Abstract
Viruses rely on hosts for their replication: thus, a critical step in the infection process is identifying a suitable host cell. Bacterial viruses, known as bacteriophages or phages, often use receptor binding proteins to discriminate between susceptible and non-susceptible hosts. By being able to evade predation, bacteria with modified or deleted receptor-encoding genes often undergo positive selection during growth in the presence of phage. Depending on the specific receptor(s) a phage uses, this may subsequently affect the bacteria’s ability to form biofilms, its resistance to antibiotics, pathogenicity, or its phenotype in various environments. In this study, we characterize the interactions between two T4-like phages, Sf22 and KRT47, and their host receptor S. flexneri outer membrane protein C (OmpC). Results indicate that these phages use a variety of surface features on the protein, and that complete resistance most frequently occurs when hosts delete the ompC gene in full, encode premature stop codons to prevent OmpC synthesis, or eliminate specific regions encoding exterior loops.
Keywords: Bacteriophage, Shigella, Phage therapy, phage-host interactions, outer membrane protein C
1. Introduction
Bacteriophages—viruses that infect bacteria, often abbreviated phages—are increasingly being applied for agricultural, industrial, and medical purposes. A benefit of phage application is their host specificity, meaning they can recognize either narrow or broad ranges of host genus, species, or subtypes [1]; however, resistance to phage is a problem when considering their use. Understanding how phages use host cell surface receptors can increase the likelihood that bacteria remain susceptible due to varying selective pressures [2]. The application of phages will likely select for the loss or modification of some receptors in the host. If the receptor and mechanism of attachment are known in advance, it may be possible to use or design phages that “kill the winner” under other control measures, for example reducing the pathogen’s virulence [3] or increasing their susceptibility to antibiotics [4–6]. Due to rising antibiotic resistance, several priority pathogens were listed for which new therapeutics are needed, making them good candidates for the development of phage therapy (WHO announcement). One genus of these pathogens is Shigella, which causes bacillary dysentery, and is responsible for approximately 267 million infections and over 200,000 deaths every year [7].
Shigella is an intracellular pathogen, which invades a layer of cells lining the gut epithelium using the type 3 secretion system (T3SS) encoded on its virulence plasmid [8,9]. In addition to the T3SS, S. flexneri requires certain outer membrane proteins to survive in and colonize the gut, including outer membrane protein C (OmpC; [10]). Of the numerous phage receptors that are known, OmpC is a common protein receptor for many phages, including members of the T4-like group [11]. These viruses have prolate icosahedral heads, long contractile tails, and genome sizes of about 165–175 k base pairs (kbp). Prior to infection, the initial contacts between T4 phages and their hosts occur through receptor binding proteins at the distal end of the phage tail fibers, either gp37 in T4 or gp38 in T2-like phages [12–14]. The structural and molecular mechanisms governing interactions between the E. coli phage T4 long tail fiber (LTF) gp37 and its receptors, OmpC and/or LPS, were recently described [15]. For gp37, the overall shape of OmpC appears to be more critical than a specific residue or loop. This is in contrast to receptors for other phages such as lambda, Ox2, Sf6, and TLS, which use LamB, OmpA, and TolC [16–19]. Phage T4 has been used in several fundamental experiments regarding phage therapy [20–24], and T4-like viruses are already being used in antibacterial products for food preparation [25,26]. These phages appear to be effective for various applications, but how bacteria may acquire resistance to these phages—and the implications thereof—is less understood.
Despite T4 being a foundational member of Tevenviridae, many phages in the subfamily encode a separate protein that binds to the end of the long tail fiber [13,27]. This receptor binding protein, denoted gp38 in phage T2, contains distinct hypervariable regions that are separated by glycine-rich motifs [12,27]. Interactions between the T2-like Salmonella virus S16 and its host have been investigated genetically and structurally, revealing it primarily uses loop 5 of OmpC rather than the entire protein, with minor variation in this loop—specifically regarding residues 227–229—conferring at least partial immunity to infection [14]. Based on an analysis of genes under positive selection in Escherichia coli, OmpC loops 4 and 5 are the regions commonly identified by both phage and contact dependent growth inhibition proteins [28]. This suggests that the T2-like, gp38-encoding phages may recognize more specific regions of their receptor, rather than the overall shape.
We previously isolated several members of the Tevenvirinae subfamily from the environment, all of which infect the genus Shigella: Sf21, Sf22, Sf23, Sf24, and Sf25 [29]. Of these, only Sf25 lacks a full-length gp38 homolog. In addition, virus Sf22 has the most clinically relevant host range, infecting all four species of Shigella—S. boydii, S. dysenteriae, S. flexneri, and S. sonnei—but not E. coli. In this work, we determined that OmpC is a critical receptor for phage Sf22 and characterized which regions are important for binding. We also identified a second phage from the environment, KRT47, which was found to use OmpC in a similar manner. Finally, we used experimental evolution to investigate how host S. flexneri gains resistance to these phages. The most common mechanism of resistance was deletion of the ompC gene or early truncation of the protein, which may result in strains with attenuated virulence.
