Abstract
Background
Whole genome sequencing (WGS) can detect variants and estimate telomere length. The clinical utility of WGS in estimating risk, progression and survival of pulmonary fibrosis patients is unknown.
Methods
In this observational cohort study, we performed WGS on 949 patients with idiopathic pulmonary fibrosis or familial pulmonary fibrosis to determine rare and common variant genotypes, estimate telomere length and assess the association of genomic factors with clinical outcomes.
Results
WGS estimates of telomere length correlated with quantitative PCR (R=0.65) and Southern blot (R=0.71) measurements. Rare deleterious qualifying variants were found in 14% of the total cohort, with a five-fold increase in those with a family history of disease versus those without (25% versus 5%). Most rare qualifying variants (85%) were found in telomere-related genes and were associated with shorter telomere lengths. Rare qualifying variants had a greater effect on telomere length than a polygenic risk score calculated using 20 common variants previously associated with telomere length. The common variant polygenic risk score predicted telomere length only in sporadic disease. Reduced transplant-free survival was associated with rare qualifying variants, shorter quantitative PCR-measured telomere lengths and absence of the MUC5B promoter (rs35705950) single nucleotide polymorphism, but not with WGS-estimated telomere length or the common variant polygenic risk score. Disease progression was associated with both measures of telomere length (quantitative PCR measured and WGS estimated), rare qualifying variants and the common variant polygenic risk score.
Conclusion
As a single test, WGS can inform pulmonary fibrosis genetic-mediated risk, evaluate the functional effect of telomere-related variants by estimating telomere length, and prognosticate clinically relevant disease outcomes.
Shareable abstract (@ERSpublications)
Whole genome sequencing yields rare and common variant genotypes and telomere length estimation to characterise disease risk and provide prognostic information regarding survival and progression for pulmonary fibrosis patients. https://bit.ly/3A8uVBe
Introduction
Idiopathic pulmonary fibrosis (IPF) is the most common fibrotic interstitial lung disease (ILD) affecting older adults. It is characterised by progressive functional decline and, in the absence of antifibrotic medication, a life expectancy of 3–5 years. Population and family-based studies have linked IPF risk to several different rare and common risk variants, which collectively involve genes that maintain the integrity of telomeres [1]. Telomeres consist of hexamer repeats (CCCTAA) located at the ends of chromosomes and serve as protective caps during cell replication. Between 5% and 25% of patients with sporadic IPF or familial pulmonary fibrosis (FPF) have rare deleterious heterozygous germline variants in one of 10 different telomere-related genes that lead to telomere shortening: TERT, TERC, RTEL1, PARN, DKC1, TINF2, NAF1, NHP2, NOP10 and ZCCHC8 [2-9]. Telomere length is inversely associated with outcome risk in patients with sporadic IPF, including worse survival [10, 11], progressive disease as measured by decline in forced vital capacity (FVC) [12], an adverse response to immunosuppressive medications [13] and increased complications after lung transplantation [14]. A polygenic score comprising multiple common, noncoding variants associated with telomere length has been causally related to IPF in mendelian randomisation studies [15].
Identification of a pathogenic telomere-related variant affects clinical management [16] and provides prognostic information for family members. However, the accurate classification of a variant can be challenging. Without functional data or genetic and clinical information from other family members, a rare variant may be considered of unknown significance per established guidelines [17]. Identifying coexistent shortened telomere length can support evidence of pathogenicity of a telomere-related variant, but telomere length is not typically reported by genetic testing laboratories and instead measured independently by flow-fluorescent in situ hybridisation (flow-FISH) or quantitative PCR (qPCR).
Whole genome sequencing (WGS) enables the identification of rare and common variants and simultaneous estimation of telomere length. In this study, we assess the utility of WGS to both identify telomere-related genetic variants and estimate telomere length. We determine the utility of estimated telomere length and genetic variants in predicting survival and rate of disease progression in a large cohort of pulmonary fibrosis patients.
Materials and methods
Study design and subjects
This retrospective cohort study was approved by the institutional review board at Columbia University Medical Center (CUMC) (IRB AAAS0753) and University of Texas Southwestern (UTSW) Medical Center (IRB 092017-007). Each subject provided written informed consent and a blood sample. The study population included patients diagnosed with IPF or FPF and recruited to participate in longitudinal observation studies through registries at UTSW and CUMC. Subjects were enrolled in the ILDs clinics at both sites and follow-up including pulmonary function testing took place as part of routine clinical treatment. Subjects with IPF from the PANTHER-IPF (NCT00957242) [18] and ACE-IPF (NCT00650091) [19] clinical trials who participated in the optional genetics sub-study were also included.
Patient diagnoses from longitudinal registries were made according to consensus guidelines [20] with multidisciplinary conference discussions at each site. Patient demographics, serial pulmonary function test results and clinical end-points were abstracted from available medical records. A positive family history was defined by having at least one third-degree relative with a history of fibrotic ILD. Clinical data from PANTHER-IPF and ACE-IPF subjects were obtained from the National Heart, Lung, and Blood Institute Biologic Specimen and Data Repository Information Coordinating Center, and did not include family history.
Measurement of telomere length
Genomic DNA was isolated from blood leukocytes from the UTSW and CUMC cohorts using Gentra reagents (Qiagen) and from PANTHER-IPF and ACE-IPF frozen blood using QIAamp DNA Blood Maxi kit (Qiagen). Leukocyte telomere length was measured by qPCR (qPCR-TL) using the RotoGene instrument (Qiagen) as previously described [21].
Terminal restriction fragment length analysis of genomic DNA isolated from peripheral blood leukocytes was performed as previously described [21]. Correlations between qPCR and Southern blot measurements were previously compared for 387 samples (Spearman rank correlation=0.83; p<2.2×10−16) [22].
Genome sequencing, variant calling and genomic analyses
Detailed methods are available in the supplementary material. WGS was performed at the Institute for Genomic Medicine according to standard protocols on Illumina’s NovaSeq 6000 platform with raw sequencing reads aligned to the hg19 reference genome. Qualifying variants (QV) were defined as rare in reference populations and predicted to be damaging by in silico tools as previously described [23]. We estimated telomere length from WGS BAM files (WGS-TL) using Telseq [24]. Principal components of ancestry were computed using FlashPCA as previously described (supplementary figure S1) [25]. Polygenic risk scores (PRSs) were calculated from 20 common single nucleotide polymorphisms (SNPs) previously associated with leukocyte telomere lengths (supplementary table S1) with higher scores predicting shorter telomere length [26]. Genomic data from consenting subjects were deposited on the Database of Genotypes and Phenotype (phs002692.v1).
