Abstract
Using genome mining and heterologous expression, we report the discovery and production of a new antimicrobial lasso peptide from species related to the Enterobacter cloacae complex. Using NMR and mass spectrometric analysis, we show that this lasso peptide, named cloacaenodin, employs a threaded lasso fold which imparts proteolytic resistance that its unthreaded counterpart lacks. Cloacaenodin has selective, low micromolar, antimicrobial activity against species related to the E. cloacae complex, including species implicated in nosocomial infections and against clinical isolates of carbapenem-resistant Enterobacterales. We further used site-directed mutagenesis to probe the importance of specific residues to the peptide’s biosynthesis, stability, and bioactivity.
Graphical Abstract
The ESKAPE pathogens are species of bacteria that are of clinical concern for their ability to evade the mechanisms of current antibiotics.1 Without new drugs and treatments, these resistant species present a threat to humanity, causing an unnecessary loss of life to infections once treatable. The last “E” in “ESKAPE” represents species of Enterobacter, which is a genus of Gram-negative γ−proteobacteria.2 While Enterobacter species commonly reside commensally in the human and animal GI tract and in the environment on decaying matter, in soil, and in sewage, certain species like E. cloacae have been the causative agents of many nosocomial outbreaks.2-4 With the emergence of Enterobacter strains displaying resistance to many last-resort antibiotics like carbapenems, these concerning pathogens call for a renewed interest in development of new treatments.3
Within this genus is the Enterobacter cloacae complex (ECC) consisting of closely related species that are commonly isolated as clinical specimens, most notably E. cloacae and E. hormaechei.4-5 Concerningly, many of these pathogens have natural resistance to β-lactam antibiotics and possess various carbapenemase genes.3 Colistin resistance has also been increasingly found in Enterobacter infections.6-12 With the rise of multidrug resistance in Enterobacter isolates, new compounds are sorely needed for future treatments.
A rapidly growing list of lasso peptides exhibit antimicrobial activity by several different mechanisms.13-24 These peptides are named after their unusual threaded shape, resembling a slipknot, and are biosynthesized from a ribosomal precursor peptide (known as A) via the action of two enzymes, a protease (known as B) and lasso cyclase (known as C).25-29 The compact, threaded structure of lasso peptides shields portions of the amide backbone, often rendering the peptides protease resistant. With advances in genomic sequencing, we can now find and predict new lasso peptides and other ribosomally synthesized and post-translationally modified peptides (RiPPs) that may not be detected from cultivation of the native species in the lab, enabling discovery of potential new drug compounds.30-35
Many lasso peptides display focused spectra of activity, presenting a route to target specific pathogens without disturbing commensal bacteria.36-38 From past investigations with lasso peptides such as ubonodin16, citrocin17, klebsidin19, microcin J25 (MccJ25)13, and capistruin14, it was noticed that these compounds often target strains that are phylogenetically similar to the producer, potentially serving as a mechanism for competition in microbial communities.39 We thus hypothesized that lasso peptide biosynthetic gene clusters (BGCs) found in pathogen-related species would be more likely to have antimicrobial activity against clinically relevant pathogens (a guilt-by-association approach), presenting an effective way to screen and prioritize genome mining hits.
Here, we report the discovery and heterologous production of a lasso peptide from members of the ECC, which we have named cloacaenodin. We solved the structure of this novel lasso peptide using solution NMR and studied its stability and structure-activity relationship via site-directed mutagenesis. Cloacaenodin showed potent and selective activity against multiple clinically relevant strains from the ECC.
Results and Discussion
Genome Mining Reveals a New Lasso Peptide from Enterobacter Species
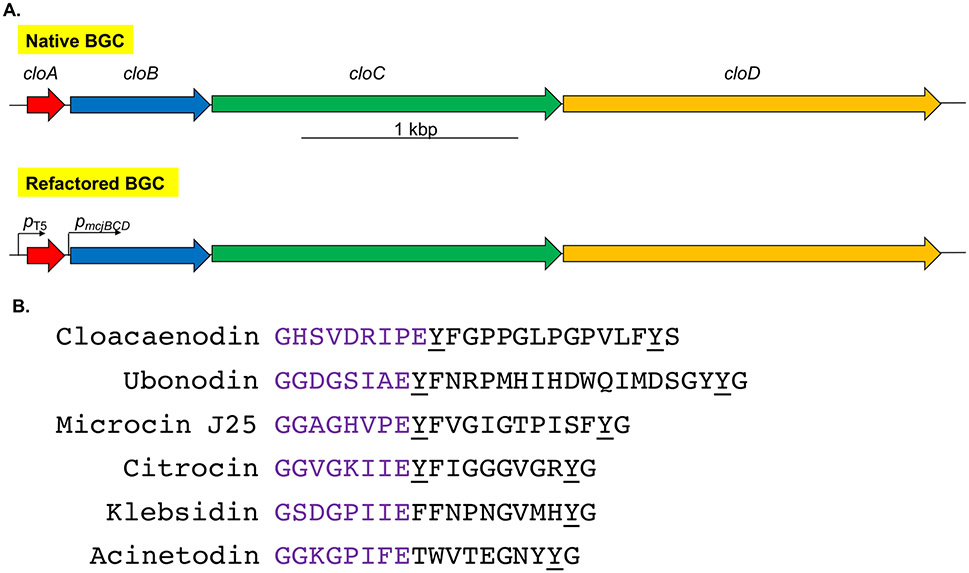
We employed our precursor-centric genome mining algorithm26 with a particular interest in putative lasso peptides with a Tyr after the ring and a Tyr in the penultimate position. The corresponding Tyr residues in the lasso peptide MccJ25 make specific hydrogen bonding contacts in the secondary channel of RNA polymerase (RNAP).40 We identified a putative lasso peptide BGC in Enterobacter hormaechei strain LB3 (on contig NZ_LFHB01000019.1), a strain that was originally identified in long beans. We later identified identical BGCs in Enterobacter cloacae strain B2 (on contig NZ_JSWY01000033.1), a strain isolated from bitter gourd, and Enterobacter kobei strain 1572712 (on contig NZ_LFHB01000019.1), a strain isolated from human sputum in China. We named this lasso peptide cloacaenodin (Figure 1), as it originates from species of the E. cloacae complex and the Latin root nodum signifies knot.
Figure 1.
Genome mining reveals a new lasso peptide from Enterobacter species. (A) The native BGC consists of a typical organization of lasso peptide genes in proteobacteria. It contains the precursor gene, cloA; the leader peptidase gene, cloB; the lasso peptide cyclase gene, cloC; and an ATP-binding cassette transporter gene for export of the mature lasso peptide, cloD. To facilitate heterologous expression, the BGC was codon-optimized and refactored into a pQE-80 vector with the cloA gene under the control of an IPTG-inducible T5 promoter, while the rest of the cluster is under the control of the constitutive pmcjBCD promoter. (B) The sequence of cloacaenodin and other known RNAP-inhibiting lasso peptides. Cloacaenodin contains a 9-membered ring (purple) and a C-terminal serine, which differs from the other lasso peptides which contain an 8-membered ring and a C-terminal glycine. Cloacaenodin contains the relatively well-conserved tyrosine residue directly after the ring, and the conserved penultimate tyrosine residue (underlined).
A protein BLAST search on the CloA protein revealed 14 distinct protein accession numbers corresponding to cloacaenodin-like precursors from other strains of Salmonella, E. coli, Citrobacter, and Enterobacter, including E. cloacae and E. hormaechei subsp. xiangfangensis (Figure S1). We queried Enterobacter cloacae and Enterobacter hormaechei in an NCBI assembly search, and found 616 total GenBank assemblies for E. cloacae and 3,754 total GenBank assemblies for E. hormaechei. Since we only observe the cloacaenodin BGC in a select few E. cloacae and E. hormaechei strains, this indicated to us that the cloacaenodin BGC is particularly rare among the currently assembled genome sequences of these two species.
From the BLAST results, we manually confirmed the presence of the B, C, and D genes (corresponding to the lasso peptide protease, cyclase, and exporter, respectively) downstream of the A gene (corresponding to the precursor). With the exception of the E. coli strains encoding a cloacaenodin-like precursor (Figure S1) which appear to have their B and C genes disrupted, the amino acid sequences of the biosynthetic enzymes all have at least a 50% amino acid identity to CloB, CloC, and CloD, with the D proteins having the highest degree of identity (Table S1-S3).
