Abstract
Ancient DNA (aDNA) samples extracted from the bone remains of six equids buried by the Vesuvius eruption in 79 AD were investigated to test pre-amplification and enzymatic repair procedures designed to enhance the rescue of nuclear genes. The extracts, which proved all positive for Equidae mtDNA amplification, proved positive only four times out of 18 when tested for single-copy Equidae nuclear genes (ɛ globin, p53 and γ interferon). Pre-amplification did not change the number of retrieved aDNA sequences but 10 times out of 14 enzymatic repair restored the amplifiability of the genes analysed, proving that repair increases the rate of successful rescue from 22 to αλµοστ 80%. These findings support the hypothesis that some of these cross-linked aDNA molecules, which are not completely separated when DNA is extracted under denaturing conditions, become homoduplex substrates for Pol I and/or T4 ligase action upon renaturation. aDNA authenticity is proved by the homology of the nucleotide sequences of loci tested to the corresponding modern Equidae sequences. Data also indicate that cross-linked homoduplex molecules selected by denaturation of the extract are repaired without any chimera formation. The general features of aDNA amplification with and without denaturation and enzymatic repair are discussed.
INTRODUCTION
Ancient DNA (aDNA) is represented by heavily damaged and highly fragmented molecules (1). Damage includes either base modification (oxidation and deamination) or base loss (apurinic and/or apyrimidinic sites) as well as single-strand DNA breaks, with either nicks, gaps or protruding ends. Interstrand cross-links have also been shown to exist at relatively high frequencies (2). All such damage interferes to different extents with common PCR procedures utilised in aDNA studies. Whereas aDNA amplification failures are quite likely due to the limited number of molecules that can be recruited for PCR reaction, the success rate is favoured by the high concentration of the primers used, making it possible to ‘fish’ the few aDNA molecules that have become denatured during the PCR procedure. However, under these conditions, there is some concern regarding the possibility of chimera or artefact formation.
Pre-amplification procedures, such as degenerated oligonucleotide–PCR (DOP–PCR) (3) and primed extension–PCR (4), have already been proposed to enhance the retrieval of aDNA molecules. However, these strategies do not eliminate the risk of chimera formation.
After initial unsuccessful attempts to use the DOP pre-amplification procedure on Pompeian and Herculanean samples, the latter were treated with the enzymatic repair protocol (5) in the search for nuclear gene sequences.
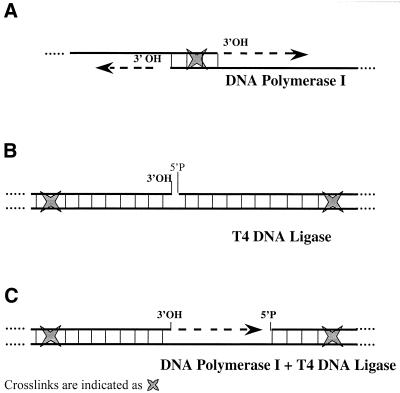
aDNA enzymatic repair can only be successful for molecules presenting unmodified 3′-OH and/or 5′-P termini which can be terminally elongated by DNA polymerase I (Fig. 1A), sealed by T4 DNA ligase (Fig. 1B) or filled in and then sealed by the concerted action of the two enzymes (Fig. 1C). More information on the DNA polymerase has became widely available over the past year and currently five complexes are known in Escherichia coli which are capable of replicating DNA using all four bases (6). A common feature to newly discovered polymerases is their ability to copy undamaged DNA with remarkably poor fidelity. In the case of aDNA molecules, unimpaired copying of aberrant bases can be prevented by using commercial Pol I polymerase for repair purposes, as it stalls at aberrant bases, thus favouring the repair of aDNA molecules containing fewer apurinic or apyrimidinic sites. This results in further improvements in aDNA sequence fidelity since the substrate of aDNA repair is the population of best conserved molecules. Nevertheless, when the reconstituted double-strand DNA molecules are submitted to selective PCR the repair strategy does not prevent chimera formation upon renaturation of separated strands (www.ipk-gatersleben.de/^blattner/aDNA/).
Figure 1.
