ABSTRACT
Timely diagnosis remains an unmet need in non-neutropenic patients at risk for aspergillosis, including those with COVID-19-associated pulmonary aspergillosis (CAPA), which in its early stages is characterized by tissue-invasive growth of the lungs with limited angioinvasion. Currently available mycological tests show limited sensitivity when testing blood specimens. Metagenomic next-generation sequencing (mNGS) to detect microbial cell-free DNA (mcfDNA) in plasma might overcome some of the limitations of conventional diagnostics. A two-center cohort study involving 114 COVID-19 intensive care unit patients evaluated the performance of plasma mcfDNA sequencing for the diagnosis of CAPA. Classification of CAPA was performed using the European Confederation for Medical Mycology (ECMM)/International Society for Human and Animal Mycoses (ISHAM) criteria. A total of 218 plasma samples were collected between April 2020 and June 2021 and tested for mcfDNA (Karius test). Only 6 patients were classified as probable CAPA, and 2 were classified as possible, while 106 patients did not fulfill CAPA criteria. The Karius test detected DNA of mold pathogens in 12 samples from 8 patients, including Aspergillus fumigatus in 10 samples from 6 patients. Mold pathogen DNA was detected in 5 of 6 (83% sensitivity) cases with probable CAPA (A. fumigatus in 8 samples from 4 patients and Rhizopus microsporus in 1 sample), while the test did not detect molds in 103 of 106 (97% specificity) cases without CAPA. The Karius test showed promising performance for diagnosis of CAPA when testing plasma, being highly specific. The test detected molds in all but one patient with probable CAPA, including cases where other mycological tests from blood resulted continuously negative, outlining the need for validation in larger studies.
KEYWORDS: CAPA, COVID-19, microbial cell-free DNA sequencing, liquid biopsy
INTRODUCTION
Severe coronavirus disease 2019 (COVID-19), resulting in pulmonary damage and systemic inflammatory burst, may promote fungal superinfections through specific immunological mechanisms such as suppression of the interferon 1 pathway (1, 2). Clinical risk factors that come with treatment of severe COVID-19, including utilization of systemic corticosteroids and anti-interleukin-6 treatment (2), may further predispose patients to fungal superinfections. In particular, COVID-19-associated pulmonary aspergillosis (CAPA) has emerged as a relevant complication in intensive care units (ICUs), affecting a median 10 to 15% of patients admitted with COVID-19-associated acute respiratory failure (ARF) (2–7). CAPA is characterized by early tissue-invasive growth in the lungs with delayed angioinvasion occurring only after several days of tissue-invasive growth, therefore mimicking the pathogenesis of invasive pulmonary aspergillosis (IPA) in other non-neutropenic patients in the ICU setting. As a result, blood biomarkers such as galactomannan often remain negative until advanced stages of the disease, for which mortality of CAPA is regularly above 80% despite appropriate antifungal treatment (8–12). Similar to other forms of IPA in non-neutropenic patients, biomarkers and mycological tests of specimens from the lower respiratory tract, optimally bronchioalveolar lavage fluid (BALF), are preferred for diagnosis of CAPA (9, 13, 14). However, BALF sampling requires bronchoscopy, which cannot always be performed safely in clinically unstable patients. In addition, other fungal diseases such as mucormycosis currently lack reliable biomarkers and are therefore often diagnosed only at autopsy (15) but can mimic IPA and also cause concomitant coinfections in up to 13% of patients with IPA (16). Hence, different and improved diagnostic assays that reliably detect and differentiate aspergillosis and mucormycosis in easily and safely obtainable samples (e.g., blood samples) are urgently needed.
Metagenomic next-generation sequencing (mNGS) to detect microbial cell-free DNA (mcfDNA) in plasma might depict a promising hypothesis-free, unbiased approach, potentially allowing earlier detection and diagnosis of fungal infections when other blood biomarkers/tests are still negative and also allowing for detection of novel pathogens (17). The application of mcfDNA sequencing has emerged as an intriguing initial diagnostic modality for suspected infectious disease when hypothesis-driven, targeted testing is not yet established (18). This strategy has the potential to enhance sensitivity while maintaining or possibly enhancing the detection range of conventional microbiological investigations (e.g., culture). The limited invasiveness of blood sampling, the potential for faster diagnosis compared to conventional testing, especially in settings of difficult to diagnose infections, and the possibility of identifying a wide range of infections rendered this method an attractive add-on to the diagnostic armamentarium across microbiology (18), although performance may differ between different mNGS platforms (19).
Karius Inc. (Redwood City, CA) commercially offers a plasma mcfDNA sequencing test (i.e., liquid biopsy), which is well established and commonly ordered in the United States. It detects, identifies and quantitates plasma mcfDNA of approximately 1,000 clinically relevant pathogens, including fungi, bacteria, DNA viruses, and eukaryotic parasites (20). Here, we evaluated the performance of plasma mcfDNA sequencing for diagnosis of CAPA in a cohort of ICU patients with COVID-19-associated respiratory failure.
MATERIALS AND METHODS
Study design.
This is a two-center cohort study including ICU patient samples obtained at the Medical University of Graz, as part of the European Confederation of Medical Mycology (ECMM) CAPA study, and Public Health Wales Microbiology Cardiff, as described before (3, 8, 9, 21). The main objective of this study was to assess whether mcfDNA sequencing detected Aspergillus directly from plasma samples, leading to improved diagnostic performance in patients with or at risk from CAPA, potentially identifying missed cases or identifying additive/other causative pathogens.
