Abstract
The thymus is an intricate organ consisting of a diverse population of thymic epithelial cells (TEC). Cortical and medullary TECs (cTECs and mTECs) and their subpopulations have distinct roles in coordinating the development and selection of functionally competent and self-tolerant T cells. Recent advances made in technologies such as single-cell RNA sequencing (scRNA-seq) have made it possible to investigate and resolve the heterogeneity in TECs. These findings have provided further understanding of the molecular mechanisms regulating TEC function and expression of tissue restricted antigens (TSAs). In this brief review, we focus on the newly characterized subsets of TECs and their diversity in relation to their functions in supporting T cell development. We also discuss recent discoveries on expression of self-antigens in the context of TEC development as well as the cellular and molecular changes occurring during embryonic development to thymic involution.
Introduction
The thymus is the primary organ responsible for the generation and maturation of T cells with antigen specificity resulting in a diverse repertoire of T cell receptors (TCRs). T cells that are unable to properly distinguish between self- and non-self-proteins risk promoting autoimmune disease. During their intra-thymic development, thymocytes are subjected to a complex selection process mediated by recognition and binding strength of their TCRs to self-peptide/MHC complexes presented by thymic epithelial cells (TEC). Cortical or cTECs are found in the cortex region of the thymus and mediate positive selection of TCRs capable of recognizing peptide/MHC complex. Following positive selection, cTECs and subsequently medullary TECs (mTECs) remove potentially autoreactive T cells that have high-affinity TCRs for self-antigens in a process termed negative selection (1). T cells with intermediate affinity are additionally redirected to a regulatory T-cell fate (2, 3). These mechanisms of deletion and diversion ensure that only thymocytes with low self-affinity will differentiate into effector T cells (Teff) and establish central tolerance. As the role of the thymus is to “educate” developing T cells against self and non-self, TECs express up to 19,293 protein-coding genes or 88% of all protein-coding genes (4, 5), which is the highest number of genes known to be expressed in any cell type. Many of these genes encode for self-proteins normally only found in differentiated cell types and thus are called tissue-restricted antigens (TRAs). At the population level, TECs express almost all protein-coding genes with only individual mature mTECS expressing 1–3% of TRAs at any given time (4–8). Additionally, mTECs have a high turnover rate (9) and several defined developmental states with varying gene expression patterns, all leading to a highly heterogeneous population.
With the ability to sequence heterogenous populations at single-cell resolution using single cell RNA sequencing (scRNA-seq) technology, we are now able to provide new insights into the functions of thymic epithelial cells and their development. While there have also been many studies done utilizing scRNA-seq on developing T cells (10), this review aims to focus on the current field of thymic epithelial cell function along with the emerging knowledge recently revealed by systems immunology approaches. Better understanding of the mechanisms in which thymic epithelial cell development and self-antigen expression is regulated can lead to insights as to why individuals have different susceptibilities to autoimmune deviations and provide better strategies for therapies involving thymic culture systems to generate T cells.
TEC heterogeneity & development
Generally, cTECs are defined as Epcam+LY51+CD45− by flow cytometry and keratin 8 (KRT8) expression whereas mTECs are defined as Epcam+UEA-1+CD45− and keratin 5 (KRT5). Early studies showed that mTECs had heterogeneous expression of MHC II and thus mTECs can be broadly divided into CD80loMHCIIlo (or mTEClo) and CD80hiMHCIIhi (or mTEChi). It was hypothesized that MHCIIhi mTECs were major players in thymic tolerance induction and MHC II genes are the prevailing contributors of genetic susceptibility to autoimmune diseases such as Type 1 Diabetes (T1D), multiple sclerosis, and rheumatoid arthritis among others (11). The mTEChi population consists of cells that have high expression of Aire and Fezf2 and early studies showed embryonic mTEClo cells gave rise to mTEChi in a reaggregate thymic organ culture (RTOC, (12). It is now appreciated that the mTEClo population includes both immature mTEC precursors as well as terminally differentiated mTECs that have previously expressed Aire (13, 14).
Recent studies using scRNA-seq and fate-mapping analyses indicate that mTEC heterogeneity, is more complex than previously thought and mTECs have recently been classified into four major subsets, termed mTEC I-IV (14, Figure 1). Table 1 resolves some of the naming variations among publications from different groups which we will refer to in this review. mTEC I is characterized by high expression of Itag6, Ly6a (15), Pdpn (16, 17) and Ccl21 (18). This population is found at the cortical-medullary junction of the thymus and production of CCL21 regulates the migration of positively selected thymocytes. The mTEC II population consists of the previously named mTEChi group expressing Aire, Fezf2, CD40, H2-Aa or CD74 and are the precursors to terminally differentiated mTEC III cells that express Pigr, Ly6d, Spink5, Ivl, and Krt10 (15) but have lost their Aire expression (18, 19). Notably, mTEC IV cells resemble tuft cells that have been described at mucosal sites, including gut and lung epithelial. Tuft cells are a type of chemosensory epithelial cell most studied for their role in controlling helminth infections by initiating type 2 mucosal immunity. When comparing transcriptional signatures of both tuft cells in the intestinal epithelium and thymus they were found to be similar and express Lrmp, Avil, Trpm5, Dclk1, Gng13, Llcam and Sox9. Thymic tuft cells are also characterized by high expression of interleukin-25 (IL-25) and are critical for the development of IL-4 producing type 2 invariant natural killer T (NKT2) in the thymus (19). Increased frequencies of ILC2 were also present in the thymus of Pou2f3−/− mice lacking tuft cells, however it is unknown whether this is linked to absence of IL-25 (15).
Figure 1. Phenotypic markers and pathways in mTEC development.
