Abstract
Background & Aims
Hepatic immune system disorder plays a critical role in the pathogenesis of acute liver injury. The intrinsic signaling mechanisms responsible for dampening excessive activation of liver macrophages are not completely understood. The Notch and Hippo-YAP signaling pathways have been implicated in immune homeostasis. In this study, we investigated the interactive cell signaling networks of Notch1/YAP pathway during acute liver injury.
Methods
Myeloid-specific Notch1 knockout (Notch1M-KO) mice and the floxed Notch1 (Notch1FL/FL) mice were subjected to lipopolysaccharide/D-galactosamine toxicity. Some mice were injected via the tail vein with bone marrow–derived macrophages transfected with lentivirus-expressing YAP. Some mice were injected with YAP siRNA using an in vivo mannose-mediated delivery system.
Results
We found that the activated Notch1 and YAP signaling in liver macrophages were closely related to lipopolysaccharide/D-galactosamine-induced acute liver injury. Macrophage/neutrophil infiltration, proinflammatory mediators, and hepatocellular apoptosis were markedly ameliorated in Notch1M-KO mice. Importantly, myeloid Notch1 deficiency depressed YAP signaling and facilitated M2 macrophage polarization in the injured liver. Furthermore, YAP overexpression in Notch1M-KO livers exacerbated liver damage and shifted macrophage polarization toward the M1 phenotype. Mechanistically, macrophage Notch1 signaling could transcriptionally activate YAP gene expression. Reciprocally, YAP transcriptionally upregulated the Notch ligand Jagged1 gene expression and was essential for Notch1-mediated macrophage polarization. Finally, dual inhibition of Notch1 and YAP in macrophages further promoted M2 polarization and alleviated liver damage.
Conclusions
Our findings underscore a novel molecular insight into the Notch1-YAP circuit for controlling macrophage polarization in acute liver injury, raising the possibility of targeting macrophage Notch1-YAP circuit as an effective strategy for liver inflammation–related diseases.
Keywords: Notch Signaling, YAP, Macrophage Polarization, Acute Liver Injury, Liver Inflammation
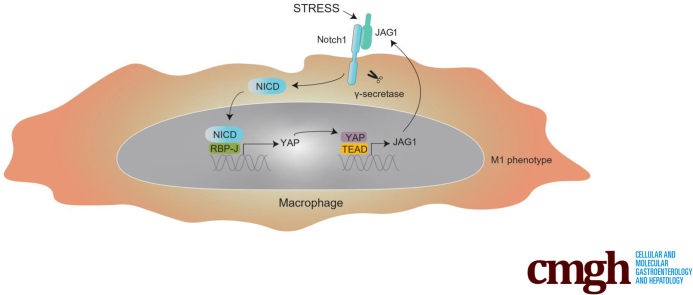
Graphical abstract
Summary.
Liver macrophages are highly plastic and adapt their phenotype according to the signals derived from the hepatic microenvironment. In this study, we identify the Notch1-YAP circuit as a key regulator of macrophage polarization and liver inflammation.
Acute liver injury is related to innate immunity dysregulation and abrupt hepatocellular injury, which contributes to high mortality and morbidity, and new treatments are desperately required.1, 2, 3, 4 Abrupt hepatocellular damage releases pathogen-associated exogenous derivates, such as pathogen-associated molecular patterns and damage-associated molecular patterns,5 which trigger the activation of inflammatory profiling, such as Kupffer cells/macrophage-derived cytokines, chemokines, reactive oxygen/nitrogen species, and death ligands. Macrophages, the important component of inflammatory profiling, dominate the production of cytokines and chemokines6 and thus are key determinants of the initiation, propagation, and resolution phases of liver injury. On the one hand, macrophage activation increases the release of oxygen free radicals and proinflammatory mediators, such as interleukin-6 (IL-6) and tumor necrosis factor α (TNF-α), which triggers the apoptotic pathway leading to the death of hepatocytes.7,8 On the other hand, distinct inflammatory stimuli and metabolic signals recruit circulating monocytes and other immune cells, exacerbating liver injury. During the progression of liver injury, macrophages, highly heterogeneous and versatile cells, can be polarized into different phenotypes in response to the local microenvironment signals.9, 10, 11 Therefore, further understanding of the macrophage phenotype switch may open up new avenues of macrophage-based therapeutic strategies for acute liver injury.
Notch signaling is highly conserved and regulates cell growth, differentiation, and survival through ligand binding and γ-secretase-mediated proteolytic cleavage that generates Notch intracellular domain (NICD). NICD translocates to the nucleus and binds to the nuclear recombinant recognition sequence binding protein at the Jκ site (RBP-J) and the nuclear effector Mastermind-like to activate transcription of canonical Notch targets including the Hairy enhancer of split (Hes) and Hes-related (Hey) family genes.12 Emerging evidence indicates that Notch signaling pathway has been implicated in regulating innate and adaptive immunity homeostasis and function.13 Recently, Notch signaling has been shown to regulate macrophage function by controlling genes involved in M1/M2 polarization. Intriguingly, these effects are different and even opposite under different pathologic conditions.14,15 It has been reported that the Notch pathway enhanced hepatic macrophage M1 activation in alcoholic steatohepatitis.16 Consistently, Notch-RBP-J signaling augmented Toll-like receptor 4 (TLR4)-induced classically M1 macrophages polarization and innate immune responses to Listeria monocytogenes.17 However, macrophage polarization is a highly dynamic process and the phenotype of polarized macrophages can be reversed under different pathologic conditions. Indeed, the activated Notch pathway was shown to facilitate M2 polarization in diffuse large B-cell lymphoma,18 whereas some data have shown that blockade of Notch pathway led to the M2-like tumor-associated macrophage.4,19 These findings suggest that Notch-mediated macrophage polarization is largely stress-dependent. Moreover, the heterogeneity of the Notch signaling outcome is evident not only in comparison between different tissues but also within the same tissue.20 There is growing evidence that the signaling diversity in cellular responses can be generated through extensive posttranslational modifications of Notch ligands and receptors, and different transcriptional regulation of Notch target genes.21 More importantly, other genes can also be regulated in parallel with those direct Notch target genes, which is accomplished by intense crosstalk with other signaling pathways, including the Hippo signaling pathway.20,21
YAP, the major transcriptional coactivator of the Hippo signaling pathway, modulates inflammation and innate immunity in liver disease, including fibrosis, nonalcoholic steatohepatitis, and fulminant hepatic failure. Recent evidence suggests that activated YAP in hepatocytes rapidly and potently promotes liver inflammation and fibrosis.22 YAP in Kupffer cells enhances the production of proinflammatory cytokines and promotes the development of nonalcoholic steatohepatitis.23 In addition, YAP has been elucidated to regulate hepatocyte proliferation in lipopolysaccharide (LPS)/D-galactosamine (D-GalN)-induced fulminant hepatic failure.24 Although previous evidence suggests the crosstalk between YAP and Notch signaling promotes severe hepatomegaly, rapid hepatocellular carcinoma initiation, and progression,25, 26, 27 it remains largely unknown as to whether and how Notch1-YAP interaction regulates macrophage polarization during acute liver injury.
In this study, we identify a previously unrecognized regulatory mechanism of the Notch1-YAP circuit in macrophage polarization. Using myeloid-specific Notch1 knockout (KO) mice combined with YAP overexpression or knockdown, we describe a bidirectional circuit whereby Notch1 and YAP directly transcriptionally regulate each other and reprogram macrophage polarization during acute liver injury. Importantly, dual inhibition of macrophage Notch1 and YAP further facilitates macrophage M2 polarization and alleviates liver damage. These results seem to prove that the Notch1-YAP circuit orchestrates proper cellular resources to reprogram macrophage polarization and liver inflammation.
Results
Macrophage Notch1 Signaling is Closely Related to LPS/D-GalN-Induced Acute Liver Injury
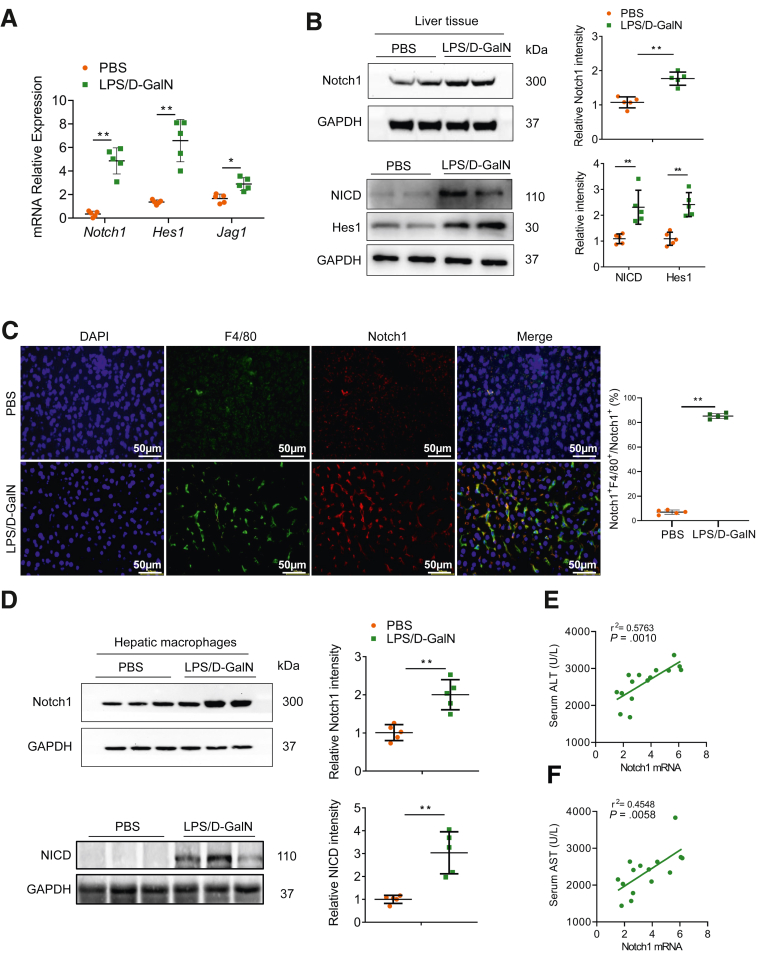
To investigate the involvement of Notch1 signaling in acute liver injury, the expressions of Notch1, Hes1, and Jag1 in liver tissues following LPS/D-GalN-induced acute liver injury were examined. As shown in Figure 1A, the Notch1 signaling was significantly activated in injured livers compared with control livers. The expressions of Notch1 and NICD were significantly increased in liver tissue after LPS/D-GalN injection (Figure 1B). Using double immunofluorescence staining, we found that the increased expression of Notch1 was primarily localized in hepatic macrophages (Figure 1C). To further determine whether LPS/D-GalN specifically influences Notch1 signaling in hepatic macrophages, we isolated hepatic macrophages from LPS/D-GalN-injured and control livers. Indeed, Western blot analysis indicated that Notch1 and NICD expression was markedly upregulated in hepatic macrophages from LPS/D-GalN-injured liver compared with control livers (Figure 1D). Importantly, the mRNA level of Notch1 in hepatic macrophages was positively correlated with the serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels (Figure 1E and F). These observations suggest that Notch1 signaling activation in hepatic macrophages is highly correlated with LPS/D-GalN-induced liver damage.
