Abstract
The availability of polygenic scores for type 2 diabetes (T2D) raises the question, whether assessing family history might become redundant. However, family history not only involves shared genetics, but also shared environment. It was the aim of this study to assess the independent and combined effects of one family risk score (FamRS) and a polygenic score (PGS) on prevalent and incident T2D risk in a population-based study from Germany (n = 3071). The study was conducted in 2004/2005 with up to 12 years of follow-up. The FamRS takes into account not only the number of diseased first grade relatives, but also age at onset of the relatives and age of participants. 256 prevalent and additional 163 incident T2D cases were recorded. Prevalence of T2D increased sharply for those within the top quantile of the PGS distribution resulting in an OR of 19.16 (p < 2 × 10–16) for the top 20% compared to the remainder of the population, independent of age, sex, BMI, physical activity and FamRS. On the other hand, having a very strong family risk compared to average was still associated with an OR of 2.78 (p = 0.001), independent of the aforementioned factors and the PGS. The PGS and FamRS were only slightly correlated (r2Spearman = 0.018). The combined contribution of both factors varied with varying age-groups, though, with decreasing influence of the PGS with increasing age. To conclude, both, genetic information and family history are relevant for the prediction of T2D risk and might be used for identification of high risk groups to personalize prevention measures.
Subject terms: Genetics, Diseases
Introduction
Type 2 diabetes (T2D) is a growing problem in modern societies. The proportion of affected individuals has been gradually increasing for the past few decades1. In addition to lifestyle factors such as obesity, disadvantageous eating behaviours, and a lack of physical activity, it has been shown that genetics plays a big role too in susceptibility to T2D2. Several studies investigated the heritability of T2D resulting in estimates between 20 and 80% with a median of 40%3–5.
This rise in prevalence needs to be addressed and halted by improving effectiveness of prevention measures. It has been suggested that those measures should be more personalized and especially target high risk groups6. Knowledge on which individual is at high genetic risk to develop T2D in the future might be one of the key points.
A cluster of T2D cases in a family can be an indication for a high genetic risk for T2D. Having first degree relatives with T2D increased the risk of individuals to develop T2D themselves by three times compared to individuals without a positive family history7. The number of affected parents, but also the age at which the parents were diagnosed greatly influenced the estimation of a person’s diabetes risk8. Therefore, we used a family risk score incorporating not only the number of affected parents and siblings, but also weights for their disease onset and takes into account the participants’ age9.
A more puristic approach to a persons’ genetic risk lies in the measurement of genotypes and calculation of genetic risk scores. With genetic information becoming more easily available, the finding that genes seem to play a role in T2D has led to studies investigating polygenic scores (PGS). Khera et al.10, showed that such a genome-wide PGS can be used to identify more than 3% of the population with a more than threefold risk for T2D than the remaining part of the general population10. Thus, such scores might help to target high-risk individuals and offer personalized prevention measures.
We have recently shown that both family history, represented as the aforementioned family risk score9, and genome-wide polygenic scores are independent predictors for the risk of stroke11 and myocardial infarction12. There are no such studies for T2D, yet.
Therefore, we aimed to investigate the independent and combined effect of a genome-wide polygenic score (PGS) and a family risk score (FamRS) on prevalent and incident T2D risk in the population-based KORA F3 study.
Methods
Study population
The KORA studies are a series of population-based studies that were conducted in the city of Augsburg, Germany, and surrounding counties. KORA F3 was performed in 2004/2005, which was the baseline visit for the present investigation. Participants were chosen randomly, stratified for 10 year age-groups and sex, resulting in 3184 participants. The analysis dataset, with available genotype data and variables of interest (family history and prevalent and incident diabetes) comprised of 3071 participants. The applied study methods included a standardized computer-assisted interview, a physical examination and a blood sampling at baseline13,14. The participants were followed-up for mortality and morbidities (including diabetes status) until 2016. The median follow-up time was 11 years with 25,363 person years.
The study was approved by the Ethics Committee of the Bavarian Medical Association (Bayerische Landesärztekammer), all research was performed in accordance with relevant guidelines/regulations and written informed consent was obtained from all participants.
Definition of diabetes
All information given in the interview at baseline (2004/2005) on diabetes and types of diabetes were validated by patient-records and/or by the patients’ general practitioners, resulting in 256 participants with type 2 diabetes. Age at diagnosis was self-declared in a standardized interview. The incident diabetes cases (n = 163) were defined as those who did not have T2D at the time of the baseline KORA F3 study (2004/2005), but who developed it in the timespan between KORA F3 and the follow-up in 2016. For incident cases, diabetes status, type of diabetes and age at diagnosis was validated by medical charge review or contactin the respionsible physician. This information was not derived for the 256 prevalent cases and was additionally missing for 280 participants. Therefore, the analysis dataset for the incident analysis comprised of 2535 participants.