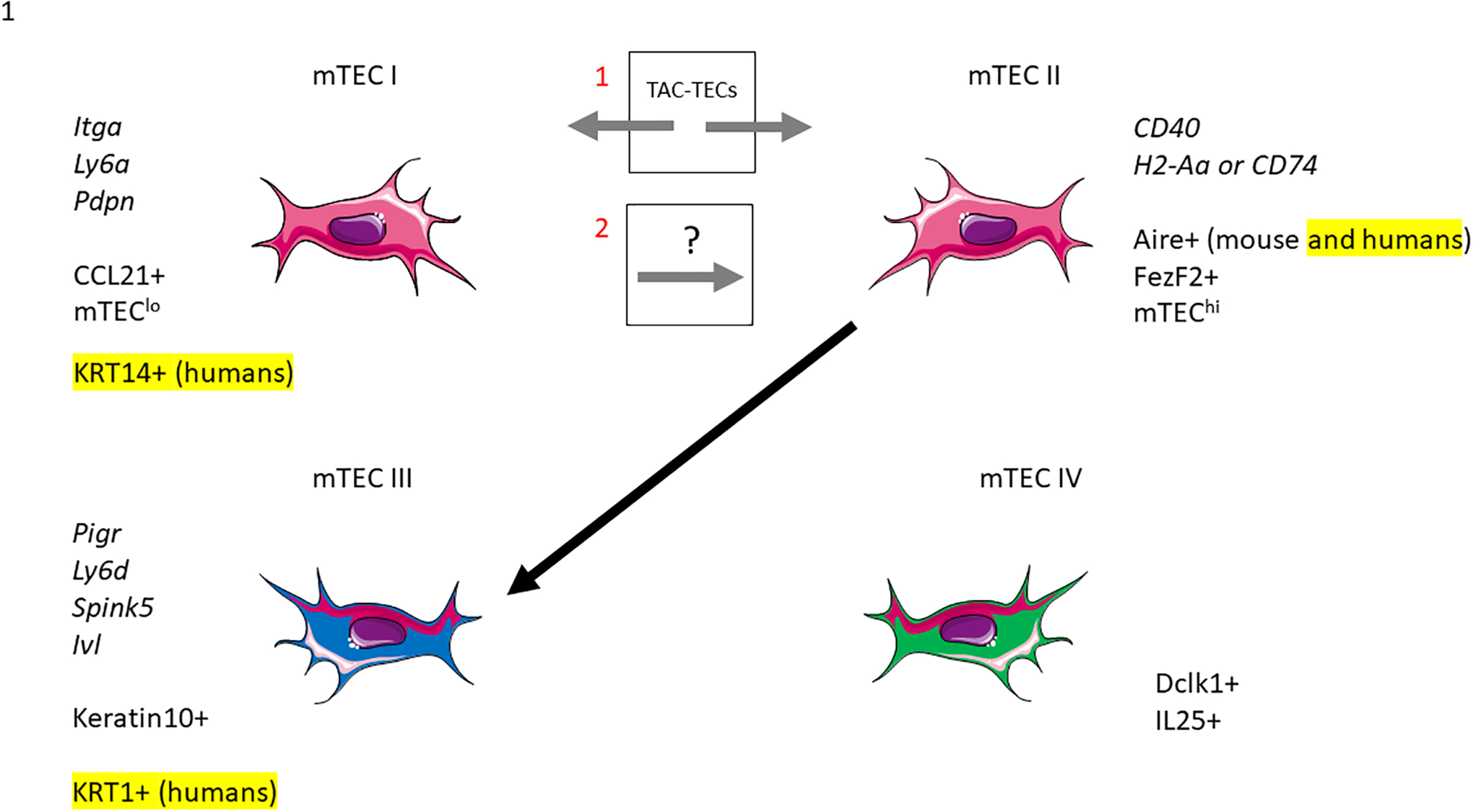
The mTEC I subset is part of the mTEClo compartment and characterized by CCL21 expression. mTEC II are the classically mature Aire+ mTEChi populations. mTEC III are the post-Aire terminally differentiated mTECs that also are found in the mTEClo compartment and mTEC IV are the newly found tuft-like cells. Additionally, there has been a newly characterized proliferating TAC-TEC subset and whether this subset is a progenitor of mTEC I and mTEC II (18), or mTEC I leads to the TAC-TEC and mTEC II populations (26) requires further study.
Table 1. mTEC subsets.
Comparison of mTEC nomenclature used by different scRNA-seq studies
| Bornstein (15) | mTEC I | mTEC II | mTEC III | mTEC IV | Identified thymic tuft-cells |
|---|---|---|---|---|---|
| Miragaia (17) | jTECs | mTEChi | mTEClo | jTECs precursors give rise to mTEChi → mTEClo | |
| Wells (18) | Ccl21-high | Aire-positive | Late-Aire | Tuft | TAC-TEC population gives rise to both Ccl21-high and Aire-positive populations |
| Lebel (36) | mTEClo | mTEChi | Post-Aire mTEC | Tuft cells | Model antigen with biased mTEChi expression supports Treg differentiation |
| Lopes (27) | CCL21+ | Aire+ | Post-Aire | Tuft-like | Self-reactive CD4+ thymocytes regulate Aire+ precursors, post-Aire and tuft-like mTEC populations |
While it was previously thought that mTEC I gives rise to the fully differentiated mTEC II population (16, 17), a more recent investigation utilizing scRNA-seq technology found a cluster of cells they termed TAC-TECs (18) as the precursor to both the Ccl21 producing mTEC I and Aire positive mTEC II populations (Figure 1). This was demonstrated using both Aire-lineage tracing and RANK-ligand transient ablation models to measure population kinetics. While TAC-TECs seem to be a precursor population, these cells did not express previously described canonical stem cell markers and were actively cycling, suggesting that they are an undifferentiated population in transition between stem cells and differentiated cells known as transit-amplifying cells (20, 21). When comparing the TAC-TEC gene signature to those of published transit-amplifying populations (21) they found a significant enrichment of previously described transit-amplifying genes (18).