Due to the producing species, this lasso peptide BGC caught our attention since we hypothesized it may have activity against members of the ECC. The gene cluster organization of ABCD (Figure 1A) all in a single operon is typical of proteobacterial lasso peptide gene clusters14, with the gene clusters of citrocin and MccJ25 being exceptions in which the A genes are transcribed divergently from the BCD operon.17, 41
Our lab has previously reported the structure and activity of lasso peptides ubonodin16 and citrocin17, which were shown to be potent inhibitors of RNAP, similar to the lasso peptides MccJ25, klebsidin, and acinetodin (Figure 1B).19,40,42-44 Due to the presence of the conserved Tyr residues, we predicted that cloacaenodin may also function by inhibiting RNAP. However, the C-terminal amino acid in cloacaenodin is serine, differing from the conserved glycine seen in other RNAP-targeting lasso peptides (Figure 1B). We were thus interested to see if and how this residue would be important to the biosynthesis and activity of this new lasso peptide. Additionally, the previous RNAP-targeting lasso peptides ubonodin, citrocin, MccJ25, acinetodin, and klebsidin have been 8-membered rings, while cloacaenodin has a 9-membered ring (Figure 1B).
Heterologous Expression of Cloacaenodin
To produce the lasso peptide, we opted for a heterologous expression strategy in E. coli, a strategy that has historically worked well for proteobacterial lasso peptide BGCs.45,46 We placed the A gene under the inducible T5 promoter in the pQE-80 plasmid with the other genes (B, C, and D) under the control of the constitutive pmcjBCD promoter (Figure 1A). This gene refactoring strategy was originally developed for MccJ2545 and has been used subsequently16,17 to allow for lasso peptide B, C, and D proteins to be already present in the cell once the A precursor is produced. To facilitate expression and eliminate any codon bias, the gene sequences were codon optimized for E. coli. The resulting codon-optimized BGC was synthesized as gBlocks and cloned into pQE-80. With this construct, we expressed cloacaenodin at room temperature in M9 minimal media overnight. Consistent with the presence of a D gene, which encodes for an ATP-binding cassette transporter that exports the lasso peptide from cells, we detected a signal that matched the expected mass of cloacaenodin in the supernatant extract through LC-MS analysis. We observed a prominent peak when extracting for the expected cloacaenodin mass and a much smaller later-eluting peak (Figure S2). Since lasso peptides are capable of unthreading47-49, we hypothesized that these peaks corresponded to the threaded and unthreaded variants of cloacaenodin.
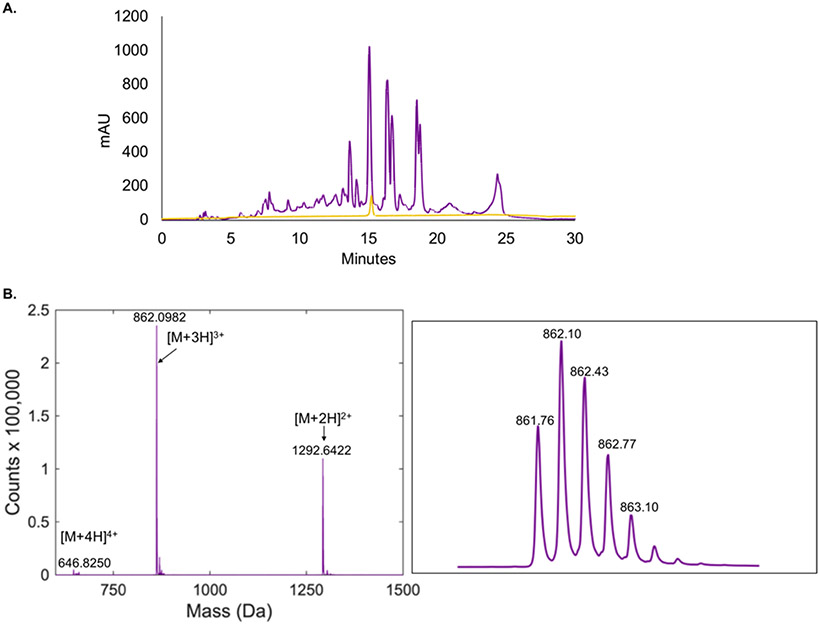
We injected the supernatant extract onto an HPLC instrument and collected the prominent peak eluting at a retention time of 15.0 min (about 60/40 water/acetonitrile) (Figure 2A). Following collection, we checked this compound using LC-MS analysis, revealing a single peak that eluted at ~10.5 minutes, containing a species consistent with the expected mass of cloacaenodin (Figure 2B). We were able to obtain a yield of ~1.1 mg/L of culture.
Figure 2.
Cloacaenodin can be biosynthesized heterologously in E. coli. (A) HPLC chromatogram of the M9 supernatant extract from E. coli heterologous expression of cloacaenodin (purple trace) and purified cloacaenodin (light orange trace). (B) Mass spectrum of purified cloacaenodin displaying the +4, +3, and +2 charge states. A close-up of the +3 charge state isotopic distribution is shown. The expected monoisotopic +3 m/z is 861.77.
We next sought to verify the threaded nature of the collected cloacaenodin sample, as the threaded structure is crucial to the bioactivity of lasso peptides.50 Heat treatment of certain lasso peptides leads to their irreversible unthreading; the unthreaded species often elutes differently than the threaded peptide.48,49 We performed a heating assay at 95 °C and noticed that after 1.5 hours ~75% of cloacaenodin now eluted at around 12.4 min instead of 10.5 min (Figure S3). Upon treatment with carboxypeptidase48,49, this later eluting peak was eliminated, and we could then observe various earlier-eluting peaks consistent with C-terminal truncations of 6, 9, or 10 amino acids (Figure S3-S4). We did not observe these truncations for the unheated cloacaenodin sample when treated with carboxypeptidase. This result supports our hypothesis that the smaller, and later eluting, peak in the supernatant extract is unthreaded cloacaenodin. This unthreaded variant in the extract may have formed due to thermal unthreading during expression and purification, exposure to organic solvents methanol and acetonitrile during purification, or both. To ensure that the sample we collected from the HPLC instrument remained stable, we froze the sample immediately upon collection until lyophilization, such that the peptide was minimally kept in solution.
Knowing that cloacaenodin was susceptible to unthreading, we tested how it would unthread at 37 °C, physiological temperature, in pure water. After a period of 72 hours, we observed that the sample of 18 μM cloacaenodin remained ~83% threaded based on relative peak area detected via LC-MS (Figure S5). We note that it is unusual for lasso peptides to unthread at this temperature, but, in terms of bioactivity, this degree of unthreading would still provide a majority of threaded peptide to exert its function.
We performed MS/MS analysis via collision-induced dissociation (CID) on both the threaded and unthreaded conformers of cloacaenodin. We observed similar fragments for both conformers, but found that the unthreaded conformer was more prone to fragmentation than the threaded conformer, consistent with previous observations that threaded lasso peptides are resistant to fragmentation.14,16,50 Certain fragmentation ions were only observable for the unthreaded conformer (Figure S6).
NMR Structure of Cloacaenodin
We next sought to determine the structure of cloacaenodin in water using 2D NMR analysis, a technique that has been successfully used to show the threaded shape of lasso peptides.51 We were concerned about cloacaenodin unthreading during the NMR acquisition, which we addressed by keeping the sample at 4 °C throughout acquisition. LC-MS analysis of the sample prior to and following NMR data collection confirmed that cloacaenodin remained threaded (Figure S7). As an additional control, 1D NMR spectra were acquired in between each 2D spectrum acquisition to ensure stability of the sample (Figure S8).
