Abstract
The Wnt-β-catenin signal transduction pathway is essential for embryonic development and adult tissue homeostasis. Wnt signaling converts TCF from a transcriptional repressor to an activator in a process facilitated by the E3 ligase XIAP. XIAP-mediated monoubiquitylation of the transcriptional corepressor Groucho (also known as TLE) decreases its affinity for TCF, thereby allowing the transcriptional coactivator β-catenin to displace it on TCF. Through a genome-scale screen in cultured Drosophila melanogaster cells, we identified the deubiquitylase USP47 as a positive regulator of Wnt signaling. We found that USP47 was required for Wnt signaling during Drosophila and Xenopus laevis development, as well as in human cells, indicating evolutionary conservation. In human cells, knockdown of USP47 inhibited Wnt reporter activity, and USP47 acted downstream of the β-catenin destruction complex. USP47 interacted with TLE3 and XIAP but did not alter their amounts; however, knockdown of USP47 enhanced XIAP-mediated ubiquitylation of TLE3. USP47 inhibited ubiquitylation of TLE3 by XIAP in vitro in a dose-dependent manner, suggesting that USP47 is the deubiquitylase that counteracts the E3 ligase activity of XIAP on TLE. Our data suggest a mechanism by which regulated ubiquitylation and deubiquitylation of TLE enhances the ability of β-catenin to cycle on and off TCF, thereby helping to ensure that the expression of Wnt target genes continues only as long as the upstream signal is present.
Introduction
The Wnt-β-catenin signaling pathway plays an essential role in animal development and in maintenance of homeostasis in adult tissues (1, 2). Mutations that disrupt the Wnt pathway result in human diseases, including cancer. The critical effector of the canonical Wnt pathway is the transcriptional coactivator β-catenin. In the absence of Wnt ligands, β-catenin is maintained at low amounts due to constitutive proteasome-dependent degradation driven by a destruction complex composed of the tumor suppressor adenomatous polyposis coli (APC), the scaffold protein Axin, and the kinases casein kinase 1α (CK1α) and glycogen synthase kinase 3 (GSK3) (3, 4). The binding of Wnt ligands to the coreceptors low-density lipoprotein receptor 5 or 6 (LRP5/6) and Frizzled (Fzd), inhibits the destruction complex, resulting in stabilization of β-catenin. Subsequently, β-catenin translocates to the nucleus, where its binding to transcription factors of the T cell factor and lymphocyte enhancer factor (TCF/LEF) family initiates a Wnt transcriptional program (1).
In the absence of a Wnt ligand, the transcriptional corepressors Groucho (Gro) in invertebrates and Transducin-like enhancer (TLE) in vertebrates form a complex with TCF/LEF to inhibit the transcription of Wnt target genes (5). The process by which β-catenin competes with Gro or TLE (Gro/TLE) binding to TCF/LEF is proposed to be a regulated event that requires the formation of a large molecular weight nuclear complex or “enhanceosome” (1, 6). Our laboratory previously identified an E3 ubiquitin ligase, X-linked inhibitor of apoptosis (XIAP), as a positive regulator of Wnt signaling (7, 8). Wnt signaling recruits XIAP to TCF/LEF to promote ubiquitylation of Gro/TLE that is bound to TCF/LEF. Ubiquitylated Gro/TLE exhibits decreased affinity for TCF/LEF, thereby allowing β-catenin to effectively compete for TCF/LEF binding.
Our previous work suggests the presence of a deubiquitylase (DUB) that acts on Gro/TLE to reverse the action of XIAP and “recharge” Gro/TLE for reassembly of the Gro/TLE-TCF/LEF complex (7). Here, we provide evidence that the deubiquitylase ubiquitin-specific protease 47 (USP47) is an evolutionarily conserved positive regulator of Wnt signaling that reverses the action of XIAP by deubiquitylating Gro/TLE in cells and in vitro. Together, our data highlight a mechanism by which XIAP and USP47 promote and oppose Gro/TLE ubiquitylation, respectively, enhancing the ability of β-catenin to cycle on and off TCF, thereby promoting the β-catenin–dependent regulation of Wnt target genes.
Results
RNAi screen identifies Usp47 as a positive regulator of Wg signaling in Drosophila
We previously identified XIAP as an E3 ligase that enhances signaling induced by Wingless (Wg), the best characterized Wnt homolog in flies (7). Using a similar approach, we performed a genome-scale RNA interference (RNAi)-based screen in Drosophila melanogaster cells to identify deubiquitylases that regulate Wg signaling (fig. S1A). Briefly, plasmids containing cDNAs corresponding to predicted and experimentally verified deubiquitylases from the Drosophila Gene Collection (Releases 1 and 2) were used as PCR templates to generate cDNAs for in vitro transcription of double-stranded RNA (dsRNA) (9). Knockdowns were performed by the addition of dsRNAs to Drosophila S2R+ cells, a Wg-sensitive isolate of the S2 cell line stably expressing the Wg-responsive luciferase reporter TOPflash (10, 11). Using this approach, we found a significant reduction in Wg-induced signaling upon knockdown of Usp47 (also called Ubp64E), which encodes the Drosophila homolog of the vertebrate deubiquitylase USP47 (fig. S1B).
Usp47 is required for Wg signaling in Drosophila
During development, Wg signaling controls patterning of the dorsoventral boundary of the Drosophila wing. Disruption of Wg signaling results in the loss of specification of sensory neurons at this boundary (12). To determine whether Usp47 is required for Wg signaling in vivo, we depleted Usp47 transcripts by RNAi-mediated knockdown in third instar larval wing imaginal discs (the precursors of the adult wing). Selective knockdown in the posterior compartment of the wing disc was achieved using the hedgehog (hh)-Gal4 driver, and effects on Wg signaling were assayed by examining expression of the Wg target gene senseless (sens) along the dorsoventral boundary. In contrast to the knockdown of the control gene yellow (y), knockdown of Usp47 in the posterior compartment of the wing disc markedly reduced sens expression (Fig. 1). To rule out off-target effects, these experiments were repeated with a different Usp47 RNAi construct and led to a similar reduction in sens expression (Fig. 1A). Notably, RNAi-mediated knockdown of Usp47 did not decrease wg expression, even though sens reporter (Scar-Sens) expression was decreased (fig. S2). These findings indicate that the reduction in sens expression resulting from Usp47 depletion was not due to a reduction in Wg. Consistent with these results, Shi et al. reported that Usp47 depletion in the wing disc resulted in decreased accumulation of Armadillo (the Drosophila homolog of β–catenin) and attenuated expression of sens (13). Together, these findings provide in vivo evidence supporting the role of Usp47 as a positive regulator of Wg signaling.
Fig. 1. Usp47 is required for the expression of the Wg target gene sens in the Drosophila wing imaginal disc.
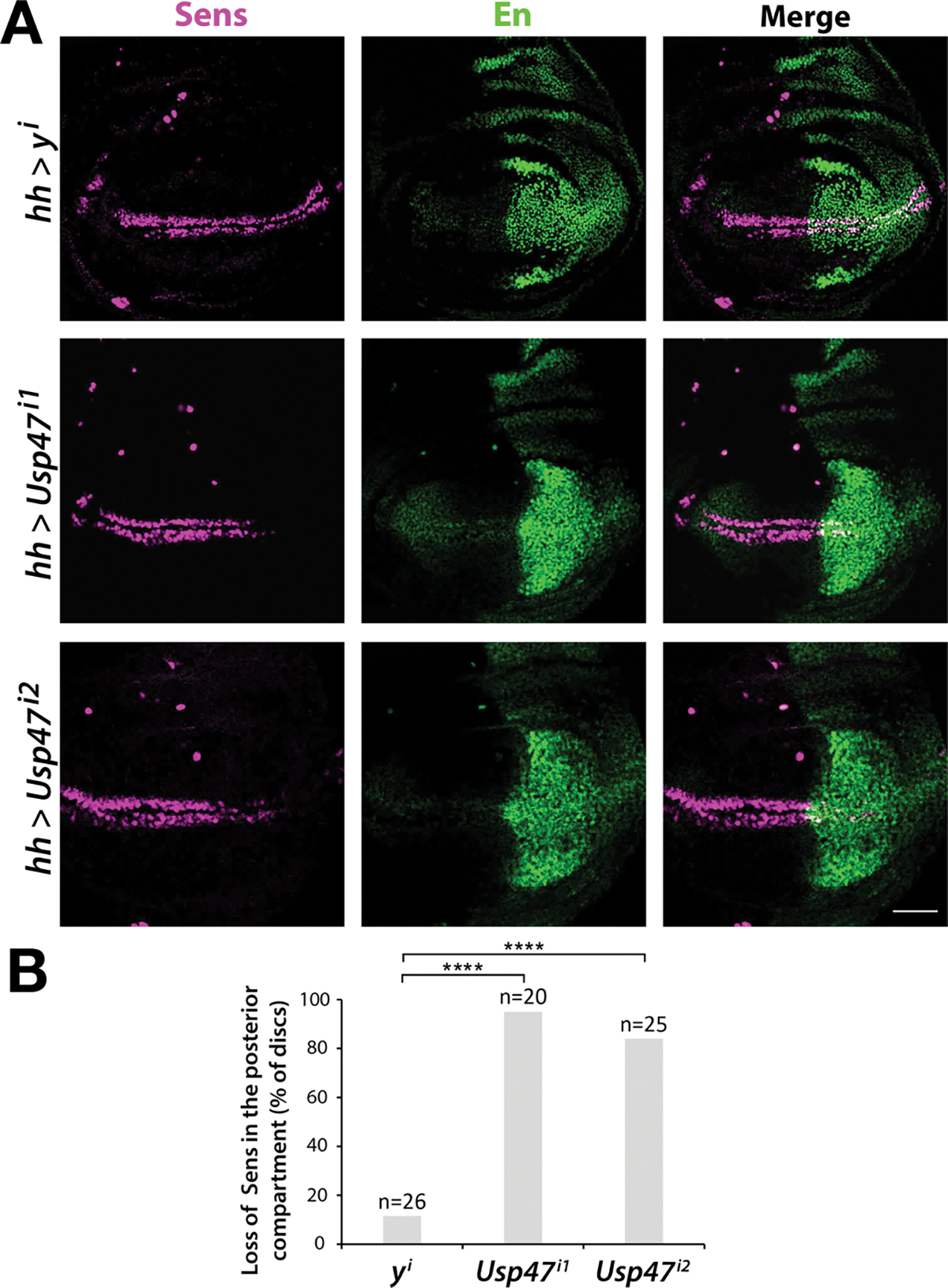
(A) Immunofluorescence staining for Sens and En in third instar wing imaginal discs in which Usp47 was knocked down by RNAi in the posterior compartment (marked by En) using the hh-Gal4 driver. Usp47 knockdown was performed in two independent RNAi lines (i1 and i2). RNAi-mediated knockdown of y is a negative control. Scale bar, 50 μm. (B) The percentage of discs that showed decreased Sens, as determined by visualization of partial or complete loss of Sens stripes in the posterior wing disc. **** p < 0.0001. Significance was assessed using two-tailed Student’s t-test versus y control. Significance was not corrected for multiple comparisons. N = 20–25 discs per group.
