Abstract
Introduction
Healthcare-associated infections (HAIs) and antimicrobial resistance (AMR) are major public health threats in upper- and lower-middle-income countries. Electronic health records (EHRs) are an invaluable source of data for achieving different goals, including the early detection of HAIs and AMR clusters within healthcare settings; evaluation of attributable incidence, mortality, and disability-adjusted life years (DALYs); and implementation of governance policies. In Italy, the burden of HAIs is estimated to be 702.53 DALYs per 100,000 population, which has the same magnitude as the burden of ischemic heart disease. However, data in EHRs are usually not homogeneous, not properly linked and engineered, or not easily compared with other data. Moreover, without a proper epidemiological approach, the relevant information may not be detected. In this retrospective observational study, we established and engineered a new management system on the basis of the integration of microbiology laboratory data from the university hospital “Policlinico Tor Vergata” (PTV) in Italy with hospital discharge forms (HDFs) and clinical record data. All data are currently available in separate EHRs. We propose an original approach for monitoring alert microorganisms and for consequently estimating HAIs for the entire period of 2018.
Methods
Data extraction was performed by analyzing HDFs in the databases of the Hospital Information System. Data were compiled using the AREAS-ADT information system and ICD-9-CM codes. Quantitative and qualitative variables and diagnostic-related groups were produced by processing the resulting integrated databases. The results of research requests for HAI microorganisms and AMR profiles sent by the departments of PTV from 01/01/2018 to 31/12/2018 and the date of collection were extracted from the database of the Complex Operational Unit of Microbiology and then integrated.
Results
We were able to provide a complete and richly detailed profile of the estimated HAIs and to correlate them with the information contained in the HDFs and those available from the microbiology laboratory. We also identified the infection profile of the investigated hospital and estimated the distribution of coinfections by two or more microorganisms of concern. Our data were consistent with those in the literature, particularly the increase in mortality, length of stay, and risk of death associated with infections with Staphylococcus spp, Pseudomonas aeruginosa, Klebsiella pneumoniae, Clostridioides difficile, Candida spp., and Acinetobacter baumannii. Even though less than 10% of the detected HAIs showed at least one infection caused by an antimicrobial resistant bacterium, the contribution of AMR to the overall risk of increased mortality was extremely high.
Conclusions
The increasing availability of health data stored in EHRs represents a unique opportunity for the accurate identification of any factor that contributes to the diffusion of HAIs and AMR and for the prompt implementation of effective corrective measures. That said, artificial intelligence might be the future of health data analysis because it may allow for the early identification of patients who are more exposed to the risk of HAIs and for a more efficient monitoring of HAI sources and outbreaks. However, challenges concerning codification, integration, and standardization of health data recording and analysis still need to be addressed.
Introduction
Hospital-acquired infections (HAIs) represent a major public health concern. In Europe, HAIs affect more than 90,000 patients in acute-care hospitals daily, thus resulting in approximately 4.5 million cases per year [1]. A study published in 2021 evaluated the incidence, attributable deaths, and burden of the most significant HAIs in Italy, which is estimated at 702.53 disability-adjusted life years (DALYs) per 100,000 population [2]. For comparison, in 2017 in Italy, the value of DALYs for ischemic heart disease, which is the major cause of death in the country and in the world, was 749 (697–805) DALYs per 100,000 population, thus confirming that HAIs represent a major public health issue.
Antimicrobial resistance (AMR) is a significant factor that affects HAIs. Data from the AR-ISS national antibiotic resistance surveillance system by the Italian National Institute of Health states that in 2019 in Italy, the resistance rates to the main classes of antibiotics for the eight alert microorganisms under surveillance (namely, Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter species) were still high or even increasing compared with those of the previous years, [3, 4] thus placing Italy almost at the bottom of the league in Europe [5].
In 2018, 16,539 isolates of multidrug-resistant (MDR) Escherichia coli, which is by far the most frequently isolated species among the eight alert microorganisms, were reported in Italy. This number increased to 18,866 in 2019. Isolates of resistant Staphylococcus aureus were the second most represented species, with 8,581 isolates in 2018 and 9,939 in 2019, followed by K. pneumoniae with 5,913 isolates in 2018 and 7,782 isolates in 2019 [6].
With regard to the specific resistance profile, the resistance rate of Escherichia coli to third-generation cephalosporins remained stable (around 30%), whereas a decreasing trend over the last five years (2015–2019) was observed for the resistance rate of fluoroquinolones (from 44.4% in 2015 to 40.7% in 2019). Concerning carbapenem-resistant K. pneumoniae, an increase was observed from 26.8% in 2018 to 28.5% in 2019. However, by considering only laboratories that participated in the surveillance system in both 2018 and 2019, a decrease from 26.4% to 22.7% was observed; therefore, this result is linked to the higher number of data collected by a greater number of laboratories that joined the surveillance network in Italy. On the other hand, carbapenem resistance was confirmed to be very low for Escherichia coli (0.4%), showed a decrease in P. aeruginosa species (13.7%), and was stable with regard to Acinetobacter (79.2%). Among gram-negative bacteria, 36.2% of K. pneumoniae isolates in 2019 were MDR (resistant to third-generation cephalosporins, aminoglycosides, and fluoroquinolones). By contrast, only 11.7% of Escherichia coli were MDR.
