Abstract
Decades of work on the spatiotemporal organization of mammalian DNA Replication Timing (RT) continues to unveil novel correlations with aspects of transcription and chromatin organization but, until recently, mechanisms regulating RT and the biological significance of the RT program had been indistinct. We now know that the RT program is both influenced by and necessary to maintain chromatin structure, forming an epigenetic positive feedback loop. Moreover, the discovery of specific cis-acting elements regulating mammalian RT at both the domain and the whole chromosome level has revealed multiple cell type specific and developmentally regulated mechanisms of RT control. We review recent evidence for diverse mechanisms employed by different cell types to regulate their RT programs and the biological significance of RT regulation during development.
Keywords: DNA Replication Timing, Nuclear Organization, transcription
Intro
The faithful duplication of a cell’s genome once per cell cycle is critical to preserve cell identity, genome integrity and progression through the cell cycle. Two mutually exclusive steps of DNA replication origin licensing in which the Mcm replicative helicase is first loaded onto DNA (G1 phase), followed by origin firing (S-phase), ensure that the entire genome is replicated once and only once (Costa & Diffley, 2022). During S-phase, origins fire at varying time points, giving rise to a distinct temporal order in which segments of the genome replicate, termed the replication timing (RT) program. The RT program is developmentally regulated and aberrations in RT are observed in diseased states like cancer and progeria (Miura et al., 2019; Rivera-Mulia, Sasaki, et al., 2019). The RT program is highly correlated with transcription and features of genome architecture. It is thus an ongoing challenge to understand how these highly correlated structural and functional properties of chromosomes are causally related.
This Current Opinions article is the fourth installment in a review series we have written each 7 years on the relationship between replication timing control, nuclear organization, and transcriptional regulation. In our last review, we discussed the contribution of chromatin-conformation capture methods, which confirmed cytogenetic evidence for spatial segregation of early and late replicating chromatin and segmentation of replication units into structural domains, allowing the alignment of microscopic features with molecular maps of chromosome folding (Rivera-Mulia & Gilbert, 2016a; Ryba et al., 2010). Since then, new methods that measure the contact frequency as well as the distance of loci to nuclear landmarks and computational methods to delineate neighborhoods within the nucleus have confirmed RT as a function that closely reflects sub-nuclear position (Vouzas & Gilbert, 2021); late replicating chromatin preferentially resides near the nuclear lamina, while early replicating chromatin typically resides in the interior of the nucleus, with the earliest replicating regions frequently near nuclear speckles (Chen et al., 2018; Chen & Belmont, 2019; van Schaik et al., 2020; Wang et al., 2021). Although these features are closely correlated, they have been uncoupled in many experimental systems (Dileep et al., 2019; Lu et al., 2010; Peycheva et al., 2022), so causal links between them remain a major challenge. Here, we focus on recent progress the field has made in discovering and understanding the major regulators of RT genome wide (Rif1), chromosome wide (ASARs), or locally (ERCEs), as well as how RT can modulate nuclear function.
Replication Timing and the Epigenome: chicken or the egg
In our 2002 Current Opinions review, we proposed that chromatin could both influence and be influenced by RT, forming a positive feedback loop, now commonly known in epigenetics as the “chicken or the egg” problem (Gilbert, 2002). Indeed, much evidence for the influence of chromatin on RT has accumulated over these two decades, including recent studies demonstrating new roles for DNA methylation and histone marks in RT (Caballero et al., 2022; Du et al., 2021; Rechem et al., 2021; Stow et al., 2022; Takebayashi et al., 2021; Wang et al., 2022). Much more difficult to demonstrate has been whether there is a role for RT in dictating chromatin structure. Since chromatin is assembled at the replication fork, it was reasonable to postulate that the RT of a domain could have a direct influence on nascent chromatin assembly. Indeed, viral plasmids replicating early/late during S phase assemble hyper/hypo acetylated chromatin (Lande-Diner et al., 2009). However, there had previously been no means to globally disrupt RT and evaluate its necessity. Recently, such a global disruption, mediated by abrupt depletion of RIF1 (Figure 1A), a known regulator of RT (Alavi et al., 2021), was shown to lead directly (in the first S phase) to the redistribution of both active and repressive histone marks. This was followed by alterations in genome architecture and transcription that accumulated gradually with each aberrant S phase (K. N. Klein et al., 2021). It will now be important to use tools, such as ChOR-seq (Petryk et al., 2021), to identify mechanisms for maintaining histone marks at replication forks that differ when replication takes place at different times and locations in the nucleus.
Figure 1: Rif1 Coordinates the RT program of hESCs.
