Abstract
Urinary tract infections (UTIs) are predominantly caused by uropathogenic Escherichia coli (E. coli). There is rapid increase in antimicrobial resistance in UTIs, also declared as a serious health threat by World Health Organization (WHO). Present study was designed to investigate the antimicrobial resistance status with specific focus on ESBLs and carbapenemases in local uropathogenic E. coli (UPEC) isolates. E. coli isolates were characterized from patients of all ages visiting diagnostic laboratories for urine examination. Demographic data was also recorded for each patient. Antibiograms were developed to observe antibiotic resistance in UPEC using Kirby Bauer disc diffusion technique. Double Disc Synergy test (DDST) was used for phenotypic ESBL test. ESBLs and carbapenemases genes were detected in UPEC using PCR. The PCR results were confirmed by sequencing. The UPEC isolates under study exhibited 78%, 77%, 74%, 72% and 55% resistance against cefotaxime, amoxicillin, erythromycin, ceftriaxone and cefixime, respectively. Resistance against colistin and meropenem was observed in 64% and 34% isolates, respectively. Phenotypic DDST identified 48% isolates as ESBLs producers. Genotypic characterization identified 70%, 74.4% and 49% prevalence of CTXM-1, TEM-1 and CTXM-15 genes respectively. One isolate was observed exhibiting co-existence of all ESBL genes. TEM-1 + CTXM-1 and TEM-1 + CTXM-1 + CTXM-15 + OXA-1 gene patterns were dominant among ESBLs. For carbapenem-resistance, 14% isolates indicated the presence of KPC whereas GES and VIM was detected in 7% and 3.4% isolates, respectively. In conclusion, our results present a high prevalence of extensively drug resistant UPEC isolates with a considerable percentage of ESBL producers. These findings propose the need of continuous surveillance for antimicrobial resistance and targeted antimicrobial therapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11274-023-03565-9.
Keywords: Extended spectrum beta lactamases, Extensively drug resistant, Uropathogenic, Carbapenemase
Introduction
Urinary tract infections (UTIs) are among the most prevalent infectious diseases reaching the overall incidence of 18/1000 in general population (Sohail et al. 2015). In Pakistan, antibiotic-resistant UTI’s are the most reported clinical infections and uropathogenic Escherichia coli (E. coli) is considered responsible for approximately 70–90% UTIs (Larramendy et al. 2020; Kahlmeter and Poulsen 2012; Tandogdu and Wagenlehner 2016; Foxman 2014). Upper UTI (pyelonephritis) is a major cause of hypertension, renal insufficiency and end-stage renal failure in children (Nader and Alejandro 2021). Early treatment of UTI with an effective antibiotic is essential for preventing long-term consequences. Treatment delay increases the risk of renal scarring and progressive kidney damage (Baziboroun et al. 2018; Terlizzi et al. 2017).
Urinary tract infections are mostly associated with increased resistance to antimicrobial agents. The emergence of multidrug resistance in UPEC may be attributed to frequent and injudicious use of antibiotics without culture sensitivity testing (Adamus-Białek et al. 2018). Additionally, extended spectrum beta-lactamases (ESBLs) are the resistance mechanisms evolved by Gram-negative bacteria, causing burden on clinical therapeutics and resulting in higher morbidity with delayed treatment and isolates are increasing at exponential rate (Shilpakar et al. 2021). ESBLs are plasmid mediated beta-lactamase genes, capable of hydrolyzing 3rd and 4th generation cephalosporins, and aztreonam (Yengui et al. 2022; Jalal et al. 2023). ESBLs are designated main drug-resistant pathogens and major variations have been detected globally in ESBL-producing isolates (Shilpakar et al. 2021; Doi et al. 2017). World Health Organization (WHO) designated ESBL-producing enteric bacteria/enterobacteriaceae as the supreme critical group for research (Larramendy et al. 2020). Plasmids harbor the genes for ESBLs that are extremely mobile and might contain genes conferring resistance to other antimicrobial groups like quinolones and aminoglycosides (Ugbo et al. 2020). SHV, TEM and CTX-M are the most common families among ESBLs (Mahamat et al. 2019; Ur Rahman et al. 2018).
In Enterobacteriaceae, increasing trend of carbapenem resistance has been reported globally from health care associated and community settings (Tilahun et al. 2021; Macareño-Castro et al. 2022). The rising incidence of carbapenem resistance is alarming. Antibiotic resistance is disseminated by different methods like ESBLs and carbapenemases, AmpC β-lactamases and porin loss. Increasing incidence of antibiotic resistance is associated with few treatment options (Tilahun et al. 2021).
Colistin is utilized as a last-resort antibiotic for the therapy of Enterobacteriaceae that are characterized as producers of carbapenemases and ESBLs. Colistin-resistant E. coli strains have become prevalent due to colistin use in farm animals (Macareño-Castro et al. 2022; Dhaouadi et al. 2020).
There are limited treatment strategies for the management of multidrug-resistant (MDR) strains and the diversity of virulence factors in E. coli (Liu et al. 2016). It is, therefore, necessary to investigate the drug resistance profiles of UPEC strains. There are very few studies on UPEC isolates from Faisalabad region (Haghighatpanah and Mojtahedi 2019; Bashir et al. 2012, 2011). There is no latest data available on UPEC locally in community settings regarding antibiotic resistance patterns. The study will provide information about circulating resistance genes in the community.
Material and methods
Study design
A total of 540 urine samples were collected from September 2018 to February 2020 from four clinical laboratories (Millat Laboratory, National Laboratory, Khidmat-E-Khalaq Laboratory and Meezan Laboratory) in Faisalabad city, Pakistan. Sterile screw-capped, wide cover containers were used to collect urine. One isolate was collected from each patient, avoiding sample repetition from the same patient. Inclusion criteria were (i) Outpatients, (ii) all age groups (iii) both genders and (iv) Positive urine culture with presumptive E. coli characterization. Exclusion criteria were (i) Inpatients and uropathogens other than E. coli.
Isolation and identification of UPEC from clinical samples
Urine samples were initially streaked on MacConkey agar (Acumedia, Michigan, US) and Cysteine Lactose Electrolyte Deficient (C.L.E.D.) agar (Hi Media laboratory Pvt Ltd, Mumbai, India) (MacConkey agar served as a selective as well as a differential culture medium. MacConkey agar was used for the isolation of Gram-negative uropathogens while C.L.E.D. agar was used for the differentiation of uropathogens based on color morphology. All media were prepared according to the manufacturer’s instructions. Urine samples were streaked on media using sterile cotton swabs. Media plates were incubated at 37 ºC overnight. Next day, plates were observed for growth. The colony of interest was picked from media containing selected pathogen using a sterile loop and mixed in 3% H2O2 on the slide for catalase test. Air bubbles and water were observed on a slide for a positive result (Habeeb et al. 2013). Triple sugar iron test is used for the characterization of various pathogens. This test is based on the pathogen’s ability for fermentation of sugars, H2S production and gas production. For TSI agar (BIOCHEM chemopharma, France), colony inoculation was performed by stabbing through the center of the medium to the bottom of the test tube and then streaking was also performed on the slant. Test tubes were incubated at 37 ºC for 18 h.
Molecular confirmation of UPEC
DNA was extracted from characterized UPEC isolates by using the conventional phenol–chloroform method (Choudhary et al. 2022). Briefly, 1.5 ml of bacterial culture was centrifuged at 10,000 rpm for 10 min. The pellet was washed in 1 ml of 10 mM Tris–HCl [pH 8.0] and re-suspended in 10% SDS followed by heating at 65 °C for 1–2 h. This bacterial suspension was mixed vigorously with chloroform: isoamyl alcohol (24:1), centrifuged at 10,000 rpm for 5 min. The supernatant containing DNA was separated. This step was repeated three times. Finally, sodium acetate (3 M; pH 4.8) and chilled isopropanol was added, mixed and stored at -20 °C overnight. This DNA preparation was centrifuged at 10,000 rpm for 10 min and supernatant was discarded. The DNA pellet was treated with 70% ethanol, the pellet was air dried and suspended in 10 mM Tris–HCl [pH 8.0]. Molecular identification was carried out using Polymerase Chain Reaction (PCR) by targeting uidA gene encoding β-glucuronidase (Table 2). The identified UPEC isolates were stored at − 20 °C as 30% glycerol stocks.
