Abstract
Background and purpose
To improve cone-beam computed tomography (CBCT), deep-learning (DL)-models are being explored to generate synthetic CTs (sCT). The sCT evaluation is mainly focused on image quality and CT number accuracy. However, correct representation of daily anatomy of the CBCT is also important for sCTs in adaptive radiotherapy. The aim of this study was to emphasize the importance of anatomical correctness by quantitatively assessing sCT scans generated from CBCT scans using different paired and unpaired dl-models.
Materials and methods
Planning CTs (pCT) and CBCTs of 56 prostate cancer patients were included to generate sCTs. Three different dl-models, Dual-UNet, Single-UNet and Cycle-consistent Generative Adversarial Network (CycleGAN), were evaluated on image quality and anatomical correctness. The image quality was assessed using image metrics, such as Mean Absolute Error (MAE). The anatomical correctness between sCT and CBCT was quantified using organs-at-risk volumes and average surface distances (ASD).
Results
MAE was 24 Hounsfield Unit (HU) [range:19-30 HU] for Dual-UNet, 40 HU [range:34-56 HU] for Single-UNet and 41HU [range:37-46 HU] for CycleGAN. Bladder ASD was 4.5 mm [range:1.6–12.3 mm] for Dual-UNet, 0.7 mm [range:0.4–1.2 mm] for Single-UNet and 0.9 mm [range:0.4–1.1 mm] CycleGAN.
Conclusions
Although Dual-UNet performed best in standard image quality measures, such as MAE, the contour based anatomical feature comparison with the CBCT showed that Dual-UNet performed worst on anatomical comparison. This emphasizes the importance of adding anatomy based evaluation of sCTs generated by dl-models. For applications in the pelvic area, direct anatomical comparison with the CBCT may provide a useful method to assess the clinical applicability of dl-based sCT generation methods.
Keywords: Deep-Learning, Synthetic computed tomography, Cone-beam computed tomography, Average surface distance, Anatomical comparison, Volume difference
1. Introduction
In radiotherapy, a treatment plan is generally made on a computed tomography (CT) image, known as the planning CT (pCT). To verify patient position prior to treatment, a cone-beam CT (CBCT) is often used to correct the patients position based on bony anatomy or target volume with reference to the pCT [1]. The anatomy visible on the pCT and daily CBCT can vary due to internal organ position and volume differences. Particularly in the pelvic region, internal bladder and rectum motions were found to be major sources of geometric uncertainty [2]. The influence of these anatomical differences on the dose can be minimized by treatment couch corrections and plan adaptations. To adapt the plan to daily anatomical changes, Plan Of the Day (PotD) and Online Plan Adaptation can be used [3], [4], [5]. However, CBCT image quality is often insufficient to enable dose calculations, due to inaccurate CT numbers and imaging artifacts. To improve CBCT quality and accuracy of CT numbers, several studies have proposed synthetic CT (sCT) generation via deep-learning (DL) [6], [7], [8], [9], [10], [11], [12], [13], [14], [15]. These dl-models can be grouped into paired- and unpaired-data models. Paired-data models are trained and validated with paired input and reference images that share geometrical information [16]. Unpaired dl-models are trained and validated with images from a target and a source domain that do not require shared information [17]. The performance of these two categories of dl-models varies. For example, Rossi et al. concluded that the paired model outperformed the unpaired ones based on image metrics, whereas on visual inspection of the anatomical differences between sCT and CBCT the unpaired models performed better [18].
To use sCTs for clinical applications such as PotD selection, sCTs require calibrated CT numbers, image quality comparable to pCT and anatomical features identical to the CBCT. To evaluate sCT image quality, the deformed pCT (pCT_deformed), deformably registered to the CBCT, is often used as ground truth. This evaluation is in most studies based on CT value-based image metrics such as Mean Absolute Error (MAE) and Root-Mean-Squared-Error (RMSE) between the ground truth and sCT [7], [8]. To evaluate the correspondence of structural features the structural similarity index measure (SSIM) between sCT and ground truth is often used [19]. However, this method is sensitive to registration errors.
Deformable registration in the pelvic area can be more prone to registration errors, since the daily anatomical changes in the pelvic area are generally larger compared to areas such as head-and-neck, [20]. In these regions, a direct evaluation on the CBCT would be desirable. However, as CT value-based image metrics cannot be used on CBCT due to artifacts and inaccurate CT numbers, a contour based evaluation might be preferred. Therefore, the present study used the contours of the CBCT and sCT to compare the anatomical correctness of sCT. The aim of this study was to emphasize the importance of anatomical correctness by quantitatively assessing sCT scans generated from CBCT scans using different paired and unpaired deep learning models.
