Abstract
Arthur et al. leverage different types of big data, either generated in house from cohorts of healthy aging and COVID-19, or downloaded from the ever-increasing public data archives, to disentangle the distinct cellular and proteomic mechanisms of COVID-19 and aging.
News and views
The last few decades have seen an explosive rise in the proportion of the world’s population living into old age, which presents healthcare-related challenges that are epitomized by the greater susceptibility of older adults to severe COVID-19. There is also an explosion in big data and an ever-increasing richness of public data repositories archiving measurements of biological samples. Can we harness research advances in translational approaches, omic data generation, data sharing, and computational techniques to advance our understanding of the aging process and its role in disease? In this issue, Arthur et al. [1] prove that we can, identifying COVID-19 specific signatures of immune subsets of CD8+ T cells and B cells, and elevated plasma proteins derived from liver- and lung and decreased skeletal muscle-derived proteins (Figure 1). Further, with this study, Arthur et al. illuminate a general path for more such advances across other fields.
Figure 1. Advanced studies combine data generation and data mining in translational studies.
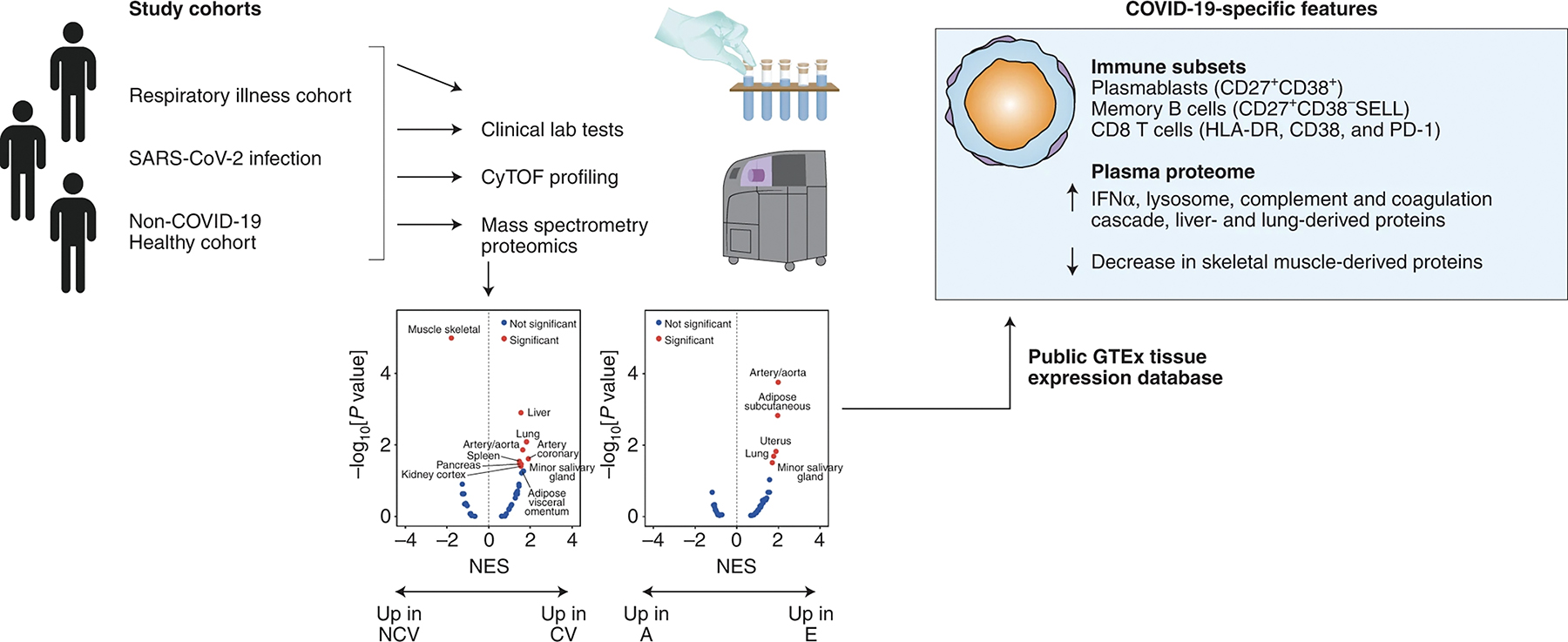
Newly generated data from Arthur et al. was combined with public data resources to elucidate a profile of COVID-19 responses distinct from aging.