2. Results and discussion
2.1. Two T4-like Shigella phages rely on OmpC as a receptor
Bacteriophages often employ a two-step process of attachment to their host. For example, Shigella phage Sf6 recognizes its S. flexneri host by binding initially to lipopolysaccharide, followed by interactions with one of at least two membrane proteins—outer membrane protein (Omp) A and/or OmpC [30]. Mechanistic studies of interactions between other Shigella phages and their hosts are scarce, but we hypothesized these membrane proteins would be utilized by other phages as well.
To investigate this hypothesis, we tested a library of phages isolated from the environment on S. flexneri Y strains lacking the OmpA (A−), OmpC (C−), or both OmpA and OmpC (A−/C−) genes. Of these, two phages were found to require OmpC (Table 1). Phage Sf22 was previously described in [29], while KRT47 was more recently isolated as described in the Materials and Methods. Plaque formation was completely restored by complementation of wild-type OmpC, suggesting that this protein is necessary and sufficient for optimal host recognition and infection by these phages. As reported in [29], phage Sf22 can infect all four species of Shigella. The LPS of these species range from smooth to rough, suggesting LPS is not a critical receptor for either phage. Between all Shigella species, the OmpC protein sequences share upwards of 93% sequence identity, with the few regions of variation mapping to the extracellular loops 2, 4, 5, and 7 (Fig. 1).
Table 1.
Efficiency of plating of Sf22 and KRT47 on S. flexneri with or without outer membrane proteins A or C.
| Host | |||||
|---|---|---|---|---|---|
| Phage | S. flexneri | ompA | ompA− + pOmpA | ompC | ompC− + pOmpC |
| Sf22 | 1.0 | 1.0 ± 0.3 | 1.2 ± 0.3 | <10−9 | 1.1 ± 0.2 |
| KRT47 | 1.0 | 1.0 | 1.0 | <10−9 | 0.9 ± 0.1 |
Fig. 1.
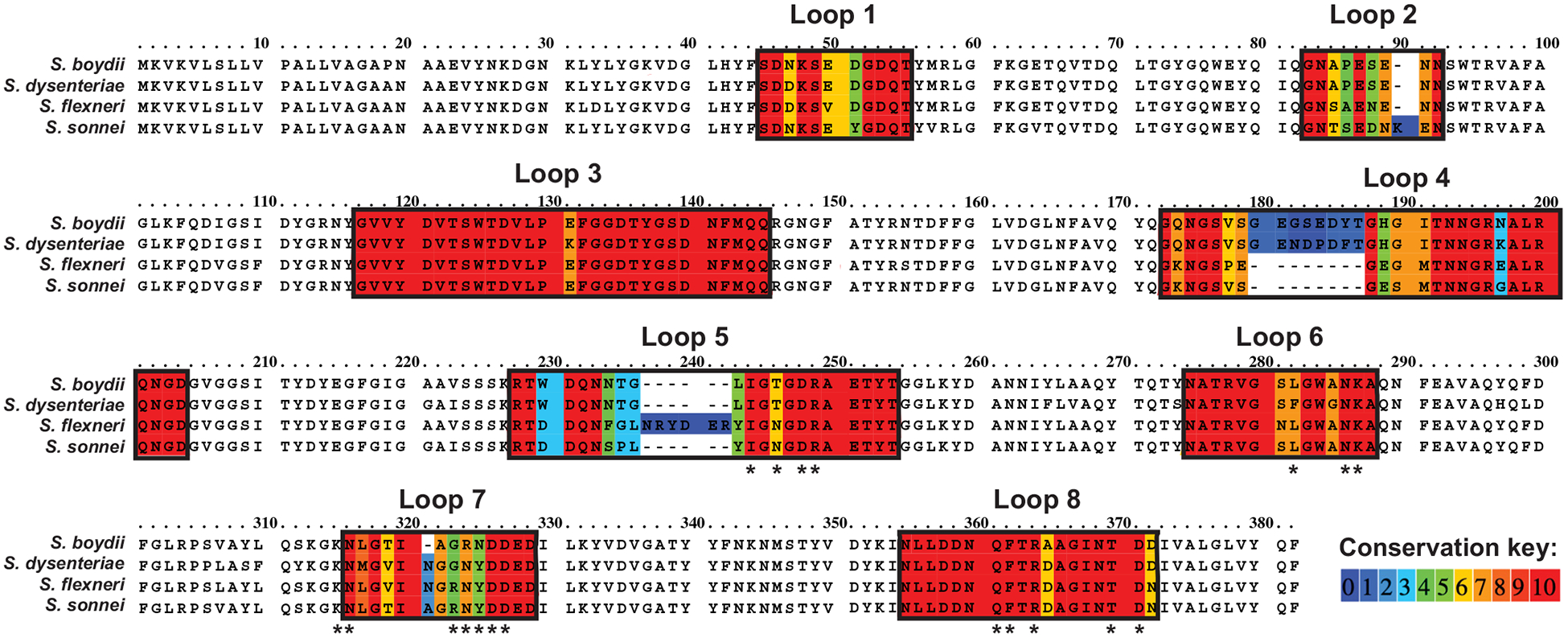
Alignments of OmpC proteins across all known species of Shigella; color key indicates conservation of amino acids. * indicates residues chosen for mutagenesis.