2. Materials and methods
2.1. Data
This study retrospectively included pCT and CBCT images of 56 randomly selected prostate cancer patients that received radiotherapy treatment between February 2020 and January 2021 at the Catharina Hospital, the Netherlands. This research was conducted on anonymized patient data and, according to Dutch law, this research was approved under so-called non-WMO legislation (medical research law waiver). pCT and CBCT images were acquired on a Philips big bore CT scanner (Philips, Eindhoven, Netherlands) and on different X-ray Volumetric Imaging (XVI) v5.0.4 systems (Elekta AB, Stockholm, Sweden), respectively. Average interval between pCT and the selected CBCT scan was 20 days (range: 2–56 days). CBCT scans were recorded with a small field-of-view (FOV), resulting in an image of 270x270 pixels with pixel size of 1.00x1.00 mm. The number of axial slices was 128 slices with a slice thickness of 1.00 mm. The pCT image size was 512x512 pixels with pixel size of 1.17x1.17 mm and slice thickness of 3.00 mm.
2.2. Image preprocessing
The pCTs were rigidly registered to the corresponding CBCT (pCT_rigid), with an iterative adaptive stochastic gradient descent optimization, and deformed (pCT_deformed) with a deformable 3D B-spline image registration, part of the Elastix registration toolbox (v5.0.0) [21]. Images were normalized (UNet [0–1], Cycle-consistent Generative Adversarial Network (CycleGAN)[-1/1]) and cropped to 256x256 pixels around the center of the image.
2.3. Deep-learning models
Three commonly used and publicly available distinctive dl-models were used in this study, namely Dual-UNet, Single-UNet and CycleGAN [7], [8], [9]. Dual-UNet was based on the architecture as proposed by Chen et al. [8]. The model was trained with Keras and Tensorflow v.2.6 in Python (v3.8). Dual-UNet was trained with CBCT and pCT_rigid as paired input and pCT_deformed as ground truth. Loss functions and hyperparameters were adopted from the optimized model in Chen et al. [8]. The Single-UNet model was based on the architecture as proposed by Jin et al. [9]. The first difference between the Dual-UNet and the Single-UNet model was an additional skip connection between input and output. The second difference was training of the model, the Single-UNet was paired trained with just the CBCT as input and pCT_deformed was set as ground truth. Loss functions and hyperparameters were adopted from the optimized model in Jin et al. [9]. The CycleGAN architecture was based on a study by Maspero et al. [7]. The implementation of the model was based on Zhu et al. and trained with Tensorflow (v2.1.0) in Python v3.8. (10). CycleGAN was trained and validated with unpaired input CBCT and ground truth pCT_rigid. Loss functions and hyperparameters were adopted from the optimized model in Maspero et al. [7].
2.4. Training of the dl-models
First the data was randomly divided in training/validation/test sets of 31/16/9 patients respectively, which was a ratio of 55 %/29 %/16 %. Preliminary analyses performed on a subset of the scans (data not shown) investigated the minimum amount of data necessary to train the UNet models and showed no improvement after including 26 paired CBCT/CT scans. Residual errors in image registration that were caused by changes in bladder and rectum filling, complicated the paired training approach in Dual-UNet and Single-UNet. To reduce training errors due to mismatch in image registration of paired datasets, images were excluded from training and validation when the image registration noticeably failed (for example, at the outer slices or in cases where the bladder size differ too much to resolve). This resulted in 2236 training, 1073 validation and 1416 test 2D images. Since CycleGAN was unpaired, slice selection was not necessary. This is an advantage of unpaired training, since CycleGAN is less sensitive to training errors caused by registration errors. The dataset was divided into sets of 3928 training, 2024 validation and 1416 test 2D images.
Models were trained on a RTX A5000 GPU (NVIDIA Corporation, Santa Clara, United States) for 200 epoch with a batch size of 40 in UNet and a batch size of 7 in CycleGAN. Out of the 200 epochs, the final model was selected based on the lowest validation loss, in order to minimize the risk of overfitting.
2.5. Evaluation
To evaluate the performance of the three models, common image quality metrics were calculated between sCT and pCT_deformed: MAE, ME, peak signal-to-noise ratio (PSNR), SSIM and RMSE [19]. These metrics were calculated within the external, bladder and rectal contours of the pCT_deformed.
In addition to the image quality metrics, the anatomical agreement between the sCT and input CBCT was evaluated. This method was based on organs-at-risk (OAR) and bone contours. OAR selected for evaluation were rectum and bladder. Contours were created by delineation of femoral joint, bladder and rectum. The observer-related variabilities in contouring were minimized with the use of an edge detection brush in Raystation V9.1 (RaySearch laboratories AB, Stockholm, Sweden). Extra attention was paid to the interface between prostate and bladder, where the contrast difference was sometimes too low for the edge detection tool to work reliably. In those cases the bladder contour was consistently delineated by one observer in all sCTs and on the corresponding CBCTs.