Figure 1.
Myeloid Notch1 signaling is closely related to LPS/D-GalN-induced liver injury. (A) The expressions of Notch1, Hes1, and Jag1 in the liver tissue of wild-type mice subjected to PBS or LPS (50 μg/kg) combined with D-GalN (700 mg/kg) for 5 hours (n = 5 mice/group). (B) Western blot analysis of Notch1 and NICD expression in liver tissue from wild-type mice treated with PBS or LPS/D-GalN injection (n = 5 mice/group). (C) Immunofluorescence images of staining with antibodies against F4/80 (green) and Notch1 (red). Nuclei were labeled with DAPI (blue). n = 5 mice/group. Scale bar, 50 μm. (D) Hepatic macrophages were isolated from mice after PBS or LPS/D-GalN injection. Notch1 and NICD expression was examined by immunoblotting. The correlation between serum ALT (E) or AST levels (F) and Notch1 expression in hepatic macrophages after LPS/D-GalN injection (n = 15 mice). The correlation coefficient was calculated by the Pearson correlation test. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Myeloid Notch1 Deficiency Alleviates LPS/D-GalN-Induced Liver Injury and Inflammation
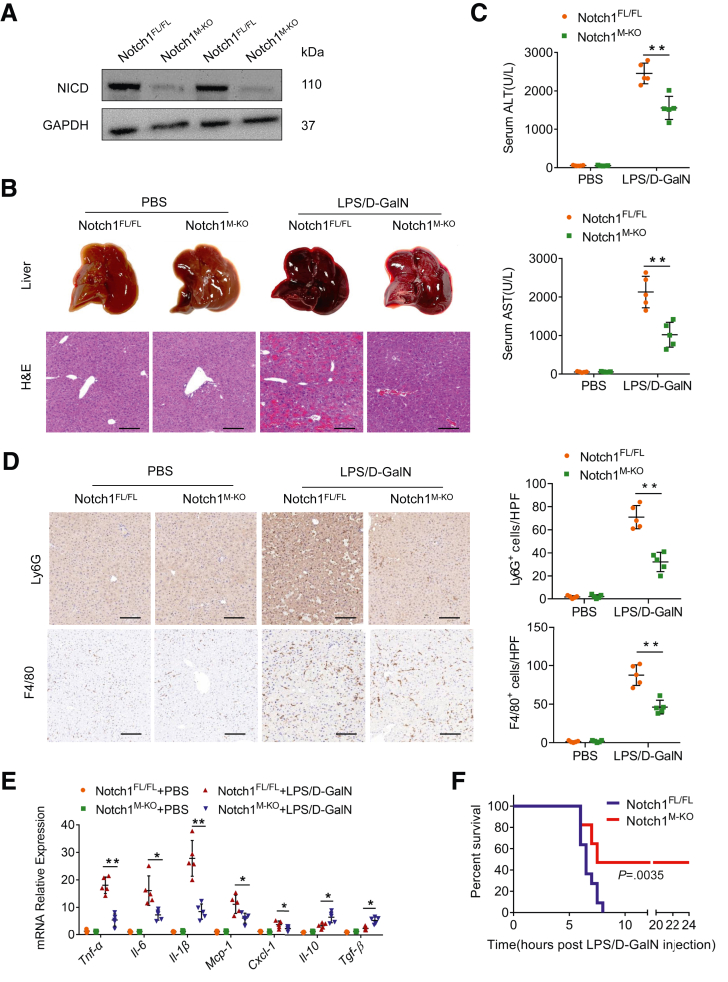
To determine whether macrophage Notch1 signaling may play a critical role in LPS/D-GalN-induced acute liver injury, we generated myeloid-specific Notch1-deficient (Notch1M-KO) and Notch1-proficient (Notch1FL/FL) mice and subjected them to LPS/D-GalN toxicity. We isolated hepatic macrophages and confirmed that Notch1M-KO mice showed Notch1 deficiency in hepatic macrophages compared with Notch1FL/FL mice (Figure 2A). Compared with Notch1FL/FL mice, Notch1M-KO mice showed decreased histologic damage after LPS/D-GalN injection (Figure 2B). Hepatocellular damage, reflected by serum ALT and AST levels, was also significantly decreased in Notch1M-KO mice (Figure 2C). Moreover, myeloid Notch1 disruption decreased the infiltration of F4/80+ macrophages and Ly6G+ neutrophils (Figure 2D), accompanied by reduced expression of proinflammatory cytokines (Tnf-α, Il-6, and Il-1β) and chemokines (Mcp-1 and Cxcl-1), and increased anti-inflammatory cytokines (Il-10 and Tgf-β) in damaged livers (Figure 2E). To determine whether myeloid Notch1 disruption exerts sustained protective effects and prevents mortality, animal survival was assessed after the LPS/D-GalN challenge. Clearly, increased animal survival was observed in Notch1M-KO mice, as compared with that in Notch1FL/FL mice (Figure 2F).
Figure 2.
Myeloid-specific deletion of Notch1 alleviates LPS/D-GalN-induced liver inflammation. (A) The NICD expression was detected by Western blot assay in hepatic macrophages from the Notch1FL/FL and Notch1M-KO livers. (B) The representative gross appearance of the collected livers and histologic staining (H&E) of liver sections from Notch1FL/FL and Notch1M-KO mice after PBS or LPS/D-GalN injection (n = 5 mice/group). Scale bars, 100 μm. (C) Liver function in serum samples was evaluated by serum ALT and AST levels (IU/L) (n = 5 samples/group). (D) Immunohistochemistry staining and quantification of Ly6G+ neutrophils and F4/80+ macrophages in liver sections (n = 5 mice/group). Scale bars, 40 μm. (E) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of proinflammatory cytokines (Tnf-α, Il-6, and Il-1β), chemokines (Mcp-1 and Cxcl-1), and anti-inflammatory cytokines (Il-10 and Tgf-β) in liver tissues (n = 5 samples/group). (F) Kaplan-Meier survival curve comparing percent survival of Notch1FL/FL and Notch1M-KO mice treated with LPS (50 μg/kg) and D-GalN (700 mg/kg) (n = 9–11 mice/group). Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Myeloid Notch1 Deficiency Reduces Hepatocellular Apoptosis in LPS/D-GalN-Induced Liver Injury
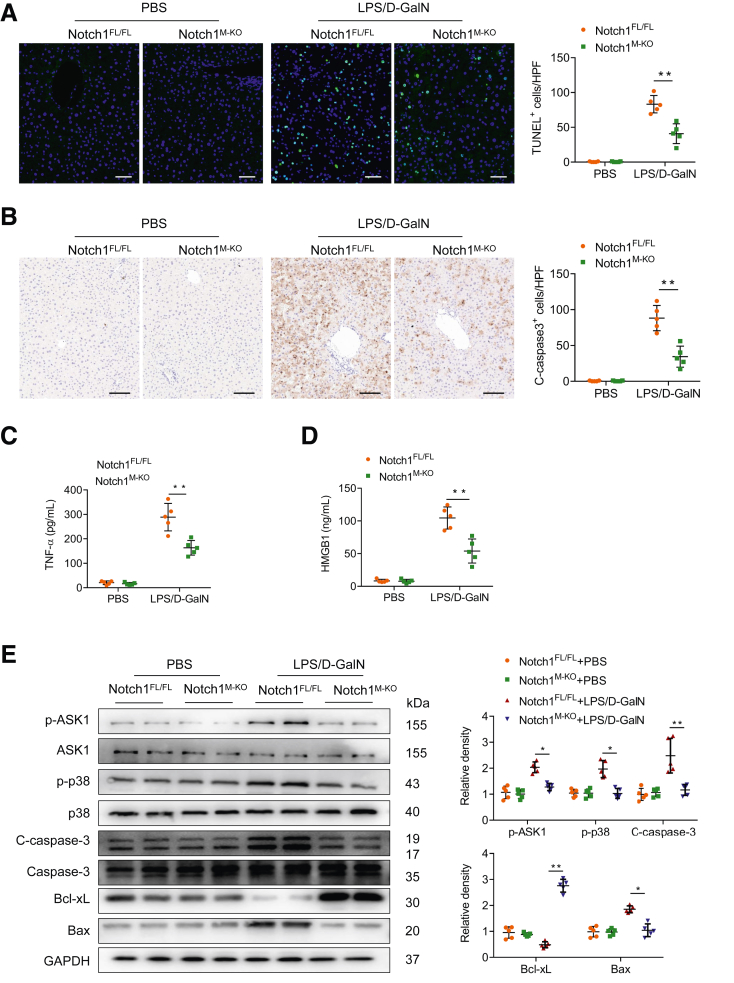
As apoptosis of hepatocytes is the major culprit underlying acute liver injury, we next analyzed the effect of myeloid Notch1 signaling on hepatocellular apoptosis by TUNEL staining. Livers in Notch1M-KO mice revealed a decreased frequency of apoptotic TUNEL+ cells compared with Notch1FL/FL livers (Figure 3A). These data were confirmed by reduced activation of caspase-3 in Notch1M-KO mice compared with that in Notch1FL/FL mice (Figure 3B). Enzyme-linked immunosorbent assay (ELISA) assay showed that myeloid Notch1 disruption significantly reduced serum TNF-α (Figure 3C) and HMGB1 (Figure 3D) production. Furthermore, Western blot analysis revealed that disruption of myeloid Notch1 downregulated p-ASK1, p-p38, and C-caspase-3 expression in Notch1M-KO livers compared with Notch1FL/FL control livers (Figure 3E), which was accompanied by increased the prosurvival Bcl-xL and reduced the proapoptotic expression of Bax in Notch1M-KO livers (Figure 3E).
Figure 3.
