Abstract
Spike (S) protein, a homotrimeric glycoprotein, is the most important antigen target for SARS-CoV-2 vaccines. A complete simulation of the advanced structure of this homotrimer during subunit vaccine development is the most likely method to improve its immunoprotective effects. In this study, preparation strategies for the S protein receptor-binding domain, S1 region, and ectodomain trimer nanoparticles were designed using ferritin nanoparticle self-assembly technology. The Bombyx mori baculovirus expression system was used to prepare three nanoparticle vaccines with high expression levels recorded in silkworms. The results in mice showed that the nanoparticle vaccine prepared using this strategy could induce immune responses when administered via both the subcutaneous administration and oral routes. Given the stability of these ferritin-based nanoparticle vaccines, an easy-to-use and low-cost oral immunization strategy can be employed in vaccine blind areas attributed to shortages of ultralow-temperature equipment and medical resources in underdeveloped areas. Oral vaccines are also promising candidates for limiting the spread of SARS-CoV-2 in domestic and farmed animals, especially in stray and wild animals.
Keywords: SARS-CoV-2 vaccine, Ferritin nanoparticle, Spike, Broad spectrum
Graphical Abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) has swept the globe for a couple of years. According to a World Health Organization (WHO) report, more than 400 million confirmed cases and 5 million deaths were reported as of February 28, 2022 (https://covid19.who.int/). The COVID-19 pathogen has been identified and named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. It is another highly pathogenic human coronavirus that was discovered after SARS-CoV (2003) and MERS-CoV (2012).
To cope with the COVID-19 pandemic, the deployment of effective physical protective measures and large investments in medical and health resources is indispensable. The development of an efficacious vaccine is of paramount importance. The World Health Organization maintains a document that includes most SARS-CoV-2 vaccines under development (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines). According to this file, > 300 candidate vaccines are currently being developed.
Preclinical studies of vaccines targeting SARS-CoV and MERS-CoV have identified, the spike (S) protein, which is responsible for receptor binding and membrane fusion, as a major antigenic target for coronavirus vaccines [2], [3], [4]. Studies of SARS-CoV-2 vaccines have confirmed these findings. Hence, most current vaccine candidates against SARS-CoV-2 target the full-length or partial S protein [5]. Similar to HIV gp160 and influenza hemagglutinin, S glycoprotein is a class I viral fusion protein that exists as a homotrimer with multiple glycosylation sites. Antigens with higher order structure similar to the native conformation are more conducive to the induction of effective immune protection [6], [7], [8]. Studies on S-based subunit vaccines have demonstrated that polymeric protein vaccines (dimers or trimers) can induce the production of more neutralizing antibodies than monomer vaccines [9], [10]. Early vaccine development strategies involving subunit vaccines, particularly trimeric subunit vaccines, have not been extensively examined. However, with the investment in more scientific tools and scientific research groups, an increasing number of studies have reported research strategies for trimeric subunit vaccines [11], [12], [13]. Kanekiyo et al. constructed a self-assembling synthetic nanoparticle vaccine with an influenza hemagglutinin trimer that improved the potency and breadth of influenza virus immunity at a lower immune dose [14]. This strategy provides an important reference for the development of trimeric S protein vaccines. The baculovirus expression system is an important subunit vaccine production platform. Various S protein-based vaccine development strategies established using this system have demonstrated certain application prospects [15], [16]. Additionally, mucosal vaccination is considered a safe and effective method for inducing durable systemic and mucosal immunity, thereby providing protection against SARS-CoV-2 [17]. Therefore, oral immunization strategies that can induce mucosal immunity have been used in previous vaccine evaluation studies on antigens with special structures or handling, such as nanoparticles, and mediate effective protection [18], [19], [20].
In the present study, using the self-assembly characteristics of Helicobacter pylori ferritin, we developed three SARS-CoV-2 nanoparticle vaccines containing receptor-binding domain (RBD), S1, and ectodomain (ECD) trimers. The BmNPV baculovirus expression system reBmBac was used for production [21]. Nanoparticle vaccines prepared using silkworms can induce protective immunity in mice via both subcutaneous and oral administration. This result indicates that depending on the stability of ferritin nanoparticles, nanovaccines could be used to develop effective vaccination programs for implementation in underdeveloped areas. Furthermore, with reference to epidemic prevention strategies for rabies and influenza, animal vaccines against SARS-CoV-2 can also be developed using this method. Disruption of the transmission chains in animals, especially pets and farm animals, is imperative [22]. The effectiveness was confirmed by detecting the specific antibody titers and the production of neutralizing antibodies induced by these three nanovaccines. Of these, the ECD-trimer nanovaccine, which demonstrated the most significant levels of neutralizing antibodies, was speculated to be suitable for further development as a candidate vaccine.
2. Materials and methods
2.1. Reagents
Rabbit anti-SARS-CoV-2 spike ECD polyclonal IgG, rabbit anti-SARS-CoV-2 spike S1 polyclonal IgG, and rabbit anti-SARS-CoV-2 spike RBD polyclonal IgG were purchased from Sino Biological (Beijing, CN). HRP-conjugated goat anti-rabbit IgG was purchased from ZSGB-BIO (Beijing, CN). CF568-labeled goat anti-rabbit IgG (H+L) was purchased from Biotium Inc. (Hayward, CA, USA). Luciferase assay system was obtained from Promega (Madison, WI, USA). Goat anti-rabbit IgG (whole-molecule) gold antibody was purchased from Sigma Aldrich (St. Louis, MO, USA).
2.2. Plasmids, bacterial strains and cell lines
A defective rescue BmNPV-BES reBmBac vector was constructed and maintained in our laboratory [21]. The pET-28a (+) vector was used in this study. The recombinant BmNPV transfer vector, pBmPH (GenBank submission ID: 2426173), was constructed and maintained in our laboratory. BmN cells were preserved in our laboratory and cultured in TC100 insect cell culture medium (Applichem, Darmstadt, Germany) with 10% fetal bovine serum (FBS, Gibco, USA) at 27 °C. HEK-293T cells were obtained from ATCC and cultured in DMEM (Thermo Fisher, Waltham, MA, USA) with 10% FBS at 37 °C. Escherichia coli strains Top10 and BL21 (DE3) were cultured in Luria-Bertani (LB) medium.
