Abstract
The Gulf pipefish Syngnathus scovelli has emerged as an important species for studying sexual selection, development, and physiology. Comparative evolutionary genomics research involving fishes from Syngnathidae depends on having a high-quality genome assembly and annotation. However, the first S. scovelli genome assembled using short-read sequences and a smaller RNA-sequence dataset has limited contiguity and a relatively poor annotation. Here, using PacBio long-read high-fidelity sequences and a proximity ligation library, we generate an improved assembly to obtain 22 chromosome-level scaffolds. Compared to the first assembly, the gaps in the improved assembly are smaller, the N75 is larger, and our genome is ~95% BUSCO complete. Using a large body of RNA-Seq reads from different tissue types and NCBI's Eukaryotic Annotation Pipeline, we discovered 28,162 genes, of which 8,061 are non-coding genes. Our new genome assembly and annotation are tagged as a RefSeq genome by NCBI and provide enhanced resources for research work involving S. scovelli.
Data description
This article presents a resource (genome assembly) that marks a technological improvement compared to the one previously published in the article, “The genome of the Gulf pipefish enables understanding of evolutionary innovations” [1].
A de novo genome assembly is evaluated based on three primary criteria: accuracy or correctness, completeness, and contiguity [2, 3]. Typically, the correctness of a genome is one of the most challenging features to measure. However, with modern, long-read sequencing technologies, the orientation of the contigs and the gene order of an assembly are highly accurate [4–6]. On the other hand, completeness and contiguity are easier to measure [6–8] yet more challenging to achieve, especially in non-model organisms. The Gulf pipefish (Syngnathus scovelli, NCBI:txid161590, fishbase ID: 3306) genome is an essential resource for the study of comparative genomics, evolutionary developmental biology, and other related topics [1, 9–15]. Given the technological constraints when it was initially sequenced, the first version of the S. scovelli genome is highly accurate and mostly complete, but it leaves considerable room for improvement with respect to contiguity [1]. Here, with the use of third-generation sequencing technology, including PacBio High Fidelity (Hi-Fi) long reads from circular consensus sequences (CCS) and Hi-C proximity ligation from Phase Genomics, we produced a nearly complete chromosome-scale genome assembly that not only improves completeness and accuracy but is also the most contiguous genome yet produced for the genus Syngnathus (Table 1).
Table 1.
Contiguity metrics from QUAST for various Syngnathus species.
| Metrics | S. acus | S. rostellatus | S. typhle | S. floridae | S. scovelli _v1 | S. scovelli _v2 |
|---|---|---|---|---|---|---|
| Number of contigs | 87 | 8,935 | 526 | 6,895 | 886 | 526 |
| Largest contig | 28,444,102 | 856,273 | 9,665,359 | 61,807,209 | 23,505,159 | 30,098,933 |
| Total length | 324,331,233 | 280,208,023 | 313,958,489 | 303,298,972 | 305,995,683 | 431,750,762 |
| Reference length | 324,331,233 | 324,331,233 | 324,331,233 | 324,331,233 | 324,331,233 | 324,331,233 |
| GC (%) | 43.46 | 43.08 | 43.29 | 43.63 | 42.95 | 45.00 |
| Reference GC (%) | 43.46 | 43.46 | 43.46 | 43.46 | 43.46 | 43.46 |
| N50 | 14,974,571 | 88,962 | 3,046,963 | 7,845,045 | 12,400,093 | 17,337,441 |
| NG50 | 14,974,571 | 70,018 | 3,012,268 | 7,783,711 | 11,493,655 | 20,118,474 |
| N75 | 11,896,884 | 34,357 | 1,098,273 | 21,150 | 8,458,319 | 13,347,818 |
| NG75 | 11,896,884 | 15,229 | 998,421 | 17,023 | 7,908,134 | 15,901,424 |
| L50 | 8 | 812 | 30 | 5 | 10 | 10 |
| LG50 | 8 | 1,092 | 32 | 6 | 11 | 7 |
| L75 | 14 | 2,068 | 72 | 1,160 | 17 | 17 |
| LG75 | 14 | 3,492 | 79 | 2,003 | 19 | 12 |
For NGx and LGx calculations, S. acus was used as the reference species. All the Sygnathus genomes (except S. scovelli) were last accessed from NCBI on 2022-July-26.