Cross-link locations necessary for DNA polymerase I (A), T4 ligase (B) and the concerted action of the two enzymes (C) to allow repair of aDNA molecules, which then become available for specific PCR analysis.
In order to circumvent the problem, we took advantage of the presence of interstrand cross-links, which prevent strand separation by allowing homoduplex molecules to be selected as unique substrates for repair. In our experiments, aDNA molecules were first denatured during the extraction procedure, following the hypothesis that their extremely low concentration would then prevent renaturation from occurring, hence the de novo formation of homo- and heteroduplexes. Thus only the cross-linked homoduplex molecules, which were partially denatured and not completely separated, could reanneal and the latter class of molecules would be the preferential substrate for repair enzymes under our experimental conditions. Figure 1 shows the model we propose to explain the role that cross-links could play in repairing aDNA after denaturation; the different panels present the cross-link locations necessary to allow the two enzymes, DNA polymerase I and T4 ligase, to carry on repair either alone or in combination.
Given that exogenous DNA contamination is a common problem in aDNA investigation, to avoid this problem and at the same time assess the feasibility and fidelity of aDNA enzymatic repair, we investigated the DNA extracted from the skeletal remains of six equids, five of which were found in the stable of the ‘Casti Amanti’ house located in Via dell’Abbondanza, Insula 12, Regio IX, Pompeii. The sixth equid was found in Herculaneum, the other city buried by the Vesuvius eruption of 79 AD. Animal skeletal remains prove to be a more suitable source of material for studying aDNA repair since problems related to human DNA contamination are minimised. Furthermore, it must be borne in mind that once an archaeological site becomes the object for aDNA studies, the analysis of aDNA extracted from animal skeletal remains found in the site is extremely important in order to authenticate the site itself in terms of suitable conditions for nucleic acid preservation. Previous studies on human remains (7,8) and on a young Barbary macaque (9) stored in Terme del Sarno showed the presence of amplifiable aDNA and supported the hypothesis that burial conditions in Pompeii are favourable for aDNA preservation. In addition, independent approaches such as histological and histochemical analysis for monitoring the presence of aDNA in osteocytes have already been carried out on two of these equids (10). There is no possibility that these bones could derive from present-day animals.
MATERIALS AND METHODS
Bone sampling
The five remains from the stable of the ‘Casti Amanti’ house in Pompeii will be referred to as CAV1, 2, 3, 4 and 5, corresponding to skeletons D, A, B, C and E, respectively (11). The sixth corresponds to a skeleton found in Herculaneum and will be referred as CAVH. A small fragment (∼0.2–0.3 g) was cut from the metacarpal bones. Samples were then mechanically ground to a fine powder with a vibration machine supplied with sterile zirconium oxide abrasive. Guidelines regarding the pre-treatment of samples have already been published (7).
aDNA extraction
The extraction method is the same as that already published (12), albeit with some minor changes: a phenyl-thiacyl-thiazolium-bromide solution (13,14) was used and a denaturing step was introduced by heating the DNA samples to 94°C for 10 min. Modern equine DNA extraction was performed following the standard phenol–chloroform procedure (15).
Genetic analysis
For mitochondrial DNA analysis, the aDNA extracts were investigated for a 146-bp mtDNA 16S rRNA gene fragment with primers designed from Equus asinus (gi1805746) and Equus caballus (gi577571) mtDNA GenBank sequences.
For single-copy genes, the following primers were used: Equidae ɛ globin (ECU70986, AFI40616), 199-bp fragment: 5′-GGCAATCCCCAAGTCAAGG-3′, 5′-CAAACTAAAAAGCAAAAAA-3′; Equidae p53 (gi 1150384, gi 1389674 and gi 1020152), 130-bp fragment: 5′-TGGGGTCTCTAGGAGGGTCT-3′, 5′-TGGGGATAACTGGTCTGCTC-3′; E.caballus γ interferon (gi 488946), 232-bp fragment: 5′-ATGGTGGGCCTCTTTTCCTGGATA-3′, 5′-TGCTCTTTTGAATGACCTGGTTAT-3′.
