Abstract
Allergic diseases such as allergic rhinitis (AR), allergic asthma (AAS), atopic dermatitis (AD), food allergy (FA), and eczema are systemic diseases caused by an impaired immune system. Accompanied by high recurrence rates, the steadily rising incidence rates of these diseases are attracting increasing attention. The pathogenesis of allergic diseases is complex and involves many factors, including maternal-fetal environment, living environment, genetics, epigenetics, and the body’s immune status. The pathogenesis of allergic diseases exhibits a marked heterogeneity, with phenotype and endotype defining visible features and associated molecular mechanisms, respectively. With the rapid development of immunology, molecular biology, and biotechnology, many new biological drugs have been designed for the treatment of allergic diseases, including anti-immunoglobulin E (IgE), anti-interleukin (IL)-5, and anti-thymic stromal lymphopoietin (TSLP)/IL-4, to control symptoms. For doctors and scientists, it is becoming more and more important to understand the influencing factors, pathogenesis, and treatment progress of allergic diseases. This review aimed to assess the epidemiology, pathogenesis, and therapeutic interventions of allergic diseases, including AR, AAS, AD, and FA. We hope to help doctors and scientists understand allergic diseases systematically.
Subject terms: Immunological disorders, Immunological disorders
Introduction
Allergic diseases are systemic disorders caused by an impaired immune system. Different allergic diseases, including AR, AAS, AD, FA and eczema, are caused by complex interactions between genetic and environmental factors. Allergic diseases are listed by the World Health Organization (WHO) as one of the top three disorders to be prevented and controlled in the 21st century. An allergic disease, whilst a systemic disease, can also manifest as different local maladies, all of which may lead to anaphylactic shock in severe cases. The incidence of allergic diseases is high, bringing much suffering to patients. It is estimated that nearly 500 million and 300 million individuals worldwide have AR and AAS, respectively,1 with an increasing number of cases. For AAS, mortality rates in women and men are 90 and 170 per million individuals, respectively. About 96% of asthma deaths occur in low-and-middle-income countries.2 It is currently estimated that FA affects 1–10% of the total population.3 The global prevalence rate of AD is 8%,4,5 with a lifetime prevalence reaching 20%.6 In 2019, there were 171.17 million patients worldwide with AD.7
Due to the different sites of allergic diseases, the clinical and pathological manifestations also differ. In AR, after stimulation by allergens, including airborne dust mites associated with fecal particles, cockroach remains, pet dander, molds and pollens, inflammatory cells such as mast cells, CD4+ T cells, B cells, macrophages and eosinophils infiltrate the lining of the nasal cavity, with infiltration into the nasal mucosa. T helper 2 (Th2) cells promote the release of immunoglobulin and cytokines, including interleukin (IL)-3, IL-4, IL-5, and IL-13; meanwhile, IgE is also produced by plasma cells. There is, however, still some uncertainty around the source of IgE production. Follicular helper T (Tfh) cells are a subpopulation of CD4+ T-effector cells, and in recent years it has been discovered that the key cells regulating IgE production are not Th2 cells, but Tfh cells. Allergens cross-link IgE that interact with mast cells, which further induces the release of multiple mediators (including histamine and leukotrienes), promotes arteriole dilation and vascular permeability, and causes pruritus, runny nose, mucus secretion, and pulmonary smooth muscle contraction.8 Over the next 4–8 hours, the released mediators and cytokines induce subsequent cellular inflammatory reactions (late inflammatory response), leading to the recurrence of symptoms, often nasal congestion, which generally persist.8,9 The immunopathological profiles of AR and AAS are very similar in terms of eosinophil, mast cell and Th2 cell infiltration. Although structural changes in airway remodeling are well characterized in AAS, they may also occur in AR. There are also pathophysiological differences between AR and AAS. In the AAS disease, mucosal pathological alterations comprise epithelial hyperplasia, goblet cell metaplasia and increased mucus generation. In the submucosal layer, smooth muscle hypertrophy, collagen accumulation and large mucus glands prevail, leading to airway narrowing and enhanced mucus generation during an asthma attack,10 with symptoms such as difficulty breathing, wheezing, chest pain, and coughing.11 The pathogenesis of AD is mainly reflected by a complex interplay between epidermal barrier dysfunction, abnormal skin microbiota and dysregulated type 2 T cell immunity.12,13 The above pathogenesis induces a series of pathological manifestations such as filamentous aggregation; weak skin barrier due to protein shortage promoting inflammatory reactions and T cell infiltration; S. aureus colonization or infection disrupting the skin barrier and inducing an inflammatory response as well as the development of epidermal edema (“spongy sclerosis”); local Th2 immune reactions further reducing the barrier function, promoting dysregulation that favors Staphylococcus species, especially S. aureus, which triggers pruritus. FA is an IgE-dependent type I hypersensitivity to a specific food allergen. Its pathological process is divided into two stages: in the allergic sensitization stage, the initial exposure to the allergen results in tolerance breakdown, with subsequent generation of specific IgE, vasoactive substances and allergic response mediators such as histamine and platelet activating factor.14 During the provocation phase, degranulation of effector cells, such as mast cells, induces allergic inflammation, and serotonin or 5-hydroxytryptamine is released in large amounts, resulting in acute gastrointestinal symptoms, including diarrhea. Besides mast cells, an allergen also reacts with sensitized basophils in the circulation, triggering a life-threatening systemic reactions featuring multiple-organ and system involvements, hypotension and shock.15 After repeated exposure to food allergens, persistent allergic inflammatory reactions occur and tissue mast cells increase, resulting in persistent gastrointestinal reactions.16
The pathogenesis of allergic diseases is complex, involving many factors such as genetics, epigenetics, environmental factors, microecology and the body’s immune function. Their recurrence rate is high, which brings great pain to and imposes a severe financial burden on patients. Therefore, this manuscript comprehensively analyzes allergic diseases, from a brief introduction of their history to their mechanism and treatment, hoping to provide not only a systematic understanding of such diseases, but also a reference for clinical doctors and scientists.
A brief history of allergic diseases
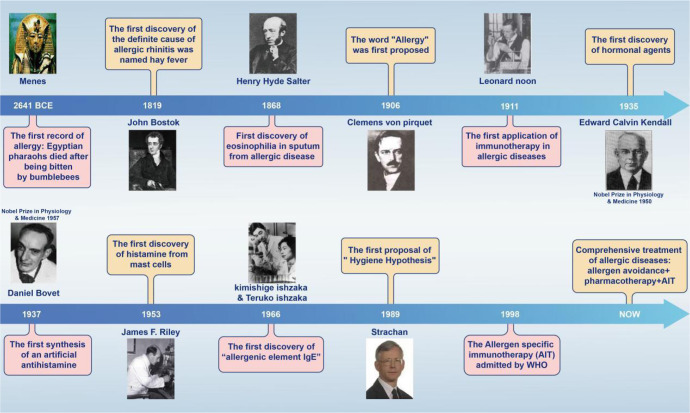
Since ancient times, people have always been drawn to allergic diseases, studying their pathogenesis and developing related treatments. The earliest recorded allergic reaction in human history was the death of the Egyptian pharaoh Menes after being bitten by a bumblebee around 2641 BCE.17 Theories on the actual causes and diagnosis of allergic diseases were further developed in the 19th century, precisely in 1819, when the British physician John Bostock, at the Royal Society of Reported Medicine, attributed summer eye and nose discomfort to hay, naming the condition “hay fever”.18 Later in 1868, Eosinophilia was first observed by Henry Hyde Salter, in the sputum of a patient with an allergic disease.19
The pathogenesis and treatment of allergic diseases have made rapid progress in the 20th century. The word “allergy” was coined by Clemens von Pirquet in 1906,20,21 which is considered to be the beginning of modern allergy science. In 1911, Leonard Noon was the first to be successful in the treatment of pollen-associated AR with low-dose flower infusion, setting a precedent for immunotherapy.22 Edward Calvin Kendall discovered the adrenocortical hormone and determined its structure and physiological effects in 1935, earning the Nobel Prize in Physiology or Medicine in 1950.23 Daniel Bovet synthesized antihistamines in 1937 and earned the Nobel Prize in Physiology or Medicine in 1957, which brought hope in the treatment of allergic diseases and has been in clinical use to this day.24,25 In 1953, James F. Riley was the first to discover that histamine in the human body mainly comes from mast cell granules.26 Up to this point in history, basic treatment methods for allergic diseases had been established, but no breakthrough had been made in mechanistic research. In terms of pathogenesis, the Ishzaka couple discovered in 1966 that the reactive hormone in the serum of patients with allergic diseases was IgE,27 providing a new experimental tool and concept for serological research. In 1989, the epidemiologist Strachan proposed the “hygiene hypothesis” on the basis that “compared with an only child, children in large families have a lower risk of developing pollen allergy and eczema”.28 The hygiene hypothesis has laid a solid foundation for studying the pathogenesis of allergic diseases from the perspectives of modern immunology, microecology, and antibiotic application. Current guidelines suggest a combined application of allergen avoidance, pharmacotherapy, and/or allergen-specific immunotherapy (AIT)29,30 (Fig. 1).
Fig. 1.
Timeline of major findings related to allergic diseases. Allergic reactions in Western countries were first recorded in 2641 BCE, when the Egyptian Pharaoh Menes died after being bitten by a bumblebee. In 1911, Leonard Noon published an article in The Lancet, reporting a clinical paper treating pollinosis by subcutaneous injection of grass pollen extract, which marked the beginning of modern immunotherapy. In 1966, the Japanese scientist Ishizaka and his wife discovered IgE, which led to a leap in the understanding of immediate allergy. Following their discovery, IgE became a new indicator for the diagnosis of allergy
Mechanism
Genetics and epigenetics
Gene-environment interactions in allergy diseases
Allergic disease is a complex disorder, whose etiology and development may involve genetic and environmental factors. Although the innate and adaptive immune systems are critical in regulating the adaptation to the external microenvironment,31 allergic diseases are considered a major cause of immune dysfunction caused by the interactions of multiple genes and the external environment in cells.32,33
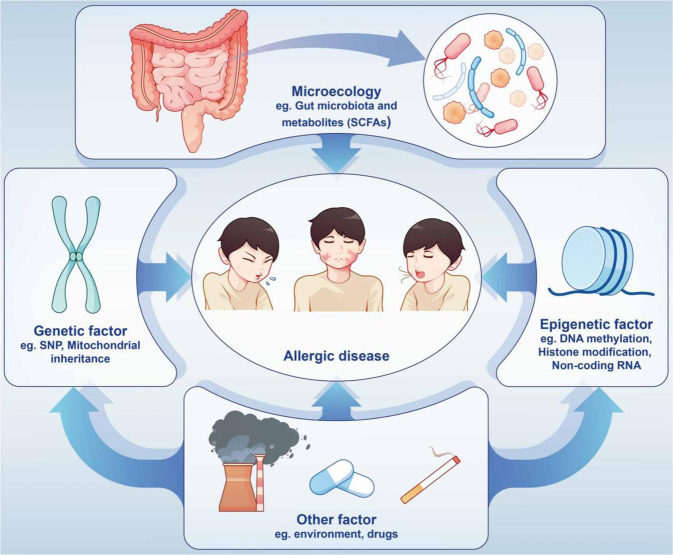
The research boom in genetics and epigenetics has substantially promoted research progress for allergic diseases. The application of genome-wide association studies (GWASs), single nucleotide polymorphism (SNP) analysis, and epigenome-wide association studies (EWASs) has laid a solid foundation for exploring the genetics of allergic diseases. Epigenetic studies mainly focus on DNA methylation, post-translational histone modifications, and non-coding RNAs. Epigenetics can explain the occurrence and development of allergic diseases in the external environment from various aspects, elucidate the mechanism of immune response plasticity in allergic disorders, and even provide diagnostic biomarkers and therapeutic targets for allergic disorders34,35 (Fig. 2).
Fig. 2.
Allergic diseases are caused by a variety of factors. External factors include changes in gut microbiota and metabolites, drugs, and air pollution. Internal influencing factors include genetic and epigenetic changes
SNPs and related GWASs in allergic diseases
When the Human Genome Project was completed in 2001,36 scientists were surprised to find that most genome sequence variations involve SNPs. SNP diversity can be found throughout different regions of the genome,37 including introns, exons, promoters, enhancers and intergenic regions, with SNPs considered as the basis of DNA sequence variation.38
Hereditary studies
Both genetics and the environment are critical to the etiology and development of allergic diseases, and it is difficult to distinguish their independent roles in allergic diseases when they are combined. Twins’ studies can be used to separate genetics from environmental factors, providing clues to the genetic component of allergic diseases. In fact, research into the genetics of allergic disease also began in twins. Twin studies revealed that FA had high heritability, and GWAS and candidate gene studies indicated marked associations of the human leukocyte antigen(HLA)-DR and HLA-DQ region with genetic variants in multiple genes, including Filaggrin (FLG), the HLA locus, and Forkhead Box Protein P3 (FOXP3).39 Peanut allergy in 82% of identical twins far exceeds the 20% concordance rate observed in dizygotic twins,40,41 further supported by the fact that children whose parents or siblings have peanut allergy are 7 times more likely to develop the disease compared with children without family risk factors. In general, heritability estimates for FA are as high as 81%,42 while the heritability of AR is estimated at approximately 91%.42 Twin studies reveal that about 25% of phenotypic variation in asthma severity can be explained by genetic factors; for example, RAD50- IL13 on chromosome 5q and the ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3)- gasdermin B (GSDMB) locus on chromosome 17q21 were found to be associated with asthma severity.43 Besides, the concordance rate for AD in identical twins is about 80%, which is remarkably elevated compared with the 20% found in fraternal twins.44
SNP and GWAS analyses in allergic diseases
To date, SNPs in no less than 34 loci and 46 genes are considered to have AD risk in different populations around the world.45 Loss-of-function mutations in FLG, which encodes filaggrin (a skin barrier protein), are considered the most important genetic risk factor for AD, although variants affecting skin and systemic immune reactions also play critical roles.46 In an AAS disease study, after analysis by GWAS, Sarnowski et al. recently detected five genetic variants related to age at onset in 5,462 asthma cases, at or around recombinant Cylindromatosis (CYLD) on 16q12 (rs1861760), IL1RL1 on 2q12 (rs10208293), HLA-DQA1 on 6p21 (rs9272346), IL33 on 9p24 (rs928413) and GSDMA on 17q12 (rs9901146), with the last four also showing associations with susceptibility to allergic diseases. Recombinant Mucin 5 Subtype AC (MUC5AC) is considered an essential factor in the natural barrier function of the airway and has a potential association with moderate to severe asthma.47 Studies have shown that SNP changes of ORMDL3 and the TSLP promoter gene are involved in AAS;48,49 polymorphisms in the alpha chain coding region of IL-4 receptor are also associated with AAS.50 Susceptibility-related GWAS data showed that asthma-related IL-33 genes are all associated with asthma and AR.51,52 Changes in IL-4 gene single nucleotide polymorphisms can increase the risk of AR;52 individuals with the Vitamin D (1,25- dihydroxyvitamin D3) receptor (VDR) rs2228570 CC and vitamin D-binding protein (VDBP) rs7041 GG genotypes have a high risk of asthma progression.53 In FA analysis, peanut allergy is clearly associated with specific locus changes in the HLA-DR and HLA-DQ genes.54 Molecular genetic analysis of the GG, GA, and AA genotypes of the IL-13 R130Q gene polymorphism revealed markedly elevated incidence rates of the GA and AA genotypes in comparison with healthy control individuals.55 Besides, the serpin B serpin (SERPINB) and cytokine gene clusters increase the risk of any FA, as well as the C11orf30/LRRC32 locus.56
In the study of AR, the largest GWAS revealed 20 novel loci associated with AR risk,57 including IL7R at 5p13.2 and SH2B adaptor protein 3 (SH2B3) on chromosome 12q24.12, which separately participate in V(D)J recombination of T cell and B cell receptors,58 blood eosinophil count59 and T cell activation pathways.60 Furthermore, we noted that AR risk loci have important effects on innate and adaptive immune responses. Loci near C-X-C chemokine receptor type 5 (CXCR5) on 11q23 and Fc Fragment of IgE Receptor Ig (FCER1G) on 1q23.3 separately encode chemokine receptor in B cells and follicular T cells61 and the γ chain of IgE receptor.62 Broad-complex,tramtrack and bric-a-brac and cap’n’collar homology 2 (BACH2) on 6q15 is critical to the induction immunomodulatory function of memory B and T cells,63,64 Leukocyte tyrosine kinase (LTK) and TYRO3 protein tyrosine kinase (TYRO3) modulate Th2-type immune responses; RAR-related orphan receptor A (RORA) regulates the development and inflammatory response of Th2 innate lymphocytes, and tumor necrosis factor (ligand) superfamily, member 11 (TNFSF11) is involved in dendritic cell activation of T cells. The Viral protein R binding protein (VPRBP) gene controls T cell proliferation and contributes to V(D)J recombination in B cells.65–68 Surprisingly, the frequencies of the tumor necrosis factor-α (TNF-α) and MRPL4 genes are starkly elevated in AR cases in Han individuals69 (Table 1).
Table 1.
Summary of genomic loci of allergic disease
| Chromosome | Gene | SNP (Variant) | Possible Allergic Mechanism | Related disease | References |
|---|---|---|---|---|---|
| 6p21.32 | HLA-DR, -DQ | rs7192, rs9275596 | Antigen-specific immune response | FA | 529 |
| 18q21.3 | SERPINB | rs12123821 | Immunological regulation or epithelial barrier function | FA | 56 |
| 19p13 | MRPL4 | rs8111930 | Involved in inflammatory adhesion process | AR | 530 |
| 11q13 | C11orf30/LRRC32 | rs2155219 | Epithelial barrier function, regulatory | AR | 531 |
| T-cell function, and immune tolerance | |||||
| 4p14 | TLR6 | rs3860069 | Pattern recognition receptors in innate immunity | AR | 531 |
| 5q31.1 | IL4 | rs2243250 | Promotes isotype class switching from IgM to IgE, promote the differentiation of T cells to a Th2 cell phenotype | AR with Asthma | 532 |
| 5q31.1 | IL13 | rs20541 | |||
| 5q22.1 | TSLP | rs1898671 | Epithelial barrier function, regulatory | AR | 531 |
| T-cell function, and immune tolerance | |||||
| 4q27 | IL2 | rs17454584/a | Immune regulatory effects. | AR | 54 |
| 3p21.2 | VPRBP | rs62257549 | Involved in T cell proliferation and V(D)J recombination in B cell development. | AR | 54 |
| 11q12-13 | FCER1B | rs569108, rs512555 | IgE cross-links allergen-bound receptors | AR | 533 |
| 13q14.11 | TNFSF11 | rs7328203 | Involved in hematopoiesis and downstream of T cell receptor activation. | AR | 54 |
| 12q24.12 | SH2B3 | rs35350651 | |||
| 3p21.2 | VPRBP | rs62257549 | Involved in T cell proliferation and V(D)J recombination in B cell development | AR | 54 |
| 15q22.2 | RORA | rs1051906 | Involved in development of natural helper cell. | AR | 54 |
| 10q24.32 | ACTR1A; TMEM180 | rs35597970 | Involved in regulating TLR-4 and cytokine signaling | AR | 54 |
| 5p13.2 | IL7R | rs7717955 | V(D)J recombination of B and T cell receptors;T cell subtypes have different levels of IL-7R on the cell surface | AR | 54 |
| 1p31.1 | LRRIQ3; NEGR1 | rs2815765 | Cell adhesion | AR | 54 |
| 17q21.2 | GSDMB | rs7216389 | Terminal differentiation of epithelial cells | AAS | 534 |
| 5p13.3 | CD14 (-159 C/T) | rs2569190 | Monocyte activity | AAS | 535 |
| 17q11.2 | NOS2 | rs10459953 | Expressed on T cells, macrophages, epithelial cells, mast cells, eosinophils, and neutrophils in response to inflammatory stimuli | AAS | 536 |
| Xp11.23 | FOXP3 | rs3761548,rs3761549 | Transcription factor of Treg cell | AAS | 537 |
| 5q31 | KIF3A | rs11740584, rs2299007 | Increased transepidermal water loss (TEWL) | AD | 538 |
| 21q22.23 | UBASH3A | rs11203203 | A phosphatase that regulates the T cell receptor (TCR) signaling pathway | AD | 539 |
| 1q12 | FLG | rs7927894 | Epidermal barrier | AD | 540 |
| 5q21.3 | TSLP | rs11466749 | Genes encoding alarmins produced by keratinocytes | ||
| 17q21.2 | ORMDL3 | rs7216389 | Epidermal barrier | ||
| 6p21.3 | GPSM3 | rs176095 | AD | 541 | |
| 11p15.4 | NLRP10 | rs878860 | Encodes a protein that belongs to the NALP protein family but lacks the leucine-rich repeat region | AD | 541 |
| 3p21.33 | GLB1 | rs6780220 | Encodes β-galactosidase-1 | AD | 541 |
| 3q13.2 | CCDC80 | rs12634229 | Encodes a protein involved in the induction of C/EBPα and peroxisome proliferator-activated receptor γ (PPARγ) | AD | 541 |
| 7p22 | CARD11 | rs4722404 | An protein for T-cell receptor (TCR) and B-cell receptor (BCR) signaling | AD | 541 |
| 10q21.2 | ZNF365 | rs10995251 | Encodes a T-cell anergy-associated transcription factor | AD | 541 |
| 20q13 | CYP24A1-PFDN4 | rs16999165 | Encodes a mitochondrial cytochrome P450 superfamily enzyme | AD | 541 |
Mitochondrial inheritance in allergic disease
Maternal inheritance is considered the most critical player in allergic disease occurrence. Most offspring mitochondria are inherited from the mother, and mitochondrial inheritance is tightly associated with asthma occurrence.70–73 Mitochondrial DNA (mtDNA) variants are significantly associated with allergic disorders, including AD and asthma. A report demonstrated that in 69 mtDNA variants, the rs28357671 locus of the MT-ND6 gene was significantly associated with mitochondrial function genes in allergic diseases, including: NLR Family Member X1 (NLRX1), oculocutaneous albinism II (OCA2) and coiled-coil-helix-coiled-coil-helix domain containing 3 (CHCHD3).74 Interestingly, the genetic cause of asthma in females may be associated with a dysfunction of mitochondrial MT-ND2 and MT-RNR2 genes, while in males, mutations in the mitochondrial cytochrome-b (CYB) gene leads to changes in reactive oxygen species (ROS) and asthma development.75
Mutations affecting mitochondrial tRNA genome sequences have been observed in the placenta of asthmatic mothers and are associated with AAS;76 for example, a rare mutation in the A3243G-tRNA Leu (UUR) MELAS gene, which is thought to be associated with asthma, was found in asthmatic patients. Maternally inherited mitochondrial diseases have been reported.77 Fukuda’s team found that 9 of the 13 differentially expressed genes in allergic patients were mitochondria-related genes, including those producing cytochrome oxidases II and III, and NADH dehydrogenase.76,78,79 In addition, polymorphisms in the ADAM metallopeptidase domain 33 (ADAM33) and cytochrome b genes located on chromosome 20 have been associated with asthma susceptibility, both of which are closely associated with mitochondrial oxidative function.80 Mitochondrial haploidy and elevated serum IgE amounts are associated in Europeans,73 which may involve diverse mutations in genes that encode mitochondrial tRNAs.76 The ATP synthase mitochondrial F1 complex assembly factor 1 gene was implicated in asthma in Caucasian children.81 Some AAS is closely related to mtDNA deficiency, and alterations in more than 25 genes (ORMDL3, 2PBP2, GSDMB, PDE4D, VEGF, Wnt, MMP-12, PRKCA, JAG1, ANKRD5, TGF-β1, IL-12β, IL-10, IL-13, IL-17, IL-25, and β2-adrenergic receptors) are associated with abnormal changes in the immune system in AAS.80,82–84
Epigenetics and Epigenome-wide Association Study (EWAS)-related analysis of allergic diseases
Epigenetics are heritable features that affect gene expression without altering the DNA sequence.85 DNA methylation is an effective factor to distinguish allergic patients from healthy people.44 DNA methylation is reflected by a methyl group added to cytosine at position 5 by DNA methyltransferases to form 5-methylcytosine,86 where a cytosine nucleotide is called a CpG, followed by a guanine nucleotide.87 CpG islands typically contain more than 200 bases, of which more than 60%-80% are guanines and cytosines (G + C).88 Methylation of CpG islands at transcription start sites (TSSs) of genes leads to gene activation or repression, and is generally thought to repress gene transcription.89
A cross-sectional study found that DNA methylation of allergy-related genes in the whole blood of allergic children may be a common parameter affecting asthma, rhinitis, and eczema; a total of 21 differential CpG loci were screened, 10 of which were in the pulmonary epithelium. The sites of replication, related to acyl-CoA thioesterase 7 (ACOT7), Lectin, Mannose Binding 2 (LMAN2) and Claudin 23 (CLDN23) genes, were all derived from eosinophils.90 Thus, changes in eosinophil levels are reflected by changes in methylation, unveiling a possible mechanism for phenotypic alterations in immune response-associated features.
