Abstract
Background:
Long non-coding RNAs (lncRNAs) are RNA molecules with over 200 nucleotides that do not code for proteins, but are known to be widely expressed and have key roles in gene regulation and cellular functions. They are also found to be involved in the onset and development of various cancers, including prostate cancer (PCa). Since PCa are commonly driven by androgen regulated signaling, mainly stimulated pathways, identification and determining the influence of lncRNAs in androgen response is useful and necessary. LncRNAs regulated by the androgen receptor (AR) can serve as potential biomarkers for PCa. In the present study, gene expression data analysis were performed to distinguish lncRNAs related to the androgen response pathway.
Methods and Results:
We used publicly available RNA-seq and ChIP-seq data to identify lncRNAs that are associated with the androgen response pathway. Using Universal Correlation Coefficient (UCC) and Pearson Correlation Coefficient (PCC) analyses, we found 15 lncRNAs that have a) highly correlated expression with androgen response genes in PCa and are b) differentially expressed in the setting of treatment with an androgen agonist as well as antagonist compared to controls. Using publicly available ChIP-seq data, we investigated the role of androgen/ androgen receptor axis in regulating expression of these lncRNAs. We observed AR binding in the promoter regions of 5 lncRNAs (MIR99AHG, DUBR, DRAIC, PVT1, and COLCA1), showing the direct influence of AR on their expression and highlighting their association with the androgen response pathway.
Conclusion:
By utilizing publicly available multi-omics data and by employing in silico methods, we identified five candidate lncRNAs that are involved in the androgen response pathway. These lncRNAs should be investigated as potential biomarkers for PCa.
Keywords: androgen response genes, correlation analysis, gene expression, long noncoding RNA, multiomic data analysis, prostate cancer
Introduction:
Prostate cancer (PCa) ranks highest in estimated new cases diagnosed and second in estimated cancer-related deaths amongst non-cutaneous malignancies in American men 1. Androgen receptor (AR) drives tumor initiation and progression in many PCa patients. AR is a steroid hormone receptor that resides in cytoplasm during its inactive state. On ligand binding and activation by androgens such as testosterone or 5 dihydrotestosterone (DHT), AR transforms into a homodimer and internalizes into the nucleus and binds to DNA at the androgen response elements (AREs), where it influences activation and expression of hundreds of genes that have ARE’s in their promoter or enhancer regions 2,3. PCa patients with localized tumors are generally treated with surgery, and/or radiation treatment as primary definitive treatment approaches. However, for patients with advanced staged, metastatic PCa, androgen deprivation therapy (ADT) is the mainstay of treatment. ADT involves treatment with first and second generation anti-androgens 4. The initially favorable response to anti-androgen treatment is often followed by resistance to ADT in many patients. Castration-resistant prostate cancers (CRPCs) have more limited treatment options and poorer prognosis due to the lack of robust efficacy of the available treatments 5,6.
Various molecular mechanisms and signaling events are involved in AR signaling pathways. Understanding these molecular events can help in identifying new biomarkers. Recent studies have used next-generation sequencing technology to decipher the role of non-coding RNAs (such as microRNAs and long non-coding RNAs (lncRNAs)) in PCa initiation and disease progression7. LncRNAs are RNA molecules with more than 200 nucleotides which are not transcribed into proteins. Although they do not code for proteins, some lncRNAs interact with and regulate chromatin function by binding to and modulating the function of various proteins and altering the transcription of may target genes as well as modulating post translational changes 8. For example, lncRNAs such as HOTAIR, XIST, and AIRN are involved in regulating chromatin remodeling 9. Although there are 30,000 to 60,000 lncRNAs in the human genome, thus far, only about 100 of these have been shown to be involved in tumor progression 10. In various cancer types, including PCa, lncRNAs can function either as oncogenes or tumor suppressor genes 11–14. In PCa, there is recognition of direct or indirect interaction of lncRNAs with AR 15,16. The lncRNA PCGEM1, interacts with AR and facilitates its binding in the promoter regions of target genes 11. Over-expression of the lncRNA HOTAIR in PCa inhibits AR degradation 17. CTBP1-AS, which is overexpressed in PCa metastases, inhibits expression of CTBP1, which functions as co-repressor of AR18. PCA3 regulates the AR signal pathway 19. Additionally, LINC00844 and MALAT1 are shown to be involved in negative regulation of the AR signaling pathway 20,21.
In view of the functions of lncRNAs in AR signaling and in order to understand the androgen regulated lncRNAs, we performed detailed in silico analysis utilizing publicly available gene expression and regulation data from PCa patients and cell lines. Furthermore, we evaluated the expression pattern of these lncRNAs in pro and anti-androgen treated PCa cell lines. These analyses will provide a better understanding of the pathways and the underlying mechanism of hormonal deprivation therapy and could lead to discovery of therapeutic targets for patients with CRPCs.
In the current study, we employed multiple correlation methods with publicly available PCa next-generation sequencing data to a) identify lncRNAs that are co-expressed with androgen response pathway genes, b) assess their gene expression patterns on androgen/anti-androgen treatment, and c) investigate the regulatory effects of AR on identified lncRNAs. The study overview is shown in Fig 1. Our analysis showed that a set of lncRNAs (PART1, CBLL1-AS1, DUBR, SOC2-AS1, SLC38A4-AS1, DRAIC, PVT1, Antisense to TUBA1B (AS-TUBA1B), IDH1-AS1, NKILA, MKLN1-AS, Antisense to C6orf1 (AS-C6orf1), MIR99AHG, COLCA1, and SNHG5) are highly regulated in PCa in an androgen dependent manner. This androgen-regulated lncRNA expression was altered during the anti-androgen treatment suggesting the role of AR in regulating these lncRNAs. Thus, some of these AR regulated lncRNA, thus, may play a critical role in PCa progression and may serve as biomarkers or therapeutic targets for PCa.
Figure 1.
