Abstract
The emergence of a polybasic cleavage motif for the protease furin in SARS-CoV-2 spike has been established as a major factor for human viral transmission. The region N-terminal to that motif is extensively mutated in variants of concern (VOCs). Besides furin, spikes from these variants appear to rely on other proteases for maturation, including TMPRSS2. Glycans near the cleavage site have raised questions about proteolytic processing and the consequences of variant-borne mutations. Here, we identify that sialic acid-containing O-linked glycans on Thr678 of SARS-CoV-2 spike influence furin and TMPRSS2 cleavage and posit O-linked glycosylation as a likely driving force for the emergence of VOC mutations. We provide direct evidence that the glycosyltransferase GalNAc-T1 primes glycosylation at Thr678 in the living cell, an event that is suppressed by mutations in the VOCs Alpha, Delta, and Omicron. We found that the sole incorporation of N-acetylgalactosamine did not impact furin activity in synthetic O-glycopeptides, but the presence of sialic acid reduced the furin rate by up to 65%. Similarly, O-glycosylation with a sialylated trisaccharide had a negative impact on TMPRSS2 cleavage. With a chemistry-centered approach, we substantiate O-glycosylation as a major determinant of spike maturation and propose disruption of O-glycosylation as a substantial driving force for VOC evolution.
Short abstract
Proteolytic cleavage of S protein is critical in SARS-CoV-2 infection. GalNAc-T1-derived sialoglycans on S confer furin resistance and may be an evolutionary driver for COVID-19 variants of concern.
Introduction
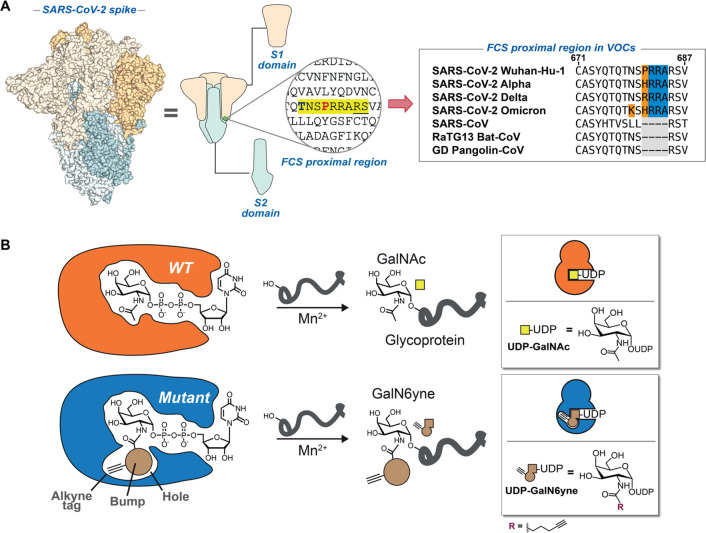
The viral surface spike protein has been the subject of intense scientific efforts to understand and curb SARS-CoV-2 transmission.1−29 Spike is a trimeric, multidomain glycoprotein (Figure 1A), with a glycan coat that plays crucial structural, immunological, and functional roles.15−28 An evolutionarily novel arginine-rich peptide sequence in SARS-CoV-2 spike has been identified as the cleavage site for the Golgi-localized convertase furin.29 This furin cleavage site (FCS) is crucial for SARS-CoV-2 transmission, as cleavage enhances receptor binding and likely the fusion activity of spike.3,30−33 On a molecular level, furin hydrolyses the peptide bond between Arg685 and Ser686, converting full-length spike (termed FL-S) into the fragments S1 and S2 in the mature protein.7,34−41 Another host protease, TMPRSS2, has been proposed to act synergistically with furin, potentially also targeting the FCS with preference to cleave before arginine.42,43
Figure 1.
Dissecting O-glycosylation on SARS-CoV-2 spike. (A) Left: SARS-CoV-2 spike model (PDB ID: 6ZGE) and (middle) its corresponding cartoon representation with the furin cleavage site (FCS) proximal region highlighted in yellow. The blue and bold T corresponds to Thr678, which is a potential glycosylation site within the FCS proximal region. Underlined R and S residues correspond to the FCS. The S1 domain (including Thr678) lies N-terminal to the FCS, and the S2 domain is C-terminal to the FCS. Right: Peptide alignment for SARS-CoV-2 variants of concern (VOCs) and related coronaviruses showing the emergence of the polybasic motif in the FCS proximal region. Highlighted in yellow is the polybasic motif of SARS-CoV-2 spike. Bold and red are amino acid changes in positions 679 and 681 in VOCs. (B) Bump-and-hole engineering allows for GalNAc-T isoenzyme-specific tagging of glycosylation substrates using the clickable substrate UDP-GalN6yne. FCS = furin cleavage site; VOCs = variants of concern.
Circulating variants of concern (VOCs) display increased proteolytic processing of spike into S1/S2.30,39,44−46 This increase has been associated with a remarkable polymorphism in the peptide stretch preceding the FCS between residues Gln675 and Pro681. Most VOCs and many minor circulating variants carry at least one mutation in that peptide region: Alpha (B.1.1.7) and Delta (B.1.617.2) display mutations at Pro681 to His and Arg, respectively, whereas all Omicron sublineages including BA.1, BA.2, and BA.5 combine P681H with the mutation N679K. Mutations in this region appear to have arisen more than once independently: Delta (P681R), Alpha (P681H), and Omicron lineages (P681H) are suggested to be on different arms of the evolutionary tree with common ancestors that do not contain mutations around the FCS, indicating that some selection pressure on this sequence must have been present during their evolution.47,48 Lower-prominence variants have featured substitutions at Gln675 and Gln677,49−53 usually to amino acids with basic functionalities (Figure 1A). In line with the cleavage enhancing effect of these mutations, preparations of VOC spike from eukaryotic expression systems contain less FL-S than WT (Wuhan/WH04/2020) spike preparations.54 While it is tempting to suggest that an increase in basic amino acids enhances furin activity simply due to extension of the polybasic cleavage site,55,56 this amino-acid-centric notion neglects the impact of post-translational modifications. Accordingly, Whittaker and colleagues observed that the P681H mutation alone does not increase furin cleavage of synthetic peptides.57 Due to the importance of spike in the viral infectious cycle, the key determinants of processing offer essential insights into the cell biology of viral maturation.
Like most surface proteins on animal viruses, SARS-CoV-2 spike is extensively coated with glycans that impact infectivity and immunogenicity of the mature virus.15−28 Among these, Asn (N)-linked glycosylation is straightforward to predict due to the existence of a peptide consensus sequence (N-X-S/T; where X = any amino acid except Pro). In contrast, the prediction of Ser/Thr-linked N-acetylgalactosamine (O-GalNAc) glycosylation, which also greatly impacts viral biology,25,58−62 is an analytical challenge due to its greater biosynthetic complexity and the lack of a peptide consensus sequence.63−73 Notably, the peptide region between Gln675 and Pro681 of SARS-CoV-2 spike harbors multiple Ser/Thr residues that may carry O-GalNAc glycans.19,26,27 Despite the analytical difficulties in understanding O-glycan biology, emerging data suggests that O-GalNAc glycosylation impacts furin-mediated spike cleavage.74 Due to the relevance of furin cleavage for viral infectivity, understanding the role of glycosylation in this process is essential.