Both participating centers collected data on demographics, underlying medical conditions, risk factors for invasive fungal infections, details on diagnostic workup, treatment, and outcome, as described before (9). For classification of CAPA, the 2020 ECMM/International Society for Human and Animal Mycoses (ISHAM) consensus criteria (13) were used. Accordingly, the patients were categorized as proven/probable/possible pulmonary and/or tracheobronchial CAPA or no evidence for CAPA.
Sample storage and measurements.
Whole blood was collected into EDTA-containing tubes between April 2020 and June 2021 and centrifuged for 10 min at 2,000 × g. The EDTA plasma was then immediately transferred to cryotubes for routine prospective testing according to local diagnostic strategies and stored at −70°C prior to being shipped on dry ice to Karius Diagnostics (Redwood City, CA) in two separate shipments: one from Graz, Austria, and one from Cardiff, United Kingdom, in June 2022. The samples were selected from CAPA cases based on the availability of plasma, which was limited in the United Kingdom due to the restricted availability of EDTA vacutainers during the pandemic. A range of plasma samples from control patients were selected to reflect the temporal range of case samples.
Liquid biopsy/mNGS.
The Karius test detects plasma mcfDNA via a Clinical Laboratory Improvement Amendments (CLIA)-validated technology (18) using sequential processes of cfDNA extraction, conversion to DNA libraries, sequencing using Illumina NextSeq 500, removal of human sequences, and alignment of remaining sequences to a curated pathogen database to identify and report any of the over 1,200 organisms included in the Karius test clinical reportable range with results significantly above background. Additional quality control steps include controls for controls for carryover, nonspecific bias, NGS quality, sample mix-up, and quantitation, as well as the automated cfDNA extraction. Once sequenced, the results are run through the Karius bioinformatics pipeline (version 3.11), which is responsible for mapping sequencing data to a curated database of genomic references, estimating the concentration for each of the detected microbes, and the applications of a series of quality control tests as listed above. We report on microbes observed at a concentration above background at statistically significant levels, in addition to other statistical tests aimed at controlling for cross-reactivity originating from homology with under-represented microbial references. For each of the reported microbes, we associated the levels observed in the >680 members of the otherwise healthy cohort studied to establish the assay’s reference interval. The metric of molecules per microliter (MPM) quantifies each organism identified by representing the number of DNA sequencing reads present per microliter of plasma.
Statistical analysis and ethics.
All statistical analyses were performed using IBM SPSS Statistics 26 (IBM, Armonk, NY) and GraphPad Prism 9 version 9.4.1. Quantitative variables are summarized by mean and standard deviation or median and interquartile range (quartiles 1 to 3) as appropriate. Categorical variables are summarized by absolute (i.e., count) and relative (i.e., percentage) frequencies.
Both centers followed local ethical requirements. This study was approved by the Medical University of Graz (EC No. 32-296 ex 19/20). In Cardiff, all samples were initially sent as part of routine diagnostic testing, with surplus material tested as a retrospective, anonymous performance evaluation with no impact on patient management, not requiring ethical approval, as confirmed previously with the local research board. The study has been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments, as well as with the International Conference on Harmonisation (ICH) Good Clinical Practice (GCP) guidelines dated July 1996 and its addendum E6 (R2) of June 2017.
RESULTS
Baseline characteristics and CAPA classification.
We included 218 plasma samples from 114 patients with underlying COVID-19-associated ARF admitted to the ICU. Patient demographics, clinical characteristics, risk factors, treatment, and outcomes are displayed in Table 1. Of the 114 patients, none were classified as proven CAPA, 6 had probable CAPA, 2 had possible CAPA (probable and possible CAPA cases summarized in Table 2), and 106 had no evidence of CAPA. In terms of other fungal infections, none of the patients was diagnosed with COVID-19-associated mucormycosis or invasive candidiasis, while one patient was diagnosed with Pneumocystis jirovecii pneumonia (PJP).
TABLE 1.
Patient demographicsa
| Characteristic | Graz (n = 79) (%) | Cardiff (n = 35) (%) |
|---|---|---|
| Age, mean (SD) (yr) | 63 (13) | 55 (18) |
| Female sex | 28 (35) | 16 (46) |
| Body mass index ≥30 kg/m2 | 21 (27) | 8 (23) |
| Underlying disease | ||
| Hematological malignancy | 13 (17) | 17 (49) |
| Solid organ transplantation | 8 (10) | 7 (20) |
| Cardiovascular disease | 52 (66) | 10 (29) |
| Pulmonary disease | 26 (33) | 9 (26) |
| Diabetes mellitus | 19 (24) | 15 (39) |
| Smoking status positive | 10 (13) | 8 (23) |
| Systemic corticosteroid therapy | 67 (85) | 18 (51) |
| ICU – respiratory support at time sample was obtained | ||
| Mechanical ventilation | 30 (38) | 25 (71) |
| Noninvasive ventilation | 15 (19) | |
| ECMO | 4 (5) | 0 (0) |
| CAPA | ||
| Possible | 0 | 2 |
| Probable | 3 | 3 |
| Proven | 0 | 0 |
| No evidenceb | 76 | 30 |
| Mortality - survival ICU discharge | 44 (56) | 23 (58) |
CAPA, COVID-19-associated pulmonary aspergillosis; ECMO, extracorporal membrane oxygenation; ICU, intensive care unit.
At time of plasma sampling for Karius test.
TABLE 2.