2.3. Recombinant BmNPV construction
Three peptide sequences of nanoparticle vaccines were designed according to the full-length sequence of the S protein (RefSeq: YP_009724390.1, original Wuhan-Hu-1 strain). The sequences of the ECD (14–1213 aa, R685N), S1 region (14–685 aa, R685N) and RBD (319–541 aa) were linked to Helicobacter pylori ferritin (RefSeq: WP_000949190.1, 5–167 aa, N19Q) using the SGG linker. The human T-cell surface glycoprotein CD5 signal peptide (RefSeq: NP_055022.2, 1–24 aa) was used as the signal peptide for the three sequences. After codon preference optimization, the three designed sequences were synthesized using GenScript (Nanjing, CN) with BamHI and EcoRI restriction sites located upstream and downstream, respectively. The ECD-ferritin sequence was inserted into the pBmPH vector via BamHI/EcoRI-mediated digestion to construct the transfer vector pBmPH-ECD-Fe. pBmPH-S1-Fe and pBmPH-RBD-Fe vectors were constructed in the same manner. The transfer vectors were then cotransfected into BmN cells with reBmBac. Recombinant BmNPV baculoviruses, reBm-ECD-Fe, reBm-S1-Fe and reBm-RBD-Fe, were obtained from the cell supernatant following cotransfection.
2.4. Expression and purification of nanoparticles
Fifth instar silkworm larvae or pupae were injected with recombinant viruses (5 μL of the supernatant, approximately 0.5 ×105 pfu) between the abdominal knobs on the backside and then reared or incubated at 25–27 °C under conditions involving 65% humidity. Larval hemolymph or pupal bodies expressing nanoparticles were harvested after 108–120 h. The crude samples were purified by gentle ultrasonic crushing and ultracentrifugation using a 30% sucrose cushion at 120,000 g for 3 h. The white transparent nanoparticle pellets were resuspended in PBS overnight. Nanoparticle concentrations were detected by ELIZA using standard samples of ECD, S1, and RBD (Sino Biological, Beijing, CN). Whether the nanoparticles were assembled successfully was observed by transmission electron microscopy (TEM). Immunoelectron microscopy (IEM) was used to identify S proteins on the surface of the nanoparticles.
2.5. Mouse study designs
Six BALB/c mice (6–8 week-old) were assigned to each group. Mice in the subcutaneous administration (SA) group were subcutaneously immunized with a 10 μg of crude purified nanoparticle vaccine ECD-Nano or an equal molar dose of S1-Nano or RBD-Nano, formulated with aluminum hydroxide adjuvant. Mice in the oral administration (OA) group were immunized with 50 μg of the crude expression product ECD-Nano or an equal molar dose of S1-Nano or RBD-Nano without adjuvant. The nanoparticle samples were purified by ultracentrifugation and diluted to an appropriate concentration for the immunization of mice. All mice were vaccinated with the aforementioned vaccines in a prime/boost manner on days 0 and 21. Sera were collected twice, on days 14 and 35, and the mice were euthanized at week 6. PBS-treated mice were used as controls.
In the safety test groups, mice were administered 50 μg of nanoparticles per day for five days continuously. Serum samples were collected on days 0 and 21. Body weight, temperature, deaths, changes in behavior, motility, and eating and drinking habits were recorded daily. Mice were euthanized and dissected on day 21. The main mouse organs (the heart, liver, spleen, lungs, and kidneys) were harvested for hematoxylin and eosin staining.
2.6. Identification of serum antibodies against the spike in mice by ELISA
Blood samples were collected from the retro-orbital plexus of mice on days 14 and 35. After coagulation at 37 °C for 2 h, blood samples were centrifuged at 2000 rpm (400 xg) for 10 min. The serum was collected and stored at − 20 °C. Recombinant ECD-His, S1-His, and RBD-His expressed and purified from E.coli BL21 (DE3) were used to coat flat-bottom 96-well plates at a final concentration of 1 μg/mL. After overnight incubation at 4 °C, plates were washed thrice with PBST, a blocking buffer containing 1% BSA in PBST was added, and the plates were incubated for 1 h at room temperature. Mouse serum samples were serially diluted and added to the plates. After 1 h of incubation at 37 °C, the plates were washed thrice, and HRP-conjugated goat anti-mouse IgG antibody was added at a dilution ratio of 1:5000. After incubation for 1 h at 37 °C, the plates were washed thrice with PBST and developed with TMB (Solarbio, Beijing, CN) for 10 min. The reaction was stopped using a stop solution (1 M H2SO4). The absorbance was measured at 450 nm using a microplate reader.
2.7. Measurement of the inhibition of binding of RBD to cell-surface ACE2
hACE2 was coexpressed with EGFP using the pEGFP-N3 vector in HEK-293T cells. Purified spike nanoparticles were incubated with PBS or antisera for 1 h. This mixture was then added to 293T-hACE2 cells and incubated for 12 h. The cells were fixed with paraformaldehyde buffer, followed by incubation with rabbit anti-SARS-CoV-2 spike RBD polyclonal IgG for 2 h at 37 °C. After washing, the cells were incubated with CF568 labeled goat anti-rabbit IgG (H+L) at a dilution ratio of 1:5000 for 2 h at 37 °C. Fluorescent images were acquired using confocal scanning microscope.
2.8. SARS-CoV-2 authentic virus neutralization assay
To assess the neutralization of SARS-CoV-2 infection in vitro, Vero cells (5 ×104) were seeded in 96-well plates and grown overnight. The SARS-CoV-2 origin strain and delta variants (approximately 100 TCID50) were preincubated with an equal volume of serum (serially diluted at a ratio of 1:2) samples obtained from immunized mice. After incubation at 37 °C for 1 h, the mixture was added to Vero cells. After three days, the cytopathogenic effects of the cells were observed under a microscope. A neutralization assay was performed using BSL-3.
2.9. Statistical analysis
All statistical analyses were performed using GraphPad Prism software (GraphPad Software Inc., San Diego, Calif., USA). All data are presented as the means ± SEM. Statistical analyses were performed using an unpaired t-test. Statistical significance was set at P < 0.05. *P < 0.05, * *P < 0.01, * **P < 0.001, * ** *P < 0.0001.