The biosynthesis of O-GalNAc glycans is initiated by the introduction of the sugar N-acetylgalactosamine (GalNAc) from the activated substrate uridine diphosphate (UDP)-GalNAc on to Ser/Thr side chains by a family of 20 GalNAc transferase (GalNAc-T1...T20) isoenzymes. GalNAc-Ts are often associated with isoenzyme-specific, decisive roles in physiological processes that are beginning to be unraveled.75−84 Understanding the substrate profiles of individual GalNAc-T isoenzymes yields insight into the regulation of such processes and can be the basis for the development of tools, diagnostics, and therapeutics. However, assigning glycosylation sites to individual GalNAc-Ts is challenging due to their complex and often overlapping interplay in the secretory pathway.85 Additionally, the initial GalNAc residue is often further elaborated, generating mature glycans containing galactose (Gal), N-acetylglucosamine (GlcNAc), and the acidic monosaccharide N-acetylneuraminic acid (Neu5Ac) as a capping structure, further complicating the analytical profiling of O-GalNAc glycoproteins by mass spectrometry (MS) glycoproteomics. Indirect methods are thus employed to establish links between GalNAc-T isoenzymes and the glycosylation sites they modify to yield insights into O-glycan biology.75,85−89 Through co-expression of the individual human GalNAc-Ts with spike in insect cells and lectin staining, Ten Hagen and colleagues found that GalNAc-T1 introduces GalNAc into recombinant spike, resulting in reduced proteolytic processing of S to S1/S2 and decreasing syncytia formation in a cellular infection model.74 Mutations at P681, including the mutation P681H, led to increased S processing related to WT spike.74 These findings are in line with earlier reports that furin cleavage of other secreted proteins can be impacted by O-glycosylation.90−94 However, the biosynthetic complexity and technical challenges associated with O-glycoproteome analysis have thus far hindered closer investigation into the molecular details behind these observations. Specifically, we currently lack knowledge on the precise glycan attachment site(s) and the impact of glycan structure on proteolysis. We also do not know yet which VOC mutations impact either O-GalNAc glycosylation, direct proteolysis, or both.
Chemical tools have provided an insight into glycobiology that is orthogonal to classical methods of molecular biology. For example, by using a tactic termed “bump-and-hole engineering”, we have developed a chemical reporter strategy for the activities of individual GalNAc-T isoenzymes in the living cell (Figure 1B).95,96 Through structure-based design, the active site of a GalNAc-T isoenzyme was expanded by mutagenesis to contain a “hole”, which is complementary to a “bump” in a chemically modified analogue of the substrate UDP-GalNAc.96 The bumped substrate “UDP-GalN6yne” contained an alkyne moiety that enabled the bioorthogonal ligation of fluorophores or biotin after transfer to a glycoprotein, allowing the profiling of the substrates of individual GalNAc-Ts.96−98 Recently, we introduced a clickable, positively charged imidazolium tag (termed ITag) that enhances MS-based analysis by increasing the charge state and improving the fragmentation-based sequencing of glycopeptides.99 Importantly, UDP-GalN6yne can be biosynthesized in the living cell through the introduction of an artificial metabolic pathway and feeding with a membrane-permeable peracetylated GalN6yne precursor (Ac4GalN6yne), allowing for the installation of a fully functional GalNAc-T bump-and-hole system.96,100 Building on the power of our chemical tools to dissect the role of O-GalNAc glycosylation, we sought to map the molecular details of glycan-mediated modulation of spike processing.
Here, with aid from this repertoire of chemical biology tools, we spotlight O-linked glycosylation as a major determinant of SARS-CoV-2 spike cleavage by the host proteases furin and TMPRSS2. We provide direct evidence by MS-glycoproteomics that identifies GalNAc-T1 as the glycosyltransferase initiating Thr678 glycosylation in the living cell. We demonstrate that the presence of elaborated glycans on Thr678 reduce proteolytic cleavage by TMPRSS2 and that a negative charge (via sialic acid) on Thr678-containing glycopeptides completely abrogates furin activity. We further confirm that mutations on Pro681 (present in major VOCs Alpha, Delta, and Omicron) impair glycosylation of Thr678 and may therefore promote proteolytic processing of spike. By emphasising O-glycosylation as a major determinant of SARS-CoV-2 spike maturation, we propose disruption of O-GalNAc glycosylation as a considerable evolutionary driver for the emergence of SARS-CoV-2 VOCs.
Results and Discussion
Establishing a protein as a GalNAc-T substrate classically features expression in cells either lacking or overexpressing the respective GalNAc-T, followed by detection by GalNAc-recognizing lectins.74,75,87−89,101 While generally powerful, identifying the modified glycosylation sites is often challenging by these methods due to the interplay and ensuing compensatory effects between GalNAc-T isoenzymes. Bump-and-hole engineering enables a direct relation to the engineered GalNAc-T isoenzyme by introduction of a GalNAc analogue which can be bioorthgonally tagged and detected by various analytical techniques (Figure 2). We incubated recombinant SARS-CoV-2 spike WT, P681R, or P681H preparations produced in human Expi293F cells with recombinant WT or bump-and-hole-engineered GalNAc-T1 ("BH" = I238A/L295A double mutant) or T2 (BH = I253A/L310A double mutant) and the bumped nucleotide sugar UDP-GalN6yne.95 We then tagged the glycosylated proteins with biotin picolyl azide by Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) and visualized glycosylation via streptavidin blot (Figure 2A). An intense, single band corresponding to the S1 fragment was observed when WT (Wuhan/WH04/2020) spike was incubated with BH-GalNAc-T1 (Figure 2B). Negligible signal was observed on preparations incubated with either BH-T2 or the corresponding WT-GalNAc-Ts. A single substitution in spike of Pro681 to either His or Arg led to near-complete abrogation of glycosylation by BH-T1. These data suggest that recombinant WT-S1 contains a dedicated GalNAc-T1 glycosylation site which is absent in recombinant unprocessed full-length protein (WT-FL-S) and obstructed upon variant-specific mutation of Pro681.
Figure 2.
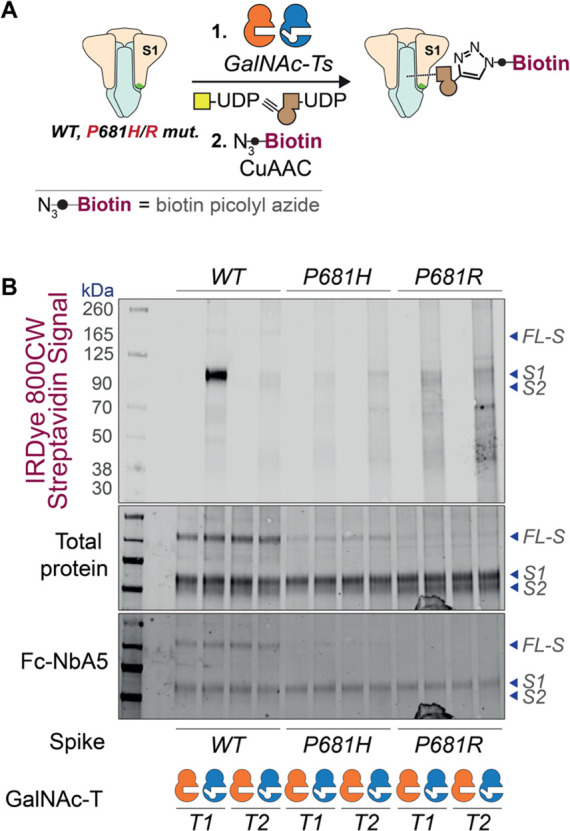
GalNAc-T isoenzyme-specific in vitro glycosylation of recombinant SARS-CoV-2 spike preparations. (A) Overview of in vitro enzymatic experiments comparing WT and BH-GalNAc-T1/T2 glycosylation with ensuing click-biotinylation of WT and P681 mutant (P681H and P681R) spike preparations. (B) Streptavidin blot of the glycosylated and biotin containing recombinant SARS-CoV-2 spike WT and P681 mutants. Visualized via IRDye 800CW-streptavidinfluorescence. Data are representative of three independent experiments. FL-S: full-length SARS-CoV-2 spike; S1: cleaved SARS-CoV-2 spike S1 domain; S2: cleaved SARS-CoV-2 spike S2 domain; Fc-NbA5: Fc-conjugated SARS-CoV-2 spike-specific nanobody binding in the receptor binding domain of the S1 region.