For P. aeruginosa, in 2019 the percentage of resistance to three or more antibiotics, including piperacillin-tazobactam, ceftazidime, carbapenems, aminoglycosides and fluoroquinolones was 13.1%, a decrease compared to previous years. However, a high and increasing percentage of multi-resistance (fluoroquinolones, aminoglycosides and carbapenems) was observed (77.3%) for Acinetobacter. For S. aureus, the proportion of methicillin-resistant (MRSA) isolates remained stable (around 34%), while significant increases were seen in the proportion of vancomycin-resistant Enterococcus faecium isolates, which stood at 21.3% in 2019. Finally, For P. aeruginosa in 2019, the percentage of resistance to three or more antibiotics, including piperacillin–tazobactam, ceftazidime, carbapenems, aminoglycosides, and fluoroquinolones, was 13.1%, which is lower than the values in previous years. However, a high and increasing percentage of multiresistance (fluoroquinolones, aminoglycosides, and carbapenems) was observed for Acinetobacter (77.3%). For Staphylococcus aureus, the proportion of methicillin-resistant Staphylococcus aureus isolates remained stable (approximately 34%), whereas significant increases were observed in the proportion of vancomycin-resistant Enterococcus faecium isolates (21.3% in 2019). Finally, for S. pneumoniae, a slight increase was observed in both the proportion of penicillin- and erythromycin-resistant isolates (11.9% and 22.4%, respectively) [4].
Catheter-associated urinary tract infections (CAUTIs) are prevalent worldwide (900,000 cases/year in the United States), whereas hospital-acquired pneumonia and blood stream infections (BSIs) are deadly and account for 67% of US annual deaths associated with HAIs. Surgical site infections are the most common and costly HAIs for surgical patients [6].
The common variables associated with HAIs are (i) invasive devices, such as intubation or CAUTIs and BSIs with vascular devices [7]; (ii); (ii) patient characteristics and conditions, such as age, immunosuppression, or comorbidities [6, 8], and (iii) intensive care units (ICUs) and high-dependency units (HDUs) [7]. For instance, the SARS-CoV-2 outbreak in 2019 resulted in a high number of patients requiring intensive care, intubation, and antimicrobials and has led the scientific community to investigate the effect of this pathogen on HAIs. Many studies are now beginning to report an alarming increase in HAIs and AMR due to COVID-19 [9, 10], particularly if we consider the increased use of drugs, the prolonged time that many COVID-19 patients have to spend in hospitals, and the difficulties of implementing AMR stewardship programs correctly during this public health emergency. The current pandemic may soon lead to further increases in HAIs and AMR [11].
Several strategies must be adopted to reduce the effect of HAIs: (i) hand hygiene; (ii) maintaining a safe, clean, hygienic hospital environment; (iii) screening and categorizing patients into risk-stratified cohorts; (iv) following patient safety guidelines; (v) antibiotic stewardship; and (vi) public health surveillance [12]. Finally, surveillance data on HAIs can be used to assess the extent, escalation, and status of infections; examine, scan, and monitor the trends of infection rates; inform alert programs; and improve performance, strategies, and competence development [13, 14]. The distribution of HAIs between hospitals and hospital wards can have significant variability; therefore, it is essential to create specific reports for healthcare settings that are under investigation to reduce bias and provide dynamic monitoring to allow proper countermeasures to be taken for the prevention and management of HAIs. EHRs are an invaluable source of information; however, most of the time, information in EHRs are stored in separate databases that are not networked.
Despite the numerous studies concerning prevalence and incidence of HAIs, and their correlation with specific risk factors, surprisingly fewer data are available on the integration of HDFs and Microbiology laboratory data. This approach is particularly useful to identify not only the correlation between risk factors associated with HAIs (both socio-demographic and hospital related), but also to evaluate their appropriate coding in the HDFs, obviously associated with the economic losses for the hospital. This retrospective observational study re-elaborates the laboratory data retrieved from different EHRs of the university hospital “Policlinico Tor Vergata” (PTV) of Rome, Italy, and integrates them with hospital discharge forms (HDFs). We suggest a novel approach for monitoring circulating alert microorganisms and for estimating the annual number of HAIs by offering a methodological tool to investigate the epidemiological aspects and possible cost implications.
Materials and methods
The conducted retrospective observational study analyzed ordinary inpatient admissions at the PTV hospital in Rome. All patients discharged from the medical and surgical wards from January 1 to December 31, 2018, were included in the analysis. Data from 2020 to 2021 have been excluded from our investigation due to possible bias introduced by the emergency situation caused by COVID-19. Moreover, the comparison with data form 2019 will be the object of further studies.
Data extraction was performed by analyzing HDFs in the databases of the Hospital Information System. These data were compiled using the AREAS-ADT information system and the ICD-9-CM classification (2007 version, which is the current standard in Italian hospitals). This newly assembled database was further processed to evaluate the quantitative and qualitative variables and the produced diagnostic-related groups (DRGs). The results of research requests for HAI and AMR evaluations were extracted from the database of the Complex Operational Unit of Microbiology.
Record linkage
First, the database was “cleaned” for individual fields of records related to personal data (patient’s name and date of birth) to achieve a semideterministic agreement between laboratory databases by using the type of sample categorized by body district of origin as keys (in addition to personal data) to identify suspected infections related to care assistance (ICAs). Second, on the basis of the patient identification number used as a deterministic key, a unique single database was created. This database shows the flow of 2018 DPR first admissions, which are linked to information of any rehospitalization and samples of alert microorganisms divided by type of infection.
Inclusion and exclusion criteria
Ordinary admissions (cod.1 variable “admission regime”) from all operating units of PTV discharged before December 31, 2018, were included in the analysis. Admissions in the nonordinary regimen (code 2-Day Hospital, code 3-Home treatment, code 4-Day Surgery with overnight stay) and patients < 18 years old were excluded because the hospital does not have a pediatric ward.