Demographics and disease characteristics of patients with probable and possible CAPAa
| Case | ECMM/ISHAM CAPA classification | EORTC/MSG host factors | Age, sex | Underlying disease | Mechanical ventilation | Systemic corticosteroids for COVID | Tocilizumab for COVID | Mycological evidence for CAPA | Antifungal therapy | Patient alive at ICU discharge |
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Probable | None | 63, m | Hypertension, coronary heart disease | No | Yes | No | BAL, GM, PCR | Isavu | No |
| Patient 2 | Probable | None | 60, m | Hypertension | Yes | Yes | No | BAL, GM, PCR, culture, LFD, serum GM | Isavu | Yes |
| Patient 3 | Probable | None | 65, m | Hypertension | Yes | No | No | Serum GM, tracheal aspiration, LFD+ | Isavu | No |
| Patient 4 | Probable | None | 58, m | JAK-2-positive essential thrombocythemia | Yes | Yes | No | NBL, culture, GM, PCR, serum GM | Vori | No |
| Patient 5 | Probable | None | 58, m | Diabetes mellitus type 2, hypertension | Yes | Yes | Yes | BAL GM, serum GM | Vori | No |
| Patient 6 | Probable | None | 53, f | Malignant neoplasm of rectum, stage 3 cervical cancer, bronchial asthma | Yes | Yes | No | BAL GM, NBL culture | L-AmB | No |
| Patient 7 | Possible | None | 52, m | None | Yes | Yes | Yes | NBL GM | Vori | Yes |
| Patient 8 | Possible | None | 72, f | Hypertension | Yes | No | No | NBL GM | Vori | Yes |
BAL, bronchoalveolar lavage; CAPA, COVID-19-associated pulmonary aspergillosis; ECMM, European Confederation for Medical Mycology; EORTC, European Organization for Research and Treatment in Cancer; f, female; GM, galactomannan; ICU, intensive care unit; Isavu, isavuconoazle; ISHAM, International Society for Human and Animal Mycoses; L-AmB, liposomal amphotericin-B; LFD, lateral flow device; m, male; MSG, Mycoses Study Group; NBL, nonbronchoscopic lavage; SOT, solid organ transplantation; TB, tuberculosis; Vori, voriconazole.
Plasma mcfDNA sequencing results and concordance with other diagnostics, clinical classification, and outcomes.
Using mcfDNA sequencing, fungal pathogens were detected in 16 plasma samples obtained from 11 patients. Molds were detected in 12 plasma samples, while other fungal pathogens were detected in 4 plasma samples. The molds detected included Aspergillus fumigatus (n = 10 samples from 6 patients), Rhizopus microsporus (n = 1), and Alternaria arborescens (n = 1). mcfDNA sequencing yielded positive results for fungal pathogens in 5 of 6 probable CAPA cases, with detection of A. fumigatus in 4 samples and R. microsporus in 1 sample. Time points and dynamics of diagnostic test results are displayed in Fig. 1. For the three probable CAPA cases from Graz, serial plasma samples were available, while samples from a single time point were available for the remaining three probable CAPA cases from Cardiff.
FIG 1.
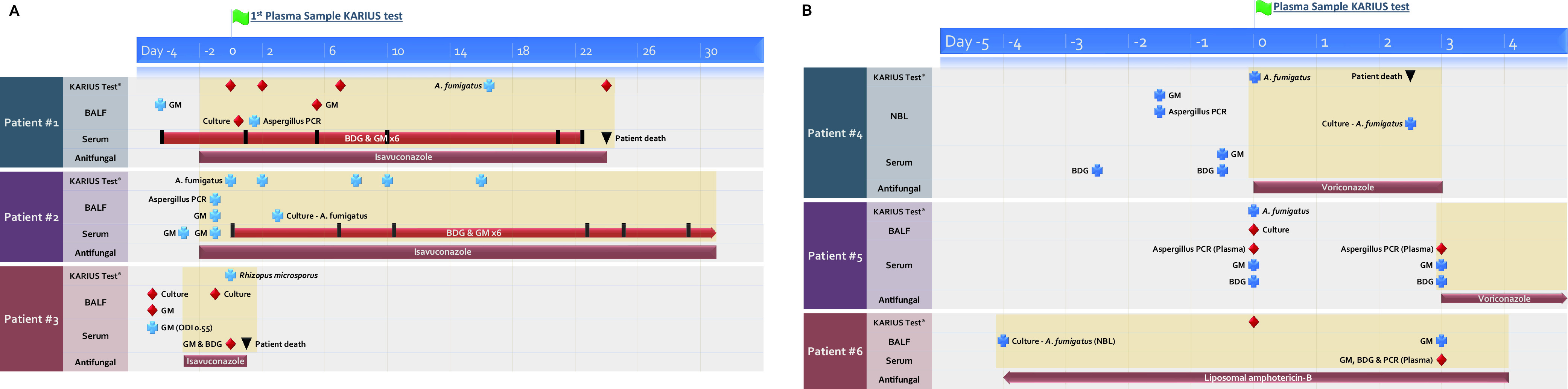
(A) Biomarker and mycological test dynamics in patients with probable COVID-19-associated pulmonary aspergillosis (CAPA) from Graz, Austria. (B) Biomarker dynamics in patients with probable CAPA from Cardiff, United Kingdom. Day 0 is defined as the day of the first Karius test. Blue plus sign, positive mycological test results (GM cutoffs >1.0 in BALF and >0.5 in serum; BDG cutoff ≥80 pg/mL); red diamond, negative mycological test results; black inverted triangle, day of patient death. Yellow background indicates diagnostic test results obtained during mold-active antifungal therapy. The red bar and arrow describe a time period of negative test results, with vertical black bars indicating the exact days of testing. BALF, bronchoalveolar lavage fluid; BDG, 1,3-β-d-glucan; GM, galactomannan; NBL, nondirected bronchial lavage; ODI, optical density index.
