Abstract
Background & Aims
Esophageal squamous cell carcinoma (ESCC) is an aggressive malignancy with a poor long-term prognosis. The molecular mechanisms underlying the initiation and progression of this tumor are largely unknown. The transcription factor GRHL3 functions as a potent tumor suppressor in SCC of skin, head, and neck. This study aims to determine whether GRHL3 also plays a role in the homeostasis of the esophageal epithelium and in the development of ESCC.
Methods
The effects of Grhl3 deletion on squamous epithelial homeostasis in embryos and adult mice were examined using immunohistochemistry, transmission electron microscopy, and real-time polymerase chain reaction. The conditionally deleted mice were subsequently used to determine susceptibility to ESCC. Whole-transcriptome sequencing (RNA-seq) was performed on ESCC in wild-type and Grhl3 deleted animals. To decipher the signaling pathways, real-time polymerase chain reaction, immunohistochemistry, analysis of chromatin immunoprecipitation sequencing, chromatin immunoprecipitation-polymerase chain reaction, and RNA seq datasets were used. Primary human samples were used to validate the findings in the mouse model.
Results
Loss of Grhl3 perturbs the proliferation-differentiation balance in the esophageal epithelium, thereby increasing the susceptibility to esophageal carcinogenesis in adult mice. Grhl3 imparts its tumor suppressor function by regulating the expression of HOPX. We have identified the Wnt/β-catenin pathway as the downstream effectors of GRHL3 and HOPX through our integrated approach using patient-derived ESCC samples and mouse models.
Conclusions
GRHL3 conveys its tumor suppressor function in ESCC through regulating its target gene HOPX, which limits Wnt/β-catenin signaling. Targeted therapies to inhibit this pathway could be a potential treatment strategy for ESCC patients with reduced GRHL3 expression.
Keywords: Transcriptional Regulation, Cancer Signaling Pathways, RNA Sequencing
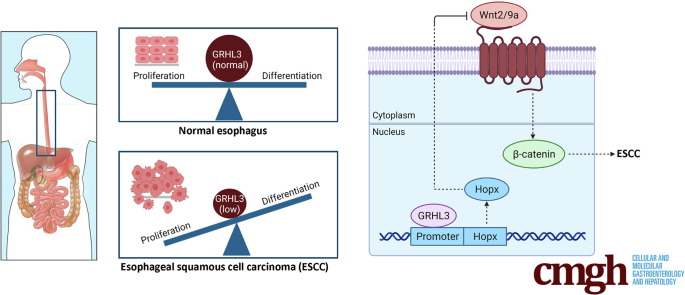
Graphical abstract
Summary.
The transcription factor GRHL3 exerts its tumor suppressor function in esophageal squamous cell carcinoma through regulating HOPX expression and Wnt signaling. The study provides a clear rationale for using targeted therapies against Wnt/β-catenin pathways in patients with reduced GRHL3 expression.
Esophageal squamous cell carcinoma (ESCC) accounts for 90% of all cases of esophageal malignancies globally. It is one of the most common and aggressive malignancies, with high prevalence in Eastern Asia, Southern and Eastern Africa, and Northern and Western Europe.1 Most patients present with locally advanced cancer requiring extensive treatment, including chemotherapy, chemoradiotherapy, and/or surgical resection.2 Even though there has been a marginal reduction in the incidence of ESCC, the medial survival of ESCC has remained unchanged at 5–13 months since the 1990s.3 The poor long-term prognosis relates to a lack of treatment options resulting from a dearth of understanding into the molecular mechanisms that underlie the initiation and progression of the tumor. Hence it is vital to identify the molecular pathogenesis of ESCC and to develop novel therapeutic targets.
Genes dysregulated in ESCC include those involved in cell cycle regulation, receptor tyrosine kinases, chromatin remodeling factors, and genes that perturb squamous cell proliferation/differentiation balance.2 The proliferating cells of the esophagus are confined to the basal cell layer, and imbalance of proliferation/differentiation induces the emergence of preneoplastic epithelium.4 One gene that maintains this balance in a variety of tissues encodes the transcription factor Grainyhead like 3 (GRHL3), shown in in vivo models to be essential for epidermal development and skin barrier formation.5,6 Grhl3 also exhibits critical suppressor function in both skin and oral epithelium, and its loss induces SCC at both sites,7,8 particularly with carcinogen exposure. In the skin, this is mediated by a Grhl3/PTEN/AKT proto-oncogenic axis,7 whereas in head and neck SCC, oncogenesis is driven by dysregulated GSK3B/C-Myc expression.8 Identification of these tissue-specific transcriptional target genes that drive malignancy in the skin and oral epithelium prompted us to investigate the consequences of loss of GRHL3 in the esophageal epithelium.
Results
Grhl3Deletion During Embryogenesis Causes Hyperproliferation of the Esophageal Epithelium
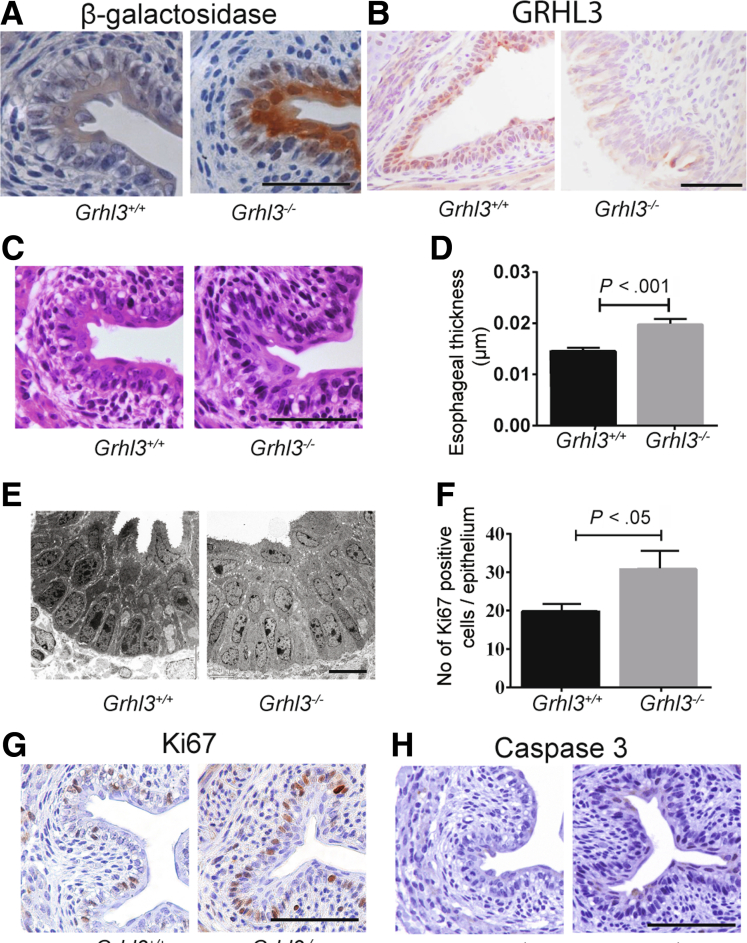
The mouse embryonic esophagus is initially lined by columnar epithelium, which converts to squamous epithelium between embryonic day (E) 10.5 and 14.5.9 During this conversion, the epithelial cells exhibit a reduction in keratin (K) 8 expression and gain in K14 expression. Previous studies in wild-type (WT) mice demonstrated expression of Grhl3 in the developing esophagus from E10.5, which persisted throughout development.10 To examine the effects of loss of Grhl3 on esophageal development, we initially focused on the constitutive Grhl3-null mice, which carry LacZ genes that report from the null alleles.11 Because these animals die at birth of skin barrier and neural tube defects, we harvested mice at E18.5 for analysis. Immunohistochemistry (IHC) detecting β-galactosidase revealed high level expression in esophageal epithelium of Grhl3-/- mice (Figure 1A), with similar staining patterns in the proximal and distal esophagus (not shown). Staining was even throughout the suprabasal layers, with almost complete sparing of the basal layer reminiscent of Grhl3 staining in the developing epidermis.12 The presence of Grhl3 protein expression in WT and its absence in KO embryonic esophagus were confirmed using IHC (Figure 1B). Histologically, the esophageal epithelium of Grhl3-/- mice was thicker compared with Grhl3+/+ mice (Figure 1C and D), with more nucleated cells in the spinous and granular layers (Figure 1E), similar to the appearance of the Grhl3-null epidermis.12 These changes were accompanied by a marked increase in the number of proliferating cells as assessed by Ki67 IHC (Figure 1F and G). No difference in the number of caspase 3-positive apoptotic cells was observed (Figure 1H).
Figure 1.
Grhl3 deletion during embryogenesis causes hyperproliferation of the esophageal epithelium. (A and B) Representative images of transverse sections of the middle esophagus of E18.5 wild-type (Grhl3+/+) and Grhl3-knockout (Grhl3-/-) mice (n = 3 each) immunohistochemically analyzed with β-galactosidase and Grhl3. Scale bar = 50 μm. (C) Representative H&E images of Grhl3+/+ and Grhl3-/- mice middle esophagus showing increased thickness of esophageal epithelium in Grhl3-/- mice. Scale bar = 50 μm. (D) Esophageal epithelial thickness was quantified from H&E images of E18.5 Grhl3+/+ and Grhl3-/- mice (n = 3 each) using ImageJ software. Three sections representing cervical, thoracic, and abdominal areas were examined, and 4 points per section were measured to calculate the average thickness of the esophagus. Statistics were calculated using unpaired, two-tailed Student t test. (E) Representative transmission electron micrograph of E18.5 Grhl3+/+ and Grhl3-/- mice middle esophagus (n = 3 each). Nucleated suprabasal squamous epithelium shown in Grhl3-/- embryo esophagus. Scale bar = 50 μm. (F) Graph shows number of Ki67 positive cells in the esophagus in E18.5 Grhl3+/+ and Grhl3-/- mice (n = 3 each), enumerated from micrographs immunohistochemically analyzed with Ki67 (representative image shown, G). Statistics were calculated using unpaired, two-tailed Student t test (∗P < .05). (H) Representative immunohistochemical images of E18.5 Grhl3+/+ and Grhl3-/- mice middle esophagus (n = 3 each) stained with caspase antibody. Scale bar = 50 μm.
