in the M. genitalium types circulating, there was no correlation between specific types and either macrolide or quinolone resistance and that when resistance occurred, it arose de novo during treatment (196). Plummer et al. (187) described a custom amplicon next-generation sequencing (NGS) approach suitable for NGS platforms such as Illumina MiSeq to detect resistance mutations and sequence types in M. genitalium associated with the 23S rRNA, parC, gyrA, and mgpB genes. They used this novel technique to characterize clinical specimens positive for M. genitalium and compared the results to those of Sanger sequencing. They concluded that their assay could provide cost-effective means of monitoring resistance mutations and genotyping for epidemiological purposes, which can be scaled according to the desired target types and numbers of specimens. More than 20 studies applying various molecular typing methods to evaluate M. genitalium in different patient populations have been published, and their results have been summarized in a recent review (197).
MANAGEMENT OF M. GENITALIUM INFECTIONS
Current CDC STI guidelines for the treatment of M. genitalium describe various regimens of doxycycline, azithromycin, and moxifloxacin, depending on whether resistance has been documented and the clinical syndrome being treated (15). The occurrence of macrolide- and fluoroquinolone-resistant infections presents a significant challenge for both laboratory diagnosis and management. To combat resistant infections, other treatment guidelines also describe various sequences and combinations of these and other antimicrobials that may differ somewhat from those in the CDC guidelines (17, 78). Some agents with limited availability, such as pristinamycin, spectinomycin, and sitafloxacin, have shown some success, and newer ones, such as omadacycline, lefamulin, gepotidacin, solithromycin, and zoliflodacin, may be potential alternatives based on in vitro activities, but there are no clinical trial data available to date to support their use (198, 199). There are also different recommendations regarding the need for a test of cure following treatment, relating to whether or not resistance mutations have been documented and the appropriate time frame to do so. Current CDC treatment guidelines provide thorough discussions of M. genitalium treatment options, so it is not necessary to delve further into treatment here.
FUTURE DIRECTIONS FOR LABORATORY DIAGNOSIS OF M. GENITALIUM INFECTIONS AND UNMET NEEDS
The much less stringent regulatory requirements for diagnostic testing for countries outside the United States have led to the proliferation of many NAATs for M. genitalium detection and testing for mutations associated with antimicrobial resistance. Although there have been several publications describing their performance in comparison to reference methods and one another, most studies have yielded fairly small numbers of specimens positive for M. genitalium. Additional studies are needed to determine which commercial NAATs are of the most value and can be recommended for use with the greatest confidence. Whether the companies developing commercial tests will decide to conduct the rigorous studies necessary to achieve FDA clearance is uncertain. However, the recent availability of three FDA-cleared automated sample-to-answer NAATs is expected to drive the more widespread utilization of molecular testing in the United States and also influence future recommendations for testing and managing such infections. Testing should be employed in research settings for studies designed to provide evidence-based indications for diagnostic testing and to improve the understanding of sequelae of symptomatic and asymptomatic infections in women, such as PID, to justify screening programs. Additional studies are also needed to improve our understanding of the contribution of M. genitalium to adverse outcomes for pregnant women and neonates. Even though culture will never be used as a diagnostic method of choice, it is important for more research and reference laboratories to attempt culture in order to acquire more clinical isolates. The availability of current isolates from different parts of the world is important so that they can be used for studies of the epidemiology, cell biology, and pathogenesis of M. genitalium infections and to assess the in vitro activities of new antimicrobial agents. There is a particular need to obtain clinical isolates that exhibit multidrug resistance in order to improve our understanding of resistance mechanisms and the clinical implications of various mutations in the QRDRs. The development of new antimicrobial agents that do not exhibit cross-resistance with existing drugs is extremely important as more infections may become untreatable should dual quinolone and macrolide resistance continue to increase in prevalence. A limited number of M. genitalium isolates have now been shown to exhibit tetracycline resistance, apparently mediated by active efflux. The extent to which such isolates occur in various patient populations is unknown but merits further investigation. The clinical significance of the 23S rRNA mutation A2062G (E. coli numbering) also needs to be investigated in order to determine whether this mutation needs to be included in macrolide resistance assays. Even though we now have the ability to detect infections within a few hours by NAATs, there are still no FDA-cleared commercially available tests in the United States that can detect macrolide or quinolone resistance, and this is a major shortcoming as it prevents the ability to perform resistance-guided therapy unless specimens are submitted to a reference laboratory and tested by a validated laboratory-developed test. The benefit of resistance-guided therapy to reduce treatment failure and help control the spread of antimicrobial resistance is extremely important. A sample-to-answer test that combines a sensitive and specific technique such as TMA with the rapid detection of macrolide and fluoroquinolone resistance in a single assay is certainly needed for diagnostic use as well as epidemiological studies. A true point-of-care test available at a reasonable cost would be especially beneficial. Finally, global surveillance to monitor changes in the prevalence of macrolide and quinolone resistance over time and investigate whether such resistance is arising de novo during treatment or as a result of clonal spread through strain genotyping will be essential.
Contributor Information
Ken B. Waites, Email: kwaites@uabmc.edu.
Romney M. Humphries, Vanderbilt University Medical Center
REFERENCES
- 1.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States. US Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. www.cdc.gov/DrugResistance/Biggest-Threats.html. [Google Scholar]
- 2.Munson E. 2017. Molecular diagnostics update for the emerging (if not already widespread) sexually transmitted infection agent Mycoplasma genitalium: just about ready for prime time. J Clin Microbiol 55:2894–2902. doi: 10.1128/JCM.00818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tully JG, Taylor-Robinson D, Cole RM, Rose DL. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288–1291. doi: 10.1016/s0140-6736(81)92461-2. [DOI] [PubMed] [Google Scholar]
- 4.Tully JG, Taylor-Robinson D, Rose DL, Cole RM, Bove JM. 1983. Mycoplasma genitalium, a new species from the human urogenital tract. Int J Syst Bacteriol 33:387–396. doi: 10.1099/00207713-33-2-387. [DOI] [Google Scholar]
- 5.Tully JG, Taylor-Robinson D, Rose DL, Furr PM, Graham CE, Barile MF. 1986. Urogenital challenge of primate species with Mycoplasma genitalium and characteristics of infection induced in chimpanzees. J Infect Dis 153:1046–1054. doi: 10.1093/infdis/153.6.1046. [DOI] [PubMed] [Google Scholar]
- 6.Aguila LKT, Patton DL, Gornalusse GG, Vojtech LN, Murnane RD, Wood GE. 2022. Ascending reproductive tract infection in pig-tailed macaques inoculated with Mycoplasma genitalium. Infect Immun 90:e00131-22. doi: 10.1128/iai.00131-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor-Robinson D, Jensen JS. 2011. Mycoplasma genitalium: from chrysalis to multicolored butterfly. Clin Microbiol Rev 24:498–514. doi: 10.1128/CMR.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yueyue W, Feichen X, Yixuan X, Lu L, Yiwen C, Xiaoxing Y. 2022. Pathogenicity and virulence of Mycoplasma genitalium: unraveling Ariadne’s thread. Virulence 13:1161–1183. doi: 10.1080/21505594.2022.2095741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGowin CL, Totten PA. 2017. The unique microbiology and molecular pathogenesis of Mycoplasma genitalium. J Infect Dis 216:S382–S388. doi: 10.1093/infdis/jix172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daley GM, Russell DB, Tabrizi SN, McBride J. 2014. Mycoplasma genitalium: a review. Int J STD AIDS 25:475–487. doi: 10.1177/0956462413515196. [DOI] [PubMed] [Google Scholar]