Statistics
We examined baseline continuous variables using a t-test or Wilcoxon signed rank sum test. Categorical variables are expressed as proportion or percentage and were compared using Chi-squared test or Fisher’s exact test as indicated. ANOVA was used to assess inter-group differences in means. Pairwise comparison testing was performed using Dunn’s test with Bonferroni correction following a significant Kruskal–Wallis test.
We created generalised additive models to test adjusted associations between telomere length and PRSs (supplementary methods). For independent predictors without nonlinear associations with telomere length, multivariate linear regression was used to estimate effect sizes. Multivariable Cox proportional hazards regression was used to estimate the association between genomic predictors and transplant-free survival, defined as time from enrolment into the registry to death or transplant, for subjects from the UTSW and CUMC registries with available genomic, clinical and outcome data. Each model was adjusted for baseline age, sex, % predicted FVC and % predicted diffusion capacity for carbon monoxide. Continuous genomic predictors were assessed as raw values and z-transformed standardised values, where hazard ratios represent change per one unit increase or one standard deviation increase, respectively. Annual FVC change was estimated for each genomic predictor using joint modelling and was restricted to registry subjects who had three or more FVC measurements that spanned >3 months. The joint model accounts for non-random dropout due to death or transplant on the slope change in FVC by including a time-to-event submodel and a mixed-effects submodel [27, 28] that includes random intercept and slope terms. A Weibull distribution was assumed for the time-to-event submodel. Both the mixed-effects and time-to-event submodels were adjusted for age, sex and study centre.
All p-values <0.05 were considered significant. All statistical analyses were performed using R statistical analysis software, version 3.6.1 (www.r-project.org).
Results
Patient characteristics
WGS of 949 unrelated subjects with IPF or FPF were included in this study (UTSW, n=626; CUMC, n=91; IPFnet, n=232) (supplementary table S2). Subjects had a median (IQR) age of 67 years (61–73 years) and were predominantly male (68%). Family history information was available for 717 subjects (76% of the total cohort). Slightly more than half (54%, 388 of 717) had a family history of pulmonary fibrosis (FPF). Of the 388 FPF probands, 60% had a clinical diagnosis of IPF and 40% had a non-IPF fibrotic ILD diagnosis.
WGS telomere length estimates
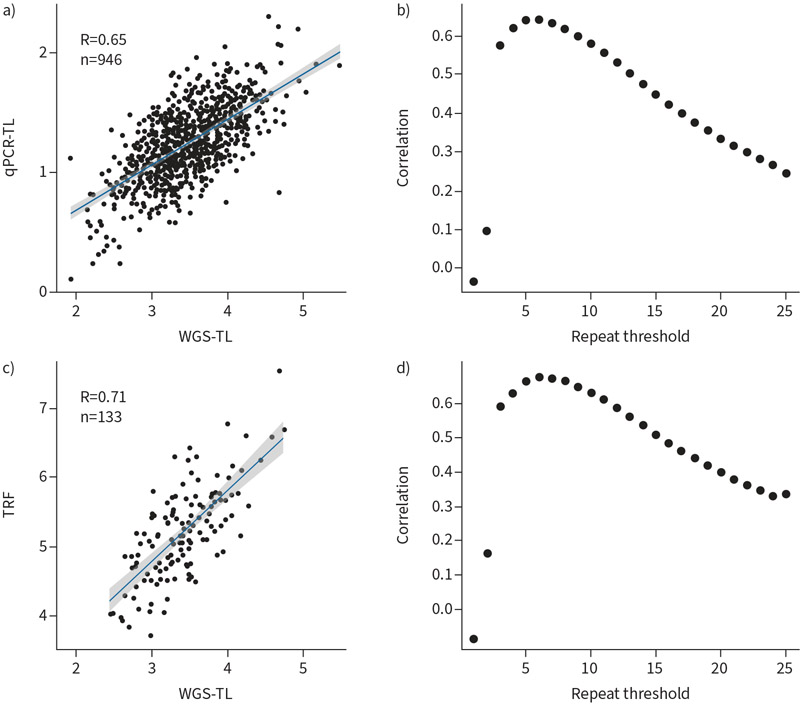
We estimated telomere length from WGS data (WGS-TL) using Telseq. Leukocyte telomere length measurement by qPCR (qPCR-TL) was available for 946 of 949 subjects (99.7%). Southern blot measurement of the telomere-containing terminal restriction fragment length was available for 133 of 949 subjects (14%). We selected a hexamer repeat threshold of six in Telseq which resulted in the best correlation between WGS-TL and qPCR-TL (R=0.65, figure 1) as well as between WGS-TL and a Southern blot measurement of telomere length (R=0.71). We used a hexamer repeat threshold of six for all Telseq estimations of telomere length. Each unit increase in Telseq was equivalent to a 1.04 kb change in telomere length by Southern blot analysis.
FIGURE 1.
Correlation of telomere length estimation by whole genome sequencing (WGS) with laboratory measurements. a) Scatterplot of telomere length measurements estimated by WGS (WGS-TL) using Telseq and measured by quantitative PCR (qPCR-TL). b) Effect of telomere length measurement correlation between WGS and qPCR methods by varying hexamer repeat threshold parameter in Telseq. Optimal repeat threshold of six shows highest correlation. c) Scatterplot of telomere measurements by WGS and by terminal restriction fragment (TRF) analysis. d) Effect of telomere length measurement correlation between WGS and Southern blot methods by varying hexamer repeat threshold parameter in Telseq. Optimal repeat threshold of six shows highest correlation.
