Abstract
Freeze tolerance, the ability of an organism to survive internal ice formation, is a striking survival strategy employed by some ectotherms living in cold environments. However, the genetic bases of this remarkable adaptation are largely unknown. The Amur sleeper (Perccottus glenii), the only known freeze-tolerant fish species, can overwinter with its entire body frozen in ice. Here, we sequenced the chromosome-level genome of the Amur sleeper and performed comparative genomic, transcriptomic, and metabolomic analyses to investigate its strategies for surviving freezing. Evolutionary analysis suggested that the Amur sleeper diverged from its closest non-cold-hardy relative about 15.07 million years ago and has experienced a high rate of protein evolution. Transcriptomic and metabolomic data identified a coordinated and tissue-specific regulation of genes and metabolites involved in hypometabolism, cellular stress response, and cryoprotectant accumulation involved in freezing and thawing. Several genes show evidence of accelerated protein sequence evolution or family size expansion were found as adaptive responses to freezing-induced stresses. Specifically, genetic changes associated with cytoskeleton stability, cryoprotectant synthesis, transmembrane transport, and neuroprotective adaptations were identified as potentially key innovations that aid in freezing survival. Our work provides valuable resources and opportunities to unveil the molecular adaptations supporting freeze tolerance in ectothermic vertebrates.
Keywords: freeze tolerance, Amur sleeper, hypometabolism, cell stress response, cytoskeleton, cryoprotectant, nerve transmission
Introduction
Ectothermic animals inhabiting seasonally or perpetually cold regions may experience low temperature exposure that could freeze their body fluids. Freeze tolerance is an amazing survival strategy, whereby some ectothermic animals tolerate the conversion of up to 82% their body water into internal ice (e.g., Hemideina maori, a tree weta; Ramløv and Westh 1993). These animals may spend prolonged periods (days to months) in a state of frozen dormancy, characterized by the cessation of vital physiological processes including heartbeat, respiration, nerve conductance, and skeletal muscle movement, followed by a return to normal life after thawing (Schmid 1982; Storey 1987, 1990). Although freeze tolerance has evolved multiple times across the animal kingdom and is exhibited in variety of taxa including insects, invertebrates, reptiles, and amphibians (Costanzo and Claussen 1990; Ramløv et al. 1992; Loomis 1995; Layne and Kefauver 1997; Bradley Shaffer et al. 2013), it is relatively rare among vertebrates compared with the common cold-tolerance strategies such as hibernation and freeze avoidance (via promoting supercooling to reduce the likelihood of freezing) (Costanzo and Lee 2013; Iwaya-Inoue et al. 2018; Mohr et al. 2020).
Natural freeze tolerance generally refers to ice formation in the extracellular spaces while resisting intercellular freezing to avoid damage to the subcellular compartments and the cytoskeleton (Costanzo and Lee 2013; Storey and Storey 2017). However, extracellular ice crystals exclude solutes, greatly elevating the osmolality of extracellular fluids, causing an outflow of intracellular water and a volume reduction for cells and organs (Storey and Storey 2020). This freezing-induced dehydration will cause molecular crowding that promotes unfavorable interactions between proteins and ions, potentially damaging macromolecules (Ramløv 2000). Cell shrinkage beyond a critical minimum volume can exert shear stress on the cytoskeleton or plasma membranes, resulting in the loss of membrane integrity upon thawing (Li et al. 2009). In addition to dehydration, freezing also exposes organisms to severe physiological stress, including hypoxia/anoxia, ischemia, and hypometabolism, all of which can generate injurious levels of reactive oxygen species (ROS) and oxidative damage to macromolecules (Storey and Storey 2017; Toxopeus and Sinclair 2018). Moreover, the reoxygenation, rehydration, and reperfusion that accompany thawing are conditions that are known to trigger a surge in ROS production (Giraud-Billoud et al. 2019).
Freeze tolerance in vertebrates has multiple components that are derived from other preexisting capabilities, such as anoxia and dehydration tolerance, which have been extensively explored in multiple terrestrial freeze-tolerant reptile (e.g., some turtles and lizards) and amphibian species, especially the wood frogs (Zhang and Storey 2012; Bradley Shaffer et al. 2013; Storey and Storey 2013, 2017; Costanzo 2019). For example, complex and coordinated cellular, molecular, and physiological adaptations that confer freezing survival include (1) mechanisms to manage extracellular ice volume and growth rate (e.g., via ice-nucleating proteins or ice-binding proteins); (2) strong metabolic rate depression coupled with selective activation of “survival” pathways to maintain cell function; and (3) the accumulation of low-molecular-weight organic compounds as cryoprotectants. However, a large gap still remains in our knowledge and understanding of the mechanisms underlying the prolonged interruption and coordinated reactivation of vital functions over a freeze/thaw cycle (Storey and Storey 2017). In particular, the genetic basis of freeze tolerance in ectothermic vertebrates remains largely unknown.
The Amur sleeper, Perccottus glenii (Odontobutidae, Perciformes), a bony fish that can overwinter with its entire body frozen in ice, is probably the only freeze-tolerant vertebrate aside from reptiles and amphibians. It is a limnophilic species native to the Amur River drainage of northeastern Asia, and has invaded European waters, leading to detrimental ecological impacts (Reshetnikov and Ficetola 2011; Xu et al. 2014). Result from freezing survival experiment of the Amur sleeper under laboratory condition showed 100% survival after 24 h of being frozen in ice at a temperature of −2 °C (Chai et al. 2020). The Amur sleeper prefers small, stagnant waterbodies (typically 40–70 cm in depth), which commonly freeze to the bottom in the winter (air temperatures generally range from −15 to −35 °C). Before freezing, this fish experiences long-term hypoxic or anoxic conditions in the ice-covered waters until its whole body is encapsulated in ice, and this frozen dormant state can be maintained for up to three/four months until warm spring (Reshetnikov 2003; Karanova 2009). The Amur sleeper can endure prolonged freezing with internal ice formation in abdominal cavity, obvious dehydration of organs, and interruption of vital physiological functions (e.g., respiration, heartbeat, blood flow, voluntary movement), and revival within a few hours after thawing. This amazing winter survival strategy meets the criteria of freeze tolerance (Storey and Storey 2017) and provides the Amur sleeper with a competitive advantage over other freshwater fishes that are unable to survive in such extreme environments, allowing this species to avoid both competitors and predators and explaining its widespread successes as an invasive species (Reshetnikov and Ficetola 2011). Since the Amur sleeper is easy to obtain and breed (Zhang, He et al. 2021), it represents a convenient model species for investigations into freeze tolerance in ectothermic vertebrates. Moreover, considering that genomic resources for freeze-tolerant reptile and amphibian species are still limited, the Amur sleeper genome may represent a superb genetic resource for in-depth studies on molecular adaptation. We hypothesized that significant genetic changes in the Amur sleeper genome combined with regulatory profiles of genes and metabolites during freezing tend to reveal core processes supporting freezing tolerance.
In this study, we generated a high-quality de novo chromosome-level genome for the Amur sleeper, as well as a de novo reference genome assembly for the closely related, but non-cold-hardy, Neodontobutis hainanensis (Lv et al. 2020). These two genomes allowed us to explore differences in the population history and evolutionary rates between the Amur sleeper and N. hainanensis. Then, we performed comparative genomic, transcriptomic, and metabolomic analyses to determine the genetic basis of freeze tolerance. Analyses revealed key processes that assist in freezing survival, and genetic changes identified as potentially evolutionary innovations were related to cytoskeleton stability, cryoprotectant synthesis, transmembrane transport and neuroprotective adaptations. Our results not only provide insights into the evolution of this complex adaptation, but also provide useful genetic resources for future studies.
Results and Discussion
Genome Characteristics
We generated the first chromosome-level genome assembly for the Amur sleeper using a combination of Nanopore long reads, BGISEQ-500 reads, and Hi-C data (supplementary table S1, Supplementary Material online). The Amur sleeper genome was estimated to be 827.25 Mb with a heterozygosity ratio of 0.65% based on k-mer analysis (supplementary fig. S1 and table S2, Supplementary Material online). Three assembly algorithms were used to obtain a high-quality Amur sleeper genome assembly, and the genome assembled by SmartDenovo was finally selected based on highest continuity and completeness (supplementary tables S3 and S4, Supplementary Material online). After removal of redundant sequences, the Amur sleeper assembly was 760.85 Mb, with a contig N50 of 4.65 Mb (supplementary table S3, Supplementary Material online). We then anchored and oriented 379 contigs (723.97 Mb, ∼95.15%) onto 22 chromosomes using Hi-C data (fig. 1A; supplementary fig. S2 and table S5, Supplementary Material online). Finally, a chromosome-level genome assembly with a contig and scaffold N50 of 4.65 Mb and 33.77 Mb, respectively, was generated (supplementary table S6, Supplementary Material online). To assess the quality of the Amur sleeper assembly, we aligned the short reads to the assembly and found that over 99% of the short reads could be mapped, which covered 98.90% of the genome assembly (supplementary table S7, Supplementary Material online). Evaluation of genome completeness based on BUSCOs identified 96.7% complete and 1.2% fragmented genes (supplementary table S8, Supplementary Material online). For comparative analyses, a de novo assembly of the N. hainanensis genome was performed and an individual with a heterozygosity ratio of 0.15% yielded a ∼848 Mb assembly containing 8,221 contigs with the N50 of 1.34 Mb (supplementary table S6, Supplementary Material online). A total of 97.21% of the short reads were mapped to the N. hainanensis assembly, and 94.9% complete BUSCO genes captured (supplementary tables S7 and S8, Supplementary Material online). The higher heterozygosity rate in the Amur sleeper genome may suggest admixture of different clades during the northwards expansion of this species (Xu et al. 2014).
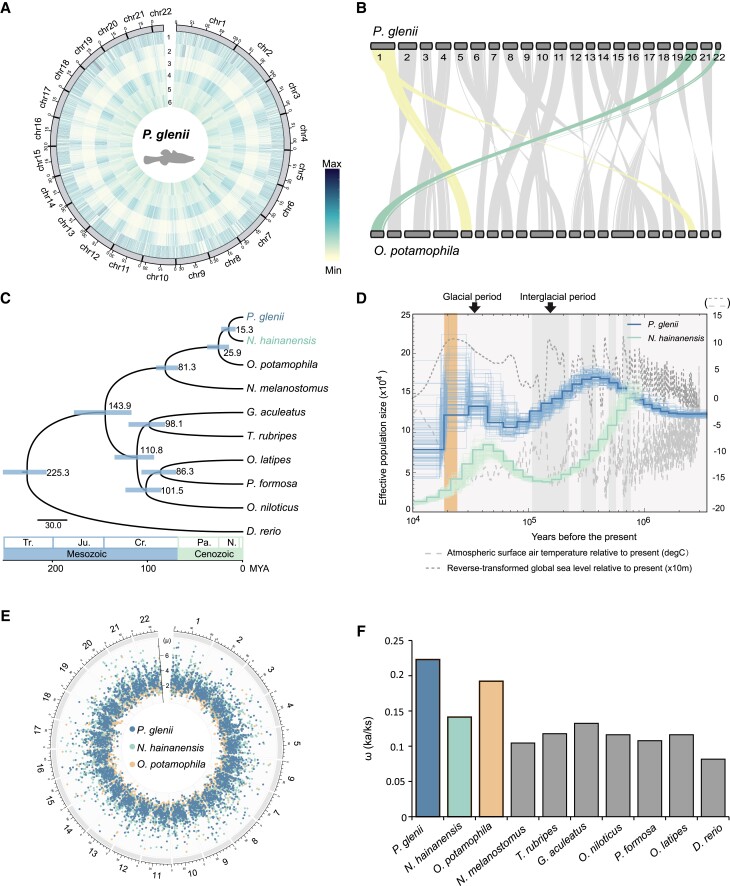
Fig. 1.