Context
Evolutionary novelties are widespread across the tree of life. However, the origin of de novo genes and their associated regulatory networks, as well as their effects on the phenotype, remain mysterious in most species. Syngnathidae is a family of teleost fishes that includes pipefishes, seahorses, and seadragons [1, 12–16]. Syngnathid fishes are known for their evolutionary novelty with respect to morphology and physiology. For instance, species in this family have variously evolved elaborate leafy appendages, male brooding structures, prehensile tails, elongated facial bones, and numerous other unusual traits [1, 12–14]. With a variety of mating systems and sex roles [12–16], the syngnathid fishes also provide an excellent study system to investigate the generality of theories on sexual selection and reproductive biology [15, 16]. Advances in comparative genomics and the evolutionary developmental biology of novel traits in syngnathids require the development of additional genomic tools. Among these are well-assembled and annotated genomes [1]. Here, we took a step in this direction by producing an improved reference genome for the Gulf pipefish.
Methods
DNA and RNA extraction
We collected S. scovelli from the Gulf of Mexico in Florida, USA (Tampa Bay), and flash froze them in liquid nitrogen. We pulverized approximately 50 mg of whole-body tissue (posterior to the urogenital opening) from a single male on liquid nitrogen, which we submitted to the University of Oregon Genomics and Cell Characterization Core Facility (UOGC3F) for high-molecular-weight DNA isolation using the PacBio Nanobind tissue kit. We submitted similar (but unpulverized) frozen tissue from the same individual fish to Phase Genomics to generate a Hi-C library using Proximo Animal (v4) technology.
In addition, we used organic extraction with TRIzol Reagent, followed by column-based binding and purification using the Qiagen RNeasy MinElute Cleanup Kit, to extract mRNA from the Brain, Eye, Gills, Muscle/Skin, Testis, Ovary, Broodpouch, and Flap tissues.
Sequencing and assembly
After the size selection of genomic DNA using the Blue Pippin (11 kb cutoff), the UOGC3F constructed a sequencing library using the SMRTbell Express Template Prep Kit 2.0. One SMRT cell was sequenced by the UOGC3F using PacBio Sequel II technology, yielding 33.39 Gb in 2.05M CCS reads (out of 6.298M Hi-Fi reads in total). We sequenced 70.4 Gb of paired-end 150 nucleotide reads (234.6 million in total) from the Hi-C library using an Illumina NovaSeq 6000 at the UOGC3F. The RNA sequencing libraries were prepared using the KAPA mRNA HyperPrep Kit. We sequenced 159 bp paired-end reads using Illumina Novaseq 6000 for each tissue from the RNA sequencing libraries for annotation.
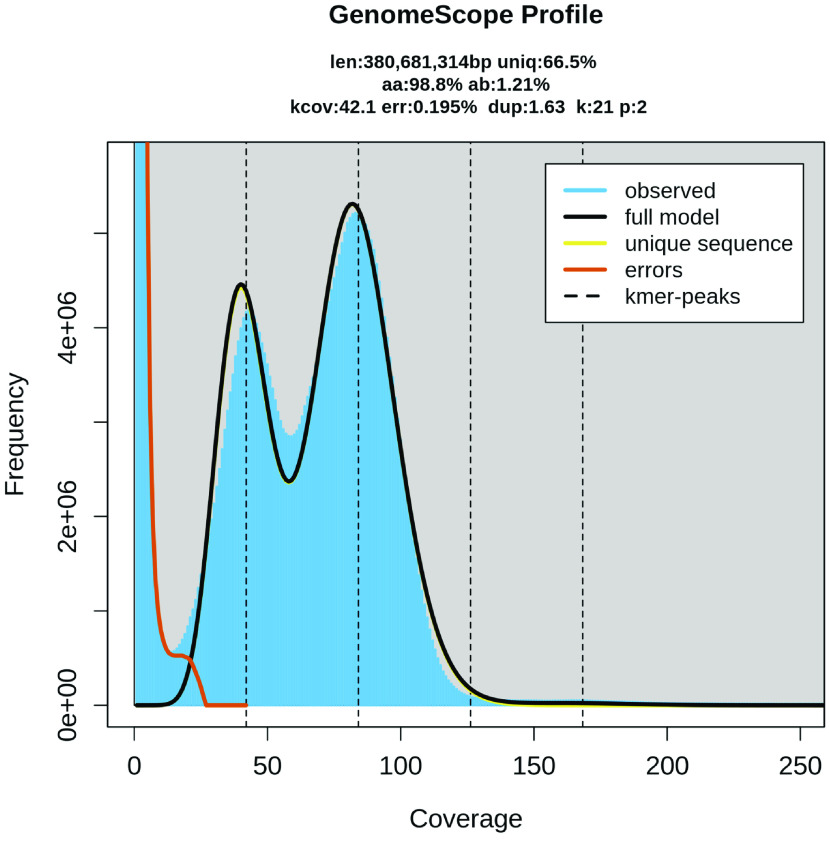
Using the Hi-Fi sequences, we estimated the genome size using genomescope2 (v2.0, RRID:SCR_017014) [17] and meryl (v2.2) [18] with a default k-mer size of 21 (Figure 1). The paired-end Hi-C reads were trimmed using trimmomatic (v0.39, RRID:SCR_011848) [19] with the parameter HEADCROP:1 to remove the first base, which was of low quality. Together with the Hi-Fi sequences, we assembled the first-pass genome assembly in Hi-C integrated mode using hifiasm (v0.16.1, RRID:SCR_021069) [18] with default parameters. The First-Pass assembly refers to the first draft consensus assembly from the Hi-Fi and Hi-C data. We extracted the consensus genome from hifiasm in fasta format and assembled the contigs into scaffolds using juicer (v1.6, RRID:SCR_017226) [20]. We used the 3D-DNA (version date: Dec 7, 2016) [21] pipeline to merely order the scaffolds. The Hi-C contact map of the ordered scaffolds was visualized using juicebox (v1.9.8, RRID:SCR_021172) with no breaking of the original contigs.
