Abstract
CDKN2A/B deletion or silencing is common across human cancer, reinforcing the general importance of bypassing its tumor suppression in cancer formation or progression. In rhabdomyosarcoma (RMS) and neuroblastoma, two common childhood cancers, the three CDKN2A/B transcripts are independently expressed to varying degrees, but one, ARF, is uniformly silenced. Although TGFβ induces certain CDKN2A/B transcripts in HeLa cells, it was unable to do so in five tested RMS lines unless the cells were pretreated with a broadly acting methyltransferase inhibitor, DZNep, or one targeting EZH2. CDKN2A/B induction by TGFβ correlated with de novo appearance of three H3K27Ac peaks within a 20 kb cis element ∼150 kb proximal to CDKN2A/B. Deleting that segment prevented their induction by TGFβ but not a basal increase driven by methyltransferase inhibition alone. Expression of two CDKN2A/B transcripts was enhanced by dCas9/CRISPR activation targeting either the relevant promoter or the 20 kb cis elements, and this “precise” manipulation diminished RMS cell propagation in vitro. Our findings show crosstalk between methyltransferase inhibition and TGFβ-dependent activation of a remote enhancer to reverse CDKN2A/B silencing. Though focused on CDKN2A/B here, such crosstalk may apply to other TGFβ-responsive genes and perhaps govern this signaling protein’s complex effects promoting or blocking cancer.
Keywords: CDKN2A/B, Cis enhancers, chromosome 9p21, EZH2, histone methyltransferase
INTRODUCTION
It has long been recognized that bypassing tumor suppressor gene activity represents a pivotal step in cancer formation and progression. This is exemplified by the CDKN2A and CDKN2B tumor suppressor genes, which lie in close proximity on chromosome 9p21 and encode three proteins that activate RB and p53, key effectors of cell cycle arrest and response to genotoxic stressors.1 Individual CDKN2A/B transcripts can be mutated,2 but the genes are more commonly co-deleted3 and can be silenced en bloc by EZH2-mediated histone methylation via Polycomb Repressive Complex 2 (PRC2)4 and promoter methylation via DNA methyltransferases.5,6
Studies of mouse eye development show that the three transcripts encoded by mouse Cdkn2a/b are not always silenced together. Expression of Arf, one of two transcripts encoded by Cdkn2a, is strictly governed in a temporally and spatially restricted pattern in later stages of embryogenesis.7–9 Tgfβ2 is required for developmentally regulated Arf expression in the mouse,10 and Tgfβ1, 2, and 3 share the capacity to drive Arf transcription in a Smad-dependent fashion in cultured mouse fibroblasts.11 This effect hinges on a non-coding, 70 kb DNA segment lying approximately 100 kb upstream of mouse Cdkna/b.12 Such studies begin to define cis enhancers engaged by an extracellular signal to drive Cdkn2a/b transcription and may provide what could be an entry point for modifying their expression in cancer cells.
Attempting to extend this to cancer models, we were initially struck by the relative paucity of human cancer cell lines in which TGFβ could induce CDKN2A/B gene expression. Of ten tested cancer cell lines (1 prostate cancer, 2 colorectal carcinoma, 1 RMS, 4 cervical cancer, and 2 osteosarcoma), induction was only observed in HeLa cells, which are known to harbor HPV E6 and E7 oncoproteins targeting p53 and RB, respectively.13,14 Similar to mouse cells, CDKN2A/B induction depended on a 20 kb cis element located approximately 110 kb from the CDKN2B transcription start site.13
To further this work, we primarily focused on rhabdomyosarcoma (RMS), a malignant soft tissue sarcoma most commonly occurring in children.15 Two major forms of RMS are recognized: a fusion-positive (FP) form typically harboring a balanced translocation generating neomorphic oncogenic fusion proteins PAX3-FOXO1 or PAX7-FOXO1, and a fusion-negative (FN) subtype often associated with RTK/RAS/MAPK pathway abnormalities.16,17 Mouse and cell culture-based models of PAX3-FOXO1 and RAS-driven RMS show the importance of CDKN2A and p53 as suppressors in both forms of the disease.18–20 Nucleic acid sequencing rarely shows deleterious mutations in CDKN2A/B, RB1, or p53 genes,17 and CDKN2A/B copy-number loss has been reported in only approximately 10% of the cases.17,21 As such, RMS formation or progression may depend on silencing CDKN2A/B transcription, creating a potentially reversible dependency.
We now report a previously unrecognized role for a methyltransferase acting through a 20 kb cis enhancer to govern TGFβ-dependent induction of CDKN2A/B transcripts in RMS cells. Using models of fusion-positive and negative RMS, we demonstrate the capacity to restore expression of two of the three CDKN2A/B transcripts with pharmacological and molecular approaches that have the potential to be more precisely applied as therapeutic strategies.
RESULTS
Assessment of CDKN2A/B Regulation in RMS
CDKN2A and CDKN2B genes, located at human chromosome 9p21, represent an unusual genomic locus that controls both RB and p53 activities. Conserved in mammals and many other amniotes,22 CDKN2A transcription can yield two different transcripts with first exons separated by over 10 kb (Fig. 1A). One of the CDKN2A transcripts and CDKN2B encode closely related members of the INK4 family proteins, p16INK4A and p15INK4B, that block cyclin D-dependent kinases 4 and 6.23,24 CDKN2A produces a second transcript from exon 1β, and it is translated in an alternate reading frame to generate p14ARF (or p19Arf in the mouse).25 This protein is thought to primarily activate p53 by sequestering MDM2,26 but we and others have shown the mouse version also exerts p53-independent effects that can control cell proliferation8 and suppress Pdgfrβ expression during late stages of eye development,9 activities essential for normal vision.7,8
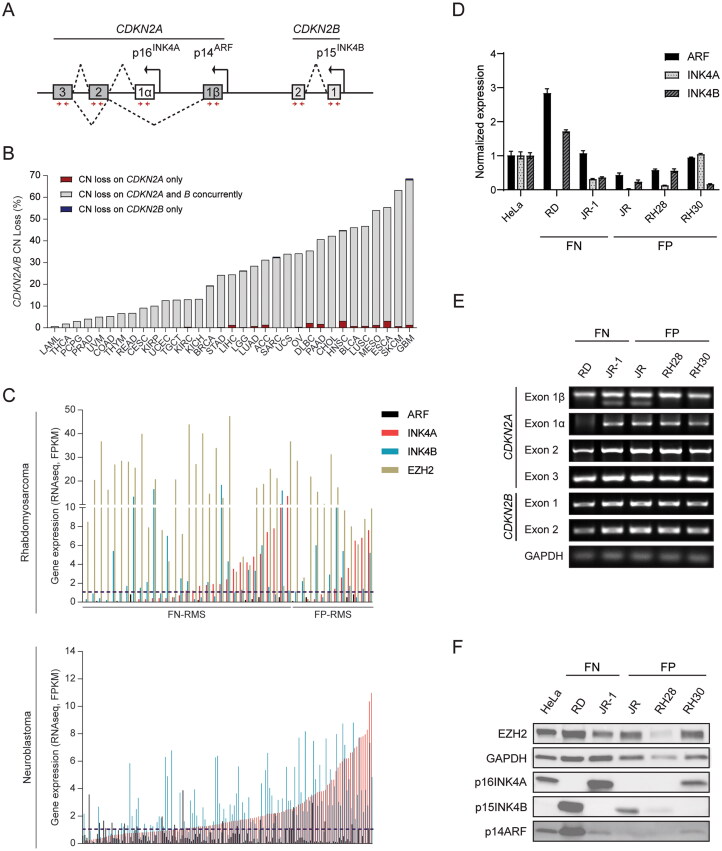
FIG 1.