3. Results
3.1. Preparation of self-assembling nanoparticle vaccines
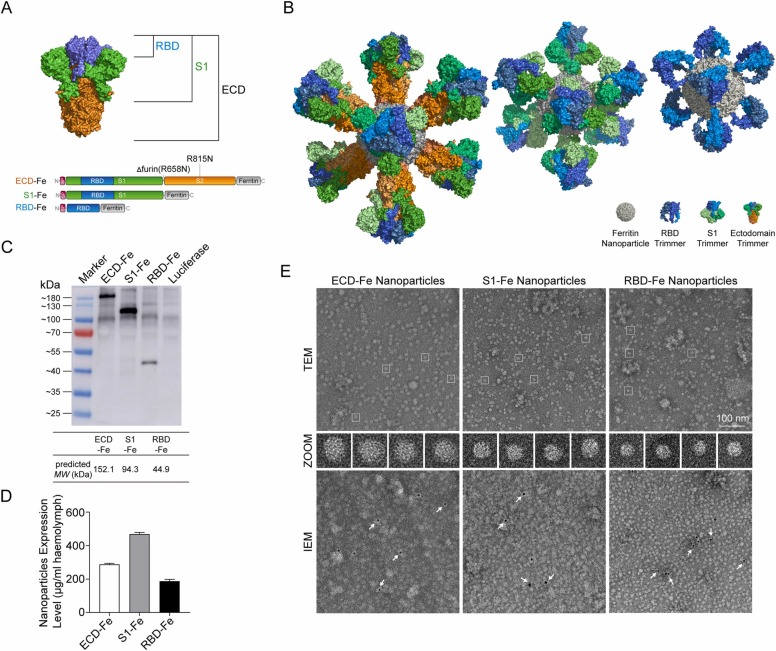
The S glycoprotein nanovaccines expressing recombinant baculoviruses were prepared using the reBmBac baculovirus expression system (BES). The RBD, S1 and ECD sequences of the S protein were fusion expressed with Helicobacter pylori ferritin at the N-terminus ( Fig. 1A). The fusion subunits produced in silkworms self-assembled into 24-mer nanoparticles. S trimers were displayed on the nanoparticle surface (Fig. 1B). Western blot analysis showed that the fusion proteins were successfully expressed (Fig. 1C), and electrophoresis showed that the molecular weight of each fusion protein was slightly higher than the predicted value. Considering the influence of posttranslational modifications, the size of the expression product was determined to be correct. The expression levels of the nanoparticles RBD-Fe, S1-Fe and ECD-Fe were 187, 468 and 287 µg/mL in larval hemolymph, respectively (Fig. 1D). After purification by ultracentrifugation, the nanoparticles were concentrated by ten-folds. Transmission electron microscopy (TEM) and immunoelectron microscopy (IEM) confirmed the self-assembly of three versions of nanoparticles: ECD-Nano, S1-Nano, and RBD-Nano (Fig. 1E).
Fig. 1.
Construction and expression of nanoparticle vaccines. (A) Schematic diagram of the vaccine components RBD-Fe, S1-Fe and ECD-Fe. Fe: Ferritin, RBD: receptor-binding domain, ECD: ectodomain. (B) Schematic diagram of the nanoparticles of the three vaccines. PyMOL software was used for visualization[23]. (C) Western blot analysis of the products prepared in silkworm using an anti-RBD antibody. (D) ELISA of the S proteins was performed to evaluate the expression levels. ECD-Fe with 287 µg/mL of larval hemolymph, S1-Fe with 468 µg/mL of larval hemolymph, and RBD-Fe with 187 µg/mL of larval hemolymph; n = 3; error bar, SEM. (E) TEM image and two-dimensional (2D) reconstruction confirmed the successful assembly of each nanoparticle, and IEM confirmed the S protein on the surface of the nanoparticle. Arrows, gold nanoparticles labeled with secondary antibody.
3.2. Immunogenicity of ECD-Nano, S1-Nano and RBD-Nano in mice
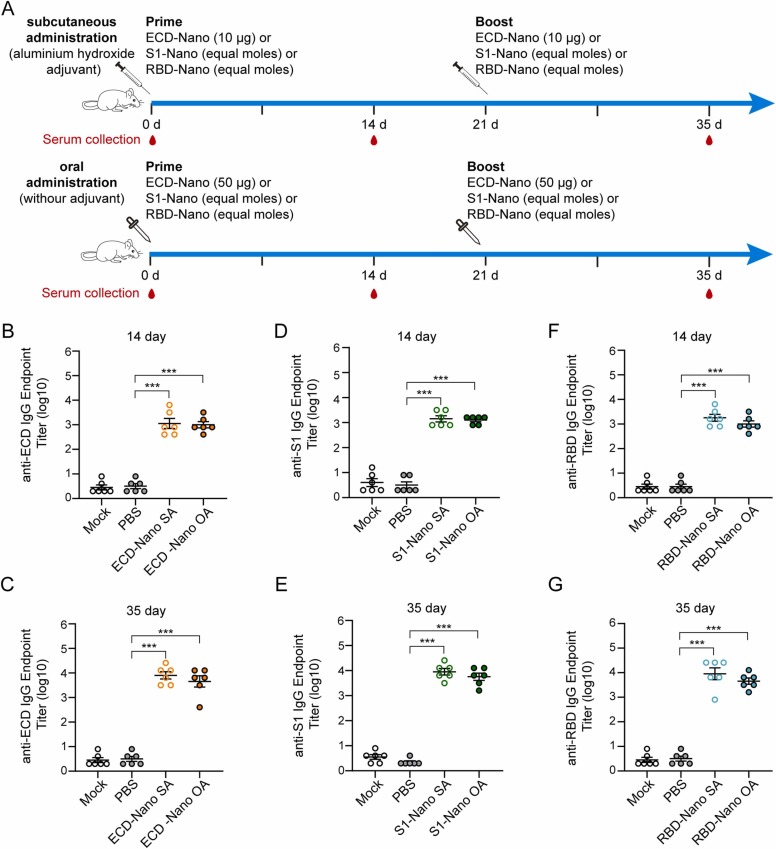
To evaluate the immunogenicity of the three nanoparticles, BALB/c mice were immunized twice at intervals of three weeks. Mice in the subcutaneous administration groups (SA) were immunized with 10 μg of ECD-Nano or equal moles of S1-Nano and RBD-Nano. In proportion, mice in the oral administration groups (OA) were immunized with 50 μg of ECD-Nano or equal moles of S1-Nano and RBD-Nano ( Fig. 2A). Sera were collected 14 d after priming and boosting. The titers of ECD-specific IgG, S1-specific IgG and RBD-specific IgG in the sera were detected by ELIZA using the corresponding recombinant proteins of ECD, S1, or RBD, which were produced and purified in E. coli (Fig. 2B-G). The geometric mean titers (GMTs) of specific IgG, whose production was elicited by ECD nanoparticle vaccines, were 1132 and 1006 when the vaccines were administered via subcutaneous and oral routes, respectively, 14 days after priming. The GMTs reached 8072 and 4528 following 35 days after priming, respectively. The GMTs of S1-specific IgG in the S1 Nano-immunized groups were 1425 and 1270 when the vaccine was administered via subcutaneous and oral routes on day 14 and 9057 and 5701 on day 35, respectively. The GMTs of RBD-specific IgG in the other two groups were 1794 and 1006 when the vaccines were administered via the subcutaneous and oral routes after priming and 9057 and 4528 after boosting, respectively. The antibody titers in the OA group were slightly lower than those in the SA group; however, the best samples reached 104.