For the bone evaluation, Dice Similarity Coefficient (DSC), Average Surface Distance (ASD) and 95th percentile Hausdorff distance (HD95) between bone contours on CBCT and sCT were calculated [22], [23]. For OAR evaluation, OAR volume differences, HD95 and ASD were calculated between contours on CBCT and sCT. ASD was calculated by averaging the minimum Euclidian distance between contour voxel of CBCT and sCT and the minimum Euclidian distance between contour voxel of sCT and CBCT.
A Wilcoxon Signed-Rank (WSR) test was used to test whether the dl-models were statistically different. Prior to the WSR test, however, a Friedman test was used to determine whether the WSR test was allowed, which was necessary as more than two groups were compared. The Friedman test, which is a non-parametric alternative to a repeated measures ANOVA, was used with a significance level of p < 0.05 [24]. If the Friedman test showed an overall significant difference, a WSR test was used to compare each pair of two dl-models mutually [25]. This way, the WSR test was used three times. Therefore, the significance level of the WSR test was corrected for multiple testing to p < 0.017.
3. Results
3.1. Visual inspection
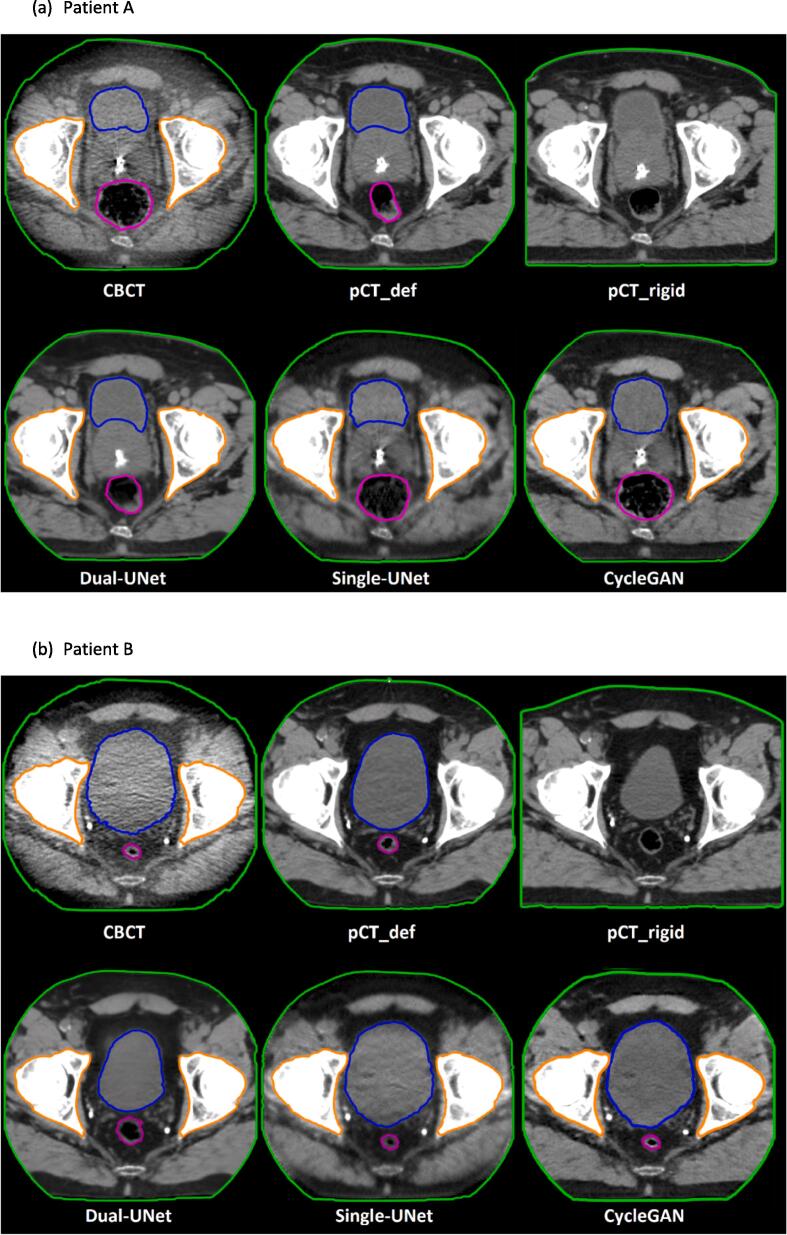
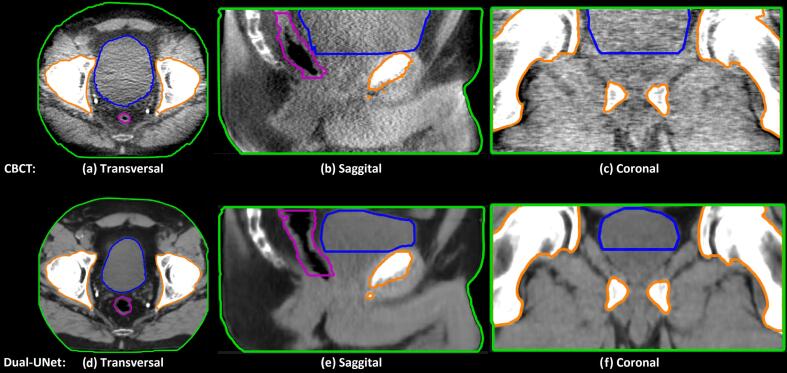
Fig. 1 shows typical examples of CBCT, sCT and CT images of two patients, showing that the image quality of the sCTs generated by the three different dl-models was improved compared to the CBCTs. Apart from the apparent image quality differences, Fig. 1 also demonstrated that the Dual-UNet resembled the pCT more in terms of anatomical features (shape and position), rather than the input CBCT. CycleGAN and Single-UNet, on the other hand, better preserved the anatomical features as present in the CBCT. For example, the bladder size was decreased significantly more by the Dual-UNet and therefore more comparable to the bladder size in the pCT_rigid (Fig. 1). The bladder volume difference for one patient is shown in more detail in Fig. 2. This example showed a smaller bladder size on the sCT generated by Dual-UNet than the bladder size of CBCT, which was 268 cm3 compared to 125 cm3, respectively.
