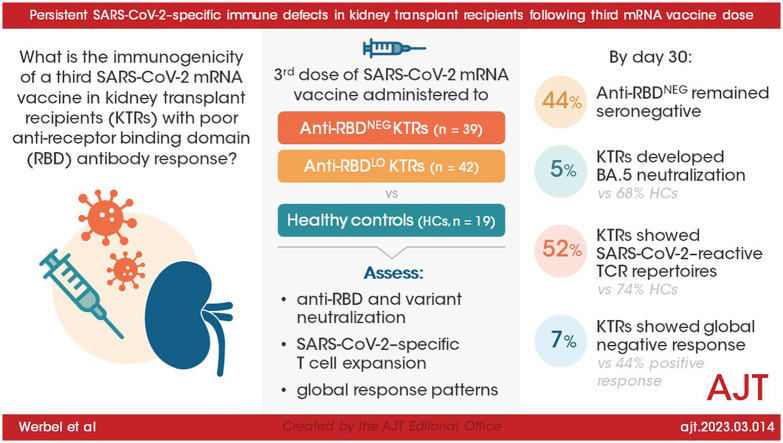
Abstract
Kidney transplant recipients (KTRs) show poorer response to SARS-CoV-2 mRNA vaccination, yet response patterns and mechanistic drivers following third doses are ill-defined. We administered third monovalent mRNA vaccines to n = 81 KTRs with negative or low-titer anti-receptor binding domain (RBD) antibody (n = 39 anti-RBDNEG; n = 42 anti-RBDLO), compared with healthy controls (HCs, n = 19), measuring anti-RBD, Omicron neutralization, spike-specific CD8+%, and SARS-CoV-2–reactive T cell receptor (TCR) repertoires. By day 30, 44% anti-RBDNEG remained seronegative; 5% KTRs developed BA.5 neutralization (vs 68% HCs, P < .001). Day 30 spike-specific CD8+% was negative in 91% KTRs (vs 20% HCs; P = .07), without correlation to anti-RBD (rs = 0.17). Day 30 SARS-CoV-2–reactive TCR repertoires were detected in 52% KTRs vs 74% HCs (P = .11). Spike-specific CD4+ TCR expansion was similar between KTRs and HCs, yet KTR CD8+ TCR depth was 7.6-fold lower (P = .001). Global negative response was seen in 7% KTRs, associated with high-dose MMF (P = .037); 44% showed global positive response. Of the KTRs, 16% experienced breakthrough infections, with 2 hospitalizations; prebreakthrough variant neutralization was poor. Absent neutralizing and CD8+ responses in KTRs indicate vulnerability to COVID-19 despite 3-dose mRNA vaccination. Lack of neutralization despite CD4+ expansion suggests B cell dysfunction and/or ineffective T cell help. Development of more effective KTR vaccine strategies is critical. (NCT04969263)
Keywords: SARS-CoV-2, kidney transplant, vaccination, immunogenicity, antibody, clinical trial
Graphical abstract
1. Introduction
Kidney transplant recipients (KTRs) demonstrate poorer humoral1 and cellular immunogenicity2 , 3 following primary mRNA SARS-CoV-2 vaccination and endure higher rates of vaccine breakthrough.4 Neutralizing antibody (nAb) is the best available correlate of protection against SARS-CoV-2 infection,5 approximated by the clinically accessible anti-receptor binding domain (anti-RBD) antibody biomarker.6 High levels of nAb, however, are required for KTRs to neutralize Omicron subvariants.7 , 8 Associations with anti-RBD response in KTRs are well defined, including the negative impact of immunosuppressive regimens containing MMF.9, 10, 11 Anti-RBD level has also emerged as a powerful predictor of response to additional vaccine doses,12, 13, 14 with the potential to identify subgroups at higher risk for COVID-19 breakthrough15, 16, 17 and the need for immunoprophylactic interventions.
The determinants and clinical impact of T cell responses induced by SARS-CoV-2 vaccines are less well delineated, in part owing to use of varying assays and metrics across studies. Additionally, discordance between antibody and T cell response has been reported in 0% to 50% of transplant recipients.18, 19, 20, 21 These patterns of humoral and/or cellular anti-SARS-CoV-2 immune responses and their underlying mechanistic drivers remain incompletely characterized. It is therefore uncertain whether immunoprotection against COVID-19 is achieved among KTRs following full (ie, 3-dose) vaccination, particularly among vulnerable KTRs who do not develop high-level anti-RBD.
Given these knowledge gaps, we enrolled a homogenous KTR cohort with poor anti-RBD response following 2-dose mRNA vaccination in a clinical trial to determine the effects of third vaccination on (1) anti-RBD and variant neutralization, (2) SARS-CoV-2–specific T cell expansion using 2 complementary assays, and (3) global patterns of immune responses as compared with healthy controls (HCs). Clinical and immunological associations with vaccine breakthroughs were recorded.
2. Methods
2.1. Participants and design
2.1.1. Study background and design
The COVID-19 Protection After Transplant (CPAT) trials were funded by the National Institutes of Health to investigate the safety and immunogenicity of SARS-CoV-2 vaccination strategies in solid organ transplant recipients. The single-arm, open-label trial described herein began August 10, 2021 to test immune responses to additional (third) homologous mRNA vaccination in KTRs who failed to respond to 2 prior mRNA vaccinations. “Failure to respond” was defined as negative (<0.8 U/mL, anti-RBDNEG) or low-titer (0.8 to 50 U/mL, anti-RBDLO) on the Roche Elecsys anti-SARS-CoV-2 S assay; this threshold was chosen given the minimal probability of neutralizing ancestral SARS-CoV-222 , 23 (Supplement).
Participants included adult, kidney-only recipients on stable calcineurin inhibitor-based immunosuppression, without major graft dysfunction or organ rejection within 6 months; full criteria are listed at ClinicalTrials.gov (NCT04969263), and the study flow diagram is presented in Supplementary Figure S1. The primary immunogenicity outcome was day 30 anti-RBD, stratified by day 0 serostatus (anti-RBDNEG/anti-RBDLO), given anticipated differential responses.14 , 24 Secondary outcomes included SARS-CoV-2 variant neutralization and cellular responses. Safety outcomes included reactogenicity and alloimmune events. Serial monitoring for SARS-CoV-2 infection occurred via polymerase chain reaction testing of nasal swabs and anti-nucleocapsid antibody testing at days 30, 90, 180, and 365; symptom screening occurred at each visit, and continuous for-cause testing was performed via clinical teams. This trial was approved by the Johns Hopkins University IRB (IRB00288774); participants provided written informed consent.
2.1.2. Healthy control (HC) cohort
In a separate, single-center prospective cohort of adult health care workers undergoing mRNA vaccination, samples were collected on day 0 and day 30 following third mRNA vaccine doses (Emory Vaccine Center, IRB#00002061). Third vaccines were administered October 2021 to November 2021, overlapping the CPAT study period; participants provided informed consent.
2.2. Antibody and neutralization assays
2.2.1. Anti-RBD antibody
Anti-RBD was measured using the semiquantitative Roche Elecsys anti-SARS-CoV-2 S pan-immunoglobulin electrochemiluminesence immunoassay. Anti-RBD in U/mL correlates ∼1:1 with World Health Organization binding antibody units. Per manufacturer, <0.8 U/mL was reported as negative (lower limit of quantification 0.4 U/mL). All samples were up-front diluted 1:50 to avoid prozone (“hook”) effects and then serially diluted until 2 equivalent signals (±10%) were obtained, with the first value utilized.
2.2.2. ACE2 inhibition assays (surrogate neutralization)
Neutralization was measured using the Meso Scale Discovery, which quantifies plasma inhibition of ACE2 binding to the full-length SARS-CoV-2 spike protein. ACE2 Meso Scale Discovery V-PLEX SARS-CoV-2 Panels 23/25/27/29/32 pre-coated with spike expressing mutations corresponding to SARS-CoV-2 variants were incubated with participant plasma; human ACE2 protein conjugated with light-emitting label was then added. If the plasma fully bound spike and blocked ACE2 binding, no light was emitted during the stimulation phase (100% inhibition; full neutralization). However, if there was no binding of spike by plasma, ACE2 was fully bound and illuminated during plate activation (0% inhibition; no neutralization). In vaccinated solid organ transplant recipients, ≥20% to 25% ACE2 inhibition (%ACE2i) on this high-throughput assay is associated with live virus nAb, including vs Omicron subvariants.25 , 26
2.2.3. Live virus neutralization
Live ancestral, Delta, and Omicron BA.1 nAb was assessed in a subset of KTRs (Supplementary Section 2.2, Fig. S2). VeroE6-TMPRSS2 cells were cultured and incubated in transport media from SARS-CoV-2–infected patients27 for RNA extraction and sequencing. The viral titer of VeroE6-TMPRSS2 cells was determined using 50% tissue culture infectious dose assays.28 nAb levels were determined using 2-fold plasma dilutions29 with the addition of 1 × 104 tissue culture infectious dose/mL virus. Samples were incubated at 37˚C for 2 days (or until complete cytopathic effect); cells were then fixed, incubated, and stained. nAb titer (NT50) was calculated as the highest serum dilution that eliminated the cytopathic effect in 50% wells; area under the curve (AUC) was calculated using GraphPad Prism to provide a continuous measure of nAb. (+)nAb was defined as >1:20 NT50 and high-level nAb as >1:160 NT50.