- 11.Jensen JS. 2004. Mycoplasma genitalium: the aetilogical agent of urethritis and other sexually transmitted diseases. J Eur Acad Dermatol Venereol 18:1–11. doi: 10.1111/j.1468-3083.2004.00923.x. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Mancuso M, Williams JA, Van Der Pol B, Fortenberry JD, Jia Q, Myers L, Martin DH. 2014. Extensive variation and rapid shift of the MG192 sequence in Mycoplasma genitalium strains from patients with chronic infection. Infect Immun 82:1326–1334. doi: 10.1128/IAI.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daubenspeck JM, Totten AH, Needham J, Feng M, Balish MF, Atkinson TP, Dybvig K. 2020. Mycoplasma genitalium biofilms contain poly-GlcNAc and contribute to antibiotic resistance. Front Microbiol 11:585524. doi: 10.3389/fmicb.2020.585524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Getman D, Jiang A, O’Donnell M, Cohen S. 2016. Mycoplasma genitalium prevalence, coinfection, and macrolide antibiotic resistance frequency in a multicenter clinical study cohort in the United States. J Clin Microbiol 54:2278–2283. doi: 10.1128/JCM.01053-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Workowski KA, Bachmann LH, Chan PA, Johnston CM, Muzny CA, Park I, Reno H, Zenilman JM, Bolan GA. 2021. Sexually transmitted infections treatment guidelines, 2021. MMWR Recommend Rep 70:1–187. doi: 10.15585/mmwr.rr7004a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latimer RL, Vodstrcil LA, Plummer EL, Doyle M, Murray GL, Fairley CK, Bodiyabadu K, Read TRH, Kaiser M, Mokany E, Guy R, Chow EPF, Bradshaw C. 2022. The clinical indications for testing women for Mycoplasma genitalium. Sex Transm Infect 98:277–285. doi: 10.1136/sextrans-2020-054818. [DOI] [PubMed] [Google Scholar]
- 17.Jensen JS, Cusini M, Gomberg M, Moi H, Wilson J, Unemo M. 2022. 2021 European guideline on the management of Mycoplasma genitalium infections. J Eur Acad Dermatol Venereol 36:641–650. doi: 10.1111/jdv.17972. [DOI] [PubMed] [Google Scholar]
- 18.Xuan Y, Wei LX, Hong X, Zhu XY, Dong SH, Yan QY, Wang LH, Wang B. 2021. A meta-analysis on the infection rates on Mycoplasma genitalium in the genitourinary tract of different populations in China. Zhonghua Liu Xing Bing Xue Za Zhi 42:335–342. doi: 10.3760/cma.j.cn112338-20200530-00791. (In Chinese.) [DOI] [PubMed] [Google Scholar]
- 19.Sonnenberg P, Ison CA, Clifton S, Field N, Tanton C, Soldan K, Beddows S, Alexander S, Khanom R, Saunders P, Copas AJ, Wellings K, Mercer CH, Johnson AM. 2015. Epidemiology of Mycoplasma genitalium in British men and women aged 16-44 years: evidence from the third National Survey of Sexual Attitudes and Lifestyles (Natsal-3). Int J Epidemiol 44:1982–1994. doi: 10.1093/ije/dyv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torrone EA, Kruszon-Moran D, Philips C, Morris MR, Bowden KE, Papp J, Bachmann LH, Weinstock H, Kersh EN. 2021. Prevalence of urogenital Mycoplasma genitalium infection, United States, 2017 to 2018. Sex Transm Dis 48:e160–e162. doi: 10.1097/OLQ.0000000000001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumann L, Cina M, Egli-Gany D, Goutaki M, Halbeisen FS, Lohrer GR, Ali H, Scott P, Low N. 2018. Prevalence of Mycoplasma genitalium in different population groups: systematic review and meta-analysis. Sex Transm Infect 94:255–262. doi: 10.1136/sextrans-2017-053384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahlangu MP, Muller EE, Venter JME, Maseko DV, Kularatne RS. 2019. The prevalence of Mycoplasma genitalium and association with human immunodeficiency virus infection in symptomatic patients, Johannesburg, South Africa, 2007-2014. Sex Transm Dis 46:395–399. doi: 10.1097/OLQ.0000000000000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Waites KB, Van Der Pol B, Aaron KJ, Hook EW, III, Geisler WM. 2019. Mycoplasma genitalium infections with macrolide and fluoroquinolone resistance-associated mutations in heterosexual African American couples in Alabama. Sex Transm Dis 46:18–24. doi: 10.1097/OLQ.0000000000000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dionne-Odom J, Geisler WM, Aaron KJ, Waites KB, Westfall AO, Van Der Pol B, Xiao L. 2018. High prevalence of multidrug-resistant Mycoplasma genitalium in human immunodeficiency virus-infected men who have sex with men in Alabama. Clin Infect Dis 66:796–798. doi: 10.1093/cid/cix853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manhart LE, Geisler WM, Bradshaw CS, Jensen JS, Martin DH. 2022. Weighing potential benefits and harms of Mycoplasma genitalium testing and treatment approaches. Emerg Infect Dis 28:e220094. doi: 10.3201/eid2808.220094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soni S, Alexander S, Verlander N, Saunders P, Richardson D, Fisher M, Ison C. 2010. The prevalence of urethral and rectal Mycoplasma genitalium and its associations in men who have sex with men attending a genitourinary medicine clinic. Sex Transm Infect 86:21–24. doi: 10.1136/sti.2009.038190. [DOI] [PubMed] [Google Scholar]
- 27.Read TRH, Murray GL, Danielewski JA, Fairley CK, Doyle M, Worthington K, Su J, Mokany E, Tan LT, Lee D, Vodstrcil LA, Chow EPF, Garland SM, Chen MY, Bradshaw CS. 2019. Symptoms, sites, and significance of Mycoplasma genitalium in men who have sex with men. Emerg Infect Dis 25:719–727. doi: 10.3201/eid2504.181258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis SC, Kent CK, Klausner JD, Rauch L, Kohn R, Hardick A, Gaydos CA. 2008. Prevalence of rectal Trichomonas vaginalis and Mycoplasma genitalium in male patients at the San Francisco STD clinic, 2005-2006. Sex Transm Dis 35:797–800. doi: 10.1097/OLQ.0b013e318177ec39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latimer RL, Shilling HS, Vodstrcil LA, Machalek DA, Fairley CK, Chow EPF, Read TR, Bradshaw CS. 2020. Prevalence of Mycoplasma genitalium by anatomical site in men who have sex with men: a systematic review and meta-analysis. Sex Transm Infect 96:563–570. doi: 10.1136/sextrans-2019-054310. [DOI] [PubMed] [Google Scholar]
- 30.Chow EPF, Lee D, Bond S, Fairley CK, Maddaford K, Wigan R, Fehler G, Lange SA, De Petra V, Bissessor M, Bradshaw CS, Howden BP, Hocking JS, Williamson DA, Chen MY. 2021. Nonclassical pathogens as causative agents of proctitis in men who have sex with men. Open Forum Infect Dis 8:ofab137. doi: 10.1093/ofid/ofab137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bissessor M, Tabrizi SN, Bradshaw CS, Fairley CK, Hocking JS, Garland SM, Twin J, Poljak M, Peel J, Chen MY. 2016. The contribution of Mycoplasma genitalium to the aetiology of sexually acquired infectious proctitis in men who have sex with men. Clin Microbiol Infect 22:260–265. doi: 10.1016/j.cmi.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Horner PJ, Taylor-Robinson D. 2011. Association of Mycoplasma genitalium with balanoposthitis in men with non-gonococcal urethritis. Sex Transm Infect 87:38–40. doi: 10.1136/sti.2010.044487. [DOI] [PubMed] [Google Scholar]
- 33.Nye MB, Harris AB, Pherson AJ, Cartwright CP. 2020. Prevalence of Mycoplasma genitalium infection in women with bacterial vaginosis. BMC Womens Health 20:62. doi: 10.1186/s12905-020-00926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oakeshott P, Aghaizu A, Hay P, Reid F, Kerry S, Atherton H, Simms I, Taylor-Robinson D, Dohn B, Jensen JS. 2010. Is Mycoplasma genitalium in women the “new chlamydia?” A community-based prospective cohort study. Clin Infect Dis 51:1160–1166. doi: 10.1086/656739. [DOI] [PubMed] [Google Scholar]
- 35.Keane FE, Thomas BJ, Gilroy CB, Renton A, Taylor-Robinson D. 2000. The association of Mycoplasma hominis, Ureaplasma urealyticum and Mycoplasma genitalium with bacterial vaginosis: observations on heterosexual women and their male partners. Int J STD AIDS 11:356–360. doi: 10.1258/0956462001916056. [DOI] [PubMed] [Google Scholar]
- 36.Manhart LE, Critchlow CW, Holmes KK, Dutro SM, Eschenbach DA, Stevens CE, Totten PA. 2003. Mucopurulent cervicitis and Mycoplasma genitalium. J Infect Dis 187:650–657. doi: 10.1086/367992. [DOI] [PubMed] [Google Scholar]
- 37.Lillis RA, Martin DH, Nsuami MJ. 2019. Mycoplasma genitalium infections in women attending a sexually transmitted disease clinic in New Orleans. Clin Infect Dis 69:459–465. doi: 10.1093/cid/ciy922. [DOI] [PubMed] [Google Scholar]
- 38.Taylor BD, Darville T, Haggerty CL. 2013. Does bacterial vaginosis cause pelvic inflammatory disease? Sex Transm Dis 40:117–122. doi: 10.1097/OLQ.0b013e31827c5a5b. [DOI] [PubMed] [Google Scholar]