We further characterized the effects of Usp47 depletion in the wing by combining CRISPR-based mutagenesis with tissue-specific expression of Cas9 and Usp47 single guide RNAs (sgRNAs). By comparison to ebony control sgRNAs, expression of Usp47 sgRNAs in the wing using the vestigial B (vgB)-Gal4 driver resulted in increased notches at the wing periphery, a hallmark of Wg signaling inactivation. To rule out potential off-target effects, we used vgB-Gal4 to express sgRNAs that target different parts of the Usp47 coding region, which also resulted in aberrant notching at the wing margin (fig. S3). Furthermore, isolation of Usp47 null alleles using CRISPR-mediated mutagenesis (see Materials and Methods) resulted in nearly complete pupal lethality, revealing that Usp47 is essential during development. Together, these findings support the conclusion that Usp47 is a positive regulator of Wg signaling in Drosophila.
Usp47 is required for Wnt signaling in Xenopus embryos and is present throughout development
To assess whether Usp47 plays a similar role in the Wnt pathway in vertebrates, we examined if knockdown or overexpression of Usp47 disrupted anterior-dorsal structure formation in Xenopus laevis embryos, a process mediated by Wnt signaling (14). Knockdown of Usp47 by dorsal injection of a usp47-targeting morpholino resulted in severely ventralized embryos, consistent with Wnt pathway inhibition (Fig. 2A). The mutant phenotype was rescued by co-injection with mouse usp47 mRNA, demonstrating the specificity of the injected usp47-targeting morpholino (Fig. 2A).
Fig. 2. Loss and gain of Usp47 perturbs axis formation in Xenopus embryos.
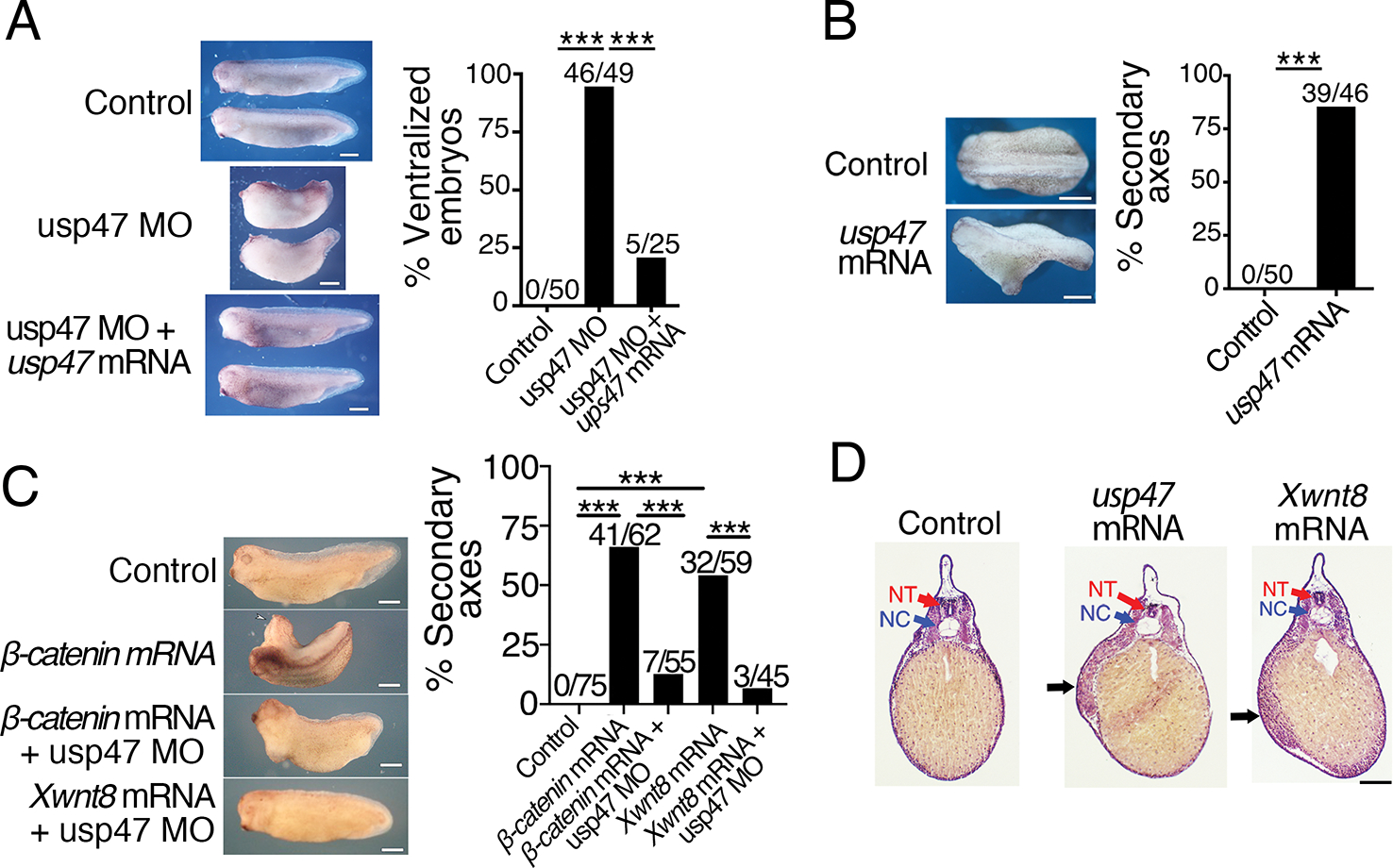
(A) Xenopus embryos at the 4-cell stage were injected dorsally with control morpholino, usp47 Morpholino (MO), or usp47 MO plus usp47 mRNA, and the dorsal-anterior index (DAI) was determined as previously described (53). The percentage of ventralized embryos (DAI ≤ 2) was quantified, and absolute numbers are shown above the bars in the graph. Representative embryos are shown. n=25–50 embryos per treatment group in each of 3 independent experiments. ***p < 0.0005. Significance was assessed using Chi-square test with Bonferroni correction. Scale bar, 1 mm. (B) 4-cell embryos were injected ventrally with control or usp47 mRNA. The percentage of embryos with secondary axis formation was quantified, and absolute numbers are shown above bars in the graph. Representative embryos are shown. n= 45–50 embryos per treatment group in each of 3 independent experiments. ***p < 0.005. Significance was assessed using Chi-square test. Scale bar, 1 mm. (C) Xenopus embryos were injected with control MO, β-catenin mRNA, XWnt8 mRNA, β-catenin mRNA plus usp47 MO, or XWnt8 mRNA plus usp47 MO. The percentage of embryos with secondary axis formation was quantified, and the absolute numbers are shown above bars in the graph. Representative embryos are shown. n= 45–75 embryos per treatment group in each of 3 independent experiments. **p < 0.0003. Significance was assessed using Chi-square test with Bonferroni correction. Scale bar, 1 mm. (D) Embryos injected with control, usp47, or Xwnt8 mRNA were fixed, sectioned, and stained with hematoxylin and eosin. Black arrows point to ectopic mesoderm. NT, neural tube; NC, notochord. Scale bar, 200 μm. Images are representative of n > 5 embryos per treatment group.
Overexpression of Usp47 by ventral injection of usp47 mRNA resulted in partial axis duplication, consistent with Wnt pathway activation (Fig. 2B). Conversely, knockdown of Usp47 by injection of a usp47-targeting morpholino suppressed secondary axis formation due to overexpression of β-catenin or XWnt8 mRNA (Fig. 2C). To examine the duplicated axes in more detail, usp47-injected embryos were sectioned and stained with hematoxylin and eosin (H&E). The H&E staining revealed muscle tissue in the secondary trunk axis that is identical to the phenotype induced by injection of low amounts of Xwnt8 mRNA (Fig. 2D).
To determine the timing of Usp47 production and its localization during Xenopus development, we performed reverse transcription-polymerase chain reaction (RT-PCR). The RT-PCR analysis revealed that usp47 is present in the unfertilized egg, gastrula, neurula, and late tailbud stages (fig. S4A). In situ hybridization analysis revealed a pattern of usp47 staining similar to that reported for β-catenin (15) (fig. S4B–H). usp47 staining was localized to the animal half of the early embryo up to the gastrula stage (fig. S4B–E). At the neurula stage, usp47 was localized to the anterior and posterior ends of the embryo (fig. S4F), whereas at the tailbud stage, usp47 staining was localized to the branchial arches, eye, and posterior end of the embryo (fig. S4G). At the tadpole stage, usp47 staining was localized primarily to the head and spinal cord (fig. S4H). Taken together, these data suggest that Usp47 is present throughout early developmental stages and is required for Xenopus development.