Figure 1.
Estimated genome size of Syngnathus scovelli based on k-mer analysis using Meryl and Genomescope.
Assessment of completeness and contiguity
To compare the completeness and contiguity of the latest version of the S. scovelli genome against the other Syngnathus genomes (Figure 2), we downloaded the genome assemblies of S. acus (GCA_024217435.2), S. rostellatus (GCA_901007895.1) [22], S. typhle (GCA_901007915.1) [22], and S. floridae (GCA_010014945.1) from NCBI. We used Benchmarking Universal Single-Copy Orthologs (BUSCO v5.2.2, RRID:SCR_015008) [23] in genome mode with the actinopterygii_odb10 database (as of 2021-02-19) to evaluate the completeness of the genome. Also, we used a k-mer-based assessment using Merqury (v2020-01-29, RRID:SCR_004231. [24]) to estimate the completeness and the base error rate. We then used the Quality Assessment Tool (QUAST v5.0.2, RRID:SCR_001228) [25] to estimate Nx and Lx statistics for our assembly.
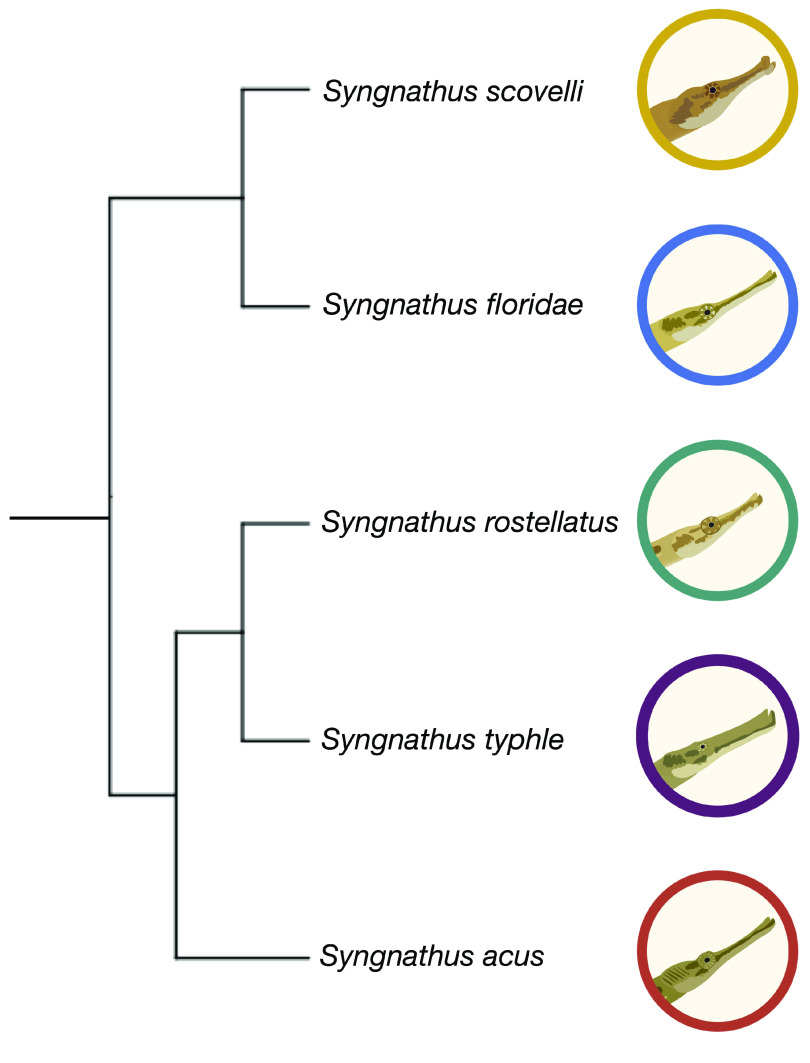
Figure 2.