- 39.Olson E, Gupta K, Van Der Pol B, Galbraith JW, Geisler WM. 2021. Mycoplasma genitalium infection in women reporting dysuria: a pilot study and review of the literature. Int J STD AIDS 32:1196–1203. doi: 10.1177/09564624211030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brihmer C, Kallings I, Nord CE, Brundin J. 1987. Salpingitis; aspects of diagnosis and etiology: a 4-year study from a Swedish capital hospital. Eur J Obstet Gynecol Reprod Biol 24:211–220. doi: 10.1016/0028-2243(87)90020-7. [DOI] [PubMed] [Google Scholar]
- 41.Haggerty CL. 2008. Evidence for a role of Mycoplasma genitalium in pelvic inflammatory disease. Curr Opin Infect Dis 21:65–69. doi: 10.1097/QCO.0b013e3282f3d9ac. [DOI] [PubMed] [Google Scholar]
- 42.Falk L, Fredlund H, Jensen JS. 2005. Signs and symptoms of urethritis and cervicitis among women with or without Mycoplasma genitalium or Chlamydia trachomatis infection. Sex Transm Infect 81:73–78. doi: 10.1136/sti.2004.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen CR, Manhart LE, Bukusi EA, Astete S, Brunham RC, Holmes KK, Sinei SK, Bwayo JJ, Totten PA. 2002. Association between Mycoplasma genitalium and acute endometritis. Lancet 359:765–766. doi: 10.1016/S0140-6736(02)07848-0. [DOI] [PubMed] [Google Scholar]
- 44.Taylor BD, Zheng X, O’Connell CM, Wiesenfeld HC, Hillier SL, Darville T. 2018. Risk factors for Mycoplasma genitalium endometritis and incident infection: a secondary data analysis of the T Cell Response against Chlamydia (TRAC) study. Sex Transm Infect 94:414–420. doi: 10.1136/sextrans-2017-053376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haggerty CL, Totten PA, Astete SG, Lee S, Hoferka SL, Kelsey SF, Ness RB. 2008. Failure of cefoxitin and doxycycline to eradicate endometrial Mycoplasma genitalium and the consequence for clinical cure of pelvic inflammatory disease. Sex Transm Infect 84:338–342. doi: 10.1136/sti.2008.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis J, Horner PJ, White PJ. 2020. Incidence of pelvic inflammatory disease associated with Mycoplasma genitalium infection: evidence synthesis of cohort study data. Clin Infect Dis 71:2719–2722. doi: 10.1093/cid/ciaa419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bjartling C, Osser S, Persson K. 2010. The association between Mycoplasma genitalium and pelvic inflammatory disease after termination of pregnancy. BJOG 117:361–364. doi: 10.1111/j.1471-0528.2009.02455.x. [DOI] [PubMed] [Google Scholar]
- 48.Lis R, Rowhani-Rahbar A, Manhart LE. 2015. Mycoplasma genitalium infection and female reproductive tract disease: a meta-analysis. Clin Infect Dis 61:418–426. doi: 10.1093/cid/civ312. [DOI] [PubMed] [Google Scholar]
- 49.Oliphant J, Azariah S. 2016. Pelvic inflammatory disease associated with Chlamydia trachomatis but not Mycoplasma genitalium in New Zealand. Sex Health 13:43–48. doi: 10.1071/SH14238. [DOI] [PubMed] [Google Scholar]
- 50.Hay PE, Kerry SR, Normansell R, Horner PJ, Reid F, Kerry SM, Prime K, Williams E, Simms I, Aghaizu A, Jensen J, Oakeshott P. 2016. Which sexually active young female students are most at risk of pelvic inflammatory disease? A prospective study. Sex Transm Infect 92:63–66. doi: 10.1136/sextrans-2015-052063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cina M, Baumann L, Egli-Gany D, Halbeisen FS, Ali H, Scott P, Low N. 2019. Mycoplasma genitalium incidence, persistence, concordance between partners and progression: systematic review and meta-analysis. Sex Transm Infect 95:328–335. doi: 10.1136/sextrans-2018-053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen CR, Nosek M, Meier A, Astete SG, Iverson-Cabral S, Mugo NR, Totten PA. 2007. Mycoplasma genitalium infection and persistence in a cohort of female sex workers in Nairobi, Kenya. Sex Transm Dis 34:274–279. doi: 10.1097/01.olq.0000237860.61298.54. [DOI] [PubMed] [Google Scholar]
- 53.Rajkumari N, Kaur H, Roy A, Gupta N, Dhaliwal LK, Sethi S. 2015. Association of Mycoplasma genitalium with infertility in North Indian women. Indian J Sex Transm Dis AIDS 36:144–148. doi: 10.4103/0253-7184.167141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alfarraj DA, Somily AM. 2017. Isolation of Mycoplasma genitalium from endocervical swabs of infertile women. Saudi Med J 38:549–552. doi: 10.15537/smj.2017.5.18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adesola Ajani T, Oluwasola TAO, Ajani MA, Bakare RA. 2017. The prevalence of, and risk factors for, Mycoplasma genitalium infection among infertile women in Ibadan: a cross-sectional study. Int J Reprod Biomed 15:613–618. [PMC free article] [PubMed] [Google Scholar]
- 56.Peipert JF, Zhao Q, Schreiber CA, Teal S, Turok DK, Natavio M, Cordon S, Daggy J. 2021. Intrauterine device use, sexually transmitted infections, and fertility: a prospective cohort study. Am J Obstet Gynecol 225:157.e1–157.e9. doi: 10.1016/j.ajog.2021.03.011. [DOI] [PubMed] [Google Scholar]
- 57.Rekha S, Nooren M, Kalyan S, Mohan M, Bharti M, Monika R, Anita S, Kiran M, Vandana N. 2019. Occurrence of Mycoplasma genitalium in the peritoneal fluid of fertile and infertile women with detailed analysis among infertile women. Microb Pathog 129:183–186. doi: 10.1016/j.micpath.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Idahl A, Jurstrand M, Olofsson JI, Fredlund H. 2015. Mycoplasma genitalium serum antibodies in infertile couples and fertile women. Sex Transm Infect 91:589–591. doi: 10.1136/sextrans-2015-052011. [DOI] [PubMed] [Google Scholar]
- 59.Jurstrand M, Jensen JS, Magnuson A, Kamwendo F, Fredlund H. 2007. A serological study of the role of Mycoplasma genitalium in pelvic inflammatory disease and ectopic pregnancy. Sex Transm Infect 83:319–323. doi: 10.1136/sti.2007.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashshi AM, Batwa SA, Kutbi SY, Malibary FA, Batwa M, Refaat B. 2015. Prevalence of 7 sexually transmitted organisms by multiplex real-time PCR in Fallopian tube specimens collected from Saudi women with and without ectopic pregnancy. BMC Infect Dis 15:569. doi: 10.1186/s12879-015-1313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peuchant O, Le Roy C, Desveaux C, Paris A, Asselineau J, Maldonado C, Chene G, Horovitz J, Dallay D, de Barbeyrac B, Bebear C. 2015. Screening for Chlamydia trachomatis, Neisseria gonorrhoeae, and Mycoplasma genitalium should it be integrated into routine pregnancy care in French young pregnant women? Diagn Microbiol Infect Dis 82:14–19. doi: 10.1016/j.diagmicrobio.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 62.Scoullar MJL, Boeuf P, Peach E, Fidelis R, Tokmun K, Melepia P, Elijah A, Bradshaw CS, Fehler G, Siba PM, Erskine S, Mokany E, Kennedy E, Umbers AJ, Luchters S, Robinson LJ, Wong NC, Vallely AJ, Badman SG, Vallely LM, Fowkes FJI, Morgan C, Pomat W, Crabb BS, Beeson JG, Healthy Mothers Healthy Babies Study Team . 2021. Mycoplasma genitalium and other reproductive tract infections in pregnant women, Papua New Guinea, 2015-2017. Emerg Infect Dis 27:894–904. doi: 10.3201/eid2703.201783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smullin CP, Green H, Peters R, Nyemba D, Qayiya Y, Myer L, Klausner J, Joseph Davey D. 2020. Prevalence and incidence of Mycoplasma genitalium in a cohort of HIV-infected and HIV-uninfected pregnant women in Cape Town, South Africa. Sex Transm Infect 96:501–508. doi: 10.1136/sextrans-2019-054255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perin J, Coleman JS, Ronda J, Neibaur E, Gaydos CA, Trent M. 2021. Maternal and fetal outcomes in an observational cohort of women with Mycoplasma genitalium infections. Sex Transm Dis 48:991–996. doi: 10.1097/OLQ.0000000000001569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trent M, Coleman JS, Hardick J, Perin J, Tabacco L, Huettner S, Ronda J, Felter-Wernsdorfer R, Gaydos CA. 2018. Clinical and sexual risk correlates of Mycoplasma genitalium in urban pregnant and non-pregnant young women: cross-sectional outcomes using the baseline data from the Women’s BioHealth Study. Sex Transm Infect 94:411–413. doi: 10.1136/sextrans-2017-053367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frenzer C, Egli-Gany D, Vallely LM, Vallely AJ, Low N. 2022. Adverse pregnancy and perinatal outcomes associated with Mycoplasma genitalium: systematic review and meta-analysis. Sex Transm Infect 98:222–227. doi: 10.1136/sextrans-2021-055352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Averbach SH, Hacker MR, Yiu T, Modest AM, Dimitrakoff J, Ricciotti HA. 2013. Mycoplasma genitalium and preterm delivery at an urban community health center. Int J Gynaecol Obstet 123:54–57. doi: 10.1016/j.ijgo.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramazanzadeh R, Khodabandehloo M, Farhadifar F, Rouhi S, Ahmadi A, Menbari S, Fallahi F, Mirnejad R. 2016. A case-control study on the relationship between Mycoplasma genitalium infection in women with normal pregnancy and spontaneous abortion using polymerase chain reaction. Osong Public Health Res Perspect 7:334–338. doi: 10.1016/j.phrp.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahimkhani M, Mordadi A, Gilanpour M. 2018. Detection of urinary Chlamydia trachomatis, Mycoplasma genitalium and human papilloma virus in the first trimester of pregnancy by PCR method. Ann Clin Microbiol Antimicrob 17:25. doi: 10.1186/s12941-018-0276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Contini C, Rotondo JC, Magagnoli F, Maritati M, Seraceni S, Graziano A, Poggi A, Capucci R, Vesce F, Tognon M, Martini F. 2018. Investigation on silent bacterial infections in specimens from pregnant women affected by spontaneous miscarriage. J Cell Physiol 234:100–107. doi: 10.1002/jcp.26952. [DOI] [PubMed] [Google Scholar]