Rare deleterious QVs
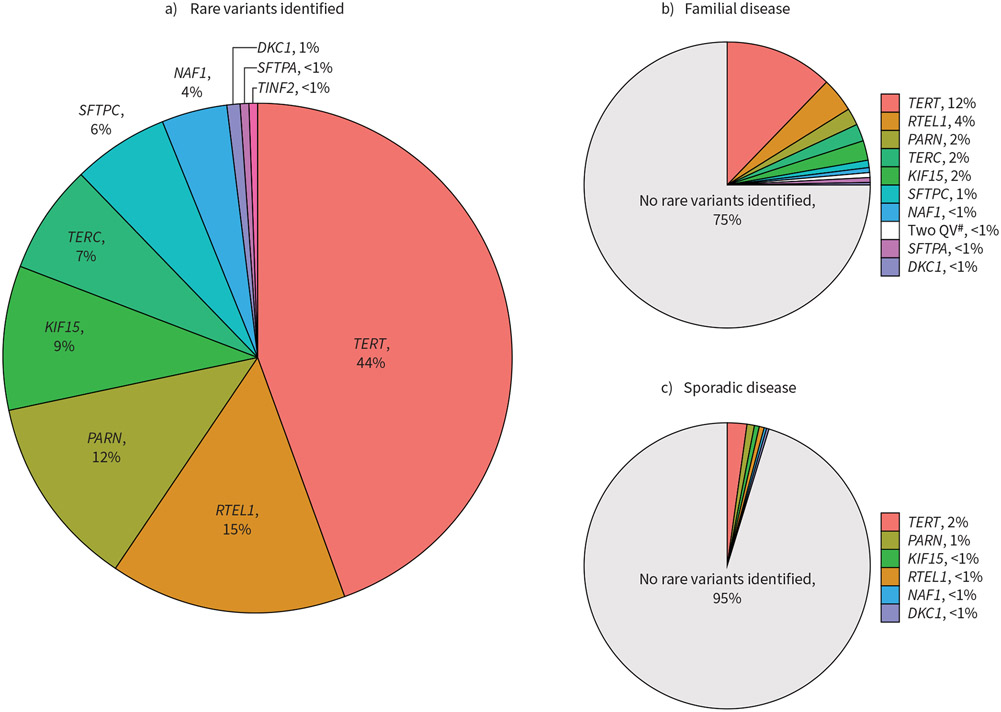
We identified carriers of rare deleterious QVs in known telomere-related (TERT, TERC, RTEL1, PARN, DKC1, TINF2, NAF1 and ZCCHC8) and non-telomere (SFTPC, SFTPA1/2 and KIF15) genes associated with adult-onset pulmonary fibrosis (table 1, supplementary table S3) [2-5, 8, 9, 23, 29-31]. A QV was defined as exceedingly rare in reference databases with a consensus in silico prediction of a damaging effect on the protein product (supplementary methods). Overall, we found that 14% of subjects (133 of 949) carried a rare QV (figure 2a, supplementary table S4). Most carriers only carried one QV, although three subjects carried two QVs (TINF2/SFTPC, PARN/SFTPC, TERT/KIF15). Most QVs (85%, 115 of 136) were found in telomere-related genes, with a smaller portion (15%, 21 of 136) found in non-telomere genes. The highest numbers of telomere-related QVs were found in TERT (n=60), RTEL1 (n=21), PARN (n=16) and TERC (n=10), with smaller numbers found in NAF1 (n=5), DKC1 (n=2) and TINF2 (n=1). No QVs were found in ZCCHC8. Rare QVs in non-telomere genes were found in KIF15 (n=12), SFTPC (n=8) and SFTPA1/2 (n=1).
TABLE 1.
Clinical and genomic characteristics of cohort
| All | Telomere-related QV carrier |
Non-telomere QV carrier |
No QV | p-value | |
|---|---|---|---|---|---|
| Subjects, n | 949 | 115# | 21# | 816 | |
| Age (years), median (IQR) | 67 (60–74) | 61 (55–67) | 59 (51–67) | 68 (62–74) | <0.0001 |
| Male | 646 (68%) | 69 (60%) | 16 (76%) | 563 (69%) | ns |
| Ethnicity | |||||
| White | 808 (85%) | 104 (90%) | 17 (81%) | 690 (84%) | ns |
| Black | 23 (3%) | 1 (1%) | 1 (5%) | 21 (3%) | ns |
| Hispanic | 67 (7%) | 7 (6%) | 3 (14%) | 57 (7%) | ns |
| Asian | 51 (5%) | 3 (3%) | 0 | 48 (6%) | ns |
| Familial disease ¶ | 388 (54%) | 84 (86%) | 15 (83%) | 291 (48%) | <0.0001 |
| IPF diagnosis | 795 (84%) | 81 (70%) | 17 (81%) | 699 (86%) | 0.00057 |
| Telomere length | |||||
| WGS-TL, mean±sd | 3.47±0.5 | 3.03±0.5 | 3.53±0.6 | 3.5±0.5 | <0.0001 |
| qPCR-TL <10th percentile+ | 385 (41%) | 91 (79%) | 6 (29%) | 288 (35%) | <0.0001 |
| MUC5B rs35705950 MAF, (95% CI) | 0.33 (0.31–0.35) | 0.26 (0.21–0.32) | 0.45 (0.30–0.61) | 0.34 (0.31–0.36) | 0.010 |
Data presented as n (%), unless otherwise indicated. Bold text indicates statistical significance. QV: qualifying variant; IPF: idiopathic pulmonary fibrosis; WGS-TL: whole genome sequencing-estimated telomere length; qPCR-TL: quantitative PCR-measured telomere length; MAF: minor allele frequency; ns: not significant.
three individuals carried both a telomere-related QV and a non-telomere QV (SFTPC/TINF2, SFTPC/PARN, KIF15/TERT) and these individuals were counted in each category
number of subjects and proportion for each category restricted to the subjects with known family histories (all, n=717; telomere-related QV carrier, n=98; non-telomere QV carrier, n=18; no QV, n=603)
number of subjects and proportion for each category restricted to the subjects with known qPCR (all, n=946; telomere-related QV carrier, n=115; non-telomere QV carrier, n=21; no QV, n=814).
FIGURE 2.
Rare variant landscape in idiopathic pulmonary fibrosis/familial pulmonary fibrosis cohort. a) All qualifying variants (QVs) found in overall cohort. QVs represent likely deleterious coding variants predicted to affect protein function. b) Prevalence of QVs in patients with familial disease. c) Prevalence of QVs in patients without a family history of fibrotic interstitial lung disease. #: two individuals with two QVs (SFTPC/TINF2, SFTPC/PARN).
The diagnostic yield of finding any rare deleterious QV was much higher in patients with familial disease compared to those without (25% versus 5%, p<0.0001) (figure 2b, c). Carriers of rare QVs in telomere-related and non-telomere genes were significantly younger than those without QVs (age difference (95% CI) of −5.8 years (−7.9– −3.6 years) and −9.2 years (−14.5– −4.0 years), respectively) after adjusting for multiple comparison testing (table 1, supplementary table S3). There was no enrichment of telomere-related QVs in any ethnicity. The overall MUC5B promoter SNP (rs35705950) minor allele frequency (MAF) was 0.33, similar to previous descriptions in other cohorts with familial and sporadic IPF [32]. We found that patients with rare telomere-related QVs had a lower MUC5B MAF when compared to those without (0.26 versus 0.34, p=0.02), consistent with a prior report [11]. There was no difference in MUC5B MAF amongst subjects with WGS-TL above versus below the median (0.31 versus 0.35, p=0.15).