Definition and calculation of the FamRS
In order to obtain the FamRS, participants were asked the following questions for both parents and all siblings in a standardized interview: “Does or did your … (e.g. father) have one of the following diseases?” with diabetes as one of the diseases in the list. No specification of diabetes subtypes was made. If the question for diabetes was answered with yes, it was followed-up by the question, if it was “before the age of 60”, “at the age of 60 or later” or “age unknown” to obtain an age-dependent weight. The FamRS score was then obtained by comparing this weighted observed number of affected family members with the number that would have been expected giving the number of relatives and age of the respective participant. More details and the algorithm can be found in9,15 and the Supplementary Material.
The calculated FamRS was then used both as a continuous variable as well as a categorical variable. For the categorical variable, the following categories were applied9: “average risk” for FamRS ≤ 0.5, “positive risk” for 0.5 < FamRS ≤ 1, “strong positive risk” for 1 < FamRS ≤ 2 and “very strong positive risk” for FamRS > 2. For most analyses, categories “positive risk” and “strong positive risk” were joined due to low numbers of participants in the “positive risk” group.
Genotyping and calculation of the PGS
In the KORA-F3 study, genotypes were derived using the Illumina Omni 2.5 and Illumina Omni Express array. Genotypes were imputed using the HRC reference panel16 and the Michigan imputation Server17. With that method, about 40 million SNPs over the whole genome are available for each of the KORA-F3 study participants. The PGS for T2D that was used in this study was developed and validated by Khera et al.10 in more than 400,000 European samples (UK Biobank). The weights for the 6,917,436 Mio SNPs included in this PGS were derived from the Polygenic Score (PGS) Catalog18 as score number “PGS000014” and calculated using PGS-Calc (https://github.com/lukfor/pgs-calc) by summing up the product of all imputed genotype scores with their respective weights.
Statistical analysis
Several logistic regression models were performed with both prevalent and incident T2D as outcome. FamRS and PGS were analyzed individually using three adjustment models each: (a) unadjusted model, (b) adjusted for sex, age, BMI and physical activity (PA) and c) additionally adjusting for PGS (in the FamRS model) or FamRS (in the PGS model). All covariates were assessed at the baseline visit. Since FamRS is highly right-skewed, it is not considered as a continuous variable, but was divided into three categories (average risk, positive or strong positive (FamRS 2) or very strong positive risk (FamRS 3)). PGS was considered as a continuous variable with the effect size given for one standard-deviation (sd) increase (1 sd of PGS = 0.123). Since a non-linear relationship of PGS with the prevalence of T2D was observed, the ORs are also given for several upper thresholds of the PGS (20%, 10%, 5% or 1%). Logistic regression models were also stratified for 10-year age-groups. Due to lower numbers of cases in these subgroups, these models are not adjusted for any other covariates except FamRS and PGS, which were included mutually. Further, FamRS was only divided into two categories in this analysis: average and above average (FamRS > 0.5). Since there were only 6 prevalent cases in the 35–44 y group, this age-group was omitted from this analysis. To account for the limited follow-up time for those, who died during follow-up, inverse probability of censoring weighting was applied for the incident analyses using package riskRegression in R. For each participant, the inverse of the probability of not being censored is calculated using a Cox-model. This is then used as weights in the logistic regression models.
To evaluate best thresholds for discrimination, ROC curves were used and AUC calculated (using function pROC in R). The predictive capacity of PGS and FamRS was determined by continuous Net Reclassification Index (NRI) and Integrated Discrimination Improvement (IDI) using the function improveProb in package Hmisc. Both adjustment models—adjusted for age, sex, BMI, physical activity plus an additional model including either PGS or FamRS – were used as baseline models to compare with. All analyses were performed using the programme R version 4.1.0. p-values < 0.05 were considered to be statistically significant.
Results
Population characteristics
Descriptive statistics can be found in Table 1. The KORA-F3 analysis dataset consists of 3071 participants, 1575 of which (51.2%) are female, in the age range 35–84 years (mean age 57.4 years) and mean BMI of 27.66. 51.7% were physically active (defined as regular activity for ≥ 1 h per week). At baseline, there were 256 participants with T2D (8.3%), 10 with type 1 diabetes and 9 other or unclear types. From the 2535 participants without prevalent T2D included in the Follow-up analysis, 163 developed T2D after the baseline visit. 35 of those participants with incident T2D and 264 without incident T2D died within the Follow-up period. Both absolute and relative numbers of T2D increased with age at baseline (Supplementary Table S1). The average age at onset (i.e. age at diagnosis as given in the interview) for participants with T2D at baseline was 58 years. Average age at baseline for those participants was 67 years. For incident T2D cases, average age of diagnosis was 67.5 years.
Table 1.