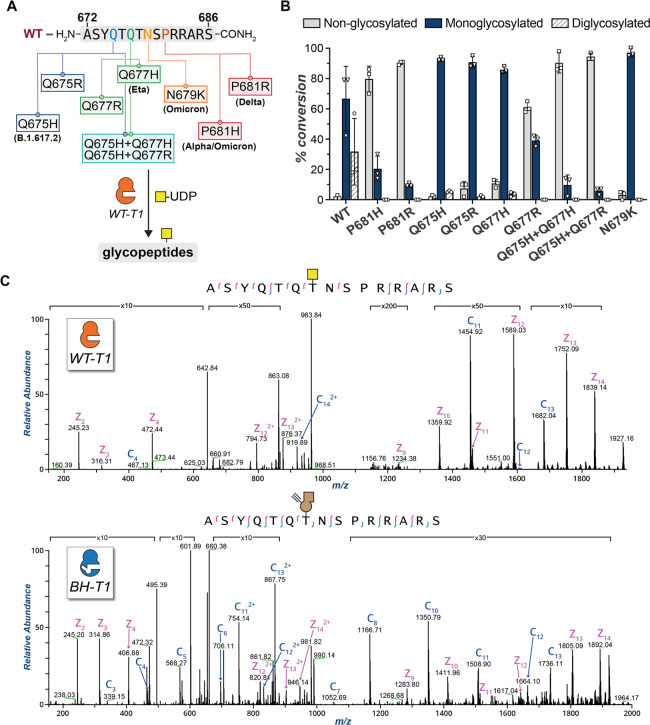
We then used a panel of synthetic peptides to study the effect of spike mutations on GalNAc-T1-mediated glycosylation. The peptide panel included variant-related mutations at the major hotspots: Gln675, Gln677, Asn679, and Pro681 (Figure 3A). GalNAc-T1 generated both mono- and diglycopeptides from a WT (Wuhan) substrate peptide (Figure 3B). Consistent with the work by Ten Hagen and colleagues,74 notable reductions in glycosylation were observed for P681H and P681R peptides with approximately 80 and 90% of non-glycosylated peptide remaining in the reaction mixture, respectively. These results validated the importance of a Pro in position +3 for GalNAc-T1 glycosylation.102 Single mutations at positions 675, 677, and 679, including N679K found in Omicron, reduced the amount of diglycosylation but largely retained monoglycosylation with almost complete consumption of the starting material. In contrast, the combination of two mutations at positions 675 and 677 showed a critical reduction of glycosylation. This was evidenced by reactions with the double mutant peptides Q675H + Q677H and Q675H + Q677R resulting in 9.7 and 5.7% conversion, respectively. These data confirmed the dependence of GalNAc-T1 on Pro681 but indicate that only certain VOC mutations impact O-GalNAc glycosylation. Mass spectrometry with electron-transfer dissociation (ETD) fragmentation revealed that monoglycopeptides are exclusively GalNAc-modified on Thr678, while diglycopeptides are modified at Thr676 and Thr678, indicating a hierarchy of sites where Thr678 is glycosylated first (Figure 3C and Supporting Figure 1). When BH-engineered GalNAc-T1 and UDP-GalN6yne were used in an in vitro glycosylation assay with WT spike-derived peptides, we observed a similar trend of glycosylating Thr678 and Thr676 sequentially, confirming that BH-T1 recapitulates the substrate specificity of WT-T1 (Figure 3C and Supporting Figure 1).
Figure 3.
Evaluation of GalNAc-T1-mediated glycosylation on synthetic peptides. (A) Peptide panel of the FCS proximal region in SARS-CoV-2 spike including WT and nine mutant peptides. All 10 peptides were subjected to enzymatic glycosylation with WT-T1 in the presence of the sugar donor UDP-GalNAc. The results are depicted in the bar chart in Figure 3B. (B) In vitro glycosylation results with recombinant WT-GalNAc-T1 and UDP-GalNAc assessed by LC-MS. Data are means ± SD from three independent replicates. (C) Tandem MS (ETD) spectra for WT (top) and BH- (bottom) GalNAc-T1 glycosylation of the WT peptide H-ASYQTQTNSPRRARS-NH2in vitro. Synthetic peptides were run on an Orbitrap Eclipse (Thermo) and subjected to ETD, followed by manual validation and hand curation. Legend: c ions are indicated in blue and z ions in pink. Yellow square = GalNAc. Brown alkyne = GalN6yne. WT-T1 = WT-GalNAc-T1. BH-T1 = BH-GalNAc-T1.
Having established a link between VOC mutations and glycosylation, we sought to rule out an immunological implication of the corresponding (glyco-)peptides that might impact any mechanistic deductions. Peptides WT, P681H, and WT-GalNAc (Figure 3A; “WT-GalNAc” corresponds to the glycosylation product of the WT peptide carrying an O-GalNAc on Thr678), were evaluated in peripheral blood mononuclear cells (PBMC) from n = 48 SARS-CoV-2 vaccinated individuals for T cell interferon-gamma (IFN-g) secretion using an enzyme-linked immunosorbent spot (ELISpot) assay.103 As shown in Supporting Figure 2, the median [IQR] response to the spike protein was 33.5 [16.7–69] spot forming cells (SFC) per million PBMC and to the combined pool of M and N proteins was 10 [0–29] SFC/million PBMC. The median [IQR] response to the peptide WT-GalNAc was 0 [0–3.7] in n = 44 individuals, compared to the peptide P681H 0 [0–3.5] in n = 21 and WT 10 [0–27] in n = 3 individuals. These results indicate that neither (glyco-)peptide is a T cell target in vaccinated individuals.
GalNAc-T Selective MS-Glycoproteomics Analysis Allows O-Glycosite and Glycan Composition Investigation in Vitro and in Engineered Cells
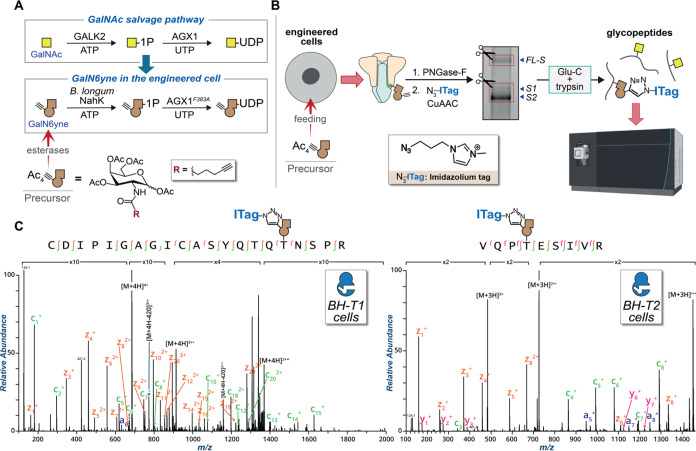
The complex dynamics of GalNAc-T isoenzymes in the secretory pathway requires new approaches to assigning their activities to specific glycosylation sites in living cells. Furthermore, the FCS-adjacent region lacks cleavage sites of the proteases most commonly used in MS sample preparation, resulting in large glycopeptides that hamper MS analyses. The use of specialized chemical tools can address these shortcomings and report on GalNAc-T activity in the secretory pathway while offering a bioorthogonal handle to aid MS analysis. We stably transfected Expi293F cells with constructs for both SARS-CoV-2 spike (Wuhan) and either WT- or BH-versions of GalNAc-T1 or T2, along with the biosynthetic machinery to generate UDP-GalN6yne in the cell from a membrane-permeable precursor (Ac4GalN6yne) that was introduced via cell feeding (Figure 4A).100 Spike samples were isolated and derivatized by CuAAC with an azide-functionalized imidazolium group containing a permanent positive charge (ITag-azide).99,104−106 This treatment introduced GalN6yne in an isoenzyme-specific fashion while endowing glycopeptides with an additional positive charge that facilitates MS analysis.99 The separated FL-S and S1/S2 fractions were subjected to in-gel digestion and analyzed by tandem MS. While collisional fragmentation (i.e., higher-energy collision dissociation, HCD) allows for the determination of monosaccharide compositions and naked peptide backbone sequences, this technique does not allow for the localization of O-glycans to their glycosites. The energy associated with collisional dissociation methods breaks the most labile bonds, which in the case of glycopeptides, are the glycosidic linkages between monosaccharides and the connection of the glycan to the peptide itself. To resolve site information in O-glycopeptides, electron-based dissociation methods must be employed; commonly, this involves electron-transfer dissociation (ETD).72,107 Thus, we used high intensity collision-induced dissociation (HCD) to obtain naked peptide sequences and glycan compositions and then used the ITag-containing GalN6yne diagnostic ion to trigger ETD fragmentation of the peptide backbone (Figure 4B).73,99,108,109 Through both computational (Byonic, ProteinMetrics) and manual validation, we found that Thr678 carried ITag-modified GalN6yne in both FL-S and S1 samples exclusively in cells expressing BH-T1, but not BH-T2 or any WT-GalNAc-Ts (Figure 4C and Supplementary Data 1). The additional positive charge of the ligated ITag permitted straightforward ETD fragmentation of a 21-amino-acid glycopeptide. In contrast, the corresponding glycopeptide in samples expressing WT-T1 could not be unambiguously sequenced, highlighting the ability of chemical tools to help advance site-specific O-glycoproteomics. We further found that both BH-T1 and BH-T2 glycosylated Thr323, a previously detected glycosylation site that had thus far not been associated with any GalNAc-T isoenzyme (Figure 4C and Supporting Figure 3). These glycosylation annotations were recapitulated through in vitro glycosylation of recombinantly expressed spike with recombinantly expressed soluble constructs of BH-GalNAc-T1 and BH-GalNAc-T2, followed by CuAAC ligation of ITag-azide and MS-glycoproteomics analysis (SI and Supporting Figure 4). Our data directly proves that GalNAc-T1 glycosylates Thr678 in the living cell.