In addition, methylation plays an important role in AD pathogenesis. The TSLP gene promoter is hypomethylated in the damaged skin of AD patients.91 Methylation is not specific to DNA, but is closely related to disease. Demethylation of histone H3 residues in the FOXP3 gene promoter region and hypermethylation of histone H3 residues in the RORC gene promote the differentiation of Th0 cells towards a regulatory T (Treg) phenotype. Conversely, these events cause Treg deficiency, one of the hallmarks of AD pathogenesis.34,92–95 Furthermore, CpG hypermethylation in IL-4 is negatively correlated with serum total IgE levels, explaining the role of Th2 immunity in AD.96 Enhanced hypermethylation of S100 calcium binding protein A5 (S100A5) was found in the epidermal part of lesions in AD cases in comparison with healthy individuals.97 Hypomethylation was observed in Recombinant Keratin 6 A (KRT6A) in keratinocytes, and methylation of cg07548383 in FLG also elevates AD risk.98
DNA methylation is also critical for AAS pathogenesis, occurrence, and development. An EWAS detected a total of 40,892 CpG sites methylated in important genes C-C motif chemokine ligand 26 (CCL26, a chemokine) and mucin 5 AC (MUC5AC, a mucin with airway defense function) among AAS patients compared with control cases.99 Chromosome 17q12-q21 hypermethylation contributes to asthma pathogenesis, with regulatory effects on all five protein-coding genes of this region, including IKAROS family zinc finger 3 (Aiolos) (IKZF3), zona pellucida-binding protein 2 (ZPBP2), ORMDL3, gasdermin A (GSDMA) and GSDMB.100 The STAT5A gene is hypermethylated in the 17q21.2 region and has been linked to increased Th1 responses and reduced infiltration of eosinophils in the airway epithelium.101 In childhood asthma, cg23602092 gene methylation status was linked to asthma symptoms,102 and hypomethylation of arachidonate 15-lipoxygenase (ALOX15) gene 17p13.2 and periostin, osteoblast specific factor (POSTN) gene 13q13.3 in nasal epithelial cells is associated with increased Th2 function.103 Methylation sites in multiple white blood cell (WBC) genes show significant associations with total IgE amounts, with the two most significant genes (ACOT7 and ZFPM1) associated with asthma.104 In adults, WNT2 gene hypermethylation in the 7q31.2 region in blood specimens is involved in neutrophilic asthma,105 and ORMDL3 hypermethylation in endobronchial airway epithelial cells contributes to asthma.106 Following hypermethylation at CpG sites, FOXP3 (12q15) and interferon-γ (IFN-γ) (Xp11.23) lead to altered T cell function and repressed Treg and T effector cell-related genes in blood.107 In adolescents, interleukin-5 receptor alpha (IL-5RA) (3p26.2) hypomethylation in blood was linked to asthma.108
In the AR disease, DNA methylation levels are tightly associated with CD4+ T cell amounts. DNA hypermethylation may downregulate IFN-γ in AR cases,109 while DNA hypomethylation increases the mRNA amounts of IL-13 and IgE.110 Alterations in hypermethylation at CpG sites in the melatonin receptor 1 A gene may be caused by paternal genetic variations in AR.111
DNA methylation might also contribute to FA pathogenesis. Reports have shown differences in DNA methylation in some mitogen-activated protein kinase (MAPK) signaling genes, e.g., human leukocyte antigen (HLA)-DQB1 and the Treg-specific demethylation region (TSDR) of FOXP3. Differential genetic DNA methylation might also contribute to FA diagnosis.39 In a pilot study of cow’s milk protein (CMA, milk allergy), hypermethylation was found in the DEXH (Asp-Glu-X-His) box polypeptide 58 (Dhx58), zinc finger protein 81 (ZNF281) and HtrA serine peptidase 2 (HTRA2) regions.112 Maternal peanut allergy also induces epigenetic changes in the IL-4 promoter in the offspring, which is associated with Th2 immune response (production of IL-4 and IgE).113 In a study of identical monozygotic (MZ) twins, the distance between peanut allergy and nonallergy in methylation profiles containing 12 DNAm signatures was reduced compared with randomly paired individuals without genetic relationships, indicating peanut allergy-associated DNAm signatures might be linked to genetic factors.114
Histone modifications
The DNA is packaged into an organized chromatin structure formed by a core histone protein consisting of H2A, H2B, H3 and H4.115 Post-translational histone modifications mainly comprise acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, and adenosine diphosphate (ADP) ribosylation of core histone tails, which reflect the epigenetic inheritance of many diseases, including AAS.115 Histone acetyltransferase (HAT)-mediated histone acetylation often loosens chromatin structure, facilitating access to transcription factors that induce gene expression. Conversely, histone deacetylation by histone deacetylases (HDACs) also leads to gene silencing. Higher levels of histone acetylation are generally associated with increased gene transcriptional activity and expression. Whether histone methylation is transcriptionally permissive or repressive depends largely on the number of methyl groups added and the position of the target amino acid residue in the histone tail.34,116–118
In adult asthmatic patients, lysine 18 acetylation of histone 3 (H3K18) and lysine 9 trimethylation of histone 3 (H3K9me3) are elevated in epithelial cells, and acetylation of H3K18 increases ΔNp63 (a p63 splice variant), epidermal growth factor receptor (EGFR) and signal transducer and activator of transcription 6 (STAT6) mRNA amounts.119 An imbalance of HAT and HDAC underlies impaired gene expression and is a determinant of asthma.120 HATs and HDACs have opposite functions, as the acetylation function of HATs promotes gene expression, while the deacetylation function of HDACs is responsible for gene silencing. In children with asthma, H3 acetylation of the FOXP3 gene contributes to Treg differentiation, and H3 histone acetylation critically affects the IL-13 gene promoter.121 Stefanowicz and collaborators assessed gene-specific alveolar epithelial histone acetylation and methylation statuses in asthma and healthy control cases, and found increased levels of H3K18ac and H3K9me3 in asthmatic patients.119 Acetylation of C-C Motif Chemokine 8 (CCL8), a neutrophil activator found in macrophages, as well as H3K18, results in elevated secreted amounts of this activator in airway smooth muscle.122 In asthma, the levels of CCR4 and CCL5 are high. CCR4 controls Th2 cell infiltration, while CCL5 is a leukocyte chemokine, and a single nucleotide polymorphism of CCR4 and CCL5 dimethylation (H3K4me2) is associated with Th2 differentiation.123 Resistance to steroid therapy in AAS has emerged, mainly due to IL-17A-induced steroid resistance resulting from decreased HDAC2 activity.123 Increased enrichment of transcriptionally active H3ac and H4ac histone markers found in AAS cases are associated with IL-13 upregulation in CD4+ T cells.121
In AR, increased HDAC activity may be involved in the pathogenetic mechanism by elevating pro-inflammatory cytokine amounts and reducing anti-inflammatory cytokine levels. Early responses are characterized by increased IL-4 expression,124 H3K9 acetylation, and H3K4 trimethylation at the IL4 locus.125 A study showed HDAC1 upregulation in nasal epithelial cells from AR patients,126 and IL-4 increased HDAC1 expression, leading to nasal epithelial barrier dysfunction. HDAC1 inhibition promotes the master regulators of T cell function, including IL-10 and CCL8, and prevents excessive activation of immune cells.127 Histone acetylation is also critical for AD pathogenesis. In AD pathogenesis, the demethylation, acetylation, and methylation of the H3 residue in the FOXP3 promoter gene region, along with the hypermethylation of the RORC gene and the methylation of the H3 residue, promote the regulation of Th0 cells. The differentiation of Tregs,34,92,94,95 thereby reduce the levels of histone acetylation at Th1 and regulatory sites.128
Non-coding RNAs in allergic diseases
Long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides in length; functional RNAs that are not translated include micro-RNAs (miRNAs), small interfering RNAs (siRNAs), lncRNAs and Pivi-interacting RNAs (piRNAs). They are essential signaling and regulatory tools that affect transcriptional processes and may also alter gene expression post-transcriptionally, with critical roles in the development of allergic diseases. We take microRNAs as an example to explain their important roles in allergic diseases.
miRNA-21, high expression of miRNA155, and low levels of Let-7a were detected in peripheral blood specimens from asthmatic children. These markers can be used for the diagnosis and prognosis of childhood asthma;129 up-regulation of miR-126 in peripheral circulation is related to immune imbalance and is considered a biomarker for asthma diagnosis.130
MicroRNAs have critical functions in the development of allergic diseases. MiR-19b reduced airway remodeling and inflammation as well as oxidative stress by downregulating TSLP to inhibit Stat3 signaling in mice with experimental asthma.131 MMP-16 and ATG7 via miR-192-5p molecules reduce airway inflammation and remodeling.132 MiR-221 can control the enhanced airway smooth muscle cell proliferation in severe asthma cases.133 The circular RNA (circHIPK3) contributes to smooth muscle cell proliferation and airway remodeling in asthma patients via miR-326/ stromal interaction molecule 1 (STIM1) signaling.134 MiR-130a-3p and miR-142-5p mediate lung macrophage polarization and are associated with airway remodeling.135 MiR-155 and miR-221 are closely associated with the regulation of Th2 responses and airway smooth muscle hyperproliferation in asthmatic patients.133,136 Vascular endothelial growth factor A (VEGF-A) amounts are elevated in sputum and serum samples from asthmatics, and has-miR-15a is associated with VEGF-A downregulation in CD4+ T cells.137 MiR-21 downregulates IL-3, IL-5 and IL-12, and inhibits IFN-γ and IL-12 production by dendritic cells, and reduces IFN-γ biosynthesis in CD4+ T cells.138 MiR-21 overexpression was also associated with the differentiation of Th2 cells in vitro. In granulocyte-infiltrating asthma, miR-221-3p in epithelial cells and sputum was inversely associated with airway eosinophilia.139 Downregulated miR-28-5p and miR-146a/b activate blood CD8+ T cells in severe asthma.140 MiR-223-3p, miR-142-3p, and miR-629-3p are involved in severe neutrophilic cellular asthma.141 MiR-126 induces Th2-type eosinophilic asthma,142 and miR-23-27-24 regulates T cell function and differentiation; meanwhile, miR-24 and miR-27 suppress Th2 cell differentiation, leading to IL-4 cytokines.143 Recently published reports revealed miR-200a is involved in asthma pathogenesis via phosphatidylinositol 3 kinase (PI3K)/ RAC-alpha serine/threonine-protein kinase (AKT) signaling.144–146 In childhood asthma, the miR-29c/B7-H3 axis controls the differentiation of Th2/Th17 cells, and the above microRNA studies might point to novel research directions for developing treatments for AAS.147
In AR, aberrantly expressed circulating lnc-NEAT1 and miR-125a were associated with Th2 cell percentage and symptoms in pediatric AR.148 In AD, Liew et al. found decreased expression of miR-335 in AD lesions compared with healthy control skin.149 Nuclear factor kappa-B (NF-κB) (p65) is a critical modulator of inflammatory immune response, and miR-124 is associated with inflammatory response and may constitute a new effector and regulator of NF-κB.150,151 In diseased skin, miR-124 is downregulated in AD patients.152 In macrophages, miR-155 targets IL-13Rα1.153 In dendritic cells (DCs), miR-221 knockdown or miR-155 overexpression promotes apoptosis, while miR-155 overexpression in mDCs enhances the production of IL-12p70.154 The study of microRNA would bring new hope in the treatment and understanding of allergic diseases.
Cell signaling pathways play critical roles in allergic diseases
Allergic diseases are immune disorders caused by an imbalance of the immunity, in which immune cells play an important role. Signaling pathways are important in intercellular signaling. This review provides a systematic review of the signaling pathways involved in allergic diseases from the nucleus to the cell membrane, in the hope of laying a solid foundation for the study of allergic diseases.
Notch signaling pathway
In the 1910s, the Notch gene was detected in Drosophila melanogaster with notched wings.155,156 Notch signaling is highly conserved. In mammals, NOTCH has four paralogs, including NOTCH1-4, with redundancy and distinct roles.157 Human Notch1-4 genes map to chromosomes 9, 1, 19 and 6, respectively. The NOTCH receptor undergoes three cleavages and is transferred to the nuclear compartment to regulate target genes transcriptionally. Notch signaling is divided into the canonical and non-canonical pathways with complex functions, but the pathway is now well known.158 It is mainly involved in diverse molecular events across species, including tissue functional damage and repair; abnormal Notch pathway might lead to different pathological processes.
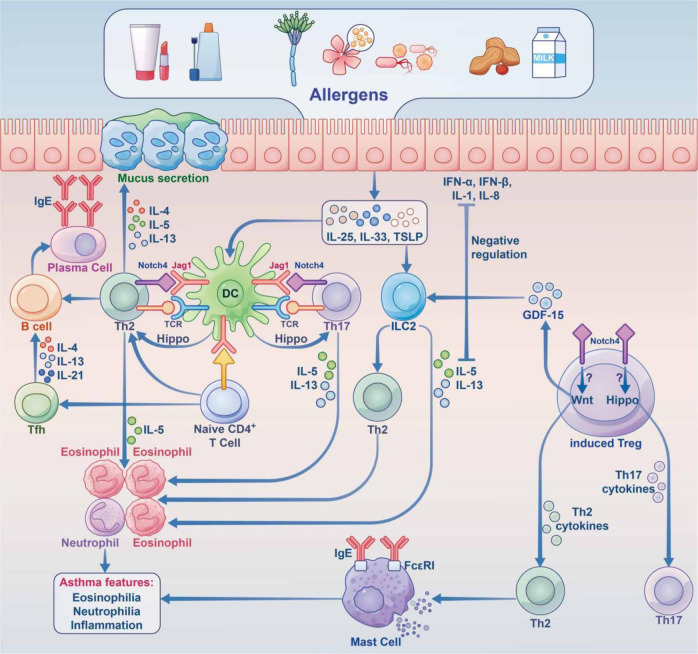
In AAS, eosinophilic asthma is dominated by Th2-type immune responses, and Notch signaling upregulates the key transcription factor Gata3.159,160 NOTCH4 is known to be critical for asthma development (Fig. 3). Repeated allergen exposure induces Tregs that produce high amounts of Notch4, which activates downstream Wnt and Hippo pathways, thereby promoting the transformation of iTregs into Th2 and Th17 cells, and exacerbating AAS.160,161
Fig. 3.
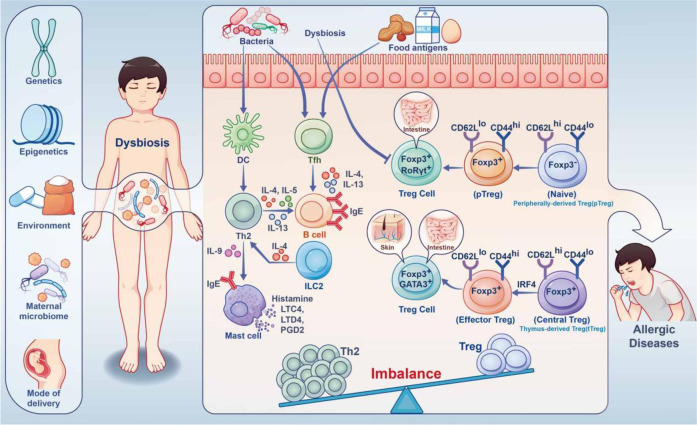
Immune imbalance caused by dysbiosis under the combined effect of gene environment in IgE-related FA. During childhood, the human microbiota is influenced by a combination of the maternal microbiome, mode of delivery, genetics, epigenetics, environment, etc. Dysbiosis resulting from aberrant damage to the gut microbiota early in life impairs Treg differentiation. This results in imbalance of Treg and Th2 cells. Food allergens and the microbiota promote T follicular helper (Tfh) responses to induce B cells, which produce large amounts of IgE through IL-4, IL-13 cytokines, causing allergic reactions
In other allergic diseases, such as AR, FA, and AD, the exact underpinning mechanism remains unclear, Notch signaling also plays important roles. In a study of AR, the serum amounts of Notch1 and Jagged1 (Jag1) in AR patients were significantly increased, which was confirmed in mouse experiments. In this study, Notch signaling could downregulate Foxp3 expression and inhibit Treg differentiation, thereby promoting AR occurrence and development.162 In a FA study, blocking the Notch signaling pathway could suppress Th2 polarization, increase Th1 cell differentiation and promote Th1/Th2 balance in a mouse model, thereby preliminarily verifying that blocking the Notch signaling pathway inhibits ovalbumin (OVA)-induced FA.163 In addition, Tfh cell production and function are dependent on Notch signaling, Notch receptors 1 and 2 are required for Tfh cell production, and Notch signaling enhances the production of type 2 cytokines in Tfh cells.164 Administration of a Notch signaling inhibitor inhibits IgE-mediated proliferation of intestinal mucosal mast cells (MMCs) in mice with food hypersensitivity, thereby attenuating allergic diarrhea and anaphylaxis.165 In AD, the epidermis of AD patients exhibits a marked deficiency in Notch receptors, leading to the upregulation of alarm TSLP, which triggers Th2-associated responses as well as TSLP and IL-31-related pruritus.166,167 Keratinocyte-produced TSLP and granulocyte-macrophage colony stimulating factor (GM-CSF) induce macrophage and DC activation, drive Th2 polarization, and promote eosinophil and mast cell infiltration, thereby enhancing the immune response.168,169
JAK/STAT signaling pathway
Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling represents a relatively simple membrane-nucleus pathway that mainly upregulates diverse key modulators involved in cancer and inflammatory processes. The well-conserved JAK/STAT pathway consists of ligand-receptor complexes, JAK and STAT. JAK consists of four cytoplasmic tyrosine kinases, i.e., JAK1, JAK2, JAK3 and TYK2. STAT proteins comprise STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b and STAT6. Each cytokine requires interaction with specific receptors on its target cells to activate this pathway, and these receptors contain associated intracellular domains composed of JAK family members.170 JAKs are inactive before exposure to cytokines, which induce JAK activation through phosphorylation by binding to their receptors.171–173 Activated JAKs phosphorylate the receptors on specific tyrosine moieties in the intracellular tail.174 STATs at receptor sites are also phosphorylated by JAKs.175,176 STATs are phosphorylated and translocated into the nuclear compartment to transcriptionally upregulate specific genes,177 often leading to cell proliferation or differentiation.
JAK/STAT signaling is highly involved in the differentiation of Th cell subsets; Th1 cell differentiation is controlled by the IFN-γ/STAT1 and IL-12/STAT4 signaling pathways.178,179 Meanwhile, Th2 cell differentiation is modulated by IL-2/STAT5 and IL-4/STAT6 signaling.180,181 The differentiation of Th17 cells mainly requires the involvement of STAT3/STAT4 signaling induced by IL-6 or IL-23.182 In allergic diseases, the JAK/STAT pathway is critical for cell proliferation and differentiation. In both rat and human bronchial smooth muscle cells, IL-13 induces JAK1-STAT6 signaling, which regulates Ras Homolog Family Member A (RhoA) activation that promotes smooth muscle contraction.183,184 IL-4 and IL-13 induce STAT6 in target cells with JAK involvement, and gene-targeted knockout mouse assays revealed STAT6 contributes to IgE synthesis, bronchial hyperresponsiveness and airway remodeling upon allergen sensitization.185,186 Further type 2 asthma-related cytokines, including IL-5 and TSLP, signal through JAK-dependent pathways. The JAK signaling pathway is critical for the differentiation of naive precursors into CD4+ Th2 cells, and the key cytokines involved are IL-2 and IL-4, which bind to cytokine receptors coupled to JAK1 and JAK3, respectively, then induce STAT5 and STAT6.187
In AAS, cytokine receptors, e.g., IL-4, IL-5, IL-13, IL-31, and TSLP, promote JAK/STAT signaling activation.188,189 In AD, Th2 immune enhancement induced by JAK/STAT signaling downstream of multiple cytokines, including IL-4, IL-5 and IL-13, is considered an essential pathogenic pathway.190 It was demonstrated that the JAK-STAT pathway regulates inflammatory processes and induces changes in the natural skin barrier, increasing TEWL (transepidermal water loss) by upregulating IFN-γ, IL-31, IL-23, and IL-22.190 STAT3 is one of the factors responsible for IL-23 expression induced by IL-6 from DCs, which is critical for Th17 lymphocyte differentiation and cellular memory, leading to a disruption of epithelial barrier integration.191
NF-κB/MAPK signaling pathway
By 2022, NF-κB has been known for 36 years. NF-κB (nuclear factor) is a protein factor with gene transcriptional regulation, which is present in almost all nucleated cells. When cells are stimulated by inflammatory mediators, NF-κB protein is activated in the cytoplasm and enters the nucleus to regulate the expression of various inflammatory factors, playing an important role in allergic diseases.
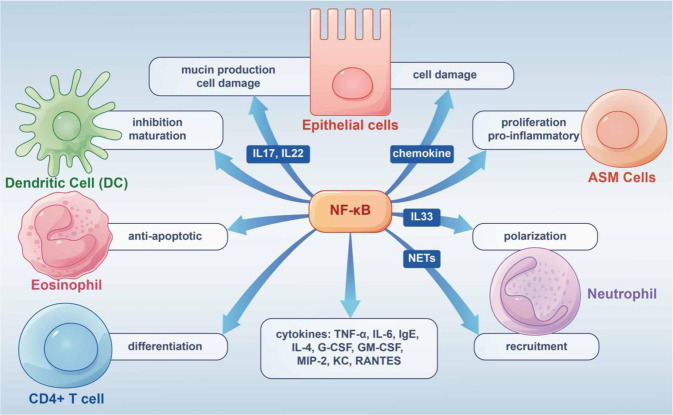
In AAS, the NF-κB/MAPK pathway controls inflammatory and immune responses by regulating TNF-α and IL-6.192–194 A 2019 study demonstrated that after nuclear translocation of phosphorylated P65, inhibited NF-κB/MAPK signaling may modulate IgE and IL-4 production.195 Enhanced NF-κB nuclear binding or production was also found in inflammatory cells collected from induced sputum in asthmatic patients.196 Besides, experiments have shown enhanced NF-κB activation in airway tissues and inflammatory cells challenged by allergens such as ovalbumin (OVA) and house dust mite (HDM) extract.197–199 In addition, the studies related to increased Tfh cells in allergic diseases, NF-κB deficiency led to a decrease in CXCR5 (Tfh cells expressing chemokine receptors) in mice and a consequent decrease in the number of Tfh cells.200 All these studies suggest that NF-κB signaling is essential in the cellular immune response of allergic diseases, especially AAS. The following focuses on the functions of NF-κB in immune cells in allergic diseases (Fig. 4).
Fig. 4.
Graphical summary of NF-κB pathway’s role in allergic diseases. The NF-κB pathway is highly involved in the occurrence and development of allergic diseases by acting on different cells and releasing inflammatory factors
Epithelial Cells
Epithelial cells play a key role in airway diseases and constitute a critical interface between the body and the environment.201 The epithelium coordinates responses to diverse invasive injuries via the production of multiple immunomodulators and inflammatory factors controlled by NF-κB. The latter mediators mostly comprise chemoattractants that induce inflammatory cell infiltration affecting epithelial function; among these, TSLP synthesized by bronchial epithelial cells is essential in Th2 responses that trigger allergic airway inflammation.201 The neutrophil-associated proteins S100A8 and S100A9 induce mucin production in airway epithelial cells through toll-like receptor 4 (TLR4)-related NF-κB pathway activation during AAS attacks.202
In isolated mouse airway epithelial cells, NF-kB activation upregulates IL-6, granulocyte colony-stimulating factor (G-CSF), GM-CSF, macrophage inflammatory Protein 2 (MIP-2), keratinocyte-derived chemokine (KC), and RANTES. NF-κB activation in mouse epithelial cells also leads to the recruitment of neutrophils for innate immune response.203 In the OVA-induced model of allergic diseases, NF-κB activation leads to airway inflammation, goblet cell hyperplasia, and induced expression inflammatory cytokines, including IL-15, IL-10, and IL-9.204
Airway Smooth Muscle (ASM) Cells, Neutrophils and Eosinophils
In AAS, acute contraction of ASM is the main factor that causes bronchospasm. ASM cells contribute to persistent histological alterations in the airway wall, ASM and inflammatory cells are important players in inflammation,205 in which thrombin and IL-1α stimulate NF-κB signaling in ASM cells.206 IL-8 over-secretion by ASM cells may increase NF-κB’s binding to the IL-8 promoter.207
AAS includes eosinophilic, neutrophilic, oligogranulocytic and mixed granulocytic types, based on the type of inflammatory infiltrating cells. Of these, neutrophil infiltration is an important cause of AAS exacerbation and resistance to hormone therapy.208 Th17 lymphocytes are critical for neutrophilic asthma, and are the major producers of IL-17A, IL-17F and IL-22, whose amounts are elevated in the airways of severe steroid-refractory asthma cases.209 NF-κB signaling is associated with IL-17 and/or IL-22-related production of epithelial mucin and ASM cell proliferation.210–212 IL-33 promotes neutrophil polarization via c-Jun N-terminal kinase and NF-κB-related pathways. NETs induce CXCL1, CXCL2, and CXCL8 expression in airway cells through TLR4/NF-κB signaling, thereby recruiting neutrophils to inflammatory sites.213 In neutrophilic asthma (NA) mice, NETs trigger the expression of chemokines by airway and alveolar epithelial cells that promote the recruitment of more neutrophils through the TLR4/NF-κB pathway, leading to epithelial cell damage.214
Airway inflammation in eosinophilic allergic asthma features infiltrated and activated eosinophils, and co-culture of epithelial cells with mast cells or eosinophils induce NF-κB-dependent cytokine production by airway epithelial cells.215,216 NF-κB signaling is critical in the survival of eosinophils, exerting an anti-apoptotic effect through autocrine TNF-α.217 NF-κB suppressors on the other hand, including MG-132, reduce eosinophil amounts and alleviate allergic inflammation.218
Dendritic cells and Lymphocytes
DCs interconnect innate and adaptive immune systems, with crucial roles in promoting immune defense and maintaining immune tolerance. Previous reports have established NF-κB signaling involvement in DC development, with NF-κB suppression preventing DC maturation associated with the upregulation of MHC and co-stimulatory molecules.219
In eosinophilic allergic asthma, naive T cells differentiate and mature into Th2 cells, which biosynthesize IL-4, IL-5 and IL-13 with NF-κB involvement, and stimulate B lymphocytes to produce immunoglobulin E (IgE).220 During CD4+ T cell differentiation, IL-6 and TGF-β are highly involved in Th17 cell differentiation.221 NF-κB signaling regulates antigen-presenting cell function and controls CD4+ T cell differentiation into Th effector cells.222,223 However, the present study is still inconclusive.
Hippo signaling and allergic disease
Hippo signaling was first discovered in Drosophila and is a highly conserved pathway.224 It mainly comprises the cascade kinase cascade transcription molecule mammalian STE20-like kinase 1/2 (MST1/2), WW domain of Sav family containing protein 1 (SAV1), and MOB kinase activator 1 (MOB1), with large tumor suppressor 1/2 (LATS1/2) upstream and Yes-associated protein (YAP)/effector molecules with PDZ- binding motif (TAZ) downstream. When Hippo signaling is not activated, unphosphorylated YAP undergoes nuclear translocation and interacts with TEAD, thereby triggering the transcription of target genes. After Hippo signaling activation, TAOK induces MST1/2 phosphorylation, and phosphorylated MST1/2 interacts with SAV1 for MST1/2-SAV1 complex formation. With activated MOB1, the latter complex phosphorylates LATS1/2. In turn, LATS1/2 phosphorylation triggers YAP activation, causing YAP capture by 4-3-3 proteins in the cytosol or degradation by SCFβ-TRCP E3 ubiquitin ligase-mediated ubiquitin-proteasome signaling.225
In AAS, Hippo signaling mainly induces cell differentiation. The Notch4 protein mediates immune tolerance and leads to Treg dysfunction, thereby promoting allergic airway inflammation.226 After alveolar macrophage engulfment of allergens and particulate pollutants, Jag1 is highly expressed on alveolar macrophages, thereby activating Notch on CD4+ T cells and promoting inflammation associated with Th2 and Th17 effector T (Teff) cells;227,228 at the same time, alveolar macrophages secrete a large amount of IL-6, which promote the expression of Notch4 on induced regulatory T (iTreg) cells, thereby activating Hippo signaling, which further exacerbates Th17 cell-induced inflammation (Fig. 5).161 In other allergic diseases, including AR, AD and FA, no associations with Hippo signaling have been reported.
Fig. 5.