2.3. Cellular analyses and methodology
2.3.1. SARS-CoV-2 spike-specific CD8+ memory T cell response
Peripheral blood mononuclear cells (PBMCs) from HLA-A∗02:01+ KTRs (n = 33) were isolated and analyzed by flow cytometry for SARS-CoV-2 spike-specific CD8+ T cell responses using HLA-peptide pentamers (Supplement, Section 2.3). Cells were washed and stained with 4 biotinylated MHC class I pentamers corresponding to immunodominant SARS-CoV-2 spike protein epitopes (FIAGLIAIV, LITGRLQSL, YLQPRTFLL, RLQSLQTYV).30 , 31 Spike-specific CD8+ T cell frequency (staining for ≥1 spike-specific epitope) was evaluated out of total memory CD8+ T cells (gated on CD3+CD4−CD8+ cells, excluding naïve CCR7+CD45RA+ T cells). Positive spike-specific CD8+ T cell response threshold was ≥0.03% (above HLA-A∗02-negative HC staining background).
2.3.2. Immunosequencing of SARS-CoV-2–associated T cell repertoires
SARS-CoV-2–associated T cell repertoires were assessed via TCR sequencing in n = 65 KTRs and n = 19 HCs using the Adaptive Biotechnologies immunoSEQ Assay platform.32 , 33 PBMCs were isolated on days 0 and 30, frozen, and sent to Adaptive for high-resolution immunosequencing. The abundance of each unique TCRβ CDR3 sequence was quantified (defining the overall TCR clonal repertoire) before and after vaccination. The set of detected TCR clones was then compared against a library of ∼5,000 “high-confidence” public clones recognizing epitopes across the SARS-CoV-2 genome found to be enriched in COVID-19 convalescent patient samples (vs prepandemic controls) using multiplex identification of antigen-specific TCRs32 to reduce potential cross-reactive TCRs.33 The same machine-learning algorithm as the clinically available FDA T-Detect COVID Test (https://www.fda.gov/media/146481/download) was applied to provide a binary classifier (T-MAP COVID), reporting whether TCR repertoires were SARS-CoV-2–reactive: ie, T-MAP “positive,” “negative,” or “indeterminate” (insufficient TCR sequences to permit classification).
TCR repertoire components were then individually evaluated: (1) breadth, the proportion of unique clones reacting to SARS-CoV-2 out of all unique TCRs (ie, diversity of SARS-CoV-2–reactive clones) and (2) depth, the proportion of all productive TCR templates that react to SARS-CoV-2 of all detected TCRs (ie, total number of SARS-CoV-2–reactive clones). These metrics were reported for CD4+ and CD8+ compartments against both spike-specific and nonspike cognate regions (eg, nucleocapsid) identified via multiplex identification of antigen-specific TCRs.
2.4. Statistical analysis
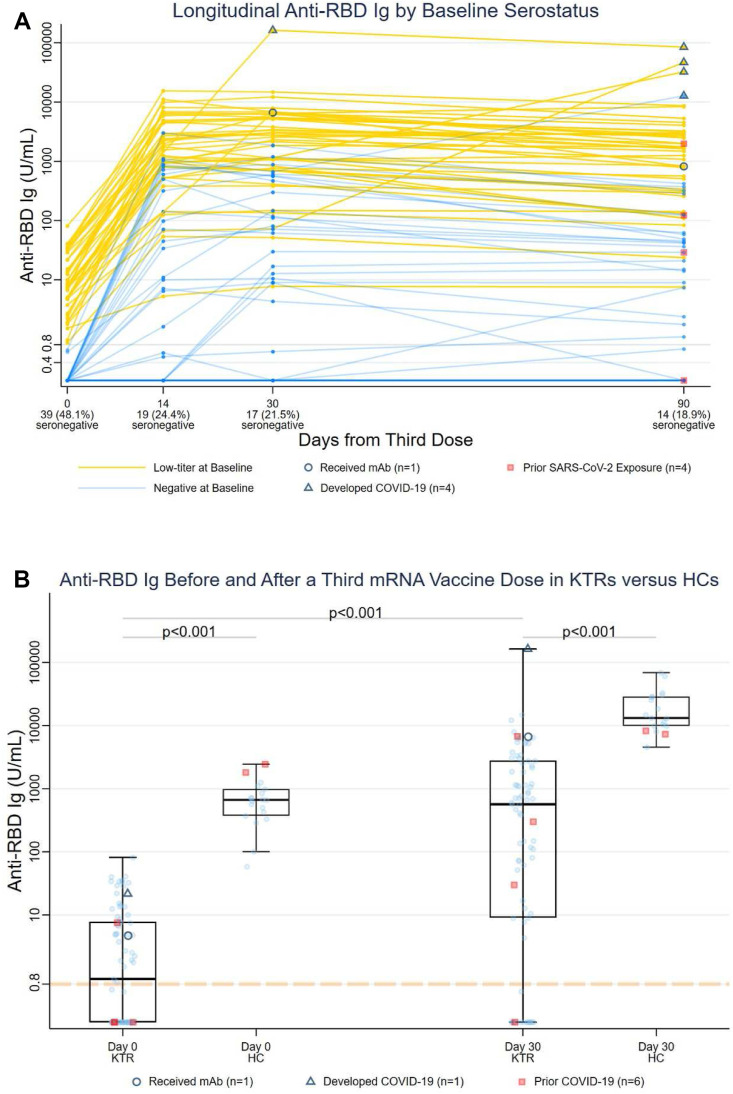
KTR characteristics were compared between (1) anti-RBDNEG and anti-RBDLO and (2) anti-RBDNEG KTRs who remained seronegative and anti-RBDNEG KTRs who seroconverted on day 30, using Fisher exact and Wilcoxon rank-sum testing (categorical and continuous variables, respectively). Day 30 anti-RBD, neutralization, and T cell responses were compared between KTRs and HCs using Wilcoxon rank-sum testing. Anti-RBD half-life for KTRs with day 30 anti-RBD 500 was estimated via exponential decay modeling. Participants who developed incident COVID-19 (for all outcomes; triangles, Fig. 1 A) or received monoclonal antibody (mAb) (for humoral outcomes; open circles, Fig. 1A) were excluded from immunogenicity analyses but included in data visualization.
Figure 1.
(A) Anti-receptor binding domain (anti-RBD) titers in KTRs following a third mRNA vaccine dose, stratified by day 0 anti-RBD level. Blue trajectories represent anti-RBDNEG (n = 39) and yellow trajectories represent anti-RBDLO low-titer (n = 42). Anti-RBD titers are represented in units/mL on the logarithmic scale. Triangles represent participants who developed incident COVID-19 (n = 4), and circles represent participants receiving monoclonal antibody (mAb) (n = 1). Squares represent participants with a history of COVID-19 prior to third vaccination. (B) Comparison of anti-receptor binding domain (anti-RBD) titers between KTRs and HCs before and 30 days after a third mRNA vaccine dose. Anti-RBD titers are represented in units/mL on the logarithmic scale. Triangles represent participants who developed incident COVID-19 (n = 1), and circles represent participants receiving monoclonal antibody (mAb) (n = 1) before day 30. Squares (n = 6) represent participants with a history of COVID-19 prior to third vaccination. HC, healthy control; KTR, kidney transplant recipient; RBD, receptor binding domain.
Associations with day 30 anti-RBD were assessed using (1) Poisson regression with robust variance estimator (RVE) for anti-RBD >2500 U/mL (potential threshold for Omicron BA.1 neutralization7 , 34) and (2) negative binomial regression with RVE for continuous anti-RBD. Based on published literature,1 multivariable models included high-dose mycophenolate (MMF; >1000 mg mycophenolate mofetil or >720 mg mycophenolic acid, daily), transplant vintage, and post hoc inclusion of day 30 CD4+ TCR breadth given mechanistic plausibility and exploratory data analysis.
The proportions of participants with SARS-CoV-2–reactive repertoires ([+]T-MAP) on day 0 and day 30 was compared used McNemar’s and Fisher exact testing. Associations of baseline characteristics with day 30 (+)T-MAP were assessed using Poisson regression with RVE; indeterminate repertoires were excluded from comparative analyses. Differences in TCR breadth and depth from day 0 to day 30 were analyzed by Wilcoxon rank-sum and matched-pairs signed-rank test as appropriate.
