ABSTRACT
To explore the presence of extrachromosomal circular DNA (eccDNA) in the anterior capsule of the lens in the eyes of patients with cataract and with high myopia. Circle-Seq was performed to identify differences in the eccDNA and gene expression between the anterior capsule of the lens of patients with simple nuclear cataract (C, n = 6 cases) and patients with nuclear cataract along with high myopia (HM, n = 6 cases). The expression of eccDNA was confirmed using routine quantitative polymerase chain reaction. The eccDNA ranked in C and HM ranged in length from 0.017 kb − 9.9 Mb with two distinctive peaks detected at 0.2 kb and 0.5 kb, while eccDNA that were differentially expressed ranged in size from 0.05 kb − 57.8 kb with two distinctive peaks observed at 0.1 kb and 0.5 kb. Only 2.5% of the eccDNA in C and 2% in HM were>25 kb in size. The gene-rich chromosomes contributed to more number of eccDNA/Mb, while several well-known high myopia candidate genes, including catenin delta 2 (CTNND2) and ubiquitin-like with PHD, exhibited significantly increased levels of eccDNA in the anterior capsule of the lens in patients with high myopia. This study highlighted the topologic analysis of the anterior capsule of eyes with high myopia, which is an emerging direction for research and clinical applications. These findings suggested that eccDNA was commonly detected in eyes with high myopia and cataracts, and the candidate genes for high myopia identified in previous studies were also observed in the eccDNA.
KEYWORDS: Extrachromosomal circular DNA, high myopia, cataract, Circle-seq, anterior capsule of the lens
Introduction
Extrachromosomal circular DNA (eccDNA) are distinct from the 22 linear autosomal pairs and a pair of sex-determining chromosomes, and have become an area of intense research interest. EccDNA can originate from chromosome sequences and have high heterogeneity in origin. Furthermore, increasing reports have confirmed that eccDNA are more prevalent in human tissues than previously anticipated [1–5], and play an important role in the pathogenesis of tumours [1,6,7]. Recent studies have revealed that the formation of oncogene-carrying eccDNA is a potent and frequent mechanism by which genes are amplified and that such eccDNA can increase intratumoral genetic heterogeneity [1,8]. Subsequently, Koche et al. discovered that eccDNA drives oncogenic genome remodelling in neuroblastoma [9], providing a new direction for modern cancer genomics. In addition, the known functions of eccDNA include sponging of transcription factors, ageing, intercellular communication, use in liquid biopsy, and stimulation of innate immune pathways [10]. However, the distribution and function of eccDNA in eye diseases, especially in high myopia and cataracts, remains unclear.
EccDNA, ranging from hundreds to thousands of base pairs in size, were first discovered by Alix Bassel and Yasuo Hoota in 1964 [11]. However, eccDNA were ignored by the scientific community at the time, as a meaningless piece of junk outside the chromosome. Later in 2014, Mischel et al. found that the resistance of glioblastoma to EGFR inhibitors was caused by the reversible loss of the EGFR-VIII gain-of-function mutation in eccDNA [12]. Subsequently, the field of eccDNA research grew, benefiting from the developments in high-throughput sequencing, that have revolutionized molecular and genetic studies [13]. One such method, Circle-Seq [14], is a gentle circular DNA purification protocol that allows for the isolation and detection of the pool of endogenous eccDNA within a cell population by high-throughput sequencing, and can aid in research on eye diseases.
High myopia, defined as myopia exceeding 6.00 dioptres or axis length≥26 mm, extensively damages the visual function of the human eye [15]. Currently, 163 million people, i.e., 2.7% of the world’s total population suffer from high myopia. A significant increase in the prevalence of this condition is predicted to occur globally, and is estimated to affect nearly a billion people by the year 2050 [16,17]. Several studies have shown that high myopia is characterized by genetic heterogeneity [18,19], and hence, it is important to elucidate the role of genes in the pathogenesis of high myopia. Several genes associated with the pathogenesis of high myopia located on the chromosomes have been identified. However, whether these genes could be present as extrachromosomal remains largely unknown.
Hence, in this study, we aimed to investigate whether eccDNA are common in the anterior capsule of the lens of the human eye. In particular, we focused on portrait distribution and circularization of eccDNA and discussing eccDNA potential function or mechanism based on bioinformatics analysis. We also compared the size profiles of the anterior capsule of the lens between patients with simple nuclear cataract and those with high myopia. Finally, we explored whether the identified candidate gene for high myopia was located extrachromosomally.
Materials and methods
Acquisition of patient samples
Informed consent was obtained from all participants included in the study. All procedures performed in the studies involving human participants were in accordance with the ethical standards of the Institutional and National Research Committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Anterior lens capsule samples (two groups) from patients with simple nuclear cataract (C, n = 6 cases) and patients nuclear cataract along with high myopia (HM, n = 6 cases) were selected based on slit lamp examination preoperatively. Each patient underwent conventional cataract phacoemulsification treatment combined with intraocular lens implantation. During continuous circular capsulorhexis (CCC), the anterior capsule (5.5 mm approximately) was collected into a cryopreservation tube and stored at − 80 ºC. Inclusion criteria: C: simple nuclear cataract, normal axial length (22 mm-24 mm); HM: high myopia combined with simple nuclear cataract, axial length>26 mm. Exclusion criteria: patients with other eye diseases, such as age-related macular degeneration, diabetic retinopathy, retinal vein occlusion, Retinal artery occlusion, uveitis, strabismus, amblyopia, etc; and patients with systemic diseases, such as hypertension, diabetes, coronary heart disease, and autoimmune disease, etc. The patient information has been presented in Table 1.