Myeloid-specific deletion of Notch1 alleviates LPS/D-GalN-induced hepatocellular apoptosis. (A) Representative TUNEL staining images and quantification of TUNEL+ cells in liver sections from the Notch1FL/FL and Notch1M-KO mice treated with PBS or LPS/D-GalN injection (n = 5 mice/group). Scale bars, 20 μm. (B) Immunohistochemistry staining and quantification of cleaved caspase-3 (C-caspase-3) positive cells in liver sections (n = 5 mice/group). Scale bars, 40 μm. ELISA analysis of serum TNF-α (C) and HMGB1 (D) levels in the Notch1FL/FL and Notch1M-KO mice (n = 5 samples/group). (E) Western blot analysis and relative density ratio of p-ASK1, ASK1, p-p38, p38, C-caspase-3, caspase-3, Bcl-xL, and Bax in the Notch1FL/FL and Notch1M-KO livers. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Myeloid Notch1 Deficiency Depresses YAP Signaling and Reprograms Macrophage Polarization
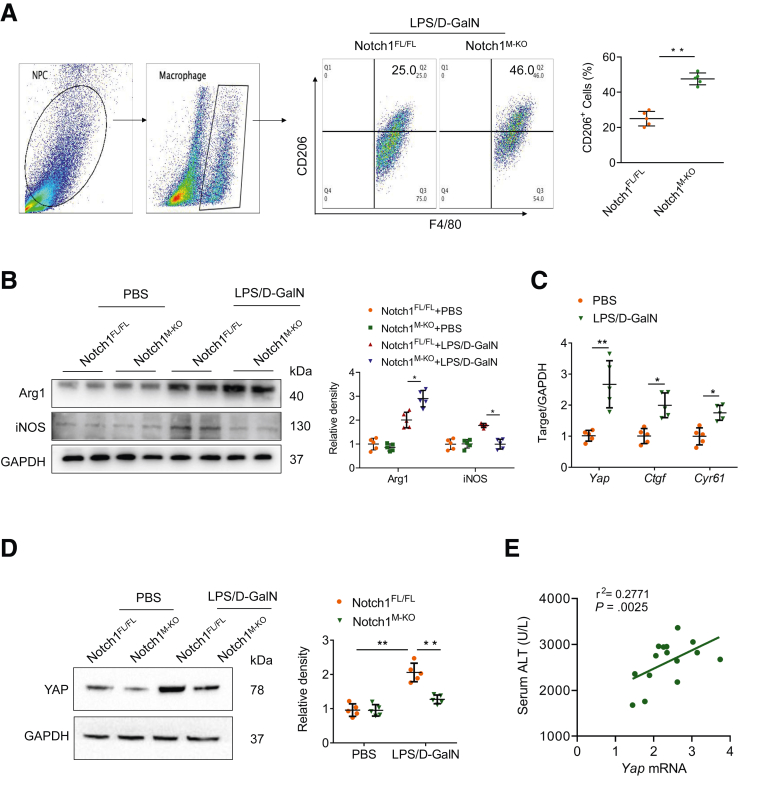
We next investigated whether and how Notch1 signaling regulates macrophage polarization in LPS/D-GalN-induced acute liver injury. Flow cytometry analysis indicated that disruption of myeloid Notch1 signaling exhibited a higher frequency of CD206 positive macrophages after LPS/D-GalN injection (Figure 4A). Moreover, Western blot analysis revealed that the disturbance of myeloid Notch1 signaling upregulated M2 markers (Arg1) and downregulated M1 markers (iNOS) expression in liver macrophages (Figure 4B). These data suggest that Notch1 is essential for controlling macrophage polarization in LPS/D-GalN-induced liver inflammation.
Figure 4.
Myeloid Notch1 deficiency depresses YAP signaling and reprograms macrophage polarization. (A) Flow cytometry analysis of liver macrophages isolated from Notch1FL/FL and Notch1M-KO mice (n = 5 mice/group). (B) The Arg1 and iNOS expression were detected by Western blot assay in liver macrophages from Notch1FL/FL and Notch1M-KO mice treated with PBS or LPS/D-GalN injection. (C) Quantitative reverse-transcriptase polymerase chain reaction analysis of Yap, Ctgf, and Cyr61 in liver macrophages (n = 5 samples/group). (D) The expression of YAP in liver macrophages from Notch1M-KO and Notch1FL/FL mice, as measured by Western blot analysis. (E) The correlation between serum ALT levels and Yap mRNA expression in liver macrophages after LPS/D-GalN injection (n = 15 mice). The correlation coefficient was calculated by the Pearson correlation test. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Previous studies indicated that the interconnection between the Notch and YAP signaling had been proposed to influence inflammation.20 We then asked whether there is putative crosstalk between Notch1 and YAP signaling in regulating macrophage polarization. Unlike phosphate-buffered saline (PBS)-treated control subjects, LPS/D-GalN injection markedly increased mRNA levels coding for YAP and YAP target genes including Ctgf and Cyr61 (Figure 4C). Consistently, Western blot analysis revealed an increased YAP protein level in hepatic macrophages after LPS/D-GalN injection (Figure 4D). Interestingly, myeloid Notch1 deficiency significantly reduced YAP expression (Figure 4D). Additionally, serum ALT level was positively correlated with Yap mRNA levels in hepatic macrophages after LPS/D-GalN injection (r2 = 0.2771; P = .0025) (Figure 4E). These results suggest that YAP may be a critical downstream factor participating in Notch1-mediated macrophage polarization and liver inflammation.
YAP is Critical for Notch1-Mediated Macrophage Polarization and Liver Inflammation
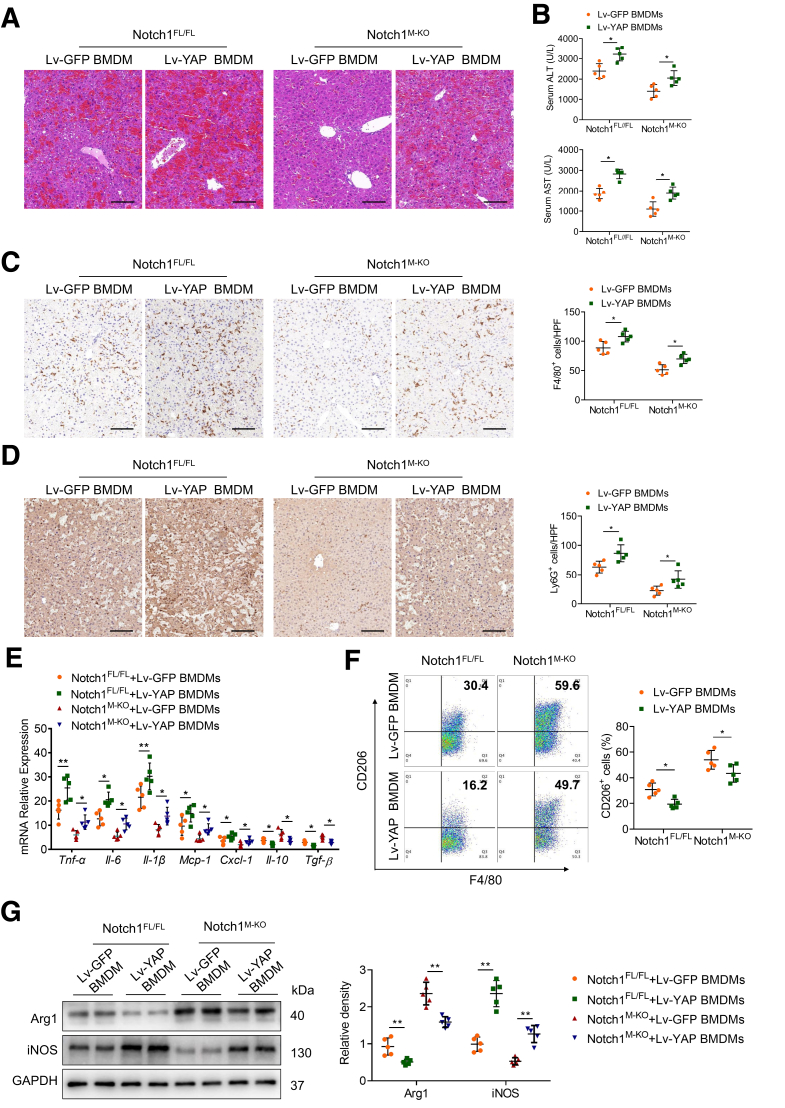
Given that myeloid-specific Notch1 deficiency depressed the YAP pathway, we then examined whether YAP contributed to Notch1-mediated macrophage polarization and liver inflammation. Bone marrow–derived macrophages (BMDMs) from Notch1FL/FL and Notch1M-KO mice were transfected with lentivirus expressing YAP (Lv-YAP) or GFP control (Lv-GFP), and adoptively transferred into Notch1FL/FL and Notch1M-KO mice, respectively. As shown in Figure 5, Lv-YAP transduction increased YAP expression in BMDMs, and the infused engineered BMDMs traveled into the liver after the LPS/D-GalN challenge. As expected, YAP overexpression aggravated LPS/D-GalN-induced liver damage (Figure 6A) and elevated serum ALT and AST levels in Notch1M-KO mice transferred with Lv-YAP-transfected BMDMs (Figure 6B). Moreover, unlike livers treated with Lv-GFP-transfected control cells, YAP overexpression increased the infiltration of F4/80+ macrophages (Figure 6C) and Ly6G+ neutrophils (Figure 6D) in Notch1M-KO mice transferred with Lv-YAP-transfected BMDMs, which was accompanied by augmented expressions of proinflammatory cytokines and chemokines including Tnf-α, Il-6, Il-1β, Mcp-1, and Cxcl-1, and decreased expressions of anti-inflammatory cytokines including Il-10 and Tgf-β (Figure 6E). Consistent with these results, flow cytometry analysis showed that YAP overexpression in the Notch1M-KO liver resulted in the decreased CD206+ macrophages (Figure 6F), which was further confirmed by downregulated M2 marker (Arg1) and upregulated M1 marker (iNOS) expression in liver macrophages (Figure 6G). These results demonstrate that YAP is required for Notch1-mediated M1 macrophage polarization and liver inflammatory response.