Supporting this hypothesis, Dhalla et al also identified a proliferating mTEC cluster that could act as a bipotent mTEC progenitor that differentiated into both Aire+ mTEC II and CCL21+ mTEC I lineages (22). Cells in the proliferating cluster upregulated genes involved in proliferation such as Mik67, and expressed Aire, suggesting it could represent the proliferating mTEChi population previously reported (22–24). Analysis of the same data using RNA velocity, a different trajectory method that relies on pre- and post-spliced RNA reads, produced conflicting results that indicated rather than differentiating into CCL21+ mTEC I, the proliferating mTEC cluster seemed to be derived from the mTEC I population (25). Another study conducted scRNA-seq of mouse TECs throughout the 1st year of year of life also identified a cluster equivalent to proliferating TAC-TECs. However, their diffusion psuedotime analysis suggested that the CCL21+ mTEC I could bifurcate into the two mTEC trajectories progressing towards Aire+ mTEC II and the proliferating mTEC cluster (26) (Figure 1).
While mTECs control thymocyte selection, CD4+ thymocytes also control the cellularity of Aire-positive mTECs by activating RANK and CD40 to induce the NF-κB signaling pathway. This reciprocal signaling between thymocytes and mTECs is referred to as thymic crosstalk. In a recent study using high through-put RNA-sequencing, Lopes et al show that self-reactive CD4+ thymocytes induce in CD80lo mTECs pivotal transcriptional regulators to control the composition, including precursors of mTEC II (Aire+), mTEC III (post-Aire cells), and mTEC IV (tuft-like mTECs) (27). They also showed upregulation in expression of TRAs, chemokines, cytokines, and adhesion molecules involved in T-cell development suggesting that self-reactive thymocytes are inducers of T-cell tolerance by controlling the developmental transcriptional programs of mTEC subsets (27).
Self-antigen expression regulation
TECs express many self-proteins normally found only in differentiated cell types and thus these genes are called tissue-restricted antigens (TRAs). While peripheral tissues have tight spatio-temporal control of gene expression during various stages of development, the diverse expression of tissue specific genes observed in the thymus is referred to as promiscuous gene expression (PGE) (4, 6, 28). In mTECs, the transcription factor Autoimmune Regulator (Aire) positively regulates 3980 TRAs whereas Aire-independent genes consist of around 3947 TRAs. Of the AIRE regulated genes, approximately 533 are directly dependent on AIRE for their expression (Aire-dependent), while expression of the remaining genes is enhanced in the presence of AIRE (Aire-enhanced) (4). Aire-dependent TRAs as generally characterized by an enrichment of H3K27me3 histone mark (trimethylation of lysine-27 of histone H3) indicating a repressive chromatin state (4). Furthermore, loss-of-function mutations in Aire causes the autoimmune polyglandular syndrome type-1 (APS-1) which is marked by thymic export of self-reactive Teff cells (29). Many of these tissue specific genes are known targets of Aire and the syndrome is characterized by severe organ-specific autoimmunity affecting parathyroid chief cells, steroidogenic cells of the adrenal cortex, pancreatic β-cells, gastic parietal cells, skin melanocytes, hepatocytes, gonads, and the lung. Recently, Fezf2 was identified as a transcription factor capable of inducing TRA expression independently of Aire (30), suggesting other potential factors are likely to contribute to the regulation of TRAs in mTECs.
Despite expression of almost all protein-coding genes at the population level, TRA expression at single-cell resolution is heterogeneous with individual mature mTECs only expressing 1–3% of TRAs at a time (4–8). On average, in a single cell TRAs accounted for approximately 10% of all genes expressed (17) and TRA repertoires are not enriched for any particular peripheral tissue (4, 5, 8, 17). While Aire-dependent genes have a smaller frequency of genes expressed and have higher mean expression levels (4, 5, 8, 17), Fezf2-induced TRAs have the same frequency as non-TRA expression but similar mean expression levels when compared to Aire-dependent genes (17). Additionally, Aire expression is only found in the mTEC II populations while interestingly Fezf2 was detected in both mTEClo and mTEChi populations (12, 18). While Aire expression is dependent on RANK signaling, both lymphotoxin β receptor (LTβR) and RANK signaling have been implicated in Fezf2 expression (30, 31). Wells et al reported conflicting results showing that Fezf2 expression was generally independent of RANK (18), therefore which signals induces Fezf2 expression needs further study. While Aire-KO and Fezf2-KO mice both develop autoimmunity in several peripheral target organs and show defective clonal deletion of autoreactive thymocytes (30, 32), the expression of exact target genes controlled by Aire and Fezf2 has also remained incompletely defined.
Accumulating evidence using scRNA-seq to investigate regulation of self-antigen co-expression points to PGE expression being coordinated by mTEC developmental stages (17, 22, 33). For example, Derbinski et al, compared expression of a small set of self-antigens between CD80lo and CD80hi mTECs and found that the CD80hi population had upregulated expression of both Aire-dependent and Aire-independent TRAs (28). More recently, several groups performed scRNA-seq and found mTECs tend to have higher expression of TRAs as they mature suggesting that the heterogeneity of self-antigen expression tend to reflect mTEC maturation trajectory (17, 22, 33). Dhalla et al went further to sort and compare total “unselected” mTECs in addition to mTECs expressing specific TRAs, namely Tspan8 and GP2 protein, and found the pre-selected self-antigens tended to cluster within mTEC subpopulations (22). This was also reproducible when comparing different mouse strains strongly suggesting that PGE in mTECs is regulated under an ordered and not random process (22). Another study also utilizing scRNA-seq of mTECs from Aire-KO mice found altered heterogeneity of mTECs and aberrant expression of CTLA-4 which was not found in wildtype (WT) mice (34). The ectopically expressed CTLA-4 was found to remove CD80/CD86 ligands expressed on thymic dendritic cells which attenuated their ability to provide co-stimulatory signals and present self-antigens transferred from mTECs, leading to impaired Treg production and autoimmunity in Aire-KO mice (34). Overall, it appears that mTECs shift through heterogenous patterns of promiscuous gene expression throughout development to eventually cover all protein-coding self-antigens.