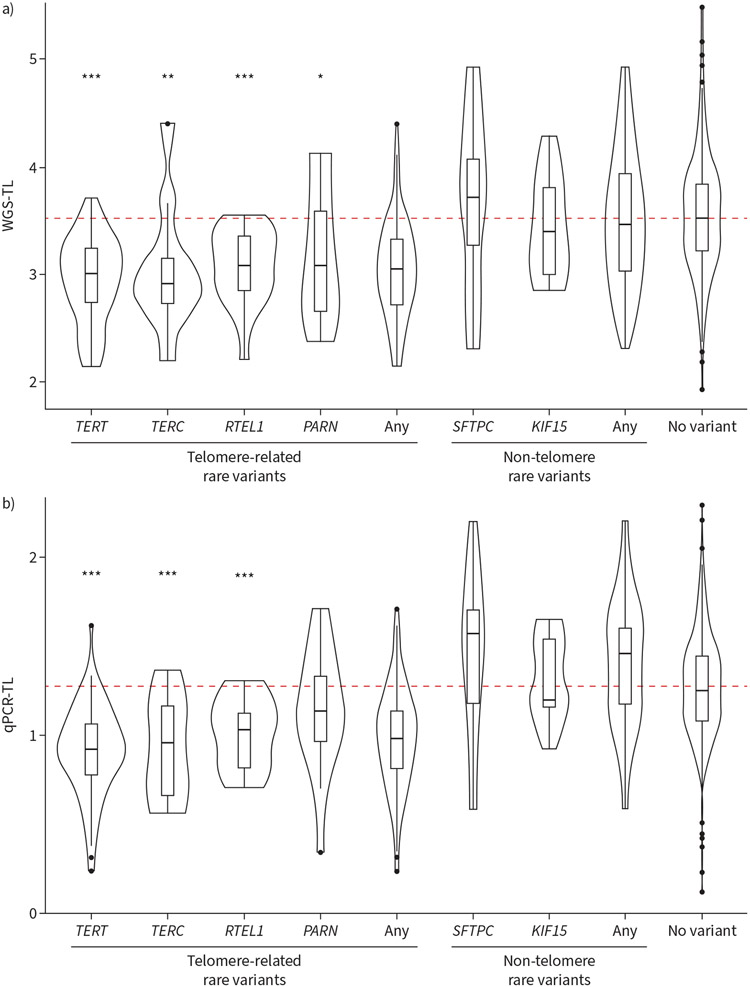
To investigate the use of WGS as a single platform to determine the biological consequence of rare telomere-related QVs, we compared WGS-TL and qPCR-TL amongst QV carriers (figure 3). Individuals carrying QVs in TERT (p<0.0001), TERC (p=0.008), RTEL1 (p<0.0001) and PARN (p=0.02) had significantly lower WGS-TL estimates compared to non-QV carriers. Individuals carrying NAF1, TINF2 and DKC1 QVs did not have significantly shorter WGS-TL although these analyses were limited by small numbers. Similarly, individuals carrying QVs in TERT, TERC and RTEL1 had significantly lower qPCR-TL measurements compared to non-QV carriers. Telomere length in carriers of telomere-related QVs was consistently short in stratified analysis by missense versus protein-truncating mutations (supplementary figure S2) and by known American College of Medical Genetics and Genomics (ACMG) classification [17] as reported in the ClinVar database (supplementary figure S3) [33]. Notably, of the telomere-related QVs, 41 were classified as variants of uncertain significance and 36 were not previously reported in ClinVar; both groups showed significantly shorter telomere lengths than non-QV carriers. All identified QVs were submitted to ClinVar.
FIGURE 3.
Telomere length stratified by rare variant carrier status. Telomere length estimated by a) whole genome sequencing (WGS-TL) and measured by b) quantitative PCR (qPCR-TL) stratified by rare variant carrier status. Rare qualifying variants were identified using WGS by predefined criteria and grouped into telomere-related and non-telomere genes. Violin plots with overlaying bar plots signifying median and interquartile range. Telomere lengths of TERT, TERC, PARN and RTEL1 rare variant carriers are significantly shorter than those belonging to subjects without any rare variant. *: p<0.05; **: p<0.01; ***: p<0.001.
Genomic determinants of telomere length
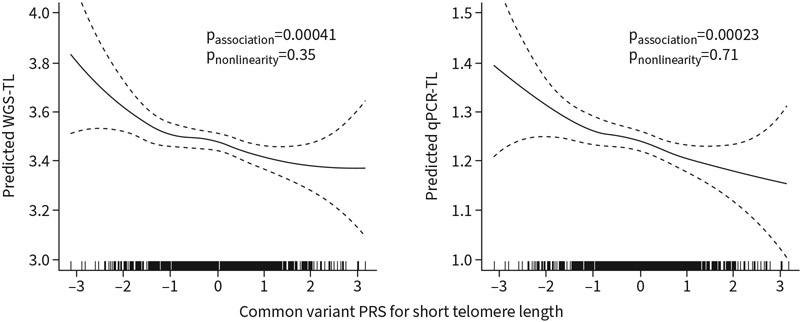
We used multivariate linear regression models to estimate the effect of age, sex and ancestry principal components, as well as rare and common variants, on WGS-TL (table 2). Older age was independently associated with shorter WGS-TL. Inheritance of rare telomere-related QVs was associated with shorter WGS-TL (p<0.0001) in adjusted models. We extracted genotypes of 20 SNPs (supplementary table S1) previously linked to telomere length [26] and calculated a PRS, with higher scores predicting shorter telomere length. We found that this PRS was associated with shorter WGS-TL in both adjusted generalised additive models and linear regression models (table 2, figure 4). We found similar associations between this PRS and shorter qPCR-TL. Associations between the PRS and WGS-TL or qPCR-TL did not meet evidence of nonlinearity in generalised additive models.
TABLE 2.