- 71.Napierala Mavedzenge S, Weiss HA. 2009. Association of Mycoplasma genitalium and HIV infection: a systematic review and meta-analysis. AIDS 23:611–620. doi: 10.1097/QAD.0b013e328323da3e. [DOI] [PubMed] [Google Scholar]
- 72.Sasaki Y, Honda M, Makino M, Sasaki T. 1993. Mycoplasmas stimulate replication of human immunodeficiency virus type 1 through selective activation of CD4+ T lymphocytes. AIDS Res Hum Retroviruses 9:775–780. doi: 10.1089/aid.1993.9.775. [DOI] [PubMed] [Google Scholar]
- 73.Roxby AC, Yuhas K, Farquhar C, Bosire R, Mbori-Ngacha D, Richardson BA, Totten PA, John-Stewart G. 2019. Mycoplasma genitalium infection among HIV-infected pregnant African women and implications for mother-to-child transmission of HIV. AIDS 33:2211–2217. doi: 10.1097/QAD.0000000000002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Latimer RL, Vodstrcil L, De Petra V, Fairley CK, Read TR, Williamson D, Doyle M, Chow EP, Bradshaw C. 2020. Extragenital Mycoplasma genitalium infections among men who have sex with men. Sex Transm Infect 96:10–18. doi: 10.1136/sextrans-2019-054058. [DOI] [PubMed] [Google Scholar]
- 75.Tully JG, Rose DL, Baseman JB, Dallo SF, Lazzell AL, Davis CP. 1995. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J Clin Microbiol 33:1851–1855. doi: 10.1128/jcm.33.7.1851-1855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor-Robinson D, Gilroy CB, Horowitz S, Horowitz J. 1994. Mycoplasma genitalium in the joints of two patients with arthritis. Eur J Clin Microbiol Infect Dis 13:1066–1069. doi: 10.1007/BF02111830. [DOI] [PubMed] [Google Scholar]
- 77.Bjornelius E, Jensen JS, Lidbrink P. 2004. Conjunctivitis associated with Mycoplasma genitalium infection. Clin Infect Dis 39:e67–e69. doi: 10.1086/423809. [DOI] [PubMed] [Google Scholar]
- 78.Soni S, Horner P, Rayment M, Pinto-Sander N, Naous N, Parkhouse A, Bancroft D, Patterson C, Fifer H. 2019. British Association for Sexual Health and HIV national guideline for the management of infection with Mycoplasma genitalium (2018). Int J STD AIDS 30:938–950. doi: 10.1177/0956462419825948. [DOI] [PubMed] [Google Scholar]
- 79.Falk L, Enger M, Jensen JS. 2015. Time to eradication of Mycoplasma genitalium after antibiotic treatment in men and women. J Antimicrob Chemother 70:3134–3140. doi: 10.1093/jac/dkv246. [DOI] [PubMed] [Google Scholar]
- 80.Golden MR, Workowski KA, Bolan G. 2017. Developing a public health response to Mycoplasma genitalium. J Infect Dis 216:S420–S426. doi: 10.1093/infdis/jix200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sweeney EL, Whiley DM, Murray GL, Bradshaw CS. 2022. Mycoplasma genitalium: enhanced management using expanded resistance-guided treatment strategies. Sex Health 19:248–254. doi: 10.1071/SH22012. [DOI] [PubMed] [Google Scholar]
- 82.Hamasuna R, Osada Y, Jensen JS. 2007. Isolation of Mycoplasma genitalium from first-void urine specimens by coculture with Vero cells. J Clin Microbiol 45:847–850. doi: 10.1128/JCM.02056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jensen JS, Hansen HT, Lind K. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J Clin Microbiol 34:286–291. doi: 10.1128/jcm.34.2.286-291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamasuna R, Osada Y, Jensen JS. 2005. Antibiotic susceptibility testing of Mycoplasma genitalium by TaqMan 5′ nuclease real-time PCR. Antimicrob Agents Chemother 49:4993–4998. doi: 10.1128/AAC.49.12.4993-4998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jensen JS, Orsum R, Dohn B, Uldum S, Worm AM, Lind K. 1993. Mycoplasma genitalium: a cause of male urethritis? Genitourin Med 69:265–269. doi: 10.1136/sti.69.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Furr PM, Taylor-Robinson D. 1984. Microimmunofluorescence technique for detection of antibody to Mycoplasma genitalium. J Clin Pathol 37:1072–1074. doi: 10.1136/jcp.37.9.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor-Robinson D. 1989. Genital mycoplasma infections. Clin Lab Med 9:501–523. doi: 10.1016/S0272-2712(18)30615-2. [DOI] [PubMed] [Google Scholar]
- 88.Moller BR, Taylor-Robinson D, Furr PM. 1984. Serological evidence implicating Mycoplasma genitalium in pelvic inflammatory disease. Lancet i:1102–1103. doi: 10.1016/s0140-6736(84)92511-x. [DOI] [PubMed] [Google Scholar]
- 89.Baseman JB, Cagle M, Korte JE, Herrera C, Rasmussen WG, Baseman JG, Shain R, Piper JM. 2004. Diagnostic assessment of Mycoplasma genitalium in culture-positive women. J Clin Microbiol 42:203–211. doi: 10.1128/JCM.42.1.203-211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clausen HF, Fedder J, Drasbek M, Nielsen PK, Toft B, Ingerslev HJ, Birkelund S, Christiansen G. 2001. Serological investigation of Mycoplasma genitalium in infertile women. Hum Reprod 16:1866–1874. doi: 10.1093/humrep/16.9.1866. [DOI] [PubMed] [Google Scholar]
- 91.Svenstrup HF, Fedder J, Kristoffersen SE, Trolle B, Birkelund S, Christiansen G. 2008. Mycoplasma genitalium, Chlamydia trachomatis, and tubal factor infertility—a prospective study. Fertil Steril 90:513–520. doi: 10.1016/j.fertnstert.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 92.Palmer HM, Gilroy CB, Claydon EJ, Taylor-Robinson D. 1991. Detection of Mycoplasma genitalium in the genitourinary tract of women by the polymerase chain reaction. Int J STD AIDS 2:261–263. doi: 10.1177/095646249100200407. [DOI] [PubMed] [Google Scholar]
- 93.Jensen JS, Uldum SA, Sondergard-Andersen J, Vuust J, Lind K. 1991. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J Clin Microbiol 29:46–50. doi: 10.1128/jcm.29.1.46-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eastick K, Leeming JP, Caul EO, Horner PJ, Millar MR. 2003. A novel polymerase chain reaction assay to detect Mycoplasma genitalium. Mol Pathol 56:25–28. doi: 10.1136/mp.56.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jensen JS, Borre MB, Dohn B. 2003. Detection of Mycoplasma genitalium by PCR amplification of the 16S rRNA gene. J Clin Microbiol 41:261–266. doi: 10.1128/JCM.41.1.261-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoshida T, Deguchi T, Ito M, Maeda S, Tamaki M, Ishiko H. 2002. Quantitative detection of Mycoplasma genitalium from first-pass urine of men with urethritis and asymptomatic men by real-time PCR. J Clin Microbiol 40:1451–1455. doi: 10.1128/JCM.40.4.1451-1455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huppert JS, Mortensen JE, Reed JL, Kahn JA, Rich KD, Hobbs MM. 2008. Mycoplasma genitalium detected by transcription-mediated amplification is associated with Chlamydia trachomatis in adolescent women. Sex Transm Dis 35:250–254. doi: 10.1097/OLQ.0b013e31815abac6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wroblewski JK, Manhart LE, Dickey KA, Hudspeth MK, Totten PA. 2006. Comparison of transcription-mediated amplification and PCR assay results for various genital specimen types for detection of Mycoplasma genitalium. J Clin Microbiol 44:3306–3312. doi: 10.1128/JCM.00553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hardick J, Giles J, Hardick A, Hsieh Y-H, Quinn T, Gaydos C. 2006. Performance of the Gen-Probe transcription-mediated amplification research assay compared to that of a multitarget real-time PCR for Mycoplasma genitalium detection. J Clin Microbiol 44:1236–1240. doi: 10.1128/JCM.44.4.1236-1240.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deguchi T, Yoshida T, Yokoi S, Ito M, Tamaki M, Ishiko H, Maeda S. 2002. Longitudinal quantitative detection by real-time PCR of Mycoplasma genitalium in first-pass urine of men with recurrent nongonococcal urethritis. J Clin Microbiol 40:3854–3856. doi: 10.1128/JCM.40.10.3854-3856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jensen JS, Bjornelius E, Dohn B, Lidbrink P. 2004. Use of TaqMan 5′ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J Clin Microbiol 42:683–692. doi: 10.1128/JCM.42.2.683-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jurstrand M, Jensen JS, Fredlund H, Falk L, Molling P. 2005. Detection of Mycoplasma genitalium in urogenital specimens by real-time PCR and by conventional PCR assay. J Med Microbiol 54:23–29. doi: 10.1099/jmm.0.45732-0. [DOI] [PubMed] [Google Scholar]