Characteristics of the KORA F3 study (n = 3,071).
| Variable | |
|---|---|
| Age (in years) | 57.4 ± 12.88 [46.0, 57.0, 67.0] |
| Women | 1575 (51.2%) |
| Anthropometric measurements | |
| Weight (in kg) | 77.93 ± 15.00 [66.90,76.80,87.60] |
| BMI | 27.66 ± 4.61 [24.42, 27.13, 30.28] |
| Height (in cm) | 167.7 ± 9.46 [160.5, 167.4, 174.6] |
| Waist circumference (in cm) | 94.93 ± 13.15 [85.6, 95.10, 103.60] |
| Hip circumference (in cm) | 106.9 ± 8.98 [100.9, 105.6, 111.8] |
| Waist-Hip-Ratio | 0.8867 ± 0.085 [0.8240, 0.8890, 0.9470] |
| Lifestyle | |
| Physically active | 1589 (51.7%) |
| Categorizations of physical activity | |
| Regularly > 2 h per week | 688 (22.4%) |
| Regularly active for ~ 1 h per week | 901 (29.3%) |
| Irregularly active for ~ 1 h per week | 460 (15%) |
| Little to no physical activity | 1,022 (33.3%) |
| Smoking categories | |
| Regular smoker | 490 (16.0%) |
| Irregular smoker | 52 (1.7%) |
| Ex-smoker | 1078 (35.1%) |
| Never-smoker | 1283 (41.8%) |
| Family history for diabetes | |
| T2D in any parent | 777 (25.5%) |
| Categorization of Family Risk score FamRS | |
| Average | 2582 (84.7%) |
| Positive FamRS | 343 (11.2%) |
| Strong positive FamRS | 125 (4.1%) |
| Blood values | |
| Glucose (mg/dl) | 107.2 ± 32.27 [91.0, 100.0, 113.0] |
| HbA1c-values (%) | 5.369 ± 0.54 [5.100, 5.300, 5.500] |
| Total cholesterol (mg/dl) | 218.3 ± 39.94 [191.0, 216.0, 243.0] |
| HDL cholesterol (mg/dl) | 58.77 ± 17.15 [46.00, 56.00, 69.00] |
| LDL cholesterol (mg/dl) | 128 ± 32.63 [105, 127, 148] |
| Triglycerides (mg/dl) | 165.2 ± 126.05 [88.0, 136.0, 201.0] |
| Diabetes | |
| Number of prevalent Type 2 cases, n (%) | 256 (8.3%) |
| Number of incident Type 2 cases, n (%) | 163 (5.3%) |
Continuous variables are shown as mean ± SD and [25%, 50%, 75%] percentile; Categorical variable in n (%).
Distribution of the FamRS
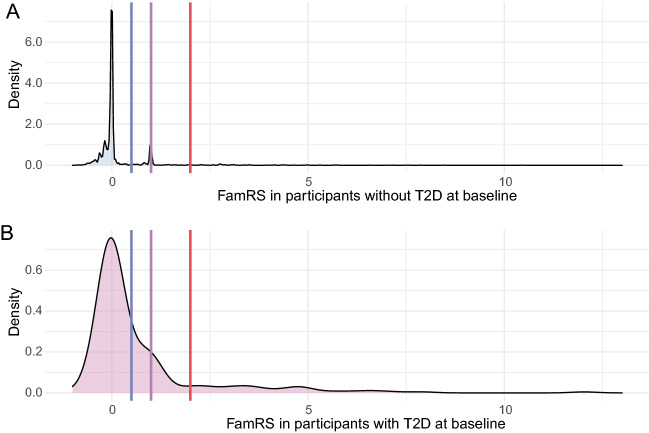
Supplementary Fig. S1 shows the increasing proportion of T2D for increasing number of relatives affected by diabetes. The FamRS distribution markedly differs between participants with or without T2D at baseline with a high peak at 1 for non-cases and a shift to higher values for cases (Fig. 1). For the incident cases, there is an overrepresentation of peaks above zero compared to those participants not developing diabetes during follow-up (Supplementary Fig. S2). Supplementary Table S1 further shows an increase of prevalent and incident T2D over the FamRS risk categories. Supplementary Fig. S3 gives the distribution of the FamRS in T2D cases stratified in 10-year age-groups, showing a decreasing trend for increasing age. Therefore, FamRS seems to be highest for younger participants with T2D.
Figure 1.
Density plot showing the FamRS distribution in participants without (lightblue, panel A) and with T2D (pink, panel B) at baseline; the lines depict the thresholds of the FamRS categories: positive family risk (blue line), strong positive family risk (purple), very strong positive family risk (red line).
