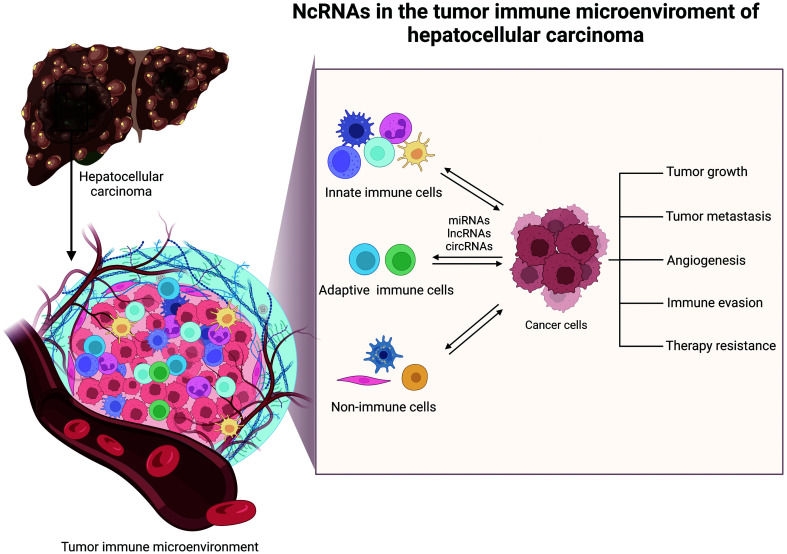
Abstract
Hepatocellular carcinoma (HCC) is one of the most prevalent malignancies. It has high mortality and poor clinical outcomes, but the molecular mechanisms in the pathogenesis of HCC are not understood. The tumor immune microenvironment (TIME) is a highly intricate system with distinct populations of innate and adaptive immune cells, as well as other stromal cells. They interact and evolve with tumor cells to influence tumor growth, migration, invasion, immune evasion, and response to therapy. Emerging evidence has shown noncoding RNAs (ncRNAs) are prominent regulators of TIME in HCC. In this review, we elaborate on the functions and molecular mechanisms of ncRNAs in remodeling TIME of HCC and discuss their diagnostic and therapeutic potential for HCC treatment.
Keywords: ncRNAs, TIME, HCC, Biomarker, Therapeutic strategy
Graphical abstract
Introduction
Hepatocellular carcinoma (HCC), is the predominant form of primary liver cancer, the fifth most common malignancy, and the fourth leading cause of cancer-related death globally.1–3 Major risk factors for HCC have been well established and include hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, abnormalities of lipid metabolism, excessive alcohol consumption, intake of dietary toxins like aristolochic acid or aflatoxin B1, diabetes.3 HCC is a biologically complex and highly heterogeneous disease, and the detailed mechanisms underlying hepatocarcinogenesis are still poorly understood. In recent decades, various preventive and therapeutic approaches have been approved and widely applied in HCC management, including antihepatitis vaccine, surgical resection, liver transplantation, and systemic treatment, etc.4,5 Notably, cancer immunotherapies have achieved pronounced clinical benefits, however, a large proportion of the immunotherapies still remain ineffective.6 Considering that worldwide mortality from HCC is continuously increasing, it is important to improve our understanding of the molecular pathogenesis of HCC, while novel diagnostic/prognostic biomarkers and therapeutic strategies are urgently needed to deal with this major public health concern.
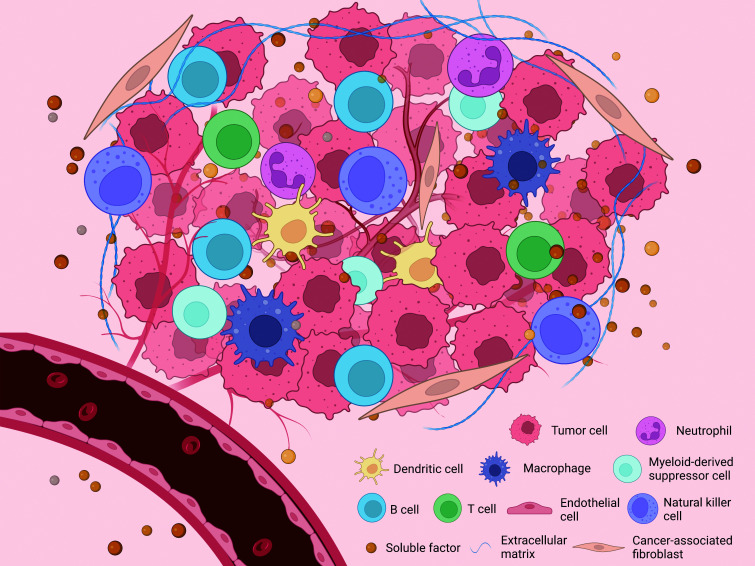
It is now clear that tumor formation and progression involve the co-evolution of neoplastic cells and surrounding stromal components. In recent years, the TIME has received significant attention as it is recognized to closely interact and co-evolve with tumor cells, affecting tumor growth, metastasis, immune escape, and the efficacy of immunotherapy. The TIME of HCC is a highly intricate and integrated system that consists of diverse cellular and noncellular components. The cellular components comprise immune cells including macrophages, neutrophils, myeloid-derived suppressor cells (MDSCs), natural killer (NK) cells, dendritic cells (DCs), T cells, B cells, cancer stem cells (CSCs), hepatic stellate cells (HSCs), vascular cells, cancer-associated fibroblasts (CAFs), and other stromal cells. The noncellular parts include the extracellular matrix and abundant soluble factors (e.g., cytokines, chemokines, growth factors) (Fig. 1). All these components dynamically interact to foster an immunosuppressive TIME. Many studies have revealed that TIME has a critical role in regulating immune evasion and the development of HCC.6–9 However, the detailed molecular mechanisms underlying TIME reprogramming in HCC are not understood.
Fig. 1. Key players in the TIME of HCC.
The TIME of HCC is a highly sophisticated system consisting of diverse cellular and noncellular components. The cellular components comprise various immune cells (macrophages, neutrophils, myeloid-derived suppressor cells, natural killer cells, dendritic cells, T cells, B cells), endothelial cells, cancer-associated fibroblasts, and other kinds of stromal cells. The noncellular counterparts include the extracellular matrix and diverse soluble factors secreted by both tumor cells and stromal cells. HCC, hepatocellular carcinoma; TIME, tumor immune microenvironment.
Noncoding RNAs (ncRNAs) refer to transcripts with no or minimal protein-coding ability. In the human genome, less than 2% of the transcripts encode proteins, while the remaining 98% are transcribed into different species of ncRNAs. ncRNAs can be classified into two major categories based on their molecular structure, including linear RNAs and circular RNAs (circRNAs). The linear RNAs can be broadly divided into two groups by their length, small noncoding RNAs (sncRNAs, <200 nt) and long noncoding RNAs (lncRNAs, >200 nt). sncRNAs consist of microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), transfer RNA-derived small RNAs (tsRNAs), and PIWI-interacting RNAs (piRNAs).10,11 Emerging evidence has shown that ncRNAs can reprogram TIME, which has profound influences on HCC tumorigenesis and progression. In this review, we systematically discuss the functional roles and molecular mechanisms of ncRNAs within the TIME of HCC, and discuss the diagnostic/therapeutic potential of ncRNAs in HCC treatment.