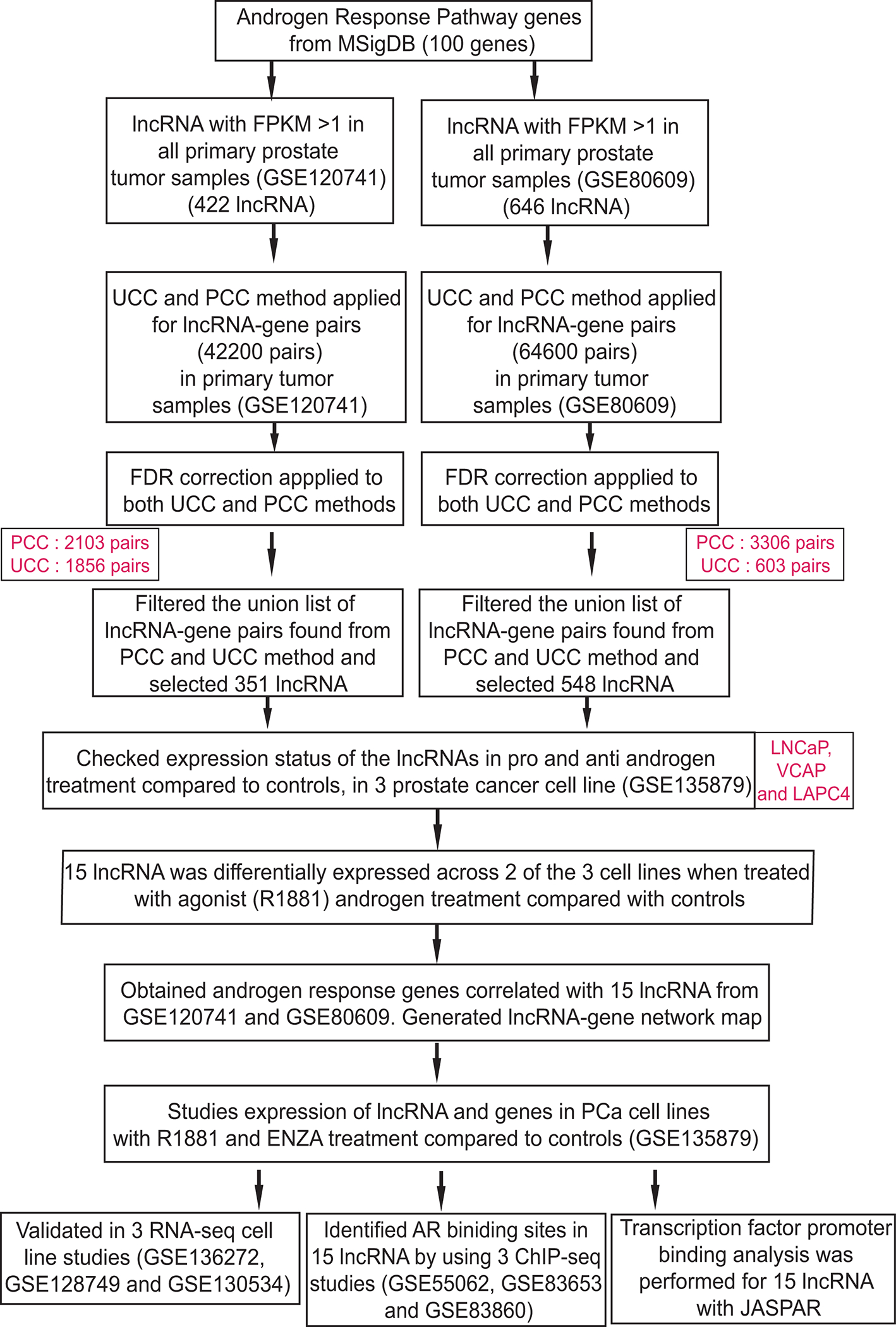
The workflow of the study.
Materials and Methods:
Data collection:
We searched RNA-sequencing data related to primary PCa tissues and androgen-treated PCa cells in the NCBI Gene expression Omnibus database (GEO) (https://www.ncbi.nlm.nih.gov/gds/). In total, 6 RNA-seq and 3 ChIP-seq studies were considered in our study. RNA sequencing data from GSE120741 and GSE80609 included 92 and 37 PCa samples respectively 22, 23. The transcriptome sequencing study from GSE135879 includes PCa cell lines (VCaP, LNCaP, and LAPC4) treated with DMSO, enzalutamide (ENZA, an anti-androgen), or metribolone (R1881, a synthetic androgen agonist) in duplicate 24. The remaining three studies (GSE136272, GSE128749, and GSE130534) were selected as validation datasets. GSE136272 includes VCaP and LNCaP cells treated with DMSO or R1881 in triplicate 25. GSE128749 dataset includes LAPC4 cells treated with ethanol (n=3) or R1881 (n=2), and LNCaP cells treated with ethanol or R1881 in triplicate 26. GSE130534 dataset includes the LNCaP cells treated with DMSO or ENZA in triplicate 27.
We also selected three sets of AR ChIP-seq data (GSE55062, GSE83653, and GSE83860) for PCa cell lines to analyze AR-mediated gene regulation. GSE55062 is comprised of ChIP-seq data for VCaP cells treated with AR (n=2) or IgG (n=1) antibody 28. The GSE83653 Chip-seq study included data for VCaP cells treated with Ab108341 (n=1) or no (n=1) antibody 29. The GSE83860 study included data for LNCaP cells treated with anti-AR (n=2) or no (n=2) antibody 30.
Table 1 provides detailed account of samples considered for analysis from above mentioned RNA-seq and ChIP-seq studies.
Table 1.
Summary of RNA-seq and ChIP-seq data considered in this study.
| GEO accession number | Source | Condition | Platform | Method | Sample size and treatment groups |
|---|---|---|---|---|---|
| GSE120741 | Tissue | PCa samples | Illumina HiSeq 2500 (Homo sapiens) | RNA-seq | 92 |
| GSE80609 | Tissue | PCa samples | Illumina HiSeq 2000 (Homo sapiens) | RNA-seq | 37 |
| GSE135879 | Cell line | VCaP (GSM4037036, GSM4037037, GSM4037038 GSM4037039, GSM4037040 and GSM4037041), LNCaP (GSM4037030, GSM4037031 GSM4037032, GSM4037033 GSM4037034, and GSM4037035) and LAPC4 (GSM4037042, GSM4037043 GSM4037044, GSM4037045 GSM4037046, and GSM4037047) | Illumina HiSeq 2500 (Homo sapiens) | RNA-seq |
VCaP, LNCaP and LAPC4 cell lines: DMSO, R1881, and ENZA in duplicates. |
| GSE136272 | Cell line | VCaP (GSM4043846, GSM4043847, GSM4043848, GSM4043849, GSM4043850 and GSM4043851) and LNCaP (GSM4043805, GSM4043806, GSM4043807, GSM4043808, GSM4043809 and GSM4043810) | Illumina HiSeq 2500 (Homo sapiens) | RNA-seq |
VCaP and LNCaP cell lines: DMSO and R1881 in triplicates. |
| GSE128749 | Cell line | LAPC4 (GSM3684385, GSM3684386, GSM3684387, GSM3684388 and GSM3684389) and LNCaP (GSM3684391, GSM3684392, GSM3684393, GSM3684394, GSM3684395 and GSM3684396) | Illumina HiSeq 2000 (Homo sapiens) | RNA-seq |
LAPC4 and VCaP cell lines: For LAPC4 cell line, ethanol (n=3) and R1881 (n=2) For LNCaP cells, ethanol and R1881 in triplicate. |
| GSE130534 | Cell line | LNCaP (GSM3741911, GSM3741912, GSM3741913, GSM3741914, GSM3741915 and GSM3741916) | Illumina NextSeq 500 (Homo sapiens) | RNA-seq |
LNCaP cell line: DMSO: 3 ENZA: 3 |
| GSE55062 | Cell line | VCaP (AR-Vehicle: GSM1328945, GSM1328947; IgG: GSM1328983) | Illumina HiSeq 1000 (Homo sapiens) | Chip-Seq |
VCaP Cell line: Control: 2 IgG: 1 |
| GSE83653 | Cell line | VCaP (AR: GSM2612456; Input DNA: GSM2612458) | Illumina HiSeq 2500 | Chip-Seq |
VCaP cell line: Control: 1 Ab108341: 1 |
| GSE83860 | Cell line | LNCaP (AR-DMSO: GSM2219856, GSM2219857; Input DNA: GSM2219876, GSM2219877) | Illumina HiSeq 2000 (Homo sapiens) | Chip-Seq |
LNCaP Cell line:Control: 2 Anti-AR: 2 |
Androgen response related genes were acquired from MSigDB’s HALLMARK_ANDROGEN_RESPONSE gene set (https://www.gsea-msigdb.org/gsea/msigdb/) 31. There were 100 genes that were highly related to the androgen response pathway. These genes included cytokines, transcription factors, cell differentiation markers, oncogenes, and tumor suppressors that are involved in the androgen response. These were identified based on clustering and included the biologically relevant genes that were coordinately expressed in that particular HALLMARK pathway.