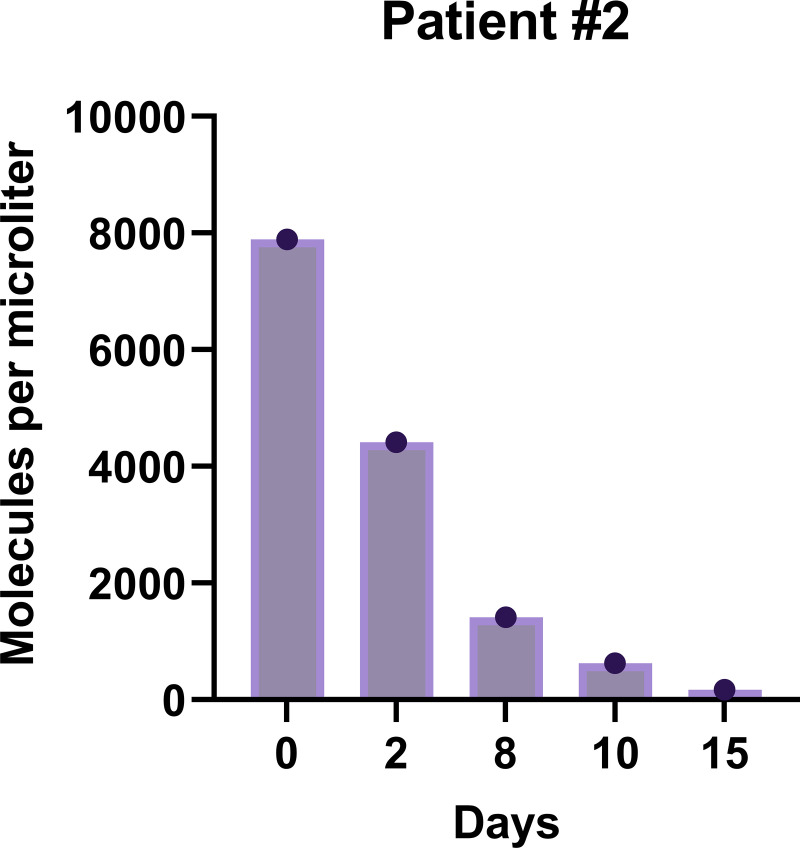
More detailed comparison of the probable CAPA cases showed that mcfDNA sequencing detected A. fumigatus DNA (MPM = 263) in plasma of patient 1, while serum galactomannan (GM) and 1,3-β-d-glucan (BDG) were continuously negative, as was BALF culture, but BALF GM and BALF Aspergillus PCR were positive. For patients 2 and 4, evidence for CAPA was derived from both blood and respiratory samples. Patient 2 had positive mcfDNA sequencing results from 5 longitudinal samples, for which MPM values over time are displayed in Fig. 2. The patient responded well to antifungal therapy and survived. In patient 4, mcfDNA sequencing yielded A. fumigatus DNA with MPM of 201. In patient 5, mcfDNA sequencing detected A. fumigatus DNA (MPM = 69) in plasma, despite serial negative results by Aspergillus PCR, while serum GM was positive. In patient 3, with a one-time slight positive serum GM and one-time positive Aspergillus lateral flow device test result from tracheal aspiration, mcfDNA sequencing detected R. microsporus (MPM = 4,419), indicating a potential mixed mold infection. This patient was treated for suspected CAPA with isavuconazole for 10 days, before R. microsporus was detected via mcfDNA sequencing, and the patient died 4 days later (no autopsy performed).
FIG 2.
Quantitative microbial cell-free DNA sequencing results for Aspergillus fumigatus from patient 2, who survived CAPA.
In patient 6 with probable CAPA, mcfDNA sequencing resulted negative. In this patient, mycological evidence for CAPA was exclusively obtained from BALF (nonbronchoscopic lavage culture and BALF GM), while all blood tests (Aspergillus PCR, GM, and BDG) were continuously negative. Overall sensitivity for mcfDNA sequencing detection of mold DNA in patients with probable CAPA was therefore 83% (5 of 6) and 67% (4 of 6) for detection of Aspergillus spp. DNA. Serum GM resulted positive in 4 of 6 cases and serum BDG in 2 of 6.
In terms of specificity, liquid biopsy detected DNA of molds (A. fumigatus [n = 2; MPM = 38 and 223]; A. arborescens [MPM = 151]) in three patients without CAPA or other clinically diagnosed mold infection. In one patient with detection of A. fumigatus (MPM = 38), there was, however, a chest computed tomography scan describing a 1.3-cm nodular lesion apical in the lower left lung lobe. Therefore, despite negative serum GM levels, the patient received antifungal treatment with posaconazole without further evaluation of the nodule (i.e., no BALF was obtained). mcfDNA sequencing was positive for A. fumigatus DNA in plasma, despite ongoing antifungal treatment. A. fumigatus DNA was also detected in a patient with asthma as underlying disease but no clinical suspicion of CAPA. A. arborescens DNA was detected in a patient with liver cirrhosis and consistently negative fungal biomarkers.
mcfDNA sequencing was also negative in two cases that were diagnosed with and appropriately treated for probable CAPA more than 1 month before the plasma sample for mcfDNA sequencing was obtained (i.e., all mycological tests were negative at the time the samples were obtained, and the patients did not fulfill CAPA criteria at the time of sampling). In addition, mcfDNA sequencing resulted negative in a patient who developed probable CAPA 1 week later (bronchoscopy with mycological evidence in respiratory tract). Overall, specificity of mcfDNA sequencing for detection of mold DNA in patients without CAPA was therefore 97% (103 of 106). mcfDNA sequencing also resulted negative in both patients with possible CAPA (one received anti-mold therapy at the time of sampling).