NcRNAs and innate immune cells in TIME
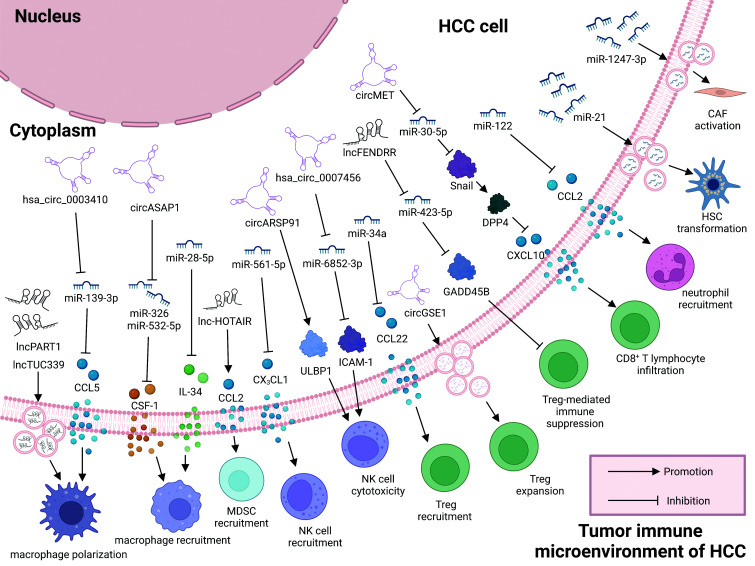
The TIME of HCC is a complex ecosystem that has various types of innate immune cells and adaptive immune cells, both have established roles in host defense against tumors through diverse mechanisms. Innate immune cells establish the body’s first line of defense against tumors, set up by macrophages, neutrophils, MDSCs, NK cells, and DCs, which recognize and act on tumor cells nonspecifically to maintain homeostasis of the host. However, under pathological conditions like cancer, the immune responses of these cells are often disturbed by TIME, which may fuel tumor growth and progression. Recently, extensive studies have indicated that ncRNAs exert a vital role in regulating the differentiation, activation, recruitment, and function of various innate immune cells during the pathogenesis of HCC, which will be discussed in the section below (Table 1, Fig. 2).12–26
Table 1. ncRNAs involved in the regulation of innate immune cells.
| NcRNA | Expression in HCC | Related immune cell | Target molecules/pathways | Function in TIME | Impact on HCC | Reference |
|---|---|---|---|---|---|---|
| miR-28-5p | Downregulated | TAMs | IL-34/FAK/ERK1/2 | Promote TAM recruitment and infiltration into HCC tissue | Promote angiogenesis, tumor growth, and metastasis | 12 |
| circASAP1 | Upregulated | TAMs | miR-326/miR-532-5p/CSF-1 | Promote TAM infiltration | Promote HCC growth and metastasis | 13 |
| hsa_circ_0110102 | Downregulated | TAMs | miR-580-5p/PPARα/CCL2 | Inhibit macrophage activation and infiltration | Inhibit HCC growth and metastasis | 14 |
| lncRNA LINC00662 | Upregulated | TAMs | miR-15a/16/107/WNT3A/Wnt/β-catenin | Promote M2 macrophage polarization | Promote tumor growth and metastasis | 15 |
| lncRNA PART1 | Upregulated | TAMs | miR-372-3p/TLR4 axis | Promote M2 macrophage polarization | Promote HCC cell proliferation, EMT, and metastasis | 16 |
| lncRNA TUC339 | Upregulated | TAMs | NA | Promote macrophage activation, M2 polarization, and pro-tumorigenic activity | Promote HCC progression | 17 |
| hsa_circ_0003410 | Upregulated | TAMs | miR-139-3p/CCL5 | Recruit and polarize M2 macrophages | Promotes HCC tumor growth and metastasis | 18 |
| miR-223 | Downregulated | Neutrophils | NA | Attenuate neutrophil maturation and activation | Inhibit HCC progression | 19 |
| miR-122 | Downregulated | Neutrophils | CCL2 | Inhibit recruitment of neutrophils | Inhibit tumor progression | 20 |
| miR-561-5p | Upregulated | NKs | CX3CL1/ CX3CR1+/STAT3 | Inhibit CX3CR1+ NK-cell infiltration and activation | Promote pulmonary metastasis | 22 |
| circRNA UHRF1 | Upregulated | NKs | miR-449c-5p/TIM-3 | Induce NK-cell exhaustion and promote NK-cell dysfunction | Promote immune evasion and resistance to anti-PD1 therapy | 23 |
| lncRNA GAS5 | Downregulated | NKs | miR-544/RUNX3 | Enhance the killing effect of NK cells | Inhibit immune evasion and tumor progression | 24 |
| circRNA hsa_circ_0007456 | Downregulated | NKs | miR-6852-3p/ICAM-1 | Strengthen the cytotoxicity of NK cells | Inhibit immune evasion and inhibit tumor growth | 25 |
| circRNA ARSP91 | Downregulated | NKs | ULBP1 | Strengthen the cytotoxicity of NK cells | Enhance innate immune surveillance, suppress HCC proliferation | 26 |
| lncRNA HOTAIR | Upregulated | MDSCs | CCL2 | Promote recruitment of MDSCs | Promote tumor growth and metastasis | 21 |
HCC, hepatocellular carcinoma; MDSCs, myeloid-derived suppressor cells; NA, not available; ncRNAs, noncoding RNAs; NKs, natural killer cells; TAMs, tumor-associated macrophages; TIME, tumor immune microenvironment.
Fig. 2. ncRNA-mediated regulation of TIME. ncRNAs (miRNAs/lncRNAs/circRNAs) regulate the development, activation, recruitment, and cellular function of multiple cell types within TIME of HCC by diverse mechanisms.
CAF, cancer-associated fibroblast; circ, circular RNA; HCC, hepatocellular carcinoma; HSC, hepatic stellate cell; lnc, long noncoding RNA; MDSC, myeloid-derived suppressor cell; NK, natural killer; Treg, regulatory T cell; TIME, tumor immune microenvironment.
NcRNAs and macrophages
Macrophages are the major component of the innate immune cells within TIME. It is acknowledged that macrophages largely originate from circulating bone marrow-derived monocytes.27 Macrophages are a highly plastic and heterogeneous cell population whose phenotypes and functions are regulated by the surrounding microenvironment. In response to specific microenvironmental stimuli, macrophages generally polarize into two phenotypes, classically activated macrophages (M1) and alternatively activated macrophages (M2). M1 macrophages elicit pro-inflammatory effects and have an antitumorigenic role. Conversely, M2 macrophages enhance anti-inflammatory response and have pro-tumorigenic functions. Most tumor-associated macrophages (TAMs) in the tumor have an M2 phenotype. Under the inductions of various signaling molecules in TIME, TAMs are recruited to the primary and metastatic tumor tissues where they suppress the immune response by secreting a plethora of pro-tumorigenic proteases, cytokines, chemokines, and growth factors, and promote tumor growth, migration, invasion, angiogenesis, and immunosuppression.28,29
An increasing number of studies show the extensive involvement of ncRNAs in macrophage recruitment and polarization in multiple cancer types, including HCC. For example, in HCC, miR-28-5p deficiency promotes the expression of interleukin (IL)-34, and activates FAK and ERK1/2 signaling in macrophages, leading to enhanced recruitment and infiltration of macrophages into HCC tumor sites.12 Similarly in another study, highly expressed circASAP1 in HCC cells functions as a competitive endogenous RNA (ceRNA) that sponges miR-326 and miR-532-5p, alleviating the repression of CSF-1 expression. CSF-1, as a potent chemoattractant, survival, and differentiation factor for macrophages,30 positively modulates TAM infiltration to HCC tumor bed, which is considered to contribute to HCC growth and metastasis.13 Another circRNA hsa_circ_0110102, which is markedly downregulated in HCC cell lines, triggers macrophage activation and hepatic infiltration via miR-580-5p/PPARα/CCL2 pathway, while increasing the production and release of pro-inflammatory cytokines COX-2/PGE2 from macrophages, and ultimately enhancing HCC cell proliferation, migration, and invasion.14
Uncontrolled macrophage polarization is commonly implicated in HCC progression, and deregulation of ncRNAs plays an essential role in mediating M1/M2 macrophage polarization. As an example, LINC00662 induces macrophage M2 polarization in a paracrine manner to potentiate HCC tumor growth and metastasis. Mechanistic studies reveal that LINC00662 acts as a ceRNA for miR-15a/16/107 to stimulate WNT3A expression and secretion from HCC cells. WNT3A then activates Wnt/β-catenin pathway in macrophages, triggering their polarization toward the M2 subtype.15 It is also reported that lncRNA PART1 is transferred from HCC cells to surrounding macrophages via HCC cell-derived extracellular vesicles (EVs) that triggers macrophage polarization toward the M2 subtype by targeting miR-372-3p/TLR4 axis.16 Similarly, TUC339, a lncRNA enriched in HCC-secreted exosomes, is transmitted from HCC cells to peri-tumor macrophages and greatly affects macrophage polarization and activity. Overexpression of TUC339 in human macrophage cell lines THP-1 contributes to M2 phenotype, polarization, and decreased phagocytic activity, decreased pro-inflammatory cytokine (IL1β and TNF-α) production, reduced costimulatory molecule expression, and augmented viability of macrophages, therefore diminishing the antitumor immune response against tumor cells.17 Cao et al.18 found that upregulated_circ_0003410 in HCC cells promoted HCC tumor growth and metastasis by elevating the ratio of M2/M1 macrophage. Mechanistically, hsa_circ_0003410 stimulates the expression of CCL5 by competitively binding miR-139-3p to recruit and polarize M2 macrophages. Many other ncRNAs have been shown to change M1/M2 macrophage polarization, such as lncRNA MALAT1,31 lncRNA TP73-AS1,32 hsa_circ_0074854,33 which promote M2 polarization, and lncRNA cox-2,34 lncRNA GAS5,35 which inhibit M2 polarization.