Associations between day 30 spike-specific TCR expansion and anti-RBD were assessed by linear fit and Spearman rank. Participants with undetectable SARS-CoV-2 TCRs were assigned values of 1 × 10-6 for analytical and visualization purposes and excluded on sensitivity analysis. Among KTRs, day 30 response patterns were assessed across 2 binary dimensions, (+)/(-)anti-RBD and (+)/(-)T-MAP, with comparison of participant characteristics.
Modeling outputs are presented in the style of Louis and Zeger35, lower 95% CIPoint Estimateupper 95% CI. Two-sided α < 0.05 denotes statistical significance. Analyses were performed using Stata/SE_17.0.
3. Results
3.1. Study population
After screening, n = 81 KTRs (n = 39 anti-RBDNEG, n = 42 anti-RBDLO) were administered a third homologous vaccine dose (22 Moderna mRNA-1273, 59 Pfizer-BNT162b2) at median (interquartile range [IQR]) 167 (149-177) days after dose 2. Demographics, laboratory, and transplant factors were similar between anti-RBDNEG and anti-RBDLO (Table 1 ).
Table 1.
Demographic and transplant characteristics of KTRs, by day 0 anti-RBD level.
| Total (N = 81) | anti-RBDNEG (N = 39) | anti-RBDLO (N = 42) | P | |
|---|---|---|---|---|
| Demographics | ||||
| Age (y), median (IQR) | 66 (57, 73) | 65 (56, 73) | 66 (57, 74) | .99 |
| Female sex, no. (%) | 27 (33) | 17 (44) | 10 (24) | .10 |
| Race, no. (%) | .75 | |||
| White | 49 (60) | 22 (56) | 27 (64) | |
| Black/African American | 24 (30) | 12 (31) | 12 (29) | |
| Asian | 4 (5) | 2 (5) | 2 (5) | |
| Other | 4 (5) | 3 (8) | 1 (2) | |
| Hispanic/Latino ethnicity, no. (%) | 2 (2) | 2 (5) | 0 (0) | .23 |
| BMI (kg/m2), median (IQR) | 26.2 (23.4, 31.2) | 27.5 (23.2, 32.0) | 25.8 (23.4, 31.1) | .53 |
| Medical comorbidities | ||||
| Diabetes, no. (%) | 27 (33) | 13 (33) | 14 (33) | >.99 |
| HCV infection, no. (%) | 5 (6) | 1 (3) | 4 (10) | .36 |
| Lung disease, no. (%) | 25 (31) | 14 (36) | 11 (26) | .47 |
| Cardiovascular disease, no. (%) | 80 (99) | 38 (97) | 42 (100) | .48 |
| Autoimmune disease, no. (%) | 12 (15) | 7 (18) | 5 (12) | .54 |
| Transplant history and immunosuppression | ||||
| Years since transplant, median (IQR) | 5.4 (2.1, 10.5) | 5.0 (2.0, 9.2) | 5.7 (3.2, 10.9) | .23 |
| Indication for most recent kidney transplantation, no. (%) | ||||
| Diabetes | 13 (16) | 6 (15) | 7 (17) | >.99 |
| Hypertension | 28 (35) | 16 (41) | 12 (29) | .25 |
| FSGS | 6 (7) | 5 (13) | 1 (2) | .10 |
| Cystic kidney disease | 11 (14) | 3 (8) | 8 (19) | .20 |
| Living donor, no. (%) | 35 (43) | 14 (36) | 21 (50) | .26 |
| DSA positive at baseline, no. (%) (n = 79) | 14 (17) | 7 (18) | 7 (18)a | >.99 |
| Baseline Immunosuppressant, no. (%) | ||||
| Mycophenolate mofetil | 56 (69) | 27 (69) | 29 (69) | >.99 |
| Total daily dose (mg), median (IQR) | 1000 (500, 1000) | 1000 (500, 1000) | 1000 (500, 1000) | .95 |
| Mycophenolic acid | 8 (10) | 5 (13) | 3 (7) | .47 |
| Total daily dose (mg), median (IQR) | 720 (540, 900) | 810 (720, 1440) | 540 (270, 810) | .16 |
| High-dose mycophenolate | 14 (17) | 9 (23) | 5 (12) | .37 |
| Prednisone | 75 (93) | 36 (92) | 39 (93) | >.99 |
| Total daily dose (mg), median (IQR) | 5 (5, 5) | 5 (5, 5) | 5 (5, 5) | .94 |
| Tacrolimus | 75 (93) | 38 (97) | 37 (88) | .20 |
| Cyclosporine | 4 (5) | 1 (3) | 3 (7) | .62 |
| Triple IS, no. (%)b | 58 (72) | 29 (74) | 29 (69) | .63 |
| COVID-19 and vaccination history | ||||
| Prior SARS-CoV-2 infectionc, no. (%) | 4 (5) | 3 (8) | 1 (2) | .35 |
| Days between second and third dose, median (IQR) | 167 (149, 177) | 159 (141, 174) | 169.5 (152, 183) | .10 |
| Vaccine manufacturer, no. (%) | .46 | |||
| Pfizer-BioNTech (BNT162b2) | 59 (73) | 30 (77) | 29 (69) | |
| Moderna (mRNA-1273) | 22 (27) | 9 (23) | 13 (31) | |
| Laboratory results | ||||
| Creatinine (mg/dL), median (IQR) | ||||
| Day 0 (Baseline) | 1.2 (1, 1.5) | 1.2 (1, 1.5) | 1.2 (1.1, 1.5) | .59 |
| Day 30 | 1.2 (1, 1.5) | 1.3 (1, 1.5) | 1.2 (1.1, 1.5) | .78 |
| Estimated GFR (ml/min/1.73m2), median (IQR) | ||||
| Day 0 | 58 (46, 73) | 57 (49, 72) | 59.5 (46, 74) | .91 |
| Day 30 | 59 (46, 72) | 55 (46, 72) | 60.5 (45, 73) | .82 |
| Baseline ALC (K/cu mm), median (IQR) | 1.01 (0.69, 1.47) | 1.01 (0.66, 1.55) | 1.00 (0.7, 1.37) | .82 |
| Baseline Total IgG (mg/dL), median (IQR) | 859 (737, 1057) | 863 (755, 1008) | 834 (732, 1086) | .89 |
| Baseline CD4+ T cell count, median (IQR)d | 172 (114, 225) | 183 (111, 228) | 171 (114, 220) | .56 |
| Recipient CMV IgG positive, no. (%) (n=72) | 38 (53) | 18 (53) | 20 (51) | >.99 |
ALC, absolute lymphocyte count; BMI, body mass index; CMV, cytomegalovirus; DSA, donor-specific antibody; FSGS, focal segmental glomerulosclerosis; GFR, glomerular filtration rate; IgG, immunoglobulin G; HCV, hepatitis C virus; IQR, interquartile range; IS, immunosuppressant. Continuous outcomes compared by Wilcoxon rank-sum testing and categorical variables were compared by Fisher exact testing.
Donor HLA typing unavailable for n = 1 participant and day 0 recipient DSA screening missing for n = 1 participant.
Any combination of 3 immunosuppressants at day 0 (calcineurin inhibitor, antimetabolite, corticosteroid).
By positive prior molecular testing or reactive anti-nucleocapsid antibody at enrollment.
T cell subtyping performed on n = 34 KTRs (16 anti-RBDNEG, 18anti-RBDLO)
The HC cohort included n = 19 persons, median (IQR) age 29 (28-35) years, all of whom received third homologous monovalent BNT162b2 vaccination median (IQR) 269 (261-277) days after dose 2 (Supplement). Two showed evidence of prior COVID-19 by day 0 anti-nucleocapsid testing.
3.2. Antibody and neutralization
3.2.1. Binding antibody responses
Among 79 KTRs (excluding n = 2 who developed COVID-19 or received mAb), median (IQR) day 30 anti-RBD titer was 561 (8.9-2567.5) U/mL (Fig. 1A), as compared with 13 170 (9915-28 755) U/mL in HCs (23-fold lower, P < .001; Fig. 1B). Day 30 median (IQR) anti-RBD was >270-fold higher in anti-RBDLO vs anti-RBDNEG KTRs: 2438.5 (740.3-5352.5)U/mL vs 9.0 (<0.4-147)U/mL (P < .001), respectively. In KTRs, anti-RBD decreased 38% by day 90, with an estimated half-life of 71 days.
Among anti-RBDNEG KTRs, 17/39 (44%) remained seronegative on day 30 (vs 0 HCs). Demographic, immunosuppressant, and vaccination factors were similar among KTRs who did vs did not seroconvert (Supplementary Table S1). Persistently seronegative KTRs demonstrated lower median (IQR) IgG levels (779 [714-881] vs 965 [846-1128] mg/dL, P = .012) and absolute lymphocyte counts (0.70 [0.59-1.36] vs 1.16 [0.93-1.57] K/mm3, P = .035), with a trend toward lower CD4+ T cell counts (120 [98-146] vs 223 [147-258] cells/μL, P = .07) (Supplementary Table S1).