The roles of the hippo and Notch pathways in AAS. Under stimulation by allergens, epithelial cells synthesize large amounts of proinflammatory cytokines (IL-25, IL-33, TSLP, etc.), thereby acting on innate lymphocytes (ILC2 cells) and DCs. Jag1 on DCs interacts with Notch receptors on T cells for Notch pathway induction. Notch transforms induced Tregs into Th2 and Th17 cells. Naive CD4 + cells affect Tfh cell class switch recombination by secreting IL-5, thus acting on B cells to induce plasma cells, which produce IgE. At the same time, Th2 cells secrete IL-4 and others to activate B cells to synthesize IgE, which interacts with IgE receptors on mast cells. In case the allergen invades the body again, it directly cross-links with IgE on the cell surface and releases a variety of active mediators, which trigger the clinical symptoms of asthma. Th17 cells are activated through the Hippo pathway; Th2 cells are activated through the Wnt pathway, and GDF-15 molecules are stimulated to act on ILC2 cells to enhance the expression of IL-13, although this remains controversial. Notch converts induced Tregs into Th2 and Th17 cells via hippo pathway-dependent mechanisms. IL interleukin, TSLP thymic stromal lymphopoietin, ILC2 group 2 innate lymphoid cell, DC dendritic cell, Jag1 jagged1
The newly discovered Hippo pathway plays a critical role in the immune function of the body, with complex crosstalk with other signaling pathways, and is regulated by other signaling pathways (such as Notch, Wnt signaling pathway, etc.). As the study of Hippo signaling pathway continues to deepen, its important function in allergic diseases will be gradually discovered.
TOLL-like receptor (TLR) signaling pathway
The increasing prevalence of allergic diseases is not only related to changes in the modern living environment (such as pollution, low-endotoxin living environments, smoking, etc.), which may induce the disorder of immune system,229,230 but also closely related to the loss of microbial biodiversity.
TLRs represents an important group of transmembrane protein receptors that are critical for proper activation of innate immunity, are highly conserved, and comprise binding domains containing arginine-rich repeats. As molecules involved in the first line of defense, TLRs are induced by pathogen-associated molecular patterns (PAMPs), which are found in diverse pathogenic organisms and absent from the host. There are eleven known human TLRs (TLR1 to TLR11), all with functions except for TLR11.231,232 TLRs are located on and within immune and non-immune cells, respectively, and are critical for initiating adaptive immune responses, including in alveolar macrophages, mast cells, epithelial cells, neutrophils, natural killer cells, and antigen presenting cells (APCs). Different TLRs recognize various groups of molecules in diverse pathogens. For example, multiple diacyl peptides can be recognized by TLR1 and TLR6, while liposomes are recognized by TLR1/2.233 TLR5 cooperates with TLR4 to recognize bacterial flagellin.234 Studies have shown that gut microbiota regulates the activity of Th1 and Th2, thereby affecting the formation of immune tolerance in the body.235 To maintain immune homeostasis, the activation of innate immune cells requires TLR signaling molecules, and innate immunity should be activated correctly to stimulate the body’s immune response.231
The strength of the TLR signaling pathway determines the possibility of allergic diseases. Accumulation of TLR4 signaling on DC was detected in house dust extract (HDE) hypersensitivity.236,237 Bacteria belonging to the healthy lung microbiome elicit baseline TLR response, whereas those involved in asthma show stronger TLR response. Mice administered asthma-associated proteobacteria show elevated amounts of neutrophils and cytokines in comparison with animals administered commensal Provetella.238
The strength of TLR signaling does determine the occurrence of allergic reactions or its absence. Strong TLR signals are protective against allergic airway disease, while low airway amounts of TLR ligands cause airway sensitization and Th2-type immunity.239–241 Studies have shown that low-level flagellin (TLR5 ligand) can enhance OVA-associated hypersensitivity in mice, whereas a high flagellin dose protects the animals from hypersensitivity by producing CD25+ Treg-dependent regulatory DCs and T cells.242 Differential responses to TLR activation have been considered a shift from Th2-type immunity to Th1-type immunity with increasing stimulation intensity.
In AAS, alveolar macrophages are immune cells with critical roles in the clearance of immune antigens. Excessive inflammatory response of these cells, however, might induce tissue damage.243 Alveolar macrophages are important in developing tolerance to inhaled allergens by inhibiting T cell proliferation and APC function.244 Asthmatic patients have reduced monocyte and macrophage amounts in comparison with healthy control individuals.245 TLR2, 4, 5, 6, 7, 8 and 9 are expressed on macrophages. TLRs have a critical function in AAS; among the TLRs expressed by human lung cells, TLR1-5, 7 and 8 have the highest expression.246 TLR2-6 and TLR9 are highly expressed on human airway epithelial cells.247,248 Mouse alveolar macrophages highly express TLR2, 4 and 9,249 and mouse macrophages produce TLR1-7 and 9 mRNAs.250,251
Lipopeptides in Gram-positive bacterial organisms and mycoplasmas show diacylation, while lipopeptides in Gram-negative bacterial and mycobacterial species show triacylation. When TLR2 polymerizes with TLR1 or TLR6, the lipopeptide is recognized by TLR2. The TLR2/TLR6 heterodimer recognizes diacylated lipopeptides such as S-FSL1 (TLR2/6), R-FSL1 (TLR2/6/CD36) and MALP-2 (macrophage-activating lipopeptide-2). Cell wall constituents in Gram-positive bacteria, including lipoteichoic acid, also bind to the TLR2/TLR6 heterodimer.252 Phagocytic cells, airway epithelial cells, smooth muscle cells, glia, mouse bone marrow-derived mast cells, and B cells all can express the TLR2 receptor.253 In OVA-sensitized animal model, TLR2 receptor pathway induction also results in enhanced pause and increased bronchioalveolar lavage fluid (BALF) eosinophil amounts.254,255 Elevated TLR2 ligand amounts can also elevate serum IgE concentration. Though TLR6 is heterodimerized with TLR2, and the heterodimer plays an important role in AAS, TLR6 is decreased in PBMC compared with healthy controls, and is also overexpressed in severe asthma cases compared with mild asthmatic patients.256 TLR4 uses the adaptor protein TRIF via myeloid differentiation primary response gene 88 (MyD88)-dependent and MyD88-independent pathway, respectively, enhancing IRF-3 induction and IFN-β production.257 In bronchial asthma cases, it was shown that TLR4 activation of macrophages produces cytokines that affects immune balance and thus affects the Th1/Th2 balance.258 After TLR5 recognizes flagellin, it induces NF-κB signaling through MyD88 and TNF receptor associated factor 6 (TRAF6), producing cytokines to trigger an inflammatory response.245 Recently, Nawijn et al. showed that intranasal TLR2 induction by aerosolized allergens promotes allergen-specific Treg proliferation to suppress asthma in a mouse model.259 Numerous studies have shown that TLR2 stimulation by parenteral or mucosal treatment with synthetic agonists prevents APCs from triggering Th2-polarizing responses, reducing IgE antibodies and immunogenicity in a mouse model of asthma.260–263
In FA, Treg activation is an important pathway by which gut DCs and macrophages induce immune tolerance, disrupting the normal immune homeostasis of the intestine.264 All microbial pattern recognition receptors (PRRs) may be involved in food tolerance and allergen presentation. TLR2 is highly expressed by intestinal epithelial cells (IECs) and DCs, and most commensal bacteria in the intestine are gram-positive organisms, meaning they have a high ability to induce TLR2.265,266 In AR, researchers were surprised to find that TLR4 inhibits allergic response in OVA-induced AR in a mouse model.267 TLR2, 3 and 4 were highly expressed in nasal mucosa specimens from AR cases in a study of 27 healthy control individuals and 42 cases of seasonal allergic rhinitis.268 All these studies have once again confirmed that TLR is critical for the etiology and progression of allergic disorders, which is worthy of further exploration.
Wnt/β-catenin signaling
In 1982, the Wnt gene was described as integrase-1 in murine breast cancer cells and the wingless gene in Drosophila.269 Wnt signaling plays core roles in the maintenance of progenitor cells and stem cells, the differentiation of T cells, and the regulation of cellular immunity. Among the Wnt-mediated signaling pathways, the most classical Wnt/β-catenin pathway contributes to maintaining human tissue homeostasis. Wnt, which mediates extracellular signals, is a secretory glycoprotein, with 19 human Wnt proteins reported as of now. This pathway relies on β-catenin, which is activated by binding extracellular Wnt ligands to membrane receptors through autocrine and paracrine processes, inhibiting the degradation of β-catenin so that it can be stably accumulated in the cytoplasm and transferred to the nuclear compartment, where it can work together with T cell factor/lymphoenhancer binding factor to stimulate the transcription of target genes.270,271
Wnt/β-catenin contributes to airway remodeling in asthma by upregulating the tenascin C/platelet-derived growth factor receptor (PDGFR) or activating p38 MAPK and its target genes c-Myc and cyclin D1 to induce proliferation in airway smooth muscle cells.272,273 A study by Trischler et al. revealed that Wnt10b, known as the classical Wnt ligand, is highly produced by T cells in AAS, and its absence increases the activation of cultured T cells and enhances immune response in animal models.274 In addition, after mesenchymal stem cell-derived exosomes and vitamin D inhibit Wnt/β-catenin signaling, airway remodeling is reduced, thereby inhibiting chronic allergic inflammation in the airway.275,276 Upregulated Notch4 in blood Tregs from asthma patients differentiates Tregs into Th2 and Th17 T cells through a Wnt and Hippo pathway-dependent mechanism, and Wnt induction upregulates growth and differentiation factor 15 (GDF15) in Tregs, and this feedforward mechanism exacerbates inflammation.226 However, Wnt-1/β-catenin pathway induction promotes allergic airway diseases. Overexpression of Wnt1 reduces DC migration to draining lymph nodes and induces an appropriate T cell tolerance response without causing T cell proliferation.277
Related literature reported that inactivation of Wnt/β-catenin signaling could reduce nasal mucosa damage and eosinophil infiltration, decrease the infiltration of nasal mast cells and enhance red blood cell immune adhesion, thereby reducing the progression of AR.278
There are few reports on this pathway in FA and AR. Research in allergic dermatitis shows that Notch deficiency is the basis for inhibiting epidermal differentiation and skin barrier defects, thus enhancing Wnt pathway, which is very important for the proliferation of epidermis cells.279
PI3K/AKT signaling
PI3K in the lipid kinase family interacts with the PH domain of the AKT protein (also known as PKB), inducing its conformational change and AKT phosphorylation. Activated AKT is transferred from the cytosol to the plasma membrane, and subsequently induces its downstream effectors, including mammalian target of rapamycin (mTOR).280
PI3K inhibitors have been considered to have great potential in the treatment of inflammation. Current evidence shows that PI3K participates in the pathogenesis of asthma through two main mechanisms. First, PI3K increases the permeability of mouse blood vessels to enhance antigen-induced airway inflammation and high reactivity.281–283 In the OVA-induced asthma model, inhibition of PI3K110δ subtype (PI3K-δ) reduces the activation of HIF-1α in airway epithelial cells, as well as antigen-induced airway inflammatory reactions and hyperresponsiveness, by regulating vascular leakage mediated by VEGF.284 Secondly, the PI3K pathway induces airway smooth muscle cell proliferation, promotes airway smooth muscle thickening and luminal stenosis, thereby participating in airway remodeling.285 With respect to immune cells, PI3K regulates the differentiation of Th cells and eosinophils in asthma,286,287 and affects the occurrence and development of inflammation in asthma. In an animal model of AAS, it was found that PI3K and AKT activities in lung tissue are increased, as well as the expression of mTOR. After treatment with a PI3K inhibitor, some pathological manifestations of asthma (such as increased amounts of activated chemokines in eosinophils, bronchoalveolar lavage fluid IL-5 and IL-13, lung tissue eosinophilia, increased mucus secretion in the respiratory tract, airway hyperresponsiveness, etc.) are obviously inhibited, and PI3K/AKT signaling highly regulates asthma pathogenesis.288
In AR, drug development research found that the anti-allergic drug α-TCP (alpha-tocopherol) and the androgen receptor antagonist bicalutamide both play anti-inflammatory and alleviating roles in AR in animal models by inhibiting the PI3K/AKT/mTOR pathway in mast cells.289,290 A mouse model with AR shows that increased leptin enhances the expression of type II innate lymphoid cell (ILC2) transcription factor and type II cytokines through the PI3K/AKT pathway.291 For mice lacking CCR3 gene in the bone marrow, the activity of PI3K/AKT signaling was also significantly reduced, and nasal eosinophil infiltration was inhibited; in addition, serum Th2 cytokines were reduced, and the symptoms of AR in mice were alleviated.292 It was found in cell experiments that ST2/PI3K/mTOR-mediated autophagy is inhibited by IL-33 secreted by nasal epithelial cells, thereby promoting mast cell degranulation in allergic asthma.293
In the skin, dysregulated PI3K/AKT pathway might result in serious pathologies featuring unchecked cell proliferation and inflammatory response.294 In addition, PI3K/AKT signaling also modulates mast cell degranulation via miRNAs in allergic skin diseases. High-expression miR-126 induces IgE-mediated mast cell degranulation related to PI3K/AKT signaling by increasing Ca2+ influx.295
Serum chitinase 3-like 1 (CHI3L1) amounts are increased in individuals with allergic disorders and promote Th2-related immunity and the polarization of M2 macrophages through PI3K/AKT signaling in FA.296
mTOR signaling pathway
mTOR represents a serine/threonine protein kinase that belongs to the PI3K-associated protein kinase (PIKK) family.297 While associated with the above paths, mTOR signaling may serve as an upstream response to other signaling pathways. mTOR has catalytic subunits in two different complexes, including mTOR complex 1 (mTORC1) and mTORC2,298 which have different susceptibilities to rapamycin, substrates, and functions.299 Studies in allergic diseases have shown that mTOR is a key molecule for sensing the immune microenvironment and determining the function and differentiation of immune cells,300 because it regulates a variety of immune cells and limits pro-inflammatory mediators.301,302 For example, increasing evidence supports that mTOR is an important regulator of Tfh cell differentiation. The balance of Tfh and Th1 cell differentiation in vivo is regulated by IL-2 signaling through PI3K, AKT and mTOR, and both mTORC1 and mTORC2 essentially promote Tfh cell differentiation and germinal centers (GC) formation, which cannot be ignored in allergic diseases.303,304
In AAS, the progenitor cells of granulocytes originate from the bone marrow and move to blood vessels and lungs when inflammation occurs. Among them, eosinophils regulate Th2 immune response, which is related to the severity of the disease,305 while ablation of mTOR leads to Gata-1 overexpression and increases eosinophil differentiation.306 The angiogenic factor fibroblast growth factor-binding protein 1 (FGFBP1) is highly expressed in asthma models with airway remodeling features, because activating mTORC1 and signal transducer and activator of transcription 3 (STAT3) signaling pathways enhances FGFBP1 expression and secretion, thus inducing angiogenesis.307
The target protein mTOR of rapamycin is involved in the growth of keratinocytes. Studies have found IL-13 activates the mTOR signaling pathway and downregulates miR-143, followed by the downregulation of epidermal barrier related proteins. Therefore, rapamycin could treat allergic dermatitis by inhibiting mTOR.308
FcƐRI signaling pathway
Fc receptors play major roles in adaptive immunity by interacting with immunoglobulins, among which FcεRI represents a high-affinity IgE receptor found on mast cells, basophils, eosinophils and APCs.309 When IgE binds to FcεRI to trigger immunity, FcεRI aggregation induces a variety of signaling pathways to regulate the secretion of allergy-associated mediators, including histamines and leukotrienes, and induces the transcription of Th2 cytokines and tumor necrosis factor (TNF) genes,310 which leads to potentially life-threatening allergic diseases. The tetrameric form of FcεRI is present in mast cells and basophils, while the trimeric form is found in other immune cells; FcεRIα, FcεRIβ and FcεRIγ (αβγ2) are encoded by the FcεR1A, FcεR1B (MS4A2) and FcεR1G genes, respectively.311,312
The pro-inflammatory effects mediated by FcεRI in different allergic disorders have the following similar mechanisms. Signal transduction in mast cells is induced by the phosphorylation of immune receptor tyrosine activation motif (ITAM) of FcεRIβ and FcεRIγ subunits by Src-protein tyrosine kinase. This results in the recruitment of tyrosine kinase Syk, which mediates the activation of some adaptor molecules (SLP76, LAT, etc.), leading to calcium mobilization.313 Therefore, dephosphorylation of tyrosine kinase activating signals downstream of the IgE-FcεRI complex may prevent allergic diseases,314 and TLR-mediated release of cytokines from mast cells depends on the expansion effect of FcεRI, which is more important in the late reactions associated with inflammation.315 There is a synergistic effect between TLR and FcεRI-mediated activation in basophils, which promotes Th2 cell differentiation and induces degranulation and cytokine release.316,317 Platelets depend on the interaction between allergens and allergen-specific IgE and FcεRI, and are directly involved in allergic asthma.318 In addition, FcεRI signaling can also activate the PI3K signaling pathway.314
In allergic diseases, sensory neurons exposed to allergens produce action potentials; the Ca2+ flux mediated by immune complexes increases, the action potentials discharge and neuropeptides are released, thereby causing pain or itching. The IgE receptor FcɛRI highly contributes to the development and remodeling of airway inflammation in allergic asthma.319 Studies in animals with experimental allergic asthma demonstrated that when the vagus nerve, which dominates the airway, senses the invasion of allergens, pain receptor neurons overexpress the immunoglobulin receptor FcɛRI and release Substance P, which drives the polarization of Th2 cells, thus triggering allergic inflammation.320
IgE-mediated FA is very common, and FcɛRI is also upregulated in the abdominal vagus nerve of mice with experimental food allergy, which promotes the skewed Th2 polarization in the intestine.321 Functional FcɛRI also exists in intestinal neurons, and stimulation of IgE antigen activates intermuscular neurons.322
NOD-like receptors signaling pathway
Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are located in the cytoplasm, and 23 species have been found in the human body to date.323,324 This pattern recognition receptor recognizes microbial compounds such as PAMPs and Damage-Associated Molecular Patterns (DAMPs) and cooperate with TLR and related pathways to trigger antibacterial immune response. The NOD-like receptor consists of the following three domains: (i) the nucleotide sequence located in the center has a domain that binds to NACHT, which drives the activation of downstream inflammatory caspase and NF-κB; (ii) the effector domain located at the N-terminal end mediates the interaction with adaptor proteins and downstream effectors and transmitting receptor excitability information; (iii) the C-terminal region is composed of leucine-rich repeats (LRRs), which constitute the microbial pattern recognition domain.325,326 NLRs recognize many pathogen-related model molecules, including microorganisms and toxins secreted by microorganisms327 which could be used for immune surveillance and host defense. The NLR signaling pathway mainly has the following functions: signal transduction, inflammasome formation, gene transcription stimulation and autophagy.328,329
After an NLR recognizes bacteria-related ligands, γ-D-glutamyl-meso- diaminopimelic acid (iE-DAP) and muramyl dipeptides (MDP), it promotes the expression of the adhesion molecule ICAM-1 on eosinophils and bronchial epithelial cells, and thus drive the cell adhesion, chemotaxis and migration of leukocytes. NLRs also induce eosinophils and bronchial epithelial cells to produce pro-inflammatory molecules such as IL-1β, IL-6, CXCL8, CCL2, CCL3, CCL4, and CCL5, thereby causing lung inflammation.330
In animal models, treatment of allergic asthma mice with NOD1 ligand induces subcutaneous fibrosis and significantly increases serum amounts of total IgE, eosinophils and the chemokine CCL5 as well as bronchoalveolar lavage fluid amounts of the Th2 cytokine IL-13.331 However, intranasal NOD2 ligand induces the expression of TSLP, IL-25 and OX40L in the lung,332 and these three molecules have been reported to promote asthma-related inflammation,333–335 blunt the production of antigen-specific CD4+Foxp3+ adaptive Tregs, and simultaneously drive CD4 T cells to produce IL-4, change the Treg/Th2 balance, block tolerance, and promote the susceptibility of airway inflammation dominated by eosinophils.332
The nucleotide-binding oligomeric domain-like receptor family Pyrin domain 3 (NLRP3) inflammasome contains NLRP3, ASC and Caspase-1, which are important constituents of the innate immune system, with critical roles in allergic disorders.331 The NLRP3-retinoid X receptor (RXR) axis drives airway epithelial cell apoptosis as well as the production of inflammatory cytokines in the lungs of asthmatic mice.336 Further study found that NLRP3 in bone marrow cells promotes the development and progression of AAS in an inflammasome-dependent manner, and RRx-001 (an inhibitor of NLRP3) could significantly decrease inflammatory cell infiltration and mucus secretion in the airway.337 PMs can cause acute exacerbation of allergic airway inflammation, activate TLR2/NF-κB/NLRP3 signaling and aggravate allergic airway inflammation.338
It was demonstrated that NOD1 and NLRP3 in AR patients are downregulated during the pollen season.339 Activation of the NLRP3/gasdermin D/IL-1β signaling pathway mediates macrophage pyroptosis and releases inflammatory mediators to local tissues, which is involved in nasal mucosa inflammation of AR.340
The microbiota plays an essential role in the occurrence and development of allergic diseases
In 1989, Strachan observed that children with more siblings in the family were less likely to develop hay fever or eczema;28 children are frequently exposed to allergens, hence the original “hygiene hypothesis” was proposed. This hypothesis is a good explanation for the phenomenon that AAS is significantly more prevalent in developed countries in comparison with underdeveloped countries. From an immunological point of view, it could be understood that the development of immune tolerance to allergens depends on the amount and degree of stimulation by microbial colonization and immune stimulating environmental signals transmitted in early life.341
With the rapid development of urbanization and industrialization, excessive use of hygiene products and antibiotics, coupled with changes in diet such as fast food, etc., would decrease microbial diversity in early life,342 resulting in impaired immune protection and the destruction of normal microorganisms.343 There is increasing evidence that the microbiota is critical for the occurrence and development of allergic disorders.344 The human microbiota mainly colonizes the gastrointestinal tract (GIT), with microorganisms also present in other body parts such as the oral cavity, nasal cavity, skin, and respiratory and reproductive tracts.345 Symbiotic microorganisms in the GIT and other organs mediate the innate and adaptive immune systems through the gut-lung and gut-skin axes. Reports have shown that many environmental factors influence the colonization, composition and metabolic activities of microbial communities in early life, thereby affecting the immune function of the body and leading to the occurrence of allergic diseases.346–349
Early-life activities and dysbiosis is tightly associated with allergic diseases
It is well established that microbial colonization starts at birth, and that microbiota composition is affected by related factors such as the prenatal and postnatal environment, which are also critical for the body’s immune function. Multiple factors, including mode of delivery,350–352 feeding choice,353 and use of antibiotics or not,354,355 can alter the composition of the gut microbiota and modulate infant tolerance to different allergens.
The use of antibiotics during pregnancy and the early postpartum period can affect the gut microbiota in normal infants and increase the risk of developing allergic diseases.356,357 Mothers exposed to antibiotics during childbirth had significantly lower microbial diversity compared with infants born to antibiotic-free mothers. The microbiota of antibiotic-exposed infants shows reduced amounts of Bacteroidetes and Bifidobacterium, alongside increased Proteus amounts. Studies have shown antibiotic utilization during pregnancy and childbirth is associated with elevated risk of AD and asthma.358–360 A study of 14,572 children, 10,220 of whom were administered antibiotics in the initial 2 years of life, revealed that early exposure to antibiotics had tight associations with childhood asthma, AR and AD.361 Studies in germ-free laboratory animals further demonstrated an interdependent association of gut microbiota with immune system development.362–365
Dysbiosis is tightly associated with allergic disease occurrence, and in some way affects the balance of immune cells in allergic diseases. Most AAS begins in childhood, and HDM, cockroach remains, pet dander, fungi and pollen are the main allergens.366 A study found lower abundances of Lachnospira, Veillonella, Faecalibacterium, and Rothia in the gut of infants are associated with higher asthma risk, and inoculating these bacteria in germ-free (GF) mice could alleviate airway inflammation and prevent the development of asthma.367 Furthermore, decreased abundance of Bifidobacterium was found in adult asthmatic patients.368 Haemophilus, Moraxella and Neisseria spp. were also observed in the airway microbial composition of asthmatic patients, and Proteus was also found in mild cases not receiving inhaled corticosteroids as well as in severe asthma cases. Actinobacteria and Klebsiella species were markedly enriched in severe asthma cases in comparison with healthy controls or mild-to-moderate asthma cases.369 In AD patients, Staphylococcus aureus colonization is an important exacerbating factor in AD pathogenesis, and gut dysbiosis is also considered an important factor in AD pathogenesis. A metagenomic analysis data showed that S. aureus constituted approximately 90% of AD skin, leading to dramatically decreased skin microbial diversity.370 In animal models, alpha-hemolysin and extracellular vesicles produced by S. aureus lead to skin barrier dysfunction and promote atopic skin inflammation.371–373 Enterotoxins secreted by staphylococci promote allergic skin inflammation by triggering substantial T cell activation, and staphylococcal delta-toxins also cause allergic skin diseases via mast cell activation.374 Clinical cohort trials have shown an early reduction in gut microbiota diversity is strongly related to elevated AD risk,375 and the presence of gut microbiota subspecies such as Clostridium Perfringens, Clostridium difficile and Faecalibacterium prausnitzii is closely associated with reduced capability of producing short-chain fatty acids (SCFAs), while L. paracasei abundance reduces the susceptibility to AD.376–378 AR is a respiratory disease that occurs in the upper respiratory tract, which includes the nose and oropharynx. Firmicutes and Actinobacteria represent key microbiota constituents in the nasal cavity of humans, while Proteobacteria, Firmicutes, and Bacteroidetes represent key phyla in the oropharynx.379–381 Microbial diversity in seasonal AR (hay fever) did not decrease, but was instead elevated during allergy season.382 Staphylococcus aureus is a microorganism involved in perennial (non-seasonal) AR.383 The etiology of the elevated incidence of FA remains undefined and may be related to the mode of delivery (natural vs. surgery) or the changes in the microbiome in early life due to antibiotic use even in low amounts.384–386
The microbiota contributes to the innate and adaptive immune systems in allergic diseases
Human mucosal tissues, such as intestinal mucosa, nasal mucosa, and other surfaces, have regular exposures to complex microbial populations comprising commensal and pathogenic organisms. The host utilizes many molecular mechanisms to mediate mucosal innate immunity for microbial homeostasis. PRRs, including TLRs and NLRs, play major roles in recognizing pathogens and inducing innate immune responses.