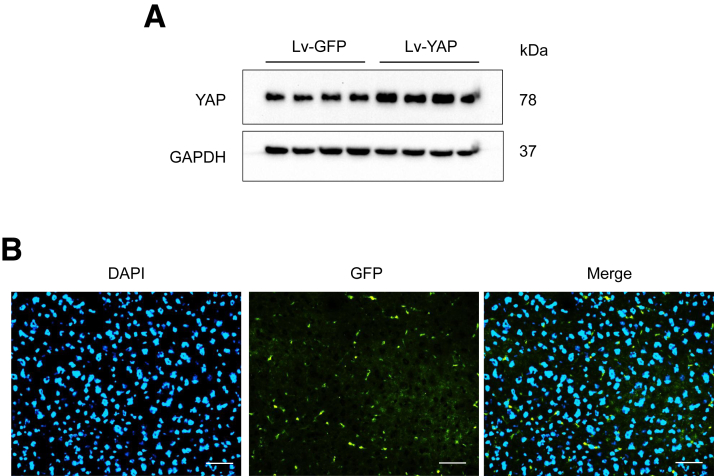
Figure 5.
BMDMs were transfected with lentivirus expressing YAP (Lv-YAP) or GFP control (Lv-GFP) and adoptively transferred into mice. (A) Western blots of YAP expression in BMDMs after Lv-YAP transduction. (B) Representative immunofluorescence staining images of the infused Lv-GFP-transfected BMDMs in the liver after the LPS/D-GalN challenge. Scale bars, 20 μm.
Figure 6.
YAP is required for Notch1-mediated macrophage polarization and liver inflammation. BMDMs from Notch1FL/FL and Notch1M-KO mice were transfected with lentivirus expressing YAP (Lv-YAP) or GFP control (Lv-GFP) and adoptively transferred into Notch1FL/FL and Notch1M-KO mice, respectively. (A) Representative H&E staining of liver sections (n = 5 mice/group). Scale bars, 100 μm. (B) The serum ALT and AST levels from the indicated groups. (C, D) Representative immunohistochemistry staining and quantification of F4/80+ macrophages and Ly6G+ neutrophils in liver sections (n = 5 mice/group). Scale bars, 40 μm. (E) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of proinflammatory cytokines (Tnf-α, Il-6, and Il-1β), chemokines (Mcp-1 and Cxcl-1), and anti-inflammatory cytokines (Il-10 and Tgf-β) in liver tissues (n = 5 samples/group). (F) Flow cytometry analysis of liver macrophages isolated from Notch1FL/FL and Notch1M-KO mice (n = 5 mice/group). (G) The expression of Arg1 and iNOS in liver macrophages from Notch1FL/FL and Notch1M-KO mice, as measured by Western blot analysis. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Activated Notch1-YAP Circuit Shifts Macrophage Polarization to M1 Phenotype In Vitro
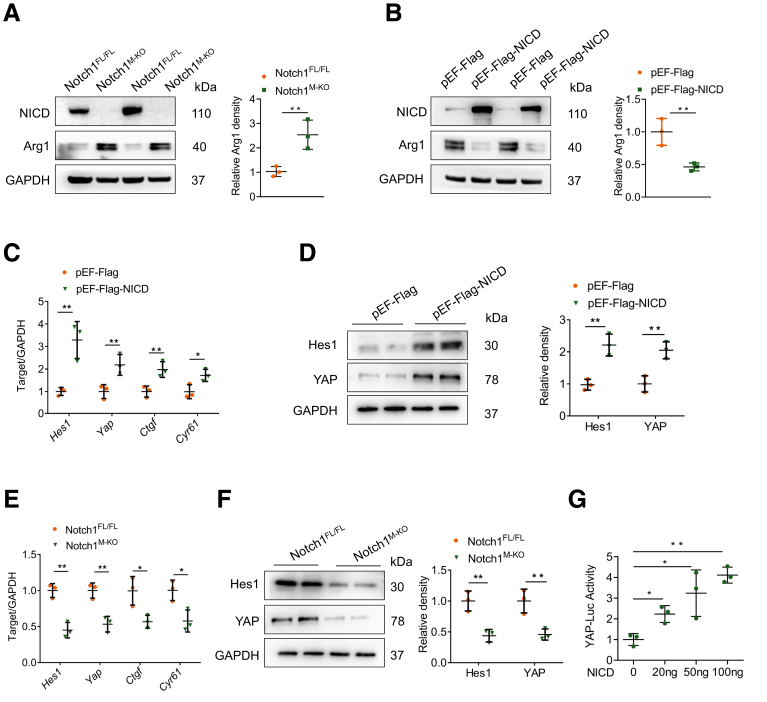
The potent regulation of myeloid Notch1 signaling on YAP activation promoted us to explore the underlying mechanisms in vitro. BMDMs were isolated from Notch1FL/FL and Notch1M-KO mice and stimulated with LPS/interferon (IFN)-γ. Western blot analysis showed that Arg1 expression was increased in Notch1M-KO BMDMs after LPS/IFN-γ stimulation (Figure 7A), whereas NICD overexpression in BMDMs with pEF-Flag-NICD plasmid transfection reduced Arg1 expression (Figure 7B). Importantly, macrophage NICD overexpression increased the mRNA levels of Hes1, Yap, and YAP signaling target genes, including Ctgf and Cyr61 (Figure 7C). Consistently, NICD overexpression elevated the protein levels of YAP and Notch signaling target Hes1 (Figure 7D). In contrast, macrophage Notch1 deficiency diminished the mRNA levels of Hes1, Yap, Ctgf, and Cyr61 (Figure 7E), accompanied by reduced protein levels of Hes1 and YAP after LPS/IFN-γ stimulation (Figure 7F). Furthermore, Notch1 stimulated YAP transcriptional activity in a dose-dependent manner, as evidenced by consistently increased YAP-luciferase reporter gene (Figure 7G).
Figure 7.
Notch1 activation upregulates YAP signaling in macrophages. (A) BMDMs were isolated from Notch1M-KO and Notch1FL/FL mice and stimulated with LPS (100 ng/mL)/IFN-γ (10 ng/mL) for 12 hours. Western blot analysis of NICD and Arg1 expression in BMDMs. (B-D) BMDMs were transfected with pEF-Flag-NICD plasmid or control vector. After 48 hours, cells were supplemented with LPS (100 ng/mL)/IFN-γ (10 ng/mL) for an additional 12 hours. (B) Western blot analysis of NICD and Arg1 expression in indicated BMDMs. (C) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of Hes1, Yap, Ctgf, and Cyr61 in indicated BMDMs. (D) Western blot analysis of Hes1 and YAP in indicated BMDMs. (E) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of Hes1, Yap, Ctgf, and Cyr61 in BMDMs from Notch1M-KO and Notch1FL/FL mice. (F) Western blot analysis of Hes1 and YAP expression in BMDMs from Notch1M-KO and Notch1FL/FL mice. (G) BMDMs were cotransfected with YAP-luciferase and pEF-Flag-NICD vectors, and the luciferase activity was measured after 48 hours. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
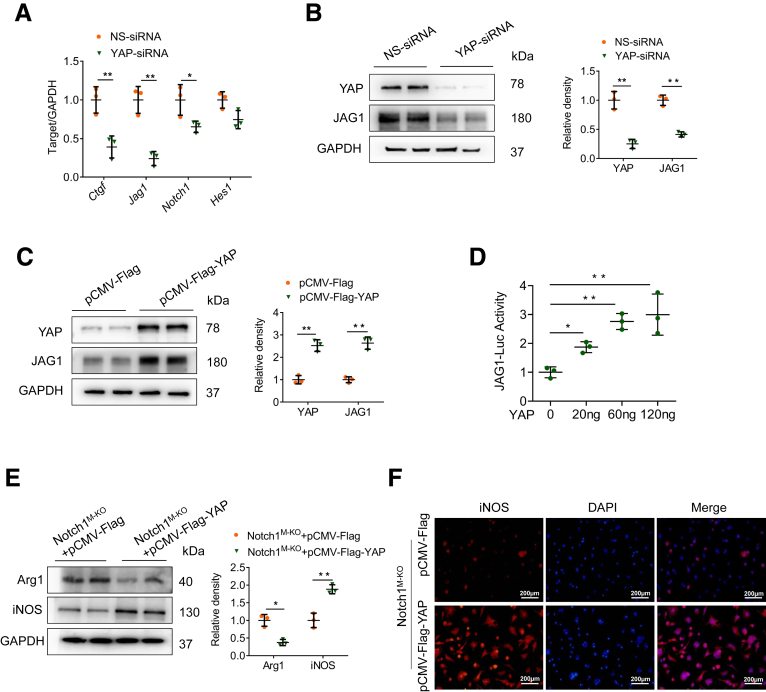
Previous evidence suggested that Notch signaling pathway may act as a functional effector downstream of the Hippo-YAP pathway in controlling liver cell fate.28 To test the hypothesis that YAP also may regulate Notch1 signaling in macrophages, BMDMs were transfected with the pCMV-Flag-YAP plasmid to overexpress YAP, or transfected with YAP siRNA to knockdown YAP followed by LPS/IFN-γ stimulation. Indeed, YAP knockdown markedly reduced the mRNA levels of YAP target gene Ctgf, and the Notch ligand gene Jag1, although there was a slight decrease in Notch1 expression (Figure 8A). Accordingly, YAP knockdown diminished the protein levels of YAP and JAG1 compared with control vector-transfected cells after LPS/IFN-γ stimulation (Figure 8B). In contrast, YAP overexpression significantly enhanced JAG1 protein expression (Figure 8C). Next, we test whether YAP also transcriptionally regulated the JAG1 activity. As expected, YAP stimulated JAG1 transcriptional activity in a dose-dependent manner, as evidenced by consistently increased JAG1-luciferase reporter gene (Figure 8D).
Figure 8.
YAP activation upregulates JAG1 in macrophages and contributes to Notch1-mediated macrophage polarization in vitro. (A, B) BMDMs were transfected with YAP-siRNA or NS-siRNA and stimulated with LPS (100 ng/mL)/IFN-γ (10 ng/mL) for 12 hours. (A) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of Ctgf, JAG1, Notch1, and Hes1. (B) Western blot analysis of YAP and JAG1 expression. (C) The expression of YAP and JAG1 in BMDMs transfected with pCMV-Flag-YAP plasmid or control vector, as measured by Western blot analysis. (D) BMDMs were cotransfected with JAG1-luciferase and pCMV-Flag-YAP vectors, and the luciferase activity was measured after 48 hours. (E, F) Notch1M-KO BMDMs were transfected with pCMV-Flag-YAP plasmid or control vector followed by LPS/IFN-γ stimulation. (E) Western blot analysis of Arg1 and iNOS expression in BMDMs. (F) Representative immunofluorescence staining of iNOS in BMDMs. Scale bars, 200 μm. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
To further examine whether YAP contributes to Notch1-mediated macrophage polarization in vitro, we isolated BMDMs from Notch1M-KO mice and transfected them with the pCMV-Flag-YAP plasmid. Obviously, YAP overexpression in Notch1M-KO BMDMs augmented iNOS but decreased Arg1 expression (Figure 8E). Consistent with these results, immunofluorescence staining also revealed that YAP overexpression increased M1 marker iNOS expression in Notch1M-KO BMDMs (Figure 8F). Collectively, these results suggest that the positive feedback loop between YAP and Notch1 signaling drives macrophage polarization toward the M1 phenotype.