For the PCR, 10 µl of aDNA extract was amplified in a 50 µl reaction mixture: 2.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl pH 9, 0.1% Triton X-100, 0.2 mM each dNTP, 0.2 µM primer, 2.5 U AmplyTaq Gold polymerase. Forty amplification cycles were performed as follows: 94°C for 1 min, 54°C (ɛ globin), 53°C (γ interferon), 55°C (p53) for 1 min and 72°C for 1 min. Amplification was started with an initial denaturation step (94°C for 6 min). Two negative controls were routinely included in each reaction, i.e. a PCR control and an extraction control. The latter was prepared at the beginning of the extraction to check for contamination during the extraction procedure. A second round of amplification was occasionally necessary for DNA sequencing.
PCR procedures were carried out in a specially designed laboratory for aDNA, which was physically separated from the extraction space. Modern DNA was manipulated in a physically separated non-aDNA laboratory.
DOP–PCR
DOP–PCR reactions were performed in 50 µl followed by ɛ globin gene amplification. The primer contained a degenerate central portion of six bases whereas the 5′ (nine bases) and 3′ (six bases) termini have a non-variable specific sequence (3).
Enzymatic repair of DNA
Escherichia coli DNA polymerase I and T4 DNA ligase were either used alone or in combination to repair different kinds of damage (Fig. 1).
Escherichia coli DNA polymerase I reaction
Aliquots (60 µl) of aDNA extracts concentrated to 10 µl volume using a Microcon 30-Millipore filter (30 kDa cut-off) were used as a template in a DNA polymerase I reaction. The 50 µl final volume reaction mixture contained 0.05 M Tris–HCl pH 7.2, 0.01 M MgSO4, 0.1 mM DTT, 50 µg/ml BSA, 0.4 mM of each dNTP and 2.5 U E.coli DNA polymerase I. The reaction was carried out in a thermal cycler for 90 min at 37°C and then incubated for 20 min at 70°C to inactivate the enzyme.
T4 DNA ligase reaction
A 10 µl aDNA extract, concentrated as described above, was used as substrate for the DNA ligase reaction. A 20 µl final volume contained 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 25 µg/ml BSA and 200 U T4 DNA ligase. The ligation reaction was performed for 1 h in a thermal cycler, with cycles between 10 and 25°C, where each temperature was held for 10 s.
Escherichia coli DNA polymerase I and T4 DNA ligase concerted reaction
An 18 µl aliquot of the DNA polymerase-treated aDNA samples was mixed with 2 µl of 10× ligase reaction buffer and 200 U T4 DNA ligase, and the ligation reaction proceeded as described above.
Aliquots of 5–10 µl of each repair reaction were successively tested with the primers selected for the PCR amplification of ɛ globin, p53 and γ interferon gene DNA fragments, respectively.
DNA sequencing
Both strands of the amplified DNA fragments, which were eluted from agarose gel slices, were directly sequenced with cycle sequencing dye terminator technology. The ɛ globin amplified DNA fragments of CAV1, CAV4 and CAV5 were also sequenced with standard 35S-labelling and polyacrylamide gel electrophoresis procedures (15). The accession numbers of GenBank deposited sequences are shown (see Figs 2–4).
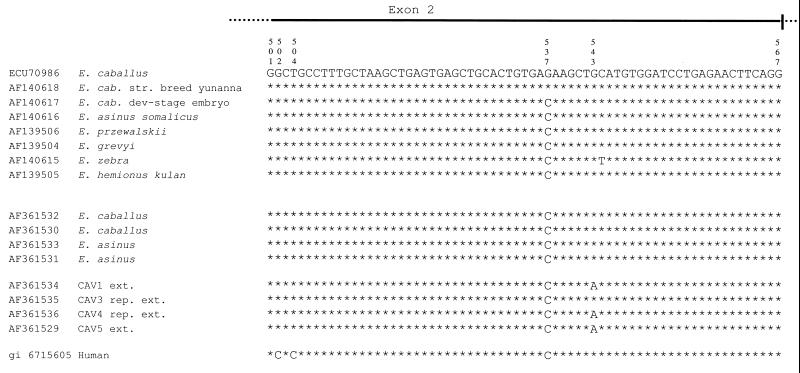
Figure 2.
