Abstract
Background:
Acute myeloid leukemia (AML) with megakaryocytic differentiation (AMkL) is a rare subtype of AML more common in children. Recent literature has identified multiple fusions associated with this type of leukemia.
Methods:
Morphology, cytogenetics, and genomic sequencing were assessed in patients from Children’s Oncology Group trials AAML0531 and AAML1031 with central-pathology review confirmed non-Down syndrome AMkL. The 5-year EFS, OS, and RR were evaluated in these AMkL subcategories.
Results:
A total of 107 cases of AMkL (5.5%) were included. Distinct fusions were identified in the majority: RBM15::MRTFA (20%), CBFA2T3::GLIS2 (16%), NUP98 (10%), KMT2A (7%), TEC::MLLT10 (2%), MECOM (1%), and FUS::ERG (1%); many of the remaining cases were classified as AMkL with (other) myelodysplasia-related changes (MRC). Very few cases had AML-associated somatic mutations. Cases with CBFA2T3::GLIS2 were enriched in trisomy 3 (p=0.015) and the RAM phenotype with associated high CD56 expression (p<.001). Cases with NUP98 fusions were enriched in trisomy 6 (p<0.001), monosomy 13/del(13q) (p<0.001), trisomy 21 (p=0.026), and/or complex karyotypes (p=0.026). While different 5-year EFS and OS were observed in the AMkL in each trial, in general, those with CBFA2T3::GLIS2 or KMT2A rearrangements had worse outcomes compared to other AMkL while those with RBM15::MRTFA or classified as AMkl-MRC fared better. AMkL with NUP98 fusions also had poor outcomes in the AAML1031 trial.
Conclusion:
Given the differences in outcomes, AMkL classification by fusions, cytogenetics, and morphology may be warranted to help in risk stratification and therapeutic options.
Keywords: acute myeloid leukemia, acute megakaryoblastic leukemia, CBFA2T3::GLIS2, NUP98 fusions, pediatric acute myeloid leukemia
INTRODUCTION
Acute myeloid leukemia (AML) with megakaryocytic differentiation (AMkL) represents less than 5% of all AML, and is defined as a leukemia with at least 20% blasts of which ≥50% are of megakaryocyte lineage.1 This leukemia has a bimodal age distribution with peaks in children less than 3 years of age and in older adults.2–4 Many childhood cases of AMkL are associated with Down syndrome (trisomy 21), which is classified as a separate entity in the World Health Organization (WHO) classification of Tumours of Haematopoietic and Lymphoid Tissues as Myeloid leukemia associated with Down syndrome.5 Excluding those with Down syndrome, AMkL accounts for 5.9–12% of AML in children, with median ages of onset of ranging from 1.4–1.8 years.6–14 Bone marrow fibrosis is a common finding in this leukemia subtype.2,15–21
Prior to more advance molecular methods, AMkL not associated with Down syndrome was most often linked to the translocation t(1;22)(p13;q13) (RBM15::MRTFA previously known as MKL1). Additional rare cases of AMkL have KMT2A (MLL) translocations.9,22–24 However, with the advent of reverse transcriptase polymerase chain reaction and next generation sequencing, additional fusion proteins in AMkL have been identified. These cryptic abnormalities include inv(16)(p13.3q24.3) (CBFA2T3::GLIS2), and t(5;11)(q35;p15) (NUP98::KMD5A).7,8,25 These four fusions are now thought to represent the most common rearrangements in AMkL.8
Childhood AMkL, excluding cases associated with Down syndrome, has an inferior overall survival compared to other categories of AML,6,12–14,26 with worse prognoses in AMkL with CBFA2T3::GLIS2, NUP98::KMD5A, and KMT2A rearrangements.8,25–27 The prognostic significance of RBM15::MRTFA is unclear with studies showing better,3,7,12 worse,13,17 or equal8,9,11 outcomes compared to fusion-negative cases. Herein, our objective was to study the morphologic, immunohistochemical, cytogenetic, and molecular features of non-Down Syndrome AMkL in cases from the Children’s Oncology Group (COG) trials AAML0531 and AAML1031 with the goal of correlating the cytogenetics and molecular classifications with outcomes.
METHODS
Patients:
Pediatric and young adults ranging from 1 month to 29.99 years of age with de novo AML and without Down syndrome were eligible for the 2 Phase III randomized COG trials and analyzed in this study. AAML0531 evaluated Gemtuzumab Ozogamicin (GO) with a dose-intensive treatment regimen; results have previously been described.28 The trial included 1022 eligible patients, enrolled between August 2006 and June 2010 from 181 participating institutions.28 AAML1031 compared standard chemotherapy with or without bortezomib and employed sorafenib in patients with high FLT3 internal tandem duplication (ITD) allelic ratios (>0.4).29 This trial risk stratified based upon minimal residual disease (MRD), FLT3 ITD allelic ratio, NPM1 mutations, CEBPα mutations, and other prognostic genetic markers. The trial included 1231 eligible patients, who enrolled between June 2011 and July 2017 from 193 participating institutions. Herein, outcome data were reviewed separately for each trial. Notably, a subset of patients included in this manuscript was previously described in pediatric AMkL/AML cohort studies.7,8,30 All participating institutions had approval by their institutional review boards (IRB) for these trials. According to institutional regulations, all patients or their parents gave written informed consent before entering this study. The studies were conducted in accordance with the Declaration of Helsinki.
Data availability statement:
The data that supports the findings of this study are available in Supporting Table S1 of this article.
Morphologic assessment:
All cases with an institutional or central pathology review (CPR) diagnosis of AMkL were identified from the AAML0531 and AAML1031 databases (n=1935). Cases were excluded from this study if CPR was not performed or the material submitted was not sufficient for diagnosis. A diagnosis of AMkL required ≥20% blasts of which ≥50% were of megakaryocyte lineage with expression of ≥1 megakaryocytic antigen by flow cytometry or immunohistochemistry. For analyses, morphologic AMkL were classified as AMkL, not otherwise specified (NOS), AMkL with genetic abnormalities, and AMkL with myelodysplasia-related changes (MRC) per the 2017 WHO hematopoietic tissue classification.1,31,32 CPR evaluated blood smears, bone marrow aspirates and biopsies, flow cytometry reports, and any immunohistochemical stains to confirm the diagnosis. Slides were re-reviewed for multilineage dysplasia and abnormal megakaryocyte maturation. If available, reticulin and collagen stains were reviewed or performed to grade bone marrow fibrosis (MF-0 to MF-3) per standardized guidelines.33,34
Cytogenetic and molecular assessment:
All cytogenetic results for this study were centrally reviewed and recorded using International System of Human Cytogenetic Nomenclature. The number of cytogenetic abnormalities and/or presence of any recurrent translocations were recorded. Screening for FLT3 ITD, NPM1, CEBPα, and WT1 mutations was performed as previously described.35–38 RNA-sequencing was performed to identify fusion transcripts using total RNA extracted from patient samples using AllPrep DNA/RNA/miRNA Universal Kit (QIAGEN, Valencia, CA, #80224), by the QIAcube system. The ribodepletion 2.0 protocol (British Columbia Genome Sciences Centre, Vancouver, BC) was employed to prepare the mRNA libraries with 75-bp strand-specific paired-end sequencing. STAR-Fusion v1.1.0 fusion detection algorithm was used, running default parameters with the pre-made GRCh37 resource library with Gencode v19 annotations (https://data.broadinstitute.org/Trinity/CTAT_RESOURCE_LIB/).39 TransABySS v1.4.10 fusion detection algorithm was established to record fusions with breakpoint reads ≥ 1, flanking pairs ≥ 2 counts, and spanning reads ≥ 2 counts.40 The dbGaP TARGET: Acute Myeloid Leukemia study (Accession: phs000465.v19.p8) displays the transcriptomic data.30
Outcome assessment and statistical analysis:
AAML0531 and AAML1031 data were current as of September 30, 2018 and June 30, 2021, respectively. EFS and OS were determined employing the Kaplan-Meier method where EFS was defined as time from study entry until failure to achieve complete remission (CR) during induction, relapse or death, and OS was defined as time from study entry to death.41 Relapse risk (RR) was calculated by cumulative incidence methods defined as time from end of induction I for patients in CR to relapse or death; deaths without a relapse were considered competing events.42 Induction I failures were defined as patients who withdrew from therapy due to a) relapse, b) persistent central nervous system disease, and/or c) refractory disease (≥20% bone marrow blasts). Any patient lost to follow-up was censored at their date of last known contact. Log-rank statistic (EFS and OS) and Gray’s statistic (RR) tested the significance of predictor variables. Potential covariates considered were age at diagnosis, morphologic classifications, presence of fibrosis, certain karyotypic abnormalities, and identified fusions. As some of the above subgroups had small numbers, the comparisons were ad hoc analyses. The chi-squared test was employed to test the significance of observed differences in proportions, Fisher’s exact test was used when data were sparse, and Student’s t-test was utilized to compare means and distributions of 2 groups. P-values <0.05 were considered statistically significant.