Fig. 2.
BALB/c mice immunized with nanoparticle vaccines produced SARS-CoV-2 ECD-S1- and RBD-specific antibodies. (A) The immunization protocols. Mice were immunized with vaccines administered via subcutaneous or oral routes. (B) and (C) Detection of ECD-specific IgG with respect to different routes of serum administration on days 14 and 35. (D) and (E) Detection of S1-specific IgG with respect to different routes of serum administration on days 14 and 35. (F) and (G) Detection of RBD-specific IgG in different routes of administration of sera from Days 14 and 35. The experiment was repeated twice, with six mice per group in each experiment; error bar, SEM; *** p < 0.001.
3.3. Nanoparticle vaccine elicits effective antibody responses in mice
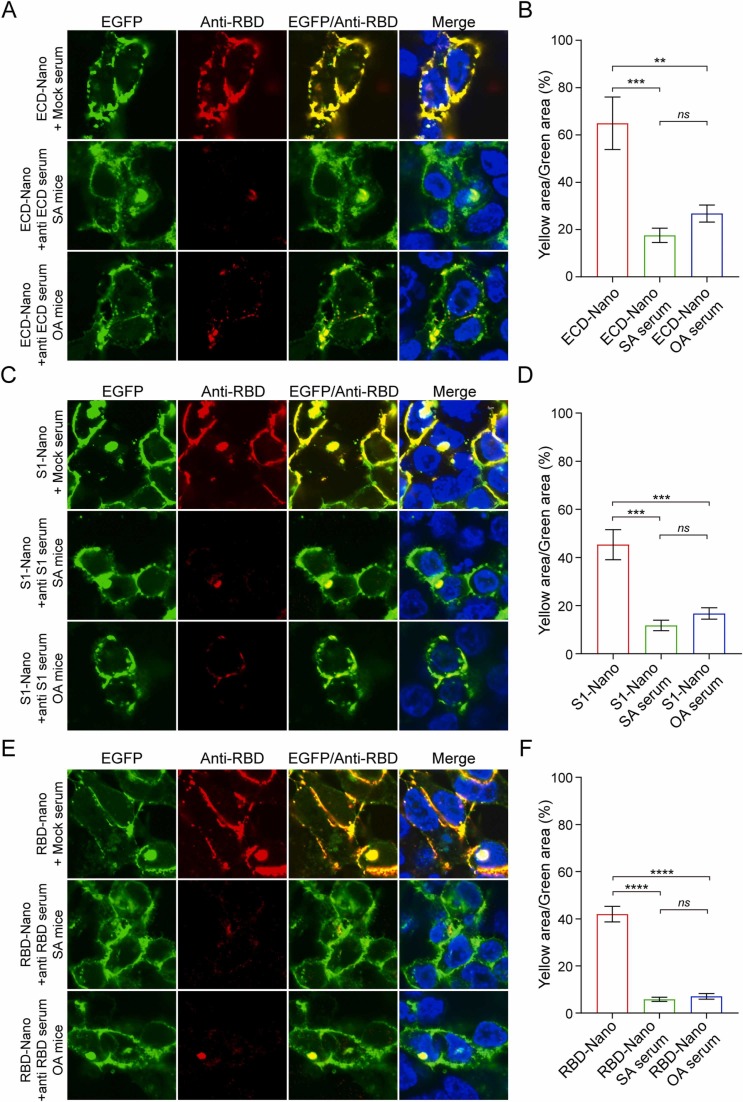
To assess whether the sera contained effective neutralizing antibodies against SARS-CoV-2 infection, we first performed confocal microscopy to detect whether specific antisera could block the interaction between nanoparticles and human angiotensin‐converting enzyme 2 (hACE2). 293T-hACE2 cells were hACE2-EGFP fusion protein-overexpressing HEK293T cells. Recombinant hACE2 was present in the cell membrane, as observed by co-localization analysis with EGFP (Fig. S1). As the confocal analysis results shown in Fig. 3, antibodies in the antisera of mice immunized with different nanovaccines could significantly inhibited the binding of RBD to hACE2 (Fig. 3).
Fig. 3.
Inhibition of the binding of S protein to hACE2. ECD-Nano (A), S1-Nano (C), and RBD-Nano (E) were added to 293T-hACE2 cells in the presence or absence of antisera at a dilution ratio of 1:10. Confocal fluorescence analysis confirmed that all three nanoparticles could specifically bind to hACE2 due to the presence of the RBD structure. After treatment with the corresponding antiserum, the colocalization signal was significantly weakened, which indicated that the antibody production induced in mice immunized with the nanoparticle vaccine had a certain neutralizing effect. The yellow area/green area of each group was analyzed and is shown in B, D and F. n = 3; error bar, SEM; ** p < 0.01, *** p < 0.001, **** p < 0.0001.
