Abstract
Non-melanoma skin cancer (NMSC) is a tumor that arises from human keratinocytes, showing abnormal control of cell proliferation and aberrant stratification. Cutaneous basal cell carcinoma (cBCC) and cutaneous squamous cell carcinoma (cSCC) are the most common sub-types of NMSC. From a molecular point of view, we are still far from fully understanding the molecular mechanisms behind the onset and progression of NMSC and to unravel targetable vulnerabilities to leverage for their treatment, which is still essentially based on surgery. Under this assumption, it is still not elucidated how the central cellular metabolism, a potential therapeutical target, is involved in NMSC progression. Therefore, our work is based on the characterization of the serine anabolism/catabolism and/or one-carbon metabolism (OCM) role in NMSC pathogenesis. Expression and protein analysis of normal skin and NMSC samples show the alteration of the expression of two enzymes involved in the serine metabolism and OCM, the Serine Hydroxy-Methyl Transferase 2 (SHMT2) and Methylen-ThetraHydroFolate dehydrogenase/cyclohydrolase 2 (MTHFD2). Tissues analysis shows that these two enzymes are mainly expressed in the proliferative areas of cBCC and in the poorly differentiated areas of cSCC, suggesting their role in tumor proliferation maintenance. Moreover, in vitro silencing of SHMT2 and MTHFD2 impairs the proliferation of epidermoid cancer cell line. Taken together these data allow us to link the central cellular metabolism (serine and/or OCM) and NMSC proliferation and progression, offering the opportunity to modulate pharmacologically the involved enzymes activity against this type of human cancer.
Subject terms: Cancer metabolism, Skin cancer
Introduction
Non-melanoma skin cancer (NMSC) represents the most common diagnosed cancer worldwide [1]. Cutaneous basal cell carcinoma (cBCC) and cutaneous squamous cell carcinoma (cSCC) are the most representative types of non-melanoma skin cancer. In general, cBCC and cSCC present different molecular pathogenesis, growth, and aggressiveness. Although there is a high incidence of these tumors [2–4], the survival is quite high because they are typically recognized in time, but the absolute number of deaths is similar to that of melanoma [5].
The cBCC is characterized by cancer cells reminiscent of the proliferative basal cells of the epidermis; preferentially arises from interfollicular stem cells, hair follicle infundibulum cells or hair follicle bulge cells, and it is the less aggressive NMSC [6, 7]; usually, cBCC shows a limited capability to invade and destroy the adjacent tissues and for these reasons it shows a low degree of malignancy [8]. Several risk factors related to cBCC insurgence have been identified, i.e. immunosuppression, genetic syndromes and skin photosensitivity (Fitzpatrick skin types I and II); but ultraviolet radiation (UV) represents the most prominent and primary risk factor for cBCC development, although the link between cBCC and UV exposure is still quite controversial [1]. Different studies demonstrate that a cumulative UV exposure determines changes in skin protein expression [9]. In fact, a chronic UV exposure leads to the increase of Reactive Oxygen Species (ROS) production, DNA damage (pyrimidine dimers) and p53-dependent apoptosis in human normal keratinocytes [10]. These molecular changes are often followed by tissue alterations, such as hyperproliferation and epidermal hyperplasia, representing the initial steps of NMSC development, which have also been proven to be fundamental in animal models of the disease [11, 12].
The cSCC develops from abnormal epidermis squamous cells and it is characterized by a strong migration and invasion capability. Tumor aggressiveness and recurrence depends on histological features such as its pathological grade and on anatomic localization. Patients with this type of cancer have a ten-year survival of about 90%, but the recurrences are very frequent. Therefore, cSCC represents the second cause of death for skin cancer after melanoma [13]. Among the risk factors for cSCC development we can mention skin photosensitivity (Fitzpatrick skin types I and II), immunosuppression, HIV, Human Papilloma Virus (HPV) infection, a chronic UV exposure and a previous story of actinic keratosis (AK) [14–17]. As well as cBCC, the chronic UV exposure is the most important environmental factor involved in cSCC insurgence; indeed, this tumor often arises on sun-exposed area [18]. In addition, different studies indicate that AK, typically manifested on skins chronically exposed to the sun-light, is a premalignant lesion that often herald cSCC development [19]. For this reason, AK is considered the most significant predictive factor for cSCC development, beneath not already identified prognostic signatures have been defined at a molecular level [20, 21]. Surgical excision is the first therapy against NMSC; however, some alternative approaches can be used, i.e. photodynamic therapy, cryotherapy, topical imiquimod 5%, and topical diclofenac sodium 3% [8, 22, 23]. SCCs also arises from squamous cells that line in the mucosal surfaces of the head and neck (HNSCC) including oral cavity, pharynx, larynx, tongue as well an in lung [24, 25]. HNSCCs are the sixth most common cancer worldwide [26]. Tobacco and alcohol are the primary risk factors, as well as