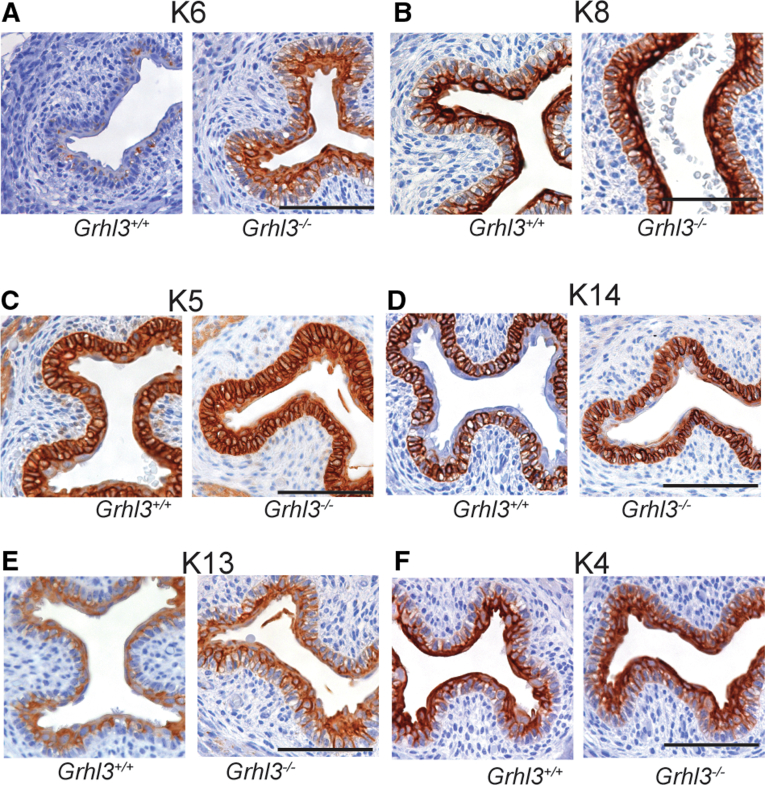
To assess the effects of loss of Grhl3 on the balance between proliferation and differentiation, we examined the expression of a variety of keratin genes. K6, a marker of hyperproliferation that is elevated in the context of epithelial damage, was markedly increased in the esophageal epithelium of Grhl3-/- mice compared with WT embryos (Figure 2A), mimicking the up-regulation we see in the Grhl3-null epidermis. Expression of K8 was also aberrant in the Grhl3-null esophagus, detectable in both basal and suprabasal layers, unlike the WT esophageal epithelium where it was mainly confined to suprabasal layers and markedly reduced expression in the cell membrane of the basal layers (Figure 2B). K5 and K14, which normally mark proliferating cells in the basal layer, and K4, a marker of differentiated suprabasal cells, remained largely unchanged, although K13, another suprabasal marker, was marginally enhanced (Figure 2C–F). Taken together, these results are indicative of a perturbation in the proliferation-differentiation equilibrium of the esophageal epithelium in the absence of Grhl3.
Figure 2.
Grhl3 deletion during embryogenesis causes perturbation of proliferation-differentiation equilibrium of esophageal epithelium. Representative immunohistochemical images of E18.5 Grhl3+/+ and Grhl3-/- mice middle esophagus (n = 3 each) stained with K6 (A), K8 (B), K5 (C), K14 (D), K13 (E), and K4 (F) antibodies. Scale bar = 50 μm.
Grhl3Deletion Is a Driver of Esophageal Squamous Cell Carcinoma
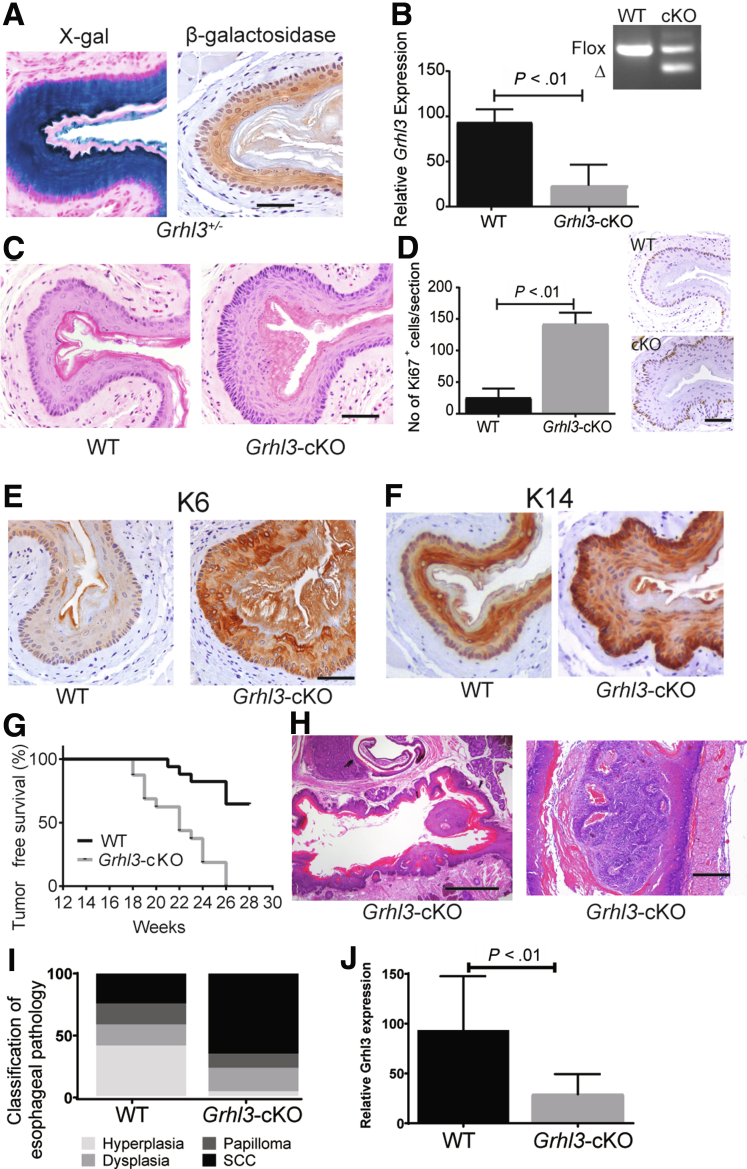
To examine Grhl3 expression in the adult esophagus, we analyzed sections from the Grhl3+/- mice with X-gal staining and β-galactosidase IHC. Robust expression was present in the suprabasal layers of the squamous epithelium, again with almost complete absence in the basal layer (Figure 3). To allow investigation of the functional role of Grhl3 in the adult esophagus, we generated a Grhl3 conditional knockout (Grhl3cKO) line using an EBV ED-L2Cre transgene. The EBV ED-L2 promoter targets expression specifically to the tongue, esophagus, and forestomach but not the skin, avoiding both neural tube and skin barrier defects that are lethal for the constitutive knockouts.13 Mice were engineered to carry a constitutively deleted allele (Grhl3-) and an undeleted floxed allele (Grhl3fl) that could be deleted with exposure to the Cre recombinase to generate the conditional null allele (Grhl3Δ). We have used this strategy previously to maximize the degree of knockdown, because the recombination efficiency induced by Cre is not 100%. This was confirmed by analysis of genomic DNA derived from the esophageal epithelium from Grhl3-/Δ/L2-Cre mice (Grhl3-cKO), which revealed a 1:1 ratio of undeleted (flox) to the deleted Grhl3 allele (Δ) at 3 months of age (Figure 3B, inset), which remained unchanged up to 12 months (not shown). Despite this, we observed a greater than 70% reduction in Grhl3 levels in the Grhl3-cKO esophageal epithelium (Figure 3B). With ageing, the Grhl3-cKO mice developed a hyperplastic esophageal epithelium, with multiple layers of nucleated cells in the spinous and granular layers (Figure 3C, qualitative observation). Similar to the Grhl3-/- embryos, the esophageal epithelium of the Grhl3-cKO mice displayed increased numbers of Ki67 positive cells (Figure 3D) and showed enhanced expression of K6 (Figure 3E). K14 expression was also perturbed, occurring at high levels in the basal layer of the Grhl3-cKO esophageal epithelium but not the WT control (Figure 3F). No substantial differences were observed in the expression patterns of K5, K4, and K13 between the WT and Grhl3-cKO mice (not shown).
Figure 3.
Grhl3 deletion is a driver of esophageal squamous cell carcinoma. (A) Transverse sections of middle esophagus of Grhl3 heterozygous mice (Grhl3+/-) showing β-galactosidase expression driving from LacZ reporter gene using X-gal staining and antibodies against β-galactosidase. Scale bar = 50 μm. (B) The mRNA expression of Grhl3 was detected by quantitative real-time PCR from esophageal epithelium collected from WT (n = 3) and Grhl3-cKO mice (n = 6). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation of replicates. Statistics were calculated using unpaired, two-tailed Student t test. (Inset) Genomic DNA from the esophageal epithelium of 12-week-old WT and Grhl3-cKO mice were analyzed using PCR to detect deletion of floxed Grhl3 allele. The undeleted (flox) band of 425 base pairs and the deleted (Δ) band of 282 base pairs are indicated. (C) Representative H&E images showing moderate hyperplasia of esophageal mucosa in Grhl3-cKO mice compared with WT mice. Scale bar = 50 μm. (D) Graph shows number of Ki67 positive cells in the esophagus in WT (n = 5) and Grhl3-cKO mice (n = 3), enumerated from micrographs immunohistochemically analyzed with Ki67 antibody (representative image shown). Statistics were calculated using unpaired, two-tailed Student t test. (E) Representative immunohistochemical images of WT and Grhl3-cKO mice esophagus (n = 3 each) stained with K6 antibody. Scale bar = 50 μm. (F) Representative immunohistochemical images of WT and Grhl3-cKO mice esophagus (n = 3 each) stained with K14 antibody. Scale bar = 50 μm. (G) Kaplan-Meier survival curve of esophageal tumor-free survival in WT (n = 10) and Grhl3-cKO mice (n = 10). Mice were euthanized upon losing 20% of their body weight. P value from the log-rank (Mantel-Cox) test is .04. (H) Representative H&E images of invasive SCC of the esophagus and occluding squamous papilloma in mid esophageal area of Grhl3-cKO mice. Scale bar = 50 μm. (I) Graph representing classification of esophageal lesions in WT (n = 10) and Grhl3-cKO mice (n = 10). (J) The mRNA expression of Grhl3 was detected by quantitative real-time PCR from esophageal tumors collected from WT (n = 10) and Grhl3-cKO mice (n = 8). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation of replicates. Statistics were calculated using unpaired, two-tailed Student t test (∗∗P < .01).