We also tested the neutralization activity against authentic SARS-CoV-2 infection in cells (Table S1). Both the original strain and the delta variants were used. Sera were diluted by two fold gradient dilution. For the original strain, only sera collected from ECD-Nano SA mice showed effective activity against VERO cells, with a geometric mean 50% neutralization titer (NT50) of 160. For the delta variants, the neutralizing titers of the ECD-Nano-induced antibody and S1-Nano-induced antibody were 1:48 and 1:32. However, owing to the stringent operating conditions of authentic viruses, this neutralization test was only a preliminary verification. During follow-up, it will be necessary to coordinate the experimental conditions for a systematic comparative evaluation to determine whether each nanoparticle vaccine can mediate induce the production of neutralizing antibodies when administered via the subcutaneous and oral routes.
3.4. Oral administration route of the nanoparticle vaccine was associated with a high level of tolerability and safety in mice
To explore the safety of oral immunization with nanoparticle vaccines, high-dose immunization experiments were performed in mice. Total immunity increased by five-fold compared to that observed in the immunogenicity experiments (Fig. S2A). After 21 days of observation, the mice were euthanized, and the serum titer and tissue sections were evaluated. At a high dose of immunization, the body temperature and behavior of mice were normal, and no pathological characteristic were observed. The GMTs of ECD-specific IgG in ECD-Nano-immunized mice, S1-specific IgG in S1-Nano-immunized mice and RBD-specific IgG in RBD-Nano-immunized mice were 2.8 × 104, 2.8 × 104, and 2.3 × 104, respectively, on day 21 (Fig. S2B). No apparent histopathological changes were observed in the organs in the three groups (Fig. S3).
4. Discussion
In this study, we designed three versions of SARS-CoV-2 nanoparticle vaccines with different spike lengths and evaluated their efficacy via two different administration routes: subcutaneous and oral. All three nanoparticle vaccines induced robust antibody responses against corresponding antigens. All neutralizing antibodies (nAbs) inhibited the binding of nanoparticles to hACE2. The nAbs collected from ECD-Nano SA-treated mice inhibited infection of cells with a nAb titer of 1:160 by the SARS-CoV-2 origin strain. ECD-Nano- and S1-Nano-induced nAbs inhibited infection by the SARS-CoV-2 delta variant in cells at titers of 1:48 and 1:32, respectively. This result preliminarily showed that the SARS-CoV-2 nanoparticle vaccine constructed based on this strategy provided broad protection against different strains. Although the orally immunized mice in this study demonstrated highly specific antibodies, the neutralization effect of the virus observed in in vitro experiments was not ideal. This may be attributed to the fact that oral administration mainly induces causes mucosal immunity, and the related mechanism still needs further in-depth examination.
Through preclinical studies on several coronaviruses, the spike protein has been identified as the best antigenic target for vaccine development [2], [3]. The SARS-CoV-2 spike protein is the best target for vaccine candidates [4], [5]. Furthermore, it is generally believed that, compared with monomers, subunit vaccines with native-structure-like trimers or at least homodimers of the spike protein are able to induce a higher production of neutralizing antibodies [10], [24]. In the development strategy of ferritin-based nanoparticle vaccines, spike subunits are distributed evenly on the nanoparticle surface as native trimer structures. The characteristics of ferritin self-assembling nanoparticles also compensate for the lower antigen density of the spike subunit [24], [25], which could improve the compatibility of the S subunit with antigen capture and presentation strategies of the host immune system [25], [26]. Ferritin-based nanoparticle vaccines can demonstrate high efficacy and broad-spectrum and persistent protection [14], [27]. Furthermore, the SARS-CoV-2 ferritin nanoparticle vaccine induces the production of highly efficient and broad-spectrum neutralizing antibodies in mice, and can provide effective immune protection for non-human primates [28], [29]. With the continuous emergence of mutant strains of SARS-CoV-2, nanoparticle vaccines designed with the S protein sequence of the newly emerged mutant strains show good immune protection in mice and non-human primates [30], [31]. Two methods link spike antigens to ferritin. One involves connecting the spike to the surface of self-assembled ferritin nanoparticles using the SpyTag-SpyCatcher (ST/SC) strategy [32]. The other involves the fusion of the spike antigen and the ferritin monomer for self-assembly [12]. Rather than the previously reported post-processing linking strategy, such as ST/SC conjugation, we adopted a fusion expression strategy in which the S subunit was linked to the N-terminus of ferritin. By exploiting the BES for high-molecular-weight protein expression, this approach simplifies the preparation of nanoparticle vaccines and improves their stability. In addition, the ECD sequence used in this study can still encode a stable S protein without the introduction of two stabilizing proline mutations [33], [34]. This suggest that ferritin-based nanoparticles may improve the stability of the recombinant S protein;however, this requires further study.
Gastrointestinal infection with SARS-CoV-2 has been confirmed [35], [36], indicating that the spike protein is resistant to the environment in the gastrointestinal tract. In addition, ferritin nanoparticles display remarkable thermal and chemical stability [37], [38]. Based on these findings, we designed an orally administered group to investigate whether nanoparticle vaccines could induce efficient antibody production in mice. These results indicate that the oral route is feasible. We believe that this nanoparticle vaccine can be used as a competitive candidate for controlling the COVID-19 pandemic in regions and countries with underdeveloped medical conditions. Many underdeveloped regions, such as Africa and Latin America, have long been vulnerable to infectious diseases [39]. According to statistical data, Africa had more than 2 million confirmed cases by the end of 2020. Ensuring that people in these areas and in other low-income countries and regions receive equal opportunities for vaccine-mediated protection is an important global challenge. Even though approved vaccines are associated with sufficient production capacity, low-temperature transportation and clean vaccination in these areas remain difficult problems to solve quickly and economically. Vaccines prepared using the ferritin nanoparticle strategy described in this study can be used as candidates to address this issue because of their stability and oral effectiveness.
In contrast, oral immunization has more advantages for immunity in animal, especially in stray or wild animals. SARS-CoV-2 can infect mammals such as nonhuman primates, cats, dogs, ferrets, minks and hamsters [40], [41], [42], [43]. The virus is transmissible between cats and ferrets [40], [44]. Widespread infections of SARS-CoV-2 have been reported in farmed mink populations, and SARS-CoV-2 has shown a trend of reverse transmission to the community [22], [45]. Consequently, some countries have decided to cull all farmed minks [46], [47]. The WHO also noted that minks appear to be a good reservoir for the disease, with a mutated strain causing infections in a dozen individuals. The transmission of SARS-CoV-2 among animals is a significant threat to farmers and the environment. Although several vaccines have been approved worldwidee, it is difficult to curtail SARS-CoV-2 infection and transmission in animals in the short term, which hinder global epidemic prevention and control.