- 103.Svenstrup HF, Jensen JS, Bjornelius E, Lidbrink P, Birkelund S, Christiansen G. 2005. Development of a quantitative real-time PCR assay for detection of Mycoplasma genitalium. J Clin Microbiol 43:3121–3128. doi: 10.1128/JCM.43.7.3121-3128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dupin N, Bijaoui G, Schwarzinger M, Ernault P, Gerhardt P, Jdid R, Hilab S, Pantoja C, Buffet M, Escande JP, Costa JM. 2003. Detection and quantification of Mycoplasma genitalium in male patients with urethritis. Clin Infect Dis 37:602–605. doi: 10.1086/376990. [DOI] [PubMed] [Google Scholar]
- 105.Xiao L, Waites KB, Wang H, Van Der Pol B, Ratliff AE, Geisler WM. 2018. Evaluation of a real-time PCR assay for detection of Mycoplasma genitalium and macrolide resistance-mediating mutations from clinical specimens. Diagn Microbiol Infect Dis 91:123–125. doi: 10.1016/j.diagmicrobio.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gaydos CA. 2017. Mycoplasma genitalium: accurate diagnosis is necessary for adequate treatment. J Infect Dis 216:S406–S411. doi: 10.1093/infdis/jix104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gaydos CA, Manhart LE, Taylor SN, Lillis RA, Hook EW, III, Klausner JD, Remillard CV, Love M, McKinney B, Getman DK. 2019. Molecular testing for Mycoplasma genitalium in the United States: results from the AMES prospective multicenter clinical study. J Clin Microbiol 57:e01125-19. doi: 10.1128/JCM.01125-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Salazar A, Espadafor B, Fuentes-Lopez A, Barrientos-Duran A, Salvador L, Alvarez M, Garcia F. 2019. Comparison between Aptima assays (Hologic) and the Allplex STI Essential assay (Seegene) for the diagnosis of sexually transmitted infections. PLoS One 14:e0222439. doi: 10.1371/journal.pone.0222439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Naidu P, Shokoples S, Martin I, Zelyas N, Singh A. 2021. Evaluation of 5 commercial assays for the detection of Mycoplasma genitalium and other urogenital mycoplasmas. Med Microbiol Immunol 210:73–80. doi: 10.1007/s00430-021-00699-1. [DOI] [PubMed] [Google Scholar]
- 110.Chernesky M, Jang D, Martin I, Speicher DJ, Clavio A, Lidder R, Ratnam S, Smieja M, Arias M, Shah A. 2020. Comparison of assays for the diagnosis of Mycoplasma genitalium and macrolide resistance mutations in self-collected vaginal swabs and urine. Sex Transm Dis 47:705–711. doi: 10.1097/OLQ.0000000000001226. [DOI] [PubMed] [Google Scholar]
- 111.Rumyantseva T, Golparian D, Nilsson CS, Johansson E, Falk M, Fredlund H, Van Dam A, Guschin A, Unemo M. 2015. Evaluation of the new AmpliSens multiplex real-time PCR assay for simultaneous detection of Neisseria gonorrhoeae, Chlamydia trachomatis, Mycoplasma genitalium, and Trichomonas vaginalis. APMIS 123:879–886. doi: 10.1111/apm.12430. [DOI] [PubMed] [Google Scholar]
- 112.Van Der Pol B, Waites KB, Xiao L, Taylor SN, Rao A, Nye M, Chavoustie S, Ermel A, Kaplan C, Eisenberg D, Chan PA, Mena L, Pacheco S, Krishnamurthy S, Mohan R, Bertuzis R, McGowin CL, Arcenas R, Marlowe EM. 2020. Mycoplasma genitalium detection in urogenital specimens from symptomatic and asymptomatic men and women by use of the cobas TV/MG test. J Clin Microbiol 58:e02124-19. doi: 10.1128/JCM.02124-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barrientos-Duran A, de Salazar A, Fuentes-Lopez A, Serrano-Conde E, Espadafor B, Chueca N, Alvarez-Estevez M, Garcia F. 2021. Comparison between Aptima assays (Hologic) and the CoBAS 6800 system (Roche) for the diagnosis of sexually transmitted infections caused by Chlamydia trachomatis, Neisseria gonorrhoeae, and Mycoplasma genitalium. Eur J Clin Microbiol Infect Dis 40:1337–1342. doi: 10.1007/s10096-020-04143-9. [DOI] [PubMed] [Google Scholar]
- 114.Pereyre S, Camelena F, Henin N, Bercot B, Bebear C. 2022. Clinical performance of four multiplex real-time PCR kits detecting urogenital and sexually transmitted pathogens. Clin Microbiol Infect 28:733.e7–733.e13. doi: 10.1016/j.cmi.2021.09.028. [DOI] [PubMed] [Google Scholar]
- 115.Chesnay A, Lancelin B, Le Brun C, Pastuszka A, Desoubeaux G, Lanotte P. 2020. Contribution of a molecular test for the diagnosis of genital infection with Trichomonas vaginalis and Mycoplasma genitalium. Ann Biol Clin (Paris) 78:623–627. doi: 10.1684/abc.2020.1589. [DOI] [PubMed] [Google Scholar]
- 116.Le Roy C, Le Hen I, Clerc M, Arfel V, Normandin F, Bebear C, de Barbeyrac B. 2012. The first performance report for the Bio-Rad Dx CT/NG/MG assay for simultaneous detection of Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium in urogenital samples. J Microbiol Methods 89:193–197. doi: 10.1016/j.mimet.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 117.Ursi D, Crucitti T, Smet H, Ieven M. 2016. Evaluation of the Bio-Rad Dx CT/NG/MG assay for simultaneous detection of Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium in urine. Eur J Clin Microbiol Infect Dis 35:1159–1163. doi: 10.1007/s10096-016-2646-4. [DOI] [PubMed] [Google Scholar]
- 118.Khatib N, Bradbury C, Chalker V, Koh GC, Smit E, Wilson S, Watson J. 2015. Prevalence of Trichomonas vaginalis, Mycoplasma genitalium and Ureaplasma urealyticum in men with urethritis attending an urban sexual health clinic. Int J STD AIDS 26:388–392. doi: 10.1177/0956462414539464. [DOI] [PubMed] [Google Scholar]
- 119.Jensen JS, Bjornelius E, Dohn B, Lidbrink P. 2004. Comparison of first void urine and urogenital swab specimens for detection of Mycoplasma genitalium and Chlamydia trachomatis by polymerase chain reaction in patients attending a sexually transmitted disease clinic. Sex Transm Dis 31:499–507. doi: 10.1097/01.olq.0000135992.98883.e4. [DOI] [PubMed] [Google Scholar]
- 120.Chernesky M, Jang D, Smieja M, Arias M, Martin I, Weinbaum B, Getman D. 2017. Urinary meatal swabbing detects more men infected with Mycoplasma genitalium and four other sexually transmitted infections than first catch urine. Sex Transm Dis 44:489–491. doi: 10.1097/OLQ.0000000000000618. [DOI] [PubMed] [Google Scholar]
- 121.Dize L, Barnes P, Jr, Barnes M, Hsieh YH, Marsiglia V, Duncan D, Hardick J, Gaydos CA. 2016. Performance of self-collected penile-meatal swabs compared to clinician-collected urethral swabs for the detection of Chlamydia trachomatis, Neisseria gonorrhoeae, Trichomonas vaginalis, and Mycoplasma genitalium by nucleic acid amplification assays. Diagn Microbiol Infect Dis 86:131–135. doi: 10.1016/j.diagmicrobio.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reinton N, Moi H, Olsen AO, Zarabyan N, Bjerner J, Tonseth TM, Moghaddam A. 2013. Anatomic distribution of Neisseria gonorrhoeae, Chlamydia trachomatis and Mycoplasma genitalium infections in men who have sex with men. Sex Health 10:199–203. doi: 10.1071/SH12092. [DOI] [PubMed] [Google Scholar]
- 123.Murray GL, Danielewski J, Bodiyabadu K, Machalek DA, Bradshaw CS, Costa AM, Birnie J, Garland SM. 2019. Analysis of infection loads in Mycoplasma genitalium clinical specimens by use of a commercial diagnostic test. J Clin Microbiol 57:e00344-19. doi: 10.1128/JCM.00344-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mobley VL, Hobbs MM, Lau K, Weinbaum BS, Getman DK, Sena AC. 2012. Mycoplasma genitalium infection in women attending a sexually transmitted infection clinic: diagnostic specimen type, coinfections, and predictors. Sex Transm Dis 39:706–709. doi: 10.1097/OLQ.0b013e318255de03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lillis RA, Nsuami MJ, Myers L, Martin DH. 2011. Utility of urine, vaginal, cervical, and rectal specimens for detection of Mycoplasma genitalium in women. J Clin Microbiol 49:1990–1992. doi: 10.1128/JCM.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shipitsyna E, Zolotoverkhaya E, Dohn B, Benkovich A, Savicheva A, Sokolovsky E, Jensen JS, Domeika M, Unemo M. 2009. First evaluation of polymerase chain reaction assays used for diagnosis of Mycoplasma genitalium in Russia. J Eur Acad Dermatol Venereol 23:1164–1172. doi: 10.1111/j.1468-3083.2009.03276.x. [DOI] [PubMed] [Google Scholar]