Different gut microbiota have different activation pathways, and some gut bacteria, e.g., Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, and Proteus vulgaris, activate HEK-293 cells through the TLR2 and TLR4 pathways.387 Flagellate bacteria, including Salmonella, Listeria, Pseudomonas, and Escherichia coli, contain flagellin, which acts by binding to TLR5.388 TLR9 functions by recognizing unmethylated CpG motifs, especially GTCGTT motifs, in the DNA of gut bacteria, including Proteus, Bacteroides and Actinobacteria, as well as Lactobacillus plantarum, etc.389 TLR signaling stimulates the maturation of innate immune cells, to properly activate APCs and initiate a moderate immune response.231 Both exposure to allergic environments and the mother’s allergic status are all factors affecting an infant’s susceptibility to allergic diseases.390 Microorganisms are present in the placenta, amniotic fluid and meconium, which are in contact with the fetus in early fetal stage; therefore, the fetus needs to develop immune tolerance during the mother-fetal period to prevent infection.391 Natural killers, DCs and macrophages in the endometrium and trophoblasts are already induced during fetal development.392,393
In allergic disorders, the microbiome exhibits a crucial function in establishing adaptive and innate immune protection. For instance, children with reduced IgG responses to specific microbial antigens than healthy counterparts are prone to allergic diseases, including asthma, AD and FD.344,394,395 In studies of babies, higher AD risk had associations with reduced levels of Proteobacteria and elevated innate inflammatory response induced by TLR-4, and Ruminococcus decrease was associated with elevated TLR2-dependent innate inflammatory response.396–398 FA in early stage is also closely associated with reduced gut microbial abundance.344
Epithelial cells
Epithelial cells on the nasal and bronchial mucosal surfaces are critical for maintaining the healthy state of respiratory mucosa. Continuous and coordinated ciliary movement enables the removal of foreign invading substances such as pollutants or allergens from surfaces. Epithelial cells also produce different cytokines and chemokines to activate inflammatory cells; meanwhile, hyperactivation of epithelial cells may trigger the onset of several disorders, including asthma and AR.399
Epithelial cells recognize PAMPs through innate PRRs, e.g., TLRs and NLRs, and such interactions affect the proliferation of epithelial cells. Microbiota-derived metabolites, including SCFAs, also have effects on epithelial cells. SCFAs, for example, stimulate inflammatory pathways by interacting with GPRs on epithelial cells in the intestine.400 P-cresol sulfate (PCS) is a microbial-derived product produced in the gut. PCS selectively reduces CCL20 production by airway epithelial cells due to uncoupling of epidermal growth factor receptor (EGFR) and Toll-like receptor 4 (TLR4) signaling, a pathway that acts distally on airway epithelial cells to reduce allergic airway responses.401
DCs
DCs, as APCs, are critical for immune responses in contact with commensal microbiota. The presence of SCFA butyrate, an end product of microbial fermentation, stimulates human monocyte-derived dendritic cell (moDC) maturation, increasing IL-10 amounts while decreasing IL-6 and IL-12 levels.402 Meanwhile, mice treated with SCFA propionate could produce new myeloid DC precursors with strong phagocytic capability but poor capability of promoting Th2 responses in the lung.
In humans, TLR9 is highly produced by macrophages and plasmacytoid DCs.403 The CpG motif is prevalent in bacteria, and the CpG motif also recognizes plasmacytoid dendritic cells (pDC) expressing TLR9 that produce pro-inflammatory cytokines, induce Th1-like immune activation patterns and activate their migration, while CpG-A oligodeoxynucleotides (ODN) induces extremely high levels of IFN-α production and CpG-B ODN induces activation of murine bone marrow-derived DCs to secrete IL-12 and IL-6.404,405
In reports assessing AAS disease models in mice and rhesus macaques, macrophage responses to TLR9 activation mainly induce Th1 type immune response, including upregulated TNFα, IL6, IL-12, IL-18, IFN-α and IFN-γ,406 resulting in the immune imbalance of Th1/2 cells.407
Macrophages
The microbiome and associated metabolites, including SCFAs, affect the function of macrophages resident in tissues. In the gut, the SCFA butyrate promotes the anti-inflammatory response of macrophages and induces Treg differentiation by activating its receptor GPR109a, which is essential in inducing tolerance to food. Besides, butyrate exerts anti-inflammatory effects on macrophages by inhibiting IL-6, IL-12 and NO production.408 In the lungs of antibiotic-treated mice, macrophages are polarized towards an M2 hypersensitivity phenotype by prostaglandin E2 (PGE 2) produced by commensal fungi.409
Lipopolysaccharide (LPS) represents a soluble cell wall constituents in common Gram-negative bacterial organisms.410 LPS can interact with complex host systems, including cellular and humoral components of the immune system, to induce the production of multiple immunomodulatory cytokines.411 It has been shown that LPS regulates lung inflammation in asthmatic mice via the TLR4 pathway in alveolar macrophages and that different doses of LPS exposure determine the type of inflammatory response.412 Improvement of AD by modulation of Th1/Th2 immune system homeostasis with LPS-activated macrophages derived from pantoid epimer (IP-PA1).413
Mast cells (MCs)
Mast cells (MCs) represent major effector cells in allergic diseases. Increasing evidence suggests that the microbiota can modulate MC function, influence MC activation through direct interactions or secreted metabolites.414 In human MCs, co-culture with Lactobacillus rhamnosus downregulates high-affinity IgE and histamine H4 receptors, while upregulating IL-8, IL-10, CCL2 and TNF-α.414 In murine experiments, Lactobacillus paracasei inhibits IgE-mediated MC activation via TLR2. However, the inhibitory effect of Lactobacillus casei is mainly through direct cell contact and not dependent on TLR or NOD1/2.414
Eosinophils
Elevated numbers of eosinophils in AD, AR and eosinophilic allergic asthma are typical symptoms for allergic diseases.415–418 Eosinophils act as major effector cells driving innate immunity in allergy and other inflammatory diseases, with an important role in clearing microbes resident in tissues.419 Meanwhile, eosinophil function is also regulated by pathogenic microorganisms; for example, Clostridium difficile stimulates the release of eosinophil-derived neurotoxins by eosinophils.420 However, when eosinophils ingest the probiotic strain Bifidobacterium, neurotoxin release is significantly reduced.420 Very interestingly, probiotics such as Lactobacillus fermentum and Lactobacillus rhamnosus, alleviate allergic inflammation involving reduced eosinophil infiltration in multiple mouse models of asthma and AD, although probiotic strains have not been shown to have a direct effect on eosinophils.421,422 NLRP12 induces allergic skin inflammation by promoting peripheral DC retention as well as neutrophil migration.423
Basophils
TLR2 was identified as the primary receptor against S. aureus,244 and intracellular NLRs also modulate microbial pathogen recognition. Intracellular NLRs, including NOD1, NOD2, NLRP1, NLRP3, NLRP4, and the interferon-inducible protein AIM2 produce a series of inflammatory molecules to resist the invasion of microorganisms.424 NOD2 is considered a major player in innate immune response to S. aureus in the skin.425 Studies have found that NOD2 expression on basophils in the peripheral blood of AD cases is markedly reduced compared with that of healthy people,426 and basophils are the main effector cells involved in Th2 polarization in allergic inflammation. Acinetobacter in the skin prevents allergic inflammation and is critical for the regulation of Th1/Th2 cell balance and anti-inflammatory responses.427
Innate Lymphoid Cells (ILCs)
ILCs, a major group of innate immune cells, are present in all parts of the respiratory tract; ILC2 cells are predominantly found in mice, while ILC3 cells are predominantly found in the human respiratory tract.428,429 Microbial signaling affects the maturation of ILC tissue-specific functions. Clostridia has been shown to stimulate ILC3 to produce IL-22, which helps to strengthen epithelial barrier and reduce intestinal permeability to dietary protein.430 ILC3 cells tolerize T-cell response and prevent IL-22 production, leading to a loss of gut bacteria.409 Furthermore, when gut macrophages release IL-1β upon microbial insult, ILC3 cells release GM-CSF and induce immune tolerance.409 TNF-β produced by ILC3 cells is essential for maintaining gut microbiota homeostasis, and IL-25 is produced by epithelial tuft cells in a microbiota-dependent manner.409 The microbiota and its metabolites induce different types of ILCs and regulate their capability of preventing allergic reactions.
Tregs
Tregs are important immune regulatory cells, which suppress allergic diseases and have critical functions in controlling the immune response. Bifidobacterium longum 35624, Clostridium fragilis, and Bacteroides fragilis all induce Tregs in the intestine, whereas other bacteria do not induce Tregs.431,432 Activation of PRRs on DCs is a critical mechanism by which gut microbial organisms mediate Treg differentiation.433 Recently, it was found that infants with reduced levels of Bifidobacterium and Faecalibacterium have elevated relative risk of asthma, characterized by elevated amounts of IL-4+ Th2 cells and reduced Treg levels,434 attenuating the adaptive immune response. In IgE-induced FA studies, dysbiosis affected the differentiation of Tregs, resulting in an imbalance of Treg and Th2 cells and leading to allergic diseases (Fig. 3).435,436
Treatment
At present, the treatment of allergic diseases mainly includes five aspects: management and therapeutic education for patients and allergen avoidance, traditional pharmacotherapy, allergen immunotherapy, biologics administration and other therapies.
Management and therapeutic education for patients and allergen avoidance
Daily management of patients with allergic diseases and educational intervention plays an essential role in managing difficult-to-treat allergic cases. It is frequent and may lead to treatment failure due to poor adherence to the prescribed treatment.437 Ensuring patient education and confidence in prescribed drug is urgently required, to achieve disease control. For example, it is necessary to inform patients regarding the correct use of intranasal spray and other pharmaceutical preparations.438 What’s more, psychosomatic aspects can also contribute to complementing topical and systemic therapies.439 Furthermore, it is also a critical issue in allergic disease management to combine the limited resources and increased digitalization. For example, mobile applications can monitor symptoms, drug use, and quality of life timely with higher efficiency, e.g., the Allergy Diary.440,441
Avoiding allergens or minimizing exposure to allergens is the first step of treatment. However, since the pathogenic allergens of different patients are often different, mainly including food allergens, aeroallergens, contact allergies, and allergens in the environment that are frequently unknown, the strategy of avoiding allergens on the basis of an allergy diagnosis is often challenging.438 Therefore, teaching patients how to avoid contacting with allergens may help achieve the best treatment effect. For example, the current standard of FA care is to avoid allergens, because there is no Food and Drug Administration (FDA) approved treatment for FA at present.442 As for AR or AD, due to their multifactorial etiologies, elimination of food or environmental allergens represents an adjuvant treatment to pharmacotherapy; thus, complete remission may not be expected after mere allergen elimination.443 Other allergen avoidance measures such as environmental intervention and use of protective devices are available to reduce exposure to causative substances for maximum effectiveness, which apply to all forms of asthma aggravated by the working environment.444 It is noteworthy that determining the individual’s relevant triggers for allergic diseases may be more difficult than diagnosing the disease itself.439 To sum up, allergic diseases normally need drugs for treatment.
Traditional pharmacotherapy
Traditional pharmacotherapy is currently an efficient and rapid treatment method, which helps improve the symptoms of most patients and enhance their quality of life. At present, there are many kinds of drug treatments for allergic diseases, with good therapeutic effects (Table 2).
Table 2.
Different Traditional pharmacotherapies and their mechanism of action and clinical application of drugs
| Traditional pharmacotherapy | Mechanism | Drugs | Stage | References | |
|---|---|---|---|---|---|
| H1-antihistamine | First-generation | Serve as neutral receptor antagonists or inverse agonists of the histamine H1 receptor, can block the action of histamine | Chlorpheniramine | Marketed | 448 |
| Diphenhydramine | Marketed | 448 | |||
| Hydroxyzine | Marketed | 448 | |||
| Doxepin | Marketed | 448 | |||
| Second-generation | Loratadine | Marketed | 446 | ||
| Cetirizine | Marketed | 446 | |||
| Fexofenadine | Marketed | 446 | |||
| Rupatadine | Marketed | 446 | |||
| Bilastine | Marketed | 446 | |||
| Astemizole | No longer approved | 447 | |||
| Terfenadine | No longer approved | 447 | |||
| Third-generation | Desloratadine | Marketed | 449 | ||
| Fexofenadine | Marketed | 449 | |||
| Levocetirizine | Marketed | 446 | |||
| Corticosteroids | Intranasal corticosteroids (INCS) | Inhibit a variety of inflammatory genes, including cytokines, inflammatory enzymes, adhesion molecules and inflammatory mediator receptors | Beclomethasone | Marketed | 438 |
| Budesonide | Marketed | 438 | |||
| Ciclesonide | Marketed | 438 | |||
| Fluticasone propionate | Marketed | 438 | |||
| Fluticasone furoate | Marketed | 438 | |||
| Mometasone furoate | Marketed | 438 | |||
| Triamcinolone acetonide | Marketed | 438 | |||
| Topical corticosteroids (TCS) | Prednicarbate | Marketed | 439 | ||
| Futicasone | Marketed | 439 | |||
| Mometasone | Marketed | 439 | |||
| Systemic corticosteroids (SCS) | Methylprednisolone | Marketed | 439 | ||
| Combination of H1-antihistamine and nasal corticosteroids | Can block the main cause of direct response and reduce inflammation through significantly different mechanisms | Azelastine hydrochloride [HCl] and fluticasone propionate | Marketed | 457 | |
| Leukotrienes (LTS) | 5-LO inhibitors prevent the synthesis of leukotriene A4 by inhibiting the catalytic activity of 5-LO, LTRAs block the cysteinyl LT (cysLT) receptor type 1 directly | Pranlukast | Marketed | 458 | |
| Montelukast | Marketed | 458 | |||
| Zafirlukast | Marketed | 458 | |||
| Other drugs | Mast cell stabilizer | Inhibit the degranulation of allergic mediators | Sodium cromoglicate | Marketed | 463 |
| Chromones | Preventive antiallergic drugs | Cromolyn sodium | Marketed | 438 | |
| Phosphodiesterase 4 inhibitors | Promote apoptosis by reducing the anti-apoptotic protein bcl-2, which can inhibit neutrophils and eosinophils in vitro | Crisaborole | Marketed | 439 | |
| Topical calcineurin inhibitors (TCI) | An inhibition of proinflammatory cytokine production by T cells and mast cells, reveal antipruritic effects which attributes to a specific effect on TRPV1 neurons in the skin | Tacrolimus | Marketed | 469 | |
| Pimecrolimus | Marketed | 469 | |||
| Other immunosuppressants | Inhibit the proliferation and differentiation of immune cells and the differentiation of T cells | Azathioprine | Marketed | 462 | |
| Cyclosporine A | Marketed | 462 | |||
| Bronchodilators | Prevent and relieve bronchoconstriction | Long‐acting muscarinic antagonist (LAMA) | Marketed | 466 | |
| Long‐acting beta‐agonist (LABA) | Marketed | 466 | |||
Website for information inquiry on Traditional clinical drugs https://www.yaozh.com/. Drugs that have been approved for listing are marked with “Marketed”
H1-antihistamines
H1-antihistamines, which serve as neutral receptor antagonists or inverse agonists of the histamine H1 receptor, can block the action of histamine.445 According to brain H1 receptor occupancy (H1RO, an indicator of antihistamines), H1‐antihistamines can be divided into the non-sedating, less-sedating and sedating groups, whose diverse chemical structures, pharmacokinetic features and potential for drug-drug and drug-food interactions make them different. First-generation H1-antihistamines have not been well studied, and because of their adverse effects, particularly sedation, they should be avoided and not recommended for use.438 New second-generation H1-antihistamines have great efficacy and safety profiles, as well as good tolerance.446 The development of second-generation H1-antihistamines occurred in the 1980s, revolutionizing allergy therapy due to no or only minimal sedative potential, including the less-sedating oral H1-antihistamines and non-sedating H1-antihistamines.447 However, because of cardiotoxic side effects, two early second-generation H1-antihistamines, i.e., astemizole and terfenadine, have now been withdrawn from the market.447,448 Third-generation oral antihistamines have improved efficacy, safety, and pharmacological and pharmacokinetic features, representing suitable candidates for the treatment of seasonal or perennial allergies, which may improve the allergic symptoms of patients.449
Corticosteroids
Corticosteroids were considered the most effective therapeutic approach for atopic disorders in the past, and could control virtually all cases of allergic diseases at high dose.450 The main role of corticosteroids is to inhibit a variety of inflammatory molecules such as cytokines, pro-inflammatory proteins, adhesion molecules and inflammatory receptors, which explain their high efficacy in complex inflammatory diseases. Corticosteroids are divided into intranasal corticosteroids (INCS), topical corticosteroids (TCS) and systemic corticosteroids (SCS).
INCS are a well-established first-line therapeutic option for adults and children with persistent or moderate-to-severe symptoms, decrease inflammation associated with AR and alleviate nasal and ocular symptoms.438,451 Their efficiency is more obvious than that of oral or intranasal antihistamines and antileukotrienes. In addition, INCS are comparable to the combination of antihistamines and antileukotrienes.452 Mechanistically, INCS exert local anti-inflammatory effects on nasal mucosal cells. New-generation inhaled corticosteroids for asthma show enhanced anti-inflammatory effects with minimal adverse effects. Undoubtedly, inhaled corticosteroids (ICS) have revolutionized asthma therapy, and are currently considered the first-line therapeutic option for all chronic asthma cases.
Topical corticosteroids (TCS) induce fewer systemic side effects, especially the recently developed topical steroids that have short half-lives, and are the first-line anti-inflammatory approach in AD.439,453 Studies have shown that topical corticosteroids used as adjunctive therapy alongside dupilumab may provide additional benefits.454 Systemic corticosteroids (SCS), one of the groups of drugs available for systemic anti-inflammatory therapy, are required for AD not sufficiently controllable with adequate topical therapies and UV light therapy. Although SCS have rapid effects, their use should be limited to 1-2 weeks because of significant risk of severe long-term side effects, as observed with methylprednisolone.439
Combination of H1-antihistamines and nasal corticosteroids
Conceptually, it is meaningful to use both nasal corticosteroids and local nasal antihistamines at the same time, because such treatment can block the main cause of direct response and reduce inflammation through significantly different mechanisms.455 A study addressed the possibility that a combination of azelastine and fluticasone (both as nasal spray) can confer significant clinical benefits in individuals with seasonal allergic rhinitis (SAR) in comparison with any drug alone.456 It has been shown that the nasal symptoms of SAR are more significantly relieved by the fixed-dose combination (FDC), containing an intranasal antihistamine (azelastine hydrochloride [HCl]) and a corticosteroid (fluticasone propionate) compared with placebo or monotherapy, and could also alleviate the nasal symptoms of perennial allergic rhinitis (PAR) more substantially than fluticasone monotherapy.457
Leukotriene receptor antagonists
Leukotrienes (LTS) are indispensable for the pathogenetic mechanisms of allergic inflammation, so inhibiting LTS represents an effective and feasible strategy in the treatment of allergic diseases, including AAS, AR, and AD. Leukotriene receptor antagonist (LTRA) is an additional treatment option for asthmatics and AR cases. In addition, some leukotriene receptor antagonists are currently used clinically for exercise-induced bronchoconstriction.458 Leukotriene is an important lipid mediator in asthma-related research.459 Leukotriene receptor antagonists improve small airway function and reduce airway inflammation in the treatment of AAS.460 In Europe, LTRAs have been approved by European Medicine Agency(EMA) only for the treatment of asthma and AR.438 Studies have shown that early intervention with a 4-week anti-leukotriene course is also beneficial for some pollen allergies.461 LTRAs are not known to cause significant congenital malformations or adverse perinatal outcomes in pregnancy safety studies. Some reports point out that LTRAs may be considered in second-line treatment of pregnant women if better treatments fail.462
Other drugs
Other drugs can also help treat allergic diseases. Mast cells can regulate angiogenesis, tissue inflammation and repair. The cells play an important role in innate and adaptive immune response, immune tolerance, and host defense. Therefore, mast cells are essential for allergic reactions. Mast cell stabilizer such as sodium cromoglicate is targeted at mast cells, which can inhibit the degranulation of allergic mediators, thus preventing allergic diseases. The side effects of sodium cromoglicate are less, and it has been clinically approved for the treatment of asthma and AR. However, the therapeutic time window of mast cell stabilizer is relatively narrow. The patient needs to be given the drug immediately before the allergen stimulation, so that the drug can play a stable and effective role.463–465
Some drugs are used for symptomatic treatment of allergic diseases. For example, most chromones can be administered as monotherapy for local symptoms. They are relatively safe drugs, but with low efficacy.438 Theophylline, a Phosphodiesterase 4 (PDE4) inhibitor, is another drug that promotes apoptosis by reducing the anti-apoptotic protein Bcl-2, which inhibits neutrophils and eosinophils in vitro. It was also noted that theophylline inhibits reactive oxygen species accumulation by neutrophils and reduces neutrophil chemotaxis.466 At present, there are four PDE4 inhibitors approved for treating human diseases, including AD and bronchial asthma.467 However, considering the associated systemic adverse effects, topical administration of inhaled PDE4 inhibitors might represent a promising alternative instead of systemic utilization, although these drugs are not recommended to use before or during pregnancy.462,468
Topical calcineurin inhibitors (TCIs) have substantial anti-inflammatory effects, inhibiting the biosynthesis of proinflammatory cytokines by T cells and mast cells, as well as antipruritic effects that are attributed to specific effects on skin Transient receptor potential vanilloid 1 (TRPV1) neurons.439 TCIs are especially useful in individuals requiring long-term treatment, and two of them have been approved for topical AD therapy; nevertheless, reports assessing the application of topical calcineurin inhibitors during pregnancy are limited.469 It is important to note that other immunosuppressants, including azathioprine and cyclosporine, do not induce congenital malformations; besides, cyclosporine is considered as first-line drug for long-term treatment of diseases.462,470
Bronchodilators, which are significant for preventing and relieving bronchoconstriction, including long‐acting muscarinic antagonist (LAMA) and long‐acting beta‐agonist (LABA) addition to ICS, sometimes have adverse effects, including tremors, palpitations and tachycardia.466,471,472
In general, H1-antihistamines are usually safe and widely used for the treatment of various allergic diseases, but some patients experience adverse effects, such as cardiotoxicity, central depression and anticholinergic effects. In addition, there are individual differences in the efficacy of antihistamines in clinical practice.473 INCS are effective in combating nasal and ocular symptoms and improving quality of life, and are safe for short-term use, but long-term safety data are lacking. Although generally well tolerated, adverse events can be observed that may lead to serious ocular complications.474 As for TCS and SCS, long-term continuous use of corticosteroids may result in local and systemic toxicity risks. LTRAs are well tolerated, non-hormonal anti-inflammatory drugs. LTRAs are primarily employed for long-term control treatment of patients with mild asthma and comorbid AR. It has also been shown that LTRAs are not associated with the risk of major congenital malformations and can be safely used during pregnancy to treat asthma.475 Nevertheless, the anti-inflammatory effect of LTRAs is not as strong as that of corticosteroids, so they are often used in combination with inhaled corticosteroids to enhance their efficacy in clinical practice. However, Some experiments have shown that LTRAs and oral H1-antihistamines have comparable effects in AR.438 In short, based on the side effects of these traditional drugs, other therapies are emerging as well, such as allergen immunotherapy (AIT) and biologics, etc.
AIT
AIT constitutes the sole disease-modifying therapy for patients with IgE-mediated inhalation allergic diseases.476 Compared with traditional pharmacotherapy, the advantage of AIT is that it is a medical intervention that can limit the natural process of the disease.477 The purpose of this therapy is to reduce the symptoms of allergic diseases by inducing tolerance to allergens.438 AIT is based on the administration of allergens that cause a given disease. Through long-term repeated exposure to specified doses of allergens, it can change the immune response and induce protective immunity, so that patients may tolerate future allergen exposure.478,479 In addition, this therapy reduces the long-term treatment cost for patients and the economic burden of allergy. From the perspective of public health, AIT plays a crucial role in allergy management.477
Application of AIT in allergic diseases
In allergic diseases, immune dysfunction is the key pathogenic factor, so the concept of inducing immune tolerance has gradually become one of the goals set for preventing and treating allergic diseases.477 In the routine use of AIT, it is recommended to take a 3-year course of treatment to achieve long-term efficacy, and long-term clinical benefits are achievable after stopping treatment.476,480 In adolescents and adults, AIT can be used to treat moderate-to-severe rhinitis and moderate asthma. In children, AIT can prevent rhinitis cases from further developing asthma symptoms.481 Researchers analyzed AR progress and changes in asthma symptoms after HDM allergen immunotherapy. The results showed that treatment of AR patients with HDM allergy drugs could indeed reduce the overall incidence rate of asthma, improve asthma symptoms and slow down the progression of asthma.482,483 Many experiments have revealed that AIT markedly reduces the symptoms in patients, changes the disease process, and improves the quality of life in allergic individuals.
Therapeutic principles of AIT in allergic diseases
The tolerance induced by AIT is related to changes in allergen-specific memory T and B cells as well as allergen-specific IgE and IgG antibody amounts. In addition, AIT has some impact on the activation threshold of mast cells, basophils and dendritic cells.438 Some cells utilize interleukin-10 (IL-10), IL-35, transforming growth factor-β (TGF-β), IL-10R, TGF-βR, cytotoxic T lymphocyte-associated antigen 4 (CTLA4), programmed cell death protein 1 (PD1) and other inhibitors to directly or indirectly alleviate the anaphylactic environment.484 Studies have shown that AIT regulates the follicular helper T (TFH) cell-to-follicular regulatory T (TFR) cell balance in allergic cases. The decrease and functional defect of TFR cells are associated with AR.485 Studies have also shown that AIT reduces the expression of CD23 on switched memory B cells, which has a positive correlation with the clinical efficacy of AIT in AR.480
Biomarkers in AIT can predict or monitor immune responses to determine efficacy as early as possible
In AIT, biomarkers can reflect some clinical or laboratory characteristics of the immune process, which is essential for monitoring the health status of patients at all times. However, there is a lack of validated genetic or blood markers that could help predict or monitor the efficacy of AIT at the individual patient level.481 In recent years, researchers have been screening for biomarkers to detect the success of AIT. Many technologies are expected to be used to examine these biomarkers, including genomics, transcriptomics, immunology, lipidomics, metabolomics, microbiology, epigenetics and proteomics.478 According to the mechanism of anaphylaxis and the development and application of the above technologies, relatively reliable candidate biomarkers for immune detection have been reported, including immunoglobulins (allergen-specific IgD, IgE and IgG4) and some cytokines or chemokines (IL-4, IL-13, IL-9, IL-25, IL-33 and TSLP). In addition, candidate biomarkers can also include specific genes or immune-related cells such as cytokine and immunoglobin transcripts, myeloid cells, innate lymphoid cells (ILC), T cells and B cells.478
The drug delivery route affects the quality of AIT
In AIT, different drug delivery routes show different side effects and final efficacy. Conventional AIT includes subcutaneous immunotherapy (SCIT) and sublingual immunotherapy (SLIT).476,486 Traditional SCIT has certain shortcomings that cannot be ignored. It requires frequent medical treatment and multiple injections, and in some rare cases, life-threatening allergic reactions and adverse events may occur.487 The typical local reaction in SCIT is redness and swelling at the injection site, and more serious systemic reactions include sneezing, nasal congestion and urticaria. Some serious symptoms often occur within 30 minutes, so individuals are usually allowed to leave the hospital 30 minutes after completing the injection.481,488 SLIT has become a particularly attractive alternative to AIT because of its rather easy administration. It usually involves allergen drops or tablets, which can be administered at home.489 Some new routes of AIT administration are also being developed in order to improve the safety and convenience of patients and maintain or even obtain better curative effects.476,477,490 Oral immunotherapy (OIT) is another SCIT option. OIT seems to have no effects on most respiratory allergens because they are easily digested in the gastrointestinal tract. Therefore, OIT is mainly used for anti-digestive food allergens, including milk, eggs, peanuts and some wheat.479 Studies have shown that OIT for IgE-mediated FA desensitizes patients who are at risk or have experienced severe allergies to peanuts, eggs and milk.477 Intralymphatic immunotherapy (ILIT) is also another better choice for SCIT. This method involves intralymphatic administration, because lymph nodes contain a large number of immune cells, so their direct contact with allergens would be faster than in SCIT and produce stronger protective IgG antibodies and immune regulation at the same time.479,491 In addition, studies have proposed epicutaneous immunotherapy (EPIT), which aims to cause fewer systemic side effects when administered through the epidermis.479
In conclusion, the current goal of improving AIT is to shorten the treatment time, improve its efficiency in order to promote the absorption and presentation of allergens, reduce side effects to improve safety, improve patient compliance, and ultimately significantly increase the utilization of this treatment method.492
Application of biologics in allergic diseases
New studies on allergic diseases are underway, especially the development and application of new biologics.441 So far, many biologics targeting Th2/1/17 inflammatory biomarkers are available, many of which are clinically applied.493 Identifying new and reliable biomarkers and clarifying novel molecular mechanisms that persist in specific reactions have become important research directions.438 We summarized some existing biologics according to differences in the mechanisms of action and target sites, providing a certain reference for follow-up studies of biologics (Table 3, Fig. 6).