High-level anti-RBD (>2500 U/mL, n = 20 [25%] KTRs vs n = 19 [100%] HCs) was associated with older transplant vintage (Ratio = 1.171.351.55 [per 5 years], P < .001) (Table 2 ) in KTRs, but not participant age or mRNA-1273 (vs BNT162b2). Anti-RBD was lower among KTRs taking high-dose MMF (Ratio = 0.040.241.70, P = .15), without reaching statistical significance.
Table 2.
Associations between clinical factors and day 30 anti-RBD level.
| Factor | >2500 U/mL ratio | P (crude) | Continuous titer ratio | P (crude) | Continuous titer ratio (adjusted) | P (adjusted) |
|---|---|---|---|---|---|---|
| Age (per 10 y) | 0.70 0.95 1.30 | .75 | 0.72 0.91 1.18 | .49 | ||
| Female sex | 0.50 1.102.43 | .82 | 0.48 1.082.42 | .85 | ||
| mRNA-1273 vaccine | 0.49 1.112.53 | .80 | 0.43 0.831.62 | .59 | ||
| Mycophenolate (n = 78) | 0.27 0.601.32 | .21 | 0.27 0.601.32 | .21 | ||
| High-dose mycophenolate | 0.04 0.24 1.70 | .15 | 0.08 0.37 1.67 | .20 | 0.020.060.18 | <.001 |
| Triple immunosuppression | 0.27 0.58 1.23 | .15 | 0.32 0.64 1.26 | .19 | ||
| Transplant vintage (per 5 y) | 1.171.351.55 | <.001 | 0.95 1.15 1.38 | .15 | 0.79 0.95 1.15 | .61 |
| Lymphocyte <1000 cell/μL | 0.85 1.904.29 | .12 | 0.91 1.78 3.49 | .09 | ||
| Absolute CD4+ count (per 100) (n = 33) | 0.59 1.06 1.92 | .85 | 0.75 0.98 1.27 | .86 | ||
| Day 0 CD4+ breadth (per 10-fold) (n = 63) | 0.92 2.57 7.23 | .07 | 0.65 1.50 3.47 | .34 | ||
| Positive day 0 T-MAP (n = 52) | 1.343.197.64 | .009 | 0.70 1.61 3.74 | .27 | ||
| Day 30 CD4+ breadth (per 10-fold) (n = 63) | 0.69 1.69 4.14 | .25 | 0.80 1.58 3.10 | .19 | 1.052.054.02 | .37 |
| Positive day 30 T-MAP (n = 55) | 1.053.4611.34 | .041 | 1.383.237.55 | .007 |
Crude univariable associations are presented for the outcomes of high-titer anti-RBD response (>2500 U/mL) and continuous anti-RBD level at day 30. An adjusted multivariable model for continuous anti-RBD response is also presented (n = 63). Bolded values represent statistical significance at the P < .05 level.
Note: All analyses excluded n = 1 participant with incident COVID-19 and n = 1 participant who received monoclonal antibody; the mycophenolate univariable analysis excluded n = 1 additional participant with inconsistent medication use during follow-up (was not prescribed high-dose mycophenolate). Sample sizes for all other univariable models are indicated next to the variable name.
3.2.2. Neutralization
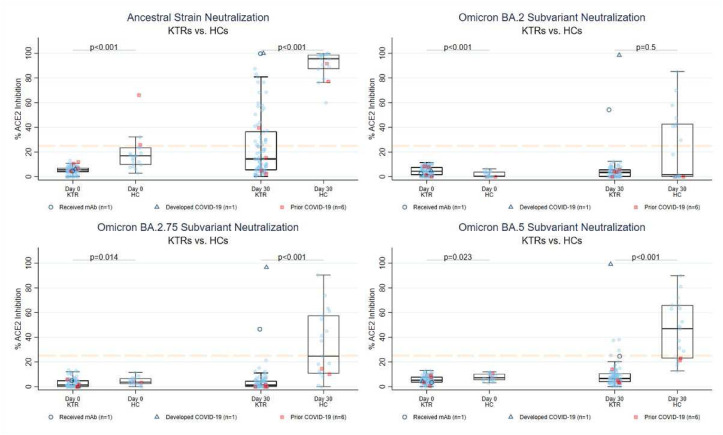
Among 79 KTRs, the proportion demonstrating ancestral strain neutralization (≥25% ACE2i) increased from 0% (n = 0) to 34% by day 30 (n = 27; 24 anti-RBDLO vs 3 anti-RBDNEG) (McNemar’s P < .001). No KTR showed Omicron subvariant neutralization on day 0 and a minimal increase by day 30: 2 (3%), 0 (0%), 0 (0%), 4 (5%) neutralized BA.1, BA.2, BA.2.75, and BA.5 spike, respectively (all anti-RBDLO; McNemar’s P > .05 all subvariants; Fig. 2 ). Of the KTRs showing BA.5 neutralization at day 30, 0/4 and 2/4 showed BQ.1.1 and XBB.1 neutralization, respectively. Confirmatory live virus testing of KTR samples on day 30 detected ancestral nAb > 1:20 in 33 (42%, 29 anti-RBDLO, median NT50 1:80), and BA.1 nAb > 1:20 in 6 (8%, all anti-RBDLO, median NT50 1:40 [low-level]) (Supplementary Fig. S2).
Figure 2.
Neutralizing capacity against SARS-CoV-2 variants before and 30 days after a third mRNA dose in KTRs and HCs. The Y axis represents percent ACE2 inhibition, ranging 0% to 100% with ≥25% consistent with neutralizing inhibition (dashed orange line). Triangles denote participants with incident COVID-19 (n = 4) and open circles denote participants receiving mAb (n = 1). Squares indicate participants with a prior history of COVID-19 (n = 4 KTRs, n = 2 HCs). HC, healthy control; KTR, kidney transplant recipient.
Among n = 19 HCs, the proportion demonstrating ancestral strain neutralization increased from 16% (n = 3) to 100% (n = 19) by day 30 (McNemar’s P < .001). No HC demonstrated Omicron subvariant neutralization on day 0, with a significant increase by day 30: 8 (42%), 9 (47%), and 13 (68%) neutralized BA.2, BA.2.75, and BA.5 spike by day 30 (McNemar’s P < .01, all subvariants). Of HCs with BA.5 neutralization on day 30, 11/13 and 12/13 showed BQ.1.1 and XBB.1 neutralization, respectively.
For each variant tested, median %ACE2i and proportion ≥25% were significantly higher on day 30 in HCs than in KTRs (P < .01 by rank-sum and Fisher exact testing, respectively, except for median BA.2 %ACE2i [P = .45]). History of prior COVID-19 did not appear associated with augmented neutralization in either group on day 30. Interestingly, the highest Omicron sublineage neutralization was observed in a KTR with breakthrough infection (see Breakthrough infections, Section 3.5).
3.3. Cellular analyses
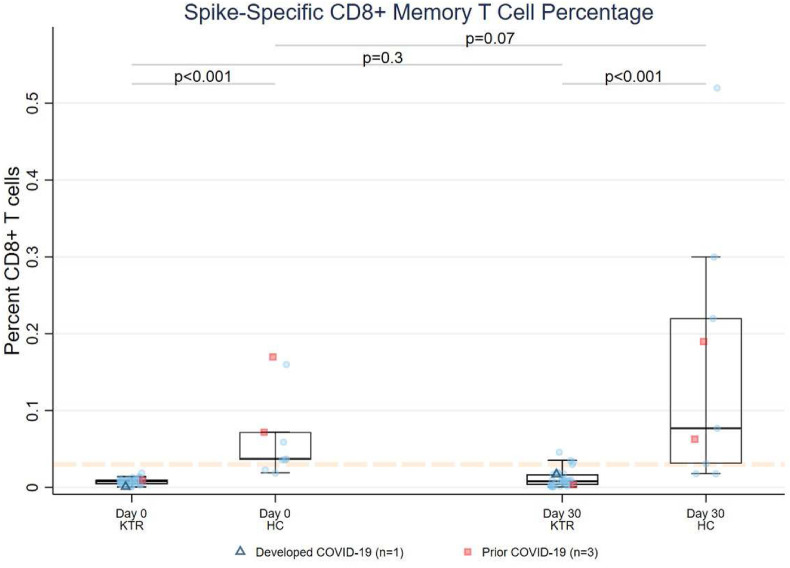
3.3.1. SARS-CoV-2 spike-specific CD8+ T cell response (pentamer staining)
Among HLA-A∗02 KTRs, 0/33 (0%) showed spike-specific CD8+ T cell response on day 0, increasing to 3/32 (9%) by day 30 (n = 2 anti-RBDLO) (McNemar’s P = .25, Fig. 3 ). In contrast, 7/9 (78%) HCs showed spike-specific CD8+ response on both day 0 and day 30. Median (IQR) CD8+% was 4.5-fold lower in KTRs than HCs on day 0 (0.0082% [0.0046-0.0098] vs 0.037% [0.036-0.072], P < .001) and 9.7-fold lower on day 30 (0.0079% [0.0031-0.014] vs 0.077% [0.031-0.22], P < .001). The change in CD8+% among KTRs from day 0 to day 30 was not significant (P = .28), although it trended toward an increase in HCs (P = .07). Day 30 CD8+ T cell response did not correlate well with anti-RBD level (KTR r s = +0.17, HC r s =-0.23, Supplementary Fig. S4), although all KTRs with positive CD8+% had positive anti-RBD (2 measurements >2500 U/mL). Both HCs with prior COVID-19 showed positive CD8+ response at day 0 and day 30, whereas the 1 KTR with prior COVID-19 showed a negative response at both time points.