- 127.Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev 23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bradshaw CS, Jensen JS, Tabrizi SN, Read TR, Garland SM, Hopkins CA, Moss LM, Fairley CK. 2006. Azithromycin failure in Mycoplasma genitalium urethritis. Emerg Infect Dis 12:1149–1152. doi: 10.3201/eid1207.051558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wood GE, Jensen NL, Astete S, Jensen JS, Kenny GE, Khosropour CM, Gillespie CW, Manhart LE, Totten PA. 2021. Azithromycin and doxycycline resistance profiles of U.S. Mycoplasma genitalium strains and their association with treatment outcomes. J Clin Microbiol 59:e00819-21. doi: 10.1128/JCM.00819-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jensen JS, Bradshaw CS, Tabrizi SN, Fairley CK, Hamasuna R. 2008. Azithromycin treatment failure in Mycoplasma genitalium-positive patients with nongonococcal urethritis is associated with induced macrolide resistance. Clin Infect Dis 47:1546–1553. doi: 10.1086/593188. [DOI] [PubMed] [Google Scholar]
- 131.Read TR, Fairley CK, Tabrizi SN, Bissessor M, Vodstrcil L, Chow EP, Grant M, Danielewski J, Garland SM, Hocking JS, Chen MY, Bradshaw CS. 2017. Azithromycin 1.5g over 5 days compared to 1g single dose in urethral Mycoplasma genitalium: impact on treatment outcome and resistance. Clin Infect Dis 64:250–256. doi: 10.1093/cid/ciw719. [DOI] [PubMed] [Google Scholar]
- 132.Jensen JS, Bradshaw C. 2015. Management of Mycoplasma genitalium infections—can we hit a moving target? BMC Infect Dis 15:343. doi: 10.1186/s12879-015-1041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tabrizi SN, Su J, Bradshaw CS, Fairley CK, Walker S, Tan LY, Mokany E, Garland SM. 2017. Prospective evaluation of ResistancePlus MG, a new multiplex quantitative PCR assay for detection of Mycoplasma genitalium and macrolide resistance. J Clin Microbiol 55:1915–1919. doi: 10.1128/JCM.02312-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Machalek DA, Tao Y, Shilling H, Jensen JS, Unemo M, Murray G, Chow EPF, Low N, Garland SM, Vodstrcil LA, Fairley CK, Hocking JS, Zhang L, Bradshaw CS. 2020. Prevalence of mutations associated with resistance to macrolides and fluoroquinolones in Mycoplasma genitalium: a systematic review and meta-analysis. Lancet Infect Dis 20:1302–1314. doi: 10.1016/S1473-3099(20)30154-7. [DOI] [PubMed] [Google Scholar]
- 135.Bercot B, Charreau I, Rousseau C, Delaugerre C, Chidiac C, Pialoux G, Capitant C, Bourgeois-Nicolaos N, Raffi F, Pereyre S, Le Roy C, Senneville E, Meyer L, Bebear C, Molina J-M, ANRS IPERGAY Study Group . 2021. High prevalence and high rate of antibiotic resistance of Mycoplasma genitalium infections in men who have sex with men: a substudy of the ANRS IPERGAY pre-exposure prophylaxis trial. Clin Infect Dis 73:e2127–e2133. doi: 10.1093/cid/ciaa1832. [DOI] [PubMed] [Google Scholar]
- 136.McIver R, Jalocon D, McNulty A, Jeoffreys NJ, Chen SC, Power M, Couldwell DL. 2019. Men who have sex with men with Mycoplasma genitalium-positive nongonococcal urethritis are more likely to have macrolide-resistant strains than men with only female partners: a prospective study. Sex Transm Dis 46:513–517. doi: 10.1097/OLQ.0000000000001009. [DOI] [PubMed] [Google Scholar]
- 137.Getman D, Cohen S, Jiang A. 23 July 2022. Distribution of macrolide resistant Mycoplasma genitalium in urogenital tract specimens from women enrolled in a US clinical study cohort. Clin Infect Dis doi: 10.1093/cid/ciac602. [DOI] [PMC free article] [PubMed]
- 138.Lau A, Bradshaw CS, Lewis D, Fairley CK, Chen MY, Kong FY, Hocking JS. 2015. The efficacy of azithromycin for the treatment of genital Mycoplasma genitalium: a systematic review and meta-analysis. Clin Infect Dis 61:1389–1399. doi: 10.1093/cid/civ644. [DOI] [PubMed] [Google Scholar]
- 139.Palich R, Gardette M, Bebear C, Caumes E, Pereyre S, Monsel G. 2021. Initial failure of pristinamycin treatment in a case of multidrug-resistant Mycoplasma genitalium urethritis eventually treated by sequential therapy. Sex Transm Dis 48:e163–e164. doi: 10.1097/OLQ.0000000000001415. [DOI] [PubMed] [Google Scholar]
- 140.Li Y, Le WJ, Li S, Cao YP, Su XH. 2017. Meta-analysis of the efficacy of moxifloxacin in treating Mycoplasma genitalium infection. Int J STD AIDS 28:1106–1114. doi: 10.1177/0956462416688562. [DOI] [PubMed] [Google Scholar]
- 141.Tickner JA, Bradshaw CS, Murray GL, Whiley DM, Sweeney EL. 2022. Novel probe-based melting curve assays for the characterization of fluoroquinolone resistance in Mycoplasma genitalium. J Antimicrob Chemother 77:1592–1599. doi: 10.1093/jac/dkac097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Couldwell DL, Tagg KA, Jeoffreys NJ, Gilbert GL. 2013. Failure of moxifloxacin treatment in Mycoplasma genitalium infections due to macrolide and fluoroquinolone resistance. Int J STD AIDS 24:822–828. doi: 10.1177/0956462413502008. [DOI] [PubMed] [Google Scholar]
- 143.Murray GL, Bodiyabadu K, Danielewski J, Garland SM, Machalek DA, Fairley CK, Jensen JS, Williamson DA, Tan LY, Mokany E, Durukan D, Bradshaw CS. 2020. Moxifloxacin and sitafloxacin treatment failure in Mycoplasma genitalium infection: association with parC mutation G248T (S83I) and concurrent gyrA mutations. J Infect Dis 221:1017–1024. doi: 10.1093/infdis/jiz550. [DOI] [PubMed] [Google Scholar]
- 144.Murray GL, Bradshaw CS, Bissessor M, Danielewski J, Garland SM, Jensen JS, Fairley CK, Tabrizi SN. 2017. Increasing macrolide and fluoroquinolone resistance in Mycoplasma genitalium. Emerg Infect Dis 23:809–812. doi: 10.3201/eid2305.161745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Murray GL, Bodiyabadu K, Vodstrcil LA, Machalek DA, Danielewski J, Plummer EL, Garland SM, Whiley DM, Sweeney EL, Bradshaw CS. 2022. parC variants in Mycoplasma genitalium: trends over time and association with moxifloxacin failure. Antimicrob Agents Chemother 66:e00278-22. doi: 10.1128/aac.00278-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hamasuna R, Le PT, Kutsuna S, Furubayashi K, Matsumoto M, Ohmagari N, Fujimoto N, Matsumoto T, Jensen JS. 2018. Mutations in ParC and GyrA of moxifloxacin-resistant and susceptible Mycoplasma genitalium strains. PLoS One 13:e0198355. doi: 10.1371/journal.pone.0198355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ando N, Mizushima D, Takano M, Mitobe M, Miyake H, Yokoyama K, Sadamasu K, Aoki T, Watanabe K, Uemura H, Yanagawa Y, Gatanaga H, Oka S. 2021. High prevalence of circulating dual-class resistant Mycoplasma genitalium in asymptomatic MSM in Tokyo, Japan. JAC Antimicrob Resist 3:dlab091. doi: 10.1093/jacamr/dlab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chua T-P, Bodiyabadu K, Machalek DA, Garland SM, Bradshaw CS, Plummer EL, Danielewski J, Vodstrcil LA, Doyle ML, Murray GL. 2021. Prevalence of Mycoplasma genitalium fluoroquinolone-resistance markers, and dual-class-resistance markers, in asymptomatic men who have sex with men. J Med Microbiol 70:e001429. doi: 10.1099/jmm.0.001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.De Baetselier I, Vuylsteke B, Reyniers T, Smet H, Van den Bossche D, Kenyon C, Crucitti T. 2022. Worryingly high prevalence of resistance-associated mutations to macrolides and fluoroquinolones in Mycoplasma genitalium among men who have sex with men with recurrent sexually transmitted infections. Int J STD AIDS 33:385–390. doi: 10.1177/09564624211070704. [DOI] [PubMed] [Google Scholar]
- 150.Li Y, Su X, Le W, Li S, Yang Z, Chaisson C, Madico G, Gong X, Reed GW, Wang B, Rice PA. 2020. Mycoplasma genitalium in symptomatic male urethritis: macrolide use is associated with increased resistance. Clin Infect Dis 70:805–810. doi: 10.1093/cid/ciz294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Waites KB, Crabb DM, Xiao L, Duffy LB, Leal SM, Jr.. 2020. In vitro activities of eravacycline and other antimicrobial agents against human mycoplasmas and ureaplasmas. Antimicrob Agents Chemother 64:e00698-20. doi: 10.1128/AAC.00698-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Khalil D, Becker CAM, Tardy F. 2017. Monitoring the decrease in susceptibility to ribosomal RNAs targeting antimicrobials and its molecular basis in clinical Mycoplasma bovis isolates over time. Microb Drug Resist 23:799–811. doi: 10.1089/mdr.2016.0268. [DOI] [PubMed] [Google Scholar]
- 153.Vodstrcil LA, Plummer EL, Doyle M, Murray GL, Bodiyabadu K, Jensen JS, Whiley D, Sweeney E, Williamson DA, Chow EPF, Fairley CK, Bradshaw CS. 2022. Combination therapy for Mycoplasma genitalium, and new insights into the utility of parC mutant detection to improve cure. Clin Infect Dis 75:813–823. doi: 10.1093/cid/ciab1058. [DOI] [PubMed] [Google Scholar]
- 154.Read TRH, Fairley CK, Murray GL, Jensen JS, Danielewski J, Worthington K, Doyle M, Mokany E, Tan L, Chow EPF, Garland SM, Bradshaw CS. 2019. Outcomes of resistance-guided sequential treatment of Mycoplasma genitalium infections: a prospective evaluation. Clin Infect Dis 68:554–560. doi: 10.1093/cid/ciy477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Durukan D, Read TRH, Murray G, Doyle M, Chow EPF, Vodstrcil LA, Fairley CK, Aguirre I, Mokany E, Tan LY, Chen MY, Bradshaw CS. 2020. Resistance-guided antimicrobial therapy using doxycycline-moxifloxacin and doxycycline-2.5 g azithromycin for the treatment of Mycoplasma genitalium infection: efficacy and tolerability. Clin Infect Dis 71:1461–1468. doi: 10.1093/cid/ciz1031. [DOI] [PubMed] [Google Scholar]