Table 2.
Primers used in this study
| Sr. No | Gene | Primer sequence (5´–3´) | Anneal. temperature | Product size (bp) | References |
|---|---|---|---|---|---|
| 1- | uidA |
F:ATCACCGTGGTGACGCATGTCGC R:CACCACGATGCCATGTTCATCTGC |
50 | 486 | Bashir et al. (2012) |
| 2- |
bla SHV-1 |
F: GGCCGCGTAGGCATGATAGA R: CCCGGCGATTTGCTGATTTC |
60 | 714 | Wajid et al. (2019) |
| 3- | bla TEM-1 |
F: CAGCGGTAAGATCCT TGAGA R: ACTCGCCGTCGTGTAGATAA |
55 | 643 | Wajid et al. (2019) |
| 4- | bla CTX-M1 |
F: AACCGTCACGCTGTTGTTAG R: TTGAGGCTGGGTGAAGTAAG |
55 | 766 | Wajid et al. (2019) |
| 5- | bla CTX-M15 |
F: CAATGTGCAGCACCAGTAA R: CGCAATATCATTGGTGGTG |
58 | 540 | Wajid et al. (2019) |
| 6- | bla CMY-2 |
F: TGGCCGTTGCCGTTATCTAC R: CCCGTTTTATGCACCCATGA |
55 | 870 | Wajid et al. (2019) |
| 7- | bla SHV-12 |
F:GGTTATTCTTATTTGTCGCTTCTT R:TACGTTACGCCACCTGGCTA |
55 | 1233 | Wajid et al. (2019) |
| 8- | OXA-1 |
F: AATGGCACCAGATTCAACTT R: CTTGGCTTTTATGCTTGATG |
55 | 640 | Li & Wang (2017) |
| 9- | bla KPC |
F:CGTCTAGTTCTGCTGTCTTG R:CTTGTCATCCTTGTTAGGCG |
55 | 538 | Farajzadeh- Sheikh et al. (2020) |
| 10- | bla VIM |
F:GTTTGGTCGCATATCGCAAC R:AATGCGCAGCACCAGGATAG |
60 | 389 | Awan et al. (2021) |
| 11 | bla GES |
F: GTTTTGCAATGTGCTCAACG R: TGCCATAGCAATAGGCGTAG |
56.65 | 371 | Poirel et al. (2011) |
Assessment of antibiotic resistance
Antibiograms of confirmed UPEC isolates were examined y following the Kirby-Bauer method (Sambrook and Fritsch 1989). Glycerol stocks were revived for obtaining fresh growth. Each isolate was inoculated in Tryptic Soy Broth at 37 °C. When turbidity was assessed according to 0.5 MacFarland standard, sterile cotton swabs were used for making a lawn culture on Mueller–Hinton Agar (MHA). Antibiotic discs were placed on the surface of inoculated MHA using sterile forceps. The plates were checked for zones of inhibition after overnight incubation at 37 °C. Antibiotic resistance was examined by using guidelines provided by the Clinical and Laboratory Standards Institute (Bauer et al. 1996; CLSI 2016). Colistin interpretation was performed according to criteria described by Gales et al. (CLSI 2013). Belonging to 10 groups, 20 antibiotics (OXOID, UK) were used in this study (Table 1). UPEC isolates were categorized as extensively drug resistant (XDR) and MDR (CLSI 2016).
Table 1.
Antibiotics used in this study
| Sr. No | Group of antibiotic | Drugs |
|---|---|---|
| 1 | Carbapenems | Meropenem(10 µg) |
| 2 | Cephalosporins | Sulbactam/ cefoperazone(105 µg) |
| Cefixime(5 µg) | ||
| Ceftriaxone(30 µg) | ||
| Cefotaxime(30 µg) | ||
| Ceftazidime(30 µg) | ||
| 3 | Quinolones and fluoroquinolones | Ofloxacin(5 µg) |
| Moxifloxacin(5 µg) | ||
| Pipemidic Acid(20 µg) | ||
| 4 | Aminoglycosides | Gentamycin(30 µg) |
| Fosfomycin(50 µg) | ||
| Tobramycin(10 µg) | ||
| 5 | Penicillins | Ampicillin/sulbactam(20 µg) |
| Amoxycillin(10 µg) | ||
| Piperacillin/ Tazobactam(110 µg) | ||
| 6 | Nitorfuran | Nitrofurantoin(300 µg) |
| 7 | Sulfonamides | Trimethoprim(5 µg) |
| 8 | Monobactams | Aztreonam(30 µg) |
| 9 | Macrolide | Erythromycin(30 µg) |
| 10 | Polymyxin | Colistin sulphate (10 µg) |
Phenotypic Assay for ESBL production
Each isolate was processed through Double Disc Synergy test (DDST) (Gales et al. 2001; Magiorakos et al. 2012). DDST was performed using five antibiotic discs (OXOID); amoxicillin-clavulanic acid (AMC), aztreonam (ATM), ceftriaxone (CRO), ceftazidime (CAZ) and cefotaxime (CTX).
AMC (20/ 10 μg) was placed at the centre of Mueller–Hinton agar plate containing inoculum of desired isolate. Other discs were placed 15 mm apart from AMC. Plate-containing discs was placed inverted in incubator at 37 ℃ overnight. The plate was observed for extension of inhibitory zone from 3rd generation cephalosporins toward AMC forming a characteristic shape denoted as “key hole”.
Molecular detection of ESBLs and carbapenemases
Molecular methods are best described for having specificity and sensitivity for the detection of resistance genes while consuming minimum time (Aruhomukama 2020). Molecular detection was performed by PCR using gene specific-primers (e-oligo, USA) (Table 2). PCR conditions were same for each primer set except annealing temperature. For ESBLs, PCR conditions were 94 °C for 5 min (initial denaturation) followed by 30 cycles of 94 °C for 30 s (denaturation), annealing (primer specific) for 30 s and 72 °C for 30 s (extension) and 72 °C for 5 min (final Extension). For carbapenems, PCR conditions were 94 °C for 5 min (initial denaturation) followed by 30 cycles of 94 °C for 30 s (denaturation), annealing (primer specific) for 40 s and 72 °C for 50 s (Extension) and 72 °C for 5 min (final extension).
PCR products were run through 1.5% (w/v) agarose gel in Tris–Borate- Ethylene diamine tetra acetic acid (TBE) buffer in an electrophoresis chamber (Thermo ELECTRON CORPORATION, USA) and detected using ultraviolet transilluminator. Voltage was adjusted to 120 V and PCR products were run for 45 min through agarose gel.
Sequencing of PCR Products
The PCR products were sequenced to check the presence of different ESBL genes in these isolates.
Statistical Analysis
The data was analyzed using GraphPad Prism 8.4.2, the Fisher’s exact test (2-tailed) value was found for the comparison of antimicrobial resistance profiles of ESBL producers and non-producer to check any significant differences between two batches of the collected isolates. P-value was calculated through Fisher’s Exact Test and was considered significant if less than 0.05.
Results
Phenotypic Identification of UPEC
Lactose fermentation and pink colonies were observed on MacConkey agar plates. On C.L.E.D. agar, bright pink colonies (with pink halo) were observed for UPEC among other uropathogens. Purified colonies were further confirmed by stabbing in TSI agar. Isolates having characteristics of yellow slant, yellow butt, gas positive(cracks/bubbles indication in agar) and H2S negative, were processed further for molecular characterization.
Phenotypic and genotypic confirmation of UPEC
There was no polymicrobial culture in urine samples. Out of 540 samples, 500 samples showed positive growth on culture media. Out of 500, 170 (34%) proved to be E. coli by phenotypic assays. Out of these presumptive 170 E. coli isolates, 89 (52.4%) exhibited the presence of uidA gene. There might be variations in uidA gene due to which 89 samples indicated presence of this gene (Clinical and Laboratory Standards Institute 2010).