Evolutionary history of the Amur sleeper (Perccottus glenii). (A) Circos plot showing the distributions of the components in the Amur sleeper genome, using 500,000 bp windows. 1, Gene frequency; 2, Density of long interspersed nuclear elements; 3, Density of long terminal repeat retrotransposons; 4, Density of DNA transposons; 5, Density of short interspersed nuclear elements; 6, Density of GC content. (B) Collinearity analysis of the Amur sleeper and Odontobutis potamophila genomes. (C) Phylogenetic tree and divergence times estimated for the Amur sleeper and nine other teleosts. Numbers near each node are the estimated divergence times, with the blue error bars indicating the 95% confidence levels. (D) Demographic histories estimated by pairwise sequentially Markovian coalescent (PSMC). Shaded orange box represents the last glacial maximum (LGM). (E) Mutation rates for the three species estimated across each genome. The number outside of the circle represent the chromosome IDs in the O. potamophila genome. µ represents the mutation rate (× 10−9 per site per year) of each window. (F) The ω (Ka/Ks) ratios of concatenated genes in ten teleosts.
A combination of de novo and homology-based prediction methods identified approximately 39.78% of the bases in the Amur sleeper genome and 45.99% of the bases in the N. hainanensis genome as repetitive sequences (supplementary table S9, Supplementary Material online). Thus, the reduced genome size of the Amur sleeper may be due to a reduction/loss in the amount of repetitive DNA. We predicted a total of 25,160 and 26,237 protein-coding genes based on combined ab initio gene annotation, homology searching and transcript mapping in the Amur sleeper and N. hainanensis genomes, respectively, and approximately 97.50% and 94.63% of which were successfully annotated using five public databases (supplementary table S10, Supplementary Material online). For genome collinearity analysis, we compared the Amur sleeper with Odontobutis potamophila, a representative confamilial species (family Odontobutidae) as our N. hainanensis genome is not at a chromosomal level. Only two chromosomal fission and fusion events were detected, indicating conserved chromosomal evolution since divergence of these two species (fig. 1B). These results reflected the high-quality of the Amur sleeper and N. hainanensis genome assemblies, as well as the accuracy of the genome annotations. Thus, these genomes will be useful genetic resources for further comparative and functional studies on freeze tolerance in ectotherms.
Population History and Evolutionary Rate
Phylogenetic analysis based on a set of 4,469 one-to-one orthologues from ten teleost species recovered the Amur sleeper and N. hainanensis as a well-supported sister relationship, with this clade recovered as sister to the monophyletic genus Odontobutis (fig. 1C; supplementary fig. S3, Supplementary Material online). The reconstructed topology is consistent with topologies inferred using the mitochondrial genome and nuclear coding genes (Li et al. 2018; Lv et al. 2020). In addition, we estimated the divergence time of the ten teleosts using MCMCtree and found that the Amur sleeper and N. hainanensis diverged about 15.30 million years ago (Ma) (95% highest probability density 8.09–23.06 Ma) in the mid-Miocene (fig. 1C).
Analysis using pairwise sequentially Markovian coalescent (PSMC) model (Li and Durbin 2011) based on whole-genome datasets revealed changes in the demographic patterns for the Amur sleeper and N. hainanensis (fig. 1D), which might be due to substantial climate oscillations, including glacial–interglacial cycles and sea level changes. We observed two population expansion events in the demographic history of the Amur sleeper (fig. 1D). The first expansion occurred earlier than ∼3 Ma and peaked ∼0.4 Ma in the largest Quaternary glaciation (0.80–0.20 Ma). This expansion timing corroborates a previous study, which suggested that the Amur sleeper might have spread from the warm south to the cold north during the Late Pliocene (2.58–3.60 Ma) (Li et al. 2018). The subsequent population decline coincided with the advent of the warm interglacial period, during which the rise in sea level caused by deglaciation resulted in a dramatic reduction in freshwater habitats. The second expansion occurred at ∼0.7 thousand years ago (Ka) and peaked the last glacial maximum (LGM, 0.26–0.19 Ka; Clark et al. 2009). Similarly, the size of the N. hainanensis population decreased sharply after ∼0.9 Ma, and a population expansion occurred at ∼0.15 Ma, followed by sharp declines predating the LGM (fig. 1D). Overall, the Amur sleeper has maintained a relatively higher and more stable effective population size than N. hainanensis over evolutionary time. This may be attributed to the strong resistance and adaptability of the Amur sleeper, which has aided its survival in extreme environments, thereby providing opportunities to expand into new ecological niches.
Because the development of freeze tolerance suggests adaptive evolution in the genome of Amur sleeper, we calculated mutation rates of the whole-genome datasets and evolutionary rates of the 4,496 one-to-one orthologues. The mutation rate across the whole Amur sleeper genome was comparable to those for the closely related species N. hainanensis and Od. potamophila (fig. 1E). Despite the similar nucleotide-level mutation rates among them, protein sequences from the Amur sleeper appear to have evolved at a higher evolutionary rate than in the other species. The Amur sleeper had a significantly higher Ka/Ks ratio (ω, nonsynonymous/synonymous substitutions ratio) (fig. 1F; supplementary fig. S4, Supplementary Material online), implying that this species may have been subject to stronger natural selection pressure.
Transcriptomic and Metabolic Profiles During Freezing and Thawing
To better understand the genetic regulatory mechanisms and metabolic adaptations important in the Amur sleeper, transcriptomes of the brain, liver, and muscle tissues, as well as the metabolomes of the liver and muscle tissues from field-collected samples in active, frozen, and thawed states were analyzed to represent the three periods during a freeze–thaw cycle, that is activity, freezing and thawing, respectively (fig. 2A and B; supplementary movies S1 and S2, Supplementary Material online). We observed obvious shrinkage in body size as well as the presence of large ice crystals in the abdominal cavity of all frozen samples (supplementary fig. S5, Supplementary Material online), suggesting significant dehydration and ice formation. Principal component analyses (PCA) of the transcriptomes identified clear differences in the three tissues among the different periods (supplementary fig. S6A, Supplementary Material online). PCAs of the liver and muscle metabolomes showed large differences between the activity and the freezing/thaw periods, but smaller differences between freezing and thawing periods (supplementary fig. S6B, Supplementary Material online). Consistent with this, we identified a substantially larger number of differentially expressed genes and significantly different metabolites between the activity and freezing/thawing periods than between the freezing and thawing periods (supplementary fig. S7A and B, Supplementary Material online). This implies substantial changes in the transcriptional and metabolomic profiles in response to freezing and thawing. However, it is important to note that this study was limited to field-collected samples which may be influenced by many other complex factors (e.g., individual and environmental differences), thus further study comparing in the field and in controlled environments is warranted.
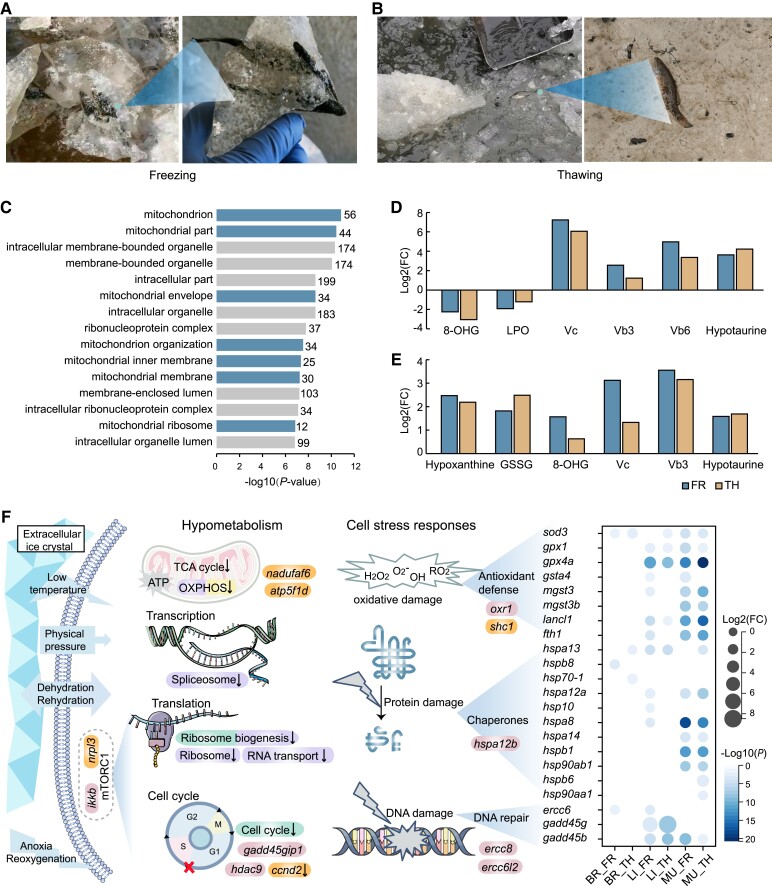
Fig. 2.
Genes and metabolites related to hypometabolism and the cell stress response. (A and B) Photographs of the Amur sleeper during freezing (A) and thawing (B). (C) The top 15 significantly enriched GO terms for downregulated genes in all three tissues (brain, liver, and muscle) during freezing compared with activity. (D and E) The log2(fold-change, FC) of metabolites related to antioxidant defense in the freezing (FR) and thawing periods (TH) relative to the activity period (AC) in the liver (D) and muscle (E) tissues. (F) Pathways and genes associated with hypometabolism and cell stress responses. The pathways related to energy-expensive cell processes that were significantly enriched in the downregulated genes in the brain (purple), liver (green), and muscle (yellow) tissues during freezing when compared with activity are shown. Genes highlighted in orange were identified as positively selected genes; genes highlighted in pink were identified as rapidly evolved genes. The heatmap shows the log2(FC) in the transcriptional expression of differentially expressed genes in FR and TH relative to AC in the brain (BR), liver (LI), muscle (MU).
Changes in Genes and Metabolites Related to Hypometabolism
Stress-induced hypometabolism is the core adaptive strategy implemented by freeze-tolerant animals to greatly decrease organismal energy demands, thereby permitting long-term survival using only their body fuel reserves (Carey et al. 2003; Storey and Storey 2013). Here, transcriptomic profiles identified 308 genes shared among all the three tissues (liver, muscle, and brain) in the Amur sleeper were downregulated during the freezing period, compared with the activity period (supplementary fig. S7A, Supplementary Material online). These downregulated genes were significantly enriched in several GO functional categories related to the mitochondria (e.g., mitochondrion, mitochondrial part, and mitochondrial protein complex) (fig. 2C). The mitochondria produce most of the adenosine triphosphate (ATP) used by cells via mitochondrial energy metabolism, including oxidative phosphorylation and tricarboxylic acid cycle reactions (Wu et al. 2007). Importantly, nadufaf6 and atp5f1d, which are components of the mitochondrial respiratory chain, were identified as positively selected genes in the Amur sleeper (fig. 2F, supplementary table S11, Supplementary Material online). The observed signal for positive selection may have implications for the regulation of ATP synthesis. KEGG analysis showed that genes downregulated in the brain and muscle tissues during the freezing period were significantly enriched in the oxidative phosphorylation pathway (fig. 2F; supplementary fig. S8A and C, Supplementary Material online). Simultaneously, the content of malonic acid, which inhibits the tricarboxylic acid cycle by affecting succinate dehydrogenase (Lu et al. 2018), increased about 3.28-fold and 6.34-fold in liver and muscle tissues, respectively, during freezing when compared with the activity period (supplementary fig. S9A and B, Supplementary Material online). The suppression of multiple mitochondrial pathways suggests a dramatic reduction in the metabolic rate in the Amur sleeper in response to freezing. Notably, given that mitochondria are also the main generators of cellular ROS, the inhibition would lead to a lower risk of ROS damage to macromolecules and organelles, including proteins, DNA, mitochondria, and cytoskeleton.