Cladogram of the five Syngnathus species in this study. This phylogeny is based on the Ultra Conserved Elements among all syngnathids [26].
Annotation using the NCBI Eukaryotic annotation pipeline
The NCBI Eukaryotic Genome Annotation Pipeline (v10.0) is an automated software pipeline identifying coding and non-coding genes, transcripts, and proteins on complete and incomplete genome submissions to NCBI. The core components of this pipeline are the RNA alignment program (STAR and Splign) and Gnomon, a gene prediction program. In this pipeline, the RNA-Seq reads from the various (Brain, Eye, Gills, Muscle/Skin, Testis, Ovary, Broodpouch, and Flap) tissues of multiple samples, including the S. scovelli individual used for Hi-Fi and Hi-C sequence data (SRR20438584–SRR20438604), were aligned to the genome. Gnomon combines the information from alignments of the transcripts and the ab initio models from a Hidden Markov Model-based algorithm to create a RefSeq annotation. This RefSeq annotation produces a non-redundant set of a predicted transcriptome and a proteome that can be used for various analyses. The Eukaryotic annotation pipeline is not publicly available; thus, we requested the staff at NCBI to annotate the S. scovelli genome.
Data validation and quality control
Assembly statistics
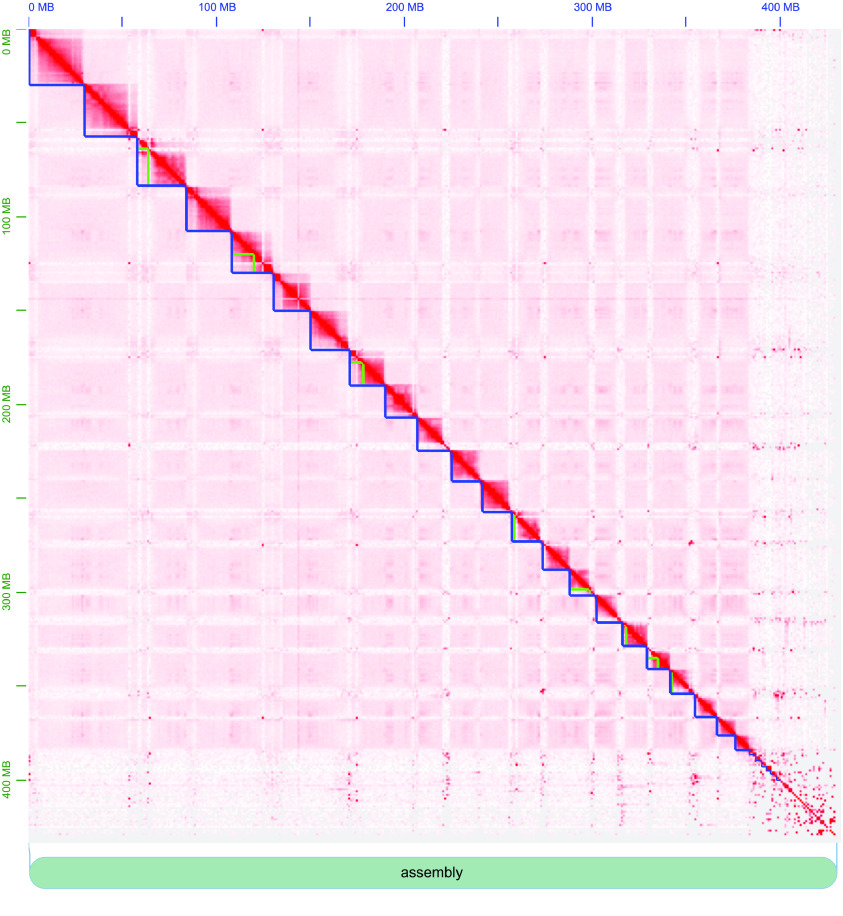
With approximately 2 million Hi-Fi reads and 234.6 million Hi-C reads, we generated the first pass consensus assembly with 585 contigs. The N50 and L50 for this assembly were 15.5 Mb and 11, respectively. We scaffolded this assembly to correct misassembles and produced a final assembly containing 526 contigs with N50 and L50 values of 17.3 Mb and 10, respectively (Table 1). This improved version of the S. scovelli genome has around three times fewer contigs compared to the original S. scovelli genome. The NG50 and NG75 are ∼1.75× and ∼2× larger, respectively, than the previous assembly, implying less fragmentation. Our new assembly reduces the number of gaps per 100 kilobase pairs (kb) from 6,837.20 Ns per 100 kb to a mere 0.27 Ns per 100 kbp, owing to the increased contiguity. This new S. scovelli genome is on par with the current best genome in the Syngnathus genus, that of S. acus, which is a complete chromosome-scale assembly. The first 22 scaffolds of the S. scovelli genome are of chromosome-scale in line with the genetic map [1] and the karyotype data [27] with a total length of around 380 Mb (Figure 3), comparable to the estimated genome size of 380 Mb (see GigaDB [28]; Table 2 and Figure 3). In addition, 88.94% of the total assembly length is captured in the 22 chromosome-scale scaffolds. For 15 of the chromosome-scale scaffolds, a single contig makes up the total length; the remaining seven are generally composed of a small number of contigs (Figure 3).