Assessment of CDKN2A/B regulation in RMS and TCGA tumors. (A) Schematic diagram showing CDKN2A/B locus, which encodes three tumor suppressor proteins p14ARF, p16INK4A, and p15INK4B. (B) Quantitative analysis showing the percentage of copy-number loss in the CDKN2A and B locus across 33 human cancers from TCGA database. (C) ARF, INK4A, INK4B, and EZH2 expression from RNA-seq data of RMS (top) and NBL (bottom) specimens in which CDKN2A/B is intact. Gene expression is presented as FPKM (fragments per kilobase of transcript per million mapped reads). (D) Quantitative analysis of mRNA expression of CDKN2A/B locus in tested RMS cell lines in comparison to HeLa cells. Gene expression of the indicated genes were normalized to the one in HeLa cells. (E–F) The analysis of (E) genome integrity of CDKN2A/B locus (PCR image) and (F) the basal levels of EZH2, p16INK4A, p15INK4B, p14ARF protein (Western blot) in tested RMS cell lines as compared to HeLa cells. The primer sets (red arrows) used for detecting genome integrity are located within the exons of CDKN2A/B and indicated in the schematic diagram A. ACC, adrenocortical carcinoma; BLCA, bladder carcinoma; BRCA, breast invasive carcinoma; CESC, cervical and endocervical cancers; CHOL, cholangiocarcinoma; COAD; colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, low-grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian cancer; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; FN, fusion-negative; FP, fusion-positive. Error bars: ±SEM.
Disruption of CDKN2A/B encoded proteins is common in human cancer. Analyzing data generated for 33 different forms of cancer in The Cancer Genome Atlas (TCGA) program of the NCI revealed that the two genes display wide ranging gene copy-number loss frequencies from very rare in LAML, THCA, and PCPG to very common in ESCA, SKCM, and GBM (Fig. 1B). In the vast majority of cases, copy-number loss involved both CDKN2A and B, implying that transcripts encoded by both are important in tumor suppression.
CDKN2A/B also must be bypassed in RMS. Our reanalysis of SNP array or next-generation DNA sequencing from 258 RMS specimens showed that 39 (15%) contained CDKN2A/B deletion, which is consistent with previous reports.17,21 Focusing on 42 RMS cases without CDKN2A/B copy-number loss, we found that a) ARF expression was essentially undetectable (FPKM < 1) in all cases of FN and FP RMS; b) INK4A and INK4B mRNA was < 2 FPKM in at least 50% of each RMS sub-type (Fig. 1C, upper panel). Statistical analysis showed that expression of each transcript subtype did not significantly correlate with expression of the others across the 42 cases (see Table S1 in the supplemental material).
Analyzing a panel of human RMS cell lines revealed similar findings. In FN (RD, JR-1) and FP (JR, RH28, RH30) RMS lines, expression of the CDKN2A/B transcripts varied, but it was mostly low when compared to HeLa cells (Fig. 1D). Moreover, only RD and JR-1 displayed RAS gene mutation, both generating the N-RASQ61H variant, but the presence of oncogenic RAS did not correlate with CDKN2A/B expression. Only the RD cell line showed deletion of one of the CDKN2A/B exons (exon 1α), which correlated with no detectable INK4A expression (Fig. 1E and F). We conclude that CDKN2A/B genes were commonly deleted in many forms of cancer, and the expression of CDKN2A/B transcripts, especially ARF and INK4B, was commonly silenced in RMS. The relatively low expression in these forms of cancer, including those with RAS mutation, supports our hypothesis for active silencing of the transcripts. We note the relatively high expression of EZH2 mRNA and protein in the RMS specimens and cell lines (Fig. 1C, upper panel, and F). This suggests that it may play a role in CDKN2A/B silencing.
To evaluate whether CDKN2A/B silencing without gene copy-number loss is found in other childhood cancers, we explored neuroblastoma (NBL), a malignant neoplasm most often arising from the adrenal glands in young children.27 Our analysis of data from 195 NBL cases revealed CDKN2A/B copy-number loss in 29%. As in RMS, NBL cases without evidence for copy-number loss showed very low ARF expression in nearly all cases, and variable silencing in INK4A and INK4B in a majority (Fig. 1C, lower panel). In the 139 NBL cases without CDKN2A/B copy-number loss, expression of ARF was again not correlated with the other two transcripts, while INK4A and INK4B showed moderate correlation in expression (r = 0.52) (see Table S1 in the supplemental material). Hence, finding ways to restore expression of these silenced tumor suppressors could have ramifications beyond RMS.
Inhibiting Methyltransferase Activity Restores CDKN2A/B Induction by TGFβ in RMS Cells
Given previous findings in the developing mouse and mouse fibroblasts10–12,28 and our studies of HeLa cells,13 we addressed whether TGFβ could boost ARF and INK4A/B expression in RMS. We confirmed that the TGFβ signaling was intact in RD cells and four additional RMS lines, as evidenced by statistically significant induction of SMAD7 mRNA following exposure to TGFβ for 72 h (Fig. 2A). Despite this, exposure to TGFβ failed to induce INK4A or ARF in any line and led to only a very small increase in INK4B expression in three of the five tested lines (Fig. 2B to D).
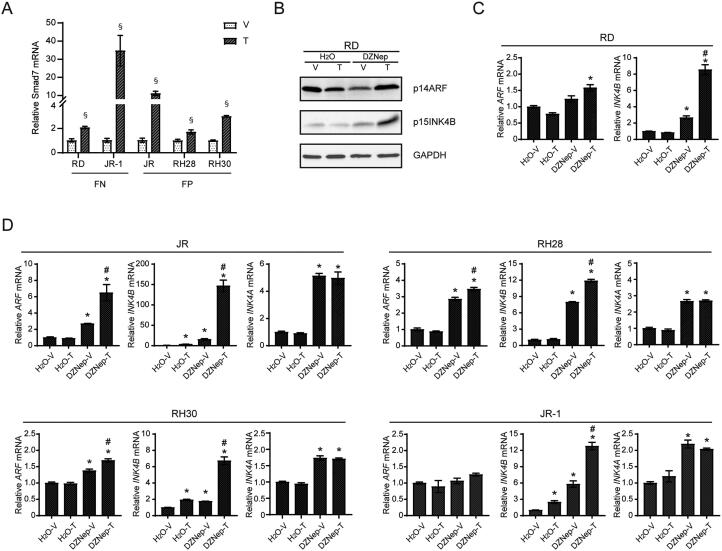
FIG 2.
Inhibition of methyltransferases with DZNep treatment on CDKN2A/B induction by TGFβ in RMS cells. (A) Quantitative analysis of Smad7 mRNA expression in tested RMS cell lines treated with vehicle or TGFβ for 72 h. (B–C) The effect of DZNep treatment on further ARF and INK4B induction by TGFβ in RD cells was determined by (B) protein levels through Western blots and (C) mRNA expression through qRT-PCR. (D) Quantitative analysis of mRNA expression of the indicated genes in tested RMS cell lines preincubated with DZNep following with TGFβ induction. For quantitative mRNA expression in panels C and D, gene expression of the indicated genes are normalized to the “H2O-V” group. V, Vehicle; T, TGFβ. Error bars: ±SEM. §P < 0.05, T vs. V; *P < 0.05, greater than “H2O-V”; #P < 0.05, greater than “DZNep-V”.
We previously showed that increased histone H3K27 acetylation in cis DNA elements correlated with TGFβ driven induction of INK4B and ARF in HeLa cells.13 H3K27 tri-methylation is known to reverse the chromatin structure effects of H3K27 acetylation,29 and EZH2 is widely recognized for its capacity to drive H3K27 methylation as part of the Polycomb Repressive Complex (PRC) 2.30 Interestingly, a Bayesian analysis pipeline was used to nominate potential RMS drivers by integrating gene copy-number and expression; genes with coordinate copy-number and expression gains were predicted to encode RMS drivers.31 EZH2 was among 25 candidates, and its importance was validated in a CRISPR/Cas9-based mini-pool screen with two RMS cell lines.31 Based on this and other reports of EZH2 in RMS,32,33 we explored whether it might contribute to CDKN2A/B silencing in this tumor.