Regarding the analysis of bacteriological data, the exclusion criteria included findings of contaminants and diagnoses of microbiological infection occurring 24 hours after hospitalization (in accordance with the definition of HAIs). In the specific case of Clostridioides difficile, which is linked to the intrahospital administration of antibiotics, samples taken during the first seven days of hospitalization were also excluded. For each detected microorganism, the dates of the first and last detection before discharge were reported.
Variables investigated
The following variables were collected from the HDFs:
Personal data (age, sex, fiscal code, etc.)
Main diagnosis (DPR) and secondary diagnoses (first to fifth) at discharge
Codes related to procedures performed during hospitalization
Length of stay (LOS)
Type of discharge
Educational qualification: low educational attainment (lower secondary school, elementary school, or no degree [ISCED 0–2]), medium educational attainment (upper secondary school [ISCED 3–4]), and high educational attainment (bachelor’s degree or other tertiary degree [ISCED 5–8])
Nationality
Marital status
The isolated HAI microorganisms in our sample can be classified into 56 families and more than 150 species. For this study, we focused our analysis on the eight most represented microorganisms in our sample, namely, A. baumannii, Escherichia coli, K. pneumoniae, P. aeruginosa, Clostridioides difficile, Candida spp., Enterococcus spp., and Staphylococcus spp.
The following variables were retrieved from the laboratory database:
Positive sample of the HAI microorganism with date of collection and department where the request was sent
Type of collected sample
Positivity of the tested AMR phenotype (MDR A. baumannii, extended-spectrum beta-lactamase Escherichia coli, K. pneumoniae with at least one AMR profile, and Staphylococcus spp with at least one AMR profile)
All data was processed to create index variables for statistics.
Statistical analysis
Data were anonymized before conducting the statistical analyses. Comparisons were performed using a two-sample t-test or Pearson’s chi-squared test as appropriate. After performing descriptive analysis (frequency, histograms, means, medians IQ, SD, etc.), univariate and multivariate logistic regression models were used to calculate the risk and estimate associations. Statistical analyses were performed using SPSS v.26.0 (IBM Corp., Armonk, NY, USA).
Ethics
This project was conducted in accordance with the Declaration of Helsinki. The project received approval from the Independent Ethics Committee of the University Hospital PTV, Rome, Italy (2022 –Protocol number 66.22) that waived the need for informed consent. To meet privacy requirements, data were anonymized by entering them in an Excel database protected by an encrypted key, which is made available only to the investigators involved in the study.
Results
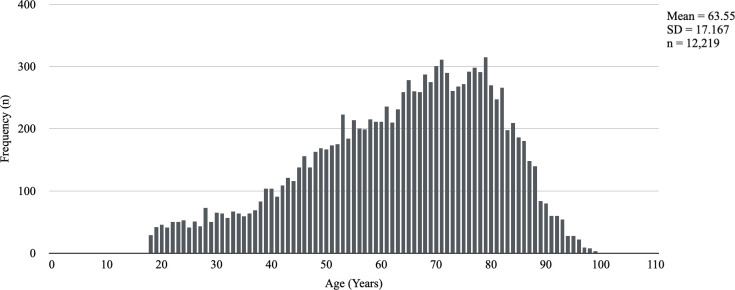
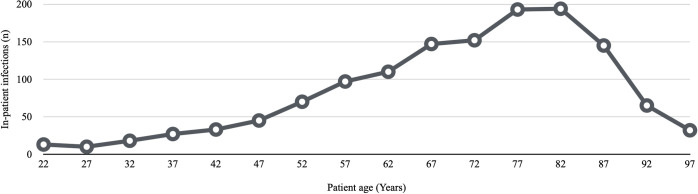
The study sample included 12,219 individuals: 6,770 were males (55.4%), and 5,449 were females (44.6%). The mean age of the patients was 63.6 ± 17.17 years (Fig 1). A total of 6,541 individuals (53.5%) were ≥ 65 years old. The average hospital stay was 9.4 ± 13.63 days, with a median of 5 days. Italian and non-Italian were the nationalities in 91.8% (11,222 individuals) and 8.2% (997 individuals) of cases, respectively. Educational level was low [15] in 76.6% of cases (9,354 individuals) and high in 23.4% of cases (2,865 individuals). The proportion of married individuals was 75.2% (9,183). A total of 1,654 individuals (13.5%) were admitted twice. Table 1 shows the data related to the descriptive analysis of the sample. Figs 1 and 2 respectively show the age distribution of the sample and inpatient infections.
Fig 1. Age distribution in the hospitalized sample.
Table 1. Descriptive analysis of the samples (n = 12,219).
| Investigated variable | N | % | Mean | SD |
|---|---|---|---|---|
| Age | 63.55 | 17.17 | ||
| Range 18–102 years | ||||
| Gender | ||||
| Male | 6,770 | 55.40 | ||
| Female | 5,449 | 44.60 | ||
| Education level | ||||
| None | 82 | 0.70 | ||
| Elementary school | 1,497 | 12.30 | ||
| Lower secondary school | 7,775 | 63.60 | ||
| Upper secondary school | 2,312 | 18.90 | ||
| Bachelor’s degree/tertiary degree | 553 | 4.50 | ||
| Marital status | ||||
| Single | 3,036 | 24.80 | ||
| Married | 9,183 | 75.20 | ||
| Nationality | ||||
| Italian | 11,222 | 91.80 | ||
| Others | 997 | 8.20 | ||
| LOS | 9.40 | 13.63 | ||
| Range 0–360 days | ||||
| Rehospitalization | ||||
| No | 10,565 | 86.50 | ||
| Yes | 1,654 | 13.50 |
Fig 2. Age distribution of inpatient infections.