Distribution of the PGS
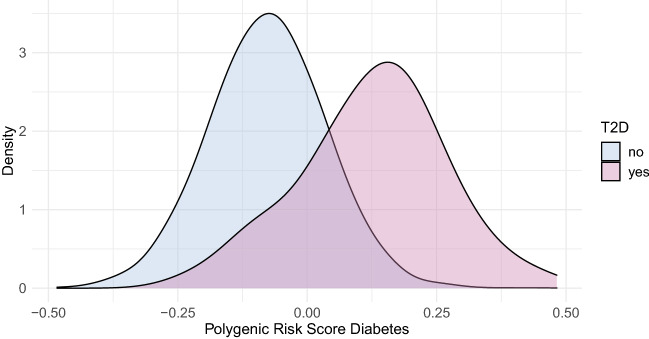
The overall distribution of the PGS in the KORA F3 study is given in Supplementary Fig. S4 together with corresponding distributions in 5 different populations from the 1000 Genomes reference population19. It closely matches the distribution of the Europeans, as expected. A clear right shift of the distribution can be observed in participants with T2D compared to those without T2D at baseline (Fig. 2), in consequence leading also to an increase of T2D cases for increasing percentile groups of the PGS (Supplementary Table S1). For incident cases, the shift is also notable but less pronounced (Supplementary Fig. S5). As was observed for the FamRS, the PGS is decreasing with age at baseline for T2D cases (Supplementary Fig. S6). Supplementary Fig. S7 depicts the prevalence of T2D in percentile groups of the PGS (in 5% groups from 0–5 to 95–100%). Up until the 75%-mark, there was no obvious difference in prevalence between the groups, with a steep increase above the 85%-percentile.
Figure 2.
Density plot showing the PGS distribution in participants without (lightblue) and with T2D (pink) at baseline.
Relationship between FamRS and PGS
The PGS was found to increase with increasing number of family members affected by T2D (Supplementary Fig. S8), which is only diluted in those with more than 6 affected family members. This group only consists of three participants, though.
The relative amount of individuals with positive to very strong positive familial risk increases with increasing PGS-percentiles (Fig. 3). The frequency of individuals with more than average FamRS is more than doubling from about 10% in the lowest 20% of the PGS distribution to 22% in the highest 20%.
Figure 3.
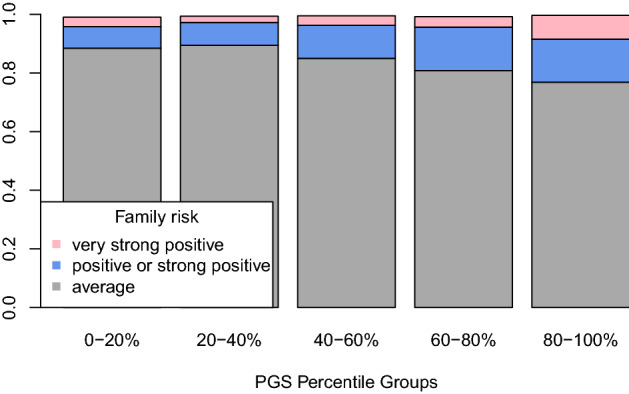
Relative amount of individuals in each FamRS category across the PGS percentile groups.
All in all, both measures are only slightly, although significantly, correlated (r2Spearman = 0.018, p = 5 × 10–14).
Results of logistic regression models
Table 2 shows the results of the logistic regression models of continuous PGS on both prevalent and incident T2D risk. The effect of the PGS was quite stable and highly significant across all models, with ORs ranging from 5.95 to 6.21 for prevalent and 1.66 and 1.68 for incident cases (per 1 sd increase). Due to the nonlinear and steep increase of T2D prevalence especially in the upper 20% of the PGS distribution (Supplementary Fig. S7), the logistic regression was repeated for several upper thresholds for PGS, with the respective remaining of the distribution as the reference category. The ORs increased from 19 to 47 for the upper 20% to 5% of the PGS distribution, even adjusted for age, sex, BMI, PA and FamRS (Table 3). For incident cases, a significant association can be observed for all presented top percentiles, with an OR of about 5 for the highest 5% of the PGS distribution compared to the remainder.
Table 2.
Results of Logistic regression models of the effect of PGS on risk of prevalent and incident diabetes.
| Outcome: Prevalent T2D | Outcome: Incident T2D | |||||
|---|---|---|---|---|---|---|
| OR* | CI (95%) | p-value | OR* | CI (95%) | p-value | |
| Unadjusted | 5.95 | [4.97–7.20] | < 2 × 10–16 | 1.66 | [1.38–1.99] | 4.49 × 10–8 |
| Adjusted for Age + Sex + BMI + Physical activity | 6.35 | [5.18–7.89] | < 2 × 10–16 | 1.68 | [1.39–2.04] | 1.08 × 10–7 |
| Adjusted for Age + Sex + BMI + Physical activity + FamRS | 6.21 | [5.06–7.74] | < 2 × 10–16 | 1.67 | [1.37–2.03] | 2.63 × 10–7 |
* OR are given for increase in 1 SD of PGS (0.123).
Table 3.
Results of Logistic regression models of the effect of PGS cut-offs on risk of prevalent and incident diabetes.
| Outcome: Prevalent T2D | Outcome: Incident T2D | |||||
|---|---|---|---|---|---|---|
| OR* | CI (95%) | p-value | OR* | CI (95%) | p-value | |
| Upper 20% of PGS | 19.16 | [13.65–27.27] | < 2 × 10–16 | 2.16 | [1.45–3.18] | 1.21 × 10–4 |
| Upper 10% of PGS | 32.09 | [22.21–47.05] | < 2 × 10–16 | 2.95 | [1.68–4.99] | 9.35 × 10–5 |
| Upper 5% of PGS | 47.03 | [29.69–76.26] | < 2 × 10–16 | 4.94 | [2.00–11.35] | 2.67 × 10–4 |
All models were adjusted for age, sex, BMI, physical activity and FamRS.