USP47 is required for Wnt signaling in cultured mammalian cells and acts downstream of the β-catenin destruction complex
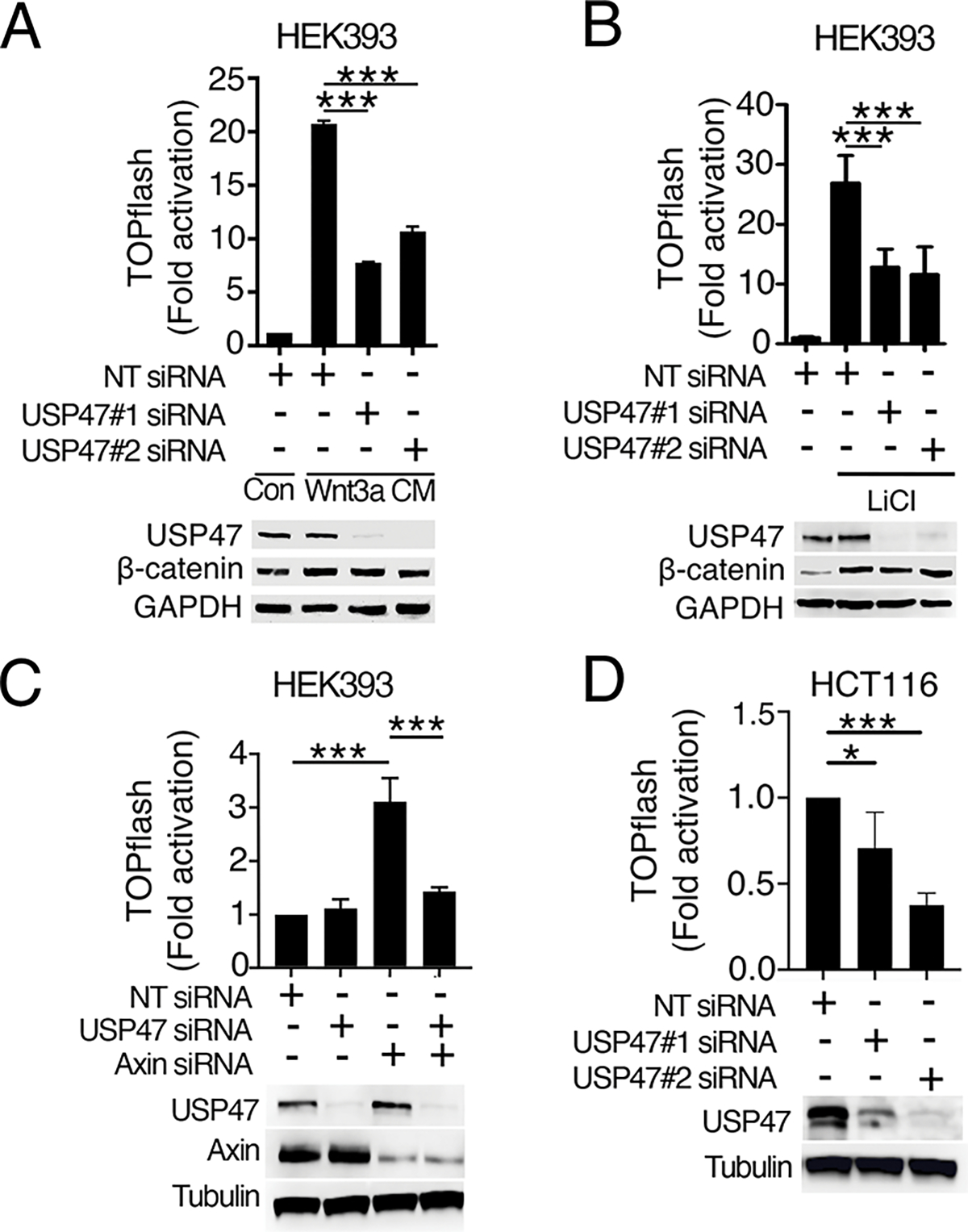
To determine if USP47 is required for Wnt signaling in mammalian cells, we tested the effects of USP47 knockdown on Wnt3a-induced pathway activation using a human cell line stably transfected with the luciferase-based Wnt reporter SuperTOPflash (HEK293STF) (16). Knockdown of USP47 with two independent short-interfering RNA (siRNA) constructs blocked Wnt3a-induced reporter activation (Fig. 3A), indicating that USP47 is required for Wnt-mediated transcriptional activation.
Fig. 3. USP47 is required for Wnt signaling in human cells and functions downstream of β-catenin stabilization.
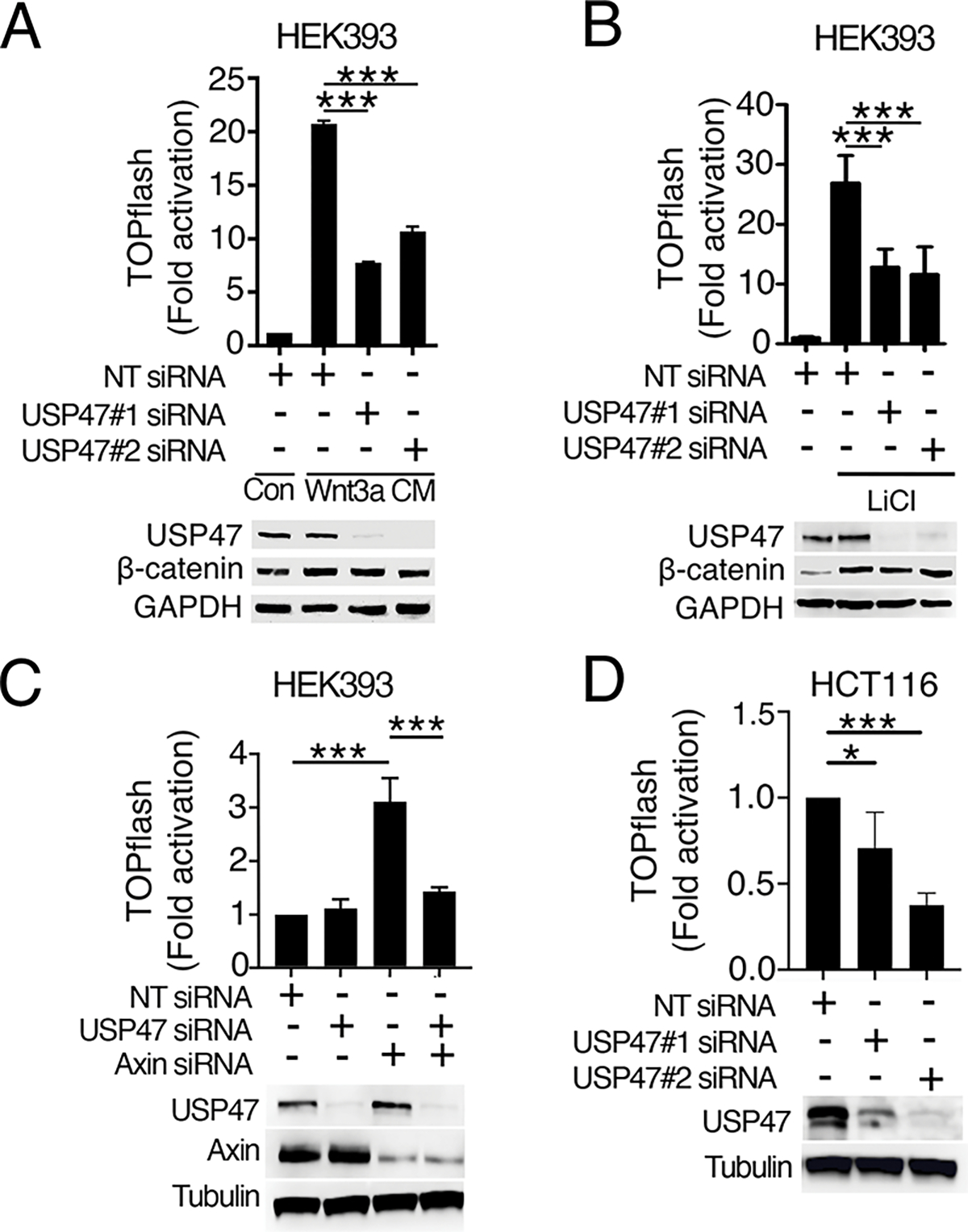
(A and B) HEK293STF cells were transfected with non-targeting (NT) control or two independent USP47 siRNAs (siRNA#1 and #2) and either treated with control conditioned media from L cells (Con) or conditioned media from L cells expressing Wnt3A (Wnt3A CM) (A) or treated with LiCl (B). TOPflash reporter activity was quantified, and cell extracts were immunoblotted for USP47 and β-catenin. GAPDH is a loading control. (C) HEK293STF cells were transfected as indicated with NT control, USP47 siRNAs, Axin siRNAs, or USP47 and Axin siRNAs. TOPflash reporter activity was quantified, and cell extracts were immunoblotted for USP47 and Axin. Tubulin is a loading control. (D) HCT116STF cells were transfected with NT control or two independent USP47 siRNAs. TOPflash reporter activity was quantified, and cell extracts were immunoblotted for USP47. Graphs show mean ± SD of TOPflash normalized to cell number and NT control. *p<0.05, ***p<0.005. Significance was assessed using two-tailed Student’s t-test versus NT or Axin knockdown. Significance was not corrected for multiple comparisons. All TOPflash results and blots are representative of at least three independent experiments. N = 3–4 wells of cells per treatment group per experiment.
Previous studies indicated that USP47 deubiquitylates β-catenin to inhibit its degradation (13). Consistent with this result, we found that knockdown of USP47 detectably reduced the amount of β-catenin (Fig. 3A, S5A). We did not, however, detect a significant effect of USP47 knockdown on β-catenin ubiquitylation (fig. S5B–C), likely due to the limited sensitivity of the assay. Inhibiting the activity of the destruction complex kinase, GSK3, with lithium blocked the decrease in β-catenin upon USP47 knockdown (Fig 3B, S5D). This suggests that USP47 stabilizes β-catenin by reversing GSK3-dependent β-catenin ubiquitylation. Even though the amount of β-catenin was unchanged with USP47 knockdown in lithium-treated HEK293STF cells (Fig 3B, S5D), Wnt reporter activity was still inhibited. Based on these results, we reasoned that USP47 must have an additional role in regulating Wnt signaling independent of its effects on β-catenin turnover and downstream of the β-catenin destruction complex.
To test whether USP47 functions downstream of the β-catenin destruction complex, we knocked down USP47 in HEK293STF cells in which the destruction complex scaffold protein, Axin, was also knocked down. Activation of the Wnt pathway upon Axin knockdown was inhibited by USP47 knockdown (Fig. 3C, S5E). Further confirmation that USP47 acts downstream of the destruction complex was obtained by siRNA-mediated knockdown of USP47 in the colorectal cancer cell line HCT116, which exhibits constitutive Wnt signaling due to a stabilizing mutation in β-catenin (17). We found that USP47 knockdown inhibited Wnt signaling in HCT116 cells (Fig. 3D) (18). These results suggest that USP47 is required for Wnt signaling downstream of the β-catenin destruction complex.