KRT47, was isolated nearly two years later, but had not been described. Its genome was sequenced and is deposited into GenBank under accession number MN781580. This phage was examined in greater detail to determine its morphology and phylogenetic relationship to other Shigella phages. Based on negative stain transmission electron microscopy and genome sequence data, this phage is also a close relative of T4. In addition, KRT47 shares 92.3% average nucleotide identity (ANI) with Sf22, where ANI is the percent nucleotide identity multiplied by the percent coverage. Like other previously described T4-like Shigella phages, KRT47 encodes a cell adhesion protein homologous to T2 gp38. This phage was also found to require OmpC for infection; however, in contrast to Sf22, phage KRT47 could infect only S. flexneri and S. sonnei. This suggests that KRT47 may either utilize a different region of OmpC, and/or it may require another component of the membrane for infection.
2.2. Bacteria gain resistance through partial or complete deletion of ompC
To determine how S. flexneri gains resistance to Sf22 or KRT47, bacteria were mixed with excess amounts of either phage and incubated overnight on agar plates. Single colonies were then transferred the following day onto a new plate covered with the same phage. For bacteria resistant to Sf22, 25 mutant strains were isolated. The ompC gene was amplified by PCR and sequenced to determine whether the gene was maintained and which mutations might confer resistance (Table 2). The gene was completely deleted in 15 mutants and truncated in one mutant. An additional seven mutants encoded wild-type OmpC, suggesting an alternative mechanism of resistance. Unfortunately, not all of ten of these survived storage during the COVID-19 lockdown: however, we were able to rescue three of them. We performed whole genome sequencing to determine alternative mechanism(s) of resistance. Two contained mutations known to affect phage infection and reproduction: 1) mutations in the lipopolysachharide synthesis pathway, and 2) a site-specific DNA recombinase, respectively. The third mutant had a synonymous mutation in a hypothetical protein, so the specific mechanism of resistance is unclear. However, a BLAST analysis of this protein shows homology to transcription factors in related Enterobacteriaceae, suggesting this mutant may be resistant to phage through gene regulation mechanisms. Two additional mutants encoded altered OmpC proteins. In one of these, 66 nucleotides corresponding to the first β-strand were deleted. Since this region is in the trimer interface, this could be either reducing the presence of OmpC in the outer membrane or reducing its ability to form trimers. In the second mutant, extracellular loop 4 gained an insertion while loops 5 and 7 were deleted in-frame, suggesting these loops might have the biggest effect on Sf22 recognition.
Table 2.
Summary of mutants isolated for resistance to Sf22 or KRT47.
| Phage | Mutation(s) in OmpC | Number of isolations |
|---|---|---|
| Sf22 | ΔompC | 15 |
| C30Δ/frameshift, V12am | 1 | |
| 12 nt insertion at 544 (loop 4) Δ10 nt at 672–682; Δ 7 nt at 688–695 (loop 5) Δ12 nt at 927–939 (loop 7) |
1 | |
| Δ66 nt at 41–107 (β-strand 1) | 1 | |
| No change | 7 | |
| KRT47 | ΔompC | 2 |
| C244T/Q81am | 5 | |
| C1089G/N363K | 1 | |
| No change | 2 |
For bacteria resistant to KRT47, ten mutant strains were isolated. In two mutants, ompC was deleted. In five mutants, a point mutation produced a stop codon at glutamine 81 (CAG to TAG), resulting in a truncated OmpC protein with only one extracellular loop. One mutant had a point mutation conferring an asparagine to lysine substitution at position 363 (Fig. 2). The remaining two mutants did not have any mutations in the ompC gene, again suggesting an alternative mechanism of resistance.
Fig. 2.