Both ncRNAs expressed in HCC cells or exosomal ncRNAs secreted by HCC cells are known to orchestrate macrophage recruitment, polarization, and activity, and macrophages reciprocally impact HCC cell behavior by regulating ncRNAs. Intercellular communication between tumor cells and microenvironmental stromal cells mediated by ncRNAs have a strong impact on HCC initiation and malignant progression. A notable example is the miR-28-5p. Specifically, TAMs induced and recruited to HCC tissues by the miR-28-5p/IL-34/FAK/ERK1/2 signaling axis suppressed the expression of miR-28-5p in HCC cells by secretion of TGF-β1, hence forming an miR-28-5p/IL-34/TAM/TGF-β1 positive feedback loop to modulate HCC growth and metastasis.12 A study by Liu et al.36 reported that miR-92a-2-5p in exosomes transported from tumor-infiltrating macrophages to HCC tumor cells increased the invasive capacity of HCC tumor cells by altering the intrinsic AR/PHLPP/p-AKT/β-catenin signaling. Likewise, RBPJ-overexpressed macrophages transmit hsa_circ_0004658 to neighboring HCC cells via shuttling exosomes, which restrains proliferation and induces apoptosis in HCC cells through the miR-499b-5p/JAM3 pathway.37
ncRNAs and neutrophils
Neutrophils, generated in the bone marrow from myeloid precursors, participate in innate immunity against cancer. Like macrophages, neutrophils have various polarization phenotypes with either tumor-suppressive or tumor-promoting immune function. Tumor-associated neutrophils (TANs) can mediate cytotoxicity toward tumor cells. Besides, TANs also promote tumor growth and metastasis by stimulating angiogenesis, orchestrating the behavior of other immune cells, and enhancing tumor cell motility, migration, and invasion.38,39 Increased neutrophil infiltration has been linked to HCC progression and poor prognosis in patients with HCC.40 Mounting evidence has indicated that ncRNAs participate in controlling the activation, polarization, recruitment, and function of neutrophils in multiple cancer types, but it remains largely unexplored in HCC.8,9 MiR-223 is abundantly expressed in neutrophil cells and serves as a modulator of neutrophils in many advanced liver diseases, including HCC and hepatitis virus infection, cirrhosis, nonalcoholic fatty liver disease, and alcohol-induced liver injury, which are important risk factors of HCC. Functionally, miR-223 has a critical role in attenuating neutrophil maturation and activation, although the exact molecular mechanism has not been clarified.19,41 In another study by Hsu et al.,20 by examining the immune cells that infiltrate hepatic parenchyma in miR-122-KO mice and control group, found that miR-122 depletion stimulated recruitment of neutrophils to the liver, driving hepatic inflammation and producing a higher level of tumor-promoting cytokines. Mechanism dissection reveals that miR-122 deficiency triggers neutrophil recruitment through upregulating CCL2. As the cellular behavior and function of neutrophils are regulated by HCC-derived exosomes, further investigation of the contents of the exosomes is warranted.42
NcRNAs and MDSCs
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of immature cells predominately originating from bone marrow precursor cells. Under pathological conditions like HCC, MDSCs and expand accumulate in TIME, and have strong immunosuppressive activity that impairs various immune responses, such as T-cell function, therefore contributing to tumorigenesis and tumor progression.43 Several studies have implicated ncRNAs in the differentiation, expansion, and immunosuppressive function of MDSCs,44–46 their contribution to developing HCC is not clear. A recent study showed that strong expression of lncRNA HOTAIR in HCC cell lines was positively associated with enhanced recruitment of MDSCs. The proportion of MDSCs in peripheral blood mononuclear cells (PBMCs) increased when they were co-cultured with HCC cells overexpressing HOTAIR. It was further confirmed that HOTAIR increased the secretion of CCL2 from HCC cells into the tumor milieu. CCL2 was a well-documented chemoattractant and was speculated to be responsible for the HOTAIR-mediated accumulation of MDSCs into the TIME.21
NcRNAs and NK cells
Natural killer (NK) cells are an indispensable part of the innate immune system and a subgroup of innate lymphoid cells. They are primarily developed in the bone marrow and migrate into the blood circulation as they mature. The status of NK-cell activation is dictated by the interactions between specific ligands and diverse activating or inhibitory receptors expressed on the NK-cell surface. NK cells have cytotoxic activity and can directly kill target cells. Beyond its cytotoxic capacity, NK cells are also producers of large amounts of cytokines, chemokines, and growth factors that contribute to innate and adaptive immune responses, such as interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), CCL3. Therefore, NK cells can also influence the immune state and function of other immune cells in TIME.47–49 Thus, NK cells mainly serve as important players in boosting antitumor immune response against tumors. NK-cell dysfunction can lead to severe immune deficiency that allows tumor cells to escape immune surveillance and thrive in the TIME. It is noteworthy that NK-cell dysfunction has been reported in the TIME of HCC,50 but the mechanism underlying the abnormal behavior of NK cells in HCC is unknown.
To the best of our knowledge, studies describing the role of ncRNAs in mediating NK-cell activity, including infiltration, activation, exhaustion, and function during HCC progression are increasing. Chen et al.22 found that upregulation of miR-561-5p expression suppressed CX3CR1+ NK-cell infiltration and activation by targeting CX3CL1, thereby promoting HCC tumorigenesis and pulmonary metastasis. miR-561-5p in HCC tumor cells reduced secretion of CX3CL1, a chemokine known to be associated with lymphocyte migration. Loss of CX3CL1 in TIME interfered with chemotaxis and activation of CX3CR1+ NK cells by inactivating STAT3 signaling in NK cells. As CX3CR1+ NK cells have strong tumor-killing activity, inhibiting the infiltration and cytotoxic activity of CX3CR1+ NK cells enabled HCC cells to escape immune surveillance, and promoted HCC proliferation and metastasis. A recent study reported that HCC cell-derived exosomal circUHRF1 contributed to immune evasion and resistance to anti-PD1 immunotherapy in HCC by inducing NK-cell exhaustion and suppressed IFN-γ and TNF-α production in NK cells. circUHRF1-mediated exhaustion and dysfunction of NK cells was attributed to increased expression of inhibitory receptor TIM-3 in NK cells.23
In HCC, the cytotoxicity of NK cells is governed by multiple ncRNAs. Low expression of the lncRNA GAS5 in the NK cells of HCC patients inhibits cytotoxic activity and accelerates tumor metastasis. Mechanistic studies show that lncRNA GAS5 deficiency inhibited RUNX3 expression in NK cells by upregulating miR-544, which suppressed the NCR1/NKp46 axis. NKp46 is a stimulatory receptor on the NK cell surface, and its inactivation impairs the killing activity of NK cells. The evidence indicates that the miR-544/RUNX3/NCR1/NKp46 pathway accounted for the GAS5-mediated regulation of NK-cell cytotoxicity.24,51 Another study reported that downregulated hsa_circ_0007456 in HCC cells reduced the cytotoxicity of NK cells toward tumor cells, which promoted immune evasion and aggressiveness of HCC. To be specific, hsa_circ_0007456 deficiency restored miR-6852-3p level and interfered with the expression of ICAM-1. ICAM-1 was reported to regulate the adhesion of cancer cells and NK cells. Hsa_circ_0007456-mediated interference of ICAM-1 decreased the susceptibility of HCC cells to NK cytolysis.25–53 Similarly, circRNA ARSP91 was found to enhance the cytotoxicity of NK cells toward HCC cells by upregulating UL16 binding protein 1, an NKG2D ligand that activates stimulatory receptors associated with the tumor-killing function of NK cells.26,54,55 Many other ncRNAs are also known to modulate the cytotoxicity of NK cells against HCC cells, such as miR-615-5p,56 miR-146a,57 miR-30c-1,58 and miR-506.59 The available evidence supports a role of ncRNAs in the regulation of NK cells during the development of HCC. Some of the ncRNAs may be potential therapeutic targets to enhance the efficacy of NK cell-based anticancer immunotherapy in the treatment of HCC.