Because of the disruption in the proliferation/differentiation balance observed in the esophageal epithelium of the Grhl3-cKO mice, we postulated that loss of Grhl3 may be a forerunner of esophageal carcinoma. Therefore, we exposed cohorts of Grhl3-cKO mice (n = 10) and WT controls (n = 10) to the chemical carcinogen 4-nitroquinolene-1 oxide (4-NQO), which has been reported previously to induce esophageal cancer.14 Mice ingested 4-NQO in drinking water for 16 weeks and were then followed for an additional 12 weeks where 4-NQO was removed from the water. At 26 weeks, 100% of the Grhl3-cKO mice exhibited substantial weight loss that necessitated euthanasia, compared with only 40% of WT controls (Figure 3G). Large occluding SCCs were observed in 65% of the Grhl3-cKO mice (Figure 3H), whereas 20% of the WT mice had smaller non-occluding SCCs, and none of the WT mice had large occluding SCCs. Histologic scoring of all lesions revealed a marked skewing toward SCC, papillomas, and dysplasia in the Grhl3-cKO mice, whereas WT mice displayed predominantly hyperplasia of the esophageal epithelium (Figure 3I). The levels of Grhl3 expressed in tumors generated in Grhl3-cKO mice showed 70% reduction compared with the WT tumors, mirroring the reduction in the premalignant esophageal epithelium (Figure 3J).
Loss of GRHL3 Induces Transcriptome Changes in Esophageal SCCs
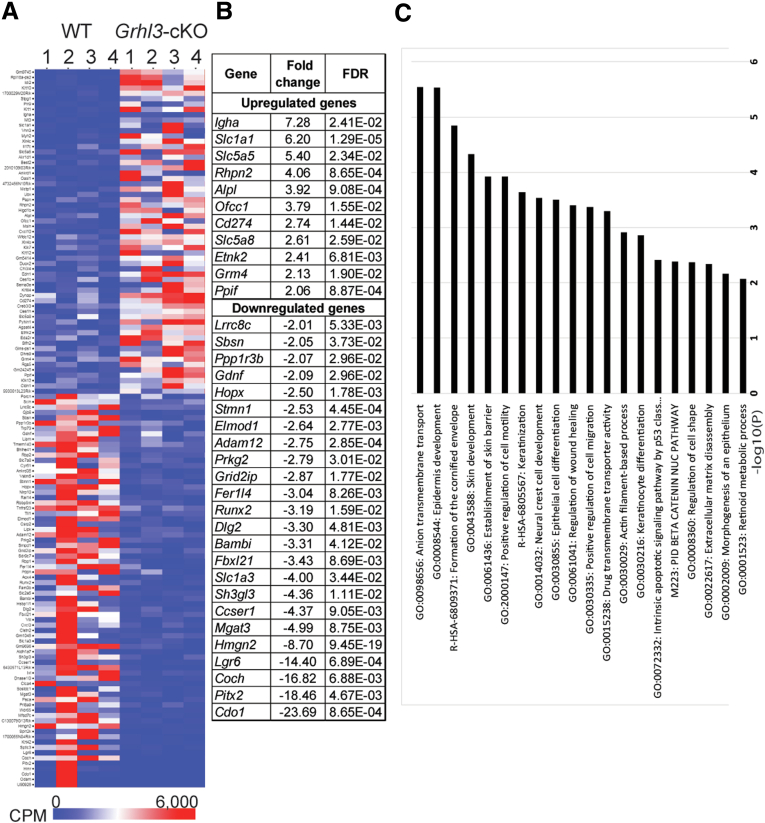
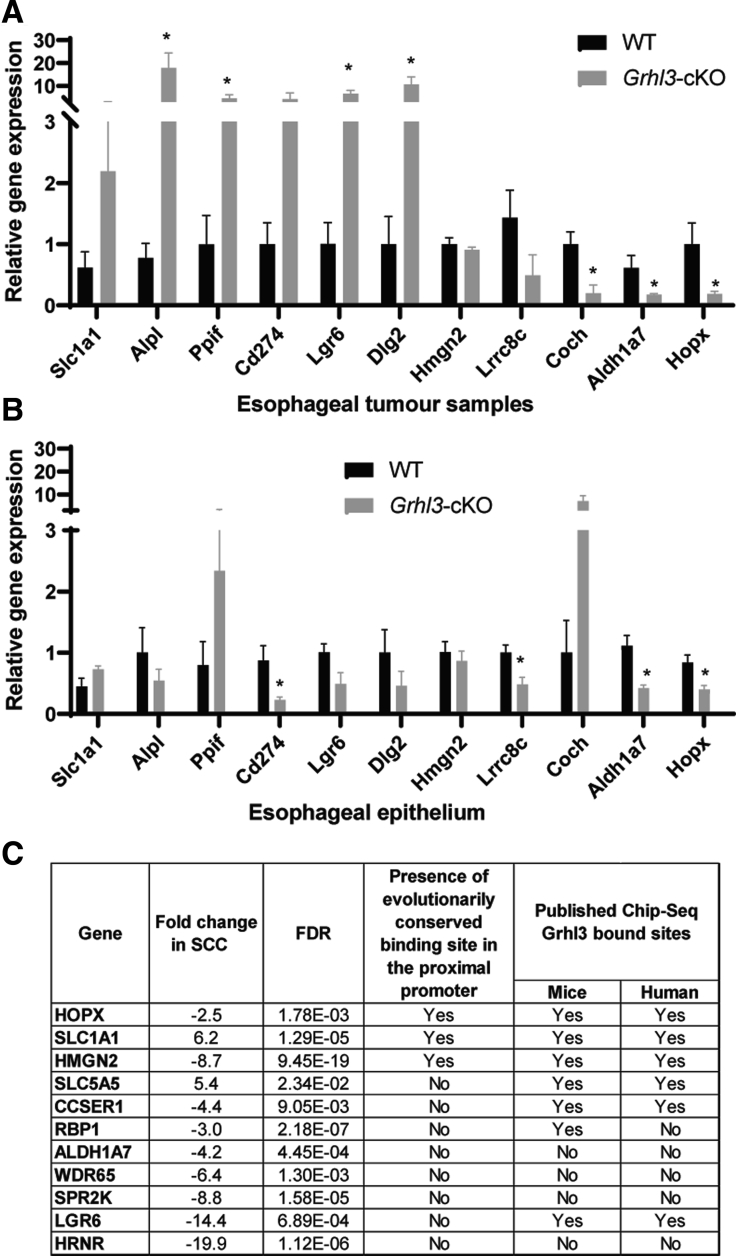
To identify the pathways contributing to esophageal SCCs with Grhl3 deletion, we performed transcriptome analysis in tumors derived from WT and Grhl3-cKO mice (n = 4). We identified 136 differentially expressed genes in the Grhl3-cKO compared with WT tumors (Figure 4A and B), 61 of which showed enhanced expression and 75 that were down-regulated (Table 1). Gene ontology analysis revealed links with expected biological and pathologic processes including epidermis development, skin barrier formation, cell migration, keratinocyte differentiation, apoptosis, and regulation of cell shape but also unanticipated changes in Wnt/β-catenin signaling (Figure 4C). To confirm the validity of our transcriptome-wide data, we performed quantitative real-time polymerase chain reaction (Q-RT-PCR) on a selection of differentially expressed genes implicated in carcinogenesis in esophageal SCCs derived from WT and Grhl3-cKO mice (Figure 5A). A strong correlation was observed between these data and the transcriptome data in almost all instances, with genes including Slc1a1, Lgr6, and Dlg2 all displaying marked up-regulation in the tumors from Grhl3-cKO mice, and Hopx, Coch, and Aldh1a7 being significantly down-regulated in these cancers. To determine which of these changes in expression could be attributed to loss of Grhl3, as opposed to effects induced by the 4-NQO exposure, we examined gene expression in the esophageal epithelium from WT and Grhl3-cKO mice that were not treated with the chemical carcinogen (Figure 5B). Surprisingly, many of the marked changes we had observed in the tumors were lost, or even reversed, in the untreated epithelium. For example, expression of Lgr6 and Dlg2 was down-regulated in the Grhl3-cKO epithelium, and Coch was up-regulated. Therefore, we focused our attention on genes that displayed concordant results in the 2 tissues, a prerequisite of driver mutations for cancer that could function cooperatively with additional mutations induced by the carcinogen.
Figure 4.
Loss of GRHL3 induces transcriptome changes in esophageal SCC. (A) Heat map of RNA-seq transcriptome analysis showing differential gene expression in ESCC from WT and Grhl3-cKO mice. Sixty-one genes were up-regulated, and 75 genes were down-regulated (false discovery rate [FDR] <0.05) in Grhl3-cKO ESCC compared with its WT counterparts. (B) List of genes that are differentially expressed more than 2-fold in Grhl3-cKO mice compared with WT mice. (C) Graph shows enriched ontology clusters in differentially expressed genes in Grhl3-cKO mice using Metascape analysis.
Table 1.