As a first step to investigate whether methyltransferase activity might render RMS cell lines “nonresponsive” with respect to CDKN2A/B induction by TGFβ, we tested 3-deazaneplanocin A (DZNep), which inhibits S-adenosylhomocysteine synthesis,34 decreases EZH2 expression,35 and broadly blocks histone methylation.36 RD cells exposed to DZNep for 7 days displayed lower EZH2 expression and H3K27me3 (Fig. 3A, left panel). DZNep modestly increased basal expression of ARF or INK4B at protein or mRNA level, and it more substantially boosted the ability of TGFβ to further elevate expression of these genes (Fig. 2B and C).
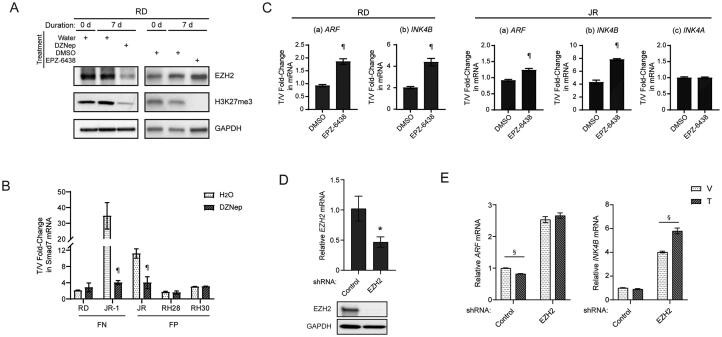
FIG 3.
The effects of HMT inhibitor specific to EZH2 on restoration of CDKN2A/B induction by TGFβ in RMS cells. (A) Representative Western blots for the indicated proteins using lysate from RD cells incubated with or without HMT inhibitors for 7 days. (B) Quantitative analysis of Smad7 mRNA expression in tested RMS cell lines preincubated with DZNep following with TGFβ induction. (C) Quantitative analysis of mRNA expression of the indicated genes in RD and JR cells preincubated with EPZ-6438 following with TGFβ induction. For quantitative mRNA expression in panels B and C, gene expression of the indicated genes are presented as fold change over expression in vehicle treated cells. (D) Quantitative analysis of EZH2 expression following targeted shRNA knockdown through qRT-PCR (top) and Western blotting (bottom) in RD cells. (E) Quantitative analysis of mRNA expression of the indicated genes with RD cells stably expressing either shRNA control or shRNA targeting EZH2, followed by vehicle or TGFβ treatment for 72 h. V, Vehicle; T, TGFβ. Error bars: ±SEM. ¶P < 0.05, “HMT inhibitor” vs. “control”; *P < 0.05, Control vs EZH2 shRNA; §P < 0.05, T vs. V.
We found similarities and differences when we extended these studies to the other RMS cell lines in our panel (Fig. 2D). Consistent with RD cells, DZNep pretreatment boosted the expression of INK4A and INK4B transcripts in all lines to varying degrees, and ARF in three of them. TGFβ induced ARF and INK4B after pretreatment with DZNep to a degree that was greater than (JR and RH30) or similar (RH28, JR-1) to that achieved by TGFβ alone. We found no evidence for TGFβ-dependent induction of INK4A, with or without DZNep pretreatment, similar to our studies of HeLa cells.13 Of note, DZNep did not enhance TGFβ induction of SMAD7; indeed, in JR-1 and JR cells, it seemed to impair a robust induction of this gene by DZNep alone (Fig. 3B).
We considered whether the effect of DZNep could be mimicked by a histone methyltransferase inhibitor that is specific to EZH2 and its deposition of H3K27me3, previously implicated as a regulator of Cdkn2a/b4 and an oncogenic driver in RMS,32,33 as mentioned above. We utilized EPZ-6438, which has 35-fold selectivity for EZH2 over EZH1 and > 4500-fold selectivity relative to other tested histone methyltransferases.37 Pretreatment of RD cells with EPZ-6438, at a dose that blocks H3K27me3 deposition without altering EZH2 protein level (Fig. 3A, right panel), significantly enhanced ARF and INK4B induction by TGFβ (Fig. 3C, left panel). In JR cells, we also observed the improved induction of ARF and INK4B, and as in the DZNep studies, the induction of INK4A was not changed by EPZ-6438 pretreatment (Fig. 3C, right panel). Finally, shRNA-mediated EZH2 knockdown in RD cells enhanced basal ARF and INK4B expression, and modestly boosted INK4B induction by TGFβ (Fig. 3D and E). Taken together, we conclude that TGFβ induction of ARF and INK4B can be restored or further augmented in some RMS cell lines by preexposure to methyltransferase inhibitors, including one specific to EZH2. We note the relatively greater effect on INK4B than ARF, and the inability of TGFβ to induce INK4A even though methyltransferase inhibition can elevate its basal level. This indicates that the molecular basis controlling expression of each transcript may still contain distinct elements. The relatively greater effect of DZNep versus EZH2-specific inhibition or knockdown suggests that other methyltransferases may contribute to render CDKN2A/B in RMS cells “nonresponsive” to TGFβ.
Distant Enhancers Are Required to Restore TGFβ-Mediated, CDKN2A/B Induction in RMS Cells
We previously defined a 20 kb cis element required for TGFβ-dependent induction of ARF and INK4B in HeLa cells, and TGFβ fostered H3K27 acetylation at three peaks within this region13 (Fig. 4A). In contrast to HeLa cells, TGFβ did not measurably change the very low level of H3K27Ac at these sites in either RD or JR cells (black versus gray line; Fig. 4B and C, top panels). Preincubation with DZNep showed a slight increase in H3K27Ac at these peaks in both cell lines, but subsequent exposure to TGFβ dramatically enhanced it in three peaks (red and blue lines, respectively; Fig. 4B and C, top panels), as in HeLa cells.13
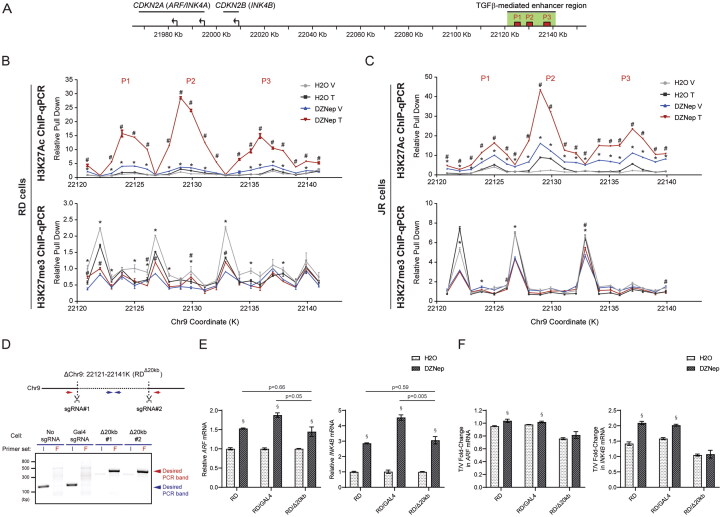
FIG 4.