RNA-Seq data analysis:
For the selected RNA-sequencing studies, raw fastq files were downloaded by using fastq-dump from sratoolkit (https://github.com/ncbi/sra-tools). The adapter sequences and low-quality reads were removed using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), followed by quality control analysis using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The trimmed fastq files were mapped using Tophat v2.1 32, and were aligned to hg38.gtf file obtained from ENSEMBL (https://ftp.ensembl.org/pub/release-108/gtf/homo_sapiens/). The procured sam files were converted to bam files and were sorted by SAMtools 33. For quantification of each gene, we used the HTseq-count from the HTSeq library34. To perform the differential gene expression analysis from the gene count files between conditions and their respective log fold-change, we utilized the DESeq2 (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html) R package. For each study, values for fragments per kilobase of transcript per million mapped reads (FPKM) were obtained by using the Cuffquant and Cuffnorm function from Cufflinks (https://cole-trapnell-lab.github.io/cufflinks/).
Chip-seq data analysis:
Raw sequence reads were trimmed using Trim_Galore. Trimmed reads were aligned to the human reference genome (hg38) using the Burrows-Wheeler Aligner (bwa mem)35. Aligned reads were sorted, and duplicate reads were marked using Picard (http://broadinstitute.github.io/picard). BED files were generated from BAM files using bamToBed 36. MACS2 was used to call AR peaks compared to control 37.
Statistical Analysis:
Statistical analyses were performed using Python, MATLAB, and R (version 4.1.1). Universal Correlation Coefficient (UCC)38 and Pearson Correlation Coefficient (PCC) methods were applied to identify lncRNA-gene pairs. The coefficient Δ was applied to detect functional relationships between the set of lncRNAs and the set of 100 genes identified in the pathway. Compared with Pearson’s correlation coefficient, which measures the strength of linearity, the universal correlation coefficient (Δ) is used as a general correlation coefficient that measures the strength of any functional dependence38. The universal correlation coefficient uses values such as 0 and 1 for representing any functional dependencies. Such as, 1 for total dependence of any functional form as the size increases, and approaches 0 for total independence. It can also take on negative values for dependence of multivalued relation form and minimizes around −1/2 for total dependence of multivalued relation of types as sample size increases. This general correlation coefficient Δ has a limiting normal distribution with known mean and standard deviation under the null hypothesis that the two variables are independent of each other, which allows the computation of a p-value for each observed value of coefficient Δ. In this application, the observed values of Δ < 0.2433 were associated with FDR adjusted p-values > 0.01, which served as our cut-off for eliminating pairs from further investigation.
Any gene-lncRNA pair with a Pearson correlation coefficient (PCC) >0.5 or <−0.5 and FDR adjusted P-value <0.01 was selected. From the DESeq2 output (https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html), lncRNA and genes with log fold change >|1| and p-value < 0.05 were considered as differentially expressed.
Transcription factor promoter binding analysis:
Promoter sequences of the selected lncRNAs were obtained from NCBI Gene (https://www.ncbi.nlm.nih.gov/gene). Based on the strand orientation of each lncRNA, the sequence was collected from 1500 bp prior to and 500 bp after the promoter start site. The sequences of lncRNA were given as input in JASPAR to identify the transcription factor promoter binding to the androgen response element (ID: MA0007.2) (http://jaspar.genereg.net/)39. JASPAR uses complex computational methods involving sequence consensus-based models, position frequency matrices (PFM), and deep learning models to predict the Transcription Factor Binding sites (TFBS), to identify Transcription Factor - DNA interactions.
Visualization and Network Analysis:
All plots were made using a heat map and the ggplot2 library in R. The gene-lncRNA interaction network analysis was performed using Cytoscape 40. The IGV (Interactive Genome Viewer) was used to visualize and annotate the results from ChIP-Seq data analysis41. Venn diagrams were made using InteractiVenn (http://www.interactivenn.net/) 42.
Results:
RNA sequencing data analysis identified lncRNAs associated with the androgen response pathway:
The current study was designed to utilize publicly available, next-generation sequencing data to identify lncRNAs associated with the androgen response pathway in PCa (Fig 1). We processed two RNA-seq studies [GSE120741 and GSE80609] comprising prostate tumor tissue samples. As we were interested in androgen response, we considered only lncRNAs expressed in all primary PCa tumors from GSE80609 and GSE120741 which yielded 646 and 422 lncRNA respectively. To understand lncRNA-gene interactions, each of these was subjected to UCC and PCC analysis with 100 androgen response genes obtained from MSigDB. PCC analysis of GSE80609 and GSE120741 led to identification of 3306 and 2103 lncRNA-gene pairs respectively, with correlation coefficients of >= 0.5 or <= −0.5 and FDR-adjusted p-values <0.01. UCC analysis of GSE80609 and GSE120741 led to identification of 603 and 1856 lncRNA-gene pairs respectively, with Δ > 0.2433 associated and FDR-adjusted p-values < 0.01 (Fig 2A). On making a union list of FDR corrected PCC and UCC method gene-lncRNA pairs for each study, we found interactions that included 548 and 351 lncRNAs in GSE80609 and GSE120741 respectively.
Figure 2.