Figure 3.
SARS-CoV-2 spike-specific CD8+ memory T cell responses before and after a third mRNA vaccine dose in KTRs and HCs. Flow cytometric data (epitope staining) are presented for HLA-A∗02 participants. The dashed orange line represents background staining threshold (<0.03%). Triangles denote participants who developed COVID-19 (n = 1) and squares indicate participants with prior history of COVID-19 (n = 3). HC, healthy control; KTR, kidney transplant recipient.
3.3.2. SARS-CoV-2 T cell repertoire analysis (TCR sequencing)
SARS-CoV-2–reactive TCR repertoires ([+]T-MAP) were detected in 10/52 (19%) KTRs on day 0, increasing to 27/52 (52%) by day 30 (McNemar’s P < .001), after excluding participants with indeterminate repertoires (n = 12). Day 30 reactive repertoires were ∼2-fold more frequent in anti-RBDLO 18/28 (64%) vs anti-RBDNEG 9/24 (38%), P = .09. In contrast, among HCs, 6/19 (32%) and 14/19 (74%) had (+)T-MAP on day 0 and day 30, respectively (McNemar’s P < .001, Fisher exact P = .11 vs day 30 KTR%).
Among KTRs, demographics factors were similar between (+) and (-)T-MAP on day 30, apart from older transplant vintage in (+)T-MAP (median [IQR] 8.1 [4.9-13.3] vs 4.9 [2.2-8.8] years, P = .04, Supplementary Table S4). No demographic or transplant factors were significantly associated with (+)T-MAP on day 30, apart from anti-RBDNEG status (Ratio = 0.300.551.00; P = .048) (Supplementary Table S3).
Among KTRs, total spike-specific TCR breadth (“unique clones”) increased 2-fold from 1.90 × 10-5 to 3.90 × 10-5 (P < .001) and depth (“total clones”) 2.9-fold from 6.9 × 10-6 to 1.99 × 10-5 (P < .001) between day 0 and day 30 (Supplementary Table S5, Supplementary Fig. S6); these measures were highly correlated (r > 0.9, Supplementary Fig. S5). TCR response was more prominent in the CD4+ compartment; spike-specific CD4+ breadth increased 1.47 × 10-5 to 2.62 × 10-5; P < .001, whereas spike-specific CD8+ breadth expansion was more limited (<1.0 × 10-6 to 1.89 × 10-6; P = .002). Notably, all dimensions of the spike-specific TCR repertoire at day 30 were 2- to 5-fold greater in anti-RBDLO vs anti-RBDNEG participants, eg, spike-specific CD4+ breadth of 3.67 × 10-5 vs 1.39 × 10-5 (P = .026). As expected, there was no significant increase in nonspike TCRs between day 0 and day 30 (median breadth 2.12 × 10-5 to 2.17 × 10-5, P = .37; median depth 9.68 × 10-6 to 9.55 × 10-6, P = .25; [Supplementary Table S5, Supplementary Fig. S6]).
Similar repertoire changes were observed in HCs between day 0 and day 30, with significant expansion of spike-specific TCRs, particularly CD4+ (P < .001 for breadth, depth), without significant nonspike TCR expansion (P > .05 for breadth, depth). Interestingly, there was no difference in day 30 spike-specific CD4+ measures between HCs and KTRs (CD4+ breadth 2.52 × 10-5 vs 2.62 × 10-5, P = .63; Supplementary Table S5). Spike-specific CD8+ measures, however, were all significantly greater in HCs vs KTRs, particularly CD8+ depth (7.6-fold higher on day 30, P = .001).
Day 30 spike-specific CD8+ TCR breadth (r s = 0.44, P = .01) and depth (r s = 0.53, P = .001) positively correlated with spike-specific CD8+% by MHC-pentamer staining, though this relationship was primarily driven by HCs (Supplementary Fig. S7).
Multivariable modeling of day 30 anti-RBD level in KTRs incorporating TCR measures revealed a positive association of day 30 spike-specific CD4+ T cell breadth (aRatio = 1.052.054.02 [per 1 log], P = .037) and a highly significant negative association of high-dose MMF (aRatio=0.020.060.18, P < .001) after accounting for transplant vintage. On sensitivity analysis excluding participants with 0 TCR breadth, the point estimate for high-dose MMF was similar (aRatio = 0.030.070.16, P < .001), whereas CD4+ breadth point estimate increased (aRatio = 5.3111.5124.92 [per 1 log], P < .001).
3.4. Response patterns after full vaccination: humoral and cellular correlations
3.4.1. Categorization of anti-RBD and T cell responses
Response patterns on day 30 were characterized using dichotomous categories of (+)/(-)anti-RBD and (+)/(-)T-MAP (n = 55 KTRs; excluding n = 8 with indeterminate T-MAP, n = 1 with incident COVID-19, and n = 1 with incident mAb). Global negative response (-)anti-RBD/(-)T-MAP was seen in 4 (7%) participants vs global positive response (+)anti-RBD/(+)T-MAP in 22 (40%). Discordant responses were seen in 29 (53%) participants: 24 (44%) with (+)anti-RBD/(-)T-MAP and 5 (9%) with (-)anti-RBD/(+)T-MAP (Supplementary Table S6). High-dose MMF was used in 3/4 (75%) with global negative responses, as compared with 20% (range 13% to 27%) of participants with other patterns (Supplementary Table S6, Fisher exact P = .037). Age and other demographic features were similar across response patterns apart from oldest transplant vintage in persons with (-)anti-RBD/(+)T-MAP (P = .046). As all HCs showed (+)anti-RBD at day 30 (74% global positive response).
3.4.2. Association of TCR repertoire expansion and anti-RBD response
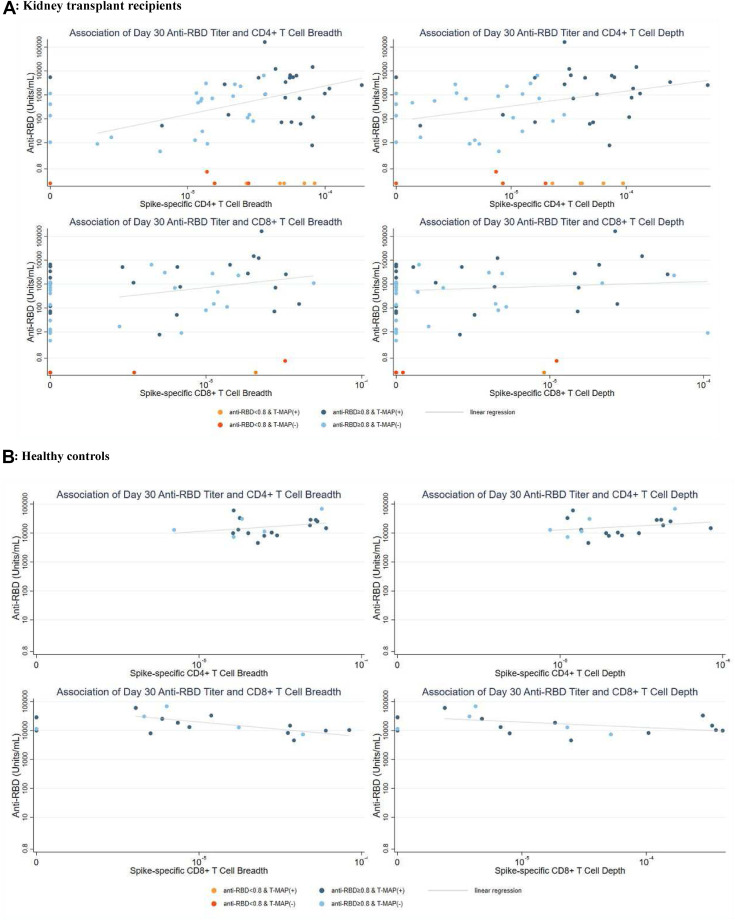
Among KTRs with (+)anti-RBD on day 30, there was a positive correlation between spike-specific CD4+ TCR breadth and anti-RBD on day 30 (Fig. 4 , r s = 0.34, P = .02); a similar association was observed with spike-specific CD4+ T cell depth (Fig. 4, r s = 0.34, P = .02). Correlations with CD4+ breadth (r s = 0.41, P = .007) and depth (rs = 0.41, P = .008) were similar after excluding KTRs with negative TCR values. In contrast, among KTRs with (-)anti-RBD on day 30, spike-specific CD4+ TCR responses varied widely. Additionally, spike-specific CD4+ TCR breadth on day 30 was similar between anti-RBDNEG KTRs who did vs those who did not seroconvert (P = .11, data not shown). Correlations between day 30 anti-RBD level and CD8+ TCR breadth (P = .06) or depth (P = .05) were not statistically significant (Fig. 4).