Investigation of TRA expression by mTEC subset show very few TRAs are differentially expressed in the mTEC I population (17) and mTEC II expresses significantly elevated levels of Aire-enhanced and Aire-dependent genes. While mTEC III had been regarded as a passive step towards mTEC death, this population has been found to express the highest number of TRAs, including Aire-unaffected TRAs and similar numbers of Aire-enhanced and Aire-dependent TRAs compared to mTEC II (17). Similarly, Fezf2-dependent TRA expression also increases as the mTEC II population undergos maturation to mTEC III (17). Supporting this finding, Wells et al also show that TRA expression peaks well after initiation of Aire expression and was maintained even after Aire expression decreased (18). Overall, Miragaia and Wells findings both suggest that while Aire is critical for inducing TRA expression, it is not necessary for maintaining it (17, 18). Interestingly, Miragaia et al observed little divergence of the TRA repertoires between each stage of maturation (17). This suggests that the TRAs encountered by thymocytes anywhere in the medulla does not depend largely on the maturation stage of the surrounding mTECs. Specifically, mTEC I are found in the cortex-medulla junction, mTEC II at the periphery of the medulla and mTEC III towards the center of the medulla (14). However, these observations are based on mRNA quantities which does not correlate linearly with protein quantities. A recent study using trans-omics analysis, combining transcriptomic and proteomic analysis, further identified signature molecules at both the mRNA and protein level that functionally and developmentally characterize cTECs and mTECs (35). Proteomic profiling of cTECs and mTECs needs further investigation and will be useful as an unbaised and powerful tool to gain insight into the distinct machinery of protein processing and peptide presentation to shape the self-tolerant TCR repertoire in T cells.
Recent evidence is now suggesting that AIRE regulates TRA expression by first directly regulating mTEC differentiation which subsequently regulates TRA expression. This was recently shown using scRNA-seq of mTECs from Aire-augmented mice, where Nishijima et al found Aire-augmented and Aire-KO mice both had altered mTEC heterogeneity and that many of the same TRAs were downregulated in both groups of mice (33). One hypothesis for this finding is that given the expression of TRAs is dependent on mTEC maturation status, perturbations of this process in either Aire-KO or Aire-augmented mice might account for the downregulation of TRAs. However, the authors did suggest that there was a possibility that feedback mechanisms initiated by Aire overexpression may also be playing a role in transcriptional suppression of TRAs.
Overall, identification of direct Aire targets has been hampered by low and promiscuous expression of TRAs. Using ATAC-seq, ChIP-seq and a reporter gene assay, the authors were able to identify CCl25 as a canonical target downstream of Aire (33). While Ccl25-deficient mice showed some inflammatory changes in the salivary gland and kidney, they did not show obvious signs of autoimmunity. Ccl25 also did not affect expression of TRAs and differentiation programs in mTECs so how reduced Ccl25 develops organ-specific autoimmune disease awaits further study. In another study using transgenic mice, expression of model TRAs were directed under either the C-reactive protein (CRP) locus, whose Aire-independent expression is preferentially detected in the mTEC I subset, or the Insulin2 (Ins2) locus to restrict expression to Aire+ mTEC II subsets. Using these models of antigen expression, it was found that mTEC I supported TCRαβ+ CD8αα intraepithelial lymphocyte development whereas mTEC II restricted antigen preferentially induces Treg differentiation to impact control of infections agents and tumor growth (36).
About a decade ago, Aire-regulated genes were shown to colocalize in chromosomal clusters (37, 38). Since then, it has been accepted that AIRE’s ability to recruit transcription factors to regions of closed chromatin would induce remodeling thus facilitating the co-expression of neighboring genes (29). The discovery that AIRE binds super enhancers supports this idea (39), providing a model that that explains both intra- and inter-chromosomal co-expression patterns of Aire regulated TRAs (8). Maragaia et al further investigated the gene clustering effect at the single cell level to determine how it changes in mTEC subsets and how it affects both Aire-enhanced and Aire-dependent TRAs (17). Overall, they found that genomic clustering tendency preferentially affects a minority of Aire-dependent TRAs and this effect seems to be established only in mTEC II and III, while limited in the mTEC I population (17).
From embryo to thymic involution
During mouse embryogenesis, an early rudiment of the thymus is evident in mouse gestation by embryonic day (E) 10–11. As development progresses, hematopoietic progenitors migrate to the thymus triggering the organ to double in size daily until birth. This formation and subsequent TEC differentiation is critically dependent on the expression of the transcription factor, Foxn1 (40–42) and mutations in the FOXN1 gene are characterized by T-cell immunodeficiency, congenital alopecia, and nail dystrophy (43). Bipotent progenitors of cTECs and mTECs arise as early as day 12.5 (44), however in an embryonic thymus, cTECs arise earlier whereas later in life mTECs are the predominant population in the adult thymus (45). mTEC I and mTEC II subpopulations become detectable at E18.5 and can be the most proliferative cells in the TECs. mTEC III can be found in the 4-week-old thymus and mTEC IV cells are present on neonatal day 6. Molecular features of TECs also differ between embryonic and adult thymus. For example, while in the adult thymus cTECs and mTECs can be separated based on their expression of keratin-5 and kertin-8 respectively (46, 47), in the early embryonic thymus a keratin 5- and keratin 8-positive TEC subset is the dominant population (48, 49) making analysis of TEC populations by this approach difficult.
Recent studies using scRNA-seq has helped provide new insights into the developmental pathway of the thymus both at embryonic stage and up until adulthood. One study characterizing the developmental dynamics of the embryonic thymus both in vivo and in vitro provided a single-cell transcriptional framework for thymus organogenesis (50). The study sampled thymic lobes beginning at embryonic day (E)12.5 and each day until birth, and they found Aire-expressing cells as early as E13.5. They also confirmed that mTEC III and IV were only found in the adult thymus (51) and not at birth (50). Additionally, they found that expression of autoimmune-implicated genes may begin during embryogenesis. Overall, their findings showed cellular heterogeneity of TECs dynamically changes with progression of thymus organogenesis, providing useful markers for developmental trajectories. Furthermore, they sequenced short term in vitro fetal thymic organ cultures (FTOCs) and found their transcriptome was comparable to in vivo development of thymii at similar timepoints (50). This provides evidence that in vitro thymic organ cultures are a physiologically relevant model and may be useful for future studies dissecting perturbations to thymic organ or T cell development. As an example, they showed that exogenous retinoic acid did not change the overall landscape of TEC compartments (50).