RESULTS
Patient Characteristics:
A total of 107 AMkL were confirmed by CPR in these trials, accounting for 5.5% of all AML cases which underwent CPR. Table 1 lists the patient demographics of these cases. There were no significant differences in age, gender, race, or ethnicity in AMkL patients between AAML0531 and AAML1031 (Supplemental Table S2). The median age at diagnosis of AMkL was 1.45 years (range 0.08–15.10 years, interquartile range 1.20 years), significantly younger than patients diagnosed with other subtypes of AML in these trials (p<0.001).
Table 1.
Comparison of pathologic findings between subtypes of AMkL
| Subtype | All cases | AMkL, NOS | AMkL, unknown cytogenetics | AMkL with RBM15::MRTFA | AMkL with CBFA2T3::GLIS2 | AMkL with NUP98 fusion | AMkL with KMT2A fusion | AMkL with MECOM fusion | AMkL with FUS::ERG | AMkL with TEC::MLLT10 | AMkL with (other) MRC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cases | 107 | 11 | 5 | 21 | 17 | 11 | 8 | 1 | 1 | 2 | 30 |
| AAML0531:AAML1031 | 48:59 | 4:7 | 3:2 | 7:14 | 7:10 | 3:8 | 3:5 | 0:1 | 1:0 | 0:2 | 20:10 ^ |
| Age Range (years) | 0.08–15.10 | 0.53–6.17 | 0.84–2.79 | 0.08–2.49# | 0.77–2.07 | 0.44–13.23 | 0.87–6.25 | 0.80 | 3.54 | 1.50–12.67 | 0.11–15.10 |
| Mean | 2.12 | 1.91 | 1.57 | 0.91 | 1.40 | 3.01 | 3.00 | 0.80 | 3.54 | 7.08 | 2.64 |
| Median | 1.45 | 1.95 | 1.18 | 0.61 | 1.35 | 2.25 | 3.08 | 0.80 | 3.54 | 7.08 | 1.48 |
| Male:Female | 48:59 | 5:6 | 0:5 | 6:15 | 7:10 | 10:1@ | 4:4 | 0:1 | 0:1 | 0:2 | 16:14 |
| Fibrosis, any | 30/39 (77%) | 3/3 (100%) | 2/2 (100%) | 8/8 (100%) | 3/4 (75%) | N/A | 4/4 (100%) | N/A | 0/1 (0%) | N/A | 10/17 (59%) |
| Fibrosis, MF-2 or MF-3 | 18/39 (46%) | 1/3 (33%) | 2/2 (100%) | 5/8 (63%) | 0/4 (0%) | N/A | 4/4 (100%) | N/A | 0/1 (0%) | N/A | 6/17 (35%) |
| Abnormal megakaryocytes | 23/43 (53%) | 4/4 (100%) | 1/2 (50%) | 2/6 (33%) | 3/6 (50%) | 2/3 (67%) | 1/3 (33%) | N/A | 1/1 (100%) | N/A | 9/18 (50%) |
| Multilineage dysplasia | 2/45 (4%) | 0/4 (0%) | 0/2 (0%) | 0/5 (0%) | 0/6 (0%) | 0/4 (0%) | 0/3 (0%) | N/A | 1/1 (100%) | N/A | 1/20 (5%) |
| Karyotype | |||||||||||
| Normal | 20/100 (20%) | 7/11 (64%)* | N/A | 2/20 (10%) | 6/17 (35%) | 0/11 (0%) | 3/8 (38%) | N/A | 0/1 (0%) | 1/2 (50%) | 1/30 (3%) |
| Trisomy 3 | 7/100 (7%) | 0/11 (0%) | N/A | 0/20 (0%) | 4/17 (24%)* | 1/11 (9%) | 2/8 (25%) | N/A | 0/1 (0%) | 0/2 (0%) | 0/30 (0%) |
| Del(5q) / monosomy 5 | 3/100 (3%) | 0/11 (0%) | N/A | 0/20 (0%) | 0/17 (0%) | 1/11 (9%) | 0/8 (0%) | N/A | 0/1 (0%) | 0/2 (0%) | 2/30 (7%) |
| Trisomy 6 | 16/100 (16%) | 0/11 (0%) | N/A | 3/20 (15%) | 0/17 (0%) | 7/11 (64%)* | 3/8 (38%) | N/A | 0/1 (0%) | 0/2 (0%) | 3/30 (10%) |
| Monosomy 7/del(7q) | 5/100 (5%) | 0/11 (0%) | N/A | 0/20 (0%) | 0/17 (0%) | 0/11 (0%) | 1/8 (13%) | N/A | 0/1 (0%) | 0/2 (0%) | 4/30 (13%)* |
| Trisomy 8 | 11/100 (11%) | 0/11 (0%) | N/A | 0/20 (0%) | 0/17 (0%) | 2/11 (18%) | 2/8 (25%) | N/A | 0/1 (0%) | 0/2 (0%) | 7/30 (23%)* |
| Del(9q) | 7/100 (7%) | 0/11 (0%) | N/A | 0/20 (0%) | 0/17 (0%) | 2/11 (18%) | 0/8 (0%) | N/A | 0/1 (0%) | 0/2 (0%) | 5/30 (17%)* |
| Monosomy 13/del(13q) | 9/100 (9%) | 0/11 (0%) | N/A | 0/20 (0%) | 0/17 (0%) | 8/11 (73%)* | 0/8 (0%) | N/A | 0/1 (0%) | 0/2 (0%) | 1/30 (3%) |
| Any chr13 structural ab. | 17/100 (17%) | 0/11 (0%) | N/A | 3/20 (15%) | 0/17 (0%) | 9/11 (82%)* | 0/8 (0%) | N/A | 0/1 (0%) | 0/2 (0%) | 5/30 (17%) |
| Trisomy 19 | 16/100 (16%) | 1/11 (9%) | N/A | 4/20 (20%) | 1/17 (6%) | 1/11 (9%) | 3/8 (38%) | N/A | 0/1 (0%) | 0/2 (0%) | 6/30 (20%) |
| Trisomy 21 | 25/100 (25%) | 1/11 (9%) | N/A | 4/20 (20%) | 3/17 (18%) | 6/11 (55%)* | 4/8 (50%) | N/A | 0/1 (0%) | 0/2 (0%) | 7/30 (23%) |
| Complex (≥3 abnormalities) | 49/100 (49%) | 0/11 (0%)& | N/A | 6/20 (30%) | 1/17 (6%)& | 9/11 (82%)* | 4/8 (50%) | N/A | 0/1 (0%) | 1/2 (50%) | 28/30 (93%)* |
| Notable Immunophenotype | |||||||||||
| RAM phenotype | 18/101 (18%) | 2/11 (18%) | 0/4 (0%) | 0/19 (0%) | 16/17 (94%)* | 0/11 (0%) | 0/8 (0%) | 0/1 (0%) | 0/1 (0%) | 0/2 (0%) | 0/27 (0%) |
| CD56 | 24/47 (51%) ^ | 3/4 (75%) | 1/3 (33%) | 0/4 (0%) | 17/17 (100%)* | 0/2 (0%) | 1/3 (33%) | N/A | N/A | N/A | 2/14 (14%) |
Ab = aberration; AMkL = acute myeloid leukemia with megakaryocytic differentiation; MRC = myelodysplasia-related changes (includes multilineage dysplasia and MDS-related karyotype)
Significantly different between AAML0531 and AAML1031
Significantly younger in age than other subtypes
Significantly more males than females compared to other subtypes
Significantly more common than other subtypes
Significantly less common than other subtypes
Leukemia morphologic, immunophenotypic, and fusion-based classifications:
By morphology, megakaryoblasts are large cells with high nuclear-to-cytoplasmic ratios, round to slightly irregular nuclear contours, fine chromatin, prominent nucleoli (sometimes multiple) and basophilic cytoplasm sometimes having pseudopod or bleb formation (Supplemental Figure S1). Table 1 details the clinical, pathologic, and cytogenetic features associated with the fusion-based classifications (see also Supplemental Tables S3 and S4). A total of 61 cases (57%) had defined fusions including 21 with RBM15::MRTFA, 17 with CBFA2T3::GLIS2, 11 with NUP98 fusions, 8 with KMT2A fusions, 2 with TEC::MLLT10, 1 with a MECOM fusion, and 1 with FUS::ERG (Figure 1). An additional 30 cases (28%) without the aforementioned fusions had either multilineage dysplasia (n=1) or MDS-related cytogenetic abnormalities (n=29), qualifying them for a diagnosis of AMkL-MRC using 2017 WHO criteria. Lastly, 11 cases were classified as AML-NOS, and 5 could not be classified due to unknown cytogenetics.