The nanoparticle vaccine used in this study has a low production cost and induces the production of high titers of specific antibodies when administered via the oral route. It can be used as a human oral vaccine similar to OPV. Furthermore, it is an effective scheme for the large-scale immunization of pets and farmed animals, especially stray or wild animals, to eliminate intermediate sources of infection and avoid mutations caused by the long-term transmission of the virus in animals. This is of great significance for the prevention and control of the global epidemic and the promotion of immunization programs. Therefore, BESs are suitable for the expression of eukaryotic proteins. Many BES-based SARS-CoV-2 vaccine development studies have confirmed that BES is also suitable for S protein [9]. The silkworm BES used in this study can be used to prepare nanoparticle vaccines in silkworms but not in cells. It has the advantages of a low production cost and high expression. Typically, the expression of the five silkworm pupae is equivalent to that observed in one liter of insect cell culture medium [48]. The recombinant baculovirus strains used in this study also have the potential to further increase the expression level by two-five-fold via a series of optimization measures [21], [49]. The advantages of low cost, high expression of BmNPV BES, and effectiveness of oral nanoparticle vaccines provide sufficient guarantee that high-dose oral immunization can control the spread of SARS-CoV-2 in animals.
The results of the high-dose immunity experiment preliminarily verified the safety of oral immunization using the nanoparticle vaccine developed in this study. Continuous high-dose immunization induced efficient specific immunity in mice, and the antibody titer was higher than that in the injection group. This suggests that subjects had a high level of tolerance to the immune dose and frequency of the oral route in practical applications. This finding demonstrates ample scope to explore the optimization of oral vaccine immunization programs.
In preliminary in vitro virus neutralization experiment, we found that ECD-Nano induced production of antibodies showed a significant neutralization effect. This may be because the ECD contains both S1 and S2 extracellular regions, which can induce the production of a richer variety of neutralizing antibodies and are more effective in preventing the virus from entering cells. The structure of the S protein of SARS-CoV-2 is similar to that of SARS-CoV and MERS-CoV. The RBD of S1 is responsible for receptor recognition and binds to ACE2, which is the preferred antigen in many subunit vaccine development strategies. The HR region of S2 is responsible for membrane fusion, mediating the entry of viruses into cells. The sequence is relatively conserved, rendering it a promising target for the development of fusion, inhibitors against SARS-CoV-2 infection [50]. However, the S protein of SARS-CoV-2 has specific characteristics. The RBD conformation changes dynamically between the exposed and hidden states,rendering it easier to evade immune surveillance against RBD [6], [7]. A furin cleavage site is present between S1/S2, which can mediate preactivation caused by the proprotein-converting enzyme, rendering it easier for the virus to enter the host cell. In other words, SARS-CoV-2 can enter host cells when S2 is directly exposed [6]. Some studies have also shown that antibodies against the RBD or HR region alone can inhibit the entry of SARS-CoV-2 into cells [24]. Therefore, vaccines containing both RBD and HR provide better immune protection. The ECD-Nano and S1-Nano vaccines used in this study showed more antigenic epitopes than RBD-Nano. Their structures were also similar to the natural trimer of the S protein. Therefore, they may be suitable candidates for vaccines.
Ethics approval and consent to participate
All experimental animals were approved by the Animal Ethics Committee of Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (License: SYXK (JING) 2019–0046). All experimental procedures and animal care strictly followed the guidelines of Institute of Animal Sciences and Beijing Administration Office of Laboratory Animals.
Consent for publication
Not applicable.
Conflict of Interest Statement
All authors disclosed no relevant relationships.
Funding
This work was supported by National Natural Sciences Foundation of China (No. 32002236, 32072796 and 31670156), The National Key Research and Development Program of China (No. 2022YFD13007), the National Transgenic Major Project of China (2019ZX08010–004) and The Agricultural Science and Technology Innovation Program .
CRediT authorship contribution statement
X.L. Y.L. and Z.Z. designed the research; X.L. Y.L. and Z.Z. supervised the project; X.L. wrote the main manuscript text; X.L., H.S., X.G., Y.S., and J.L. completed animal experiments. X.L., Y.Y., D.L. and Z.Z. completed protein expression and purification. J.J. and X.L. completed the virus neutralization experiment. All authors reviewed the manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.procbio.2023.03.026.
Appendix A. Supplementary material
Supplementary material
.
Data availability
No data was used for the research described in the article.