The mean age was 68.3 ± 15.771 years in the infection group and 63.0 ± 17.243 years in the noninfection group (t = 10.781; p < 0.001). The mean LOS was 31.03 ± 25.610 days in the infection group and 6.71 ± 7.865 days in the noninfection group (t = 74.683; p < 0.001).
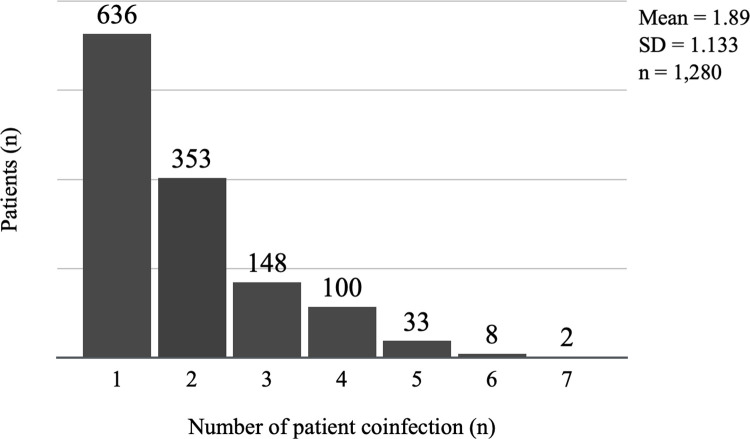
In this retrospective analysis, 1,352 individuals (11.1%) presented one or more suspected ICAs. The number of reported HAI microorganisms (including Candida spp., Proteus, Aspergillus spp.) was 3,549, thus resulting in an average of 2.62 microorganism reported per positive patient. Among these, 2,938 microorganisms (67.6%) are regarded alert microorganisms for AMR, thus resulting in an average of 1.77 AMR alert microorganism reported per positive patient. Concurrent infections from different microorganisms are very frequent, and approximately 50% of infected individuals tested positive for more than one microorganism during their period of hospitalisation (Fig 3).
Fig 3. Distribution of concurrent infections on the sample.
Univariate analysis for the risk of infection showed statistically significant associations with age, LOS, marital status, and low level of education (Table 2). However, there was no significant association between sex and nationality. The binary logistic regression approach was used for multivariate analysis, which showed a persistent association of HAIs with age and LOS in the hospital (Table 2).
Table 2. Association between sociodemographic and outcome variables for infections with HAI associated microorganisms: a binary logistic regression analysis.
| Univariate analysis | Multivariate analysis forward stepwise method | |||||||
|---|---|---|---|---|---|---|---|---|
| Investigated variable | OR | 95% CI | p | OR | 95% CI | p | ||
| lower | upper | lower | upper | |||||
| Age (decade) | 1.217 | 1.174 | 1.262 | <0.001 | 1.178 | 1.126 | 1.233 | <0.001 |
| Gender (male) | 0.980 | 0.875 | 1.098 | = 0.731 | ||||
| Education level | 0.766 | 0.705 | 0.832 | <0.001 | ||||
| Marital status (single) | 1.361 | 1.184 | 1.566 | <0.001 | ||||
| Nationality (italian) | 0.835 | 0.670 | 1.040 | = 0.107 | ||||
| LOS (days) | 1.146 | 1.138 | 1.153 | <0.001 | 1.144 | 1.137 | 1.152 | <0.001 |
Concerning wards, the analysis showed a higher risk of infection associated with medical intervention than with surgical intervention (OR = 1.603; 95% CI = 1.430–1.797; p < 0.001). The risk increased among those over the age of 65 (OR = 1.872; 95% CI = 1.616–2.169; p<0.001), whereas the risk was not statistically significant among those under the age of 65 (OR = 1.185; 95% CI = 0.984–1.428; p = 0.074). Table 3 presents the frequency distributions.
Table 3. Distribution of HAI microorganisms by ward type.
| Inpatient ward | Presence of infection | Total | ||
|---|---|---|---|---|
| n | % | |||
| All patients | Medical | 780 | 13.5% | 5,775 |
| Surgical | 572 | 8.9% | 6,444 | |
| Total | 1,352 | 11.1% | 12,219 | |
| Patients with age ≥ 65 years | Medical | 544 | 16.9% | 3,226 |
| Surgical | 324 | 9.8% | 3,315 | |
| Total | 868 | 13.3% | 6,541 | |
| Patients with age < 65 years | Medical | 236 | 9.3% | 2,549 |
| Surgical | 248 | 7.9% | 3,129 | |
| Total | 484 | 8.5% | 5,678 | |
ICUs had a higher risk of infection in all patients (OR = 3.071; 95% CI = 2.539–3.715; p<0.001). Considering age, the risk for those under 65 was higher (OR = 5.478; 95% IC = 4.036–7.435; p < 0.001) than for those over 65 (OR = 2.094; 95% CI = 1.639–2.675; p < 0.001).
Table 4 lists the frequency of infections per site. Sociodemographic and outcome variables were analyzed for the eight microorganisms under investigation and are presented in Table 5.