Equidae ɛ globin genes. Sequence alignment from nucleotides 501–567 of the Equidae ɛ globin gene exon 2. The eight sequences at the top are from GenBank. Four sequences (E.caballus AF361532 and AF361530; E.asinus AF361533 and AF361531) were newly determined from modern animals. Four sequences are from ancient Pompeii Equidae: CAV1 AF361534 and CAV5 AF361529 sequences were determined directly from amplified extracts whereas CAV3 AF361535 and CAV4 AF361536 sequences come from the amplification of repaired extracts. The bottom line shows the human sequence of the homologous fragment.
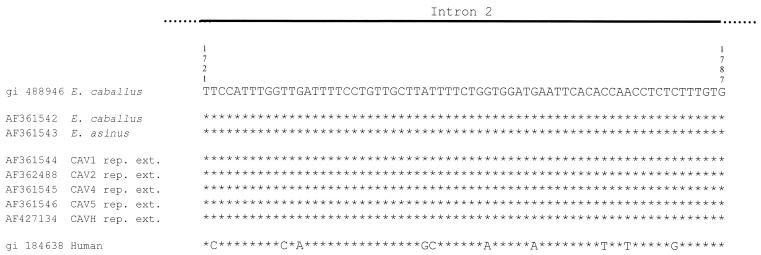
Figure 4.
The Equidae γ interferon gene. Sequence alignment from nucleotides 1721–1787 of Equidae γ interferon gene intron 2. Top, E.caballus GenBank DNA sequence; middle, newly determined sequences from modern E.caballus AF361542 and E.asinus AF361543; bottom, ancient Equidae sequences from repaired DNA extracts (AF361544, AF362488, AF361545, AF361546 and AF427134). The bottom line shows the human γ interferon sequence, which contains 10 substitutions out of 67 nt with respect to the Equidae sequences.
Sequence alignment
Sequences are aligned either against modern homologous sequences present in GenBank or the sequences from modern E.caballus and E.asinus DNA samples determined in our laboratory. MegAlign software (DNASTAR Inc.) and the Clustal method for pairwise alignment were used.
RESULTS
The DNA extracted from skeletal remains of six equids was first investigated for the presence of the mtDNA 16S rRNA gene fragment. The extracts were further investigated for the amplification of nuclear gene fragments encoding ɛ globin, p53 and γ interferon (Table 1). The specificity of all selected Equidae primers was tested by checking for the absence of cross-amplification with human DNA. Table 1 shows that the extracts exhibit a different amplifiability pattern, most likely related to the degree of preservation. However, when evaluating the latter, not only are there differences among bones from the same burial place (7,8) but also within the same bone. These observations are in line with the differences found in the amplifiability pattern among the Casti Amanti and Herculaneum equine extracts: (i) CAV1 and CAV5 prove amplifiable for mtDNA, ɛ globin and p53 loci, but not for the γ interferon locus; (ii) CAV2, CAV3, CAV4 and CAVH are positive for mtDNA and negative for all three single-copy genes tested.
Table 1. PCR amplifications: aDNA extracts.
| mtDNA | Single-copy genes | |||
|---|---|---|---|---|
| ɛ globin | p53 | γ interferon | ||
| CAV1 | + | + | + | – |
| CAV2 | + | – | – | – |
| CAV3 | + | – | – | – |
| CAV4 | + | – | – | – |
| CAV5 | + | + | + | – |
| CAVH | + | – | – | – |
Since the retrieval of single-copy genes, by means of DOP–PCR of extracts previously negative to specific PCR also proved unsuccessful, all extracts were subjected to DNA enzymatic repair procedures and then further tested for amplification of all three single-copy genes selected in our experiments (Table 2). In order to establish which kind of single-strand DNA breaks could be used for repair after aDNA denaturation, the two enzymes, E.coli DNA polymerase I and T4 DNA ligase, were used alone or in combination. Only the CAV4 p53 locus proved amplifiable after repair either using DNA polymerase I or T4 DNA ligase independently (data not shown), indicating the presence in these aDNA extracts of molecules with 3′ termini or single-strand nicks with ends which can be recruited for repair purposes. The other five showed successful amplification only when repaired by the concerted action of the two enzymes, indicating the prevalence of gapped recruitable molecules in these samples.