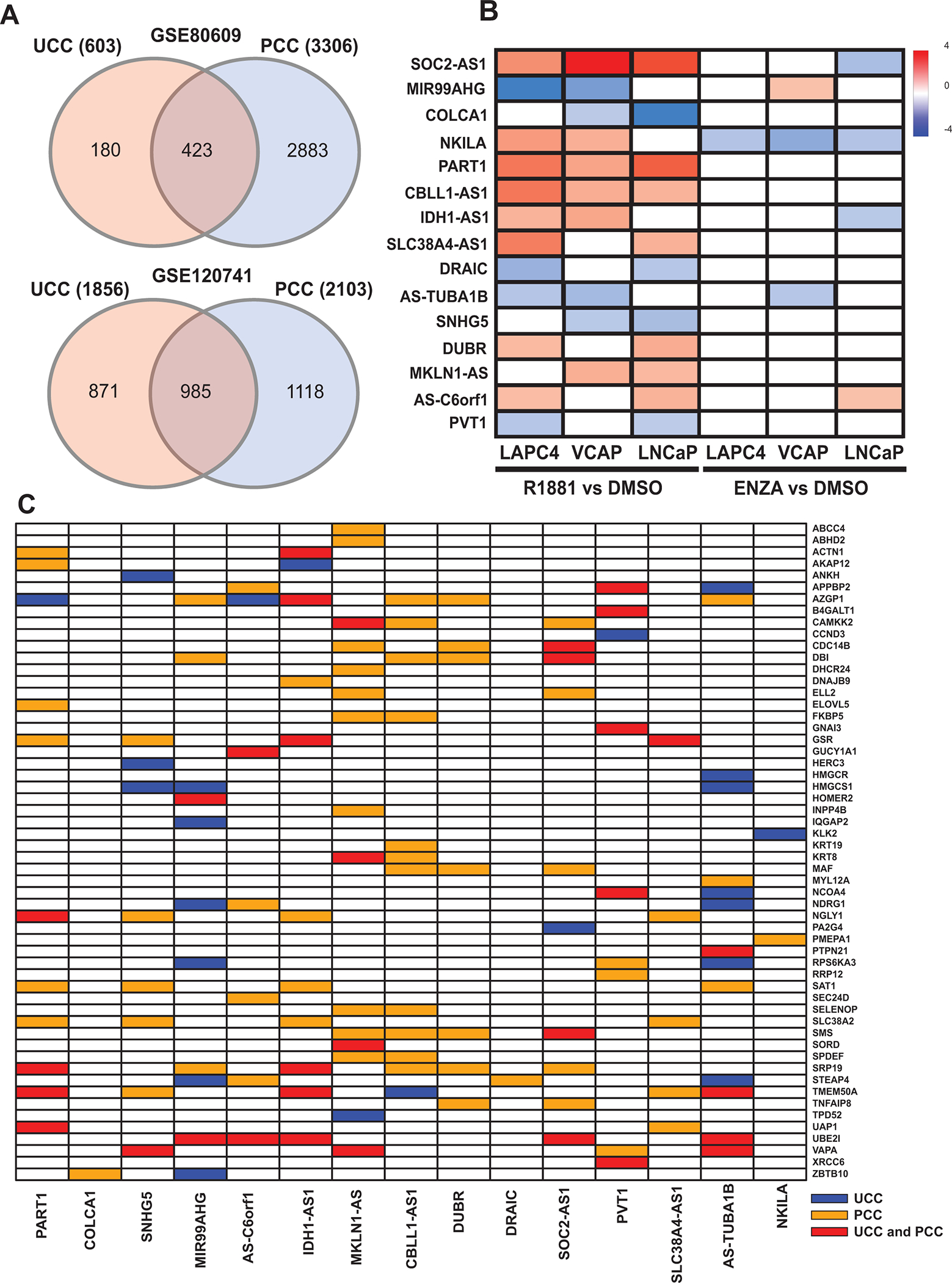
Gene expression and statistical analysis identified lncRNA-gene pairs. (A) Venn diagram depicting the statistical method by which lncRNA-gene pairs were selected (Top- GSE80609 and Bottom – GSE120741). (B) Heatmap showing the expression pattern of 15 lncRNAs with R1881 treatment compared with control and ENZA treatment compared with control for VCaP, LNCaP and LAPC4 cell lines (GSE135879). (C) Heatmap representing the statistical method that identified 15 lncRNA and 56 androgen response genes. (lncRNA: Long non-coding RNA, ENZA: enzalutamide)
To analyze the expression pattern of these lncRNAs after androgen or anti-androgen treatment of PCa cells, we compared them with differentially expressed lncRNAs from GSE135879 RNA-seq data. Our initial analysis led to identification of 249, 354, and 419 differentially expressed lncRNAs in LNCaP, LAPC4 and VCaP cells, respectively, after R1881 (androgen agonist) treatment when compared with control; 119, 40, and 92 differentially expressed lncRNAs were evident after ENZA treatment on comparison with controls for LNCaP, LAPC4, and VCaP cells, respectively.
Comparative analysis of 548 and 351 lncRNAs from two studies [GSE80609 and GSE120741] and differentially expressed lncRNAs on R1881 treatment compared to control, from GSE135879 led to selection of 15 lncRNAs. Only lncRNAs differentially expressed in at least 2 of 3 cell lines were selected (Fig 2B). These included PART1, PVT1, COLCA1, SNHG5, MIR99AHG, IDH1-AS1, MKLN1-AS, CBLL1-AS1, DUBR, DRAIC, SOC2-AS1, NKILA, Antisense to TUBA1B (AS-TUBA1B), Antisense to C6orf1 (AS-C6orf1), and SLC38A4-AS1. Of these, 9 lncRNAs were significantly (p-value > 0.05) upregulated on R1881 treatment compared to control (PART1, IDH1-AS1, MKLN1-AS1, CBLL1-AS1, DUBR, SOC2-AS1, SLC38A4-AS1, AS-C6orf1 and NKILA); the others (COLCA1, SNHG5, MIR99AHG, DRAIC, AS-TUBA1B, and PVT1) were significantly down-regulated on R1881 treatment. Of the 15 lncRNAs, 4 lncRNAs (NKILA, SOC2-AS1, AS-TUBA1B and IDH1-AS1) showed significant down-regulation and 2 lncRNAs (MIR99AHG and AS-C6orf1) showed significant up regulation on ENZA treatment compared to controls, in at least 1 of the 3 cell lines (Fig 2B). Statistical methods identifying 15 lncRNAs and their interacting 56 androgen-related genes obtained from GSE80609 and GSE120741 are presented in Fig 2C.
lncRNA-gene network analysis to understand gene expression patterns on treatment with R1881 or ENZA:
We assessed the effects of androgen and anti-androgen treatment on 15 lncRNAs and their interacting partners (i.e., androgen response pathway genes) using transcriptome sequencing data from GSE135879. Using Cytoscape, we generated lncRNA-gene network comprised of 71 nodes and 126 edges. Nodes represented 15 lncRNAs and 56 protein-coding androgen response pathway genes; edges were drawn based on the gene expression correlation between the lncRNA and androgen response genes. The centrality of each node (such as degree, betweenness, and closeness) was obtained using Cytoscape’s NetworkAnalyzer feature. Network analysis showed that MKLN1-AS1 had the highest correlation with androgen response genes (degree = 15). This was followed by AS-TUBA1B, CBLL1-AS1, PART1, IDH1-AS1, MIR99AHG and SOC2-AS1 with degrees of 14, 12, 11, 11, 11 and 10, respectively. PVT1, SNHG5, DUBR, AS-C6orf1and SLC38A4-AS1 correlated with at least 5 androgen-response pathway genes.