ncRNAs and DCs
DCs are considered the most efficient antigen-presenting cells with a key role in linking innate and adaptive immune system. They take up and process antigens, converting them into peptides that are presented to T cells by major histocompatibility complex molecules that trigger activation and protective immune responses. In the tumor milieu, normal DC activity is disturbed and often has immunosuppressive and tolerogenic effects that boosts the malignant progression of tumors.60,61 Studies of dendritic cells in HCC are relatively scarce despite considerable evidence showing they have a critical role in many malignancies. ncRNAs have been shown to regulate the development, differentiation, recruitment, and function of DCs in many cancers, but their regulatory roles in the pathogenesis of HCC have not been extensively studies.8,9,62 In recent years, several ncRNAs were reported to mediate the infiltration of dendritic cells in HCC. For example, Wu et al.63 found that the expression level of lncRNA ASB16-AS1 was negatively correlated with tumor-infiltrating neutrophils in HCC, as shown by CIBERSORT, TIMER, xCell, quanTIseq, EPIC and MCP-counter.63 The CIBERSORT algorithm confirmed that that MIR210HG was negatively correlated with, and LINC01224 was positively correlated, with DC infiltration.64 The detailed molecular mechanisms were not described in the studies.
ncRNAs and adaptive immune cells in TIME
Despite being prominent regulators for many types of innate immune cells within TIME of HCC, emerging evidence has revealed that ncRNAs also participate in the regulation of adaptive immune cells, including various T and B cell subgroups (Table 2, Fig. 2).65–73
Table 2. ncRNAs involved in the regulation of adaptive immune cells.
| NcRNA | Expression in HCC | Related immune cell | Target molecules/pathway | Function in TIME | Impact on HCC | Reference |
|---|---|---|---|---|---|---|
| lncRNA NEAT1 | Upregulated | CD8+ T cells | miR-155/Tim-3 | Induce CD8+ T cells apoptosis and dampen its cytolysis activity against HCC cells | Promote immune evasion and tumor progression | 65 |
| lncRNA lnc-Tim3 | Upregulated | CD8+ T cells | Tim-3/Bat3/Lck/ZAP70/AP-1/NF-AT1 and Bat3/p300/p53/RelA | Stimulate CD8+ T-cell exhaustion | Promote immunosuppression and tumor growth | 66 |
| circRNA circMET | Upregulated | CD8+ T lymphocytes | miR-30-5p/Snail/DPP4/CXCL10 | Stimulate CD8+ lymphocyte infiltration | Enhance immunosuppression | 67 |
| microRNA-132 | Upregulated | Th17 | SNIP1 | Promote Th17 differentiation and function | Promote HCC cell migration and EMT | 68 |
| microRNA-34a | Downregulated | Tregs | CCL22 | Suppress Treg recruitment | Enhance immune surveillance, suppress tumor growth, and metastasis | 69 |
| lncRNA EGFR | Upregulated | Tregs | EGFR/AP-1/NF-AT1 | Stimulate Treg differentiation, inhibit CTL activity | Promote immunosuppression and HCC growth | 70 |
| circRNA circGSE1 | Upregulated | Tregs | miR-324-5p/TGFBR1/Smad3/FOXP3 axis | Induce the expansion of Tregs | Promote immune escape, enhance tumor growth and metastasis | 71 |
| lncRNA FENDRR | Downregulated | Tregs | miR-423-5p/GADD45B | Inhibit Treg infiltration | Suppress immune escape and tumor growth | 72 |
| lncRNA LINC00261 | Downregulated | B cells | miR105-5p/SELL | Promote B-cell dysfunction | Promote HCC progression | 73 |
HCC, hepatocellular carcinoma; NA, not available; ncRNA, noncoding RNA; Th, T-helper cell; TIME, tumor immune microenvironment; Treg, regulatory T cells.
ncRNAs and T cells
T lymphocytes are the primary effector cells in cellular immunity and include subsets with distinct roles in immunity and immune-mediated pathologies.74 Cytotoxic T cells (CTLs) kill and eradicate malignant cells.75 T-helper (Th) cells are differentiated from CD4+ T cells and have subpopulations with either pro- or antitumorigenic activity in the tumor milieu.76 Regulatory T cells (Tregs) are differentiated from CD4+ T cells and suppress antitumor responses of other immune cells, with immunosuppressive activity in the TIME. Infiltration of a large number of Tregs into tumor tissue is often associated with poor prognosis.77 Additionally, In the setting of HCC, recent studies have shown that substantial changes in the expression profiles of ncRNAs occur during T-cell development, activation, and differentiation, indicating a crucial role of ncRNAs in regulating T-cell activity.
Increasing evidence shows the pivotal role of ncRNAs in mediating the antitumor response of CTLs against malignant HCC cells. For example, lncRNA NEAT1 was shown to contribute to the immune escape of HCC by affecting the antitumor activity of CD8+ T cells. The lncRNA NEAT1 was significantly upregulated in the PBMCs of HCC patients, and overexpression of lncRNA NEAT1 induced CD8+ T-cell apoptosis and impaired the cytolysis of HCC cells via regulating miR-155/Tim-3 signaling. Tim-3 is an inhibitory immune checkpoint receptor expressed on T cells; its activation enforces T-cell exhaustion and induces T-cell apoptosis and dysfunction.65,78 The lncRNA lnc-Tim3 was shown to stimulate CD8+ T-cell exhaustion by targeting Tim-3, and was linked to immunosuppression and malignant behavior in HCC. Mechanistically, lnc-Tim3 competitively bound to Tim-3 in CD8+ T cells, resulting in release of Bat3 from the C-terminal end of Tim-3 and accumulation of the catalytically inactive form of Lck, which suppressed downstream T-cell signaling (ZAP70/AP-1/NF-AT1 signaling) and endogenous cytokine production (IL2/IFN-γ). The released Bat3 formed a complex with p300, which increased its nuclear translocation and enhanced p300-dependent p53 and RelA transcriptional activation of anti-apoptosis genes and promoted survival of Tim-3+ exhausted CD8+ T cells. The dual mechanism contributed to CD8+ T-cell exhaustion.66 In addition, the circRNA circMET is preferentially expressed in HCC tumors and associated with poor clinical outcomes. CircMET overexpression hinders CD8+ T-cell infiltration in HCC tissues through the miR-30-5p/Snail/DPP4/CXCL10 axis, which enhances the immunosuppressive properties of TIME that favor HCC cell survival and metastasis.67
Th cell differentiation results from regulation of genes and involves transcription factors, including STAT3, RUNX-1, and others.79 However, ncRNAs are also emerging as important regulators of Th cell differentiation. Feng et al.68 observed that overexpression of miR-132 promoted Th17 differentiation and production of IL22 and IL17 possibly by targeting of the downstream protein SNIP1. IL22 activated hepatic stellate cells (HSCs), which then promoted HCC cell migration and epithelial-mesenchymal transition (EMT).68
ncRNAs are also implicated in the modulation of Tregs in HCC. For example, HBV infection-activated TGF-β signaling suppresses the expression of microRNA-34a, resulting in increased production of CCL22, which facilitates recruitment of CD4+CD25+ Tregs into the TIME. Sustained activation of TGF-β-miR-34a-CCL22 axis promotes the development of intrahepatic venous metastasis in HCC patients via generating an immunosuppressive TIME that favors tumor cell survival and dissemination.69 ncRNAs have also been found to participate in the differentiation of Tregs during HCC development, as shown by lncRNA lnc-EGFR. Lnc-EGFR is highly expressed in Tregs of HCC patients and is positively correlated with HCC immune evasion and tumor growth. Lnc-EGFR specifically binds to epithelial growth factor receptor (EGFR) and stabilizes it by blocking its ubiquitination by c-CBL. Persistent activation of EGFR triggers a. downstream signaling cascade (RAS/ERK/AP-1/NF-AT1). It is important to note that the NF-AT transcription factors are widely expressed in a variety of leukocytes, including T cells, and regulate genes involved in lymphocyte development. lnc-EGFR-activated AP-1/NF-AT1 signaling has been shown to stimulate Treg differentiation, as shown by an increased ratio of Tregs in CD4+ T cells and in TIME. Intriguingly, the AP-1/NF-AT1 complex enhanced transcription of lnc-EGFR, EGFR, and Foxp3 by binding to their promoters, thus forming a forward-feedback loop in Tregs that impaired antitumor immunity and promoted HCC progression.70 Similarly, exosomal circGSE1 from HCC cells promoted immune escape, tumor growth, and metastasis by promoting Treg differentiation and proliferation by regulating an miR-324-5p/TGFBR1/Smad3/FOXP3 axis.71 In addition, Yu et al.72 reported that poorly expressed lncRNA FENDRR in HCC cells acted an miR-423-5p sponge to downregulate GADD45B, enhance the immune-suppressive activity of Tregs, and allow HCC cells to escape from immune surveillance.72 Taken together, the studies underscore the crucial role of ncRNAs in T cell-mediated immunosuppression and might inspire immunotherapy.