Table 2. PCR amplifications: aDNA extracts repaired with DNA polymerase I and T4 ligase.
| CAV1a | γ interferon | + |
| CAV2 | ɛ globin | – |
| p53 | – | |
| γ interferon | + | |
| CAV3 | ɛ globin | + |
| p53 | – | |
| γ interferon | – | |
| CAV4a | ɛ globin | + |
| p53 | + | |
| γ interferon | + | |
| CAV5a | γ interferon | + |
| CAVH | ɛ globin | – |
| p53 | + | |
| γ interferon | + |
aThese samples were also tested with DNA polymerase I and T4 ligase independently as discussed in the text.
+, successful amplification; –, unsuccessful amplification.
The repair procedure successfully retrieved γ interferon DNA from five equids with the exception of CAV3, which was positive for ɛ globin locus only. Refractoriness to repair, as also shown by the CAV2 ɛ globin and p53 loci, by the CAV3 γ interferon locus and by the CAVH ɛ globin locus, could be due either to the presence in the aDNA molecules of damage which cannot be repaired by the two enzymes or to cross-links within the fragment selected by the primers.
The amplification products were then sequenced and aligned against modern homologous sequences present in GenBank and against sequences obtained from modern E.caballus and E.asinus DNA samples. The latter were determined in a separate laboratory not dedicated to aDNA.
The most significant difference observed between modern and ancient Equidae ɛ globin sequences (Fig. 2) is the G→A transition in position 543, which corresponds to a silent substitution from CUG to CUA, both of which are L (Leu) codons. As confirmed both by cycle dye terminator and standard sequencing procedures conducted on different extracts, the results indicate a homozygous state. Such a transition can be detected in CAV1 and CAV5 extracts and in the CAV4 repaired extract, whereas in the CAV3 repaired extract, the G is conserved. The G which is present at nucleotide position 537 in the two upper E.caballus sequences shown in Figure 2 corresponds to C in all other listed sequences, i.e. the GAG codon changes to GAC and corresponds to the amino acid substitution E (Glu)→D (Asp), both polar and negatively charged. In the human sequence, two Cs at nucleotide positions 502 and 504 give rise to a codon change from GCU to CCC and to the amino acid substitution A (Ala)→P (Pro).
Figures 3 and 4 show the alignments of the p53 and γ interferon gene sequences. The data show that none of the single genes tested is preferentially lost or retrieved, indicating that aDNA damage is randomly present.
Figure 3.
The Equidae p53 gene. Sequence alignment from nucleotides 418–481 of the Equidae p53 gene intron 6. Top, two E.caballus sequences from GenBank; middle, sequences newly obtained from modern E.caballus AF361540 and E.asinus AF361541; bottom, sequences from ancient Equidae without and with DNA repair. The bottom line shows the homologous human sequence gi 4731629. The E.asinus sequence gi 1020152 was excluded from the comparison since it contains as many as 13 indels (insertion/deletion) out of the 65 nt being investigated.
DISCUSSION
The results presented in this paper demonstrate that the aDNA molecules extracted from Pompeii and Herculaneum animal remains contain 3′-OH and 5′-P termini which are suitable for enzymatic repair, and that aDNA denaturation before repair greatly enhances the retrieval of aDNA, thus increasing the number of loci that can be investigated in each extract.
The model we propose to explain these findings is based on the hypothesis that the presence of interstrand cross-links, a form of damage reported to occur in aDNA molecules, physically prevents strand separation when aDNA is subjected to denaturation. Consequently, among the homoduplexes that survive denaturation there may be molecules, which are recruitable substrates for DNA enzymatic repair as they present unmodified 5′-P and 3′-OH ends. In all cases, gene-specific PCR will only be successful when cross-links are located outside the region targeted for specific amplification. The formation of other substrate molecules suitable for repair, both in hetero- and homoduplex configurations, is most likely prevented by the very low amount of aDNA molecules, which make the aDNA separated strands extremely diluted. Thus, double-helix reconstitution would appear fairly implausible.