Nodes were colored based on the directionality of gene regulation, with red symbolizing up-regulation and blue representing down-regulation. Network plots showed the effects of R1881 and ENZA treatment on lncRNA-gene networks (Fig 3A and 3B). However, AS-TUBA1B and AS-C6orf1 showed similar expression on treatment with R1881 and ENZA compared to controls. In the validation datasets (GSE136272, GSE128749, and GSE130534), 9 lncRNA showed similar results as those for Figure 3 (Supplementary Figure 1). When treated with R1881 compared to control (GSE136272 and GSE128749), 6 lncRNAs (IDH1-AS1, PART1, CBLL1-AS1, NKILA, DUBR and SOC2-AS1) showed significant upregulation and 3 lncRNAs (COLCA1, DRAIC and MI99AHG) showed significant down regulation. When LNCAP cells were treated with ENZA (GSE130534), none of the lncRNAs showed differential regulation in comparison with controls.
Figure 3.
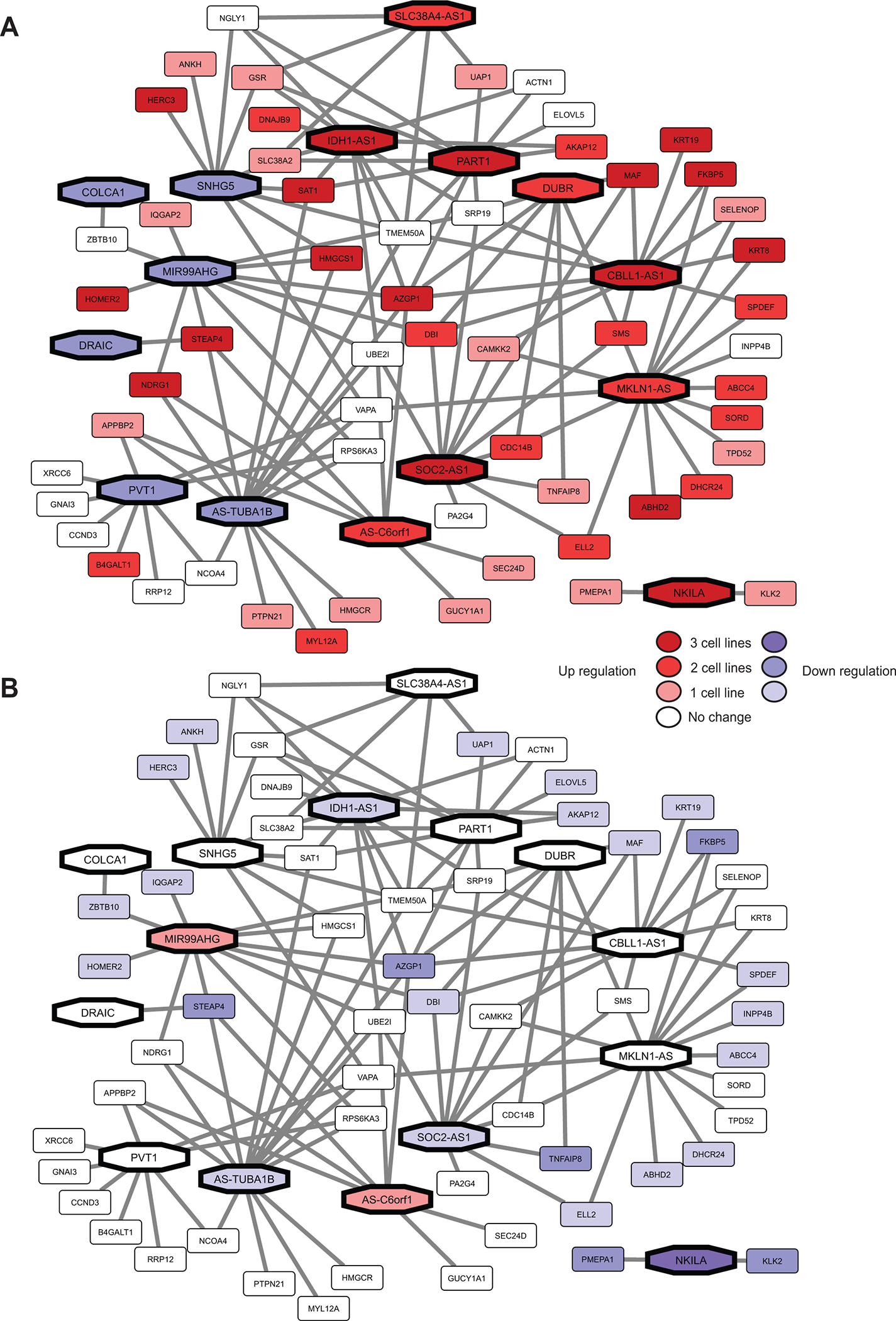
Network analysis of the selected lncRNA-gene pairs, which consists of 15 lncRNA and 56 androgen response genes. The expression pattern of 3 cell lines treated with (A) R1881 treatment compared to control and (B) ENZA compared with control. The darker the color represent of regulation (p<0.05) in more cell lines. (lncRNA: Long non-coding RNA, ENZA: enzalutamide)
ChIP sequencing data revealed a direct regulatory influence of androgen on lncRNA expression.
To elucidate the direct regulatory effect of AR on these 15 lncRNAs, we analyzed three independent AR ChIP-seq datasets (GSE55062, GSE83653, and GSE83860). From all three studies, an AR binding site was evident in the genomic region of MIR99AHG. In 2 of 3 studies, AR binding sites were present in the promoter regions of COLCA1, DRAIC, DUBR, and PVT1 (Fig 4).
Figure 4.