virus infection [27–30]. Similarly to cSCCs, HNSCC incidence is continuously rising, and it is expected to increase by 30% by 2030. Unlike cSCCs, one of the main factors affecting the low survival rate of HNSCC, is the high percentage of diagnosis at advanced stage, being the recurrence rate of about 50% during the first 2 years from diagnosis. For this reason, several clinical and/or pathological parameters have been indicated to predict prognosis, recurrence, and survival, and to drive the therapeutic choice [31–37]. Notably, cSCCs and HNSCCs display hallmarks of solid tumors mildly responsive to systemic immunotherapy [38–45]. Thus, immunotherapy with PD-1 blockade has emerged as the latest standard-of-care treatment strategy developed for advanced and metastatic SCCs. A crucial step of NMSC and HNSCC development is the alteration of normal epidermal cell proliferation; indeed, several studies report that cancer cells can adapt their metabolism to achieve this effect [46–52]. The term one-carbon metabolism (OCM) refers to a complex metabolic pathway involved in the generation of one-carbon units (hydroxymethyl groups) used by the cells for the biosynthesis of fundamental anabolic precursors, for cellular redox homeostasis and for methylation reactions [53]. The importance of OCM in cancer was recognized when it was observed that a diet poor of folate in children with acute leukaemia was able to reduce the number of their leukaemic cells but, at date, the involvement of this metabolism in the pathogenesis of different cancer types is still under evaluation [54]. In general, the alteration of serine and OCM is strongly associated with different types of cancer with a high rate of cellular proliferation [55]. The OCM reactions have a pivotal role in the production of pyrimidine and purine nucleotides (DNA and RNA synthesis), S-adenosylmethionine (SAM), necessary for DNA methylation and to produce reduced GSH for ROS scavenging [56]. Closely related to OCM is the serine biosynthesis pathway, an alternative route of glycolysis in which glucose is converted in serine through the action of three enzymes: phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1) and phosphoserine phosphatase (PSPH) [57]. After the generation of serine form glucose, the amino acid is metabolized both in the cytosol and in the mitochondria through the activity of Serine Hydroxy-Methyl Transferase enzymes (SHMT1 and SHMT2 enzymes, respectively), thus representing the major source of one-carbon units. In general, the interconnection of serine anabolic/catabolic reactions and OCM are a crucial metabolic hub for cellular energy production, regulation of proliferation rate, and cellular differentiation and for these reasons it is essential to describe a possible way to reprogram these metabolisms in cancer. This work lays the foundation for identifying the link between serine and/or OCM metabolism and abnormal proliferation of NMSC, of the most aggressive (cSCC), trying to identify possible metabolic vulnerabilities against this type of cancer.
Results
Serine and OCM enzymes are differentially expressed in NMSC
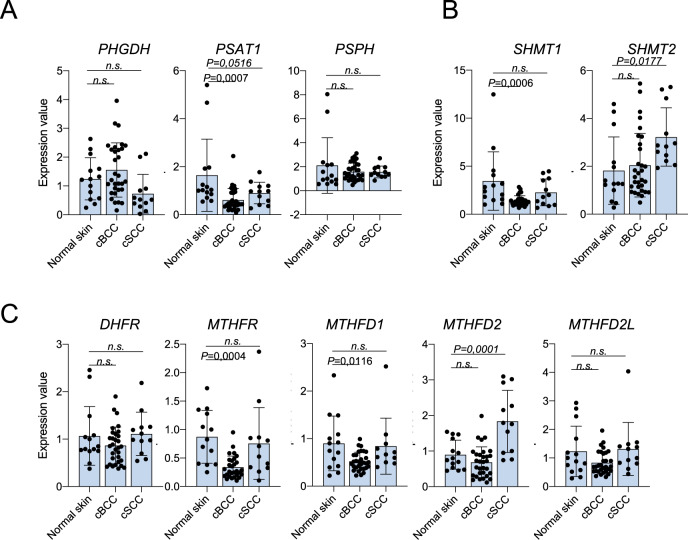
Although the involvement of serine metabolism and OCM in the proliferation and progression of different human malignancies [46, 57] is, at date, quite well described, its pathogenic role in NMSC has yet to be investigated. We analyzed the expression of the enzymes involved in serine de novo biosynthesis (PHGDH, PSAT1 and PSPH), the enzymes involved in serine anabolism/catabolism reactions in the cytoplasm and in the mitochondria (SHMT1 and SHMT2, respectively) and the cytoplasmic and mitochondrial enzymes involved in the formation of the cofactor tetrahydrofolate (THF), DHFR, MTHFR, MTHFD1, MTHFD2, MTHFD2L. The mRNA expression was analyzed using collected human samples of normal skin (n = 14), cBCC (n = 31) and cSCC (n = 12), Fig. 1. mRNA levels, evaluated through RT-qPCR (Fig. 1A), showed the mitochondrial enzymes SHMT2 and MTHFD2 differentially expressed in tumor samples of cSCC, the most aggressive NMSC. While in cBCC we observed the reduced expression of PSAT1, SHMT1, MTHFR, MTHFD1. To expand these results, we interrogated a publicly available dataset (GSE7553) of cBCC (n = 15), cSCC (n = 11) and normal skin (n = 4) as the healthy control. The mRNA expression values are represented by boxplots in Supplementary Fig. S1A–C. The small number of samples did not allow us to find striking changes in NMSC gene expression for many of the enzymes analyzed, compared to their expression in normal skin, except for two genes whose expression is significantly reduced in cBCC (SHMT1 P = 0,046 and MTHFD2L P = 0,0004). Only one gene expression was increased in cSCC (PSPH P = 0,0295) compared to normal skin. These data indicate a possible involvement of serine and OCM metabolism in NMSC.
Fig. 1. Expression analysis of serine de novo biosynthesis, serine anabolism/catabolism and one-carbon metabolism enzymes in human samples of normal skin, cBCC and cSCC.
A Expression analysis through RTqPCR of serine biosynthesis, B SHMT1 and SHMT2 and C one-carbon metabolism enzymes in a cohort of human samples collected from IDI-IRCCS -normal skin (n = 14), cBCC (n = 31), cSCC (n = 12). The p-value was obtained using Student’s t test. Values were considered significant when the p-value < 0.05, n.s. not significant.