- 156.Clinical and Laboratory Standards Institute. 2011. Methods for antimicrobial susceptibility testing of human mycoplasmas. Approved guideline, CLSI document M43-A. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 157.Waites KB, Crabb DM, Xiao L, Duffy LB. 2017. In vitro activities of gepotidacin (GSK2140944) and other antimicrobial agents against human mycoplasmas and ureaplasmas. Antimicrob Agents Chemother 61:e01064-17. doi: 10.1128/AAC.01064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jensen JS, Norgaard C, Scangarella-Oman N, Unemo M. 2020. In vitro activity of the first-in-class triazaacenaphthylene gepotidacin alone and in combination with doxycycline against drug-resistant and -susceptible Mycoplasma genitalium. Emerg Microbes Infect 9:1388–1392. doi: 10.1080/22221751.2020.1775498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gosse M, Lysvand H, Pukstad B, Nordbo SA. 2016. A novel SimpleProbe PCR assay for detection of mutations in the 23S rRNA gene associated with macrolide resistance in Mycoplasma genitalium in clinical samples. J Clin Microbiol 54:2563–2567. doi: 10.1128/JCM.01233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Braam JF, van Marm S, Severs TT, Belousov Y, Mahoney W, Kusters JG. 2018. Sensitive and specific assay for the simultaneous detection of Mycoplasma genitalium and macrolide resistance-associated mutations. Eur J Clin Microbiol Infect Dis 37:2137–2144. doi: 10.1007/s10096-018-3350-3. [DOI] [PubMed] [Google Scholar]
- 161.Touati A, Peuchant O, Jensen JS, Bebear C, Pereyre S. 2014. Direct detection of macrolide resistance in Mycoplasma genitalium isolates from clinical specimens from France by use of real-time PCR and melting curve analysis. J Clin Microbiol 52:1549–1555. doi: 10.1128/JCM.03318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Twin J, Jensen JS, Bradshaw CS, Garland SM, Fairley CK, Min LY, Tabrizi SN. 2012. Transmission and selection of macrolide resistant Mycoplasma genitalium infections detected by rapid high resolution melt analysis. PLoS One 7:e35593. doi: 10.1371/journal.pone.0035593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kristiansen GQ, Lisby JG, Schonning K. 2016. A 5′ nuclease genotyping assay for identification of macrolide-resistant Mycoplasma genitalium in clinical specimens. J Clin Microbiol 54:1593–1597. doi: 10.1128/JCM.00012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Salado-Rasmussen K, Jensen JS. 2014. Mycoplasma genitalium testing pattern and macrolide resistance: a Danish nationwide retrospective survey. Clin Infect Dis 59:24–30. doi: 10.1093/cid/ciu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wold C, Sorthe J, Hartgill U, Olsen AO, Moghaddam A, Reinton N. 2015. Identification of macrolide-resistant Mycoplasma genitalium using real-time PCR. J Eur Acad Dermatol Venereol 29:1616–1620. doi: 10.1111/jdv.12963. [DOI] [PubMed] [Google Scholar]
- 166.Jensen JS. 2012. Protocol for the detection of Mycoplasma genitalium by PCR from clinical specimens and subsequent detection of macrolide resistance-mediating mutations in region V of the 23S rRNA gene. Methods Mol Biol 903:129–139. doi: 10.1007/978-1-61779-937-2_8. [DOI] [PubMed] [Google Scholar]
- 167.Drud ST, Njuguna P, Ebeyan S, Erskine S, Holm M, Johansson SC, Tan LY, Jensen JS. 2020. Evaluation of the ResistancePlus MG FleXible assay for detection of wild-type and 23S rRNA-mutated Mycoplasma genitalium strains. J Clin Microbiol 58:e01900-19. doi: 10.1128/JCM.01900-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Manhart LE, Jensen JS. 2020. Quinolone resistance-associated mutations in Mycoplasma genitalium: not ready for prime time. Sex Transm Dis 47:199–201. doi: 10.1097/OLQ.0000000000001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tabrizi SN, Tan LY, Walker S, Twin J, Poljak M, Bradshaw CS, Fairley CK, Bissessor M, Mokany E, Todd AV, Garland SM. 2016. Multiplex assay for simultaneous detection of Mycoplasma genitalium and macrolide resistance using PlexZyme and PlexPrime technology. PLoS One 11:e0156740. doi: 10.1371/journal.pone.0156740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Le Roy C, Henin N, Bebear C, Pereyre S. 2017. Evaluation of a commercial multiplex quantitative PCR (qPCR) assay for simultaneous detection of Mycoplasma genitalium and macrolide resistance-associated mutations in clinical specimens. J Clin Microbiol 55:978–979. doi: 10.1128/JCM.02168-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Su JP, Tan LY, Garland SM, Tabrizi SN, Mokany E, Walker S, Bradshaw CS, Read T, Murray GL. 2018. Evaluation of the SpeeDx ResistancePlus MG diagnostic test for Mycoplasma genitalium on the Applied Biosystems 7500 Fast quantitative PCR platform. J Clin Microbiol 56:e01245-17. doi: 10.1128/JCM.01245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pitt R, Cole MJ, Fifer H, Woodford N. 2018. Evaluation of the Mycoplasma genitalium Resistance Plus kit for the detection of M. genitalium and mutations associated with macrolide resistance. Sex Transm Infect 94:565–567. doi: 10.1136/sextrans-2017-053366. [DOI] [PubMed] [Google Scholar]
- 173.Le Roy C, Bebear C, Pereyre S. 2020. Clinical evaluation of three commercial PCR assays for the detection of macrolide resistance in Mycoplasma genitalium. J Clin Microbiol 58:e01478-19. doi: 10.1128/JCM.01478-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Le Roy C, Pereyre S, Henin N, Bebear C. 2017. French prospective clinical evaluation of the Aptima Mycoplasma genitalium CE-IVD assay and macrolide resistance detection using three distinct assays. J Clin Microbiol 55:3194–3200. doi: 10.1128/JCM.00579-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Murray GL, Danielewski J, Bradshaw CS, Williamson DA, Birnie J, Su JP, De Petra V, Tan LY, Wee R, Machalek DA, Read TRH, Garland SM. 2020. Performance of the ResistancePlus MG diagnostic test for Mycoplasma genitalium using samples collected with Hologic Aptima specimen collection kits. J Med Microbiol 69:244–248. doi: 10.1099/jmm.0.001140. [DOI] [PubMed] [Google Scholar]
- 176.Shedko ED, Khayrullina GA, Goloveshkina EN, Akimkin VG. 2021. Clinical evaluation of commercial PCR assays for antimicrobal [sic] resistance in Mycoplasma genitalium and estimation of resistance-mediated mutation prevalence in Moscow and Moscow region. Eur J Clin Microbiol Infect Dis 40:1413–1418. doi: 10.1007/s10096-021-04170-0. [DOI] [PubMed] [Google Scholar]
- 177.Sweeney EL, Mhango LP, Ebeyan S, Tan LY, Bletchly C, Nimmo GR, Whiley DM. 2020. Evaluation of the ResistancePlus MG FleXible cartridge for near-point-of-care testing of Mycoplasma genitalium and associated macrolide resistance mutations. J Clin Microbiol 58:e01897-19. doi: 10.1128/JCM.01897-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fernandez-Huerta M, Salmeron P, Silgado A, Espasa M, Pumarola T, Tulsiani-Drud S, Barbera MJ, Hoyos-Mallecot Y, Serra-Pladevall J. 2020. Clinical evaluation of the ResistancePlus MG FleXible test on the GeneXpert Infinity-48s instrument: a near-patient assay for simultaneous detection of Mycoplasma genitalium and macrolide resistance. Diagn Microbiol Infect Dis 97:115062. doi: 10.1016/j.diagmicrobio.2020.115062. [DOI] [PubMed] [Google Scholar]
- 179.Murray GL, Doyle M, Bodiyabadu K, Vodstrcil LA, Garland SM, Danielewski J, Machalek DA, McGuinness C, Plummer EL, De Petra V, Williamson DA, Bradshaw CS. 2021. Evaluation of ResistancePlus MG FleXible, a ‘near-patient’ test for the detection of Mycoplasma genitalium and macrolide resistance mutations, using freshly collected clinical samples. J Med Microbiol 70:e001271. doi: 10.1099/jmm.0.001271. [DOI] [PubMed] [Google Scholar]
- 180.Le Roy C, Bebear C, Pereyre S. 2021. Performance of three commercial molecular diagnostic assays for the simultaneous detection of Mycoplasma genitalium and macrolide resistance. J Clin Microbiol 59:e00020-21. doi: 10.1128/JCM.00020-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tagg KA, Jeoffreys NJ, Couldwell DL, Donald JA, Gilbert GL. 2013. Fluoroquinolone and macrolide resistance-associated mutations in Mycoplasma genitalium. J Clin Microbiol 51:2245–2249. doi: 10.1128/JCM.00495-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Deguchi T, Yasuda M, Horie K, Seike K, Kikuchi M, Mizutani K, Tsuchiya T, Yokoi S, Nakano M, Hoshina S. 2015. Drug resistance-associated mutations in Mycoplasma genitalium in female sex workers, Japan. Emerg Infect Dis 21:1062–1064. doi: 10.3201/eid2106.142013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ong JJ, Magooa MP, Chikandiwa A, Kelly H, Didelot M-N, Muller EE, Maseko V, Segondy M, Delany-Moretlwe S, Kularatne R, Mayaud P, HIV in Africa Research Partnership Study Group . 2019. Prevalence and antimicrobial resistance of Mycoplasma genitalium infection among women living with human immunodeficiency virus in South Africa: a prospective cohort study. Clin Infect Dis 69:873–876. doi: 10.1093/cid/ciz045. [DOI] [PubMed] [Google Scholar]