After birth the thymus continues to expand in size, although at a reduced rate, until peaking in size at ~4 weeks of age. It is then maintained in the adult mouse until puberty, around 8 weeks, after which it begins to decline (23). While TECs display considerable proliferative potential (23), it remains unclear why thymic involution occurs. Thymic involution refers to the diminished TEC cellularity and turnover, disrupted thymic architecture, decreased thymic output and reduced T cell function that occurs in an age-dependent manner (52–54). Aging, infections, pregnancy, stress, and other processes can all cause thymic involution, or atrophy (55–57). Involution and reduced T cell output has been linked to age-related incidence of cancer, infection, and autoimmunity (58). While reduced levels of the transcription factor, Foxn1 contributes to thymic involution (59), genetic manipulation of cell-cycle regulators such as cyclins and E2F transcription factors can maintain thymic mass in aged mice (60, 61).
Another group performing transcriptome sequencing of neonatal TECs after birth included skin epithelial cells for comparison. They found that cell cycle progression is differentially regulated in TECs and skin epithelial cells and that many positive regulators of cell division are repressed in TECs relative to skin epithelial cells which could be related to thymic involution (62). Also supporting this hypothesis, Ki et al, showed thymic involution is associated with downregulation of cell-cycle genes in the CD80lo mTEC subset including decreased E2F3 activity in cTEC and CD80lo mTEC cells (51). Another study investigating transcriptional diversity of TECs through development also found increased proliferation and ribosomal biogenesis in fetal TEC and displayed diminishing expression with age (63). In this study, they showed that genes controlling ribosomal biogenesis and cell cycling in fetal stages in TEC development were targets of Myc which also had an age-correlated decline in expression (63). Overall, this further established Myc as a possible therapeutic target in modulating thymic regeneration and function.
Human thymus
While the thymus has been extensively studied using animal models, human immunity cannot be understood without further investigation into human TEC heterogeneity and development. The study of human fetal thymus organogenesis has been mainly limited to morphological descriptions (64). Thymus organogenesis initiates from the third pharyngeal pouches during week 6 of gestation (65). During week 7, thymic and parathyroird primordium contains undifferentiated bi-potent thymic epithelial progenitor cells (TEPCs) (64) and at week 8, the differentiation of TECs occurs. After week 12, the cortical and medullary epithelial regions are distinct and CD4+ and CD8+ single-positive T lymphocytes appear (65).
To further characterize the molecular profiles of TECs and their interactions with developing thymocytes using human samples, two groups took single-cell transcriptomic approaches. Park et al, sequenced thymii spanning from embryonic, fetal, pediatric, and adult stages (66) whereas Zeng et al, analyzed thymii from the thymus, AGM (aorta-gonad-mesonephros) region, liver and blood of human embryos and fetuses (67). Park et al, found TEC populations that were conserved from mouse to human included PSMB11-positive cTECs, KRT14-positive mTEC I, Aire-expressing mTEC II, and KRT1-expressing mTEC III. mTEC IV tuft-like cells were found and enriched but were not specifically expressing DCLK1 or POU2F3. Additionally, there were two populations specific to humans which included MYOD1- and MYOG- expressing myoid cells, TEC(myo)s and NEUROD1-, NEUROG1- and CHGA-expressing TEC(neuro)s, that resemble neuroendocrine cells (66).
Another study investigated the transcriptional landscape of early thymic epithelial cell development and potential cell-cell interactions during early thymus organogenesis by using cells from multiple hemogenic and hematopoietic sites spanning embryonic and fetal stages. In human thymic primordia, TECs are relatively rare, accounting for about 1% of total cells (67, 68). Similarly in mice, few mature mTECs were detected during the early embryonic stages evidenced in the absence of Aire (67, 69–71) whereas cTECs proliferated and developed at a significantly faster rate than mTECs. Using prediction methods to calculate cellular interactions (72) they found that interactions between TECs and mesenchymal cells, as well as those between TECs and endothelial cells were the strongest, suggesting that cross talk between each type of stromal cell was important for early thymus organogenesis (67). Additionally, both embryonic and fetal early thymic progenitors (ETPs) had intensive interactions with TECs (67), consistent with the role of TECs in the hematopoietic progenitor-seeding thymus and T cell development.
The study of human thymus development has been limited; however, a recent study was able to isolate and expand clonogenic CD49f expressing mTEC and cTECs which could repopulate whole-organ scaffolds to reconstitute an anatomic phenocopy of the human thymus. The authors showed that the repopulated thymus scaffolds were able to support mature T cell development in vivo after transplantation into humanized immuno-deficient mice (73). Additionally, scRNA-seq analysis of the expanded cells in culture provided gene signatures useful for dissecting the complexity and properties of these thymic clonogenic cells. Further development of this organ reconstruction system and other ex vivo tools (73–76) will be valuable to address the roles of TECs and other human stromal cells (e.g. dendritic cells) during both organogenesis and thymopoesis.
Conclusions
Unbiased transcriptomic analysis has been powerful in advancing our understanding of TECs. Global gene expression analysis identified promiscuous gene expression in mTECs (4, 6, 28, 77) and single-cell RNA-sequencing has revealed enormous diversity in mTEC subpopulations, including the novel tuft-like cell population (13, 15). Recent discoveries have contributed to a new appreciation of TEC diversity to provide multiple thymic microenvironments supporting different stages of thymocyte development. With new tools to study cell heterogeneity, we can further understand the mechanisms contributing to their development and identify new markers for each population. Overall, research in TEC function contributing to thymocyte development and their molecular characteristics during development will provide better understanding of the etiology of multiple T-cell-related diseases. These insights will improve strategies for T-cell based immunotherapies in cancer and autoimmune diseases.