Clinical and genomic predictors of telomere length in multivariate linear regression model stratified by family history
| Variable | Multivariable analysis | |||||
|---|---|---|---|---|---|---|
| Total cohort (n=949) |
Familial disease (n=388) |
Sporadic disease (n=329) |
||||
| β±SE | p-value | β±SE | p-value | β±SE | p-value | |
| WGS-estimated telomere length | ||||||
| Age (years) | −0.006±0.002 | 0.00012 | −0.006±0.002 | 0.0065 | −0.009±0.003 | 0.0020 |
| Male sex | 0.05±0.03 | 0.12 | 0.02±0.05 | 0.76 | 0.10±0.07 | 0.13 |
| PC of ancestry | ||||||
| PC1 | −0.22±0.07 | 0.0038 | −0.08±0.32 | 0.79 | −0.24±0.10 | 0.020 |
| PC2 | 0.17±0.10 | 0.10 | 0.16±0.19 | 0.41 | 0.40±0.20 | 0.040 |
| Any rare telomere-related QV | −0.52±0.05 | <0.0001 | −0.56±0.06 | <0.0001 | −0.40±0.13 | 0.0030 |
| Common variant PRS for short telomere length (standardised)# | −0.05±0.02 | 0.00038 | −0.05±0.02 | 0.053 | −0.07±0.03 | 0.0081 |
| qPCR-measured telomere length | ||||||
| Age (years) | −0.004±0.0009 | <0.0001 | −0.004±0.001 | 0.0016 | −0.003±0.002 | 0.07 |
| Male sex | −0.02±0.02 | 0.31 | −0.04±0.03 | 0.15 | 0.04±0.04 | 0.27 |
| PC of ancestry | ||||||
| PC1 | −0.21±0.04 | <0.0001 | −0.25±0.18 | 0.16 | −0.22±0.06 | 0.0001 |
| PC2 | 0.08±0.06 | 0.18 | 0.02±0.11 | 0.85 | 0.11±0.11 | 0.32 |
| Any rare telomere-related QV | −0.21±0.03 | <0.0001 | −0.34±0.03 | <0.0001 | −0.23±0.08 | 0.0026 |
| Common variant PRS for short telomere length (standardised)# | −0.03±0.009 | 0.00042 | −0.01±0.01 | 0.32 | −0.05±0.02 | 0.00068 |
Analyses are adjusted for all variables listed (age, male sex, PC1, PC2, presence of rare telomere-related QV, common variant PRS). Bold text indicates statistical significance. WGS: whole genome sequencing; PC: principal component; QV: qualifying variant; PRS: polygenic risk score; qPCR: quantitative PCR.
standardised scores are z-transformed, β estimates are for every 1 sd increase in PRSs above mean.
FIGURE 4.
Telomere length polygenic risk score (PRS) association with telomere length. Generalised additive model fit using z-transformed standardised common variant PRSs (see Methods) to predict whole genome sequencing-estimated telomere length (WGS-TL) and quantitative PCR-measured telomere length (qPCR-TL). Model is adjusted for age, sex, first two principal components of ancestry and presence of any rare telomere-related variant. Associations estimated using nonparametric locally weighted smoothing spline. p-values for association calculated using ANOVA for parametric effects. ANOVA for nonparametric effects used to estimate probability of nonlinearity.
The presence of any telomere-related rare variant was associated with a 10-fold greater effect on WGS-TL than one standard deviation increase in the common variant PRS (β= −0.52 versus β= −0.05, respectively; p<0.0001 versus p=0.00038, respectively). We repeated the analysis stratified by family history, restricting it to the 717 subjects with available family history data (table 2). We found that for both familial and sporadic disease, the presence of a rare telomere-related QV was associated with shorter WGS-TL (β= −0.56, p<0.0001 and β= −0.40, p=0.0030, respectively). Conversely, the common variant PRS was associated with WGS-TL only for those with sporadic disease (β= −0.07, p=0.0081), and not for those with familial disease. We found similar independent associations of age, rare telomere-related QVs and the common variant PRS with qPCR-TL stratified by family history.
Genomic determinants of clinical outcomes
We assessed the clinical utility of WGS-derived genomic characteristics on transplant-free survival using a subset of the UTSW and CUMC registry cohorts that had available data for baseline pulmonary function tests and clinical outcome (death or transplant) (n=474, supplementary table S5, supplementary figure S4). In fully adjusted models accounting for age, sex, ancestry principal components and baseline lung function, each standard deviation increase in qPCR-TL was associated with reduced rates of transplant or mortality (HR 0.82, 95% CI 0.73–0.93, p=0.0018) (table 3). In contrast, WGS-TL was not predictive of survival (HR 0.95, 95% CI 0.85–1.07, p=0.42 for each standard deviation increase). The presence of any rare telomere-related QV was associated with worse transplant-free survival (HR 1.62, 95% CI 1.16–2.26, p=0.0043), whereas each standard deviation increase of the common variant PRS, predicting shorter telomere length, trended towards worse transplant-free survival (HR 1.13, 95% CI 0.99–1.32, p=0.058). The presence of the MUC5B rs35705950 minor allele was associated with improved survival (HR 0.79, 95% CI 0.62–0.99, p=0.049).
TABLE 3.
Genomic factors association with reduced transplant-free survival
| Model | HR per 1 unit increase (95% CI) |
HR per 1 sd increase (95% CI) |
p-value |
|---|---|---|---|
| Baseline+WGS-TL # | 0.91 (0.73–1.14) | 0.95 (0.85–1.07) | 0.42 |
| Baseline+qPCR-TL (ln[T/S]) # | 0.52 (0.35–0.79) | 0.82 (0.73–0.93) | 0.0018 |
| Baseline+rare telomere-related QV (present vs absent) | 1.62 (1.16–2.26) | – | 0.0043 |
| Baseline+common variant PRS for short telomere length ¶ | 2.27 (0.97–5.27) | 1.13 (0.99–1.27) | 0.058 |
| Baseline+MUC5Brs35705950 SNP (GG vs GT/TT) | 0.79 (0.62–0.99) | – | 0.049 |
The analysis was restricted to registry subjects with available outcome and baseline data (n=474). Baseline variables include age, sex, % predicted forced vital capacity, % predicted diffusion capacity of the lung for carbon monoxide and first two principal components of ancestry. Hazard estimates are for the continuous variables listed in addition to baseline variables. Bold text indicates statistical significance. WGS-TL: whole genome sequencing-estimated telomere length; qPCR-TL: quantitative PCR-measured telomere length; QV: qualifying variant; PRS: polygenic risk score.
β estimates for telomere length shown for one unit or 1 sd increase
β estimates for PRSs shown for one unit or 1 sd increase (higher score predicting shorter telomere length).