To assess whether USP47 could be acting in the nucleus to regulate Wnt signaling, we determined the localization pattern of endogenous USP47 in the mouse intestine (fig. S6A). We found that USP47 was predominantly nuclear throughout the small intestine and in colon crypts. Immunostaining of USP47 in HEK293T and RKO human cell lines further confirmed the strong nuclear localization pattern of USP47 (fig. S6B). In addition, we did not observe any change in USP47 localization upon Wnt stimulation (fig. S6C). To determine whether depletion of USP47 affected the accumulation of nuclear β-catenin upon Wnt stimulation, we performed single-cell analysis with two independent HEK293T clonal lines that have greatly reduced amounts of USP47 due to CRISPR-Cas9 editing (fig. S6D). We found that depletion of USP47 did not affect the accumulation of nuclear β-catenin upon Wnt3a treatment for the two cell lines when compared to their parental control (fig. S6E).
USP47 interacts with XIAP and TLE3 but does not affect their stability
To determine the nuclear role of USP47 in the Wnt pathway, we identified potential USP47 binding partners. USP47 coimmunoprecipitated with XIAP when tagged versions of both proteins were overexpressed (Fig. 4A) in HEK293 cells. Because our previous studies demonstrated that XIAP ubiquitylates TLE3 (7), we sought to determine whether USP47 interacts with TLE3. We found that overexpressed XIAP and TLE3 coimmunoprecipitated with endogenous USP47 (Fig. 4B). Coimmunoprecipitation of endogenous USP47, XIAP, and TLE3 from HEK293T cells further confirmed their interactions (Fig. 4C). To determine whether all three proteins can form a complex, we performed a competitive binding assay. We found that TLE3 inhibited the interaction between USP47 and XIAP, suggesting that TLE3 and USP47 compete for binding to XIAP and that the three proteins do not form a heterotrimeric complex (Fig. 4D, S7A).
Fig. 4. USP47 interacts with XIAP and TLE3.
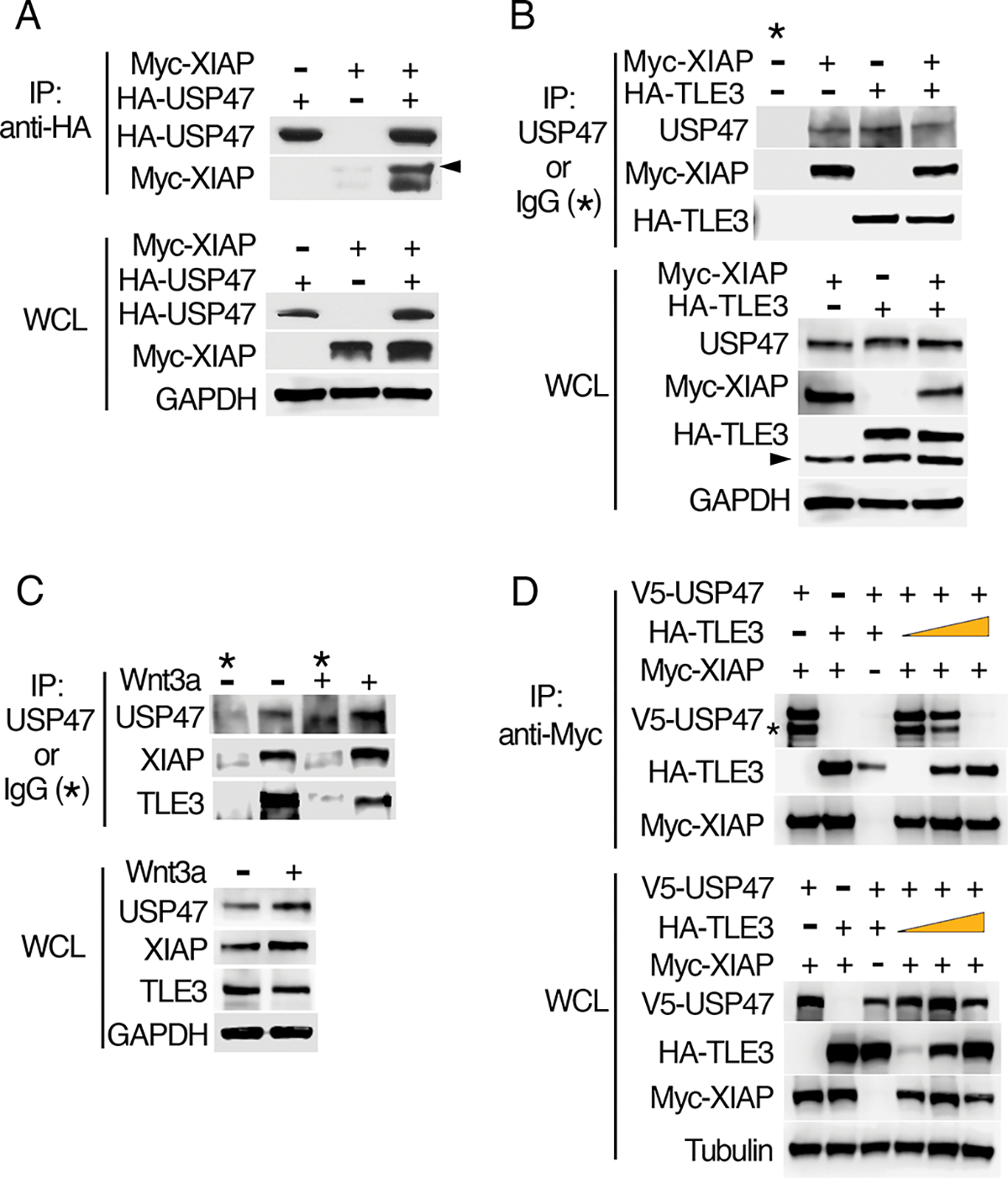
(A) HEK293 cells were transfected with plasmids encoding Myc-XIAP and HA-USP47as indicated. HA immunoprecipitates (IP) and whole-cell lysates (WCL) were immunoblotted for HA and Myc. Arrowhead indicates autoubiquitylated Myc-XIAP. GAPDH is a loading control. (B) HEK293 cells were transfected with plasmids encoding Myc-XIAP and HA-TLE3 as indicated, and cell extracts were immunoprecipitated with an antibody recognizing endogenous USP47 or with IgG (negative control). Co-immunoprecipitated Myc-XIAP and HA-TLE3 were detected by immunoblotting for Myc and HA, respectively. Arrowhead indicates nonspecific band. (C) HEK293 cells were incubated in the presence or absence of purified recombinant Wnt3a, and cell extracts were immunoprecipitated with a USP47-specific antibody or IgG (negative control). Co-immunoprecipitated, endogenous XIAP and TLE3 were detected by immunoblotting. (D) HEK293 cells were transfected with the indicated expression constructs, and Myc-XIAP was immunoprecipitated. Co-immunoprecipitated V5-USP47 and HA-TLE3 were detected by immunoblotting for V5 and HA, respectively. Asterisk marks proteolytic fragment of USP47 occasionally observed during the immunoprecipitation step. All immunoblots are representative of at least three independent experiments.
Because deubiquitylating enzymes are known to stabilize their partner E3 ligases by counteracting their autoubiquitylation, we sought to determine whether USP47 affects steady-state amounts of XIAP. We found that siRNA-mediated knockdown of USP47 had no observable effect on the amounts of XIAP (Fig. 5A, S7B). Similarly, knockdown of USP47 did not noticeably affect the amounts of TLE3. We next tested the possibility that USP47 is a substrate of XIAP. We found that neither XIAP overexpression (Fig. 5B, S7C) nor knockdown (Fig. 5C, S7D) resulted in an observable change in USP47 amounts, suggesting that XIAP does not affect the stability of USP47. Finally, we demonstrated that the mobility of USP47 did not change in an in vitro ubiquitylation assay with XIAP, demonstrating that USP47 is not likely to be a substrate of XIAP in cells (Fig. 5D).
Fig. 5. USP47 and XIAP do not affect one another’s stability.
(A) HEK293 cells were transfected with non-targeting (NT) control or two independent USP47 siRNAs (USP47 #1 and #2). Whole-cell lysate was collected and immunoblotted for endogenous USP47, TLE3, and XIAP. GAPDH is loading control. (B) HEK293 cells were transfected as indicated with vector control (Con) or Myc-XIAP and treated with purified recombinant Wnt3a prior to immunoblotting for USP47 and XIAP. (C) HEK293 cells transfected with NT control or two independent XIAP siRNAs (XIAP #1 and #2) were incubated in the presence or absence of Wnt3a and cell extracts were immunoblotted for USP47 and XIAP. (D) In vitro–translated USP47 was incubated in an in vitro ubiquitylation assay with recombinant proteins as indicated and USP47 was detected by immunoblotting. All immunoblots are representative of at least three independent experiments.
USP47 deubiquitylates TLE3 in cells and in vitro
We previously showed that XIAP ubiquitylates TLE3 to reduce its affinity for TCF/LEF (7). We speculated that USP47 acts as a deubiquitylase for TLE3 to “recharge” the system, thereby allowing for rebinding of TLE3 to TCF/LEF. To test whether USP47 acts as a deubiquitylase for TLE3, we knocked down USP47 and overexpressed tagged forms of TLE3 (Myc-TLE3), ubiquitin (His6-Ub), and XIAP (FLAG-XIAP) in HEK293 cells. In the presence of overexpressed XIAP, monoubiquitylated TLE3 species, which are readily detected by immunoblotting (7), increased upon USP47 knockdown (Fig. 6A). To more directly determine whether USP47 deubiquitylates TLE3, we performed an in vitro ubiquitylation assay with ubiquitylated TLE3 and recombinant USP47 protein. USP47 decreased the amount of ubiquitylated TLE3 in vitro in a dose-dependent manner (Fig. 6B). To test how deubiquitylation of TLE3 by USP47 alters TLE3 binding to TCF, we performed an in vitro binding assay and found that TLE3 was pulled down with TCF4, but ubiquitylated TLE3 was not (Fig. 6C), consistent with our previous results (7). In agreement with the greater affinity of deubiquitylated TLE3 for TCF4, the addition of USP47 increased the total amount of TLE3 that was pulled down with TCF4. These results indicate that USP47 directly deubiquitylates TLE3 to oppose the activity of XIAP (Fig. 7).