References
- 1.Abramson J, Anderson G, Thymic Epithelial Cells (2017), doi: 10.1146/annurev-immunol. [DOI] [PubMed] [Google Scholar]
- 2.Lio CWJ, Hsieh CS, A Two-Step Process for Thymic Regulatory T Cell Development. Immunity. 28, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tai X, Erman B, Alag A, Mu J, Kimura M, Katz G, Guinter T, McCaughtry T, Etzensperger R, Feigenbaum L, Singer DS, Singer A, Foxp3 Transcription Factor Is Proapoptotic and Lethal to Developing Regulatory T Cells unless Counterbalanced by Cytokine Survival Signals. Immunity. 38, 1116–1128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, Heger A, Ponting CP, Holländer GA, Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Kuchler R, Huber W, Kyewski B, Steinmetz LM, Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol. 16, 933–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derbinski J, Schulte A, Kyewski B, Klein L, Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Journal of Immunology. 196, 2915–2922 (2016). [PubMed] [Google Scholar]
- 7.Villasenor Jennifer, Besse Whitney, Benoist Christophe, Mathis Diane, Ectopic expression of peripheral-tissue antigens in the thymic epithelium: Probabilistic, monoallelic, misinitiated. Proceedings of the National Academy of Sciences. 105, 15854–15859 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meredith M, Zemmour D, Mathis D, Benoist C, Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol. 16, 942–949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray D, Abramson J, Benoist C, Mathis D, Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. Journal of Experimental Medicine. 204, 2521–2528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Lan Y, Liu B, Zhang H, Hu H, T Cell Development: Old Tales Retold By Single-Cell RNA Sequencing. Trends Immunol. 42 (2021), pp. 165–175. [DOI] [PubMed] [Google Scholar]
- 11.Tsai S, Santamaria P, MHC class II polymorphisms, autoreactive T-cells, and autoimmunity. Front Immunol. 4 (2013), doi: 10.3389/fimmu.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, McConnell FM, Scott HS, Penninger JM, Jenkinson EJ, Lane PJL, Anderson G, RANK signals from CD4+3- inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. Journal of Experimental Medicine. 204, 1267–1272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, Parent A. v., Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS, Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature. 559, 627–631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzger TC, Khan IS, Gardner JM, Mouchess ML, Johannes KP, Krawisz AK, Skrzypczynska KM, Anderson MS, Lineage Tracing and Cell Ablation Identify a Post-Aire-Expressing Thymic Epithelial Cell Population. Cell Rep. 5, 166–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Tóth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann VS, Abramson J, Amit I, Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 559 (2018), pp. 622–626. [DOI] [PubMed] [Google Scholar]
- 16.Onder L, Nindl V, Scandella E, Chai Q, Cheng HW, Caviezel-Firner S, Novkovic M, Bomze D, Maier R, Mair F, Ledermann B, Becher B, Waisman A, Ludewig B, Alternative NF-κB signaling regulates mTEC differentiation from podoplanin-expressing presursors in the cortico-medullary junction. Eur J Immunol. 45, 2218–2231 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Miragaia RJ, Zhang X, Gomes T, Svensson V, Ilicic T, Henriksson J, Kar G, Lönnberg T, Single-cell RNA-sequencing resolves self-antigen expression during mTEC development. Sci Rep. 8 (2018), doi: 10.1038/s41598-017-19100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells KL, Miller CN, Gschwind AR, Wei W, Phipps JD, Anderson MS, Steinmetz LM, Combined transient ablation and single cell rna sequencing reveals the development of medullary thymic epithelial cells. Elife. 9, 1–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, v Parent A, Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS, Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature. 559, 627–631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray D, Abramson J, Benoist C, Mathis D, Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. Journal of Experimental Medicine. 204, 2521–2528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basak O, Krieger TG, Muraro MJ, Wiebrands K, Stange DE, Frias-Aldeguer J, Rivron NC, van de Wetering M, van Es JH, van Oudenaarden A, Simons BD, Clevers H, Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc Natl Acad Sci U S A. 115, E610–E619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhalla F, Baran‐Gale J, Maio S, Chappell L, Holländer GA, Ponting CP, Biologically indeterminate yet ordered promiscuous gene expression in single medullary thymic epithelial cells. EMBO J. 39 (2020), doi: 10.15252/embj.2019101828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray DHD, Seach N, Ueno T, Milton MK, Liston A, Lew AM, Goodnow CC, Boyd RL, Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells (2006), doi: 10.1182/blood-2006-02. [DOI] [PubMed] [Google Scholar]
- 24.Wong K, Lister NL, Barsanti M, Lim JMC, Hammett M. v., Khong DM, Siatskas C, Gray DHD, Boyd RL, Chidgey AP, Multilineage potential and self-renewal define an epithelial progenitor cell population in the adult thymus. Cell Rep. 8, 1198–1209 (2014). [DOI] [PubMed] [Google Scholar]
- 25.la Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, P. v. Kharchenko, RNA velocity of single cells. Nature. 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baran-Gale J, Morgan MD, Maio S, Dhalla F, Calvo-Asensio I, Deadman ME, Handel AE, Maynard A, Chen S, Green F, Sit R. v., Neff NF, Darmanis S, Tan W, May AP, Marioni JC, Ponting CP, Holländer GA, Ageing compromises mouse thymus function and remodels epithelial cell differentiation. Elife. 9, 1–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes N, Boucherit N, Santamaria JC, Provin N, Charaix J, Ferrier P, Giraud M, Irla M, Thymocytes trigger self-antigen-controlling pathways in immature medullary thymic epithelial stages. Elife. 11 (2022), doi: 10.7554/ELIFE.69982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derbinski J, Gäbler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B, Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. Journal of Experimental Medicine. 202, 33–45 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathis D, Benoist C, Aire. Annu Rev Immunol. 27 (2009), pp. 287–312. [DOI] [PubMed] [Google Scholar]