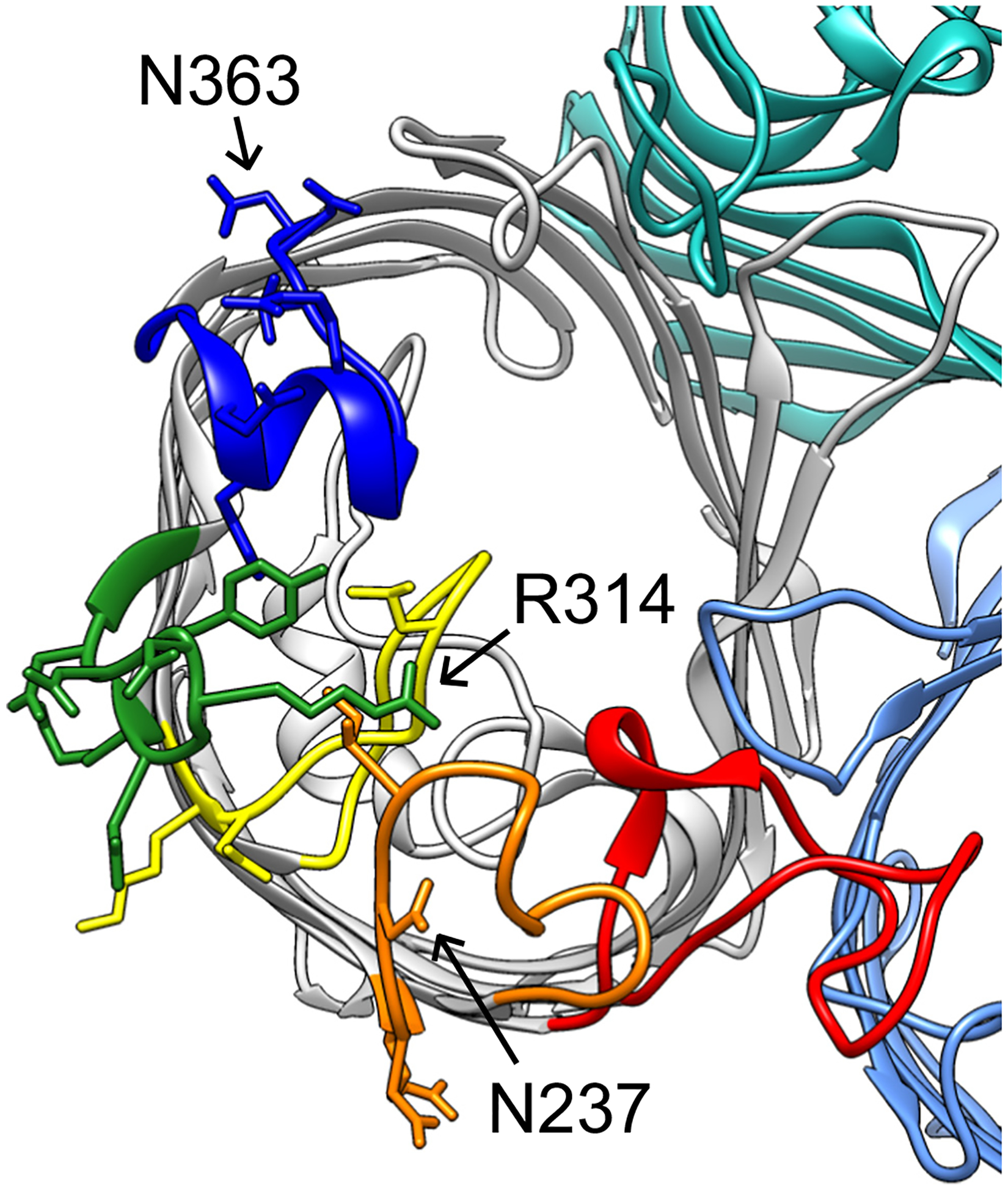
Structure of OmpC (PDB ID 2J1N), with extracellular loops four through eight of one monomer corresponding to red, orange, yellow, green, and blue, respectively. Visible side chains represent residues targeted for site-directed mutagenesis; N237, R314, and N363 are indicated by arrows.
2.3. Two additional single substitutions in OmpC confer resistance
To further determine the specific region recognized by Sf22 and KRT47, a series of mutations were made at the conserved residues of variable loops 5 and 7, plus the conserved loops 6 and 8. As shown in Fig. 2, these regions are away from the dimeric or trimeric interfaces of the porin, so they are unlikely to affect any inter-subunit interactions. Mutations in loop 4 were not generated, as these were more likely to interfere with these interactions.
Based on the efficiency of plating data illustrated in Fig. 3, only two of these mutations blocked infection by both phages. First, substituting asparagine 237 to threonine reduced efficiency of plating to below 10−6, which was the most dramatic reduction observed. Interestingly, this site is a threonine in S. boydii and S. dysenteriae, which Sf22 can naturally infect, yet the presence of this residue in the S. flexneri OmpC eliminates infectivity. The second substitution, arginine 314 to glutamate, reduced efficiency of plating to below 10−2. In both cases, these changes create a more negatively-charged surface in the same region (Fig. 2, arrows). The additional negative charge in this region may be sufficient to block recognition by the phage tail proteins. Conversely, the phage may use electrostatics of the opposite charge on the other side of the β-barrel, as the asparagine 363 to lysine similarly conferred resistance to infection. To ensure that these observed differences were due to phage interactions and not mislocalization of OmpC to the membrane, we fractionated outer membranes from strains expressing wild-type or mutant ompC genes. This was done by using a modified version of method 1 as described in [31]. We then performed mass spectrometry analysis on these outer membrane fractions and, using OmpA abundance as a reference, compared the ratio of OmpC to OmpA for each mutant strain to that of the unmodified OmpC. We chose to focus on two variants that demonstrated a severe phenotype (N237T and R314E), and two variants that demonstrated no phenotype (N237A and R314A). The results of this experiment indicate that all OmpC proteins reach the membrane, although N237T is reduced compared to the other four which were highly similar in signal. While the wild-type OmpC was present at a ratio of 0.594 ± 0.155 OmpC/OmpA, the N237A mutant was reduced approximately 20-fold to 0.012 ± 0.006. Conversely, the other mutant OmpC proteins were present at levels of 0.407 ± 0.052 (N237A), 0.407 ± 0.118 (R314E), and 0.455 ± 0.049 (R314A). Therefore, since R314E is localized to the membrane at wild type levels, it is very likely the substitution that is affecting phage attachment. For, N237T, the mutation may still affect phage binding, but the 20-fold reduction makes it difficult to determine the relative contribution to the phenotype.