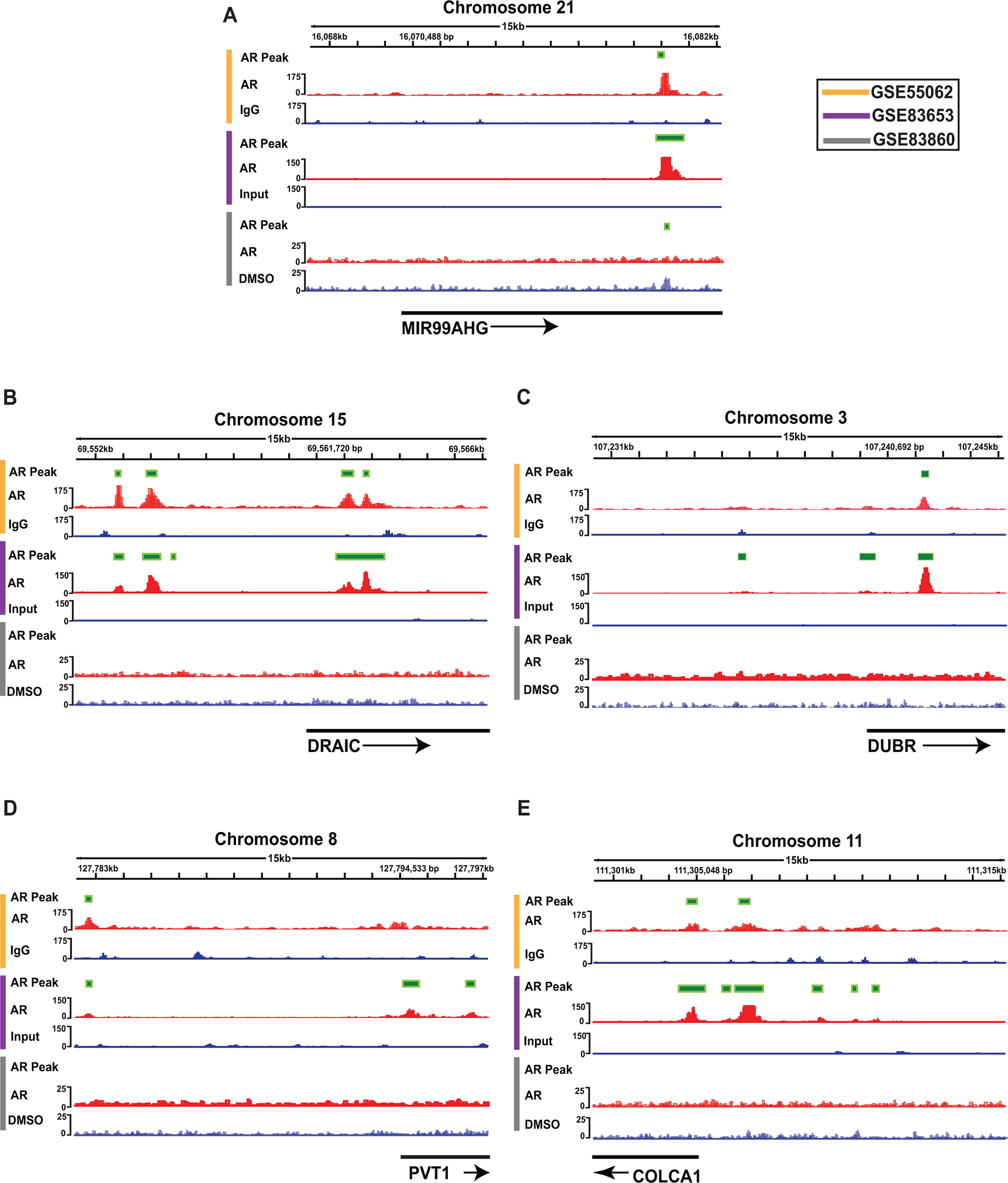
ChIP-Seq analysis for AR binding in the locus of androgen response regulated lncRNAs. ChIP-seq analysis revealed that AR binding sites were present in 5 lncRNAs in at least 2 studies that were assessed. ChIP-Seq Analysis of (A) MIR99AHG, (B) DRAIC, (C) DUBR, (D) PVT1, and (E) COLCA1. The color codes on the side of the images represent the study from which the results were obtained. (lncRNA: Long non-coding RNA, ENZA: enzalutamide, ChIP-seq: chromatin immunoprecipitation sequencing)
In silico promoter analysis to evaluate the AR regulatory influence on lncRNA gene expression.
Using the JASPAR database, 15 lncRNA promoter regions were analyzed for AR binding sites. The results are presented in Supplementary Table 1. In JASPAR analysis, COLCA1, MIR99AHG, IDH1-AS1, CBLL1-AS1, DUBR, and DRAIC had scores greater than 10. COLCA1 had the highest score of 18.1, followed by DRAIC, CBLL1-AS1, IDH1-AS1, MIR99HG, and DUBR. These scores suggest that the mentioned lncRNAs could have a putative binding site of AR (MA0007.2).
Discussion:
To understand the gene expression and molecular events in complex diseases such as cancers of various types, next generation sequencing is being widely applied. A meta-analysis with RNA sequencing and comparative transcriptomics using bioinformatics approach are useful to assess the changes in various disease conditions and after treatments. RNA-sequencing is one of the commonly used technologies and the generated data are made available for analysis as they are deposited in public repositories 43. These publicly available data has allowed in-depth analyses and interpretation of these data by researchers, leading to various discoveries and better understanding of cancer. The ability of RNA-seq to identify the expression patterns of genes other than protein-coding genes is an added advantage. However we understand that lncRNAs have lower expression levels compared to protein-coding transcripts, which can make them more difficult to detect in RNA-seq experiments44. There are tools such as FANTOM-CAT, which are widely used to understand the biological functions of lncRNA45. In order to understand the expression and regulation of lncRNAs in PCa, we utilized these publicly available RNA-sequencing data in the current study. We used various RNA-seq studies involving human PCa and the effects of R1881 and ENZA on PCa cells. To identify the lncRNA involved in Androgen response pathway, we used HALLMARK_ANDROGEN_RESPONSE gene set from Molecular Signatures Database (MSigDB). The Hallmark gene sets from MSigDB are collection of gene sets that have been curated to represent various biological pathways, processes, and states. The putative HALLMARK_ANDROGEN_RESPONSE gene set includes genes that are known to be activated or repressed in response to androgen signaling, such as androgen receptor (AR) and its target genes. The gene set was curated from the literature and from publicly available gene expression data sets. While the HALLMARK_ANDROGEN_RESPONSE may not be exhaustive, the gene set used in our analysis helped to identify and better understand the regulation of lncRNAs in androgen response in PCa. Our comparative analysis performed here led to identification of lncRNAs that were altered with the treatment of an androgen agonist (R1881) or antagonist (ENZA). Furthermore, ChIP-Seq data analysis confirmed binding of AR to the promoters of several androgen regulated lncRNAs. Although previous studies established that various lncRNAs are involved in the androgen response and in PCas 46,47, our investigation led to the identification of lncRNAs that are targets of the androgen –androgen receptor axis.
Herein, we used two statistical approach to analyze the RNA-sequencing data, namely the PCC and UCC methods. PCC is widely used in identifying the strength of linear relationships; UCC method is used to capture general or nonlinear relationships.