- 30.Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, Kodama T, Takayanagi H, Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell. 163, 975–987 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Cosway EJ, Lucas B, James KD, Parnell SM, Carvalho-Gaspar M, White AJ, Tumanov A. v., Jenkinson WE, Anderson G, Redefining thymus medulla specialization for central tolerance. Journal of Experimental Medicine. 214, 3183–3195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson G, Jenkinson WE, Co-ordination of intrathymic self-representation. Nat Immunol. 16, 895–896 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Nishijima H, Matsumoto M, Morimoto J, Hosomichi K, Akiyama N, Akiyama T, Oya T, Tsuneyama K, Yoshida H, Matsumoto M, Aire Controls Heterogeneity of Medullary Thymic Epithelial Cells for the Expression of Self-Antigens. The Journal of Immunology. 208, 303–320 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Morimoto J, Matsumoto M, Miyazawa R, Yoshida H, Tsuneyama K, Matsumoto M, Aire suppresses CTLA-4 expression from the thymic stroma to control autoimmunity. Cell Rep. 38 (2022), doi: 10.1016/j.celrep.2022.110384. [DOI] [PubMed] [Google Scholar]
- 35.Apavaloaei A, Brochu S, Dong M, Rouette A, Hardy M-P, Villafano G, Murata S, Melichar HJ, Perreault C, PSMB11 Orchestrates the Development of CD4 and CD8 Thymocytes via Regulation of Gene Expression in Cortical Thymic Epithelial Cells. The Journal of Immunology. 202, 966–978 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Lebel MÈ, Coutelier M, Galipeau M, Kleinman CL, Moon JJ, Melichar HJ, Differential expression of tissue-restricted antigens among mTEC is associated with distinct autoreactive T cell fates. Nat Commun. 11 (2020), doi: 10.1038/s41467-020-17544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derbinski J, Pinto S, Rö S, Hexel K, Kyewski B, “Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism” (2008), (available at www.pnas.org/cgi/content/full/). [DOI] [PMC free article] [PubMed]
- 38.Johnnidis JB, Venanzi ES, Taxman DJ, P-Y Ting J, Benoist CO, Mathis DJ, “Chromosomal clustering of genes controlled by the aire transcription factor” (2005), (available at www.pnas.orgcgidoi10.1073pnas.0502670102). [DOI] [PMC free article] [PubMed]
- 39.Bansal K, Yoshida H, Benoist C, Mathis D, The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol. 18, 263–273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nehls Michael, Pfeifer Dietmar, Schorpp Michael, Hedrich Hans, Boehm Thomas, “New member of the winged-helix protein family disrupted in mouse and rat nude mutations” (1994). [DOI] [PubMed]
- 41.Blackburn CC, Augustine CL, Li R, Harvey RP, Malint MA, Boydt RL, Miller JFAP, Morahan G, “The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors (nude mice/thymus)” (1996), (available at https://www.pnas.org). [DOI] [PMC free article] [PubMed]
- 42.Corbeaux T, Hess I, Swann JB, Kanzler B, Haas-Assenbaum A, Boehm T, Thymopoiesis in mice depends on a Foxn1-positive thymic epithelial cell lineage. Proc Natl Acad Sci U S A. 107, 16613–16618 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pignata C, Fiore M, Guzzetta V, Castaldo A, Sebastio G, Porta F, Guarino A, “Congenital Alopecia and Nail Dystrophy Associated With Severe Functional T-cell Immunodeficiency in Two Sibs” (1996). [DOI] [PubMed]
- 44.Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ, Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature. 441, 988–991 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Wang HX, Pan W, Zheng L, Zhong XP, Tan L, Liang Z, He J, Feng P, Zhao Y, Qiu YR, Thymic Epithelial Cells Contribute to Thymopoiesis and T Cell Development. Front Immunol. 10 (2020), doi: 10.3389/fimmu.2019.03099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohigashi I, Kozai M, Takahama Y, “Development and developmental potential of cortical thymic epithelial cells” (2016). [DOI] [PubMed]
- 47.Lucas B, Mccarthy NI, Baik S, Cosway E, James KD, Parnell SM, White AJ, Jenkinson WE, Anderson G, “Control of the thymic medulla and its influence on abT-cell development” (2016). [DOI] [PMC free article] [PubMed]
- 48.Klug DB, Carter C, Gimenez-Conti IB, Richie ER, Cutting Edge: Thymocyte-Independent and Thymocyte-Dependent Phases of Epithelial Patterning in the Fetal Thymus. The Journal of Immunology. 169, 2842–2845 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Klug DB, Carter C, Crouch E, Roop D, Conti CJ, Richie ER, “Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment” (1998), (available at www.pnas.org.). [DOI] [PMC free article] [PubMed]
- 50.Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R, A Single-Cell Transcriptomic Atlas of Thymus Organogenesis Resolves Cell Types and Developmental Maturation. Immunity. 48, 1258–1270.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ki S, Park D, Selden HJ, Seita J, Chung H, Kim J, Iyer VR, Ehrlich LIR, Global transcriptional profiling reveals distinct functions of thymic stromal subsets and age-related changes during thymic involution. Cell Rep. 9, 402–415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chinn IK, Blackburn CC, Manley NR, Sempowski GD, Changes in primary lymphoid organs with aging. Semin Immunol 24 (2012), pp. 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haynes L, Maue AC, Effects of aging on T cell function. Curr Opin Immunol. 21 (2009), pp. 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikolich-Žugich J, Li G, Uhrlaub JL, Renkema KR, Smithey MJ, Age-related changes in CD8 T cell homeostasis and immunity to infection. Semin Immunol. 24 (2012), pp. 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dooley J, Liston A, Molecular control over thymic involution: From cytokines and microRNA to aging and adipose tissue. Eur J Immunol. 42 (2012), pp. 1073–1079. [DOI] [PubMed] [Google Scholar]