Dual Inhibition of Notch1-YAP Circuit Attenuates LPS/D-GalN-Induced Liver Injury
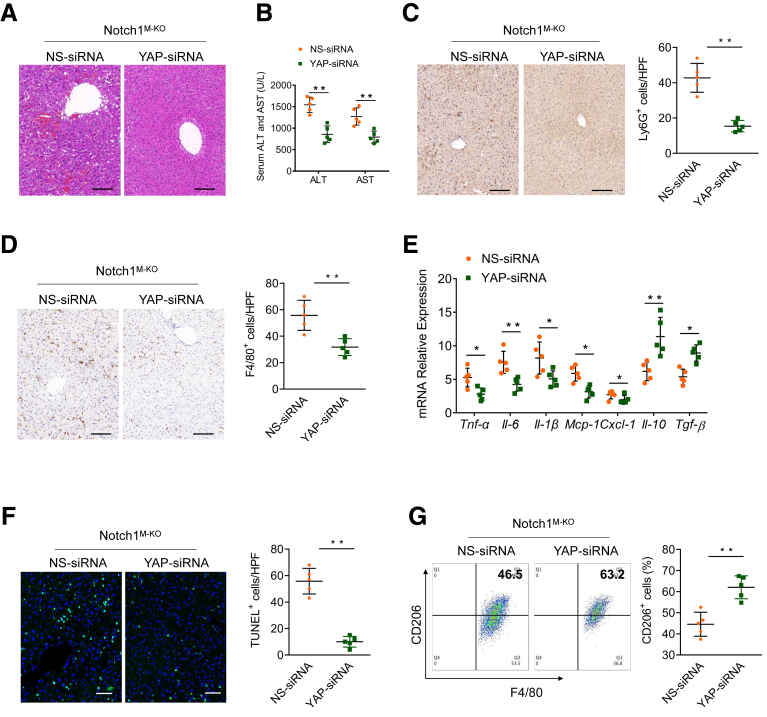
Having demonstrated that the positive Notch1-YAP circuit facilitates M1 macrophage polarization, we speculated that simultaneous inhibition of the Notch1 and YAP pathways may further alleviate inflammatory responses and liver damage. To confirm this hypothesis, we disrupted YAP in Notch1FL/FL and Notch1M-KO livers using a YAP siRNA with an in vivo mannose-mediated delivery system that enhances delivery to cells expressing a mannose-specific membrane receptor to macrophages as previously described.29,30 We found that inhibition of YAP alone decreased LPS/D-GalN-induced liver injury, as manifested by decreased serum ALT and AST levels, intrahepatic macrophage/neutrophil infiltration, hepatocellular apoptosis, and proinflammatory mediators (Figure 9). Importantly, histologic analysis of the livers showed that YAP knockdown in Notch1M-KO liver further decreased liver damage compared with Notch1M-KO mice with nonspecific (NS) siRNA treatment (Figure 10A). Similarly, serum ALT and AST levels were significantly decreased in Notch1M-KO mice with YAP siRNA treatment (Figure 10B). Moreover, YAP knockdown reduced the infiltration of Ly6G+ neutrophils and F4/80+ macrophages in the Notch1M-KO livers (Figure 10C and D). Consistent with these changes, YAP knockdown reduced the expressions of proinflammatory cytokines and chemokines including Tnf-α, Il-6, Il-1β, Mcp-1, and Cxcl-1, and increased the expressions of anti-inflammatory cytokines including Il-10 and Tgf-β (Figure 10E), and also inhibited hepatocellular apoptosis (Figure 10F). Furthermore, flow cytometry analysis indicated that YAP knockdown increased CD206+ macrophages compared with NS siRNA treatment in the Notch1M-KO livers (Figure 10G). Taken together, these results demonstrate that dual inhibition for silencing the Notch1-YAP circuit in macrophages is more effective for the alleviation of LPS/D-GalN-induced liver injury.
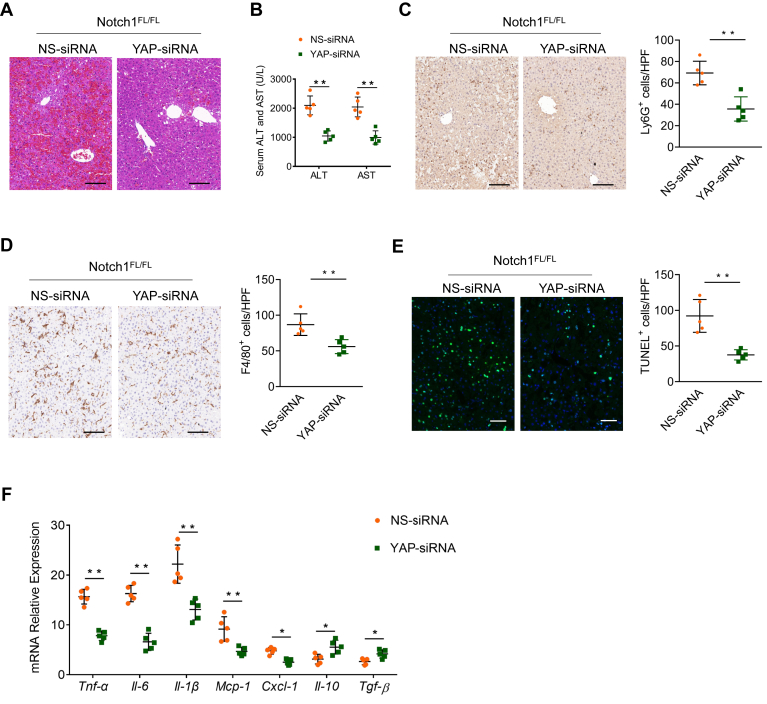
Figure 9.
Inhibition of YAP in macrophages attenuates LPS/D-GalN-induced liver injury. Notch1FL/FL mice were injected by the tail vein with YAP-siRNA or NS control siRNA mixed with mannose-conjugated polymers at 6 hours before LPS/D-GalN treatment (n = 5 mice/group). (A) Representative H&E staining of liver sections. (B) Serum ALT and AST levels (U/L). (C, D) Immunohistochemical staining and quantification of Ly6G+ neutrophils and F4/80+ macrophages in liver sections. Scale bars, 40 μm. (E) Representative TUNEL-staining images and quantification of TUNEL+ cells in liver sections. Scale bars, 20 μm. (F) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of proinflammatory cytokines (Tnf-α, Il-6, and Il-1β), chemokines (Mcp-1 and Cxcl-1), and anti-inflammatory cytokines (Il-10 and Tgf-β) in liver tissues. Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Figure 10.
Dual inhibition of Notch1-YAP circuit attenuates LPS/D-GalN-induced liver inflammation and damage. Notch1M-KO mice were injected by the tail vein with YAP-siRNA or NS control siRNA mixed with mannose-conjugated polymers at 6 hours before LPS/D-GalN treatment. (A) Representative H&E staining of liver tissue. (B) The hepatocellular function was evaluated by serum ALT and AST levels (U/L). (C, D) Immunohistochemical staining and quantification of Ly6G+ neutrophils and F4/80+ macrophages in liver sections (n = 5 mice/group). Scale bars, 40 μm. (E) Quantitative reverse-transcriptase polymerase chain reaction–assisted detection of proinflammatory cytokines (Tnf-α, Il-6, and Il-1β), chemokines (Mcp-1 and Cxcl-1), and anti-inflammatory cytokines (Il-10 and Tgf-β) in liver tissues (n = 5 samples/group). (F) Representative TUNEL-staining images and quantification of TUNEL+ cells in liver sections (n = 5 mice/group). Scale bars, 20 μm. (G) Flow cytometry analysis of liver macrophages isolated from Notch1M-KO mice with YAP-siRNA or NS-siRNA treatment (n = 5 mice/group). Data are presented as the mean ± standard deviation. ∗P < .05, ∗∗P < .01.
Discussion
Liver macrophages are highly plastic and adapt their phenotype according to the signals derived from the hepatic microenvironment. They play central roles in liver homeostasis and injury recognition, including regulating inflammation, fibrosis, and hepatocarcinogenesis. Notch and Hippo-YAP signaling pathways have been implicated in immune homeostasis. However, these signals may mount diverse and even opposite biologic effects that are highly context-specific in a cell-type-dependent manner. In the present study, using myeloid-specific Notch1 KO mice combined with YAP overexpression or knockdown, we identified Notch1-YAP circuit as a key regulator of macrophage polarization. We demonstrated that macrophage Notch1 and YAP signaling pathways were closely related to LPS/D-GalN-induced acute liver injury. Also, macrophage Notch1 signaling could transcriptionally activate YAP gene expression, reciprocally, YAP transcriptionally upregulated the Notch ligand JAG1 gene expression, thus forming a bidirectional Notch1-YAP signaling circuit. Moreover, the activated Notch1-YAP circuit in hepatic macrophages facilitated M1 macrophage polarization and contributed to LPS/D-GalN-induced acute liver injury. Finally, silencing the Notch1-YAP circuit by simultaneously inhibiting Notch1 and YAP pathways in macrophages was more effective to alleviate LPS/D-GalN-induced liver injury. Collectively, these findings highlight the importance of Notch1-YAP circuit in liver inflammation and provide evidence and rationale for combination therapies cotargeting Notch1 and YAP in macrophages.