The amplification of the γ interferon fragment from aDNA of CAV1, CAV 4 and CAV5, which was tested after independent treatment with Pol I and T4 ligase, did not show positive results. However, the same treatment for CAV4 p53 proved PCR positive both after Pol I and T4 ligase alone. These results indicate that in CAV4 there is more heterogeneous damage, thus conserving the termini intact. Since current literature claims that each archaeological site presents peculiar burial conditions that may affect aDNA preservation, the characteristics of the Vesuvius pyroclastic materials that covered Pompeii and Herculaneum, and the temperature value, coupled with almost 2000 years of unchanged burial conditions, may have played a role in modifying the spectrum of known biochemical damage, favouring one or more kinds of damage with respect to the others.
Our model is supported by the perfect matching of nucleotide sequences obtained from the amplification of denatured-repaired aDNA with the homologous modern nucleotide sequence of Equidae genes and this finding further supports the absence of heteroduplex. While these data prove the authenticity of the DNA, they also show that enzymatic repair does not interfere with the genetic analysis of aDNA. Furthermore, the high degree of homology observed between ancient and extant DNA sequences supports the argument that the subsequent amplification of intact molecules selected during the first rounds of the reaction could be favoured by pausing of DNA polymerase I on abasic sites.
The G→A transition on position 543 of the ɛ globin gene found in CAV1 and CAV5 extracts and in the CAV4-repaired extract (Fig. 2) cannot be interpreted as a consequence of depurination because this occurrence could not only affect the G in position 543 in three different individuals. However, it cannot be interpreted as a DNA polymerase mistake since both strands of amplified fragments unequivocally show the presence of this mutation. This mutation, which consists of a silent Leu substitution, could be interpreted as a breed-specific nucleotide polymorphism. The analysis of a much larger sample would establish whether or not the A haplotype is absent in extant equids. Although data on the horse a gene complex exist in the literature (16), analogous information has not yet been produced to attempt to draw a picture of the evolution of the equid ɛ globin locus.
One limit of the approach described in this work is that the physical locations of the cross-links along the aDNA molecule, as shown in Figure 1, impose an upper limit to the size of amplifiable fragment. However, this constraint could be theoretically bypassed if the θ DNA polymerase, an enzyme shown to be able to repair cross-links (6,17) is used after denaturation. In fact, if cross-links were removed from the homoduplex molecules that survived denaturation, the size of the amplifiable fragment could also be expanded.
By way of a final consideration, the yeast extraordinary array of repair DNA polymerase from α to i, where different enzymes repair different kinds of lesions in different ways, may became a possible kit of new reagents in the repair repertoire of archaeological DNA investigation.
To conclude, we propose that aDNA denaturation, followed by enzymatic repair, should be routinely introduced in aDNA investigations before specific PCR. This method makes it possible to greatly increase both the number of assays which can be performed in each extract and to extend the genetic investigation to a number of nuclear loci which previously were almost inaccessible. Thus, the increase of the amount of amplifiable molecules by enzymatic repair proves a new strategy for successfully analysing aDNA nuclear genes.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Professor P. G. Guzzo, Archaeological Superintendent of Pompeii, and Dr A. M. Ciarallo for granting access to the collection of remains. We are grateful to Professor E. Giulotto, University of Pavia, for providing us with modern horse DNA (E.caballus) and Dr A. Genovese, Faculty of Veterinary Science, University of Naples Federico II and Dr F. Landolfi for providing us with horse (E.caballus) and donkey (E.asinus) blood samples. The stimulating cultural support of the P.I.T.A.G.O.R.A. Institute is greatly appreciated. We are grateful to Dr P. Gorman for editing the manuscript. This research is financed in part by the National Research Council (CNR 99.03736.PF36) and by the Campania Regional Authorities L.R. 41-1996 and 1997 to M.C. and in part by MURST PRIN-99 to A.C.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +39 081 5665879; Fax: +39 081 5667547; Email: antonino.cascino@unina2.it AF361529–AF361546, AF362488, AF427134, AF427135