In the liver and brain of the Amur sleeper, genes that were downregulated during the freezing period, compared with the activity period, were also enriched in pathways related to transcription, translation, and cell division (e.g., spliceosome, ribosome, cell cycle) (fig. 2F; supplementary fig. S8A, Supplementary Material online), all of which are energy-expensive cell processes (Storey and Storey 2004). Studies in wood frogs and the western painted turtle have shown that cell cycle suppression is a general feature of freeze tolerance (Zhang and Storey 2012; Bradley Shaffer et al. 2013). Remarkably, the gene ccnd2, which encodes the G1/S specific cyclin D2, was positively selected in the Amur sleeper and is downregulated in the freezing period (fig. 2F; supplementary table S11, Supplementary Material online). It has been reported that inhibition of ccnd2 expression arrests the cell cycle in the G1 phase (Xiao et al. 2021). In addition, Gadd45gip1 and hdac9, which are associated with the G1/S transition of the cell cycle (Li and Durbin 2011), were also identified as rapidly evolved genes (fig. 2F; supplementary table S12, Supplementary Material online). Moreover, a number of negative regulators of mTORC1, including deptor, ddit4, tsc1, and akt1 (Coronel et al. 2022), were significantly upregulated during the freezing period compared with the activity period (supplementary fig. S10, Supplementary Material online), indicating a strong depression of mTORC1 activity during freezing. Previous studies have suggested that reductions in mTORC1 activity may lead to the subsequent inhibition of protein translation and the cell cycle in hibernating animals (Logan et al. 2019; Dias et al. 2021). Furthermore, two mTOR pathway repressor, nrpl3 and ikkb (Lee et al. 2007; Baldassari et al. 2016), were identified as a positively selected gene and a rapidly evolving gene, respectively (fig. 2F; supplementary tables S11 and S12, Supplementary Material online). Thus, genes that experienced marked genetic alterations may participate in the suppression of energy-expensive cell processes in the Amur sleeper during freezing period, although their putative functions still await further verification. Collectively, our results suggest that hypometabolism is an adaptive response in the Amur sleeper to support freezing, yielding a minimization of energetic needs and ROS generation.
Changes in Genes and Metabolites Associated With Cellular Stress Responses
Both freezing and thawing have important consequences, including physical ice damage, anoxia/reoxygenation, dehydration/rehydration, and ischemia/reperfusion, all of which can expose cells and organs to quick bursts of ROS and severe oxidative stress (Storey and Storey 2017; Zhang, Gupta et al. 2021). Therefore, an integrated suite of cellular stress responses is of paramount importance to freeze-tolerant species. Core elements of the cellular stress responses include antioxidant defense, protein stabilization by chaperones, and DNA damage repair (Kültz 2005). Our metabolomic analysis of the liver and muscle tissues showed substantial increase in the content of the antioxidants vitamin C (Vc), Vb3, Vb6 and hypotaurine during the freezing and thawing periods, with the most pronounced changes observed for Vc content (a 150.2-fold increase) in the liver and Vb3 (a 11.7-fold increase) in the muscle during freezing (fig. 2D and E; supplementary figs. S11 and S12, Supplementary Material online). The metabolites glutathione oxidized (GSSG), hypoxanthine, and 8-hydroxyguanosine (8-OHG), which serve as markers of oxidative stress (Joanisse and Storey 1996; Hira et al. 2014), were notably increased in muscle tissue during the freezing and thawing periods (fig. 2E; supplementary fig. S12, Supplementary Material online). In contrast, levels of 8-OHG and total lipid peroxidation (LPO) in liver tissue decreased significantly, probably due to the observed hypometabolism and increases in antioxidant levels that suppress ROS (fig. 2D; supplementary fig. S11, Supplementary Material online, and see below). In fact, the liver as the center of glucose homeostasis and lipid metabolism plays a vital role in coping with freezing imposed stresses, likely is more prone to oxidative stress and requires enhanced antioxidant defenses, chaperones, and DNA repair mechanisms to maintain cellular homeostasis. Additionally, the observed hypometabolism could also contribute to the low levels of oxidative stress.
Our transcriptome profiles also identified a set of genes crucial for antioxidation that were upregulated in the freezing and thawing periods. For example, gene encoding the core antioxidant enzyme superoxide dismutase (sod3) was strongly upregulated in all three tissues, whereas the glutathione peroxidases genes (gpx1, gpx4a) were upregulated in the liver and muscle tissues. Other enzymes or proteins that have crucial antioxidant capacity, including the glutathione S-transferase (GST) isozymes (gsta4, mgst3, mgst3b) and ferritin (fth1) (Tsuji et al. 2000), were also significantly upregulated in a tissue- and context- (activity, freezing, and thawing) specific manner (fig. 2F). These regulation profiles can reflect perturbations from homeostasis and cellular damage in each of the organ to generate tissue-specific functions to cope with the different stresses, as previously documented in multiple other freeze-tolerant organisms (Toxopeus and Sinclair 2018; Wu et al. 2018). Collectively, these cellular stress responses by enzymatic and non-enzymatic antioxidants to address freezing/thawing support the hypothesis of a preparative mechanism against oxidative stress (POS) (Giraud-Billoud et al. 2019) whereby organisms enhance antioxidant defenses during a number of stresses (e.g., freezing/thawing, anoxia/reoxygenation, torpor/arousal, etc.), and suggest that tissue-specific regulation of the antioxidant system is a part of the adaptive mechanisms that allow Amur sleeper to survive prolonged freezing and recover after thawing with the avoidance of oxidative damage.
Heat shock proteins are the best-known protein chaperones that promote the refolding of denatured or misfolded proteins and prevent the denaturation and aggregation of unfolded proteins (King and MacRae 2015). Our transcriptomic profiles revealed that a number of HSP genes (including hspa13, hspb8, hsp70-1, hspa12a, hsp10, hspa8, hspa14, hspb1, hsp90ab1, hspb6, and hsp90aa1) were significantly upregulated during freezing and thawing, especially in the muscle (fig. 2F). Moreover, genes involved in DNA damage repair, including ercc6, gadd45g, and gadd45b, were also upregulated in a tissue-dependent manner. Some cellular stress response-associated genes (shc1, oxr1, hspa12b, ercc8, and ercc6l2) experienced positive selection and rapid evolution in the Amur sleeper (fig. 2F; supplementary tables S11 and S12, Supplementary Material online). Shc1 and oxr1 have been shown to be important for oxidative stress protection (Koch et al. 2008; Sanada et al. 2014), whereas ercc8 and ercc6l2 are essential factors in the transcription-coupled repair (TCR) pathway that performs DNA excision repair (Fousteri and Mullenders 2008; Tummala et al. 2018). Overall, genetic modifications and regulation profiles of the stress associated genes and metabolites provide strong evidence for the involvement of multiple cellular defense strategies in the Amur sleeper during freezing and thawing. Further studies are needed to define the function of these genes and proteins in cellular stress responses during freezing and thawing.
Changes in Genes Associated With the Cytoskeleton
The cytoskeleton, which comprises actin filaments (microfilament, MF), microtubules (MT), and intermediate filaments (IF), not only help control cell shape, bear external forces, and maintain the stability of internal cell structures, but also enables cells to carry out essential functions such as division and movement (Alberts et al. 2002; Fletcher and Mullins 2010). During freezing, the cytoskeleton is subjected to direct physical stress from extracellular ice, substantial changes in cell volume, and the presence of destructive ROS (Storey and Storey 2017; Ou et al. 2018). Importantly, multimeric cytoskeletal components quickly depolymerize at near-freezing temperatures, resulting in catastrophic functional impairment (Ou et al. 2018). Therefore, adaptive modifications of cytoskeletal-related genes are likely necessary to mitigate freeze-associated cytoskeletal damage. As expected, genomic comparisons showed that 285 gene families were significantly expanded in the Amur sleeper compared with other teleosts (supplementary table S13, Supplementary Material online). Several of the GO terms significantly enriched in these expanded gene families are associated with microtubules (e.g., microtubule associated complex, microtubule organizing center attachment site, LINC complex) (fig. 3A). Notably, three expanded gene families (kntc2, spc24, and haus3) are components of the kinetochore-microtubule attachments that ensure proper chromosome segregation during cell division, which is crucial for cellular viability (Cheeseman and Desai 2008; Umbreit et al. 2012; Zhang et al. 2019) (fig. 3B; supplementary fig. S13, Supplementary Material online). This is consistent with a requirement for stable kinetochore-microtubule attachments at low temperature, which confer resistance to microtubule depolymerization during cold exposure whereas unattached microtubules are depolymerized (Margolis et al. 1990).
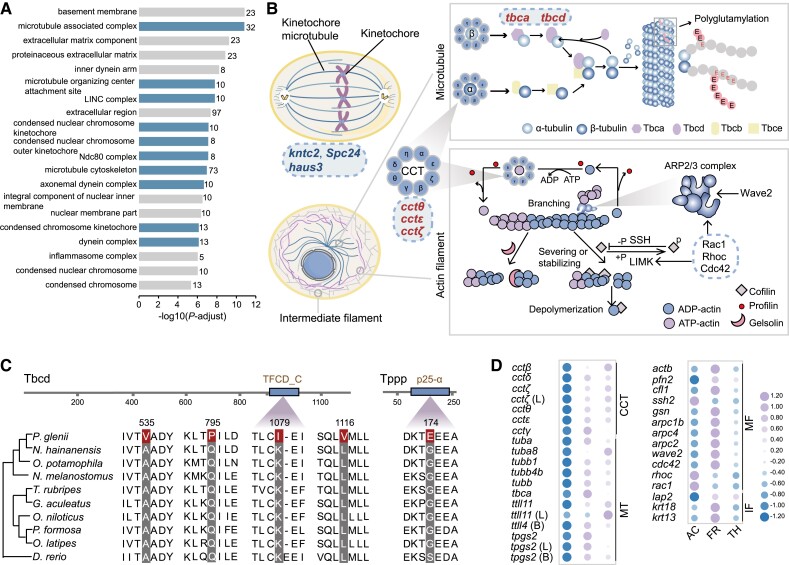
Fig. 3.
Changes in genes related to the cytoskeleton. (A) The top 20 significantly enriched GO terms for the expanded gene families are shown for cellular components (GO terms related to the microtubule are shown in blue). (B) Schematic showing cytoskeletal composition, microtubules in mitosis, microtubule assembly, and the regulation of the actin cytoskeleton. Rapidly evolving genes are marked in red and expanded gene families are marked in blue. (C) Alignments of the positively selected genes tbcd and tppp from several teleosts. Positively selected sites are highlighted in red. (D) Changes in the expression levels of genes related to the cytoskeleton during three periods (AC, activity; FR, freezing; TH, thawing). Purple represents higher expression levels, and blue represents lower expression levels. L and B represent liver and brain tissue, respectively (CCT, CCT complex; MT, microtubule; MF, microfilament; IF, intermediate filament).
Across all identified rapidly evolving genes, the most significantly overrepresented KEGG pathways were involved in the regulation of the actin cytoskeleton (supplementary fig. S14, Supplementary Material online). Of particular interest, five chaperone genes (cctθ, cctζ, cctɛ, tbca, and tbcd) exhibited significant signatures of elevated rates of evolution (fig. 3B; supplementary table S12, Supplementary Material online). The genes cctθ, cctζ, and cctɛ encode different subunits of the chaperonin-containing t-complex polypeptide 1 (CCT complex) (Leitner et al. 2012) and are essential for the biogenesis of actin and tubulin, which are required for the assembly of actin filaments and microtubules (Llorca et al. 2001). The genes tbca and tbcd encode tubulin-specific chaperones (TBCs) that bring together α- and β-tubulin subunits to form the assembly-competent heterodimers, which are required for the biogenesis of tubulin (Tian and Cowan 2013). Tbcd was further identified as being positively selected and having four sites with evidence of positively selection, with one of them located in the TFCD-C domain (fig. 3C). Moreover, the gene tppp encoding tubulin polymerization-promoting protein, a microtubule regulatory protein that promotes the incorporation of tubulin heterodimers into growing microtubule filaments (Hlavanda et al. 2002), was also identified as a positively selected gene with a mutation (G174E) in the p25-alpha domain (fig. 3C). Tppp is a multiple effector of the microtubule organization that not only protects the microtubules from cold depolymerization via its microtubule bundling activity, but also increases the level of acetylation by inhibiting the activity of histone deacetylase 6 (Tőkési et al. 2010), rendering microtubules more mechanically stable (Xu et al. 2017). As the rapidly evolving genes and positively selected genes identified in the Amur sleeper are known to be important for cytoskeletal stability, especially in tubulin biogenesis, this suggests that these genes have been co-opted to prevent irreparable structural damage to the cytoskeletal system and maintain normal function over the freeze/thaw cycle.