References
- 1.Coronaviridae V. Study group of the international committee on taxonomy of the species severe acute respiratory syndrome-related coronavirus: classifying 2019-ncov and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 5.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X., Cao D., Kong L., Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 2020;11(1):3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., Guo X.V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.Y., Kwong P.D., Sodroski J.G., Yin M.T., Sheng Z., Huang Y., Shapiro L., Ho D.D. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 9.Tian J.H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Logue J., Portnoff A.D., Norton J., Guebre-Xabier M., Zhou B., Jacobson K., Maciejewski S., Khatoon R., Wisniewska M., Moffitt W., Kluepfel-Stahl S., Ekechukwu B., Papin J., Boddapati S., Jason Wong C., Piedra P.A., Frieman M.B., Massare M.J., Fries L., Bengtsson K.L., Stertman L., Ellingsworth L., Glenn G., Smith G. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021;12(1):372. doi: 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., Yang M., Li Y., Cheng H., Yuan Y., Zhang W., Ke C., Wong G., Qi J., Qin C., Yan J., Gao G.F. A universal design of betacoronavirus vaccines against covid-19, MERS and SARS. Cell. 2020;182(3):722–733. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce M.G., Chen W.H., Sankhala R.S., Hajduczki A., Thomas P.V., Choe M., Martinez E.J., Chang W.C., Peterson C.E., Morrison E.B., Smith C., Chen R.E., Ahmed A., Wieczorek L., Anderson A., Case J.B., Li Y., Oertel T., Rosado L., Ganesh A., Whalen C., Carmen J.M., Mendez-Rivera L., Karch C.P., Gohain N., Villar Z., McCurdy D., Beck Z., Kim J., Shrivastava S., Jobe O., Dussupt V., Molnar S., Tran U., Kannadka C.B., Soman S., Kuklis C., Zemil M., Khanh H., Wu W., Cole M.A., Duso D.K., Kummer L.W., Lang T.J., Muncil S.E., Currier J.R., Krebs S.J., Polonis V.R., Rajan S., McTamney P.M., Esser M.T., Reiley W.W., Rolland M., de Val N., Diamond M.S., Gromowski G.D., Matyas G.R., Rao M., Michael N.L., Modjarrad K. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021;37(12) doi: 10.1016/j.celrep.2021.110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell A.E., Zhang K., Sanyal M., Tang S., Weidenbacher P.A., Li S., Pham T.D., Pak J.E., Chiu W., Kim P.S. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 2021;7(1):183–199. doi: 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Huang B., Zhu Y., Tan W., Zhu M. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell Mol. Immunol. 2021;18(3):749–751. doi: 10.1038/s41423-021-00643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanekiyo M., Wei C.J., Yassine H.M., McTamney P.M., Boyington J.C., Whittle J.R., Rao S.S., Kong W.P., Wang L., Nabel G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499(7456):102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Oosten L., Altenburg J.J., Fougeroux C., Geertsema C., van den End F., Evers W.A.C., Westphal A.H., Lindhoud S., van den Berg W., Swarts D.C., Deurhof L., Suhrbier A., Le T.T., Torres Morales S., Myeni S.K., Kikkert M., Sander A.F., de Jongh W.A., Dagil R., Nielsen M.A., Salanti A., Sogaard M., Keijzer T.M.P., Weijers D., Eppink M.H.M., Wijffels R.H., van Oers M.M., Martens D.E., Pijlman G.P. Two-Component nanoparticle vaccine displaying glycosylated spike s1 domain induces neutralizing antibody response against SARS-CoV-2 variants. mBio. 2021;12(5) doi: 10.1128/mBio.01813-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goepfert P.A., Fu B., Chabanon A.L., Bonaparte M.I., Davis M.G., Essink B.J., Frank I., Haney O., Janosczyk H., Keefer M.C., Koutsoukos M., Kimmel M.A., Masotti R., Savarino S.J., Schuerman L., Schwartz H., Sher L.D., Smith J., Tavares-Da-Silva F., Gurunathan S., DiazGranados C.A., de Bruyn G. Safety and immunogenicity of SARS-CoV-2 recombinant protein vaccine formulations in healthy adults: interim results of a randomised, placebo-controlled, phase 1-2, dose-ranging study. Lancet Infect. Dis. 2021;21(9):1257–1270. doi: 10.1016/S1473-3099(21)00147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mudgal R., Nehul S., Tomar S. Prospects for mucosal vaccine: shutting the door on SARS-CoV-2. Hum. Vaccin. Immunother. 2020;16(12):2921–2931. doi: 10.1080/21645515.2020.1805992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellier B., Saura A., Lujan L.A., Molina C.R., Lujan H.D., Klatzmann D. A thermostable oral SARS-CoV-2 vaccine induces mucosal and protective immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.837443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammadi G., Sotoudehnia Koranni Z., Jebali A. The oral vaccine based on self-replicating RNA lipid nanoparticles can simultaneously neutralize both SARS-CoV-2 variants alpha and delta. Int. Immunopharmacol. 2021;101 doi: 10.1016/j.intimp.2021.108231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kar S., Devnath P., Emran T.B., Tallei T.E., Mitra S., Dhama K. Oral and intranasal vaccines against SARS-CoV-2: current progress prospects advantages and challenges. Immun. Inflamm. Dis. 2022;10(4) doi: 10.1002/iid3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Wei Y., Li Y., Li H., Yang X., Yi Y., Zhang Z. A highly efficient and simple construction strategy for producing recombinant baculovirus bombyx mori nucleopolyhedrovirus. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0152140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., Bouwmeester-Vincken N., Harders F., Hakze-van der Honing R., Wegdam-Blans M.C.A., Bouwstra R.J., GeurtsvanKessel C., van der Eijk A.A., Velkers F.C., Smit L.A.M., Stegeman A., van der Poel W.H.M., Koopmans M.P.G. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.W.L. DeLano, Pymol: An open-source molecular graphics tool, CCP4 Newsletter on protein crystallography 40(1) 82–92.