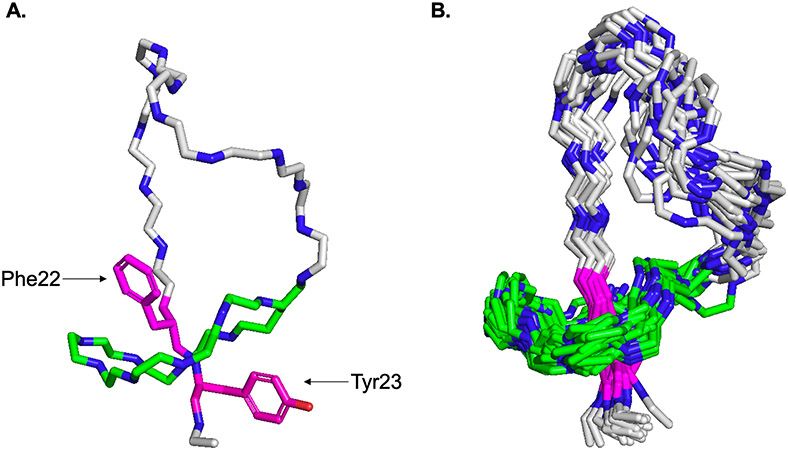
We acquired TOCSY at a mixing time of 80 ms (Figure S9) along with two NOESY spectra: one at 150 ms (Figure S10) and one at 300 ms (Figure S11). The TOCSY and 300 ms NOESY were used together to assign proton shifts (Table S8). We note that this NMR analysis was particularly challenging due to the presence of five prolines in the sequence. In addition to the prolines disrupting the NOESY walk during the assignment of residues, these prolines can lead to multiple isomers of cloacaenodin due to cis/trans isomerization as observed previously in MccJ25.52 We found multiple possibilities in our assignments of Pro17 and Gly18 (Table S8), and due to this ambiguity in assigning the chemical shifts for these protons, we chose to leave these residues undefined in our model building. The chemical shifts for the amide protons of Phe22 and Tyr23 (9.210 ppm and 10.320 ppm respectively) are markedly downshifted from the BMRB average; the unique chemical environment experienced by these residues suggest that they may be the amino acids passing through the isopeptide bonded ring. The wide dispersion of the amide proton chemical shifts in the spectra (6.364 ppm – 10.320 ppm) provides further support for the lassoed structure of cloacaenodin as it has been previously demonstrated that the amide proton shifts for unthreaded lasso peptides are generally in a narrower range.51
The 150 ms NOESY spectrum was integrated for through-space distance restraints. From the NOESY spectrum, we observed interactions between protons from Phe22 with Asp5, Arg6, Pro8, and Glu9, as well as between protons from Tyr23 with Gly1, His2, Asp5, Ile7, Pro8, and Glu9 (Table S9). These interactions indicated that Phe22 and Ty23 likely function as the bulky steric lock residues to keep the peptide threaded. Our assignments as well as through-space NOEs from integrated peaks and explicit distance constraints around the isopeptide bond (calculated from the rubrivinodin crystal structure PDB 5OQZ) 53 were given to CYANA 2.154 using the automated mode, where all prolines were presumed trans.
The top 20 structures calculated by CYANA show Phe22 and Tyr23 as the upper and lower steric lock residues respectively (Figure 3A), similar to the Phe19 and Tyr20 steric locks of MccJ2550,55,56. Cloacaenodin has a loop region of 13 amino acids, which is the second largest loop observed after ubonodin, and a short tail of only two amino acids (Figure 3B). This large loop and short tail structure is similar to the RNAP-inhibiting lasso peptides MccJ25, ubonodin, citrocin, klebsidin, and acinetodin, but with cloacaenodin being unique with its C-terminal serine and 9-membered ring (Figure S12).
Figure 3.
NMR structure of cloacaenodin (A) Representative cloacaenodin structure is shown. Carbons from ring residues are colored in green. Carbons from loop and tail residues are colored in grey. The steric locks Phe22 and Tyr23 are displayed in magenta (oxygen for Tyr23 sidechain is in red). (B) 20 lowest energy structures are overlaid. These model structures have been deposited to the Protein Data Bank under PDB code 8DYN.
Threaded cloacaenodin resists protease cleavage
One of the advantages of lasso peptides is that they are often resistant to proteolysis. We previously established that cloacaenodin was resistant to C-terminal proteolysis by the exopeptidase carboxypeptidase (Figure S3-S4), and we wanted to see if this peptide would be resistant to endopeptidases.
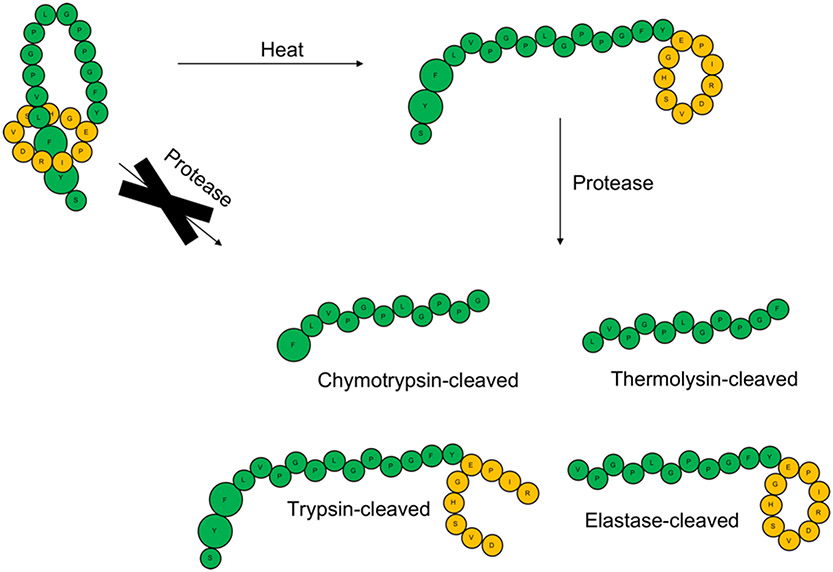
The sequence of cloacaenodin contains residues cleavable by trypsin, chymotrypsin, elastase, and thermolysin. As expected, when we tested a sample of unthreaded cloacaenodin (generated by heating), we observed fragments consistent with cleavage of the unthreaded peptide both in its linear and ring segments. In stark contrast, under the same conditions, the threaded peptide remained resistant to proteolysis by each of these four proteases (Figure 4, Figure S13-S16). This is consistent with the reported proteolytic resistance of previously discovered lasso peptides and demonstrates that the threaded structure of cloacaenodin is essential to its proteolytic stability.57-59
Figure 4.
Threaded cloacaenodin is resistant to proteolysis while the unthreaded conformer is proteolyzed into the major products shown. Loop/tail residues are colored green and ring residues are colored orange. Phe22 and Tyr23 are enlarged to denote their role as the steric lock residues for cloacaenodin. The LC-MS data underlying this schematic are shown in Figure S13-S16.
Cloacaenodin has antimicrobial activity against multiple Enterobacter strains
After characterizing the structure and proteolytic resistance of cloacaenodin, we next tested if cloacaenodin could inhibit bacterial growth. As a preliminary test of antimicrobial activity, we cloned a version of our expression plasmid where the cloD gene was removed, so that the lasso peptide could not be exported by the E. coli upon IPTG-induced expression. We found that E. coli XL-1 Blue colonies did not appear on LB agar when IPTG was added to the plate (Figure S17), indicative of intracellular cloacaenodin toxicity.
Of the antimicrobial lasso peptides characterized to date, most target strains that are phylogenetically or environmentally related to the producing strain.60 Therefore we chose to test strains that were members of the ECC as well as strains that may reside in the same environmental niche as Enterobacter strains. We acknowledge that there are different proposed names for some of the strains we tested, such as the re-classification of E. amnigenus and E. nimipressuralis to the genus Lelliottia.61
We used spot-on-lawn assays in M63 agar against a panel of bacteria using purified cloacaenodin dissolved in water (Figure S18A, Table S4A). Cloacaenodin had no observed activity against E. coli MG1655, B. subtilis 168, Salmonella enterica serovar Newport, Klebsiella aerogenes ATCC 13048, Enterobacter hormaechei ATCC 700323, and Enterobacter kobei BAA-260. However, we observed zones of inhibition in the low micromolar range when we tested cloacaenodin against E. cloacae ATCC 13047, E. mori DSM 26271, E. asburiae DSM 17506, E. amnigenus ATCC 33072, and E. nimipressuralis DSM 18955 (Table 1). E. cloacae is the most clinically relevant strain that we found activity against, as this pathogen is a frequent cause of nosocomial infections around the globe.2 The other strains that we found as susceptible to cloacaenodin are known to be causative agents for certain plant diseases or occasional human pathogens.62-65
Table 1.