The key strategy here is the combination and integration of traditional clinical laboratory values, immune cell subsets, plasma proteomic profiles, and existing big data resources to identify relevant pathways which distinguish effects of aging from COVID-19 (moderate from severe) and other respiratory illnesses. By comparing healthy populations across different age groups with patients with COVID-19 and other non-COVID-19 respiratory illnesses, this study found, as has been reported before for this infection [2–5], that COVID-19 patients with severe disease were more likely to be older and male. In clinical laboratory testing, patients in the COVID-19 and respiratory illness cohorts had more neutrophils and lower red blood cell counts compared to age-matched controls, while no statistical differences in the monocyte or platelet counts were observed. Other effects specific to COVID-19 included decreased albumin and calcium, as well as biomarkers of kidney function correlating with disease severity.
The authors used multiparameter mass cytometry (CyTOF) assays of primary peripheral blood mononuclear cells to identify immune cell populations and major subpopulations affected by aging and respiratory infection. Reduced proportions of naive cells in aging were noted, as has been well recognized across the studies of aging in the immune system [6–8], and which were further decreased with pulmonary infection (not exclusively to SARS-CoV-2). Both COVID-19 and other patient groups showed increased proportions of B cells compared to age-matched healthy controls. Specific to COVID-19 were an increase in CD27+CD38+ plasmablasts and a decrease in CD27+CD38-SELL memory B cells. Clustering on established T cell markers identified 12 subsets of CD4 T cells and 10 subsets of CD8 T cells with both age-specific differences and COVID-19 disease-specific changes. CD8+ T cells in particular show an increase in aging of a subpopulation of CD8+ T cells expressing the cytotoxicity marker GZMK. Notably, compared to age-matched healthy controls, COVID-19 patients showed reduced levels of CD4 T cells, and patients with moderate COVID-19 infection showed an increase in CD8 T cells. Effector subsets of CD8 T cells expressing granzyme molecules were also increased compared to age-matched controls and other respiratory infections. COVID-19 patients specifically showed an increase in CD8+ T cells expressing HLA-DR, CD38, and PD-1, which the authors suggest may arise from effector memory subpopulations. CyTOF profiles differentiate 11 subsets of NK cells which differ between healthy and infected (not exclusively to SARS-CoV-2). One NK cell cluster was found to decrease with age (CD56+CD57-SELL+ cluster). Analysis of myeloid lineages showed decreases in classical myeloid cells with a cluster of HLA-DRlow myeloid cells in pulmonary infection (not exclusively to SARS-CoV-2), suggesting an immunosuppressive population. The overall implications of these differences in immune cell populations are to disentangle COVID-19 specific changes from effects of other infections and of aging. Recognizing these specialized cell subsets highlights their roles and suggests pathways critical to advance our understanding of COVID-19 disease mechanisms and responses.
To map the plasma proteomes from the COVID-19 and the healthy aging cohort, the aptamer-based SOMAscan technology was used to detect and quantify ~4700 proteins. Since different plasma types had been collected for the two cohorts used in this study, a direct comparison was not possible. Instead, the authors devised a strategy to identify first those proteins that change due to COVID-19 or due to aging. In a subsequent step, the true, i.e., age-adjusted, COVID-19 associated differences in the plasma proteome were mapped. After removal of those proteins which showed aging-associated changes, this approach identified 337 up- and 421 down-regulated proteins respectively, that change specifically in response to COVID-19. The importance of accounting for age as a confounder was underscored by the fact that the most upregulated pathways observed in COVID-19 patients, namely matrisome and ECM glycoproteins, were also identified as being upregulated with age.
The subsequent Gene Set Enrichment Analysis (GSEA) identified interferon alpha response, lysosome, complement and coagulation cascade, and IL2/STAT6 and IL6/STAT3 signaling as being COVID-19 specific upregulated pathways, even after accounting for age as a confounder. This age-adjusted analysis suggests pathways for investigation of biomarkers or interventions truly focused towards COVID-19.
Not all classical plasma proteins were represented in the current iteration of the SOMAscan platform and hence it was not surprising to note that many of the plasma proteins previously described in the COVID-19 context [9–12] were not recapitulated by Arthur et al. While missing out on certain bona fide plasma proteins, the current iteration of the SOMAscan platform is an excellent tool to identify a wide range of tissue-specific proteins [13] and hence, their aging- and COVID-19-associated dysregulation could be readily mapped in this study. To this end, Arthur et al. take advantage of the GTEx tissue expression database, a public RNA-seq resource, to identify the likely tissue sources of the proteins in the plasma identified by SOMAscan. For example, the COVID-19 patients showed a significant increase in liver- and lung-derived proteins, and a concomitant decrease in skeletal muscle-derived proteins, while aging was associated with increases in arterial and subcutaneous adipose tissue proteins. This use of big data repository information enriches the current study and emphasizes the benefits of FAIR principles (the Findability, Accessibility, Interoperability, and Reuse of digital assets) [14] in support of their conclusions.