Fig. 6. USP47 deubiquitylates TLE3.
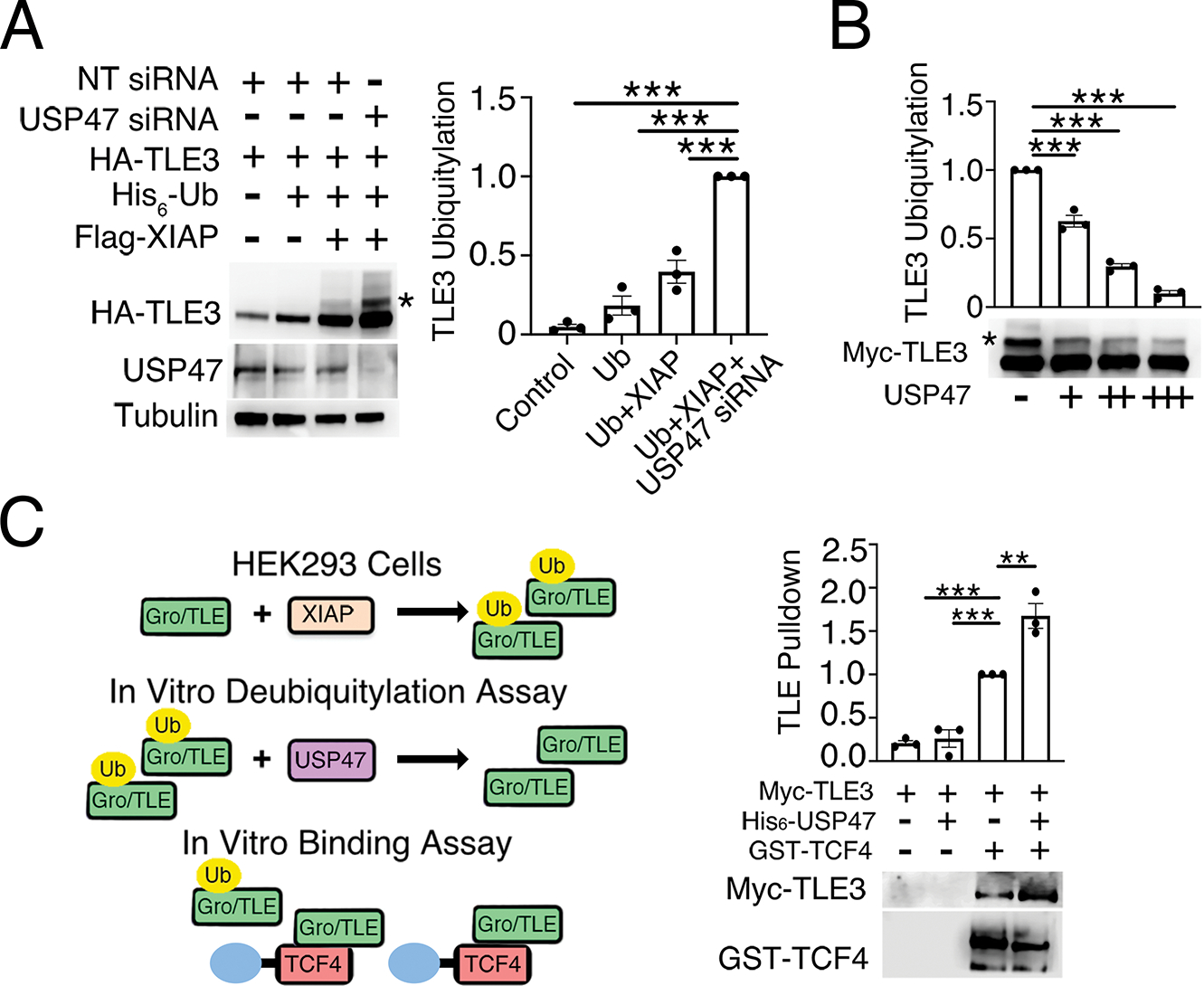
(A) HEK293 cells were transfected with the indicated expression constructs, and whole-cell lysates were immunoblotted for HA and USP47. Tubulin is loading control. Ubiquitylated TLE3 was quantified and normalized to non-targeting (NT) control. Graph shows mean ± SEM of three independent replicates. ***p<0.005 (two-tailed Student’s t-test) versus Ub plus XIAP. (B) Ubiquitylated Myc-TLE3 was incubated with increasing amounts of recombinant USP47 protein in an in vitro deubiquitylation assay. Ubiquitylated TLE3 was detected by immunoblotting for Myc, quantified, and normalized to buffer control. Graph shows mean ± SEM of three independent experiments. ***p<0.005 (two-tailed Student’s t-test) versus buffer control. Significance was not corrected for multiple comparisons. (C) Ubiquitylated Myc-TLE3 was isolated from HEK293 cells and incubated with recombinant His-tagged USP47 protein in an in vitro deubiquitylation reaction followed by a binding assay performed with purified GST-TCF4. Bound Myc-TLE3 was eluted with sample buffer and detected by immunoblotting for Myc. Quantification of Myc-TLE3 immunoblots was normalized to buffer control. Graph shows mean ± SEM of three independent experiments. **p<0.01, ***p<0.005 (two-tailed Student’s t-test) versus no USP47 control. Significance was not corrected for multiple comparisons. All immunoblots are representative of at least three independent experiments.
Fig. 7. Model for XIAP and USP47 in regulating Gro/TLE ubiquitylation in the Wnt pathway.
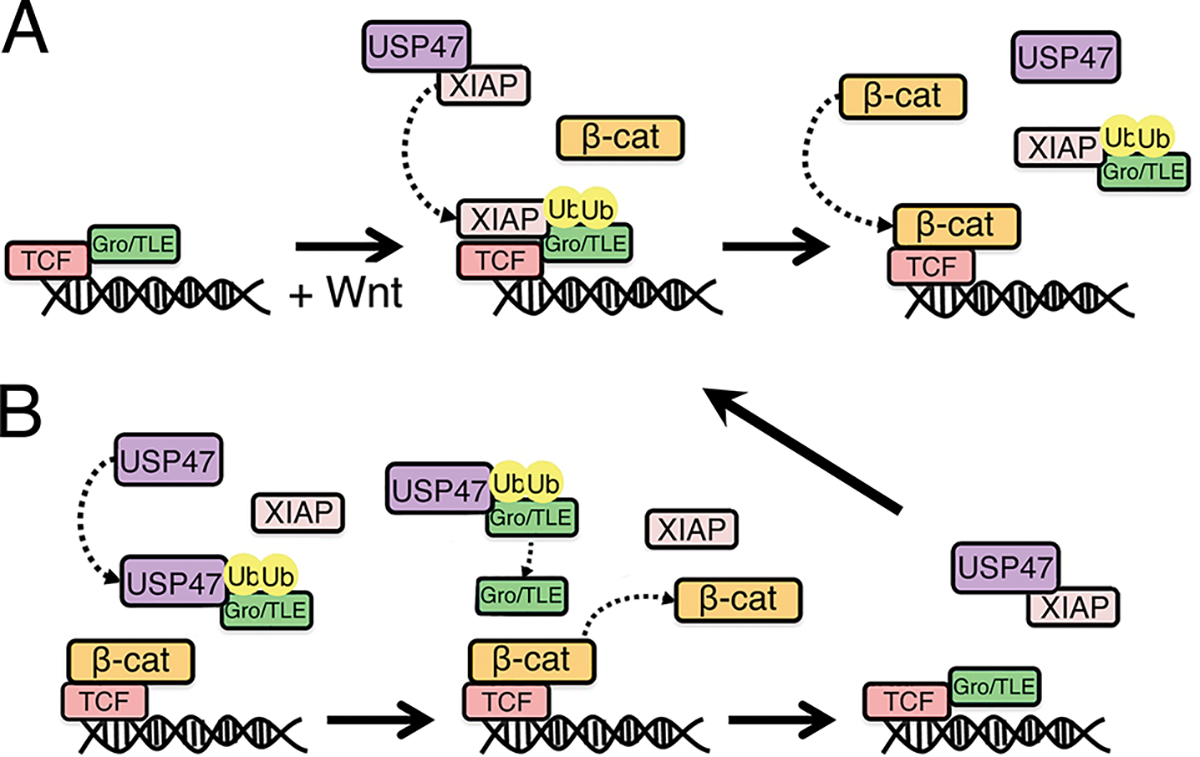
(A) Upon Wnt signaling, XIAP ubiquitylates Gro/TLE, which promotes Gro/TLE dissociation from TCF to allow binding of β-catenin to TCF. (B) USP47 deubiquitylates Gro/TLE, thereby allowing it to displace β-catenin and rebind to TCF, setting up a cycle of β-catenin binding to and dissociation from TCF.
High USP47 correlates with reduced survival in pancreatic cancer patients
USP47 is abundant in several types of cancer (19). Using the cBioPortal in The Cancer Genome Atlas (20, 21), we found that high amounts of USP47 were significantly associated with reduced survival of patients with pancreatic cancer (fig. S8). Recurrent inactivating mutations in RNF43, a ubiquitin ligase that targets Wnt receptors, have also been found in pancreatic cancer (22). Future studies will focus on whether high amounts of USP47 potentiate Wnt signaling in pancreatic cancers with RNF43 mutations.
Discussion
Here, we showed that USP47 is an evolutionarily conserved positive regulator of the Wnt pathway. USP47 was previously reported to deubiquitylate β-catenin, thereby increasing its stability (13). We observed no noticeable effect on β-catenin ubiquitylation upon knockdown of USP47, raising the possibility that multiple deubiquitylases may be involved in regulating this process under normal conditions or that others can compensate when USP47 is reduced or absent. Our findings indicate that USP47 also acts in the nucleus to regulate Wnt-mediated transcription by catalyzing the deubiquitylation of Gro/TLE. We previously proposed a model in which Wnt signaling promotes recruitment of XIAP to Gro/TLE-TCF/LEF; subsequently, XIAP ubiquitylates Gro/TLE to facilitate its dissociation and allow β-catenin binding to TCF/LEF (Fig. 7A). Furthermore, we showed that this activity of XIAP was required to cycle β-catenin on and off TCF to promote Wnt signaling (7). Our current study suggests that USP47 is the deubiquitylase that counteracts the E3 ligase activity of XIAP (Fig. 7B). Given that USP47 opposes the activity of XIAP, a positive Wnt regulator, by deubiquitylating its substrate, Gro/TLE, the finding that USP47 promotes Wnt signaling was unexpected. We speculate that by mediating the deubiquitylation of Gro/TLE, USP47 “recharges” Gro/TLE, the ubiquitylated form of which does not bind TCF, to dislodge β-catenin from TCF, thereby establishing a cycle of β-catenin binding to and dissociating from TCF/LEF. Unlike that for XIAP, there is no evidence that USP47 is recruited to TCF in response to Wnt signaling, and we favor a model in which USP47 acts in a recycling step for Gro/TLE. We show that although XIAP, USP47, and TLE3 can form dimeric complexes with one another, they are unable to form a stable trimeric complex. Furthermore, we cannot distinguish whether XIAP and USP47 have overlapping binding sites on TLE3 or whether the binding of XIAP or USP47 induces a conformational change in TLE3 that precludes the binding of USP47 or XIAP, respectively.