Table 4. Frequency of alert microorganisms over the site of infection.
| Bacteremia / sepsis | Respiratory tract | Gastrointestinal tract | Urinary tract | Surgical site | Other sites | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Staphylococcus spp | 473 | 50.2 | 95 | 16.4 | 0 | 0.0 | 16 | 1.6 | 66 | 14.7 | 95 | 21.3 |
| Klebsiella pneumoniae | 98 | 10.4 | 65 | 11.2 | 41 | 39.4 | 97 | 9.6 | 45 | 10.0 | 20 | 4.5 |
| Enterococcus spp | 98 | 10.4 | 29 | 5.0 | 0 | 0.0 | 243 | 24.1 | 122 | 27.2 | 81 | 18.1 |
| Candida spp | 68 | 7.2 | 93 | 16.1 | 2 | 1.9 | 305 | 30.3 | 47 | 10.5 | 96 | 21.5 |
| Escherichia coli | 47 | 5.0 | 30 | 5.2 | 3 | 2.9 | 169 | 16.8 | 41 | 9.2 | 30 | 6.7 |
| Acinetobacter baumannii | 33 | 3.5 | 59 | 10.2 | 22 | 21.2 | 26 | 2.6 | 25 | 5.6 | 19 | 4.3 |
| Pseudomonas aeruginosa | 21 | 2.2 | 69 | 12.0 | 1 | 1.0 | 56 | 5.6 | 29 | 6.5 | 22 | 4.9 |
| Clostridioides difficile | 0 | 0.0 | 0 | 0.0 | 33 | 31.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Vibrions | 0 | 0.0 | 0 | 0.0 | 1 | 1.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Proteus | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 33 | 3.3 | 29 | 6.5 | 0 | 0.0 |
| Aspergillus | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 18 | 4.0 |
| Others | 105 | 11.1 | 138 | 23.9 | 1 | 0.8 | 63 | 6.3 | 44 | 9.8 | 66 | 14.8 |
| Total | 943 | 100.0 | 578 | 100.0 | 104 | 100.0 | 1,008 | 100.0 | 448 | 100.0 | 447 | 100.0 |
Table 5. Intrahospital decease and infections: a binary logistic regression analysis.
| Univariate Analysis | Multivariate analysis forward stepwise method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Investigated variable | OR | 95% CI | p | OR | 95% CI | p | ||||
| lower | upper | lower | upper | |||||||
| Staphylococcus spp | 7.801 | 6.306 | 9.651 | <0.001 | 2.948 | 2.249 | 3.864 | <0.001 | ||
| Pseudomonas aeruginosa | 9.106 | 6.584 | 12.595 | <0.001 | 2.435 | 1.648 | 3.600 | <0.001 | ||
| Klebsiella pneumoniae | 9.334 | 7.084 | 12.299 | <0.001 | 2.640 | 1.884 | 3.700 | <0.001 | ||
| Escherichia coli | 3.108 | 2.203 | 4.384 | <0.001 | ||||||
| Enterococcus spp | 4.835 | 3.797 | 6.157 | <0.001 | ||||||
| Clostridioides difficile | 9.748 | 4.828 | 19.681 | <0.001 | 4.841 | 2.164 | 10.830 | <0.001 | ||
| Candida spp | 8.589 | 6.891 | 10.707 | <0.001 | 3.131 | 2.368 | 4.141 | <0.001 | ||
| Acinetobacter baumannii | 12.558 | 8.816 | 17.889 | <0.001 | 2.443 | 1.585 | 3.764 | <0.001 | ||
A binary logistic regression analysis was conducted to provide a risk calculation between infections and intrahospital death that accounted for 26% (278/1,352) of patients with at least one HAI in our study. Table 5 shows the results. All reported microorganisms were significantly associated with an increased risk of mortality in the univariable analysis, and this association was confirmed in the multivariate analysis, except for Escherichia coli and Enterococcus spp. Multivariate analysis was probably influenced by the high rate of concurrent infections.
Regarding the antibiotic resistance spectra, all isolated microorganisms were tested for resistance phenotypes. Overall, 141 patients showed at least 1 infection with AMR phenotype; for the 4 AMR microorganisms considered in this study, we first evaluated the association with intrahospital death and then evaluated the effect of these microorganisms when considering the mortality associated with an infection caused by the same microorganism without an AMR profile (Table 6).
Table 6. Univariate analysis for intrahospital decease associated with AMR infections and non-AMR infections.
| Non-AMR | AMR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Investigated variable | OR | 95% CI | p | n | OR | 95% CI | p | n | |||
| lower | upper | lower | upper | ||||||||
| Staphylococcus spp | 6.563 | 5.197 | 8.287 | <0.001 | 676 | 18.275 | 11.355 | 29.412 | <0.001 | 70 | |
| Klebsiella pneumoniae | 8.282 | 6.022 | 11.391 | <0.001 | 303 | 12.539 | 7.520 | 20.908 | <0.001 | 63 | |
| Escherichia coli | 2.899 | 2.013 | 4.176 | <0.001 | 310 | 6.171 | 2.441 | 15.600 | <0.001 | 9 | |
| Acinetobacter baumannii | 9.699 | 6.457 | 14.571 | <0.001 | 156 | 33.997 | 15.627 | 73.963 | <0.001 | 28 | |
In addition, we evaluated the percentage of HAIs for which coding was missing from the DRGs. In 407 cases (30.1%), coding for the diagnosis of infection by HAIs was not performed. Coding in medical wards, surgical wards, and ICUs was lacking in 27.0%, 36.8%, and 26.4% of cases, respectively (Table 7).