Our transcriptomic analyses further clarified the dynamic rearrangement of cytoskeleton in the Amur sleeper. Multiple genes associated with three components of the cytoskeleton (MT, MF, and IF) exhibited significant fluctuations in expression levels in freezing and thawing periods, especially in the muscle tissue. Genes encoding the subunits of the CCT complex (cctβ, cctδ, cctζ, cctθ, cctɛ and cctγ) were significantly upregulated during freezing and thawing (fig. 3D), indicating the potential reassembly of dissociated cytoskeletal monomers. Similarly, the upregulation of the CCT complex to prevent cold-induced actin depolymerization was also reported in two species of insects (Kayukawa and Ishikawa 2009; Zhang et al. 2011). With respect to microtubules, genes encoding α-tubulin (tuba), β-tubulin (tubb1, tubb4b, tubb) and tubulin-folding cofactor (tbca) were upregulated in muscle during freezing and thawing (fig. 3D). This upregulation likely contributes to the maintenance of the tubulin pool, which becomes more important at low temperatures due to cold-induced microtubules destabilization (Des Marteaux et al. 2018). In addition, genes involved in the post-translational modification of tubulins (i.e., tpgs2, ttll11, and ttll4) (Wang et al. 2022) also exhibited tissue-specific patterns of elevation (fig. 3D). Such alterations may help to promote microtubule cold-stability. For more cold-resistant actin filaments, genes encoding actin (actb) and actin-binding proteins profilin (pfn2), gelsolin (gsn), cofilin (cfl1), and the arp2⁄3 complex (arpc1b, arpc4 and arpc2) (Pollard 2016) were significantly upregulated in muscle tissue during freezing and decreased upon thawing. Moreover, the upstream regulators of cofilin and the arp2⁄3 complex, such as ssh2, wave2, cdc42, rac1, and rhoc (Campellone and Welch 2010; Mizuno 2013), were also differentially expressed, supporting dynamic rearrangements of the actin cytoskeleton (fig. 3B and D). Several genes that participate in the assembly or post-translational modification of intermediate filaments, such as krt13, krt18, and lap2 (Naetar et al. 2017; Wu et al. 2017), were significantly differentially expressed in muscle tissue during freezing and thawing (fig. 3D). Real-time quantitative PCR (RT-qPCR) analysis of these genes recovered expression patterns consistent with those determined using the transcriptome analysis (supplementary fig. S15A and B, Supplementary Material online). We speculate that these specific regulatory and genetic changes play important roles in cytoskeleton stability to avoid the harmful effects associated with freezing and thawing. These results provide evidence for adaptive modifications of the cytoskeletal components in maintaining cell viability over a freeze/thaw cycle.
Cryoprotectants and Transmembrane Transporters
A well-known strategy for freezing survival is the accumulation of large quantities of low-molecular-weight cryoprotectants, which not only reduce ice formation but also limit cell shrinkage via osmic effects (Storey and Storey 1986, 1996b). Sugars (e.g., glucose and trehalose), polyhydric alcohols (e.g., glycerol and sorbitol) and amino acids (e.g., proline) are common colligative (concentration-dependent) cryoprotectants found in cold-hardy amphibians and insects, and urea contributes in some cases in amphibians (Storey and Storey 2017; Toxopeus and Sinclair 2018). In fact, cryoprotectants may be interchangeable and function on a colligative basis, or they may perform non-colligatively, with each cryoprotectant molecule having a unique cryoprotective role, such as protecting membrane or/and protein stability (Toxopeus et al. 2019; Kennedy et al. 2020). Our metabolomic analyses identified a variety of putative new cryoprotectants that showed distinct fluctuations in content in the muscle and liver of the Amur sleeper. Unexpectedly, glucose was the most abundant sugar in both the liver and muscle tissue, but only showed an apparent increase (3.6-fold) in the muscle during the freezing period compared with the activity period (fig. 4A). Myo-inositol and glycerol were the most abundant polyhydric alcohols in both tissues but with no significant fluctuations among the three periods. In contrast, arabitol increased significantly between the activity and freezing periods in muscle (5.8-fold; fig. 4A), indicating an obvious freeze response. Despite a global repression of amino acid metabolism, the aspartic acid content of muscle increased significantly (7.88-fold) in the freezing period compared with the activity period (fig. 4A), suggesting that aspartic acid potentially plays a protective role. The patterns observed in the Amur sleeper, wherein some putative cryoprotectants accumulated independent of freezing stimulation, are likely indicative of a seasonal acquisition, consistent with the variation of glucose and glycol levels in amphibians (Layne and Jones 2001; Irwin and Lee 2003; Niu et al. 2018).
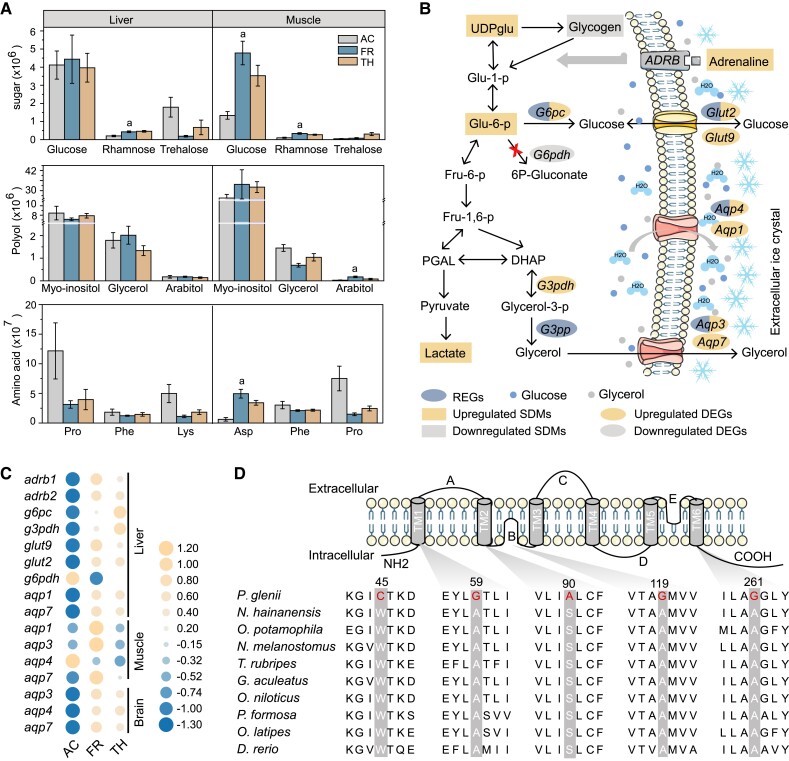
Fig. 4.
Putative cryoprotectants and their movement with water. (A) The content of three most abundant sugar, polyhydric alcohols (polyols), and amino acids in the liver and muscle during freezing (AC, activity; FR, freezing; TH, thawing). Lowercase letters above the bar correspond to significant increases in freezing compared with activity. Error bars represent the mean ± SD (n = 6). (B) Schematic showing the glucose and glycerol biosynthetic pathways, as well as the movement of water and cryoprotectants. (C) Patterns of differential expression among cryoprotectant-associated genes. Orange represents higher expression levels, and blue represents lower expression levels. (D) Alignment of multiple aqp4 amino acid sequences from the Amur sleeper and other nine teleosts. Specific mutations are shown in red.
During freezing, glycogenolysis is clearly the dominant pathway producing cryoprotectants glucose and polyhydric alcohols. In the liver of the Amur sleeper, we observed a sharp drop in glycogen (as much as 133-fold), in conjunction with obvious rise in UDP-glucose (6.40-fold) and glucose-6-phosphate (2.92-fold) during the freezing period compared with the activity period (fig. 4B; supplementary fig. S16, Supplementary Material online). Meanwhile, the significant lactate accumulation suggests that glucose serves as the fuel for anaerobic metabolism (supplementary fig. S16, Supplementary Material online). Adrenaline-mediated triggering of β-adrenergic signaling (the fight-or-flight response) has been linked with rapid activation of glycogenolysis (Storey and Storey 1996a). Consistent with this, the adrenaline content rose 2.20-fold; simultaneously, beta-1 adrenergic receptor (adrb1) and beta-2 adrenergic receptor (adrb2) were significantly upregulated during the freezing period compared with the activity period (supplementary fig. S16, Supplementary Material online; fig. 4B and C). Glucose-6-phosphatase (g6pc), which converts Glu-6-p to glucose in the terminal step of glucose synthesis (Foster et al. 1997), was also significantly upregulated (fig. 4B and C). In contrast, glucose 6-phosphatedehydrogenase (g6pdh) was noticeably downregulated, preventing Glu-6-p from being used for other purposes (Cowan and Storey 2001). Importantly, g6pc was identified as a rapidly evolving gene and had nine specific amino acid replacements (fig. 4B; supplementary fig. S17A, Supplementary Material online), four of which were located in the transmembrane domains (supplementary fig. S17B, Supplementary Material online). Glycerol-3-phosphate phosphatase (g3pp), a crucial enzyme for glycerol biosynthesis (Raymond 2015) was also identified as a rapidly evolving gene and had nine specific amino acid mutations (fig. 4B; supplementary fig. S18, Supplementary Material online). These genetic changes might promote glucose and glycerol synthesis in the liver; the glucose and glycerol thus produced may be exported and circulated throughout the body for cryoprotection before freezing.
The formation of ice crystals and the accumulation of cryoprotectants increase osmolality and have profound effects on cellular water content (Storey and Storey 2013). Aquaporins (AQPs) play an essential role in rapid osmoregulation, as they facilitate the diffusion of water and osmolytes across cell membranes (Hill et al. 2004). There are two subsets of the AQP family: AQP1, AQP2, AQP4, and AQP5 are permeated only by water, whereas AQP3, AQP7, and AQP9 can also be permeated by glycerol and even larger solutes (Verkman and Mitra 2000). We identified aqp3 and aqp4 as rapidly evolving genes in the Amur sleeper (fig. 4B). In particular, the amino acid sequence of aqp4 has five specific mutations that cause amino acid polarity changes, including three alanine (A) to glycine (G) substitutions at sites that were highly conserved in the corresponding proteins of other fish species (fig. 4D). Furthermore, aqp1, aqp3, aqp4, and aqp7 were generally upregulated during the freezing period across all tissues examined (fig. 4C), providing functional evidence that these AQPs are crucial for the control of ice content, cell volume, and fluid homeostasis in the Amur sleeper (fig. 4C). Moreover, the transcription levels of two glucose transporter glut2 and glut9 (Augustin et al. 2004; Mueckler and Thorens 2013) in liver tissue were increased significantly during the freezing period when compared with the activity period (fig. 4B and C), suggesting a rapid export of cryoprotective glucose during the onset of freezing. Moreover, glut2 was identified as a rapidly evolving gene in the Amur sleeper and was also upregulated during thawing (fig. 4B; supplementary table S12, Supplementary Material online), which might facilitate reuptake of glucose by hepatocytes to restore glycogen pools and mitigate hyperglycemia (Storey and Storey 1986; Mueckler and Thorens 2013). The dynamic regulation of glut2 expression is crucial for survival during freeze–thaw cycles in organisms that employ glucose as a cryoprotectant, as has been demonstrated in amphibians (Storey and Storey 1988; Rosendale et al. 2014). The expression patterns of these differentially expressed cryoprotectant and transporter genes were validated using RT-qPCR (supplementary fig. S15D and E, Supplementary Material online). Together, our results indicate that genes associated with cryoprotectant synthesis and transmembrane transporters contribute to freezing survival by facilitating cryoprotectant accumulation and osmoregulation. However, the effects of the mutations in these genes still require further investigation.