Table 1.
Patient Characteristics of C and HM.
| gender |
ages |
axial |
eye |
|||||
| Samples |
C |
HM |
C |
HM |
C |
HM |
C |
HM |
| 1 | F | M | 60 | 61 | 22.65 | 27.44 | OD | OS |
| 2 | F | M | 63 | 65 | 22.19 | 28.51 | OD | OD |
| 3 | F | F | 66 | 60 | 22.16 | 33.01 | OD | OS |
| 4 | F | F | 78 | 77 | 22.73 | 28.16 | OS | OD |
| 5 | M | F | 74 | 57 | 22.96 | 30.30 | OS | OS |
| 6 | M | F | 50 | 62 | 23.77 | 27.00 | OS | OD |
| P | 0.771 | 0.000 | ||||||
| Mean | 65.17±10.05 | 63.66±7.03 | 22.74±0.59 | 29.07±2.24 | ||||
F: female; M: male; C: simple nuclear cataract; HM: nuclear cataract complicated with high myopia;
Tissue DNA preparation and eccDNA sequencing
The Circle-Seq eccDNA sequencing service was provided by CloudSeq Biotech Inc. (Shanghai, China) by following published procedures with slight modifications [2]. The anterior capsule of the lens was mixed into one sample in every 2 cases. Specifically, three samples of each group were suspended in L1 solution (Plasmid Mini AXE; A&A Biotechnology) and supplemented with Proteinase K (Thermo Fisher Scientific, Waltham, MA, USA) prior to overnight incubation at 50°C with agitation. After lysis, samples were alkaline-treated, followed by precipitation of proteins and separation of chromosomal DNA from circular DNA through an ion exchange membrane column (Plasmid Mini AXE; A&A Biotechnology,Gdynia, Poland). The column-purified DNA were treated with FastDigest MssI (Thermo Fisher Scientific,Waltham, MA, USA) to remove mitochondrial circular DNA and incubated at 37°C for 16 h. Next, the remaining linear DNA were removed by exonuclease (Plasmid-Safe ATP-dependent DNase, Epicentre,Madison, WI, USA) at 37°C in a heating block, and the enzyme reaction was carried out continuously for seven days, adding additional ATP and DNase every 24 h (30 units/day) according to the manufacturer’s protocol (Plasmid-Safe ATP-dependent DNase, Epicentre). The eccDNA-enriched samples were used as templates for phi29 polymerase amplification reactions (REPLI-g Midi Kit, Qiagen, Hilden, Germany), amplifying eccDNA at 30°C for 46–48 h. Phi29-amplified DNA were sheared by sonication (Bioruptor, Diagenode, Philadelphia, PA), and the fragmented DNA was subjected to library preparation using NEBNext® Ultra II DNA Library Prep Kit for Illumina (New England Biolabs,Ipswich, Massachusetts, USA). Sequencing was carried out on an Illumina NovaSeq 6000 with 150 bp paired end mode, according to the manufacturer’s instructions.
Sequencing analysis of eccDNA
Paired-end reads were harvested from the Illumina NovaSeq 6000 sequencer and quality controlled by Q30. After 3’ adaptor-trimming and removal of low-quality reads using cutadapt software (v1.9.1) [20], the high-quality clean reads were aligned to the reference genome (UCSC hg19) with BWA software (v0.7.12) [21]. The Circle-map software (v1.1.4) [14] was used to detect eccDNA within all samples, and SAMtools software (v0.2) [22] was used to obtain raw soft-clipped read counts of the break point. Then, the EdgeR software (v0.6.9) [23] was used to perform normalization and differentially expressed eccDNA filter by p-value and fold change. The BedTools software (v2.27.1) [24] was used to annotate the eccDNA. Gene Ontology (GO) and pathway enrichment analyses were performed based on differentially expressed eccDNA-associated genes. IGV software (v2.4.10) [25] was used for cDNA visualization.
Validation of eccDNA by routine quantitative polymerase chain reaction (qPCR)
Four upregulated and one downregulated eccDNA in different genomic regions and chromosomes were chosen for experimental validation. Briefly, DNA was extracted from three samples of each group and treated with FastDigest MssI and exonuclease to remove mitochondrial circular DNA and linear DNA, as described above. Rolling circle amplification was then performed to increase the yield. PCR was performed to assess the candidate eccDNA level, using primers in these products with Accurate Taq Master Mix (dye plus) (Accurate Biotechnology, China). The reaction conditions were 95°C for 10 min, 40 cycles of 95°C for 10 s, 60°C for 60 s, and 95°C for 15 s. The primers for eccDNA were designed using the ‘out-facing’ strategy and are described in Table 2. The PCR products were loaded onto 1.5% agarose gels and visualized using an ultraviolet luminescent image analyser (LAS-4000 Mini; GE Healthcare Life Sciences, Pittsburgh, US). Besides, gene expression C and HM in mRNA level was verified by PCR.
Table 2.