We further examined the effect of genomic characteristics on disease progression, measured by rate of FVC decline per year using a joint model accounting for non-random dropout and adjusting for age, sex, ancestry principal components and study centre. We restricted the analysis to those subjects with three or more FVC measurements over 3 months (n=268, supplementary table S6, supplementary figure S4). Subjects with the shortest quartile (Q1) of WGS-TL had a greater rate of FVC decline (−240 mL·year−1, 95% CI −284– −196 mL·year−1, versus −188 mL·year−1, 95% CI −216– −159 mL·year−1; p=0.047) than those in the second through fourth quartiles (table 4, supplementary figure S5), as did those with the shortest quartile (Q1) of qPCR-TL (−266 mL·year−1, 95% CI −320– −212 mL·year−1, versus −187 mL·year−1, 95% CI −213– −160 mL·year−1; p=0.0087). The inheritance of a rare telomere-related QV was associated with a faster rate of FVC decline (−267 mL·year−1, 95% CI −331– −202 mL·year−1, versus −194 mL·year−1, 95% CI −220– −168 mL·year−1; p=0.039) compared to those without a telomere-related QV. Similarly, those in the first quartile (Q1) of the common variant PRS, predicted to have the shortest telomere length, had a faster rate of FVC decline (−251 mL·year−1, 95% CI −299– −204 mL·year−1, versus −190 mL·year−1, 95% CI −218– −162 mL·year−1; p=0.026). There was no effect of the MUC5B SNP on the rate of FVC decline.
TABLE 4.
Genomic factors associated with disease progression
| Variable | Subjects, n |
FVC measurements, n |
ΔFVC, mL·year−1 (95% CI) |
p-value |
|---|---|---|---|---|
| WGS-TL | ||||
| Q1# | 73 | 454 | −240 (−284– −196) | 0.047 |
| Q2–4 | 195 | 1192 | −188 (−216– −159) | |
| qPCR-TL | ||||
| Q1# | 54 | 334 | −266 (−320– −212) | 0.0087 |
| Q2–4 | 214 | 1312 | −187 (−213– −160) | |
| Rare telomere-related QV | ||||
| Present | 36 | 208 | −267 (−331– −202) | 0.039 |
| Absent | 232 | 1438 | −194 (−220– −168) | |
| Common variant PRS for short telomere length | ||||
| Q1¶ | 70 | 422 | −251 (−299– −204) | 0.026 |
| Q2–4 | 198 | 1224 | −190 (−218– −162) | |
| MUC5B rs35705950 | ||||
| Any minor (GT/TT) | 160 | 1015 | −201 (−232– −170) | 0.74 |
| Major (GG) | 108 | 631 | −209 (−248– −170) |
A joint model was used to estimate FVC progression adjusted for age, sex, study centre and first two principal components for ancestry. The analysis was restricted to subjects (n=268) with ⩾3 FVC measurements spanning at least 3 months of follow-up, with all FVC measures obtained within 5 years of enrolment. Bold text indicates statistical significance. FVC: forced vital capacity; WGS-TL: whole genome sequencing-estimated telomere length; qPCR-TL: quantitative PCR-measured telomere length; QV: qualifying variant; PRS: polygenic risk score.
the first quartile (Q1) for the WGS-TL and qPCR-TL represents those with the shortest telomere lengths
the first quartile (Q1) for the PRS represents those genetically predicted to have the shortest telomere lengths.
Discussion
WGS yields a wealth of data regarding rare and common genetic variants, as well as genomic structural elements such as telomere repeats. As the cost of WGS decreases, this modality will be more accessible for clinical applications. Because rare variants, common variants and telomere length can all inform disease risk and clinical progression, pulmonary fibrosis patients may benefit from this technology. Here, we validate telomere length estimated by WGS (WGS-TL) against two independent measures of telomere length. We confirm that WGS-TL is independently associated with rare deleterious QVs in telomere-related genes and with common variants previously linked to telomere length by genome-wide association studies (GWAS). In addition, we have found that WGS-TL, rare QVs and the common variant PRS for short telomere length are all associated with disease progression.
We estimated telomere length from WGS data using Telseq, which estimates the quantity of telomere hexamers (TTAGGG) after adjusting for guanine/cytosine (GC) content. Prior studies have used Telseq with its default threshold of seven hexamer repeats per 100 base pairs [34]. We leveraged three pulmonary fibrosis cohorts with qPCR-TL and found improved correlation when using a Telseq threshold of six hexamer repeats per 150 base pairs. Correlation between WGS-TL and qPCR-TL remained robust (R⩾0.6) when higher thresholds (10 repeats per 150 base pairs) were used, comparable to default Telseq parameters. Biases in estimating WGS-TL may exist depending on depth of coverage, sample quality and the sequencing platform used [35]. Because telomere repeats are found in unmapped, repetitive regions of the genome where quality control measures are less rigorous, there may be variability in WGS-TL estimates for different samples, especially when sequenced and analysed on different platforms. In this study, all WGS sequencing and analysis was performed on the same platform to minimise variability. The correlation between the WGS-TL and Southern blot measurement (R=0.71) was less robust than the correlation we previously reported between qPCR-TL and Southern blot measured length (R=0.83) [22]. Despite these limitations, we have found that WGS-TL offers corroborating evidence of variant pathogenicity and has utility in predicting disease progression. In contrast, WGS-TL was not predictive of transplant-free survival, consistent with a recent study that found that WGS-TL did not predict survival after lung transplantation [36]. Thus, techniques that directly measure telomere length, such as qPCR, may offer more discriminatory power in prognosticating survival in patients with pulmonary fibrosis [13, 37-40].
The genetic architecture of pulmonary fibrosis is composed of both rare variants and common variants, which have marked differences in contributing to disease risk. The stratification of patients by family history accentuates several distinctions. Patients with a family history of fibrotic ILD are five times more likely to have inherited a rare likely pathogenic variant (25% versus 5%). Similarly, most patients with rare QVs in the telomere-related or non-telomere-related genes report having a family member with fibrotic ILD (86% and 83%, respectively). While the effect of rare QVs in the telomere-related genes have significant effects on telomere length for patients regardless of family history, the effect of the common variant PRS is notable only for those without a family history. These common SNPs may serve as the genetic basis for the unexplained telomere shortening in sporadic cases without identifiable rare telomerase mutations [21]. Thus, this study is consistent with a recent mendelian randomisation study [15] that used a polygenic score to implicate telomere shortening in the development of IPF. While the MUC5B promoter SNP is associated with a genetic predisposition to developing IPF [32] and early radiographically apparent interstitial lung abnormalities [41], we found that this SNP was not associated with disease progression but was associated with improved survival, consistent with a prior report [42].
