Summary
RNA interference (RNAi)‐based technologies are starting to be commercialized as a new approach for agricultural pest control. Horizontally transferred genes (HTGs), which have been transferred into insect genomes from viruses, bacteria, fungi or plants, are attractive targets for RNAi‐mediated pest control. HTGs are often unique to a specific insect family or even genus, making it unlikely that RNAi constructs targeting such genes will have negative effects on ladybugs, lacewings and other beneficial predatory insect species. In this study, we sequenced the genome of a red, tobacco‐adapted isolate of Myzus persicae (green peach aphid) and bioinformatically identified 30 HTGs. We then used plant‐mediated virus‐induced gene silencing (VIGS) to show that several HTGs of bacterial and plant origin are important for aphid growth and/or survival. Silencing the expression of fungal‐origin HTGs did not affect aphid survivorship but decreased aphid reproduction. Importantly, although there was uptake of plant‐expressed RNA by Coccinella septempunctata (seven‐spotted ladybugs) via the aphids that they consumed, we did not observe negative effects on ladybugs from aphid‐targeted VIGS constructs. To demonstrate that this approach is more broadly applicable, we also targeted five Bemisia tabaci (whitefly) HTGs using VIGS and demonstrated that knockdown of some of these genes affected whitefly survival. As functional HTGs have been identified in the genomes of numerous pest species, we propose that these HTGs should be explored further as efficient and safe targets for control of insect pests using plant‐mediated RNA interference.
Keywords: horizontally transferred genes, green peach aphid, whitefly, seven‐spotted ladybug, RNA interference, virus‐induced gene silencing
Introduction
Aphids, whiteflies and other hemipteran insect pests cause considerable damage to agricultural crops. Although current control strategies, in particular chemical pesticides, provide some level of protection, insects continuously develop new tolerance or resistance mechanisms (Bass et al., 2014; Koch et al., 2018). Genetically engineered Bt crops have been shown to be effective in controlling specific insect pests, without long‐term environmental persistence (Mendelsohn et al., 2003; Sears et al., 2001; Sujii et al., 2013; Tian et al., 2015). However, there has been only limited success in the development of Bt toxins that target hemipteran pests (Chougule and Bonning, 2012; Lu et al., 2010; Pessoa et al., 2016; Sujii et al., 2013). Thus, there is a need to develop novel and environmentally friendly transgenic plant approaches for the control of phloem‐feeding insects such as aphids and whiteflies.
RNA interference (RNAi)‐based gene expression silencing has emerged as a novel and powerful strategy for agricultural pest control. As phloem feeders, hemipteran pests are less likely to take up surface sprays of RNAi constructs, which can be deployed against chewing herbivores such as Colorado potato beetles (Leptinotarsa decemlineata; Rodrigues et al., 2021). However, plant‐mediated RNAi has been effective for targeting hemipteran gene expression (Chung et al., 2021). Several classes of aphid genes have been successfully inhibited by plant‐mediated RNAi, including C002 (Coleman et al., 2015; Mutti et al., 2006; Pitino et al., 2011), receptor of activated kinase C (Rack‐1) (Coleman et al., 2015; Pitino et al., 2011), MpPInt01 and MpPInt02 (Coleman et al., 2015), Mp55 (Elzinga et al., 2014); and those encoding aquaporin, sucrase‐transglucosidase, and sugar transporters (Tzin et al., 2015), V‐ATPase E and tubulin folding cofactor D (Guo et al., 2014), serine protease (Bhatia et al., 2012), hunchback (Mao and Zeng, 2014), carboxylesterase CbE E4, juvenile hormone binding protein, calreticulin, and cathepsin, carboxylesterase (Xu et al., 2014), salivary sheath protein (Abdellatef et al., 2015), and a zinc finger protein (Xie et al., 2022). Similarly, genes that have been effectively targeted using plant‐mediated RNAi for controlling Bemisia tabaci (whiteflies) include cytochrome P450 genes cyp315a1 and cyp18a1 (Luan et al., 2013), and those encoding ecdysone receptor (EcR) and acetylcholinesterase (AchE) (Malik et al., 2016), aquaporin and α‐glucosidase (Raza et al., 2016), v‐ATPase (Ibrahim et al., 2017; Thakur et al., 2014), cyclophilin B (CypB) and heat shock protein 70 (hsp70) (Kanakala et al., 2019), and phenolic glucoside malonlyltransferase (BtPMaT1) (Xia et al., 2021).
As part of the regulatory approval process for commercialization, RNAi‐based transgenic plants should be assessed for biological risks, which include effects on non‐target insect species with similar gene sequences (Casacuberta et al., 2015). Essential genes that are potential targets for RNAi‐mediated pest control, including many of those described above for the control of aphids and whiteflies, are often highly conserved across the insect phylogeny. As it is not currently feasible to examine the genomes of all potential non‐target insects, bioinformatic analysis alone is not sufficient for the design of RNAi constructs that are unlikely to have off‐target effects on beneficial insect species.
Horizontally transferred genes (HTGs), which are often taxon‐, genus‐ or even species‐specific (Wybouw et al., 2016), are an attractive option for limiting potential off‐target effects during RNAi‐mediated pest control. HTGs, including those transferred from prokaryotes to prokaryotes, prokaryotes to eukaryotes, and eukaryotes to eukaryotes, have been described in all branches of life (Husnik and McCutcheon, 2018; Soucy et al., 2015). Importantly, the integration, expression and maintenance of HTGs in the recipient genome suggest that their presence provides a selective advantage (Soucy et al., 2015). There are many hurdles for HTGs to become functional in the recipient genome, particularly in the case of genes that are transformed from prokaryotes to eukaryotes. For instance, the presence or absence of introns, variable GC contents, codon usage preferences, and differences in transcriptional promoters can limit successful gene expression in a recipient species (Husnik and McCutcheon, 2018). Nevertheless, functional HTGs have been described in many species, including aphids and whiteflies (Chen et al., 2016; Chung et al., 2018; Luan et al., 2015; Novakova and Moran, 2012; Parker and Brisson, 2019; Sloan et al., 2014).
Prior to this study, a few HTGs with different origins have been investigated in aphid genomes. Carotenoid biosynthetic genes in aphids were shown to be horizontally transferred from a fungus, including the highly duplicated carotene desaturase (Tor) and the carotenoid cyclase‐carotenoid synthase (CarRP) (Novakova and Moran, 2012). One carotenoid desaturase gene copy was present only in a red Myzus persicae (green peach aphid) isolate, and a point mutation in this gene, which arose in a lab culture, resulted in the loss of red colour in the aphids (Moran and Jarvik, 2010). Similar red colour reduction was also observed in Acyrthosiphon pisum (pea aphid) nymphs when silencing a carotenoid desaturase in the parental aphids (Ding et al., 2020). These results indicated that carotenoid biosynthetic genes of fungal origin remain functional in the recipient aphid genomes and confer to the aphid red‐green colour polymorphism, which in turn influence their susceptibility to natural enemies (Moran and Jarvik, 2010). More recently, 2 A. pisum HTGs of bacterial origin, amiD and ldcA1, were targeted using RNAi constructs that were supplied in artificial diet (Chung et al., 2018). Enriched amidD and ldcA1 expression in A. pisum bacteriocytes, which harbour Buchnera aphidicola endosymbionts, coupled with the fact that amiD and ldcA were lost when the peptidoglycan biosynthetic genes were not present in the symbiont Buchnera genome (Smith et al., 2022), suggested that amiD and ldcA1 function in degrading bacterial peptidoglycan, thereby protecting Buchnera from host attack (Chung et al., 2018). Consistent with this hypothesis, knockdown of amiD and ldcA1 by RNAi caused a significant reduction in Buchnera abundance and inhibited aphid growth. In addition to HTGs from fungi and bacteria, HTGs that originated from viruses have also been found in aphids. For example, the cytolethal distending toxin subunit B (cdtB) found in the genome of M. persicae strain G006 was suggested to be involved in aphid resistance to a predatory wasp (Verster et al., 2019). In the pea aphid genome, two HTGs from a densovirus have been described and demonstrated to modulate the aphid wing plasticity (Parker and Brisson, 2019).
Our previous analysis of the B. tabaci MEAM1 genome identified 142 HTGs from bacteria, fungi and plants (Chen et al., 2016). Several of these genes were proposed to contribute to broad host range and insecticide resistance of whiteflies. Interestingly, a recent study found that a detoxifying gene in whiteflies, phenolic glucoside malonyltransferase (BtPMaT1), likely originated from plants and was able to neutralize host plant defensive metabolites (Xia et al., 2021). Plant‐mediated silencing of BtPMaT1 expression impairs whitefly detoxification functions and confers full whitefly resistance in tomato plants, confirming the utility of this HTG for whitefly control (Xia et al., 2021).
To further explore HTGs as potential RNAi targets for aphid control, we assembled the genome of a red, tobacco‐adapted strain of M. persicae (Ramsey et al., 2007, 2014), conducted a genome‐wide annotation of HTGs, and compared this to the HTG profiles of other aphid species. Then, using plant‐mediated virus‐induced gene silencing (VIGS), we tested the RNAi effects of these HTGs on aphid survival. Additionally, we conducted RNAi of a subset of the HTGs that have been annotated in the B. tabaci MEAM1 (Chen et al., 2016). We found that knockdown of these HTGs using plant‐mediated RNAi can significantly affect insect performance. Importantly, the knockdown of the aphid HTGs did not affect the survival of Coccinella septempunctata (seven‐spotted ladybugs) that fed on these aphids. Our results suggest that HTGs will be effective and safe targets for plant genetic engineering to control aphid populations in the field.