The TGFβ-mediated enhancer region on CDKN2A/B reactivation followed by epigenetic modification in RMS cells. (A) Diagram indicating the CDKN2A/B locus, together with the TGFβ-mediated enhancer region including enhancer peaks identified previously (denoted as P1–3)13 on the chromosome 9p21. (B-C) H3K27Ac and H3K27me3 ChIP-qPCR across the TGFβ-mediated enhancer region identified in the previous study with 1 kb resolution. The ChIP assay was performed in either (B) RD or (C) JR cells preincubated with or without DZNep for 7 days following vehicle or TGFβ treatment for another 72 h. Pull down signal is presented as relative pull down normalized against signal obtained at coordinate: 22121 k in “H2O-V” group. The data were obtained from single experiment with triplicate samples in qPCR analysis. (D) RDΔ20kb cell clones were generated through CRISPR/Cas9-mediated homologous recombination. Deletion of 20 kb region with the sgRNA pair and its further validation through PCR with assigned primer sets were followed by previous design13 and highlighted in the schematic diagram. The representative gel indicated that compared to the control strains, the deletion clones can only amplify flanking (red arrow) but not internal (blue arrow) signal, suggesting homozygous deletion of the clones. (E-F) Quantitative analysis of mRNA expression of the indicated genes in RDΔ20kb cells preincubated with or without DZNep for 7 days following vehicle or TGFβ treatment for another 72 h, compared to control strains (RD and RD/Gal4 cells). In panel E, gene expression of the indicated genes are only shown with vehicle treated cells and normalized to the one in control (H2O) group, to address the effect of DZNep treatment on baseline expression of the genes. mRNA expression is presented as T/V fold-change with expression in TGFβ treated cells over one in vehicle treated cells in panel F, to address the effect of DZNep treatment on TGFβ-mediated ARF/INK4B induction. V, Vehicle; T, TGFβ. Error bars: ±SEM. *P < 0.05, DZNep V vs H2O V; #P < 0.05, DZNep T vs DZNep V; §P < 0.05, DZNep vs H2O.
We explored whether H3K27 tri-methylation at these peaks might be altered by DZNep because very little H3K27me3 was reported in this region in HeLa cells,38 in which TGFβ induces ARF and INK4B without methyltransferase inhibition.13 ChIP-qPCR showed that H3K27me3 is mostly deposited at sites flanking P1, P2, and P3 in RD and JR cells, and the methylation decreases following DZNep but is not consistently altered by TGFβ with or without DZNep pretreatment (gray versus blue line; Fig. 4B and C, lower panel).
To further explore how the 20 kb region contributed to DZNep and TGFβ effects in RD cells, we used CRISPR/Cas9 to generate a targeted deletion of the region (ΔChr9:22121-22141K, Δ20 kb), which we confirmed to be deleted in two separate clones (Fig. 4D). Comparing these clones to parental cells and controls in which nontargeting GAL4 sgRNA was used showed that the ability of DZNep to induce the basal expression of ARF and INK4B was not affected by the deletion (Fig. 4E). In contrast, loss of the 20 kb enhancer erased the induction of these transcripts by TGFβ (Fig. 4F). We conclude that the 20 kb region contributes to TGFβ-driven induction of ARF and INK4B in RMS, but it is not required for a broadly acting histone methyltransferase inhibitor to augment their basal expression.
CRISPRa-Mediated ARF and INK4B Promoter Activation Decreases RMS Cell Accumulation
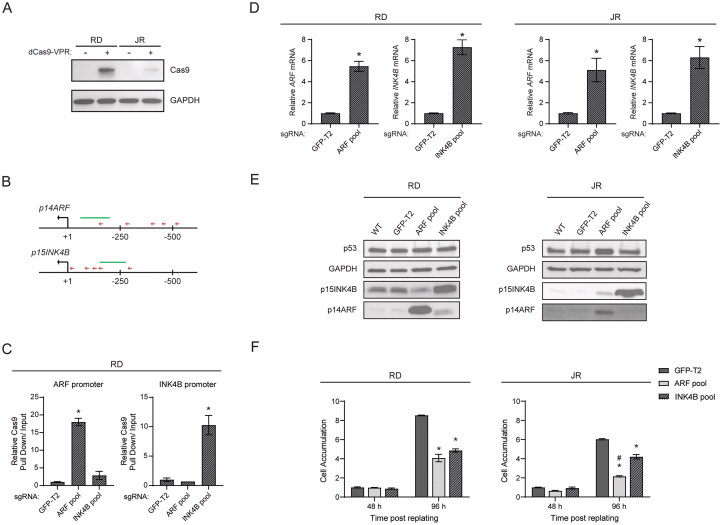
To begin to understand the potential to “precisely” reprogram CDKN2A/B expression in RMS, we explored whether we could use a CRISPR-based activation (CRISPRa) system to directly induce ARF or INK4B expression. In this system, sgRNAs guide the nuclease-dead Cas9 (dCas9) that has been modified to include a transactivation motif.39,40 We utilized dCas9-VPR, which contains reiterated VP16 motifs (VP64) and two other transactivation domains, p65 and Rta,41 and can be stably expressed using a lentiviral vector in RD and JR cells (Fig. 5A). We designed sgRNAs to target regions 40–500 bp upstream of the transcriptional start sites of ARF and INK4B (Fig. 5B), and we used a pool of lentiviral vectors to deliver those guides or a nontargeting sgRNA (GFP-T2) as a control. Five days following transduction with the aforementioned vectors, we confirmed by ChIP-qPCR that dCas9-VPR was recruited to the ARF and INK4B promoters in response to the expression of the respective sgRNA pools but not the nontargeting control (Fig. 5C). At that time point, expression analyses showed a 5- to 7-fold induction in ARF and INK4B mRNA and their respective proteins in both RD and JR cells in comparison to the nontargeting control (Fig. 5D and E).
FIG 5.
CRISPRa-mediated, promoter activation of ARF/INK4B in RMS cells. (A) Representative Western blots using anti-Cas9 antibodies to detect dCas9-VPR expression with lysate prepared from transduced RD and JR cells stably expressing dCas9-VPR. (B) A schematic diagram indicating the position of sgRNAs (red arrows), which were used for CRISPR activation of the ARF and INK4B locus, relative to the transcription starting site of corresponding genes. (C) Quantitative analysis of representative ChIP assays by using anti-Cas9 antibodies to validate the recruitment of dCas9-VPR to the promoter region of ARF and INK4B. ChIP was performed using RD cells stably expressing dCas9-VPR and sgRNAs targeting either ARF promoter (ARF pool), INK4B promoter (INK4B pool), or GFP-T2 (nontargeting control). Immunoprecipitated DNA and input DNA were amplified with primers for genomic regions (green lines) indicated in the schematic diagram B, and the pull down signal of the indicated locus is presented as relative pull down normalized against signal obtained in “GFP-T2” group. (D) Quantitative analysis of mRNA expression of the indicated genes in RD and JR cells following CRISPRa-mediated ARF/INK4B activation. Gene expression of the indicated genes are normalized to the “GFP-T2” group. (E) Representative Western blots for the indicated proteins using lysate from RD and JR cells following CRISPRa-mediated ARF/INK4B activation. (F) Cell proliferation assays of RD and JR cells following CRISPRa-mediated ARF/INK4B activation. Cells were counted at 48 and 96 h postreplating and further normalized to the “GFP-T2” group at 48 h. Error bars: ±SEM. *P < 0.05, vs GFP-T2; #P < 0.05, ARF pool vs INK4B pool.
We then tested whether restoring CDKN2A/B expression by CRISPRa could limit cell accumulation in vitro. To do this, we cultured RD-dCas-VPR and JR-dCas-VPR cells for four days in puromycin after delivering the sgRNA pool, at which point cells were harvested and replated at low density to monitor cell accumulation 48 and 96 h later. No significant difference was observed after 48 h, but the number of cells was significantly lower at 96 h (Fig. 5F). Targeting ARF had a significantly greater effect in JR cells than RD (Fig. 5F, right panel). This difference correlated with multiple common p53 mutations, such as mutp53R248W and mutp53P72R, in RD but not JR cells, and with a greater induction of p53 protein in JR (Fig. 5E). We used EdU pulse labeling to explore whether the decreased accumulation at 96 h correlated with decreased progression through S-phase. We found no difference in the percent of EdU-positive cells in the control versus experimental plates at 96 h [54, 53, and 57% positive (RD) and 57, 53, and 53% positive (JR), for GFP-T2, ARF, and INK4B pools, respectively (P > 0.05)], although the overall number of Hoechst + nuclei in the plates decreased (CR and SXS; negative data not shown). Together, our findings indicate that “precise” re-induction of native ARF/INK4B in established tumor cell lines can limit accumulation without measurably altering G1-S phase progression.