NcRNAs and B cells
The importance of T cells in tumor immune surveillance is well established, but the contribution of B cells has been studied to a much lesser extent. B cells contribute to humoral immunity by producing antibodies. Recent advances in B-cell biology have revealed that B cells participate in antigen presentation, promote T-cell responses, and release a variety of cytokines. B cell subsets have protumor or antitumor activities, including regulatory B cells with immunosuppressive activity.80,81 Recent studies have shown tumor-infiltrating B cells were associated with tumor progression and immunotherapy response in human cancers, including HCC.82–84 The regulatory role of ncRNAs during B-cell development, differentiation, apoptosis, and function have been described,8,85,86 but little is known of ncRNA-mediated B-cell regulation of the pathogenesis of HCC. A recent bioinformatics analysis revealed that the LINC00261/MiR105-5p/SELL signaling axis was involved in B-cell dysfunction and was associated with overall survival in HCC patients. Details of the molecular mechanism were not clarified.73
ncRNAs and other stromal components in the TIME
In addition to immune cells, CSCs, HSCs, CAFs, and many other stromal cells are components of the TIME in HCC.6 Evidence of the regulatory effects of ncRNAs on a variety of nonimmune cells is increasing (Table 3, Fig. 2).87–92–99
Table 3. ncRNAs involved in the regulation of other stromal cells in TIME.
| NcRNA | Expression in HCC | Related stromal cell | Target molecules/pathways | Function in TIME | Impact on HCC | References |
|---|---|---|---|---|---|---|
| lncRNA lncTCF7 | Upregulated | CSCs | TCF7/Wnt signaling | Promote self-renewal of human liver CSCs | Promote tumor propagation | 87 |
| lncRNA lnc-β-Catm | Upregulated | CSCs | EZH2/Wnt-β-catenin | Sustain liver CSC self-renewal | Promote tumor propagation | 88 |
| lncRNA LncBRM | Upregulated | CSCs | YAP signaling | Promote CSC self-renewal | Promote tumor propagation | 89 |
| lncRNA lncHDAC2 | Upregulated | CSCs | Hedgehog signaling | Promote self-renewal of liver CSCs | Promote tumor growth | 90 |
| lncRNA lncSOX4 |
Upregulated | CSCs | STAT3/SOX4 signaling | Sustain liver CSC self-renewal | Promote tumor initiation | 91 |
| lncRNA lnc-DILC | Downregulated | CSCs | IL-6/JAK2/STAT3 a signaling | Suppress self-renewal of liver CSCs | Inhibit tumor initiation and progression | 92 |
| microRNA-145 | Downregulated | HSCs | ZEB2/Wnt-β-catenin | Repress HSC activation and proliferation | Repress liver fibrosis and tumorigenesis | 93 |
| microRNA-708 | Downregulated | HSCs | ZEB1/Wnt-β-catenin | Repress HSC activation and proliferation | Repress liver fibrosis and tumorigenesis | 94 |
| lncRNA-MEG3 | Downregulated | HSCs | miR-212/SMO/Hh signaling | Inhibit HSC activation | Inhibit liver fibrosis | 95 |
| microRNA-378 | Downregulated | HSCs | Hh signaling | Limit HSC activation | Inhibit liver fibrosis | 96 |
| microRNA-21 | Upregulated | HSCs, CAFs | PTEN/PDK1/AKT signaling | Convert HSCs to CAFs | Promote HCC angiogenesis | 97 |
| microRNA-124 | Downregulated | HSCs | IQGAP1/NF-κB axis | Inhibit cytokine secretion of HSCs | Reduce inflammatory response | 98 |
| miR-1247-3p | Upregulated | CAFs | B4GALT3, β1-integrin/NF-κB axis | Induce CAF activation | Foster lung metastasis of HCC | 99 |
CAF, cancer-associated fibroblast; CSC, cancer stem cell; HCC, hepatocellular carcinoma; HSC, hepatic stellate cell; NA, not available; ncRNA, noncoding RNA; TIME, tumor immune microenvironment.
ncRNAs and CSCs
Cancer stem cells (CSCs) are a rare population of cells within the tumor bulk that share many intrinsic features with normal stem cells, such as self-renewal and differentiation. CSCs have been found to exist in many solid tumors, including HCC. The stem-cell like properties of liver CSCs may contribute to the heterogeneity, resistance to treatment, metastasis, and high rate of recurrence of HCC, which makes CSCs an attractive target for cancer therapy.100,101 Recently, increasing studies have described the ability of ncRNAs to modulate self-renewal, differentiation, and stemness of liver CSCs through activating diverse CSC-related signaling pathways, such as Wnt-β-catenin signaling, YAP signaling, Hedgehog signaling, STAT3 signaling, TGF-β signaling, or cell cycle-related signaling.101 LncRNA lncTCF7 and lnc-β-Catm, both are seen highly expressed in HCC tumor tissues and liver CSCs and correlate with poor prognosis in HCC, promote self-renewal maintenance of liver CSCs through activation of Wnt-β-catenin signaling pathway. Mechanistically, lncTCF7 recruits the SWI/SNF chromatin remodeling complex to the promoter region of target gene TCF7 to promote its transcription. TCF7 then triggers downstream Wnt signaling cascade, which primes the self-renewal of liver CSCs and tumor propagation. Lnc-β-Catm associates with EZH2 to catalyze methylation of β-catenin, thus hindering β-catenin ubiquitination and stabilizing it, allowing β-catenin to start Wnt signaling and sustain the self-renewal of liver CSCs.87,88 Similarly, lncBRM sequesters BRM to form BRG1-BAF complex, starting YAP1 signaling in liver CSCs, which drives CSC self-renewal process.89 lncHDAC2 is highly expressed in the CD13+CD133+ subset of liver CSCs, where it contributes to self-renewal maintenance by recruiting the nucleosome remodeling and deacetylase (NuRD) complex to promote PTCH1 and activate Hedgehog signaling.90 Another lncRNA lncSOX4 mediates liver CSCs self-renewal via STAT3-SOX4 signaling axis. LncSOX4 interacts with and recruits STAT3 to bind to SOX4 promoter, triggering SOX4 expression, which is required for liver CSCs self-renewal and tumor initiation.91 In addition, lncRNA-DILC is examined significantly downregulated in EPCAM+ CSCs; it abrogates IL6 transcription and abolishes STAT3 activation, thus repressing self-renewal and expansion of liver CSCs. Lnc-DILC depletion helps with HCC start and progression.92
NcRNAs and HSCs
HSCs play vital roles in the tumorigenesis and progression of HCC, largely because activation of HSCs contributes to hepatic fibrosis. HSCs can secrete a variety of bioactive contents to maintain liver inflammation and regulate tumor-associated pathways, which then trigger immunosuppression, angiogenesis, and therapy resistance of HCC. Under pathological conditions, HSCs are changed from the quiescent stage to the active stage, and the activated HSCs eventually differentiate into myofibroblast-like cells.102,103 Increasing studies have described the molecular mechanisms underlying HSC activation, and ncRNAs emerge as prominent participants in the regulation of HSC activation. For example, as the Wnt-β-catenin signaling pathway is documented to be generally hyperactivated in HSCs during liver fibrosis to orchestrate cell activation, proliferation, and maintain homeostasis,104 many ncRNAs have been revealed to regulate HSC activation via Wnt-β-catenin pathway, as exampled by microRNA-145 and microRNA-708. MicroRNA-145 and microRNA-708 are both poorly expressed in fibrotic liver tissues and activated HSCs, and their deregulations are both able to activate the Wnt-β-catenin pathway via increasing expression of ZEB2 and ZEB1, respectively. The hyperactivated Wnt-β-catenin pathway thus accelerates the activation and proliferation of HSCs.93,94 Hedgehog (Hh) signaling is another cascade activated in HSCs and regulates hepatic fibrogenesis. Hh signaling is also regulated by various ncRNAs, such as lncRNA-MEG3, microRNA-378, etc. LncRNA-MEG3 inhibits Hh signaling-mediated EMT process in HSC activation via associating with SMO protein and sponging miR-212. While microRNA-378 limits HSC activation by suppressing Gli3 expression, which is a downstream transcription factor of Hh signaling.95,96 Zhou et al.97 identified that tumor-derived exosomal miRNA-21 was internalized by HSCs and it directly targeted phosphatase and tensin homolog (PTEN), resulting in activation of PDK1/AKT signaling in HSCs, which primed the conversion from normal HSCs to CAFs and promoted angiogenesis of HCC. ncRNAs can also impair the HSC function to produce inflammatory cytokines. One such example is microRNA-124, which inhibits HSC secretion of TNF-α, IL-1β, and IL-6 by targeting the IQGAP1/NF-κB axis.98
ncRNAs and CAFs
As the most important and abundant component of the stromal cell population in TIME, CAFs are crucial players during the occurrence and malignant progression of HCC. Upon stimulation by the TIME, fibroblasts are activated and converted into CAFs. CAFs have been reported to modulate HCC progression through diverse mechanisms, including remodeling the extracellular matrix, secreting soluble factors or exosomes, and regulating the behavior of various immune cells, which can either potentiate or oppose HCC progression.105–107 Many ncRNAs are known to regulate CAF formation and activation during HCC development. For example, the abovementioned HCC cell-derived miR-21 could convert HSCs into CAFs via targeting PTEN and activating PDK1/AKT signaling cascade in HSCs. Activated CAFs release various angiogenic factors to stimulate angiogenesis in HCC tumors.97 Fang and colleagues unveil that HCC cell-derived exosomal miR-1247-3p potentiates CAFs activation to foster lung metastasis of HCC. Mechanistically, miR-1247-3p is transferred from HCC cells to fibroblasts in lung pre-metastasis niche via exosomes. MiR-1247-3p subsequently drives normal fibroblast transformation to CAFs by decreasing its target gene B4GALT3 expression to activate β1-integrin-NF-κB signaling. Activated CAFs promote stemness, EMT, chemoresistance, and tumorigenicity of HCC cells by releasing IL-6 and IL-8.99 Finally, dynamic intercellular communications mediated by exosomes are widely seen between CAFs and HCC cells and strongly affect HCC progression and therapy response.108 Therefore, as a major cargo in exosomes, ncRNAs are speculated to play important roles during the interaction, which deserves further elucidation.