The cycling of β-catenin on and off promoters has been previously speculated to provide a mechanism by which cells are able to maintain a Wnt-mediated transcriptional response (23). A similar phenomenon has been observed with other transcription factors, including Smad4, p53, and FoxO, all of which are also regulated by cycles of ubiquitylation and deubiquitylation (24–26). The alternating binding of coactivator and corepressor (often triggered by ubiquitylation) has been proposed to be a feature of promoters regulated by inducible transcription factors (particularly nuclear receptors) and may allow for potential regulatory checkpoints (27). In such a scenario, binding of the corepressor would allow the system to reset and monitor the state of pathway activation before proceeding with a further transcriptional response. This would contribute to the responsiveness of the system by ensuring that Wnt-activated gene expression only continued in the presence of the activating signal and could be rapidly shut down when the signal is no longer present. Such a mechanism would allow for more precise control of gene transcription and a more rapid reversal upon the termination of the signal.
USP47 participates in DNA damage repair, cell adhesion, and epithelial-mesenchymal transition (EMT) (28). Mechanistically, USP47 regulates multiple signaling pathways through the stabilization of its substrate proteins, including modulators of EMT [such as the Hippo pathway effector YAP, E-cadherin, SNAIL, and Special AT-rich Binding protein 1 (SATB1)], MAPK, transcription elongation factor A3 (TCEA3, a regulator of transcription and apoptosis), and β-transducin repeats–containing protein (β-TRCP), which is involved in a variety of cellular processes including Wnt signaling (29–35). Thus, USP47 may play multiple roles in regulating Wnt signaling through its effects on β-catenin, β-TRCP, and TLE3. USP47 has also been shown to deubiquitylate monoubiquitylated Polβ, and inhibition of USP47 leads to the accumulation of DNA strand breaks and decreased cell viability (36). Inhibitors of USP47 are being developed as anti-cancer agents (37). Our results predict that drugs targeting USP47 may also inhibit the Wnt pathway, thereby providing an effective strategy for targeting cancers through its effects on DNA damage repair, cell survival, and Wnt signaling.
Materials and Methods
Drosophila dsRNA Generation and S2 Cell RNAi Screen
Screening for deubiquitylase regulators of Wg signaling was performed as previously reported (7). Verified or predicted deubiquitylases (DUBs) were identified using the “Termlink” function on www.flybase.org, resulting in a list of 29 DUBs. The Drosophila Gene Collection (DGC) was used to isolate plasmid cDNA encoding each DUB. T7 and T3 RNA polymerase promoters were added to the 5’ and 3’ end of each cDNA, respectively, via PCR primers specific for the vector. Primer sequences are as follows for DUBs in pOTB7/pOT2:
Forward-5’- CAGAGATGCATAATACGACTCACTATAGGGAGATTAGGTGACACTATAGAACT-3’
Reverse-5’- CCAAGCCTTCAATTAACCCTCACTAAAGGGAGAAAGCCCGCTCATTAGGCGGGTTAAA-3’
For DUBs in pBSSK/pFlc1:
Forward 5’- CAGAGATGCATAATACGACTCACTATAGGGAGACGACTCACTATAGGGCGAAT-3’
Reverse-5’- CCAAGCCTTCAATTAACCCTCACTAAAGGGAGATTAACCCTCACTAAAGGGAACAAAA-3’.
dsRNAs were synthesized in an in vitro transcription reaction using mMessage mMachine (Ambion) according to the manufacturer’s instructions. dsRNAs were purified using the Rneasy Mini Kit (Qiagen) and were added to Drosophila S2 reporter cells stably transfected with a Wg TOPflash luciferase transcriptional reporter and a constitutively expressed lacZ gene (gift from R. Nusse, Stanford). S2 reporter cells were then incubated with dsRNAs for 72 hr. followed by incubation for 24 hr with Wg-conditioned media from a Drosophila S2 cell line (S2-Tub-wg) stably transfected to express Wg (gift from R. Nusse, Stanford). Cells were lysed in 1X Passive Lysis Buffer (Promega), and luciferase and β-galactosidase activities were measured using Steady Glo and β-Glo assays (Promega), respectively. Luciferase activity was normalized to β-galactosidase activity (a measure of cell number).
Drosophila stocks and crosses
RNAi lines for Usp47 (Usp47i1: Vienna Drosophila Resource Center [VDRC] #103743 and Usp47i2: VDRC #26027) and y (VDRC #106068) were expressed, along with UAS-Dcr-2 (BDSC #24648), in third instar larval wing imaginal discs using the hh-Gal4 (38) driver. Crosses were performed at 25°C.
Generation of mScar:T2A:sens flies
To generate a transcriptional reporter for sens expression in Drosophila, the coding sequence for the red fluorophore mScarlet was inserted 5’ of the sens coding sequence by CRISPR-assisted homology-directed repair. To avoid functional interference of Sens function by the fluorescent tag, the coding sequence of both proteins was separated by a T2A cleavage signal. CRISPR-assisted HDR was performed as previously described (39). Briefly, a donor plasmid was constructed containing homology arms of around 1 kb on either side of the insertion site and the mScar:T2A sequence. Homology arms were PCR amplified with primers sens5’armfwd GACGGTATCGATAAGCTTGATATCGAATTCcaccgatcagcagtgaaatcctctgcc, sens5’armrev CTTTATTACAGCTTCGCCCTTACTAACCATTTTGATTCTGGATCTTTACTTTTTGG,
sens3’armfwd ATGCGGTGACGTCGAGGAGAATCCTGGTCCAATGAATCACCTATCGCCGCC, and
sens3’armrev GAGCTCCACCGCGGTGGCGGCCGCTCTAGAACCTGGTGCTCCTCCTCATC from genomic DNA of nos-Cas9 flies. The mScar:T2A sequence was ordered as a gBlock from Integrated DNA Technologies and assembled with the homology arms and EcoRI and XbaI digested pBluescript plasmid by In-Fusion cloning (Takara Bio, Shiga, Japan). A sgRNA with the protospacer TAGGTGATTCATTTTGATTC was cloned into pCFD3 as previously described (39). 500 ng/μl donor plasmid and 100 ng/μl sgRNA plasmid were microinjected into nos-cas9 embryos. Progeny carrying the insertion were identified by PCR and the accuracy of the knock-in was confirmed by PCR with primers annealing outside the homology arms and subsequent Sanger sequencing.
Generation of tissue-specific CRISPR mutations in Drosophila
CRISPR target sites were identified using https://flycrispr.org/target-finder/ (40) gRNA sequences (table S1). Two sets of two guide RNAs for Usp47 and one set for the control gene ebony were cloned into the pCFD6: UAS-t::gRNA plasmid, following the protocol described at http://www.crisprflydesign.org/. Plasmids were injected by BestGene and integrated at the attP40 site. Wing-specific mutations were generated by crossing the gRNA-containing lines to vgB-Gal4; UAS-Cas9. Crosses were performed at 29°C.
Generation of germline Usp47 mutants using CRISPR/Cas9
Usp47 germline mutants described in Table S2 were generated using Usp47 gRNA line 2 (table S1). The gRNA-containing line was crossed to yw; UAS-Cas9 nos-Gal4::VP16 (Bloomington Drosophila Stock Center #54593). The Usp47 mutants isolated encode only 39 or 40 out of the total 1538 amino acids (table S2).
Drosophila immunohistochemistry
Third instar larval wing imaginal discs were dissected in PBS and fixed in 4% paraformaldehyde in PBS for 20 minutes. After fixation, discs were washed with PBS with 0.1% Triton X-100 followed by incubation in PBS with 0.1% Triton X-100 and 10% BSA for 1 hr. at room temperature. Wing discs were incubated with guinea pig anti-Senseless (41) or rabbit anti-dsRed (Clontech/TaKaRa, Cat. No. 632496, 1:500, to detect the sens reporter mScar:T2A:sens), mouse anti-Wg (4D4, Developmental Studies Hybridoma bank [DSHB], 1:500), and mouse anti-Engrailed (En) (4D9, [DSHB], 1:20) at 4°C overnight in PBS with 0.5% Triton X-100, followed by incubation with secondary antibodies (anti-guinea pig and anti-mouse Alexa Fluor 488 and 555 conjugates; ThermoFisher Scientific, 1:500) for 2 hr. at room temperature. Specimens were stained with DAPI (2 ug/ml) and mounted in Prolong Gold (Invitrogen). Fluorescent images were obtained on a Nikon A1Rsi confocal microscope.
Plasmids and purified proteins
pCS2-Myc-XIAP, pCS2-Myc-TLE3, pCS2-USP47, pCS2-HA-USP47, and pCS2-HA-Ub were generated using standard PCR-based cloning strategies. pMAL-C5E-6XTUBE was synthesized by Genscript Biotech based on the 6XTUBE sequence (42). pEBB-FLAG-XIAP was purchased from Addgene and described previously (43, 44). pTwist-CMV-BetaGlobin-V5-USP47 was purchased from Twist Biosciences. The following plasmids were generous gifts: pMT107-His-Ub (W. Tansey, Vanderbilt University), and pcDNA3-HA-TLE3 (A. Kispert, Hannover Medical School). MBP-TUBE was purified according to the manufacturer’s protocol for pMAL-based vectors (New England Biolabs). GST-TCF4 was purified as described previously (7). Recombinant His10-USP47 protein was purchased from R&D Systems. Recombinant murine Wnt3a protein was purchased from TIME Bioscience.