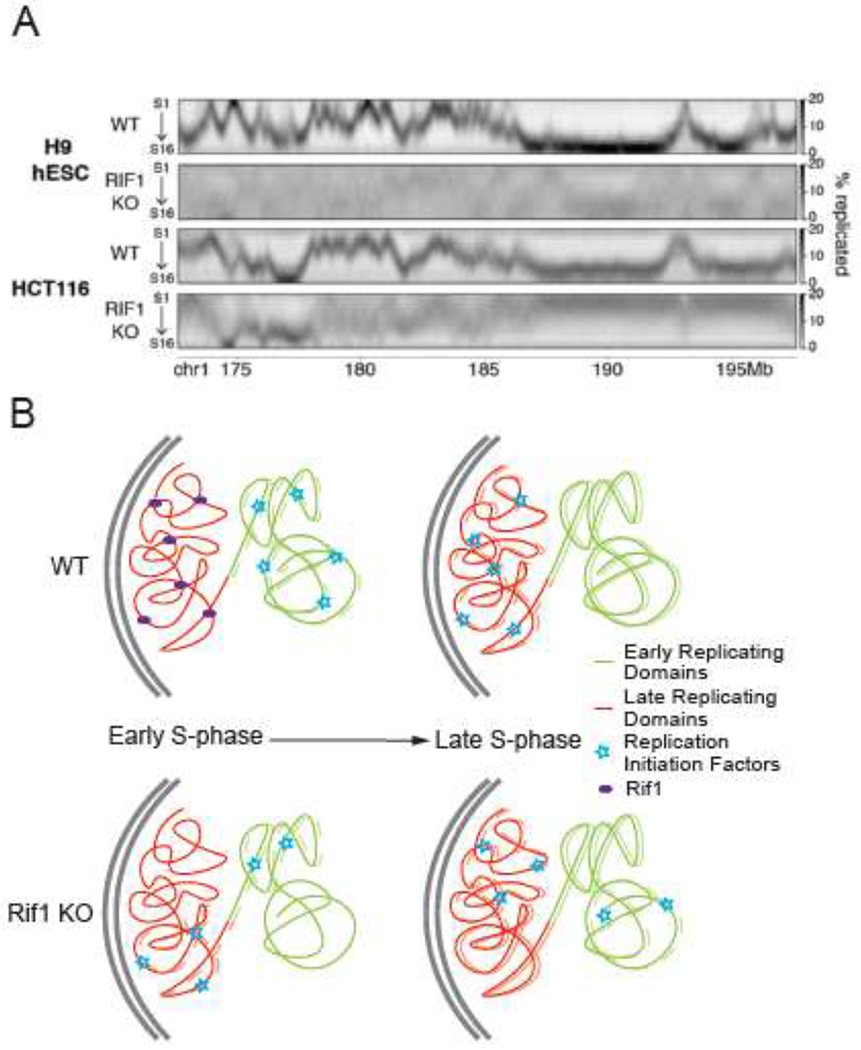
(A) Parental (WT) and RIF1 knockout (KO) high resolution Repli-seq (as described in Zhao, 2020) of a 20Mb segment of chromosome 1 in the human ESC line WA09 (H9) (top) and in human colorectal carcinoma, HCT116 cells (bottom). (from Klein et al, 2021). Nascent DNA that was replicated in each of 16 temporal windows of S phase was sequenced and the density of reads displayed as a heat map (Y-axis rows). Most cells in a WT population replicate any given genomic region within a defined temporal window of S phase. In RIF1 KO cells, either the entire genome (H9) or large segments of the genome (HCT116) are replicated stochastically. HCT116 differs from H9 in having late replicating regions that are unaffected by RIF1 KO. (B) Model of Rif1 dependent RT control. During early S-phase in WT cells Rif1 shields late replicating chromatin domains from replication initiation factors. This directs the replication initiation factors to early replicating chromatin domains. During late S-phase, Rif1 is removed from late replicating domains, allowing their access to initiation factors. KO of Rif1 allows late replicating domains to compete with early domains for limiting replication initiation factors, resulting in an increase in the heterogeneity of replication timing.
Although mechanisms by which chromatin proteins influence RT in mammalian cells are still poorly understood, based on findings in budding and fission yeast, it is generally presumed that they work by recruiting or antagonizing a limiting pool of initiation factors that activate the replicative helicases assembled in early G1 phase (He et al., 2022; Richards et al., 2022; Santos et al., 2022; Yoshida et al., 2014). In mammalian cells, most trans-acting factors that affect RT do so only in a local or domain-specific manner (Du et al., 2021; Spracklin et al., 2022; van Schaik et al., 2022). To date, the regulator with the most consistent effect on RT in species from yeast to human and with the most global effect in most mammalian cell types is RIF1. RIF1 recruits Protein Phosphatase 1 (PP1) to antagonize the phosphorylation of MCM in yeast and Drosophila and it was recently shown that mouse Rif1 also contains a PP1 binding domain that is necessary for Rif1 to properly regulate RT (Cho et al., 2022; Gnan et al., 2021; Hiraga et al., 2014; Mattarocci et al., 2014; Peace et al., 2014). Rif1’s role in RT nicely illustrates some the complexities of RT regulation in mammalian cells. First, mutation of the PP1 interacting domain did not fully account for the phenotypic effects of Rif1 gene disruption; the remaining activity is still unknown (Gnan et al., 2021). Second, Rif1 is enriched almost exclusively in late replicating chromatin but its loss results in equally profound effects of both early and late replicating regions. One explanation is that depletion of Rif1 allows late domains to compete with early domains for a limiting pool of initiation factors, resulting in more heterogeneous replication firing times genome-wide (Figure 1B), a mechanism that is familiar in yeast mutants that affect RT (He et al., 2022; Santos et al., 2022; Yoshida et al., 2014). Lastly, different cell types exhibit varied RT phenotypes in response to Rif1 loss (discussed below). Cell type specificity is a recurrent feature of RT regulatory mechanisms in mammals (discussed below) and sets them apart from yeast. Clearly, a ripe area of investigation in the future will be to unveil the myriad alternative mechanisms cells employ to regulate RT and whether they all converge downstream on pathways conserved in yeasts.