Table 7. Share of infection coding performed by ward type.
| Ward | Coding not done | Total | ||
|---|---|---|---|---|
| n | % | n | % | |
| Medical | 203 | 27.0% | 753 | 100.0% |
| Surgical | 162 | 36.8% | 440 | 100.0% |
| Intensive care unit | 42 | 26.4% | 159 | 100.0% |
| Total | 407 | 30.1% | 1,352 | 100.0% |
Discussion
The results of this study confirm the alarming data described in the literature regarding the spread of microorganisms of concern and consequent infections in acute care institutions.
Our analysis investigated a specific case study, that of PTV, a university hospital that serves as a referral for high medical specialty functions for about 1,5 million citizens living in the city of Rome and province. Our sample showed a prevalence of infections, in ordinary hospitalization, of 11.1% (1352/12219). This data is high in comparison with the most recent available data from studies across Europe [1, 16] and Italy [17] which showed a prevalence of HAIs of approximately 8.00%.
As HAIs are a multifactorial phenomenon, our study allowed us to investigate in further detail their incidence in relation to several factors both sociodemographic and associated with specific medical procedures. Moreover, while many studies focus on the incidence of infections due to a specific microorganism, our approach allowed us to investigate the phenomenon as a whole, showing interesting results especially concerning multiple HAIs acquired by the single patient. The combined approach of data from the laboratory, the clinical wards and from hospital discharge card allowed a broader view on the topic, and gave more interesting insight. First, the sociodemographic variables associated with HAIs provide us important insights, thus highlighting their role in defining the absolute risk of developing a HAIs (Table 2). Our data showed that being unmarried increased the risk of HAIs. We inferred that marital status could be an indirect indication of loneliness, which is a powerful risk factor for infectious disease susceptibility [18, 19] and immunosuppression (i.e. higher susceptibility of developing HAIs).On the other hand a low education level is associated with poor socioeconomic conditions linked to physical weakness and comorbidities [20].
Nevertheless, the association of the education level with HAI was not confirmed on the multivariate analysis.
In our study, both in univariate and multivariate analysis, age and LOS are significantly associated with HAIs, while gender is not associated in either analysis, this is supported also by the study of Golfera and colleagues [21]. However, as demonstrated in the narrative revision of Cristina and colleagues (2021) elderly patients are identified as belonging to the high-risk group for the development of healthcare-associated infections (HAI) due to the age-related immune system decline, known as immunosenescence [22].
The association of the above-mentioned variables with HAI was not confirmed on the multivariate analysis, probably because of the effect of age. We confirmed the well-known association with the increase in LOS appears in our analysis. Ohannessian and colleagues [23] estimated the change in LOS due to infection(s); by using a multistate model and the time of infection onset. Results from their study show an increase in LOS of 5.0 days (95% CI = 4.6–5.4 days). The LOS increased with the number of infected sites and was higher for patients who were discharged alive from the ICU, but no increase in LOS was found for patients presenting with late-onset HAI, after day 25 of admission [23]. This suggests that the increase in LOS is attributable to infection at the early stage of hospitalization. However in the study of Cristina and colleagues HAI in geriatric patients are responsible for longer hospital stays [22].
Concerning wards, our study showed that, the risk of HAI is higher in medical wards than surgical wards (13.5% and 8.9% of patients, respectively) (Table 3) this is also confirmed by the literature [21, 24]. Our study confirmed this data for both over 65 and under 65 year old patients. This can be explained by the greater number of elderly patients (>65 years) in medical wards than in surgical wards; however, the difference in HAI incidence was not confirmed only considering patients under 65 years. We can therefore assume that perioperative prophylaxis in surgical wards plays a role in reducing infections [25] even if, particular attention must be paid to compliance with the protocol and to its duration to avoid the promotion of AMR insurgence [16, 26, 27].
Concerning the site of infection, HAIs that are frequently observed in our study include urinary tract infections and BSIs; this fact is in accordance with the global trends described in the literature [6, 7, 28].
Intra-hospital death is frequently observed in patients with HAIs, and all analysed microorganisms are significantly associated with increased risk of mortality in the univariate analysis. In multivariate analysis there is no association with increased risk of mortality and Escherichia coli and Enterococcus spp microorganisms.
In this study we showed that approximately 50% of patients who acquired HAIs during their hospitalization tested positive for at least two or more microorganisms, therefore the multivariate analysis provided relevant insights into the effect of concurrent infections. The multivariate analysis showed that some microorganisms, such as Clostridioides difficile, occur less frequently than others. On the other hand, microorganisms such as Escherichia coli and Enterococcus spp. are more likely associated with at least one concurrent HAI, this finding probably explains why they are not associated with mortality in the multivariate analysis (Table 5).
The same approach has been used for infections caused by AMR microorganisms. The risk of intrahospital death is extremely high compared to infections from the same microorganisms with no AMR profile. A relevant example is MDR Acinetobater baumannii which increases mortality by 34 times, compared with 9 times in non-AMR cases (Table 6).
Conclusions
The phenomenon investigated represents a major challenge for public health worldwide, but few epidemiological studies have been conducted on the subject. Furthermore, epidemiological studies are usually conducted in limited areas and with only partial integration of laboratory data, often leading to the detriment of an overall picture.
The record-linkage experiment between laboratory data and HDF flows represents an innovative method from the epidemiological point of view in the assessment of HAIs, which, to the best of our knowledge, has never been fully used, particularly in situations where an integrated regional bacteriological data collection system has not been implemented.
The author considers this approach as a promising tool for deciphering the epidemiological frame of intrinsic comorbidity and is a starting point in combining environmental, clinical, social individual and organizational factors to calculate and define the risk of HAIs.