Figure 1. Fusions present in acute myeloid leukemia with megakaryocytic differentiation.
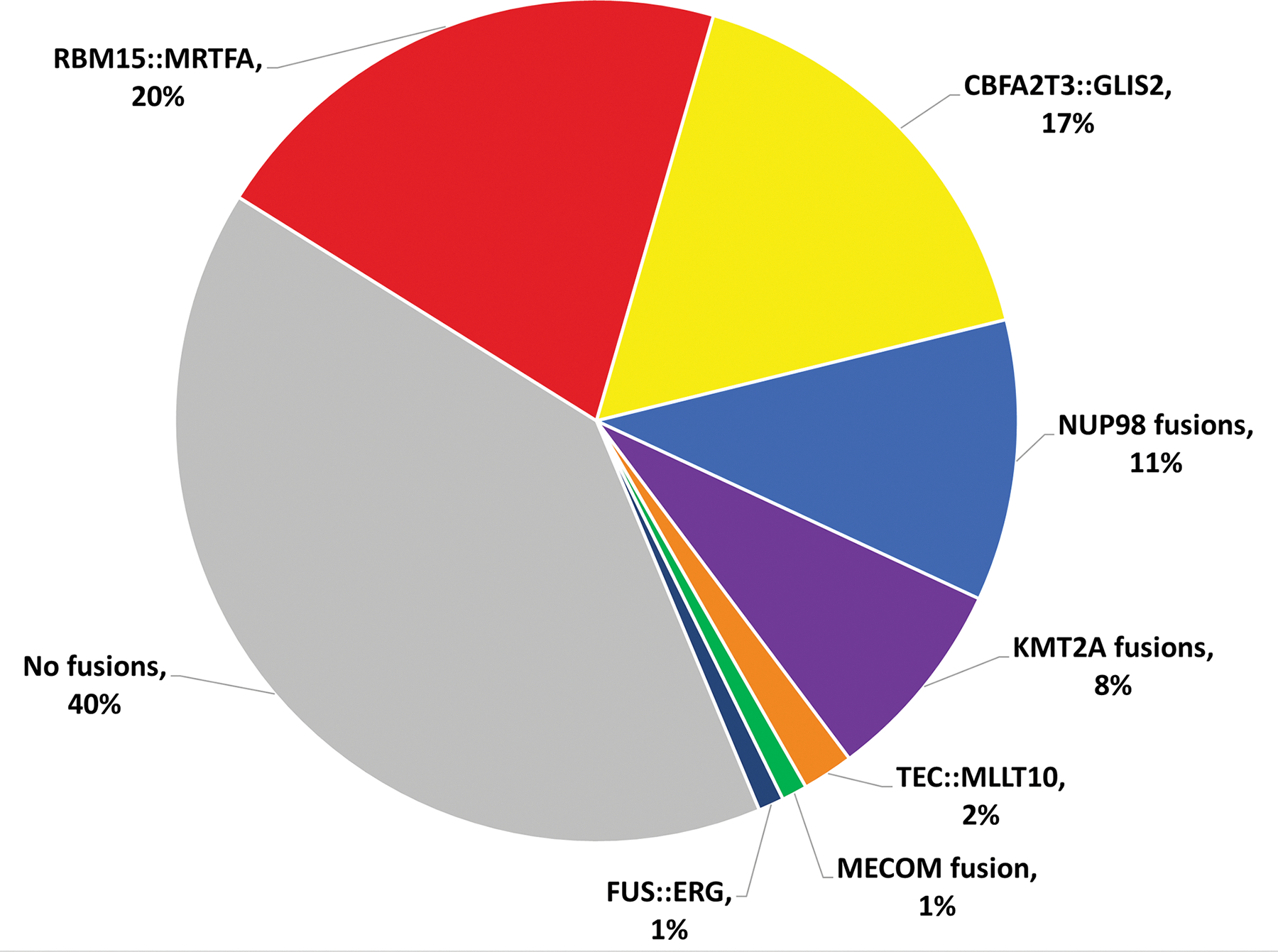
In this study, 61 of 102 patients with available cytogenetic data had identified fusion proteins.
By flow cytometry and/or immunohistochemistry, the blasts expressed ≥1 megakaryocytic antigens, including CD61 (n=90/92), CD41 (n=60/61), and CD42b (n=37/38). Other variably expressed antigens included CD13 (n=37/60), CD33 (n=72/85), CD34 (n=44/73), CD117 (n=40/60), HLA-DR (n=23/52), CD71 (n=21/24), CD4 (n=34/50), and CD7 (n=37/54). CD56 was also expressed in 24/47 (51%) tested cases, most associated with CBFA2T3::GLIS2 (p<0.001). The RAM phenotype, defined by bright CD56, dim/negative CD45 and CD38, and negative HLA-DR,43 was noted in 18/101 cases, and was significantly more common in those with CBFA2T3::GLIS2 (p<0.001).
Multilineage dysplasia was identified in 2/45 (4%) of evaluated cases, including the case with FUS::ERG. However, 23/43 (53%) marrows assessed for megakaryocytic maturation demonstrated abnormal megakaryocytes, including micromegakaryocytes and forms with separate nuclear lobes (Supplemental Figure S2). These cases were not restricted to any specific subgroup. Bone marrow aspirates in AMkL cases were often hemodilute due to marrow fibrosis. Of the 39 bone marrow biopsies with reticulin staining, 77% had at least mild fibrosis (≥MF-1) (Table 1, Supplemental Figure S2).
Cytogenetic and molecular characterization:
Of the 107 AMkL, 100 had karyotype data and 2 had FISH or molecular data that allowed classification into the categories depicted in Table 1. Normal karyotypes were identified in 18 cases (18%), but 10 of these cases (56%) had molecularly-identified cryptic translocations (6 with CBFA2T3::GLIS2, 2 with RMB15::MRTFA, and one each of KMT2A::MLLT3 and TEC::MLLT10). CBFA2T3::GLIS2 were more commonly associated with trisomy 3 compared to other classifications (p=0.015); an additional case with CBFA2T3::GLIS2 demonstrated a translocation involving chromosome 3p. The karyotypes in CBFA2T3::GLIS2 were significantly less complex (p<0.001). NUP98 translocations were present in 11 cases, including 9 with a KDM5A partner, one with a NSD1 partner, and one with a BPTF partner. NUP98 fusions were associated with trisomy 6 (p<.001), monosomy 13/del(13q) (p<0.001), and trisomy 21 (p=0.026); the karyotypes were more complex (p=0.026). Note that NUP98-rearranged AMkL with monosomy 13/del(13q) only had KDM5A fusion partners; the single case of NUP98::KDM5A without monosomy 13/del(13q) had a translocation involving 13q. Of the 8 KMT2A-rearranged AMkL, fusion partners included MLLT10 (n=3), MLLT3 (n=3), MLLT11 (n=1), and unknown (n=1). A total of 30 other cases were classified as AMkL-MRC based upon karyotype or multilineage dysplasia; 28 of these cases had complex karyotypes, and an additional case had del(7q) and trisomy 8, the former of which is a myelodysplasia-related cytogenetic abnormality. Monosomy 7/del(7q), trisomy 8, and del(9q) were more common cytogenetic abnormalities identified in these AMkL-MRC (p=0.027, p=0.016, and p=0.024 respectively).