Differentially Expressed Genes in Grhl3 cKO Esophageal Tumors Compared With WT Tumors
| No. | Gene | Expression in cKO tumor vs WT tumor | Fold-change | False discovery rate | P value |
|---|---|---|---|---|---|
| 1 | Gm9745 | 4.86 | 28.95 | 6.19E-14 | 1.72E-17 |
| 2 | Rpl10a-ps2 | 4.52 | 22.89 | 1.36E-10 | 9.49E-14 |
| 3 | Idi2 | 4.39 | 20.91 | 1.18E-11 | 5.74E-15 |
| 4 | Krt10 | 4.07 | 16.77 | 6.19E-14 | 1.43E-17 |
| 5 | 1700029M20Rik | 3.85 | 14.47 | 6.54E-07 | 7.74E-10 |
| 6 | Stpg1 | 3.60 | 12.17 | 5.91E-06 | 9.46E-09 |
| 7 | Prr9 | 3.39 | 10.50 | 5.85E-05 | 1.35E-07 |
| 8 | Krt1 | 3.12 | 8.72 | 6.66E-12 | 2.78E-15 |
| 9 | Igha | 2.86 | 7.28 | 0.02408037 | .0002295 |
| 10 | Mt3 | 2.82 | 7.06 | 4.55E-07 | 5.06E-10 |
| 11 | Slc1a1 | 2.65 | 6.26 | 1.29E-05 | 2.16E-08 |
| 12 | Vnn3 | 2.59 | 6.03 | 0.0212799 | .00019541 |
| 13 | Myh2 | 2.53 | 5.77 | 0.01216068 | 8.46E-05 |
| 14 | Xlr4c | 2.52 | 5.75 | 1.64E-06 | 2.28E-09 |
| 15 | Il17c | 2.51 | 5.70 | 0.0077887 | 4.77E-05 |
| 16 | Slc5a5 | 2.43 | 5.40 | 0.02343524 | .00021846 |
| 17 | Akr1d1 | 2.39 | 5.25 | 1.58E-05 | 2.86E-08 |
| 18 | Best2 | 2.29 | 4.89 | 0.00957711 | 6.33E-05 |
| 19 | 2010109I03Rik | 2.27 | 4.84 | 5.70E-11 | 3.57E-14 |
| 20 | Ankrd1 | 2.26 | 4.79 | 0.02390158 | .00022447 |
| 21 | Oasl1 | 2.24 | 4.71 | 5.85E-05 | 1.36E-07 |
| 22 | 4732456N10Rik | 2.14 | 4.41 | 0.0017232 | 7.07E-06 |
| 23 | Mctp1 | 2.13 | 4.37 | 1.65E-05 | 3.21E-08 |
| 24 | Uox | 2.12 | 4.35 | 0.03069151 | .00033521 |
| 25 | Pspn | 2.12 | 4.34 | 0.00666409 | 3.85E-05 |
| 26 | Rhpn2 | 2.02 | 4.06 | 0.00086482 | 2.96E-06 |
| 27 | Higd1b | 2.00 | 4.00 | 0.0201145 | .00018051 |
| 28 | Alpl | 1.97 | 3.92 | 0.00090791 | 3.41E-06 |
| 29 | Ofcc1 | 1.92 | 3.79 | 0.01549823 | .00012183 |
| 30 | Msln | 1.91 | 3.76 | 0.00021943 | 5.50E-07 |
| 31 | Cxcl10 | 1.89 | 3.70 | 0.00382155 | 1.91E-05 |
| 32 | Wfdc12 | 1.89 | 3.70 | 0.00666409 | 3.83E-05 |
| 33 | Xlr4b | 1.88 | 3.68 | 0.00088665 | 3.19E-06 |
| 34 | Klk7 | 1.88 | 3.68 | 0.00040908 | 1.17E-06 |
| 35 | Krt12 | 1.85 | 3.61 | 0.00192656 | 8.83E-06 |
| 36 | Gm5414 | 1.79 | 3.45 | 0.01008913 | 6.74E-05 |
| 37 | Duox2 | 1.78 | 3.44 | 0.02711639 | .00028295 |
| 38 | Chi3l4 | 1.76 | 3.39 | 0.02585033 | .00025716 |
| 39 | Edn1 | 1.73 | 3.31 | 2.18E-07 | 2.22E-10 |
| 40 | Ces1b | 1.65 | 3.13 | 0.02522116 | .00024914 |
| 41 | Sema3e | 1.59 | 3.00 | 0.01982114 | .00017512 |
| 42 | Krt84 | 1.49 | 2.82 | 0.01212562 | 8.35E-05 |
| 43 | Dynap | 1.48 | 2.80 | 0.00206661 | 9.63E-06 |
| 44 | Cd274 | 1.45 | 2.73 | 0.0143833 | .00010662 |
| 45 | Creb3l3 | 1.40 | 2.65 | 0.01987858 | .00017701 |
| 46 | Ces1h | 1.40 | 2.64 | 4.96E-05 | 1.07E-07 |
| 47 | Slc5a8 | 1.38 | 2.61 | 0.02594167 | .00025987 |
| 48 | Pyhin1 | 1.37 | 2.59 | 9.02E-05 | 2.20E-07 |
| 49 | Agpat4 | 1.36 | 2.57 | 1.80E-05 | 3.64E-08 |
| 50 | Etnk2 | 1.27 | 2.41 | 0.0068103 | 4.03E-05 |
| 51 | Eda2r | 1.23 | 2.34 | 0.00120149 | 4.68E-06 |
| 52 | Slfn2 | 1.21 | 2.31 | 0.00379206 | 1.87E-05 |
| 53 | Glns-ps1 | 1.17 | 2.25 | 0.0431795 | .00053468 |
| 54 | Dhrs9 | 1.10 | 2.15 | 0.01280229 | 9.17E-05 |
| 55 | Grm4 | 1.09 | 2.13 | 0.01895824 | .00016617 |
| 56 | Rgs5 | 1.07 | 2.10 | 0.00089158 | 3.29E-06 |
| 57 | Gm24245 | 1.06 | 2.09 | 0.01721095 | .00014367 |
| 58 | Ppif | 1.04 | 2.06 | 0.00088665 | 3.21E-06 |
| 59 | Klk12 | 1.00 | 2.01 | 0.02484335 | .0002385 |
| 60 | Cldn1 | 1.00 | 2.01 | 0.01531555 | .00011933 |
| 61 | 9930013L23Rik | 1.00 | 2.00 | 0.01159924 | 7.91E-05 |
| 62 | Porcn | –1.00 | 2.00 | 0.03518776 | .00040389 |
| 63 | Scin | –1.01 | 2.02 | 0.00481262 | 2.64E-05 |
| 64 | Lrrc8c | –1.02 | 2.03 | 0.00532646 | 3.00E-05 |
| 65 | Gjb6 | –1.04 | 2.05 | 0.04022195 | .00047847 |
| 66 | Sbsn | –1.04 | 2.05 | 0.03733291 | .00043697 |
| 67 | Ppp1r3b | –1.05 | 2.07 | 0.02964876 | .00031969 |
| 68 | Trp73 | –1.06 | 2.09 | 0.0017369 | 7.25E-06 |
| 69 | Gdnf | –1.06 | 2.09 | 0.02964876 | .00031742 |
| 70 | Lipm | –1.10 | 2.14 | 0.03374927 | .00037723 |
| 71 | Tmem140 | –1.11 | 2.15 | 0.00280495 | 1.37E-05 |
| 72 | Bhlhe41 | –1.13 | 2.19 | 0.01880423 | .00016221 |
| 73 | Rbp2 | –1.18 | 2.26 | 0.00481262 | 2.56E-05 |
| 74 | Slc7a8 | –1.18 | 2.27 | 0.0017232 | 7.00E-06 |
| 75 | Cyr61 | –1.23 | 2.34 | 0.02497147 | .00024146 |
| 76 | Ankrd35 | –1.30 | 2.46 | 0.00057789 | 1.81E-06 |
| 77 | Vstm5 | –1.33 | 2.51 | 0.00183842 | 8.06E-06 |
| 78 | Stmn1 | –1.34 | 2.53 | 0.0004453 | 1.33E-06 |
| 79 | Hopx | –1.34 | 2.53 | 0.00177526 | 7.53E-06 |
| 80 | Nlrp10 | –1.34 | 2.54 | 5.85E-05 | 1.38E-07 |
| 81 | Rai14 | –1.37 | 2.58 | 0.01740342 | .00014649 |
| 82 | Rbbp8nl | –1.39 | 2.63 | 0.00683881 | 4.09E-05 |
| 83 | Tnfrsf23 | –1.40 | 2.64 | 8.51E-08 | 7.70E-11 |
| 84 | Tll1 | –1.40 | 2.65 | 0.02074218 | .00018902 |
| 85 | Elmod1 | –1.40 | 2.65 | 0.0027709 | 1.32E-05 |
| 86 | Csrp2 | –1.42 | 2.68 | 0.02711639 | .00027879 |
| 87 | Lipk | –1.43 | 2.69 | 0.0143833 | .00010714 |
| 88 | Adam12 | –1.46 | 2.75 | 0.00028507 | 7.73E-07 |
| 89 | Prkg2 | –1.48 | 2.79 | 0.03014967 | .00032719 |
| 90 | Smpd1 | –1.52 | 2.86 | 8.89E-07 | 1.11E-09 |
| 91 | Grid2ip | –1.52 | 2.87 | 0.01771554 | .00015158 |
| 92 | Sdr9c7 | –1.56 | 2.94 | 0.0005923 | 1.92E-06 |
| 93 | Rbp1 | –1.57 | 2.98 | 2.18E-07 | 2.27E-10 |
| 94 | Fer1l4 | –1.61 | 3.04 | 0.008261 | 5.11E-05 |
| 95 | Pdpn | –1.62 | 3.06 | 0.01455309 | .00011136 |
| 96 | Aox4 | –1.62 | 3.08 | 0.04713894 | .00060338 |
| 97 | Runx2 | –1.68 | 3.19 | 0.01589786 | .00012784 |
| 98 | Fam3b | –1.68 | 3.20 | 0.01711759 | .0001417 |
| 99 | Slc2a5 | –1.69 | 3.24 | 0.00040908 | 1.17E-06 |
| 100 | Bambi | –1.73 | 3.31 | 0.04121757 | .00049891 |
| 101 | Hsbp1l1 | –1.74 | 3.33 | 0.02522116 | .00024596 |
| 102 | Dlg2 | –1.75 | 3.36 | 0.00481262 | 2.62E-05 |
| 103 | Fbxl21 | –1.78 | 3.43 | 0.008685 | 5.49E-05 |
| 104 | Vill | –1.87 | 3.66 | 0.00489226 | 2.72E-05 |
| 105 | Cxcl3 | –1.88 | 3.67 | 0.00025812 | 6.64E-07 |
| 106 | Clstn2 | –1.89 | 3.71 | 0.01589786 | .00012829 |
| 107 | Gm1045 | –1.94 | 3.83 | 0.0143833 | .00010697 |
| 108 | Slc1a3 | –2.01 | 4.03 | 0.03439203 | .00039237 |
| 109 | Gm9696 | –2.07 | 4.19 | 0.0005923 | 1.94E-06 |
| 110 | Aldh1a7 | –2.10 | 4.28 | 0.0004453 | 1.32E-06 |
| 111 | Sh3gl3 | –2.12 | 4.36 | 0.01108265 | 7.48E-05 |
| 112 | Ccser1 | –2.13 | 4.37 | 0.00905438 | 5.92E-05 |
| 113 | 6430571L13Rik | –2.20 | 4.58 | 0.00026295 | 6.95E-07 |
| 114 | Ivl | –2.20 | 4.59 | 4.42E-05 | 9.23E-08 |
| 115 | Dnase1l3 | –2.20 | 4.59 | 3.74E-09 | 2.87E-12 |
| 116 | Clca4 | –2.20 | 4.60 | 0.00481262 | 2.59E-05 |
| 117 | Sostdc1 | –2.25 | 4.75 | 0.0068103 | 4.02E-05 |
| 118 | Mgat3 | –2.32 | 4.99 | 0.00874882 | 5.66E-05 |
| 119 | Psca | –2.43 | 5.39 | 9.27E-13 | 3.22E-16 |
| 120 | Prl8a9 | –2.56 | 5.88 | 0.01589786 | .00012786 |
| 121 | Wdr65 | –2.70 | 6.49 | 0.0013019 | 5.16E-06 |
| 122 | Mfsd7c | –2.74 | 6.67 | 2.69E-06 | 3.93E-09 |
| 123 | C130079G13Rik | –3.02 | 8.11 | 7.76E-09 | 6.48E-12 |
| 124 | Hmgn2 | –3.13 | 8.73 | 9.45E-19 | 6.57E-23 |
| 125 | Sprr2k | –3.14 | 8.83 | 1.58E-05 | 2.75E-08 |
| 126 | 1700055N04Rik | –3.34 | 10.12 | 4.99E-06 | 7.63E-09 |
| 127 | Krt42 | –3.46 | 10.98 | 0.0166903 | .00013701 |
| 128 | Sptlc3 | –3.66 | 12.60 | 8.67E-15 | 1.21E-18 |
| 129 | Lgr6 | –3.85 | 14.43 | 0.000689 | 2.30E-06 |
| 130 | Coch | –4.07 | 16.82 | 0.00688372 | 4.17E-05 |
| 131 | Pitx2 | –4.21 | 18.46 | 0.00467098 | 2.40E-05 |
| 132 | Hrnr | –4.28 | 19.40 | 1.12E-06 | 1.48E-09 |
| 133 | Cdo1 | –4.57 | 23.69 | 0.00086482 | 3.01E-06 |
| 134 | Odam | –5.94 | 61.26 | 0.008685 | 5.50E-05 |
| 135 | U90926 | –7.50 | 180.90 | 1.65E-05 | 3.13E-08 |
| 136 | Xist | –9.55 | 750.27 | 1.49E-11 | 8.27E-15 |
NOTE. Data are from RNA-seq analysis.