Fig. 3.
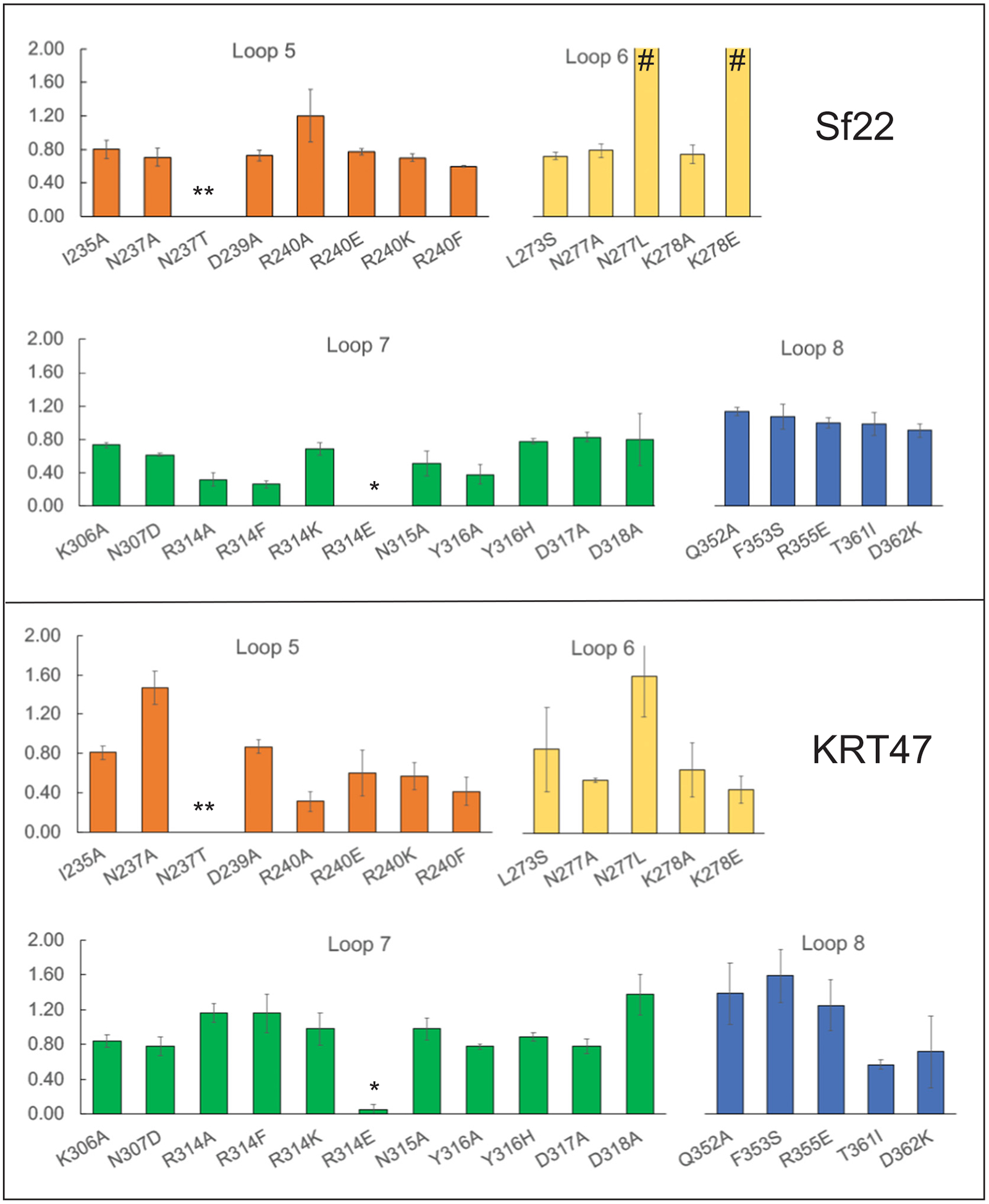
Efficiency of plating on cells expressing mutant ompC genes. Efficiency of plating ratio is on the y-axis, with mutations in OmpC represented on the x-axis. Symbols indicate: * <10−2; ** <10−6; # >2.0.
2.4. Minor differences in phage gp38 proteins may affect host range
For T2 and other T2-like viruses, it has been genetically and structurally demonstrated that gp38 interacts with OmpC. To examine which regions of the Sf22 and KRT47 gp38 protein may recognize the OmpC protein, sequences of the gp38 protein were aligned for three phages: Sf22, KRT47, and Sf24, where the latter does not require OmpC for infection. The alignment shown in Fig. 4A illustrates that these proteins exhibit the greatest differences in the C-terminal half of the protein. A model of the Sf22 gp38 protein, shown in Fig. 4B, was generated using Phyre2 [32], and the regions of these differences were mapped onto the structure. All regions of variation correspond to the five loops on the receptor-binding domain region of the protein. While this does not indicate single specific loops that may be responsible for OmpC binding, it is consistent with the hypothesis that these hypervariable segments of gp38 are responsible for determining host range [14,27].
Fig. 4.