Some lncRNAs identified by UCC and PCC method are shown to be involved in androgen response and in PCa. In PCa cells, it has been shown that knockout of PART1 lncRNA suppresses cell proliferation and apoptosis by modulating toll-like receptors 48. For LNCaP cell lines, PART1 has been shown to regulate the androgen response49. Similar to the results we found in our analysis, it has been shown that SNHG5 is downregulated by DHT 47. In PCa, IDH1-AS1 has been shown to promote tumor growth by inducing up-regulation of PAX5, but its role in the androgen response needs to be explored 50. DRAIC, localized in the cytoplasm, is a tumor suppressor that is regulated by AR and FOXA1; this lncRNA suppresses metastasis and cellular migration 51. When cells are treated with R1881, DRAIC is down-regulated 51; and we saw the same result in our analysis. Further, repression of DRAIC is required to recruit AR in the promoter region 51. SOCS2-AS1 is an androgen-inducing lncRNA that, in PCa cells, promotes cell growth and inhibits apoptosis. SOC2-AS1 promotes castration-resistant and androgen-dependent cell growth 52. Knockdown of PVT1 inhibits PCa growth, promotes apoptosis, and regulates hundreds of genes, of which 160 genes are involved in androgen repression 53,54. Also, higher levels of PVT1 relate to poorer overall and disease-free survival55. We identified other lncRNAs, including COLCA1, IRF1-AS1, MKLN1-AS, CBLL1-AS1, DUBR, MIR99AHG, and SLC38A4-AS1. These are potentially involved in the androgen response, and which may function as biomarkers. The ChIP-Seq analysis of 15 lncRNAs identified 5 lncRNAs (MIR99AHG, DUBR, DRAIC, COLCA1 and PVT1) that had an AR binding site near the genomic or promoter region. The transcriptomic and ChIP-Seq analyses point to the importance of these lncRNAs and their regulations in androgen response.
Our current study utilized only transcriptomic data. Similar comparative analysis with knockout of identified lncRNAs will provide tailored information on the regulation of androgen response genes. The current bioinformatics approach makes use of available data to identify lncRNAs involved in this response. A future direction is to experimentally validate association of these lncRNAs with androgen response genes in PCa cell lines, as well as evaluate the prognostic potential of these lncRNAs using the PCa patient RNA-seq data.
Conclusion:
By analyzing various sets of RNA-seq and ChIP-seq data, we found 15 lncRNAs that could be involved in the PCa-related androgen response pathway. Furthermore, 5 lncRNAs showed AR binding sites at their genomic or promoter regions, suggesting that these are regulated by androgen binding. We also identified some lncRNAs that could be useful as biomarkers. Future studies will focus on validation of some these androgen regulated lncRNAs and their biological functions and impact in PCa progression.
Supplementary Material
Supplementary figure 1. Validation of lncRNA-gene network findings in other RNA-seq studies. (A) Expression pattern of the lncRNA-gene network with R1881 treatment compared with control in a 3 cell line study, from 2 RNA-seq data (GSE136272 and GSE128749). The darker the color represent the regulation in more cell lines. (lncRNA: Long non-coding RNA)
Supplementary table 1. Transcription factor binding site analysis of promoter region of lncRNAs using JASPAR. The Excel file presents the results obtained from JASPAR. Each hit with a relative profile score threshold above 80% was selected and presented. (lncRNA: Long non-coding RNA)
Acknowledgments:
This study was supported by the UAB Department of Pathology, the UAB O’Neal Comprehensive Cancer Center, and the UAB Heersink School of Medicine. S.V. and U.M were supported by funding from U54 CA118948. S.V. was also supported by funding from DOD, W81XWH-19-1-0588. Dr. Donald Hill from the UAB Department of Pathology provided help in editing this manuscript.
Funding/support:
Department of Pathology, UAB. S.V. and U.M were supported by U54CA118948. S.V. was also supported by DOD funding, W81XWH-19-1-0588.
Footnotes
Conflict of Interests: The authors declare no conflict of interests.
References:
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. Jan 2021;71(1):7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Fontana F, Anselmi M, Limonta P. Molecular mechanisms and genetic alterations in prostate cancer: From diagnosis to targeted therapy. Cancer Lett. May 28 2022;534:215619. doi: 10.1016/j.canlet.2022.215619 [DOI] [PubMed] [Google Scholar]
- 3.Ozturan D, Morova T, Lack NA. Androgen Receptor-Mediated Transcription in Prostate Cancer. Cells. Mar 5 2022;11(5)doi: 10.3390/cells11050898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. Feb 2009;6(2):76–85. doi: 10.1038/ncpuro1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. Jul 23 2009;138(2):245–56. doi: 10.1016/j.cell.2009.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke JA, Guns ES, Lubik AA, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. Aug 1 2008;68(15):6407–15. doi: 10.1158/0008-5472.CAN-07-5997 [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Liu KY, Liu Q, Cao Q. Androgen Receptor-Related Non-coding RNAs in Prostate Cancer. Front Cell Dev Biol. 2021;9:660853. doi: 10.3389/fcell.2021.660853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra K, Kanduri C. Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. Noncoding RNA. Dec 3 2019;5(4)doi: 10.3390/ncrna5040054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. Nov 2011;33(11):830–9. doi: 10.1002/bies.201100084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Li M, Li J, et al. Comprehensive Characterization of Androgen-Responsive lncRNAs Mediated Regulatory Network in Hormone-Related Cancers. Dis Markers. 2020;2020:8884450. doi: 10.1155/2020/8884450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. Aug 29 2013;500(7464):598–602. doi: 10.1038/nature12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickard MR, Williams GT. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes (Basel). Jul 7 2015;6(3):484–99. doi: 10.3390/genes6030484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. Aug 6 2010;142(3):409–19. doi: 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik R, Patel L, Prensner JR, et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res. Aug 2014;12(8):1081–7. doi: 10.1158/1541-7786.MCR-14-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang G, Su Z, Liang X, Huang Y, Lan Z, Jiang X. Long non-coding RNAs in prostate tumorigenesis and therapy (Review). Mol Clin Oncol. Dec 2020;13(6):76. doi: 10.3892/mco.2020.2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taheri M, Khoshbakht T, Jamali E, Kallenbach J, Ghafouri-Fard S, Baniahmad A. Interaction between Non-Coding RNAs and Androgen Receptor with an Especial Focus on Prostate Cancer. Cells. Nov 16 2021;10(11)doi: 10.3390/cells10113198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang A, Zhao JC, Kim J, et al. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. Oct 6 2015;13(1):209–221. doi: 10.1016/j.celrep.2015.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takayama K, Horie-Inoue K, Katayama S, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. Embo j. Jun 12 2013;32(12):1665–80. doi: 10.1038/emboj.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira LB, Palumbo A, de Mello KD, et al. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer. Nov 6 2012;12:507. doi: 10.1186/1471-2407-12-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lingadahalli S, Jadhao S, Sung YY, et al. Novel lncRNA LINC00844 Regulates Prostate Cancer Cell Migration and Invasion through AR Signaling. Mol Cancer Res. Dec 2018;16(12):1865–1878. doi: 10.1158/1541-7786.MCR-18-0087 [DOI] [PubMed] [Google Scholar]