Fig. 1.
Examples of CBCT, sCT and CT images of two patients. Purple line is rectum contour, blue line is bladder contour and orange line is bone contour. For each patient, the first row shows from left to right: cone-beam CT and registered planning CT. The second row shows synthetic CTs generated by three different dl-models. The window width and window level of the shown CT images are 350 and 40 HU. The window width and window level of the shown CBCT images are 350 and −230 HU. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
A qualitative example of large anatomical differences between CBCT and sCT generated by Dual-UNet in transversal, sagittal and coronal view. Purple line is rectum contour, blue line is bladder contour and orange line is bone contour. This example shows a smaller bladder size of the sCT generated by Dual-UNet than the bladder size of CBCT, which was 268 cm3 compared to 125 cm3 respectively. The window width and window level of the shown CT images are 350 and 40 HU. The window width and window level of the shown CBCT images are 350 and −230 HU. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. CT value-based evaluation
According to the MAE, PSNR and SSIM, Dual-UNet performed best in CT value-based evaluation between the different dl-models (Table 1 and Supplementary material A). The MAE, reported as median [range], was smallest in Dual-UNet compared to Single-UNet and CycleGAN, which was respectively 24 HU [range: 19–30 HU], 40 HU [range: 34–56 HU] and 41 HU [range: 37–46 HU]. Significant difference between Dual-UNet and Single-UNet were found in MAE within external (p = 0.01), bladder (p = 0.01) and rectum (p = 0.01). This was also observed between Dual-UNet and CycleGAN. No significant difference was found between Single-UNet and Cyclegan. Significant difference was also observed in the SSIM Dual-UNet compared to the other two networks (Single-UNet and CycleGAN) (p = 0.01 and p = 0.01 respectively).
Table 1.
Image metrics calculated between sCT and pCT_deformed per dl-model (Median [range]). The best performance is indicated in bold.