Figure 3.
Visualization of contact maps from Hi-C reads for Syngnathus scovelli (v2). The first 22 primary assembly features (blue lines) sum to about 380 Mb in size, which is the estimated genome size for the species. The green lines reflect the individual contigs from the hifiasm assembly that were organized into chromosome-level scaffolds based on Hi-C contact data.
Table 2.
Contiguity metrics from QUAST for the first pass and the scaffolded assembly of S. scovelli _v2.
| Metrics | Haplotype1 | Haplotype2 | Primary consensus assembly | Scaffolded assembly |
|---|---|---|---|---|
| Number of contigs | 901 | 544 | 585 | 526 |
| Largest contig | 21,671,036 | 23,661,123 | 30,098,933 | 30,098,933 |
| Total length | 427,545,154 | 428,155,884 | 431,749,582 | 431,750,762 |
| GC (%) | 44.99 | 44.78 | 45.00 | 45.00 |
| N50 | 10,825,652 | 10,535,849 | 15,551,623 | 17,337,441 |
| N75 | 4,999,310 | 4,477,557 | 11,049,644 | 13,347,818 |
| L50 | 15 | 15 | 11 | 10 |
| L75 | 29 | 30 | 19 | 17 |
| Number of N’s per 100 kbp | 0.00 | 0.00 | 0.00 | 0.27 |
BUSCO and Merqury results
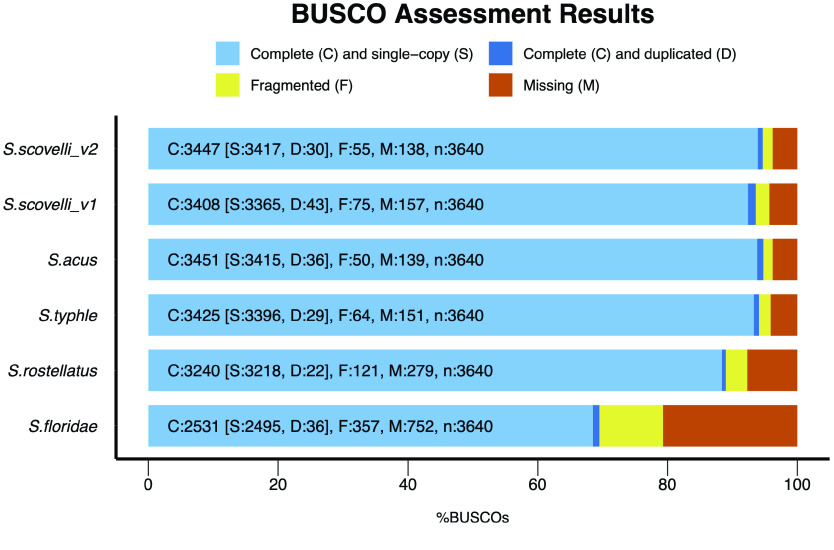
BUSCO results suggest a high degree of completeness as it found 95% of the orthologs in the Actinopterygii dataset (94.7% [S: 93.9%, D: 0.8%], F: 1.5%, M: 3.8%, n: 3,640) when run in genome mode (Figure 4) and the Merqury evaluation suggests that the genome is ∼86% complete with a quality value (QV) of 61.37 and an error rate of 7.3 × 10−5 % (see GigaDB [28] for more details; Tables 3 and 4).
Figure 4.
Comparison of BUSCO completeness among all the five Syngnathus species.