Materials and methods
Insect and plant cultures
The tobacco‐adapted M. persicae strain USDA‐Red (Ramsey et al., 2007, 2014) was maintained on Nicotiana tabacum (tobacco) plants in a growth room at 23 °C with a 16:8 h light:dark photoperiod. A B. tabaci MEAM1 colony was obtained from Angela Douglas (Cornell University) and maintained on an acylsugar‐deficient asat2‐1 mutant Nicotiana benthamiana lineage (Feng et al., 2022) at 23 °C with a 16:8 h light:dark photoperiod. A C. septempunctata colony was maintained on A. pisum on Vicia faba (faba bean). Mealybug ladybird beetle (Cryptolaemus montrouzieri) adults were purchased from Amazon (www.amazon.com). Nicotiana benthamiana wild type and asat2‐1 mutant plants (Feng et al., 2022) for aphid, whitefly and ladybug experiments were grown in Cornell Mix [by weight 56% peat moss, 35% vermiculite, 4% lime, 4% Osmocoat slow‐release fertilizer (Scotts, Marysville, OH), and 1% Unimix (Scotts)] in a Conviron growth chamber with a photosynthetic photon flux density of 200 mmol/m/s and a 16:8 h day:night photoperiod, at 23 °C with 50% relative humidity.
Sequencing, assembly and annotation of the USDA‐Red M. persicae genome
To produce a chromosome‐scale genome assembly of the USDA‐Red M. persicae strain, we combined PacBio long‐read, Illumina short‐read and Hi‐C sequencing of genomic DNA. To aid in gene prediction, the M. persicae transcriptome was sequenced using Illumina strand‐specific RNA‐Seq and PacBio Iso‐Seq platforms. Details for the sequencing, assembly and annotation of the M. persicae genome are elaborated in the Supplemental Methods.
Identification of horizontally transferred genes
To identify HTGs in M. persicae strain USDA‐Red, we compared protein sequences in the USDA‐Red genome to six databases of complete proteomes in UniProt, including archaea, bacteria, fungi, plants, metazoa (excluding proteins from species in the Arthropoda), and other eukaryotes (the remaining eukaryotes excluding fungi, plants, and metazoa). The index of horizontal gene transfer, h, was calculated by subtracting the bitscore of the best metazoan match from that of the best non‐metazoan match as described by Crisp et al. (2015). We defined candidate HTGs as those with h ≥ 30 and the bitscore of the best non‐metazoan match hit ≥100. The corresponding genome sequences of these candidates were compared against NCBI nt database to exclude contamination. We then performed phylogenetic analyses to validate HTGs. For each candidate gene, its protein sequence was compared by BLASTP against the protein databases of six taxa (archaea, bacteria, fungi, plants, metazoan and other eukaryotes). The top five hits from each taxon were extracted, and aligned with the protein sequence of the candidate gene using MUSCLE (Larkin et al., 2007). Each alignment was trimmed to exclude regions where gaps were more than 20% of sequences. Phylogenetic trees were constructed with PhyML (Guindon et al., 2009) using a JTT model with 100 bootstraps. HTGs were considered validated if the genes were monophyletic with taxa of plants, bacteria, fungi or other microorganisms.
To identify HTGs in M. persicae strain G006 (Mathers et al., 2017; Ramsey et al., 2007), we implemented de novo HTG annotation following the same protocol as for M. persicae strain USDA‐Red. We conducted a homology search using the HTGs that we found in M. persicae strain USDA‐Red and other genes previously reported in aphids (Parker and Brisson, 2019; Verster et al., 2019). The HTG homologues in other aphid species (A. pisum, Aphis glycines, Aphis gossypii, Daktulosphaira vitifoliae, Diuraphis noxia, Eriosoma lanigerum, Myzus cerasi, Pentalonia nigronervosa, Rhopalosiphum maidis, and Rhopalosiphum padi) were identified using reciprocal homology blast (Table 1; gene IDs in other aphid species are listed in Table S1). The newest‐version genomes and annotations of all aphid species were downloaded from AphidBase (https://bipaa.genouest.org/is/aphidbase/) and used in this study. In a previous publication, we described the identification of whitefly HTGs (Chen et al., 2016).
Table 1.
Horizontally transferred genes identified in aphid genomes
| ID in M. persicae USDA‐Red | Annotation | Origin | M. persicae G006 | A. pisum | A. glycines | A. gossypii | D. vitifoliae | D. noxia | E. lanigerum | M. cerasi | P. nigronervosa | R. maidis | R. padi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MPE127429 | Putative LD carboxypeptidase | Bacteria | y | y | y | y | n | y | y | y | y | y | y |
| MPE127428 | ATP binding protein | Bacteria | y | y | y | y | y | y | y | y | y | y | y |
| MPE015894 | N‐acetylmuramoyl‐L‐alanine amidase | Bacteria | y | y | n | n | n | y | n | y | n | n | n |
| MPE005086 | Cytolethal distending toxin subunit B | Viruses | y | n | n | n | n | n | n | y | n | n | n |
| MPE025920 | Non‐structural protein NS1 superfamily | Viruses | y | y | y | n | y | n | n | y | y | n | y |
| MPE014463 | Non‐structural protein NS1 superfamily | Viruess | y | n | n | n | n | n | n | n | n | n | n |
| MPE006690 | Non‐structural protein NS1 superfamily | Viruses | y | n | n | n | n | n | n | n | n | n | n |
| MPE006487 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006492 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006495 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006496 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006497 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006498 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006500 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006501 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006504 | Lycopene cyclase/phytoene synthase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE006493 | Carotenoid desaturase | Fungi | y | y | y | y | y | y | y | y | y | y | n |
| MPE006499 | Carotenoid desaturase | Fungi | y | y | y | y | y | y | y | y | y | y | n |
| MPE006502 | Carotenoid desaturase | Fungi | y | y | y | y | y | y | y | y | y | y | n |
| MPE006505 | Carotenoid desaturase | Fungi | y | y | y | y | y | y | y | y | y | y | y |
| MPE002740 | Unknown with ankyrin repeats | Plants | y | y | y | y | n | n | y | y | y | y | y |
| MPE008575 | Unknown with ankyrin repeats | Plants | y | n | n | n | n | n | n | y | n | n | n |
| MPE008576 | Unknown with ankyrin repeats | Planst | y | n | n | n | n | n | n | y | n | n | n |
| MPE010373 | Unknown with ankyrin repeats | Plants | y | y | n | n | n | n | n | y | y | y | y |
| MPE011636 | Reverse transcriptase domain‐containing protein | Plants | y | n | n | n | n | n | n | y | n | n | y |
| MPE013670 | Unknown with ankyrin repeats | Plants | y | n | n | n | n | n | n | y | n | n | n |
| MPE016359 | Unknown with ankyrin repeats | Plants | y | y | y | y | n | y | n | y | y | y | y |
| MPE018635 | Unknown with ankyrin repeats | Plants | y | n | n | n | n | n | n | n | n | n | n |
| MPE018636 | Unknown with ankyrin repeats | Plants | y | y | n | n | n | n | n | n | n | n | y |
| MPE023973 | Unknown with ankyrin repeats | Plants | y | n | y | n | n | y | n | y | n | y | y |
Design of dsRNA and cloning of target HTG sequences in the TRV‐VIGS vector
For each target gene, dsRNA fragments with a length of 150–300 bp were designed using the ERNAi website (https://www.dkfz.de/signaling/e‐rnai3/; Horn and Boutros, 2010). To reduce the chances of off‐target silencing, the designed fragments were checked against the reference genomes of the experimental species, M. persicae strain USDA‐Red (this study), B. tabaci MEAM1 (Chen et al., 2016), N. benthamiana (Bombarely et al., 2012), and C. septempunctata, using a VIGS tool (Fernandez‐Pozo et al., 2015) and local BLAST (Mount, 2007) to eliminate dsRNA fragments that contain any perfect matches of ≥19 nt for HTGs of bacterial and plant origin and >21 nt for HTGs of fungal origin to the reference genomes. The retained dsRNA fragments were cloned into the TRV‐VIGS vector (Senthil‐Kumar and Mysore, 2014) using the Invitrogen Gateway recombination cloning system (Invitrogen). Briefly, dsRNA fragments were first PCR‐amplified from aphid cDNA using primers designed based on the dsRNA fragments (Table S2). PCR products were then cloned into pDONR207 using the Gateway BP clonase, dsRNA fragments in pDONR207 were swapped into Gateway compatible TRV2 plasmid (pTRV2) by recombination using Gateway LR clonase. The final constructs were named TRV2‐GOI (gene of interest). In parallel, we obtained a construct TRV2‐PDS, which carries a dsRNA fragment targeting N. benthamiana phytoene desaturase as a positive control for expression silencing (Velasquez et al., 2009). As negative controls, we used TRV2‐GFP or TRV2‐GUS, which carry dsRNA fragments targeting Green Fluorescent Protein (GFP) or Escherichia coli beta‐glucuronidase (GUS) genes, respectively, but not sequences that are found in either the target insects or N. benthamiana (Vaghchhipawala et al., 2011), as well as a TRV2‐EV (empty vector).
Transient agrobacterium infection of N. benthamiana for TRV‐based VIGS
To infiltrate plants, plasmids (TRV1 and TRV2 carrying the gene of interest) were transformed into Agrobacterium tumefaciens and then infiltrated into N. benthamiana as previously described (Senthil‐Kumar and Mysore, 2014). Briefly, the TRV1 and TRV2‐GOI plasmids were transformed into A. tumefaciens strain GV3101. Each TRV2‐GOI Agrobacterium culture was mixed with one carrying TRV1 and adjusted to a final OD600 of 0.3 with cell suspension buffer [final concentrations at 0.01 m MES (2‐(N‐morpholino)ethanesulfonic acid), 0.01 m MgCl2, and 0.2 mm acetosyringone]. After 3 h of incubation at 23 °C, the Agrobacterium TRV mixtures were infiltrated to saturate three leaves of v4‐stage N. benthamiana using 1‐mL needleless syringes. Inoculated plants were kept in a growth chamber for 2–3 weeks, at which point plants infiltrated with TRV‐PDS showed photobleaching symptoms that indicated viral spread. The TRV‐GOI plant leaves were assayed for the presence of TRV1 and the expression of GOI dsRNA using PCR (Figure S1). Leaf samples were collected for RNA extraction using the SV Total RNA Isolation system (Promega, Fitchburg, WI) and cDNA synthesis using the High‐Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). The synthesized cDNA samples were amplified by PCR using Phusion™ High‐Fidelity DNA Polymerase (ThermoFisher Scientific, Waltham, MA) and the VIGS primers (Table S2, Figures S2 and S3). A 20 μL PCR reaction contains 4 μL betaine, 4 μL 5× High‐Fidelity buffer, 1 μL 2.5 mm dNTPs, 0.2 μL Phusion polymerase, 1 μL each of 10 mm forward and reverse primers, 1 μL cDNA and 7.8 μL of ddH2O. The PCR program was 95 °C for 3 min, followed by 25 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min, with a final step of 72 °C for 5 min. The confirmed TRV‐GOI plants were then used for aphid bioassays.