Primers used for qPCR.
| Gene | Genetic locus | Types of primers | Gene sequence |
|---|---|---|---|
| UHRF1BP1L | chr12:100507034–100507634 | 1-Forward | AGGCCTCTAATCCCAGCACT |
| 1-Reverse | CCTGTCTCAGCCTCCAGAGT | ||
| CTNND2 | chr5:11524266–11525000 | 2-Forward | CCCTCCAGGAGTTCCTGTCT |
| 2-Reverse | TGGGAAGGTGGAGGTCACAG | ||
| LAMA3 | chr18:21446269–21446595 | 3-Forward | GCATCTTGTAGTTCAGGCCA |
| 3-Reverse | AGGTGTGTGTGACTTGCTGG | ||
| ZNF776 | chr19:58258376–58258734 | 4-Forward | GCAAAGGCCTGGAGATGAAC |
| 4-Reverse | GACGCCACAATTACCTGAGC | ||
| LAMA1 | chr18:7084795–7084945 | 5-Forward | GTTGGCTAGGCCTGAGACTTG |
| 5-Reverse | GCTGGGAAGAAATCCAAAAACC | ||
| Internal reference | Carrier pGEX-5X-2 | 6-Forward | GGGCTGGCAAGCCACGTTTGGTG |
| 6-Reverse | CCGGGAGCTGCATGTGTCAGAGG |
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE207646 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207646).
Overview of eccDNA map in anterior capsular of the lens
In order to characterize eccDNA in eyes with high myopia and cataracts, we adapted the method for genome-scale detection of eccDNA (Circle-Seq) [2,26,27]. The eccDNA were purified from the anterior capsule of the lens of patients with C and HM, respectively. We chose to compare these two groups of anterior capsule of the lens as they provided a perfect contrast, allowing the exploration of the pathogenesis of high myopia. Additionally, compared with the sclera, the anterior capsule of the lens is the most readily available human eye tissue material in clinical work. We have previously confirmed that the use of these two groups can yield meaningful results to elucidate the pathogenesis of high myopia [28].
Purification, enrichment, and detection of eccDNA for the sequencing library were performed in the following manner (Figure 1); 1) gentle DNA isolation by column separation, 2) removal of remaining linear DNA by exonuclease, 3) rolling-circle amplification, and 4) sequencing and mapping of paired-end reads to the human genome to identify structural variations resulting from DNA circularization. We performed qPCR verification of the linear gene after enzymatic digestion to ensure that the linear DNA was completely removed. After image and base identification, the raw reads were harvested using an Illumina Novaseq 6000 sequencer. Subsequently, more than 120 million high-quality clean reads were compared to the human reference genome (HG19) using BWA software after the removal of joints – adaptors and low-quality reads (Supplementary Table S1). Finally, 8,219 eccDNA in 23 pairs of chromosomes, including 2708 eccDNA in C and 3648 eccDNA in HM, that had a combined length of 10.85% (335.25 Mb) of the human genome, were identified using Circle-map software in these specimens (Figure 2a, Supplementary Table S2). Most of these eccDNA were detected in three C and HM specimens, indicating that the existence of eccDNA was a common event in eyes with cataract and with high myopia (Supplementary Table S2). In addition, the eccDNA that were detected were hierarchically ranked based on significant differences identified between C and HM specimens. We assigned upregulation (uregul) and downregulation (dregul) support to eccDNA with fold change≥2.0 (HM/C, log (FC) ≥ 1.0, p ≤ 0.05). We detected 783 uregul and 460 dregul eccDNA in this study (Figure 2b, Supplementary Table S3).
Figure 1.
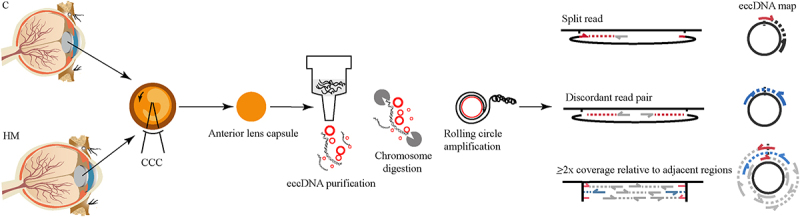
Circle-Seq method for mapping of eccDNA. OD: right eye OS: left eye CCC: Continuous circular capsulorhexis, an important operation in cataract surgery.
Figure 2.
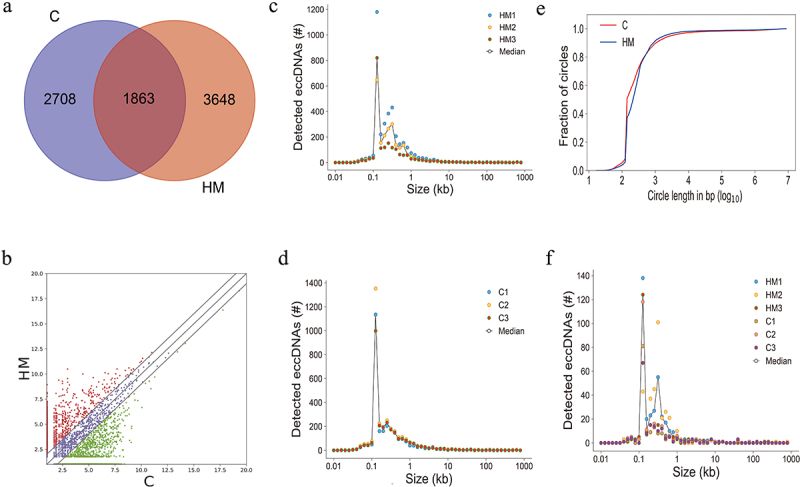
Genome-wide detection and analysis of eccDnas distribution by high throughput sequencing in HM and C samples. (a). the number of eccDnas of C and HM. (b). Scatter plots showing the differentially expressed eccDnas (fold changes ≥2 and p < 0.05). (c). Number of eccDnas relative to size in kb of each C samples. (d). Number of eccDnas relative to size in kb of each HM samples. (e). Cumulative frequency plots of HM (blue) and C (red) eccDNA in anterior capsular of the lens. (f). Number of significant differentially eccDnas relative to size in kb of HM and C samples.