Figure 4.
Uncovering the relationship between GalNAc-T1 and Thr678 by chemical tools. (A) GalNAc salvage pathway and UDP-GalN6yne biosynthesis. Expression of the kinase NahK and the pyrophosphorylase AGX1F383A permits biosynthesis of UDP-GalN6yne in engineered cells. (B) Graphical representation of the MS-glycoproteomics methodology for engineered cells: SARS-CoV-2 spike was recombinantly expressed in Expi293F cells coexpressing NahK, AGX1F383A, and either BH-GalNAc-T1 or T2. Following purification and de-N-glycosylation, ITag-azide was introduced by CuAAC, and the protein preparation subjected to in-gel digestion and MS by HCD-triggered ETD. (C) Annotated tandem MS (ETD) spectra of the major hits tagged by BH-GalNAc-T1 (left) and BH-GalNAc-T2 (right). c ions are indicated in green, z ions in orange, and y ions in pink. Data are representative from one experiment using 10 μg recombinant spike which was confirmed in a second experiment with a 2 μg preparation. Yellow square = GalNAc. Brown alkyne = GalN6yne. Ac4GalN6yne precursor = membrane-permeable peracetylated GalN6yne. BH-T1 cells = Expi293F cells cotransfected with WT SARS-CoV-2 spike and BH-GalNAc-T1. BH-T2 cells = Expi293F cells cotransfected with WT SARS-CoV-2 spike and BH-GalNAc-T2.
Elaborated O-Linked Sialoglycans on Thr678 Confer Proteolytic Resistance on SARS-CoV-2 Spike
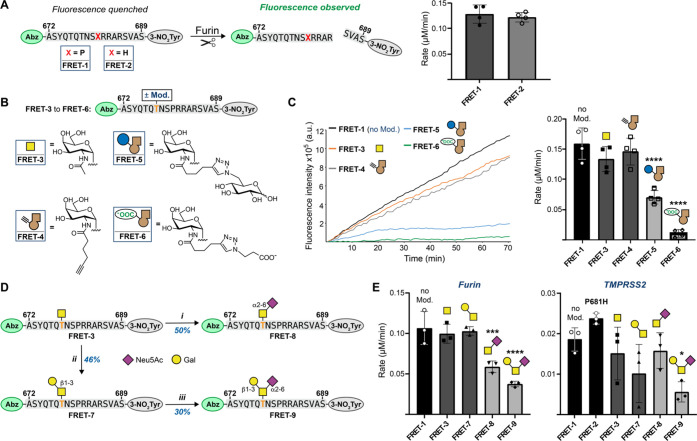
Glycosylation has the potential to modulate the proteolytic processing of a peptide depending on the distance to the cleavage site and glycan composition, as previously proposed for spike upon co-expression with GalNAc-T1.74,90,93,94 We used a direct method to investigate whether O-GalNAc glycans on Thr678 modulate cleavage by furin. To this end, we designed synthetic Förster resonance energy transfer (FRET)-active substrate peptides to assess proteolytic activity. Peptides spanning residues 672 to 689 contained N-terminal 2-aminobenzoyl (Abz) and C-terminal 3-nitrotyrosine (3-NO2Tyr) as fluorescence donor and quencher moieties, respectively. An increase in fluorescence intensity indicated proteolytic cleavage (Figure 5A).110 To test whether the P681H mutation could have any intrinsic, non-glycan-dependent impact on furin activity, we first compared non-glycosylated substrates corresponding to either WT (FRET-1) or P681H mutant spike (FRET-2). The P681H mutation had no discernible effect on the rate of furin-mediated cleavage, confirming the data by Whittaker and colleagues that the addition of a basic amino acid is not by itself a defining characteristic of spike furin cleavage enhancement in existing VOCs.57
Figure 5.
Chemical elaboration of O-glycosylation to assess proteolytic cleavage of glycopeptides. (A) Left: Experimental design and comparison of furin cleavage between WT and P681H peptide substrates. Peptides containing N-terminal 2-aminobenzoate (Abz) as a FRET donor and C-terminal 3-nitrotyrosine (3-NO2Tyr) as a FRET quencher that are separated upon proteolytic cleavage. Right: Rates of furin cleavage of glycopeptides FRET-1 and FRET-2 obtained through linear regression and normalization to control runs without furin. Data are means ± SD of four independent experiments. (B) Chemical modifications of synthetic (glyco-)peptides FRET-3 through FRET-6, generated via in vitro glycosylation and CuAAC. (C) Left: Time course of fluorescence increase upon furin cleavage reactions of 20 μM FRET-1 and FRET-3 through FRET-6 with 0.8 U/mL furin. Linear fluorescence increase is shown and normalized to the corresponding control run without furin. Right: Rates of furin cleavage of glycopeptides FRET-1 and FRET-3 through FRET-6 obtained through linear regression and normalization to control runs without furin. Data are means ± SD of four independent experiments. (D) Chemoenzymatic synthesis of glycopeptides FRET-7 through FRET-9: (i) ST6GALNAC1 (150 μg/mL), CMP-Neu5Ac (1.5 equiv), pH 7.5, 37 °C for 16 h, 46% yield; (ii) DmC1GalT1 (1 μM), UDP-Gal (1.5 equiv), pH 7.5, 37 °C for 16 h, 50% yield; (iii) ST6GALNAC2 (10 μg/mL), CMP-Neu5Ac (1.5 equiv), pH 7.5, 37 °C for 48 h, 30% yield. Bold and orange T denotes Thr678 in the glycopeptides. (E) Rates of furin (left) and TMPRSS2 (right) cleavage of (glyco-)peptides FRET-1 through FRET-3 and FRET-7 through FRET-9 obtained through linear regression and normalization to control runs without furin. Data are means ± SD of three independent experiments. Group comparison was performed via one-way ANOVA with Tukey’s multiple comparisons test, and asterisks annotate P values: *P < 0.0332 ; **P < 0.0021; ***P < 0.0002; ****P < 0.0001, compared to the nonmodified peptide (FRET-1). CMP = cytidine monophosphate; CIAP = calf intestinal alkaline phosphatase. Yellow square = GalNAc. Brown alkyne = GalN6yne. Yellow circle = Gal = galactose. Purple diamond = Neu5Ac = N-acetylneuraminic acid.