Demographic Data
Out of 89, female population was 70 (79%) whereas male population was 19 (21%). Age data indicated that No. of < 18 years patients were 5 (6%); 18–60 years were 76 (85%) and > 60 years were 8 (9%). Among ESBL producers; females were 25 (58%) whereas males were 18 (42%). Age-wise distribution among these patients was < 18 years were 3 (7%), 18–60 years were 38 (88%) and > 60 years were 2 (5%). Among non-ESBL producers; females were 45 (98%) while only one was male (2%). Age-wise distribution among these patients was < 18 years were 2 (4%), 18–60 years were 38 (83%) and > 60 years were 6 (13%).
Phenotypic confirmation of ESBL production and antibiotic resistance
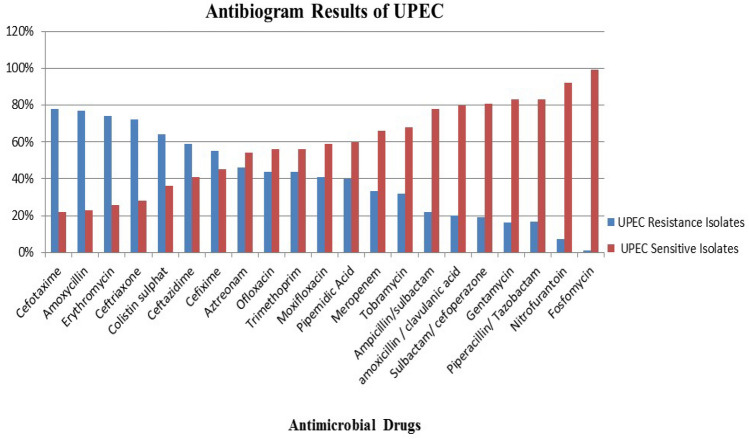
XDR UPEC isolates were 63 (71%) Out of 89, 43 (48%) isolates exhibited the presence of ESBLs by DDST. UPEC isolates indicated 78%, 77%, 74%, 72%, 59% and 55% resistance against cefotaxime, amoxicillin, erythromycin, ceftriaxone, ceftazidime and cefexime, respectively. For colistin; 50% isolates indicated intermediate zones and 14% showed resistance. Meropenem resistance was observed 22% UPEC isolates. Susceptible and intermediate UPEC isolates were 66.5% and 11.5%, respectively. The intermediately resistant isolates were considered resistant during statistical and data analysis (Fig. 1, Fig. 3, Table 3).
Fig. 1.
Graphical presentation of antibiogram of UPEC in the study
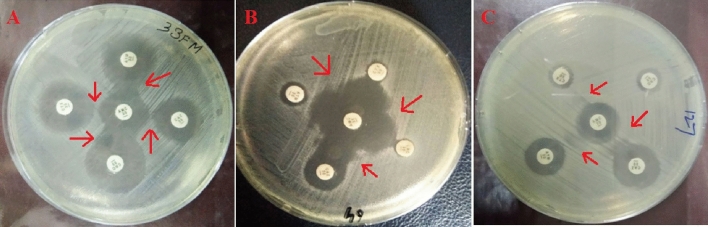
Fig. 3.
Phenotypic detection of ESBLs by DDST. Figure parts A and B indicate synergy between AMC and CTX, CAZ, ATM and CRO. Figure part C indicates synergy between AMC and CRO, CAZ and ATM
Table 3.
Comparative analysis of AMR profile of ESBL producers and non-producers
| Antimicrobial drugs | Non-ESBL producing UPEC isolates (n = 46) | ESBL producing UPEC isolates (n = 43) | p value | ||
|---|---|---|---|---|---|
| Resistant | Susceptible | Resistant | Susceptible | ||
| Meropenem | 29 (63%) | 17 (37%) | 11 (26%) | 32 (74%) | 0.0006*** |
| Fosfomycin | 0 (0%) | 46 (100%) | 1 (2%) | 42 (98%) | 0.4831 |
| Tobramycin | 17 (37%) | 29 (63%) | 13 (30%) | 30 (70%) | 0.6540 |
| Gentamycin | 7 (15%) | 39 (85%) | 9 (21%) | 34 (79%) | 0.5845 |
| Erythromycin | 45 (98%) | 1 (2%) | 43 (100%) | 0 | 1.0000 |
| Pipemidic Acid | 17 (37%) | 29 (63%) | 23 (54%) | 20 (46%) | 0.5845 |
| Moxifloxacin | 25 (54%) | 21 (46%) | 22 (51%) | 21 (49%) | 0.8332 |
| Ofloxacin | 25 (54%) | 21 (46%) | 16 (37%) | 27 (63%) | 0.1372 |
| Nitrofurantoin | 7 (15%) | 39 (85%) | 1 (2%) | 42 (98%) | 0.0593 |
| Aztreonam | 27 (59%) | 19 (41%) | 34 (79%) | 9 (21%) | 0.0435* |
| Trimethoprim | 33 (72%) | 13 (28%) | 28 (65%) | 15 (35%) | 0.6483 |
| Ceftazidime | 28 (61%) | 18 (39%) | 32 (74%) | 11 (26%) | 0.1847 |
| Ceftriaxone | 33 (72%) | 13 (28%) | 34 (79%) | 9 (21%) | 0.4689 |
| Cefexime | 34 (74%) | 12 (26%) | 34 (79%) | 9 (21%) | 0.6237 |
| Cefotaxime | 35 (76%) | 11 (24%) | 38 (88%) | 5 (12%) | 0.1709 |
| Sulbactam/ cefoperazone | 7 (15%) | 39 (85%) | 9 (21%) | 34 (79%) | 0.5845 |
| Piperacillin/ Tazobactam | 9 (20%) | 37 (80%) | 6 (13%) | 37 (87%) | 0.5763 |
| Amoxycillin | 44 (96%) | 2 (4%) | 40 (93%) | 3 (7%) | 0.6698 |
|
Ampicillin/ sulbactam |
11 (24%) | 35 (76%) | 10 (23%) | 33 (77%) | 1.0000 |
| Amoxicillin / Clavulanic acid | 9 (20%) | 37 (80%) | 7 (17%) | 36 (83%) | 0.7856 |
| Colistin sulphate | 36 (78%) | 10 (22%) | 31 (72%) | 12 (28%) | 0.6242 |
***Indicates extremely statistically significant
*Designates statistically significant
Among ESBL-producers, these isolates possessed 100%, 93%, 88%, 79% and 79% resistance against erythromycin, amoxycillin, cefotaxime, ceftriaxone and cefixime, respectively. For colistin; 30 (70%) isolates exhibited intermediate zones and 1(3%) showed resistance. For meropenem; 15% showed resistance, 10% intermediate zones and 75% showed sensitivity to meropenem.
Gene Detection according to Ambler Classification
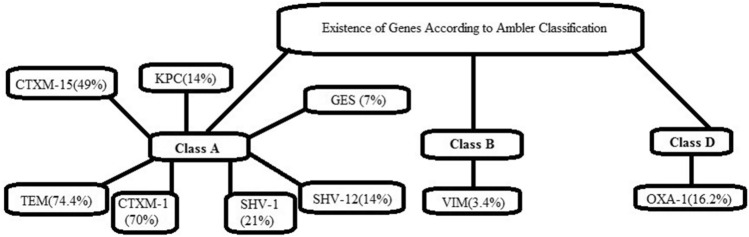
Ambler classification is based on the discrimination of resistance pattern induced by these enzymes and in most of the cases Class A and Class D is present (Aruhomukama 2020). In our study, isolates indicated the presence of Class A genes (84% ESBLs + 19% Carbapenemases) (Fig. 2).
Fig. 2.
Distribution of ESBLs and carbapenem-resistance genes on the basis of Ambler classification
Occurrence of ESBL genes
All isolates indicated the presence of gene(s) except three isolates (Table 4). One isolate indicated the presence of all the tested genes.TEM-1, CTXM-1 and CTXM-15 were present in 74.4%, 70% and 49%, respectively. SHV-1 and OXA-1 were present in 21% and 16.2% isolates respectively. SHV-12 was present in 14% isolates and CMY-2 was present in 9.3% isolates (Table 4). These genes were also co-existed in different patterns in ESBL-positive isolates. TEM-1 + CTXM-1 and TEM-1 + CTXM-1 + CTXM-15 + OXA-1 patterns indicated high prevalence followed by TEM-1 + CTXM-1 + CTXM-15 and TEM-1 + CTXM-1 + CTXM-15 + SHV-1 + OXA-1 gene patterns (Table 5).