The length distribution of eccDNA has been shown in Figure 2c,d. The eccDNA ranked in C and HM ranged in length from 0.017 kb − 9.9 Mb with two distinctive peaks at 0.2 kb and 0.5 kb (Figure 2e), while uregul and dregul eccDNA ranged in size from 0.05 kb − 57.8 kb with two distinctive peaks at 0.1 kb and 0.5 kb (Figure 2f). Interestingly, only 2.5% of the eccDNA in HM and 2% in C were>25 kb (Figure 3a,b and e), including eccDNA that were long enough to encode gene segments or even complete genes. Here, we have referred to eccDNA with lengths>25 kb as ecDNA. Around 2.5% of the significant differentially eccDNA (uregul and dregul) were ecDNA (Figure 3c,d and f) indicating that no significant differences were present between ecDNA in C or HM.
Figure 3.
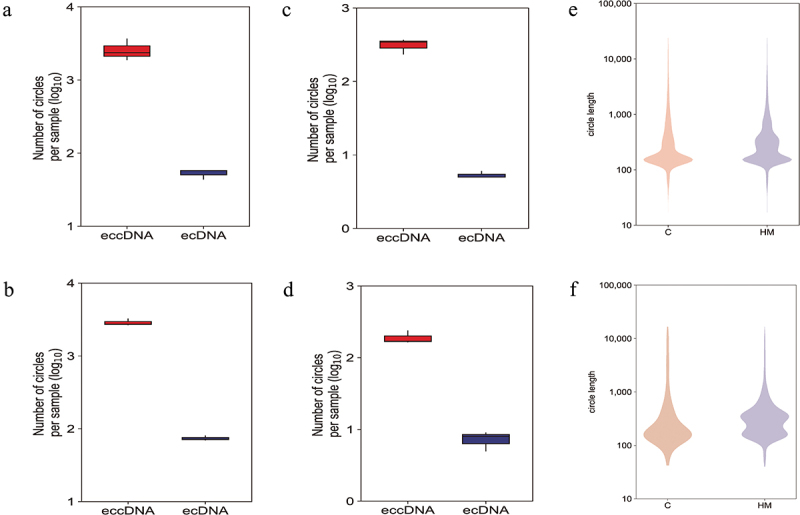
A genome-wide map of eccDnas and ecDnas in anterior capsular of the lens. (a). Number of ecDnas and eccDnas of HM samples. (b). Number of ecDnas and eccDnas of C samples. (c). Number of significant differentially eccDnas and ecDnas of HM samples. (d). Number of significant differentially eccDnas and ecDnas of C samples. (e): Size distribution of all ecDnas and eccDnas identified using Circle-seq in HM and C samples. (f). Size distribution of significant differentially eccDnas and ecDnas identified using Circle-seq in HM and C samples.
Significant differentially eccDNA were derived from all the chromosomes (Figure 4a), reproducing the oscillating length distribution observed in C and HM (Figure 4b,c). The genomic distribution of eccDNA revealed that the gene-rich chromosomes 17 and 19 contributed to a 1.6-fold and 3.3-fold higher average frequency of eccDNA/Mb than other chromosomes (Figure 5a). The genomic distribution of ecDNA revealed that the gene-rich chromosomes 17 and 19 contributed to a 2.7-fold and 5.7-fold higher average frequency of ecDNA/Mb than other chromosomes (Figure 5b). Interestingly, there were no ecDNA on chromosomes 10, 15, and 18. Notably, eccDNA and ecDNA were enriched in genic regions (Figure 5c,d). While ecDNA regularly contained all genes (80%), eccDNA mostly included a fraction of genes, indicating that DNA circularization affected the various coding and noncoding regions with yet unknown functional consequences.
Figure 4.
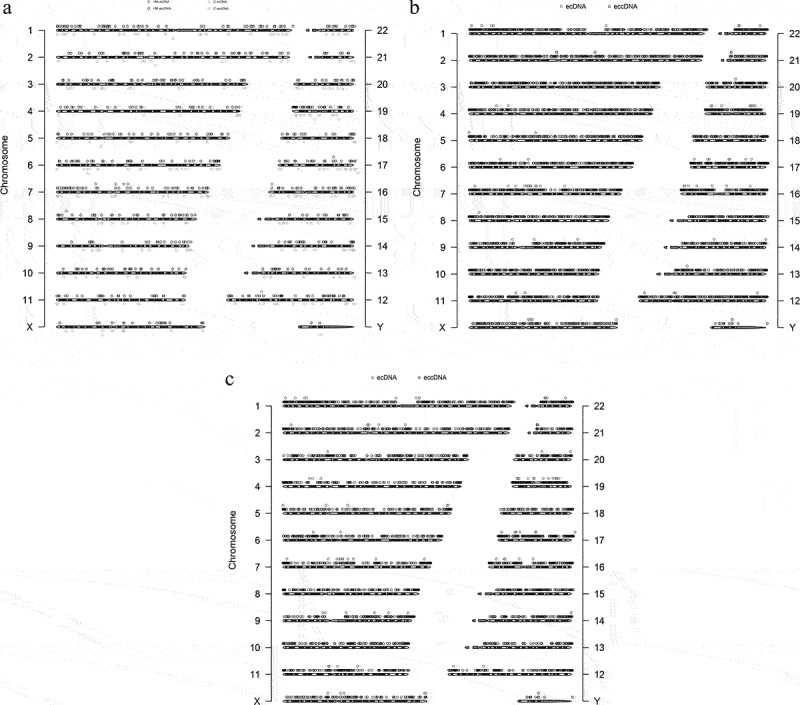
Genomic overview of eccDNA types detected anterior capsular of the lens. (a). Chromosomal map of significant differentially eccDnas and ecDnas from Circle-Seq. (b). Chromosomal map of all eccDnas and ecDnas of HM samples. (c). Chromosomal map of all eccDnas and ecDnas of C samples.