Recently, a study presented the surprising finding that Mcm6 depletion can cause a dramatic global disruption of RT in mouse B cells (Peycheva et al., 2022). The ability to disrupt RT led to the discovery that RT is necessary for the recurring AID-mediated translocations observed between early replicating sites in B cells. However, the mechanism by which a reduction in the levels of a protein complex so critical to replication initiation could result in alterations in RT, and whether this is a natural means to control RT, remains to be demonstrated and may be difficult to discern. In budding yeast, depletion of Mcm4 decreased Mcm loading at a subset of origins and only locally altered RT (Dukaj & Rhind, 2021). One model is that a lower Mcm dosage may reduce a competitive advantage of early domains for limiting factors. Interestingly, while there is evidence that Mcm density positively correlates with early replication in yeast (Dukaj & Rhind, 2021), recent maps of human Mcm double hexamers detect a much higher density of Mcm in late replicating domains (Li et al., 2023). Also, since the activated B cells in Peycheva studies have unusually rapid S phases and depletion required several days of Mcm6 shRNA expression, it will be important to rule out indirect effects of replication stress, not assessed in this study
Coordinating Replication Timing With S phase of the Cell Cycle
Replication must also be timed to ensure that the entire genome is replicated within S phase or risk entering mitosis with unreplicated DNA. This is usually attributed to a G2/M checkpoint that restrains entry into mitosis until replication is completed. However, in the latest chapter in what is emerging as one of the most impactful discoveries in the history of chromosome biology, a class of cis-acting very long non-coding RNAs (vlncRNAs) has been shown to be responsible for ensuring each chromosome pair completes synchronous replication during S phase. Numerous studies over the last century have documented severely delayed RT (DRT) and delayed mitotic condensation (DMC) in over 80% of cancers (Thayer, 2012). This phenomenon was traced to specific loci containing >200kb vlncRNAs that are mono-allelically expressed, asynchronously replicated and coat their native chromosome territory. These vlncRNAs, termed Asynchronous Replication and Autosomal RNAs (ASARs), have since been validated as being both necessary for proper RT of 4 different human chromosomes by deletions (as small as 6-8kb) within the ASAR genes (Heskett et al., 2022b), and sufficient to induce DRT/DMC by insertion into ectopic sites, and by oligonucleotide directed RNA degradation of the ectopically expressed RNA (Platt et al., 2018). Disrupting their normal regulation in 6 different human cell types and 2 mouse cell types (Breger et al., 2004) delays replication and condensation of their resident chromosome into G2 and mitosis and leads to catastrophic genome rearrangements resembling chromothripsis (Breger et al., 2005; Stoffregen et al., 2011).
The most recent pair of reports on ASARs reveal their genome-wide abundance and demonstrate that they function through interactions with abundant poorly understood RNA binding proteins. In the first study, the core characteristics of known ASARs were used to identify hundreds of potential ASAR elements genome wide, found to comprise 2.5% of the genome (Heskett et al., 2022a), remarkably identical to the percentage of randomly induced translocations that caused DRT and DMC in the pioneering studies (Thayer, 2012). A total of 8 of these elements on 4 chromosomes were shown to cause DRT and DCC phenotypes when disrupted, validating their ASAR activity (Heskett et al., 2020, 2022a). In the second study (Thayer et al., 2022), a group of RNA binding proteins were found to associate with a 7kb region within one of the ASARs. Ectopic expression of only that RBP binding region caused DRT and DMC of its resident chromosome. Moreover, depletion of any of 10 of these RNA binding proteins eliminated localization of ASARs RNAs from their chromosome territories and produced DRT/DMC aberrations across every autosome pair. Thus, ASARs constitute novel cis-acting functional elements of human chromosomes that ensure their timely replication within the confines of S-phase, preventing catastrophic genome instability. With respect to the RT program, it will be interesting to see how ensuring replication is completed during S phase is related to the temporal order of replication.
RT and transcription
The mechanistic basis for the correlation between transcription and early RT remains a major gap in our understanding. A caveat to the global correlation is that it is driven by the majority of genes that are constitutively early replicating and expressed (Rivera-Mulia et al., 2015). Genes that are developmentally regulated for both transcription and RT do not show a strong correlation between RT and transcriptional activity; 2/3 are expressed and late replicating in at least one cell type and some are even induced when switching from early to late replicating (Hiratani et al., 2010; Rivera-Mulia et al., 2015). When the RT program is disrupted genome-wide by either RIF1 (K. N. Klein et al., 2021) or Mcm (Peycheva et al., 2022) depletion, very few genes are transcriptionally altered. Asynchronously replicated and mono-allelically expressed genes regions can be randomly fixed as either early or late replicating, even for different genes within the same loci (Heskett et al., 2022a). Thus it is clear that early replication is neither necessary or sufficient for transcription and vice versa. However, as more groups have experimented with various methods to induce transcriptional activity and study its effect on RT, a high degree of context-dependence has emerged. In one case, targeting a strong acidic transcriptional activator to the promoters of late replicating genes both activated their transcription and advanced their RT (Brueckner et al., 2020; Therizols et al., 2014). In another study, insertion of a strongly expressed reporter gene did not advance RT (Hassan-Zadeh et al., 2012). Yet another study showed that activation of transcription of a long silent and late replicating gene by the Dox-inducible promoter altered sites of initiation but did not advance RT in the presence or absence of Dox, leading the authors to conclude that transcription is not sufficient to advance RT (Blin et al., 2019). In this same study, using stronger promoters to drive the same gene both led to more robust transcription and did advance RT. What is missing from all of these studies is a means to determine whether it is the transcription itself that is promoting the RT advance, or some associated step of transcriptional activation that may be promoter-specific. It is clear from every genomics survey of RT and transcription that genic transcription is neither necessary nor sufficient for early RT (Rivera-Mulia & Gilbert, 2016b), however this does not rule out steps of transcription that precede processive transcriptional elongation. Indeed, in the Hassan-Zedeh study where strong transcription was not sufficient to advance RT (Hassan-Zadeh et al., 2012), adding a binding site for the USF transcription factor nearby did elicit an RT advance. Two classical studies further illustrate this point. In one, targeting a histone acetyltransferase to a late replicating region, led to an advance in RT without any detectable activation of transcription (Goren et al., 2008). In another, targeting a transgene to a late replicating locus let to high levels of histone acetylation and advanced RT but in an orientation-dependent manner and deletion of the promoter driving transcription retained orientation-dependent histone acetylation and early RT (Lin et al., 2003). Together, these results suggest that a high degree of histone acetylation, not transcription, is what drives early replication. What is needed are systematic studies of different promoters that do or do not advance RT, to determine which steps in their transcriptional induction mechanisms are able to advance RT.
Developmental Control of DNA RT
Systematic studies of RT during cell fate changes have found that about half of the genome undergoes programmed changes in RT in at least one cell lineage (Hiratani et al., 2004, 2008, 2010; Perry et al., 2004; Rivera-Mulia et al., 2015). The mechanisms regulating these changes are an active area of pursuit and can be naively thought of as consisting of repressive mechanisms that delay RT vs. active mechanisms that advance RT.