Table 3.
Targets and clinical stages of biologics
| Biologic | Description | Clinical stage | References |
|---|---|---|---|
| Omalizumab | Recombinant humanized anti-IgE antibody | Marketed | 441 |
| Mepolizumab | IgG1 antibody against IL-5 | Marketed | 496 |
| Reslizumab | Anti-IL-5 IgG4 antibody | Marketed | 496 |
| Benralizumab | An IL-5Rα-directed cytolytic monoclonal antibody | Marketed | 496 |
| Dupilumab | Fully humanized antibody that targets IL-4Rα subunits | Marketed | 500 |
| Tralokinumab | IgG4 monoclonal antibody targeting IL-13 | Marketed | 500 |
| Lebrikizumab | Inhibit the dimerization of IL-13Rα1 and IL-4Rα | Phase III | 500 |
| Nemolizumab | Humanized monoclonal antibody targeted to block IL-31 receptor A | Marketed in Japan | 501 |
| Enokizumab | A humanized immunoglobulin G1k anti-IL-9 mAb | Phase II | 516 |
| Baricitinib | Small molecule that inhibit JAK1 and JAK2 | Marketed | 501 |
| Tofacitinib | Inhibit JAK1 and JAK3 | Marketed | 501 |
| Upadacitinib | A selective JAK1 inhibitor | Marketed | 501 |
| Ruxolitinib | Small molecule JAK1/2 inhibitor | Marketed | 505 |
| Teglarinad Chloride | Inhibitors targeting IKK complex in NF-κB pathway | No application | 542 |
| Disulfiram | Inhibitors targeting UPS in NF-κB pathway | No application | 542 |
| TPCA-1 | Small molecule IKK-β inhibitor | No application | 218 |
| AS602868 | Small molecule IKK-β inhibitor | No application | 218 |
| Idelalisib | PI3Kδ kinase inhibitor | Marketed | 542 |
| Alpelisib | PI3K inhibitor, inhibiting PI3Kα (PIK3CA) activity | Marketed | 542 |
| Copanlisib | PI3K inhibitor, inhibiting PI3Kα and PI3Kδ | Marketed in the USA | 542 |
| Duvelisib | Selective PI3K δ/γ inhibitor | Marketed | 542 |
| Rapamycin | mTOR protein specific inhibitor | Marketed | 505 |
| SAHM1 | Inhibit the formation of classical Notch transcription complex | No application | 510 |
| DAPT | γ- Secretase inhibitor, inhibit Notch 1 signaling | No application | 510 |
| Avasimibe | Inhibit the Wnt/β-catenin signaling pathway | No application | 511 |
| Resiquimod (R848) | Agonist of Toll-like receptors 7/8 (TLR7/TLR8) | No application | 512 |
| GSK2245035 | Toll-like receptor 7 agonist | Phase II | 513 |
| Tezepelumab | Monoclonal antibody against TSLP | Marketed in the USA | 514 |
| Etokimab | IgG1 antibody can neutralize IL-33 | Phase II | 515 |
| GBR830 | An antibody targeting OX40 | Phase II | 500 |
| AMG853 | A CRTH2 antagonist reduces Th2-mediated inflammation | Phase II | 189 |
| OC000459 | A CRTH2 antagonist reduces Th2-mediated inflammation | Phase III | 189 |
| BI671800 | A CRTH2 antagonist reduces Th2-mediated inflammation | Phase I | 189 |
| N6022 | Small molecule inhibitor of GSNOR | Phase II | 519 |
Website for information inquiry on biologics https://www.yaozh.com/. Biologics that have been approved for listing are marked with “ Marketed”, and unlisted biologics are inquired about their clinical research stage in allergic diseases. “ No application” indicates that no clinical study on the application of this biological agent to treat allergic diseases has been found
Fig. 6.
Application of biologics in allergic diseases. Multiple cell interactions trigger allergic reactions. Therefore, treatment with biologics aims to target the cytokines produced by various cells, with a potential impact on the interaction between cells. Some biologics exert their effects by targeting IgE, IL-5, IL-4, IL-13, IL-31, IL-9, IL-33, and TSLP, among others
Targeting IgE
Targeting IgE molecules is one of the most important methods for nipping allergic reactions in the body with lasting effects. Anti-IgE antibody treatment markedly reduces the serum amounts of free IgE molecules in allergic individuals, thus exerting a certain effect.494 Recombinant humanized anti-IgE antibodies, such as omalizumab, have been approved by the FDA for severe asthma, substantially improving clinical symptoms in patients with poor disease control.441,495
Targeting IL-5
Eosinophils are critical in asthma pathogenesis, and airway eosinophil inflammation is related to the severity of asthma. Biologics, including mepolizumab, reslizumab and benralizumab, have been developed to target IL-5 or IL-5Rα and then affect the survival and differentiation of eosinophils. Studies have shown that blocking the IL-5 receptor could indeed alleviate severe eosinophilic asthma and severe uncontrolled asthma.496–498
Targeting IL-4/IL-13
IL-4 and IL-13 represent key driving factors in type 2 immune diseases. For example, dupilumab was approved by the FDA and the EMA (EU) for adults with moderate-to-severe AD. Dupilumab is a fully humanized antibody that targets IL-4Rα subunits, thus inhibiting the IL-4/IL-13 axis. These two cytokines induce IgE production in B cells, goblet cell metaplasia, mucus production, basement membrane thickening and fibrosis. A recent experiment showed that blocking the IL-4/IL-13 pathway alleviates glucocorticoid-dependent severe asthma, moderate-to-severe uncontrolled asthma and AD. Biologics also significantly alleviate AR symptoms, especially nasal symptoms.441,496,497,499 Many biologics have been developed to target IL-4 and IL-13, inhibiting the dimerization of IL-13Rα1 and IL-4Rα or directly targeting the IL-4Rα subunits to play a role.499–502 More and more biologics that block IL-4 and IL-13 activities have been shown to alleviate nasal symptoms in patients with uncontrolled asthma and AR.503
Targeting IL-31
Monoclonal antibodies, including nemolizumab, a humanized monoclonal antibody targeting IL-31 receptor A (IL-31RA), relieve itching symptoms and IL-31 signal transduction in the pathogenesis of AD. In mice, IL-31 and its receptor IL-31RA are involved in AD-induced pruritus. In another animal experiment, the monkey scratch model, it was found that the scratch induced by IL-31 is significantly alleviated after a single injection of nemolizumab.501,504
Targeted signaling pathways
Some biologics target signaling pathways, including baricitinib, tofacitinib, upadacitinib and ruxolitinib, which target JAK/STAT signaling. Targeting JAK/STAT signaling is critical for T cell activation, and could also block the downstream pathways of many important cytokine receptors. T cells are essential for atopic inflammation. The JAK protein in cells activates STAT protein dimerization and transfer to the nucleus, thereby increasing the gene expression of inflammatory mediators. Therefore, JAK/STAT pathway suppression represents an effective and feasible therapeutic approach in AD.500,501,505 Asthma is often accompanied by inflammation, and many pro-inflammatory chemokines, cytokines, adhesion molecules, airway mucins, growth and angiogenesis factors are upregulated through Rel/Nuclear Factor-κB (NF-κB) transcription factor family.218,506 So asthma and NF-κB mediated signal transduction is inextricably linked, and a series of NF-κB signaling intermediate inhibitors have been produced, e.g., DNA oligonucleotides and DNA-peptide molecules acting as NF-κB bait sequences; in addition, small molecule inhibitors and some proteasome inhibitors affect NF-κB signal transduction.218 For example, small molecule inhibitors such as TPCA-1 and AS602868 inhibit IκB kinase (IKK)-β to block NF-κB signaling. In addition, proteasome inhibitors can block NF-κB signal transduction by targeting the 20 S proteasome, ultimately regulating eosinophil function.218,507 Some biologics regulate the signal cascade by inhibiting PI3K or any downstream target, because PI3K-mediated signals pass through IKKα phosphorylation, and protein kinase B (PKB, AKT) phosphorylation directly activates IKK, which then enters NF-κB signal cascade reaction pathway. For example, idelalisib, alpelisib, copanlisib and duvelisib are PI3K inhibitors.508 The mechanistic target of rapamycin (mTOR) pathway is also considered a signaling pathway regulating innate and adaptive immune cells. It was found that rapamycin targeting mTOR inhibits eosinophil differentiation and reduces allergic airway inflammation in mouse models.505,509 Notch signaling is also associated with the pathogenesis of allergic airway inflammation. Notch is necessary to maintain Th1 and Th2 programs. As a biological agent, stapled α-helical peptide derived from mastermind-like 1 (SAHM1) targets Notch to effectively reduce the inflammatory symptoms of mice with experimental allergic airway inflammation and accelerate recovery. In addition to being related to T cells, Notch also is critical for the differentiation of lung organs and alveoli. For example, in a mouse asthma model, intranasal administration of γ-secretase inhibitors (GSIs) could block the Notch signaling pathway and reduce allergic pulmonary edema.510 Avasimibe (Ava), as a specifically targets acetyl-CoA acetyltransferase 1 (ACAT1) inhibitor, has been proven to alleviate the disruption of the airway epithelial barrier by inhibiting the Wnt/β-catenin signaling pathway. It has good safety and may be a promising drug for the clinical treatment of AAS.511
Other biologics
Currently, the use of new therapeutic targets is also being explored. For example, Toll-like receptors are involved in the activation of innate immunity in the respiratory mucosa. Some agonists can induce cytokines, including TNF-α, IL-6 and IFN-α; such biologics are well tolerated and may not cause systemic immune activation.512,513 In the study of AAS, due to its complex pathogenesis, certain new biological targets have attracted attention from researchers, e.g., epithelial cell-derived cytokines (IL-1, IL-33, IL-25 and TSLP).496,503,514,515 In addition, IL-9 is also associated with allergic diseases. In the study of asthma, mouse models overexpressing IL-9 show enhanced eosinophilic airway inflammation, IgE production and mast cell proliferation. Therefore, biologics targeting IL-9 have also been designed.516 There are also antibodies that target the costimulatory molecule OX40 (CD134), which is critical for T cell expansion.500
Chemoattractant receptor-homologous molecule expressed on TH2 cells (CRTH2), also known as prostaglandin D2 receptor 2, is a 7-transmembrane G protein-coupled receptor expressed by Th2 cells, eosinophils and basophils. The CRTH2 receptors are involved in inducing the migration and activation of Th2 lymphocytes, eosinophils and basophils. At the same time, it is also involved in the up regulation of adhesion molecules and the release of proinflammatory Th2 cytokines such as IL-4, IL-5 and IL-13. Therefore, it plays a certain role in the course of allergic diseases. It has been proved that the number of CRTH2-positive cells is related to the severity of asthma. At present, some CRTH2 antagonists, such as AMG853, OC000459 and BI671800, have been developed to treat asthma, AD and AR. These CRTH2 inhibitors reduce the Th2-mediated inflammatory response by blocking the activation of mast cells, basophils and eosinophils.189,517,518
Nitric oxide (NO) is an important signal molecule in many physiological processes, such as maintenance of vascular tone and endothelial barrier function, immune defense and apoptosis. NO can also regulate cell function through post-translational modification of proteins. NO can react with glutathione to form S-nitrosoglutathione (GSNO), which effectively transduces NO signal. GSNO concentration can be regulated by GSNO-reductase (GSNOR), which provides the “brake” for signal transduction. There is evidence that GSNOR polymorphisms increase the expression of GSNOR and are associated with an increased risk of asthma. Inhibition of GSNOR can lead to the preservation of endogenous GSNO and limit eosinophilic inflammation, mucus production and airway hyper reactivity (AHR). N6022 is the first small molecule inhibitor of GSNOR in the treatment of asthma. Some studies have shown that in the mouse asthma model, N6022 significantly reduces airway hyperreactivity and shows a strong anti-inflammatory effect.519–521
Biologics-targeted therapies target the endotypes of allergic diseases based on pathogenesis, reducing the occurrence of adverse events and improving the efficiency of treatment, and are considered promising therapeutic approaches. However, many biologics have not been developed far enough and have not been better evaluated. The response to treatment varies greatly from individual to individual. Therefore, the dose and duration of treatment need to be better defined and the details need to be further optimized. In addition, the relatively expensive price of biologics also limits their application to some extent.522–524
Other therapies
Different treatment options have varying degrees of side effects; thus, a growing number of alternatives therapies have also been developed. In addition to the aforementioned treatment methods, other therapies also offer different therapeutic effects in the treatment of allergic diseases, including antibacterial and antimycotic therapies, phototherapy, early introduction therapy, circadian regulation therapy and so on.
The microbiota and allergic diseases are closely related. For example, Staphylococcus aureus is the main cause of AD, and Malassezia furfur is also associated with skin immune response and barrier function. Therefore, antibacterial and antimycotic therapies have also become an option for treating allergic diseases.439 Phototherapy has immunosuppressive and immunomodulatory effects, which can inhibit the effects of anaphylaxis and histamine release triggered by mast cell antigens. Therefore, rhinophototherapy is considered a promising non-invasive alternative treatment option for perennial or seasonal AR cases, especially low-level laser therapy (LLLT).525,526 However, it has been demonstrated that photochemotherapy has certain carcinogenicity. In addition, in AD, oral psoralen plus ultraviolet A (PUVA) as a type of phototherapy also has many side effects. Therefore, PUVA for AD treatment has been abandoned to a large extent, and it is recommended to apply combination therapy.439 In patients with FA, early consumption is more beneficial than delayed administration. A trial showed that early introduction of eggs combined with appropriate eczema treatment is practical, and may effectively reduce the odds of egg allergy in high-risk infants.527 One of the important research fields to further examine new methods to prevent or treat modern allergic diseases is to understand the relationship between circadian biology and allergy; correspondingly, it is suggested to develop a lifestyle in which the endogenous biological clock is consistent with the environmental cycle, or undergo appropriate therapeutic interventions to prevent or treat allergic disease in the future.528
Acknowledgements
We thank Professor Qianfei Wang (Beijing Institute of genomics, Chinese Academy of Sciences) and Professor Qianben Wang (Department of Pathology, Duke University School of Medicine, Durham, USA) for critical comments and advice on the manuscript. This work was supported by the General project of National Natural Science Foundation of China (No. 82174243, 81973715 and 82204948), National Key R&D Program of China (2020YFC2003100), Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (No: ZYYCXTD-C-202001), General project of Beijing Natural Science Foundation (No.7202110).
Author contributions
J.W., Y.Z., L.H., H.Z., J.L., L.W., H.Z. and T.W. wrote the manuscript and created the figures and table. J.W., Y.Z. and Q.W. provided the conceptual idea. L.C. edited the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ji Wang, Yumei Zhou
References
- 1.Bousquet J, Van Cauwenberge P, Khaltaev N, Aria Workshop G, World Health O. Allergic rhinitis and its impact on asthma. J. Allergy Clin. Immunol. 2001;108:S147–S334. doi: 10.1067/mai.2001.118891. [DOI] [PubMed] [Google Scholar]
- 2.Meghji J, Mortimer K, Jayasooriya S, Marks GB. Lung health in LMICs: tackling challenges ahead – Authors’ reply. Lancet. 2021;398:490. doi: 10.1016/S0140-6736(21)01252-6. [DOI] [PubMed] [Google Scholar]
- 3.Kattan JD, Wang J. Allergen component testing for food allergy: ready for prime time? Curr. Allergy Asthma Rep. 2013;13:58–63. doi: 10.1007/s11882-012-0311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsoi LC, et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J. Allergy Clin. Immunol. 2020;145:1406–1415. doi: 10.1016/j.jaci.2019.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lloyd-Lavery A, et al. What’s new in atopic eczema? An analysis of systematic reviews published in 2016. Part 2: epidemiology, aetiology and risk factors. Clin. Exp. Dermatol. 2019;44:370–375. doi: 10.1111/ced.13853. [DOI] [PubMed] [Google Scholar]
- 6.Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat. Rev. Dis. Prim. 2018;4:1. doi: 10.1038/s41572-018-0001-z. [DOI] [PubMed] [Google Scholar]
- 7.Dong WL, et al. The prevalence and year lived with disability of atopic dermatitis in China: Findings from the global burden of disease study 2019. World Allergy Organ J. 2021;14:100604. doi: 10.1016/j.waojou.2021.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small, P. et al. In J. Otolaryngol. S5-S27 (2007).
- 9.Dykewicz MS, Hamilos DL. Rhinitis and sinusitis. J. Allergy Clin. Immunol. 2010;125:S103–S115. doi: 10.1016/j.jaci.2009.12.989. [DOI] [PubMed] [Google Scholar]
- 10.Fahy JV. Type 2 inflammation in asthma–present in most, absent in many. Nat. Rev. Immunol. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mims JW. Asthma: definitions and pathophysiology. Int Forum Allergy Rhinol. 2015;5(Suppl 1):S2–S6. doi: 10.1002/alr.21609. [DOI] [PubMed] [Google Scholar]
- 12.Feld M, et al. The pruritus- and TH2-associated cytokine IL-31 promotes growth of sensory nerves. J. Allergy Clin. Immunol. 2016;138:500–508 e524. doi: 10.1016/j.jaci.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Wilson SR, et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013;155:285–295. doi: 10.1016/j.cell.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinthrajah RS, et al. Diagnosis of food allergy. Pediatr. Clin. North Am. 2015;62:1393–1408. doi: 10.1016/j.pcl.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deschildre A, Lejeune S. How to cope with food allergy symptoms? Curr. Opin. Allergy Clin. Immunol. 2018;18:234–242. doi: 10.1097/ACI.0000000000000447. [DOI] [PubMed] [Google Scholar]
- 16.Chinthrajah RS, Hernandez JD, Boyd SD, Galli SJ, Nadeau KC. Molecular and cellular mechanisms of food allergy and food tolerance. J. Allergy Clin. Immunol. 2016;137:984–997. doi: 10.1016/j.jaci.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krombach JW, Kampe S, Keller CA, Wright PM. Pharaoh Menes’ death after an anaphylactic reaction–the end of a myth. Allergy. 2004;59:1234–1235. doi: 10.1111/j.1398-9995.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 18.Ramachandran M, Aronson JK. John Bostock’s first description of hayfever. J. R. Soc. Med. 2011;104:237–240. doi: 10.1258/jrsm.2010.10k056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjermer L. History and future perspectives of treating asthma as a systemic and small airways disease. Respir. Med. 2001;95:703–719. doi: 10.1053/rmed.2001.1148. [DOI] [PubMed] [Google Scholar]
- 20.Huber B. [100 years of allergy: Clemens von Pirquet - his idea of allergy and its immanent concept of disease] Wien. Klin. Wochenschr. 2006;118:573–579. doi: 10.1007/s00508-006-0701-3. [DOI] [PubMed] [Google Scholar]
- 21.German AI. [Munchener Medizinische Wochenschrift/24 July 1906: Allergy by Clemens v. Pirquet, Vienna] MMW Munch. Med Wochenschr. 1978;120:474. [PubMed] [Google Scholar]
- 22.Noon L. Prophylactic inoculation against hay fever. Int Arch. Allergy Appl Immunol. 1953;4:285–288. doi: 10.1159/000228032. [DOI] [PubMed] [Google Scholar]
- 23.de Herder WW. Heroes in endocrinology: Nobel Prizes. Endocr. Connect. 2014;3:R94–R104. doi: 10.1530/EC-14-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WB S. The uses and abuses of antihistamine drugs. Bull. N. Y Acad. Med. 1951;27:309–324. [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles S, Shear NH. Antihistamines. Clin. Dermatol. 1989;7:48–59. doi: 10.1016/0738-081X(89)90007-2. [DOI] [PubMed] [Google Scholar]
- 26.Riley JF. Editorial: heparin, histamine and mast cells. Blood. 1954;9:1123–1126. doi: 10.1182/blood.V9.12.1123.1123. [DOI] [PubMed] [Google Scholar]
- 27.Ishizaka K, I T. Biological function of gamma E antibodies and mechanisms of reaginic hypersensitivity. Clin. Exp. Immunol. 1970;6:25–42. [PMC free article] [PubMed] [Google Scholar]
- 28.DP S. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen JN, Broge L, Jacobi H. Allergy immunotherapy: the future of allergy treatment. Drug Disco. Today. 2016;21:26–37. doi: 10.1016/j.drudis.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Bousquet J LR, Malling HJ. Allergen immunotherapy: therapeutic vaccines for allergic diseases. A WHO position paper. J. Allergy Clin. Immunol. 1998;102:558–562. doi: 10.1016/S0091-6749(98)70271-4. [DOI] [PubMed] [Google Scholar]
- 31.Agache, I. & Akdis, C. A. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic diseases. J. Clin. Investig.130 (2019). [DOI] [PMC free article] [PubMed]
- 32.Palmer, L. J., C. W. In Analysis of multifactorial disease 22 (2000).