Figure 5.
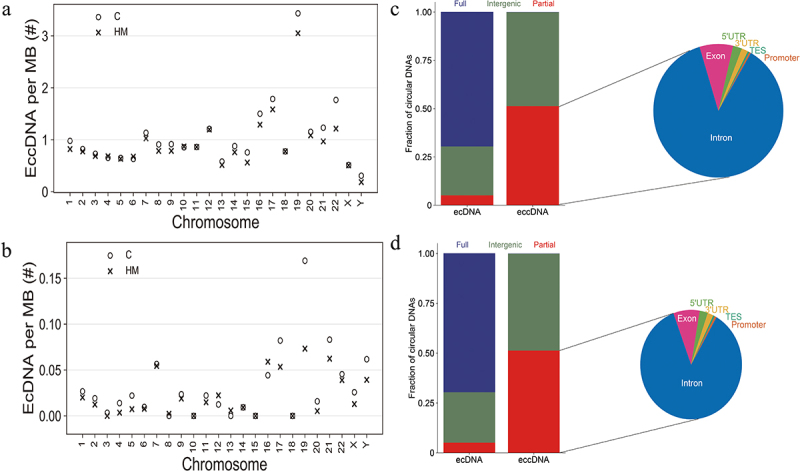
The relationship between eccDNA and genomes and chromosomes. (a). Significant differentially eccDNA frequency relative to chromosome. (b). Significant differentially ecDNA frequency relative to chromosome. (c). Fraction of genomic regions affected by eccDNA compared to ecDNA in C samples. D. Fraction of genomic regions affected by eccDNA compared to ecDNA in HM samples.
Genes associated with the eccDNA are involved in important biological pathways
Using the Circle-Seq analysis, GO enrichment analysis, and KEGG pathway enrichment analysis of eccDNA, we revealed the potential function of the genes associated with eccDNA in multiple signalling pathways and cellular processes. The GO project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). Fisher’s exact test was used to determine if there was more overlap between the gene list and the GO annotation list. We found that several well-known high myopia candidate genes, including catenin delta 2 (CTNND2, P = 0.002, fold change (FC) = 8.28), ubiquitin-like with PHD and ring finger domains 1 binding protein 1 like (UHRF1BP1L, P = 0.03, FC = 4.19), and laminin subunit alpha 1 (LAMA1, P = 0.03, FC = 3.96), exhibited significantly increased levels of eccDNA in the anterior capsule of the lens of high myopia (Supplementary Table 3). Routine PCR confirmed the results of Circle-Seq and demonstrated that the eccDNA of CTNND2 (circular CTNND2, c-CTNND2) and UHRF1BP1L (circular UHRF1BP1L, c-UHRF1BP1L) displayed higher levels of eccDNA enrichment in the anterior capsule of the lens of patients with high myopia than in patients with simple nuclear cataract (Figure 6a). In addition, we further identified the difference of gene expression C and HM in mRNA level. The results showed that the expression level of genes was consistent with that of genes carried by eccDNA (Figure 6b). The motif analysis flanking eccDNA junctional sites by high throughput sequencing showed the nucleotide composition from 10 bp upstream to 10 bp downstream of the start and end position inferred from the reference genome (hg19, Figure 6c).
Figure 6.
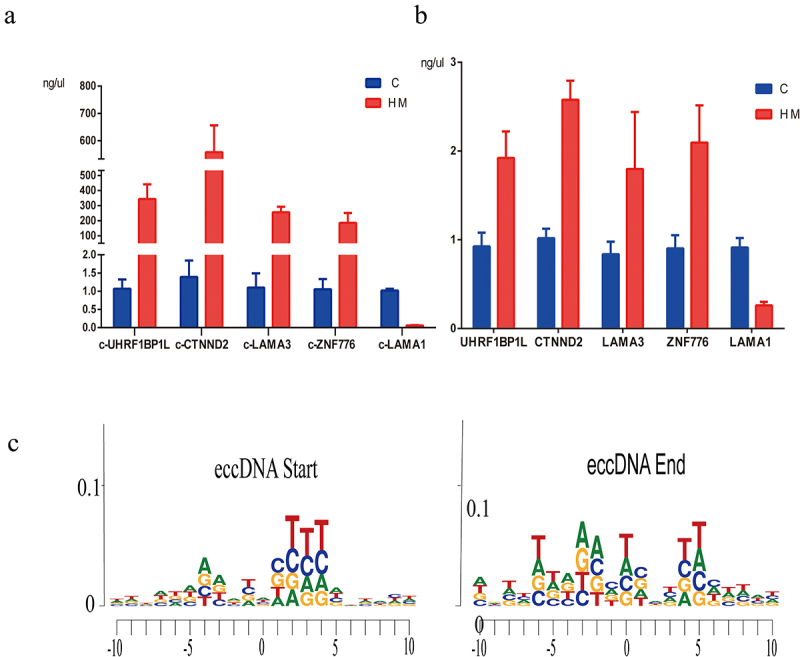
The PCR results of eccDnas and motif analysis. (a). the PCR results of 5 candidate eccDnas (c-gene) in 3 pairs of C and HM samples. (b). the PCR results of gene expression in mRNA level. (c). the motif analysis flanking eccDNA junctional sites by high throughput sequencing.