Gene Changes Correlated With Neuroprotective Adaptations
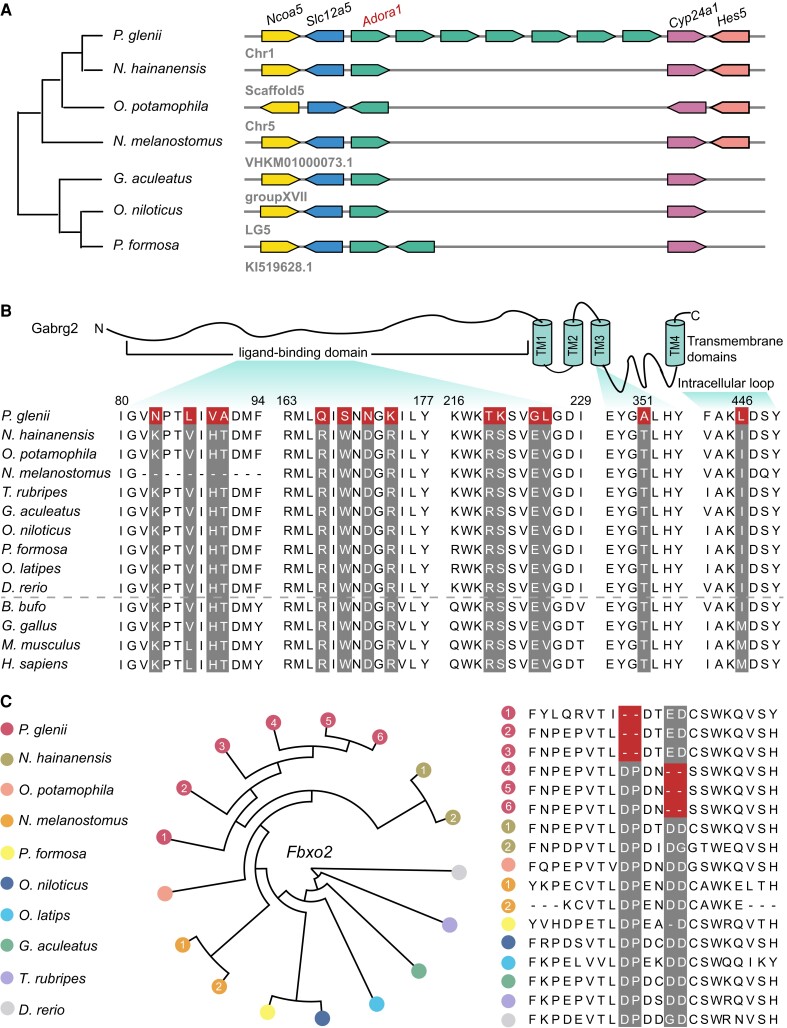
Freeze-tolerant animals endure a prolonged state of frozen dormancy with interrupted nerve transmission (Storey and Storey 2017). Thus, neuroprotective mechanisms controlling the entry to and exit from the dormant state, as well as maintaining cellular homeostasis, are vital for freezing survival, yet this subject has received almost no attention in ectothermic vertebrates. Interestingly, we found seven copies of the adenosine A1-receptor (adora) in the Amur sleeper genome. The adora genes are in a tandem array and situated between slc12a5 and cyp24a1 genes (fig. 5A). The adenosine A1-receptor is generally linked to the inhibition of the release of neurotransmitters and has its greatest inhibitory effects on the excitatory glutamatergic system (Dunwiddie and Masino 2001). It has been demonstrated that stimulation of adora in the central nervous system (CNS) plays a key role in the entrance into hibernation in mammals (Tamura et al. 2005). The observed expansion of adora might enhance the suppression of neurotransmission and thus, be critically important for the induction or maintenance of dormancy in the Amur sleeper. Moreover, we found that gabrg2, encoding the γ2 subunit of gamma-aminobutyric acid receptor type A that mediates the inhibition of the CNS by combining with gamma-aminobutyric acid (GABA), possesses a strong signal of positive selection in the Amur sleeper. Remarkably, a total of 14 positively selected sites were detected in the gabrg2 gene, which is highly conserved in human, mouse, chicken, and frog, with twelve of those sites located in the extracellular ligand-binding domain (fig. 5B). Our transcriptomic analyses further identified multiple genes (glna, slc38a2, slc38a3, pkaca, and pkcb) in the GABAergic synaptic pathway that were upregulated in the brain tissue during the freezing and thawing periods (supplementary fig. S19, Supplementary Material online). Zebrafish exposed to GABA-enhancing drugs and mice fed black-sticky-rice giant embryo containing high levels of GABA exhibited decreased levels of anxiety (Stewart et al. 2011; Jung et al. 2017). Thus, the observed mutations in gabrg2 and the upregulation of genes associated with the GABAergic synaptic pathway may have important anti-anxiety effects and facilitate the initiation of dormancy.
Fig. 5.
Expanded and positively selected genes related to nerve activity. (A) Expansion of the adenosine A1-receptor (adora) gene family. Seven copies of the adora gene are arranged in tandem in the Amur sleeper genome. (B) Schematic representation of the gabrg2 protein, and alignment of the Amur sleeper gabrg2 amino acids sequence with the corresponding sequences of 13 vertebrates, including nine teleosts (Bufo bufo, frog; Gallus gallus, chicken; Mus musculus, mouse; Homo sapiens, human). The 14 positively selected sites in the Amur sleeper gabrg2 sequence are highlighted in red. (C) F-box only protein 2 (fbxo2) gene family has a significant expansion in the Amur sleeper genome (left) compared with other teleosts. The Amur sleeper has six copies of this gene, whereas other species only have 1 or 2. Alignment of six copies of fbxo2 from Amur sleeper with the corresponding proteins from other teleosts identified two specific 2-amino acid deletions (right).
In parallel with the receptors that mediate neural inhibition, we found the metabotropic glutamate receptor 5 (mglur5), which modulates cell excitability and synaptic transmission (Niswender and Conn 2010), was also under positive selection. In addition to neuronal excitability, mglur5 also play important roles in the induction of long-lasting forms of synaptic plasticity and long-term depression (Niswender and Conn 2010). Two positively selected sites (C607L and L608T) in mglur5 were identified in the conserved dimer interface (supplementary fig. S20, Supplementary Material online), which plays a critical role in activation via a massive conformational change (Llinas del Torrent et al. 2019). The two mutations may affect its activation, and thus have implications for neural activity regulation while frozen and for reactivation upon thawing.
Autophagy is a conserved degradation pathway carried out by lysosomes, which plays a vital role in the clearance of cellular damage and nutrient recycling under stressful conditions (Settembre et al. 2013). A recent study suggested that autophagy is necessary for neuronal survival and function in the Amur sleeper during and after wintering (Gordon et al. 2020). However, oxidative stress can lead to lysosomal membrane permeabilization (LMP), resulting in leakage of hydrolases into the cytosol triggering cell death (Papadopoulos and Meyer 2017). Previous studies have provided evidence of LMP in neurons exposed to the cold (Ou et al. 2018) and established that lysophagy can clear damaged lysosomes though a selective from of macroautophagy as a critical response to LMP in the CNS (Papadopoulos and Meyer 2017). Notably, our genomic analyses found that the lysophagy related gene f-box only protein 2 (fbxo2) experienced significant expansion with an increase in copy number (to six copies) in the Amur sleeper relative to other species (which typically have one to two copies) (fig. 5C). A multiple sequence alignment identified a pair of 2-amino acids deletions specific to the Amur sleeper, with each copy of fbxo2 including one of these deletions that might affect the three-dimensional structure of the encoded proteins (fig. 5C; supplementary fig. S21, Supplementary Material online). Fbxo2 is a brain enriched glycan-binding F-box protein that functions as a subunit of the ubiquitin protein ligase complex SCF (Liu et al. 2020). It has been demonstrated that fbxo2 is recruited to damaged lysosomes and participates in their clearance, and that fbxo2-knockout mice show a delayed lysophagy progression and increased neuronal cell death after lysosomal damage (Liu et al. 2020). Our transcriptomic analyses also showed high expression levels of multiple fbxo2 copies were maintained in brain tissue in freezing and thawing periods (supplementary fig. S22, Supplementary Material online). Thus, fbxo2 gene expansion might indicate enhanced lysophagy function in the Amur sleeper to maintain neuronal homeostasis during freezing and thawing. Further functional studies on these genetic changes may yield insights into how nerve activity regulation is coordinated and maintained upon freezing and thawing.
Conclusion
Freeze tolerance, a fascinating example of a complex animal adaptation, has been extensively investigated in multiple hibernating species including reptiles, amphibians and insects. However, the genetic basis of freezing survival remains unclear. We used a multi-omics approach to characterize this adaptation in the only known freeze-tolerant fish species, the Amur sleeper. Our analyses revealed a suite of coordinated molecular adaptations in the Amur sleeper related to multiple cell/organ preservation strategies that likely mitigate the detrimental effects of freezing and thawing. Potential evolutionary innovations associated with cytoskeletal stability, cryoprotectant synthesis, transmembrane transport, and neuroprotective adaptation were identified, which may be key to the Amur sleeper's freezing survival. The identification of these genetic changes provides a framework for further functional characterization. Considering the present study only focuses on genetic changes of orthologues, future studies examining other genomic features such as species-specific genes, non-coding genes, convergent evolution, and epigenetic modifications are worth to be explored. Overall, this study not only provides useful genomic resources and insights into the molecular adaptation of freeze tolerance in ectothermic vertebrates, but also has potential implications for the development of better cryopreservation technologies and the exploration of the causes of mental diseases in biomedicine, since many genes reported here are associated with multiple neurological and neuropsychiatric disorders in humans.
Materials and Methods
Genome Sequencing and De Novo Assembly
Wild Amur sleeper and N. hainanensis individuals were collected from Heilongjiang and Guangxi Province, China, respectively. Tissues used for obtaining the genome sequences were sampled from a single individual of each species. All experiments in this study were approved by the Institutional Animal Care and Use Committee of Institute of Hydrobiology, Chinese Academy of Sciences (Approval ID: Y21304506) and were conducted in compliance with relevant guidelines. Muscle tissues from the Amur sleeper and N. hainanensis were used for DNA extraction and genome sequencing. For the Amur sleeper, a total of 70.29 Gbp of long reads and 101.02 Gbp of short reads were generated. For N. hainanensis, a total of 86.06 Gbp of long reads and 121.71 Gbp of short reads were generated. The long reads were sequenced using the PrometlON DNA sequencer on the Oxford Nanopore platform and the short reads were generated on the BGISEQ-500 platform. For Hi-C sequencing of the Amur sleeper, liver tissue was used for the extraction of DNA for library preparation. Hi-C libraries were sequenced on the Illumina NovaSeq platform, and a total of 95.60 Gbp Hi-C reads were generated (supplementary table S1, Supplementary Material online).
We adapted the KmerFreq_AR program from the SOAPdenovo2 package, which is based on k-mer distribution, to estimate genome size from about 45 Gb of BGISEQ-500 short reads filtered using fastp (Chen et al. 2018). The estimated genome size of the Amur sleeper and N. hainanensis were 827.25 Mb and 840.86 Mb, respectively (supplementary table S2, Supplementary Material online). First, Nanopore long reads of the two species were corrected using the NextCorrect modules of NextDenovo (https://github.com/Nextomics/NextDenovo). For the Amur sleeper, we used wtdbg2 (Ruan and Li 2020), Flye (Kolmogorov et al. 2019), and Smartdenovo (https://github.com/ruanjue/smartdenovo) for de novo assembly with corrected long reads due to the high heterozygosity estimated from the genome characteristics (supplementary fig. S1 and table S2, Supplementary Material online). Next, we applied three rounds of polishing using filtered short reads with Pilon1.23 (Walker et al. 2014). We also filtered the redundant contigs caused by the high heterozygosity using the script fasta2homozygous.py from Redundans (https://github.com/lpryszcz/redundans). The quality of these three genomes was assessed. Finally, Hi-C reads were aligned to the best assembly version via Bowtie2 (Langmead and Salzberg 2012), and then Lachesis (Burton et al. 2013) was used to anchor the draft genome onto 22 chromosomes. For N. hainanensis, the Nanopore reads were assembled using wtdbg2 and then three rounds of polishing using short reads were applied. To estimate the quality of the genomes from the two species, short reads were mapped back to the genome using BWA-MEM (Li and Durbin 2009). Completeness of the two genomes was evaluated using Busco v5 (Manni et al. 2021) with the actinopterygii_odb10 database.