*OR is given for those above this cut-off with the remainder of the population as the reference.
The respective results for FamRS are given in Table 4. Having a positive or strong positive family risk is significantly associated with T2D risk with an OR of 2.08, with more than double the risk (OR 4.73) for a very strong positive family risk. The ORs do not markedly change, if the model is adjusted for age, sex, BMI and PA. It is attenuated, though, but still significant, when the PGS is additionally included in the model.
Table 4.
Results of Logistic regression models of the effect of FamRS on risk of prevalent and incident diabetes
| FamRS categories | Outcome: Prevalent T2D | Outcome: Incident T2D | |||||
|---|---|---|---|---|---|---|---|
| OR* | CI (95%) | p-value | OR* | CI (95%) | p-value | ||
| Unadjusted | FamRS 2 | 2.08 | [1.45–2.92] | 4.05 × 10–5 | 1.40 | [0.85–2.19] | 0.165 |
| FamRS 3 | 4.73 | [3.04–7.20] | 1.77 × 10–14 | 3.46 | [1.85–6.09] | 5.89 × 10–5 | |
| Adjusted for Age + Sex + BMI + Physical activity | FamRS 2 | 2.47 | [1.67–3.61] | 4.30 × 10–6 | 1.60 | [0.95–2.58] | 0.062 |
| FamRS 3 | 4.33 | [2.65–6.95] | 2.17 × 10–9 | 3.74 | [1.94–6.82] | 3.49 × 10–5 | |
| Adjusted for Age + Sex + BMI + Physical activity + PGS | FamRS 2 | 1.67 | [1.01–2.71] | 0.041 | 1.45 | [0.86–2.35] | 0.149 |
| FamRS 3 | 2.78 | [1.49–5.09] | 0.001 | 3.47 | [1.79–6.35] | 1.07 × 10–4 | |
*ORs are given for FamRS categories “positive and strong positive family risk” (FamRS 2) and “very strong positive family risk” (FamRS 3) compared to average family risk.
The effect on incident diabetes was more robust with hardly a difference in ORs between the adjustment models. However, only very strong positive family risk remains significantly associated with an OR of 3.47 in the fully adjusted model. Simplifying the family history to “parents with T2D yes/no” also yields significant association with T2D risk (Supplementary Table S2). The ORs are in about the same range as the one for (strong) positive family risk (OR ranging from 2.14 to 2.68 in the prevalent models and 1.49–1.67 in the incident models), therefore missing the higher risk for those with very strong positive FamRS.
Since Supplementary Figs. S3 and S6 showed decreasing FamRS and PGS with increasing age, it was also evaluated, whether the effect of both scores changed with age. Therefore, logistic regression models were further stratified into 10y age-groups. The effect of PGS is slightly higher for those being 45–64 years at baseline than for older participants with no significant difference, though. The effect also doesn’t change, if additionally adjusted for FamRS (Supplementary Fig. S9, panel B). In contrast, the direction of interaction between age and FamRS on T2D changes, when PGS is taken into account (Supplementary Fig. S9, panel A). In line with the PGS, the OR for FamRS decreases slightly with increasing age in the unadjusted model. It increases, though, especially for the highest age-group, when PGS is adjusted for.
Discrimination and reclassification measures
When the PGS is used as a classifier for prevalent diabetes, the area under the curve (AUC) amounts to 0.869, and the calculated best threshold is estimated to be 0.016 (~ 75% percentile), (Supplementary Table S3. For incident cases, AUC is markedly lower, but still significantly different from 0.5.
For FamRS the AUC for prevalent cases is 0.617 with the best threshold at 0.109. This threshold reflects the boundary of the high peak at around 0 especially for participants without T2D (Fig. 1A). Therefore, most non-cases are captured (specificity = 0.841), but many cases missed (sensitivity = 0.384) (Supplementary Table SS3 The AUC for incident cases (0.538) does not significantly differ from 0.5.
To evaluate whether PGS or FamRS do improve reclassification compared to a model already including several explanatory variables, NRI and IDI were additionally calculated. However, this was restricted to prevalent cases, since AUCs indicated a low predictive power for incident cases.
Adding the PGS to the baseline model could correctly increase T2D probability for 80% of the prevalent cases and decrease for 83% of non-cases, respectively (Supplementary Table S4). This results in an overall NRI of 1.261. The corresponding IDI was 0.328 (p = 1.02 × 10–66). These measures were nearly identical, when FamRS was also included to the baseline model.
Adding the FamRS to the baseline model could merely ameliorate prediction for non-cases by correctly down-shifting the probability for 85% of the non-cases (NRI = 0.361, p = 3.38 × 10–9), although overall NRI and IDI were still significant (IDI = 0.025, p = 0.0002). IDI was not significant any more, when FamRS was compared to a model already including the PGS (Supplementary Table S4).