SHMT2 and MTHFD2 are differentially expressed in cSCC and BCC
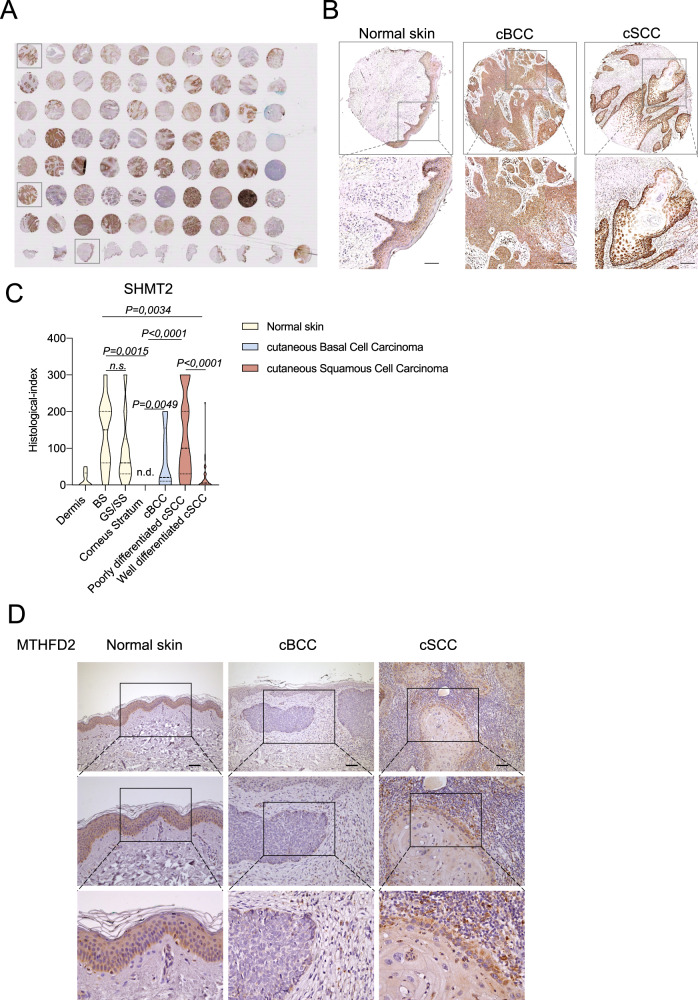
To further investigate the role of the modulated enzymes, additional NMSC samples from commercially available Tissue Micro Array (TMA), were analyzed (Fig. 2A). Specifically, 14 and 43 samples of cBCC and cSCC were analyzed for SHMT2 expression, respectively. NMSC samples were compered to 10 samples of normal skin. The immunohistochemistry performed on TMA, indicates an increased expression of SHMT2 in the basal layer of the skin compared to the upper differentiated layers. In cBCC representative samples SHMT2 was detected in all tumor cells, in contrast to cSCC representative samples where the expression of SHMT2 decreased in the differentiated area of the tumor (Fig. 2B). This evidence was confirmed by the analysis of the histological score, obtained by a combination of the SHMT2 intensity expression and the percentage of the tumor sample positive for its expression. The differentiated upper layers of human skin have a reduced SHMT2 expression compared to the basal undifferentiated layer and similarly SHMT2 is significantly less expressed in the well differentiated areas of cSCC compared to the poorly differentiated ones (Fig. 2C). Globally, SHMT2 expression in cBCC is strongly up regulated compared to the last differentiated layer of human skin, the corneum stratum.
Fig. 2. SHMT2 and MTHFD2 are differentially expressed in normal skin and Non-Melanoma skin cancer.
A Immunohistochemical staining for SHMT2 performed on Tissue Micro Array (TMA), containing cases of normal skin (n = 10), cutaneuos Basal Cell Carcinoma (n = 14) and cutaneous Squamous Cell carcinoma (n = 43). B SHMT2 expression in 3 representative samples of normal skin, cBCC and cSCC. C Histological index of SHMT2 expression in normal skin (divided into Dermis, Basal Stratum-BS-, Granulosum Stratum-GS-, Spinous Stratum-SS-, and Corneum Stratum), cBCC and cSCC (divided into Well differentiated cSCC and Poorly differentiated cSCC). D Immunohistochemical staining for MTHFD2 performed on FFPE tissues of normal skin, cutaneuos Basal Cell Carcinoma (cBCC) and cutaneous Squamous Cell carcinoma (cSCC). Scale Bar = 100 μm. The p-value was obtained using Student’s t test. Values were considered significant when the p-value < 0.05, n.s. not significant.
MTHFD2 staining shows that the enzyme is abundantly expressed in the basal and suprabasal levels of human normal skin and in poorly differentiated areas of cSCC, while it is less expressed in BCC (Fig. 2D). These data are consistent to the MTHFD2 expression at mRNA levels (Fig. 1). These observations suggest that serine catabolism/anabolism and OCM enzymes are differently involved in cBCC and cSCC.