Figure 5.
Loss of GRHL3 induces transcriptome changes in esophageal SCC. (A) The mRNA expression of selected genes by Q-RT-PCR from esophageal tumor samples collected from WT and Grhl3-cKO mice. Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation of 3–5 replicates. Statistics were calculated using multiple unpaired t test using GraphPad prism (∗P < .05). (C) The mRNA expression of selected genes by Q-RT-PCR from esophageal epithelium collected from WT and Grhl3-cKO mice. Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation of 4–8 replicates. Statistics were calculated using multiple unpaired t test using GraphPad prism (∗P < .05). (C) Table showing putative Grhl3 target genes that are differentially expressed in Grhl3-cKO mice.
HopxIs a Direct Target of GRHL3
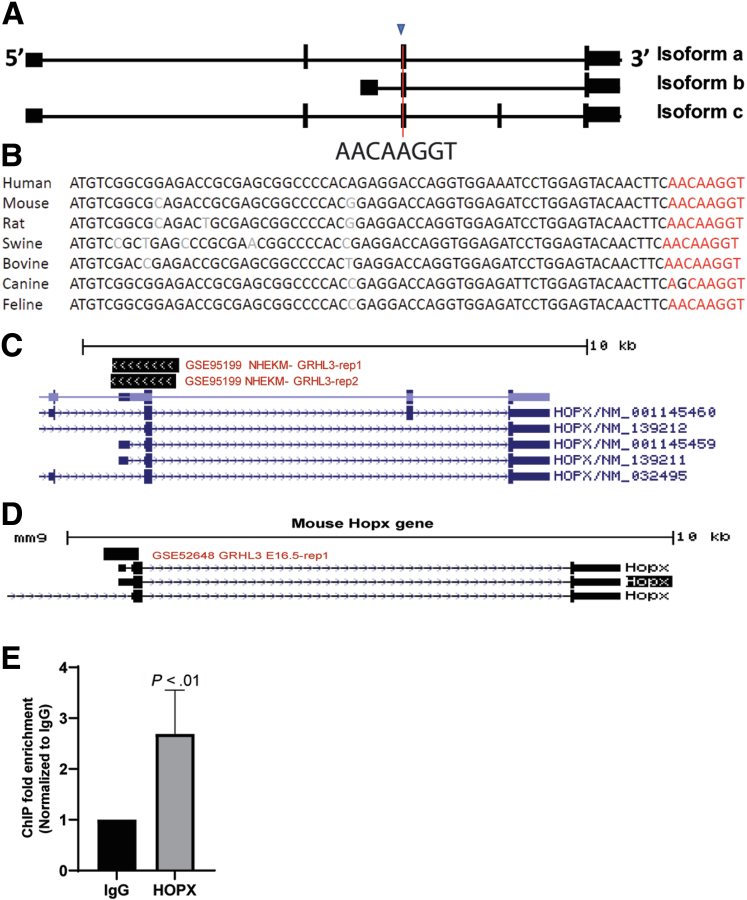
As a transcription factor, Grhl3 exerts its biological influence through the regulation of specific target genes. We have previously defined the core GRHL3 consensus DNA binding site (AACCGGTT), which is evolutionarily conserved from Drosophila to humans over 700 million years.5 Using this sequence, we developed an algorithm to identify direct GRHL3 target genes that was based on (1) the presence of a predicted GRHL3 binding site within 2 kb of the transcription start site; (2) evolutionary conservation of this site across vertebrates; (3) the presence of a chromatin immunoprecipitation (ChIP)-sequencing peak on published datasets; and (4) differential expression in WT versus Grhl3-cKO tissues (Figure 5C). This strategy has been highly successful, leading to the identification of critical GRHL3 target genes in wound repair,15 skin barrier formation,5 and epithelial cancers.7,8 Because our previous studies have shown that GRHL3 functions as a tumor suppressor in SCCs from multiple tissue types by transcriptional activation of key target genes, we focused on genes that displayed reduced expression in the absence of Grhl3 and met all other criteria. On this basis, only Hopx fulfilled the stated requirements: (1) a GRHL3 binding site was identified (Figure 6A); (2) the site was conserved (Figure 6B); and (3) ChIP-seq data identified a clear ChIP-seq peak at the 5′ end of the coding exon (Figure 6C and D). We validated the GRHL3 occupancy to the Hopx gene at the conserved site by ChIP-Q-PCR (Figure 6E).
Figure 6.
HOPX is a direct target of GRHL3. (A) Location of GRHL3 binding site in the human HOPX genome. Blue arrowhead indicates start codon. (B) Alignment of GRHL3 binding site (red) in the indicated species showing conservation of the site in multiple species. (C) ChIP-seq peak in the promoter region of human HOPX genome from GRHL3 ChIP performed on primary human keratinocytes (NHEK, GSE95199), aligned in the hg19 UCSC genome assembly. (D) ChIP-seq peak in the promoter region of mouse Hopx genome from GRHL3 ChIP performed on E16.5 mouse back skin (GSE52648), aligned in the UCSC mm9 genome assembly. (E) ChIP-Q-PCR from EPC cells demonstrating more than 2-fold enrichment over immunoglobulin G (IgG) in the binding site of HOPX promoter (n = 4 replicates). Statistics were calculated using unpaired, one-tailed Student t test.
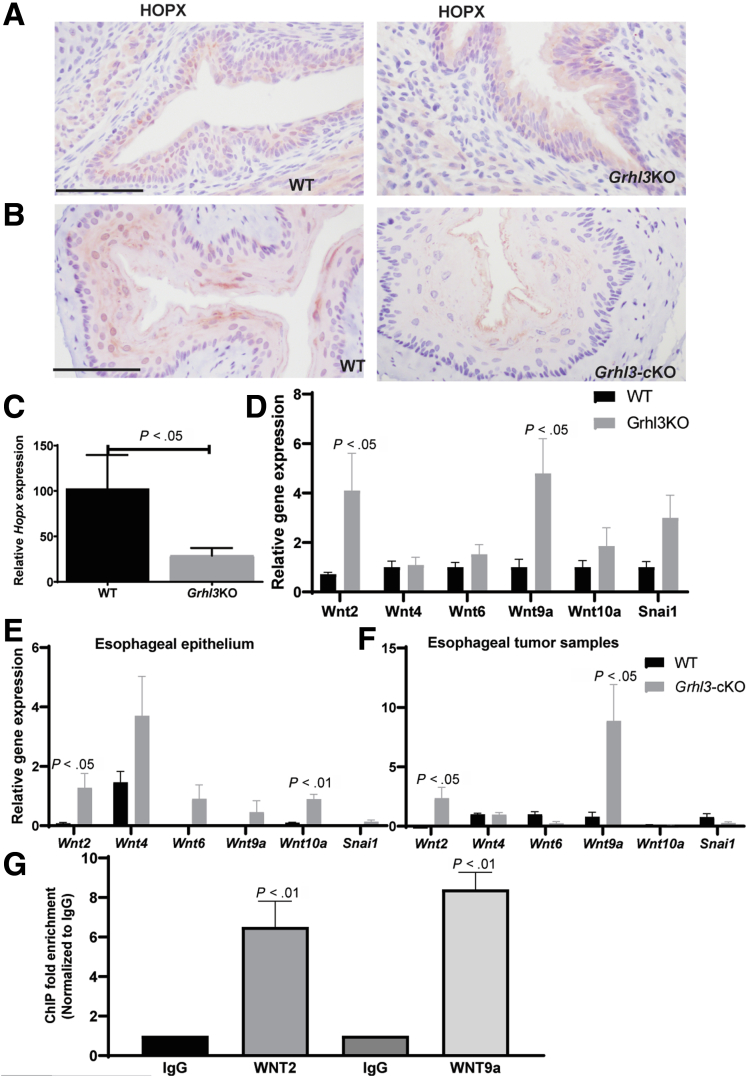
To determine the localization and expression pattern of Hopx in esophageal epithelium, we performed IHC. In WT embryos, the Hopx expression correlates with Grhl3 expression in the suprabasal cell layer and a complete absence of expression in the Grhl3-/- embryonic esophageal epithelium (Figure 7A). Similarly, there is reduced Hopx expression in the adult esophageal epithelium (Figure 7B). The expression of Hopx was reduced in the esophageal epithelium lacking Grhl3 expression (Figure 7C). Hopx has been shown to inhibit Wnt signaling in cardiomyoblasts, airway epithelium, and non-small cell lung cancer by repressing the expression of multiple Wnt ligands,16,17 and alterations in Wnt/β-catenin signaling were observed in our gene ontology analysis (Figure 4C). To examine this directly, we evaluated the expression of Wnt ligands in the esophageal epithelium of the Grhl3-null mice (Figure 7D) and Grhl3-cKO mice (Figure 7E and F) using Q-RT-PCR. Wnt2 and Wnt9a were both up-regulated in tissue from Grhl3-cKO esophageal tumors and Grhl3-null embryos, whereas all other Wnts tested were unchanged. The protein expression of Wnt9a in esophageal tumor sample is also markedly increased in Grhl3-cKO mice (data not shown). We then validated HOPX occupancy in the CPG island of the Wnt2 and Wnt9a promoters using ChIP-Q-PCR (Figure 7G).