Aphid bioassays
For each GOI, 5 or 6 infiltrated plants were used for aphid bioassays, and two aphid cages were attached to the adaxial side of fully developed young leaves on each plant. To set up the bioassays, 10 adult aphids were added to each cage and allowed to produce nymphs for 24 h. The adults were removed, and 25 newborn nymphs were scored for survival every 24 h for up to 5 days. After 5 days, 10–15 adult aphids were collected and flash‐frozen in liquid nitrogen for target gene expression analyses using quantitative reverse transcriptase‐PCR (qRT‐PCR) (Primers see Table S2, Figures S2 and S3).
To test the effects of silencing bacterial and plant‐origin HTGs on aphid reproduction, immediately after the 5th‐day survival check, the aphid cages were moved to previously uninfested leaves and five adult aphids were left in each cage to measure aphid reproduction for a week. To test the effects of dsTor and dsCarRP on aphid reproduction, one adult aphid was added to each cage for 24 h, and then the adult aphid was removed. Nymphs that were deposited by the adult aphids were collected after 7 days, the number of progeny and the survival of the nymphs was scored, and adults were collected for qRT‐PCR analysis.
Ladybug bioassays
On wildtype N. benthamiana plants, M. persicae prefer feeding on older, senescing leaves but the TRV virus tends to move to younger and new leaves. To generate enough aphids for the ladybug bioassays, we performed additional VIGS experiments and maintained a larger aphid population on asat2‐1 mutant N. benthamiana plants (Feng et al., 2022), which are deficient in acyl sugars and allow efficient M. persicae feeding on the TRV‐infected younger leaves.
For ladybug bioassays, 2‐day‐old C. septempunctata larvae were used for all experiments. Each ladybug larva was individually caged in a vented plastic cup and aphids from VIGS N. benthamiana plants were served every other day. The survival of each ladybug was tracked for 7 days, and 10 ladybugs were monitored for each of the VIGS treatments. The experiments were repeated twice using wildtype N. benthamiana and twice with asat2‐1 mutant N. benthamiana. To confirm the tri‐trophic persistence of RNAi signals, we also conducted assays using C. montrouzieri adults.
Whitefly bioassays
As B. tabaci strain MEAM1 could not survive on wildtype N. benthamiana, we used asat2‐1 mutant N. benthamiana (Feng et al., 2022) for bioassays. For each experiment, 3 or 4 TRV‐GOI infiltrated plants were used for each gene of interest, and three cages were attached to each plant. Ten young adult whiteflies (emerged within the past week) were placed in each cage. After 7 days, the survival rates of whiteflies were assessed and compared across different treatments. The surviving whiteflies were collected for q‐RT‐PCR analyses of target gene expression. The whitefly VIGS bioassays were repeated three times with similar results. Two replicates of a separate experiment were set up to collect whiteflies after 1 day for gene expression analyses.
mRNA expression and qPCR analysis
We used q‐RT‐PCR to measure the expression of target GOIs in treated aphids and whiteflies. Total RNA was extracted from 10 to 20 M. persicae using the SV Total RNA Isolation system (Promega) and RNA was reverse‐transcribed using the High‐Capacity cDNA Reverse Transcription Kits (Applied Biosystems). cDNAs were then diluted 10‐fold and used for qPCR reactions. Each qPCR reaction contained 5 μL of the PowerUp™ SYBR™ Green PCR master mix (Applied Biosystems), with 1 μL of each qPCR primer (Table S2) and 1 μL of cDNA. qPCR primers were designed to not overlap with the selected dsRNA in each of the targeted genes to avoid detecting signals from ingested dsRNAs. PCR reactions were initiated with an incubation at 95 °C for 30 s, followed by a 40 cycles of 95 °C for 5 s, 60 °C for 1 min and a melting curve was collected after the reaction. The Ct values were used to quantify and analyse gene expression according to the method (Livak and Schmittgen, 2001). Ubiquitin and/or EF1a were used as the internal control genes in each qPCR experiment for HTGs of bacterial and plant origin and RpL7 was used as an internal control for Tor and CarRP. Changes in the expression levels of each gene were calculated by comparing the ratio of the ΔΔC T values of samples from aphids and whiteflies fed on TRV2‐GOI plants to those fed on TRV2 empty vector control plants.
Data analyses and statistics
For both insect bioassays and gene expression assays, data were pooled from multiple experiments for statistical analyses. We tested for differences using the univariate analysis of variance with a fixed factor of treatments and a random factor of experiment, followed by a Bonferroni post hoc test for multiple comparison corrections using SPSS v.25 (IBM, Armonk, NY). Differences in ladybug survival were tested using the Cox mixed‐effect model (Therneau and Grambsch, 2000) followed by a Tukey post hoc test using R Studio v 1.3.959 (RStudio Team, 2020). For dsTor and dsCarRP, the aphid reproduction data were tested using ANOVA followed by the Tukey's HSD test. For gene expression data, Mann–Whitney U‐tests were used to test for differences.
Results
Chromosome‐scale assembly of the M. persicae genome
As the genome of the USDA‐Red strain of M. persicae had not been sequenced, we generated a chromosome‐scale genome assembly by combining PacBio long‐read, Illumina short‐read and Hi‐C sequencing. The assembled M. persicae genome has a total length of 383.0 Mb and consists of 331 contigs with an N50 length of 4.5 Mb. A total of 364.4 Mb (95.1% of the assembly) are clustered into six chromosomes (Figure 1), which is consistent with the commonly observed 2n = 12 karyotype of M. persicae (Blackman, 1980). To evaluate the completeness of the M. persicae genome assembly, the Illumina sequencing reads were aligned to the assembly, allowing up to three mismatches using BWA‐MEM (Li and Durbin, 2009), resulting in a mapping rate of 95.2%, indicating that most of the acquired reads were successfully assembled into the genome. Furthermore, BUSCO (Simao et al., 2015) showed that 94.2% of the core eukaryotic genes were at least partially captured by the genome assembly and 92.6% were completely captured. Combining evidence from ab initio predictions, protein homology and transcripts derived from Illumina strand‐specific RNA‐Seq and PacBio Iso‐Seq data of the USDA‐Red strain, we predicted a total of 27 430 protein‐coding genes in the M. persicae genome. The assembled genome has been deposited in GenBank under PRJNA888091.
Figure 1.
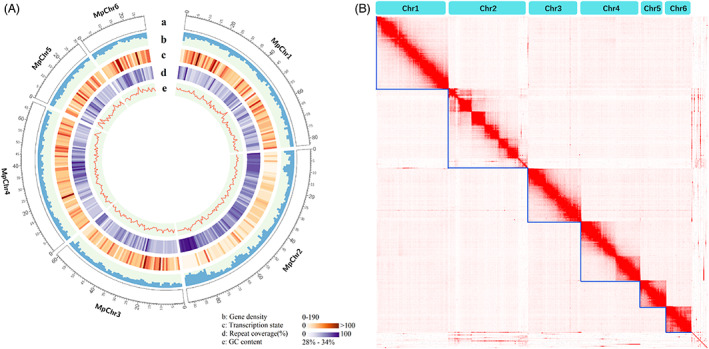
Chromosome‐scale genome assembly of a tobacco‐adapted red Myzus persicae strain. (A) M. persicae genome landscape. (a) Ideogram of the 6 M. persicae pseudochromosomes at the Mb scale. (b) Gene density is represented as number of genes per Mb. (c) Transcription state. The transcription level was estimated by read counts per million mapped reads in 1‐Mb windows. (d) Percentage of coverage of repeat sequences per Mb. (e) Guanine‐cytosine (GC) content in 1‐Mb windows. (B) Heatmap showing the frequency of HiC contacts along the M. persicae genome assembly. Blue lines indicate chromosome‐scale scaffolds.
Horizontally transferred genes (HTGs) in M. persicae str. USDA‐Red
We identified 30 genes that were predicted to be horizontally transferred from bacteria, viruses, fungi or plants into M. persicae strains USDA‐Red and G006 (Table 1 and Table S1). Genes of bacterial origin include those encoding an uncharacterized protein with a predicted ATP binding function, an N‐acetylmuramoyl‐L‐alanine amidase (amiD) and a microcin c7 self‐immunity protein, which is a homologue of the A. pisum murein tetrapeptide carboxypeptidase (ldcA1, ACYPI009109) (Table 1). Genes of viral origin were all identified based on homology search using previously reported HTGs (Parker and Brisson, 2019; Verster et al., 2019). For the non‐structural protein NS1 superfamily genes, three copies were identified in the M. persicae USDA‐Red genome, whereas either one or two copies were present in other species, including A. pisum in which these HTGs were initially identified (Parker and Brisson, 2019). Genes of fungal origin were mainly predicted to be involved in carotenoid biosynthesis, with nine copies of lycopene cyclase‐phytoene synthase and four copies of carotenoid desaturase genes (Table 1). Genes of plant origin are most similar to those from microalgae and encode uncharacterized proteins with predicted ankyrin repeats, that is, MPE002740, MPE008575, MPE008576, MPE010373, MPE013670, MPE016359, MPE018635, MPE018636, MPE023973 (Table 1). In addition, one other plant‐origin HTG, MPE011636, was identified from our de novo annotation (Table 1).