- 24.Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., Qiao Y., Zhan Y., Liu J., Zhang J., Zhang X., Peng Z., Li Y., Lin Y., Liang L., Wang G., Chen Y., Chen Q., Pan T., He X., Zhang H. NanopArticle Vaccines Based on the Receptor Binding Domain (Rbd) and Heptad Repeat (Hr) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53(6):1315–1330. doi: 10.1016/j.immuni.2020.11.015. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., Steinmetz N.F. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15(8):646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 27.Georgiev I.S., Joyce M.G., Chen R.E., Leung K., McKee K., Druz A., Van Galen J.G., Kanekiyo M., Tsybovsky Y., Yang E.S., Yang Y., Acharya P., Pancera M., Thomas P.V., Wanninger T., Yassine H.M., Baxa U., Doria-Rose N.A., Cheng C., Graham B.S., Mascola J.R., Kwong P.D. Two-component ferritin nanoparticles for multimerization of diverse trimeric antigens. ACS Infect. Dis. 2018;4(5):788–796. doi: 10.1021/acsinfecdis.7b00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyce M.G., King H.A.D., Elakhal-Naouar I., Ahmed A., Peachman K.K., Macedo Cincotta C., Subra C., Chen R.E., Thomas P.V., Chen W.H., Sankhala R.S., Hajduczki A., Martinez E.J., Peterson C.E., Chang W.C., Choe M., Smith C., Lee P.J., Headley J.A., Taddese M.G., Elyard H.A., Cook A., Anderson A., McGuckin Wuertz K., Dong M., Swafford I., Case J.B., Currier J.R., Lal K.G., Molnar S., Nair M.S., Dussupt V., Daye S.P., Zeng X., Barkei E.K., Staples H.M., Alfson K., Carrion R., Krebs S.J., Paquin-Proulx D., Karasavva N., Polonis V.R., Jagodzinski L.L., Amare M.F., Vasan S., Scott P.T., Huang Y., Ho D.D., de Val N., Diamond M.S., Lewis M.G., Rao M., Matyas G.R., Gromowski G.D., Peel S.A., Michael N.L., Bolton D.L., Modjarrad K. A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci. Transl. Med. 2022;14(632):eabi5735. doi: 10.1126/scitranslmed.abi5735. [DOI] [PubMed] [Google Scholar]
- 29.Li D., Martinez D.R., Schafer A., Chen H., Barr M., Sutherland L.L., Lee E., Parks R., Mielke D., Edwards W., Newman A., Bock K.W., Minai M., Nagata B.M., Gagne M., Douek D.C., DeMarco C.T., Denny T.N., Oguin T.H., 3rd, Brown A., Rountree W., Wang Y., Mansouri K., Edwards R.J., Ferrari G., Sempowski G.D., Eaton A., Tang J., Cain D.W., Santra S., Pardi N., Weissman D., Tomai M.A., Fox C.B., Moore I.N., Andersen H., Lewis M.G., Golding H., Seder R., Khurana S., Baric R.S., Montefiori D.C., Saunders K.O., Haynes B.F. Breadth of SARS-CoV-2 neutralization and protection induced by a nanoparticle vaccine. Nat. Commun. 2022;13(1):6309. doi: 10.1038/s41467-022-33985-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J., Thomas P.V., McMahan K., Jacob-Dolan C., Liu J., He X., Hope D., Martinez E.J., Chen W.H., Sciacca M., Hachmann N.P., Lifton M., Miller J., Powers O.C., Hall K., Wu C., Barrett J., Swafford I., Currier J.R., King J., Corbitt C., Chang W.C., Golub E., Rees P.A., Peterson C.E., Hajduczki A., Hussin E., Lange C., Gong H., Matyas G.R., Rao M., Paquin-Proulx D., Gromowski G.D., Lewis M.G., Andersen H., Davis-Gardner M., Suthar M.S., Michael N.L., Bolton D.L., Joyce M.G., Modjarrad K., Barouch D.H. Protection against SARS-CoV-2 Omicron BA.1 variant challenge in macaques by prime-boost vaccination with Ad26.COV2.S and SpFN. Sci. Adv. 2022;8(47):eade4433. doi: 10.1126/sciadv.ade4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tai W., Chai B., Feng S., Zhuang X., Ma J., Pang M., Pan L., Yang Z., Tian M., Cheng G. Development of a ferritin-based nanoparticle vaccine against the SARS-CoV-2 omicron variant. Signal Transduct. Target. Ther. 2022;7(1):173. doi: 10.1038/s41392-022-01041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang Y.F., Sun C., Zhuang Z., Yuan R.Y., Zheng Q., Li J.P., Zhou P.P., Chen X.C., Liu Z., Zhang X., Yu X.H., Kong X.W., Zhu Q.Y., Zhong Q., Xu M., Zhong N.S., Zeng Y.X., Feng G.K., Ke C., Zhao J.C., Zeng M.S. Rapid development of SARS-CoV-2 Spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano. 2021 doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- 33.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He D., Marles-Wright J. Ferritin family proteins and their use in bionanotechnology. New Biotechnol. 2015;32(6):651–657. doi: 10.1016/j.nbt.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Liu Z., Zhou X., Guo Z., Zhang J., Zhu P., Yao S., Zhu M. Ferritin nanoparticle-based SpyTag/SpyCatcher-enabled click vaccine for tumor immunotherapy. Nanomedicine. 2019;16:69–78. doi: 10.1016/j.nano.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Nkengasong J.N., Ndembi N., Tshangela A., Raji T. COVID-19 vaccines: how to ensure Africa has access. Nature. 2020;586(7828):197–199. doi: 10.1038/d41586-020-02774-8. [DOI] [PubMed] [Google Scholar]
- 40.Halfmann P.J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., Kinoshita N., Hattori S.I., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Kawaoka Y. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020;383(6):592–594. doi: 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosco-Lauth A.M., Hartwig A.E., Porter S.M., Gordy P.W., Nehring M., Byas A.D., VandeWoude S., Ragan I.K., Maison R.M., Bowen R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis transmission and response to reexposure in cats. Proc. Natl. Acad. Sci. 2020;117(42):26382–26388. doi: 10.1073/pnas.2013102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlottau K., Rissmann M., Graaf A., Schon J., Sehl J., Wylezich C., Hoper D., Mettenleiter T.C., Balkema-Buschmann A., Harder T., Grund C., Hoffmann D., Breithaupt A., Beer M. SARS-CoV-2 in fruit bats ferrets pigs and chickens: an experimental transmission study. Lancet Microbe. 2020;1(5):e218–e225. doi: 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets cats dogs and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard M., Kok A., de Meulder D., Bestebroer T.M., Lamers M.M., Okba N.M.A., Fentener van Vlissingen M., Rockx B., Haagmans B.L., Koopmans M.P.G., Fouchier R.A.M., Herfst S. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11(1):3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munnink B.B.O., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M. Jumping back and forth: anthropozoonotic and zoonotic transmission of SARS-CoV-2 on mink farms. bioRxiv. 2020 [Google Scholar]
- 46.Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect. Dis. 2021;21(1):18–19. doi: 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oreshkova N., Molenaar R.J., Vreman S., Harders F., Oude Munnink B.B., Hakze-van der Honing R.W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R.S., Tacken M.G., de Rooij M.M., Weesendorp E., Engelsma M.Y., Bruschke C.J., Smit L.A., Koopmans M., van der Poel W.H., Stegeman A. SARS-CoV-2 infection in farmed minks the Netherlands April and May 2020. Eur. Surveill. 2020;25(23) doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usami A., Ishiyama S., Enomoto C., Okazaki H., Higuchi K., Ikeda M., Yamamoto T., Sugai M., Ishikawa Y., Hosaka Y., Koyama T., Tobita Y., Ebihara S., Mochizuki T., Asano Y., Nagaya H. Comparison of recombinant protein expression in a baculovirus system in insect cells (Sf9) and silkworm. J. Biochem. 2011;149(2):219–227. doi: 10.1093/jb/mvq138. [DOI] [PubMed] [Google Scholar]
- 49.Liu X., Yang X., Mehboob A., Hu X., Yi Y., Li Y., Zhang Z. A construction strategy for a baculovirus-silkworm multigene expression system and its application for coexpression of type I and type II interferons. Microbiologyopen. 2020;9(3) doi: 10.1002/mbo3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharm. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
No data was used for the research described in the article.