In the case of regulators that delay RT, several pathways that are present in almost all cell types nonetheless vary in their importance to regulation of RT in different cell types. For example, the depletion of RIF1 has a nearly genome-wide effect on the RT program of human embryonic stem cells (hESCs), while HCT116 colon cancer cells have a substantial number of RIF1-independent domains whose late replication is regulated, at least in part, by H3K9 methyltransferases (K. N. Klein et al., 2021). Interestingly, chromatin in HCT116 that is dependent vs. independent of RIF1 for late replication have distinct chromatin composition and interaction patterns that can be found at varying abundance in different cell types (Spracklin et al., 2022). Finally, mESCs have a substantial number of Rif1-independent late replicating domains that are associated with the nuclear lamina whereas in hESCs, late replication of LADs is not RIF1-independent (Foti et al., 2016).
An exciting recent advance in understanding RT regulation is the discovery of Early Replication Control Elements (ERCEs), cis-acting elements that regulate RT, transcription and genome architecture of entire domains (Sima et al., 2019). While one ERCE can create a mid-S replicating domain, early replicating domains contain multiple redundant ERCEs in which deletion of a single ERCE can have little to no effect on RT, while deletion of all ERCEs results in late replication. ERCEs do not significantly overlap with origins of replication and may, rather, create a sub-nuclear environment that increases the probability of initiation in their vicinity. Consistently, ERCEs interact physically with each other, and are highly decorated with acetylated histones, which could create a 3D hub of active chromatin (Sima et al., 2019). It is known that RT is established in early G1 phase at the Timing Decision Point (TDP), which coincides with the re-assembly of genome architecture after each mitosis and is subsequently lost during S phase. It is tempting to speculate that the TDP is when ERCEs organize into hubs. In fact, the 3D clustering of Fkh1/2 transcriptional activators, which occurs during G1 in budding yeast, is necessary for early replication of a large fraction of the yeast genome (Ostrow et al., 2017; Zhang et al., 2019), and a pair of reporter genes capable of advancing RT in chicken DT40 cells were also found to interact (Brossas et al., 2020). Discovered in mESCs, ERCEs are sites of Oct4, Sox2 and Nanog (OSN) co-binding, all of which are master regulators of transcription in mESCs. Master regulators of transcription were also implicated in bipartite RT and transcriptional regulatory networks (TRNs), constructed through network analysis of RT and transcription changes during hESC differentiation to multiple lineages, and were shown to bind at sites within the domains that switch to early RT in the corresponding cell type (Rivera-Mulia, Kim, et al., 2019). Altogether, this suggests that the developmental regulation of RT may be triggered by master TFs that control cell fate changes and have been co-opted to create chromatin environments conducive to early replication. It is now important to directly determine whether these master TFs regulate ERCE activity.
Why would RT be regulated during development? The hypothesis that TRNs trigger RT changes combined with the recent demonstration that the epigenome can be remodeled by changes in RT provides an attractive albeit speculative model in which cell signaling during development initiates changes in master TFs, which both induce transient changes in gene expression and also create new ERCEs. ERCE reprogramming would elicit changes in RT, rapidly altering chromatin assembled throughout the domain, locking in the cell fate change (K. N. Klein et al., 2021). In this model (Figure 2) RT plays an important role in the transition from cell specification to cell determination during development. In fact, Rif1 depletion, which leads to global loss of RT and disruption of the epigenome in many cell types, and interferes with ERCE function through as yet unidentified mechanisms, has very little effect on cell growth or cellular phenotypes, suggesting that the RT program is not necessary to maintain cellular identity. Nonetheless, Rif1 null mutations are lethal in mice, whose development is arrested at various points after gastrulation (Buonomo et al., 2009), consistent with a role for RT in cell lineage determination.
Figure 2:
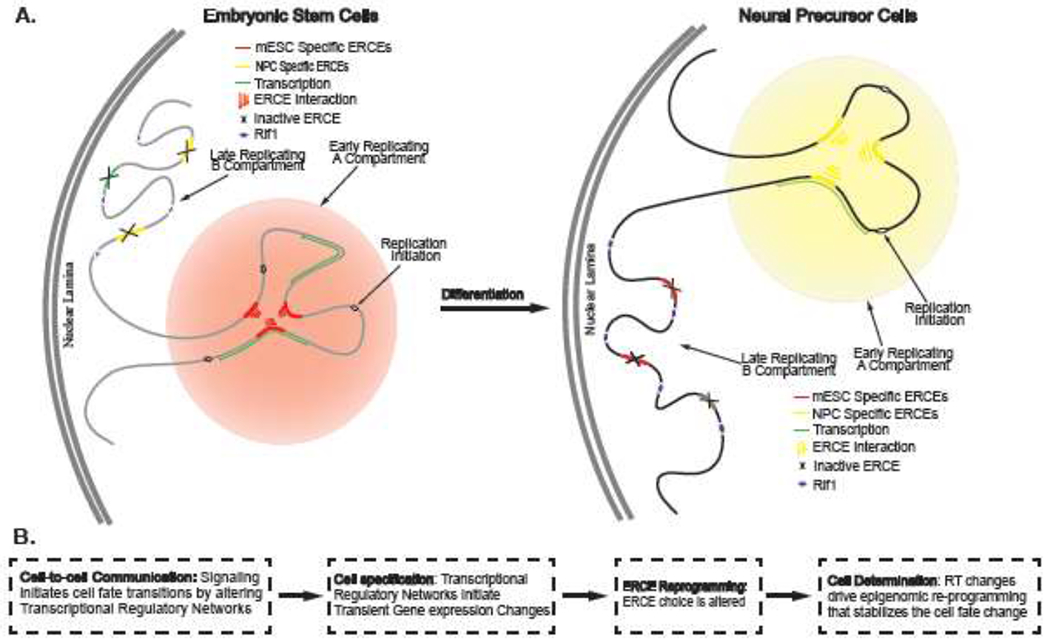
Replication Timing (RT) as a Driver of Transitions from Cell Specification to Cell Determination. (A) Given that ERCEs bind core transcriptional regulatory network (TRN) transcription factors (TFs), we propose that developmental changes in RT are driven by the differential usage of cell type specific ERCEs. ERCEs interact and may create 3D hubs that seed the assembly of sub-nuclear microenvironments promoting early replication. Changes in TRNs during differentiation, would inactivate some ERCEs and activate others, resulting dramatic shifts in domain compartmentalization and RT. (B) Cell fate transitions are initiated by cell cell communication through juxtracrine and paracrine signaling. These factors alter TRNs in cells that are competent to receive the signals. We speculate that the altered TRNs alone can initiate cell fate changes (specification) but are not initially sufficiently robust to stabilize them to fluctuations in cell signaling. Rather, TRN changes alter the set of active ERCEs, generating a new RT program that reprograms the epigenome of the cell, thus stabilizing the cell fate change.