Minimal inhibitory concentration (MIC) of cloacaenodin against susceptible strains. An asterisk indicates that the strain is a clinical isolate.
| Species | MIC in M63 media |
|---|---|
| Enterobacter cloacae ATCC 13047 | 7.5 μM (agar) / 940 nM (liquid) |
| E. nimipressuralis DSM 18955 | 1.9 μM (agar) |
| E. asburiae DSM 17506 | 3.8 μM (agar) |
| E. mori DSM 26271 | 1.9 μM (agar) |
| E. amnigenus ATCC 33072 | 500 nM (agar) / 230 nM (liquid) |
| * E. cloacae UCI102 | 1.0 μM (agar) |
| * E. ludwigii MGH216 | 1.0 μM (agar) |
| * E. kobei MGH178 | 2.0 μM (agar) |
| * E. hormaechei UCI35 | 2.0 μM (agar) |
| * E. asburiae MGH243 | 1.0 μM (agar) |
| * E. asburiae UCI193 | 1.0 μM (agar) |
The type strain of E. cloacae (ATCC 13047) tested above was isolated from human cerebrospinal fluid in 189066 before the era of antibiotics. We were motivated to test whether cloacaenodin would have activity against more recent clinical isolates. We tested a panel of 12 clinical isolates of Enterobacter using the spot-on-lawn assay (Table S4B). These isolates are part of a larger collection of carbapenem-resistant Enterobacterales (CRE) that have been extensively analyzed for their genetic mechanisms of resistance.67 In 6 of these strains, 3 of which are classified as resistant to carbapenem antibiotics, we consistently observed low micromolar values of the minimal inhibitory concentration (MIC) of cloacaenodin (Table 1, Figure S18B). In the other 6 clinical isolates (Table S4B), we observed either weak or no activity. The observation of strain-specific activity is consistent with the observed narrow-spectrum activity of ubonodin against Burkholderia strains due to transport through PupB.16,68 These assays demonstrate that cloacaenodin has the potential to serve as a potent therapeutic against Enterobacter strains that have evolved resistance to last-resort antibiotics.
We also tested the bioactivity of a sample of cloacaenodin that had been heated and then HPLC-purified to isolate the unthreaded peptide. At a concentration of 15 μM, we observed no activity of this unthreaded peptide against E. cloacae, consistent with our understanding of the essential nature of the threaded structure for activity (Figure S19).
Cloacaenodin was also tested against E. cloacae and E. amnigenus using a broth microdilution assay in M63 media. We observed an MIC value of 940 nM for E. cloacae and 230 nM for E. amnigenus from this assay, showing that cloacaenodin is more active in solution than on solid media (Table 1, Figure S20). When testing against E. cloacae, we noticed larger OD values for sub-MIC treated wells, which motivated us to see if there was a phenotypic abnormality for these cells. When looking at some of the treated bacteria under the microscope, we observed that E. cloacae cells that were treated with sub-MIC values of cloacaenodin exhibited a degree of filamentation (Figure S21). Filamentation has previously been observed for E. coli cells upon exposure to MccJ25.13
Stability and Bioactivity of Cloacaenodin Variants
To gain insight into the contribution of specific cloacaenodin residues toward its stability and bioactivity, we carried out site-directed mutagenesis on various residues, with the main results summarized in Table 2. We were intrigued by the fact that we saw a peak of unthreaded cloacaenodin in the supernatant extract (Figure S2). We reasoned that increasing the sidechain volume of the steric lock residues Phe22 and Tyr23 may retard the unthreading process. Swapping of the upper steric lock Phe22 to Trp still resulted in comparable levels of unthreaded peptide in the extract, while a Y23W variant only had the threaded peptide observable in its supernatant extract (although the yield was reduced >10-fold) (Figure S22).
Table 2.
Summary of yield, stability, and bioactivity of cloacaenodin variants. For yield, +++ signifies made at near wild-type levels, ++ signifies made at ~40-60% wild-type levels, + signifies made at ≤10% wild-type levels, all as detected by absorbance at UV-215 measured from HPLC. L signifies only detected via LC-MS. Stability was judged by ratio of threaded-to-unthreaded peptide in supernatant extract after purification, judged by LC-MS. N/A stands for not applicable. Variants that were not tested for bioactivity are listed as n.d. for no data.
| Cloacaenodin Variant |
Yield Relative to WT |
Stability Relative to WT |
Bioactivity Relative to WT |
|---|---|---|---|
| F22W | ++ | Comparable | Comparable |
| Y23W | + | More | n.d. |
| S24G | +++ | Less | Abolished |
| S24T | ++ | Comparable | Less |
| S24C | ++ | Comparable | Less |
| S24A | ++ | Comparable | Comparable |
| S24Y | ++ | Comparable | Comparable |
| 10-membered ring | L | Less | n.d. |
| G1A | Not tolerated | N/A | N/A |
| E9D | Not tolerated | N/A | N/A |
| P8A | ++ | Comparable | n.d. |
| V4P | L | More | n.d. |
| Y10A | + | Comparable | Much less |
Since cloacaenodin is unique in having a C-terminal serine compared to other lasso peptides that inhibit RNAP, we next swapped this residue to the more typical C-terminal glycine. To our surprise, when we expressed the S24G variant, we found that the majority of the peptide was now unthreaded in the supernatant extract, based on the later retention time as well as heating and carboxypeptidase assays (Figure S23). This unthreaded peptide completely lost bioactivity when tested against E. amnigenus (Figure S24). However, when expressed intracellularly without the cloD gene present, the S24G variant still inhibited E. coli cell growth, suggesting that at least some fraction of the peptide remains threaded in the cytoplasm and exerts antimicrobial activity (Figure S25). Surprised by the S24G results, we next made conservative substitutions to the S24 residue (cloacaenodin S24A, S24C, and S24T), all of which were similarly stable to wild-type and had similar or decreased bioactivity compared to wild-type (Figure S24). Given the unthreading behavior, we hypothesized swapping the S24 to a bulkier Tyr, Phe, or Trp residue may stabilize the lassoed shape, but we found that these variants still allowed unthreading. The S24Y mutant was purified by HPLC and had similar bioactivity as wild-type (Figure S24).
We next asked if the cloacaenodin biosynthetic enzymes could tolerate an increase in ring size. An increase in ring size from 9 to 10 aa was previously reported for the lasso peptide fuscanodin/fusilassin.69 We cloned a variant of cloacaenodin with an extra alanine near the middle of the ring, so that the new ring sequence would be GHSVADRIPE. We detected a single peak corresponding to the expected mass and isotopic distribution in the supernatant, eluting at about 12.1 minutes, made at about 1% of the yield as wild-type. Heating assays and carboxypeptidase assays of the extract demonstrated that this was an unthreaded peptide (Figure S26). We hypothesize that the lasso peptide is first made threaded by the B and C enzymes, but then readily unthreads upon being secreted into the supernatant and during our purification procedure (Figure S27).
While changes to the isopeptide-bonded residues are generally not tolerated for lasso peptides70,71, several reports demonstrated that certain biosynthetic enzymes could tolerate variations at these positions.23,72,73 For cloacaenodin, G1A and E9D variants were not produced, as the variants could not be detected in the supernatant or the cell pellet.
A report by Hegemann et al. demonstrated that a proline at position 8 in the 9-membered ring of the lasso peptide caulosegnin II was vital for prevention of unthreading at 95 °C.74 For cloacaenodin, we did not observe a dramatic decrease in the ratio of threaded-to-unthreaded peptide in the extract for a cloacaenodin P8A variant (Figure S28). However, swapping the Val4 residue to Pro appeared to favor the threaded peptide, suggesting that rigidification of the ring with proline can stabilize the lasso peptide against unthreading. This increased stability comes at a cost, however, as the V4P variant is only made at about 1% of the wild-type yield (Figure S29).