There was a clear paucity of common AML-associated somatic mutations. Of the 101 tested AMkL, no cases had detectable FLT3 ITD, NPM1, or CEBPα mutations. WT1 mutations were identified in 2 of 97 patients, one with NUP98::NSD1, and another with AMkL-MRC. Figure 2 depicts an oncoprint of mutations by fusion groups. More comprehensive genomic screening by whole genome and targeted exome performed in 87 of the patients demonstrated a lack of prominent somatic single nucleotide variants/indels in these patients (Supplemental Table S1); 60 AMkL (69%) lacked such mutations. Some of the more commonly mutated genes included NRAS (n=8), MYH11 (n=5), and PTPN11 (n=3).
Figure 2. Oncoprint illustrating fusions present in pediatric AMKL, and their co-operating cytogenetic abnormalities and mutations.
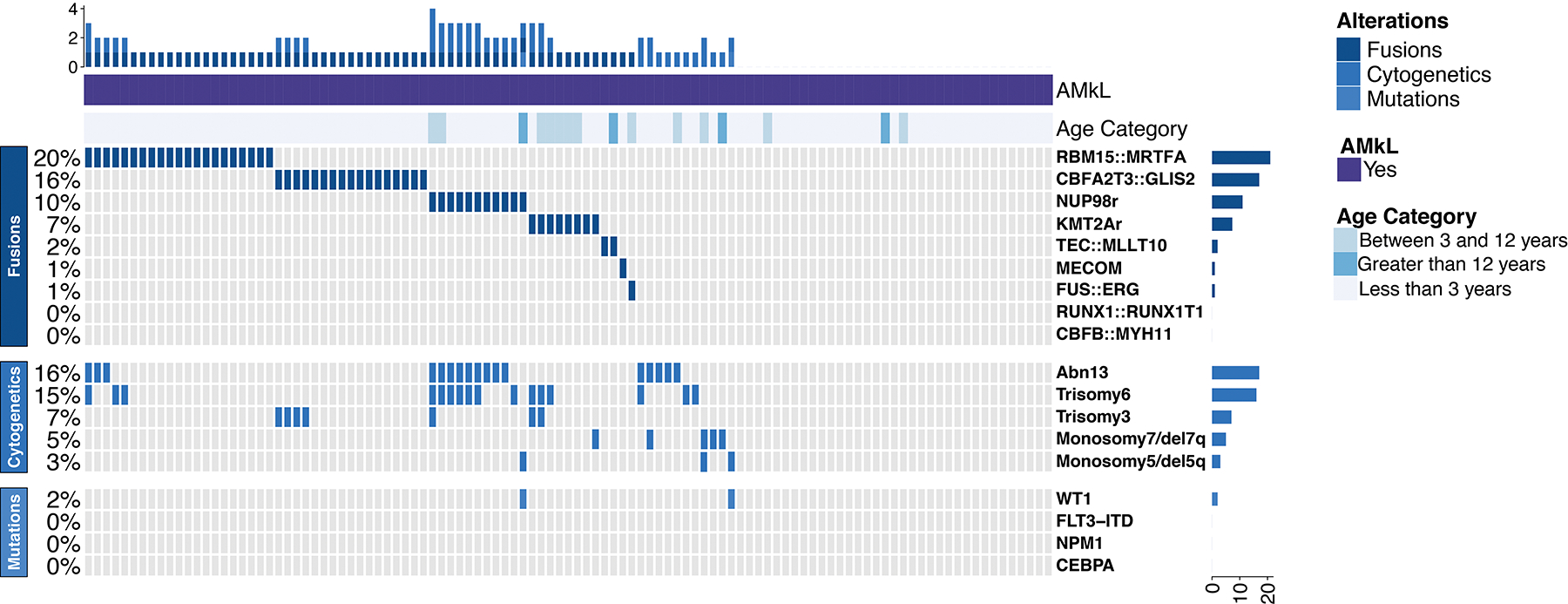
The abnormalities present in chr13 (Abn13) include del(13q), monosomy 13, trisomy 13, and chr13 translocations. Additional cytogenetic aberrations reported are copy number variations (trisomy 3, trisomy 6, monosomy 7/del7q, and monosomy5/del5q).
Outcomes:
Table 2 lists the 5-year EFS, OS, and RR for these AMkL. The 5-year EFS and OS were different between the two trials, with those treated on AAML0531 having better outcomes than those treated on AAML1031 (5-year EFS of 62 ± 14% vs 37 ± 13%, p=0.009, respectively; 5-year OS of 64 ± 14% vs 45 ± 14%, p=0.069, respectively). While AMkL compared to other FAB subtypes in AAML0531 did not show differences in 5-year EFS or OS, AMkL in AAML1031 had a worse 5-year OS than the other FAB subtypes (44 ± 14% vs 66 ± 3%, p=0.001) (Figure 3).
Table 2.
Outcome measures in pediatric AMkL per Children’s Oncology Group trial
| AAML0531 | AAML1031 | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | N | 5-year EFS | 5-year OS | 5-year RR from end of course 1b | N | 5-year EFS | 5-year OS | 5-year RR from end of course 1b |
| Morphology | ||||||||
| All M7 FAB | 48 | 62 ± 14% ^ | 64 ± 14% # | 31 ± 17% | 59 | 37 ± 13% ^ | 45 ± 14% *,# | 45 ± 16% |
| M0-M6 FAB | 816 | 53 ± 3% | 66 ± 3% | 36 ± 4% | 1012 | 47 ± 3% | 66 ± 3% * | 43 ± 4% |
| FAB M7 Karyotype | ||||||||
| Not complex | 20 | 53 ± 22% | 53 ± 22% | 55 ± 34% | 31 | 39 ± 18% | 38 ± 18% | 38 ± 22% |
| Complex | 24 | 71 ± 18% | 75 ± 17% | 24 ± 20% | 25 | 36 ± 20% | 53 ± 22% | 49 ± 26% |
| FAB M7 fusion-based classification a | ||||||||
| AMkL, NOS | 4 | - | - | - | 7 | 43 ± 37% | 42 ± 37% | 25 ± 50% |
| AMkL with RBM15::MRTFA fusion | 7 | 86 ± 26% | 86 ± 26% | 0 ± 0% | 14 | 50 ± 27% | 54 ± 14% | 38 ± 29% |
| AMkL with CBFA2T3::GLIS2 | 7 | 43 ± 37% | 43 ± 37% | - | 10 | 10 ± 19% | 10 ± 19% | 60 ± 56% |
| AMkL with NUP98 fusion | 3 | 100 ± 0% | 100 ± 0% | 0 ± 0% | 8 | 30 ± 35% | 42 ± 44% | - |
| AMkL with KMT2A fusion | 3 | - | - | 0 ± 0% | 5 | 40 ± 44% | 40 ± 44% | 25 ± 50% |
| AMkL with (other) myelodysplasia-related changes | 20 | 75 ± 20% | 80 ± 19% | 18 ± 19% | 10 | 58 ± 32% | 69 ± 30% | 35 ± 34% |
Abbreviations: AMkL, acute myeloid leukemia with megakaryocytic differentiation; EFS, event-free survival; FAB, French-American-British classification; N, number; NOS, not otherwise specified; OS, overall survival; RR, relapse risk
Excluding those with unknown cytogenetics or fusions identified in ≤2 cases (MECOM, FUS::ERG, and TEC::MLLT10)
Including only those with a complete remission
AAML1031 M7 significantly worse compared to AAML1031 M0-M6 FAB group (p=0.001)
AAML0531 M7 5-year EFS significantly better compared to AAML1031 M7 group (p=0.009)
AAML0531 M7 5-year OS better compared to AAML1031 M7 group but not significantly (p=0.069)
- 5-year estimate is undefined
Figure 3. AMkL vs other AML Kaplan–Meier curves per trial.