Genome Prediction Annotation
We combined RepeatMasker v4.06 (Tarailo-Graovac and Chen 2009) with RepeatProteinMask v4.06 for homology repeat sequence prediction by aligning the genome sequences against the RepBase library. For de novo repeat prediction, we adopted RepeatModeler v1.08 along with LTR-FINDER v1.06 (Xu and Wang 2007) based on the de novo repeat library. We used three different methods, namely, ab initio annotation, homology annotation, and transcriptome-based annotation, to predict the whole gene set for the two genomes. Briefly, Augustus (Stanke et al. 2008), Snap (Leskovec and Sosič 2016), and GeneScan (Burge and Karlin 1997) were used for de novo gene prediction based on the repeat masked genome sequences. The Augustus and Snap programs were trained with the transcript and zebrafish genome training sets, respectively. For homology-based annotation, protein sequences from Oreochromis niloticus, Oryzias latipes, Danio rerio, Anabas testudineus, Neogobius melanostomus, and Poecilla formosa were downloaded from Ensembl (supplementary table S14, Supplementary Material online) and aligned to the two genomes using the TBLASTN program. GeneWise (Birney et al. 2004) was used to identify accurate gene structures for the alignment produced by TblastN. In addition, GeMoMa (Keilwagen et al. 2016) was used for homology-based prediction with the zebrafish genome as a reference genome. For the transcriptome-based annotation, ten tissues, including muscle, skin, heart, liver, spleen, brain, eye, gonad, kidney, and blood from the same individual of Amur sleeper, and the above tissues without blood from the same individual of N. hainanensis were sampled for RNA extraction and transcriptome sequence. Finally, a total of 62.49 Gb RNA-seq reads for the Amur sleeper and 87.30 Gb for N. hainanensis were generated by BGISEQ-500 (supplementary table S15, Supplementary Material online). We also performed full-length transcriptome sequence, a total of 8,570,679 subreads with a mean length of 2,009 bp for the Amur sleeper and 16,621,922 subreads with a mean length of 1,719 bp for N. hainanensis were generated in PACBIO_SMRT platform (supplementary table S16, Supplementary Material online). BGISEQ-500 RNA-seq reads were mapped to the genomes using Hisat2 (Kim et al. 2015), and then transcripts were generated using StringTie (Pertea et al. 2015). Full-length transcriptome data were used to construct consensus sequences using IsoSeq3 (https://github.com/PacificBiosciences/IsoSeq), and subsequently mapped to the genomes with Gmap (Wu and Watanabe 2005). Both types of transcripts were then processed with PASA (Haas et al. 2003) to obtain the final results. Finally, all gene models generated from these three strategies were integrated with EVidenceModeler (EVM) (Haas et al. 2008). Functional annotations of the predicted gene sets were obtained by mapping to public functional databases, including SwissProt, NCBI-Nr, KEGG, GO, and InterPro.
Syntenic Relationship With the Odontobutis potamophila Genome
To evaluate genome collinearity of the Amur sleeper with its close relatives, we assembled and annotated the Od. potamophila genome based on raw data downloaded from NCBI (Jia et al. 2021). First, coding sequences from the 22 autosomal chromosomes of the two species were aligned by LAST v942 (Kielbasa et al. 2011). These results were then subjected to MCscan (Tang et al. 2008) to identify syntenic blocks.
Phylogenetic Analysis
In addition to the Amur sleeper and N. hainanensis, genomes of eight other teleosts, including Od. potamophila, Or. niloticus, Oryzias latipes, D. rerio, N. melanostomus, P. formosa, Gasterosteus aculeatus, and Takifugu rubripes (supplementary table S14, Supplementary Material online) were used to perform comparative genomic analyses. First, we identified orthologous gene clusters using OrthoFinder v2.3.4 (Emms and Kelly 2015) with default parameters. A total of 4,469 single-copy genes were identified in the 10 species. Protein sequences for each single-copy orthologue were aligned using MAFFT v7.310 (Katoh and Standley 2013) and the corresponding coding sequence alignments were obtained with pal2nal v14 (Suyama et al. 2006). We removed poorly aligned regions of each coding sequence alignment using Gblocks v0.91b (Talavera and Castresana 2007) with the codon model. Alignments of less than 50 codons were discarded (Wu et al. 2021). All filtered coding sequence alignments were concatenated into super-genes for each species to construct a phylogenetic tree using RAXML v8.2.4 (Stamatakis 2014) with 1,000 ultrafast bootstrap replicates. To estimate divergence times, MCMCtree from the PAML software package was performed on the inferred phylogenetic tree with D. rerio as the outgroup and fourfold degenerate sites (4D) extracted from the super-genes. We set three calibration time points (Or. niloticus–Oryzias latipes: ∼88–139 Ma; N. melanostomus–P. glenii: ∼59–89 Ma; G. aculeatus–D. rerio: ∼206–252 Ma) taken from the TimeTree database to calibrate the calculated divergence times.
Inference of Demographic History
We inferred demographic histories for the Amur sleeper and N. hainanensis by pairwise sequentially Markovian coalescent (PSMC) analysis (Li and Durbin 2011). Short reads used for the genome assemblies were aligned to the two reference genomes using BWA-MEM (Li and Durbin 2009) with default parameters. To generate consensus diploid sequences for the two individuals separately, the SAMtools mpileup with bcftools and vcfutils.pl pipeline (https://github.com/lh3/psmc) was applied. We then used the fq2psmcfa program of PSMC to convert the consensus fastq files into psmcfa format, the input file format for PSMC. Finally, the effective population history was inferred using PSMC with 100 bootstraps and plotted using the psmc_plot.pl pipeline. Generation time for the two species were set as 2 and the substitution rate (μ) were estimated by r8s (5.50e-9 per site per generation for the Amur sleeper and 6.46e-9 per site per generation for N. hainanensis).
Mutation Rate and Strength of Natural Selection
We chose four closely related species for whole-genome synteny alignment using LAST v942 (Kielbasa et al. 2011) with the Od. potamophila genome sequence used as a reference. The aligned results were submitted to the subprogram “roast” of Multiz v3 (Blanchette et al. 2004) to generate one-to-one alignment sequences. A sliding window (100 kb) along the synteny alignment was applied to estimate the mutation rate. First, branch lengths for each window were estimated using RAxML (Stamatakis 2014) based on the neutral regions (repetitive sequences, regions located within genes, or sequences 3 kb upstream/downstream of them were filtered). The mutation rates were then calculated with r8s using the estimated branch lengths and divergence time previously estimated. In addition, we calculated ω (ka/ks) ratios based on 4,469 one-to-one orthologues from the ten teleosts. The free-ratio model, allowing a separate ω for each branch of a tree, from the codeml program in PAML (Yang 2007) was run on the concatenated orthologues and for each of the orthologues.
Gene Family Expansion and Contraction
We used CAFE v5 (De Bie et al. 2006) to test for expansion and contraction of gene families in the Amur sleeper based on the results from the OrthoFinder (Emms and Kelly 2015) analyses and the estimated divergence times from MCMCtree. If the copy number of the Amur sleeper was higher or lower than that found in its close ancestral branch lineage, then we identified this gene family as a substantially expanded or contracted gene family. Any gene family, with a false discovery rate (FDR) adjusted P-value < 0.05, was thought to experience significant expansion or contraction. Functional categories and pathways in significantly expanded gene families were identified by performing GO terms enrichment analysis and KEGG pathway enrichment analysis using KOBAS (http://kobas.cbi.pku.edu.cn/). GO terms or KEGG pathways with a P-value <0.05 were considered significantly enriched.
Identification of Positively and Rapidly Evolved Genes
All one-to-one orthologous genes were used to assess the contribution of natural selection on the Amur sleeper genome by calculating the ratios (ω) of nonsynonymous substitution (dN) to synonymous substitution (dS) using the PAML4 software package (Yang 2007). The two-ratio branch model (model = 2, NSsites = 0) with null model (model = 0, NSsites = 0) was used to detect rapidly evolving genes and branch-site model (model = 2, NSsites = 2, fix_omega = 0) with null model (model = 2, NSsites = 2, fix_omega = 1) was used to detect positively selected genes, with the Amur sleeper as the foreground branch. Likelihood ratio tests (LRTs) were applied to test the significance of the differences between alterative and null models for each orthologue. We treated a gene as a rapidly evolving gene when the FDR-adjusted P-value < 0.05, and it had a higher ω ratio in the Amur sleeper. Genes with P < 0.05 were considered as a positively selected gene. Finally, we removed false positive results by manual checking. The final positively selected genes and rapidly evolving genes were then assessed for enrichment of functional categories and pathways.
Transcriptome Sequencing and Analysis
Active, frozen and thawed Amur sleeper weighted 7–12 g were collected in the autumn (September 20, 2020), winter (February 25, 2021), and early spring (March 30, 2021), respectively in the same shallow bog in Qiqihaer, Heilongjiang, China (48.30°N, 125.25°E). Active samples were collected at the water temperature at 9 °C, frozen samples were collected from the bottom of the completely frozen bog (45 cm in depth, ice temperature at −2 °C, supplementary movie S1, Supplementary Material online). Frozen samples were dissected immediately after collection without thawing as cryoprotectants kept the bodies relatively soft. The thawing samples were collected at the surface of the bog as the ice melt (water temperature at 2 °C, supplementary movies S2, Supplementary Material online). After collection, brain, muscle, and liver tissues from nine individuals (three replicates per period) were dissected immediately and snap frozen in liquid nitrogen. RNA was extracted using TRIzol (Invitrogen, USA) and used for the generation of paired-end (PE) libraries. Each library was sequenced on an Illumina HiSeq platform with 150 bp PE reads. A total of 206.81 Gb clean data were generated (supplementary table S17, Supplementary Material online). Clean reads were mapped to the reference Amur sleeper genome using Hisat2 (Kim et al. 2015). StringTie (Pertea et al. 2015) was then used to generate gene expression level counts in fragments per kilobase of transcript per million fragments mapped. PCA was performed based on the expression pattern of all genes. To identify differentially expressed genes between the different periods, the number of reads mapped to gene regions was quantified by the FeatureCounts program (Liao et al. 2014), a part of the Subread package v2.0.0 (http://subread.sourceforge.net/). The R package DESeq2 (Love et al. 2014) was used for the detection of differentially expressed genes based on the read count table generated by featureCounts. An FDR-adjusted P-value < 0.05 and a 2-fold-change > 2 was set as the level of significance. Enrichments of KEGG pathways and GO terms of the differentially expressed genes were estimated using the annotations of all identified transcripts as a background.
Metabolite Extraction, Detection, and Analysis
Muscle and liver samples from 18 individuals (six replicates per period) were used for LC–MS/MS analysis. 50 mg of each sample was homogenized with 1,000 µl of ice-cold methanol/water (70%, v/v). The homogenate was centrifuged with 12,000 rpm at 4 °C for 10 min, and 400 μl of supernatant was stored in −20 °C refrigerator overnight. Then the supernatant was centrifuged at 12,000 rpm at 4 °C for 3 min to obtain 200 μl of supernatant. The metabolites detection was carried out by a combination of non-targeted detection (Ultra-performance liquid chromatography (UPLC) and Quadrupole-Time of Flight) and widely targeted detection (UPLC and Tandem mass spectrometry (MS/MS)). PCA was performed using the statistics function prcomp within R to identify general trends in content changes. Metabolites with VIP (variable importance in projection) >= 1, absolute Log2FC (fold-change) >= 1 and FDR P-value < 0.05 were regarded as significantly different metabolites. Identified metabolites were annotated using the KEGG Compound database (http://www.kegg.jp/kegg/compound/), and then annotated metabolites were mapped to the KEGG Pathway database (http://www.kegg.jp/kegg/pathway.html).