Genomic comparisons across different aphid species (A. pisum, A. glycines, A. gossypii, D. vitifoliae, D. noxia, E. lanigerum, M. cerasi, P. nigronervosa, R. maidis, and R. padi) showed that most HTGs from M. persicae strains USDA‐Red and G006 are present in multiple other aphid species (Table 1). For instance, the fungal‐origin lycopene cyclase/phytoene synthase gene and all of its duplicates were found in all assessed aphid species. By contrast, one of the plant‐origin uncharacterized ankyrin repeat protein genes (MPE0018635) and the two additional copies of non‐structural protein gene (MPE014463 and MPE006690) were found only in M. persicae strains G006 and USDA‐Red (Table 1).
Tri‐trophic persistence of RNA
To examine the possibility of tri‐trophic persistence of RNAi signals, we used two ladybug species, C. septempunctata larvae (Figure 2) and C. montrouzieri adults (Figure S4). A dsRNA fragment (dsGFP), targeting the GFP gene, which it is not naturally present in plants, aphids or ladybugs, was expressed in N. benthamiana. The plant‐expressed dsRNA of GFP fragments were consistently detected in both ladybug species that were fed on aphids that fed on dsGFP plants (Figure 2 and Figure S4). As the aphids were removed from the plants for feeding C. septempunctata larvae and C. montrouzieri, we could rule out direct transfer of the RNA from plants to beetles. Given the observed transfer of RNA from plants to beetles via aphids, we proposed using RNAi to target HTGs that are specific to insect pests, thereby avoiding negative impacts on non‐target species, in particular beneficial insects like C. septempunctata larvae and C. montrouzieri.
Figure 2.
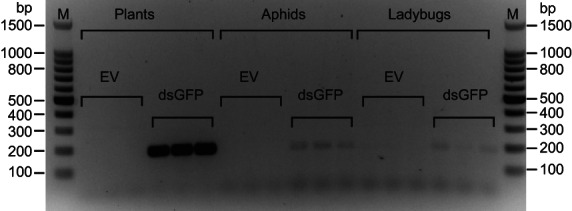
Evidence of tri‐trophic persistence of dsRNA. Agarose gel image showing the detection of dsGFP in Coccinella septempunctata ladybug larvae after consuming Myzus persicae aphids that had fed on Nicotiana benthamiana infiltrated with TRV2‐GFP. The expected amplicon size is 180 bp, which is consistent to the band size shown in the gel. PCR from cDNA samples of the following treatments are shown in each lane: Plants: plants that infiltrated with TRV2 empty vector (EV) or TRV2‐GFP vector (dsGFP); Aphids: aphids fed on EV and dsGFP VIGS plants; Ladybugs: ladybugs fed on aphids that were fed on EV and dsGFP VIGS plants. Each treatment was performed with three biological replicates (shown as three lanes), and this experiment was repeated twice. M: Promega 100 bp DNA ladder.
Silencing of M. persicae bacterial‐origin HTGs using VIGS
To evaluate the effects of silencing bacterial‐origin HTGs (amidase, ATP binding protein, and ldcA) on both plant‐feeding aphids and the ladybugs consuming these aphids, we knocked down gene expression in M. persicae using TRV VIGS. Expression of all three VIGS‐targeted genes was significantly reduced relative to aphids feeding on TRV2‐EV, TRV2‐GUS, and/or TRV2‐GFP control plants (P < 0.05) (Figure 3). For the amidase gene, we observed a significantly lower aphid survival rate as early as 24 h after the initiation of aphid feeding on VIGS plants, compared to both TRV2‐GUS and EV controls (P < 0.05). In the case of the ATP binding protein gene, aphid survival rates decreased significantly on the TRV2‐ATPb VIGS plants compared to TRV2‐GFP (only initially at 24 h post feeding) or EV plants (throughout the time period monitored, from 24 to 120 h post‐feeding) (P < 0.05). In the case of ldcA, we observed a significantly lower aphid survival rate on TRV2‐ldcA plants compared to both TRV2‐GFP and EV controls (P < 0.05). We did not observe significant effects on aphid reproduction with any of the three tested genes. Furthermore, there were no significant effects on the survival of C. septempunctata, when using either wildtype or asat2‐1 mutant N. benthamiana plants for the VIGS experiments.
Figure 3.
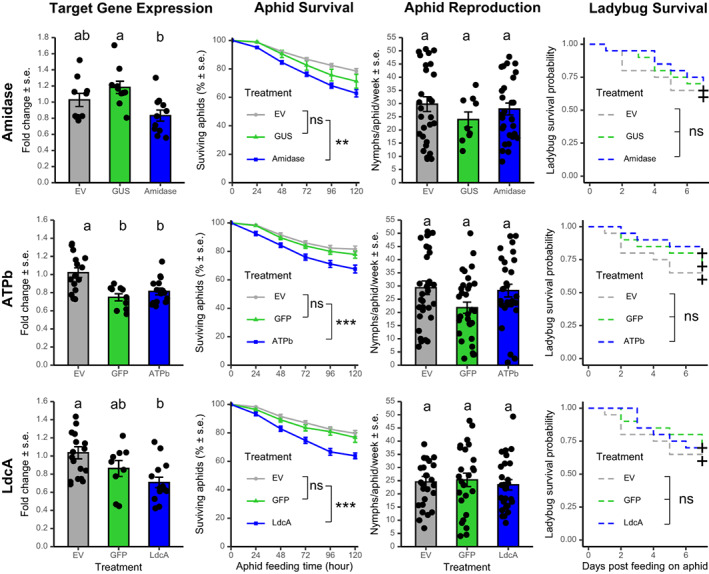
Knockdown of bacterial‐origin horizontally transferred genes reduced aphid survival with no effects on ladybug survival. Bioassay experiments were repeated at least 5 times, with 5–8 plants for each construct as biological replicates in each experiment. Dots on top of each bar graph represent individual data points collected from multiple experiments. Letters above the bars indicate significant differences between treatments by ANOVA (P < 0.05) with a block effect (experiment). Significant differences in the aphid survival between treatments are indicated for the last time point (i.e. 120 h), **P < 0.01, ***P < 0.001, ns, not significant.
VIGS‐mediated silencing of M. persicae plant‐origin HTGs
After 120 h of feeding, expression of five plant‐origin HTGs, all of which encode uncharacterized proteins with ankyrin repeat domains (Table 1), was significantly down‐regulated by VIGS in comparison to M. persicae on TRV2‐GFP and/or EV control plants (P < 0.05) (Figure 4). The effects on gene expression varied from a 20% decrease for MPE018635 to a 50% decrease for MPE013670. We observed a significantly lower aphid survival rate on the VIGS plants compared to both TRV2‐GFP and EV controls (P < 0.05) (Figure 4). The decreases in aphid survival ranged from 30% for MPE018635 to 40% for MPE010373 after 120 h of aphid feeding (Figure 4). The significant decreases in aphid survival were observed starting from 24 h after initiation of aphid feeding and continuing to decrease over time, with the exception of MPE023973, for which significant decreases were observed only starting at 96 h (Figure 4). Similar to the bacterial‐origin HTGs, we did not observe significant effects on aphid reproduction with any of these five plant‐origin HTGs. Similarly, there were no significant effects on the survival of C. septempunctata, when using either wildtype or asat2‐1 mutant N. benthamiana plants for the VIGS experiments. For three additional genes of plant origin, MPE018636, MPE002740 and MPE016359, we did not find significant effects on aphid survival. In the case of dsMPE018636 VIGS plants, we observed a significant decrease in aphid survival in only one of five VIGS experiments. However, MPE018636 expression was not significantly decreased in that experiment.
Figure 4.
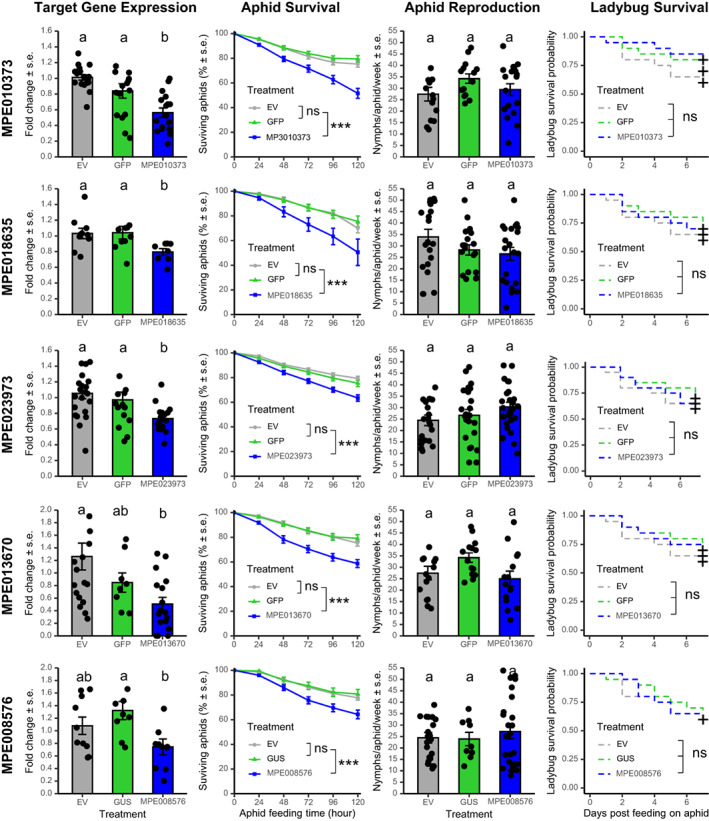
Knockdown plant‐origin horizontally transferred genes reduced aphid survival with no effects on ladybug survival. Bioassay experiments were repeated at least 5 times, with 5–8 plants for each construct as biological replicates in each experiment. Dots on top of each bar graph represent individual data points collected from multiple experiments. Letters above the bars indicate significant differences between treatments by ANOVA (P < 0.05) with a block effect (experiment). Significant differences of the aphid survival between treatments are indicated for the last time point (i.e. 120 h), **P < 0.01, ***P < 0.001, ns, not significant.