Notch signaling is a highly conserved pathway involved in cell-fate decisions and tissue homeostasis through the interaction with adjacent cells.12 Emerging evidence demonstrates that Notch signaling controls the homeostasis of several innate cell populations, and thus the deregulation of the Notch cascade received substantial attention recently in inflammatory diseases.13 Investigation of Notch signaling has focused predominantly on lymphocytes, and knowledge about the effects of Notch signaling in myeloid lineage cells is not yet fully understood.13 In macrophages, TLR ligands have been reported to activate the Notch signaling and the well-known pathway of nuclear factor-κB involved in proinflammatory responses.31 Moreover, Notch and TLR pathways cooperated to activate canonical Notch target genes Hes1 and Hey1, and to increase the production of canonical TLR-induced cytokines IL-6, TNF-α, and IL-12.32 However, it has been reported that Notch signaling also suppresses TLR-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor-κB activation.33 Additionally, disruption of myeloid Notch1 promotes RhoA/ROCK signaling and aggravates ischemia/reperfusion-induced sterile liver inflammation.34 In this study, our results revealed that the Notch1 signaling activity in hepatic macrophages was significantly upregulated, and was closely associated with LPS/D-GalN-induced liver injury. Furthermore, myeloid-specific Notch1 deficiency ameliorated LPS/D-GalN-induced liver damage and inflammation. Although reasons for these conflicting conclusions about the role of Notch1 signaling in inflammation remain to be elucidated, 1 possible explanation is that the Notch1 pathway exerts diverse and even opposite biologic effects through the different targets according to different microenvironments and pathologic states. Indeed, previous studies showed that myeloid Notch1 signaling promotes the reprogramming of mitochondrial metabolism for proinflammatory M1 macrophage activation and liver inflammation in a murine model of alcoholic steatohepatitis.16 Additionally, the Notch-RBP-J signaling and TLR signaling are integrated at the level of IFN regulatory factor 8 protein synthesis to promote inflammatory M1 macrophage polarization.17 In keeping with these studies, our data demonstrated that the blockade of Notch1 signaling facilitated M2 macrophage polarization, accounting for the reduction of LPS/D-GalN-induced liver inflammation. However, it has also been reported that Notch1 signaling pathway modulates nuclear factor-κB activation via PI3K and MAPK pathways and exhibits macrophage M2 functional polarization in systemic lupus erythematosus.35 Similarly, RBP-J, the downstream molecule of the Notch signaling, is required for M2 macrophage polarization in response to chitin and mediates function of alternatively activated macrophages.36 Overall, these findings suggest that the bifunctionality of Notch1 pathway on macrophage polarization seems to be highly context-specific. It is also possible that the activation status of Notch signaling varies in the different diseases to respond to various stimuli, in acute or chronic inflammation, and in the different stages of disease (the early or late stages), resulting in the different roles of Notch signaling in various cell types and cellular activities. Nonetheless, it is clear to conclude that Notch1 signaling-dependent macrophage polarization may play a pivotal role in inflammatory response, which could provide a potential therapeutic approach for inflammatory diseases.
Given the complexity of Notch signaling and the heterogeneity of macrophages under various environmental stimuli, specific Notch signaling network is needed to be clarified in macrophage polarization. Intriguingly, several lines of evidence support the fact that Notch and Hippo-YAP/TAZ signaling pathways are intricately linked. A mechanical interplay between Notch and YAP/TAZ has been described in various cellular contexts, which influences cell self-renewal, stem cell differentiation, cell fate decisions, epithelial-stromal interactions, morphogenesis, and inflammation.20 However, the crosstalk between Notch1 and YAP in modulating macrophage polarization is not well established. The present study identified a bidirectional overlap between Notch1 and YAP signaling in activated macrophages. Based on in vitro studies, we demonstrated that Notch1 signaling transcriptionally upregulated YAP expression, promoting M1 macrophage polarization. In turn, the increase of YAP transcriptional activity can feed Notch1 signaling by upregulating the expression of Notch ligand JAG1. Moreover, our in vivo studies revealed that YAP overexpression in macrophages aggravated LPS/D-GalN-induced liver injury in Notch1M-KO mice, which was accompanied by increased infiltration of macrophages/neutrophils, and augmented expressions of proinflammatory mediators. Collectively, our results identify YAP as an important regulator of Notch1-mediated M1 macrophage polarization and liver inflammation during LPS/D-GalN-induced acute liver injury. These findings are in agreement with a recent report that YAP activation in macrophages enhanced proinflammatory response by increasing IL-6 expression and impeded reparative response by decreasing Arg1 expression in myocardial infarction.37 A similar report has also shown that YAP-mediated Kupffer cell activation critically contributes to the production of proinflammatory cytokines and the perpetuation of liver inflammation in nonalcoholic steatohepatitis.23 In addition, the work of Ji's group showed that YAP activation in hepatic parenchymal cells (hepatocytes) attenuated hepatic damage in liver ischemia-reperfusion injury.38 The work of Camargo's group revealed that YAP signaling in biliary epithelial cells and hepatocytes is a protective rheostat and regenerative regulator in the mammalian liver.39 Our current study demonstrated that inhibition of the Notch1-YAP circuit in macrophages attenuated LPS/D-GalN-induced acute liver injury. Consistent with our results, a previous study also demonstrated that YAP aggravated inflammatory bowel disease by regulating M1/M2 macrophage polarization and that myeloid-specific knockout of YAP attenuated inflammatory bowel disease.40 Collectively, these findings add to the nuance, complexity, and variability of YAP signaling depending on environmental conditions and cell type. More importantly, our current study demonstrates that the Notch1 signaling forms a positive feedback loop with the Hippo signaling effector YAP to promote M1 macrophage polarization and liver inflammatory responses. However, further work is needed to elucidate the exact mechanisms of the intracellular signal chain between the Notch1-YAP circuit and the downstream targets for regulating macrophage polarization.
Another important implication of our results is that the myeloid Notch1-YAP circuit could be involved in the regulation of apoptotic pathways during LPS/D-GalN-induced acute liver injury. It is well documented that apoptosis of hepatocytes is a seminal feature of acute liver injury and fulminant hepatic failure.41, 42, 43 ASK1 was identified as a mitogen-activated protein kinase, which activates the c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinase pathways and is required for TNF-α-induced apoptosis.44,45 In addition, a recent report showed that ASK1 activated hepatic JNK and p38 to promote apoptosis, inflammation, and hepatic fibrosis, whereas ASK1 inhibition reduced cell death and fibrosis downstream of inflammatory signaling induced by NLR family pyrin domain containing 3.46 Consistent with these observations, our study demonstrated that myeloid Notch1 deficiency inhibited ASK1, p38, and caspase-3 activation, which was accompanied by decreased apoptotic hepatocytes in Notch1M-KO livers. Moreover, we found that the serum TNF-α levels were markedly reduced in Notch1M-KO mice compared with the Notch1FL/FL control animals. Indeed, hepatocellular apoptosis, a contributor to many acute and chronic liver diseases, can be a consequence of overactivation of the immune response and is often mediated by TNF-α production.47 Besides, HMGB1, a member of damage-associated molecular patterns, is usually released from damaged or dying cells during apoptosis in response to oxidative stress. As an early mediator of inflammation, HMGB1 provides danger signals and promotes inflammation and tissue damage during acute liver injury.29,48 In agreement with previous findings indicating that HMGB1 is intricately involved in the progression of LPS/D-GalN-induced acute liver injury,49,50 our results showed that serum HMGB1 levels were dramatically increased after LPS/D-GalN challenge, whereas myeloid Notch1 disruption significantly reduced serum HMGB1 concentration.
Various signaling pathways have been implicated in the regulation of liver homeostasis and repair, including the Notch and Hippo signaling. Abnormal Notch and Hippo signalings promote the development and/or progression of various liver diseases, including liver inflammatory diseases.21,51 The present study further demonstrated that upregulated Notch1 and YAP signalings in liver macrophages were highly correlated with LPS/D-GalN-induced liver injury. More importantly, dual inhibition of Notch1 and YAP in macrophages further promoted M2 polarization and attenuated liver injury. Our results suggest that targeting macrophage Notch1-YAP circuit might be a new opportunity for personalized medicine and the development of novel therapeutic interventions. Moreover, a benefit of combination treatment might be the advantage of lowering the dose of each drug, minimizing toxicity. Nevertheless, further cell- and context-specific in-depth understanding of Notch and Hippo-YAP signaling in liver homeostasis and disease is crucial to translate these concepts into new therapeutic strategies.
In conclusion, we uncover a novel Notch1-YAP circuit for reprogramming macrophage polarization and liver inflammation. These findings suggest that targeting this circuit in macrophages could be a promising therapeutic strategy for treating inflammatory liver diseases.
Methods
Animals
The floxed Notch1 (Notch1FL/FL) mice (Jackson Laboratory, Bar Harbor, ME) and the mice expressing Cre recombinase under the control of the Lysozyme 2 (Lyz2) promoter (LysM-Cre; Jackson Laboratory) were used to generate myeloid-specific Notch1 (Notch1M-KO) mice. Mouse genotyping was performed by using a standard protocol with primers described in the JAX genotyping protocols database. Male Notch1M-KO and Notch1FL/FL mice at 6–8 weeks of age were used in these experiments. The mice were bred in a standard environment with a 12-hour light/dark cycle. All animal protocols were approved by the Animal Care and Use Committee of Wuhan University in China (Permit No: WP20220117). The animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health.
Mouse Acute Liver Injury Model
LPS and D-GalN (Sigma-Aldrich, St. Louis, MO) were used to induce acute liver injury in mice. The mice were injected intraperitoneally with a dose of 50 μg/kg LPS plus 700 mg/kg D-GalN. The mice were sacrificed 5 hours after injection. Some animals were injected via the tail vein with YAP siRNA or NS control siRNA (2 mg/kg) (Santa Cruz Biotechnology) mixed with mannose-conjugated polymers (Polyplus transfection, Illkirch, France) at a ratio according to the manufacturer’s instructions 6 hours before LPS/D-GalN injection. Some animals were injected via the tail vein with BMDMs (5 × 106 cells in PBS/mouse) transfected with lentivirus-expressing YAP (Lv-YAP) 24 hours before LPS/D-GalN injection.
Hepatocellular Function Assay
Serum ALT and AST levels, an indicator of hepatocellular injury, were measured with Micro Glutamic-pyruvic Transaminase Assay Kit and Micro Glutamic-oxalacetic Transaminase Assay Kit (Solarbio, Beijing, China) in a standard clinical automatic analyzer (Dimension Xpand; Siemens Dade Behring, Munich, Germany).
Histology, Immunohistochemistry, Immunofluorescence Staining
Liver sections (5 μm) were stained with hematoxylin and eosin. Liver macrophage and neutrophil infiltration were detected by immunohistochemistry staining, using primary rat antimouse F4/80 Ab and primary rat antimouse Ly6G Ab (Invitrogen), respectively. Cleaved caspase-3 was detected using primary rabbit anti-mouse cleaved caspase-3 Ab (Cell Signaling Technology). The secondary biotinylated goat antirabbit IgG (Vector, Burlingame, CA) was used for immunohistochemistry staining. Macrophage NICD was detected using primary rabbit antimouse NICD (Cell Signaling Technology) and secondary biotinylated goat antirabbit IgG (Vector). F4/80 and Notch1 double-positive macrophages were identified by rat antimouse F4/80 (Bio-Rad, Hercules, CA) and mouse monoclonal antibody Notch1 (Santa Cruz Biotechnology) followed by incubating with secondary Alexa Fluor 488 AffiniPure Goat Anti-Rat IgG (H+L) or Cy3 AffiniPure Goat Anti-Mouse IgG (H+L) (Jackson Immunoresearch), respectively. The average number of positive cells was quantified by analyzing at least 10 random high-power fields per animal, with Image-Pro Plus software. Detailed antibodies are listed in Table 1.