Figure 4.
Response patterns following third mRNA vaccine doses: correlation of SARS-CoV-2 antibody and T cell responses. Scatterplot of anti-receptor binding domain (RBD) level and dimensions of SARS-CoV-2 T cell receptor expansion (spike-specific CD4+ and CD8+ breadth and depth) on the logarithmic scale at day 30 post vaccination among kidney transplant recipients (A, n = 55) and healthy controls (B, n = 19). Data points are colorized by response pattern, (+)/(-) anti- RBD and (+)/(-)T-MAP (SARS-CoV-2-reactive T cell repertoire). Trend lines visualize correlation between vaccine responses in participants with detectable signatures (i.e., (+)anti-RBD and categorizable T cell receptor repertoire).
KTRs with global positive responses at day 30 had median (IQR) anti-RBD 1499 (118-5225) U/mL, including 10 (45%) with anti-RBD >2500 U/mL, and 2 (9%) demonstrated Omicron BA.5 neutralization. In contrast, KTRs with (+)anti-RBD/(-)T-MAP (discordant pattern) demonstrated median (IQR) anti-RBD 441 (23-1124) U/mL (P = .03 vs global positive), including only 3 (10%) with anti-RBD >2500U/mL (0 showed BA.5 neutralization). Overall, anti-RBD >2500U/mL was achieved in 37% (+)T-MAP vs 11% (-)T-MAP KTRs (P = .029).
In contrast, among HCs, there was no significant correlation between CD4+ TCR measures and anti-RBD (breadth r s = 0.24 [P = .3]; depth r s = 0.20 [P = .4]). Additionally, there was no significant difference in anti-RBD level if (+)T-MAP vs (-)T-MAP (median 13 976 U/mL vs 12 885 U/mL, P = .7).
3.5. Breakthrough infections
There were 13 SARS-CoV-2 infections among KTRs (16%) at median 99 days (range 13-141) after third vaccination (Table 3 ). Four KTRs were infected before day 90, during the Delta wave, whereas most (88%) late infections occurred during the Omicron BA.1 wave. Nearly all cases (92%) were symptomatic; 2 (15%) required hospitalization without intensive care. Median (IQR) anti-RBD level preinfection was 91 (16-429) U/mL, including 3 (23%) with negative titers; none displayed preinfection BA.1 neutralization, although 2 showed Delta neutralization (1 had received mAb).
Table 3.
Clinical and immunological characteristics of 13 breakthrough SARS-CoV-2 infections.
| Age; y (decade) | Sex | Days from dose 3 to infectiona | Variant waveb | Day 0 anti-RBD level | Day 30 anti-RBD level | Preinfection anti-RBD | Pre- infection Delta %ACE2i | Pre- infection BA.1 %ACE2i | Preinfection T cell reactivity | mAb received for infection | Days from infection to sampling | Postinfection anti-RBD | Postinfection Delta %ACE2i | Postinfection BA.1 %ACE2i | COVID-19 severity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50-59 | M | 13 | Delta (Sept 2021) | 21.6 | 21.6 | 21.6 | 9.0% | 1.3% | T-MAP(-) CD8(-) | None | 18 | 163 120 | >99% | 98.1% | Mild: symptomatic, outpatient |
| 30-39 | M | 74 | Delta (Nov 2021) | 2.7 | 1145 | 1145 | 11.8% | 3.4% | T-MAP(+) | Casirivimab/ imdevimab | 15 | 32 400 | >99% | 22.0% | Mild: symptomatic, outpatient |
| 40-49 | F | 74 | Delta (Nov 2021) | <0.8 | 666 | 666 | 33.3% | 4.8% | CD8(-) | None | 22 | 12 758 | 72.9% | 19.7% | Mild: symptomatic, outpatient |
| 50-59 | M | 76 | Delta (Nov 2021) | 3.8 | 73.1 | 73.1 | 8.8% | 6.8% | T-MAP(+) | Casirivimab/imdevimab | 16 | 46 880 | >99% | 72.0% | Moderate: hospitalization, oxygen by nasal cannula |
| 70-79 | M | 95 | BA.1 (Dec 2021) | 1.9 | 2310 | 1484 | 13.5% | 3.0% | T-MAP(-) CD8(-) | None | 86 | 29 820 | 98.3% | 71.5% | Mild: symptomatic, outpatient |
| 60-69 | M | 98 | BA.1 (Dec 2021) | <0.8 | 301 | 123 | 4.5% | 5.7% | CD8(-) | Sotrovimab | 77 | 10 580 | 73.6% | 11.9% | Mild: symptomatic, outpatient |
| 70-79 | M | 98 | Delta (Nov 2021) | <0.8 | <0.8 | <0.8 | 4.5% | 8.1% | .. | Bamlanivimab/etesevimab | 86 | 3400 | 90.5% | 0% | Mild: symptomatic, outpatient |
| 40-49 | F | 99 | BA.1 (Dec 2021) | <0.8 | <0.8 | <0.8 | 4.5% | 0.0% | .. | None | .. | .. | .. | .. | Moderate: hospitalization, oxygen by nasal cannula |
| 60-69 | M | 122 | BA.1 (Jan 2022) | 2.5 | 2153 | 1079 | 6.3% | 2.2% | .. | Sotrovimab | 51 | 19 550 | 90.4% | 62.9% | Mild: symptomatic, outpatient |
| 50-59 | F | 126 | BA.1 (Dec 2021) | 4.7 | 6650c | 821c | 98.0%c | 0.0%c | T-MAP(+) | Sotrovimab | 66 | 10 910 | .. | .. | Mild: symptomatic, outpatient |
| 60-69 | F | 127 | BA.1 (Jan 2022) | <0.8 | <0.8 | <0.8 | 6.4% | 9.0% | T-MAP(+) | Sotrovimab | 61 | 6740 | 39.3% | 12.5% | Mild: symptomatic, outpatient |
| 70-79 | M | 128 | BA.1 (Dec 2021) | <0.8 | 52.6 | 23.7 | 9.6% | 6.6% | CD8(-) | None | 58 | 44 617d | >99%d | >99%d | Mild: asymptomatic, outpatient |
| 50-59 | M | 141 | BA.1 (Jan 2022) | 10.1 | 838 | 110 | 6.6% | 2.7% | T-MAP(-) CD8(-) | Sotrovimab | 46 | 12 475 | 53.1% | 15.2% | Mild: symptomatic, outpatient |
Note: Breakthrough infections identified via positive SARS-CoV-2 test or anti-nucleocapsid antibody seroconversion. Preinfection measures (anti-RBD, surrogate neutralization [%ACE2 inhibition, ≥25% consistent with neutralizing capacity], T cell reactivity [T-MAPTM TCRseq classifier and/or spike-specific CD8%]) represent last available timepoint before confirmed infection. Postinfection measures represent first timepoint following breakthrough. COVID-19 treatment was at the discretion of the primary transplant team.
Date of PCR confirmation available for 12 participants, date of symptom onset used for remaining 1 participant.
Delta wave defined as August 1 to December 1, 2021. Omicron wave (BA.1) defined as December 24, 2021 to February 1, 2022; there were no infections during period of Delta and Omicron co-circulation December 1 to December 23, 2021). Confirmatory sequencing was not performed.
Received prior casirivimab/imdevimab on day 16 post vaccination (active against the Delta variant).
Received fourth vaccine dose (monovalent mRNA booster) before postinfection sampling.
Postinfection anti-RBD and neutralization were augmented in 4 KTRs infected before day 90 (triangles, Figure 1, Figure 2), above nearly all other participants (2 also received mAb). Two participants with breakthrough were the only KTRs to demonstrate high-level BA.1 nAb on day 90 (Supplementary Fig. 2), including 1 with high-level neutralization against all Omicron subvariants including BQ.1.1 and XBB.1 (did not receive mAb). Neutralizing capacity after BA.1 infections was variable, including 3/8 KTRs showing ACE2i <25% after infection (all received mAb). Of 10 participants with preinfection SARS-CoV-2 T cell data, 6/6 (100%) had negative CD8+ response by MHC-pentamer staining and 3/7 (43%) had (-)T-MAP; 1 participant with (+)T-MAP preinfection required hospitalization.