VIGS‐mediated silencing of the fungal‐origin Tor and CarRP genes
Carotene desaturase (Tor) is required for the production of the red carotenoids that provide the colour of M. persicae strain USDA‐Red. Carotenoid cyclase–carotenoid synthase (CarRP) catalyses both the committed step for carotenoid biosynthesis and later steps involving the formation of cyclic carotenoids (Hansen and Moran, 2011; Novakova and Moran, 2012). We conducted Tor and CarRP VIGS experiments and checked their expression at a time point of 7 days after aphids were born on VIGS plants. Compared to aphids on dsGFP control plants, the expression levels of both genes were reduced by approximately 50% (P < 0.05) in aphids feeding on TRV2‐Tor and/or TRV2‐CarRP N. benthamiana plants (Figure 5A,B). No apparent macroscopic phenotypic changes (particularly loss of red colour) were observed among the groups of aphids. With only about 50% reduction in gene expression in the VIGS experiments, the lack of a visible colour change in the aphids could be due to residual carotenoid accumulation. Nevertheless, progeny production by aphids that were reared on the TRV2‐Tor or TRV2‐CarRP VIGS plants over 8 days was significantly reduced compared to the TRV2‐GFP control (Figure 5C).
Figure 5.
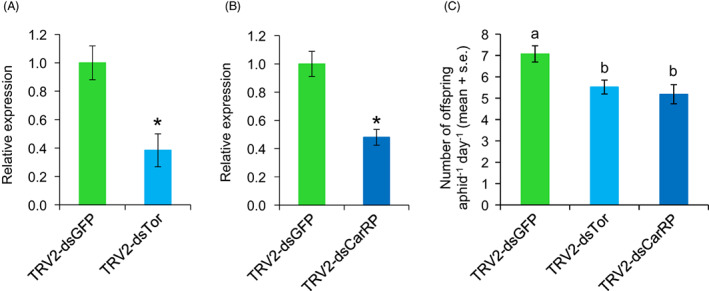
Silencing of Tor and CarRP genes reduces aphid reproduction. (A) Tor gene and (B) CarRP gene expression after 7 days exposure of Myzus persicae to TRV‐VIGS in Nicotiana benthamiana. Data shown are the mean ± standard error of 5 or 6 biological replicates, with three aphids pooled for each replicate. The Y‐axis has arbitrary units, with gene expression in the TRV‐GFP strain normalized as 1. *P < 0.05, Mann–Whitney U‐test relative to the control. (C) Reproductive output per aphid over 8 days of M. persicae reared on N. benthamiana plants with the two RNAi constructs. ANOVA results are shown, with different letters referring to treatments with significantly different expression by Tukey's HSD test.
VIGS‐mediated silencing of M. persicae viral‐origin cdtB
We tested one of the viral‐origin HTGs encoding the cytolethal distending toxin B (cdtB), which was suggested to be involved in aphid resistance to a predatory wasp (Verster et al., 2019). Somewhat unexpectedly, cdtB gene expression was consistently observed to be up‐regulated in the TRV‐cdtB plants relative to TRV‐GFP and EV controls (Figure 6a). This may be indicative of unexplained up‐regulation of cdtB expression as a compensatory response to the attempted RNA interference. We did not observe any effects on aphid survival in our plant‐mediated VIGS experiments, which were conducted in the absence of wasp predation (Figure 6b).
Figure 6.
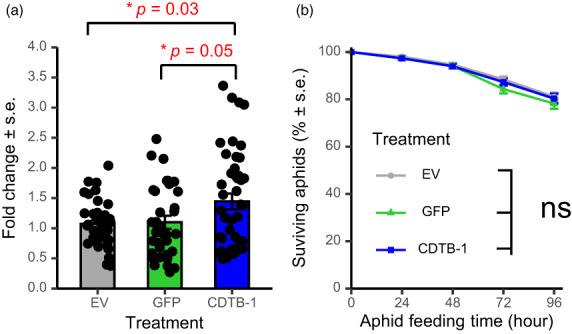
VIGS up‐regulated cdtB (virus‐origin HTG) gene expression (a), but did not affect aphid survival (b). Bioassays for were replicated 4 times with similar results. Each experiments included 5–8 plants for each construct as biological replicates. Significant differences between groups were tested using ANOVA with a block effect (experiment), followed by multiple comparisons using the Bonferroni method. *P < 0.05, ns, not significant.
VIGS‐mediated silencing of whitefly HTGs
Bemisia tabaci MEAM1 has at least 142 HTGs from bacteria and fungi (Chen et al., 2016). As VIGS targets, we chose five B. tabaci HTGs, encoding a squalene synthase (SQS, Bta11043), a diaminopimelate decarboxylase (DAPDC, Bta03593), an argininosuccinate synthase (ASS, Bta00062), a cyanate hydratase (CYN, Bta20016), and an aromatic peroxygenase (APO, Bta04808), respectively. In parallel, we chose three essential whitefly genes as positive controls, including acetylcholine esterase (AchE) and toll‐like receptor 7 (TLR7) genes, which have been targeted previously by RNAi (Malik et al., 2016), and the tubulin‐specific chaperon D gene (TBCD), which has been targeted by VIGS in aphids (Guo et al., 2014). We initially conducted VIGS experiments with wildtype N. benthamiana, but the low survival rate of B. tabaci on these plants made it impossible to reliably assess the effectiveness of gene expression silencing and interpret the results (Figure 7a). Instead, we conducted whitefly VIGS experiments with asat2‐1 mutant N. benthamiana, which allows improved whitefly reproduction and survival (Feng et al., 2022). In VIGS experiments with asat2‐1 plants, AchE and SQS expression was reduced after 1 and 7 days (Figure 7b,c), which resulted in significantly reduced whitefly survival (Figure 7d). Although TLR7 and TBCD gene expression was reduced by VIGS only on day 1 when feeding on asat2‐1 plants (Figure 7b), there was nevertheless a negative effect on whitefly survival over 7 days (Figure 7d). Two possible explanations for this observation are: (i) There was a survivor bias in that we could only measure gene expression levels in surviving whiteflies, and perhaps all whiteflies with efficient TLR7 and TBCD expression silencing were dead after 7 days; or (ii) there may be gene expression compensation at the whole‐insect level over time, but not in specific tissues that affect insect survival. Quantitative PCR of fractionated whiteflies would be necessary to determine the time course of TLR7 and TBCD expression silencing and whether VIGS primarily affects gene expression in specific tissue types.
Figure 7.
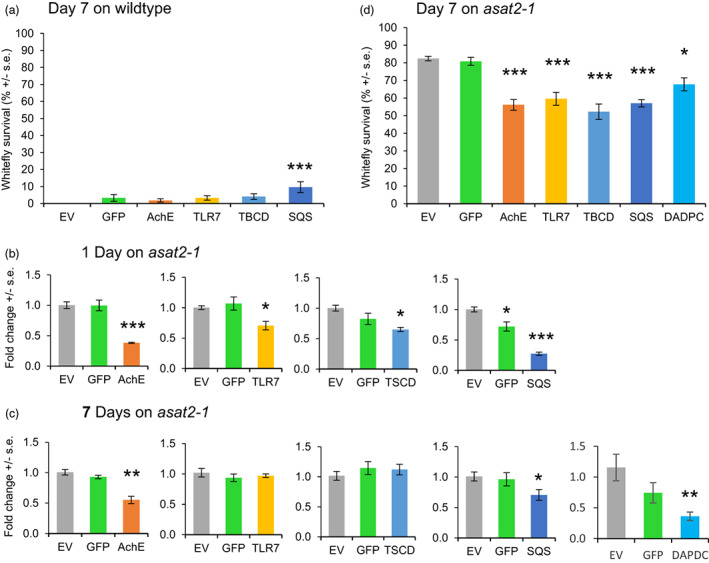
Virus induced gene silencing (VIGS) of whitefly genes using tobacco rattle virus (TRV). Bemisia tabaci (whiteflies) were placed on wildtype (a) and asat2‐1 (b–d) Nicotiana benthamiana plants infected with TRV expressing VIGS constructs, and gene expression was measured after one (b) and 7 days (c). In each experiment, the empty virus vector (EV) and GFP‐carrying virus were used as controls. Gene expression was normalized to 1 for EV control plants. Whitefly survival was assessed on TRV‐infected plants after 7 days of feeding on (a) wildtype plants or (d) asat2‐1 plants. GFP/AchE/SQS/TLR7/TBCD/DADPC: VIGS plants with TRV expressing RNA constructs targeting GFP, AchE, SQS, TLR7, TBCD, and DADPC, respectively; AchE: acetylcholinesterase, SQS: squalene synthase, TLR7: toll‐like receptor 7, TBCD: tubulin‐specific chaperon D, DADPC: diaminopimelate decarboxylase. The survival and the 7‐day qPCR experiments were repeated three times. The 1‐day qPCR experiments were repeaetd twice. Significant differences were determined using one‐way ANOVA with a fixed factor of treatments and a block effect of experiment, followed by a Dunnett's post hoc test for comparing VIGS constructs to the empty vector (EV) control. Mean ± SE of n = 3 (a), n = 9 (b), and n = 27 (c, d). *P < 0.05; **P < 0.01, ***P < 0.001.