Diagnostic/therapeutic potential of ncRNAs in HCC
One reason leading to the high mortality of HCC is that a significant percentage of patients is diagnosed at advanced stages. The diagnosis of HCC relies on serum α-fetoprotein measurement and ultrasonography imaging, etc. However, these diagnostic modalities still remain insufficient, especially for diagnosis of early-stage HCC.5 Therefore, novel biomarkers with higher sensitivity and specificity are urgently needed. Emerging evidence indicates that a myriad of ncRNAs show aberrant and tissue-specific/cell-specific expression patterns in HCC, and many are detectable and relatively stable in body fluids. These unique properties of ncRNAs make them promising noninvasive biomarkers for HCC detection. Besides, ncRNAs also display prognostic value since the expression levels of multiple ncRNAs are closely correlated with tumor stage and clinical outcomes of HCC, such as metastasis and recurrence.11 In addition, certain ncRNAs that modulate resistance are proven to be associated with treatment response, indicating their potential to predict treatment response.23,109 Of note, many ncRNAs are encapsulated in circulating exosomes, which protect them from being degraded by RNase. And exosomal ncRNA detection has the advantage of noninvasive, repeatable, and real-time tracking.110,111 Taken together, ncRNAs could serve as potential diagnostic/prognostic biomarkers, however, further efforts must validate the sensitivity and specificity of them as biomarkers.
Despite a lack of reliable biomarkers, effective therapeutic options/targets are also limited for HCC treatment.5 Recently, ncRNAs have been documented to play widespread roles in gene regulation and participate in diverse signaling cascades.112 Most important, ncRNAs function as a pivotal regulator in TIME during HCC progression by influencing the differentiation, activation, recruitment, and function of various types of cells within TIME, including diverse immune cells and many other nonimmune stromal cells. The ncRNA-mediated regulation of the TIME and cancer type-specific deregulation of ncRNAs indicate that ncRNAs are highly promising therapeutic targets for HCC treatment. To date, many approaches have been developed to target ncRNAs and govern their expression or function, including small molecule inhibitors, aptamers, antisense oligonucleotides, RNA interference, and CRISPR/Cas9 gene editing technology.113–115 RNA-based therapeutic method is still in its infancy and many difficulties and limitations have emerged during its application. For example, difficulty of using antisense oligonucleotides is to optimize their specific delivery to target cells and to augment their stability in vivo.113 One challenge of CRISPR/Cas9 is to avoid adverse off-target effects. Besides, there are concerns like whether these treatments might cause unwanted side effects, such as affecting other parts within TIME.112,114 In addition, many ncRNAs exist in regulatory feedback loops, thus, it might be difficult to modulate their expression. Overcoming these challenges will improve the efficacy of these RNA-based cancer therapies.
It is important to highlight that ncRNA-based therapy is a promising approach in HCC immunotherapy. Given their vital roles in TIME, it might be possible to modulate the immune response of multiple immune cells within TIME by manipulating the expression pattern of specific ncRNA, such as facilitating recruitment of various antitumor immune cells to the tumor site, enhancing cytotoxicity of NK cells, inhibiting the function of immunosuppressive cells, which could be effective to boost antitumor immune response and restrain immune escape, ultimately hindering tumor growth and malignant progression. In addition, targeting different ncRNAs combined with other therapeutic strategies might show significant benefit in the treatment of HCC. For example, given that circUHRF1 has been proven to drive resistance to anti-PD1 immunotherapy in HCC patients,23 the combined therapy of ncRNA-targeted drugs and anti-PD1 immunotherapy may therefore display synergistic effects in inhibiting tumor progression.
Conclusions and perspectives
TIME is an integrated system consisting of diverse cellular and noncellular parts. It closely interacts with tumors and greatly contributes to the occurrence and progression of HCC. And as summarized in this review, ncRNA is emerging as a prominent regulator in reprogramming the TIME of HCC. An impressive number of ncRNAs exhibit aberrant expression patterns in HCC, and they can modulate the development, biological behavior, and function of various cell types within the TIME, which ultimately elicit profound influences on tumorigenesis, tumor growth, metastasis, angiogenesis, and immune evasion in HCC. Of note, the current knowledge regarding the regulatory role of ncRNAs in TIME principally focuses on miRNAs and lncRNAs, but novel classes of ncRNA like circRNAs and piRNAs await further investigation. Accumulating evidence has indicated ncRNAs as important mediator in the crosstalk between TIME and neoplastic cells in various cancer types, but their mediatory role has not been elucidated in HCC. Apart from focusing on regulating ncRNAs on TIME, it is also important to dissect the exact mechanisms of how ncRNAs are dysregulated in TIME of HCC, which lets us gain a more comprehensive understanding of the complex regulatory network between TIME and ncRNAs in HCC pathogenesis. Because of the roles of ncRNAs within TIME of HCC, novel diagnostic/prognostic biomarkers and therapeutic interventions based on ncRNAs are under development for treating HCC, however, the majority are still in the experimental stages due to various limitations. Further investigations must translate those research findings into clinical applications.