CRISPR-Dependent Modifications of TGFβ-Dependent Enhancers Influence ARF and INK4B Expression in HeLa and RMS Cell Lines
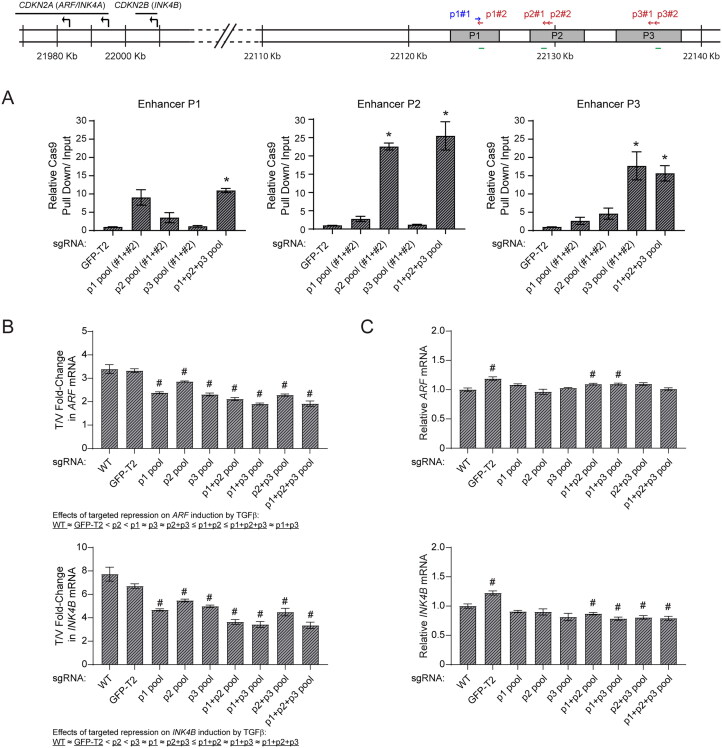
We next explored whether CDKN2A/B gene expression could be restored by localizing dCas9-VPR to the silenced remote enhancers required for TGFβ-dependent ARF and INK4B induction. As there were three distinct H3K27Ac peaks located within the 20 kb enhancer region (Fig. 4A to C), we first used HeLa cells and a CRISPR interference (CRISPRi) approach to test whether one or more were dominant with respect to CDKN2A/B induction by TGFβ. Here we employed dCas9 fused to a Kruppel-associated box (KRAB) motif (dCas9-KRAB),42 which we stably expressed in HeLa cells. We designed two sgRNAs targeting each of the three enhancer peaks, and we showed by ChIP-qPCR that their expression as either a “mini-pool” targeting one individual peak or as a pool targeting all three peaks localized dCas-KRAB to the respective enhancer (Fig. 6A). We found the induction of ARF and INK4B mRNA by TGFβ to be significantly blunted when dCas9-KRAB was targeted to any one of the enhancer peaks, but the magnitude of blunting seemed least for P2, even though dCas9-KRAB was targeted most robustly to that peak (Fig. 6A and B). Similarly, targeting P2 did not seem to add to the repressive effects of targeting P1, P3, or both on ARF and INK4B induction by TGFβ (Fig. 6B). Localizing dCas9-KRAB to each of the three enhancer peaks had a modest, if any, effect on baseline expression of ARF or INK4B mRNA (Fig. 6C), which is also consistent with our previous findings when the 20 kb region was deleted in HeLa cells.13 We conclude that, in HeLa cells, localizing a trans repressor to cis enhancer elements, particularly P1 and P3, limited ARF and INK4B mRNA induction by TGFβ without affecting basal expression.
FIG 6.
Assessment of enhancer dependency within the TGFβ-dependent region on ARF/INK4B expression through CRISPRi in HeLa cells. (A) Quantitative analysis of representative ChIP assays by using anti-Cas9 antibodies to validate the recruitment of dCas9-KRAB to the previously defined enhancer peaks (P1, P2, and P3) located within TGFβ-dependent region. ChIP was performed using HeLa cells stably expressing dCas9-KRAB and sgRNAs targeting enhancer peaks either individually (P1, P2, and P3 pool) or concurrently (P1 + P2 + P3 pool), together with the one targeting GFP-T2 as a nontargeting control. The position of designed sgRNAs (arrows) in relation to enhancer peaks and genomic regions (green lines) where primers were designed to amplify immunoprecipitated DNA and input DNA are indicated in the schematic diagrams above. Pull down signal of the indicated locus is presented as relative pull down normalized against signal obtained in the “GFP-T2” group. (B-C) Quantitative analysis of mRNA expression of the indicated genes in HeLa cells with assigned enhancer peaks silenced through CRISPR interference, following vehicle or TGFβ treatment for 72 h. In the panel B, mRNA expression is presented as T/V fold-change with expression in TGFβ treated cells over one in vehicle treated cells, in comparison to the value obtained from control CRISPRi strains with either no sgRNA (WT) or nontargeting sgRNA (GFP-T2) expression. In the panel C, gene expression of the indicated genes are only shown with vehicle treated cells and normalized to the wild-type (WT) group, to address the effect of targeted enhancer silencing on baseline expression of the genes. Error bars: ±SEM. *P < 0.05, vs GFP-T2; #P < 0.05, vs WT.
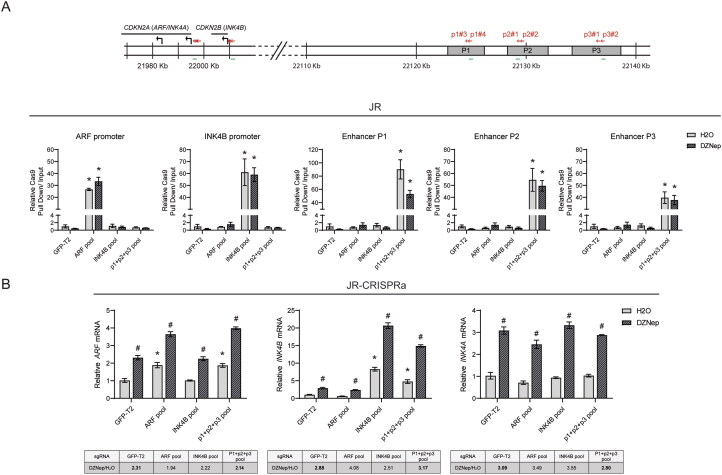
Having concluded that the individual enhancer peaks can contribute to TGFβ-driven expression of ARF and INK4B in HeLa cells, and that CRISPRa could induce these transcripts when targeted to promoter elements in RMS cell lines, we explored whether CRISPRa could achieve the same effect when targeting the silenced cis enhancers alone or following exposure to DZNep. As a first step, we confirmed that a pool of guides targeting P1, P2, and P3 could localize the dCas9-VPR protein to each enhancer element. With the notable exception of the P1 enhancer, DZNep did not substantially impede or enhance dCas9-VPR localization (Fig. 7A). Targeting dCas9-VPR to the three enhancers boosted the expression of ARF and INK4B mRNA to approximately the same degree as targeting the respective promoters (Fig. 7B, left two panels). DZNep pretreatment again elevated expression of ARF and INK4B mRNA, and CRISPRa targeting of the P1, 2, and 3 enhancers further elevated their expression. Finally, like TGFβ which fails to boost the expression of INK4A, artificially activating these enhancers did not induce INK4A expression even after DZNep exposure (Fig. 7B, right panel). This indicated that the selective action of TGFβ on ARF and INK4B depended on molecular mechanisms intrinsic to the enhancers and unrelated to signals emanating from TGFβ.