4. Discussion
In this trial designed to systematically characterize immunogenicity of third mRNA vaccines in poor anti-RBD responders, we demonstrated substantial SARS-CoV-2–specific immune deficits despite full vaccination in KTRs. The findings confirm the major impact of preceding anti-RBD serostatus on subsequent responses, with nearly half anti-RBDNEG failing to seroconvert. Although some participants with anti-RBDLO attained high anti-RBD titers, only 5% showed Omicron BA.5 neutralization (none neutralized BQ.1.1). SARS-CoV-2–specific CD4+ responses measured by TCR sequencing improved with vaccination, dovetailing with highest-level anti-RBD, to define a global positive response pattern in 40% KTRs. Yet, even in these participants, SARS-CoV-2–specific CD8+ responses by MHC-pentamer staining and TCR sequencing were limited; <10% KTRs showed spike-specific CD8+ staining, and CD8+ TCR depth was >7-fold lower vs HCs. Breakthrough infections were common, predominately occurring among KTRs with lower anti-RBD and poor neutralizing capacity, without clear relation to measures of T cell reactivity.
This trial further supports the negative association of high-dose MMF with humoral vaccine response,6 , 10 , 11 which strengthened after accounting for CD4+ TCR breadth and transplant vintage, suggesting heavy lymphocyte impairments. Given suboptimal immune responses and neutralization in many KTRs following repeated mRNA vaccination,8 , 13 , 36 exploring perivaccination MMF reduction among low alloimmune risk KTRs is of great interest, having shown safety and potentially augmented immunogenicity in small observational studies37 and a clinical trial38; a multicenter CPAT trial (NCT05077254) is currently underway.
Although persistent anti-RBD seronegativity was common, there was no clear association with standard clinical or transplant characteristics, and many showed equivalent CD4+ expansion as anti-RBD responders. Remarkably, CD4+ expansion was similar between KTRs and HCs, despite striking differences in anti-RBD and neutralization. This suggests spike-specific CD4+ T cell reactivity as necessary but not sufficient for high-level anti-RBD responses in KTRs. Coupled with findings of lower lymphocyte counts and IgG levels in persistently seronegative participants, these investigations suggest B cell dysfunction and/or qualitative T cell defect as contributors to poor antibody response.3 This may include metabolic dysfunction related to MMF,39 ineffective T follicular helper cell production, and/or costimulation.6 , 19 Investigating the state and interactions of B and T cells in KTRs with poor humoral response despite T cell reactivity is a potential avenue to delineate mechanisms of vaccine nonresponse and target augmentation strategies.
Breakthrough infections were common, predominantly among those with poor plasma neutralizing capacity. In the era of active mAbs, there was no critical illness, yet with loss of activity against newer Omicron sublineages, outcomes may not be as favorable. Delta variant infection elicited impressive humoral responses, including 1 KTR with cross-variant neutralization against BQ.1.1 and XBB.1, whereas immunogenicity following BA.1 infection was more variable, potentially related to high antigenic distance from the vaccine strain.40 Notably, several participants showed SARS-CoV-2 T cell reactivity prior to infection, including 1 participant who required hospitalization, suggesting cellular markers may not correlate as strongly with protection against COVID-19. Given poor CD8+ response and lack of correlation with anti-RBD, it is challenging to presume strong T cell immunoprotection in the absence of high-level humoral response, although this remains a critical scientific frontier.
Strengths of this trial include explicit focus on high-risk KTRs, using clinically available biomarkers and studying associations with neutralizing measures and deeper evaluation of SARS-CoV-2–associated T cell compartments. Additionally, breakthrough ascertainment was robust, leveraging serial assessment of pre- and postinfection immune responses. Limitations include smaller sample size, resulting from strict inclusion criteria and contemporaneous availability of third vaccines in the community, reducing power to detect immunological associations. Additionally, due to HLA and PBMC restrictions, T cell analyses were not performed on all participants. Although the broader SARS-CoV-2–reactive T cell repertoire was interrogated, focus was upon public/immunoprevalent epitopes, and functional capacity and metabolic state of cells were not explicitly evaluated. Furthermore, although HCs provided critical framing of poor multifactorial KTR responses, cohorts were not age- and comorbidity-matched, which may explain some variance in immunogenicity. Thus, the findings are hypothesis-generating and should be considered alongside other investigations into the varied cellular immunoprotection following vaccination and infection19 , 41 and their real-world implications for KTRs.
In summary, a third mRNA vaccine dose augmented anti-RBD in KTRs with prior detectable antibody after a 2-dose series, albeit to levels far below that of HCs; ≤5% demonstrated contemporary Omicron sublineage neutralization. Spike-reactive CD4+ T cell repertoires after vaccination correlated with highest-level anti-RBD response in KTRs yet did not fully discriminate humoral responders. High-dose MMF significantly impaired anti-RBD response, potentially via B cell dysfunction and/or ineffective CD4+ help. Paucity of neutralization and CD8+ response suggests vulnerability to infection in the majority of these high-risk KTRs in the Omicron era. Alternative vaccination strategies are needed to enhance immunoprotection in KTRs, particularly those with negative anti-RBD, including targeted immunosuppression reduction37 , 42 or exploration of alternative platforms including adjuvanted vaccines.
Funding
This work was supported by the National Institutes of Health (NIH) to CMD and DLS (U01 AI138897). Additional research support was provided by the NIH to CMD and DLS (U01 AI134591), WAW (K23 AI157893) AART (R01 AI20938), AHK (K08 AI156021), ACJ (T32 AI070081), CPL (U19 AI051731, R01 MD011682, R01 AI126322, U01 AI138909), the National Cancer Institute to ALC and SLK (U54 CA260491), the Johns Hopkins University Center for AIDS Research to WAW (P30 AI094189), the James O. Robbins Fellowship to ACJ, and the James M. Cox Foundation and the Carlos and Marguerite Mason Trust to CPL.
Disclosure
The funder of the study (US NIH) was not involved in patient recruitment, data collection, analysis, or visualization; the funder was involved in protocol design, data interpretation, manuscript writing, and the decision to submit for publication as per cooperative agreement. No agencies provided payment for the writing of this report. All authors had access to the full data in the study and accept responsibility for the decision to submit for publication.
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. Outside of the submitted work, the following authors declare: W. Werbel: AstraZeneca (consulting), Novavax (advisory board), GlobalData (consulting); A. Karaba: Roche (consulting); R. Avery: Aicuris, Astellas, Chimerix, Merck, Oxford Immunotec, Qiagen, Regeneron, Takeda/Shire (research support to institution); M. Bettinotti: CareDx, One Lambda Thermofisher (scientific advisory board); N. Rouphael: ICON, EMMES, MICRON, Krog (consulting), ARLG, TMRC, CDC, Moderna (advisory board), NIH, Doris Duke Foundation (grant review committee); C. Durand: Abbvie, GlaxoSmithKline (research support to institution), Gilead (grant review committee); D. Segev: CSL Behring (consulting), Novartis, Sanofi, Jazz Pharmaceuticals, Veloxis, Mallinckrodt, Thermo Fisher Scientific, Regeneron, AstraZeneca (honorarium, consulting).