Real-time Quantitative PCR Assay
To validate differentially expressed genes across different periods of the Amur sleeper, RT-qPCR was performed. First-strand cDNA was synthesized from 1 ug of total RNA samples using Hifair II firstst-strand cDNA Synthesis SuperMix for RT-qPCR (Yeasen, China). The RT-qPCR was performed with the Hieff qPCR SYBR® Green Master Mix (Yeasen, China) and a LightCycler 480 II Instrument (Roche, Switzerland). Six biological replicates and three reaction replicates for each group were used. Expression values were calculated using the detected threshold cycle (Ct) value using the geNorm algorithm. Non-tissue-specific reference genes elongation factor alpha (EF1α) and 18S rRNA were selected as internal controls to normalize the relative expression levels. Statistical analysis was performed using an unpaired two-tailed Student's t test.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB31000000), the National Natural Science Foundation of China (32170480, 31972866, 32102797), Youth Innovation Promotion Association, Chinese Academy of Sciences (http://www.yicas.cn), and the Young Top-notch Talent Cultivation Program of Hubei Province. This research was supported by the Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences, China. We thank Dr Baosheng Wu and Dr Chenguang Feng for advice on analyses and in particular Mr Sheng Jiang and Mrs Shuxiang Zhou for collecting the samples.
Contributor Information
Haifeng Jiang, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi, China.
Wenqi Lv, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; University of Chinese Academy of Sciences, Beijing, China.
Ying Wang, Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China; Hubei Engineering Research Center for Protection and Utilization of Special Biological Resources in the Hanjiang River Basin, School of Life Sciences, Jianghan University, Wuhan, China.
Yuting Qian, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; University of Chinese Academy of Sciences, Beijing, China.
Cheng Wang, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; University of Chinese Academy of Sciences, Beijing, China.
Ning Sun, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; University of Chinese Academy of Sciences, Beijing, China.
Chengchi Fang, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China.
David M Irwin, Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada; Banting and Best Diabetes Centre, University of Toronto, Toronto, Canada.
Xiaoni Gan, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China.
Shunping He, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China; Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, China.
Liandong Yang, The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China; Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China.
Author Contributions
L.Y., H.J., S.H., Y.W., and X.G. designed and managed the project. H.J., W.L., and C.W. collected and prepared the Amur sleeper samples. W.L., and Y.Q. performed genome assembly and gene annotation. W.L., H.J., C.W., and Y.W. conducted the bioinformatic analysis and transcriptome, metabolism analysis. H.J., W.L., D.M.I., C.F., N.S., X.G., and L.Y. wrote and revised the manuscripts.
Conflict of interest statement. The authors declare no competing interests.
Data Availability
The sequence data have been deposited in the NCBI BioProject database with accession numbers PRJNA818152 (P. glenii), PRJNA818180 (N. hainanensis). The genome assembly files are under accession numbers JALDQB000000000 (P. glenii) and JALDNG000000000 (N. hainanensis).
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. The cytoskeleton and cell behavior. In: Molecular biology of the cell. 4th ed. New York: Garland Science. [Google Scholar]
- Augustin R, Carayannopoulos MO, Dowd LO, Phay JE, Moley JF, Moley KH. 2004. Identification and characterization of human glucose transporter-like protein-9 (GLUT9)-alternative splicing alters trafficking. J Biol Chem. 279:16229–16236. [DOI] [PubMed] [Google Scholar]
- Baldassari S, Licchetta L, Tinuper P, Bisulli F, Pippucci T. 2016. GATOR1 complex: the common genetic actor in focal epilepsies. J Med Genet. 53:503–510. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. Genewise and genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley Shaffer H, Minx P, Warren DE, Shedlock AM, Thomson RC, Valenzuela N, Abramyan J, Amemiya CT, Badenhorst D, Biggar KK, et al. 2013. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 14:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268:78–94. [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu RL, Kitzman JO, Shendure J. 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 31:1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. 2010. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 11:237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. 2003. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 83:1153–1181. [DOI] [PubMed] [Google Scholar]
- Chai L, Huang P, Bao X. 2020. Tolerant ability and physiological and biochemical responses of Chinese sleeper Perccottus glenii to icing up and hypoxia environment. J Dalian Ocean Univ. 35:218–222. (Chinese with English abstract). [Google Scholar]
- Cheeseman IM, Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 9:33–46. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The last glacial maximum. Science. 325:710–714. [DOI] [PubMed] [Google Scholar]
- Coronel L, Häckes D, Schwab K, Riege K, Hoffmann S, Fischer M. 2022. p53-mediated AKT and mTOR inhibition requires RFX7 and DDIT4 and depends on nutrient abundance. Oncogene. 41:1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo JP. 2019. Overwintering adaptations and extreme freeze tolerance in a subarctic population of the wood frog, Rana sylvatica. J Com Physiol B. 189:1–15. [DOI] [PubMed] [Google Scholar]
- Costanzo JP, Claussen DL. 1990. Natural freeze tolerance in the terrestrial turtle, Terrapene carolina. J Exp Zool. 254:228–232. [Google Scholar]
- Costanzo JP, Lee RE. 2013. Avoidance and tolerance of freezing in ectothermic vertebrates. J Exp Zool. 216:1961–1967. [DOI] [PubMed] [Google Scholar]
- Cowan KJ, Storey KB. 2001. Freeze-thaw effects on metabolic enzymes in wood frog organs. Cryobiology. 43:32–45. [DOI] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 22:1269–1271. [DOI] [PubMed] [Google Scholar]
- Des Marteaux LE, Stinziano JR, Sinclair BJ. 2018. Effects of cold acclimation on rectal macromorphology, ultrastructure, and cytoskeletal stability in Gryllus pennsylvanicus crickets. J Insect Physiol. 104:15–24. [DOI] [PubMed] [Google Scholar]
- Dias IB, Bouma HR, Henning RH. 2021. Unraveling the big sleep: molecular aspects of stem cell dormancy and hibernation. Front Physiol. 12:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwiddie TV, Masino SA. 2001. The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci. 24:31–55. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. Orthofinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher DA, Mullins RD. 2010. Cell mechanics and the cytoskeleton. Nature. 463:485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JD, Pederson BA, Nordlie RC. 1997. Glucose-6-phosphatase structure, regulation, and function: an update. Proc Soc Exp Biol Med. 215:314–332. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH. 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18:73–84. [DOI] [PubMed] [Google Scholar]
- Giraud-Billoud M, Rivera-Ingraham GA, Moreira DC, Burmester T, Castro-Vazquez A, Carvajalino-Fernández JM, Dafre A, Niu C, Tremblay N, Paital B, et al. 2019. Twenty years of the ‘preparation for oxidative stress’ (POS) theory: ecophysiological advantages and molecular strategies. Comp Biochem Phys A. 234:36–49. [DOI] [PubMed] [Google Scholar]
- Gordon RY, Santalova I, Mikheeva I, Karanova M, Khutsian S. 2020. Change in the state of neurons in the Medulla Oblongata of fish Perccottus glehni during wintering (ultrastructural and biochemical studies). Cell Tissue Biol. 14:209–217. [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Shachar-Hill B, Shachar-Hill Y. 2004. What are aquaporins for? J Membrane Biol. 197:1–32. [DOI] [PubMed] [Google Scholar]
- Hira HS, Samal P, Kaur A, Kapoor S. 2014. Plasma level of hypoxanthine/xanthine as markers of oxidative stress with different stages of obstructive sleep apnea syndrome. Ann Saudi Med. 34:308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavanda E, Kovács J, Oláh J, Orosz F, Medzihradszky KF, Ovádi J. 2002. Brain-specific p25 protein binds to tubulin and microtubules and induces aberrant microtubule assemblies at substoichiometric concentrations. Biochemistry. 41:8657–8664. [DOI] [PubMed] [Google Scholar]
- Irwin JT, Lee JRE. 2003. Geographic variation in energy storage and physiological responses to freezing in the gray treefrogs Hyla versicolor and H. chrysoscelis. J Exp Biol. 206:2859–2867. [DOI] [PubMed] [Google Scholar]
- Iwaya-Inoue M, Sakurai M, Uemura M. 2018. Survival strategies in extreme cold and desiccation. Singapore: Springer. [Google Scholar]
- Jia Y, Zheng J, Liu S, Li F, Chi M, Cheng S, Gu Z. 2021. A chromosome-level genome assembly of the dark sleeper Odontobutis potamophila. Genome Biol Evol. 13:evaa271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanisse DR, Storey KB. 1996. Oxidative damage and antioxidants in Rana sylvatica, the freeze-tolerant wood frog. Am J Physiol-Reg I. 271:R545–R553. [DOI] [PubMed] [Google Scholar]
- Jung W-Y, Kim S-G, Lee J-S, Kim H-K, Son B-G, Kim J-W, Suh J-W. 2017. Effect of feeding high gamma-aminobutyric acid-containing giant embryo black sticky rice (Oryza sativa L.) on anxiety-related behavior of C57BL/6 mice. J Med Food. 20:777–781. [DOI] [PubMed] [Google Scholar]
- Karanova M. 2009. Free amino acid composition in blood and muscle of the gobi Precottus glehni at the period of preparation and completion of hibernation. J Evol Biochem Physiol. 45:67–77. [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayukawa T, Ishikawa Y. 2009. Chaperonin contributes to cold hardiness of the onion maggot Delia antiqua through repression of depolymerization of actin at low temperatures. PLoS One. 4:e8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, Hartung F. 2016. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy JR, Harley CD, Marshall KE. 2020. Drivers of plasticity in freeze tolerance in the intertidal mussel Mytilus trossulus. J Exp Biol. 223:jeb233478. [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AM, MacRae TH. 2015. Insect heat shock proteins during stress and diapause. Ann Rev Entomol. 60:59–75. [DOI] [PubMed] [Google Scholar]
- Koch OR, Fusco S, Ranieri SC, Maulucci G, Palozza P, Larocca LM, Cravero AA, Farre SM, De Spirito M, Galeotti T, et al. 2008. Role of the life span determinant P66shcA in ethanol-induced liver damage. Lab Invest. 88:750–760. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37:540–546. [DOI] [PubMed] [Google Scholar]
- Kültz D. 2005. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 67:225–257. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layne JR Jr, Jones AL. 2001. Freeze tolerance in the gray treefrog: cryoprotectant mobilization and organ dehydration. J Exp Zool. 290:1–5. [DOI] [PubMed] [Google Scholar]
- Layne JR Jr, Kefauver J. 1997. Freeze tolerance and postfreeze recovery in the frog Pseudacris crucifer. Copeia. 260–264. [Google Scholar]
- Lee D-F, Kuo H-P, Chen C-T, Hsu J-M, Chou C-K, Wei Y, Sun H-L, Li L-Y, Ping B, Huang W-C, et al. 2007. IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 130:440–455. [DOI] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, et al. 2012. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 20:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskovec J, Sosič R. (snap co-authors) 2016. Snap: a general-purpose network analysis and graph-mining library. ACM T Intell Syst Tec. 8:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Benoit JB, Lopez-Martinez G, Elnitsky MA, Lee RE, Denlinger DL. 2009. Distinct contractile and cytoskeletal protein patterns in the Antarctic midge are elicited by desiccation and rehydration. Proteomics. 9:2788–2797. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. (bwa co-authors) 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (Psmc co-authors) 2011. Inference of human population history from individual whole-genome sequences. Nature. 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, He Y, Jiang J, Liu Z, Li C. 2018. Molecular systematics and phylogenetic analysis of the Asian endemic freshwater sleepers (Gobiiformes: Odontobutidae). Mol Phylogenet Evol. 121:1–11. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. [DOI] [PubMed] [Google Scholar]