In addition, the GO analysis demonstrated that the upregulated eccDNA-related genes were mainly involved in a variety of biological processes, including positive regulation of dendritic spine development (ontology: biological process; Figure 7a), plasma membrane bounded cell projection (ontology: cellular component; Figure 7b), and ribonucleotide binding (ontology: molecular function; Figure 7c). The downregulated eccDNA-related genes were significantly associated with xenobiotic glucuronidation (ontology: biological process; Figure 7d), filamentous actin (ontology: cellular component; Figure 7e), and glucuronosyltransferase activity (ontology: molecular function; Figure 7f). Thus, the GO biological process classification indicated that most of the genes associated with the eccDNA were enriched in the cellular process (Figure 7c), and were involved in the formation of dendritic spine development (Figure 7a). This suggested that dendritic spine development was associated with the formation of pathological damage in high myopia. The KEGG pathway analysis showed the results for C and HM (Figure 8).
Figure 7.
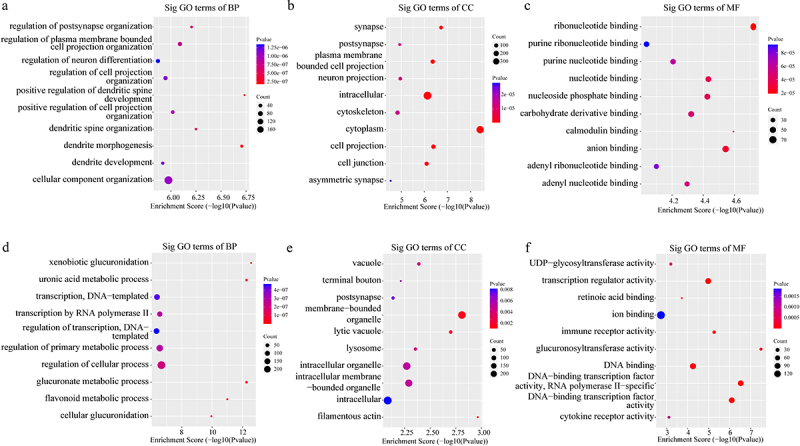
The results of the GO analysis. (a-c). Major gene ontology terms (BP, CC, MF, respectively) were significantly enriched for upregulation eccDNA-associated genes. (d-f). Major gene ontology terms (BP, CC, MF, respectively) were significantly enriched for downregulation eccDNA-associated genes. BP: biological process; CC: cellular component; MF: molecular function.
Figure 8.
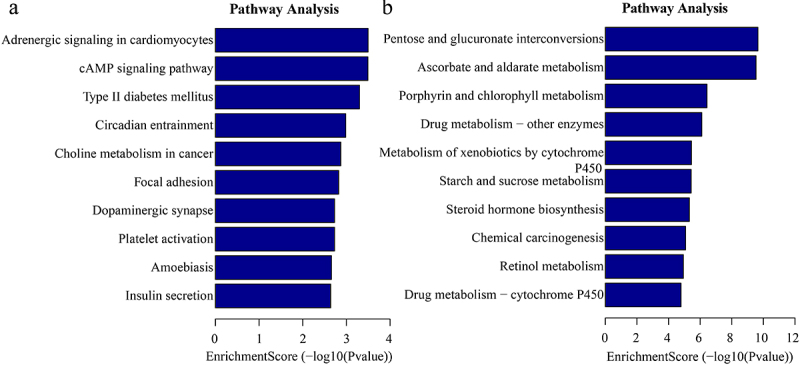
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of significant differentially eccDNA (select the top 10 by the value of p.Adjust). (a) the top ten significantly enriched pathways for the upregulation eccDNA. (b). the top ten significantly enriched pathways for the downregulation eccDNA.
Discussion
In this study, we demonstrated the presence of eccDNA in the eyeball, including the eyes with cataract and high myopia. To the best of our knowledge, this is the first systematic survey of the eye genome for eccDNA, where we have identified more than 8,000 eccDNA. Thus, this study increases the number of reported eccDNA in human somatic tissue. Our findings are likely an accurate estimate of the true set of eccDNA in cataract eyes and eyes with high myopia as the majority of the identified eccDNA (including elements<1 kb) were included in the analysis. Notably, we demonstrated that eccDNA could be mapped to any region of the human genome, and the frequency of eccDNA formation was associated with the chromosomes or coding genes and repetitive regions. This finding was in agreement with prior studies on human somatic tissue, human germline, and yeast, which reported that gene-rich chromosomes and repetitive sequences had a higher tendency to circularize and form eccDNA [2,26,29–31]. In addition, we showed that the length distribution of the eccDNA detected in cases of cataract and high myopia implied that most of the eccDNA were<1 kb with two distinctive peaks at 0.2 kb, 0.5 kb, in agreement with a previous study on human muscle tissue [2]. There were no detectable differences between the length distribution of eccDNA of C and HM, indicating that high myopia may not affect the length of DNA circularization.
Finally, the candidate genes associated with high myopia, that have been identified in previous studies, were present in the eccDNA. The GO and KEGG pathway analyses showed that the genes generating the candidate eccDNA at differential levels were mainly involved in biological processes of dendritic spine development, and may be related to high myopia; however, this has not been reported previously. Therefore, our findings provide new evidence that eccDNA elements of chromosomal origin are common in human somatic tissue and can be formed from any part of the human genome [2,3,10]. Moreover, this is the first time that the candidate genes for high myopia identified in previous studies were found on eccDNA, which provides a new perspective for the study of high myopia-related genes.