The Tyr9 sidechain in MccJ25 (corresponding to Tyr10 in cloacaenodin) was shown to hydrogen bond with RNA polymerase in a co-crystal structure.40 We purified a cloacaenodin Y10A variant but found that, despite being threaded, it had drastically reduced activity against E. amnigenus (Figure S30). When expressing this peptide in E. coli without CloD present, the E. coli cells could still grow, suggesting that the Y10A variant lacks activity due to decreased target binding as opposed to decreased transport into susceptible cells (Figure S17).
Discussion and Conclusion
This paper focuses on a novel antimicrobial lasso peptide cloacaenodin. This lasso peptide is particularly notable because of its potent antimicrobial activity against multiple members of the Enterobacter genus, including those implicated in nosocomial infections. While other lasso peptides with antimicrobial activity against Gram-negative bacteria such as klebsidin, capistruin, and citrocin have only modest potency, the MIC of cloacaenodin is in the high nanomolar to single micromolar range for the strains tested here. This potency coupled with the urgency to develop new antimicrobial interventions against the ESKAPE pathogen Enterobacter makes cloacaenodin a promising lead for antibiotic development. We noted that BGCs related to cloacaenodin are present in 5 different species of Enterobacter as well as other enterobacteria (Figure S1). Furthermore, many of the BGCs appear in genomic contexts consistent with being plasmid-borne. The mobility of the cloacaenodin BGC may be due to horizontal gene transfer between these strains which are all part of the gut microbiota. We speculate this horizontal gene transfer occurs because cloacaenodin confers a competitive advantage on the producing cells.
We were initially interested in cloacaenodin not only because of its antimicrobial potential but also because it differs in structure from other antimicrobial lasso peptides targeting Gram-negative bacteria. Specifically, cloacaenodin has a 9-membered ring and a C-terminal serine, in contrast to other characterized examples which have 8-membered rings and C-terminal glycine (Figure 1). Cloacaenodin is also unusually rich in proline residues with 5 prolines, 4 of which are in the 13 aa loop region. As we noted above, the threaded structure of cloacaenodin protects it from proteolysis (Figure 4). The high prevalence of prolines in the structure likely contributes to this proteolytic resistance as well. Proline cis/trans isomerization is known in lasso peptides75, and so in principle cloacaenodin may exist as an ensemble of up to 32 (ie 25) different conformers. In addition to the likely role of proline in engendering cloacaenodin with proteolytic resistance, this conformational flexibility may also be related to the antimicrobial activity.
Methods
Detailed materials and methods used in this study can be found in the supplementary information.
Cloacaenodin BGC Identification
The cloacaenodin BGC was found through a precursor-centric genome mining approach26, where we prioritized hits having a Tyr after the ring as well as a penultimate Tyr. Subsequent BlastP searches were conducted on the cloA gene to identify cloacaenodin-like BGCs in other strains.
Plasmid Construction
For heterologous expression in E. coli, the cloacaenodin BGC was refactored and cloned into the pQE-80 vector. Primers were used to assemble the cloA gene which is under the control of the pT5 promoter, and synthesized gBlocks were used to assemble the cloBCD genes which are under the control of the promoter for mcjBCD. Mutagenic primers were used to generate plasmids to express cloacaenodin variants.
Heterologous Expression and Purification
To generate cloacaenodin and cloacaenodin variants, we used E. coli BL-21 which had been transformed via electroporation with a plasmid to express cloacaenodin or a given variant. The transformed cells were grown at 37 °C with shaking in M9 minimal media (containing M9 salts supplemented with glucose, MgSO4, thiamine, and amino acids) with 100 μg/mL ampicillin. Once the OD600 of the culture reached ~0.2, 1 mM IPTG was added to the culture, which was then allowed to continue growing at room temperature with shaking overnight.
Following expression, the cultures were centrifuged to isolate the supernatant. The supernatant was further purified via a C8 column and eluted with methanol. The methanol extract was dried via rotary evaporation, and the dried extract was resuspended in water (1 mL/L of culture).
The resuspended extracts were purified via HPLC, and masses were verified via LC-MS analysis. Some peptide variants expressed too poorly for HPLC analysis, so they were instead only analyzed via LC-MS. The cloacaenodin Y10A variant required a second round of HPLC purification before use in antimicrobial assays. HPLC-purified peptides were lyophilized before use in later assays.
NMR Data Acquisition and Determination of Cloacaenodin Structure
We acquired NMR spectra, including TOCSY and two NOESY spectra (300 ms and 150 ms mixing time), at the Princeton University Department of Chemistry NMR Facility. We maintained the sample at 4 °C during acquisition to ensure that cloacaenodin remained threaded. Following data acquisition, MNova was used for residue assignment, and through-space distance restraints were manually chosen from the 150 ms mixing time NOESY spectrum. With these residue assignments and distance restraints, CYANA 2.1 was employed to generate 20 structures of cloacaenodin. Each of the 20 structures was then energy minimized with Avogadro.
Evaluation of Antimicrobial Activity
Purified cloacaenodin was dissolved in water and its concentration measured by determining its absorbance at 280 nm using a NanoDrop instrument. A spot-on-lawn assay was employed to evaluate antimicrobial activity against a panel of strains. A strain of interest was cultured overnight in LB media. This culture was then used to inoculate 5 mL of LB, which was grown until mid-exponential phase (OD600 ~ 0.4-0.6). The culture was then used to inoculate 10 mL of melted M63 soft agar at an inoculum of ~107 CFUs/mL. The soft agar was poured atop a plate with 10 mL of M63 agar and allowed to dry. Dissolved cloacaenodin (10 μL) was spotted on the plate at various concentrations and then allowed to dry on the plate. After incubation of the plate overnight, the plate was analyzed for zones of inhibition. The last visible spot was defined as the MIC.
For E. cloacae and E. amnigenus, a broth microdilution assay in M63 media was also used to evaluate antimicrobial activity. The same procedure above was followed, but instead of inoculating M63 soft agar, the culture was used to inoculate liquid cultures (starting OD600 of 0.0005) in a 96-well plate. The plates were then grown with shaking and their OD600 was measured after 8 hours and 16 hours. The lowest concentration with no visible bacterial growth was defined as the MIC. For some of the cultures grown in the plate at sub-MIC concentrations, microscopy was employed to view the morphology of the bacteria.
Proteolytic Susceptibility Assays
Cloacaenodin (or a variant) was incubated with carboxypeptidase B and carboxypeptidase Y at 20 °C for 16 hours in a buffer of 50 mM sodium acetate, pH 6.0. For trypsin digestion, cloacaenodin was incubated in 50 mM ammonium bicarbonate buffer with trypsin in a 1:100 trypsin:peptide weight ratio. The reaction proceeded for 30 minutes to 1 hour before quenching. For α-chymotrypsin digestion, cloacaenodin was incubated in a buffer of 100 mM Tris, 10 mM CaCl2, pH 8.0, with 0.04 mg/mL α-chymotrypsin. The reaction proceeded for 1 hour at 25 °C before quenching. For elastase digestion, cloacaenodin was incubated in a buffer of 50 mM Tris, pH 9.0, with 0.04 mg/mL elastase. The reaction proceeded for 1 hour at 25 °C before quenching. For thermolysin digestion, cloacaenodin was incubated in a buffer of 50 mM Tris, 0.5 mM CaCl2, pH 8.0 with 0.04 mg/mL thermolysin. The reaction proceeded for 1 hour at 30 °C before quenching. All reactions were quenched via addition of 1% formic acid.
Supplementary Material
Acknowledgements
We thank István Pelczer (Princeton University NMR Facility) for his help in acquiring the NMR spectra. We thank Professor Daniel Cohen (Princeton University Department of Mechanical and Aerospace Engineering) and members of the Cohen group for the use of and assistance with microscopy. We thank all members of the Link lab for helpful feedback and discussion.
This work was supported by US National Institutes of Health (NIH) grant GM107036 to A.J.L., as well as NIH grant U19AI110818 to the Broad Institute. We also acknowledge the support of the Princeton University SEAS Funds for the funding towards the Focused Research Team for Precision Antibiotics.
Footnotes
Conflict of Interest
D.V.C. and A.J.L. have applied for a patent on aspects of the antimicrobial activity of cloacaenodin.