Aside from mold pathogens, mcfDNA sequencing also detected other fungal pathogens in a subset of samples: Candida albicans (n = 2), Candida glabrata (n = 1, same patient also had C. albicans), and P. jirovecii (n = 1). In one patient in whom mcfDNA sequencing detected C. albicans, serum BDG was strongly positive at >500 pg/mL, together with a high quantitative yield in mcfDNA sequencing (MPM = 5,688); whether the patient received antifungals is unknown. In the other patient, there was no evidence of invasive Candida infection (MPM = 23 and 107, respectively), but the patient was colonized with both C. albicans and C. glabrata in BAL and had also C. albicans detected in urine. P. jirovecii was detected in one solid organ transplant recipient (MPM = 30,112) who had a serum BDG level of 160 pg/mL on the day of plasma sample and a positive result for P. jirovecii in the same-day sputum PCR. Thus, the patient was clinically diagnosed with PJP. Despite targeted PJP treatment, the patient died after 6 days at the ICU.
DISCUSSION
Timely diagnosis of invasive mold diseases remains an unmet need, particularly in non-neutropenic patients at risk for IPA, where the disease in its early stages is primarily characterized by tissue-invasive growth in the lungs (22). The currently available fungal biomarkers and conventional mycological tests, such as GM (9) and BDG (23), show limited sensitivity when testing blood due to this pathophysiological feature (9). Here, we evaluated whether mcfDNA sequencing performed in 218 plasma samples from a cohort of 114 patients could aid the early detection of CAPA and overcome some of the limitations of established tests. Testing plasma samples from patients admitted with COVID-19-associated ARF to ICUs in Graz and Cardiff, we found that mcfDNA sequencing holds promise for diagnosis of CAPA, as a specific test (97% specificity) resulting positive for mold DNA in five of six patients with probable CAPA.
Previously, plasma mcfDNA sequencing has shown promise for diagnosing invasive mold infections, detecting the same fungus identified from the biopsy tissue at the genus level in seven of nine patients with proven mold disease (17). In addition, the test was able to detect Aspergillus lentulus, a cryptic Aspergillus spp. potentially requiring alternative antifungal therapy that was wrongly identified as A. fumigatus by conventional methods (17). Evaluating the clinical impact of mcfDNA sequencing for fungi, bacteria, viruses, and parasites, a recent analyses evaluated 82 Karius mcfDNA tests from a mixed cohort of adults and children, most of whom were immunocompromised (18). Among those cases, the Karius test results led to positive impact on clinical patient management in six cases (7.3%) and negative impact in three cases (3.7%) (18). Of note, in three of the six cases in which the Karius test had a positive clinical impact, the test led to early diagnosis of fungal infections (invasive candidiasis, mucormycosis, IPA) that had been missed by conventional methods but were later confirmed (18). In one additional mucormycosis case, the test was also able to identify the causative Mucorales genus but also wrongly identified a number of bacterial pathogens, leading to escalation of antibacterial coverage that was in retrospect deemed unnecessary, leading to negative assessment of the clinical utility (18).
Our study represents the largest study to date to evaluate the diagnostic performance of the Karius test in plasma for fungal infections. In a cohort of 114 ICU patients with severe COVID-19, we found that the test was highly specific for confirming CAPA (97% specificity), while detecting A. fumigatus in four of six cases with probable CAPA, including a case where all other blood biomarkers and tests for aspergillosis had resulted negative. Kinetics of quantitative mcfDNA test results may also hold significant value, as shown in one of those patients who responded to antifungal treatment and survived, where MPM values for A. fumigatus decreased markedly over time.
An important added benefit of mcfDNA sequencing is its potential to also detect rare fungal pathogens that may be missed or misclassified by conventional diagnostics, such as cryptic Aspergillus spp. (17) or members of the Mucorales order (18). This was also shown in our study, where plasma mcfDNA detected R. microsporus in a patient that had been diagnosed with probable CAPA 10 days earlier and had been on isavuconazole treatment. Despite isavuconazole being active against Mucorales (24, 25), the patient died a few days later, and the comparably high MPM value indicates a mixed fungal infection that was missed by conventional diagnostics. However, this could not be ultimately proven, because there was no further evaluation performed during clinical routine to rule out mixed fungal infection, nor was autopsy performed. Indeed, coinfections with Aspergillus and Mucorales may occur relatively frequently. Recently coinfections have been reported in 13% of patients with probable IPA (16) and also in 7% (26 of 378) of patients with COVID-19-associated mucormycosis (26). For the latter, coinfection with initial diagnosis of CAPA, potentially triggering inappropriate treatment for mucormycosis, was a major independent risk factor for mortality (26). In addition, mcfDNA sequencing also detected P. jirovecii in a patient with PJP pneumonia and C. albicans in a patient with suspected Candida infection (also high MPM value), outlining the broad potential of this test for detecting fungal infections and highlighting the benefits over current species/genus-specific assays. While other fungal biomarker tests can provide a broad detection range (e.g., BDG), the Karius system also provides a species-level identification that could be critical to administering appropriate antifungal therapy. In light of a loaded antifungal pipeline with several new antifungal classes entering late-stage clinical development (27, 28), accurate detection of causative fungal species will become even more important in the near future for enabling optimal targeted treatment.