The proportion of preventable HAIs is high, and infections associated with certain procedures can be prevented by reducing unnecessary interventions, choosing safer equipment, adopting aseptic patient care measures, and observing proper hand washing techniques [12–14]. The cost of HAIs is high to the patient and the facility in both health and economic terms; hence, there is a need to adopt safe care practices that prevent or control transmission, particularly of alert microorganisms. Control programs need to be implemented at different levels (national, regional, and local) to ensure the implementation of measures that have proven effective in minimizing the risk of infections, but this cannot be achieved without improving epidemiological knowledge by conducting prospective studies and improving the transmission of information in administrative flows.
Limitations
The weaknesses of this study includes its lack of systematic data collection and record linkage with environmental monitoring (surfaces, fomites, air, water, medical devices, etc.) and medical surgical procedures/devices. We are currently working to integrate these types of data into the system. In addition, we need to integrate key data from personnel attitudes regarding hand hygiene and other behaviours in daily activities. Microbiological data is integrated in our database but only for patients who tested positive for HAIs, thus resulting in the lack of information about the total number of microbiological tests conducted. Some variables associated with HAIs, except LOS in hospitals, had an unclear causal relationship with the risk of infection because of the observational nature of the study. A prospective study could be useful for assessing this bidirectional relationship.
Data Availability
The data are subjected to the Personal Data Competent Authority, and we can not make them available on a public repository. Fellow researchers can request access to the data sending a request to the Independent Ethical Committee (comitato.etico@ptvonline.it) or to Fondazione PTV Policlinico Tor Vergata Hospital Directorate (direzione.sanitaria@ptvonline.it).
Funding Statement
The authors received no specific funding for this work.
References
- 1.Suetens C, Latour K, Kärki T, Ricchizzi E, Kinross P, Moro ML, et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance [Internet]. 2018. Nov 15 [cited 2021 Nov 15];23(46). Available from: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2018.23.46.1800516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordino V, Vicentini C, D’Ambrosio A, Quattrocolo F, Zotti CM, Novati R, et al. Burden of healthcare-associated infections in Italy: incidence, attributable mortality and disability-adjusted life years (DALYs) from a nationwide study, 2016. J Hosp Infect. 2021. Jul;113:164–71. doi: 10.1016/j.jhin.2021.04.023 [DOI] [PubMed] [Google Scholar]
- 3.Monasta L, Abbafati C, Logroscino G, Remuzzi G, Perico N, Bikbov B, et al. Italy’s health performance, 1990–2017: findings from the Global Burden of Disease Study 2017. Lancet Public Health. 2019. Dec;4(12):e645–57. doi: 10.1016/S2468-2667(19)30189-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epicentro. Italy’s antimicrobial resistance surveillance system (AR-ISS) report dati 2019; 2020 [cited 2022 Feb 22]. Database: Archivio dei rapporti delle sorveglianze AR-ISS e CPE [internet].Available from:: https://www.epicentro.iss.it/antibiotico-resistenza/ar-iss/RIS-1_2020.pdf.
- 5.European Centre for Disease Prevention and Control. Country summaries antimicrobial resistance in the EU/EEA (EARS-Net) Annual Epidemiological Report 2019; 2020 [cited 2022 Feb 22]. Database: ECDC [internet]. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/Country%20summaries-AER-EARS-Net%20202019.pdf.
- 6.Liu JY, Dickter JK. Nosocomial Infections. Gastrointest Endosc Clin N Am. 2020. Oct;30(4):637–52. [DOI] [PubMed] [Google Scholar]
- 7.Stewart S, Robertson C, Pan J, Kennedy S, Dancer S, Haahr L, et al. Epidemiology of healthcare-associated infection reported from a hospital-wide incidence study: considerations for infection prevention and control planning. J Hosp Infect. 2021. Aug;114:10–22. doi: 10.1016/j.jhin.2021.03.031 [DOI] [PubMed] [Google Scholar]
- 8.Deng Y, Zheng Z, Cheng S, Lin Y, Wang D, Yin P, et al. The factors associated with nosocomial infection in elderly hip fracture patients: gender, age, and comorbidity. Int Orthop. 2021. Dec;45(12):3201–9. doi: 10.1007/s00264-021-05104-3 [DOI] [PubMed] [Google Scholar]
- 9.O’Toole RF. The interface between COVID-19 and bacterial healthcare-associated infections. Clin Microbiol Infect. 2021. Jun;S1198743X21002974. doi: 10.1016/j.cmi.2021.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantón R, Gijón D, Ruiz-Garbajosa P. Antimicrobial resistance in ICUs: an update in the light of the COVID-19 pandemic. Curr Opin Crit Care. 2020. Oct;26(5):433–41. doi: 10.1097/MCC.0000000000000755 [DOI] [PubMed] [Google Scholar]
- 11.Afshinnekoo E, Bhattacharya C, Burguete-García A, Castro-Nallar E, Deng Y, Desnues C, et al. COVID-19 drug practices risk antimicrobial resistance evolution. Lancet Microbe. 2021. Apr;2(4):e135–6. doi: 10.1016/S2666-5247(21)00039-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque M, McKimm J, Sartelli M, Dhingra S, Labricciosa FM, Islam S, et al. Strategies to Prevent Healthcare-Associated Infections: A Narrative Overview. Risk Manag Healthc Policy. 2020. Sep;Volume 13:1765–80. doi: 10.2147/RMHP.S269315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridelberg M, Nilsen P. Using surveillance data to reduce healthcare–associated infection: a qualitative study in Sweden. J Infect Prev. 2015. Sep;16(5):208–14. doi: 10.1177/1757177415588380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bruin JS, Seeling W, Schuh C. Data use and effectiveness in electronic surveillance of healthcare associated infections in the 21st century: a systematic review. J Am Med Inform Assoc. 2014. Sep 1;21(5):942–51. doi: 10.1136/amiajnl-2013-002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UNESCO Institute for Statistics. International standard classification of education: ISCED 2011 [Internet]. Montreal, Quebec: UNESCO Institute for Statistics; 2012 [cited 2021 Nov 15]. Available from: http://www.uis.unesco.org/Education/Documents/isced-2011-en.pdf
- 16.Cassini A, Plachouras D, Eckmanns T, Abu Sin M, Blank HP, Ducomble T, et al. Burden of Six Healthcare-Associated Infections on European Population Health: Estimating Incidence-Based Disability-Adjusted Life Years through a Population Prevalence-Based Modelling Study. Harbarth S, editor. PLOS Med. 2016. Oct 18;13(10):e1002150. doi: 10.1371/journal.pmed.1002150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Italian ministry of health. Studio di prevalenza italiano sulle infezioni correlate all’assistenza e sull’uso di antibiotici negli ospedali per acuti—protocollo ECDC [internet]. Università di Torino; 2018 [cited 2022 Feb 22]. Available from: https://www.salute.gov.it/imgs/C_17_pubblicazioni_2791_allegato.pdf.