| Contour | dl-model | Mean Absolute Error [HU] | Peak Signal-to-Noise Ratio [dB] | Structural Similarity Index Measure [-] |
|---|---|---|---|---|
| External | Dual-UNet | 25 [19–30] | 39.4 [39.0–40.8] | 0.74 [0.68–0.79] |
| Single-UNet | 40 [34–56] | 36.4 [35.0–37.33] | 0.65 [0.52–0.70] | |
| CycleGAN | 41 [37–46] | 37.3 [36.6–37.8] | 0.67 [0.59–0.70] | |
| Bladder | Dual-UNet | 19 [13–29] | 40 [39–45] | – |
| Single-UNet | 40 [30–66] | 37 [34–39] | – | |
| CycleGAN | 37 [26–56] | 38 [36–40] | – | |
| Rectum | Dual-UNet | 18 [12–23] | 41 [37–45] | – |
| Single-UNet | 50 [32–95] | 36 [32–39] | – | |
| CycleGAN | 41 [27–55] | 37 [33–40] | – |
3.3. Anatomical comparison of bones
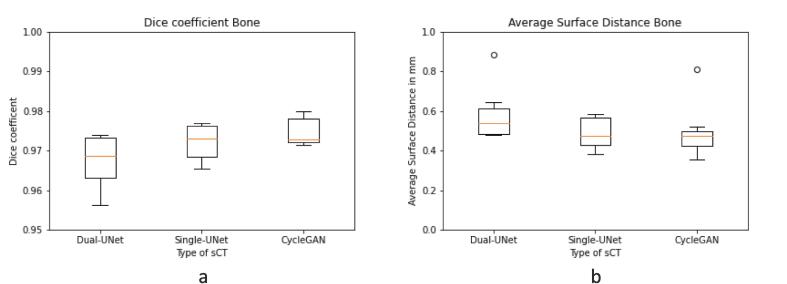
The median DSC in bone contours between CBCT and sCT, were 0.97 [range: 0.96–0.97] for Dual-UNet, 0.97 [range: 0.97–0.98] for Single-UNet and 0.97 [range: 0.95–0.98] for CycleGAN (Fig. 3a). The median ASD was 0.5 mm [range: 0.5–0.9 mm] for Dual-UNet, 0.5 mm [range: 0.4–0.6 mm] for Single-UNet and 0.5 mm [range: 0.4–0.8 mm] for GycleGAN. There was no significant difference in DSC and ASD found between the three DL models.
Fig. 3.
Comparison of bone contours of CBCT and sCT per dl-model. (a) Dice coefficient. (b) ASD in mm. Horizontal lines in boxes are medians, dots are outliers.
3.4. Anatomical comparison of OAR
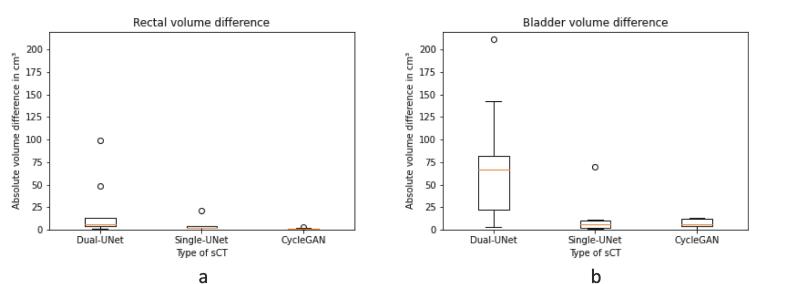
In contrast to the anatomical comparison of the bones, significant differences between dl-models in the anatomical comparison of OAR were found. Absolute rectum volume differences were largest in Dual-UNet, then Single-UNet, and smallest in CycleGAN, with median 6.5 cm3 [range: 1.4–98.9 cm3], 1.9 cm3 [range: 0.2–21.3 cm3] and 0.8 cm3 [range: 0.1–2.8 cm3], respectively (Fig. 4a). This resulted in a significant difference between Dual-UNet and CycleGAN (p < 0.01). The absolute bladder volume differences between the dl-models were larger in Dual-UNet than Single-UNet and CycleGAN, respectively 67 cm3 [range: 3–220 cm3], 6 cm3 [range: 1–70 cm3] and 6 cm3 [range: 0–13 cm3] (Fig. 4b). This resulted in a significant difference between Dual-UNet and CycleGAN (p < 0.01).
Fig. 4.
OAR absolute volume differences between CBCT and sCT in cm3 per dl-model. (a) Rectal absolute volume difference. (b) Bladder absolute volume difference. Horizontal lines in boxes are medians, dots are outliers.
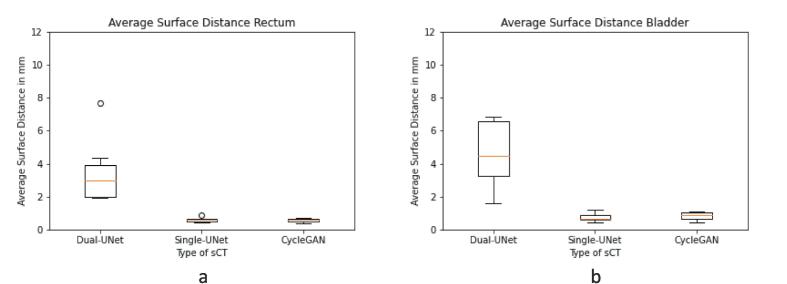
The ASD of the rectum contour between CBCT and sCT was largest in Dual-UNet and small in Single-UNet and CycleGAN, respectively 3.0 mm [range: 1.9–7.7 mm], 0.6 mm [range: 0.45–0.87 mm] and 0.6 mm [range: 0.4–0.7 mm] (Fig. 5a). This resulted in a significant difference between Dual-UNet, Single-UNet (p = 0.01) and CycleGAN (p = 0.01). In the bladder contours, the ASD was largest in Dual-UNet and small in Single-UNet and CycleGAN, respectively 4.5 mm [range: 1.6–12.3 mm], 0.7 mm [range: 0.4–1.2 mm] and 0.9 mm [range: 0.4–1.1 mm] (Fig. 5b). This resulted in a significant difference between Dual-UNet and Single-UNet (p = 0.01) or CycleGAN (p = 0.01).