Of Origins and Timing: wherein lies the regulation?
It is clear that initiation site usage and the timing of firing are independently regulated from yeast to human, but the extent to which an origin has an intrinsic tendency to initiate at a particular time is much less clear. In yeasts, which have very efficient origins, disruption of timing mechanisms results in the same origins firing at alternative times during S phase (Ostrow et al., 2017; Zhang et al., 2019). The same is true in mammals; when regions replicate at different times, the most efficient origins within those regions are typically the same sites (Besnard et al., 2012; Peycheva et al., 2022). Moreover, conditions can be found in which timing is maintained but origin specification is lost or in which timing is lost, but origin specification maintained (Dimitrova et al., 2002; Peycheva et al., 2022). It is important to keep in mind that initiation site usage is highly heterogeneous in mammalian cells, with initiation within any ~50kb zone occurring in less than 10% of S phases and at any particular site within each zone at a frequency decidedly less than that (K. Klein et al., 2017). Replication timing is also heterogeneous, but less so, with the majority of sequences replicating within 25% of S phase at least 50% of the time (Dileep & Gilbert, 2018; Zhao et al., 2020). Finally, ERCEs clearly are necessary for RT but do not overlap with detectable replication origins (Sima et al., 2019). Altogether, it is clear that replication timing is regulated independent of origin specification. Thus, it came as a surprise when it was recently reported that an 800bp deletion of a single origin site upstream of the Myc gene caused a shift in RT from early to middle replicating (Peycheva et al., 2022). One possibility is that, since Myc is in a constitutively early region (early in all cell types), regulation of RT may be different in constitutive vs. developmentally-regulated RT domains. Nonetheless, there are other origins in this region so it is puzzling that deletion of one origin would render all other origins ineffective. Indeed, insertions containing origins also require additional activities to significantly advance RT (Brossas et al., 2020; Hassan-Zadeh et al., 2012). In fact, an early study demonstrated that, inserting an early origin into a late replicating site advanced its RT, but deletion of the origin sequences themselves showed that the RT advancing activity was distinct from the origin sequences, demonstrating that such sequences can reside close together (Lin et al., 2003). Thus, an alternative possibility is that the deleted region containing one of the Myc locus origins will be found to contain an independent element regulating RT.
Conclusions and Future Perspectives
In the last 7 years, the field has taken decisive steps in discovering and understanding some of the major regulators of RT in mammalian cells. RT can be controlled genome wide by factors like Rif1, chromosome wide by ASARs, and domain wide by ERCEs, LADs or certain flavors of heterochromatin (combinations of histone marks, histone variants and DNA methylation). These factors are developmentally regulated; Rif1, LADs and chromatin flavors vary in their importance for late replication control in different cell types, while ERCEs may control early replication through cell type specific master TFs of cell identity. It has also become clear that RT regulates the assembly of the epigenome, completing the self-reinforcing positive feedback loop we proposed in this series of Current Opinions articles over 20 years ago, and solidifying RT as an epigenomic regulator (Gilbert, 2002; Hiratani & Gilbert, 2009). Finally, the regulation of whole chromosome replication by ASARs has unveiled a central role for a huge fraction of genomic dark matter in the regulation of chromosome replication and genome stability.
In the next 7 years, we expect to see great progress in elucidating the mechanisms by which ERCEs and ASARs control RT and whether their varied functions in regulating replication, architecture and transcription are related or separable. In the case of ASARs, in addition to understanding how they interact with numerous previously uncharacterized RNPs to ensure timely chromosome replication, it will be critical to understand whether replicating in a timely fashion is related to replicating in the proper temporal order. To fully understand the structure of ERCEs and their mechanism of developmental control of RT, it is critical to study ERCEs both in their native context, and to understand them sufficiently so as to build ERCEs from minimal DNA sequences. The spatio-temporal mechanisms by which RT is established at the TDP is also an important area of pursuit. Modern tools to study nuclear organization may reveal how sub-nuclear location and RT are related. Finally, we envision RT becoming an integral part of developmental biology; in particular, how the interplay between induced changes in RT and the resulting epigenetic reprogramming may drive cell fate transitions is a ripe area of research.
Acknowledgements
This work was supported by National Institute of Health (NIH) grant GM083337 (D.M.G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: none
Conflict of Intrest
The authors declare no conflict of interest.