Table 3.
k-mer based assembly evaluation for completeness using Merqury.
| Assembly | k-mer set used | solid k-mers in the assembly | Total solid k-mers in the read set | Completeness (%) |
|---|---|---|---|---|
| S. scovelli _v2 | all | 272,969,166 | 318,487,563 | 85.708 |
Table 4.
k-mer based quality evaluation using Merqury.
| Assembly | k-mers uniquely found only in the assembly | k-mers found in both the assembly and the read set | QV | Error rate |
|---|---|---|---|---|
| S. scovelli_v2 | 6,614 | 431,737,882 | 61.3697 | 7.29504 × 10−7 |
Consistent with the BUSCO contiguity metrics, the genome is on par with S. acus for completeness, which is also around 95% complete. Missing genes make up the majority of the remaining 5% of genes. We identified genes likely to be truly missing from the S. scovelli genome and more broadly from members of Syngnathidae (including the seahorses, genus Hippocampus along with Syngnathus) by confirming their absence across the BUSCO results from the present assembly, four additional members of the genus Syngnathus, and six additional Hippocampus publicly available assemblies (see GigaDB [28] for additional details). Of the missing BUSCO genes, 83 are shared among all the species of Syngnathus, and 38 are missing from both genera (see GigaDB [28] for additional details). Future work could profitably explore these missing genes, as some may be related to the interesting novel traits in syngnathid fishes.
Annotation results
After masking about 43% of the genome, the annotations resulted in the prediction of about 28,162 genes, of which 8,061 are non-coding genes (see GigaDB [28]; Tables 5 and 6). The 28,162 genes produce about 59,938 transcripts, of which 47,846 are mRNA, and the rest is made up of other types of RNAs such as tRNA, lncRNA, and others. Out of the 20,101 coding genes, 18,616 had a protein with an alignment covering 50% or more of the query against the UniProtKB curated protein set, and 9,152 had an alignment covering 95% or more of the query.
Table 5.
Gene and Feature Statistics from NCBI Eukaryotic Pipeline.
| Feature | S. scovelli_v2 |
|---|---|
| Genes and pseudogenes | 29,062 |
| protein-coding | 20,101 |
| non-coding | 8,061 |
| Transcribed pseudogenes | 0 |
| Non-transcribed pseudogenes | 887 |
| genes with variants | 10,398 |
| Immunoglobulin/T-cell receptor gene segments | 9 |
| other | 4 |
| mRNAs | 47,846 |
| fully-supported | 47,491 |
| with >5% ab initio | 89 |
| partial | 39 |
| with filled gap(s) | 0 |
| known RefSeq | 0 |
| model RefSeq | 47,846 |
| non-coding RNAs | 12,092 |
| fully-supported | 7,318 |
| with >5% ab initio | 0 |
| partial | 5 |
| with filled gap(s) | 0 |
| known RefSeq | 0 |
| model RefSeq | 10,741 |
| pseudo transcripts | 0 |
| fully-supported | 0 |
| with >5% ab initio | 0 |
| partial | 0 |
| with filled gap(s) | 0 |
| known RefSeq | 0 |
| model RefSeq | 0 |
| CDSs | 47,855 |
| fully-supported | 47,491 |
| with >5% ab initio | 115 |
| partial | 39 |
| with major correction(s) | 144 |
| known RefSeq | 0 |
| model RefSeq | 47,846 |
Table 6.
Detailed Feature Lengths from NCBI Eukaryotic Pipeline.
| Feature | Count | Mean length (bp) | Median length (bp) | Min length (bp) | Max length (bp) |
|---|---|---|---|---|---|
| Genes | 28,166 | 11,149 | 4,361 | 56 | 677,970 |
| All transcripts | 59,938 | 3,654 | 2,773 | 56 | 106,526 |
| mRNA | 47,846 | 3,907 | 3,042 | 204 | 98,797 |
| misc_RNA | 2,018 | 3,844 | 2,824 | 138 | 22,974 |
| tRNA | 1,351 | 74 | 73 | 71 | 87 |
| lncRNA | 5,304 | 3,880 | 1,632 | 112 | 106,526 |
| snoRNA | 117 | 123 | 126 | 62 | 319 |
| snRNA | 378 | 142 | 141 | 56 | 196 |
| rRNA | 2,920 | 1,228 | 154 | 118 | 4,380 |
| Single-exon | 514 | 2,381 | 1,944 | 358 | 21,617 |
| coding | 514 | 2,381 | 1,944 | 358 | 21,617 |
| CDSs | 47,846 | 2,373 | 1,617 | 96 | 97,746 |
| Exons | 277,161 | 325 | 142 | 2 | 38,823 |
| coding | 260,368 | 299 | 140 | 2 | 38,823 |
| non-coding | 27,774 | 515 | 152 | 9 | 36,521 |
| Introns | 247,597 | 1,355 | 160 | 30 | 611,280 |
| coding | 235,861 | 1,207 | 152 | 30 | 611,280 |
| non-coding | 22,579 | 2,911 | 304 | 30 | 498,241 |
Reuse potential
The new version of the S. scovelli genome opens doors to more accurate results by enhancing the comparative genome data analysis and facilitating the creation of robust tools for molecular genetic studies. We generated the original version of the genome to focus on the genetic mechanisms underlying the unique body plan among pipefishes and seahorses. This genome version takes us one step closer to uncovering these evolutionary mysteries and aids in answering other unknown features, such as the effects of sexual selection and mate choice systems on genome evolution.