The simultaneous addition of PGS and FamRS to the baseline model enhances the prediction for both prevalent cases (NRIevents = 0.579) and non-cases (NRInon-events = 0.665), which leads to an overall NRI of 1.244 and IDI of 0.336 (p = 1.47 × 10–66).
Discussion
In this study we found a strong association for both, a genome-wide polygenic score and family history, represented by a weighted family risk score, with the risk of prevalent and incident T2D in a population-based study from Southern Germany. Both scores were only slightly correlated with each other and were found to be independent predictors for T2D.
The PGS was found to be clearly right-shifted for prevalent and to a lesser extent incident T2D cases. The ORs were almost unaffected by any of the adjustments. While the finding of this association itself is in line with previous studies10,20, the extent of the effect size markedly differs. Mars et al.21 evaluated PGSs in different populations resulting in a pooled OR of 1.66 per 1 sd increase in mixed European populations, considering incident and prevalent cases jointly, while we found an OR of around 6 for prevalent and 1.66 in incident cases. In our study, the association was found to be non-linear, with a sharp increase of risk for the upper 20–25% of the distribution, which corresponds to the best discriminating cutoff. Also for these highest risk groups, a marked difference to the literature could be observed: For the top 5% vs. remainder, both Khera et al.10 and Mahajan et al.20,22 observed an OR of 2.75, while our study resulted in an adjusted OR of 47 for the same group. One explanation could be difference in prevalence (~ 2% in10, ~ 8% in KORA-F3) or definition/validation of Type 2 diabetes cases.
PGS was also found to be a valuable predictor for T2D with an AUC of 0.869 for prevalent and 0.613 for incident cases. It also significantly improves reclassification in addition to an already adjusted model and correctly shifts the probabilities for the majority of cases and non-cases to higher respectively lower values. Also here, reported AUCs for PGS were markedly lower (0.64–0.6620).
The FamRS was found to be significantly associated with both prevalent and incident T2D, supporting results of previous studies, which defined family history as any first-degree relative with T2D8 or by also taking into account the number of affected relatives23. The occurrence of T2D in either one of the parents was significantly associated with T2D risk with an OR of about 2.4 for prevalent and 1.6 for incident cases, even in the fully adjusted model. This is comparable to a Dutch study24, which found a HR of 2.2 for parental T2D, also after adjustment for lifestyle factors and obesity. When only looking at parental T2D, the risk discrimination between those with (strong) positive and very strong positive family risk would have been missed. A “very strong positive family risk” (FamRS > 2) occurs, if there are two or more T2D cases in a family with an age of onset before the age of 60. With such a measure, someone with very high familial risk might be identified, but a high fraction of individuals at risk might be missed, especially in small families. This is reflected in the prediction and reclassification measures. Although FamRS can significantly improve prediction, it is a better measure for correctly down-classifying non-cases than up-classifying cases.
However, family history scores taking into account family structure and age of onset, as the FamRS we used, have been shown to be more efficient than only looking at positive family history yes/no or even only disease status in parents9,11,12,25.
Unlike PGS, the effect size of FamRS is not stable across the different adjustment models for prevalent T2D. Although there is no difference from the unadjusted model to the model adjusting for age, sex, BMI and PA, adjustment for the PGS leads to an attenuation of the OR for FamRS risk categories, but it is still significantly associated. This means that a part of the information of FamRS is likely to be of genetic nature, while another part is independent from the PGS.
Cornelis et al.26 estimated by simulation and comparison with empirical data that about one third of the association of parental T2D with T2D risk is due to shared environment, while the remainder is due to shared genetics. This finding is supported by our data. Similar results have already been found for myocardial infarction12,27 and stroke11. For T2D, however, such data are sparse. Chatterjee et al.28 predicted—based on a theoretical model and observed SNP effect size distribution from GWAS—that family history, defined as presence of any affected first-grade relative, hardly improves polygenic risk prediction. In this theoretical framework, however, family history was assumed to represent only shared genetics. In UK-Biobank, however, including the family history of each of the relatives separately in a model improved prediction accuracy of polygenic risk scores29.
In this context, it should be noted that a recall bias cannot be excluded for the determination of the FamRS. It has been shown that both older individuals and those not affected from Diabetes were less accurate reporting the disease status of relatives than younger individuals and individuals with T2D30. This recall bias cannot play a role for incident cases, though, for which the FamRS was also shown to be significantly associated. In these incident models, the effect sizes for FamRS stay quite stable over different adjustment models. Especially for the very strong family risk group, there is also no decrease in effect size from the prevalent to the incident models. This is different for the PGS.