Figure 7.
Reduced expression of HOPX activates Wnt signaling pathway in esophageal epithelium. (A) Representative immunohistochemical images of middle esophagus from WT E18.5 and Grhl3KO embryo esophagus (n = 3 each) stained with HOPX antibody. Scale bar = 50 μm. (B) Representative immunohistochemical images of middle esophagus from WT and Grhl3-cKO adult esophagus (n = 3 each) stained with HOPX antibody. Scale bar = 50 μm. (C) The mRNA expression of Hopx was detected by Q-RT-PCR from esophagus collected from WT (n = 3) and Grhl3KO mice (n = 6). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation. Statistics were calculated using unpaired, two-tailed Student t test. (D) The mRNA expression of indicated genes by Q-RT-PCR from esophagus collected from WT (n = 4) and Grhl3KO mice (n = 4). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation. Statistics were calculated using multiple unpaired t test using GraphPad prism. (E) The mRNA expression of indicated genes by Q-RT-PCR from esophageal epithelium collected from WT (n = 6) and Grhl3-cKO mice (n = 6). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation. Statistics were calculated using multiple unpaired t test using GraphPad Prism. (F) The mRNA expression of indicated genes by Q-RT-PCR from esophageal tumors collected from WT (n = 5–6) and Grhl3-cKO mice (n = 5–6). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation. Statistics were calculated using multiple unpaired t test using GraphPad Prism. (G) ChIP-Q-PCR from EPC cells demonstrating more than 4-fold enrichment over immunoglobulin G (IgG) in WNT2 and WNT9a promoter (n =3 replicates). Statistics were calculated using unpaired, one-tailed Student t test.
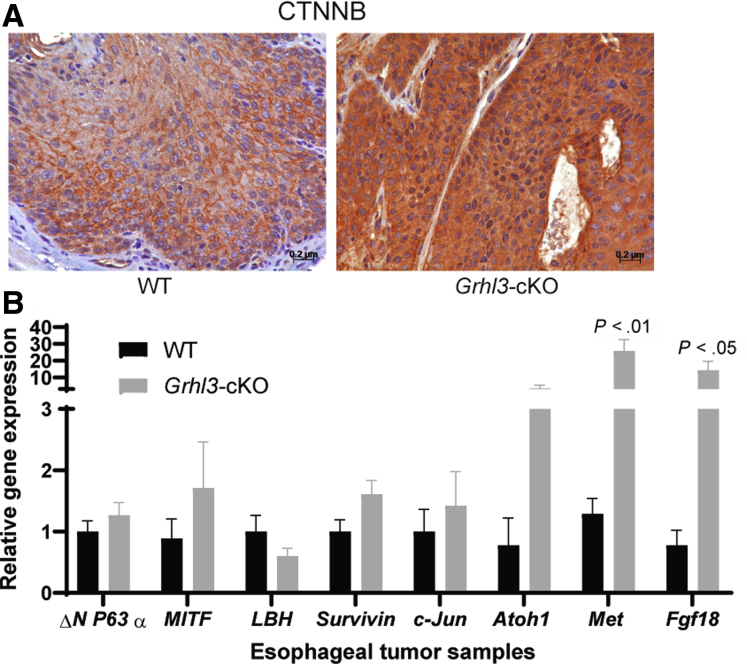
β-catenin immunohistochemical analysis of esophageal SCCs revealed a marked increase in expression in Grhl3-cKO compared with WT tumors (Figure 8A), suggesting Wnt-β catenin signaling is active specifically in these cancers, in support of our RNA-seq pathway enrichment analysis. We examined several known Wnt pathway target genes altered in other cancers in esophageal tumor samples.18,19 Among them, Met and Fgf18 were significantly up-regulated in the ESCCs derived from Grhl3-cKO mice compared with WT controls (Figure 8B).
Figure 8.
Wnt pathway is dysregulated esophageal SCC samples. (A) Representative immunohistochemical images of WT and Grhl3-cKO mice esophageal tumors (n = 3 each) stained with CTNNB antibody. Scale bar = 50 μm. (B) The mRNA expression of indicated genes by Q-RT-PCR from esophageal tumors collected from WT (n = 8–9) and Grhl3-cKO mice (n = 8–9). Relative gene expression was calculated using HPRT as the housekeeping gene. Data were represented as mean ± standard deviation. Statistics were calculated using multiple unpaired t test using GraphPad Prism.
GRHL3andHOPXExpression Is Reduced in Patient-Derived Esophageal SCC Samples
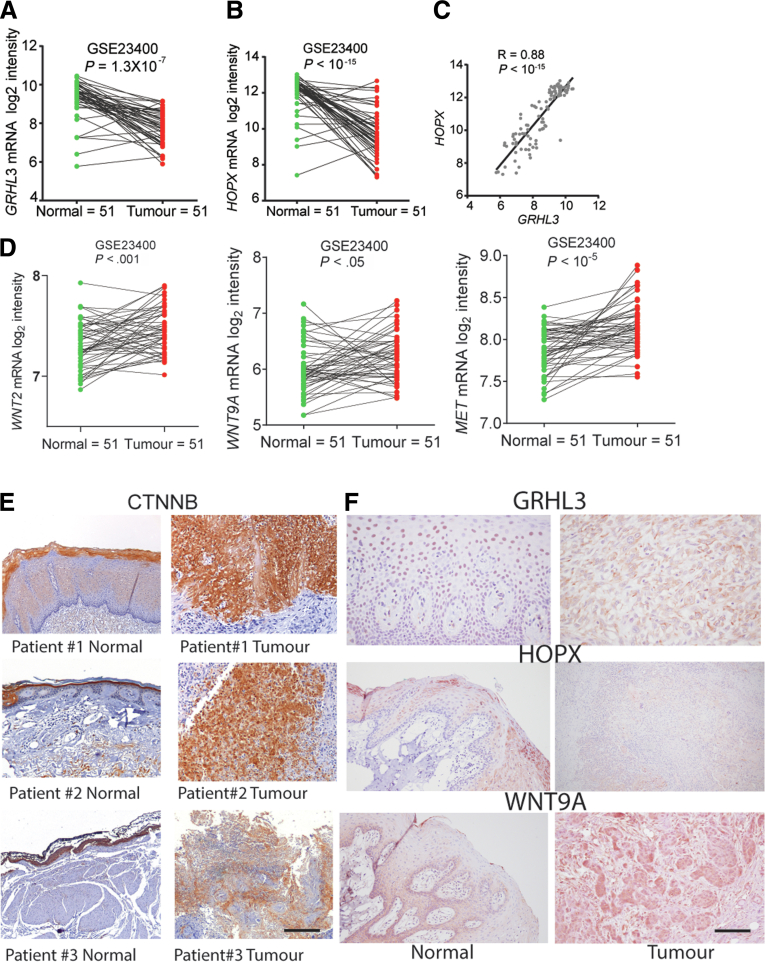
The genome-wide gene expression in the tumors from 51 ESCC cases to their matched normal tissue samples were studied using Affymetrix microarray.20 The mRNA expression of GRHL3 was 2.9-fold down-regulated in ESCC samples compared with normal matched controls (Figure 9A, GSE23400).20,21 The expression of HOPX mRNA was also significantly down-regulated in ESCC patient samples (Figure 9B), and the correlation between GRHL3 and HOPX mRNA levels (r value 0.88) suggested that they were inter-related (Figure 9C). Similar to mice SCC, WNT2, WNT9A, and MET were up-regulated in these ESCC samples compared with normal matched controls (Figure 9D). However, fibroblast growth factor 18 was unchanged between normal and tumor samples (data not shown). The increased expression of β-catenin in human patient samples indicates this pathway is active in oral squamous cell carcinoma (Figure 9E). These findings support our hypothesis that loss of GRHL3 leads to down-regulation of HOPX and subsequent activation of the Wnt/β-catenin pathway. We had validated the expression of GRHL3, HOPX, and WNT9a in human samples (Figure 9E).
Figure 9.