FIG 7.
CRISPRa-mediated activation of TGFβ-dependent enhancers on ARF/INK4B expression in JR cells. (A) Quantitative analysis of representative ChIP assays by using anti-Cas9 antibodies to validate the recruitment of dCas9-VPR to the assigned genomic regions. ChIP was performed using CRISPRa strains, including sgRNAs targeting the promoter regions of ARF and INK4B (ARF pool and INK4B pool), remote enhancers required for ARF/INK4B induction by TGFβ (P1 + P2 + P3 pool), or GFP-T2 (nontargeting control), followed by treatment of DZNep or H2O. The position of designed sgRNAs (red arrows) for CRISPR activation and the genomic regions (green lines) where primers were designed to amplify immunoprecipitated DNA and input DNA are indicated in the schematic diagrams above, and the pull down signal of the indicated locus is presented as relative pull down normalized against signal obtained in the nontargeting control group with treatment of H2O (GFP-T2 with H2O). (B) Quantitative analysis of mRNA expression of the indicated genes in JR cells with assigned promoter or enhancer regions activated through CRISPRa, following DZNep or H2O treatment for 7 days. Gene expression of the indicated genes are normalized to the “GFP-T2 with H2O” group. To evaluate whether DZNep treatment facilitates CRISPRa-mediated gene activation, the value of “DZNep/H2O fold-change” with expression in DZNep treated cells over one in H2O treated cells is calculated and listed as the table below the column chart. Error bars: ±SEM. *P < 0.05, vs “GFP-T2 with H2O”; #P < 0.05, DZNep vs H2O.
DISCUSSION
The role that TGFβ plays in cancer is confusing in that many papers have shown its capacity to either promote or arrest cancer formation or progression. Our findings shed some new light on this problem. The mouse Arf transcript, encoded by Cdkn2a orthologous to human CDKN2A, is essential for vascular remodeling in the primary vitreous space during eye development.8 In addition to developing cancer, Arf null mice are blind.7,8 The expression of Arf was initially thought to be silenced during mouse embryo development, but it is actually exquisitely controlled in a spatially and temporally restricted pattern that is distinct from Ink4a expression.7–9 TGFβ2 is essential for Arf expression during eye development,10 and a distal cis regulatory element residing in the gene desert flanking Cdkn2a and Cdkn2b is required for TGFβ-driven Arf induction.12 The mouse TGFβ-ARF signaling pathway, including a requirement for a distant, cis enhancer, is conserved in human Hela cells, but TGFβ does not directly induce CDKN2A/B transcripts in most human cancer cell lines we tested.13 We now show that blocking histone methyltransferase activity using either broadly acting or EZH2-specific inhibitors restores TGFβ responsiveness. The effect was specific to ARF and INK4B, not INK4A, and INK4B induction was greatest. We narrowed the cis regulatory element to a 20 kb region in which TGFβ activates a de novo enhancer evidenced by new H3K27 acetylation marks. This same enhancer element was found in HeLa cells which shows conservation of this regulatory region across different cell lineages. Methyltransferase inhibition and EZH2 knockdown boosted basal expression of all three CDKN2A/B transcripts, but TGFβ and directly targeting the enhancers with dCas-VPR only induced ARF and INK4B. Other recent papers have addressed crosstalk between histone or DNA methylation and TGFβ effects on cancer,43–45 but ours is the first to implicate histone methylation of a remote enhancer in governing anticancer effects of the TGFβ pathway.
The noncoding segment of DNA flanking CDKN2A and CDKN2B has been implicated in a number of human diseases. For example, genome-wide association studies link SNPs to risk for coronary artery disease,46–48 type 2 diabetes mellitus,49,50 and Alzheimer’s disease.51,52 As introduced above, the knockout of a 70 kb noncoding mouse DNA segment orthologous to the coronary artery disease risk interval increased weight gain and lowered survival when animals were fed a high fat/high calorie diet.53 Subsequently, those animals were shown to display an ocular developmental defect reminiscent of a childhood disease, persistent hyperplastic primary vitreous (PHPV) (sometimes known as persistent fetal vasculature).12 The closest mouse and human protein-coding genes are CDKN2A and CDKN2B, residing approximately 60 kb from the most prominent SNPs,47 and mouse Cdkn2a/b transcripts are lower in the heart and the primary vitreous in mice lacking the CAD risk interval.12,53 Loss of Arf expression in that model, like direct Arf knockout in the mouse, directly leads to primary vitreous expansion by derepressing Pdgfrb expression,54 which results in excess vascular endothelial cell proliferation.9
The cis enhancer we characterized in HeLa and RMS cells lies in a large gene desert increasingly implicated in gene regulation. The first insights for this stemmed from finding a long noncoding RNA, CDKN2BAS/ANRIL that is transcribed from the ARF promoter in the opposite direction.55 Early mechanistic studies showed that ANRIL localized the PRC2 complex to silence CDKN2A/B expression.56 However, the aforementioned 70 kb deletion in the mouse included the lncRNA and further repressed Cdkn2a/b, which suggested the existence of cis enhancers in the region. Our and others’ reports support that concept. For example, a recent paper provided bioinformatics and functional evidence for super enhancer clusters within a topologically associating domain that spans that region.38 Detailed mechanistic studies show the interdependence of a distinct subset of these enhancers which support the expression of all three CDKN2A/B transcripts.38 When deleted or silenced by a CRISPR/Cas9-based approach, EZH2 is enriched at the ARF, INK4A, and INK4B promoters, and their expression is silenced. Our findings further this work by showing that specific enhancer elements (P1, P2, and P3) can be engaged by TGFβ in RMS cells (like HeLa cells), but only after blocking histone methylation. Publicly available 4 C data from HeLa cells38 does not support the physical interaction of this 20 kb region to the CDKN2A/B locus, which is consistent with an inactivated status of the 20 kb enhancers without TGFβ.13,38 Further experiments will be needed to prove whether TGFβ leads to conformational changes bringing the enhancers to INK4B and ARF promoters and to define mechanisms that exclude INK4A induction. We anticipate that such studies can help clarify the discordant expression of these tumor suppressors in cancers, their capacity to be induced therapeutically, and how sequence variants in the noncoding region may influence phenotypes as disparate as type 2 diabetes and Alzheimer’s disease.
Our findings may also have relevance for human cancer biology. The 9p21 region is among the most commonly deleted genes in all of human cancer, and as we showed, most deletions disrupt CDKN2A and CDKN2B coding sequence. That the region is so commonly deleted underscores its general importance as a barrier to cancer formation and/or progression, but it also raises the question of how CDKN2A/B is bypassed when not deleted. Indeed, for RMS and neuroblastoma, ARF is essentially universally silenced, and most cases have no or low expression of INK4B and/or INK4A, as well. Pharmacologically restoring their expression is not a new concept. Inhibiting DNA and histone methylation has been explored in many human cancers, including RMS.57,58 However, those approaches have not substantially influenced clinical care, perhaps because the beneficial effects of such manipulations are likely to be dictated by the underlying state of the genes (e.g., deleted or intact) and by the epigenetic state of key enhancers. With that in mind, we predict that EZH2 inhibitors, or a more broadly acting methyltransferase inhibitor, might be more effective in cases in which (a) CDKN2A/B genes are intact, (b) ARF/INK4B expression is low, (c) p53 and RB1 genes are intact, (d) cis enhancers show H3K27 tri-methylation, and (e) there is evidence for endogenous TGFβ signaling. These variables could realistically be developed into a multianalyte predictive test using molecular or pathological tools that are readily available. We also recognize that merely boosting expression of INK4B or ARF may not be sufficient to fully activate either RB1 or p53-dependent cell cycle arrest, senescence, or apoptosis. Additional mechanistic and preclinical studies combining “smart” epigenetic treatments with low-dose cytotoxic agents may reveal more effective and tolerable therapies than the maximally intensive cytotoxic approaches used for RMS today.15
MATERIALS AND METHODS
Datasets for Gene Copy-Number and Expression
Copy-number variation (CNV) data across many types of human cancers were obtained from the National Cancer Institute (NCI) TCGA program through the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Custom PERL scripts were then generated to compile and calculate the frequency of copy-number loss on CDKN2A and B locus, individually or concurrently, among 33 different cancer types.