In vitro modulation of the mitochondrial enzymes SHMT2 and MTHFD2, acts on cSCC proliferation
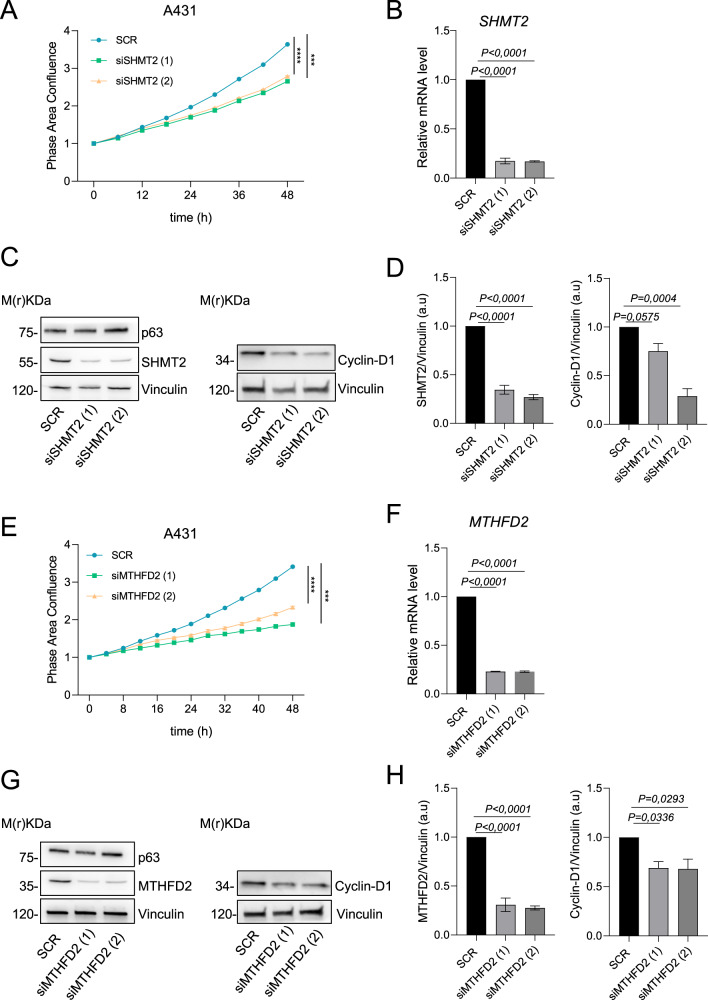
Next, we focused the attention on the SHMT2 and MTHFD2, the two mitochondrial enzymes significantly modulated at mRNA and protein level in cSCCs. The mitochondrial enzymes SHMT2 and MTHFD2 are involved in fundamental metabolic pathways, serine and THF metabolism, respectively. To characterize their functional role in cancer, we decided to modulate their expression in A431 cells, using two transient and specific siRNAs. The evaluation of cellular proliferation was made using Incucyte® Live-Cell analysis. The confluence curves show a significant reduction of A431 growth after in vitro silencing of SHMT2 and MTHFD2 and confirms the possible involvement of these enzymes in controlling cSCC proliferation (Fig. 3A–H). As control, by RT-qPCR and western blot, we confirmed SHMT2 and MTHFD2 knockdown (Fig. 3B–F, C–G). In line with the growth curve results, western blots confirmed the reduction in the expression of cyclin D1, after 72 hours silencing of SHMT2 and MTHFD2 (Fig. 3C, D, G, H). These data globally confirm the involvement of serine metabolism and OCM in NMSC proliferation, serving as hypothetical targets against this tumor.
Fig. 3. SHMT2 and MTHFD2 silencing effects on A431 proliferation.
A Confluence analysis of A431 cells after SHMT2 silencing. B SHMT2 expression analysis through qPCR after silencing. C Western blot analysis after SHMT2 silencing, one representative experiment of 3 is shown. D Relative ratio of SHMT2 and Cyclin-D1 expression normalized to that of Vinculin. E Confluence analysis of A431 cells after MTHFD2 silencing. F MTHFD2 expression analysis through qPCR after silencing. G Western blot analysis after MTHFD2 silencing, one representative experiment of 3 is shown in H. Relative ratio of MTHFD2 and Cyclin-D1 expression normalized to that of Vinculin. The p-value was obtained using Student’s t test and One-Way Anova. Values were considered significant when the p-value < 0.05, n.s. not significant.
Discussion
Human epidermis is a multilayer stratified epithelium, consisting of keratinocytes and melanocytes, which represent the most common cell types of the tissue. In particular, the physiological keratinocyte proliferation, differentiation and stratification are essential for normal epidermis formation and homeostasis [58]. Alterations of this phenomenon and the insurgence of others such as the epithelial-to-mesenchymal transition are responsible of different skin disorders, including NMSC [59]. A strict metabolic regulation is crucial for normal cell fate and our work focus on the importance of this metabolic control in NMSC. Serine and OCM represent the major source of anabolic precursors for cellular energy production, DNA methylation and redox balance [60–62]. SHMT2 and MTHFD2 enzymes are two essential turning points of serine and OCM, respectively. SHMT2 catalyzes, in the mitochondrial compartment, the reversible conversion of serine and THF to glycine and 5,10-methylen-THF to guarantee the one-carbon units for the THF cycle [63], while the mitochondrial enzyme MHFD2 is responsible of the 10-formylTHF production through two different reactions catalyzed by the dehydrogenase and cyclohydrolase domains [64].