Abbreviations
- CAF
cancer-associated fibroblast
- ceRNA
competitive endogenous RNA
- circRNA
circular RNA
- CSC
cancer stem cell
- CTL
cytotoxic T cell
- DC
dendritic cell
- EMT
epithelial-mesenchymal transition
- EV
extracellular vesicle
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- Hh signaling
Hedgehog signaling
- HSC
hepatic stellate cell
- lncRNA
long noncoding RNA
- MDSC
myeloid-derived suppressor cell
- miRNA
microRNA
- NA
not available
- ncRNA
noncoding RNA
- NK cell
natural killer cell
- PBMC
peripheral blood mononuclear cell
- piRNA
PIWI-interacting RNA
- sncRNA
small noncoding RNA
- snoRNA
small nucleolar RNA
- TAM
tumor-associated macrophage
- TAN
tumor-associated neutrophil
- Th cell
T-helper cell
- TIME
tumor immune microenvironment
- Treg
regulatory T cell
- tsRNA
transfer RNA-derived small RNA
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 6.Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18(8):525–543. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Liu Q, Liao Q. Long noncoding RNA: a dazzling dancer in tumor immune microenvironment. J Exp Clin Cancer Res. 2020;39(1):231. doi: 10.1186/s13046-020-01727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Xie Y, Luo Y. The Role of Long Non-Coding RNAs in the Tumor Immune Microenvironment. Front Immunol. 2022;13:851004. doi: 10.3389/fimmu.2022.851004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179(5):1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 12.Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z, Cao Y, et al. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63(5):1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 13.Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2020;72(3):906–922. doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARα/CCL2 pathway. Aging (Albany NY) 2021;13(8):11969–11987. doi: 10.18632/aging.202900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian X, Wu Y, Yang Y, Wang J, Niu M, Gao S, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol Oncol. 2020;14(2):462–483. doi: 10.1002/1878-0261.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Che J, Xu L, Yang W, Zhou W, Zhou C. Tumor-derived extracellular vesicles containing long noncoding RNA PART1 exert oncogenic effect in hepatocellular carcinoma by polarizing macrophages into M2. Dig Liver Dis. 2022;54(4):543–553. doi: 10.1016/j.dld.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int J Mol Sci. 2018;19(10):2958. doi: 10.3390/ijms19102958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao P, Ma B, Sun D, Zhang W, Qiu J, Qin L, et al. hsa_circ_0003410 promotes hepatocellular carcinoma progression by increasing the ratio of M2/M1 macrophages through the miR-139-3p/CCL5 axis. Cancer Sci. 2022;113(2):634–647. doi: 10.1111/cas.15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye D, Zhang T, Lou G, Liu Y. Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med. 2018;50(9):1–12. doi: 10.1038/s12276-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–2883. doi: 10.1172/jci63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujisaka Y, Iwata T, Tamai K, Nakamura M, Mochizuki M, Shibuya R, et al. Long non-coding RNA HOTAIR up-regulates chemokine (C-C motif) ligand 2 and promotes proliferation of macrophages and myeloid-derived suppressor cells in hepatocellular carcinoma cell lines. Oncol Lett. 2018;15(1):509–514. doi: 10.3892/ol.2017.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EB, Zhou ZJ, Xiao K, Zhu GQ, Yang Y, Wang B, et al. The miR-561-5p/CX(3)CL1 Signaling Axis Regulates Pulmonary Metastasis in Hepatocellular Carcinoma Involving CX(3)CR1(+) Natural Killer Cells Infiltration. Theranostics. 2019;9(16):4779–4794. doi: 10.7150/thno.32543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang P, Xiang L, Chen W, Li S, Huang S, Li J, et al. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun. 2019;25(2):99–109. doi: 10.1177/1753425919827632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021;12(1):94. doi: 10.1038/s41419-020-03334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17(1):388–397. doi: 10.3892/ol.2018.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17(12):887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 28.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193(6):727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol. 2020;318(3):C649–c663. doi: 10.1152/ajpcell.00510.2018. [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Huang ZB, Liao CJ, Hu XW, Li SL, Qi M, et al. LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-β1 signaling. Cell Signal. 2020;75:109738. doi: 10.1016/j.cellsig.2020.109738. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int J Nanomedicine. 2021;16:2803–2818. doi: 10.2147/ijn.S284560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. 2018;119(3):2951–2963. doi: 10.1002/jcb.26509. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Li FY, Zhao W, Gao ZK, Shen B, Xu H, et al. Long non-coding RNA GAS5 overexpression inhibits M2-like polarization of tumour-associated macrophages in SMCC-7721 cells by promoting PTEN expression. Int J Exp Pathol. 2020;101(6):215–222. doi: 10.1111/iep.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu G, Ouyang X, Sun Y, Xiao Y, You B, Gao Y, et al. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death Differ. 2020;27(12):3258–3272. doi: 10.1038/s41418-020-0575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Zhang J, Li P, Li T, Zhou Z, Wu H. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 2022;13(1):32. doi: 10.1038/s41419-021-04345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16(7):431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 39.Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol. 2019;16(10):601–620. doi: 10.1038/s41571-019-0222-4. [DOI] [PubMed] [Google Scholar]
- 40.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13(3):159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut. 2021;70(4):784–795. doi: 10.1136/gutjnl-2020-322526. [DOI] [PubMed] [Google Scholar]
- 42.Han Q, Zhao H, Jiang Y, Yin C, Zhang J. HCC-Derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells. 2019;8(6):558. doi: 10.3390/cells8060558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19(2):108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Zhao S, Sui H, Liu H, Yao M, Su Y, et al. MicroRNAs/LncRNAs Modulate MDSCs in Tumor Microenvironment. Front Oncol. 2022;12:772351. doi: 10.3389/fonc.2022.772351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leija Montoya G, González Ramírez J, Sandoval Basilio J, Serafín Higuera I, Isiordia Espinoza M, González González R, et al. Long Non-coding RNAs: Regulators of the Activity of Myeloid-Derived Suppressor Cells. Front Immunol. 2019;10:1734. doi: 10.3389/fimmu.2019.01734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Safarzadeh E, Asadzadeh Z, Safaei S, Hatefi A, Derakhshani A, Giovannelli F, et al. MicroRNAs and lncRNAs-A New Layer of Myeloid-Derived Suppressor Cells Regulation. Front Immunol. 2020;11:572323. doi: 10.3389/fimmu.2020.572323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu SY, Fu T, Jiang YZ, Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19(1):120. doi: 10.1186/s12943-020-01238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19(3):200–218. doi: 10.1038/s41573-019-0052-1. [DOI] [PubMed] [Google Scholar]
- 49.Cózar B, Greppi M, Carpentier S, Narni-Mancinelli E, Chiossone L, Vivier E. Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 2021;11(1):34–44. doi: 10.1158/2159-8290.Cd-20-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung PS, Jang JW. Natural Killer Cell Dysfunction in Hepatocellular Carcinoma: Pathogenesis and Clinical Implications. Int J Mol Sci. 2018;19(11):3648. doi: 10.3390/ijms19113648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai CB, Mager DL. Role of runt-related transcription factor 3 (RUNX3) in transcription regulation of natural cytotoxicity receptor 1 (NCR1/NKp46), an activating natural killer (NK) cell receptor. J Biol Chem. 2012;287(10):7324–7334. doi: 10.1074/jbc.M111.306936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong JU, Uong TNT, Chung WK, Nam TK, Ahn SJ, Song JY, et al. Effect of irradiation-induced intercellular adhesion molecule-1 expression on natural killer cell-mediated cytotoxicity toward human cancer cells. Cytotherapy. 2018;20(5):715–727. doi: 10.1016/j.jcyt.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen C, et al. MiRNA-296-3p-ICAM-1 axis promotes metastasis of prostate cancer by possible enhancing survival of natural killer cell-resistant circulating tumour cells. Cell Death Dis. 2013;4(11):e928. doi: 10.1038/cddis.2013.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113(15):3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 55.Huergo-Zapico L, Acebes-Huerta A, López-Soto A, Villa-Álvarez M, Gonzalez-Rodriguez AP, Gonzalez S. Molecular Bases for the Regulation of NKG2D Ligands in Cancer. Front Immunol. 2014;5:106. doi: 10.3389/fimmu.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahmoon MA, Youness RA, Gomaa AI, Hamza MT, Waked I, El Tayebi HM, et al. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017;35(2-3):76–87. doi: 10.1080/08977194.2017.1354859. [DOI] [PubMed] [Google Scholar]