To address CDKN2A/B deletion in RMS, 258 RMS specimens containing SNP array or whole-genome sequencing (WGS) data were compiled from three sources [NCI (n = 138), the Children’s Oncology Group (COG) (n = 102), and the University of Texas Southwestern (UTSW) (n = 26)]. Note that eight COG cases were also found in the NCI dataset, and the duplicates were excluded. CDKN2A/B expression was extracted through RNA-seq data from 42 RMS specimens downloaded from The European Genome-phenome Archive (EGA) with accession number EGAD00001000878. For CDKN2A/B regulation in neuroblastoma, 195 cases containing both WGS and RNA-seq data were gathered from the NCI Gabriella Miller Kids’ First program. Genomics and expression data analysis was performed as previously described.31
Cell Culture and Treatment
Rhabdomyosarcoma cell lines (RD, JR-1, JR, RH28, and RH30) and HeLa cells, validated by STR testing, either provided by P. Houghton or obtained from ATCC and previously reported,22 were cultivated in RPMI with 10% FBS supplemented with penicillin/streptomycin. Cell manipulation with TGFβ or vehicle control was performed as previously described.13 For inhibition of histone methylation, 3-deazaneplanocin A (DZNep; Selleckchem) or EPZ-6438 (tazemetostat; Selleckchem) was added to the medium to the final concentration 1 µM (except 5 µM DZNep in RD cells57 before or with the addition of TGFβ.
Western Blotting
Western blotting was performed as previously described9 with primary antibodies directed against p14ARF (#53640, Santa Cruz), p15INK4B (#271791, Santa Cruz), H3K27me3 (#9733, Cell Signaling), EZH2 (#39933, Active Motif), Cas9 (#SAB4200701, Sigma-Aldrich), HSC70 (#7298, Santa Cruz), and GAPDH (#47724, Santa Cruz). Chemiluminescent detection of proteins was visualized through the ChemiDoc XRS + imaging system (Bio-Rad).
Quantitative RT-PCR
Total RNA extraction and cDNA preparation were performed as previously described.12 qPCR analysis was performed using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) and the CFX96/CFX Opus 96 (Bio-Rad) for real-time PCR detection. RT-qPCR primer sequences are listed in Supplementary Table S2. Results are pooled from three individual samples.
ChIP-qPCR
Chromatin immunoprecipitation (ChIP) was carried out as described previously.11 Antibodies against H3K27Ac (#4729, Abcam), RNAPII (#47701, Santa Cruz), and Cas9 (#C15310258, Diagenode) were used for immunoprecipitation. Quantitative PCR analysis was performed as described above using primers listed in Supplementary Table S2.
CRISPR Based Experiments
CRISPR-based genomic deletion in RD cells, including the lentiviral expression vectors used, sgRNA design for targeted 20 kb deletion, and primer sets for PCR deletion screen, was carried out as previously described.13
For CRISPR activation (CRISPRa) and interference (CRISPRi), either dCas9-VPR (Edit-R CRISPRa lentiviral hCMV-Blast-dCas9-VPR, Horizon) or dCas9-KRAB (Lenti-dCas9-KRAB-blast, Addgene) expression vectors were stably expressed in tested cell lines through lentiviral transduction followed by blasticidin selection. Five or six individual sgRNA oligonucleotide sequences were designed using established algorithms59 to target either the human ARF/INK4B promoters within 40–500 bp upstream of the TSS of each gene or the center of the predicted enhancer region (P1, P2, P3). The sgRNA oligonucleotides, purchased from Sigma-Aldrich, were subcloned into the lentiviral expression vector, CROPseq-Guide(MS2)-Puro (Addgene), and lentivirus stocks were produced as described.60 To analyze how CDKN2A/B expression was affected by CRISPR/Cas9-based modification, cells were transduced with all sgRNAs targeting the region of interest and then selected with puromycin for 4 days. In cell accumulation assays, 5 × 104 cells per well were seeded in a six-well plate following transduction and selection and counted in three technical replicates at 48 and 96 h. The sgRNA oligonucleotide and PCR primer sets are listed in Supplementary Table S2.
Statistical Analysis
Quantitative data are presented as a bar graph with mean ± SEM for three individual representative experiments unless otherwise noted. Statistical significance was determined by Student’s t test. Gene expression correlation among CDKN2A/B transcripts is determined by correlation coefficient (r) pairwise and presented as correlation matrix listed in Supplementary Table S1.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge technical assistance provided by J. Aguayo and J. Liu, and many helpful comments by other members of the Skapek laboratory.
Funding Statement
We acknowledge financial support from the Cancer Prevention and Research Institute of Texas [RP180319] and the National Cancer Institute Cancer Center Support Grant [P30CA142543].
SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed online at https://doi.org/10.1080/10985549.2023.2186074
DATA AVAILABILITY STATEMENT
Copy-number variation (CNV) data were obtained from the National Cancer Institute (NCI) TCGA program using the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). CDKN2A/B expression in RMS and neuroblastoma was extracted through RNA-seq data from 42 specimens downloaded from The European Genome-phenome Archive (EGA) with accession number EGAD00001000878 (for RMS), and from 195 cases NCI Gabriella Miller Kids’ First program (https://commonfund.nih.gov/kidsfirst) (for neuroblastoma).