We hypothesized that an increase of furin processing in VOC mutant spike may not stem directly from recognition of the bare peptide sequence but rather a decreased capacity of GalNAc-T1 to introduce O-GalNAc glycans to peptides with mutations proximal to the furin recognition site. We thus tested whether glycosylation of furin substrate peptides impacts proteolytic cleavage. FRET reporter peptides carrying GalNAc, the simplest O-glycan, (FRET-3) or its alkyne-containing analogue GalN6yne (FRET-4) were generated by chemical and chemoenzymatic synthesis, respectively. Glycosylation with the single monosaccharides alone did not substantially impact the furin cleavage rate compared to the WT peptide FRET-1 (Figure 5C and Supporting Figure 5). We speculated that elaboration of GalNAc to larger or charged glycans might introduce additional structural constraints on furin recognition. To test this notion, the alkyne tag present on GalN6yne gave an opportunity to modify the biophysical properties of glycopeptides in a straightforward fashion, enabling synthetic efforts to furnish glycopeptides with specific additional groups or functionalities. We reacted the alkyne-equipped glycopeptide FRET-4 with two organic azides under CuAAC conditions: to evaluate the impact of a larger glycan, we used 6-azido-6-deoxy-glucose yielding pseudodisaccharide FRET-5, while 3-azido-propionic acid introduced an additional acidic functionality to investigate the impact of a negative charge on furin cleavage in glycopeptide FRET-6. Both click-elaborated glycopeptides displayed a significant reduction in furin cleavage (Figure 5C and Supporting Figure 5). FRET-5 exhibited an 80% decrease in the rate of furin cleavage with respect to FRET-1, which is attributable to the relative steric expansion. Strikingly, FRET-6, which carried a smaller, negatively charged modification, resulted in a 93% rate reduction, almost completely abrogating furin activity. While these modifications are not naturally occurring, we concluded that the elaboration of O-glycans on Thr678, especially with negatively charged modifications, severely impedes the activity of furin.
Sialic acid, a common capping monosaccharide of O-glycans, is negatively charged under physiological pH. With the results above, we reasoned that in naturally occurring O-glycans, the presence of elaborated glycans, especially sialic-acid-containing ones, might modulate furin cleavage. We chemoenzymatically synthesized a set of spike-derived glycopeptides carrying common elaborated mammalian O-glycans to test on our cleavage assay. First, Drosophila melanogaster C1GalT1 was used to extend FRET-3 with β3-linked galactose to give Core-1 O-GalNAc disaccharide FRET-7.111 α2,6-Linked sialic acid was introduced into FRET-3 and FRET-7 using the enzymes ST6GALNAC1 and ST6GALNAC2, respectively, to yield the sialoglycopeptides FRET-8 and FRET-9 (Figure 5D).112 While the uncharged disaccharide in FRET-7 only had a marginal (3.5% decrease) effect on the furin rate compared to the parental peptide FRET-1, the presence of a sialic acid led to a striking 45% reduction of furin rate in glycopeptide FRET-8 and a 65% reduction in glycopeptide FRET-9 (Figure 5E). We concluded that the elaboration of O-glycans on Thr678 with negatively charged sialic acid residues severely hampers furin activity.
To our knowledge, in contrast to furin,90 TMPRSS2 has not been comprehensively probed for cleavage of glycopeptide substrates. We sought to establish whether O-GalNAc glycans could modulate TMPRSS2 activity in a similar fashion to furin. Recombinant TMPRSS2 was subjected to our selection of (glyco-)peptide FRET substrates FRET-1, FRET-3, and FRET-7 to FRET-9 (Figure 5E). While all glycans somewhat impacted TMPRSS2 activity with respect to the non-glycosylated peptide FRET-1, the trisaccharide-containing sialoglycopeptide FRET-9 (70% rate reduction) impeded cleavage more drastically than all other (glyco-)peptides.
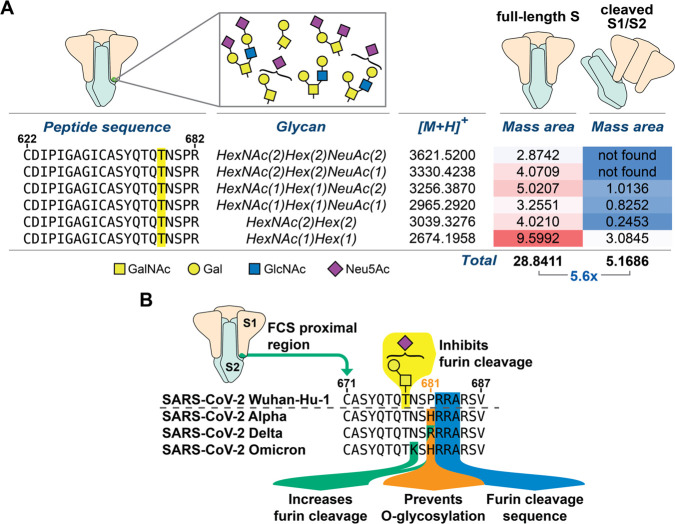
Our in vitro glycosylation experiments suggested that S1 contains the only available GalNAc-T1 substrate on WT spike after secretion from human cell culture (Figure 2B). Such behavior could be explained by the presence of elaborated, sialylated O-glycans on recombinant FL-S, which would both prevent furin cleavage and block access by GalNAc-T1 in vitro. This would suggest an enrichment of sialylated O-glycans on FL-S relative to processed S1. After SDS-PAGE, in-gel digestion allowed us to directly test this notion by individually cutting out the bands corresponding to the cleaved (S1/S2) and uncleaved (FL-S) fractions from the same recombinant WT-spike preparation. We subjected the FL-S and S1/S2 gel bands to MS-glycoproteomics analysis, searching for both simple and elaborated O-GalNAc glycans in each of the fractions. By calculating the intact masses of various expected O-glycopeptides in recombinant spike and then obtaining the associated extracted ion chromatograms (XICs), we found that over 5-fold higher glycopeptide signal is present in FL-S when compared to cleaved S1/S2 fractions from the same spike preparation (Figure 6A and Supplementary Data 2). Furthermore, when accounting for sialic-acid-containing glycopeptides only, an ∼8-fold higher abundance was observed for FL-S relative to S1/S2 (Supplementary Data 2).
Figure 6.
Extracted ion abundance for O-glycopeptides from FL-S or S1/S2 recombinant spike fractions. (A) Mass areas were normalized against their corresponding base peaks and multiplied by a factor of 10 000. All data analysis was performed in Thermo XCalibur software from extracted ion chromatograms and hand-curated. Highlighted T corresponds to Thr678, which was found to be modified by diverse O-glycans. Data are from one experiment. (B) Summary of VOC mutations within the FCS proximal region and their established impact on glycosylation and proteolysis.