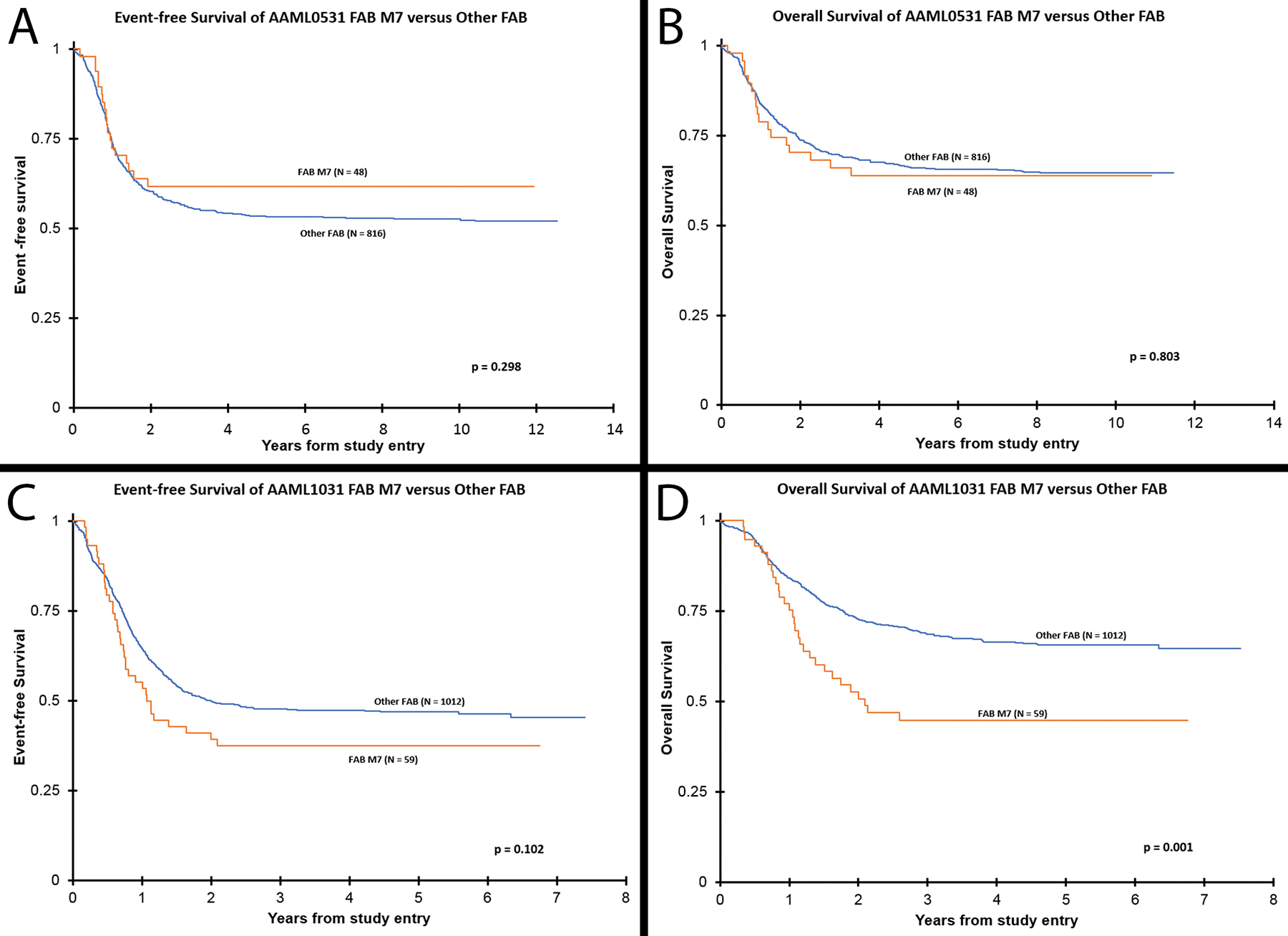
(A) Event-free survival and (B) overall survival of all cases of AMkL (FAB M7) versus all other centrally-reviewed AML FAB subtypes from trial AAML0531. As shown, M7 cases have relatively equivalent EFS and OS compared to other FAB subgroups. (C) EFS and (D) OS of all cases of AMkL versus all other centrally reviewed AML FAB subtypes from trial AAML1031; as shown AMkL have decreased 5-year OS compared to the other FAB subgroups. In general, AMkL on AAML0531 had better outcomes than AMkL on AAML1031.
To study the outcomes of AMkL by their fusion classification, the following cases were excluded due to low numbers: 5 with unknown cytogenetics, 2 with TEC::MLLT10, 1 with FUS::ERG, and 1 with a MECOM fusion. The outcomes from the remaining subgroups were determined (Table 2 and Figure 4). In general, cases of AMkL-MRC without recurrent fusions and those with RBM15::MRTFA had better outcomes than the other subgroups, while those with CBFA2T3::GLIS2 or KMT2A fusions had worse outcomes. AMkL, NOS (i.e. AMkL without identified fusions, complex karyotypes, or cytogenetic abnormalities that define myelodysplasia-related changes) had variably decreased EFS and OS, especially when compared with the more favorable AMkL-MRC and AMkL with RBM15::MRTFA. Interestingly, complex karyotypes in general (not considering the presence of any fusion proteins) did not lead to significantly different survival rates. Additionally, AMkL with NUP98 fusions had different outcomes in the trials, with increased EFS and OS in the AAML0531 trial compared to those in the AAML1031 trial, though the number of AMkL with NUP98 fusions in the AAML0531 trial was quite small (n=3).
Figure 4. AMkL subgroup Kaplan–Meier curves per trial.
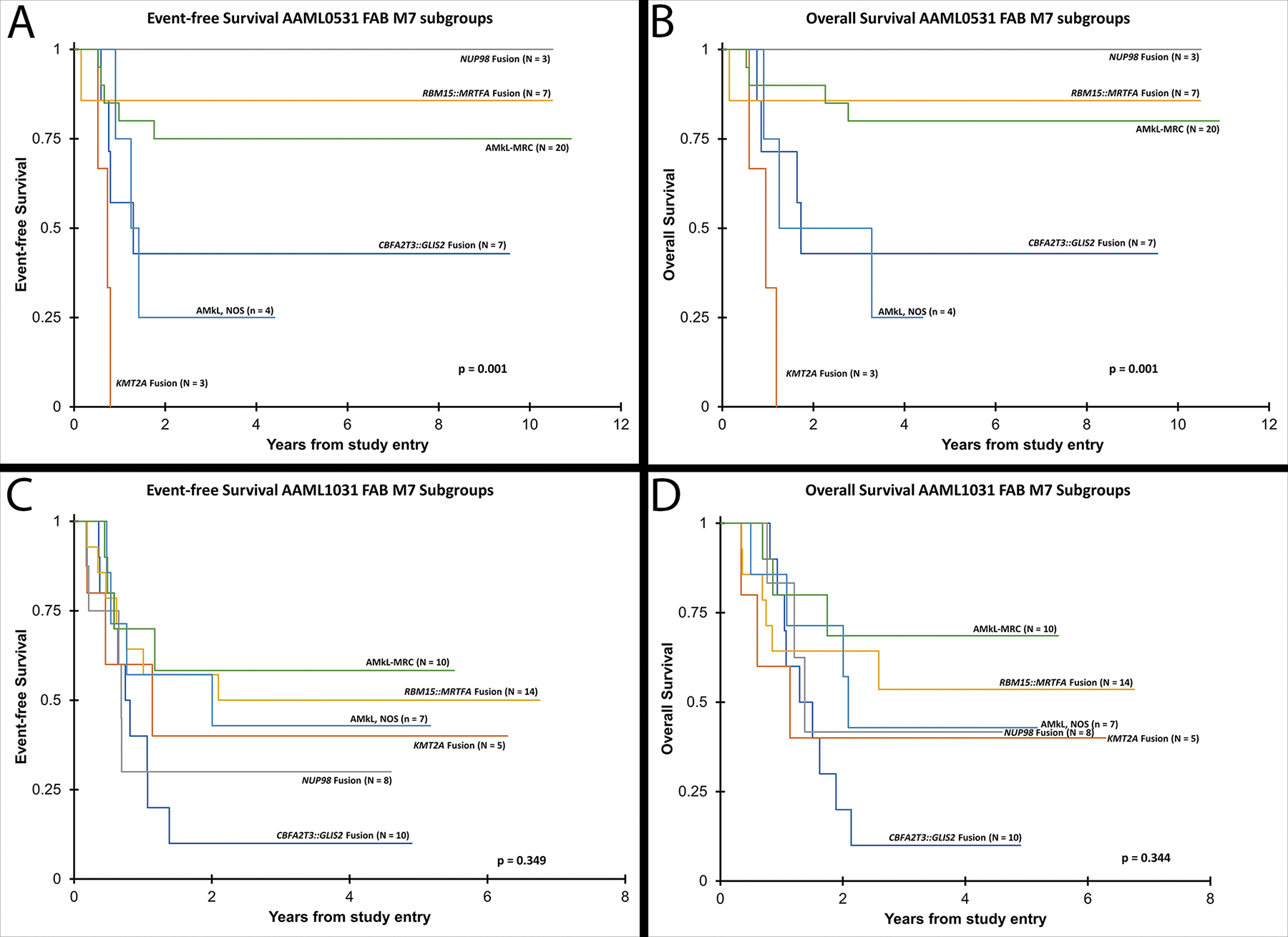
(A) Event-free survival and (B) overall survival of major AMkL subgroups in trial AAML0531 show statistically significant different survivals depending on the identified fusion, other myelodysplasia-related changes (MRC), or not otherwise specified (NOS). (C) Event-free survival and (D) overall survival of major AMkL subgroups in trial AAML1031 did not show statistically significant different survivals depending on the subcategory.