Discussion
Identification of HTGs in insect pests
With the increasing number of available genome sequences, HTGs have been identified in all branches of life, including a broad range of agricultural insect pests such as aphids and whiteflies (Husnik and McCutcheon, 2018; Soucy et al., 2015). Many of those HTGs have been shown to be expressed and functional after the incorporation into the recipient genomes (Husnik and McCutcheon, 2018; Wybouw et al., 2016), providing the recipient species benefits such as nutrition, protection and adaptation to environmental stresses (Husnik and McCutcheon, 2018; Soucy et al., 2015). HTGs can help the recipient insects to digest plant cell wall components that would otherwise be non‐digestible (Kirsch et al., 2014; Pauchet et al., 2014; Pauchet and Heckel, 2013; Zhao et al., 2014). For example, the pectin‐degrading polygalacturonases found in herbivorous beetles are ancient HTGs from fungi that help beetles to digest cell wall components such as pectin, cellulose and hemicellulose (Kirsch et al., 2014). HTGs can also help recipient insects detoxify plant defensive metabolites (Daimon et al., 2008). For instance, mulberry latex, such as 1,4‐dideoxy‐1,4‐imino‐D‐arabinitol (D‐AB1) and 1‐deoxynojirimycin (DNJ), are highly toxic to non‐mulberry specialist insects, but present no harm to the mulberry specialist silkworm, Bombyx mori. A beta‐fructofuranosidase gene that was horizontally transferred from bacteria is highly expressed in the B. mori gut and helps in digesting plant‐defensives alkaloidal sugars (Daimon et al., 2008). In sap‐feeding insects, HTGs from bacteria have been found to complement the function of their obligate endosymbiont bacteria in the biosynthesis of essential nutrients, such as essential amino acids and vitamins (Husnik et al., 2013; Luan et al., 2015; Sloan et al., 2014). Some HTGs have been proposed to be involved in protecting recipient insects from pathogen or predators (Verster et al., 2019). For example, the cdtB gene of bacteriophage origin in the vinegar flies and aphids may function in defending these insects against natural enemies (Verster et al., 2019). As these nutritional and protective functions shown above are critical to insect development, survival and reproduction, and most of them are species‐ or genus‐specific, those HTGs in insect pests should be further explored as efficient and specific targets for the control of pest species.
HTGs in aphids and their potential for aphid control
Many HTGs were described in aphid genomes as aphid genomic resources started becoming available (Ding et al., 2020; International Aphid Genomics Consortium, 2010; Moran and Jarvik, 2010; Nikoh et al., 2010; Nikoh and Nakabachi, 2009; Parker and Brisson, 2019; Verster et al., 2019). Notably, aphids acquired carotenoid biosynthesis genes, which determine body colour through horizonal transfer from fungi. The absence of visible changes in aphid body colour after Tor and CarRP transcriptional silencing indicates that the observed 50% reduction in gene expression (Figure 5) does not cause a complete loss of carotenoid production. Additionally, aphid body colour is influenced by the environment, including abiotic (e.g. temperature) and biotic environmental (e.g. predation and endosymbionts) factors (Tsuchida, 2016). Thus, the colour changes from the knockdown of Tor and CarRP could be concealed by other factors that were not a focus in our RNAi experiments.
Aphid body colour further influences the ecological interactions between aphids and their predators, such as wasps (Moran and Jarvik, 2010). Due to reduced survival and/or reproduction, these predator interactions can be costly for aphids, (Nelson, 2007). Consistent with previous research, our plant‐mediated VIGS experiments showed negative effects on aphid performance, in this case progeny production. Although Tor and CarRP are believed to be dispensable during aphid development, the changes in Tor and CarRP gene expression could be perceived by the aphids as a metabolic disturbance, thereby incurring the observed reproductive costs.
We used plant‐mediated VIGS to determine that three HTGs of bacteria origin influence M. persicae survival. Transiently knocking down ATPb, amiD and LdcA expression significantly reduced aphid survival (Figure 3). This result for amiD and LdcA in M. persicae is consistent with an investigation of these two genes in A. pisum, in which knocking down amiD and ldcA by dsRNAs in artificial diet reduced aphid growth (Chung et al., 2018). In the case of ATPb, the VIGS construct caused no significant expression knockdown relative to the GFP control but there was nevertheless a decrease in aphid survival. There are several possible explanations for this: (i) Expression silencing is tissue‐localized and we do not observe a significant effect when analysing whole insects, (ii) Transient silencing and/or compensatory gene expression may be occurring, which can nevertheless have longer‐term negative effects, (iii) The constructs may be targeting multiple genes, with significant effects not being observed when analysing an individual gene, or (iv) the GFP construct may have unexpected negative effects on the expression of some aphid genes.
All of the plant‐derived HTGs that we identified in M. persicae encode uncharacterized proteins with ankyrin repeat domains. The ankyrin repeat domains consists of ~33 amino acid tandem motifs, which have been well documented in protein–protein interaction studies (Sedgwick and Smerdon, 1999). Since the first report of ankyrin repeat proteins in yeast (Saccharomyces cerevisiae), fruit flies (Drosophila melanogaster) and nematodes (Caenorhabditis elegans) (Breeden and Nasmyth, 1987), ankyrin repeat proteins have been identified in numerous organisms, ranging from viruses and bacteria to humans (Sedgwick and Smerdon, 1999). Bacterial ankyrin repeat proteins are mainly found in species that are obligately or facultatively associated with eukaryotic hosts (Siozios et al., 2013). Different ankyrin repeat proteins have been identified that regulate host‐pathogen and host‐symbiont interactions (Kumagai et al., 2007; Nguyen et al., 2014; Pan et al., 2008). Aphids are well known for their interactions with associated obligate and facultative symbionts, as well as being vectors for many plant viruses, and expression of horizontally acquired ankyrin repeat proteins was detected previously in aphid bacteriocyte and/or gut transcriptomes (Duncan et al., 2016; Feng et al., 2018). The functions of these proteins in mediating interactions between aphids and their symbionts will require further investigation. Nevertheless, our results show that these horizontally transferred ankyrin repeat proteins are important for aphid survival (Figure 4).
Selection of non‐specific controls for RNAi experiments
In several previous insect RNAi experiments, researchers chose a fragment of GFP as the negative control (Dong et al., 2022; Jain et al., 2022; Pitino et al., 2011; Tariq et al., 2019; Ye et al., 2019; Yoon et al., 2020), presumably because this sequence is not present in their target insects and plants. However, our experiments show that a GFP fragment may lead to non‐specific changes in the expression of some targeted genes. For instance, we observed that our target gene expression was similarly repressed by the negative control and TRV‐GFP (e.g. the results for ATPb in Figure 3). This was not the case when we used TRV‐GUS as the non‐specific negative control, as in the case of the amidase gene in Figure 3 and MPE008576 in Figure 4. Therefore, gene fragments used as negative controls for RNAi experiments will need to be cautiously selected and tested for their effects on the specific target genes of interest.
HTGs in whiteflies
In the whitefly genome, 64 genes were predicted to be horizontally transferred from bacteria and 78 genes were predicted to be horizontally transferred from fungi (Chen et al., 2016). Here, we have targeted five whitefly HTGs, which were chosen based on their predicted metabolic functions, using plant‐mediated VIGS and demonstrated that knocking down HTGs can significantly reduce whitefly survivorship (Figure 7). These results suggest that HTGs could be targets for the purpose of whitefly control.
Given that horizontal transfer of functionally expressed microbial genes into insect germlines is rare on an evolutionary timescale, there is likely a selective advantage to having these genes expressed in whiteflies. This was confirmed by the observation that VIGS of both SQS and DAPDC reduced whitefly survival relative to control plants (Figure 7d). Transient expression knockdown of SQS and DAPDC, as well as other horizontally transferred genes, will enable future research to study the functions of these genes in whitefly metabolism. Due to their importance for whitefly survival, as well their absence in beneficial insects such as ladybugs and lacewings, horizontally transferred genes also are attractive targets for controlling whiteflies by RNA interference.
Avoiding negative effects in beneficial species
Despite the reduction in aphid reproduction and/or survival when HTGs were targeted by RNAi, we did not observe any negative effects on the survival of C. septempunctata larvae that were feeding on these aphids. This is consistent with the lack of target sequence homology in the beetles, which do not have the HTGs. However, it is also possible that there are indirect negative effects on predatory insects feeding on aphids that were targeted with RNAi constructs. For instance, silencing cathepsin L gene expression in M. persicae reduced their protein content and made the less suitable as prey for C. septempunctata (Rauf et al., 2019).
Conclusions
The well‐established N. benthamiana VIGS system, which allows rapid cloning of target‐specific constructs (Liu et al., 2002), will allow screening of other horizontally transferred genes to identify those that are most suitable for hemipteran pest control. Such assays will be facilitated by the use of asat2 mutant N. benthamiana, which improves aphid growth and allows B. tabaci survival (Feng et al., 2022). For the majority of tested M. persicae and B. tabaci HTGs, knocking down expression by means of plant‐mediated VIGS decreased survival and/or reproduction. Importantly, no negative effects were observed on C. septempunctata. Together, our results indicate that HTGs have potential as efficient and biologically safe targets for hemipteran pest control by plant‐mediated RNA interference.
Conflict of interest
The authors declare that they have no conflicts of interest related to this research.
Author contributions
GJ, HF and VT designed experiments. HF, VT, SS, SH and FA: cloned genes, constructed virus vectors, measured gene expression, and conducted insect bioassays. HF, WC and GJ analysed data. HF and GJ wrote the manuscript. ZF, TU and GJ obtained funding and provided other essential resources. All authors approved the final manuscript.
Supporting information
Figure S1 Examples of planta expression of dsRNAs that targeting horizontally transferred genes.
Figure S2 Carotenoid desaturase nucleotide sequence alignment.
Figure S3 Carotenoid cyclase synthase nucleotide alignment.
Figure S4 Tri‐trophic persistence of dsRNA using a dsRNA targeting GFP.
Table S1 Homologous annotation of HTGs across aphid species.
Table S2 Primers used for aphid and whitefly VIGS.
Acknowledgements
This research funded through the United Stated Department of Agriculture awards 2021‐67013‐33565 and 2021‐67014‐342357 to GJ, agreement HR0011‐17‐2‐0053 from the Defence Advanced Research Projects Agency (DARPA) Insect Allies Program with the Boyce Thompson Institute, a scholarship from the International Research Support Initiative Program of the Pakistan Higher Education Commission to SH 1‐8/HEC/HRD/2020/10897, and the Binational Agricultural Research and Development Fund award FI‐471‐2012 to VT.