Data availability
Proposals to access de-identified data from the CPAT trials can be submitted to the CPAT trials data coordinating center (contact: christinedurand@jhmi.edu), with transfer approved on an individual basis via formal data use agreement.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajt.2023.03.014.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Boyarsky B.J., Werbel W.A., Avery R.K., et al. Antibody response to 2-dose SARS-CoV-2 mRNA vaccine series in solid organ transplant recipients. JAMA. 2021;325(21):2204–2206. doi: 10.1001/jama.2021.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yahav D., Rahamimov R., Mashraki T., et al. Immune response to third dose BNT162b2 COVID-19 vaccine among kidney transplant recipients-a prospective study. Transpl Int. 2022;35 doi: 10.3389/ti.2022.10204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rincon-Arevalo H., Choi M., Stefanski A.L., et al. Impaired humoral immunity to SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients and dialysis patients. Sci Immunol. 2021;6(60) doi: 10.1126/sciimmunol.abj1031. [DOI] [PubMed] [Google Scholar]
- 4.Sun J., Zheng Q., Madhira V., et al. Association between immune dysfunction and COVID-19 breakthrough infection after SARS-CoV-2 vaccination in the US. JAMA Intern Med. 2022;182(2):153–162. doi: 10.1001/jamainternmed.2021.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert P.B., Donis R.O., Koup R.A., Fong Y., Plotkin S.A., Follmann D. A Covid-19 milestone attained – a correlate of protection for vaccines. N Engl J Med. 2022;387(24):2203–2206. doi: 10.1056/NEJMp2211314. [DOI] [PubMed] [Google Scholar]
- 6.Charmetant X., Espi M., Benotmane I., et al. Infection or a third dose of mRNA vaccine elicits neutralizing antibody responses against SARS-CoV-2 in kidney transplant recipients. Sci Transl Med. 2022;14(636) doi: 10.1126/scitranslmed.abl6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar D., Hu Q., Samson R., et al. Neutralization against Omicron variant in transplant recipients after three doses of mRNA vaccine. Am J Transplant. 2022;22(8):2089–2093. doi: 10.1111/ajt.17020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaba A.H., Johnston T.S., Aytenfisu T.Y., et al. A fourth dose of COVID-19 vaccine does not induce neutralization of the omicron variant among solid organ transplant recipients with suboptimal vaccine response. Transplantation. 2022;106(7):1440–1444. doi: 10.1097/TP.0000000000004140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manothummetha K., Chuleerarux N., Sanguankeo A., et al. Immunogenicity and risk factors associated with poor humoral immune response of SARS-CoV-2 vaccines in recipients of solid organ transplant: a systematic review and meta-analysis. JAMA Netw Open. 2022;5(4) doi: 10.1001/jamanetworkopen.2022.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantauskaite M., Müller L., Kolb T., et al. Intensity of mycophenolate mofetil treatment is associated with an impaired immune response to SARS-CoV-2 vaccination in kidney transplant recipients. Am J Transplant. 2022;22(2):634–639. doi: 10.1111/ajt.16851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell J., Chiang T.P., Alejo J.L., et al. Effect of mycophenolate mofetil dosing on antibody response to SARS-CoV-2 vaccination in heart and lung transplant recipients. Transplantation. 2022;106(5):e269–e270. doi: 10.1097/TP.0000000000004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caillard S., Thaunat O., Benotmane I., Masset C., Blancho G. Antibody response to a fourth messenger RNA COVID-19 vaccine dose in kidney transplant recipients: a case series. Ann Intern Med. 2022;175(3):455–456. doi: 10.7326/L21-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Midtvedt K., Vaage J.T., Heldal K., Munthe L.A., Lund-Johansen F., Åsberg A. Fourth dose of the SARS-CoV-2 vaccine in kidney transplant recipients with previously impaired humoral antibody response. Am J Transplant. 2022;22(11):2704–2706. doi: 10.1111/ajt.17091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benotmane I., Gautier G., Perrin P., et al. Antibody response after a third dose of the mRNA-1273 SARS-CoV-2 vaccine in kidney transplant recipients with minimal serologic response to 2 doses. JAMA. 2021;326(11):1063–1065. doi: 10.1001/jama.2021.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alejo J.L., Chiang T.P.Y., Bowles Zeiser L., et al. Incidence and severity of COVID-19 among vaccinated solid organ transplant recipients during the omicron wave. Transplantation. 2022;106(9):e413–e415. doi: 10.1097/TP.0000000000004226. [DOI] [PubMed] [Google Scholar]
- 16.Caillard S., Chavarot N., Bertrand D., et al. Occurrence of severe COVID-19 in vaccinated transplant patients. Kidney Int. 2021;100(2):477–479. doi: 10.1016/j.kint.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tau N., Yahav D., Schneider S., Rozen-Zvi B., Abu Sneineh M., Rahamimov R. Severe consequences of COVID-19 infection among vaccinated kidney transplant recipients. Am J Transplant. 2021;21(8):2910–2912. doi: 10.1111/ajt.16700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall V.G., Ferreira V.H., Ierullo M., et al. Humoral and cellular immune response and safety of two-dose SARS-CoV-2 mRNA-1273 vaccine in solid organ transplant recipients. Am J Transplant. 2021;21(12):3980–3989. doi: 10.1111/ajt.16766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrezenmeier E., Rincon-Arevalo H., Stefanski A.L., et al. B and T cell responses after a third dose of SARS-CoV-2 vaccine in kidney transplant recipients. J Am Soc Nephrol. 2021;32(12):3027–3033. doi: 10.1681/ASN.2021070966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sattler A., Schrezenmeier E., Weber U.A., et al. Impaired humoral and cellular immunity after SARS-CoV-2 BNT162b2 (tozinameran) prime-boost vaccination in kidney transplant recipients. J Clin Invest. 2021;131(14) doi: 10.1172/JCI150175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havlin J., Svorcova M., Dvorackova E., et al. Immunogenicity of BNT162b2 mRNA COVID-19 vaccine and SARS-CoV-2 infection in lung transplant recipients. J Heart Lung Transplant. 2021;40(8):754–758. doi: 10.1016/j.healun.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Departments of Research & Development and Product Management, for Roche Diagnostics Solutions, Core Lab. Correlation between Elecsys® Anti-SARS-CoV-2 S assay results and the detection of functional SARS-CoV-2 neutralizing antibodies. Memo. February. 2021;8 [Google Scholar]
- 23.Resman Rus K., Korva M., Knap N., Avšič Županc T., Poljak M. Performance of the rapid high-throughput automated electrochemiluminescence immunoassay targeting total antibodies to the SARS-CoV-2 spike protein receptor binding domain in comparison to the neutralization assay. J Clin Virol. 2021;139 doi: 10.1016/j.jcv.2021.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werbel W.A., Boyarsky B.J., Ou M.T., et al. Safety and immunogenicity of a third dose of SARS-CoV-2 vaccine in solid organ transplant recipients: a case series. Ann Intern Med. 2021;174(9):1330–1332. doi: 10.7326/L21-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaba A.H., Zhu X., Liang T., et al. A third dose of SARS-CoV-2 vaccine increases neutralizing antibodies against variants of concern in solid organ transplant recipients. Am J Transplant. 2022;22(4):1253–1260. doi: 10.1111/ajt.16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaba A.H., Kim J.D., Chiang T.P.Y., et al. Neutralizing activity and 3-month durability of tixagevimab and cilgavimab prophylaxis against Omicron sublineages in transplant recipients. Am J Transplant. 2023;23(3):423–428. doi: 10.1016/j.ajt.2022.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein S.L., Pekosz A., Park H.S., et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest. 2020;130(11):6141–6150. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaecher S.R., Touchette E., Schriewer J., Buller R.M., Pekosz A. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J Virol. 2007;81(20):11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carreño J.M., Alshammary H., Tcheou J., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 30.Shomuradova A.S., Vagida M.S., Sheetikov S.A., et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53(6):1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberhardt V., Luxenburger H., Kemming J., et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597(7875):268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klinger M., Pepin F., Wilkins J., et al. Multiplex identification of antigen-specific T cell receptors using a combination of immune assays and immune receptor sequencing. PLOS ONE. 2015;10(10) doi: 10.1371/journal.pone.0141561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder T.M., Gittelman R.M., Klinger M., et al. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv. 2020 doi: 10.1101/2020.07.31.20165647. [DOI] [Google Scholar]
- 34.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185(3):457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis T.A., Zeger S.L. Effective communication of standard errors and confidence intervals. Biostatistics. 2009;10(1):1–2. doi: 10.1093/biostatistics/kxn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osmanodja B., Ronicke S., Budde K., et al. Serological response to three, four and five doses of SARS-CoV-2 vaccine in kidney transplant recipients. J Clin Med. 2022;11(9) doi: 10.3390/jcm11092565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrezenmeier E., Rincon-Arevalo H., Jens A., et al. Temporary antimetabolite treatment hold boosts SARS-CoV-2 vaccination-specific humoral and cellular immunity in kidney transplant recipients. JCI Insight. 2022;7(9) doi: 10.1172/jci.insight.157836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kho M.M.L., Messchendorp A.L., Frölke S.C., et al. Alternative strategies to increase the immunogenicity of COVID-19 vaccines in kidney transplant recipients not responding to two or three doses of an mRNA vaccine (RECOVAC): a randomised clinical trial. Lancet Infect Dis. 2023;23(3):307–319. doi: 10.1016/S1473-3099(22)00650-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson E., Roznik K., Karaba A.H., et al. Lipid-oxidizing B cells enable successful vaccine response despite immunosuppression. Immunity. Posted online April. 2022;22 doi: 10.2139/ssrn.4090935. [DOI] [Google Scholar]
- 40.Reynolds C.J., Pade C., Gibbons J.M., et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science. 2022;377(6603) doi: 10.1126/science.abq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira V.H., Solera J.T., Hu Q., et al. Homotypic and heterotypic immune responses to Omicron variant in immunocompromised patients in diverse clinical settings. Nat Commun. 2022;13(1):4489. doi: 10.1038/s41467-022-32235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connolly C.M., Chiang T.P., Boyarsky B.J., et al. Temporary hold of mycophenolate augments humoral response to SARS-CoV-2 vaccination in patients with rheumatic and musculoskeletal diseases: a case series. Ann Rheum Dis. 2022;81(2):293–295. doi: 10.1136/annrheumdis-2021-221252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proposals to access de-identified data from the CPAT trials can be submitted to the CPAT trials data coordinating center (contact: christinedurand@jhmi.edu), with transfer approved on an individual basis via formal data use agreement.