- 56.Lynch HE, Goldberg GL, Chidgey A, van den Brink MRM, Boyd R, Sempowski GD, Thymic involution and immune reconstitution. Trends Immunol. 30 (2009), pp. 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ventevogel MS, Sempowski GD, Thymic rejuvenation and aging. Curr Opin Immunol. 25 (2013), pp. 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer DB, The effect of age on thymic function. Front Immunol. 4 (2013), doi: 10.3389/fimmu.2013.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, Xiao S, Manley NR, Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood. 113, 567–574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Puebla ML, Lacava M, Miliani De Marval PL, Jorcano JL, Richie ER, Conti CJ, “Animal Model Cyclin D2 Overexpression in Transgenic Mice Induces Thymic and Epidermal Hyperplasia whereas Cyclin D3 Expression Results Only in Epidermal Hyperplasia” (2000). [DOI] [PMC free article] [PubMed]
- 61.Scheijen B, Bronk M, van der Meer T, de Jong D, Bernards R, High Incidence of Thymic Epithelial Tumors in E2F2 Transgenic Mice. Journal of Biological Chemistry. 279, 10476–10483 (2004). [DOI] [PubMed] [Google Scholar]
- 62.St-Pierre C, Brochu S, Vanegas JR, Dumont-Lagacé M, Lemieux S, Perreault C, Transcriptome sequencing of neonatal thymic epithelial cells. Sci Rep. 3 (2013), doi: 10.1038/srep01860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cowan JE, Malin J, Zhao Y, Seedhom MO, Harly C, Ohigashi I, Kelly M, Takahama Y, Yewdell JW, Cam M, Bhandoola A, Myc controls a distinct transcriptional program in fetal thymic epithelial cells that determines thymus growth. Nat Commun. 10 (2019), doi: 10.1038/s41467-019-13465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farley AM, Morris LX, Vroegindeweij E, Depreter MLG, Vaidya H, Stenhouse FH, Tomlinson SR, Anderson RA, Cupedo T, Cornelissen JJ, Clare BC, Dynamics of thymus organogenesis and colonization in early human development. Development (Cambridge). 140, 2015–2026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haynes BF, Heinly CS, “Early Human T Cell Development: Analysis of the Human Thymus at the Time of Initial Entry of Hematopoietic Stem Cells into the Fetal Thymic Microenvironment,” (available at http://rupress.org/jem/article-pdf/181/4/1445/1106635/1445.pdf). [DOI] [PMC free article] [PubMed]
- 66.Park JE, Botting RA, Conde CD, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, Wilbrey-Clark A, Roberts K, Kedlian VR, Ferdinand JR, He X, Webb S, Maunder D, Vandamme N, Mahbubani KT, Polanski K, Mamanova L, Bolt L, Crossland D, de Rita F, Fuller A, Filby A, Reynolds G, Dixon D, Saeb-Parsy K, Lisgo S, Henderson D, Vento-Tormo R, Bayraktar OA, Barker RA, Meyer KB, Saeys Y, Bonfanti P, Behjati S, Clatworthy MR, Taghon T, Haniffa M, Teichmann SA, A cell atlas of human thymic development defines T cell repertoire formation. Science (1979). 367 (2020), doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng Y, Liu C, Gong Y, Bai Z, Hou S, He J, Bian Z, Li Z, Ni Y, Yan J, Huang T, Shi H, Ma C, Chen X, Wang J, Bian L, Lan Y, Liu B, Hu H, Single-Cell RNA Sequencing Resolves Spatiotemporal Development of Pre-thymic Lymphoid Progenitors and Thymus Organogenesis in Human Embryos. Immunity. 51, 930–948.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Seach N, Hammett M, Chidgey A, Isolation, characterization, and reaggregate culture of thymic epithelial cells. Methods in Molecular Biology. 945, 251–272 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Venanzi ES, Gray DHD, Benoist C, Mathis D, Lymphotoxin Pathway and Aire Influences on Thymic Medullary Epithelial Cells Are Unconnected. The Journal of Immunology. 179, 5693–5700 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Zuklys S, Balciunaite G, Agarwal A, Fasler-Kan E, Palmer E, Holländer GA, Normal Thymic Architecture and Negative Selection Are Associated with Aire Expression, the Gene Defective in the Autoimmune-Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy (APECED). The Journal of Immunology. 165, 1976–1983 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Nishikawa Y, Hirota F, Yano M, Kitajima H, Miyazaki JI, Kawamoto H, Mouri Y, Matsumoto M, Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. Journal of Experimental Medicine. 207, 963–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramilowski JA, Goldberg T, Harshbarger J, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, Forrest ARR, A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 6 (2015), doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campinoti S, Gjinovci A, Ragazzini R, Zanieri L, Ariza-McNaughton L, Catucci M, Boeing S, Park JE, Hutchinson JC, Muñoz-Ruiz M, Manti PG, Vozza G, Villa CE, Phylactopoulos DE, Maurer C, Testa G, Stauss HJ, Teichmann SA, Sebire NJ, Hayday AC, Bonnet D, Bonfanti P, Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds. Nat Commun. 11 (2020), doi: 10.1038/s41467-020-20082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan Y, Tajima A, Goh SK, Geng X, Gualtierotti G, Grupillo M, Coppola A, Bertera S, Rudert WA, Banerjee I, Bottino R, Trucco M, Bioengineering thymus organoids to restore thymic function and induce donor-specific immune tolerance to allografts. Molecular Therapy. 23, 1262–1277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chung B, Montel-Hagen A, Ge S, Blumberg G, Kim K, Klein S, Zhu Y, Parekh C, Balamurugan A, Yang OO, Crooks GM, Engineering the human thymic microenvironment to support thymopoiesis in vivo. Stem Cells. 32, 2386–2396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hun M, Barsanti M, Wong K, Ramshaw J, Werkmeister J, Chidgey AP, Native thymic extracellular matrix improves in vivo thymic organoid T cell output, and drives in vitro thymic epithelial cell differentiation. Biomaterials. 118, 1–15 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Anderson Mark S., Venanzi Emily S., Klein Ludger, Chen Zhibin, Berzins Stuart P., Turley Shannon J., von Boehmer Harald, Bronson Roderick, Dierich Andrée, Benoist Christophe, Mathis Diane, Projection of an Immunological Self Shadow Within the Thymus by the Aire Protein. Science (1979). 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]