- Liu EA, Schultz ML, Mochida C, Chung C, Paulson HL, Lieberman AP. 2020. Fbxo2 mediates clearance of damaged lysosomes and modifies neurodegeneration in the Niemann-Pick C brain. JCI Insight. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas del Torrent C, Casajuana-Martin N, Pardo L, Tresadern G, Pérez-Benito L. 2019. Mechanisms underlying allosteric molecular switches of metabotropic glutamate receptor 5. J Chem Inf Model. 59:2456–2466. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martín-Benito J, Gómez-Puertas P, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. 2001. Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J Structural Biol. 135:205–218. [DOI] [PubMed] [Google Scholar]
- Logan SM, Wu C-W, Storey KB. 2019. The squirrel with the lagging eIF2: global suppression of protein synthesis during torpor. Comp Biochem Phys A. 227:161–171. [DOI] [PubMed] [Google Scholar]
- Loomis S. 1995. Freezing tolerance of marine invertebrates. Oceanogr Mar Biol. 33:337–350. [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Zhao Y, Gao X, Wu J, Zhou H, Tang P, Wei Q, Wei Z. 2018. Effect of tricarboxylic acid cycle regulator on carbon retention and organic component transformation during food waste composting. Bioresour Technol. 256:128–136. [DOI] [PubMed] [Google Scholar]
- Lv W, Jiang H, Bo J, Wang C, Yang L, He S. 2020. Comparative mitochondrial genome analysis of Neodontobutis hainanensis and Perccottus glenii reveals conserved genome organization and phylogeny. Genomics. 112:3862–3870. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simao FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Rauch CT, Pirollet F, Job D. 1990. Specific association of STOP protein with microtubules in vitro and with stable microtubules in mitotic spindles of cultured cells. EMBO J. 9:4095–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K. 2013. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal. 25:457–469. [DOI] [PubMed] [Google Scholar]
- Mohr SM, Bagriantsev SN, Gracheva EO. 2020. Cellular, molecular, and physiological adaptations of hibernation: the solution to environmental challenges. Ann Rev Cell and Dev Biol. 36:315–338. [DOI] [PubMed] [Google Scholar]
- Mueckler M, Thorens B. 2013. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 34:121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naetar N, Ferraioli S, Foisner R. 2017. Lamins in the nuclear interior− life outside the lamina. J Cell Sci. 130:2087–2096. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. 2010. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol. 50:295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YG, Wang JJ, Men SK, Zhao YF, Lu SS, Tang XL, Chen Q. 2018. Urea and plasma ice-nucleating proteins promoted the modest freeze tolerance in Pleske's High altitude frog Nanorana pleskei. J Comp Physiol B. 188:599–610. [DOI] [PubMed] [Google Scholar]
- Ou J, Ball JM, Luan Y, Zhao T, Miyagishima KJ, Xu Y, Zhou H, Chen J, Merriman DK, Xie Z, et al. 2018. iPSCs from a hibernator provide a platform for studying cold adaptation and its potential medical applications. Cell. 173:851–863.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos C, Meyer H. 2017. Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and lysophagy. Curr Biol. 27:R1330–R1341. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 2016. Actin and actin-binding proteins. Cold Spring Harbor Perspect Biol. 8:a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramløv U-B. 2000. Aspects of natural cold tolerance in ectothermic animals. Hum Reprod. 15:26–46. [DOI] [PubMed] [Google Scholar]
- Ramløv H, Bedford J, Leader J. 1992. Freezing tolerance of the New Zealand alpine weta, Hemideina maori Hutton [Orthoptera; Stenopelmatidae]. J Ther Biol. 17:51–54. [Google Scholar]
- Ramløv H, Westh P. 1993. Ice formation in the freeze-tolerant alpine weta Hemideina-maori Hutton (Orthoptera, Stenopelmatidae). Cryo-Lett. 14:169–176. [Google Scholar]
- Raymond JA. 2015. Two potential fish glycerol-3-phosphate phosphatases. Fish Physiol Biochem. 41:811–818. [DOI] [PubMed] [Google Scholar]
- Reshetnikov AN. 2003. The introduced fish, rotan (Perccottus glenii), depresses populations of aquatic animals (macroinvertebrates, amphibians, and a fish). Hydrobiologia. 510:83–90. [Google Scholar]
- Reshetnikov AN, Ficetola GF. 2011. Potential range of the invasive fish rotan (Perccottus glenii) in the Holarctic. Biol Invasions. 13:2967–2980. [Google Scholar]
- Rosendale AJ, Lee RE Jr, Costanzo JP. 2014. Effect of physiological stress on expression of glucose transporter 2 in liver of the wood frog. Rana sylvatica. J Exp Zool Part A. 321:566–576. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada Y, Asai S, Ikemoto A, Moriwaki T, Nakamura N, Miyaji M, Zhang-Akiyama Q-M. 2014. Oxidation resistance 1 is essential for protection against oxidative stress and participates in the regulation of aging in Caenorhabditis elegans. Free Radical Res. 48:919–928. [DOI] [PubMed] [Google Scholar]
- Schmid WD. 1982. Survival of frogs in low-temperature. Science. 215:697–698. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 14:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 24:637–644. [DOI] [PubMed] [Google Scholar]
- Stewart A, Wu N, Cachat J, Hart P, Gaikwad S, Wong K, Utterback E, Gilder T, Kyzar E, Newman A, et al. 2011. Pharmacological modulation of anxiety-like phenotypes in adult zebrafish behavioral models. Prog Neuropsychopharmacol Biol Psychiatry. 35:1421–1431. [DOI] [PubMed] [Google Scholar]
- Storey KB. 1987. Organ-specific metabolism during freezing and thawing in a freeze-tolerant frog. Am J Physiol. 253:R292–R297. [DOI] [PubMed] [Google Scholar]
- Storey KB. 1990. Life in a frozen state: adaptive strategies for natural freeze tolerance in amphibians and reptiles. Am J Physiol. 258:R559–R568. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. 1986. Freeze tolerant frogs: cryoprotectants and tissue metabolism during freeze–thaw cycles. Can J Zool. 64:49–56. [Google Scholar]
- Storey KB, Storey JM. 1988. Freeze tolerance in animals. Physiol Rev. 68:27–84. [DOI] [PubMed] [Google Scholar]
- Storey JM, Storey KB. 1996a. β-Adrenergic, hormonal, and nervous influences on cryoprotectant synthesis by liver of the freeze-tolerant wood frog Rana sylvatica. Cryobiology. 33:186–195. [Google Scholar]
- Storey KB, Storey JM. 1996b. Natural freezing survival in animals. Annu Rev Ecol Evol S. 27:365–386. [Google Scholar]
- Storey KB, Storey JM. 2004. Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev. 79:207–233. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. 2013. Molecular biology of freezing tolerance. Compr Physiol. 3:1283–1308. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. 2017. Molecular physiology of freeze tolerance in vertebrates. Physiol Rev. 97:623–665. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. 2020. Mitochondria, metabolic control and microRNA: advances in understanding amphibian freeze tolerance. BioFactors. 46:220–228. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Shintani M, Nakamura A, Monden M, Shiomi H. 2005. Phase-specific central regulatory systems of hibernation in Syrian hamsters. Brain Res. 1045:88–96. [DOI] [PubMed] [Google Scholar]
- Tang HB, Bowers JE, Wang XY, Ming R, Alam M, Paterson AH. 2008. Perspective - synteny and collinearity in plant genomes. Science. 320:486–488. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 25:4–10. [DOI] [PubMed] [Google Scholar]
- Tian G, Cowan NJ. 2013. Tubulin-specific chaperones: components of a molecular machine that assembles the α/β heterodimer. Methods cell biol. 115:155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tőkési N, Lehotzky A, Horváth I, Szabó B, Oláh J, Lau P, Ovádi J. 2010. TPPP/P25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J Biol Chem. 285:17896–17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toxopeus J, Koštál V, Sinclair BJ. 2019. Evidence for non-colligative function of small cryoprotectants in a freeze-tolerant insect. Proc Biol Sci. 286:20190050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toxopeus J, Sinclair BJ. 2018. Mechanisms underlying insect freeze tolerance. Biol Rev. 93:1891–1914. [DOI] [PubMed] [Google Scholar]
- Tsuji Y, Ayaki H, Whitman SP, Morrow CS, Torti SV, Torti FM. 2000. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol Cell Biol. 20:5818–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala H, Dokal AD, Walne A, Ellison A, Cardoso S, Amirthasigamanipillai S, Kirwan M, Browne I, Sidhu JK, Rajeeve V, et al. 2018. Genome instability is a consequence of transcription deficiency in patients with bone marrow failure harboring biallelic ERCC6L2 variants. P Nati Acad Sci U S A. 115:7777–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, Davis TN. 2012. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci U S A. 109:16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkman A, Mitra AK. 2000. Structure and function of aquaporin water channels. Am J Physiol-Renal. 278:F13–F28. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Paudyal SC, Kang Y, Owa M, Liang F-X, Spektor A, Knaut H, Sánchez I, Dynlacht BD. 2022. Regulators of tubulin polyglutamylation control nuclear shape and cilium disassembly by balancing microtubule and actin assembly. Cell Res. 32:190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BS, Feng CG, Zhu CL, Xu WJ, Yuan Y, Hu ML, Yuan K, Li YX, Ren YD, Zhou Y, et al. 2021. The genomes of two billfishes provide insights into the evolution of endothermy in teleosts. Mol Biol Evol. 38:2413–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lu C, Dong X, Zhang Z, Yang M, Xu H. 2017. Proteomics analysis of zebrafish brain following chronically exposed to bisphenol A. Toxicol Environ Chem. 99:469–481. [Google Scholar]
- Wu CW, Tessier SN, Storey KB. 2018. Stress-induced antioxidant defense and protein chaperone response in the freeze-tolerant wood frog Rana sylvatica. Cell Stress Chaperon. 23:1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- Wu F, Yang F, Vinnakota KC, Beard DA. 2007. Computer modeling of mitochondrial tricarboxylic acid cycle, oxidative phosphorylation, metabolite transport, and electrophysiology. J Biol Chem. 282:24525–24537. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Dai Y, Li L, Geng F, Xu Y, Wang J, Wang S, Zhao J. 2021. Tetrahydrocurcumin ameliorates Alzheimer's Pathological phenotypes by inhibition of microglial cell cycle arrest and apoptosis via Ras/ERK signaling. Biomed Pharmacother. 139:111651. [DOI] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, Nachury MV. 2017. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science. 356:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35:W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yin W, Chen A, Li J, Lei G, Fu C. 2014. Phylogeographical analysis of a cold-temperate freshwater fish, the Amur sleeper (Perccottus glenii) in the Amur and liaohe river basins of northeast Asia. Zool Sci. 31:671–679. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gupta A, Storey KB. 2021. Freezing stress adaptations: critical elements to activate Nrf2 related antioxidant defense in liver and skeletal muscle of the freeze tolerant wood frogs. Com Biochem Phys B. 254:110573. [DOI] [PubMed] [Google Scholar]
- Zhang Y, He B, Cao D, Bai Q, He L. 2021. Reproductive biology and artificial propagation of Amur Sleeper (Perccottus glenii). Chin J Fish. 34:77–81. [Google Scholar]
- Zhang J, Storey KB. 2012. Cell cycle regulation in the freeze tolerant wood frog, Rana sylvatica. Cell Cycle. 11:1727–1742. [DOI] [PubMed] [Google Scholar]
- Zhang G, Storey JM, Storey KB. 2011. Chaperone proteins and winter survival by a freeze tolerant insect. J Insect Physiol. 57:1115–1122. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhuang R, Ye Q, Zhuo J, Chen K, Lu D, Wei X, Xie H, Xu X, Zheng S. 2019. High expression of human AugminComplex submit 3 indicates poor prognosis and associates with tumor progression in hepatocellular carcinoma. J Cancer. 10:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data have been deposited in the NCBI BioProject database with accession numbers PRJNA818152 (P. glenii), PRJNA818180 (N. hainanensis). The genome assembly files are under accession numbers JALDQB000000000 (P. glenii) and JALDNG000000000 (N. hainanensis).