This study comes with some important limitations. First, despite this being one of the largest studies to date evaluating the Karius test for diagnosis of fungal disease, sample size and in particular the number of cases with probable/proven CAPA or other mold infection was limited. Although our results were in line with previous reports, larger studies are needed to determine the performance of the Karius test in cases with invasive fungal diseases (IFDs). Several factors may influence the performance of this diagnostic approach when applied in clinical routine that need to be assessed in large and representative cohorts. Furthermore, in all but one case in which mcfDNA sequencing resulted negative 7 days before a patient developed CAPA, plasma samples for the mcfDNA sequencing were obtained when CAPA diagnosis was already established, limiting interpretation of whether or not fungal DNA might have been measurable before detection of any other evidence for CAPA.
In conclusion, this study showed encouraging results of the Karius test and its potential to become a valuable, highly specific, diagnostic add-on in settings of poor blood biomarker performance, such as the ICU setting and other settings with non-neutropenic patients at risk for invasive pulmonary mold infections. Further studies including more longitudinal sample time points and optimally including time points before patients develop invasive infection are needed to fully assess the potential of mNGS as a more sensitive diagnostic tool compared to conventional methods. Further studies are also needed to evaluate the potential real-world impact of the Karius test and to define optimal ways of utilization alongside conventional microbiological methods. Improved access to testing is required to demonstrate clinical utility given that the management of deadly IFDs is time critical (29), particularly with the emergence of antifungal resistance (30). Until faster turnaround time can be achieved, the test may primarily serve a role as adjunct/confirmatory test, while established tests with faster turnaround time, such as galactomannan and culture, may guide patient management.
ACKNOWLEDGMENTS
Karius tests were provided by Karius Diagnostics (Redwood City, CA), and the company also paid for shipping of the samples. They had no role in the study design, data collection, analysis, interpretation, decision to publish, writing of the manuscript, or decision to submit the manuscript for publication. The study was also supported by an investigator initiated research grant by Gilead sciences (M.H.).
M.H. received research funding from Gilead, Astellas, MSD, Euroimmune, Scynexis, F2G, and Pfizer. P.L.W. performed diagnostic evaluations and received meeting sponsorship from Bruker, Dynamiker, and Launch Diagnostics; received speaker fees, expert advice fees, and meeting sponsorship from Gilead; received speaker and expert advice fees from F2G; received speaker fees MSD and Pfizer; and received speaker fees and performed diagnostic evaluations for Associates of Cape Cod and IMMY. Juergen Prattes received research funding from MSD and Pfizer outside this work. The other authors declare no conflict of interest.
Contributor Information
Martin Hoenigl, Email: hoeniglmartin@gmail.com.
Juergen Prattes, Email: juergen.prattes@medunigraz.at.
Kimberly E. Hanson, University of Utah
REFERENCES
- 1.Feys SG, Khan SM, Choi M, Boeckx S, Chatelain B, Cunha D, Davaveye C, Hermans Y, Hertoghs G, Humblet Baron M, Jacobs S, Lagrou C, Marcelis K. 2022. Lung epithelial and myeloid immunity in influenza- and COVID-19-associated pulmonary aspergillosis: An observational study. Lancet Resp Med 10:1147–1159. 10.1016/S2213-2600(22)00259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoenigl M, Seidel D, Sprute R, Cunha C, Oliverio M, Goldman GH, Ibrahim AS, Carvalho A. 2022. COVID-19-associated fungal infections. Nat Microbiol 7:1127–1140. 10.1038/s41564-022-01172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prattes J, Wauters J, Giacobbe DR, Salmanton-García J, Maertens J, Bourgeois M, Reynders M, Rutsaert L, Van Regenmortel N, Lormans P, Feys S, Reisinger AC, Cornely OA, Lahmer T, Valerio M, Delhaes L, Jabeen K, Steinmann J, Chamula M, Bassetti M, Hatzl S, Rautemaa-Richardson R, Koehler P, Lagrou K, Hoenigl M. 2022. Risk factors and outcome of pulmonary aspergillosis in critically ill coronavirus disease 2019 patients: a multinational observational study by the European Confederation of Medical Mycology. Clin Microbiol Infect 28:580–587. 10.1016/j.cmi.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gangneux JD, Fekkar E, Luyt A, Botterel CE, De Prost N, Tadié J-M, Reizine F, Houzé S, Timsit J-F, Iriart X, Riu-Poulenc B, Sendid B, Nseir S, Persat F, Wallet F, Le Paper P, Canet E, Novara A, Manai M, Cateau E, Thille AW, Brun S, Cohen Y, Alanio A, Mégarbane B, Cornet M, Terzi N, Lamhaut L, Sabourin E, Desoubeaux G, Ehrmann S, Hennequin C, Voiriot G, Nevez G, Aubron EC, Letscher-Bru V, Meziani F, Blaize M, Mayaux J, Monsel A, Boquel F, Robert-Gangneax F, Le Tulzo Y, Sequin P, Guegan H, Autier B, Lesouhaitier M, Pelletier R, Belaz S, Bonnal C, et al. 2022. Fungal infections in mechanically ventilated COVID-19 patients in the ICU during the 1 first wave: the French multicenter MYCOVID study. Lancet Resp Med 10:180–190. 