The enzymatic activity of SHMT2 and MTHFD2 is often deregulated in different types of human cancer affecting the cellular mechanism above-mentioned (DNA methylation and synthesis, redox balance, and protein synthesis) [65, 66]. Recent studies have led to the development of chemical inhibitors specifically targeting SHMT2 and MTHFD2, potentially promising as treatments to impair proliferation and aggressiveness of cancer cells [67]. To date, nothing is known about the involvement of these enzymes in NMSC progression; our preliminary bioinformatic analysis showed a trend to the de-regulation of the genes involved in serine and OCM in NMSC, if compared to normal skin (Supplementary Fig. 1A–C). However, RT-qPCR analysis on a larger number of samples increased the level of this evidence since SHMT2 and MTHFD2 are significantly up-regulated in cSCC, the more aggressive NMSC (Fig. 1A, C). The analysis of additional samples of NMSC on TMA confirms that SHMT2 is more expressed in the proliferative layer of human skin compared to the differentiated ones (Fig. 2B, C). Interestingly, SHMT2 is expressed in all cBCC cells, in contrast to cSCC, where SHMT2 is significantly downregulated in the more differentiated areas of the tumor (Fig. 2C). In addition, similarly to SHMT2, also MTHFD2 is more expressed in proliferating cells of cSCC (Fig. 2D), suggesting that these cells rely on this metabolism to sustain their growth. Finally, the in vitro modulation of SHMT2 and MTHFD2 determines the reduction of A431 cells ability to undergo to a complete cellular confluence (Fig. 3A, E) and the decrease of the proliferation markers cyclin D1 (Fig. 3C, D, G, H). Taken together these data show the fundamental role of serine and OCM, in particular of the mitochondrial enzymes SHMT2 and MTHFD2, in NMSC pathogenesis suggesting hypothetical metabolic vulnerabilities on which to act in the treatment of these types of tumors.
Material and methods
Cell culture, transfection, and proliferation assay
A431 cells was cultured in DMEM (Corning, Cat. No 10-102-CVR). For the Phase Area Confluence analysis, Incucyte® Live-Cell Analysis (Essen BioScience) was used. For siRNA-mediated knockdown experiments, 2,5 × 105 cells were seeded in 60 mm plates and transfected with specific siRNAs (Supplementary Table 2) using Lipofectamine RNAiMAX transfection reagent (Invitrogen). The cells were collected 72 h after transfection.
Immunohistochemical staining and histological scoring
TMA sections (purchased from US Biomax, Cat. No. SK801c, Rockville, MD, USA) were dewaxed for 2 h at 60 °C, then treated with Bio-Clear (Bio Optica) and rehydrated with alcohol scale and ddH2O. Immunohistochemical staining was performed using UltraTek HRP anti-polyvalent (DAB) (ScyTek laboratories, Cat. No. AMF080). Samples were boiled at 95 °C for 15 min in sodium citrate buffer at pH 6.0 for antigen retrieval. Anti-SHMT2 antibody was incubated (1:100, Sigma Cat. No. HPA020549) for 1 h. Anti-MTHFD2 antibody was incubated (1:500, Abcam Cat. No. ab151447) for 1 h. Sections were counterstained with Mayer’s haematoxylin (Bioptica, Cat. No. 05-06002E).
Histological index was visually analyzed in a blinded manner by a pathologist using a semi-quantitative method. NMSC cases were analyzed for staining intensity: 0 (not detected), 1 + (weak), 2 + (intermediate), and 3 + (strong). For each case, the histological “H-score” (0–300) was calculated by multiplying the percentage of positive cells (0–100%) by the intensity (0–3).
Immunoblotting
A431 cells were lysed with Ripa lysis buffer (50 mM Tris-cl pH 7.4; 150 mM NaCl; 1% NP40; 0.25% Na-deoxycholate; 1 mM AEBSF; and 1 mM DTT). The total protein extracts (20 µg) were separated using SDS polyacrylamide gels and transferred on PVDF membrane, using Trans-Blot Turbo Transfer System. Then, the membranes were incubated over-night using the following antibodies: anti-SHMT2 (Sigma, Cat. No. HPA020549, 1:500), anti-CycD1 (Santa Cruz, Cat. No. sc-8396 1:500), anti-MTHFD2 (Abcam Cat. No. ab151447 1:1000), anti-p63-α (Cell Signalling, Cat. No. 13109, 1:500) and anti-vinculin (Sigma, Cat. No. V9131 1:10000). The western blot signals were captured using Alliance™ Q9-ATOM Light.
RNA extraction and RT-qPCR
A431 cells were lysed in RLT lysis buffer (Qiagen). The Total RNA was isolated and purified using the RNeasy Mini Kit (Qiagen). The RNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific). Total RNA was used for cDNA synthesis with a SensiFAST™ cDNA synthesis kit (Bioline). The qPCR was performed with a Fast SYBR™ Green Master Mix (Applied Biosystems) in an QuantStudio Real-Time PCR Systems (Applied Biosystems) using appropriate qPCR primers (Supplementary Table 1). TBP was used as housekeeping genes for data normalization. The expression of each gene was defined by the threshold cycle (Ct), and relative expression levels were calculated by using the 2 − ΔΔCt method.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0 software (San Diego, CA, USA); Student’s t test was used for all the analysis except for confluence analysis in which One-way Anova statistical analysis was used. Values of P < 0.05 and P < 0.01 were considered statistically significant.
Supplementary information
Supplementary Figure 1 and Supplementary Table 1 and 2
Acknowledgements
We thank Dr Artem Smirnov, Dr Anna Maria Lena and Dr Carlo Ganini for discussion and support.
Author contributions
EC designed research; AC performed most of the experiments, bioinformatical analysis, qPCR and immunohistochemical staining; AZ performed the experiments on A431 cells; MM, LF and FR collected the human samples and extracted the relative RNA; GT performed the IHC analysis of MTHFD2; EC, GM and AC discussed and analyzed the data; AC and EC wrote the paper.