Acknowledgements
We are truly grateful for the dedicated efforts of Emily Rose and her students at Valdosta State University, who collected the S. scovelli samples crucial for this work (Florida Fish and Wildlife Conservation Commission Permit: SAL-17-0182-E, SAL-18-0182-E). We also thank Jeff Bishop and Tina Arredondo from the University of Oregon (UO) GC3F for library preparation and sequencing assistance. We want to acknowledge the staff at Phase Genomics for their helpful Hi-C technical support. We are grateful to Mike Coleman and Mark Allen for assisting with Talapas Supercomputer Cluster at UO and Benji Oswald with Research Computing and Data Services at UI. We greatly appreciate the support of the NCBI staff for the Eukaryotic Annotation Pipeline. We thank Jacelyn Shu for her pipefish illustrations. We are grateful for the helpful comments and review by Sven Winter and Yue Song on the manuscript.
Funding Statement
This work was funded by National Science Foundation (NSF) Grant 2015419 to WAC and AGJ and Grant 1953170 to AGJ. We also acknowledge the startup funds provided by the University of Idaho to AGJ.
Data Availability
The genome is available on NCBI with the assembly accession number GCA_024217435.2. The genome is annotated via the NCBI eukaryotic genome annotation pipeline, and the annotation report release (100) is available here. Several smaller contigs and contaminant microbes were removed in the annotation pipeline yielding a more robust genome assembly. The sequence identifier for the chromosome-level scaffolds is available in the GigaDB [28]. The NCBI Bioproject accession number is PRJNA851781, the raw Hi-Fi sequence accession is SRR19820733, the Hi-C sequence accession is SRR22219025, and the RNA-Seq sequence files from various tissues are SRR20438584–SRR20438604. Additional data is available in the GigaDB [28].
Declarations
List of abbreviations
BUSCO: Benchmarking Universal Single-Copy Orthologs; CCS: Circular Consensus Sequence; Gb: Giga basepair; Hi-Fi: High-Fidelity; Mb: Mega basepair; NCBI: National Center for Biotechnology Information; not: nucleotide; QUAST: Quality Assessment Tool; QV: Quality Value; SMRT: Single Molecule Real Time; University of Oregon Genomics and Cell Characterization Core Facility (UOGC3F).
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was funded by National Science Foundation (NSF) Grant 2015419 to WAC and AGJ and Grant 1953170 to AGJ. We also acknowledge the startup funds provided by the University of Idaho to AGJ.
Authors’ contributions
Author contributions, described using the CRedIT taxonomy are as follows:
Conceptualization: BR, CMS, SB, WAC, AGJ; Methodology: BR, CMS, SB, BDJ, EB; Software: BR, CMS, HH, MC; Validation: BR, CMS; Formal Analysis: BR, CMS; Investigation: BR, CMS; Resources: MC, BDJ, EB, MM; Data Curation: BR, CMS, MC; Writing – Original Draft Preparation: BR, CMS, AGJ; Writing – Review & Editing: BR, CMS, AGJ; Visualization: BR, CMS; Supervision: WAC, AGJ; Project Administration: CMS, SB, WAC, AGJ; Funding Acquisition: WAC, AGJ.