10.1016/S2213-2600(21)00442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssen NAF, Nyga R, Vanderbeke L, Jacobs C, Ergün M, Buil JB, van Dijk K, Altenburg J, Bouman CSC, van der Spoel HI, Rijnders BJA, Dunbar A, Schouten JA, Lagrou K, Bourgeois M, Reynders M, van Regenmortel N, Rutsaert L, Lormans P, Feys S, Debavaye Y, Tamion F, Costa D, Maizel J, Dupont H, Chouaki T, Nseir S, Sendid B, Brüggemann RJM, van de Veerdonk FL, Wauters J, Verweij PE. 2021. Multinational observational cohort study of COVID-19-associated pulmonary aspergillosis. Emerg Infect Dis 27:2892–2898. 10.3201/eid2711.211174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Permpalung N, Chiang TP, Massie AB, Zhang SX, Avery RK, Nematollahi S, Ostrander D, Segev DL, Marr KA. 2022. COVID-19 associated pulmonary aspergillosis in mechanically ventilated patients. Clin Infect Dis 74:83–91. 10.1093/cid/ciab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kariyawasam RM, Dingle TC, Kula BE, Vandermeer B, Sligl WI, Schwartz IS. 2022. Defining COVID-19-associated pulmonary aspergillosis: systematic review and meta-analysis. Clin Microbiol Infect 28:920–927. 10.1016/j.cmi.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prattes J, Wauters J, Giacobbe DR, Lagrou K, Hoenigl M; ECMM-CAPA Study Group. 2021. Diagnosis and treatment of COVID-19 associated pulmonary apergillosis in critically ill patients: results from a European confederation of medical mycology registry. Intensive Care Med 47:1158–1160. 10.1007/s00134-021-06471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Autier B, Prattes J, White PL, Valerio M, Machado M, Price J, Egger M, Gangneux JP, Hoenigl M. 2022. Aspergillus lateral flow assay with digital reader for the diagnosis of COVID-19-associated pulmonary aspergillosis (CAPA): a multicenter study. J Clin Microbiol 60:e0168921. 10.1128/JCM.01689-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellière S, Dudoignon E, Voicu S, Collet M, Fodil S, Plaud B, Chousterman B, Bretagne S, Azoulay E, Mebazaa A, Dépret F, Mégarbane B, Alanio A. 2022. Combination of mycological criteria: a better surrogate to identify COVID-19 associated pulmonary aspergillosis patients and evaluate prognosis? J Clin Microbiol 60:e0216921. 10.1128/jcm.02169-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giacobbe DR, Prattes J, Wauters J, Dettori S, Signori A, Salmanton-García J, Maertens J, Bourgeois M, Reynders M, Rutsaert L, Van Regenmortel N, Lormans P, Feys S, Klimko N, Shadrivova O, Cornely OA, Rautemaa-Richardson R, Koehler P, Lagrou K, Bassetti M, Hoenigl M. 2022. Prognostic impact of bronchoalveolar lavage fluid galactomannan and Aspergillus culture results on survival in COVID-19 intensive care unit patients: a post hoc analysis from the European Confederation of Medical Mycology (ECMM) COVID-19-Associated Pulmonary Aspergillosis Study. J Clin Microbiol 60:e0229821. 10.1128/jcm.02298-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ergün M, Brüggemann RJM, Alanio A, Dellière S, van Arkel A, Bentvelsen RG, Rijpstra T, van der Sar-van der Brugge S, Lagrou K, Janssen NAF, Buil JB, van Dijk K, Melchers WJG, Reijers MHE, Schouten JA, Wauters J, Cordey A, Soni S, White PL, van de Veerdonk FL, Verweij PE. 2021. Aspergillus test profiles and mortality in critically-ill COVID-19 patients. J Clin Microbiol 59:e0122921. 10.1128/JCM.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koehler P, Bassetti M, Chakrabarti A, Chen SCA, Colombo AL, Hoenigl M, Klimko N, Lass-Florl C, Oladele RO, Vinh DC, Zhu LP, Boll B, Bruggemann R, Gangneux JP, Perfect JR, Patterson TF, Persigehl T, Meis JF, Ostrosky-Zeichner L, White PL, Verweij PE, Cornely OA, European Confederation of Medical Mycology, International Society for Human Animal Mycology, Asia Fungal Working Group, INFOCUS LATAM/ISHAM Working Group, ISHAM Pan Africa Mycology Working Group, European Society for Clinical Microbiology, Infectious Diseases Fungal Infection Study Group, ESCMID Study Group for Infections in Critically Ill Patients, Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy, Medical Mycology Society of Nigeria, Medical Mycology Society of China Medicine Education Association, Infectious Diseases Working Party of the German Society for Haematology and Medical Oncology, Association of Medical Microbiology, Infectious Disease Canada . 2021. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect Dis 21:e149–e162. 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenks JP, Frank J, Speiss B, Mehta SR, Boch T, Buchheidt D, Hoenigl M. 2020. Performance of the bronchoalveolar lavage fluid Aspergillus galactomannan lateral flow assay with cube reader for diagnosis of invasive pulmonary aspergillosis: a multicenter cohort study. Clin Infect Dis 73:e1737–e1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoenigl M, Seidel D, Carvalho A, Rudramurthy SM, Arastehfar A, Gangneux JP, Nasir N, Bonifaz A, Araiza J, Klimko N, Serris A, Lagrou K, Meis JF, Cornely OA, Perfect JR, White PL, Chakrabarti A. 2022. The emergence of COVID-19 associated mucormycosis: a review of cases from 18 countries. Lancet Microbe 3:e543–e552. 