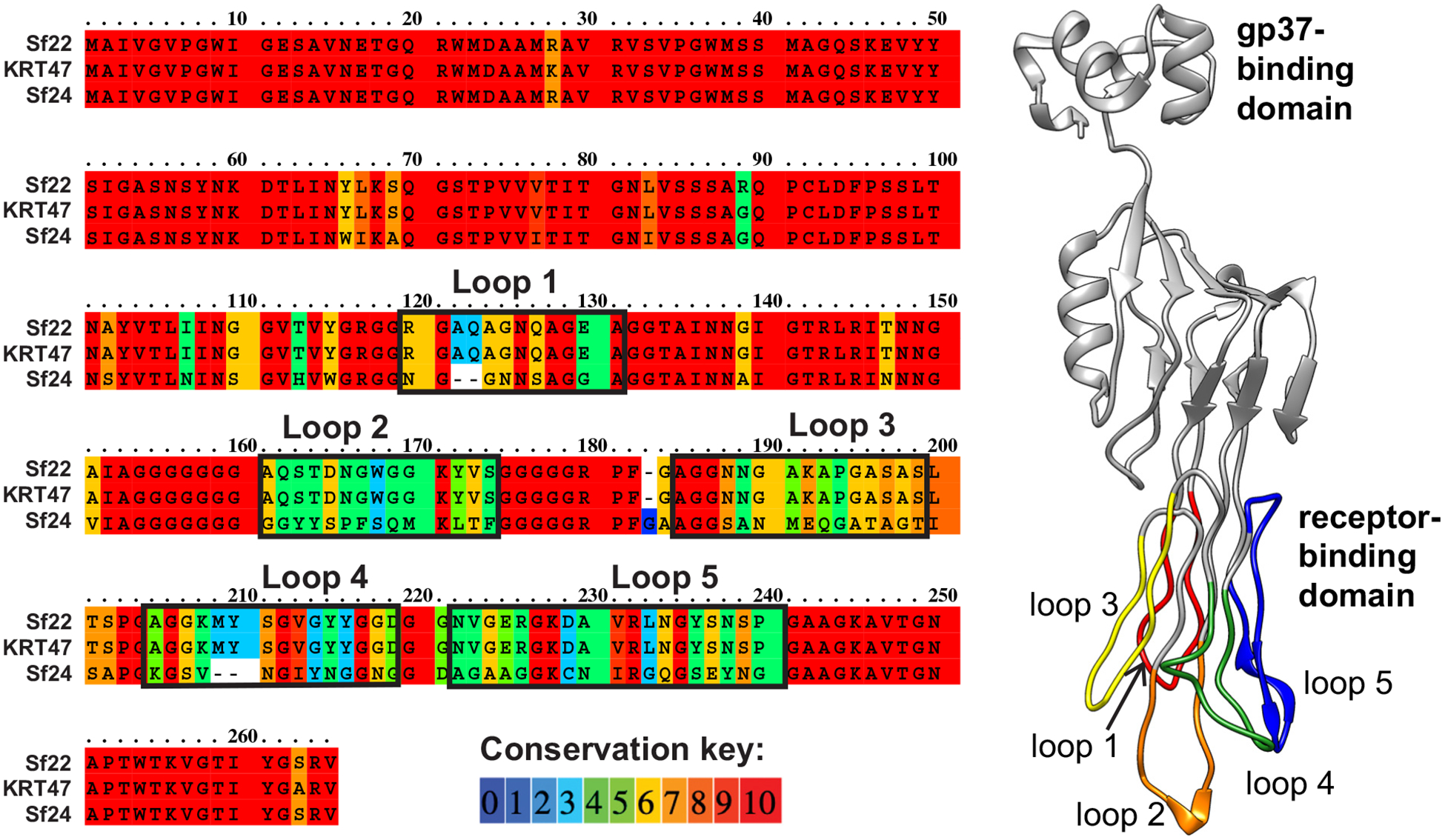
A) Alignment of gp38 proteins from Sf22, KRT47, and Sf24. B) Model of gp38 from Sf22, with variable loops highlighted.
2.5. OmpC-utilizing phages may reduce pathogenicity if not bacterial load
Like many species of bacteria, Shigella are becoming increasingly resistant to antibiotics [33]. Bacteriophages are a vast resource for combating antibiotic-resistant pathogens, and phage therapy is an active area of research [34]. Multiple phages can be combined in “cocktails” to target all possible strains of a given pathogen, eliminating the need to find and match a phage with the specific strain in an infected person [2] and reducing the chances of phage resistance mutants arising. Outside the clinical setting, bacteriophages may be applied to drinking water to reduce enteric bacteria in the environment or upon consumption [20]. Though phages can be highly effective in both settings, their increased usage requires careful investigation because of additional effects on virulence, microbial communities, and gene flow [35].