Fig. 5.
ASD in mm between OAR contours of CBCT and sCT of different dl-Model Types. (a) Rectum contours. (b) Bladder contours. Horizontal lines in boxes are medians, dots are outliers.
4. Discussion
In this study, the anatomical correctness of sCT scans generated from CBCT scans was quantitatively assessed for paired and unpaired dl-models on prostate cancer patients with varying bladder and rectum filling. Although Dual-UNet performed best in standard image quality measures, like MAE and SSIM, the preservation of CBCT anatomy was worst in Dual-UNet. This confirmed that Dual-UNet generated sCTs with the highest image quality upon visual inspection, in terms of contrast, noise and sharpness. However, by visual inspection it could also be observed that the anatomy of the CBCT was not adequately preserved in the sCTs generated by Dual-UNet. This emphasizes the importance of adding anatomy based evaluation of sCTs generated by dl-models.
Three different dl-models were evaluated on sCT generation from CBCT images of prostate cancer patients: paired Dual-UNet, paired Single-UNet and unpaired CycleGAN. All three dl-models resulted in sCTs with a reduced amount of noise compared to the original CBCT scan. Compared to the model proposed in Chen et al. our Dual-UNet performed better in terms of MAE and PSNR [8]. For the CycleGAN model, our MAE was lower than the MAE reported by Kurz et al. and comparable to the MAE reported by Eckl et al. [11], [26]. Dual-UNet performed best in the standard image metrics MAE, PSNR and SSIM (Table 1), however, CycleGAN and Single-UNet performed best in preserving anatomical features. The present finding that the best performance in SSIM did not correspond with the highest anatomical correctness is in agreement with the findings of Rossi et al. [18].
In the bone DSC and ASD comparison, no significant differences between the different dl-models were found. The ASD of the bone contours were smaller than found in segmentation studies [27], [28]. Since the contours used in the present study are within the precision of dl-segmentation model ranges, the observed significant ASD differences between the three different dl-models were due to dl-model differences in their capability to preserve of CBCT anatomy.
Significant rectal volume differences were found between the different dl-model types. The rectal volume difference between contours on Dual-UNet and CBCT was around three times larger than the rectal volume difference between contours generated by manual delineation and dl-segmentation models on MRI and CT [29], [30], [31]. The rectal volume difference in Single-UNet and CycleGAN was, however, in the range of these dl-segmentation models [29], [30], [31]. The rectum volume difference in Dual-UNet was up to 99 cm3, in Single-UNet up to 4 cm3 and in CycleGAN up to 3 cm3. The rectal volume difference was depending on the CBCT and pCT anatomy difference, caused by difference in rectal filling between the pCT and daily CBCT. Even larger volume differences up to 221 cm3 were found in the bladder, which led to a HD95 of 29 mm in Dual-UNet (Supplementary material B). The HD95 difference in Dual-UNet was around three times higher than in dl-segmentation studies [29], [30], [32]. This HD95 is a clinically relevant difference, since for example Planning Target Volume (PTV) margins of 5 mm are used in treatment planning. This emphasizes that the observed differences between dl-models could have a clinically relevant impact on the estimated bladder volumes and position.
To include OAR locations, the ASD was calculated. The ASD of the rectum and bladder contours of the sCTs generated by the Single-UNet and CycleGAN were within the range of dl-segmentation studies for male pelvic ASDs [27], [30], [33], [34]. However, the Dual-UNet resulted in ASDs of four times the reported autocontouring ASD values. These ASDs between Dual-UNet and CBCT were in the range of clinically used PTV margins. OAR volume differences and ASD were larger in Dual-UNet when the CBCT and pCT anatomy difference was larger due to rectal and bladder filling.
In further research, if auto-contouring on CBCT is improved, auto-contoured based ASD and volume comparison could even further improve the anatomical comparison of dl-models. With the use of auto-contouring, dl-models could be improved on anatomical preservation by adding the ASD and volume comparison to the loss functions of the dl-models. The influence on the OAR dose could be investigated, with respect to the organ boundary location in the sCTs, and therefore the location of OAR contours.