This is the first study to demonstrate that the common variant PRS of short telomere length offers prognostic value, because it is predictive of telomere length and disease progression. This is also the first study to directly compare the effect of individual rare telomere-related QVs to common variants with regard to telomere length. We found that the effect size of one rare telomere-related QV on telomere length was approximately 10 times the effect of one standard deviation change in the common variant PRS. In fact, the effect of a single rare telomere-related QV exceeded the theoretical maximum PRS, which was not obtained by any individual in our cohorts. This finding is consistent with the paradigm of an inverse relationship between variant allele frequency and phenotype effect size. All rare QVs, by definition, have a population-based allele frequency of <0.0005, whereas the common variants have an allele frequency that ranges between 0.01 and 0.9.
Determining the pathogenicity of rare variants is particularly problematic for a late-onset lethal disorder, because it is often challenging to test for co-segregation or de novo variants. The lack of mutational hot spots and the large number of “private” mutations further complicates determination of pathogenicity. In addition, assays of protein function are laborious and not widely available. Short telomere length of circulating blood cells has been used as a surrogate functional assay but requires a fresh blood draw and send-out to a reference laboratory specialising in Flow-FISH. Additionally, there is uncertainty regarding the optimal cut-off informing short telomere length by Flow-FISH and there can be divergent results for granulocytes and lymphocyte subsets. Thus, a single test that can simultaneously detect telomere gene rare QVs and offer supporting evidence of pathogenicity through the estimation of telomere length is particularly attractive. The strict bioinformatic-driven approach used in this study screens for rare QVs using consensus in silico tools to assess pathogenicity and differs from previous studies [11, 43] that focused on ACMG classification [17] or used looser bioinformatic filters. We found that rare QVs in telomere-related genes were predictive of short telomere length and relevant clinical outcomes, validating our approach.
Our study has several limitations. The retrospective nature of our study did not allow for standardisation of follow-up, nor protocolisation of treatment. Furthermore, we restricted our clinical analyses to cohorts with longitudinal follow-up to focus on clinically relevant outcomes, thus limiting our sample size. As such we were not able to adjust for all clinically relevant variables, such as medications. Owing to data limitations, we were unable to assess WGS-TL for a large control cohort to derive age-adjusted values. Nevertheless, we confirmed that older age was a significant predictor of shorter WGS-TL and accounted for this factor by including age as a covariate. The measurement of repeat elements like telomere length from WGS data may be influenced by DNA isolation techniques, sequencing platform and analysis pipeline, potentially affecting reproducibility across cohorts and platforms. In this study, we included the IPFnet cohort, for which DNA was isolated using a different protocol, and found it to have comparable WGS-TL to other cohorts. This study did not attempt to examine the clinical impact of returning genetic findings to patients and their medical providers. We recognise that genetic testing is complicated by several factors, including inheritance pattern (autosomal dominant, autosomal recessive or X-linked), incomplete penetrance and variable expressivity, unknown genotype–phenotype correlations, impact of environmental exposures, potential phenocopies and genetic anticipation due to telomere shortening. Lastly, our study consisted of a predominantly non-Hispanic White population and future studies with recruitment of diverse ethnicities will be needed to generalise findings for all individuals.
In summary, we have found that WGS offers a robust estimate of telomere content and provides information relevant to patient care. Simultaneous detection of rare telomere-related variants and estimation of telomere length can offer preliminary evidence for pathogenicity. Ultimately, clinical classification of a variant as pathogenic or likely pathogenic requires multiple lines of evidence in accordance with ACMG guidelines, and implementation of genetic testing should only be done by those with expertise in clinical genetics and pulmonary fibrosis. Pre-test genetic counselling is needed because incidental findings are frequently found in exome or genome sequencing. Identification of rare pathogenic variants has implications not only for the person in whom it is found, but also for family members. Once confirmed, all first-degree family members should receive genetic counselling and cascade genetic testing should be offered. Rare deleterious variants in telomere-related genes predict worse survival and faster FVC decline, thus providing a rationale for earlier institution and sustained treatment with antifibrotic medications capable of slowing the progression of disease. Furthermore, WGS technology allows for simultaneous SNP genotyping and compilation of a PRS, which can predict disease progression. Thus, WGS offers multidimensional characterisation of disease risk and prognostic outcomes for pulmonary fibrosis patients.
Supplementary Material
Support statement:
National Institutes of Health grant support include R01HL093096 (C.K. Garcia), K23HL148498 and UL1TR001105 (C.A. Newton), Stony Wold-Herbert Fund and T32HL105323 (D. Zhang). Funding information for this article has been deposited with the Crossref Funder Registry.
Footnotes
Conflict of interest: D. Zhang has received consulting fees from Boehringer Ingelheim and is supported by grants from the Stony Wold-Herbert Fund and the NHLBI (HL105323). C.A. Newton has received consulting fees from Boehringer Ingelheim and is supported by a career development award from the NHLBI. I. Noth has received consulting fees from Boehringer Ingelheim, Genentech and Sanofi-Aventis. F.J. Martinez reports grants from the NIH/NHLBI, Afferent/Merck, Bayer, Biogen, Nitto, Patara/Respivant, Promedior/Roche and Veracyte; consulting fees from Abvie, Boehringer Ingelheim, BMS, Bridge Biotherapeutics, CSL Behring, DevPro, Genentech, IQVIA, Lung Therapeutics, Sanofi, Shionogi, twoXAR and Veracyte; travel support from Boehringer Ingelheim, CSL Behring and Patara/Respivant; participation on advisory boards for Biogen and Boehringer Ingelheim; and honoraria from United Therapeutics. D. Goldstein is an employee of Actio Biosciences; La Jolla, CA, USA. C.K. Garcia reports support from NIH, the Department of Defense and Boehringer Ingelheim outside the scope of this work, lecture honoraria from Three Lakes Foundation and Stanford University, and other support from AstraZeneca. B. Wang, G. Povysil and G. Raghu have nothing to disclose.