Funding
This work was mainly supported by Ministry of Health and IDI-IRCCS, Grant RF-2019-12368888 RicercaCorrente 2022 and European Union-NextGenerationEU through the Italian Ministry of University and Research under PNRR-M4C2-I1.3 Project PE_00000019 "HEAL ITALIA" (to EC). Partially supported by LazioInnova, Grant A0375-2020-36568 UTV-IDI (to EC). Funding from the Associazione Italiana Ricerca sul Cancro (AIRC) under IG Grant 22206 to EC is also gratefully acknowledged.
Data availability
For Serine biosynthesis and One Carbon metabolism enzymes expression at mRNA level a public database available through GEO datasets (https://www.ncbi.nlm.nih.gov/gds) was analyzed (GSE7553).
Competing interests
The authors declare no competing interests.
Ethics statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Bioethics Committee (Protocol No. 552/1, December 14, 2018). Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-023-01398-x.
References
- 1.Didona D, Paolino G, Bottoni U, Cantisani C. Non Melanoma Skin Cancer Pathogenesis Overview. Biomedicines. 2018;6:6. doi: 10.3390/biomedicines6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ragaini BS, Blizzard L, Newman L, Stokes B, Albion T, Venn A. Temporal trends in the incidence rates of keratinocyte carcinomas from 1978 to 2018 in Tasmania, Australia: a population-based study. Discov Oncol. 2021;12:30. doi: 10.1007/s12672-021-00426-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geller AC, Swetter SM. Reporting and registering nonmelanoma skin cancers: A compelling public health need. Br J Dermatol. 2012;166:913–5. doi: 10.1111/j.1365-2133.2012.10911.x. [DOI] [PubMed] [Google Scholar]
- 4.Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012;166:1069–80. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- 5.Shalhout SZ, Emerick KS, Kaufman HL, Miller DM. Immunotherapy for Non-melanoma Skin Cancer. Curr Oncol Rep. 2021;23:125. doi: 10.1007/s11912-021-01120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson SC, Eberl M, Vagnozzi AN, Belkadi A, Veniaminova NA, Verhaegen ME, et al. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell. 2015;16:400–12. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mancini M, Cappello A, Pecorari R, Lena AM, Montanaro M, Fania L, et al. Involvement of transcribed lncRNA uc.291 and SWI/SNF complex in cutaneous squamous cell carcinoma. Discov Oncol. 2021;12:14. doi: 10.1007/s12672-021-00409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apalla Z, Nashan D, Weller RB, Castellsagué X. Skin Cancer: Epidemiology, Disease Burden, Pathophysiology, Diagnosis, and Therapeutic Approaches. Dermatol Ther. 2017;7:5–19. doi: 10.1007/s13555-016-0165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gloster HM, Brodland DG. The epidemiology of skin cancer. Dermatol Surg. 1996;22:217–26. doi: 10.1111/j.1524-4725.1996.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 10.Ouhtit A, Konrad Muller H, Gorny A, Ananthaswamy HN. UVB-induced experimental carcinogenesis: dysregulation of apoptosis and p53 signalling pathway. Redox Rep. 2000;5:128–9. doi: 10.1179/135100000101535447. [DOI] [PubMed] [Google Scholar]
- 11.Bootorabi F, Manouchehri H, Changizi R, Barker H, Palazzo E, Saltari A, et al. Zebrafish as a Model Organism for the Development of Drugs for Skin Cancer. Int J Mol Sci. 2017;18:1550. doi: 10.3390/ijms18071550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo I, Sartor E, Fagotto L, Colombo A, Tiso N, Alaibac M. The Zebrafish model in dermatology: an update for clinicians. Discov Oncol. 2022;13:48. doi: 10.1007/s12672-022-00511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corchado-Cobos R, García-Sancha N, González-Sarmiento R, Pérez-Losada J, Cañueto J. Cutaneous Squamous Cell Carcinoma: From Biology to Therapy. Int J Mol Sci. 2020;21:2956. doi: 10.3390/ijms21082956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brogden DRL, Khoo CC, Kontovounisios C, Pellino G, Chong I, Tait D, et al. Anal squamous cell carcinoma in a high HIV prevalence population. Discov Oncol. 2021;12:3. doi: 10.1007/s12672-021-00397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choquet H, Ashrafzadeh S, Kim Y, Asgari MM, Jorgenson E. Genetic and environmental factors underlying keratinocyte carcinoma risk. JCI Insight. 2020;5:e134783. doi: 10.1172/jci.insight.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun M, Ji H, Xu N, Jiang P, Qu T, Li Y. Real-world data analysis of immune checkpoint inhibitors in stage III-IV adenocarcinoma and squamous cell carcinoma. BMC Cancer. 2022;22:762. doi: 10.1186/s12885-022-09843-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Q, Jun T, Wang B, Patel VG, Mellgard G, Zhong X, et al. Clinical factors associated with outcome in solid tumor patients treated with immune-checkpoint inhibitors: a single institution retrospective analysis. Discov Oncol. 2022;13:73. doi: 10.1007/s12672-022-00538-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol. 2014;810:120–40. doi: 10.1007/978-1-4939-0437-2_7. [DOI] [PubMed] [Google Scholar]
- 19.Calzavara-Pinton P, Ortel B, Venturini M. Non-melanoma skin cancer, sun exposure and sun protection. G Ital Dermatol Venereol. 2015;150:369–78. [PubMed] [Google Scholar]