Here, we have identified the receptor for two Shigella phages in the T4 family, which are viruses that have been investigated for biocontrol and are currently being applied in various settings [20–26]. We subsequently determined how Shigella may become resistant to these phages. In both cases, deleting ompC or producing a truncated protein appears to be the primary mechanism of resistance, as single substitutions alone seem to be ineffective based on an analysis of mutant OmpC proteins. Since OmpC is necessary for Shigella to effectively colonize and persist in the human gut, using these phages may force the bacteria to either maintain its pathogenicity but be susceptible to the phage; or to develop phage resistance but lose its pathogenicity. Similar studies have demonstrated that bacteria must “choose” between antibiotic resistance or phage resistance [6]. Combining phages with antibiotics is one way to increase the lifespan of antibiotics, and to make treatments more effective. Alternatively, or in addition, using phages that target receptors necessary for pathogenesis may be another option for treatment. In either case, applying these phages in clinical settings or to products for human consumption may reduce the risk and burden of Shigella infections.
3. Materials and methods
3.1. Bacteriophage, bacteria, and plasmids
Bacteriophage Sf22 has been previously described [29]. Bacteriophage KRT47 was isolated on June 20, 2018 from the Red Cedar River in East Lansing, MI, using similar techniques as those used for Sf22 isolation. Single plaques were picked and purified twice by subsequent plating. Genomic DNA was purified from a high-titer stock via phenol-chloroform extraction as described in [29] and sequenced by the Massachusetts General Hospital Center for Computational and Integrative Biology (MGH CCIB). Genome annotation was conducted as in [29].
The S. flexneri knockout strains ompA, ompC, and ompA−/C− have been described in the PE577 background [30]. The ompC gene was cloned with its regulatory micF sequence {Andersen, 1989 #30} by amplifying PE577 genomic DNA, with a 5′ primer introducing a BamHI site and a 3′ primer introducing a HindIII site. The amplified gene was digested with BamHI and HindIII and ligated into pACYC184 digested with the same enzymes. The sequence was confirmed by Sanger sequencing and expression was confirmed by SDS-PAGE. To alter specific residues of OmpC, site-directed mutagenesis was performed by the primer-mediated QuikChange method (Agilent) with the following modification: VeraSeq high-fidelity enzyme (Enzymatics) was used for 20 cycles prior to DpnI digestion. Mutations were again confirmed by Sanger sequencing.
3.2. Host range determination and efficiency of plating
Initial host range assays were performed by dipping a toothpick into a high-titer phage stock and stabbing the toothpick into LB agar plates overlaid with test bacteria. Plates were incubated at 37 °C overnight. A clearing around the stab was indicative of a positive growth score. If the clearing was small, additional plaque assays were conducted to confirm growth on the host. Initial receptor assays were performed using the same toothpick method on S. flexneri PE577, OmpA, OmpC, and OmpA−/C− knockout strains. Stabs that resulted in no clearing on the knockout strains were selected for further efficiency of plating analysis.
To quantify the efficiency of plating, dilutions of high-titer phage stocks were plated using the double agar overlay method on various cell types. Efficiency of plating was calculated as the titer on the test bacteria divided by the titer on the permissive strain, S. flexneri PE577. All experiments used three biological replicates of each host strain.
3.3. Mass spectrometry analysis of outer membrane fractions
To determine abundance of outer membrane proteins A and C, we performed mass spectroscopy analysis of outer membrane fractions generated by a modified version of method 1 as described in [31]. In brief we created spheroplasts from 25 mL of bacterial culture, collected the outer membrane fraction, then TCA precipitated the proteins. These samples were separated by SDS-PAGE and the resulting single bands were excised for all proteins. The gel slices were proteolytically digested with trypsin and analyzed by LC/MS/MS at the RTSF Mass Spectrometry Core facility at Michigan State University. Three biological replicates were analyzed for each variant, with the averages and standard deviations across these three replicates reported in the text.
Acknowledgements
This work was supported by the National Institutes of Health GM110185 and National Science Foundation CAREER Award 1750125 to K.N.P. and by startup funds from the University of Florida to S.M.D.