It has been shown that the heritability for T2D is twice as high in patients with age at onset 35–60 years (h2 = 0.69) compared to patients with onset up to 75 years (h2 = 0.31)31. Since the age of onset was about 10 years higher for incident cases in our study than for prevalent cases, the drop in effect sizes for the PGS can likely be explained by that. This assumption is further strengthened by the finding that effect sizes decrease for increasing 10y age-groups. However, confidence intervals overlap to a high degree, which advises against overinterpretations. However, previous studies32,33 have also found that polygenic scores play a greater role for those developing T2D at a younger age. For FamRS, however, there is only a drop in effect sizes for increasing age-groups, if it is unadjusted for the PGS. The drop cannot be observed any more, if it is adjusted for PGS, therefore getting rid of the “shared genetics” component. For higher age, the “shared environment” component, which also partly results in acquired habits and lifestyle, might thus play a bigger role in the FamRS-variable in older participants.
In conclusion, can and should PGS and FamRS be used to identify individuals with high T2D risk? Both scores have been shown to be quite independent from each other, and contribute to risk prediction in varying extent, depending primarily on age of onset. Even though a healthy lifestyle and especially having a normal body weight is beneficial for all in terms of T2D risk, independent of the genetic risk34, it has been shown that those being at high genetic risk defined by either high PGS35, but also high PGS and positive family history36 benefit most from adhering to a healthy lifestyle.
Supplementary Information
Author contributions
C.L.: designed the research. E.D. and C.L.: performed data analysis and drafted the manuscript. L.F. and S.S.: managed genotyping data and calculated the polygenic risk score. C.G., A.P., F.K., H.G.: conceived, designed and oversaw conduction of the KORA-F3 study. L.F., S.S., F.K., H.G.: contributed to interpretation of the data and critical revision of the manuscript. All authors have read and approved the manuscript.
Funding
The KORA study was initiated and financed by the Helmholtz Zentrum München – GermanResearch Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. This project is also funded by the Bavarian State Ministry of Health and Care through the research project DigiMed Bayern (www.digimed-bayern.de). The funding agencies had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
Data availability
The datasets generated and/or analysed during the current study are not publicly available due to restrictions from Helmholtz Zentrum München in accordance with the informed consent given by the study participants, but are available upon request from the corresponding author Prof. Lamina for any researcher based on a standard agreement on data provision within the KORA Research Platform using the digital tool KORA.PASST (https://helmholtz-muenchen.managed-otrs.com/external/c/requestprojectapplication).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Harald Grallert and Claudia Lamina.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-31496-w.
References
- 1.Thibault V, et al. Factors that could explain the increasing prevalence of type 2 diabetes among adults in a Canadian province: A critical review and analysis. Diabetol. Metab. Syndr. 2016;8(1):1–10. doi: 10.1186/S13098-016-0186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuschieri S. The genetic side of type 2 diabetes—A review. Diabetes Metab. Syndr. Clin. Res. Rev. 2019;13(4):2503–2506. doi: 10.1016/j.dsx.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Poulsen P, Ohm Kyvik K, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—A population-based twin study. Diabetologia. 1999;42(2):139–145. doi: 10.1007/S001250051131. [DOI] [PubMed] [Google Scholar]
- 4.J. B. Meigs, L. A. Cupples, & P. W. F. Wilson, Parental transmission of type 2 diabetes the Framingham offspring study, 2000. [Online]. http://diabetesjournals.org/diabetes/article-pdf/49/12/2201/365516/11118026.pdf. [DOI] [PubMed]
- 5.Willemsen G, et al. The concordance and heritability of type 2 diabetes in 34,166 twin pairs from international twin registers: The discordant twin (DISCOTWIN) consortium. Twin Res. Hum. Genet. 2015;18(6):762–771. doi: 10.1017/THG.2015.83. [DOI] [PubMed] [Google Scholar]
- 6.Wareham NJ. Personalised prevention of type 2 diabetes. Diabetologia. 2022;65(11):1796–1803. doi: 10.1007/S00125-022-05774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Florez JC, Hirschhorn J, Altshuler D. The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu. Rev. Genomics Hum. Genet. 2003;4:257–291. doi: 10.1146/ANNUREV.GENOM.4.070802.110436. [DOI] [PubMed] [Google Scholar]
- 8.Scott RA, et al. The link between family history and risk of type 2 diabetes is not explained by anthropometric, lifestyle or genetic risk factors: The EPIC-InterAct study. Diabetologia. 2013;56(1):60–69. doi: 10.1007/s00125-012-2715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RR, et al. Usefulness of cardiovascular family history data for population-based preventive medicine and medical research (the Health Family Tree Study and the NHLBI Family Heart Study) Am. J. Cardiol. 2001;87(2):129–135. doi: 10.1016/s0002-9149(00)01303-5. [DOI] [PubMed] [Google Scholar]