In conclusion, although Dual-UNet performed best in standard image quality measures, the contour based anatomical feature comparison with the CBCT showed Dual-UNet performed worst on anatomical comparison. This emphasizes the importance of adding anatomy based evaluation of sCTs generated by dl-models. For applications in the pelvic area, where accurate deformable registration is not always guaranteed, a direct comparison of the anatomical structure with the input CBCT may provide a useful method to assess the clinical applicability of DL based sCT generation methods.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank dr. D.C. Rijkaart (radiation oncologist at Catharina hospital Eindhoven Netherlands) for assistance with delineation and review of the contours used in this study. Yvonne de Hond was financially supported by a research grant by Elekta (grant number SOW_20210426, Elekta Ltd., Crawley, UK).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2023.100416.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ding G.X., Duggan D.M., Coffey C.W., Deeley M., Hallahan D.E., Cmelak A., et al. A study on adaptive IMRT treatment planning using kV cone-beam CT. Radiother Oncol. 2007;85:116–125. doi: 10.1016/j.radonc.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 2.Fokdal L., Honoré H., Høyer M., Meldgaard P., Fode K., Von der Maase H. Impact of changes in bladder and rectal filling volume on organ motion and dose distribution of the bladder in radiotherapy for urinary bladder cancer. Int J Radiat Oncol Biol Phys. 2004;59:436–444. doi: 10.1016/j.ijrobp.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 3.Lutkenhaus L.J., De Jong R., Geijsen E.D., Visser J., Van Wieringen N., Bel A. Potential dosimetric benefit of an adaptive plan selection strategy for short-course radiotherapy in rectal cancer patients. Radiother Oncol. 2016;119:525–530. doi: 10.1016/j.radonc.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Murthy V., Master Z., Adurkar P., Mallick I., Mahantshetty U., Bakshi G., et al. 'Plan of the day' adaptive radiotherapy for bladder cancer using helical tomotherapy. Radiother Oncol. 2011;99:55–60. doi: 10.1016/j.radonc.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 5.De Jong R., Crama K., Visser J., Van Wieringen N., Wiersma J., Geijsen E., et al. Online adaptive radiotherapy compared to plan selection for rectal cancer: quantifying the benefit. Radiat Oncol. 2020;15:162. doi: 10.1186/s13014-020-01597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spadea M.F., Maspero M., Zaffino P., Seco J. Deep learning based synthetic-CT generation in radiotherapy and PET: A review. Med Phys. 2021;48:6537–6566. doi: 10.1002/mp.15150. [DOI] [PubMed] [Google Scholar]
- 7.Maspero M., Houweling A.C., Savenije M.H.F., van Heijst T.C.F., Verhoeff J.J.C., Kotte A.N.T.J., et al. A single neural network for cone-beam computed tomography-based radiotherapy of head-and-neck, lung and breast cancer. Phys Imaging Radiat Oncol. 2020;14:24–31. doi: 10.1016/j.phro.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L., Liang X., Shen C., Jiang S., Wang J. Synthetic CT generation from CBCT images via deep learning. Med Phys. 2020;47:1115–1125. doi: 10.1002/mp.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin K.H., McCann M.T., Froustey E., Unser M. Deep Convolutional Neural Network for Inverse Problems in Imaging. IEEE Trans Image Process. 2017;26:4509–4522. doi: 10.1109/TIP.2017.2713099. [DOI] [PubMed] [Google Scholar]
- 10.Liang X., Chen L., Nguyen D., Zhou Z., Gu X., Yang M., et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab22f9. [DOI] [PubMed] [Google Scholar]
- 11.Kurz C., Maspero M., Savenije M.H.F., Landry G., Kamp F., Pinto M., et al. CBCT correction using a cycle-consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab4d8c. [DOI] [PubMed] [Google Scholar]
- 12.Harms J., Lei Y., Wang T., Zhang R., Zhou J., Tang X., et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. 2019;46:3998–4009. doi: 10.1002/mp.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Chen X., Zhu J., Yang B., Wei R., Xiong R., et al. A two-step method to improve image quality of CBCT with phantom-based supervised and patient-based unsupervised learning strategies. Phys Med Biol. 2022;67 doi: 10.1088/1361-6560/ac6289. [DOI] [PubMed] [Google Scholar]
- 14.Yuan N., Rao S., Chen Q., Sensoy L., Qi J., Rong Y. Head and neck synthetic CT generated from ultra-low-dose cone-beam CT following Image Gently Protocol using deep neural network. Med Phys. 2022;49:3263–3277. doi: 10.1002/mp.15585. [DOI] [PubMed] [Google Scholar]
- 15.Rusanov B., Hassan G.M., Reynolds M., Sabet M., Kendrick J., Rowshanfarzad P., et al. Deep learning methods for enhancing cone-beam CT image quality toward adaptive radiation therapy: A systematic review. Med Phys. 2022;49:6019–6054. doi: 10.1002/mp.15840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. 2015 Medical Image Computing and Computer-Assisted Intervention -- MICCAI. 2015;234-41. https://doi.org/10.1007/978-3-319-24574-4_28.