References
- 1.Mathai SK, Newton CA, Schwartz DA, et al. Pulmonary fibrosis in the era of stratified medicine. Thorax 2016; 71: 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrovski S, Todd JL, Durheim MT, et al. An exome sequencing study to assess the role of rare genetic variation in pulmonary fibrosis. Am J Respir Crit Care Med 2017; 196: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kropski JA, Mitchell DB, Markin C, et al. A novel dyskerin (DKC1) mutation is associated with familial interstitial pneumonia. Chest 2014; 146: e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley SE, Gable DL, Wagner CL, et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci Transl Med 2016; 8: 351ra107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gable DL, Gaysinskaya V, Atik CC, et al. ZCCHC8, the nuclear exosome targeting component, is mutated in familial pulmonary fibrosis and is required for telomerase RNA maturation. Genes Dev 2019; 33: 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benyelles M, O’Donohue MF, Kermasson L, et al. NHP2 deficiency impairs rRNA biogenesis and causes pulmonary fibrosis and Hoyeraal-Hreidarsson syndrome. Hum Mol Genet 2020; 29: 907–922. [DOI] [PubMed] [Google Scholar]
- 7.Kannengiesser C, Manali ED, Revy P, et al. First heterozygous NOP10 mutation in familial pulmonary fibrosis. Eur Respir J 2020; 55: 1902465. [DOI] [PubMed] [Google Scholar]
- 8.Stuart BD, Choi J, Zaidi S, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet 2015; 47: 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsakiri KD, Cronkhite JT, Kuan PJ, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA 2007; 104: 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart BD, Lee JS, Kozlitina J, et al. Effect of telomere length on survival in patients with idiopathic pulmonary fibrosis: an observational cohort study with independent validation. Lancet Respir Med 2014; 2: 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dressen A, Abbas AR, Cabanski C, et al. Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study. Lancet Respir Med 2018; 6: 603–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton CA, Oldham JM, Ley B, et al. Telomere length and genetic variant associations with interstitial lung disease progression and survival. Eur Respir J 2019; 53: 1801641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton CA, Zhang D, Oldham JM, et al. Telomere length and use of immunosuppressive medications in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2019; 200: 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton CA, Kozlitina J, Lines JR, et al. Telomere length in patients with pulmonary fibrosis associated with chronic lung allograft dysfunction and post-lung transplantation survival. J Heart Lung Transplant 2017; 36: 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duckworth A, Gibbons MA, Allen RJ, et al. Telomere length and risk of idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease: a Mendelian randomisation study. Lancet Respir Med 2021; 9: 285–294. [DOI] [PubMed] [Google Scholar]
- 16.Newton CA, Batra K, Torrealba J, et al. Telomere-related lung fibrosis is diagnostically heterogeneous but uniformly progressive. Eur Respir J 2016; 48: 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Idiopathic Pulmonary Fibrosis Clinical Research Network, Raghu G, Anstrom KJ, et al. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med 2012; 366: 1968–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noth I, Anstrom KJ, Calvert SB, et al. A placebo-controlled randomized trial of warfarin in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2012; 186: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghu G, Remy-Jardin M, Myers JL, et al. Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care Med 2018; 198: e44–e68. [DOI] [PubMed] [Google Scholar]
- 21.Cronkhite JT, Xing C, Raghu G, et al. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med 2008; 178: 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz de Leon A, Cronkhite JT, Katzenstein AL, et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS One 2010; 5: e10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D, Povysil G, Kobeissy PH, et al. Rare and common variants in KIF15 contribute to genetic risk of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2022; 206: 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Z, Mangino M, Aviv A, et al. Estimating telomere length from whole genome sequence data. Nucleic Acids Res 2014; 42: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron-Christie S, Wolock CJ, Groopman E, et al. Exome-based rare-variant analyses in CKD. J Am Soc Nephrol 2019; 30: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Stoma S, Lotta LA, et al. Genome-wide association analysis in humans links nucleotide metabolism to leukocyte telomere length. Am J Hum Genet 2020; 106: 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogan JW, Laird NM. Model-based approaches to analysing incomplete longitudinal and failure time data. Stat Med 1997; 16: 259–272. [DOI] [PubMed] [Google Scholar]
- 28.Crowther MJ, Abrams KR, Lambert PC. Flexible parametric joint modelling of longitudinal and survival data. Stat Med 2012; 31: 4456–4471. [DOI] [PubMed] [Google Scholar]
- 29.Nogee LM, Dunbar AE 3rd, Wert SE, et al. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med 2001; 344: 573–579. [DOI] [PubMed] [Google Scholar]
- 30.Nathan N, Giraud V, Picard C, et al. Germline SFTPA1 mutation in familial idiopathic interstitial pneumonia and lung cancer. Hum Mol Genet 2016; 25: 1457–1467. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Kuan PJ, Xing C, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet 2009; 84: 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seibold MA, Wise AL, Speer MC, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med 2011; 364: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landrum MJ, Chitipiralla S, Brown GR, et al. ClinVar: improvements to accessing data. Nucleic Acids Res 2020; 48: D835–DD44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhindsa RS, Mattsson J, Nag A, et al. Identification of a missense variant in SPDL1 associated with idiopathic pulmonary fibrosis. Commun Biol 2021; 4: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taub MA, Conomos MP, Keener R, et al. Novel genetic determinants of telomere length from a trans-ethnic analysis of 109,122 whole genome sequences in TOPMed. bioRxiv 2020; preprint [ 10.1101/74910]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alder JK, Sutton RM, Iasella CJ, et al. Lung transplantation for idiopathic pulmonary fibrosis enriches for individuals with telomere-mediated disease. J Heart Lung Transplant 2022; 41: 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ley B, Newton CA, Arnould I, et al. The MUC5B promoter polymorphism and telomere length in patients with chronic hypersensitivity pneumonitis: an observational cohort-control study. Lancet Respir Med 2017; 5: 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Planas-Cerezales L, Arias-Salgado EG, Buendia-Roldan I, et al. Predictive factors and prognostic effect of telomere shortening in pulmonary fibrosis. Respirology 2019; 24: 146–153. [DOI] [PubMed] [Google Scholar]
- 39.Snetselaar R, van Batenburg AA, van Oosterhout MFM, et al. Short telomere length in IPF lung associates with fibrotic lesions and predicts survival. PLoS One 2017; 12: e0189467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai J, Cai H, Li H, et al. Association between telomere length and survival in patients with idiopathic pulmonary fibrosis. Respirology 2015; 20: 947–952. [DOI] [PubMed] [Google Scholar]
- 41.Hunninghake GM, Hatabu H, Okajima Y, et al. MUC5B promoter polymorphism and interstitial lung abnormalities. N Engl J Med 2013; 368: 2192–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peljto AL, Zhang Y, Fingerlin TE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA 2013; 309: 2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borie R, Tabeze L, Thabut G, et al. Prevalence and characteristics of TERT and TERC mutations in suspected genetic pulmonary fibrosis. Eur Respir J 2016; 48: 1721–1731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.