Table 1.
List of Antibodies Used in This Study
| Antibodies | Company | Catalog number | Dilution |
|---|---|---|---|
| Anti-NICD | Cell Signaling Technology | Cat#: 4147S | 1:1000 |
| Anti-Hes1 | Abmart | Cat#: T55649S | 1:1000 |
| Anti-Notch1 | ABclonal | Cat#: A7636 | 1:500 |
| Anti-Notch1 | Santa Cruz Biotechnology | Cat#: sc-376403 | 1:50 |
| Anti-GAPDH | ABclonal | Cat#: AC001 | 1:1000 |
| Anti-F4/80 | Santa Cruz Biotechnology | Cat#: sc-52664 | 1:50 |
| Anti-Ly6G | Cell Signaling Technology | Cat#: 87048 | 1:50 |
| F4/80, PE/Cyanine7 | BioLegend | Cat#: 123114 | 1:100 |
| CD206, APC | Invitrogen | Cat#: 17-2061-82 | 1:100 |
| Anti-Arginase1 | Santa Cruza Biotechnology | Cat#: sc-271430 | 1:500 |
| Anti-iNOS | Abmart | Cat#: T55993S | 1:1000 |
| Anti-YAP | ABclonal | Cat#: A11265 | 1:500 |
| Anti-C-caspase-3 | Cell Signaling Technology | Cat#: 9664S | 1:1000 |
| Anti-Caspase-3 | Cell Signaling Technology | Cat#: 9662S | 1:1000 |
| Anti-p-ASK1 | Affinity Biosciences | Cat#: AF3477 | 1:500 |
| Anti-ASK1 | Affinity Biosciences | Cat#: AF6477 | 1:500 |
| Anti-p-P38 | Cell Signaling Technology | Cat#: 4511S | 1:1000 |
| Anti-P38 | Cell Signaling Technology | Cat#: 8690S | 1:1000 |
| Anti-Bcl-xL | Cell Signaling Technology | Cat#: 2764S | 1:1000 |
| Anti-Bax | Cell Signaling Technology | Cat#: 2772S | 1:1000 |
| Anti-JAG1 | Cell Signaling Technology | Cat#: 70109T | 1:1000 |
| Alexa Fluor 488 AffiniPure Goat Anti-Rat IgG (H+L) | Jackson Immunoresearch | Cat#: 112-545-003 | 1:50 |
| Cy3 AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson Immunoresearch | Cat#:111-165-003 | 1:50 |
TUNEL Assay
Apoptotic cells were identified with an apoptosis detection kit (S7110, EMD Millipore Co, Merck KGaA, Darmstadt, Germany). Cells with nuclear-positive staining by fluorescent antibodies for DNA fragmentation were visualized directly by fluorescence microscopy and counted.
Western Blot Analysis
Protein was extracted from liver tissue or cells with ice-cold protein lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate, 1% Triton-100). Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL). Equal amounts of proteins (30–50 μg) were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked with 5% skim milk. The membranes were incubated with primary antibodies overnight at 4°C, followed by the corresponding secondary antibodies. The used primary antibodies included anti-NICD, anti-Hes1, anti-JAG1, anti-YAP, anti-p-p38, anti-p38, antiapoptosis signal-regulating kinase 1 (ASK1), anti-p-ASK1, anti-Bcl-xL, anti-Bax, anti-Caspase-3, anti-cleaved-Caspase-3, anti-Arg1, anti-iNOS, and anti-Notch1. Protein levels were quantified with Image Lab Software and normalized to the loading control GADPH. Detailed antibodies are listed in Table 1.
Isolation of Hepatic Macrophages and BMDMs
Primary hepatic macrophages were isolated as described.29 In brief, mouse livers were perfused in situ with 37°C warmed HBSS solution, followed by a collagenase buffer (collagenase type IV, Sigma-Aldrich). Perfused livers were dissected and teased through 70 μm nylon mesh cell strainers (BD Biosciences). The nonparenchymal cells were separated from hepatocytes and layered onto a 50%/25% 2-step Percoll gradient (Sigma-Aldrich). After centrifugation, hepatic macrophages in the middle layer were collected and allowed to adhere to cell culture plates in DMEM with 10% FBS, 10 mM HEPES, 2 mM GlutaMax, 100 U/mL penicillin, and 100 μg/mL streptomycin for 15 min at 37°C. Murine primary BMDMs were isolated from mice.29 In brief, bone marrow cells were harvested from the femurs and tibias of Notch1FL/FL and Notch1M-KO mice and cultured in DMEM supplemented with 10% FBS and 20% L929-conditioned medium.
Lentiviral Vector Construction
The 293T cells were cultured in DMEM supplemented with 10% FBS and were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells were cotransfected with p-Lv-YAP, psPAX2, and pCMV-VSV-G (Addgene, plasmid #12260 and #8454, MA) using FuGENE HD transfection reagent (Promega) to package the lentiviruses according to the manufacturer’s instructions. The viral vector-containing supernatant was collected and filtered through a 0.45-μm filter after 48 hours posttransfection. GFP lentivirus (Lv-GFP, Addgene plasmid #174171) was used as a control.
Plasmid Constructs
NICD was produced by polymerase chain reaction amplification. The NICD fragments that were amplified from mouse BMDM cDNA were cloned into the EcoRI/XbaI site of pEF-Flag-N. The pCMV-Flag-YAP plasmid and empty vector were kindly provided by Dr. Bin Zhao (Zhejiang University, China).
In Vitro Transfection
BMDMs (1 × 106/well) were transfected with the pEF-Flag-NICD plasmid or pCMV-Flag-YAP plasmid. After 48 hours, cells were supplemented with 100 ng/mL LPS and 10 ng/mL IFN-γ for an additional 12 hours.
Quantitative Reverse-Transcriptase Polymerase Chain Reaction Analysis
Total RNA was extracted from liver tissue or cell cultures using Trizol Reagent (Takara) according to the manufacturer’s instructions. Reverse transcription was performed using 1000 ng of total RNA in the first-strand cDNA synthesis reaction with PrimeScript RT reagent Kit (Takara). Reverse-transcriptase polymerase chain reaction was performed using an ABI 7900 sequence detector (Invitrogen). Reverse-transcriptase polymerase chain reaction was performed using SYBR Premix Ex Taq (Takara) and values were calculated by their ratios to the housekeeping gene Gapdh. Primer sequences are provided in Table 2 for the amplification of Ctgf, Cyr61, JAG1, Tnf-α, Il-6, Il-1β, Il-10, Tgf-β, Mcp-1, Cxcl-1, Notch1, Hes1, Yap, and Gapdh.
Table 2.
List of Primers Used in This Study
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| Ctgf | CACCTAAAATCGCCAAGCCTG | AGTTCGTGTCCCTTACTTCCTG |
| Cyr61 | ACCGCTCTGAAAGGGATCTG | TGTTTACAGTTGGGCTGGAAG |
| JAG1 | AGAAGTCAGAGTTCAGAGGCGTCC | AGTAGAAGGCTGTCACCAAGCAAC |
| TNF-α | CCTCTCTCTAATCAGCCCTCTG | GAGGACCTGGGAGTAGATGAG |
| IL-6 | TCCTACCCCAATTTCCAATGCT | TAACGCACTAGGTTTGCCGA |
| IL-1β | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| Notch1 | GGATCACATGGACCGATTGC | ATCCAAAAGCCGCACGATAT |
| Yap | TACTGATGCAGGTACTGCGG | TCAGGGATCTCAAAGGAGGAC |
| Hes1 | CTTGGATTTTAGGAGAGACTT | GCATGGTCAGTCACTTAATAC |
| MCP-1 | GAAGGAATGGGTCCAGACAT | ACGGGTCAACTTCACATTCA |
| CXCL-1 | TGGCTGGGATTCACCTCAAGAACA | TTTCTGAACCAAGGGAGCTTCAGG |
| Gapdh | CATCACTGCCACCCAGAAGACTG | ATGCCAGTGAGCTTCCCGTTCAG |
| IL-10 | TGAATTCCCTGGGTGAGAAG | TGGCCTTGTAGACACCTTGG |
| Tgf-β | TGCGCTTGCAGAGATTAAAA | CTGCCGTACAACTCCAGTGA |
Flow Cytometry Analysis
Cell surface expression analysis of liver macrophages was performed using the antimurine CD206 (BD Bioscience, San Diego, CA) and F4/80-FITC (Biomedicals, Augst, Switzerland). Cells were incubated for 15 minutes on ice with anti-CD16/CD32 monoclonal antibody (Biolegend, San Diego, CA) to reduce NS binding. Cells were washed in buffer, incubated with each antibody for 30 minutes, and subjected to flow cytometry using the FACS Calibur (BD Biosciences). Detailed antibodies are listed in Table 1.
Luciferase Assays
The YAP promoter: luciferase reporter plasmid (YAP luciferase) was constructed in pGL3 luciferase vector (Promega) according to the manufacturer’s instructions. The JAG1 promoter: luciferase reporter plasmid (JAG1 luciferase) was constructed in pGL3 luciferase vector (Promega) according to the manufacturer’s instructions. BMDMs were cotransfected with pGL3-YAP-luciferase and pEF-Flag-NICD, or pGL3-JAG1-luciferase and pCMV-Flag-YAP vectors. After 48 hours, the cells were lysed with Passive Lysis Buffer, and the transcriptional activity was measured using a luciferase assay system (Promega).
ELISA
Murine serum and BMDM culture supernatants were harvested for cytokine analysis. ELISA kits were used to measure TNF-α and HMGB1 levels (eBiosciences, CA).
Statistical Analysis
Data are expressed as mean ± standard deviation and analyzed by Permutation t-test and Pearson correlation. Two-sided P values less than .05 were considered statistically significant. Multiple group comparisons were performed using 1-way analysis of variance followed by Bonferroni post hoc test. For Kaplan-Meier survival curves, statistical comparison was determined by log-rank analyses. All analyses were performed by SPSS version 17.0 (SPSS, Chicago, IL).