Supplementary Information
Detailed experimental methods and materials , including details on cloacaenodin BGC identification, cloacaenodin heterologous expression, NMR analysis, antimicrobial assays, and protease assays, are included in the supplementary information (PDF).
References
- (1).Pendleton JN; Gorman SP; Gilmore BF Clinical Relevance of the ESKAPE Pathogens. Expert Rev Anti-Infe 2013, 11 (3), 297–308. 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- (2).Davin-Regli A; Pagès J-M Enterobacter aerogenes and Enterobacter cloacae; Versatile Bacterial Pathogens Confronting Antibiotic Treatment. Front Microbiol 2015, 6, 392. 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Annavajhala MK; Gomez-Simmonds A; Uhlemann A-C Multidrug-Resistant Enterobacter cloacae Complex Emerging as a Global, Diversifying Threat. Front Microbiol 2019, 10, 44. 10.3389/fmicb.2019.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mezzatesta ML; Gona F; Stefani S Enterobacter cloacae Complex: Clinical Impact and Emerging Antibiotic Resistance. Future Microbiol 2012, 7 (7), 887–902. 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- (5).Davin-Regli A; Lavigne J-P; Pagès J-M Enterobacter Spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin Microbiol Rev 2019, 32 (4). 10.1128/cmr.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bialvaei AZ; Kafil HS Colistin, Mechanisms and Prevalence of Resistance. Curr Med Res Opin 2015, 31 (4), 707–721. 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- (7).Nation RL; Li J Colistin in the 21st Century. Curr Opin Infect Dis 2009, 22 (6), 535–543. 10.1097/qco.0b013e328332e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zong Z; Feng Y; McNally A Carbapenem and Colistin Resistance in Enterobacter: Determinants and Clones. Trends Microbiol 2021, 29 (6), 473–476. 10.1016/j.tim.2020.12.009. [DOI] [PubMed] [Google Scholar]
- (9).Norgan AP; Freese JM; Tuin PM; Cunningham SA; Jeraldo PR; Patel R Carbapenem- and Colistin-Resistant Enterobacter cloacae from Delta, Colorado, in 2015. Antimicrob Agents Ch 2016, 60 (5), 3141–3144. 10.1128/aac.03055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zeng K-J; Doi Y; Patil S; Huang X; Tian G-B Emergence of the Plasmid-Mediated mcr-1 Gene in Colistin-Resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Ch 2016, 60 (6), 3862–3863. 10.1128/aac.00345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kang KN; Klein DR; Kazi MI; Guérin F; Cattoir V; Brodbelt JS; Boll JM Colistin Heteroresistance in Enterobacter cloacae Is Regulated by PhoPQ-Dependent 4-Amino-4-Deoxy-l-Arabinose Addition to Lipid A. Mol Microbiol 2019, 111 (6), 1604–1616. 10.1111/mmi.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Liao W; Cui Y; Quan J; Zhao D; Han X; Shi Q; Wang Q; Jiang Y; Du X; Li X; Yu Y High Prevalence of Colistin Resistance and mcr-9/10 Genes in Enterobacter Spp. in a Tertiary Hospital over a Decade. Int J Antimicrob Ag 2022, 59 (5), 106573. 10.1016/j.ijantimicag.2022.106573. [DOI] [PubMed] [Google Scholar]
- (13).Salomón RA; Farías RN Microcin 25, a Novel Antimicrobial Peptide Produced by Escherichia coli. J Bacteriol 1992, 174 (22), 7428–7435. 10.1128/jb.174.22.7428-7435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Knappe TA; Linne U; Zirah S; Rebuffat S; Xie X; Marahiel MA Isolation and Structural Characterization of Capistruin, a Lasso Peptide Predicted from the Genome Sequence of Burkholderia thailandensis E264. J Am Chem Soc 2008, 130 (34), 11446–11454. 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- (15).Iwatsuki M; Uchida R; Takakusagi Y; Matsumoto A; Jiang C-L; Takahashi Y; Arai M; Kobayashi S; Matsumoto M; Inokoshi J; Tomoda H; Ōmura S Lariatins, Novel Anti-Mycobacterial Peptides with a Lasso Structure, Produced by Rhodococcus jostii K01-B0171. J Antibiot 2007, 60 (6), 357–363. 10.1038/ja.2007.48. [DOI] [PubMed] [Google Scholar]
- (16).Cheung-Lee WL; Parry ME; Zong C; Cartagena AJ; Darst SA; Connell ND; Russo R; Link AJ Discovery of Ubonodin, an Antimicrobial Lasso Peptide Active against Members of the Burkholderia cepacia Complex. Chembiochem 2020, 21 (9), 1335–1340. 10.1002/cbic.201900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cheung-Lee WL; Parry ME; Cartagena AJ; Darst SA; Link AJ Discovery and Structure of the Antimicrobial Lasso Peptide Citrocin. J Biol Chem 2019, 294 (17), 6822–6830. 10.1074/jbc.ra118.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Tsunakawa M; Hu S-L; Hoshino Y; Detlefson DJ; Hill SE; Furumai T; White RJ; Nishio M; Kawano K; Yamamoto S; Fukagawa Y Oki T Siamycins I and II, New Anti-HIV Peptides: I. Fermentation, Isolation, Biological Activity and Initial Characterization. J Antibiot 1995, 48 (5), 433–434. 10.7164/antibiotics.48.433. [DOI] [PubMed] [Google Scholar]
- (19).Metelev M; Arseniev A; Bushin LB; Kuznedelov K; Artamonova TO; Kondratenko R; Khodorkovskii M; Seyedsayamdost MR; Severinov K Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. ACS Chem Biol 2017, 12 (3), 814–824. 10.1021/acschembio.6b01154. [DOI] [PubMed] [Google Scholar]
- (20).Metelev M; Tietz JI; Melby JO; Blair PM; Zhu L; Livnat I; Severinov K; Mitchell DA Structure, Bioactivity, and Resistance Mechanism of Streptomonomicin, an Unusual Lasso Peptide from an Understudied Halophilic Actinomycete. Chem Biol 2015, 22 (2), 241–250. 10.1016/j.chembiol.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Gavrish E; Sit CS; Cao S; Kandror O; Spoering A; Peoples A; Ling L; Fetterman A; Hughes D; Bissell A; Torrey H; Akopian T; Mueller A; Epstein S; Goldberg A; Clardy J; Lewis K Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2. Chem Biol 2014, 21 (4), 509–518. 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Stariha LM; McCafferty DG Discovery of the Class I Antimicrobial Lasso Peptide Arcumycin. Chembiochem 2021, 22 (16), 2632–2640. 10.1002/cbic.202100132. [DOI] [PubMed] [Google Scholar]
- (23).Li Y; Ducasse R; Zirah S; Blond A; Goulard C; Lescop E; Giraud C; Hartke A; Guittet E; Pernodet J-L; Rebuffat S Characterization of Sviceucin from Streptomyces Provides Insight into Enzyme Exchangeability and Disulfide Bond Formation in Lasso Peptides. ACS Chem Biol 2015, 10 (11), 2641–2649. 10.1021/acschembio.5b00584. [DOI] [PubMed] [Google Scholar]