Conclusion
Our data strongly indicate that elaborated, negatively charged O-GalNAc glycans on Thr678 of SARS-CoV-2 spike have a supressing effect on proteolytic cleavage. Such glycans are produced on lung epithelial cells which express GalNAc-T1,74 suggesting that glycosylation is a physiologically relevant modification that could restrict the maturation (by proteolysis) of spike in WT SARS-CoV-2.113 The propensity of SARS-CoV-2 variants of concern to outcompete each other has been linked to both increased infectivity and immune escape. Within the evolutionary trajectory to the Alpha, Delta, and Omicron variants, notable changes in the amino acid sequence proximal to the FCS indicated that proteolytic cleavage is gradually enhanced, congruent with their increased infectivity. Mutations of Pro681 have been detected in early variants such as Alpha (P681H). We found that this mutation did not intrinsically increase the rate of cleavage by both furin and TMPRSS2 but impacted O-glycosylation as a restricting factor for spike processing.45 Notably, the analogous mutation found on the more transmissible Delta variant (P681R) has been linked to an increase in furin cleavage,45 suggesting an evolutionary trajectory that convolves suppression of O-glycosylation with increasing intrinsic furin recognition. This trend is further underlined by the mutations found in Omicron, which features both the P681H and the N679K mutations: in contrast to P681H and consistent with mapped amino acid preferences of GalNAc-T1,114 the N679K mutation does not substantially impact glycosylation, but it leads to enhanced furin cleavage of synthetic peptides.45 These Omicron mutations therefore act synergistically and have likely evolved to both suppress glycosylation and intrinsically enhance furin cleavage. Since the closest relatives to SARS-CoV-2, the strains RaTG13 Bat-CoV and GD Pangolin-CoV, share the exact same peptide sequence with WT (Wuhan) spike but without an FCS (Figure 1), we speculate that O-glycosylation might have been a remnant of ancestral strains that is being lost due to selective pressure toward increased furin cleavage, apparently without directly mutating the Thr678 residue.116
The presence of O-GalNAc glycans adjacent to proteolytic cleavage sites has been found to impact processing of secreted proteins.90−94 The glycosylation site at Thr678 of spike is not in direct proximity of the FCS, potentially explaining why a single GalNAc residue is not sufficient to modulate furin activity and only minimally impacts TMPRSS2 activity. The necessity for the glycan to be elaborated or sialylated to reveal the suppressing effect upon the rates of cleavage further highlights the need for accurate glycan tracing techniques.115 Tuning the chemical properties of glycopeptides in a straightforward fashion by CuAAC was pivotal in enabling an initial understanding of the substrate-activity relationship of furin. This strategy informed the targeted synthesis of elaborated O-glycopeptide substrates, providing a convenient method to fine-tune substrate scope in a time- and resource-efficient manner. Chemical tools thus yielded insights into glycosyltransferase specificity, improved the efficacy of detection by detectability by MS, and allowed the exploration of protease substrate specificity, showcasing the power of such tools in biomedical discovery.
Acknowledgments
We thank David Briggs, Sarah Maslen, Neil McDonald, Phil Walker, Steve Gamblin, and Nicola O’Reilly for valuable support and David Bauer, Theresa Zeisner, Jennifer Milligan, and John Diffley for helpful discussions. We thank Kelly Ten Hagen and Nadine Samara for providing crucial insights. We thank Junwon Choi and Carolyn R. Bertozzi for UDP-GalN6yne. The Legacy study was approved by London Camden and Kings Cross Health Research Authority (HRA) Research and Ethics committee (REC) IRAS number 286469 and sponsored by University College London. For the purposes of Open access, the author has applied a CC-BY copyright to any author accepted version of a manuscript arising from this submission.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.2c01349.
Detailed experimental procedures and additional supporting data including additional annotated spectra, interferon-gamma ELIspot results, fluorescence traces for protease cleavage experiments, characterization of synthetic (glyco-)peptides, and additional references (PDF)
Data analysis of glycoproteomics hits from engineered cells (XLSX)
Data analysis for the extracted ion abundance for O-glycopeptides from FL-S or S1/S2 spike fractions (XLSX)
Transparent Peer Review report available (PDF)
Author Contributions
E.G.-R., M.Z.-H., G.B.-T., and A.M. contributed equally to this work.
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (CC2127, CC1283, and CC2112), the UK Medical Research Council (CC2127, CC1283, and CC2112), and the Wellcome Trust (CC2127, CC1283, and CC2112). This work was funded by the UK Biotechnology and Biological Sciences Research Council (BB/V008439/1 to E.G.-R., L.D.V., and B.S., BB/V014862/1 to G.B.-T. and B.S., and BB/M028836/1 to S.L.F.), the Engineering and Physical Sciences Research Council (EP/S005226/1 to S.L.F.), the Wellcome Trust (218304/Z/19/Z to A.M. and B.S.), and the European Research Council (788231 to S.L.F.). M.Z.-H. was supported by a Crick-HEI studentship funded by the Department of Chemistry at Imperial College London and the Francis Crick Institute. S.A.M. is supported by the Yale Science Development Fund and NIGMS R35 GM147039. K.E.M. is supported by a Yale Endowed Postdoctoral Fellowship. R.J.W. receives support from Rosetrees (M926) and the Wellcome Trust (203135 and 222574). R.H.-G. thanks ARAID, the Agencia Estatal de Investigación (AEI, BFU2016-75633-P and PID2019-105451GB-I00), and Gobierno de Aragón (E34_R17 and LMP58_18 to R.H.-G.) with FEDER (2014–2020) funds for “Building Europe from Aragón” for financial support. The PBMC portion of this work was supported jointly by the National Institute for Health Research (NIHR) University College London Hospitals Department of Health’s NIHR Biomedical Research Centre (BRC) and core funding from the Francis Crick Institute, which receives its funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust. E.C.W. is supported by the BRC’s funding scheme. Plasmids used herein were generated by Moremen et al.117 for the PSI Materials Repository related to human and bacterial glycosylation enzymes (Glyco-enzyme clone collection) and generated through the support of NIH grant RR005351.
The authors declare the following competing financial interest(s): S.A.M. is a consultant for InterVenn Biosciences and Arkuda Therapeutics. The other authors declare no conflict of interest.
Supplementary Material
References
- Hoffmann M.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira R. A.; Guzman H. V.; Boopathi S.; Baker J. L.; Poma A. B. Characterization of structural and energetic differences between conformations of the SARS-CoV-2 spike protein. Materials (Basel). 2020, 13 (23), 5362. 10.3390/ma13235362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D. J.; et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330. 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A. G.; et al. Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2. Nat. Commun. 2021, 12, 837. 10.1038/s41467-021-21006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D. J.; et al. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2022586118. 10.1073/pnas.2022586118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B.; et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020, 370, 203–208. 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541. 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. L.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y.; et al. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.; et al. A multiscale coarse-grained model of the SARS-CoV-2 virion. Biophys. J. 2021, 120, 1097–1104. 10.1016/j.bpj.2020.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A. G.; et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020, 27, 763–767. 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A.; Supekar N. T.; Gleinich A. S.; Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbalm K. E.; Tiemeyer M.; Wells L.; Aoki K.; Zhao P. Glycomics-informed glycoproteomic analysis of site-specific glycosylation for SARS-CoV-2 spike protein. STAR Protoc. 2020, 1, 100214. 10.1016/j.xpro.2020.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; et al. O-Glycosylation Landscapes of SARS-CoV-2 Spike Proteins. Front. Chem. 2021, 9, 689521. 10.3389/fchem.2021.689521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison A. M.; et al. Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S. Chem. Sci. 2022, 13, 386–395. 10.1039/D1SC04832E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.; Qin S.; Dai L.; Tian Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target. Ther. 2021, 6, 396. 10.1038/s41392-021-00809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. D.; et al. Site-Specific Steric Control of SARS-CoV-2 Spike Glycosylation. Biochemistry 2021, 60, 2153–2169. 10.1021/acs.biochem.1c00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Wang M.; Yu D.; Zhang X. Variations in SARS-CoV-2 Spike Protein Cell Epitopes and Glycosylation Profiles During Global Transmission Course of COVID-19. Front. Immunol. 2020, 11, 565278. 10.3389/fimmu.2020.565278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant O. C.; Montgomery D.; Ito K.; Woods R. J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020, 10, 14991. 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L.; et al. Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020, 6, 1722–1734. 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; et al. Inhibition of SARS-CoV-2 viral entry upon blocking N-and O-glycan elaboration. Elife 2020, 9, e61552. 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C.; et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv 2020; 2020.07.29.227462. 10.1101/2020.07.29.227462. [DOI]