GRHL3 and HOPX expression is reduced in patient-derived esophageal SCC samples leading to activation of Wnt signaling pathway. (A) GRHL3 mRNA expression in 51 oral squamous cell carcinoma (OSCC) samples compared with its own paired normal tissue, calculated from GSE23400. Statistics were calculated using unpaired, two-tailed Student t test, and P values are indicated on the graph. (B) HOPX mRNA expression in 51 OSCC samples compared with its own paired normal tissue, calculated from GSE23400. Statistics were calculated using unpaired, two-tailed Student t test, and P values are indicated on the graph. (C) Correlation between GRHL3 and HOPX mRNA expression in patient samples calculated from GSE23400. Pearson r method using GraphPad prism is used to calculate the correlation between the mRNA expression. (D) WNT2, WNT9A, and MET mRNA expression in 51 OSCC samples compared with its own paired normal tissue, calculated from GSE23400. Statistics were calculated using unpaired, two-tailed Student t test, and P values are indicated on the graph. (E) Representative immunohistochemical images of 3 human patient esophageal samples immunohistochemically stained with CTNNB. Scale bar = 50 μm. (F) Representative immunohistochemical images of human patient esophageal samples immunohistochemically stained with GRHL3, HOPX, and WNT9A. Scale bar = 50 μm.
Discussion
Using Grhl3 deleted mouse models and primary human samples we have identified molecular pathways involved in the pathogenesis of ESCCs, which offer insights for potential novel therapeutics and biomarkers. Grhl3 is robustly expressed in the esophageal epithelium of embryo and adult mice, and its reduced expression dysregulates the proliferation/differentiation balance and increases the propensity to ESCCs with exposure to the carcinogen 4-NQO. Because it is predominantly expressed in the suprabasal layers, the role of GRHL3 in controlling proliferation has been extensively studied in the epidermis. Using single cell RNA-seq, Lin et al22 have shown that differentiation of keratinocytes in the epidermis is a gradualistic process, and loss of GRHL3 induces emergence of epidermal stem cells with proliferative potential by suppressing Wnt signaling.
Tissue-specific transcriptional control of GRHL3 in established in skin cancer and head and neck squamous cell carcinoma, and the signaling pathways7,8 induced by loss of Grhl3 in mice differed markedly between the epidermis and oral epithelium. Adding to this complexity of transcriptional control by GRHL3, we identified increased Wnt/β-catenin signaling in ESCCs, induced by differential regulation of one of the known Grhl3 target genes, HOPX. HOPX was initially reported as a differentiation marker, and its expression is markedly suppressed in a variety of cancers and is shown to inhibit metastatic progression.17,23,24 Even though epigenetic regulation including DNA hypermethylation is reported as a mechanism of reduced expression of HOPX in nasopharyngeal carcinoma,23 it does not play a role in HOPX expression in lung adenocarcinoma and human epidermal keratinocytes.17,25 This holds true in the case of ESCCs, where its expression is transcriptionally regulated by GRHL3.
Our integrated approach using patient-derived ESCC samples and mouse models identified the Wnt/β-catenin pathway as the downstream signaling target of GRHL3 and HOPX. One of the up-regulated Wnt ligands, Wnt2, in both mice and human ESCCs is significantly associated with poor clinical outcomes in patients and has been shown to activate canonical Wnt/β-catenin signaling pathway in esophageal cancer cells.26 Wnt2 has been shown to be secreted by tumor fibroblasts, and it is probable that reduced expression of HOPX removes the transcriptional control leading to enhanced expression of these secretory ligands. Through whole-genome analysis, it has been shown that HOPX regulates multiple Wnt genes, and in tissues, with Hopx deletion there is an expansion of Wnt signaling.16 We propose that the tumor suppressor function of HOPX observed in multiple cancers is due to its ability to repress Wnt signaling, which is based on the evidence from ESCC. The presence of stem cells in the esophagus is still under debate; however, using organoids, it has been shown that a non-quiescent stem cell population resides in the basal epithelium of mice esophagus, and Wnt has been shown to regulate esophageal self-renewal.27 It is possible that these stem cells are activated with reduced expression of GRHL3 and HOPX through Wnt signaling leading to ESCC.
Our work identified a molecular signature in ESCCs that provides a clear rationale for using targeted therapies in this cancer. Several Wnt antibodies (vantictumab, inhibitor of frizzled receptor) and β-catenin inhibitors (PRI-724) are already phase 1 clinical trials in solid cancers28 and are promising agents for ESCC. There are challenges in inhibiting the Wnt pathway, because this pathway plays an important role in the maintenance of stem cells and regeneration of tissues and organs. Specific inhibitors targeting cancer stem cells are required to overcome side effects; nevertheless, ESCC is a potential solid cancer that could benefit from Wnt/β-catenin inhibition in select patients with low GRHL3 expression levels.
Methods
Experimental Animals
All experiments were preapproved by the AMREP Animal Ethics Committee. The generation and genotyping of Grhl3+/– have been described previously.11 To generate Grhl3-cKO mice, transgenic B6. Cg-Tg(ED-L2-cre) 267Jkat/Nci13 were purchased from NCI Mouse Repository and crossed with Grhl3+/– mice. The resultant L2Cre+/Grhl3+/- mice were crossed with Grhl3fl/fl mice to provide the Grhl3Δ/–/L2Cre+ experimental animals (Grhl3-cKO). Both male and female mice were used in the experiments presented here.
Esophageal tumors were induced in 5-month-old Grhl3-cKO mice through the administration of 50 μg/mL 4-NQO (Sigma-Aldrich) in drinking water for 16weeks, followed by reversion to regular water and monitoring for 24 weeks.8 All animals underwent weekly oral cavity examination and were euthanized by cervical dislocation when exhibiting significant weight loss or at week 24. A complete autopsy was performed on all animals, and histopathologic lesions in the esophagus were scored by a certified pathologist.
Immunohistochemistry
For IHC, tissues were collected and fixed in 4% paraformaldehyde overnight and analyzed as described previously.7 Antibodies used for IHC and the dilutions used are shown in Table 2.
Table 2.
Details of Antibodies Used for Immunohistochemistry and Their Concentration
| Antibodies | Catalogue no. | Concentration | Species |
|---|---|---|---|
| β-galactosidase | ab9361 | 1:1000 | Chicken polyclonal |
| Ki67 | ab6667 | 1:100 | Rabbit monoclonal |
| K6 | PRB-169P | 1:500 | Rabbit polyclonal |
| K8 | MABT329M | 1:50 | Rat monoclonal |
| K5 | PRB-160P | 1:100 | Rabbit polyclonal |
| K14 | PRB-155P | 1:2000 | Rabbit polyclonal |
| K4 | ab9004 | 1:400 | Mouse monoclonal |
| K13 | ab92551 | 1:1000 | Mouse monoclonal |
| CTNNB | CST9562 | 1:300 | Rabbit polyclonal |
| HOPX | PA5-114744 | 1:100 | Rabbit polyclonal |
| WNT9A | PA5-96871 | 1:100 | Rabbit polyclonal |
| GRHL3 | HPA059960 | 1:100 | Rabbit polyclonal |
RNA Preparation and Quantitative Real-Time Polymerase Chain Reaction
For gene expression analysis, esophagus from E18.5 Grhl3+/+ and Grhl3–/– embryos and esophageal epithelium from WT and Grhl3-cKO mice were homogenized in Trizol (Invitrogen), and RNA was extracted according to the manufacturer’s instructions. Esophageal tumors were macro-dissected from the slides after staining with Histogene staining solution (Thermo Fisher Scientific). RNA was extracted using RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. Quantitative real-time PCR was carried out as described previously,11 with hypoxanthine-guanine phosphoribosyltransferase serving as the internal control in all experiments. A Student t test was used to determine statistical differences in expression levels, with P values <.05 considered significant, and the results were analyzed using GraphPad Prism. The error bars in all expression analyses represent the standard deviation. The primers used for Q-RT-PCR are shown in Table 3.
Table 3.
Primer Sequences for Quantitative Real-time Polymerase Chain Reaction
| Species | Gene | Sense | Antisense |
|---|---|---|---|
| Mouse | Grhl3 | AAGGAAGATGTCGAATGAACTTG | TCGTCCTCATTACTGTAGGGAAA |
| Mouse | Slc1a1 | GGATGCCATGTTGGACCTGA | CCCGCTTGGTTTTGTACTGC |
| Mouse | Alpl | TCGGAACAACCTGACTGACC | GTCAATCCTGCCTCCTTCCA |
| Mouse | Ppif | CCGACGAGAACTTCACACTGA | ATGCTTGCCATCTAGCCAGTC |
| Mouse | Cd274 | CGCCTGCAGATAGTTCCCAA | AGCCGTGATAGTAAACGCCC |
| Mouse | Lgr6 | CTGTCCGCTGACTGCTCC | ACTGAGGTCTAGGTAAGCCGT |
| Mouse | Dlg2 | TCCTTAGCAGCACATGCCC | TCGATACTTCTTTACGTTAGTCCG |
| Mouse | Hmgn2 | GCGAGGTTGTCTGCTAAACC | GCGAGGTTGTCTGCTAAACC |
| Mouse | Lrrc8c | CCCCCAGAGATTAATGTGGCT | GAACTCGGTCACCGGAATCA |
| Mouse | Coch | GAGGGAGCGGTTCCCATTC | ATGCTGGACACTGACGCATA |
| Mouse | Aldh1a7 | TTTGGCTGTCCCTGTCCAAT | ACCATGTTCGCCCAGTTCTC |
| Mouse | Hopx | GCCCCAGTGTAAAGAAATGGT | GTGACGGATCTGCACTCTGA |
| Mouse | ΔNP63α | GAGCAGCCTTGACCAGTCTC | GGTTCGTGTACTGTGGCTCA |
| Mouse | MITF | GCAAGAGGGAGTCATGCAGT | AGAACTGCTGCTCTTCAGAGGT |
| Mouse | LBH | GATCGGCTGAGATGACCGAG | ATGGGTCCGGAAAGATCTGA |
| Mouse | Survivin | AGAACAAAATTGCAAAGGAGACCA | CTGGGATGCGTGGCTTAGAT |
| Mouse | c-Jun | GGGAGCATTTGGAGAGTCCC | TTTGCAAAAGTTCGCTCCCG |
| Mouse | Atoh1 | GTGCGATCTCCGAGTGAGAG | GGGATAAGCCCCGAACAACA |
| Mouse | Met | TCTGGGAGCTCATGACGAGA | CTTCGTACAAGGCGTCTGGA |
| Mouse | FGF18 | TGGGGAAGCCTGATGGTACT | CCCTTGGGGTAACGCTTCAT |
| Mouse | Wnt2 | CTCTCGGTGGAATCTGGCTC | CCTGTAGCTCTCATGTACCACC |
| Mouse | Wnt4 | CAGGAAGGCCATCTTGACACAC | GTCTTTACCTCGCAGGAGCC |
| Mouse | Wnt6 | ACTGGGGGTTCGAGAATGTC | TCTCTCGGATGTCCTGCTGC |
| Mouse | Wnt9a | GCCTACTTCGGGCTGACG | GGTCGCAGGCCTTGTAGTG |
| Mouse | Wnt10a | CCGAGAGCCTCACAGAGACA | GTTCTCCATCACCGCCTGC |
| Mouse | Snai1 | AAGATGCACATCCGAAGCCA | ATGGCTTCTCACCAGTGTGG |
Transmission Electron Microscopy
Ultrathin (∼80 nm) sections of the transverse plane of each esophagus were examined with a Hitachi H-7500 transmission electron microscope operated at 80 kv. Digital images were captured with a Gatan multi-scan camera using the software digital micrograph.