Table 4.
Occurrence of ESBL genes
| ESBL genes | Isolate no. (%) |
|---|---|
| CTXM-1 | 30 (70%) |
| TEM-1 | 32 (74.4%) |
| CTXM-15 | 21 (49%) |
| CMY-2 | 4 (9.3%) |
| SHV-12 | 6 (14%) |
| SHV-1 | 9 (21%) |
| OXA-1 | 7 (16.2%) |
Table 5.
Co-existence of ESBL genes
| Gene Pattern | Isolate no. (%) |
|---|---|
| TEM-1 + CTXM-1 | 5 (12%) |
| TEM-1 + CTXM-1 + CTXM-15 | 4 (9.3%) |
| TEM-1 + CTXM-15 | 2 (5%) |
| TEM-1 + CTXM-1 + CTXM-15 + CMY-2 + SHV-12 + SHV-1 | 2 (5%) |
| TEM-1 + CTXM-1 + CTXM-15 + CMY-2 | 1 (2.3%) |
| TEM-1 + CTXM-1 + SHV-12 | 1 (2.3%) |
| TEM-1 + CTXM-1 + SHV-1 | 2 (5%) |
| TEM-1 + CTXM-1 + CTXM-15 + SHV-1 | 2 (5%) |
| TEM-1 + CTXM-1 + CTXM-15 + OXA-1 | 5 (12%) |
| TEM-1 + CTXM-1 + CTXM-15 + SHV-1 + OXA-1 | 3 (7%) |
| TEM-1 + CTXM-1 + SHV-1 + OXA-1 | 1 (2.3%) |
| TEM-1 + CTXM-1 + OXA-1 | 2 (5%) |
| TEM-1 + CTXM-1 + CTXM-15 + SHV-1 + OXA-1 + VIM | 1 (2.3%) |
Molecular detection of carbapenem-resistance
Klebsiella pneumoniae carbapenemase (KPC), Verona integron-encoded MBL (VIM) and Guiana extended spectrum (GES) carbapenems (Noster et al. 2021) were detected in this study. Two (7%) isolates indicated the presence of GES and VIM was detected in one (3.4%) isolate. KPC was detected in four (14%) isolates. Co-existence was not observed for carbapenem resistance genes Figs. 4 and 5.
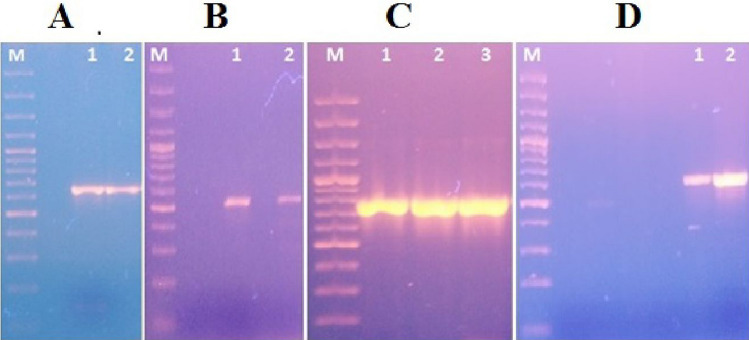
Fig. 4.
Genotypic detection of ESBL genes (TEM-1, CTXM-15, CTXM-1, OXA-1) by PCR Lane M indicates sizes (in base pairs) of 3000, 2000, 1500, 1200, 1000, 900, 800, 700, 600, 500, 400, 300, 200, 100 (Cat no. SM0323, Thermo Fisher Scientific, USA) Part A (Lane 1&2) represents the amplification of TEM-1 gene (643 bp) Part B (Lane 1&2) represents the amplification of CTXM-15 gene (540 bp) Part C (Lane1, 2 and 3) represents the amplification of CTXM-1 gene (766 bp) Part D (Lane 1 and 2) represents the amplification of OXA-1 (640 bp)
Fig. 5.
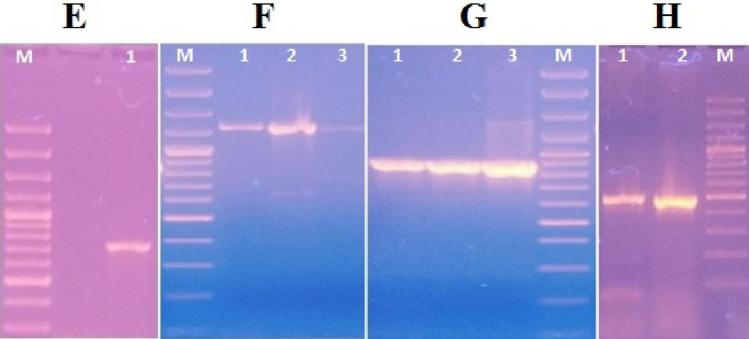
Genotypic detection of ESBL genes (SHV-1, SHV-12, CMY-2, uidA) by PCR Lane M indicates sizes (in base pairs) of 3000, 2000, 1500, 1200, 1000, 900, 800, 700, 600, 500, 400, 300, 200, 100 (Cat no. SM0323, Thermo Fisher Scientific, USA) Part E (Lane 1) represents the amplification of SHV-1 (714 bp) Part F (Lane 1, 2 and 3) represents the amplification of SHV-12 (1233 bp) Part G (Lane 1, 2 and 3) represents the amplification of CMY-2 (870 bp) Part H (Lane 1 and 2) represents the amplification of uidA (486 bp)
Sequenced PCR Products
Sequences were aligned with protein databases using blastx on NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences represented significant alignment with their related proteins (WP_240093217.1, AKE33354.1, ACG58887.1, WP_063859857.1, NPI00138.1, and AFR79065.1). Sequences were 99%, 100%, 83%, 96%, 99%, and 97% identical respectively.
Interpretation of Statistical Analysis
The comparative antimicrobial resistance between ESBL producers and ESBL non producers was found extremely significant by statistical analysis for aztreonam, amoxicillin, erythromycin, cefixime and trimethoprim. Ceftazidime indicated very significant association. Nitrofurantoin, ofloxacin and cefotaxime indicated statistical significance.