- 21.Dai X, Liu L, Liang Z, Guo K, Xu S, Wang H. Silencing of lncRNA MALAT1 inhibits cell cycle progression via androgen receptor signaling in prostate cancer cells. Pathol Res Pract. Apr 2019;215(4):712–721. doi: 10.1016/j.prp.2019.01.011 [DOI] [PubMed] [Google Scholar]
- 22.Stelloo S, Nevedomskaya E, Kim Y, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun. Nov 21 2018;9(1):4900. doi: 10.1038/s41467-018-07270-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun SJ, Kim SK, Kim J, et al. Transcriptomic features of primary prostate cancer and their prognostic relevance to castration-resistant prostate cancer. Oncotarget. Dec 29 2017;8(70):114845–114855. doi: 10.18632/oncotarget.22296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyquist MD, Corella A, Mohamad O, et al. Molecular determinants of response to high-dose androgen therapy in prostate cancer. JCI Insight. Oct 3 2019;4(19)doi: 10.1172/jci.insight.129715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brzezinka K, Nevedomskaya E, Lesche R, et al. Characterization of the Menin-MLL Interaction as Therapeutic Cancer Target. Cancers (Basel). Jan 14 2020;12(1)doi: 10.3390/cancers12010201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poluri RTK, Beauparlant CJ, Droit A, Audet-Walsh E. RNA sequencing data of human prostate cancer cells treated with androgens. Data Brief. Aug 2019;25:104372. doi: 10.1016/j.dib.2019.104372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Handle F, Prekovic S, Helsen C, et al. Drivers of AR indifferent anti-androgen resistance in prostate cancer cells. Sci Rep. Sep 24 2019;9(1):13786. doi: 10.1038/s41598-019-50220-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. Jun 12 2014;510(7504):278–82. doi: 10.1038/nature13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bose R, Karthaus WR, Armenia J, et al. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature. Jun 29 2017;546(7660):671–675. doi: 10.1038/nature22820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malinen M, Niskanen EA, Kaikkonen MU, Palvimo JJ. Crosstalk between androgen and pro-inflammatory signaling remodels androgen receptor and NF-kappaB cistrome to reprogram the prostate cancer cell transcriptome. Nucleic Acids Res. Jan 25 2017;45(2):619–630. doi: 10.1093/nar/gkw855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. doi:doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. Apr 25 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. Nov 1 2011;27(21):2987–93. doi: 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. Jan 15 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. Jul 15 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. Mar 15 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu N, Huang X, Huang S. A measure of general functional dependence between two continuous variables. Communications in Statistics - Theory and Methods. 2017/05/03 2017;46(9):4327–4352. doi: 10.1080/03610926.2015.1081951 [DOI] [Google Scholar]
- 39.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. Jan 7 2022;50(D1):D165–D173. doi: 10.1093/nar/gkab1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. Nov 2003;13(11):2498–504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. Jan 2011;29(1):24–6. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. May 22 2015;16:169. doi: 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. Jan 2009;10(1):57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atkinson SR, Marguerat S, Bähler J. Exploring long non-coding RNAs through sequencing. Seminars in Cell & Developmental Biology. 2012/04/01/ 2012;23(2):200–205. doi: 10.1016/j.semcdb.2011.12.003 [DOI] [PubMed] [Google Scholar]
- 45.Imada EL, Sanchez DF, Collado-Torres L, et al. Recounting the FANTOM CAGE-Associated Transcriptome. Genome Res. Jul 2020;30(7):1073–1081. doi: 10.1101/gr.254656.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. Nov 2017;108(11):2107–2114. doi: 10.1111/cas.13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan X, Huang W, Yang S, et al. Identification of androgen-responsive lncRNAs as diagnostic and prognostic markers for prostate cancer. Oncotarget. Sep 13 2016;7(37):60503–60518. doi: 10.18632/oncotarget.11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun M, Geng D, Li S, Chen Z, Zhao W. LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol Chem. Mar 28 2018;399(4):387–395. doi: 10.1515/hsz-2017-0255 [DOI] [PubMed] [Google Scholar]
- 49.Lin B, White JT, Ferguson C, et al. PART-1: a novel human prostate-specific, androgen-regulated gene that maps to chromosome 5q12. Cancer Res. Feb 15 2000;60(4):858–63. [PubMed] [Google Scholar]
- 50.Zhang N, Li Z, Bai F, Zhang S. PAX5-induced upregulation of IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy. Cell Death Dis. Sep 30 2019;10(10):734. doi: 10.1038/s41419-019-1932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakurai K, Reon BJ, Anaya J, Dutta A. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res. May 2015;13(5):828–38. doi: 10.1158/1541-7786.MCR-15-0016-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Misawa A, Takayama K, Urano T, Inoue S. Androgen-induced Long Noncoding RNA (lncRNA) SOCS2-AS1 Promotes Cell Growth and Inhibits Apoptosis in Prostate Cancer Cells. J Biol Chem. Aug 19 2016;291(34):17861–80. doi: 10.1074/jbc.M116.718536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Videira A, Beckedorff FC, daSilva LF, Verjovski-Almeida S. PVT1 signals an androgen-dependent transcriptional repression program in prostate cancer cells and a set of the repressed genes predicts high-risk tumors. Cell Commun Signal. Jan 11 2021;19(1):5. doi: 10.1186/s12964-020-00691-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Li C, Mudd A, Gu X. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Biosci Biotechnol Biochem. Dec 2017;81(12):2301–2306. doi: 10.1080/09168451.2017.1387048 [DOI] [PubMed] [Google Scholar]
- 55.Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in cancer: A review and meta-analysis. Clin Chim Acta. Nov 2017;474:1–7. doi: 10.1016/j.cca.2017.08.038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Validation of lncRNA-gene network findings in other RNA-seq studies. (A) Expression pattern of the lncRNA-gene network with R1881 treatment compared with control in a 3 cell line study, from 2 RNA-seq data (GSE136272 and GSE128749). The darker the color represent the regulation in more cell lines. (lncRNA: Long non-coding RNA)
Supplementary table 1. Transcription factor binding site analysis of promoter region of lncRNAs using JASPAR. The Excel file presents the results obtained from JASPAR. Each hit with a relative profile score threshold above 80% was selected and presented. (lncRNA: Long non-coding RNA)