DISCUSSION
In this study, 107 cases of central pathology confirmed AMkL from two COG trials (AAML0531 and AAML1031) were categorized into subgroups based upon morphology/dysplasia, cytogenetics, and molecular features. While there was a paucity of genetic mutations, these AMkL harbored a variety of fusions which helped to subclassify the leukemias; such subclassifications are helpful in determining outcomes.
AMkL is a rarer subtype of AML in children. The diagnosis of AMkL may be challenging, as bone marrow aspirate smears are often hemodilute and/or aparticulate due to marrow fibrosis. Such marrow fibrosis can also lead to suboptimal specimens for flow cytometry, cytogenetics, and molecular testing, with falsely decreased blast percentages. Bone marrow core biopsies may be helpful in characterizing the blast percentage and immunophenotypes of these leukemias. While only performed in a subset of our cohort (n=39), 77% of core biopsies with reticulin staining had ≥MF-1 fibrosis, while 46% had MF-2 or MF-3 fibrosis, concordant with prior studies.15,21 Prominent fibrosis has been reported in AMkL with RBM15::MRTFA,16–20 but this is one of the first reports detailing a high rate of fibrosis in those with KMT2A fusions. In one adult study, fibrosis did not correlate with survival.44
Previous reports have shown inferior outcomes in AMkL compared to other AML subtypes, with 4- and 5-year EFS ranging from 36.6–51% and 41–47%, respectively, and 4- and 5-year OS ranging from 56–58.6% and 49–60%.8,9,13,14,26 In this study, the 48 AMkL cases on AAML0531 had equivalent to slightly better 5-year EFS and OS, while the 59 AMkL cases on AAML1031 had equivalent to decreased 5-year EFS and OS compared to these prior studies. The different outcomes of AMkL in these two current COG trials is not surprising due to the differences in therapy in these trials. AAML0531 randomized the enrolled patients to receive standard 5-course chemotherapy with or without two doses of GO. While there were more AMkL patients on the no-GO Arm than on the GO Arm (n=30 vs 18, respectively), this difference was not statistically significant; however, the GO arm generally had significantly improved EFS and decreased RR compared to the no-GO Arm.28 In AAML1031, the patients who were classified as low risk (having favorable cytogenetic/molecular features or uninformative cytogenetic/molecular features) but with negative MRD at end of induction (including 30 of the AMkL in this series), received 4-course chemotherapy with or without bortezomib. Notably, these 30 AMkL did not have favorable cytogenetic/molecular features as there were no cases of t(8;21) (RUNX1::RUNX1T1), inv(16)/t(16;16) (CBFB::MYH11), NPM1, and CEBPα in our AMkL cohort. Getz et al. reported that the reduced cytarabine exposure in these low-risk patients without favorable cytogenetics led to reduced disease-free and overall survival.45
In this study, we subclassified the AMkL into separate cohorts based upon pathology, cytogenetic, and involved fusion proteins. Our results suggest that genotype and not phenotype defines the outcome of these cases. Despite the small numbers of cases in these subcategories and different outcomes in the two COG trials, we showed a poor prognosis of AMkL with CBFA2T3::GLIS2 and KMT2A fusions. Previous reports have confirmed the decreased outcomes of CBFA2T3::GLIS2 with 4–5-year EFS ranging from 8–33% and 4–5-year OS of ranging from 14–38%,7,8,25,27,46 though some of those reports did notably include a subset of patients in this study. Similarly, AMkL with KMT2A fusions have reported poor prognoses, with 5-year EFS ranging from 27–28.5% and 5-year OS ranging from 27–32.4%.7,9,27
CBFA2T3::GLIS2 is cryptic, as it cannot be detected by routine karyotype. In fact, of the 17 CBFA2T3::GLIS2 cases in this series, none had karyotypic evidence of the fusion and 6 had normal karyotypes. This fusion’s surprisingly high association with trisomy 3 and lack of complex karyotypes also suggests that it is especially important to screen for cryptic fusions in these cases. Other clues come from its immunophenotype, as CBFA2T3::GLIS2 is highly associated with bright CD56 expression and the RAM phenotype. Only 18 of 101 cases in this study displayed the RAM phenotype, 16 of which had CBFA2T3::GLIS2, concordant with previous studies.47
The most common fusion identified in this series was RBM15::MRTFA. These 21 RBM15::MRTFA cases were significantly younger in age (mean of 0.91 years), consistent with prior studies showing RBM15::MRTFA patients are amongst the youngest with AMkL.7–9 Our study confirmed that RBM15::MRTFA AMkL generally have a more favorable outcome than other subgroups of AMkL.7,8,12,27
In our cohort, NUP98 fusions were seen with KMD5A, NSD1, and BPTF partner genes. Concordant with literature, the most common fusion partner was KMD5A, a partner often associated with AMkL.7,8,25–27 Some karyotypic findings, including trisomy 6, monosomy 13/del 13(q), and trisomy 21, were significantly increased in the cases with NUP98 fusions. NUP98::KDM5A with trisomy 21, monosomy 13/del 13(q), trisomy 6, and complex karyotypes have previously been reported,7,8,26 but this is the first study to highlight the significant increases in these abnormalities. Specifically, all 9 NUP98::KDM5A had structural chromosome 13 abnormalities, with 8 having monosomy 13 (n=2) or del(13q) (n=6), and the remaining case having a translocation involving 13q; notably, one case each with monosomy 13 and del(13q) also demonstrated translocations involving 13q.
The 5-year EFS and OS of AMkL with NUP98 rearrangements were quite different between the AAML0531 and AAML1031 trials. Only 3 patients had AMkL with NUP98 rearrangements in the AAML0531 trial, all of whom have survived without relapse, although this favorable survival may be influenced by small numbers. In contrast, the 8 NUP98-rearranged AMkL patients on AAML1031 (7 partnered with KDM5A) had reduced EFS and OS, more consistent with prior studies. Two AMkL cohorts studied by de Rooij et al.7,27 demonstrated that patients with NUP98::KDM5A had 5-year EFS and OS ranging from 22–25% and 22–35%, respectively, though notably both not statistically significant. However, with a different cohort, de Rooij et al.8 found this translocation to be an independent predictor of poor outcome with 4-year EFS and OS both of 36%. Hara et al.26 found similarly worse EFS. Two of the de Rooij et al. cohorts7,8 included a subset of patients described in this study.