- 18.Liotta G, Marazzi MC, Orlando S, Palombi L. Is social connectedness a risk factor for the spreading of COVID-19 among older adults? The Italian paradox. Abete P, editor. PLOS ONE. 2020. May 21;15(5):e0233329. doi: 10.1371/journal.pone.0233329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanguas J, Pinazo-Henandis S, Tarazona-Santabalbina FJ. The complexity of loneliness. Acta Bio Medica Atenei Parm. 2018. Jun 7;89(2):302–14. doi: 10.23750/abm.v89i2.7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnelly JP, Lakkur S, Judd SE, Levitan EB, Griffin R, Howard G, et al. Association of Neighborhood Socioeconomic Status With Risk of Infection and Sepsis. Clin Infect Dis. 2018. Jun 1;66(12):1940–7. doi: 10.1093/cid/cix1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golfera M, Toscano F, Cevenini G, Marco MFD, Porchia BR, Serafini A, et al. Predicting Healthcare-associated Infections: are Point of Prevalence Surveys data useful? J Prev Med Hyg. 2022. Jun;63(2):E304–9. doi: 10.15167/2421-4248/jpmh2022.63.2.1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cristina ML, Spagnolo AM, Giribone L, Demartini A, Sartini M. Epidemiology and Prevention of Healthcare-Associated Infections in Geriatric Patients: A Narrative Review. Int J Environ Res Public Health. 2021. May;18(10):5333. doi: 10.3390/ijerph18105333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohannessian R, Gustin MP, Bénet T, Gerbier-Colomban S, Girard R, Argaud L, et al. Estimation of Extra Length of Stay Attributable to Hospital-Acquired Infections in Adult ICUs Using a Time-Dependent Multistate Model*: Crit Care Med. 2018. Jul;46(7):1093–8. doi: 10.1097/CCM.0000000000003131 [DOI] [PubMed] [Google Scholar]
- 24.Chernet AZ, Dasta K, Belachew F, Zewdu B, Melese M, Ali MM. Burden of Healthcare-Associated Infections and Associated Risk Factors at Adama Hospital Medical College, Adama, Oromia, Ethiopia. Drug Healthc Patient Saf. 2020. Oct;Volume 12:177–85. doi: 10.2147/DHPS.S251827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prospero E, Barbadoro P, Marigliano A, Martini E, D’Errico MM. Perioperative antibiotic prophylaxis: improved compliance and impact on infection rates. Epidemiol Infect. 2011. Sep;139(9):1326–31. doi: 10.1017/S0950268810002505 [DOI] [PubMed] [Google Scholar]
- 26.Cicuttin E, Sartelli M, Scozzafava E, Tartaglia D, Cremonini C, Brevi B, et al. Antibiotic Prophylaxis in Torso, Maxillofacial, and Skin Traumatic Lesions: A Systematic Review of Recent Evidence. Antibiotics. 2022. Jan 21;11(2):139. doi: 10.3390/antibiotics11020139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothe, Kathrin M.D.; Münster, Nathalie; Hapfelmeier, Alexander Ph.D.; Ihbe-Heffinger, Angela Ph.D.; Paepke, Stefan M.D.; Niemeyer, Markus M.D., Ph.D.; et al., Plastic and Reconstructive Surgery: January 31, 2022—Volume—Issue—10.1097/PRS.0000000000008900 doi: 10.1097/PRS.0000000000008900 Does the Duration of Perioperative Antibiotic Prophylaxis Influence the Incidence of Postoperative Surgical-Site Infections in Implant-Based Breast Reconstruction in Women with Breast Cancer? A Retrospective Study. [DOI] [PubMed]
- 28.Kollef, Marin H. MD1; Torres, Antoni MD, PhD2; Shorr, Andrew F. MD, MPH, MBA3; Martin-Loeches, Ignacio MD, PhD4; Micek, Scott T. PharmD. Nosocomial Infection, Critical Care Medicine. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are subjected to the Personal Data Competent Authority, and we can not make them available on a public repository. Fellow researchers can request access to the data sending a request to the Independent Ethical Committee (comitato.etico@ptvonline.it) or to Fondazione PTV Policlinico Tor Vergata Hospital Directorate (direzione.sanitaria@ptvonline.it).