- 33.Agache, I. et al. Emerging concepts and challenges in implementing the exposome paradigm in allergic diseases and asthma: a Practall document. Allergy (2019). [DOI] [PubMed]
- 34.Potaczek DP, et al. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017;9:539–571. doi: 10.2217/epi-2016-0162. [DOI] [PubMed] [Google Scholar]
- 35.Tost JO. & France clinical reviews in allergy and immunology a translational perspective on epigenetics in allergic diseases. J. Allergy Clin. Immunol. 2018;142:715–726. doi: 10.1016/j.jaci.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Willey DL. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 37.Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet. 2010;58:250–264. doi: 10.1034/j.1399-0004.2000.580402.x. [DOI] [PubMed] [Google Scholar]
- 38.MarkSonkn Single nucleotide polymorphisms: from the evolutionary past. Nature (2001). [DOI] [PubMed]
- 39.Demirdag Y, Bahna S. The role of genetics in food allergy. Expert Rev. Clin. Immunol. 2022;18:401–411. doi: 10.1080/1744666X.2022.2054414. [DOI] [PubMed] [Google Scholar]
- 40.Sicherer SH, et al. Genetics of peanut allergy: a twin study. J. Allergy Clin. Immunol. 2000;106:53–56. doi: 10.1067/mai.2000.108105. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Zhang S, Tsai HJ, Hong X, Wang X. Genetic and environmental contributions to allergen sensitization in a Chinese twin study. Clin. Exp. Allergy. 2010;39:991–998. doi: 10.1111/j.1365-2222.2009.03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol. Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raby BA. Asthma severity, nature or nurture: genetic determinants. Curr. Opin. Pediatr. 2019;31:1. doi: 10.1097/MOP.0000000000000758. [DOI] [PubMed] [Google Scholar]
- 44.Choi, B. Y., Han, M., Kwak, J. W. & Kim, T. H. Genetics and Epigenetics in Allergic Rhinitis. (2021). [DOI] [PMC free article] [PubMed]
- 45.Yazd NKK, Patel RR, Dellavalle RP, Dunnick CA. Genetic risk factors for development of atopic dermatitis: a systematic review. Curr. Dermatol. Rep. 2017;6:297–308. doi: 10.1007/s13671-017-0199-0. [DOI] [Google Scholar]
- 46.Brown SJ, Elias MS, Bradley M. Genetics in atopic dermatitis: historical perspective and future prospects. Acta Derm. Venereol. 2020;100:adv00163. doi: 10.2340/00015555-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoettler N, Ober C. Genetic architecture of moderate-to-severe asthma mirrors that of mild asthma. J. Allergy Clin. Immunol. 2019;144:1521–1523. doi: 10.1016/j.jaci.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Agache I, Akdis C, Jutel M, Virchow JC. Untangling asthma phenotypes and endotypes. Allergy. 2012;67:835–846. doi: 10.1111/j.1398-9995.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- 49.Andiappan AK, Yang YS, Lee B, Suri BK, Chew FT. Functional variants of 17q12-21 are associated with allergic asthma but not allergic rhinitis. J. allergy Clin. Immunol. 2015;137:758–766.e753. doi: 10.1016/j.jaci.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 50.Beghé B, Barton S, Rorke S, Peng Q, Holloway JW. Polymorphisms in the interleukin-4 and interleukin-4 receptor α chain genes confer susceptibility to asthma and atopy in a Caucasian population. Clin. Exp. Allergy. 2003;33:1111–1117. doi: 10.1046/j.1365-2222.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- 51.Bønnelykke K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat. Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 52.Jiang F, Yan A. IL-4 rs2243250 polymorphism associated with susceptibility to allergic rhinitis: a meta-analysis. Biosci. Rep. 2021;41:BSR20210522. doi: 10.1042/BSR20210522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, Xu Y. Association between vitamin D receptor gene polymorphism rs2228570 and allergic rhinitis. Pharmacogenomics Pers. Med. 2020;ume 13:327–335. doi: 10.2147/PGPM.S262402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnelykke K, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat. Genet. 2013;45:902–906. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashley, S. E. et al. Genetic variation at the Th2 immune gene IL13 is associated with IgE‐mediated paediatric food allergy. Clin. Exp. Allergy (2017). [DOI] [PubMed]
- 56.Marenholz I, et al. Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat. Commun. 2017;8:1056. doi: 10.1038/s41467-017-01220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waage J, et al. Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis. Nat. Genet. 2018;50:1072–1080. doi: 10.1038/s41588-018-0157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consortium, T. E. G. A. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet.advance online publication (2015). [DOI] [PMC free article] [PubMed]
- 59.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 60.Mori T, et al. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-gamma-producing T cells. J. Immunol. 2014;193:1728–1736. doi: 10.4049/jimmunol.1303243. [DOI] [PubMed] [Google Scholar]
- 61.León B, et al. Regulation of T(H)2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat. Immunol. 2013;13:681–690. doi: 10.1038/ni.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lawrence T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinnakasu, R. et al. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. (2016). [DOI] [PubMed]
- 64.Roychoudhuri, R. et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat. Immunol. [DOI] [PMC free article] [PubMed]
- 65.Rothlin, C. TAM receptors and pleiotropic inhibitors of the innate immune response. Cell131 (2007). [DOI] [PubMed]
- 66.Kassmeier MD, et al. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J. 2014;31:945–958. doi: 10.1038/emboj.2011.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamblet CE, Makowski SL, Tritapoe JM, Pomerantz JL. NK cell maturation and cytotoxicity are controlled by the intramembrane aspartyl protease SPPL3. J. Immunol. 2016;196:2614. doi: 10.4049/jimmunol.1501970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Galibert LA. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 69.Xin W, Yuan Z, Zheng F, Luo Z. The association between polymorphisms in the MRPL4 and TNF-α genes and susceptibility to allergic rhinitis. Plos One. 2013;8:e57981. doi: 10.1371/journal.pone.0057981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveti, J. F., Kercsmar, C. M. & Susan, R. Pre- and perinatal risk factors for asthma in inner city African-American Children. Am. J. Epidemiol., 570. [DOI] [PubMed]
- 71.Litonjua AA, Carey VJ, Burge HA. Parental history and the risk for childhood asthma. Does mother confer more risk than father? Am. J. Respir. Crit. Care Med. 1998;158:176–181. doi: 10.1164/ajrccm.158.1.9710014. [DOI] [PubMed] [Google Scholar]
- 72.Maternal history, sensitization to allergens, and current wheezing, rhinitis, and eczema among children in Costa Rica. Pediat. Pulmonol.33 (2002). [DOI] [PubMed]
- 73.Raby BA, et al. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. J. Allergy Clin. Immunol. 2007;120:351–358. doi: 10.1016/j.jaci.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 74.Jang H, Kim M, Hong JY, Cho HJ, Kim KW. Mitochondrial and nuclear mitochondrial variants in allergic diseases. Allergy Asthma Immunol. Res. 2020;12:877. doi: 10.4168/aair.2020.12.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flaquer, A., Heinzmann, A., Rospleszcz, S., Mailaparambil, B. & Grychtol, R. Association study of mitochondrial genetic polymorphisms in asthmatic children. Mitochondrion14 (2014). [DOI] [PubMed]
- 76.Zifa E, et al. Mitochondrial genetic background plays a role in increasing risk to asthma. Mol. Biol. Rep. 2012;39:4697–4708. doi: 10.1007/s11033-011-1262-8. [DOI] [PubMed] [Google Scholar]
- 77.Qian L, Mehrabi NE, Athari SM, Athari SS. Mitochondria signaling pathways in allergic asthma. J. Investig. Med. 2022;70:863–882. doi: 10.1136/jim-2021-002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Polonikov AV, et al. Polymorphism -930A > G of the cytochrome b gene is a novel genetic marker of predisposition to bronchial asthma. Ter. Arkh. 2009;81:5–31. [PubMed] [Google Scholar]
- 79.Fukuda T, Mochida S, Fukushima Y, Makino S. Detection of allergen-induced genes in peripheral blood mononuclear cells of patients with allergic asthma using subtractive hybridization. J. Allergy Clin. Immunol. 1995;96:1076–1082. doi: 10.1016/S0091-6749(95)70193-1. [DOI] [PubMed] [Google Scholar]
- 80.Xu YD, et al. The early asthmatic response is associated with glycolysis, calcium binding and mitochondria activity as revealed by proteomic analysis in rats. Respir. Res. 2010;11:107. doi: 10.1186/1465-9921-11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schauberger EM, et al. Identification of ATPAF1 as a novel candidate gene for asthma in children. J. Allergy Clin. Immunol. 2011;128:753–760.e711. doi: 10.1016/j.jaci.2011.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raby BA, et al. Importin-13 genetic variation is associated with improved airway responsiveness in childhood asthma. Respir. Res. 2009;10:67. doi: 10.1186/1465-9921-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Silverman EK, et al. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J. Allergy Clin. Immunol. 2003;112:870–876. doi: 10.1016/S0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 84.Reddy PH. Mitochondrial dysfunction and oxidative stress in asthma: implications for mitochondria-targeted antioxidant therapeutics. Pharmaceuticals. 2011;4:429. doi: 10.3390/ph4030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Devries A, Vercelli D. Epigenetic mechanisms in asthma. Ann. Am. Thorac. Soc. 2016;13(Suppl 1):S48. doi: 10.1513/AnnalsATS.201507-420MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moen, E. L. et al. New themes in the biological functions of 5‐methylcytosine and 5‐hydroxymethylcytosine. Immunol. Rev.263 (2015). [DOI] [PMC free article] [PubMed]
- 87.Neidhart, M. DNA methylation and complex human disease (Oxford: Academic Press, 2016).
- 88.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-Wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 89.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 90.Xu, C. J., Gruzieva, O., Qi, C., Esplugues, A. & Koppelman, G. H. Shared DNA methylation signatures in childhood allergy: the MeDALL study. J. Allergy Clin. Immunol. (2020). [DOI] [PMC free article] [PubMed]
- 91.Luo Y, Zhou B, Zhao M, Tang J, Lu Q. Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin. Exp. Dermatol. 2013;39:48–53. doi: 10.1111/ced.12206. [DOI] [PubMed] [Google Scholar]
- 92.Harb H, Renz H. Update on epigenetics in allergic disease. J. Allergy Clin. Immunol. 2015;135:15–24. doi: 10.1016/j.jaci.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 93.Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY. Epigenetic regulation of gene expression in keratinocytes. J. Investig. Dermatol. 2012;132:2505–2521. doi: 10.1038/jid.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nedoszytko B, et al. The role of regulatory T cells and genes involved in their differentiation in pathogenesis of selected inflammatory and neoplastic skin diseases. Part III: polymorphisms of genes involved in Tregs’ activation and function. Postepy Dermatol. Allergol. 2017;34:517–525. doi: 10.5114/pdia.2017.67053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kehrmann, J. et al. Impact of 5-aza-2’-deoxycytidine and epigallocatechin-3-gallate for induction of human regulatory T cells. Immunology142 (2014). [DOI] [PMC free article] [PubMed]
- 96.Boorgula MP, et al. Replicated methylation changes associated with eczema herpeticum and allergic response. Clin. Epigenetics. 2019;11:122. doi: 10.1186/s13148-019-0714-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmidt, A. D. & Strong, C. D. G. Current understanding of epigenetics in atopic dermatitis. Exp. Dermatol.30 (2021). [DOI] [PMC free article] [PubMed]
- 98.Ziyab AH, Karmaus W, Holloway JW, Zhang H, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J. Eur. Acad. Dermatol. Venereol. 2013;27:e420–e423. doi: 10.1111/jdv.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nicodemus-Johnson, J. et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. Jci Insight1 (2016). [DOI] [PMC free article] [PubMed]
- 100.Moussette S, Tuwaijri AA, Kohan-Ghadr HR, Elzein S, Naumova AK. Role of DNA methylation in expression control of the IKZF3-GSDMA region in human epithelial cells. Plos ONE. 2017;12:e0172707. doi: 10.1371/journal.pone.0172707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stefanowicz, D. et al. DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PLoS ONE7 (2012). [DOI] [PMC free article] [PubMed]
- 102.Hong et al. Nasal DNA methylation is associated with childhood asthma. [DOI] [PMC free article] [PubMed]
- 103.Yang, I. V. et al. The nasal methylome and childhood atopic asthma. J. Allergy Clin. Immunol. (2017). [DOI] [PMC free article] [PubMed]
- 104.Ivana et al. DNA methylation and childhood asthma in the inner city. J. Allergy Clin. Immunol. (2015). [DOI] [PMC free article] [PubMed]
- 105.Gunawardhana LP, et al. Characteristic DNA methylation profiles in peripheral blood monocytes are associated with inflammatory phenotypes of asthma. Epigenetics. 2014;9:1302–1316. doi: 10.4161/epi.33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nicodemus-Johnson J, et al. Genome-wide methylation study identifies an il-13-induced epigenetic signature in asthmatic airways. Am. J. Respir. Crit. Care Med. 2016;193:376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scott RR, et al. Asthma discordance in twins is linked to epigenetic modifications of T cells. PLoS ONE. 2012;7:e48796. doi: 10.1371/journal.pone.0048796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arathimos R, et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin. Epigenetics. 2017;9:112. doi: 10.1186/s13148-017-0414-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bayrak Degirmenci P, et al. Allergic Rhinitis and Its Relationship with IL-10, IL-17, TGF-β, IFN-γ, IL 22, and IL-35. Dis. Markers. 2018;2018:9131432. doi: 10.1155/2018/9131432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li, J. Y. et al. Association between DNA hypomethylation at IL13 gene and allergic rhinitis in house dust mite‐sensitized subjects. Clin. Exp. Allergy46 (2016). [DOI] [PubMed]
- 111.Sarnowski, C. et al. DNA methylation within melatonin receptor 1 A (MTNR1A) mediates paternally transmitted genetic variant effect on asthma plus rhinitis. J. Allergy Clin. Immunol., 748-753 (2016). [DOI] [PubMed]
- 112.Petrus, N. C. M. et al. Cow’s milk allergy in Dutch children: an epigenetic pilot survey. Clin. Transl. Allergy6 (2016). [DOI] [PMC free article] [PubMed]
- 113.Song, Y. et al. Maternal allergy increases susceptibility to offspring allergy in association with TH2-biased epigenetic alterations in a mouse model of peanut allergy. J. Allergy Clin. Immunol. (2014). [DOI] [PMC free article] [PubMed]
- 114.Zhou, X., Han, X., Lyu, S. C., Bunning, B. J. & Nadeau, K. C. Targeted DNA methylation profiling reveals epigenetic signatures in peanut allergy. JCI insight6, 143058. [DOI] [PMC free article] [PubMed]
- 115.Salam MT. Asthma epigenetics. Adv. Exp. Med. Biol. 2014;795:183. doi: 10.1007/978-1-4614-8603-9_11. [DOI] [PubMed] [Google Scholar]
- 116.Bilal AA, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. 2018;14:39. doi: 10.1186/s13223-018-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Angiolilli et al. The acetyl code in rheumatoid arthritis and other rheumatic diseases. [DOI] [PubMed]
- 118.Kaniskan, H. Ü., Martini, M. L. & Jin, J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. (2018). [DOI] [PMC free article] [PubMed]
- 119.Stefanowicz D, et al. Elevated H3K18 acetylation in airway epithelial cells of asthmatic subjects. Respir. Res. 2015;16:95. doi: 10.1186/s12931-015-0254-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kabesch M, Adcock IM. Epigenetics in asthma and COPD. Biochimie. 2012;94:2231–2241. doi: 10.1016/j.biochi.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 121.Harb, H. et al. Childhood allergic asthma is associated with increased IL-13 and FOXP3 histone acetylation. J. Allergy Clin. Immunol. (2015). [DOI] [PubMed]
- 122.Clifford, R. L., Patel, J. K., John, A. E., Tatler, A. L. & Knox, A. J. CXCL8 Histone H3 acetylation is dysfunctional in airway smooth muscle in asthma: regulation by BET. AJP Lung Cell. Mol. Physiol.308, ajplung.00021.02015 (2015). [DOI] [PMC free article] [PubMed]
- 123.Brook PO, Perry MM, Adcock IM, Durham AL. Epigenome-modifying tools in asthma. Epigenomics. 2015;7:1017–1032. doi: 10.2217/epi.15.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-γ loci accompany Th1/Th2 differentiation. J. Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 125.Wang Y, Lv L, Zang H, Gao Z, Zhou X. Regulation of Trek1 expression in nasal mucosa with allergic rhinitis by specific immunotherapy. Cell Biochem. Funct. 2015;33:23–28. doi: 10.1002/cbf.3075. [DOI] [PubMed] [Google Scholar]
- 126.Cho JS, Kang JH, Han IH, Um JY, Lee HM. Antiallergic effects of trichostatin A in a murine model of allergic rhinitis. Clin. Exp. Otorhinolaryngol. 2015;8:243–249. doi: 10.3342/ceo.2015.8.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu, H., Shan, L., Schiller, P.W., Mai, A. & Peng, T. Histone deacetylase-3 activation promotes tumor necrosis factor-α (TNF-α) expression in cardiomyocytes during Lipopolysaccharide stimulation. J. Biol. Chem. (2010). [DOI] [PMC free article] [PubMed]
- 128.Alashkar Alhamwe, B. et al. Decreased histone acetylation levels at Th1 and regulatory loci after induction of food allergy. Nutrients12 (2020). [DOI] [PMC free article] [PubMed]
- 129.Karam, R. A. & Elrahman, D. A. Differential expression of miR-155 and Let-7a in the plasma of childhood asthma: potential biomarkers for diagnosis and severity. Clin. Biochem. (2019). [DOI] [PubMed]
- 130.Tian M, et al. Changes in circulating microRNA-126 levels are associated with immune imbalance in children with acute asthma. Int J. Immunopathol. Pharm. 2018;32:2058738418779243. doi: 10.1177/2058738418779243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ye L, Yan M, Jian W, Jin ML. Effects of microRNA-19b on airway remodeling, airway inflammation and degree of oxidative stress by targeting TSLP through the Stat3 signaling pathway in a mouse model of asthma. Oncotarget. 2017;8:47533–47546. doi: 10.18632/oncotarget.17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lou L, Tian M, Chang J, Li F, Zhang G. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed. Pharmacother. 2019;122:109692. doi: 10.1016/j.biopha.2019.109692. [DOI] [PubMed] [Google Scholar]
- 133.Perry, M. M., Baker, J. E., Gibeon, D. S., Adcock, I. M. & Chung, K. F. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am. J. Respir. Cell. Mol. Biol., 7–17 (2014). [DOI] [PMC free article] [PubMed]
- 134.Lin J, Feng X, Zhang J. Circular RNA circHIPK3 modulates the proliferation of airway smooth muscle cells by miR-326/STIM1 axis. Life Sci. 2020;255:117835. doi: 10.1016/j.lfs.2020.117835. [DOI] [PubMed] [Google Scholar]
- 135.Shi, J. et al. miR‐142‐5p and miR‐130a‐3p regulate pulmonary macrophage polarization and asthma airway remodeling. Immunol. Cell Biol.98 (2020). [DOI] [PubMed]
- 136.Malmh Ll,C, et al. MicroRNA-155 is essential for TH2-mediated allergen-induced eosinophilic inflammation in the lung. J. Allergy Clin. Immunol. 2014;133:1429–1438.e1427. doi: 10.1016/j.jaci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 137.Nakano T, et al. Lower levels of hsa-mir-15a, which decreases VEGFA, in the CD4+ T cells of pediatric patients with asthma. J. Allergy Clin. Immunol. 2013;132:1224–1227.e1212. doi: 10.1016/j.jaci.2013.06.041. [DOI] [PubMed] [Google Scholar]
- 138.Lu TX, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 2011;187:3362–3373. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kan, Z. et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. AJP Lung Cell. Mol. Physiol.315, ajplung.00567.02017- (2018). [DOI] [PubMed]
- 140.Tsitsiou E, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J. Allergy Clin. Immunol. 2012;129:95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 141.Tm A, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum - ScienceDirect. J. Allergy Clin. Immunol. 2016;137:1433–1446. doi: 10.1016/j.jaci.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 142.Collison A, Herbert C, Siegle JS, Mattes J, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm. Med. 2011;11:29. doi: 10.1186/1471-2466-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cho, S. et al. miR-23∼27∼24 clusters control effector T cell differentiation and function. J. Exp. Med. (2016). [DOI] [PMC free article] [PubMed]
- 144.Ghosh, J. A. & Ulaganathan Resveratrol attenuates experimental allergic asthma in mice by restoring inositol polyphosphate 4 phosphatase (INPP4A). Int. Immunopharmacol. (2012). [DOI] [PubMed]
- 145.Xia H, Li Y, Lv X. MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer. Int J. Oncol. 2016;49:1325–1333. doi: 10.3892/ijo.2016.3628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Lin YJ, et al. Tumor hypoxia regulates Forkhead Box C1 to promote lung cancer progression. Theranostics. 2017;7:1177–1191. doi: 10.7150/thno.17895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang X, et al. The role of miR-29c/B7-H3 axis in children with allergic asthma. J. Transl. Med. 2018;16:218. doi: 10.1186/s12967-018-1590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu X, et al. Aberrant expressions of circulating lncRNA NEAT1 and microRNA-125a are linked with Th2 cells and symptom severity in pediatric allergic rhinitis. J. Clin. Lab Anal. 2022;36:e24235. doi: 10.1002/jcla.24235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wen, C. L., Sundaram, G. M., Shan, Q., Guang, L. G. & Sampath, P. Belinostat resolves skin barrier defects in atopic dermatitis by targeting the dysregulated miR-335:SOX6 axis. J. Allergy Clin. Immunol.146 (2020). [DOI] [PubMed]
- 150.O’Connell RM, Rao DS, Baltimore D. microRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 2011;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 151.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-κB. Immunol. Rev. 2012;246:205–220. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 152.Yang, Z. et al. MicroRNA-124 alleviates chronic skin inflammation in atopic eczema via suppressing innate immune responses in keratinocytes. Cell. Immunol., S0008874917301235 (2017). [DOI] [PubMed]
- 153.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The Interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1) J. Biol. Chem. 2011;286:1786. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.C, L. et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. blood (2011). [DOI] [PMC free article] [PubMed]
- 155.Metz CW, Bridges CB. Incompatibility of Mutant Races in Drosophila. Proc. Natl Acad. Sci. USA. 1917;3:673–678. doi: 10.1073/pnas.3.12.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mohr OL. Character changes caused by mutation of an entire region of a chromosome in Drosophila. Genetics. 1919;4:275–282. doi: 10.1093/genetics/4.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou B, et al. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct. Target Ther. 2022;7:95. doi: 10.1038/s41392-022-00934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tindemans I, et al. Increased surface expression of NOTCH on memory T cells in peripheral blood from patients with asthma. J. Allergy Clin. Immunol. 2019;143:769–771.e763. doi: 10.1016/j.jaci.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 160.Tindemans I, et al. Notch signaling in T cells is essential for allergic airway inflammation, but expression of the Notch ligands Jagged 1 and Jagged 2 on dendritic cells is dispensable. J. Allergy Clin. Immunol. 2017;140:1079–1089. doi: 10.1016/j.jaci.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 161.Hammad H, Lambrecht BN. Wnt and Hippo pathways in regulatory T cells: a NOTCH above in asthma. Nat. Immunol. 2020;21:1313–1314. doi: 10.1038/s41590-020-0797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jiao WE, et al. Notch signaling promotes development of allergic Rhinitis by suppressing Foxp3 expression and Treg cell differentiation. Int Arch. Allergy Immunol. 2019;178:33–44. doi: 10.1159/000493328. [DOI] [PubMed] [Google Scholar]
- 163.Jiang S, Han S, Chen J, Li X, Che H. Inhibition effect of blunting Notch signaling on food allergy through improving T(H)1/T(H)2 balance in mice. Ann. Allergy Asthma Immunol. 2017;118:94–102. doi: 10.1016/j.anai.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 164.Dell’Aringa M, Reinhardt RL. Notch signaling represents an important checkpoint between follicular T-helper and canonical T-helper 2 cell fate. Mucosal Immunol. 2018;11:1079–1091. doi: 10.1038/s41385-018-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Honjo A, et al. Pharmacologic inhibition of Notch signaling suppresses food antigen-induced mucosal mast cell hyperplasia. J. Allergy Clin. Immunol. 2017;139:987–996.e910. doi: 10.1016/j.jaci.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 166.Ziegler SF, Artis D. Sensing the outside world: TSLP regulates barrier immunity. Nat. Immunol. 2010;11:289–293. doi: 10.1038/ni.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Niebuhr M, Mamerow D, Heratizadeh A, Satzger I, Werfel T. Staphylococcal alpha-toxin induces a higher T cell proliferation and interleukin-31 in atopic dermatitis. Int Arch. Allergy Immunol. 2011;156:412–415. doi: 10.1159/000323905. [DOI] [PubMed] [Google Scholar]
- 168.Liu YJ. Thymic stromal lymphopoietin: master switch for allergic inflammation. J. Exp. Med. 2006;203:269–273. doi: 10.1084/jem.20051745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yoo J, et al. Spontaneous atopic dermatitis in mice expressing an inducible thymic stromal lymphopoietin transgene specifically in the skin. J. Exp. Med. 2005;202:541–549. doi: 10.1084/jem.20041503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stark GR, Darnell JE., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-L. [DOI] [PubMed] [Google Scholar]
- 172.Wilks AF, et al. Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol. Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kawamura M, et al. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc. Natl Acad. Sci. USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Argetsinger LS, et al. Identification of JAK2 as a growth hormone receptor-associated tyrosine kinase. Cell. 1993;74:237–244. doi: 10.1016/0092-8674(93)90415-M. [DOI] [PubMed] [Google Scholar]
- 175.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 176.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 177.Schindler C, Darnell JE., Jr. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev. Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 178.Lighvani AA, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc. Natl Acad. Sci. USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Afkarian M, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat. Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 180.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4’s role in Th2 differentiation and cell expansion. J. Immunol. 2001;166:7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 181.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/S1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 182.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 183.Chiba Y, Goto K, Matsusue K, Kimura S, Misawa M. Identification and characterization of rat RhoA gene promoter. J. Pharm. Sci. 2010;112:467–472. doi: 10.1254/jphs.09346SC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chiba Y, Goto K, Misawa M. Interleukin-13-induced activation of signal transducer and activator of transcription 6 is mediated by an activation of Janus kinase 1 in cultured human bronchial smooth muscle cells. Pharm. Rep. 2012;64:454–458. doi: 10.1016/S1734-1140(12)70788-0. [DOI] [PubMed] [Google Scholar]
- 185.Kuperman DA, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 2002;8:885–889. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 186.Kuperman DA, et al. Dissecting asthma using focused transgenic modeling and functional genomics. J. Allergy Clin. Immunol. 2005;116:305–311. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 187.Ruterbusch M, Pruner KB, Shehata L, Pepper M. In vivo CD4(+) T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu Rev. Immunol. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- 188.Hsieh YY, et al. JAK-1 rs2780895 C-related genotype and allele but not JAK-1 rs10789166, rs4916008, rs2780885, rs17127114, and rs3806277 are associated with higher susceptibility to asthma. Genet Test. Mol. Biomark. 2011;15:841–847. doi: 10.1089/gtmb.2011.0002. [DOI] [PubMed] [Google Scholar]
- 189.Howell MD, Fitzsimons C, Smith PA. JAK/STAT inhibitors and other small molecule cytokine antagonists for the treatment of allergic disease. Ann. Allergy Asthma Immunol. 2018;120:367–375. doi: 10.1016/j.anai.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 190.Leung DY, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J. Allergy Clin. Immunol. 2014;134:769–779. doi: 10.1016/j.jaci.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bao L, Zhang H, Chan LS. The involvement of the JAK-STAT signaling pathway in chronic inflammatory skin disease atopic dermatitis. Jakstat. 2013;2:e24137. doi: 10.4161/jkst.24137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Paplińska M, et al. Expression of eotaxins in the material from nasal brushing in asthma, allergic rhinitis and COPD patients. Cytokine. 2012;60:393–399. doi: 10.1016/j.cyto.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 193.Shieh YH, et al. Zerumbone enhances the Th1 response and ameliorates ovalbumin-induced Th2 responses and airway inflammation in mice. Int Immunopharmacol. 2015;24:383–391. doi: 10.1016/j.intimp.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 194.Gao, G. M. The progress of eotaxin and the association with ashtma. Int. J. Immunol. (2006).