In the present study, a distribution map of ecDNA (>25 kb) in the chromosome has also been generated. Unlike prior studies [1,2], we identified that ecDNA accounted for a small portion of all eccDNA in eyes with cataract and high myopia. With an average gene size of 27 kb in the human genome, many of these ecDNAs may be large enough to contain full genes [32], as human somatic cells contain Mb-sized ring chromosomes [33,34]. This phenomenon suggests that eccDNA may be capable of gene delivery. Indeed, multiple studies have confirmed that eccDNA have transcription ability [2,9].
The differences in the length and chromosome distribution of eccDNA in cases of high myopia were smaller than cataract, but we still identified more than 1,000 eccDNA with statistical differences. With the development of high-throughput sequencing technology, knowledge of the biological properties and functions of eccDNA has broadened, including cell development and communication, ageing, adaptive evolution, molecular sponges, tumour occurrence and heterogeneity, drug resistance, genome instability, and lipid biopsy [10]. Sin et al. explored the presence of eccDNA in the plasma of pregnant women and provided a new direction for prenatal diagnosis [5]. In our study, after gene annotation of eccDNA using BedTools, we found several previously identified candidate genes related to high myopia, such as LAMA1, UHRFlBPlL, and CTNND2, that were successfully validated using routine PCR. LAMA1 encodes one of the alpha 1 subunits of laminin that make up a major component of the basement membrane and have been implicated in a wide variety of biological processes including cell adhesion, differentiation, migration, signalling, neurite outgrowth and metastasis. Qingjiong Zhang found that early onset high myopia with midline alopecia areata could be caused not only by mutations of the COL18A1 gene but also by mutations in the LAMA1 gene [35] UHRFlBPlL, also known as BLTP3B or SHIP164, enables GARP complex binding activity and protein homodimerization activity. In 2013, HAWTHORNE et al. discovered UHRFlBPlL is associated with high myopia though a genetic association study of the MYP3 locus in a family-based high-grade myopia cohort [36CTNND2 encodes an adhesive junction-associated protein of the armadillo/beta-catenin superfamily and is implicated in brain and eye development and cancer formation [37]. The results of several studies have indicated that polymorphisms in the CTNND2 gene are associated with the pathogenesis of high myopia [37–40]. High myopia is prevalent in a population-specific manner. In 2013, PAN et al. found that the prevalence of high myopia in Chinese Singaporeans was significantly higher than that in Indian and Malaysian ethnic groups [41]. An additional risk of myopia in children of parents with myopia has been confirmed [42], indicating that genetic factors play an important role in the pathogenesis of high myopia. Till date, more than 150 genes and over 20 chromosomal loci have been identified to be associated with myopia. However, the candidate gene locus of high myopia located outside the chromosome has not yet been identified. Our findings may provide new ideas and methods for studying sites of high myopia-related genes.
In conclusion, we identified the genome-wide presence of eccDNA and ecDNA in patients with high myopia and cataract. The eccDNA map generated in this study revealed features of eccDNA (e.g., genome-wide distribution in patients with high myopia) and identified the common as well as selective characteristics of eccDNA and their functional implications. This comprehensive eccDNA profiling provides a solid basis for the determination of potential functional roles of genes in eccDNA in high myopia. Future studies are required to explore the potential mechanism leading to the different levels of eccDNA in eyes of patients with cataract and high myopia and to determine the regulatory mechanisms involved in the formation of eccDNA in patients with high myopia.
Supplementary Material
Acknowledgments
The author thanks NewCore BiodataStudio in Shanghai for sequencing date analysis; Jing Sun and Yang Cai for important clinical and scientific suggestions. No honorarium, grant, or other form of payment was given to anyone to produce the manuscript. Each author listed on the manuscript has seen and approved the submission of this version of the manuscript and takes full responsibility for the manuscript.