- 57.Xu D, Han Q, Hou Z, Zhang C, Zhang J. miR-146a negatively regulates NK cell functions via STAT1 signaling. Cell Mol Immunol. 2017;14(8):712–720. doi: 10.1038/cmi.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong J, Liu R, Zhuang R, Zhang Y, Fang L, Xu Z, et al. miR-30c-1* promotes natural killer cell cytotoxicity against human hepatoma cells by targeting the transcription factor HMBOX1. Cancer Sci. 2012;103(4):645–652. doi: 10.1111/j.1349-7006.2012.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su Z, Ye X, Shang L. MiR-506 Promotes Natural Killer Cell Cytotoxicity against Human Hepatocellular Carcinoma Cells by Targeting STAT3. Yonsei Med J. 2019;60(1):22–29. doi: 10.3349/ymj.2019.60.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Worbs T, Hammerschmidt SI, Förster R. Dendritic cell migration in health and disease. Nat Rev Immunol. 2017;17(1):30–48. doi: 10.1038/nri.2016.116. [DOI] [PubMed] [Google Scholar]
- 61.Giovanelli P, Sandoval TA, Cubillos-Ruiz JR. Dendritic Cell Metabolism and Function in Tumors. Trends Immunol. 2019;40(8):699–718. doi: 10.1016/j.it.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Scalavino V, Liso M, Serino G. Role of microRNAs in the Regulation of Dendritic Cell Generation and Function. Int J Mol Sci. 2020;21(4):1319. doi: 10.3390/ijms21041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L, Liao W, Wang X, Zhao Y, Pang J, Chen Y, et al. Expression, prognosis value, and immune infiltration of lncRNA ASB16-AS1 identified by pan-cancer analysis. Bioengineered. 2021;12(2):10302–10318. doi: 10.1080/21655979.2021.1996054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu ZK, Wu KF, Zhang RY, Kong LM, Shang RZ, Lv JJ, et al. Pyroptosis-Related LncRNA Signature Predicts Prognosis and Is Associated With Immune Infiltration in Hepatocellular Carcinoma. Front Oncol. 2022;12:794034. doi: 10.3389/fonc.2022.794034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol. 2019;110:1–8. doi: 10.1016/j.biocel.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 66.Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018;9(5):478. doi: 10.1038/s41419-018-0528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19(1):92. doi: 10.1186/s12943-020-01213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng R, Cui Z, Liu Z, Zhang Y. Upregulated microRNA-132 in T helper 17 cells activates hepatic stellate cells to promote hepatocellular carcinoma cell migration in vitro. Scand J Immunol. 2021;93(5):e13007. doi: 10.1111/sji.13007. [DOI] [PubMed] [Google Scholar]
- 69.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22(3):291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. doi: 10.1038/ncomms15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113(6):1968–1983. doi: 10.1111/cas.15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, et al. Long Non-coding RNA FENDRR Acts as a miR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids. 2019;17:516–529. doi: 10.1016/j.omtn.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song H, Huang XF, Hu SY, Lu LL, Yang XY. The LINC00261/MiR105-5p/SELL axis is involved in dysfunction of B cell and is associated with overall survival in hepatocellular carcinoma. PeerJ. 2022;10:e12588. doi: 10.7717/peerj.12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dong C. Cytokine Regulation and Function in T Cells. Annu Rev Immunol. 2021;39:51–76. doi: 10.1146/annurev-immunol-061020-053702. [DOI] [PubMed] [Google Scholar]
- 75.Halle S, Halle O, Förster R. Mechanisms and Dynamics of T Cell-Mediated Cytotoxicity In Vivo. Trends Immunol. 2017;38(6):432–443. doi: 10.1016/j.it.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 76.Basu A, Ramamoorthi G, Albert G, Gallen C, Beyer A, Snyder C, et al. Differentiation and Regulation of T(H) Cells: A Balancing Act for Cancer Immunotherapy. Front Immunol. 2021;12:669474. doi: 10.3389/fimmu.2021.669474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27(1):109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson AC. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol Res. 2014;2(5):393–398. doi: 10.1158/2326-6066.Cir-14-0039. [DOI] [PubMed] [Google Scholar]
- 79.Shui X, Chen S, Lin J, Kong J, Zhou C, Wu J. Knockdown of lncRNA NEAT1 inhibits Th17/CD4(+) T cell differentiation through reducing the STAT3 protein level. J Cell Physiol. 2019;234(12):22477–22484. doi: 10.1002/jcp.28811. [DOI] [PubMed] [Google Scholar]
- 80.Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- 81.Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol. 2020;20(5):294–307. doi: 10.1038/s41577-019-0257-x. [DOI] [PubMed] [Google Scholar]
- 82.Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66(2):342–351. doi: 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Petitprez F, de Reyniès A, Keung EZ, Chen TW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 84.Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 85.Li J, Wan Y, Ji Q, Fang Y, Wu Y. The role of microRNAs in B-cell development and function. Cell Mol Immunol. 2013;10(2):107–112. doi: 10.1038/cmi.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao C, Nemazee D, Gonzalez-Martin A. MicroRNA control of B cell tolerance, autoimmunity and cancer. Semin Cancer Biol. 2020;64:102–107. doi: 10.1016/j.semcancer.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 88.Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, et al. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23(7):631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 89.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, et al. The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J Hepatol. 2019;70(5):918–929. doi: 10.1016/j.jhep.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 91.Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598. doi: 10.1038/ncomms12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Sun W, Shen W, Xia M, Chen C, Xiang D, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64(6):1283–1294. doi: 10.1016/j.jhep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 93.Zhou DD, Wang X, Wang Y, Xiang XJ, Liang ZC, Zhou Y, et al. MicroRNA-145 inhibits hepatic stellate cell activation and proliferation by targeting ZEB2 through Wnt/β-catenin pathway. Mol Immunol. 2016;75:151–160. doi: 10.1016/j.molimm.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 94.Yang J, Tao Q, Zhou Y, Chen Q, Li L, Hu S, et al. MicroRNA-708 represses hepatic stellate cells activation and proliferation by targeting ZEB1 through Wnt/β-catenin pathway. Eur J Pharmacol. 2020;871:172927. doi: 10.1016/j.ejphar.2020.172927. [DOI] [PubMed] [Google Scholar]
- 95.Yu F, Geng W, Dong P, Huang Z, Zheng J. LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212. Cell Death Dis. 2018;9(10):1014. doi: 10.1038/s41419-018-1068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, et al. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun. 2016;7:10993. doi: 10.1038/ncomms10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37(1):324. doi: 10.1186/s13046-018-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang J, Xu C, Wu M, Wu Y, Jia X, Zhou C, et al. MicroRNA-124 inhibits hepatic stellate cells inflammatory cytokines secretion by targeting IQGAP1 through NF-κB pathway. Int Immunopharmacol. 2021;95:107520. doi: 10.1016/j.intimp.2021.107520. [DOI] [PubMed] [Google Scholar]
- 99.Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9(1):191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaiser J. The cancer stem cell gamble. Science. 2015;347(6219):226–229. doi: 10.1126/science.347.6219.226. [DOI] [PubMed] [Google Scholar]
- 101.Lee TK, Guan XY, Ma S. Cancer stem cells in hepatocellular carcinoma - from origin to clinical implications. Nat Rev Gastroenterol Hepatol. 2022;19(1):26–44. doi: 10.1038/s41575-021-00508-3. [DOI] [PubMed] [Google Scholar]
- 102.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14(7):397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 103.Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev. 2017;121:27–42. doi: 10.1016/j.addr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Monga SP. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology. 2015;148(7):1294–1310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fang Z, Xu J, Zhang B, Wang W, Liu J, Liang C, et al. The promising role of noncoding RNAs in cancer-associated fibroblasts: an overview of current status and future perspectives. J Hematol Oncol. 2020;13(1):154. doi: 10.1186/s13045-020-00988-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Song M, He J, Pan QZ, Yang J, Zhao J, Zhang YJ, et al. Cancer-Associated Fibroblast-Mediated Cellular Crosstalk Supports Hepatocellular Carcinoma Progression. Hepatology. 2021;73(5):1717–1735. doi: 10.1002/hep.31792. [DOI] [PubMed] [Google Scholar]
- 108.Yang X, Li Y, Zou L, Zhu Z. Role of Exosomes in Crosstalk Between Cancer-Associated Fibroblasts and Cancer Cells. Front Oncol. 2019;9:356. doi: 10.3389/fonc.2019.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fornari F, Pollutri D, Patrizi C, La Bella T, Marinelli S, Casadei Gardini A, et al. In Hepatocellular Carcinoma miR-221 Modulates Sorafenib Resistance through Inhibition of Caspase-3-Mediated Apoptosis. Clin Cancer Res. 2017;23(14):3953–3965. doi: 10.1158/1078-0432.Ccr-16-1464. [DOI] [PubMed] [Google Scholar]
- 110.Abdelrahman MM, Fawzy IO, Bassiouni AA, Gomaa AI, Esmat G, Waked I, et al. Enhancing NK cell cytotoxicity by miR-182 in hepatocellular carcinoma. Hum Immunol. 2016;77(8):667–673. doi: 10.1016/j.humimm.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 111.Wang W, Hao LP, Song H, Chu XY, Wang R. The Potential Roles of Exosomal Non-Coding RNAs in Hepatocellular Carcinoma. Front Oncol. 2022;12:790916. doi: 10.3389/fonc.2022.790916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11(2):125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacology & therapeutics. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.Can-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]