References
- Alavi S, Ghadiri H, Dabirmanesh B, Moriyama K, Khajeh K, & Masai H (2021). G-quadruplex binding protein Rif1, a key regulator of replication timing. In Journal of biochemistry (Vol. 169, Issue 1). 10.1093/jb/mvaa128 [DOI] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin J-M, & Lemaitre J-M (2012). Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nature Structural & Molecular Biology, 19(8), 837–844. 10.1038/nsmb.2339 [DOI] [PubMed] [Google Scholar]
- Blin M, Le Tallec B, Nähse V, Schmidt M, Brossas C, Millot GA, Prioleau MN, & Debatisse M (2019). Transcription-dependent regulation of replication dynamics modulates genome stability. Nature Structural and Molecular Biology, 26(1), 58–66. 10.1038/s41594-018-0170-1 [DOI] [PubMed] [Google Scholar]
- Breger KS, Smith L, & Thayer MJ (2005). Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Human Molecular Genetics, 14(19), 2813–2827. 10.1093/hmg/ddi314 [DOI] [PubMed] [Google Scholar]
- Breger KS, Smith L, Turker MS, & Thayer MJ (2004). Ionizing radiation induces frequent translocations with delayed replication and condensation. Cancer Research, 64(22), 8231–8238. 10.1158/0008-5472.CAN-04-0879 [DOI] [PubMed] [Google Scholar]
- Brossas C, Valton A, Venev SV, Chilaka S, Counillon A, Laurent M, Goncalves C, Duriez B, Picard F, Dekker J, & Prioleau M (2020). Clustering of strong replicators associated with active promoters is sufficient to establish an early-replicating domain. The EMBO Journal, 39(21), 1–21. 10.15252/embj.201899520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner L, Zhao PA, Schaik T, Leemans C, Sima J, Peric-Hupkes D, Gilbert DM, & Steensel B (2020). Local rewiring of genome–nuclear lamina interactions by transcription. The EMBO Journal, 39(6), 1–17. 10.15252/embj.2019103159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SBC, Wu Y, Ferguson D, & Lange T. De. (2009). Mammalian Rif1 contributes to replication stress survival and homology-directed repair. Journal of Cell Biology, 187(3). 10.1083/jcb.200902039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero M, Ge T, Rebelo AR, Seo S, Kim S, Brooks K, Zuccaro M, Kanagaraj R, Vershkov D, Kim D, Smogorzewska A, Smolka M, Benvenisty N, West SC, Egli D, Mace EM, & Koren A (2022). Comprehensive analysis of DNA replication timing across 184 cell lines suggests a role for MCM10 in replication timing regulation. Human Molecular Genetics, 31(17), 2899–2917. 10.1093/hmg/ddac082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, & Belmont AS (2019). Genome organization around nuclear speckles. In Current Opinion in Genetics and Development (Vol. 55). 10.1016/j.gde.2019.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, Steensel B. Van, Ma J, & Belmont AS (2018). Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. Journal of Cell Biology, 217(11). 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C-Y, Seller CA, & O’Farrell PH (2022). Temporal control of late replication and coordination of origin firing by self-stabilizing Rif1-PP1 hubs in Drosophila. Proceedings of the National Academy of Sciences, 119(26). 10.1073/pnas.2200780119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, & Diffley JFX (2022). The Initiation of Eukaryotic DNA Replication. In Annual Review of Biochemistry (Vol. 91). 10.1146/annurev-biochem-072321-110228 [DOI] [PubMed] [Google Scholar]
- Dileep V, & Gilbert DM (2018). Single-cell replication profiling to measure stochastic variation in mammalian replication timing. Nature Communications, 9(1). 10.1038/s41467-017-02800-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Wilson KA, Marchal C, Lyu X, Zhao PA, Li B, Poulet A, Bartlett DA, Rivera-Mulia JC, Qin ZS, Robins AJ, Schulz TC, Kulik MJ, McCord RP, Dekker J, Dalton S, Corces VG, & Gilbert DM (2019). Rapid Irreversible Transcriptional Reprogramming in Human Stem Cells Accompanied by Discordance between Replication Timing and Chromatin Compartment. Stem Cell Reports, 13(1). 10.1016/j.stemcr.2019.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, & Gilbert DM (2002). Mammalian nuclei become licensed for DNA replication during late telophase. Journal of Cell Science, 115(1), 51–59. 10.1242/jcs.115.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Smith GC, Luu PL, Ferguson JM, Armstrong NJ, Caldon CE, Campbell EM, Nair SS, Zotenko E, Gould CM, Buckley M, Chia KM, Portman N, Lim E, Kaczorowski D, Chan CL, Barton K, Deveson IW, Smith MA, … Clark SJ (2021). DNA methylation is required to maintain both DNA replication timing precision and 3D genome organization integrity. Cell Reports, 36(12). 10.1016/j.celrep.2021.109722 [DOI] [PubMed] [Google Scholar]
- Dukaj L, & Rhind N (2021). The capacity of origins to load mcm establishes replication timing patterns. PLoS Genetics, 17(3). 10.1371/journal.pgen.1009467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, Buness A, Klein FA, Huber W, Johnstone E, Loos R, Bertone P, Gilbert DM, Manke T, Jenuwein T, & Buonomo SCB (2016). Nuclear Architecture Organized by Rif1 Underpins the Replication-Timing Program. Molecular Cell, 61(2), 260–273. 10.1016/j.molcel.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2002). Replication timing and transcriptional control: Beyond cause and effect. Current Opinion in Cell Biology, 14(3), 377–383. 10.1016/S0955-0674(02)00326-5 [DOI] [PubMed] [Google Scholar]