After AMkL with recurrent fusions has been separated, the two main remaining subgroups are AMkL-MRC and AMkL, NOS. Our study is one of the first to include these classifications in AMkL. In general, AML with complex karyotypes, myelodysplasia-defining cytogenetic abnormalities, and/or morphologic dysplasia have poorer outcomes compared to AML, NOS or AML with certain favorable recurrent cytogenetic abnormalities.32,48,49 In this study, cases with complex karyotypes, but not having known gene rearrangements were classified as AMkL-MRC, including 28 with complex karyotypes, 1 with myelodysplasia-related cytogenetic abnormalities lacking a complex karyotype, and 1 with multilineage dysplasia. Similar to prior reports, monosomy 5/del(5q) and monosomy 7/del(7q) were not commonly identified in these pediatric cases of AMkL (though this group was still significantly enriched in cases with monosomy7/del(7q)) and the most common gains included trisomies 8, 19, and 21.2,9 Interestingly, these 30 AMkL-MRC cases had relatively increased 5-year EFS and OS compared to the other subcategories. It may be that purifying the AMkL-MRC cohort, excluding specific fusion products, leads to better prognoses. Notably, in the upcoming WHO hematolymphoid tumor 5th edition50 and International Consensus Classification (ICC) of myeloid neoplasms and acute leukemias,51 the AML-MRC category is removed in favor of AML, myelodysplasia-related (AML-MR, WHO 5th edition) and AML with myelodysplasia-related cytogenetic abnormalities or AML with myelodysplasia-related gene mutations (ICC), categories depicting cases of AML with specific cytogenetic and molecular abnormalities but removing cases with only morphologic dysplasia. In this study, such categorization would have reclassified only 1 case of AMkL from the AML-MRC category to AMkL, NOS.
Conversely, cases without defined fusion products and relatively normal karyotypes, classified as AMkL, NOS in this study had relatively poor outcomes. Our cohort of 11 cases may be considered more of a pure group than similar cohorts in past studies, as many cases with cryptic fusions can have normal karyotypes.
The relatively small numbers of AMkL in these COG trials could lead to some false associations. Additionally, the 2 trials could not be combined for outcomes analysis due to their statistically significant differences in 5-year EFS and OS. Considering the small size of these subgroups, the comparisons performed herein are more ad hoc analyses and exploratory in nature. Larger scale studies of similar cohorts are recommended to confirm our findings.
In conclusion, full cytogenetic and RNA fusion testing is recommended for all AMkL to enable classification similar to that presented here. The identification of recurrent fusion proteins should prevent classification into the AMkL, NOS and AMkL-MRC categories, even in the cases with complex karyotypes. Such subgroups can aid in future prognostication and development of therapeutic strategies especially in those with poor outcomes.
Supplementary Material
Supplemental Figure 1. Acute myeloid leukemia with megakaryocytic differentiation morphology. A-B) Aspirate smears of AML, NOS demonstrate megakaryoblasts as large cells with cytoplasmic pseudopods or bleb formation. They have round, slightly irregular nuclei with fine reticular chromatin and 1–3 nucleoli. Rare cytoplasmic granules can be seen. C) AML with RBM15::MRTFA has blasts in which the differential diagnosis may include a small round blue cell tumor. D) AML with KMT2A rearrangements, classically associated with monocytic differentiation, may also have distinctly megakaryoblastic cells.
Supplemental Figure 2. Acute myeloid leukemia with megakaryocytic differentiation morphology on bone marrow core biopsies. A) Abnormal megakaryocytic maturation is often present in AMkL, including micromegakaryocytes and forms with separate nuclear lobes. B) On a core biopsy, the megakaryoblasts are large with smooth chromatin and prominent nucleoli. C) Low power may demonstrate increased reticulin fibrosis, here myelofibrosis score 3 (MF-3) (reticulin stain).
ACKNOWLEDGMENTS
This research was supported by COG Chairs Grant number U10CA098543, NCTN Network Group Operations Center Grant (U10CA180886), Statistics and Data Center Grant Number U10CA098413, and NCTN Statistics and Data Center Grant U10CA180899 from the National Cancer Institute, National Institutes of Health; St. Baldrick’s Foundation; Seattle Children’s Hospital Mark Alan Bomgardner Endowment (K.M.C); and Canada Research Chair in Pediatric Oncology Supportive Care (L.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviation key:
- AMkL
Acute myeloid leukemia with megakaryocytic differentiation
- AML
Acute myeloid leukemia
- COG
Children’s Oncology Group
- CR
Complete remission
- EFS
Event-free survival
- FAB
French-American-British
- GO
Gemtuzumab Ozogamicin
- ICC
International Consensus Classification
- IRB
Institutional review board
- ITD
Internal tandem duplications
- MF
Myelofibrosis
- MRC
Myelodysplasia-related changes
- NOS
Not otherwise specified
- OS
Overall survival
- RR
Relapse risk
- WHO
World Health Organization
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors report no financial interests or potential conflicts of interest.
REFERENCES
- 1.Arber DA, Brunning RD, Orazi A, et al. Acute myeloid leukaemia, NOS. In: Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2017:156–168. [Google Scholar]
- 2.Dastugue N, Lafage-Pochitaloff M, Pages MP, et al. Cytogenetic profile of childhood and adult megakaryoblastic leukemia (M7): a study of the Groupe Francais de Cytogenetique Hematologique (GFCH). Blood. 2002;100(2):618–626. [DOI] [PubMed] [Google Scholar]
- 3.Duchayne E, Fenneteau O, Pages MP, et al. Acute megakaryoblastic leukaemia: a national clinical and biological study of 53 adult and childhood cases by the Groupe Francais d’Hematologie Cellulaire (GFHC). Leuk Lymphoma. 2003;44(1):49–58. [DOI] [PubMed] [Google Scholar]
- 4.Giri S, Pathak R, Prouet P, Li B, Martin MG. Acute megakaryocytic leukemia is associated with worse outcomes than other types of acute myeloid leukemia. Blood. 2014;124(25):3833–3834. [DOI] [PubMed] [Google Scholar]
- 5.Arber DA, Baumann I, Niemeyer CM, Brunning RD, Porwit A. Myeloid proliferations associated with Down syndrome. In: Swerdlow SH, Campo E, Harris NL, et al. , eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2017:122–128. [Google Scholar]
- 6.Barnard DR, Alonzo TA, Gerbing RB, Lange B, Woods WG, Children’s Oncology G. Comparison of childhood myelodysplastic syndrome, AML FAB M6 or M7, CCG 2891: report from the Children’s Oncology Group. Pediatr Blood Cancer. 2007;49(1):17–22. [DOI] [PubMed] [Google Scholar]
- 7.de Rooij JD, Hollink IH, Arentsen-Peters ST, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27(12):2280–2288. [DOI] [PubMed] [Google Scholar]
- 8.de Rooij JD, Masetti R, van den Heuvel-Eibrink MM, et al. Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: a retrospective intergroup study. Blood. 2016;127(26):3424–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inaba H, Zhou Y, Abla O, et al. Heterogeneous cytogenetic subgroups and outcomes in childhood acute megakaryoblastic leukemia: a retrospective international study. Blood. 2015;126(13):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karol SE, Coustan-Smith E, Cao X, et al. Prognostic factors in children with acute myeloid leukaemia and excellent response to remission induction therapy. Br J Haematol. 2015;168(1):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maarouf N, Mahmoud S, Khedr R, et al. Outcome of Childhood Acute Megakaryoblastic Leukemia: Children’s Cancer Hospital Egypt 57357 Experience. Clin Lymphoma Myeloma Leuk 2019;19(3):e142–e152. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien MM, Cao X, Pounds S, et al. Prognostic features in acute megakaryoblastic leukemia in children without Down syndrome: a report from the AML02 multicenter trial and the Children’s Oncology Group Study POG 9421. Leukemia. 2013;27(3):731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweitzer J, Zimmermann M, Rasche M, et al. Improved outcome of pediatric patients with acute megakaryoblastic leukemia in the AML-BFM 04 trial. Ann Hematol. 2015;94(8):1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teyssier AC, Lapillonne H, Pasquet M, et al. Acute megakaryoblastic leukemia (excluding Down syndrome) remains an acute myeloid subgroup with inferior outcome in the French ELAM02 trial. Pediatr Hematol Oncol 2017;34(8):425–427. [DOI] [PubMed] [Google Scholar]