- 17.Zhu JY, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. IEEE International Conference on Computer Vision (ICCV). 2017;2242-51. https://doi.org/10.1109/ICCV.2017.244.
- 18.Rossi M., Cerveri P. Comparison of Supervised and Unsupervised Approaches for the Generation of Synthetic CT from Cone-Beam CT. Diagnostics. 2021;11:1435. doi: 10.3390/diagnostics11081435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Bovik A.C., Sheikh H.R., Simoncelli E.P. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 20.Boulanger M., Nunes J.C., Chourak H., Largent A., Tahri S., Acosta O., et al. Deep learning methods to generate synthetic CT from MRI in radiotherapy: A literature review. Phys Med. 2021;89:265–281. doi: 10.1016/j.ejmp.2021.07.027. [DOI] [PubMed] [Google Scholar]
- 21.Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 22.Dice L.R. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;3:297–302. doi: 10.2307/1932409. [DOI] [Google Scholar]
- 23.Birsan T., Tiba D. One hundred years since the introduction of the set distance by dimitrie pompeiu. CSMO 2005: System Modeling and Optimization. 2006;199:35–39. doi: 10.1007/0-387-33006-2_4. [DOI] [Google Scholar]
- 24.Hoffman J.I.E. Analysis of Variance. II. More Complex Forms. Academic Press. 2019:419–441. [Google Scholar]
- 25.Wilcoxon F. Individual Comparisons by Ranking Methods. Biometrics. 1945;1:80–83. doi: 10.2307/3001968. [DOI] [Google Scholar]
- 26.Eckl M., Hoppen L., Sarria G.R., Boda-Heggemann J., Simeonova-Chergou A., Steil V., et al. Evaluation of a cycle-generative adversarial network-based cone-beam CT to synthetic CT conversion algorithm for adaptive radiation therapy. Phys Med. 2020;80:308–316. doi: 10.1016/j.ejmp.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Tong N., Gou S., Chen S., Yao Y., Yang S., Cao M., et al. Multi-task edge-recalibrated network for male pelvic multi-organ segmentation on CT images. Phys Med Biol. 2021;66 doi: 10.1088/1361-6560/abcad9. [DOI] [PubMed] [Google Scholar]
- 28.Amjad A., Xu J., Thill D., Lawton C., Hall W., Awan M.J., et al. General and custom deep learning autosegmentation models for organs in head and neck, abdomen, and male pelvis. Med Phys. 2022;49:1686–1700. doi: 10.1002/mp.15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuisma A., Ranta I., Keyriläinen J., Suilamo S., Wright P., Pesola M., et al. Validation of automated magnetic resonance image segmentation for radiation therapy planning in prostate cancer. Phys Imaging Radiat Oncol. 2020;13:14–20. doi: 10.1016/j.phro.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Y., Wang T., Tian S., Fu Y., Patel P., Jani A.B., et al. Male pelvic CT multi-organ segmentation using synthetic MRI-aided dual pyramid networks. Phys Med Biol. 2021;66 doi: 10.1088/1361-6560/abf2f9. [DOI] [PubMed] [Google Scholar]
- 31.Olsson C.E., Suresh R., Niemelä J., Akram S.U., Valdman A. Autosegmentation based on different-sized training datasets of consistently-curated volumes and impact on rectal contours in prostate cancer radiation therapy. Phys Imaging Radiat Oncol. 2022;22:67–72. doi: 10.1016/j.phro.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbani N., Baudier T., Rit S., Franco F.D., Okoli F., Jaouen V., et al. Deep learning-based segmentation in prostate radiation therapy using Monte Carlo simulated cone-beam computed tomography. Med Phys. 2022;49:6930–6944. doi: 10.1002/mp.15946. [DOI] [PubMed] [Google Scholar]
- 33.Schreier J., Genghi A., Laaksonen H., Morgas T., Haas B. Clinical evaluation of a full-image deep segmentation algorithm for the male pelvis on cone-beam CT and CT. Radiother Oncol. 2020;145:1–6. doi: 10.1016/j.radonc.2019.11.021. [DOI] [PubMed] [Google Scholar]
- 34.Walker Z., Bartley G., Hague C., Kelly D., Navarro C., Rogers J., et al. Evaluating the Effectiveness of Deep Learning Contouring across Multiple Radiotherapy Centers. Phys Imaging Radiat Oncol. 2022;24:121–128. doi: 10.1016/j.phro.2022.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.