- 195.Yuan F, et al. JAX2, an ethanol extract of Hyssopus cuspidatus Boriss, can prevent bronchial asthma by inhibiting MAPK/NF-κB inflammatory signaling. Phytomedicine. 2019;57:305–314. doi: 10.1016/j.phymed.2018.12.043. [DOI] [PubMed] [Google Scholar]
- 196.Hart LA, Krishnan VL, Adcock IM, Barnes PJ, Chung KF. Activation and localization of transcription factor, nuclear factor-kappaB, in asthma. Am. J. Respir. Crit. Care Med. 1998;158:1585–1592. doi: 10.1164/ajrccm.158.5.9706116. [DOI] [PubMed] [Google Scholar]
- 197.Tully JE, et al. Epithelial NF-κB orchestrates house dust mite-induced airway inflammation, hyperresponsiveness, and fibrotic remodeling. J. Immunol. 2013;191:5811–5821. doi: 10.4049/jimmunol.1301329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poynter ME, et al. NF-kappa B activation in airways modulates allergic inflammation but not hyperresponsiveness. J. Immunol. 2004;173:7003–7009. doi: 10.4049/jimmunol.173.11.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Poynter ME, Irvin CG, Janssen-Heininger YM. Rapid activation of nuclear factor-kappaB in airway epithelium in a murine model of allergic airway inflammation. Am. J. Pathol. 2002;160:1325–1334. doi: 10.1016/S0002-9440(10)62559-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Oh H, Ghosh S. NF-κB: roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 2013;252:41–51. doi: 10.1111/imr.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gras D, Chanez P, Vachier I, Petit A, Bourdin A. Bronchial epithelium as a target for innovative treatments in asthma. Pharm. Ther. 2013;140:290–305. doi: 10.1016/j.pharmthera.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 202.Kang JH, Hwang SM, Chung IY. S100A8, S100A9 and S100A12 activate airway epithelial cells to produce MUC5AC via extracellular signal-regulated kinase and nuclear factor-κB pathways. Immunology. 2015;144:79–90. doi: 10.1111/imm.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cheng DS, et al. Airway epithelium controls lung inflammation and injury through the NF-kappa B pathway. J. Immunol. 2007;178:6504–6513. doi: 10.4049/jimmunol.178.10.6504. [DOI] [PubMed] [Google Scholar]
- 204.Sheller JR, et al. Nuclear factor kappa B induction in airway epithelium increases lung inflammation in allergen-challenged mice. Exp. Lung Res. 2009;35:883–895. doi: 10.3109/01902140903019710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Koziol-White CJ, Panettieri RA., Jr. Airway smooth muscle and immunomodulation in acute exacerbations of airway disease. Immunol. Rev. 2011;242:178–185. doi: 10.1111/j.1600-065X.2011.01022.x. [DOI] [PubMed] [Google Scholar]
- 206.Tran T, et al. Stimulus-dependent glucocorticoid-resistance of GM-CSF production in human cultured airway smooth muscle. Br. J. Pharm. 2005;145:123–131. doi: 10.1038/sj.bjp.0706174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.John AE, Zhu YM, Brightling CE, Pang L, Knox AJ. Human airway smooth muscle cells from asthmatic individuals have CXCL8 hypersecretion due to increased NF-kappa B p65, C/EBP beta, and RNA polymerase II binding to the CXCL8 promoter. J. Immunol. 2009;183:4682–4692. doi: 10.4049/jimmunol.0803832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Radermecker C, Louis R, Bureau F, Marichal T. Role of neutrophils in allergic asthma. Curr. Opin. Immunol. 2018;54:28–34. doi: 10.1016/j.coi.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 209.Chien JW, et al. Increased IL-17A secreting CD4+ T cells, serum IL-17 levels and exhaled nitric oxide are correlated with childhood asthma severity. Clin. Exp. Allergy. 2013;43:1018–1026. doi: 10.1111/cea.12119. [DOI] [PubMed] [Google Scholar]
- 210.Fujisawa T, et al. NF-κB mediates IL-1β- and IL-17A-induced MUC5B expression in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2011;45:246–252. doi: 10.1165/rcmb.2009-0313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dragon S, Hirst SJ, Lee TH, Gounni AS. IL-17A mediates a selective gene expression profile in asthmatic human airway smooth muscle cells. Am. J. Respir. Cell Mol. Biol. 2014;50:1053–1063. doi: 10.1165/rcmb.2012-0267OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chang Y, et al. Th17-associated cytokines promote human airway smooth muscle cell proliferation. FASEB J. 2012;26:5152–5160. doi: 10.1096/fj.12-208033. [DOI] [PubMed] [Google Scholar]
- 213.Wan R, et al. Neutrophil extracellular traps amplify neutrophil recruitment and inflammation in neutrophilic asthma by stimulating the airway epithelial cells to activate the TLR4/ NF-κB pathway and secrete chemokines. Aging. 2020;12:16820–16836. doi: 10.18632/aging.103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wenzel SE, et al. Bronchoscopic evaluation of severe asthma. Persistent inflammation associated with high dose glucocorticoids. Am. J. Respir. Crit. Care Med. 1997;156:737–743. doi: 10.1164/ajrccm.156.3.9610046. [DOI] [PubMed] [Google Scholar]
- 215.Wong CK, et al. Induction of adhesion molecules upon the interaction between eosinophils and bronchial epithelial cells: involvement of p38 MAPK and NF-kappaB. Int. Immunopharmacol. 2006;6:1859–1871. doi: 10.1016/j.intimp.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 216.Wang CB, et al. Induction of IL-6 in co-culture of bronchial epithelial cells and eosinophils is regulated by p38 MAPK and NF-kappaB. Allergy. 2005;60:1378–1385. doi: 10.1111/j.1398-9995.2005.00884.x. [DOI] [PubMed] [Google Scholar]
- 217.Kankaanranta H, et al. Tumour necrosis factor-α regulates human eosinophil apoptosis via ligation of TNF-receptor 1 and balance between NF-κB and AP-1. PLoS ONE. 2014;9:e90298. doi: 10.1371/journal.pone.0090298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Edwards MR, et al. Targeting the NF-kappaB pathway in asthma and chronic obstructive pulmonary disease. Pharm. Ther. 2009;121:1–13. doi: 10.1016/j.pharmthera.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rescigno M, Martino M, Sutherland CL, Gold MR, Ricciardi-Castagnoli P. Dendritic cell survival and maturation are regulated by different signaling pathways. J. Exp. Med. 1998;188:2175–2180. doi: 10.1084/jem.188.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pelaia G, et al. Cellular mechanisms underlying eosinophilic and neutrophilic airway inflammation in asthma. Mediators Inflamm. 2015;2015:879783. doi: 10.1155/2015/879783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ahern PP, Izcue A, Maloy KJ, Powrie F. The interleukin-23 axis in intestinal inflammation. Immunol. Rev. 2008;226:147–159. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- 222.Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity. 2002;16:257–270. doi: 10.1016/S1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 223.O’Keeffe M, et al. Distinct roles for the NF-kappaB1 and c-Rel transcription factors in the differentiation and survival of plasmacytoid and conventional dendritic cells activated by TLR-9 signals. Blood. 2005;106:3457–3464. doi: 10.1182/blood-2004-12-4965. [DOI] [PubMed] [Google Scholar]
- 224.Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tang W, et al. Hippo signaling pathway and respiratory diseases. Cell Death Disco. 2022;8:213. doi: 10.1038/s41420-022-01020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Harb H, et al. A regulatory T cell Notch4-GDF15 axis licenses tissue inflammation in asthma. Nat. Immunol. 2020;21:1359–1370. doi: 10.1038/s41590-020-0777-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kumar P, et al. Soluble OX40L and JAG1 Induce Selective Proliferation of Functional Regulatory T-Cells Independent of canonical TCR signaling. Sci. Rep. 2017;7:39751. doi: 10.1038/srep39751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fermino ML, et al. Galectin-3 negatively regulates the frequency and function of CD4(+) CD25(+) Foxp3(+) regulatory T cells and influences the course of Leishmania major infection. Eur. J. Immunol. 2013;43:1806–1817. doi: 10.1002/eji.201343381. [DOI] [PubMed] [Google Scholar]
- 229.Begin P, Nadeau KC. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin. Immunol. 2014;10:27. doi: 10.1186/1710-1492-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Palmer LJ, et al. Independent inheritance of serum immunoglobulin E concentrations and airway responsiveness. Am. J. Respir. Crit. Care Med. 2000;161:1836–1843. doi: 10.1164/ajrccm.161.6.9805104. [DOI] [PubMed] [Google Scholar]
- 231.Gangloff SC, Guenounou M. Toll-like receptors and immune response in allergic disease. Clin. Rev. Allergy Immunol. 2004;26:115–125. doi: 10.1007/s12016-004-0006-0. [DOI] [PubMed] [Google Scholar]
- 232.Takeda K, Akira S. Roles of Toll-like receptors in innate immune responses. Genes Cells. 2001;6:733–742. doi: 10.1046/j.1365-2443.2001.00458.x. [DOI] [PubMed] [Google Scholar]
- 233.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 234.Mizel SB, Honko AN, Moors MA, Smith PS, West AP. Induction of macrophage nitric oxide production by Gram-negative flagellin involves signaling via heteromeric Toll-like receptor 5/Toll-like receptor 4 complexes. J. Immunol. 2003;170:6217–6223. doi: 10.4049/jimmunol.170.12.6217. [DOI] [PubMed] [Google Scholar]
- 235.Riiser A. The human microbiome, asthma, and allergy. Allergy Asthma Clin. Immunol. 2015;11:35. doi: 10.1186/s13223-015-0102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.McAlees JW, et al. Distinct Tlr4-expressing cell compartments control neutrophilic and eosinophilic airway inflammation. Mucosal Immunol. 2015;8:863–873. doi: 10.1038/mi.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Thomas SY, et al. MyD88-dependent dendritic and epithelial cell crosstalk orchestrates immune responses to allergens. Mucosal Immunol. 2018;11:796–810. doi: 10.1038/mi.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Larsen JM, et al. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology. 2015;144:333–342. doi: 10.1111/imm.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Karvonen AM, et al. Quantity and diversity of environmental microbial exposure and development of asthma: a birth cohort study. Allergy. 2014;69:1092–1101. doi: 10.1111/all.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Remot A, et al. Bacteria isolated from lung modulate asthma susceptibility in mice. ISME J. 2017;11:1061–1074. doi: 10.1038/ismej.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Eisenbarth SC, et al. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J. Exp. Med. 2002;196:1645–1651. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Shim JU, et al. Flagellin suppresses experimental asthma by generating regulatory dendritic cells and T cells. J. Allergy Clin. Immunol. 2016;137:426–435. doi: 10.1016/j.jaci.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 243.Holt PG, et al. Downregulation of the antigen presenting cell function(s) of pulmonary dendritic cells in vivo by resident alveolar macrophages. J. Exp. Med. 1993;177:397–407. doi: 10.1084/jem.177.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Strickland DH, Thepen T, Kees UR, Kraal G, Holt PG. Regulation of T-cell function in lung tissue by pulmonary alveolar macrophages. Immunology. 1993;80:266–272. [PMC free article] [PubMed] [Google Scholar]
- 245.Lun SW, Wong CK, Ko FW, Hui DS, Lam CW. Expression and functional analysis of toll-like receptors of peripheral blood cells in asthmatic patients: implication for immunopathological mechanism in asthma. J. Clin. Immunol. 2009;29:330–342. doi: 10.1007/s10875-008-9269-1. [DOI] [PubMed] [Google Scholar]
- 246.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J. Immunol. 2002;168:554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 247.Sha Q, Truong-Tran AQ, Plitt JR, Beck LA, Schleimer RP. Activation of airway epithelial cells by toll-like receptor agonists. Am. J. Respir. Cell Mol. Biol. 2004;31:358–364. doi: 10.1165/rcmb.2003-0388OC. [DOI] [PubMed] [Google Scholar]
- 248.Armstrong L, et al. Expression of functional toll-like receptor-2 and -4 on alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 2004;31:241–245. doi: 10.1165/rcmb.2004-0078OC. [DOI] [PubMed] [Google Scholar]
- 249.Fernandez S, Jose P, Avdiushko MG, Kaplan AM, Cohen DA. Inhibition of IL-10 receptor function in alveolar macrophages by Toll-like receptor agonists. J. Immunol. 2004;172:2613–2620. doi: 10.4049/jimmunol.172.4.2613. [DOI] [PubMed] [Google Scholar]
- 250.Applequist SE, Wallin RP, Ljunggren HG. Variable expression of Toll-like receptor in murine innate and adaptive immune cell lines. Int Immunol. 2002;14:1065–1074. doi: 10.1093/intimm/dxf069. [DOI] [PubMed] [Google Scholar]
- 251.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Long EM, Millen B, Kubes P, Robbins SM. Lipoteichoic acid induces unique inflammatory responses when compared to other toll-like receptor 2 ligands. PLoS ONE. 2009;4:e5601. doi: 10.1371/journal.pone.0005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fuchs B, Braun A. Modulation of asthma and allergy by addressing toll-like receptor 2. J. Occup. Med. Toxicol. 2008;3(Suppl. 1):S5. doi: 10.1186/1745-6673-3-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Redecke V, et al. Cutting edge: activation of Toll-like receptor 2 induces a Th2 immune response and promotes experimental asthma. J. Immunol. 2004;172:2739–2743. doi: 10.4049/jimmunol.172.5.2739. [DOI] [PubMed] [Google Scholar]
- 255.Akdis CA, et al. Inhibition of T helper 2-type responses, IgE production and eosinophilia by synthetic lipopeptides. Eur. J. Immunol. 2003;33:2717–2726. doi: 10.1002/eji.200323329. [DOI] [PubMed] [Google Scholar]
- 256.Chun E, et al. Toll-like receptor expression on peripheral blood mononuclear cells in asthmatics; implications for asthma management. J. Clin. Immunol. 2010;30:459–464. doi: 10.1007/s10875-009-9363-z. [DOI] [PubMed] [Google Scholar]
- 257.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 258.Siwiec J, et al. [Evaluation of Th1/Th2 lymphocyte balance and lipopolysaccharide receptor expression in asthma patients] Pneumonol. Alergol. Pol. 2009;77:123–130. [PubMed] [Google Scholar]
- 259.Nawijn MC, et al. TLR-2 activation induces regulatory T cells and long-term suppression of asthma manifestations in mice. PLoS ONE. 2013;8:e55307. doi: 10.1371/journal.pone.0055307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhou C, Kang XD, Chen Z. A synthetic Toll-like receptor 2 ligand decreases allergic immune responses in a mouse rhinitis model sensitized to mite allergen. J. Zhejiang Univ. Sci. B. 2008;9:279–285. doi: 10.1631/jzus.B0730029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Patel M, et al. TLR2 agonist ameliorates established allergic airway inflammation by promoting Th1 response and not via regulatory T cells. J. Immunol. 2005;174:7558–7563. doi: 10.4049/jimmunol.174.12.7558. [DOI] [PubMed] [Google Scholar]
- 262.Taylor RC, Richmond P, Upham JW. Toll-like receptor 2 ligands inhibit TH2 responses to mite allergen. J. Allergy Clin. Immunol. 2006;117:1148–1154. doi: 10.1016/j.jaci.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 263.Fuchs B, et al. A Toll-like receptor 2/6 agonist reduces allergic airway inflammation in chronic respiratory sensitisation to Timothy grass pollen antigens. Int Arch. Allergy Immunol. 2010;152:131–139. doi: 10.1159/000265534. [DOI] [PubMed] [Google Scholar]
- 264.Kim KS, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351:858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 265.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Xu H, Shu H, Zhu J, Song J. Inhibition of TLR4 inhibits allergic responses in murine allergic rhinitis by regulating the NF-kappaB pathway. Exp. Ther. Med. 2019;18:761–768. doi: 10.3892/etm.2019.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Fransson M, et al. Up-regulation of Toll-like receptors 2, 3 and 4 in allergic rhinitis. Respir. Res. 2005;6:100. doi: 10.1186/1465-9921-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 270.Liu J, et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target Ther. 2022;7:3. doi: 10.1038/s41392-021-00762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Matsuda-Hirose H, et al. Selective inhibition of beta-Catenin/Co-Activator cyclic AMP response element-binding protein-dependent signaling prevents the emergence of hapten-induced atopic dermatitis-like dermatitis. Ann. Dermatol. 2019;31:631–639. doi: 10.5021/ad.2019.31.6.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kwak HJ, et al. The Wnt/beta-catenin signaling pathway regulates the development of airway remodeling in patients with asthma. Exp. Mol. Med. 2015;47:e198. doi: 10.1038/emm.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Jia XX, et al. Wnt/beta-catenin signaling pathway regulates asthma airway remodeling by influencing the expression of c-Myc and cyclin D1 via the p38 MAPK-dependent pathway. Exp. Ther. Med. 2019;18:3431–3438. doi: 10.3892/etm.2019.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Trischler J, et al. Immune modulation of the T cell response in asthma through Wnt10b. Am. J. Respir. Cell Mol. Biol. 2016;54:584–593. doi: 10.1165/rcmb.2014-0425OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Song J, Zhu XM, Wei QY. MSCs reduce airway remodeling in the lungs of asthmatic rats through the Wnt/β-catenin signaling pathway. Eur. Rev. Med Pharm. Sci. 2020;24:11199–11211. doi: 10.26355/eurrev_202011_23608. [DOI] [PubMed] [Google Scholar]
- 276.Huang Y, Wang L, Jia XX, Lin XX, Zhang WX. Vitamin D alleviates airway remodeling in asthma by down-regulating the activity of Wnt/beta-catenin signaling pathway. Int Immunopharmacol. 2019;68:88–94. doi: 10.1016/j.intimp.2018.12.061. [DOI] [PubMed] [Google Scholar]
- 277.Reuter S, et al. The Wnt/beta-catenin pathway attenuates experimental allergic airway disease. J. Immunol. 2014;193:485–495. doi: 10.4049/jimmunol.1400013. [DOI] [PubMed] [Google Scholar]
- 278.Li J, Xue K, Zheng Y, Wang Y, Xu C. RORA overexpression alleviates nasal mucosal injury and enhances red blood cell immune adhesion function in a mouse model of allergic rhinitis via inactivation of the Wnt/beta-catenin signaling pathway. Int Arch. Allergy Immunol. 2019;180:79–90. doi: 10.1159/000500637. [DOI] [PubMed] [Google Scholar]
- 279.Leung DYM, Berdyshev E, Goleva E. Cutaneous barrier dysfunction in allergic diseases. J. Allergy Clin. Immunol. 2020;145:1485–1497. doi: 10.1016/j.jaci.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Tasioudi KE, et al. Immunohistochemical and molecular analysis of PI3K/AKT/mTOR pathway in esophageal carcinoma. APMIS. 2015;123:639–647. doi: 10.1111/apm.12398. [DOI] [PubMed] [Google Scholar]
- 281.Choi YH, Jin GY, Li LC, Yan GH. Inhibition of protein kinase C delta attenuates allergic airway inflammation through suppression of PI3K/Akt/mTOR/HIF-1 alpha/VEGF pathway. PLoS ONE. 2013;8:e81773. doi: 10.1371/journal.pone.0081773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zou W, Ding F, Niu C, Fu Z, Liu S. Brg1 aggravates airway inflammation in asthma via inhibition of the PI3K/Akt/mTOR pathway. Biochem. Biophys. Res. Commun. 2018;503:3212–3218. doi: 10.1016/j.bbrc.2018.08.127. [DOI] [PubMed] [Google Scholar]
- 283.Lee KS, et al. Phosphoinositide 3-kinase-delta inhibitor reduces vascular permeability in a murine model of asthma. J. Allergy Clin. Immunol. 2006;118:403–409. doi: 10.1016/j.jaci.2006.04.041. [DOI] [PubMed] [Google Scholar]
- 284.Kim SR, et al. HIF-1alpha inhibition ameliorates an allergic airway disease via VEGF suppression in bronchial epithelium. Eur. J. Immunol. 2010;40:2858–2869. doi: 10.1002/eji.200939948. [DOI] [PubMed] [Google Scholar]
- 285.Burgess JK, et al. Dual ERK and phosphatidylinositol 3-kinase pathways control airway smooth muscle proliferation: differences in asthma. J. Cell Physiol. 2008;216:673–679. doi: 10.1002/jcp.21450. [DOI] [PubMed] [Google Scholar]
- 286.Huang L, et al. OX40L induces helper T cell differentiation during cell immunity of asthma through PI3K/AKT and P38 MAPK signaling pathway. J. Transl. Med. 2018;16:74. doi: 10.1186/s12967-018-1436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Stark AK, Sriskantharajah S, Hessel EM, Okkenhaug K. PI3K inhibitors in inflammation, autoimmunity and cancer. Curr. Opin. Pharm. 2015;23:82–91. doi: 10.1016/j.coph.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhao Y, et al. PI3K-AKT-mTOR signaling pathway: the intersection of allergic asthma and cataract. Pharmazie. 2019;74:598–600. doi: 10.1691/ph.2019.9080. [DOI] [PubMed] [Google Scholar]
- 289.Wu G, et al. Anti-allergic function of α-Tocopherol is mediated by suppression of PI3K-PKB activity in mast cells in mouse model of allergic rhinitis. Allergol. Immunopathol. 2020;48:395–400. doi: 10.1016/j.aller.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 290.Zhang Y, et al. Bicalutamide, an androgen receptor antagonist, effectively alleviate allergic rhinitis via suppression of PI3K-PKB activity. Eur. Arch. Otorhinolaryngol. 2023;280:703–711. doi: 10.1007/s00405-022-07538-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Zeng Q, Luo X, Tang Y, Liu W, Luo R. Leptin regulated ILC2 Cell through the PI3K/AKT pathway in allergic rhinitis. Mediators Inflamm. 2020;2020:4176082. doi: 10.1155/2020/4176082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Yuan J, et al. Gene knockdown of CCR3 reduces eosinophilic inflammation and the Th2 immune response by inhibiting the PI3K/AKT pathway in allergic rhinitis mice. Sci. Rep. 2022;12:5411. doi: 10.1038/s41598-022-09467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Nian JB, et al. Epithelial cells expressed IL-33 to promote degranulation of mast cells through inhibition on ST2/PI3K/mTOR-mediated autophagy in allergic rhinitis. Cell Cycle. 2020;19:1132–1142. doi: 10.1080/15384101.2020.1749402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Karagianni, F., Pavlidis, A., Malakou, L. S., Piperi, C. & Papadavid, E. Predominant role of mTOR signaling in skin diseases with therapeutic potential. Int. J. Mol. Sci.23 (2022). [DOI] [PMC free article] [PubMed]
- 295.Bao Y, et al. MicroRNA-126 accelerates IgE-mediated mast cell degranulation associated with the PI3K/Akt signaling pathway by promoting Ca(2+) influx. Exp. Ther. Med. 2018;16:2763–2769. doi: 10.3892/etm.2018.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kim EG, et al. Chitinase 3-like 1 contributes to food allergy via M2 macrophage polarization. Allergy Asthma Immunol. Res. 2020;12:1012–1028. doi: 10.4168/aair.2020.12.6.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 298.Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10:31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Delgoffe GM, Powell JD. mTOR: taking cues from the immune microenvironment. Immunology. 2009;127:459–465. doi: 10.1111/j.1365-2567.2009.03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev. Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zou H, et al. MTOR-mediated autophagy is involved in the protective effect of ketamine on allergic airway inflammation. J. Immunol. Res. 2019;2019:5879714. doi: 10.1155/2019/5879714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Preite S, Huang B, Cannons JL, McGavern DB, Schwartzberg PL. PI3K orchestrates T follicular helper cell differentiation in a context dependent manner: implications for autoimmunity. Front Immunol. 2018;9:3079. doi: 10.3389/fimmu.2018.03079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wang P, et al. The Regulatory Effects of mTOR Complexes in the Differentiation and Function of CD4(+) T Cell Subsets. J. Immunol. Res. 2020;2020:3406032. doi: 10.1155/2020/3406032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Jacobsen EA, et al. Allergic pulmonary inflammation in mice is dependent on eosinophil-induced recruitment of effector T cells. J. Exp. Med. 2008;205:699–710. doi: 10.1084/jem.20071840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zhu C, et al. mTOR complexes differentially orchestrates eosinophil development in allergy. Sci. Rep. 2018;8:6883. doi: 10.1038/s41598-018-25358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chen X, et al. Poly-L-arginine promotes asthma angiogenesis through induction of FGFBP1 in airway epithelial cells via activation of the mTORC1-STAT3 pathway. Cell Death Dis. 2021;12:761. doi: 10.1038/s41419-021-04055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Jia QN, Zeng YP. Rapamycin blocks the IL-13-induced deficiency of epidermal barrier related proteins via upregulation of miR-143 in HaCaT Keratinocytes. Int. J. Med. Sci. 2020;17:2087–2094. doi: 10.7150/ijms.45765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Méndez-Enríquez E, Hallgren J. Mast cells and their progenitors in allergic asthma. Front Immunol. 2019;10:821. doi: 10.3389/fimmu.2019.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Turner H, Kinet JP. Signalling through the high-affinity IgE receptor Fc epsilonRI. Nature. 1999;402:B24–B30. doi: 10.1038/35037021. [DOI] [PubMed] [Google Scholar]
- 311.Amo G, et al. FCERI and histamine metabolism gene variability in selective responders to NSAIDS. Front Pharm. 2016;7:353. doi: 10.3389/fphar.2016.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kanagaratham C, El Ansari YS, Lewis OL, Oettgen HC. IgE and IgG antibodies as regulators of mast cell and basophil functions in food allergy. Front Immunol. 2020;11:603050. doi: 10.3389/fimmu.2020.603050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Shin JS, Greer AM. The role of FcεRI expressed in dendritic cells and monocytes. Cell Mol. Life Sci. 2015;72:2349–2360. doi: 10.1007/s00018-015-1870-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Suber J, Iweala OI. Strategies for mast cell inhibition in food allergy. Yale J. Biol. Med. 2020;93:719–731. [PMC free article] [PubMed] [Google Scholar]
- 315.Suurmond J, Dorjée AL, Knol EF, Huizinga TW, Toes RE. Differential TLR-induced cytokine production by human mast cells is amplified by FcɛRI triggering. Clin. Exp. Allergy. 2015;45:788–796. doi: 10.1111/cea.12509. [DOI] [PubMed] [Google Scholar]
- 316.Suurmond J, et al. Activation of human basophils by combined toll-like receptor- and FcεRI-triggering can promote Th2 skewing of naive T helper cells. Eur. J. Immunol. 2014;44:386–396. doi: 10.1002/eji.201343617. [DOI] [PubMed] [Google Scholar]
- 317.Pellefigues, C. IgE Autoreactivity in atopic dermatitis: paving the road for autoimmune diseases? Antibodies9 (2020). [DOI] [PMC free article] [PubMed]
- 318.Pitchford SC, et al. Allergen induces the migration of platelets to lung tissue in allergic asthma. Am. J. Respir. Crit. Care Med. 2008;177:604–612. doi: 10.1164/rccm.200702-214OC. [DOI] [PubMed] [Google Scholar]
- 319.Guo W, et al. IgE aggravates the senescence of smooth muscle cells in abdominal aortic aneurysm by upregulating LincRNA-p21. Aging Dis. 2019;10:699–710. doi: 10.14336/AD.2018.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Crosson T, et al. FcεR1-expressing nociceptors trigger allergic airway inflammation. J. Allergy Clin. Immunol. 2021;147:2330–2342. doi: 10.1016/j.jaci.2020.12.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Liang H, et al. Vagal activities are involved in antigen-specific immune inflammation in the intestine. J. Gastroenterol. Hepatol. 2011;26:1065–1071. doi: 10.1111/j.1440-1746.2011.06710.x. [DOI] [PubMed] [Google Scholar]
- 322.Yashiro, T. et al. Pathophysiological roles of neuro-immune interactions between enteric neurons and mucosal mast cells in the gut of food allergy mice. Cells10 (2021). [DOI] [PMC free article] [PubMed]
- 323.Chandler, C. E., Harberts, E. M. & Ernst, R. K. Pathogen sensing: toll-like receptors and NODs (Innate Immunity). Ency. Microbiol., 443–456 (2019).