Discussion
Urinary tract infection has become a significant problem due to emergence of MDR pathogens in human population (Godambe et al. 2017). E. coli is a major cause of infections in newborns and all age groups in both hospital and community settings (Aurilio et al. 2022). Among UPEC isolates, high resistance rates were observed in amoxicillin, cefotaxime, ceftriaxone, ceftazidime and cefixime indicating 77%, 78%, 72%, 59% and 55% resistant isolates respectively in this study. Our study is in accordance with the reports from different centers indicating resistance against 3rd generation cephalosporins and penicillins in Pakistan (Coura et al. 2021; Wu et al. 2021; Ahmed et al. 2022). ESBL producing UPEC bring about resistance against broad-spectrum antibiotics like 3rd generation cephalosporins due to the empiric use of these antibiotics for UTIs (Akhtar et al. 2021). In the present study, 64% resistance was observed in for colistin sulphate. Colistin sulphate, fosfomycin, nitrofurantoin, polymyxin B, and chloramphenicol proved promising for Paediatric patients of UTIs at the Institute of Child Health, Lahore, Pakistan (Nasir, et al. 2021). Another study from Children Hospital Lahore, indicated high susceptibilities for colistin (84.8%), nitrofurantoin (64.7%) and meropenem (54%) (White 2021). Uropathogenic gram negative bacteria were 22.8% that indicated resistance against colistin from patients at Peshawar, Pakistan (Iqbal et al. 2021). In this study, significant ESBL prevalence (48%) was found by DDST Naeem et al. reported 57.89% ESBL-positive E. coli isolates recovered from tertiary care centres of Peshawar and Islamabad (Mir et al. 2022). From Karachi, Nasir et al. reported 66.2% ESBL-producing E. coli isolates retrieved from urology department of Jinnah postgraduate medical center (Arif et al. 2022). Khan and Bari reported 92% ESBL-positive E. coli from urine samples at Qazi Hussain Ahmed Medical Complex Nowshera (Naeem et al. 2021). A study was conducted by Samin et al. at Diabetes Hospital Peshawar and Nishter Hospital Multan, ESBL-positive E. coli isolates were recovered from 81.4% subjects (Nasir et al. 2021)..Percentages of TEM-1 and CTXM-1 were 74.4% and 70% in this study. CTX-M enzymes are the most prevalent ESBLs and declared pandemic. TEM enzymes have been studied well and contribute 90% resistance in E. coli reviewed by Hussain et al. Khan and Bari 2021). TEM gene has also been found mainly (66.6%) in hospital aerosols from Tai'an City, China (Samin et al. 2021). There is significant proportion (49%) of isolates containing CTXM-15 in present investigation. Latest research from different countries around the globe also indicated a high prevalence of CTXM-15 (Hussain et al. 2021; Wu et al. 2020; Hassuna et al. 2020; Legese et al. 2022; Carvalho et al. 2021). High prevalence of CTXM-15 is responsible for clonal dissemination of ST-131 armed with ESBL genes and virulence factors. ST-131 also dominantly colonizes human gut(Surgers et al. 2019) CTXM-15 gene was also found significantly in companion animals in Italy, Germany and Europe Companion animals have become reservoires of CTXM-15 gene in Italy, Germany and Europe. ST131 clone of UPEC remained responsible for this gene most of the time. ST131 is responsible for MDR infections globally (Hassuna et al. 2020). Others also investigated ST131 and found predominance of CTXM-15 (Surgers et al. 2019; Mazumder et al. 2020). Co-existence of TEM-1 and CTXM-1 was present in 12% isolates in our case. A meta-analysis from Pakistan indicated abundance of CTXM-1 and co-existence of this gene TEM (Abdelrahim et al. 2021). In our study co-existence of CTXM-15 with TEM-1 and CTXM-1 was 9.3%, benefitting bacteria to escape pressure of multiple antibiotic treatments thus worsening the situation. In Philippines, Samples from pig farms indicated 4.17% co-existence of these genes (Alqasim et al. 2020).
Antibiotic usage in poultry is transferring ESBLs to humans at community level (Bilal et al. 2021; Gundran et al. 2019).
In our study SHV-1 was found in few isolates (21%) and SHV-12 was also found in low percentage (14%). The prevalence was in accordance with different studies performed around the globe. Pandit et al. detected 4.8% SHV in MDR UPEC isolates (Falgenhauer et al. 2019) In Upper Egypt, 59.7% isolates indicated ESBL-production from 62% MDR UPEC. SHV was co-existed with CTX-M in 10% isolates and co-existence of SHV with TEM was present in 6.25% isolates (Hussain et al. 2021). Nwafia et al. detected SHV in 2.86% E. coli isolates recovered from both community and hospital. SHV was also coexisted with CTX-M in 20% isolates and in TEM/CTX-M in 14.29% isolates (Kim et al. 2021).SHV was detected in high prevalence (39.44%) among drug users (Nahar et al. 2022). SHV enzymes are detected frequently in Klebsiella pneumoniae outbreaks than E. coli (Khan and Bari 2021). In this study, CMY-2 gene was present in 9.3% isolates ESBL genes are not unique for UPEC and these genes are found in E. coli derived from different sources. CMY type is predominantly detected type of plasmid- mediated AmpC and 64 plasmid- mediated variants exist of CMY type. Multidrug resistance emerges due to plasmid-mediated AmpC β-lactamase production(Pandit et al. 2020).CMY-2 (n = 2) wasdetected in Enterobacteriaceae isolated from sepsis patients(Wu et al. 2020). CMY-2 was detected in 3.7% carbapenem resistant E. coli isolates recovered from tertiary-care center in Lebanon(Nwafia et al. 2019). In Mexico, 4.5% isolates indicated presence of CMY-variants in paediatric patients (Barani et al. 2021). Poultry meat has been declared a main source of ESBL/AmpC-producing E. coli and K. pneumonia (Muriuki et al. 2022) ESBL/AmpC-producing E. coli recovered from calves were at high frequency as compared to cows/ manure pits at farms in Quebec, Canada (Dagher et al. 2018).
Prevalence of OXA-1 was 16.2% in this study. The result is in accordance with reports on extraintestinal E. coli (Carvalho et al. 2021) and female patients with UTI episode (Merida-Vieyra et al. 2020). OXA-1 was also detected in fecal samples of healthy individualsin India (Kurittu et al. 2021) and USA (Massé et al. 2021). It is the indication of E. coli being a reservoir of antimicrobial resistance genes. There are reports about co-existence of OXA-1 gene with NDM-5 and CTXM-variants /only with NDM-5 (Zeng et al. 2021; Chandran et al. 2017; Rubin et al. 2020). In this study, OXA-1 co-existed with different genes. TEM-1 + CTXM-1 + CTXM-15 + OXA-1 pattern was present in 12% isolates. Virulent MDR E. coli isolates that produce ESBLs, are the main reason for UTI severity and it may lead to acute kidney injury, considered an important risk factor for diabetic patients (Nasir et al. 2021; Hao et al. 2022). Three isolates did not indicate presence of any gene which might be due to the production of different enzymes.
In our study, 34% UPEC isolates indicated resistance against meropenem and presence of carbapenemase genes is another alarming finding. KPC gene was detected in 14% isolates. From Pakistan, recent reports indicated the presence of NDM in pediatric patients (Sun et al. 2018; Corbellini et al. 2022) and along with other carbapenemase genes from hospital-settings [81 whereas NDM was not detected in our study. In Islamabad, occurrence of carbapenem-resistances genes has been detected along with ESBLs in sewage indicating circulation of genes in community (Naziri et al. 2020). TEM-1 + CTXM-1 + CTXM-15 + SHV-1 + OXA-1 + VIM pattern was observed in one isolate in our study and only that isolate indicated presence of VIM gene. GES was present in 7% isolates in this study. Systematic review analysis indicated UTIs the most reported infection in Pakistan and KPC was prominently detected in E. coli (31.67%). E. coli indicated significant resistance for antibiotics used as first line therapy (Abdelrahim et al. 2021). Statistical analysis indicated association between ESBL producing and non-ESBL producing UPEC isolates. Statistical significance was investigated on E. coli isolates obtained from UTI patients in Iran. Statistically significant difference was found for imipenem, gentamycin, levofloxacin, ciprofloxacin, Tetracycline, ampicillin and ceftazidime (Aslam et al. 2021). In Palestine, statistical analysis indicated no significance for chloramphenicol and aminoglycosides than other drugs in human clinical isolates. Both ESBL and non-ESBL E. coli isolates were susceptible to imipenem (Yasmin et al. 2022). Gharavi et al. did not find significant effect of ESBL expression for Imipenem, Meropenem, Nitrofurantoin, and Amikacin in E. coli isolates (Abrar et al. 2018). In this study, there is need to do more work on drug resistance genes of different drug groups on molecular level. There is need to explore virulence factors pertaining to UPEC in isolates recovered in this study, which can help in assessment of the virulence potential of XDR isolates and this can be added in our future projects.
Conclusion
In conclusion, the data indicated the higher prevalence of UTIs in females as compared to males. Antibiotic sensitivity analysis identified Fosfomycin and Nitrofurantoin as the most effective antimicrobials, and Cefotaxime and Amoxicillin as the least effective ones. ESBL genes were present in more than 40% UPEC and co-existed in different combinations. Some isolates also indicated the phenotypic and genotypic occurrence of carbapenem resistance genes. This situation is quite alarming, therefore, it is important to emphasize that antibiotic prescription should only be based on clinical diagnosis of urine culture. Antibiotic usage in animal husbandry should also be monitored. Routes of transmission should be identified to prevent transmission from animals to humans and vice versa. Over the counter drug availability is another main problem for generating drug resistance among pathogens. Antibiotics spread in the environment through different routs could lead to emergence of XDR and MDR pathogens. According to our knowledge, it is first study indicating the presence of KPC in UPEC as well as co-existence of different ESBL genes in UPEC from Faisalabad region.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge the support provided by Government College University, Faisalabad and National Institute for Biotechnology & Genetic Engineering College (NIBGE-C), for completion of this research project.