References
- Abdellatef, E. , Will, T. , Koch, A. , Imani, J. , Vilcinskas, A. and Kogel, K.H. (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae . Plant Biotechnol. J. 13, 849–857. [DOI] [PubMed] [Google Scholar]
- Bass, C. , Puinean, A.M. , Zimmer, C.T. , Denholm, I. , Field, L.M. , Foster, S.P. , Gutbrod, O. et al. (2014) The evolution of insecticide resistance in the peach potato aphid, Myzus persicae . Insect Biochem. Mol. Biol. 51, 41–51. [DOI] [PubMed] [Google Scholar]
- Bhatia, V. , Bhattacharya, R. , Uniyal, P.L. , Singh, R. and Niranjan, R.S. (2012) Host generated siRNAs attenuate expression of serine protease gene in Myzus persicae . PLoS One, 7, e46343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, R.L. (1980) Chromosome‐numbers in the Aphididae and their taxonomic significance. Syst. Entomol. 5, 7–25. [Google Scholar]
- Bombarely, A. , Rosli, H.G. , Vrebalov, J. , Moffett, P. , Mueller, L.A. and Martin, G.B. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant‐microbe biology research. Mol. Plant Microbe Interact. 25, 1523–1530. [DOI] [PubMed] [Google Scholar]
- Breeden, L. and Nasmyth, K. (1987) Similarity between cell‐cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila . Nature, 329, 651–654. [DOI] [PubMed] [Google Scholar]
- Casacuberta, J.M. , Devos, Y. , du Jardin, P. , Ramon, M. , Vaucheret, H. and Nogue, F. (2015) Biotechnological uses of RNAi in plants: risk assessment considerations. Trends Biotechnol. 33, 145–147. [DOI] [PubMed] [Google Scholar]
- Chen, W. , Hasegawa, D.K. , Kaur, N. , Kliot, A. , Pinheiro, P.V. , Luan, J. , Stensmyr, M.C. et al. (2016) The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 14, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chougule, N.P. and Bonning, B.C. (2012) Toxins for transgenic resistance to hemipteran pests. Toxins (Basel), 4, 405–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S.H. , Jing, X. , Luo, Y. and Douglas, A.E. (2018) Targeting symbiosis‐related insect genes by RNAi in the pea aphid‐Buchnera symbiosis. Insect Biochem. Mol. Biol. 95, 55–63. [DOI] [PubMed] [Google Scholar]
- Chung, S.H. , Feng, H. and Jander, G. (202) Engineering pest tolerance through plant‐mediated RNA interference. Curr. Opin. Plant Biol. 60, 102029. [DOI] [PubMed] [Google Scholar]
- Coleman, A.D. , Wouters, R.H. , Mugford, S.T. and Hogenhout, S.A. (2015) Persistence and transgenerational effect of plant‐mediated RNAi in aphids. J. Exp. Bot. 66, 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp, A. , Boschetti, C. , Perry, M. , Tunnacliffe, A. and Micklem, G. (2015) Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 16, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon, T. , Taguchi, T. , Meng, Y. , Katsuma, S. , Mita, K. and Shimada, T. (2008) Beta‐fructofuranosidase genes of the silkworm, Bombyx mori: insights into enzymatic adaptation of B. mori to toxic alkaloids in mulberry latex. J. Biol. Chem. 283, 15271–15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B.Y. , Niu, J. , Shang, F. , Yang, L. , Zhang, W. , Smagghe, G. and Wang, J.J. (2020) Parental silencing of a horizontally transferred carotenoid desaturase gene causes a reduction of red pigment and fitness in the pea aphid. Pest Manag. Sci. 76, 2423–2433. [DOI] [PubMed] [Google Scholar]
- Dong, Y. , Wu, M. , Zhang, Q. , Fu, J. , Loiacono, F.V. , Yang, Y. , Wang, Z. et al. (2022) Control of a sap‐sucking insect pest by plastid‐mediated RNA interference. Mol. Plant, 15, 1176–1191. [DOI] [PubMed] [Google Scholar]
- Duncan, R.P. , Feng, H. , Nguyen, D.M. and Wilson, A.C.C. (2016) Gene family expansions in aphids maintained by endosymbiotic and nonsymbiotic traits. Genome Biol. Evol. 8, 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga, D.A. , De Vos, M. and Jander, G. (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact. 27, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H. , Wang, L. , Wuchty, S. and Wilson, A.C.C. (2018) microRNA regulation in an ancient obligate endosymbiosis. Mol. Ecol. 27, 1777–1793. [DOI] [PubMed] [Google Scholar]
- Feng, H. , Acosta‐Gamboa, L. , Kruse, L.H. , Tracy, J.D. , Chung, S.H. , Fereira, A.R.N. , Shakir, S. et al. (2022) Acylsugars Protect Nicotiana benthamiana Against Insect Herbivory and Desiccation. Plant Mol. Biol. 109, 505–522. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Pozo, N. , Rosli, H.G. , Martin, G.B. and Mueller, L.A. (2015) The SGN VIGS tool: user‐friendly software to design virus‐induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant, 8, 486–488. [DOI] [PubMed] [Google Scholar]
- Guindon, S. , Delsuc, F. , Dufayard, J.F. and Gascuel, O. (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137. [DOI] [PubMed] [Google Scholar]
- Guo, H.Y. , Song, X.G. , Wang, G.L. , Yang, K. , Wang, Y. , Niu, L.B. , Chen, X.Y. et al. (2014) Plant‐generated artificial small RNAs mediated aphid resistance. PLoS One, 9, e97410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, A.K. and Moran, N.A. (2011) Aphid genome expression reveals host‐symbiont cooperation in the production of amino acids. Proc. Natl Acad. Sci. USA, 108, 2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, T. and Boutros, M. (2010) E‐RNAi: a web application for the multi‐species design of RNAi reagents–2010 update. Nucleic Acids Res. 38, W332–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik, F. and McCutcheon, J.P. (2018) Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 16, 67–79. [DOI] [PubMed] [Google Scholar]
- Husnik, F. , Nikoh, N. , Koga, R. , Ross, L. , Duncan, R.P. , Fujie, M. , Tanaka, M. et al. (2013) Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell, 153, 1567–1578. [DOI] [PubMed] [Google Scholar]
- Ibrahim, A.B. , Monteiro, T.R. , Cabral, G.B. and Aragao, F.J.L. (2017) RNAi‐mediated resistance to whitefly (Bemisia tabaci) in genetically engineered lettuce (Lactuca sativa). Transgenic Res. 26, 613–624. [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium (2010) Genome sequence of the pea aphid Acyrthosiphon pisum . PLoS Biol. 8, e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R.G. , Fletcher, S.J. , Manzie, N. , Robinson, K.E. , Li, P. , Lu, E. , Brosnan, C.A. et al. (2022) Foliar application of clay‐delivered RNA interference for whitefly control. Nat. Plants, 8, 535–548. [DOI] [PubMed] [Google Scholar]
- Kanakala, S. , Kontsedalov, S. , Lebedev, G. and Ghanim, M. (2019) Plant‐mediated silencing of the whitefly Bemisia tabaci cyclophilin B and heat shock protein 70 impairs insect development and virus transmission. Front. Physiol. 10, 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch, R. , Gramzow, L. , Theissen, G. , Siegfried, B.D. , Ffrench‐Constant, R.H. , Heckel, D.G. and Pauchet, Y. (2014) Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: key events in the evolution of herbivory in beetles. Insect Biochem. Mol. Biol. 52, 33–50. [DOI] [PubMed] [Google Scholar]
- Koch, R.L. , Hodgson, E.W. , Knodel, J.J. , Varenhorst, A.J. and Potter, B.D. (2018) Management of insecticide‐resistant soybean aphids in the upper midwest of the United States. J. Integr. Pest Manag. 9, 1–7. [Google Scholar]
- Kumagai, H. , Hakoyama, T. , Umehara, Y. , Sato, S. , Kaneko, T. , Tabata, S. and Kouchi, H. (2007) A novel ankyrin‐repeat membrane protein, IGN1, is required for persistence of nitrogen‐fixing symbiosis in root nodules of Lotus japonicus . Plant Physiol. 143, 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.L. , Schiff, M. and Dinesh‐Kumar, S.P. (2002) Virus‐induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Wu, K. , Jiang, Y. , Xia, B. , Li, P. , Feng, H. , Wyckhuys, K.A. et al. (2010) Mirid bug outbreaks in multiple crops correlated with wide‐scale adoption of Bt cotton in China. Science, 328, 1151–1154. [DOI] [PubMed] [Google Scholar]
- Luan, J.B. , Ghanim, M. , Liu, S.S. and Czosnek, H. (2013) Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 43, 740–746. [DOI] [PubMed] [Google Scholar]
- Luan, J.B. , Chen, W. , Hasegawa, D.K. , Simmons, A.M. , Wintermantel, W.M. , Ling, K.S. , Fei, Z. et al. (2015) Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap‐feeding insects. Genome Biol. Evol. 7, 2635–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H.J. , Raza, A. , Amin, I. , Scheffler, J.A. , Scheffler, B.E. , Brown, J.K. and Mansoor, S. (2016) RNAi‐mediated mortality of the whitefly through transgenic expression of double‐stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 6, 38469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, J.J. and Zeng, F.R. (2014) Plant‐mediated RNAi of a gap gene‐enhanced tobacco tolerance against the Myzus persicae . Transgenic Res. 23, 145–152. [DOI] [PubMed] [Google Scholar]
- Mathers, T.C. , Chen, Y. , Kaithakottil, G. , Legeai, F. , Mugford, S.T. , Baa‐Puyoulet, P. , Bretaudeau, A. et al. (2017) Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 18, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn, M. , Kough, J. , Vaituzis, Z. and Matthews, K. (2003) Are Bt crops safe? Nat. Biotechnol. 21, 1003–1009. [DOI] [PubMed] [Google Scholar]
- Moran, N.A. and Jarvik, T. (2010) Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science, 328, 624–627. [DOI] [PubMed] [Google Scholar]
- Mount, D.W. (2007) Using the Basic Local Alignment Search Tool (BLAST). CSH Protoc. 2007, pdb.top17. [DOI] [PubMed] [Google Scholar]
- Mutti, N.S. , Park, Y. , Reese, J.C. and Reeck, G.R. (2006) RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum . J. Insect Sci. 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, E.H. (2007) Predator avoidance behavior in the pea aphid: costs, frequency, and population consequences. Oecologia, 151, 22–32. [DOI] [PubMed] [Google Scholar]