REFERENCES
- 1.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Flanders TY, Pollock PM, Hayward NK.. The CDKN2A (p16) gene and human cancer. Mol Med. 1997;3:5–20. doi: 10.1007/BF03401664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim WY, Sharpless NE.. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Gu X, Su I-h, Bottino R, Contreras JL, Tarakhovsky A, Kim SK.. Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 6.Kusy S, Larsen C-J, Roche J.. p14ARF, p15INK4b and p16INK4a methylation status in chronic myelogenous leukemia. Leuk Lymphoma. 2004;45:1989–1994. doi: 10.1080/10428190410001714025. [DOI] [PubMed] [Google Scholar]
- 7.Martin AC, Thornton JD, Liu J, Wang X, Zuo J, Jablonski MM, Chaum E, Zindy F, Skapek SX.. Pathogenesis of persistent hyperplastic primary vitreous in mice lacking the Arf tumor suppressor gene. Invest Ophthalmol Vis Sci. 2004;45:3387–3396. doi: 10.1167/iovs.04-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKeller RN, Fowler JL, Cunningham JJ, Warner N, Smeyne RJ, Zindy F, Skapek SX.. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc Natl Acad Sci U S A. 2002;99:3848–3853. doi: 10.1073/pnas.052484199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva RLA, Thornton JD, Martin AC, Rehg JE, Bertwistle D, Zindy F, Skapek SX.. Arf-dependent regulation of Pdgf signaling in perivascular cells in the developing mouse eye. Embo J. 2005;24:2803–2814. doi: 10.1038/sj.emboj.7600751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman-Anderson NE, Zheng Y, McCalla-Martin AC, Treanor LM, Zhao YD, Garfin PM, He T-C, Mary MN, Thornton JD, Anderson C, et al. Expression of the Arf tumor suppressor gene is controlled by Tgfβ2 during development. Development. 2009;136:2081–2089. doi: 10.1242/dev.033548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Zhao YD, Gibbons M, Abramova T, Chu PY, Ash JD, Cunningham JM, Skapek SX.. Tgfβ signaling directly induces Arf promoter remodeling by a mechanism involving Smads 2/3 and p38 MAPK. J Biol Chem. 2010;285:35654–35664. doi: 10.1074/jbc.M110.128959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y, Devitt C, Liu J, Iqbal N, Skapek SX.. Arf induction by Tgfβ is influenced by Sp1 and C/ebpβ in opposing directions. PLoS One. 2013;8:e70371. doi: 10.1371/journal.pone.0070371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-T, Xu L, Bennett L, Hooks JC, Liu J, Zhou Q, Liem P, Zheng Y, Skapek SX.. Identification of de novo enhancers activated by TGFβ to drive expression of CDKN2A and B in HeLa cells. Mol Cancer Res. 2019;17:1854–1866. doi: 10.1158/1541-7786.MCR-19-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werness B, Levine A, Howley P.. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 15.Skapek SX, Ferrari A, Gupta AA, Lupo PJ, Butler E, Shipley J, Barr FG, Hawkins DS.. Rhabdomyosarcoma. Nat Rev Dis Primers. 2019;5:1. doi: 10.1038/s41572-018-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saab R, Spunt SL, Skapek SX.. Chapter 7 – Myogenesis and rhabdomyosarcoma: the Jekyll and Hyde of skeletal muscle. In: Dyer MA, editors. Current topics in developmental biology. Vol. 94. San Diego (CA): Academic Press; 2011. p. 197–234. [DOI] [PubMed] [Google Scholar]
- 17.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM.. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65:4490–4495. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 19.Linardic CM, Naini S, Herndon JE, II, Kesserwan C, Qualman SJ, Counter CM.. The PAX3-FKHR fusion gene of rhabdomyosarcoma cooperates with loss of p16INK4A to promote bypass of cellular senescence. Cancer Res. 2007;67:6691–6699. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y-X, Finckenstein FG, Abdueva DA, Shahbazian V, Chung B, Weinberg KI, Triche TJ, Shimada H, Anderson MJ.. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68:6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–724. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil J, Peters G.. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 23.Hannon GJ, Beach D.. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 24.Serrano M, Hannon GJ, Beach D.. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 25.Quelle DE, Zindy F, Ashmun RA, Sherr CJ.. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. [DOI] [PubMed] [Google Scholar]
- 26.Michael D, Oren M.. The p53–Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/S1044-579X(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 27.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA.. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Devitt C, Liu J, Mei J, Skapek SX.. A distant, cis-acting enhancer drives induction of Arf by Tgfβ in the developing eye. Dev Biol. 2013;380:49–57. doi: 10.1016/j.ydbio.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calo E, Wysocka J.. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y.. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 31.Xu L, Zheng Y, Liu J, Rakheja D, Singleterry S, Laetsch TW, Shern JF, Khan J, Triche TJ, Hawkins DS, et al. Integrative Bayesian analysis identifies rhabdomyosarcoma disease genes. Cell Rep. 2018;24:238–251. doi: 10.1016/j.celrep.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, Giordano A.. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8:172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 33.Marchesi I, Fiorentino FP, Rizzolio F, Giordano A, Bagella L.. The ablation of EZH2 uncovers its crucial role in rhabdomyosarcoma formation. Cell Cycle. 2012;11:3828–3836. doi: 10.4161/cc.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glazer RI, Knode MC, Tseng CKH, Haines DR, Marquez VE.. 3-deazaneplanocin A: a new inhibitor of S-adenosylhomocysteine synthesis and its effects in human colon carcinoma cells. Biochem Pharmacol. 1986;35:4523–4527. doi: 10.1016/0006-2952(86)90774-4. [DOI] [PubMed] [Google Scholar]
- 35.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RKM, Tan PBO, Liu ET, Yu Q.. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA.. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Scott MP, Chesworth R, Moyer MP, Copeland RA, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farooq U, Saravanan B, Islam Z, Walavalkar K, Singh AK, Jayani RS, Meel S, Swaminathan S, Notani D.. An interdependent network of functional enhancers regulates transcription and EZH2 loading at the INK4a/ARF locus. Cell Rep. 2021;34:108898. doi: 10.1016/j.celrep.2021.108898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R.. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannigan A, Smith P, Kalna G, Lo Nigro C, Orang’ C, O'Brien DI, Shah R, Syed N, Spender LC, Herrera B, et al. Epigenetic downregulation of human disabled homolog 2 switches TGF-β from a tumor suppressor to a tumor promoter. J Clin Invest. 2010;120:2842–2857. doi: 10.1172/JCI36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S-H, Kim O, Kim H-J, Hwangbo C, Lee J-H.. Epigenetic regulation of TGF-β-induced EMT by JMJD3/KDM6B histone H3K27 demethylase. Oncogenesis. 2021;10:17. doi: 10.1038/s41389-021-00307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinchure OS, Sharma V, Tabasum S, Ghosh S, Singh RP, Sarkar C, Kulshreshtha R.. Polycomb complex mediated epigenetic reprogramming alters TGF-β signaling via a novel EZH2/miR-490/TGIF2 axis thereby inducing migration and EMT potential in glioblastomas. Int J Cancer. 2019;145:1254–1269. doi: 10.1002/ijc.32360. [DOI] [PubMed] [Google Scholar]
- 46.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 47.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann H-E, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 50.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emanuele E, Lista S, Ghidoni R, Binetti G, Cereda C, Benussi L, Maletta R, Bruni AC, Politi P.. Chromosome 9p21.3 genotype is associated with vascular dementia and Alzheimer’s disease. Neurobiol Aging. 2011;32:1231–1235. doi: 10.1016/j.neurobiolaging.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Züchner S, Gilbert JR, Martin ER, Leon-Guerrero CR, Xu P-T, Browning C, Bronson PG, Whitehead P, Schmechel DE, Haines JL, et al. Linkage and association study of late-onset Alzheimer disease families linked to 9p21.3. Ann Hum Genet. 2008;72:725–731. doi: 10.1111/j.1469-1809.2008.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA.. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widau RC, Zheng Y, Sung CY, Zelivianskaia A, Roach LE, Bachmeyer KM, Abramova T, Desgardin A, Rosner A, Cunningham JM, et al. p19Arf represses platelet-derived growth factor receptor β by transcriptional and posttranscriptional mechanisms. Mol Cell Biol. 2012;32:4270–4282. doi: 10.1128/MCB.06424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I.. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 56.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y.. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciarapica R, Carcarino E, Adesso L, De Salvo M, Bracaglia G, Leoncini PP, Dall’Agnese A, Verginelli F, Milano GM, Boldrini R, et al. Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS. BMC Cancer. 2014;14:139. doi: 10.1186/1471-2407-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarnowski M, Tkacz M, Czerewaty M, Poniewierska‑Baran A, Grymuła K, Ratajczak MZ.. 5‑Azacytidine inhibits human rhabdomyosarcoma cell growth by downregulating insulin‑like growth factor 2 expression and reactivating the H19 gene product miR‑675, which negatively affects insulin‑like growth factors and insulin signaling. Int J Oncol. 2015;46:2241–2250. doi: 10.3892/ijo.2015.2906. [DOI] [PubMed] [Google Scholar]
- 59.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E.. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Y, Xu L, Hassan M, Zhou X, Zhou Q, Rakheja D, Skapek SX.. Bayesian modeling identifies PLAG1 as a key regulator of proliferation and survival in rhabdomyosarcoma cells. Mol Cancer Res. 2020;18:364–374. doi: 10.1158/1541-7786.MCR-19-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Copy-number variation (CNV) data were obtained from the National Cancer Institute (NCI) TCGA program using the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). CDKN2A/B expression in RMS and neuroblastoma was extracted through RNA-seq data from 42 specimens downloaded from The European Genome-phenome Archive (EGA) with accession number EGAD00001000878 (for RMS), and from 195 cases NCI Gabriella Miller Kids’ First program (https://commonfund.nih.gov/kidsfirst) (for neuroblastoma).