Author contributions
B.E. wrote the manuscript and prepared the figures. Y. S. and A. H. conceived the idea, improved and finalized the manuscript and supervised the whole project. M.Q. and A. A. helped during experimental work.
Funding
The research project was supported jointly by Department of Bioinformatics & Biotechnology, Government College University, Faisalabad and National Institute for Biotechnology & Genetic Engineering College (NIBGE-C), PIEAS. The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Code availability
There was no software or code developed during this study, so this is not applicable.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declared no financial or non-financial competing interests.
Ethics approval
This study was reviewed and approved by the Department Ethical Committee of GCUF. The letter is provided along with the uploaded documents (Ref. No: GCUF/BNB/2018/1177b).
Consent of participants
Informed consent from patients was not required by the Ethical Committee, because there was no contact with patients and all data were de-identified.
Consent to publication
No personal details of participants are included in this study, so consent to publish is not required by the participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Asma Haque, Email: asma@gcuf.edu.pk.
Yasra Sarwar, Email: yasrasarwar@yahoo.com.
References
- Abdelrahim SS, Fouad M, Abdallah N, Ahmed RF, Zaki S. Comparative study of CTX-M-15 producing Escherichia coli ST131 clone isolated from urinary tract infections and acute diarrhoea. Infect Drug Resist. 2021;14:4027–4038. doi: 10.2147/IDR.S325669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrar S, Hussain S, Khan RA, et al. Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: first systematic meta-analysis report from Pakistan. Antimicrob Resist Infect Control. 2018;7:26. doi: 10.1186/s13756-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamus-Białek W, Baraniak A, Wawszczak M, et al. The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Mol Biol Rep. 2018;45(5):1055–1065. doi: 10.1007/s11033-018-4254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Khalid H, Mushtaq M, et al. The Molecular characterization of virulence determinants and antibiotic resistance patterns in human bacterial uropathogens. Antibiotics. 2022;11(4):516. doi: 10.3390/antibiotics11040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar SMH, Sattar A, Hayat K, et al. Uropathogens spectrum and antibiotic susceptibility in sexually active females of age group 20 to 40 years. RMJ. 2021;46(1):14–17. [Google Scholar]
- Alqasim A, Jaffal AA, Alyousef AA. Prevalence and molecular characteristics of sequence type 131 clone among clinical uropathogenic Escherichia coli isolates in Riyadh. Saudi Arabia Saudi J Biol Sci. 2020;27(1):296–302. doi: 10.1016/j.sjbs.2019.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif A, Ullah I, Ullah O, Zaman R. Identification of colistin resistance and its bactericidal activity against uropathogenic gram negative bacteria from Hayatabad Medical Complex Peshawar. Pak J Med Sci. 2022;38(4):981–986. doi: 10.12669/pjms.38.4.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruhomukama D. Review of phenotypic assays for detection of extended-spectrum β-lactamases and carbapenemases: a microbiology laboratory bench guide. Afr Health Sci. 2020;20(3):1090–1108. doi: 10.4314/ahs.v20i3.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam F, Lodhi H, Bashir R, et al. Prevalence of New Delhi Metallo-βLactamase-1 (blaNDM-1) Gene in Children from Tertiary Care Hospital of Pakistan. Pakistan J Zool. 2021;54(3):1455–1458. doi: 10.17582/journal.pjz/20201021081020. [DOI] [Google Scholar]
- Aurilio C, Sansone P, Barbarisi M, et al. Mechanisms of action of carbapenem resistance. Antibiotics. 2022;11(3):421. doi: 10.3390/antibiotics11030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan AB, Yan A, Sarwar Y, et al. Detection of synergistic antimicrobial resistance mechanisms in clinical isolates of Pseudomonas aeruginosa from post-operative wound infections. Appl Microbiol Biotechnol. 2021;105(24):9321–9332. doi: 10.1007/s00253-021-11680-6. [DOI] [PubMed] [Google Scholar]
- Barani A, Bafroee AST, Jabalameli L. Abundance of extended-spectrum β-lactamase genes among intestinal Escherichia coli strains from drug users. Arch Microbiol. 2021;203(6):3245–3255. doi: 10.1007/s00203-021-02316-4. [DOI] [PubMed] [Google Scholar]
- Bashir S, Sarwar Y, Ali A, et al. Multiple drug resistance patterns in various phylogenetic groups of uropathogenic E. coli isolated from Faisalabad region of Pakistan. Braz J Microbiol. 2011;42(4):1278–1283. doi: 10.1590/S1517-83822011000400005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir S, Haque A, Sarwar Y, Ali A, Anwar M1. Virulence profile of different phylogenetic groups of locally isolated community acquired uropathogenic E. coli from Faisalabad region of Pakistan. Ann Clin Microbiol Antimicrob. 2012 doi: 10.1186/1476-0711-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1996;45(4):493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- Baziboroun M, Bayani M, Poormontaseri Z, Shokri M, Biazar T. Prevalence and antibiotic susceptibility pattern of extended spectrum beta lactamases producing Escherichia coli isolated from outpatients with Urinary Tract Infections in Babol, Northern of Iran. Curr Issues Pharm Med Sci. 2018;31(2):61–64. doi: 10.1515/cipms-2018-0013. [DOI] [Google Scholar]
- Bilal H, Khan MN, Rehman T, et al. Antibiotic resistance in Pakistan: a systematic review of past decade. BMC Infect Dis. 2021;21:244. doi: 10.1186/s12879-021-05906-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho I, Carvalho JA, Martínez-Álvarez S, et al. Characterization of ESBL-Producing Escherichia coli and Klebsiella pneumoniae Isolated from clinical samples in a Northern Portuguese Hospital: predominance of CTX-M-15 and high genetic diversity. Microorganisms. 2021;9(9):1914. doi: 10.3390/microorganisms9091914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran SP, Sarkar S, Diwan V, et al. Detection of virulence genes in ESBL producing, quinolone resistant commensal Escherichia coli from rural Indian children. J Infect Dev Ctries. 2017;11(5):387–392. doi: 10.3855/jidc.8574. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Tomar A, Ansari K, et al. Antimicrobial activity of certain drugs against the different isolates found in bovine fecal samples. J Pharmacogn Phytochem. 2022;11(3):186–192. [Google Scholar]
- Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing twentieth informational supplement, M100–S20. Wayne: CLSI; 2010. [Google Scholar]
- CLSI . Performance standards for antimicrobial susceptibility testing twenty-third informational supplement. Wayne PA: CLSI document M100–S23 Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- CLSI . Performance standards for antimicrobial susceptibility testing twenty-third informational supplement. 26. Wayne PA: CLSI document M100S Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- Corbellini S, Scaltriti E, Piccinelli G, et al. Genomic characterisation of Escherichia coli isolates co-producing NDM-5 and OXA-1 from hospitalised patients with invasive infections. J Glob Antimicrob Resist. 2022;28:136–139. doi: 10.1016/j.jgar.2021.12.018. [DOI] [PubMed] [Google Scholar]
- Coura FM, Savini VMD, Xavier RGC, et al. Virulence genes profile and antimicrobial susceptibility of community-acquired bacterial urinary tract infections in a Brazilian hospital. Curr Microbiol. 2021;78(11):3913–3923. doi: 10.1007/s00284-021-02650-2. [DOI] [PubMed] [Google Scholar]
- Dagher C, Salloum T, Alousi S, Arabaghian H, Araj GF, Tokajian S. Molecular characterization of Carbapenem resistant Escherichia coli recovered from a tertiary hospital in Lebanon. PLoS ONE. 2018;13(9):e0203323. doi: 10.1371/journal.pone.0203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaouadi S, Soufi L, Hamza A, et al. Co-occurrence of mcr-1 mediated colistin resistance and β-lactamase-encoding genes in multidrug-resistant Escherichia coli from broiler chickens with colibacillosis in Tunisia. J Glob Antimicrob Resist. 2020;22:538–545. doi: 10.1016/j.jgar.2020.03.017. [DOI] [PubMed] [Google Scholar]