REFERENCES
- 1.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Paabo S. (1989) Ancient DNA: Extraction, characterization, molecular cloning and enzymatic amplification. Proc. Natl Acad. Sci. USA, 86, 1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pusch C.M., Graeme,J.N., Bachmann,L. and Scholz,M. (2000) Degenerate oligonucleotide-primed preamplification of ancient DNA allows the retrieval of authentic DNA sequences. Anal. Biochem., 279, 118–122. [DOI] [PubMed] [Google Scholar]
- 4.Satoh K., Takahashi,K., Itoh,Y. and Kobayashi,R. (1998) Typing of DNA using the primer extension preamplification (PEP) method—studies of readability of typing and deletion limits. Nippon Hoigaku Zasshi, 52, 184–190. [PubMed] [Google Scholar]
- 5.Pusch C.M., Giddings,I. and Scholz,M. (1998) Repair of degraded duplex DNA from prehistoric samples using Escherichia coli DNA polymerase I and T4 DNA ligase. Nucleic Acids Res., 26, 857–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman M.F. and Tippin,B. (2000) The expanding polymerase universe. Nat. Rev. Mol. Cell Biol., 1, 101–109. [DOI] [PubMed] [Google Scholar]
- 7.Cipollaro M., Di Bernardo,G., Galano,G., Galderisi,U., Guarino,F.M., Angelini,F. and Cascino,A. (1998) Ancient DNA in human bone remains from Pompeii archaeological site. Biochem. Biophys. Res. Commun., 247, 901–904. [DOI] [PubMed] [Google Scholar]
- 8.Cipollaro M., Di Bernardo,G., Forte,A., Galano,G., Galderisi,U., Guarino,F.M., Angelini,F. and Cascino,A. (1999) Ancient DNA analysis of human bone remains from the Caius Iulius Polybius house in Pompeii. Croat. Med. J., 40, 392–397. [PubMed] [Google Scholar]
- 9.Bailey J.F., Henneberg,M., Colson,I.B., Ciarallo,A., Hedges,R.E.M. and Sykes,B. (1999) Monkey business in Pompeii-unique find of a juvenile Barbary macaque skeleton in Pompeii identified using osteology and ancient DNA techniques. Mol. Biol. Evol., 16, 1410–1414. [DOI] [PubMed] [Google Scholar]
- 10.Guarino F.M., Angelini,F., Odierna,G., Bianco,M.R., Di Bernardo,G., Forte,A., Cascino,A. and Cipollaro,M. (2000) Detection of DNA in ancient bones by histochemical methods. Biotech. Histochem., 75, 110–117. [DOI] [PubMed] [Google Scholar]
- 11.Cocca T., Lorizio,R., Capaldo,L., Calamo,A. and Genovese,A. (1995) The equids in the ‘Casti Amanti’ house stable in Pompeii: first observations. In National Research Council, University of Catania and Consorzlo Catonia ricerche, Proceedings of 1st International Congress on: Science and Technology for the Safeguard of Cultural Heritage in the Mediterranean Basin. Luxograph S.r.l., Palermo, Vol. 2, pp. 1577–1581.
- 12.Hoss M. and Paabo S. (1993) DNA extraction from Pleistocene bones by a silica-based purification method. Nucleic Acids Res., 21, 3913–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poinar H.N., Hofreiter,M., Spaulding,W.G., Martin,P.S., Stankiewicz,B.A., Bland,H., Evershed,R.P., Possnert,G. and Paabo,S. (1998) Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science, 281, 402–406. [DOI] [PubMed] [Google Scholar]
- 14.Vasan S., Zhang,X., Kapurniotu,A., Bernhagen,J., Teichberg,S., Basgen,J., Wagle,D., Shih,D., Terlecky,I., Bucala,R., Cerami,A., Egan,J. and Ulrich,P. (1996) An agent cleaving glucose-derived protein crosslinks in vitro and in vivo.Nature, 382, 275–278. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Oakenfull E.A. and Clegg,J.B. (1988) Phylogenetic relationships within the genus Equus and the evolution of α and θ globin genes. J. Mol. Evol., 47, 772–783. [DOI] [PubMed] [Google Scholar]
- 17.Sharief F.S., Vojta,P.J., Ropp,P.A. and Coopeland,W.C. (1999) Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eight human DNA polymerase. Genomics, 59, 90–96. [DOI] [PubMed] [Google Scholar]