- 15.Athale UH, Razzouk BI, Raimondi SC, et al. Biology and outcome of childhood acute megakaryoblastic leukemia: a single institution’s experience. Blood. 2001;97(12):3727–3732. [DOI] [PubMed] [Google Scholar]
- 16.Cairney AE, McKenna R, Arthur DC, Nesbit ME, Jr., Woods WG. Acute megakaryoblastic leukaemia in children. Br J Haematol. 1986;63(3):541–554. [DOI] [PubMed] [Google Scholar]
- 17.Carroll A, Civin C, Schneider N, et al. The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia: a Pediatric Oncology Group Study. Blood. 1991;78(3):748–752. [PubMed] [Google Scholar]
- 18.Chan WC, Carroll A, Alvarado CS, et al. Acute megakaryoblastic leukemia in infants with t(1;22)(p13;q13) abnormality. Am J Clin Pathol. 1992;98(2):214–221. [DOI] [PubMed] [Google Scholar]
- 19.Lion T, Haas OA. Acute megakaryocytic leukemia with the t(1;22)(p13;q13). Leuk Lymphoma. 1993;11(1–2):15–20. [DOI] [PubMed] [Google Scholar]
- 20.Lion T, Haas OA, Harbott J, et al. The translocation t(1;22)(p13;q13) is a nonrandom marker specifically associated with acute megakaryocytic leukemia in young children. Blood. 1992;79(12):3325–3330. [PubMed] [Google Scholar]
- 21.Ribeiro RC, Oliveira MS, Fairclough D, et al. Acute megakaryoblastic leukemia in children and adolescents: a retrospective analysis of 24 cases. Leuk Lymphoma. 1993;10(4–5):299–306. [DOI] [PubMed] [Google Scholar]
- 22.Borkhardt A, Haas OA, Strobl W, et al. A novel type of MLL/AF10 fusion transcript in a child with acute megakaryocytic leukemia (AML-M7). Leukemia. 1995;9(10):1796–1797. [PubMed] [Google Scholar]
- 23.Buchanan J, Tirado CA. A t(16;21)(p11;q22) in Acute Myeloid Leukemia (AML) Resulting in Fusion of the FUS/TLS and ERG Genes: A Review of the Literature. J Assoc Genet Technol. 2016;42(1):24–33. [PubMed] [Google Scholar]
- 24.Morerio C, Rapella A, Tassano E, Rosanda C, Panarello C. MLL-MLLT10 fusion gene in pediatric acute megakaryoblastic leukemia. Leuk Res. 2005;29(10):1223–1226. [DOI] [PubMed] [Google Scholar]
- 25.Gruber TA, Larson Gedman A, Zhang J, et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell. 2012;22(5):683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara Y, Shiba N, Ohki K, et al. Prognostic impact of specific molecular profiles in pediatric acute megakaryoblastic leukemia in non-Down syndrome. Genes Chromosomes Cancer. 2017;56(5):394–404. [DOI] [PubMed] [Google Scholar]
- 27.de Rooij JD, Branstetter C, Ma J, et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 2017;49(3):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamis AS, Alonzo TA, Meshinchi S, et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol. 2014;32(27):3021–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aplenc R, Meshinchi S, Sung L, et al. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: a report from the Children’s Oncology Group. Haematologica. 2020;105(7):1879–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolouri H, Farrar JE, Triche T Jr, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arber DA, Brunning RD, Le Beau MM, et al. Acute myeloid leukaemia with recurrent genetic abnormalities. In: Swerdlow SH, Campo E, Harris NL, et al. , eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2017:130–149. [Google Scholar]
- 32.Arber DA, Brunning RD, Orazi A, et al. Acute myeloid leukaemia with myelodysplasia-related changes. In: Swerdlow SH, Campo E, Harris NL, et al. , eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2017:150–152. [Google Scholar]
- 33.Thiele J, Kvasnicka HM, Orazi A, et al. Primary myelofibrosis. In: Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2017:44–50. [Google Scholar]
- 34.Thiele J, Kvasnicka HM, Facchetti F, Franco V, van der Walt J, Orazi A. European consensus on grading bone marrow fibrosis and assessment of cellularity. Haematologica. 2005;90(8):1128–1132. [PubMed] [Google Scholar]
- 35.Brown P, McIntyre E, Rau R, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood. 2007;110(3):979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho PA, Alonzo TA, Gerbing RB, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood. 2009;113(26):6558–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho PA, Zeng R, Alonzo TA, et al. Prevalence and prognostic implications of WT1 mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood. 2010;116(5):702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meshinchi S, Woods WG, Stirewalt DL, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001;97(1):89–94. [DOI] [PubMed] [Google Scholar]
- 39.Haas BJ, Dobin A, Li B, Stransky N, Pochet N, Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson G, Schein J, Chiu R, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7(11):909–912. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 42.Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. 2nd ed. Hoboken, NJ: John Wiley & Sons, Inc.; 2002. [Google Scholar]
- 43.Eidenschink Brodersen L, Alonzo TA, Menssen AJ, et al. A recurrent immunophenotype at diagnosis independently identifies high-risk pediatric acute myeloid leukemia: a report from Children’s Oncology Group. Leukemia. 2016;30(10):2077–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niino D, Tsuchiya T, Tomonaga M, Miyazaki Y, Ohshima K. Clinicopathological features of acute megakaryoblastic leukaemia: Relationship between fibrosis and platelet-derived growth factor. Pathol Int. 2013;63(3):141–149. [DOI] [PubMed] [Google Scholar]
- 45.Getz KD, Alonzo TA, Sung L, et al. Cytarabine dose reduction in patients with low-risk acute myeloid leukemia: A report from the Children’s Oncology Group. Pediatr Blood Cancer. 2022;69(1):e29313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masetti R, Pigazzi M, Togni M, et al. CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood. 2013;121(17):3469–3472. [DOI] [PubMed] [Google Scholar]
- 47.Pardo LM, Voigt AP, Alonzo TA, et al. Deciphering the Significance of CD56 Expression in Pediatric Acute Myeloid Leukemia: A Report from the Children’s Oncology Group. Cytometry B Clin Cytom. 2020;98(1):52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arber DA, Stein AS, Carter NH, Ikle D, Forman SJ, Slovak ML. Prognostic impact of acute myeloid leukemia classification. Importance of detection of recurring cytogenetic abnormalities and multilineage dysplasia on survival. Am J Clin Pathol. 2003;119(5):672–680. [DOI] [PubMed] [Google Scholar]
- 49.Weinberg OK, Seetharam M, Ren L, et al. Clinical characterization of acute myeloid leukemia with myelodysplasia-related changes as defined by the 2008 WHO classification system. Blood. 2009;113(9):1906–1908. [DOI] [PubMed] [Google Scholar]
- 50.Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36(7):1703–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arber DA, Orazi A, Hasserjian RP, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Acute myeloid leukemia with megakaryocytic differentiation morphology. A-B) Aspirate smears of AML, NOS demonstrate megakaryoblasts as large cells with cytoplasmic pseudopods or bleb formation. They have round, slightly irregular nuclei with fine reticular chromatin and 1–3 nucleoli. Rare cytoplasmic granules can be seen. C) AML with RBM15::MRTFA has blasts in which the differential diagnosis may include a small round blue cell tumor. D) AML with KMT2A rearrangements, classically associated with monocytic differentiation, may also have distinctly megakaryoblastic cells.
Supplemental Figure 2. Acute myeloid leukemia with megakaryocytic differentiation morphology on bone marrow core biopsies. A) Abnormal megakaryocytic maturation is often present in AMkL, including micromegakaryocytes and forms with separate nuclear lobes. B) On a core biopsy, the megakaryoblasts are large with smooth chromatin and prominent nucleoli. C) Low power may demonstrate increased reticulin fibrosis, here myelofibrosis score 3 (MF-3) (reticulin stain).
Data Availability Statement
The data that supports the findings of this study are available in Supporting Table S1 of this article.