- 184.Conway RJH, Cook S, Malone C, Bone S, Hassan-Ibrahim MO, Soni S. 2020. Clearance of Mycoplasma genitalium infection with moxifloxacin in the presence of quinolone resistance-associated mutations. Sex Transm Dis 47:197–198. doi: 10.1097/OLQ.0000000000001095. [DOI] [PubMed] [Google Scholar]
- 185.Sweeney EL, Lowry K, Ebeyan S, Lundgren M, Whiley DM. 2020. Evaluation of the SpeeDx MG parC (beta) PCR assay for rapid detection of Mycoplasma genitalium quinolone resistance-associated mutations. J Clin Microbiol 58:e01432-20. doi: 10.1128/JCM.01432-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shipitsyna E, Khusnutdinova T, Budilovskaya O, Shedko E, Goloveshkina E, Khayrullina G, Krysanova A, Shalepo K, Savicheva A, Unemo M. 16 June 2022. Performance of the first commercial dual resistance assay, AmpliSens Mycoplasma genitalium-ML/FQ-Resist-FL, for detection of potential macrolide and quinolone resistance-associated mutations and prevalence of M. genitalium resistance mutations in St. Petersburg, Russia. Sex Transm Infect doi: 10.1136/sextrans-2021-055249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Plummer EL, Murray GL, Bodiyabadu K, Su J, Garland SM, Bradshaw CS, Read TRH, Tabrizi SN, Danielewski JA. 2020. A custom amplicon sequencing approach to detect resistance associated mutations and sequence types in Mycoplasma genitalium. J Microbiol Methods 179:106089. doi: 10.1016/j.mimet.2020.106089. [DOI] [PubMed] [Google Scholar]
- 188.Sweeney EL, Bradshaw CS, Murray GL, Whiley DM. 2022. Individualised treatment of Mycoplasma genitalium infection—incorporation of fluoroquinolone resistance testing into clinical care. Lancet Infect Dis 22:e267–e270. doi: 10.1016/S1473-3099(21)00629-0. [DOI] [PubMed] [Google Scholar]
- 189.Sweeney EL, Tickner J, Bletchly C, Nimmo GR, Whiley DM. 2020. Genotyping of Mycoplasma genitalium suggests de novo acquisition of antimicrobial resistance in Queensland, Australia. J Clin Microbiol 58:e00641-20. doi: 10.1128/JCM.00641-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pineiro L, Idigoras P, Cilla G. 2019. Molecular typing of Mycoplasma genitalium-positive specimens discriminates between persistent and recurrent infections in cases of treatment failure and supports contact tracing. Microorganisms 7:609. doi: 10.3390/microorganisms7120609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kokotovic B, Friis NF, Jensen JS, Ahrens P. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J Clin Microbiol 37:3300–3307. doi: 10.1128/JCM.37.10.3300-3307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ma L, Martin DH. 2004. Single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene among Mycoplasma genitalium strains from clinical specimens. J Clin Microbiol 42:4876–4878. doi: 10.1128/JCM.42.10.4876-4878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hjorth SV, Bjornelius E, Lidbrink P, Falk L, Dohn B, Berthelsen L, Ma L, Martin DH, Jensen JS. 2006. Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J Clin Microbiol 44:2078–2083. doi: 10.1128/JCM.00003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ma L, Taylor S, Jensen JS, Myers L, Lillis R, Martin DH. 2008. Short tandem repeat sequences in the Mycoplasma genitalium genome and their use in a multilocus genotyping system. BMC Microbiol 8:130. doi: 10.1186/1471-2180-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cazanave C, Charron A, Renaudin H, Bebear C. 2012. Method comparison for molecular typing of French and Tunisian Mycoplasma genitalium-positive specimens. J Med Microbiol 61:500–506. doi: 10.1099/jmm.0.037721-0. [DOI] [PubMed] [Google Scholar]
- 196.Dumke R, Rust M, Glaunsinger T. 2019. MgpB types among Mycoplasma genitalium strains from men who have sex with men in Berlin, Germany, 2016-2018. Pathogens 9:12. doi: 10.3390/pathogens9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dumke R. 2022. Molecular tools for typing Mycoplasma pneumoniae and Mycoplasma genitalium. Front Microbiol 13:904494. doi: 10.3389/fmicb.2022.904494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bradshaw CS, Jensen JS, Waites KB. 2017. New horizons in Mycoplasma genitalium treatment. J Infect Dis 216:S412–S419. doi: 10.1093/infdis/jix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.van der Schalk TE, Braam JF, Kusters JG. 2020. Molecular basis of antimicrobial resistance in Mycoplasma genitalium. Int J Antimicrob Agents 55:105911. doi: 10.1016/j.ijantimicag.2020.105911. [DOI] [PubMed] [Google Scholar]
- 200.Kirkconnell B, Weinbaum B, Santos K, Le Nguyen T, Vinluan B, Astete S, Wood GE, Totten PA, Getman DK. 2019. Design and validation of transcription-mediated-amplification nucleic acid amplification tests for Mycoplasma genitalium. J Clin Microbiol 57:e00264-19. doi: 10.1128/JCM.00264-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tabrizi SN, Costa AM, Su J, Lowe P, Bradshaw CS, Fairley CK, Garland SM. 2016. Evaluation of the Hologic Panther transcription-mediated amplification assay for detection of Mycoplasma genitalium. J Clin Microbiol 54:2201–2203. doi: 10.1128/JCM.01038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Salado-Rasmussen K, Tolstrup J, Sedeh FB, Larsen HK, Unemo M, Jensen JS. 2022. Clinical importance of superior sensitivity of the Aptima TMA-based assays for Mycoplasma genitalium detection. J Clin Microbiol 60:e02369-21. doi: 10.1128/jcm.02369-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shah A, Jang D, Martin I, Speicher DJ, Lidder R, Clavio A, Ratnam S, Smieja M, Chernesky M. 2021. Workflow and throughput of commercial assays to detect Mycoplasma genitalium and macrolide resistance-mediating mutations. Sex Transm Dis 48:e132–e134. doi: 10.1097/OLQ.0000000000001360. [DOI] [PubMed] [Google Scholar]
- 204.Stafford IA, Hummel K, Dunn JJ, Muldrew K, Berra A, Kravitz ES, Gogia S, Martin I, Munson E. 2021. Retrospective analysis of infection and antimicrobial resistance patterns of Mycoplasma genitalium among pregnant women in the southwestern USA. BMJ Open 11:e050475. doi: 10.1136/bmjopen-2021-050475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Romano SS, Jensen JS, Lowens MS, Morgan JL, Chambers LC, Robinson TS, Totten PA, Soge OO, Golden MR, Manhart LE. 2019. Long duration of asymptomatic Mycoplasma genitalium infection after syndromic treatment for nongonococcal urethritis. Clin Infect Dis 69:113–120. doi: 10.1093/cid/ciy843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chernesky M, Jang D, Martin I, Arias M, Shah A, Smieja M, Ratnam S, Getman D, Schachter J. 2019. Mycoplasma genitalium, Chlamydia trachomatis, and Neisseria gonorrhoeae detected with Aptima assays performed on self-obtained vaginal swabs and urine collected at home and in a clinic. Sex Transm Dis 46:e87–e89. doi: 10.1097/OLQ.0000000000001004. [DOI] [PubMed] [Google Scholar]
- 207.de Salazar A, Barrientos-Duran A, Espadafor B, Fuentes-Lopez A, Chueca N, Garcia F. 2021. Macrolide and fluoroquinolone resistance of Mycoplasma genitalium in southern Spain, 2018–2019. Sex Transm Infect 97:8–10. doi: 10.1136/sextrans-2019-054386. [DOI] [PubMed] [Google Scholar]
- 208.Herrmann B, Malm K. 2021. Comparison between Abbott m2000 RealTime and Alinity m STI systems for detection of Chlamydia trachomatis, Neisseria gonorrhoeae, and Mycoplasma genitalium. Eur J Clin Microbiol Infect Dis 40:2217–2220. doi: 10.1007/s10096-020-04135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.de Jong AS, Rahamat-Langendoen JC, van Alphen P, Hilt N, van Herk C, Pont S, Melchers W, van de Bovenkamp J. 2016. Large two-centre study into the prevalence of Mycoplasma genitalium and Trichomonas vaginalis in the Netherlands. Int J STD AIDS 27:856–860. doi: 10.1177/0956462415596496. [DOI] [PubMed] [Google Scholar]
- 210.Pereyre S, Laurier Nadalie C, Bebear C, Investigator Group . 2017. Mycoplasma genitalium and Trichomonas vaginalis in France: a point prevalence study in people screened for sexually transmitted diseases. Clin Microbiol Infect 23:122.e1–122.e7. doi: 10.1016/j.cmi.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 211.Waites KB, Bébéar C. 2023. Mycoplasma and ureaplasma. In Carroll KC, Pfaller MA (ed), Manual of clinical microbiology, 13th Edition. ASM Press, Washington, DC. [Google Scholar]