- Gnan S, Flyamer IM, Klein KN, Castelli E, Rapp A, Maiser A, Chen N, Weber P, Enervald E, Cardoso MC, Bickmore WA, Gilbert DM, & Buonomo SCB (2021). Nuclear organisation and replication timing are coupled through RIF1–PP1 interaction. Nature Communications, 12(1). 10.1038/s41467-021-22899-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, & Cedar H (2008). DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes and Development. 10.1101/gad.468308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Zadeh V, Chilaka S, Cadoret J-C, Ma MK-W, Boggetto N, West AG, & Prioleau M-N (2012). USF Binding Sequences from the HS4 Insulator Element Impose Early Replication Timing on a Vertebrate Replicator. PLoS Biology, 10(3), e1001277. 10.1371/journal.pbio.1001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Petrie MV, Zhang H, Peace JM, & Aparicio OM (2022). Rpd3 regulates single-copy origins independently of the rDNA array by opposing Fkh1-mediated origin stimulation. Proceedings of the National Academy of Sciences, 119(40). 10.1073/pnas.2212134119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heskett MB, Smith LG, Spellman P, & Thayer MJ (2020). Reciprocal monoallelic expression of ASAR lncRNA genes controls replication timing of human chromosome 6. RNA, 26(6), 724–738. 10.1261/RNA.073114.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heskett MB, Vouzas AE, Smith LG, Yates PA, Boniface C, Bouhassira EE, Spellman PT, Gilbert DM, & Thayer MJ (2022a). Epigenetic control of chromosome-associated lncRNA genes essential for replication and stability. Nature Communications, 13(1), 6301. 10.1038/s41467-022-34099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heskett MB, Vouzas AE, Smith LG, Yates PA, Boniface C, Bouhassira EE, Spellman PT, Gilbert DM, & Thayer MJ (2022b). Epigenetic control of chromosome-associated lncRNA genes essential for replication and stability. Nature Communications, 13(1), 6301. 10.1038/s41467-022-34099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Alvino GM, Chang FJ, Lian HY, Sridhar A, Kubota T, Brewer BJ, Weinreich M, Raghuraman MK, & Donaldson AD (2014). Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7- mediated phosphorylation of the MCM complex. Genes and Development, 28(4), 372–383. 10.1101/gad.231258.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, & Gilbert DM (2009). Replication timing as an epigenetic mark. In Epigenetics (Vol. 4, Issue 2). 10.4161/epi.4.2.7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Leskovar A, & Gilbert DM (2004). Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.0406687101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, Rathjen PD, & Gilbert DM (2010). Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Research. 10.1101/gr.099796.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schübeler D, & Gilbert DM (2008). Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biology, 6(10), 2220–2236. 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KN, Zhao PA, Lyu X, Sasaki T, Bartlett DA, Singh AM, Tasan I, Zhang M, Watts LP, Hiraga SI, Natsume T, Zhou X, Baslan T, Leung D, Kanemaki MT, Donaldson AD, Zhao H, Dalton S, Corces VG, & Gilbert DM (2021). Replication timing maintains the global epigenetic state in human cells. Science, 372(6540). 10.1126/science.aba5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K, Wang W, Borrman T, Chan S, Zhang D, Weng Z, Hastie A, Chen C, Gilbert DM, & Rhind N (2017). Genome-Wide Identification of Early-Firing Human Replication Origins by Optical Replication Mapping. BioRxiv, 214841. 10.1101/214841 [DOI] [Google Scholar]
- Lande-Diner L, Zhang J, & Cedar H (2009). Shifts in Replication Timing Actively Affect Histone Acetylation during Nucleosome Reassembly. Molecular Cell, 34(6), 767–774. 10.1016/j.molcel.2009.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dong J, Wang W, Yu D, Fan X, Hui YC, Lee CSK, Lam WH, Alary N, Yang Y, Zhang Y, Zhao Q, Chen C-L, Tye B-K, Dang S, & Zhai Y (2023). The human pre-replication complex is an open complex. Cell, 186(1), 98–111.e21. 10.1016/j.cell.2022.12.008 [DOI] [PubMed] [Google Scholar]
- Lin CM, Fu H, Martinovsky M, Bouhassira E, & Aladjem MI (2003). Dynamic Alterations of Replication Timing in Mammalian Cells. Current Biology, 13(12), 1019–1028. 10.1016/S0960-9822(03)00382-8 [DOI] [PubMed] [Google Scholar]
- Lu J, Li F, Murphy CS, Davidson MW, & Gilbert DM (2010). G2 phase chromatin lacks determinants of replication timing. Journal of Cell Biology, 189(6), 967–980. 10.1083/jcb.201002002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarocci S, Shyian M, Lemmens L, Damay P, Altintas DM, Shi T, Bartholomew CR, Thomä NH, Hardy CFJ, & Shore D (2014). Rif1 Controls DNA replication timing in yeast through the PP1 Phosphatase Glc7. Cell Reports, 7(1). 10.1016/j.celrep.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Miura H, Takahashi S, Poonperm R, Tanigawa A, Takebayashi S ichiro, & Hiratani I (2019). Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization. Nature Genetics, 51(9), 1356–1368. 10.1038/s41588-019-0474-z [DOI] [PubMed] [Google Scholar]
- Ostrow AZ, Kalhora R, Gana Y, Villwocka SK, Linke C, Barberisb M, Chena L, & Aparicioa OM (2017). Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.1612422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace JM, Ter-Zakarian A, & Aparicio OM (2014). Rif1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PloS One, 9(5), e98501. 10.1371/journal.pone.0098501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P, Sauer S, Billon N, Richardson WD, Spivakov M, Warnes G, Livesey FJ, Merkenschlager M, Fisher AG, & Azuara V (2004). A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle, 3(12). 10.4161/cc.3.12.1346 [DOI] [PubMed] [Google Scholar]
- Petryk N, Reverón-Gómez N, González-Aguilera C, Dalby M, Andersson R, & Groth A (2021). Genome-wide and sister chromatid-resolved profiling of protein occupancy in replicated chromatin with ChOR-seq and SCAR-seq. In Nature Protocols (Vol. 16, Issue 9). 10.1038/s41596-021-00585-3 [DOI] [PubMed] [Google Scholar]
- Peycheva M, Neumann T, Malzl D, Nazarova M, Schoeberl UE, & Pavri R (2022). DNA replication timing directly regulates the frequency of oncogenic chromosomal translocations. Science, 377(6612). 10.1126/science.abj5502 [DOI] [PubMed] [Google Scholar]