- Doi Y, Iovleva A, Bonomo RA. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J Travel Med. 2017 doi: 10.1093/jtm/taw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim-Saraie HS, Nezhad NZ, Heidari H, et al. Detection of antimicrobial susceptibility and integrons among extended-spectrum β-lactamase producing Uropathogenic Escherichia coli Isolates in Southwestern Iran. Oman Med J. 2018;33(3):218–223. doi: 10.5001/omj.2018.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgenhauer L, Imirzalioglu C, Oppong K, et al. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol. 2019;9:3358. doi: 10.3389/fmicb.2018.03358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajzadeh-Sheikh A, Savari M, Hosseini Nave H, et al. Frequency and molecular epidemiology of class A ESBLs producing Enteroinvasive Escherichia coli (EIEC) isolates among patients with diarrhea. Gastroenterol Hepatol Bed Bench. 2020;13(1):77–85. [PMC free article] [PubMed] [Google Scholar]
- Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28(1):1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Gales AC, Reis AO, Jones RN, et al. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol. 2001;39(1):183–190. doi: 10.1128/JCM.39.1.183-190.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharavi MJ, Zarei J, Roshani-Asl P, et al. Comprehensive study of antimicrobial susceptibility pattern and extended spectrum beta-lactamase (ESBL) prevalence in bacteria isolated from urine samples. Sci Rep. 2021;11:578. doi: 10.1038/s41598-020-79791-0021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godambe LP, Bandekar J, Shashidhar R, et al. Species specific PCR based detection of Escherichia coli from Indian foods. 3 Biotech. 2017;7(1):130. doi: 10.1007/s13205-017-0784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundran RS, Cardenio PA, Salvador RT, et al. Prevalence, antibiogram, and resistance profile of extended-spectrum β-lactamase-producing Escherichia coli isolates from Pig Farms in Luzon. Philippines Microb Drug Resist. 2019;26(2):160–168. doi: 10.1089/mdr.2019.0019. [DOI] [PubMed] [Google Scholar]
- Habeeb MA, Sarwar Y, Ali A, Salman M, Haque A. Rapid emergence of ESBL producers in E. coli causing urinary and wound infections in Pakistan. Pak J Med Sci. 2013;29(2):540–544. doi: 10.12669/pjms.292.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighatpanah M, Mojtahedi A. Characterization of antibiotic resistance and virulence factors of Escherichia coli strains isolated from Iranian in patients with urinary tract infections. Infect Drug Resist. 2019;12:2747–2754. doi: 10.2147/IDR.S219696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Zeng Z, Xiao X, et al. Genomic and phenotypic characterization of a colistin-resistant Escherichia coli Isolate co-harboring blaNDM-5, blaOXA-1, and blaCTX-M-55 isolated from urine. Infect Drug Resist. 2022;15:1329–1343. doi: 10.2147/IDR.S355010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassuna NA, Khairalla AS, Farahat EM, Hammad AM, Abdel-Fattah M. Molecular characterization of extended-spectrum β lactamase- producing E. coli recovered from community-acquired urinary tract infections in upper Egypt. Sci Rep. 2020;10:2772. doi: 10.1038/s41598-020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain HI, Aqib AI, Seleem MN, et al. Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Microb Pathog. 2021;158:105040. doi: 10.1016/j.micpath.2021.105040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z, Mumtaz MZ, Malik A. Extensive drug-resistance in strains of Escherichia coli and Klebsiella pneumoniae isolated from paediatric urinary tract infections. J Taibah Univ Med Sci. 2021;16(4):565–574. doi: 10.1016/j.jtumed.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal NA, Al-Ghamdi AM, Momenah AM, et al. Prevalence and antibiogram Pattern of Klebsiella pneumoniae in a Tertiary Care Hospital in Makkah, Saudi Arabia: An 11-Year Experience. Antibiotics. 2023;12(1):164. doi: 10.3390/antibiotics12010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlmeter G, Poulsen HO. Antimicrobial susceptibility of Escherichia coli from community-acquired urinary tract infections in Europe: the ECO·SENS study revisited. Int J Antimicrob Agents. 2012;39(1):45–51. doi: 10.1016/j.ijantimicag.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Khan H, Bari F. Extended spectrum Beta lactamases (ESBLs): preference for age and gender; a hospital-based study. Microb Infect Dis. 2021;2(2):326–332. doi: 10.21608/MID.2020.41773.1061. [DOI] [Google Scholar]
- Kim H, Kim YA, Seo YH, Lee H, Lee K. Prevalence and molecular epidemiology of extended-spectrum-β-lactamase (ESBL)-producing Escherichia coli from multiple sectors of poultry industry in Korea. Antibiotics. 2021;10(9):1050. doi: 10.3390/antibiotics10091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurittu P, Khakipoor B, Aarnio M, et al. Plasmid-Borne and Chromosomal ESBL/AmpC Genes in Escherichia coli and Klebsiella pneumonia in Global Food Products . Front Microbiol. 2021;12:592291. doi: 10.3389/fmicb.2021.592291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larramendy S, Deglaire V, Dusollier P, et al. Risk factors of extended-spectrum beta-lactamases-producing escherichia coli community acquired urinary tract infections: a systematic review. Infect Drug Resist. 2020;13:3945–3955. doi: 10.2147/IDR.S269033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legese MH, Asrat D, Aseffa A, Hasan B, Mihret A, Swedberg G. Molecular epidemiology of extended-spectrum beta-lactamase and AmpC producing Enterobacteriaceae among sepsis patients in Ethiopia: a prospective multicenter study. Antibiotics. 2022;11(2):131. doi: 10.3390/antibiotics11020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Wang M, Li X. ST37 Klebsiella pneumoniae: development of carbapenem resistance in vivo during antimicrobial therapy in neonates. Future Microbiol. 2017;12(10):891–904. doi: 10.2217/fmb-2016-0165. [DOI] [PubMed] [Google Scholar]
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- Macareño-Castro J, Solano-Salazar A, Dong LT, et al. Fecal microbiota transplantation for carbapenem-Resistant Enterobacteriaceae: a systematic review. J Infect. 2022;84(6):749–759. doi: 10.1016/j.jinf.2022.04.028. [DOI] [PubMed] [Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Mahamat OO, Lounnas M, Hide M, et al. High prevalence and characterization of extended-spectrum ß-lactamase producing Enterobacteriaceae in Chadian hospitals. BMC Infect Dis. 2019;19:205. doi: 10.1186/s12879-019-3838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé J, Lardé H, Fairbrother JM, et al. Prevalence of antimicrobial resistance and characteristics of Escherichia coli isolates from fecal and manure pit samples on dairy farms in the Province of Québec. Canada. Front Vet Sci. 2021;8:654125. doi: 10.3389/fvets.2021.654125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R, Abdullah A, Ahmed D, Hussain A. High prevalence of blaCTX-M-15 Gene among extended-spectrum β-lactamase-producing Escherichia coli isolates causing extraintestinal infections in Bangladesh. Antibiotics. 2020;9(11):796. doi: 10.3390/antibiotics9110796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merida-Vieyra J, De Colsa-Ranero A, Calderón-Castañeda Y, Aquino-Andrade A. Detection of CMY-type beta-lactamases in Escherichia coli isolates from paediatric patients in a in a tertiary care hospital in Mexico. Antimicrob Resist Infect Control. 2020;9:168. doi: 10.1186/s13756-020-00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir N, Khurshid R, Zafar A, et al. Frequency and susceptibility profile of pathogens causing urinary tract infections in pediatric population. P J M H S. 2022;16(2):142–144. doi: 10.53350/pjmhs22162142. [DOI] [Google Scholar]