10.1016/s2666-5247(21)00237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aerts RB, Bevers S, Beuselinck K, Schauwvlieghe A, Lagrou K, Lagrou K, Maertens J. 2022. Mucorales PCR to track down Aspergillus and Mucorales coinfections in at-risk hematology patients: a case-control study. Front Cell Infect Microbiol 12:1080921. 10.3389/fcimb.2022.1080921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong DK, Blauwkamp TA, Kertesz M, Bercovici S, Truong C, Banaei N. 2018. Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn Microbiol Infect Dis 92:210–213. 10.1016/j.diagmicrobio.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Hogan CA, Yang S, Garner OB, Green DA, Gomez CA, Dien Bard J, Pinsky BA, Banaei N. 2021. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis 72:239–245. 10.1093/cid/ciaa035. [DOI] [PubMed] [Google Scholar]
- 19.Schulz E, Grumaz S, Hatzl S, Gornicec M, Valentin T, Huber-Kraßnitzer B, Kriegl L, Uhl B, Deutsch A, Greinix H, Krause R, Neumeister P. 2022. Pathogen detection by metagenomic next-generation sequencing during neutropenic fever in patients with hematological malignancies. Open Forum Infect Dis 9:ofac393. 10.1093/ofid/ofac393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD, Cheung A, Rogers ZN, Meshulam-Simon G, Huijse L, Balakrishnan S, Quinn JV, Hollemon D, Hong DK, Vaughn ML, Kertesz M, Bercovici S, Wilber JC, Yang S. 2019. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 4:663–674. 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 21.White PL, Dhillon R, Cordey A, Hughes H, Faggian F, Soni S, Pandey M, Whitaker H, May A, Morgan M, Wise MP, Healy B, Blyth I, Price JS, Vale L, Posso R, Kronda J, Blackwood A, Rafferty H, Moffitt A, Tsitsopoulou A, Gaur S, Holmes T, Backx M. 2021. A national strategy to diagnose COVID-19 associated invasive fungal disease in the ICU. Clin Infect Dis 73:e1634–e1644. 10.1093/cid/ciaa1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson GR, III, Young JH. 2021. Aspergillus infections. N Engl J Med 385:1496–1509. 10.1056/NEJMra2027424. [DOI] [PubMed] [Google Scholar]
- 23.Egger M, Prüller F, Krause R, Prattes J, Hoenigl M. 2022. Utility of serum 1,3-β-d-glucan testing for diagnosis and prognostication in COVID-19-associated pulmonary aspergillosis. Microbiol Spectr 10:e0137322. 10.1128/spectrum.01373-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenks JD, Salzer HJ, Prattes J, Krause R, Buchheidt D, Hoenigl M. 2018. Spotlight on isavuconazole in the treatment of invasive aspergillosis and mucormycosis: design, development, and place in therapy. Drug Des Devel Ther 12:1033–1044. 10.2147/DDDT.S145545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson GR, III, Garcia-Diaz J, Miceli MH, Nguyen MH, Ostrosky-Zeichner L, Young JH, Fisher CE, Clark NM, Greenberg RN, Spec A, Kovanda L, Croos-Dabrera R, Kontoyiannis DP. 2022. Systemic antifungal therapy with isavuconazonium sulfate or other agents in adults with invasive mucormycosis or invasive aspergillosis (non-fumigatus): a multicentre, non-interventional registry study. Mycoses 65:186–198. 10.1111/myc.13412. [DOI] [PubMed] [Google Scholar]
- 26.Özbek L, Topçu U, Manay M, Han Esen B, Bektas SN, Aydın S, Özdemir B, Seidel D, Hoenigl M, Ergonul O. 2022. A meta-analysis of 556 individual COVID-19-associated mucormycosis cases: learning from the pandemic. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4174291.
- 27.Hoenigl M, Sprute R, Egger M, Arastehfar A, Cornely OA, Krause R, Lass-Flörl C, Prattes J, Spec A, Thompson GR, III, Wiederhold N, Jenks JD. 2021. The antifungal pipeline: fosmanogepix, ibrexafungerp, olorofim, opelconazole, and rezafungin. Drugs 81:1703–1729. 10.1007/s40265-021-01611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoenigl M, Sprute R, Arastehfar A, Perfect JR, Lass-Flörl C, Bellmann R, Prattes J, Thompson GR, III, Wiederhold NP, Al Obaidi MM, Willinger B, Arendrup MC, Koehler P, Oliverio M, Egger M, Schwartz IS, Cornely OA, Pappas PG, Krause R. 2022. Invasive candidiasis: investigational drugs in the clinical development pipeline and mechanisms of action. Expert Opin Investig Drugs 31:795–812. 10.1080/13543784.2022.2086120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slavin MA, Chen YC, Cordonnier C, Cornely OA, Cuenca-Estrella M, Donnelly JP, Groll AH, Lortholary O, Marty FM, Nucci M, Rex JH, Rijnders BJA, Thompson GR, Verweij PE, White PL, Hargreaves R, Harvey E, Maertens JA. 2021. When to change treatment of acute invasive aspergillosis: an expert viewpoint. J Antimicrob Chemother 77:16–23. 10.1093/jac/dkab317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rößler S, Bader O, Stölzel F, Sommer U, Spiess B, Geibel S, Buchheidt D, Groß U, Baretton G, Jacobs E, Ostrosky-Zeichner L. 2017. Progressive dispersion of azole resistance in Aspergillus fumigatus: fatal invasive aspergillosis in a patient with acute myeloid leukemia infected with an A. fumigatus strain with a cyp51A TR(46) Y121F M172I T289A allele. Antimicrob Agents Chemother 61:e00270-17. 10.1128/AAC.00270-17. [DOI] [PMC free article] [PubMed] [Google Scholar]