- (24).Shao M; Ma J; Li Q; Ju J Identification of the Anti-Infective Aborycin Biosynthetic Gene Cluster from Deep-Sea-Derived Streptomyces sp. SCSIO ZS0098 Enables Production in a Heterologous Host. Mar Drugs 2019, 17 (2), 127. 10.3390/md17020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Montalbán-López M; Scott TA; Ramesh S; Rahman IR; van Heel AJ; Viel JH; Bandarian V; Dittmann E; Genilloud O; Goto Y; Burgos MJG; Hill C; Kim S; Koehnke J; Latham JA; Link AJ; Martínez B; Nair SK; Nicolet Y; Rebuffat S; Sahl H-G; Sareen D; Schmidt EW; Schmitt L; Severinov K; Süssmuth RD; Truman AW; Wang H; Weng J-K; van Wezel GP; Zhang Q; Zhong J; Piel J; Mitchell DA; Kuipers OP; van der Donk WA New Developments in RiPP Discovery, Enzymology and Engineering. Nat Prod Rep 2021, 38 (1), 130–239. 10.1039/d0np00027b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Maksimov MO; Pelczer I; Link AJ Precursor-Centric Genome-Mining Approach for Lasso Peptide Discovery. Proc Natl Acad Sci USA 2012, 109 (38), 15223–15228. 10.1073/pnas.1208978109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Duquesne S; Destoumieux-Garzón D; Zirah S; Goulard C; Peduzzi J; Rebuffat S Two Enzymes Catalyze the Maturation of a Lasso Peptide in Escherichia coli. Chem Biol 2007, 14 (7), 793–803. 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- (28).Yan K-P; Li Y; Zirah S; Goulard C; Knappe TA; Marahiel MA; Rebuffat S Dissecting the Maturation Steps of the Lasso Peptide Microcin J25 in Vitro. Chembiochem 2012, 13 (7), 1046–1052. 10.1002/cbic.201200016. [DOI] [PubMed] [Google Scholar]
- (29).Cheung-Lee WL; Link AJ Genome Mining for Lasso Peptides: Past, Present, and Future. J Ind Microbiol Biotech 2019, 46 (9–10), 1371–1379. 10.1007/s10295-019-02197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kloosterman AM; Cimermancic P; Elsayed SS; Du C; Hadjithomas M; Donia MS; Fischbach MA; van Wezel GP; Medema MH Expansion of RiPP Biosynthetic Space through Integration of Pan-Genomics and Machine Learning Uncovers a Novel Class of Lanthipeptides. PLOS Biol 2020, 18 (12), e3001026. 10.1371/journal.pbio.3001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Agrawal P; Khater S; Gupta M; Sain N; Mohanty D RiPPMiner: A Bioinformatics Resource for Deciphering Chemical Structures of RiPPs Based on Prediction of Cleavage and Cross-Links. Nucleic Acids Res 2017, 45 (W1), W80–W88. 10.1093/nar/gkx408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kloosterman AM; Shelton KE; van Wezel GP; Medema MH; Mitchell DA RRE-Finder: A Genome-Mining Tool for Class-Independent RiPP Discovery. mSystems 2020, 5 (5), e00267–20. 10.1128/msystems.00267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhong Z; He B; Li J; Li Y-X Challenges and Advances in Genome Mining of Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs). Synthetic Syst Biotechnology 2020, 5 (3), 155–172. 10.1016/j.synbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).de los Santos ELC NeuRiPP: Neural Network Identification of RiPP Precursor Peptides. Sci Rep 2019, 9 (1), 13406. 10.1038/s41598-019-49764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Russell AH; Truman AW Genome Mining Strategies for Ribosomally Synthesised and Post-Translationally Modified Peptides. Comput Struct Biotechnology J 2020, 18, 1838–1851. 10.1016/j.csbj.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li Y; Rebuffat S The Manifold Roles of Microbial Ribosomal Peptide–Based Natural Products in Physiology and Ecology. J Biol Chem 2020, 295 (1), 34–54. 10.1074/jbc.rev119.006545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rebuffat S Ribosomally Synthesized Peptides, Foreground Players in Microbial Interactions: Recent Developments and Unanswered Questions. Nat Prod Rep 2022, 39 (2), 273–310. 10.1039/d1np00052g. [DOI] [PubMed] [Google Scholar]
- (38).Cao L; Do T; Link AJ Mechanisms of Action of Ribosomally Synthesized and Posttranslationally Modified Peptides (RiPPs). J Ind Microbiol Biot 2020, 48 (3–4), kuab005. 10.1093/jimb/kuab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Granato ET; Meiller-Legrand TA; Foster KR The Evolution and Ecology of Bacterial Warfare. Curr Biol 2019, 29 (11), R521–R537. 10.1016/j.cub.2019.04.024 [DOI] [PubMed] [Google Scholar]
- (40).Braffman NR; Piscotta FJ; Hauver J; Campbell EA; Link AJ; Darst SA Structural Mechanism of Transcription Inhibition by Lasso Peptides Microcin J25 and Capistruin. Proc Natl Acad Sci USA 2019, 116 (4), 1273–1278. 10.1073/pnas.1817352116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Solbiati JO; Ciaccio M; Farías RN; González-Pastor JE; Moreno F; Salomón RA Sequence Analysis of the Four Plasmid Genes Required To Produce the Circular Peptide Antibiotic Microcin J25. J Bacteriol 1999, 181 (8), 2659–2662. 10.1128/jb.181.8.2659-2662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mukhopadhyay J; Sineva E; Knight J; Levy RM; Ebright RH Antibacterial Peptide Microcin J25 Inhibits Transcription by Binding within and Obstructing the RNA Polymerase Secondary Channel. Mol Cell 2004, 14 (6), 739–751. 10.1016/j.molcel.2004.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Adelman K; Yuzenkova J; La Porta A; Zenkin N; Lee J; Lis JT; Borukhov S; Wang MD; Severinov K Molecular Mechanism of Transcription Inhibition by Peptide Antibiotic Microcin J25. Mol Cell 2004, 14 (6), 753–762. 10.1016/j.molcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- (44).Kirsch SH; Haeckl FPJ; Müller R Beyond the Approved: Target Sites and Inhibitors of Bacterial RNA Polymerase from Bacteria and Fungi. Nat Prod Rep 2022, 39 (6), 1226–1263. 10.1039/d1np00067e. [DOI] [PubMed] [Google Scholar]
- (45).Pan SJ; Cheung WL; Link AJ Engineered Gene Clusters for the Production of the Antimicrobial Peptide Microcin J25. Protein Expres Purif 2010, 71 (2), 200–206. 10.1016/j.pep.2009.12.010. [DOI] [PubMed] [Google Scholar]
- (46).Hegemann JD; Zimmermann M; Zhu S; Klug D; Marahiel MA Lasso Peptides from Proteobacteria: Genome Mining Employing Heterologous Expression and Mass Spectrometry. Biopolymers 2013, 100 (5), 527–542. 10.1002/bip.22326. [DOI] [PubMed] [Google Scholar]
- (47).Koos JD; Link AJ Heterologous and in Vitro Reconstitution of Fuscanodin, a Lasso Peptide from Thermobifida fusca. J Am Chem Soc 2019, 141 (2), 928–935. 10.1021/jacs.8b10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Allen CD; Chen MY; Trick AY; Le DT; Ferguson AL; Link AJ Thermal Unthreading of the Lasso Peptides Astexin-2 and Astexin-3. ACS Chem Biol 2016, 11 (11), 3043–3051. 10.1021/acschembio.6b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Hegemann JD Factors Governing the Thermal Stability of Lasso Peptides. Chembiochem 2020, 21 (1–2), 7–18. 10.1002/cbic.201900364. [DOI] [PubMed] [Google Scholar]
- (50).Wilson K-A; Kalkum M; Ottesen J; Yuzenkova J; Chait BT; Landick R; Muir T; Severinov K; Darst SA Structure of Microcin J25, a Peptide Inhibitor of Bacterial RNA Polymerase, Is a Lassoed Tail. J Am Chem Soc 2003, 125 (41), 12475–12483. 10.1021/ja036756q. [DOI] [PubMed] [Google Scholar]
- (51).Xie X; Marahiel MA NMR as an Effective Tool for the Structure Determination of Lasso Peptides. Chembiochem 2012, 13 (5), 621–625. 10.1002/cbic.201100754. [DOI] [PubMed] [Google Scholar]
- (52).Jeanne Dit Fouque K; Hegemann JD; Zirah S; Rebuffat S; Lescop E; Fernandez-Lima F Evidence of Cis/Trans-Isomerization at Pro7/Pro16 in the Lasso Peptide Microcin J25. J Am Soc Mass Spectr 2019, 30 (6), 1038–1045. 10.1007/s13361-019-02134-5. [DOI] [PubMed] [Google Scholar]