References
- 1.Small CM, Bassham S, Catchen J et al. The genome of the Gulf pipefish enables understanding of evolutionary innovations. Genome Biol., 2016; 17(1): 1–23. doi: 10.1186/s13059-016-1126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alhakami H, Mirebrahim H, Lonardi S. . A comparative evaluation of genome assembly reconciliation tools. Genome Biol., 2017; 18(1): 1–14. doi: 10.1186/s13059-017-1213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dida F, Yi G. . Empirical evaluation of methods for de novo genome assembly. PeerJ Comput. Sci., 2021; 7: e636. doi: 10.7717/peerj-cs.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox EJ, Reid-Bayliss KS, Emond MJ et al. Accuracy of next generation sequencing platforms. Next Gener. Seq. Appl., 2014; 1: 1000106. doi: 10.4172/jngsa.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu T, Chitnis N, Monos D et al. Next-generation sequencing technologies: An overview. Human Immunol., 2021; 82(11): 801–811. doi: 10.1016/j.humimm.2021.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Sohn J-I, Nam J-W. . The present and future of de novo whole-genome assembly. Brief. Bioinform., 2018; 19(1): 23–40. doi: 10.1093/bib/bbw096. [DOI] [PubMed] [Google Scholar]
- 7.Ekblom R, Wolf JBW. . A field guide to whole-genome sequencing, assembly and annotation. Evol. Appl., 2014; 7(9): 1026–1042. doi: 10.1111/eva.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wajid B, Serpedin E. . Do it yourself guide to genome assembly. Brief. Funct. Genom., 2016; 15(1): 1–9. doi: 10.1093/bfgp/elu042. [DOI] [PubMed] [Google Scholar]
- 9.Jones AG, Avise JC. . Microsatellite analysis of maternity and the mating system in the Gulf pipefish Syngnathus scovelli, a species with male pregnancy and sex-role reversal. Mol. Ecol., 1997; 6(3): 203–213. doi: 10.1046/j.1365-294x.1997.00173.x. [DOI] [PubMed] [Google Scholar]
- 10.Ratterman NL, Rosenthal GG, Jones AG. . Sex recognition via chemical cues in the sex-role-reversed gulf pipefish (Syngnathus scovelli). Ethology, 2009; 115(4): 339–346. doi: 10.1111/j.1439-0310.2009.01619.x. [DOI] [Google Scholar]
- 11.Begovac PC, Wallace RA. . Stages of oocyte development in the pipefish, Syngnathus scovelli . J. Morphol., 1988; 197(3): 353–369. doi: 10.1002/jmor.1051970309. [DOI] [PubMed] [Google Scholar]
- 12.Paczolt KA, Jones AG. . Post-copulatory sexual selection and sexual conflict in the evolution of male pregnancy. Nature, 2010; 464(7287): 401–404. doi: 10.1038/nature08861. [DOI] [PubMed] [Google Scholar]
- 13.Haase D, Roth O, Kalbe M et al. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: evidence from deep transcriptome sequencing. Biol. Lett., 2013; 9(2): 20130044. doi: 10.1098/rsbl.2013.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mobley KB, Small CM, Jones AG. . The genetics and genomics of Syngnathidae: pipefishes, seahorses and seadragons. J. Fish Biol., 2011; 78(6): 1624–1646. doi: 10.1111/j.1095-8649.2011.02967.x. [DOI] [PubMed] [Google Scholar]
- 15.Whittington CM, Griffith OW, Qi W et al. Seahorse brood pouch transcriptome reveals common genes associated with vertebrate pregnancy. Mol. Biol. Evol., 2015; 32(12): 3114–3131. doi: 10.1093/molbev/msv177. [DOI] [PubMed] [Google Scholar]
- 16.Whittington CM, Friesen CR. . The evolution and physiology of male pregnancy in syngnathid fishes. Biol. Rev., 2020; 95(5): 1252–1272. doi: 10.1111/brv.12607. [DOI] [PubMed] [Google Scholar]
- 17.Ranallo-Benavidez TR, Jaron KS, Schatz MC. . GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun., 2020; 11(1): 1–10. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurk S, Walenz BP, Rhie A et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res., 2020; 30(9): 1291–1305. doi: 10.1101/gr.263566.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. . Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014; 30(15): 2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng H, Concepcion GT, Feng X et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods, 2021; 18(2): 170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand NC, Shamim MS, Machol I et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst., 2016; 3(1): 95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth O, Solbakken MH, Tørresen OK et al. Evolution of male pregnancy associated with remodeling of canonical vertebrate immunity in seahorses and pipefishes. Proc. Natl. Acad. Sci. USA, 2020; 117(17): 9431–9439. doi: 10.1073/pnas.1916251117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seppey M, Manni M, Zdobnov EM. . BUSCO: assessing genome assembly and annotation completeness. In: Gene Prediction. Springer, 2019; pp. 227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 24.Rhie A, Walenz BP, Koren S et al. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol., 2020; 21(1): 1–27. doi: 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurevich A, Saveliev V, Vyahhi N et al. QUAST: quality assessment tool for genome assemblies. Bioinformatics, 2013; 29(8): 1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longo SJ, Faircloth BC, Meyer A et al. Phylogenomic analysis of a rapid radiation of misfit fishes (Syngnathiformes) using ultraconserved elements. Mol. Phylogenet. Evol., 2017; 113: 33–48. doi: 10.1016/j.ympev.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Vitturi R, Libertini A, Campolmi M et al. Conventional karyotype, nucleolar organizer regions and genome size in five Mediterranean species of Syngnathidae (Pisces, Syngnathiformes). J. Fish Biol., 1998; 52(4): 677–687. doi: 10.1111/j.1095-8649.1998.tb00812.x. [DOI] [Google Scholar]
- 28.Ramesh B, Small CM, Healey H et al. Supporting data for “Improvements to the Gulf Pipefish Syngnathus scovelli Genome”. GigaScience Database, 2023; 10.5524/102353. [DOI] [PMC free article] [PubMed] [Google Scholar]