- Platt EJ, Smith L, & Thayer MJ (2018). L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. The Journal of Cell Biology, 217(2), 541–553. 10.1083/jcb.201707082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechem C Van, Ji F, Chakraborty D, Black JC, Sadreyev RI, & Whetstine JR (2021). Collective regulation of chromatin modifications predicts replication timing during cell cycle. Cell Reports, 37(1). 10.1016/j.celrep.2021.109799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards L, Das S, & Nordman JT (2022). Rif1-Dependent Control of Replication Timing. Genes, 13(3). 10.3390/genes13030550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, Schulz TC, Menendez L, Kulik MJ, Dalton S, Gabr H, Kahveci T, & Gilbert DM (2015). Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Research, 25(8), 1091–1103. 10.1101/gr.187989.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, & Gilbert DM (2016a). Replication timing and transcriptional control: Beyond cause and effect - part III. Current Opinion in Cell Biology, 40, 168–178. 10.1016/j.ceb.2016.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, & Gilbert DM (2016b). Replication timing and transcriptional control: Beyond cause and effect - part III. In Current Opinion in Cell Biology (Vol. 40). 10.1016/j.ceb.2016.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Kim S, Gabr H, Chakraborty A, Ay F, Kahveci T, & Gilbert DM (2019). Replication timing networks reveal a link between transcription regulatory circuits and replication timing control. Genome Research, 29(9), 1415–1428. 10.1101/gr.247049.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Sasaki T, Trevilla-Garcia C, Nakamichi N, Knapp DJHF, Hammond CA, Chang BH, Tyner JW, Devidas M, Zimmerman J, Klein KN, Somasundaram V, Druker BJ, Gruber TA, Koren A, Eaves CJ, & Gilbert DM (2019). Replication timing alterations in leukemia affect clinically relevant chromosome domains. Blood Advances, 3(21). 10.1182/bloodadvances.2019000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, & Gilbert DM (2010). Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Research, 20(6). 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MM, Johnson MC, Fiedler L, & Zegerman P (2022). Global early replication disrupts gene expression and chromatin conformation in a single cell cycle. Genome Biology, 23(1), 217. 10.1186/s13059-022-02788-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, Bartlett DA, Zhao PA, Washburn BK, Nora EP, Kraft K, Mundlos S, Bruneau BG, Ljungman M, Fraser P, … Gilbert DM (2019). Identifying cis Elements for Spatiotemporal Control of Mammalian DNA Replication. Cell, 176(4), 816–830.e18. 10.1016/j.cell.2018.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G, Abdennur N, Imakaev M, Chowdhury N, Pradhan S, Mirny L, & Dekker J (2022). Diverse silent chromatin statesmodulate genome compartmentalization and loop extrusion barriers. Nature Structural & Molecular Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffregen EP, Donley N, Stauffer D, Smith L, & Thayer MJ (2011). An autosomal locus that controls chromosomewide replication timing and mono-allelic expression. Human Molecular Genetics, 20(12), 2366–2378. 10.1093/hmg/ddr138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow EC, Simmons JR, An R, Schoborg TA, Davenport NM, & Labrador M (2022). A Drosophila insulator interacting protein suppresses enhancer-blocking function and modulates replication timing. Gene, 819. 10.1016/j.gene.2022.146208 [DOI] [PubMed] [Google Scholar]
- Takebayashi SI, Ryba T, Wimbish K, Hayakawa T, Sakaue M, Kuriya K, Takahashi S, Ogata S, Hiratani I, Okumura K, Okano M, & Ogata M (2021). The temporal order of DNA replication shaped by mammalian DNA methyltransferases. Cells, 10(2). 10.3390/cells10020266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MJ (2012). Mammalian chromosomes contain cis-acting elements that control replication timing, mitotic condensation, and stability of entire chromosomes. BioEssays : News and Reviews in Molecular, Cellular and Developmental Biology, 34(9), 760–770. 10.1002/bies.201200035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MJ, Heskett MB, Smith LG, Spellman P, & Yates PA (2022). ASAR RNAs control replication through interactions with dozens of RNA binding proteins. Biorxiv. 2022.06.04.494840; doi: 10.1101/2022.06.04.494840 [DOI] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, & Bickmore WA (2014). Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science, 346(6214), 1238–1242. 10.1126/science.1259587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik T, Manzo SG, Vouzas AE, Liu NQ, Teunissen H, de Wit E, Gilbert DM, & van Steensel B (2022). Dynamic chromosomal interactions and control of heterochromatin positioning by Ki-67. EMBO Reports, e55782. 10.15252/embr.202255782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik T, Vos M, Peric-Hupkes D, Celie PHN, & van Steensel B (2020). Cell cycle dynamics of lamina-associated DNA. EMBO Reports, 21(11). 10.15252/embr.202050636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouzas AE, & Gilbert DM (2021). Mammalian DNA replication timing. Cold Spring Harbor Perspectives in Biology, 13(7). 10.1101/cshperspect.a040162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Huang Y, Cheng E, Liu X, Zhang Y, Yang J, Young JTF, Brown GW, Yang X, & Shang Y (2022). LSD1 is required for euchromatic origin firing and replication timing. Signal Transduction and Targeted Therapy, 7(1), 102. 10.1038/s41392-022-00927-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Zhang R, van Schaik T, Zhang L, Sasaki T, Peric-Hupkes D, Chen Y, Gilbert DM, van Steensel B, Belmont AS, & Ma J (2021). SPIN reveals genome-wide landscape of nuclear compartmentalization. Genome Biology, 22(1). 10.1186/s13059-020-02253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Bacal J, Desmarais D, Padioleau I, Tsaponina O, Chabes A, Pantesco V, Dubois E, Parrinello H, Skrzypczak M, Ginalski K, Lengronne A, & Pasero P (2014). The Histone Deacetylases Sir2 and Rpd3 Act on Ribosomal DNA to Control the Replication Program in Budding Yeast. Molecular Cell, 54(4), 691–697. 10.1016/j.molcel.2014.04.032 [DOI] [PubMed] [Google Scholar]
- Zhang H, Petrie MV, He Y, Peace JM, Chiolo IE, & Aparicio OM (2019). Dynamic relocalization of replication origins by fkh1 requires execution of ddk function and cdc45 loading at origins. ELife, 8, 1–20. 10.7554/eLife.45512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao PA, Sasaki T, & Gilbert DM (2020). High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells. Genome Biology, 21(1), 1–20. 10.1186/s13059-020-01983-8 [DOI] [PMC free article] [PubMed] [Google Scholar]