RNA-seq Sample Preparation and Sequencing
Qiagen RNeasy micro kits were used to extract RNA from esophageal tumors from WT and Grhl3-cKO mice treated with 4-NQO. Specifically, tumors were macro-dissected from frozen cryosections to avoid contamination with normal esophageal tissue. RNA was quantified using Qubit RNA fluorometry (Invitrogen), and RNA integrity was assessed with the Agilent Bioanalyzer 2100 (Agilent Technologies). Illumina’s TruSeq stranded mRNA chemistry was used to produce libraries that were poly-A selected to remove structural RNA, which was performed by Micromon (Monash University, Melbourne, Australia). The libraries were sequenced to produce at least 20 million paired-end 75 base pair reads per sample using Illumina NextSeq500 in High-Output mode. The FastQC software (S. Andrews, FastQC: A Quality Control Tool for High Throughput Sequence Data. 2015. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used to assess the quality of the raw sequence data, and all samples passed the quality control. Sequences were then mapped to the mouse reference genome (Grcm38, Ensembl) using RNAsik-pipe/1.4.7 (https://github.com/MonashBioinformaticsPlatform/RNAsik-pipe/tree/1.5.3), and gene-level counts were obtained by the feature Counts tool.29 Further analysis was carried out using the edgeR30 package configured Degust software (https://zenodo.org/record/3501067#.YYi4E3JxVhE). Counts per million were calculated for each gene to standardize for differences in library size, and filtering was carried out to retain genes with a baseline expression level of at least 1.0 CPM in 3 or more samples.
ChIP-Q-PCR
Approximately 50 million EPC cells were cross-linked for 10 minutes in 0.75% formaldehyde at room temperature. After quenching cross-linking for 5 minutes with 125 mmol/L glycine and washing the cells in cold phosphate-buffered saline, the cells were lysed in 50 mmol/L Tris-HCl, pH 8.1,1% sodium dodecyl sulfate, 10 mmol/L EDTA, and protease inhibitor (added freshly). Chromatin was sheared to 200–500 base pair fragments using the S220 Covaris sonicator (105 peak incident power; duty factor 7; 200 cycles/burst; 40 seconds; USA). The sheared DNA was immunoprecipitated with anti-Grhl3Ab (Sigma HPA059960-100μL), HOPX Polyclonal Ab (PA5-114744, Invitrogen), or Rabbit IgG antibody (∼10 μg chromatin, 0.5 μg antibody for ChIP-Q-PCR) overnight. Protein-A agarose beads were added to each reaction for 1 hour at 4°C the following day. The magnetic beads were then washed in a series of buffers including a low salt buffer (20 mmol/L Tris-HCl, pH 8.1, 150 mmol/L NaCl, 2 mmol/L EDTA, 0.1% sodium dodecyl sulfate, 1% Triton X-100), a high salt buffer for 15 minutes (20 mmol/L Tris-HCl, pH 8.1, 500 mmol/L NaCl, 2 mmol/L EDTA, 0.1% sodium dodecyl sulfate, 1% Triton X-100), a LiCl buffer (10 mmol/L Tris-HCl, pH 8.1, 0.25 mol/L LiCl, 1 mmol/L EDTA, 1% NP-40, 1% deoxycholic acid), and finally with TE buffer, with each wash lasting for 5 minutes. Complexes were eluted in 1% sodium dodecyl sulfate, 0.1 mol/L NaHCO3. DNA was uncross-linked, and either Q-PCR was performed for regions of interest (primer sequences in Table 4). For ChIP-Q-PCR experiments, Ct values for immunoprecipitation (either specific antibody or immunoglobulin G) were normalized to input. Fold-change enrichment between specific antibody (Grhl3 or HOPX) and immunoglobulin G is shown for ChIP-Q-PCR data. The data included are representative of ChIPs from at least 3 replicates and are the mean ± standard deviation.
Table 4.
Primer Sequences for Chromatin Immunoprecipitation Polymerase Chain Reaction
| Species | Gene | Sense | Antisense |
|---|---|---|---|
| Human | HOPX | GCAGAAGCGATGGGAGATCAT | CACAGAGGACCAGGTGGAAA |
| Human | WNT2 | CCGAGAGGGGCGTTCATATT | CTGGCCTTTATCGCTCGCTG |
| Human | WNT9a | GGAACTGACTTACAGGGGGC | AAGTCCAGAGGGCAAGTGTG |
All authors had access to the study data and had reviewed and approved the final manuscript.
Acknowledgments
The authors thank Monash bioinformatics platform, animal facility and histopathology platform for the assistance in the experiments. They acknowledge the Victorian biobank for providing human patient samples.
CRediT Authorship Contributions
Smitha Rose Georgy, BVSc, MVSc, PhD, MANZCVS Diplomate ACVP (Conceptualization: Lead; Funding acquisition: Equal; Investigation: Lead; Methodology: Lead)
Diar Riyanti Rudiatmoko (Investigation: Supporting)
Alana Auden (Methodology: Supporting; Validation: Supporting)
Darren Partridge (Investigation: Supporting; Methodology: Supporting; Validation: Supporting)
Tariq Butt (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting)
Seema Srivastava (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting)
Nick Wong (Data curation: Supporting; Software: Supporting)
Marina Rose Carpinelli (Formal analysis: Supporting)
Dijina Swaroop (Methodology: Supporting; Software: Supporting)
Mirjana Bogeski (Data curation: Supporting; Validation: Supporting),
Stephen M. Jane (Conceptualization: Lead; Formal analysis: Equal; Supervision: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by Cancer Australia’s Priority-driven Collaborative Cancer Research Scheme in conjunction with the National Health and Medical Research Council project grant and The Garnett Passé and Rodney Williams Memorial Foundation Research Training Fellowship to SRG.
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Smyth E.C., Lagergren J., Fitzgerald R.C., et al. Oesophageal cancer. Nat Rev Dis Primers. 2017;3 doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Njei B., McCarty T.R., Birk J.W. Trends in esophageal cancer survival in United States adults from 1973 to 2009: a SEER database analysis. J Gastroenterol Hepatol. 2016;31:1141–1146. doi: 10.1111/jgh.13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcolea M.P., Greulich P., Wabik A., et al. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol. 2014;16:615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ting S.B., Caddy J., Hislop N., et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- 6.Ting S.B., Wilanowski T., Cerruti L., et al. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003;370(Pt 3):953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darido C., Georgy S.R., Wilanowski T., et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Georgy S.R., Cangkrama M., Srivastava S., et al. Identification of a novel proto-oncogenic network in head and neck squamous cell carcinoma. J Natl Cancer Inst. 2015;107:1–13. doi: 10.1093/jnci/djv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W.Y., Slack J.M., Tosh D. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Dev Biol. 2005;284:157–170. doi: 10.1016/j.ydbio.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 10.Auden A., Caddy J., Wilanowski T., et al. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Ting S.B., Wilanowski T., Auden A., et al. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat Med. 2003;9:1513–1519. doi: 10.1038/nm961. [DOI] [PubMed] [Google Scholar]
- 12.Yu Z., Lin K.K., Bhandari A., et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa H., Wang T.C., Zukerberg L., et al. The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach. Oncogene. 1997;14:1185–1190. doi: 10.1038/sj.onc.1200937. [DOI] [PubMed] [Google Scholar]
- 14.Tang X.H., Knudsen B., Bemis D., et al. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10(Pt 1):301–313. doi: 10.1158/1078-0432.ccr-0999-3. [DOI] [PubMed] [Google Scholar]
- 15.Caddy J., Wilanowski T., Darido C., et al. Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev Cell. 2010;19:138–147. doi: 10.1016/j.devcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain R., Li D., Gupta M., et al. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348:aaa6071. doi: 10.1126/science.aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung W.K., Zhao M., Liu Z., et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boon E.M., van der Neut R., van de Wetering M., et al. Wnt signaling regulates expression of the receptor tyrosine kinase met in colorectal cancer. Cancer Res. 2002;62:5126–5128. [PubMed] [Google Scholar]
- 19.Thakur R., Mishra D.P. Pharmacological modulation of beta-catenin and its applications in cancer therapy. J Cell Mol Med. 2013;17:449–456. doi: 10.1111/jcmm.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su H., Hu N., Yang H.H., et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17:2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes D.R., Yu J., Shanker K., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Z., Jin S., Chen J., et al. Murine interfollicular epidermal differentiation is gradualistic with GRHL3 controlling progression from stem to transition cell states. Nat Commun. 2020;11:5434. doi: 10.1038/s41467-020-19234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren X., Yang X., Cheng B., et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun. 2017;8 doi: 10.1038/ncomms14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yap L.F., Lai S.L., Patmanathan S.N., et al. HOPX functions as a tumour suppressor in head and neck cancer. Sci Rep. 2016;6 doi: 10.1038/srep38758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J.M., Sim S.M., Kim H.Y., et al. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol. 2010;89:537–546. doi: 10.1016/j.ejcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Fu L., Zhang C., Zhang L.Y., et al. Wnt2 secreted by tumour fibroblasts promotes tumour progression in oesophageal cancer by activation of the Wnt/beta-catenin signalling pathway. Gut. 2011;60:1635–1643. doi: 10.1136/gut.2011.241638. [DOI] [PubMed] [Google Scholar]
- 27.DeWard A.D., Cramer J., Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 30.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]