- Nguyen, M.T.H.D. , Liu, M. and Thomas, T. (2014) Ankyrin‐repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 23, 1635–1645. [DOI] [PubMed] [Google Scholar]
- Nikoh, N. and Nakabachi, A. (2009) Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh, N. , McCutcheon, J.P. , Kudo, T. , Miyagishima, S.Y. , Moran, N.A. and Nakabachi, A. (2010) Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 6, e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova, E. and Moran, N.A. (2012) Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus. Mol. Biol. Evol. 29, 313–323. [DOI] [PubMed] [Google Scholar]
- Pan, X. , Luhrmann, A. , Satoh, A. , Laskowski‐Arce, M.A. and Roy, C.R. (2008) Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science, 320, 1651–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, B.J. and Brisson, J.A. (2019) A laterally transferred viral gene modifies aphid wing plasticity. Curr. Biol. 29, e2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauchet, Y. and Heckel, D.G. (2013) The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacteria through horizontal gene transfer. Proc. Biol. Sci. 280, 20131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauchet, Y. , Kirsch, R. , Giraud, S. , Vogel, H. and Heckel, D.G. (2014) Identification and characterization of plant cell wall degrading enzymes from three glycoside hydrolase families in the cerambycid beetle Apriona japonica . Insect Biochem. Mol. Biol. 49, 1–13. [DOI] [PubMed] [Google Scholar]
- Pessoa, R. , Rossi, G.D. and Busoli, A.C. (2016) Transgenic cotton‐fed Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) affects the parasitoid Encarsia desantisi Viggiani (Hymenoptera: Aphelinidae) development. Neotrop. Entomol. 45, 102–106. [DOI] [PubMed] [Google Scholar]
- Pitino, M. , Coleman, A.D. , Maffei, M.E. , Ridout, C.J. and Hogenhout, S.A. (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS One, 6, e25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, J.S. , Wilson, A.C. , De Vos, M. , Sun, Q. , Tamborindeguy, C. , Winfield, A. , Malloch, G. et al. (2007) Genomic resources for Myzus persicae: EST sequencing, SNP identification, and microarray design. BMC Genomics, 8, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, J.S. , Elzinga, D.A. , Sarkar, P. , Xin, Y.R. , Ghanim, M. and Jander, G. (2014) Adaptation to nicotine feeding in Myzus persicae . J. Chem. Ecol. 40, 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf, I. , Asif, M. , Amin, I. , Naqvi, R.Z. , Umer, N. , Mansoor, S. and Jander, G. (2019) Silencing cathepsin L expression reduces Myzus persicae protein content and the nutritional value as prey for Coccinella septempunctata . Insect Mol. Biol. 28, 785–797. [DOI] [PubMed] [Google Scholar]
- Raza, A. , Malik, H.J. , Shafiq, M. , Amin, I. , Scheffler, J.A. , Scheffler, B.E. and Mansoor, S. (2016) RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): potential technology for the control of whitefly. PLoS One, 11, e0153883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, T.B. , Mishra, S.K. , Sridharan, K. , Barnes, E.R. , Alyokhin, A. , Tuttle, R. , Kokulapalan, W. et al. (2021) First sprayable double‐stranded RNA‐based biopesticide product targets proteasome subunit beta type‐5 in Colorado potato beetle (Leptinotarsa decemlineata). Front. Plant Sci. 12, 728652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2020) RStudio: Integrated Development for R. Boston, MA: RStudio. http://www.rstudio.com/ [Google Scholar]
- Sears, M.K. , Hellmich, R.L. , Stanley‐Horn, D.E. , Oberhauser, K.S. , Pleasants, J.M. , Mattila, H.R. , Siegfried, B.D. et al. (2001) Impact of Bt corn pollen on monarch butterfly populations: a risk assessment. Proc. Natl Acad. Sci. USA, 98, 11937–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick, S.G. and Smerdon, S.J. (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316. [DOI] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2014) Tobacco rattle virus‐based virus‐induced gene silencing in Nicotiana benthamiana . Nat. Protoc. 9, 1549–1562. [DOI] [PubMed] [Google Scholar]
- Simao, F.A. , Waterhouse, R.M. , Ioannidis, P. , Kriventseva, E.V. and Zdobnov, E.M. (2015) BUSCO: assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics, 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- Siozios, S. , Ioannidis, P. , Klasson, L. , Andersson, S.G. , Braig, H.R. and Bourtzis, K. (2013) The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One, 8, e55390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, D.B. , Nakabachi, A. , Richards, S. , Qu, J. , Murali, S.C. , Gibbs, R.A. and Moran, N.A. (2014) Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap‐feeding insects. Mol. Biol. Evol. 31, 857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.E. , Li, Y. , Perreau, J. and Moran, N.A. (2022) Elucidation of host and symbiont contributions to peptidoglycan metabolism based on comparative genomics of eight aphid subfamilies and their Buchnera. PLoS Genet. 18, e1010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy, S.M. , Huang, J. and Gogarten, J.P. (2015) Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. [DOI] [PubMed] [Google Scholar]
- Sujii, E.R. , Togni, P.H. , de A Ribeiro, P. , de A Bernardes, T. , Milane, P.V. , Paula, D.P. , Pires, C.S. et al. (2013) Field evaluation of Bt cotton crop impact on nontarget pests: cotton aphid and boll weevil. Neotrop. Entomol. 42, 102–111. [DOI] [PubMed] [Google Scholar]
- Tariq, K. , Ali, A. , Davies, T.G.E. , Naz, E. , Naz, L. , Sohail, S. , Hou, M.L. et al. (2019) RNA interference‐mediated knockdown of voltage‐gated sodium channel (MpNav) gene causes mortality in peach‐potato aphid, Myzus persicae . Sci. Rep. 9, 5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, N. , Upadhyay, S.K. , Verma, P.C. , Chandrashekar, K. , Tuli, R. and Singh, P.K. (2014) Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v‐ATPase A gene. PLoS One, 9, e87235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau, T.M. and Grambsch, P.M. (2000) Modeling Surivival Data: Extending the Cox Model. New York, NY: Springer‐Verlag. [Google Scholar]
- Tian, J.C. , Yao, J. , Long, L.P. , Romeis, J. and Shelton, A.M. (2015) Bt crops benefit natural enemies to control non‐target pests. Sci. Rep. 5, 16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T. (2016) Molecular basis and ecological relevance of aphid body colors. Curr. Opin. Insect Sci. 17, 74–80. [DOI] [PubMed] [Google Scholar]
- Tzin, V. , Yang, X. , Jing, X. , Zhang, K. , Jander, G. and Douglas, A.E. (2015) RNA interference against gut osmoregulatory genes in phloem‐feeding insects. J. Insect Physiol. 79, 105–112. [DOI] [PubMed] [Google Scholar]
- Vaghchhipawala, Z. , Rojas, C.M. , Senthil‐Kumar, M. and Mysore, K.S. (2011) Agroinoculation and agroinfiltration: simple tools for complex gene function analyses. Methods Mol. Biol. 678, 65–76. [DOI] [PubMed] [Google Scholar]
- Velasquez, A.C. , Chakravarthy, S. and Martin, G.B. (2009) Virus‐induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 10, 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster, K.I. , Wisecaver, J.H. , Karageorgi, M. , Duncan, R.P. , Gloss, A.D. , Armstrong, E.E. , Price, D.K. et al. (2019) Horizontal transfer of bacterial cytolethal distending toxin B genes to insects. Mol. Biol. Evol. 36, 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw, N. , Pauchet, Y. , Heckel, D.G. and Van Leeuwen, T. (2016) Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol. Evol. 8, 1785–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J. , Guo, Z. , Yang, Z. , Han, H. , Wang, S. , Xu, H. , Yang, X. et al. (2021) Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell, 184, e1617. [DOI] [PubMed] [Google Scholar]
- Xie, X. , Shang, F. , Ding, B.Y. , Yang, L. and Wang, J.J. (2022) Assessment of a zinc finger protein gene (MPZC3H10) as potential RNAi target for green peach aphid Myzus persicae control. Pest Manag. Sci. 78, 4956–4962. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Duan, X. , Lv, Y. , Zhang, X. , Nie, Z. , Xie, C. , Ni, Z. et al. (2014) Silencing of an aphid carboxylesterase gene by use of plant‐mediated RNAi impairs Sitobion avenae tolerance of Phoxim insecticides. Transgenic Res. 23, 389–396. [DOI] [PubMed] [Google Scholar]
- Ye, C. , Jiang, Y.D. , An, X. , Yang, L. , Shang, F. , Niu, J.Z. and Wang, J.J. (2019) Effects of RNAi‐based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci. Rep. 9, 3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J.S. , Tian, H.G. , McMullen, J.G. , Chung, S.H. and Douglas, A.E. (2020) Candidate genetic determinants of intraspecific variation in pea aphid susceptibility to RNA interference. Insect Biochem. Mol. Biol. 103408, 103408. [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Doucet, D. and Mittapalli, O. (2014) Characterization of horizontally transferred beta‐fructofuranosidase (ScrB) genes in Agrilus planipennis . Insect Mol. Biol. 23, 821–832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Examples of planta expression of dsRNAs that targeting horizontally transferred genes.
Figure S2 Carotenoid desaturase nucleotide sequence alignment.
Figure S3 Carotenoid cyclase synthase nucleotide alignment.
Figure S4 Tri‐trophic persistence of dsRNA using a dsRNA targeting GFP.
Table S1 Homologous annotation of HTGs across aphid species.
Table S2 Primers used for aphid and whitefly VIGS.