Funding Statement
This work was supported by the [Autonomous and open project of Tianjin Key Laboratory of Retinal Function and Diseases] under Grant[number 2020tjswmm002]; [Funded by Tianjin Key Medical Discipline (Specialty) Construction Project] under Grant [number TJYXZDXK-037A]; [Tianjin Education Commission Scientific Research Project (Natural Science)] under Grant [number 2022ZD059]; [Tianjin Science and Technology Plan Project (subject)] under Grant [number 21KPHDRC00140];[The Science&Technology Development Fund of Tianjin Education Commission for Higher Education] under Grant [number 2020KJ179];[Talent program of Tianjin medical university eye hospital] under Grant [number YDYYRCXM-B2018-01LC].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
GEO Series accession number GSE207646 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207646)
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2023.2192324
References
- [1].Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moller HD, Mohiyuddin M, Prada-Luengo I, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018;9:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shibata Y, Kumar P, Layer R, et al. Extrachromosomal microDnas and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pennisi E. Circular DNA throws biologists for a loop. Science. 2017;356:996. [DOI] [PubMed] [Google Scholar]
- [5].Sin STK, Jiang P, Deng J, et al. , Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proceedings of the National Academy of Sciences of the United States of America. 2020. Jan 21;117:1658–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Verhaak RGW, Bafna V, Mischel PS. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer. 2019;19(5):283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kumar P, Kiran S, Saha S, et al. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. 2020;6:eaba2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].deCarvalho AC, Kim H, Poisson LM, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nature Genet. 2018;50:708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koche RP, Rodriguez-Fos E, Helmsauer K, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nature Genet. 2020;52:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Paulsen T, Kumar P, Koseoglu MM, et al. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 2018;34:270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hotta Y, Bassel A , Molecular size and circularity of DNA in cells of mammals and higher plants. Proceedings of the National Academy of Sciences of the United States of America. 1965. Feb;53;356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nathanson DA, Gini B, Mottahedeh J, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salmaninejad A, Motaee J, Farjami M, et al. Next-generation sequencing and its application in diagnosis of retinitis pigmentosa. Ophthalmic Genet. 2019;40:393–402. [DOI] [PubMed] [Google Scholar]
- [14].Prada-Luengo I, Krogh A, Maretty L, et al. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinf. 2019;20:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saw SM, Gazzard G, Shih-Yen EC, et al. Myopia and associated pathological complications. Ophthalmic & Physiological Optics: The Journal of the British College of Ophthalmic Opticians. 2005;25(5):381–391. [DOI] [PubMed] [Google Scholar]
- [16].Holden BA, Fricke TR, Wilson DA, et al. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmol. 2016;123:1036–1042. [DOI] [PubMed] [Google Scholar]
- [17].Wong YL, Saw SM. Epidemiology oF pathologic myopia in Asia and worldwide. Asia-Pac J Ophthalmol. 2016;5(6):394–402. [DOI] [PubMed] [Google Scholar]
- [18].Young TL, Metlapally R, Shay AE. Complex trait genetics of refractive error. Arch Ophtalmol. 2007;125(1):38–48. [DOI] [PubMed] [Google Scholar]
- [19].Chen CY, Scurrah KJ, Stankovich J, et al. Heritability and shared environment estimates for myopia and associated ocular biometric traits: the genes in myopia (GEM) family study. Hum Genet. 2007;121(3–4):511–520. DOI: 10.1007/s00439-006-0312-0 [DOI] [PubMed] [Google Scholar]
- [20].Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):3. [Google Scholar]
- [21].Li H, Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Quinlan AR, Hall IM. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Moller HD, Parsons L, Jorgensen TS, et al. , Extrachromosomal circular DNA is common in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2015. Jun 16;112:E3114–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moller HD, Bojsen RK, Tachibana C, et al. Genome-wide purification of extrachromosomal circular DNA from eukaryotic cells. J Vis Exp. 2016;e54239. DOI: 10.3791/54239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wen K, Zhang Y, Li Y, et al. Comprehensive analysis of transcriptome-wide m 6 a methylome in the anterior capsule of the lens of high myopia patients. Epigenetics. 2020;16(9):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gresham D, Usaite R, Germann SM, et al. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proceedings of the National Academy of Sciences of the United States of America, 2010. Oct 26;107:18551–18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Turner DJ, Miretti M, Rajan D, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nature Genet. 2008;40:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Moller HD, Larsen CE, Parsons L, et al. Formation of Extrachromosomal Circular DNA from Long Terminal Repeats of Retrotransposons in Saccharomyces cerevisiae. G3: Genes | Genomes | Genetics. 2015;G3(6):453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. [DOI] [PubMed] [Google Scholar]
- [33].Tumer Z, Harboe TL, Blennow E, et al. Molecular cytogenetic characterization of ring chromosome 15 in three unrelated patients. Am J Med Genet A. 2004;130A(4):340–344. [DOI] [PubMed] [Google Scholar]
- [34].Guilherme RS, Meloni VF, Kim CA, et al. Mechanisms of ring chromosome formation, ring instability and clinical consequences. BMC Med Gene. 2011;12:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang P, Jia X, Xiao X, et al. An early diagnostic clue for COL18A1- and LAMA1-associated diseases: high myopia with alopecia areata in the cranial midline. Front Cell Dev Biol. 2021;9:644947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hawthorne F, Feng S, Metlapally R, et al. Association mapping of the high-grade myopia MYP3 locus reveals novel candidates UHRF1BP1L, PTPRR, and PPFIA2. Invest Ophthalmol Visual Sci. 2013;54(3):2076–2086. DOI: 10.1167/iovs.12-11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lu Q, Aguilar BJ, Li M, et al. Genetic alterations of δ-catenin/nprap/neurojungin (CTNND2): functional implications in complex human diseases. Hum Genet. 2016;135(10):1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li YJ, Goh L, Khor CC, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmol. 2011;118(2):368–375. DOI: 10.1016/j.ophtha.2010.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lu B, Jiang D, Wang P, et al. Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Visual Sci. 2011;52(11):8258–8261. DOI: 10.1167/iovs.11-7914 [DOI] [PubMed] [Google Scholar]
- [40].Yu Z, Zhou J, Chen X, et al. Polymorphisms in the CTNND2 gene and 11q24.1 genomic region are associated with pathological myopia in a Chinese population. Ophthalmol J int d’ophtalmologie Int j ophthalmol Zeitschrift fur Augenheilkunde. 2012;228(2):123–129. [DOI] [PubMed] [Google Scholar]
- [41].Pan CW, Zheng YF, Anuar AR, et al. Prevalence of refractive errors in a multiethnic Asian population: the Singapore epidemiology of eye disease study. Invest Ophthalmol Visual Sci. 2013;54(4):2590–2598. DOI: 10.1167/iovs.13-11725 [DOI] [PubMed] [Google Scholar]
- [42].Xiang F, He M, Morgan IG. The impact of parental myopia on myopia in Chinese children: population-based evidence. Optom Vis Sci. 2012;89:1487–1496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GEO Series accession number GSE207646 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207646)