While this is an elegant and informative study, no one study can address all the questions we want to answer. It is not yet clear which of the specific immune cell and protein changes identified may directly influence COVID19 disease severity, which mechanisms may mediate these effects, and whether new therapeutic approaches may be identified. Critically important, the authors excluded obesity from their analysis. Although understandable, as removal of this potential confounder significantly reduced the complexity of the analysis, this will be an essential gap to address as obesity is recognized as a critical susceptibility factor to severe COVID-19 [5]. Future proteomic studies will have to include carefully designed age-, sex-, and BMI-matched cohorts. In addition, plasma proteomics itself is notoriously complex due to the extreme differential of protein abundances in plasma, clouding detection of the thousands of tissue leakage proteins. As such, SOMAscan, which is able to reliably detect >4700 proteins, has advanced into a veritable plasma proteomic platform to detect these tissue leakage proteins in particular. Notably, the non-plasma focused selection of detectable proteins in SOMAscan may thus underrepresent essential aspects of the immune system, whose modulation is a key role of blood. Thus, sample sparing LC/MS-based plasma proteomics and/or antibody-based cytokine assays should be considered for a comprehensive and immune-relevant plasma proteome mapping, with the additional advantage that very little additional sample is needed for these assays. Additional in-depth proteomic profiles across translational studies will also enrich data resources for comparison.
In summary, the integrated approach presented in this paper including patient disease status and clinical laboratory data, newly generated in-depth multidimensional data, and incorporating existing big data resources is a real trifecta and paves the road for best practices across translational studies far beyond COVID-19 and aging studies.
ADD ACKNOWLEDGEMENTS (E.G. FUNDING SOURCES)
Our Laboratories are supported in part by US National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) Human Immunology Project Consortium (HIPC) awards U19AI118608, U19 AI 089992; and awards to RRM (AI127865, AI142624, DA043337, AG055362) and to HS (AI 152179-02, AG 071858).
Footnotes
ADD COMPETING INTERESTS STATEMENT
The authors report no relevant competing interests.
Contributor Information
Ruth R. Montgomery, Department of Internal Medicine, Yale School of Medicine, New Haven, CT
Hanno Steen, Department of Pathology, Boston Children’s Hospital and Harvard Medical School, Boston, MA; Precision Vaccines Program, Boston Children’s Hospital, Boston, MA.
References
- 1.Arthur, Cellular and plasma proteomic determinants of COVID-19 and non-COVID-19 pulmonary diseases relative to healthy aging. Nat. Aging, 2021. in press. [DOI] [PubMed] [Google Scholar]
- 2.Mathew D, et al. , Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science, 2020. 369(6508). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laing AG, et al. , A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med, 2020. 26(10): p. 1623–1635. [DOI] [PubMed] [Google Scholar]
- 4.Lucas C, et al. , Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature, 2020. 584(7821): p. 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palaiodimos L, et al. , Severe obesity, increasing age and male sex are independently associated with worse in-hospital outcomes, and higher in-hospital mortality, in a cohort of patients with COVID-19 in the Bronx, New York. Metabolism, 2020. 108: p. 154262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thome JJ, et al. , Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell, 2014. 159(4): p. 814–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw AC, Goldstein DR, and Montgomery RR, Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol, 2013. 13: p. 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolich-Zugich J, The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol, 2018. 19(1): p. 10–19. [DOI] [PubMed] [Google Scholar]
- 9.Messner CB, et al. , Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst, 2020. 11(1): p. 11–24 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stukalov A, et al. , Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature, 2021. [DOI] [PubMed] [Google Scholar]
- 11.Shu T, et al. , Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity, 2020. 53(5): p. 1108–1122 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen B, et al. , Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell, 2020. 182(1): p. 59–72 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filbin MR, et al. , Plasma proteomics reveals tissue-specific cell death and mediators of cell-cell interactions in severe COVID-19 patients. bioRxiv, 2020. [Google Scholar]
- 14.Wilkinson MD, et al. , The FAIR Guiding Principles for scientific data management and stewardship. Sci Data, 2016. 3: p. 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]