- 20.Werner RN, Sammain A, Erdmann R, Hartmann V, Stockfleth E, Nast A. The natural history of actinic keratosis: a systematic review. Br J Dermatol. 2013;169:502–18. doi: 10.1111/bjd.12420. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Jin Y, Gong B, Li L, Zhao Q. Pan-cancer analysis of necroptosis-related gene signature for the identification of prognosis and immune significance. Discov Oncol. 2022;13:17. doi: 10.1007/s12672-022-00477-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fania L, Samela T, Moretta G, Ricci F, Dellambra E, Mancini M, et al. Attitudes among dermatologists regarding non-melanoma skin cancer treatment options. Discov Oncol. 2021;12:31. doi: 10.1007/s12672-021-00421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Moloney M, Tran A, Zheng S, Rogers J. Local control comparison of early-stage non-melanoma skin Cancer (NMSC) treated by superficial radiotherapy (SRT) and external beam radiotherapy (XRT) with and without dermal image guidance: a meta-analysis. Discov Oncol. 2022;13:129. doi: 10.1007/s12672-022-00593-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatti V, Fierro C, Annicchiarico-Petruzzelli M, Melino G, Peschiaroli A. ΔNp63 in squamous cell carcinoma: defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol Oncol. 2019;13:981–1001. doi: 10.1002/1878-0261.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nomori H, Shiraishi A, Honma K, Shoji K, Otsuki A, Cong Y, et al. Differences between lung adenocarcinoma and squamous cell carcinoma in histological distribution of residual tumor after induction chemoradiotherapy. Discov Oncol. 2021;12:36. doi: 10.1007/s12672-021-00431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piotrowski I, Zhu X, Saccon TD, Ashiqueali S, Schneider A, de Carvalho Nunes AD, et al. miRNAs as Biomarkers for Diagnosing and Predicting Survival of Head and Neck Squamous Cell Carcinoma Patients. Cancers. 2021;13:3980. doi: 10.3390/cancers13163980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castellano LRC, et al. Implications and Emerging Therapeutic Avenues of Inflammatory Response in HPV+ Head and Neck Squamous Cell Carcinoma. Cancers. 2022;14:5406. doi: 10.3390/cancers14215406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galizia D, Minei S, Maldi E, Chilà G, Polidori A, Merlano MC. How Risk Factors Affect Head and Neck Squamous Cell Carcinoma (HNSCC) Tumor Immune Microenvironment (TIME): Their Influence on Immune Escape Mechanisms and Immunotherapy Strategy. Biomedicines. 2022;10:2498. doi: 10.3390/biomedicines10102498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindele A, Holm A, Nylander K, Allard A, Olofsson K. Mapping human papillomavirus, Epstein–Barr virus, cytomegalovirus, adenovirus, and p16 in laryngeal cancer. Discov Oncol. 2022;13:18. doi: 10.1007/s12672-022-00475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Prim. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bisheshar SK, van der Kamp MF, de Ruiter EJ, Ruiter LN, van der Vegt B, Breimer GE, et al. The prognostic role of tumor associated macrophages in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oral Oncol. 2022;135:106227. doi: 10.1016/j.oraloncology.2022.106227. [DOI] [PubMed] [Google Scholar]
- 32.Ranganath K, Feng AL, Franco RA, Varvares MA, Faquin WC, Naunheim MR, et al. Molecular Biomarkers of Malignant Transformation in Head and Neck Dysplasia. Cancers. 2022;14:5581. doi: 10.3390/cancers14225581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li K, Wu G, Fan C, Yuan H. The prognostic significance of primary tumor size in squamous cell carcinoma of the penis. Discov Oncol. 2021;12:22. doi: 10.1007/s12672-021-00416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Chen J, Shi Y, Fang X, Tang Z. MAPK Signaling Pathway in Oral Squamous Cell Carcinoma: Biological Function and Targeted Therapy. Cancers. 2022;14:4625. doi: 10.3390/cancers14194625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song D, Wei Y, Hu Y, Chen X, Zheng Y, Liu M, et al. Identification of prognostic biomarkers associated with tumor microenvironment in ceRNA network for esophageal squamous cell carcinoma: a bioinformatics study based on TCGA database. Discov Oncol. 2021;12:46. doi: 10.1007/s12672-021-00442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Beeraka NM, Xie L, Dong L, Liu J, Wang L. Co-expression of High-mobility group box 1 protein (HMGB1) and receptor for advanced glycation end products (RAGE) in the prognosis of esophageal squamous cell carcinoma. Discov Oncol. 2022;13:64. doi: 10.1007/s12672-022-00527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smirnov A, Anemona L, Novelli F, Piro CM, Annicchiarico-Petruzzelli M, Melino G, et al. p63 Is a Promising Marker in the Diagnosis of Unusual Skin Cancer. Int J Mol Sci. 2019;20:5781. doi: 10.3390/ijms20225781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL, Bruno TC. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer. 2023;23:173–88. doi: 10.1038/s41568-022-00531-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv Y, Lin W. Comprehensive analysis of the expression, prognosis, and immune infiltrates for CHDs in human lung cancer. Discov Oncol. 2022;13:29. doi: 10.1007/s12672-022-00489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel R, Chang ALS. Immune Checkpoint Inhibitors for Treating Advanced Cutaneous Squamous Cell Carcinoma. Am J Clin Dermatol. 2019;20:477–82. doi: 10.1007/s40257-019-00426-w. [DOI] [PubMed] [Google Scholar]