Footnotes
Accession numbers
The genome of bacteriophage KRT47 has been deposited in NCBI GenBank as accession number MN781580. All representations of OmpC are based on PDB ID 2J1N.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].de Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE, Molecular and evolutionary determinants of bacteriophage host range, Trends Microbiol. 27 (2019) 51–63. [DOI] [PubMed] [Google Scholar]
- [2].Chan BK, Abedon ST, Loc-Carrillo C, Phage cocktails and the future of phage therapy, Future Microbiol. 8 (2013) 769–783. [DOI] [PubMed] [Google Scholar]
- [3].Chan BK, Brown K, Kortright KE, Mao S, Turner PE, Extending the lifetime of antibiotics: how can phage therapy help? Future Microbiol. 11 (2016) 1105–1107. [DOI] [PubMed] [Google Scholar]
- [4].Escobar-Paramo P, Gougat-Barbera C, Hochberg ME, Evolutionary dynamics of separate and combined exposure of Pseudomonas fluorescens SBW25 to antibiotics and bacteriophage, Evol. Appl 5 (2012) 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang QG, Buckling A, Phages limit the evolution of bacterial antibiotic resistance in experimental microcosms, Evol. Appl 5 (2012) 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE, Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa, Sci. Rep 6 (2016) 26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].G.B.D.D.D. Collaborators, Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016, Lancet Infect. Dis 18 (2018) 1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sansonetti PJ, Kopecko DJ, Formal SB, Involvement of a plasmid in the invasive ability of shigella flexneri, Infect. Immun 35 (1982) 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schroeder GN, Hilbi H, Molecular pathogenesis of shigella spp.: controlling host cell signaling, invasion, and death by type III secretion, Clin. Microbiol. Rev 21 (2008) 134–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bernardini ML, Sanna MG, Fontaine A, Sansonetti PJ, OmpC is involved in invasion of epithelial cells by shigella flexneri, Infect. Immun 61 (1993) 3625–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bertozzi Silva J, Storms Z, Sauvageau D, Host receptors for bacteriophage adsorption, FEMS Microbiol. Lett (2016) 363. [DOI] [PubMed] [Google Scholar]
- [12].Montag D, Riede I, Eschbach ML, Degen M, Henning U, Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution, J. Mol. Biol 196 (1987) 165–174. [DOI] [PubMed] [Google Scholar]
- [13].Montag D, Hashemolhosseini S, Henning U, Receptor-recognizing proteins of T-even type bacteriophages. The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb, J. Mol. Biol 216 (1990) 327–334. [DOI] [PubMed] [Google Scholar]
- [14].Dunne M, Denyes JM, Arndt H, Loessner MJ, Leiman PG, Klumpp J, Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition, Structure 26 (1573–1582) (2018), e1574. [DOI] [PubMed] [Google Scholar]
- [15].Islam MZ, Fokine A, Mahalingam M, Zhang Z, Garcia-Doval C, van Raaij MJ, Rossmann MG, Rao VB, Molecular anatomy of the receptor binding module of a bacteriophage long tail fiber, PLoS Pathog 15 (2019), e1008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Charbit A, Gehring K, Nikaido H, Ferenci T, Hofnung M, Maltose transport and starch binding in phage-resistant point mutants of maltoporin. Functional and topological implications, J. Mol. Biol 201 (1988) 487–496. [DOI] [PubMed] [Google Scholar]
- [17].Morona R, Tommassen J, Henning U, Demonstration of a bacteriophage receptor site on the Escherichia coli K12 outer-membrane protein OmpC by the use of a protease, Eur. J. Biochem 150 (1985) 161–169. [DOI] [PubMed] [Google Scholar]
- [18].Porcek NB, Parent KN, Key residues of S. Flexneri OmpA mediate infection by bacteriophage Sf6, J. Mol. Biol 427 (2015) 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].German GJ, Misra R, The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage, J. Mol. Biol 308 (2001) 579–585. [DOI] [PubMed] [Google Scholar]
- [20].Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brussow H, In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy, Antimicrob. Agents Chemother 48 (2004) 2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bruttin A, Brussow H, Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy, Antimicrob. Agents Chemother 49 (2005) 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weiss M, Denou E, Bruttin A, Serra-Moreno R, Dillmann ML, Brussow H, In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli, Virology 393 (2009) 16–23. [DOI] [PubMed] [Google Scholar]
- [23].Sarker SA, McCallin S, Barretto C, Berger B, Pittet AC, Sultana S, Krause L, Huq S, Bibiloni R, Bruttin A, Reuteler G, Brussow H, Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh, Virology 434 (2012) 222–232. [DOI] [PubMed] [Google Scholar]
- [24].Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, Salamon P, Youle M, Rohwer F, Bacteriophage adhering to mucus provide a non-host-derived immunity, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 10771–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mai V, Ukhanova M, Reinhard MK, Li M, Sulakvelidze A, Bacteriophage administration significantly reduces shigella colonization and shedding by shigella-challenged mice without deleterious side effects and distortions in the gut microbiota, Bacteriophage 5 (2015), e1088124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Soffer N, Woolston J, Li M, Das C, Sulakvelidze A, Bacteriophage preparation lytic for shigella significantly reduces shigella sonnei contamination in various foods, PLoS One 12 (2017), e0175256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM, The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage’s host specificity, Genome Biol. Evol 3 (2011) 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R, Genes under positive selection in Escherichia coli, Genome Res 17 (2007) 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Doore SM, Schrad JR, Dean WF, Dover JA, Parent KN, Shigella phages isolated during a dysentery outbreak reveal uncommon structures and broad species diversity, J. Virol 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Parent KN, Erb ML, Cardone G, Nguyen K, Gilcrease EB, Porcek NB, Pogliano J, Baker TS, Casjens SR, OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in shigella, Mol. Microbiol 92 (2014) 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Thein M, Sauer G, Paramasivam N, Grin I, Linke D, Efficient subfractionation of gram-negative bacteria for proteomics studies, J. Proteome Res 9 (2010) 6135–6147. [DOI] [PubMed] [Google Scholar]
- [32].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis, Nat. Protoc 10 (2015) 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Puzari M, Sharma M, Chetia P, Emergence of antibiotic resistant shigella species: a matter of concern, J. Infect. Public Health 11 (2018) 451–454. [DOI] [PubMed] [Google Scholar]
- [34].Gordillo Altamirano FL, Barr JJ, Phage therapy in the postantibiotic era, Clin. Microbiol. Rev 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Meaden S, Koskella B, Exploring the risks of phage application in the environment, Front. Microbiol 4 (2013) 358. [DOI] [PMC free article] [PubMed] [Google Scholar]