- 10.Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hämmerle M, et al. A family and a genome-wide polygenic risk score are independently associated with stroke in a population-based study. Stroke. 2022;53(7):2331–2339. doi: 10.1161/STROKEAHA.121.036551. [DOI] [PubMed] [Google Scholar]
- 12.Schnitzer F, et al. Association between a polygenic and family risk score on the prevalence and incidence of myocardial infarction in the KORA-F3 study. Atherosclerosis. 2022;352:10–17. doi: 10.1016/j.atherosclerosis.2022.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Löwel H, Döring A, Schneider A, Heier M, Thorand B, Meisinger C. The MONICA Augsburg surveys—Basis for prospective cohort studies. Gesundheitswesen. 2005;67(Suppl 1):S13–18. doi: 10.1055/s-2005-858234. [DOI] [PubMed] [Google Scholar]
- 14.Holle R, Happich M, Löwel H, Wichmann HE, MONICA/KORA Study Group KORA—A research platform for population based health research. Gesundheitswesen. 2005;67(Suppl 1):S19–25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 15.Lamina C, et al. Correlation between a positive family risk score and peripheral artery disease in one case-control and two population-based studies. Atherosclerosis. 2014;237(1):243–250. doi: 10.1016/j.atherosclerosis.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48(10):1284–1287. doi: 10.1038/NG.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert SA, et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021;53(4):420–425. doi: 10.1038/S41588-021-00783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udler MS, McCarthy MI, Florez JC, Mahajan A. Genetic risk scores for diabetes diagnosis and precision medicine. Endocr. Rev. 2019;40(6):1500–1520. doi: 10.1210/ER.2019-00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mars N, et al. Genome-wide risk prediction of common diseases across ancestries in one million people. Cell Genomics. 2022;2(4):100118. doi: 10.1016/J.XGEN.2022.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50(11):1505–1513. doi: 10.1038/S41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hariri S, Yoon PW, Qureshi N, Valdez R, Scheuner MT, Khoury MJ. Family history of type 2 diabetes: A population-based screening tool for prevention? Genet. Med. 2006;8(2):102–108. doi: 10.1097/01.gim.0000200949.52795.df. [DOI] [PubMed] [Google Scholar]
- 24.Abbasi A, et al. Maternal and paternal transmission of type 2 diabetes: Influence of diet, lifestyle and adiposity. J. Int. Med. 2011;270(4):388–396. doi: 10.1111/J.1365-2796.2011.02347.X. [DOI] [PubMed] [Google Scholar]
- 25.Feng R, McClure LA, Tiwari HK, Howard G. A new estimate of family disease history providing improved prediction of disease risks. Stat. Med. 2009;28(8):1269. doi: 10.1002/SIM.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornelis MC, Zaitlen N, Hu FB, Kraft P, Price AL. Genetic and environmental components of family history in type 2 diabetes. Hum. Genet. 2015;134(2):259. doi: 10.1007/S00439-014-1519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tada H, et al. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur. Heart J. 2016;37(6):561–567. doi: 10.1093/eurheartj/ehv462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet. 2013;45(4):400–405. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hujoel MLA, Loh P-R, Neale BM, Price AL. Incorporating family history of disease improves polygenic risk scores in diverse populations. Cell Genomics. 2022;2(7):100152. doi: 10.1016/J.XGEN.2022.100152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bensen JT, et al. Accuracy of proband reported family history: The NHLBI family heart study (FHS) Genet. Epidemiol. 1999;17:141–150. doi: 10.1002/(SICI)1098-2272(1999)17:2. [DOI] [PubMed] [Google Scholar]
- 31.Almgren P, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia study. Diabetologia. 2011;54(11):2811–2819. doi: 10.1007/S00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- 32.Kong X, Xing X, Zhang X, Hong J, Yang W. Early-onset type 2 diabetes is associated with genetic variants of β-cell function in the Chinese Han population. Diabetes. Metab. Res. Rev. 2020;36(2):e3214. doi: 10.1002/dmrr.3214. [DOI] [PubMed] [Google Scholar]
- 33.Piko P, Werissa NA, Fiatal S, Sandor J, Adany R. Impact of genetic factors on the age of onset for type 2 diabetes mellitus in addition to the conventional risk factors. J. Pers. Med. 2021;11(1):1–17. doi: 10.3390/jpm11010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnurr TM, et al. Obesity, unfavourable lifestyle and genetic risk of type 2 diabetes: A case-cohort study. Diabetologia. 2020;63(7):1324–1332. doi: 10.1007/S00125-020-05140-5. [DOI] [PubMed] [Google Scholar]
- 35.Ye Y, Chen X, Han J, Jiang W, Natarajan P, Zhao H. Interactions between enhanced polygenic risk scores and lifestyle for cardiovascular disease, diabetes, and lipid levels. Circ. Genomic Precis. Med. 2021;14(1):E003128. doi: 10.1161/CIRCGEN.120.003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding M, et al. Additive and multiplicative interactions between genetic risk score and family history and lifestyle in relation to risk of type 2 diabetes. Am. J. Epidemiol. 2020;189(5):445–460. doi: 10.1093/AJE/KWZ251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available due to restrictions from Helmholtz Zentrum München in accordance with the informed consent given by the study participants, but are available upon request from the corresponding author Prof. Lamina for any researcher based on a standard agreement on data provision within the KORA Research Platform using the digital tool KORA.PASST (https://helmholtz-muenchen.managed-otrs.com/external/c/requestprojectapplication).