- Sanda M.; Morrison L.; Goldman R. N-and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 2021, 93, 2003–2009. 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztain T.; et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021, 13, 963–968. 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A. G.; Benton D. J.; Roustan C.; Borg A.; Hussain S.; Martin S. R.; Rosenthal P. B.; Skehel J. J.; Gamblin S. J. Nat. Comm. 2022, 13, 1178. 10.1038/s41467-022-28768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak A. J.; Yu A.; Ke Z.; Briggs J. A. G.; Voth G. A. Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core. Nat. Commun. 2022, 13, 1002. 10.1038/s41467-022-28654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 2020, 78, 779–784.e5. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao B.; Olvera de la Cruz M. Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano 2020, 14, 10616–10623. 10.1021/acsnano.0c04798. [DOI] [PubMed] [Google Scholar]
- Coutard B.; et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020, 176, 104742. 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örd M.; Faustova I.; Loog M. The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci. Reports 2020, 10, 16944. 10.1038/s41598-020-74101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S.; et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020, 5, 92. 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker G. R. SARS-CoV-2 spike and its adaptable furin cleavage site. Lancet Microbe 2021, 2, e488–e489. 10.1016/S2666-5247(21)00174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T. P.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Johnson B. A.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa G.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLOS Pathog. 2021, 17, e1009246. 10.1371/journal.ppat.1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essalmani R. Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity. J. Virol. 2022, 96, e00128-22. 10.1128/jvi.00128-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J. M.; et al. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discovery 2014, 4, 1310–1325. 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39, 110829. 10.1016/j.celrep.2022.110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B.; Jaimes J. A.; Whittaker G. R.. Intrinsic furin-mediated cleavability of the spike S1/S2 site from SARS-CoV-2 variant B.1.529 (Omicron). bioRxiv, 2022; 2022.04.20.488969. 10.1101/2022.04.20.488969. [DOI]
- Peacock T. P.; et al. The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. bioRxiv, 2020; 2020.09.30.318311. 10.1101/2020.09.30.318311. [DOI]
- Viana R.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nat. 2022, 603, 679–686. 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha L. B.; Foster C.; Rawlinson W.; Tedla N.; Bull R. A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. 10.1002/rmv.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli A.; et al. Role of Q675H Mutation in Improving SARS-CoV-2 Spike Interaction with the Furin Binding Pocket. Viruses 2021, 13, 2511. 10.3390/v13122511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins 2021, 89, 569–576. 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.; Wang W. Fast-spreading SARS-CoV-2 variants: challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target. Ther. 2021, 6, 226. 10.1038/s41392-021-00644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P.; et al. Occurrence of a substitution or deletion of SARS-CoV-2 spike amino acid 677 in various lineages in Marseille, France. Virus Genes 2022, 58, 53–58. 10.1007/s11262-021-01877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A.; Basiouni S.; Parvin R.; Hafez H. M.; Shehata A. A. Evolutionary insights into the furin cleavage sites of SARS-CoV-2 variants from humans and animals. Arch. Virol. 2021, 166, 2541–2549. 10.1007/s00705-021-05166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A. G.; et al. Evolution of the SARS-CoV-2 spike protein in the human host. Nat. Commun. 2022, 13, 1178. 10.1038/s41467-022-28768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P.; Brunak S.; Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004, 17, 107–112. 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Remacle A. G.; et al. Substrate Cleavage Analysis of Furin and Related Proprotein Convertases: A COMPARATIVE STUDY. J. Biol. Chem. 2008, 283, 20897–20906. 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B.; et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 2022, 25, 103589. 10.1016/j.isci.2021.103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall H. H.; Nielsen M. A. I.; King-Smith S.; de Haan N.; Bagdonaite I. Global functions of O-glycosylation: promises and challenges in O-glycobiology. FEBS J. 2021, 288, 7183–7212. 10.1111/febs.16148. [DOI] [PubMed] [Google Scholar]
- Simon E. J.; Linstedt A. D. Site-specific glycosylation of Ebola virus glycoprotein by human polypeptide GalNAc-transferase 1 induces cell adhesion defects. J. Biol. Chem. 2018, 293, 19866–19873. 10.1074/jbc.RA118.005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen M. B.; et al. An innate antiviral pathway acting before interferons at epithelial surfaces. Nat. Immunol. 2016, 17, 150–158. 10.1038/ni.3319. [DOI] [PubMed] [Google Scholar]
- Stone J. A.; Nicola A. V.; Baum L. G.; Aguilar H. C. Multiple Novel Functions of Henipavirus O-glycans: The First O-glycan Functions Identified in the Paramyxovirus Family. PLOS Pathog. 2016, 12, e1005445. 10.1371/journal.ppat.1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I.; et al. Global Mapping of O-Glycosylation of Varicella Zoster Virus, Human Cytomegalovirus, and Epstein-Barr Virus *. J. Biol. Chem. 2016, 291, 12014–12028. 10.1074/jbc.M116.721746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager K. T.; Narimatsu Y.; Joshi H. J.; Clausen H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- Čaval T.; de Haan N.; Konstantinidi A.; Vakhrushev S. Y. Quantitative characterization of O-GalNAc glycosylation. Curr. Opin. Struct. Biol. 2021, 68, 135–141. 10.1016/j.sbi.2020.12.010. [DOI] [PubMed] [Google Scholar]
- Kotwal G. J.; et al. Role of Glycosylation in Transport of Vesicular Stomatitis Virus Envelope Glycoprotein. A new class of mutant defective in glycosylation and transport of G protein. J. Biol. Chem. 1986, 261, 8936–8943. 10.1016/S0021-9258(19)84472-1. [DOI] [PubMed] [Google Scholar]
- Chernykh A.; Kawahara R.; Thaysen-Andersen M. Towards structure-focused glycoproteomics. Biochem. Soc. Trans. 2021, 49, 161–186. 10.1042/BST20200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.; et al. Advances in mass spectrometry-based glycoproteomics. Electrophoresis 2018, 39, 3104–3122. 10.1002/elps.201800272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Huang J.; Li L. Recent advances in mass spectrometry (MS)-based glycoproteomics in complex biological samples. Trends Analyt. Chem. 2019, 118, 880–892. 10.1016/j.trac.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams J. L.; et al. Recent advances in glycoinformatic platforms for glycomics and glycoproteomics. Curr. Opin. Struct. Biol. 2020, 62, 56–69. 10.1016/j.sbi.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Khoo K. H. Advances toward mapping the full extent of protein site-specific O-GalNAc glycosylation that better reflects underlying glycomic complexity. Curr. Opin. Struct. Biol. 2019, 56, 146–154. 10.1016/j.sbi.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Malaker S. A.; et al. The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 7278–7287. 10.1073/pnas.1813020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Angarita V.; Malaker S. A. Mucinomics as the Next Frontier of Mass Spectrometry. ACS Chem. Biol. 2021, 16, 1866–1883. 10.1021/acschembio.1c00384. [DOI] [PubMed] [Google Scholar]