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All authors had access to the study data and had reviewed and approved the final manuscript.
Acknowledgments
The authors thank Linhao Xing (Wuhan University School of Basic Medical Sciences) for helping with the graphical abstract.
CRediT Authorship Contributions
Yan Yang (Data curation: Lead; Writing – original draft: Lead)
Ming Ni (Data curation: Lead)
Ruobin Zong (Formal analysis: Supporting)
Mengxue Yu (Data curation: Equal)
Yishuang Sun (Data curation: Supporting)
Jiahui Li (Data curation: Supporting)
Pu Chen (Conceptualization: Lead; Funding acquisition: Lead)
Changyong Li (Conceptualization: Lead; Funding acquisition: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the National Key Research and Development Program of China (No.2018YFA0109000), the Wuhan Municipal Science and Technology Bureau (2020020601012210), and the Natural Science Foundation of Hubei Province (2017CFB620).
Contributor Information
Pu Chen, Email: puchen@whu.edu.cn.
Changyong Li, Email: lichangyong@whu.edu.cn.
References
- 1.Bernal W., Lee W.M., Wendon J., et al. Acute liver failure: a curable disease by 2024? J Hepatol. 2015;62(Suppl 1):S112–S120. doi: 10.1016/j.jhep.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Possamai L.A., Thursz M.R., Wendon J.A., et al. Modulation of monocyte/macrophage function: a therapeutic strategy in the treatment of acute liver failure. J Hepatol. 2014;61:439–445. doi: 10.1016/j.jhep.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Bernal W., Auzinger G., Dhawan A., et al. Acute liver failure. Lancet. 2010;376:190–201. doi: 10.1016/S0140-6736(10)60274-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhou D., Huang C., Lin Z., et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Strnad P., Tacke F., Koch A., et al. Liver: guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 6.Weigert A., von Knethen A., Fuhrmann D., et al. Redox-signals and macrophage biology. Mol Aspects Med. 2018;63:70–87. doi: 10.1016/j.mam.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Brenner C., Galluzzi L., Kepp O., et al. Decoding cell death signals in liver inflammation. J Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 8.Ilyas G., Zhao E., Liu K., et al. Macrophage autophagy limits acute toxic liver injury in mice through down regulation of interleukin-1beta. J Hepatol. 2016;64:118–127. doi: 10.1016/j.jhep.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y.Y., Li X.F., Meng X.M., et al. Macrophage phenotype in liver injury and repair. Scand J Immunol. 2017;85:166–174. doi: 10.1111/sji.12468. [DOI] [PubMed] [Google Scholar]
- 10.Tacke F., Zimmermann H.W. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol. 2014;60:1090–1096. doi: 10.1016/j.jhep.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Ye L., He S., Mao X., et al. Effect of hepatic macrophage polarization and apoptosis on liver ischemia and reperfusion injury during liver transplantation. Front Immunol. 2020;11:1193. doi: 10.3389/fimmu.2020.01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radtke F., Fasnacht N., Macdonald H.R. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Radtke F., MacDonald H.R., Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 14.Thapa B., Lee K. Metabolic influence on macrophage polarization and pathogenesis. BMB Rep. 2019;52:360–372. doi: 10.5483/BMBRep.2019.52.6.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W., Liu Y., Chen J., et al. The Notch signaling pathway regulates macrophage polarization in liver diseases. Int Immunopharmacol. 2021;99 doi: 10.1016/j.intimp.2021.107938. [DOI] [PubMed] [Google Scholar]
- 16.Xu J., Chi F., Guo T., et al. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J Clin Invest. 2015;125:1579–1590. doi: 10.1172/JCI76468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H., Zhu J., Smith S., et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y.H., Cai K., Xu P.P., et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct Target Ther. 2021;6:10. doi: 10.1038/s41392-020-00437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y.C., He F., Feng F., et al. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 2010;70:4840–4849. doi: 10.1158/0008-5472.CAN-10-0269. [DOI] [PubMed] [Google Scholar]
- 20.Totaro A., Castellan M., Di Biagio D., et al. Crosstalk between YAP/TAZ and Notch signaling. Trends Cell Biol. 2018;28:560–573. doi: 10.1016/j.tcb.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler F., Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mooring M., Fowl B.H., Lum S.Z.C., et al. Hepatocyte stress increases expression of Yes-associated protein and transcriptional coactivator with PDZ-binding motif in hepatocytes to promote parenchymal inflammation and fibrosis. Hepatology. 2020;71:1813–1830. doi: 10.1002/hep.30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song K., Kwon H., Han C., et al. Yes-associated protein in Kupffer cells enhances the production of proinflammatory cytokines and promotes the development of nonalcoholic steatohepatitis. Hepatology. 2020;72:72–87. doi: 10.1002/hep.30990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Liu L., Zhang S., et al. Long noncoding RNA NEAT1 suppresses hepatocyte proliferation in fulminant hepatic failure through increased recruitment of EZH2 to the LATS2 promoter region and promotion of H3K27me3 methylation. Exp Mol Med. 2020;52:461–472. doi: 10.1038/s12276-020-0387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv Y., Kim K., Sheng Y., et al. YAP controls endothelial activation and vascular inflammation through TRAF6. Circ Res. 2018;123:43–56. doi: 10.1161/CIRCRESAHA.118.313143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W., Khan S.K., Gvozdenovic-Jeremic J., et al. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J Clin Invest. 2017;127:137–152. doi: 10.1172/JCI88486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Song X., Liao H., et al. Overexpression of Mothers against decapentaplegic homolog 7 activates the Yes-associated protein/Notch cascade and promotes liver carcinogenesis in mice and humans. Hepatology. 2021;74:248–263. doi: 10.1002/hep.31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yimlamai D., Christodoulou C., Galli G.G., et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C., Sheng M., Lin Y., et al. Functional crosstalk between myeloid Foxo1-beta-catenin axis and Hedgehog/Gli1 signaling in oxidative stress response. Cell Death Differ. 2021;28:1705–1719. doi: 10.1038/s41418-020-00695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu S.S., Lau C.M., Barham W.J., et al. Macrophage-specific RNA interference targeting via "click", mannosylated polymeric micelles. Mol Pharm. 2013;10:975–987. doi: 10.1021/mp300434e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palaga T., Buranaruk C., Rengpipat S., et al. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur J Immunol. 2008;38:174–183. doi: 10.1002/eji.200636999. [DOI] [PubMed] [Google Scholar]
- 32.Hu X., Chung A.Y., Wu I., et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q., Wang C., Liu Z., et al. Notch signal suppresses Toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor kappaB activation. J Biol Chem. 2012;287:6208–6217. doi: 10.1074/jbc.M111.310375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu L., Yue S., Jiang L., et al. Myeloid Notch1 deficiency activates the RhoA/ROCK pathway and aggravates hepatocellular damage in mouse ischemic livers. Hepatology. 2018;67:1041–1055. doi: 10.1002/hep.29593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W., Xu W., Xiong S. Blockade of Notch1 signaling alleviates murine lupus via blunting macrophage activation and M2b polarization. J Immunol. 2010;184:6465–6478. doi: 10.4049/jimmunol.0904016. [DOI] [PubMed] [Google Scholar]
- 36.Foldi J., Shang Y., Zhao B., et al. RBP-J is required for M2 macrophage polarization in response to chitin and mediates expression of a subset of M2 genes. Protein Cell. 2016;7:201–209. doi: 10.1007/s13238-016-0248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mia M.M., Cibi D.M., Abdul Ghani S.A.B., et al. YAP/TAZ deficiency reprograms macrophage phenotype and improves infarct healing and cardiac function after myocardial infarction. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Lu T., Zhang C., et al. Activation of YAP attenuates hepatic damage and fibrosis in liver ischemia-reperfusion injury. J Hepatol. 2019;71:719–730. doi: 10.1016/j.jhep.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepe-Mooney B.J., Dill M.T., Alemany A., et al. Single-cell analysis of the liver epithelium reveals dynamic heterogeneity and an essential role for YAP in homeostasis and regeneration. Cell Stem Cell. 2019;25:23–38. doi: 10.1016/j.stem.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X., Li W., Wang S., et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. 2019;27:1176–1189. doi: 10.1016/j.celrep.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C., Jin H., Wang Y., et al. Critical role of OX40 in drug-induced acute liver injury. Br J Pharmacol. 2020;177:3183–3196. doi: 10.1111/bph.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y., Xue W., Zhang W., et al. Histone methyltransferase G9a protects against acute liver injury through GSTP1. Cell Death Differ. 2020;27:1243–1258. doi: 10.1038/s41418-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehrke N., Hovelmeyer N., Waisman A., et al. Hepatocyte-specific deletion of IL1-RI attenuates liver injury by blocking IL-1 driven autoinflammation. J Hepatol. 2018;68:986–995. doi: 10.1016/j.jhep.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Saitoh M., Nishitoh H., Fujii M.,T., et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tobiume K., Matsuzawa A., Takahashi T., et al. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2:222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuster-Gaul S., Geisler L.J., McGeough M.D., et al. ASK1 inhibition reduces cell death and hepatic fibrosis in an Nlrp3 mutant liver injury model. JCI Insight. 2020;5 doi: 10.1172/jci.insight.123294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaufmann T., Jost P.J., Pellegrini M., et al. Fatal hepatitis mediated by tumor necrosis factor TNFalpha requires caspase-8 and involves the BH3-only proteins Bid and Bim. Immunity. 2009;30:56–66. doi: 10.1016/j.immuni.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsung A., Sahai R., Tanaka H., et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med. 2005;201:1135–1143. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kusakabe J., Hata K., Miyauchi H., et al. Complement-5 inhibition deters progression of fulminant hepatitis to acute liver failure in murine models. Cell Mol Gastroenterol Hepatol. 2021;11:1351–1367. doi: 10.1016/j.jcmgh.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W., Sun L., Deng Y., et al. Synergistic effects of antibodies against high-mobility group box 1 and tumor necrosis factor-alpha antibodies on D-(+)-galactosamine hydrochloride/lipopolysaccharide-induced acute liver failure. FEBS J. 2013;280:1409–1419. doi: 10.1111/febs.12132. [DOI] [PubMed] [Google Scholar]
- 51.Russell J.O., Camargo F.D. Hippo signalling in the liver: role in development, regeneration and disease. Nat Rev Gastroenterol Hepatol. 2022;19:297–312. doi: 10.1038/s41575-021-00571-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All authors had access to the study data and had reviewed and approved the final manuscript.