- Muriuki CW, Ogonda LA, Kyanya C, et al. Phenotypic and genotypic characteristics of uropathogenic Escherichia coli Isolates from Kenya. Microb Drug Resist. 2022;28(1):31–38. doi: 10.1089/mdr.2020.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader S, Alejandro H. Urinary tract infections in children: epidemiology and risk factors. Waltham, MA: UpToDate; 2021. pp. 1–19. [Google Scholar]
- Naeem S, Bilal H, Muhammad H, et al. Detection of blaNDM-1 gene in ESBL producing Escherichia coli and Klebsiella pneumoniae isolated from urine samples. J Infect Dev Ctries. 2021;15(4):516–522. doi: 10.3855/jidc.12850. [DOI] [PubMed] [Google Scholar]
- Nahar S, Urmi UL, Ali T, et al. ESBL genes, blaTEM, blaOXA, and blaSHV in poultry gut bacteria: an endemic public health burden in Bangladesh: ESBL genes in poultry gut bacteria. Bangladesh Med Res Counc Bull. 2022;47(2):165–174. doi: 10.3329/bmrcb.v47i2.57775. [DOI] [Google Scholar]
- Nasir F, M. Khan I, Kashif S,, et al. Prevalence of ESBLs secreting and carbapenem-resistant E. coli from urinary tract infection. RMJ. 2021;46(3):518–521. [Google Scholar]
- Nasir F, Khan MI, Kashif S, et al. Prevalence of ESBLs secreting and carbapenem-resistant E. coli from urinary tract infection. Rawal Med J. 2021;46(3):518–521. [Google Scholar]
- Naziri Z, Derakhshandeh A, Borchaloee AS, et al. Treatment failure in urinary tract infections: a warning witness for virulent multi-drug resistant esbl- producing Escherichia coli. Infect Drug Resist. 2020;13:1839–1850. doi: 10.2147/IDR.S256131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosheen S, Bukhari NI, Junaid K, et al. Phylogenetic diversity and mutational analysis of New Delhi Metallo-β-lactamase (NDM) producing E. coli strains from pediatric patients in Pakistan. Saudi. J Biol Sci. 2021;28(10):5875–5883. doi: 10.1016/j.sjbs.2021.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noster J, Thelen P, Hamprecht A. Detection of multidrug-resistant Enterobacterales-from ESBLs to Carbapenemases. Antibiotics. 2021;10(9):1140. doi: 10.3390/antibiotics10091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwafia IN, Ohanu ME, Ebede SO, Ozumba UC. Molecular detection and antibiotic resistance pattern of extended-spectrum beta-lactamase producing Escherichia coli in a tertiary Hospital in Enugu. Nigeria Ann Clin Microbiol Antimicrob. 2019;18:41. doi: 10.1186/s12941-019-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit R, Awal B, Shrestha SS, Joshi G, Rijal BP, Parajuli NP. Extended-spectrum β-lactamase (ESBL) genotypes among multidrug-resistant uropathogenic Escherichia coli clinical isolates from a teaching hospital of Nepal. Interdiscip Perspect Infect Dis. 2020;2020:6525826. doi: 10.1155/2020/6525826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Walsh TR, Cuvillier V, Nordmann P, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Rubin J, Mussio K, Xu Y, et al. Prevalence of antimicrobial resistance genes and integrons in commensal gram-negative bacteria in a college community. Microb Drug Resist. 2020;26(10):1227–1235. doi: 10.1089/mdr.2019.0279. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch MT. Molecular cloning. USA: Cold Spring Harbor Laboratory Press; 1989. p. 2. [Google Scholar]
- Samin KA, Malik S, Sadiq S, Rasheeq T, Sajid NK, Iqbal W. Acute Kidney injury is a risk factor among type 2 diabetic patients after UTI due to extended-spectrum beta-lactamase producing organisms. Pak J Med Health Sci. 2021;15(5):1718–1720. doi: 10.53350/pjmhs211551718. [DOI] [Google Scholar]
- Shilpakar A, Ansari M, Rai KR, Rai G, Rai SK. Prevalence of multidrug-resistant and extended-spectrum beta-lactamase producing Gram-negative isolates from clinical samples in a tertiary care hospital of Nepal. Trop Med Health. 2021;49(1):23. doi: 10.1186/s41182-021-00313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M, Khurshid M, Saleem HG, et al. Characteristics and antibiotic resistance of urinary tract pathogens isolated from Punjab Pakistan. Jundishapur J Microbiol. 2015;8(7):e19272. doi: 10.5812/jjm.19272v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xu J, He F. Draft genome sequence of an NDM-5, CTX-M-15 and OXA-1 co-producing Escherichia coli ST167 clinical strain isolated from a urine sample. J Glob Antimicrob Resist. 2018;14:284–286. doi: 10.1016/j.jgar.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Surgers L, Boersma P, Girard PM, et al. Molecular epidemiology of ESBL-producing E. coli and K. pneumoniae: establishing virulence clusters. Infect Drug Resist. 2019;12:119–127. doi: 10.2147/IDR.S179134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandogdu Z, Wagenlehner FME. Global epidemiology of urinary tract infections. Curr Opin Infect Dis. 2016;29(1):73–79. doi: 10.1097/QCO.0000000000000228. [DOI] [PubMed] [Google Scholar]
- Tayh G, Elmanama A, Selmi R, et al. Antibiotic resistance profile and molecular characterization of extraintestinal pathogenic Escherichia coli (ExPEC) from human clinical samples in gaza strip, palestine. Lett Appl Microbiol. 2022 doi: 10.1093/lambio/ovac033. [DOI] [PubMed] [Google Scholar]
- Terlizzi ME, Gribaudo G, Maffei ME. UroPathogenic Escherichia coli (UPEC) Infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol. 2017;8:1566. doi: 10.3389/fmicb.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilahun M, Kassa Y, Gedefie A, Ashagire M. Emerging carbapenem-resistant Enterobacteriaceae infection its epidemiology and novel treatment options: a review. Infect Drug Resist. 2021;14:4363–4374. doi: 10.2147/IDR.S337611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugbo EN, Anyamene CO, Moses IB, et al. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among extended spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae of clinical origin. Gene Rep. 2020;21:100909. doi: 10.1016/j.genrep.2020.100909. [DOI] [Google Scholar]
- Ur Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J. The growing functional and genetic diversity of extended spectrum beta-lactamases. Biomed Res Int. 2018;2018:9519718. doi: 10.1155/2018/9519718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajid M, Awan AB, Saleemi MK, et al. Multiple Drug resistance and virulence profiling of Salmonella enterica serovars typhimurium and enteritidis from poultry farms of Faisalabad. Pakistan Microb Drug Resist. 2019;25(1):133–142. doi: 10.1089/mdr.2018.0121. [DOI] [PubMed] [Google Scholar]
- White RT. Escherichia Coli: placing resistance to third-generation cephalosporins and fluoroquinolones in australia and new zealand into perspective. Microbiol Aust. 2021;42:104–110. doi: 10.1071/MA21031. [DOI] [Google Scholar]
- Wu B, Qi C, Wang L, et al. Detection of microbial aerosols in hospital wards and molecular identification and dissemination of drug resistance of Escherichia coli. Environ Int. 2020;137:105479. doi: 10.1016/j.envint.2020.105479. [DOI] [PubMed] [Google Scholar]
- Wu D, Ding Y, Yao K, Gao W, Wang Y. Antimicrobial resistance analysis of clinical Escherichia coli isolates in Neonatal Ward. Front Pediatr. 2021 doi: 10.3389/fped.2021.670470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin S, Karim A-M, Lee S-H, Zahra R. Temporal variation of meropenem resistance in E coli Isolated from Sewage Water in Islamabad Pakistan. Antibiotics. 2022;11(5):635. doi: 10.3390/antibiotics11050635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengui M, Trabelsi R, Khannous L, et al. Rapid detection of beta-lactamases genes among Enterobacterales in urine samples by using real-time PCR. Biomed Res Int. 2022;2022:8612933. doi: 10.1155/2022/8612933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Xiao S, Gu F, et al. Antimicrobial resistance and molecular epidemiology of uropathogenic Escherichia coli isolated from female patients in Shanghai, China. Front Cell Infect Microbiol. 2021;11:653983. doi: 10.3389/fcimb.2021.653983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
There was no software or code developed during this study, so this is not applicable.