- Cioce A.; Malaker S. A.; Schumann B. Generating orthogonal glycosyltransferase and nucleotide sugar pairs as next-generation glycobiology tools. Curr. Opin. Chem. Biol. 2021, 60, 66–78. 10.1016/j.cbpa.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; et al. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, 2109905118. 10.1073/pnas.2109905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen K. G.; Fritz T. A.; Tabak L. A. All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 2003, 13, 1R–16R. 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Revoredo L.; et al. Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology 2016, 26, 360–376. 10.1093/glycob/cwv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T. A.; et al. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 2011, 286, 14493–14507. 10.1074/jbc.M111.218701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. L.; Tran D. T.; Tabak L. A. Members of the GalNAc-T family of enzymes utilize distinct Golgi localization mechanisms. Glycobiology 2018, 28, 841–848. 10.1093/glycob/cwy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T. A.; et al. The Lectin Domain of the Polypeptide GalNAc Transferase Family of Glycosyltransferases (ppGalNAc Ts) Acts as a Switch Directing Glycopeptide Substrate Glycosylation in an N- or C-terminal Direction, Further Controlling Mucin Type O-Glycosylation *. J. Biol. Chem. 2013, 288, 19900–19914. 10.1074/jbc.M113.477877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaman E.-M.; Carter D. R. F.; Brooks S. A. GALNTs: master regulators of metastasis-associated epithelial-mesenchymal transition (EMT). Glycobiology 2022, 32, 556. 10.1093/glycob/cwac014. [DOI] [PubMed] [Google Scholar]
- Wandall H. H.; Nielsen M. A. I.; King-Smith S.; de Haan N.; Bagdonaite I. Global functions of O-glycosylation: promises and challenges in O-glycobiology. FEBS J. febs. 2021, 288, 7183 10.1111/febs.16148. [DOI] [PubMed] [Google Scholar]
- Pratt M. R.; et al. Deconvoluting the Functions of Polypeptide N-α-Acetylgalactosaminyltransferase Family Members by Glycopeptide Substrate Profiling. Chem. Biol. 2004, 11, 1009–1016. 10.1016/j.chembiol.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bennett E. P.; et al. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu Y.; et al. Exploring regulation of protein O-glycosylation in isogenic human HEK293 cells by differential O-glycoproteomics. Mol. Cell Proteomics 2019, 18, 1396–1409. 10.1074/mcp.RA118.001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C.; et al. Mining the O-glycoproteome using zinc-finger nuclease–glycoengineered SimpleCell lines. Nat. Methods 2011, 8, 977–982. 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Schjoldager K. T. B. G.; et al. Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 9893–9898. 10.1073/pnas.1203563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakhrushev S. Y.; et al. Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 2013, 12, 932–944. 10.1074/mcp.O112.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram Schjoldager K. T-B.; et al. A Systematic Study of Site-specific GalNAc-type O-Glycosylation Modulating Proprotein Convertase Processing. J. Biol. Chem. 2011, 286, 40122–40132. 10.1074/jbc.M111.287912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarsky K.; Kingsley D.; Krieger M. Use of a mutant cell line to study the kinetics and function of O-linked glycosylation of low density lipoprotein receptors. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 4335–4339. 10.1073/pnas.85.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K.; et al. Polypeptide GalNAc-transferase T3 and Familial Tumoral Calcinosis: Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 2006, 281, 18370–18377. 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Schjoldager K. T-B. G.; et al. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 2010, 285, 36293–36303. 10.1074/jbc.M110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci V. S.; et al. Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 5520–5525. 10.1073/pnas.1402218111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.; et al. Engineering Orthogonal Polypeptide GalNAc-Transferase and UDP-Sugar Pairs. J. Am. Chem. Soc. 2019, 141, 13442–13453. 10.1021/jacs.9b04695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann B.; et al. Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells. Mol. Cell 2020, 78, 824–834.e15. 10.1016/j.molcel.2020.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce A.; Malaker S. A.; Schumann B. Generating orthogonal glycosyltransferase and nucleotide sugar pairs as next-generation glycobiology tools. Curr. Opin. Chem. Biol. 2021, 60, 66–78. 10.1016/j.cbpa.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce A.; et al. Optimization of Metabolic Oligosaccharide Engineering with Ac4GalNAlk and Ac4GlcNAlk by an Engineered Pyrophosphorylase. ACS Chem. Biol. 2021, 16, 1961–1967. 10.1021/acschembio.1c00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle B.; et al. Benefits of Chemical Sugar Modifications Introduced by Click Chemistry for Glycoproteomic Analyses. J. Am. Soc. Mass Spectrom. 2021, 32, 2366–2375. 10.1021/jasms.1c00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce A. Cell-specific Bioorthogonal Tagging of Glycoproteins. Nat. Commun. 2022, 13, 6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K.-H.; et al. GALNT14 promotes lung-specific breast cancer metastasis by modulating self-renewal and interaction with the lung microenvironment. Nat. Commun. 2016, 7, 13796. 10.1038/ncomms13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Rivas M.; Lira-Navarrete E.; Gerken T. A.; Hurtado-Guerrero R. Polypeptide GalNAc-Ts: from redundancy to specificity. Curr. Opin. Struct. Biol. 2019, 56, 87–96. 10.1016/j.sbi.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler A.; et al. Adaptive immunity and neutralizing antibodies against SARS-CoV-2 variants of concern following vaccination in patients with cancer: the CAPTURE study. Nat. Cancer 2021, 2, 1321–1337. 10.1038/s43018-021-00274-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittel I.; Galan M. C. Chemo-enzymatic synthesis of imidazolium-tagged sialyllactosamine probes. Bioorg. Med. Chem. Lett. 2015, 25, 4329–4332. 10.1016/j.bmcl.2015.07.049. [DOI] [PubMed] [Google Scholar]
- Sittel I.; Galan M. C. Imidazolium-labeled glycosides as probes to harness glycosyltransferase activity in human breast milk. Org. Biomol. Chem. 2017, 15, 3575–3579. 10.1039/C7OB00550D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittel I.; Tran A. T.; Benito-Alifonso D.; Galan M. C. Combinatorial ionic catch-and-release oligosaccharide synthesis (combi-ICROS). Chem. Commun. 2013, 49, 4217–4219. 10.1039/c2cc37164b. [DOI] [PubMed] [Google Scholar]
- Ince D.; Lucas T. M.; Malaker S. A. Current strategies for characterization of mucin-domain glycoproteins. Curr. Opin. Chem. Biol. 2022, 69, 102174. 10.1016/j.cbpa.2022.102174. [DOI] [PubMed] [Google Scholar]
- Riley N. M.; Malaker S. A.; Bertozzi C. R. Electron-Based Dissociation Is Needed for O-Glycopeptides Derived from OpeRATOR Proteolysis. Anal. Chem. 2020, 92, 14878–14884. 10.1021/acs.analchem.0c02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley N. M.; Malaker S. A.; Driessen M. D.; Bertozzi C. R. Optimal Dissociation Methods Differ for N- A nd O-Glycopeptides. J. Proteome Res. 2020, 19, 3286–3301. 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. T.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp3 papain-like protease. Biochem. J. 2021, 478, 2517–2531. 10.1042/BCJ20210244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ramírez A. M.; et al. Structural basis for the synthesis of the core 1 structure by C1GalT1. Nat. Commun. 2022, 13, 2398. 10.1038/s41467-022-29833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong G.; et al. A facile chemoenzymatic synthesis of SARS-CoV-2 glycopeptides for probing glycosylation functions. Chem. Commun. 2021, 57, 6804–6807. 10.1039/D1CC02790E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N.; et al. The Human Lung Glycome Reveals Novel Glycan Ligands for Influenza A Virus. Sci. Rep. 2020, 10, 5320. 10.1038/s41598-020-62074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T. A.; et al. Emerging Paradigms for the Initiation of Mucin-type Protein O-Glycosylation by the Polypeptide GalNAc Transferase Family of Glycosyltransferases. J. Biol. Chem. 2011, 286, 14493–14507. 10.1074/jbc.M111.218701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu M. N.; Lokugamage K. G.; Plante J. A. QTQTN motif upstream of the furin-cleavage site plays a key role in SARS-CoV-2 infection and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 2022, 1119, e2205690119 10.1073/pnas.2205690119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.; Pristera N.; Wang W.; Zhang X.; Wu Q. Effect of sialylated O-glycans in pro–brain natriuretic peptide stability. Clin. Chem. 2010, 56, 959–966. 10.1373/clinchem.2009.140558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreman K. W.; et al. Expression system for structural and functional studies of human glycosylation enzymes. Nat. Chem. Biol. 2017, 14, 156–162. 10.1038/nchembio.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.