Cell lines and transfections
HEK293 cell lines were purchased from the American Type Culture Collection. Wg-secreting cells were purchased from the Drosophila Genomics Resource Center. The following cell lines were gifts: HEK293STF (J. Nathans, Johns Hopkins University) HCT116 WTKO (R. Coffey, Vanderbilt University), and S2 reporter cells (R. Nusse, Stanford University). HEK293 cell lines were cultured in DMEM plus 10% FBS and antibiotics. HCT116 cells were cultured in McCoy’s 5a Medium plus 10% FBS. Drosophila S2 cells were cultured in Schneider’s medium plus 10% FBS. DNA transfections were performed with CaCl2, or Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. CaCl2 transfections were performed as follows. To transfect an individual well of a 12-well plate, 1 μg of DNA was mixed with 6.1 μl of CaCl2 and sterile water for a final volume of 50 μl and added to a tube containing 50 μl of 2x HBS (280mM NaCl, 10mM KCl, 1.5 mM Na2HPO4-2H2O, 12 mM Dextrose (D-Glucose), and 50 mM HEPES), mixed by bubbling air, and added dropwise to the cells. siRNA transfections were performed using Dharmafect-1 (HEK293) or Dharmafect-2 (HCT116). The following siRNAs were used:
NT siRNA: 5’- UAAGGCUAUGAAGAGAUACUU-3’
USP47 siRNA#1: 5’-CGAGAGAAGCUUAGUGAAAUU-3’
USP47 siRNA#2: 5’-GCUUAUAAGAUGAUGGAUUUU-3’
hAXIN1 siRNA#1: 5’-GCGUGGAGCCUCAGAAGUUUU-3’
hAXIN1 siRNA#2: 5’-CCGAGGAGAAGCUGGAGGAUU-3’
Axin siRNA knockdowns were performed by combining hAXIN1 siRNA#1 and #2. Similarly, unless otherwise stated, USP47 siRNA knockdowns were performed by combining USP47 siRNA#1 and #2.
Generation of USP47 CRISPR-Cas9 edited clones
Guide RNAs were designed using publicly available prediction tools at https://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design (45). The following guide sequences were used to generate the USP47–83 and USP47–118 clones:
USP47 gRNA#1: 5’-GGAACCTTTGACTTGGTGTG-3’
USP47 gRNA#2: 5’-GAAGATGTGGCCAACAAAGT-3’
pCas-Guide plasmids (OriGene Technologies) containing USP47 gRNA#1 and USP47 gRNA#2 were co-transfected into HEK293T cells, GFP positive cells FACS sorted, and immunoblotting performed of individual clones to assess USP47 protein levels as previously described (46). Were unable to obtain clones that showed complete knockout of USP47 protein as assessed by immunoblotting, which may reflect the fact that USP47 is an essential gene. Alternatively, the difficulty of obtaining cells with complete knockout of USP47 in HEK293 cells may be attributed to its pseudotriploid nature (47). Regardless, several clones were identified that showed significantly reduced amounts of USP47 protein by immunoblotting, including USP47–83 and USP47–118, which were further expanded for use.
Immunoblotting, Immunoprecipitation, and GST Pull-downs
For immunoblots, cells were lysed in non-denaturing lysis buffer (NDLB) (50 mM Tris-Cl (pH 7.4), 300 mM NaCl, 5 mM EDTA, 1% Triton X-100) with 1 mM PMSF and soluble fractions were obtained. Immunoblot intensities were measured using ImageJ (National Institute of Health). For co-immunoprecipitations, cells were lysed in NDLB with 1mM PMSF and PhosStop phosphatase inhibitor cocktail tablets (Roche). Lysates were diluted to 1 mg/ml with lysis buffer and incubated with antibody overnight with rotation at 4°C followed by the addition of Protein A/G beads (Santa Cruz) for 2 hr. Beads were then washed 5X with NDLB. Bound proteins were eluted from beads with protein sample buffer and analyzed by SDS-PAGE/immunoblotting. For competition binding assay, increasing amounts of HA-TLE3 was co-transfected (50ng, 500ng, 5000ng) with V5-USP47 (5000ng) and/or Myc-XIAP (5000ng). Control empty vector was added so the total amount of DNA was consistent across samples. For in vitro binding assays, GST-TCF4 (8 μg) was diluted in NDLB and incubated with ubiquitylated Myc-TLE3 (see Ubiquitylation assays) treated with or without recombinant His10-USP47 protein (3.84 μg) for 1 hr with rotation at 4°C. Glutathione beads (Thermo Scientific) were added for 2 hr with rotation at 4°C. Beads were washed seven times for 5 min at 4°C in NDLB, and proteins eluted with sample buffer and analyzed by SDS-PAGE followed by immunoblotting.
Ubiquitylation assays
In vitro ubiquitylation assays were performed in 10μl reactions using the Ubiquitin Conjugation Initiation Kit (R&D Systems) and USP47 (R&D Systems). Increasing amounts of USP47 were added (0.32 μg, 0.64 μg, or 0.96 μg). Ubiquitylated Myc-TLE3 was prepared by transfection of HEK293FT cells with HA-Ub, Myc-TLE3, and FLAG-XIAP. Whole cell lysate was collected and used as a substrate. USP47 in vitro ubiquitylation assays were performed in 20 μl reactions using the Ubiquitin Thioester/Conjugation Initiation Kit (Boston Biochem) and the following: 1μM UbcH5a (Boston Biochem); 2.5 μg GST-XIAP, and 1 mM DTT. USP47 protein, generated by the TNT Transcription/Translation System (Promega), was used as a substrate. Reactions were carried out at 30°C for 90 min and were terminated by the addition of sample buffer. Reaction products were resolved by SDS-PAGE and visualized by immunoblotting. For the tandem Ub-binding entity (TUBE) assay (48), cells were treated with 10 μM of the proteasome inhibitor MG132 (Sigma-Aldrich) or dimethyl sulfoxide (DMSO) for 4 hr. and lysed in NDLB with 1mM PMSF and PhosStop phosphatase inhibitor cocktail tablets (Roche). Lysates were diluted to 1 mg/ml with lysis buffer and incubated with 50 μg of MBP-TUBE protein O/N with rotation at 4°C followed by the addition of amylose resin (NEB) for 2 hr. Resin was then washed five times with 0.1% TBS-Tween. Bound proteins were eluted from the resin with protein sample buffer and analyzed by SDS-PAGE/immunoblotting.
Immunofluorescence and single-cell analysis
For immunofluorescence, cells were seeded on a black, clear-bottom 96-well imaging plate (Greiner 655090) and allowed to adhere overnight. Cells were then treated for 6 hr. with increasing concentrations of Wnt3a (R&D 5036-WN). Cells were then fixed with 4% PFA/sucrose and permeabilized in 0.2% Triton X-100/PBS and blocked with 2.5% BSA/PBS for 1 hr. at room temperature. Cells were then stained with primary antibody for USP47 (Bethyl IHC-00235, 1:500) and β-catenin (BD Biosciences 610154, 1:1000) overnight at 4°C. Washes were performed 3X with PBST followed by incubation with AlexaFluor 488/546 (Invitrogen A11001/A11035, 1:1000) for 2 hr. at room temperature protected from light. Cells were stained for DAPI for 30 min. and imaged using a PerkinElmer CLS high-content microscope. Segmentation was performed using Harmony software as previously described (49), and nuclear and total USP47 and β-catenin were measured.
Reporter Assays
For cell-based luciferase assays, cells were plated and transfected with siRNA or DNA, as described above. Wnt3a-conditioned medium (from L cells stably transfected to express Wnt3a; CRL-2467, ATCC) or control medium (from L cells; CRL-2648, ATCC ) or recombinant murine Wnt3a (TIME Bioscience) was added to HEK293STF or HCT116 WTKO cells stably transfected with a luciferase-based Wnt reporter 24 hr. after transfection. Cells were lysed 24 hr. after Wnt3a treatment with 1X Passive Lysis Buffer (Promega), and luciferase activity was measured with Steady Glo (Promega). Luciferase activities were normalized to viable cell numbers using the CellTiter-Glo assay (Promega). All assays were performed in triplicate. All graphs were made using Prism 4 (GraphPad Software, Inc.). Statistical analysis was performed using the two-tailed, unpaired student’s t-test. A value of p<0.05 is considered statistically significant.
Xenopus laevis studies
Xenopus embryos were fertilized in vitro, dejellied, cultured, and injected with 25 ng of Morpholino and/or 2 ng of mRNA as previously described (50). Morpholinos with the following sequences were purchased from Gene Tools:
Standard Control MO: 5’-CCTCTTACCTCAGTTACAATTTATA-3’
USP47 MO: 5’-GCTGACTCTCTTCTCCAGGCCTCAT-3’
Capped Xwnt8 and USP47 mRNA were generated using mMessage mMachine (Ambion) according to the manufacturer’s instructions. Animal caps were excised from stage 9 embryos, cultured until stage 11, and RT-PCRs of USP47, Chordin, and ODC transcripts were performed as described (51). In situ hybridization analysis was performed as described (52) using a probe against Xenopus USP47 and using SP6 polymerase for the antisense strand. For whole embryo sectioning and staining, embryos were formalin-fixed, processed, embedded into paraffin, and stained with hematoxylin and eosin for histological analysis. Statistical analysis was performed using Chi-square test with Bonferroni correction. All experiments involving the use of Xenopus embryos was approved by the Institutional Animal Care and Use Committee (IACUC) at Vanderbilt University and were performed in accordance with their policies and guidelines.
Supplementary Material
Acknowledgments:
We thank Jeremy Nathans (Johns Hopkins University), Robert Coffey (Vanderbilt University), Roel Nusse (Stanford University), William Tansey (Vanderbilt University), and Andreas Kinspert (Hannover Medical School) for reagents; Heather Kroh (Vanderbilt University) for technical help; Laura A. Lee (Vanderbilt University Medical Center) for critical reading of the manuscript.
Funding:
This work was supported by the National Institutes of Health (NIH) Grants: T32CA009213 to CRC; 5T32GM008554 to SK; T32CA00959228 to VHN; 5F30ES016504 to AJH; R35GM136233 to YA; R35GM122516 to EL; R01CA219189 to DLR; R01CA244188 to YA, EL, and DJR; DK103126 and R35GM147128 to CAT; K08 CA240901-01A1 and R01CA272875 to VLW. Additional support include the Lombardi Comprehensive Cancer Center grant to DLR, American Cancer Society 133934-CSDG-19-216-01-TBG and RSG-22-084-01-MM grants to VLW, and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) SFB1324 to MB.