- 41.Qin Q, Jun T, Wang B, Patel VG, Mellgard G, Zhong X, et al. Clinical factors associated with outcome in solid tumor patients treated with immune-checkpoint inhibitors: a single institution retrospective analysis. Discov Oncol. 2022;13:73. doi: 10.1007/s12672-022-00538-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W-H, Wang Y, Yan C, Du W-D, Al-Aroomi MA, Zheng L, et al. CC chemokine receptor 7 promotes macrophage recruitment and induces M2-polarization through CC chemokine ligand 19&21 in oral squamous cell carcinoma. Discov Oncol. 2022;13:67. doi: 10.1007/s12672-022-00533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Y, Chen J, Shi Y, Fang X, Tang Z. MAPK Signaling Pathway in Oral Squamous Cell Carcinoma: Biological Function and Targeted Therapy. Cancers. 2022;14:4625. doi: 10.3390/cancers14194625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagata Y, Yamamoto S, Kato K. Immune checkpoint inhibitors in esophageal cancer: Clinical development and perspectives. Hum Vaccin Immunother. 2022;18:2143177. doi: 10.1080/21645515.2022.2143177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meliante PG, Barbato C, Zoccali F, Ralli M, Greco A, de Vincentiis M, et al. Programmed Cell Death-Ligand 1 in Head and Neck Squamous Cell Carcinoma: Molecular Insights, Preclinical and Clinical Data, and Therapies. Int J Mol Sci. 2022;23:15384. doi: 10.3390/ijms232315384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman AC, Maddocks ODK. Serine and Functional Metabolites in Cancer. Trends Cell Biol. 2017;27:645–57. doi: 10.1016/j.tcb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Ling R, Chen G, Tang X, Liu N, Zhou Y, Chen D. Acetyl-CoA synthetase 2(ACSS2): a review with a focus on metabolism and tumor development. Discov Oncol. 2022;13:58. doi: 10.1007/s12672-022-00521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smirnov A, Anemona L, Montanaro M, Mauriello A, Annicchiarico-Petruzzelli M, Campione E, et al. Transglutaminase 3 is expressed in basal cell carcinoma of the skin. Eur J Dermatol. 2019;29:477–83. doi: 10.1684/ejd.2019.3636. [DOI] [PubMed] [Google Scholar]
- 49.Pecorari R, Bernassola F, Melino G, Candi E. Distinct interactors define the p63 transcriptional signature in epithelial development or cancer. Biochem J. 2022;479:1375–92. doi: 10.1042/BCJ20210737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frezza V, Fierro C, Gatti E, Peschiaroli A, Lena AM, Petruzzelli MA, et al. Np63 promotes IGF1 signalling through IRS1 in squamous cell carcinoma. Aging. 2018;10:4224–40. doi: 10.18632/aging.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smirnov A, Lena AM, Cappello A, Panatta E, Anemona L, Bischetti S, et al. ZNF185 is a p63 target gene critical for epidermal differentiation and squamous cell carcinoma development. Oncogene. 2019;38:1625–38. doi: 10.1038/s41388-018-0509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smirnov A, Panatta E, Lena A, Castiglia D, di Daniele N, Melino G, et al. FOXM1 regulates proliferation, senescence and oxidative stress in keratinocytes and cancer cells. Aging. 2016;8:1384–97. doi: 10.18632/aging.100988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattaini KR, Sullivan MR, vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. 2016;214:249–57. doi: 10.1083/jcb.201604085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic. N. Engl J Med. 1948;238:787–93. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 55.Ganini C, Amelio I, Bertolo R, Candi E, Cappello A, Cipriani C, et al. Serine and one-carbon metabolisms bring new therapeutic venues in prostate cancer. Discov Oncol. 2021;12:45. doi: 10.1007/s12672-021-00440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–8. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–83. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Candi E, Schmidt R, Melino G. The cornified envelope: A model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 59.Genenger B, Perry JR, Ashford B, Ranson M. A tEMTing target? Clinical and experimental evidence for epithelial-mesenchymal transition in the progression of cutaneous squamous cell carcinoma (a scoping systematic review) Discov Oncol. 2022;13:42. doi: 10.1007/s12672-022-00510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maddocks ODK, Labuschagne CF, Adams PD, Vousden KH. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol Cell. 2016;61:210–21. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labuschagne CF, van den Broek NJF, Mackay GM, Vousden KH, Maddocks ODK. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–58. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 62.Cappello A, Mancini M, Madonna S, Rinaldo S, Paone A, Scarponi C, et al. Extracellular serine empowers epidermal proliferation and psoriasis-like symptoms. Sci Adv. 2022;8:eabm7902. doi: 10.1126/sciadv.abm7902. [DOI] [PubMed] [Google Scholar]
- 63.Giardina G, Paone A, Tramonti A, Lucchi R, Marani M, Magnifico MC, et al. The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells. FEBS J. 2018;285:3238–53. doi: 10.1111/febs.14610. [DOI] [PubMed] [Google Scholar]
- 64.Shin M, Momb J, Appling DR. Human mitochondrial MTHFD2 is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase. Cancer Metab. 2017;5:11. doi: 10.1186/s40170-017-0173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li AM, Ducker GS, Li Y, Seoane JA, Xiao Y, Melemenidis S, et al. Metabolic Profiling Reveals a Dependency of Human Metastatic Breast Cancer on Mitochondrial Serine and One-Carbon Unit Metabolism. Mol Cancer Res. 2020;18:599–611. doi: 10.1158/1541-7786.MCR-19-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang C, Zhang J, Liao M, Yang Y, Wang Y, Yuan Y, et al. Folate-mediated one-carbon metabolism: a targeting strategy in cancer therapy. Drug Discov Today. 2021;26:817–25. doi: 10.1016/j.drudis.2020.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 and Supplementary Table 1 and 2
Data Availability Statement
For Serine biosynthesis and One Carbon metabolism enzymes expression at mRNA level a public database available through GEO datasets (https://www.ncbi.nlm.nih.gov/gds) was analyzed (GSE7553).