- (53).Jeanne Dit Fouque K; Lavanant H; Zirah S; Hegemann JD; Fage CD; Marahiel MA; Rebuffat S; Afonso C General Rules of Fragmentation Evidencing Lasso Structures in CID and ETD. Analyst 2018, 143 (5), 1157–1170. 10.1039/c7an02052j. [DOI] [PubMed] [Google Scholar]
- (54).Güntert P; Buchner L Combined Automated NOE Assignment and Structure Calculation with CYANA. J Biomol NMR 2015, 62 (4), 453–471. 10.1007/s10858-015-9924-9. [DOI] [PubMed] [Google Scholar]
- (55).Bayro MJ; Mukhopadhyay J; Swapna GVT; Huang JY; Ma L-C; Sineva E; Dawson PE; Montelione GT; Ebright RH Structure of Antibacterial Peptide Microcin J25: A 21-Residue Lariat Protoknot. J Am Chem Soc 2003, 125 (41), 12382–12383. 10.1021/ja036677e. [DOI] [PubMed] [Google Scholar]
- (56).Rosengren KJ; Clark RJ; Daly NL; Göransson U; Jones A; Craik DJ Microcin J25 Has a Threaded Sidechain-to-Backbone Ring Structure and Not a Head-to-Tail Cyclized Backbone. J Am Chem Soc 2003, 125 (41), 12464–12474. 10.1021/ja0367703. [DOI] [PubMed] [Google Scholar]
- (57).Hegemann JD; Zimmermann M; Xie X; Marahiel MA Caulosegnins I–III: A Highly Diverse Group of Lasso Peptides Derived from a Single Biosynthetic Gene Cluster. J Am Chem Soc 2013, 135 (1), 210–222. 10.1021/ja308173b. [DOI] [PubMed] [Google Scholar]
- (58).Hegemann JD; Zimmermann M; Zhu S; Steuber H; Harms K; Xie X; Marahiel MA Xanthomonins I-III: A New Class of Lasso Peptides with a Seven-Residue Macrolactam Ring. Angew Chem Int Edit 2014, 53 (8), 2230–2234. 10.1002/anie.201309267. [DOI] [PubMed] [Google Scholar]
- (59).Son S; Jang M; Lee B; Hong Y-S; Ko S-K; Jang J-H; Ahn JS Ulleungdin, a Lasso Peptide with Cancer Cell Migration Inhibitory Activity Discovered by the Genome Mining Approach. J Nat Prod 2018, 81 (10), 2205–2211. 10.1021/acs.jnatprod.8b00449. [DOI] [PubMed] [Google Scholar]
- (60).Tan S; Moore G; Nodwell J Put a Bow on It: Knotted Antibiotics Take Center Stage. Antibiotics (Basel) 2019, 8 (3), 117. 10.3390/antibiotics8030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Brady C; Cleenwerck I; Venter S; Coutinho T; de Vos P Taxonomic Evaluation of the Genus Enterobacter Based on Multilocus Sequence Analysis (MLSA): Proposal to Reclassify E. nimipressuralis and E. amnigenus into Lelliottia Gen. Nov. as Lelliottia nimipressuralis Comb. Nov. and Lelliottia amnigena Comb. Nov., Respectively, E. gergoviae and E. pyrinus into Pluralibacter Gen. Nov. as Pluralibacter gergoviae Comb. Nov. and Pluralibacter pyrinus Comb. Nov., Respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia Gen. Nov. as Kosakonia cowanii Comb. Nov., Kosakonia radicincitansx Comb. Nov., Kosakonia oryzae Comb. Nov. and Kosakonia arachidis Comb. Nov., Respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis Nom. Nov., Cronobacter helveticus Comb. Nov. and Cronobacter pulveris Comb. Nov., Respectively, and Emended Description of the Genera Enterobacter and Cronobacter. Syst Appl Microbiol 2013, 36 (5), 309–319. 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- (62).Zhu B; Lou M-M; Xie G-L; Wang G-F; Zhou Q; Wang F; Fang Y; Su T; Li B; Duan Y-P Enterobacter mori sp. nov., associated with bacterial wilt on Morus alba L. Int J Syst Evol Micr 2011, 61 (Pt 11), 2769–2774. 10.1099/ijs.0.028613-0. [DOI] [PubMed] [Google Scholar]
- (63).Koth K; Boniface J; Chance EA; Hanes MC Enterobacter asburiae and Aeromonas hydrophila: Soft Tissue Infection Requiring Debridement. Orthopedics 2012, 35 (6), e996–9. 10.3928/01477447-20120525-52. [DOI] [PubMed] [Google Scholar]
- (64).Westerfeld C; Papaliodis GN; Behlau I; Durand ML; Sobrin L Enterobacter amnigenus Endophthalmitis. Retin Cases Brief Rep 2009, 3 (4), 409–411. 10.1097/icb.0b013e31818a46c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Xue Y; Hu M; Chen S; Hu A; Li S; Han H; Lu G; Zeng L; Zhou J Enterobacter asburiae and Pantoea ananatis Causing Rice Bacterial Blight in China. Plant Dis 2021, 105 (8), 2078–2088. 10.1094/pdis-10-20-2292-re. [DOI] [PubMed] [Google Scholar]
- (66).Ren Y; Ren Y; Zhou Z; Guo X; Li Y; Feng L; Wang L Complete Genome Sequence of Enterobacter cloacae subsp. cloacae Type Strain ATCC 13047. J Bacteriol 2010, 192 (9), 2463–2464. 10.1128/jb.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Salamzade R; Manson AL; Walker BJ; Brennan-Krohn T; Worby CJ; Ma P; He LL; Shea TP; Qu J; Chapman SB; Howe W; Young SK; Wurster JI; Delaney ML; Kanjilal S; Onderdonk AB; Bittencourt CE; Gussin GM; Kim D; Peterson EM; Ferraro MJ; Hooper DC; Shenoy ES; Cuomo CA; Cosimi LA; Huang SS; Kirby JE; Pierce VM; Bhattacharyya RP; Earl AM Inter-Species Geographic Signatures for Tracing Horizontal Gene Transfer and Long-Term Persistence of Carbapenem Resistance. Genome Med 2022, 14 (1), 37. 10.1186/s13073-022-01040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Do T; Thokkadam A; Leach R; Link AJ Phenotype-Guided Comparative Genomics Identifies the Complete Transport Pathway of the Antimicrobial Lasso Peptide Ubonodin in Burkholderia. ACS Chem Biol 2022. 10.1021/acschembio.2c00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).DiCaprio AJ; Firouzbakht A; Hudson GA; Mitchell DA Enzymatic Reconstitution and Biosynthetic Investigation of the Lasso Peptide Fusilassin. J Am Chem Soc 2019, 141 (1), 290–297. 10.1021/jacs.8b09928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Pavlova O; Mukhopadhyay J; Sineva E; Ebright RH; Severinov K Systematic Structure-Activity Analysis of Microcin J25. J Biol Chem 2008, 283 (37), 25589–25595. 10.1074/jbc.m803995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Knappe TA; Linne U; Robbel L; Marahiel MA Insights into the Biosynthesis and Stability of the Lasso Peptide Capistruin. Chem Biol 2009, 16 (12), 1290–1298. 10.1016/j.chembiol.2009.11.009. [DOI] [PubMed] [Google Scholar]
- (72).Liu T; Ma X; Yu J; Yang W; Wang G; Wang Z; Ge Y; Song J; Han H; Zhang W; Yang D; Liu X; Ma M Rational Generation of Lasso Peptides Based on Biosynthetic Gene Mutations and Site-Selective Chemical Modifications. Chem Sci 2021, 12 (37), 12353–12364. 10.1039/d1sc02695j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Zimmermann M; Hegemann JD; Xie X; Marahiel MA The Astexin-1 Lasso Peptides: Biosynthesis, Stability, and Structural Studies. Chem Biol 2013, 20 (4), 558–569. 10.1016/j.chembiol.2013.03.013. [DOI] [PubMed] [Google Scholar]
- (74).Hegemann JD; Fage CD; Zhu S; Harms K; Di Leva FS; Novellino E; Marinelli L; Marahiel MA The Ring Residue Proline 8 Is Crucial for the Thermal Stability of the Lasso Peptide Caulosegnin II. Mol Biosyst 2016, 12 (4), 1106–1109. 10.1039/c6mb00081a. [DOI] [PubMed] [Google Scholar]
- (75).Schröder HV; Zhang Y; Link AJ Dynamic Covalent Self-Assembly of Mechanically Interlocked Molecules Solely Made from Peptides. Nat Chem 2021, 13 (9), 850–857. 10.1038/s41557-021-00770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.