Footnotes
Competing interests: EL and DJR are co-founders of StemSynergy Therapeutics, a company that seeks to develop inhibitors of major signaling pathways (including the Wnt pathway) for the treatment of cancer. The remaining authors declare that they have no competing interests.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Requests for reagents should be directed to the Lead Contact, Ethan Lee (ethan.lee@vanderbilt.edu).
References and Notes
- 1.Anthony CC, Robbins DJ, Ahmed Y, Lee E, Nuclear Regulation of Wnt/β-Catenin Signaling: It’s a Complex Situation. Genes (Basel) 11, 886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A, Lee E, The way Wnt works: Components and mechanism. Growth Factors 31, 1–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P, The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol 9, 207–210 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X, Beta-Trcp couples Beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proceedings of the National Academy of Sciences 96, 6273–6278 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A, Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395, 604–608 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Fiedler M, Graeb M, Mieszczanek J, Rutherford TJ, Johnson CM, Bienz M, An ancient Pygo-dependent Wnt enhanceosome integrated by Chip/LDB-SSDP. eLife 4, e09073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson Alison J., Wallace Heather A., Freeman Tanner J., Beauchamp RD, Lee Laura A., Lee E, XIAP Monoubiquitylates Groucho/TLE to Promote Canonical Wnt Signaling. Molecular Cell 45, 619–628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng VH, Hang BI, Sawyer LM, Neitzel LR, Crispi EE, Rose KL, Popay TM, Zhong A, Lee LA, Tansey WP, Huppert S, Lee E, Phosphorylation of XIAP at threonine 180 controls its activity in Wnt signaling. J Cell Sci 131, jcs210575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, Yu C, Carlson J, George R, Celniker S, Rubin GM, The Drosophila Gene Collection: Identification of Putative Full-Length cDNAs for 70% of D. melanogaster Genes. Genome Research 12, 1294–1300 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korinek V, Constitutive Transcriptional Activation by a beta -Catenin-Tcf Complex in APC−/− Colon Carcinoma. Science 275, 1784–1787 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Yanagawa S-I, Lee J-S, Ishimoto A, Identification and Characterization of a Novel Line of Drosophila Schneider S2 Cells That Respond to Wingless Signaling. Journal of Biological Chemistry 273, 32353–32359 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Benjumea FJ, Cohen SM, Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 121, 4215–4225 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Shi J, Liu Y, Xu X, Zhang W, Yu T, Jia J, Liu C, Deubiquitinase USP47/UBP64E Regulates β-Catenin Ubiquitination and Degradation and Plays a Positive Role in Wnt Signaling. Molecular and Cellular Biology 35, 3301–3311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heasman J, Patterning the early Xenopus embryo. Development 133, 1205–1217 (2006). [DOI] [PubMed] [Google Scholar]
- 15.DeMarais RTMAA, The armadillo homologs beta-catenin and plakoglobin are differentially expressed during early development of Xenopus laevis. Developmental Biology 153, 337–346 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, Nathans J, Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 116, 883–895 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Ilyas M, Tomlinson IPM, Rowan A, Pignatelli M, Bodmer WF, β-Catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci U S A 94, 10330–10334 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, Jernigan KK, Meyers KC, Hang BI, Waterson AG, Kim K, Melancon B, Ghidu VP, Sulikowski GA, LaFleur B, Salic A, Lee LA, Miller DM, Lee E, Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat Chem Biol 6, 829–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvestrini VC, Thomé CH, Albuquerque D, de Souza Palma C, Ferreira GA, Lanfredi GP, Masson AP, Delsin LEA, Ferreira FU, de Souza FC, de Godoy LMF, Aquino A, Carrilho E, Panepucci RA, Covas DT, Faça VM, Proteomics analysis reveals the role of ubiquitin specific protease (USP47) in Epithelial to Mesenchymal Transition (EMT) induced by TGFβ2 in breast cells. Journal of Proteomics 219, 103734 (2020). [DOI] [PubMed] [Google Scholar]
- 20.N. The Cancer Genome Atlas Research, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z, GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, Eshleman JR, Goggins MG, Wolfgang CL, Canto MI, Schulick RD, Edil BH, Choti MA, Adsay V, Klimstra DS, Offerhaus GJ, Klein AP, Kopelovich L, Carter H, Karchin R, Allen PJ, Schmidt CM, Naito Y, Diaz LA Jr., Kinzler KW, Papadopoulos N, Hruban RH, Vogelstein B, Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci U S A 108, 21188–21193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sierra J, Yoshida T, Joazeiro CA, Jones KA, The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes & Development 20, 586-600 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W, Mono- Versus Polyubiquitination: Differential Control of p53 Fate by Mdm2. Science 302, 1972–1975 (2003). [DOI] [PubMed] [Google Scholar]
- 25.van der Horst A, de Vries-Smits AMM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BMT, FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8, 1064–1073 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S, FAM/USP9x, a Deubiquitinating Enzyme Essential for TGFβ Signaling, Controls Smad4 Monoubiquitination. Cell 136, 123–135 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Perissi V, Rosenfeld MG, Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol 6, 542–554 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Pan K, Fu J, Xu W, Role of Ubiquitin-Specific Peptidase 47 in Cancers and Other Diseases. Frontiers in Cell and Developmental Biology 9, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sako-Kubota K, Tanaka N, Nagae S, Meng W, Takeichi M, Minus end–directed motor KIFC3 suppresses E-cadherin degradation by recruiting USP47 to adherens junctions. Molecular Biology of the Cell 25, 3851–3860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashton-Beaucage D, Lemieux C, Udell CM, Sahmi M, Rochette S, Therrien M, The Deubiquitinase USP47 Stabilizes MAPK by Counteracting the Function of the N-end Rule ligase POE/UBR4 in Drosophila. PLOS Biology 14, e1002539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi B-J, Park S-A, Lee S-Y, Cha YN, Surh Y-J, Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci Rep 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bufalieri F, Infante P, Bernardi F, Caimano M, Romania P, Moretti M, Lospinoso Severini L, Talbot J, Melaiu O, Tanori M, Di Magno L, Bellavia D, Capalbo C, Puget S, De Smaele E, Canettieri G, Guardavaccaro D, Busino L, Peschiaroli A, Pazzaglia S, Giannini G, Melino G, Locatelli F, Gulino A, Ayrault O, Fruci D, Di Marcotullio L, ERAP1 promotes Hedgehog-dependent tumorigenesis by controlling USP47-mediated degradation of βTrCP. Nat Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L, Dong L, Wang Y, Liu L, Long H, Li H, Li J, Yang X, Liu Z, Duan G, Dai X, Lin Z, Reversible regulation of SATB1 ubiquitination by USP47 and SMURF2 mediates colon cancer cell proliferation and tumor progression. Cancer Letters 448, 40–51 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Pan B, Yang Y, Li J, Wang Y, Fang C, Yu F-X, Xu Y, USP47-mediated deubiquitination and stabilization of YAP contributes to the progression of colorectal cancer. Protein & Cell 11, 138–143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou X, Xia J, Feng Y, Cui L, Yang Y, Yang P, Xu X, USP47-Mediated Deubiquitination and Stabilization of TCEA3 Attenuates Pyroptosis and Apoptosis of Colorectal Cancer Cells Induced by Chemotherapeutic Doxorubicin. Front. Pharmacol. 12, 713322–713322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons Jason L., Dianova Irina I., Khoronenkova Svetlana V., Edelmann Mariola J., Kessler Benedikt M., Dianov Grigory L., USP47 Is a Deubiquitylating Enzyme that Regulates Base Excision Repair by Controlling Steady-State Levels of DNA Polymerase β. Molecular Cell 41, 609–615 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Suresh Kumar KG, Goldenberg SJ, Mattern MR, Nicholson B, Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Med. Chem. Lett. 3, 789–792 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanimoto H, Itoh S, ten Dijke P, Tabata T, Hedgehog Creates a Gradient of DPP Activity in Drosophila Wing Imaginal Discs. Molecular Cell 5, 59–71 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Port F, Chen HM, Lee T, Bullock SL, Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM, Highly Specific and Efficient CRISPR/Cas9-Catalyzed Homology-Directed Repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolo R, Abbott LA, Bellen HJ, Senseless, a Zn Finger Transcription Factor, Is Necessary and Sufficient for Sensory Organ Development in Drosophila. Cell 102, 349–362 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, Tanaka K, Saeki Y, Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat Commun 9, 524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Ubiquitin Protein Ligase Activity of IAPs and Their Degradation in Proteasomes in Response to Apoptotic Stimuli. Science 288, 874–877 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Lewis J, Burstein E, Reffey SB, Bratton SB, Roberts AB, Duckett CS, Uncoupling of the Signaling and Caspase-inhibitory Properties of X-linked Inhibitor of Apoptosis. Journal of Biological Chemistry 279, 9023–9029 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE, Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32, 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bylund L, Kytölä S, Lui WO, Larsson C, Weber G, Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet Genome Res 106, 28–32 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Hjerpe R, Aillet F, Lopitz‐Otsoa F, Lang V, England P, Rodriguez MS, Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin‐binding entities. EMBO reports 10, 1250–1258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabel CR, Alizadeh E, Robbins DJ, Ahmed Y, Lee E, Thorne CA, Single-Cell Analyses Confirm the Critical Role of LRP6 for Wnt Signaling in APC-Deficient Cells. Dev Cell 49, 827–828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng HB, Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol 36, 657–662 (1991). [PubMed] [Google Scholar]
- 51.Cselenyi CS, Jernigan KK, Tahinci E, Thorne CA, Lee LA, Lee E, LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3’s phosphorylation of -catenin. Proceedings of the National Academy of Sciences 105, 8032–8037 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harland RM, In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 36, 685–695 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Kao KR, Elinson RP, The entire mesodermal mantle behaves as Spemann’s organizer in dorsoanterior enhanced Xenopus laevis embryos. Developmental Biology 127, 64–77 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Requests for reagents should be directed to the Lead Contact, Ethan Lee (ethan.lee@vanderbilt.edu).