- 324.Wicherska-Pawłowska, K., Wróbel, T. & Rybka, J. Toll-Like Receptors (TLRs), NOD-like receptors (NLRs), and RIG-I-like receptors (RLRs) in innate immunity. TLRs, NLRs, and RLRs ligands as immunotherapeutic agents for hematopoietic diseases. Int. J. Mol. Sci.22 (2021). [DOI] [PMC free article] [PubMed]
- 325.Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: versatile cytosolic sentinels. Physiol. Rev. 2015;95:149–178. doi: 10.1152/physrev.00009.2014. [DOI] [PubMed] [Google Scholar]
- 326.Kim YK, Shin JS, Nahm MH. NOD-like receptors in infection, immunity, and diseases. Yonsei Med J. 2016;57:5–14. doi: 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Gurung P, Kanneganti TD. Immune responses against protozoan parasites: a focus on the emerging role of Nod-like receptors. Cell Mol. Life Sci. 2016;73:3035–3051. doi: 10.1007/s00018-016-2212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat. Immunol. 2011;12:121–128. doi: 10.1038/ni.1985. [DOI] [PubMed] [Google Scholar]
- 329.Travassos LH, Carneiro LA, Girardin S, Philpott DJ. Nod proteins link bacterial sensing and autophagy. Autophagy. 2010;6:409–411. doi: 10.4161/auto.6.3.11305. [DOI] [PubMed] [Google Scholar]
- 330.Tsang, M. S., Hou, T., Chan, B. C. & Wong, C. K. Immunological roles of NLR in allergic diseases and its underlying mechanisms. Int. J. Mol. Sci. 22 (2021). [DOI] [PMC free article] [PubMed]
- 331.Wong CK, et al. NOD-like receptors mediated activation of eosinophils interacting with bronchial epithelial cells: a link between innate immunity and allergic asthma. Cell Mol. Immunol. 2013;10:317–329. doi: 10.1038/cmi.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Duan W, et al. Innate signals from Nod2 block respiratory tolerance and program T(H)2-driven allergic inflammation. J. Allergy Clin. Immunol. 2010;126:1284–1293.e1210. doi: 10.1016/j.jaci.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhou B, et al. Thymic stromal lymphopoietin as a key initiator of allergic airway inflammation in mice. Nat. Immunol. 2005;6:1047–1053. doi: 10.1038/ni1247. [DOI] [PubMed] [Google Scholar]
- 334.Tamachi T, et al. IL-25 enhances allergic airway inflammation by amplifying a TH2 cell-dependent pathway in mice. J. Allergy Clin. Immunol. 2006;118:606–614. doi: 10.1016/j.jaci.2006.04.051. [DOI] [PubMed] [Google Scholar]
- 335.Jember AG, Zuberi R, Liu FT, Croft M. Development of allergic inflammation in a murine model of asthma is dependent on the costimulatory receptor OX40. J. Exp. Med. 2001;193:387–392. doi: 10.1084/jem.193.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lv J, et al. Heme oxygenase-1 protects airway epithelium against apoptosis by targeting the proinflammatory NLRP3-RXR axis in asthma. J. Biol. Chem. 2018;293:18454–18465. doi: 10.1074/jbc.RA118.004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Ma M, Li G, Qi M, Jiang W, Zhou R. Inhibition of the inflammasome activity of NLRP3 attenuates HDM-induced allergic asthma. Front Immunol. 2021;12:718779. doi: 10.3389/fimmu.2021.718779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Dai MY, et al. Particulate matters induce acute exacerbation of allergic airway inflammation via the TLR2/NF-κB/NLRP3 signaling pathway. Toxicol. Lett. 2020;321:146–154. doi: 10.1016/j.toxlet.2019.12.013. [DOI] [PubMed] [Google Scholar]
- 339.Bogefors J, et al. Nod1, Nod2 and Nalp3 receptors, new potential targets in treatment of allergic rhinitis? Allergy. 2010;65:1222–1226. doi: 10.1111/j.1398-9995.2009.02315.x. [DOI] [PubMed] [Google Scholar]
- 340.Zhou H, et al. Activation of NLRP3 inflammasome contributes to the inflammatory response to allergic rhinitis via macrophage pyroptosis. Int Immunopharmacol. 2022;110:109012. doi: 10.1016/j.intimp.2022.109012. [DOI] [PubMed] [Google Scholar]
- 341.Haspeslagh E, Heyndrickx I, Hammad H, Lambrecht BN. The hygiene hypothesis: immunological mechanisms of airway tolerance. Curr. Opin. Immunol. 2018;54:102–108. doi: 10.1016/j.coi.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Sbihi H, et al. Thinking bigger: how early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy. 2019;74:2103–2115. doi: 10.1111/all.13812. [DOI] [PubMed] [Google Scholar]
- 343.Coyte KZ, Rao C, Rakoff-Nahoum S, Foster KR. Ecological rules for the assembly of microbiome communities. PLoS Biol. 2021;19:e3001116. doi: 10.1371/journal.pbio.3001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Peroni DG, Nuzzi G, Trambusti I, Di Cicco ME, Comberiati P. Microbiome composition and its impact on the development of allergic diseases. Front Immunol. 2020;11:700. doi: 10.3389/fimmu.2020.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Dekaboruah E, Suryavanshi MV, Chettri D, Verma AK. Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020;202:2147–2167. doi: 10.1007/s00203-020-01931-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sugahara H OT, Hashikura N, Abe F, Xiao JZ. Differences in folate production by bifidobacteria of different origins. Biosci. Microbiota Food Health. 2015;34:87–93. doi: 10.12938/bmfh.2015-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Rowland I, et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 2018;57:1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Gomez de Agüero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 349.Francino MP. Early development of the gut microbiota and immune health. Pathogens. 2014;3:769–790. doi: 10.3390/pathogens3030769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Stokholm J, et al. Cesarean section changes neonatal gut colonization. J. Allergy Clin. Immunol. 2016;138:881–889 e882. doi: 10.1016/j.jaci.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 351.Shaterian N, Abdi F, Ghavidel N, Alidost F. Role of cesarean section in the development of neonatal gut microbiota: a systematic review. Open Med. 2021;16:624–639. doi: 10.1515/med-2021-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Kim G, et al. Delayed establishment of gut microbiota in infants delivered by Cesarean section. Front. Microbiol. 2020;11:2099. doi: 10.3389/fmicb.2020.02099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Ma J, et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci. Rep. 2020;10:15792. doi: 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Zhong H, et al. Impact of probiotics supplement on the gut microbiota in neonates with antibiotic exposure: an open-label single-center randomized parallel controlled study. World J. Pediatr. 2021;17:385–393. doi: 10.1007/s12519-021-00443-y. [DOI] [PubMed] [Google Scholar]
- 355.Yousuf EI, et al. Persistence of suspected probiotic organisms in preterm infant gut microbiota weeks after probiotic supplementation in the NICU. Front. Microbiol. 2020;11:574137. doi: 10.3389/fmicb.2020.574137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Dierikx TH, et al. The influence of prenatal and intrapartum antibiotics on intestinal microbiota colonisation in infants: a systematic review. J. Infect. 2020;81:190–204. doi: 10.1016/j.jinf.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 357.Milliken S, Allen RM, Lamont RF. The role of antimicrobial treatment during pregnancy on the neonatal gut microbiome and the development of atopy, asthma, allergy and obesity in childhood. Expert Opin. Drug Saf. 2019;18:173–185. doi: 10.1080/14740338.2019.1579795. [DOI] [PubMed] [Google Scholar]
- 358.Panduru, M. et al. Antibiotics administration during last trimester of pregnancy is associated with atopic dermatitis - a cross-sectional study. Sciendo (2020). [DOI] [PubMed]
- 359.Stensballe LG, Simonsen J, Jensen SM, Bonnelykke K, Bisgaard H. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. J. Pediatr. 2013;162:832–838 e833. doi: 10.1016/j.jpeds.2012.09.049. [DOI] [PubMed] [Google Scholar]
- 360.Geng M, et al. Prenatal low-dose antibiotic exposure and children allergic diseases at 4 years of age: a prospective birth cohort study. Ecotoxicol. Environ. Saf. 2021;225:112736. doi: 10.1016/j.ecoenv.2021.112736. [DOI] [PubMed] [Google Scholar]
- 361.Aversa Z, et al. Association of Infant antibiotic exposure with childhood health outcomes. Mayo Clin. Proc. 2021;96:66–77. doi: 10.1016/j.mayocp.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Moreau MC DR, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 1978;21:532–539. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Fulde M, H M. Maturation of the enteric mucosal innate immune system during the postnatal period.pdf. Immunol. Rev. 2014;260:21–34. doi: 10.1111/imr.12190. [DOI] [PubMed] [Google Scholar]
- 364.Wesemann DR, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Sansonetti PJ. To be or not to be a pathogen: that is the mucosally relevant question. Mucosal Immunol. 2011;4:8–14. doi: 10.1038/mi.2010.77. [DOI] [PubMed] [Google Scholar]
- 366.Holgate ST. Innate and adaptive immune responses in asthma. Nat. Med. 2012;18:673–683. doi: 10.1038/nm.2731. [DOI] [PubMed] [Google Scholar]
- 367.Arrieta MC, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 368.Hevia A, et al. Allergic patients with long-term asthma display low levels of Bifidobacterium adolescentis. PLoS ONE. 2016;11:e0147809. doi: 10.1371/journal.pone.0147809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Huang YJ, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J. Allergy Clin. Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Yao Y, et al. Identification of staphylococcal protein A in infected atopic dermatitis lesions. J. Invest. Dermatol. 2010;130:2502–2504. doi: 10.1038/jid.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Niebuhr M, Scharonow H, Gathmann M, Mamerow D, Werfel T. Staphylococcal exotoxins are strong inducers of IL-22: a potential role in atopic dermatitis. J. Allergy Clin. Immunol. 2010;126:1176–1183 e1174. doi: 10.1016/j.jaci.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 373.Hong SW, et al. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 2011;66:351–359. doi: 10.1111/j.1398-9995.2010.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Ando T, et al. Mast cells are required for full expression of allergen/SEB-induced skin inflammation. J. Invest. Dermatol. 2013;133:2695–2705. doi: 10.1038/jid.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Abrahamsson TR, et al. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 2012;129:434–440. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 376.Marrs T, Flohr C. The role of skin and gut microbiota in the development of atopic eczema. Br. J. Dermatol. 2016;175(Suppl. 2):13–18. doi: 10.1111/bjd.14907. [DOI] [PubMed] [Google Scholar]
- 377.Penders J, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J. Allergy Clin. Immunol. 2013;132:601–607 e608. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 378.Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J. Allergy Clin. Immunol. 2016;137:852–860. doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 379.Bassis CM TA, Young VB, Pynnonen MA. The nasal cavity microbiota of healthy adults. Microbiome. 2014;2:27. doi: 10.1186/2049-2618-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Liu CM PL, et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015;1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Lemon KP, et al. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio. 2010;1:e00129–00110. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Choi CH, et al. Seasonal allergic rhinitis affects sinonasal microbiota. Am. J. Rhinol. Allergy. 2014;28:281–286. doi: 10.2500/ajra.2014.28.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Shiomori T, Yoshida S, Miyamoto H, Makishima K. Relationship of nasal carriage of Staphylococcus aureus to pathogenesis of perennial allergic rhinitis. J. Allergy Clin. Immunol. 2000;105:449–454. doi: 10.1067/mai.2000.104256. [DOI] [PubMed] [Google Scholar]
- 384.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin. Exp. Allergy. 2008;38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 385.Hirsch AG, et al. Early-life antibiotic use and subsequent diagnosis of food allergy and allergic diseases. Clin. Exp. Allergy. 2017;47:236–244. doi: 10.1111/cea.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Sanchez-Valverde F, et al. The impact of caesarean delivery and type of feeding on cow’s milk allergy in infants and subsequent development of allergic march in childhood. Allergy. 2009;64:884–889. doi: 10.1111/j.1398-9995.2008.01931.x. [DOI] [PubMed] [Google Scholar]
- 387.Erridge C, Duncan SH, Bereswill S, Heimesaat MM. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLoS ONE. 2010;5:e9125. doi: 10.1371/journal.pone.0009125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Kant R, de Vos WM, Palva A, Satokari R. Immunostimulatory CpG motifs in the genomes of gut bacteria and their role in human health and disease. J. Med. Microbiol. 2014;63:293–308. doi: 10.1099/jmm.0.064220-0. [DOI] [PubMed] [Google Scholar]
- 390.Aagaard K, et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6:237ra265. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Chu DM, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016;8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N. Engl. J. Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 393.Koga K, Mor G. Toll-like receptors at the maternal-fetal interface in normal pregnancy and pregnancy disorders. Am. J. Reprod. Immunol. 2010;63:587–600. doi: 10.1111/j.1600-0897.2010.00848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Christmann BS, et al. Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol. 2015;136:1378–1386 e1371-1375. doi: 10.1016/j.jaci.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017;66:515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 396.West CE, et al. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin. Exp. Allergy. 2015;45:1419–1429. doi: 10.1111/cea.12566. [DOI] [PubMed] [Google Scholar]
- 397.Yiu JH, Dorweiler B, Woo CW. Interaction between gut microbiota and toll-like receptor: from immunity to metabolism. J. Mol. Med. 2017;95:13–20. doi: 10.1007/s00109-016-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Sun L, Liu W, Zhang LJ. The role of toll-like receptors in skin host defense, psoriasis, and atopic dermatitis. J. Immunol. Res. 2019;2019:1824624. doi: 10.1155/2019/1824624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat. Med. 2012;18:684–692. doi: 10.1038/nm.2737. [DOI] [PubMed] [Google Scholar]
- 400.Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18:2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Wypych TP, et al. Microbial metabolism of l-tyrosine protects against allergic airway inflammation. Nat. Immunol. 2021;22:279–286. doi: 10.1038/s41590-020-00856-3. [DOI] [PubMed] [Google Scholar]
- 402.Nastasi C, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci. Rep. 2015;5:16148. doi: 10.1038/srep16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Müller T, Hamm S, Bauer S. TLR9-mediated recognition of DNA. Handb. Exp. Pharmacol. 2008;183:51–70. doi: 10.1007/978-3-540-72167-3_3. [DOI] [PubMed] [Google Scholar]
- 404.Hartmann G, Weiner GJ, Krieg AM. CpG DNA: a potent signal for growth, activation, and maturation of human dendritic cells. Proc. Natl Acad. Sci. USA. 1999;96:9305–9310. doi: 10.1073/pnas.96.16.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Krug A, et al. Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur. J. Immunol. 2001;31:2154–2163. doi: 10.1002/1521-4141(200107)31:7<2154::AID-IMMU2154>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 406.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev. Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 407.Choudhury BK, et al. In vivo role of p38 mitogen-activated protein kinase in mediating the anti-inflammatory effects of CpG oligodeoxynucleotide in murine asthma. J. Immunol. 2002;169:5955–5961. doi: 10.4049/jimmunol.169.10.5955. [DOI] [PubMed] [Google Scholar]
- 408.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl Acad. Sci. USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 410.Bortolatto J, et al. Toll-like receptor 4 agonists adsorbed to aluminium hydroxide adjuvant attenuate ovalbumin-specific allergic airway disease: role of MyD88 adaptor molecule and interleukin-12/interferon-gamma axis. Clin. Exp. Allergy. 2008;38:1668–1679. doi: 10.1111/j.1365-2222.2008.03036.x. [DOI] [PubMed] [Google Scholar]
- 411.O’Hara AM, Moran AP, Würzner R, Orren A. Complement-mediated lipopolysaccharide release and outer membrane damage in Escherichia coli J5: requirement for C9. Immunology. 2001;102:365–372. doi: 10.1046/j.1365-2567.2001.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Dong L, Li H, Wang S, Li Y. Different doses of lipopolysaccharides regulate the lung inflammation of asthmatic mice via TLR4 pathway in alveolar macrophages. J. Asthma. 2009;46:229–233. doi: 10.1080/02770900802610050. [DOI] [PubMed] [Google Scholar]
- 413.Yoshida A, Kohchi C, Inagawa H, Nishizawa T, Soma G. Improvement of allergic dermatitis via regulation of the Th1/Th2 immune system balance by macrophages activated with lipopolysaccharide derived from Pantoea agglomerans (IP-PA1) Anticancer Res. 2009;29:4867–4870. [PubMed] [Google Scholar]
- 414.Siebenhaar F, Redegeld FA, Bischoff SC, Gibbs BF, Maurer M. Mast cells as drivers of disease and therapeutic targets. Trends Immunol. 2018;39:151–162. doi: 10.1016/j.it.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 415.Simon D, Braathen LR, Simon H-U. Eosinophils and atopic dermatitis. Allergy. 2004;59:561–570. doi: 10.1111/j.1398-9995.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- 416.Fitzhugh DJ, Lockey RF. Allergen immunotherapy: a history of the first 100 years. Curr. Opin. Allergy Clin. Immunol. 2011;11:554–559. doi: 10.1097/ACI.0b013e32834c3134. [DOI] [PubMed] [Google Scholar]
- 417.Price DB, et al. Blood eosinophil count and prospective annual asthma disease burden: a UK cohort study. Lancet Respir. Med. 2015;3:849–858. doi: 10.1016/S2213-2600(15)00367-7. [DOI] [PubMed] [Google Scholar]
- 418.Woodruff PG, et al. Relationship between airway inflammation, hyperresponsiveness, and obstruction in asthma. J. Allergy Clin. Immunol. 2001;108:753–758. doi: 10.1067/mai.2001.119411. [DOI] [PubMed] [Google Scholar]
- 419.Rosenberg HF, Masterson JC, Furuta GT. Eosinophils, probiotics, and the microbiome. J. Leukoc. Biol. 2016;100:881–888. doi: 10.1189/jlb.3RI0416-202R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Hosoki K, et al. Differential activation of Eosinophils by ‘Probiotic’ Bifidobacterium bifidum and ‘Pathogenic’ Clostridium difficile. Int. Arch. Allergy Immunol. 2010;152(Suppl. 1):83–89. doi: 10.1159/000312131. [DOI] [PubMed] [Google Scholar]
- 421.Choi CY, et al. Anti-inflammatory potential of a heat-killed Lactobacillus strain isolated from Kimchi on house dust mite-induced atopic dermatitis in NC/Nga mice. J. Appl. Microbiol. 2017;123:535–543. doi: 10.1111/jam.13515. [DOI] [PubMed] [Google Scholar]
- 422.Wang X, et al. Oral administration of Lactobacillus paracasei L9 attenuates PM2.5-induced enhancement of airway hyperresponsiveness and allergic airway response in murine model of asthma. PLoS ONE. 2017;12:e0171721. doi: 10.1371/journal.pone.0171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Arthur JC, et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J. Immunol. 2010;185:4515–4519. doi: 10.4049/jimmunol.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Michels KR, Lukacs NW, Fonseca W. TLR activation and allergic disease: early life microbiome and treatment. Curr. Allergy Asthma Rep. 2018;18:61. doi: 10.1007/s11882-018-0815-5. [DOI] [PubMed] [Google Scholar]
- 425.Hruz P, et al. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc. Natl Acad. Sci. USA. 2009;106:12873–12878. doi: 10.1073/pnas.0904958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Wong CK, Chu IM, Hon KL, Tsang MS, Lam CW. Aberrant expression of bacterial pattern recognition receptor NOD2 of basophils and microbicidal peptides in atopic dermatitis. Molecules. 2016;21:471. doi: 10.3390/molecules21040471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Fyhrquist N, et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J. Allergy Clin. Immunol. 2014;134:1301–1309 e1311. doi: 10.1016/j.jaci.2014.07.059. [DOI] [PubMed] [Google Scholar]
- 428.Montaldo E, et al. Human innate lymphoid cells. Immunol. Lett. 2016;179:2–8. doi: 10.1016/j.imlet.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 429.Bal SM, et al. IL-1beta, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 2016;17:636–645. doi: 10.1038/ni.3444. [DOI] [PubMed] [Google Scholar]
- 430.Stefka AT, et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl Acad. Sci. USA. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Konieczna P, et al. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS ONE. 2013;8:e62617. doi: 10.1371/journal.pone.0062617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Lyons A, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin. Exp. Allergy. 2010;40:811–819. doi: 10.1111/j.1365-2222.2009.03437.x. [DOI] [PubMed] [Google Scholar]
- 433.Konieczna P, et al. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: potential role for myeloid and plasmacytoid dendritic cells. Gut. 2012;61:354–366. doi: 10.1136/gutjnl-2011-300936. [DOI] [PubMed] [Google Scholar]
- 434.Fujimura KE, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Koizumi, S. I. & Ishikawa, H. Transcriptional regulation of differentiation and functions of effector T regulatory cells. Cells8 (2019). [DOI] [PMC free article] [PubMed]
- 436.Rachid R, Stephen-Victor E, Chatila TA. The microbial origins of food allergy. J. Allergy Clin. Immunol. 2021;147:808–813. doi: 10.1016/j.jaci.2020.12.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Eicher L, et al. A systematic review of factors influencing treatment adherence in chronic inflammatory skin disease - strategies for optimizing treatment outcome. J. Eur. Acad. Dermatol Venereol. 2019;33:2253–2263. doi: 10.1111/jdv.15913. [DOI] [PubMed] [Google Scholar]
- 438.Bousquet J, et al. Allergic rhinitis. Nat. Rev. Dis. Prim. 2020;6:95. doi: 10.1038/s41572-020-00227-0. [DOI] [PubMed] [Google Scholar]
- 439.Wollenberg A, et al. ETFAD/EADV Eczema task force 2020 position paper on diagnosis and treatment of atopic dermatitis in adults and children. J. Eur. Acad. Dermatol. Venereol. 2020;34:2717–2744. doi: 10.1111/jdv.16892. [DOI] [PubMed] [Google Scholar]
- 440.Bousquet J, et al. Treatment of allergic rhinitis using mobile technology with real-world data: The MASK observational pilot study. Allergy. 2018;73:1763–1774. doi: 10.1111/all.13406. [DOI] [PubMed] [Google Scholar]
- 441.Meng Y, Wang C, Zhang L. Recent developments and highlights in allergic rhinitis. Allergy. 2019;74:2320–2328. doi: 10.1111/all.14067. [DOI] [PubMed] [Google Scholar]
- 442.Tordesillas L, Berin MC, Sampson HA. Immunology of food allergy. Immunity. 2017;47:32–50. doi: 10.1016/j.immuni.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 443.Katoh N, et al. Japanese guidelines for atopic dermatitis 2020. Allergol. Int. 2020;69:356–369. doi: 10.1016/j.alit.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 444.Nakamura Y, et al. Japanese guidelines for adult asthma 2020. Allergol. Int. 2020;69:519–548. doi: 10.1016/j.alit.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 445.Kawauchi, H., Yanai, K., Wang, D. Y., Itahashi, K. & Okubo, K. Antihistamines for allergic rhinitis treatment from the viewpoint of nonsedative properties. Int. J. Mol. Sci.20 (2019). [DOI] [PMC free article] [PubMed]
- 446.Brozek JL, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines: 2010 revision. J. Allergy Clin. Immunol. 2010;126:466–476. doi: 10.1016/j.jaci.2010.06.047. [DOI] [PubMed] [Google Scholar]
- 447.Wang XY, Lim-Jurado M, Prepageran N, Tantilipikorn P, Wang de Y. Treatment of allergic rhinitis and urticaria: a review of the newest antihistamine drug bilastine. Ther. Clin. Risk Manag. 2016;12:585–597. doi: 10.2147/TCRM.S105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Estelle F, Simons R. M.D. Advances in H 1 -Antihistamines. N. Engl. J. Med. 2004;351:2203–2217. doi: 10.1056/NEJMra033121. [DOI] [PubMed] [Google Scholar]
- 449.Casale TB, Oppenheimer JJ. Next generation antihistamines: therapeutic rationale, accomplishments and advances. Expert Opin. Investig. Drugs. 2002;11:807–817. doi: 10.1517/13543784.11.6.807. [DOI] [PubMed] [Google Scholar]
- 450.Barnes, P. J. Therapeutic strategies for allergic diseases. Nature402 (1999). [DOI] [PubMed]
- 451.Wu EL, et al. Epistaxis risk associated with intranasal corticosteroid sprays: a systematic review and meta-analysis. Otolaryngol. Head. Neck Surg. 2019;161:18–27. doi: 10.1177/0194599819832277. [DOI] [PubMed] [Google Scholar]
- 452.Rodrigo GJ, Neffen H. Efficacy of fluticasone furoate nasal spray vs. placebo for the treatment of ocular and nasal symptoms of allergic rhinitis: a systematic review. Clin. Exp. Allergy. 2011;41:160–170. doi: 10.1111/j.1365-2222.2010.03654.x. [DOI] [PubMed] [Google Scholar]
- 453.Bieber T. Atopic dermatitis: an expanding therapeutic pipeline for a complex disease. Nat. Rev. Drug Disco. 2022;21:21–40. doi: 10.1038/s41573-021-00266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Blauvelt A, et al. Long-term management of moderate-to-severe atopic dermatitis with dupilumab and concomitant topical corticosteroids (LIBERTY AD CHRONOS): a 1-year, randomised, double-blinded, placebo-controlled, phase 3 trial. Lancet. 2017;389:2287–2303. doi: 10.1016/S0140-6736(17)31191-1. [DOI] [PubMed] [Google Scholar]
- 455.Kaliner MA. A novel and effective approach to treating rhinitis with nasal antihistamines. Ann. Allergy Asthma Immunol. 2007;99:383–391. doi: 10.1016/S1081-1206(10)60560-1. [DOI] [PubMed] [Google Scholar]
- 456.Hampel FC, et al. Double-blind, placebo-controlled study of azelastine and fluticasone in a single nasal spray delivery device. Ann. Allergy Asthma Immunol. 2010;105:168–173. doi: 10.1016/j.anai.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 457.Segall N, Prenner B, Lumry W, Caracta CF, Tantry SK. Long-term safety and efficacy of olopatadine-mometasone combination nasal spray in patients with perennial allergic rhinitis. Allergy Asthma Proc. 2019;40:301–310. doi: 10.2500/aap.2019.40.4233. [DOI] [PubMed] [Google Scholar]
- 458.Shim JS, Kim MH, Kim MH, Cho YJ, Chun EM. Risk of neuropsychiatric diseases according to the use of a leukotriene receptor antagonist in middle-aged and older adults with asthma: a nationwide population-based study using health claims data in Korea. J. Allergy Clin. Immunol. Pr. 2021;9:4290–4297. doi: 10.1016/j.jaip.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 459.Hamri, S. et al. Convenient approach for the synthesis of ONO-LB-457, a potent leukotriene B4 receptor antagonist. Tetrahedron77 (2021).
- 460.Kim JH, et al. Airway mechanics after withdrawal of a leukotriene receptor antagonist in children with mild persistent asthma: Double-blind, randomized, cross-over study. Pediatr. Pulmonol. 2020;55:3279–3286. doi: 10.1002/ppul.25085. [DOI] [PubMed] [Google Scholar]
- 461.Osada T, Okano M. Japanese cedar and cypress pollinosis updated: new allergens, cross-reactivity, and treatment. Allergol. Int. 2021;70:281–290. doi: 10.1016/j.alit.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 462.Pfaller B, Bendien S, Ditisheim A, Eiwegger T. Management of allergic diseases in pregnancy. Allergy. 2022;77:798–811. doi: 10.1111/all.15063. [DOI] [PubMed] [Google Scholar]
- 463.Lin P, et al. A phosphatase-mimetic nano-stabilizer of mast cells for long-term prevention of allergic disease. Adv. Sci. 2021;8:2004115. doi: 10.1002/advs.202004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Xu H, Bin NR, Sugita S. Diverse exocytic pathways for mast cell mediators. Biochem Soc. Trans. 2018;46:235–247. doi: 10.1042/BST20170450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Jackson, C. W., Pratt, C. M., Rupprecht, C. P., Pattanaik, D. & Krishnaswamy, G. Mastocytosis and mast cell activation disorders: clearing the air. Int. J. Mol. Sci.22 (2021). [DOI] [PMC free article] [PubMed]
- 466.Sze E, Bhalla A, Nair P. Mechanisms and therapeutic strategies for non-T2 asthma. Allergy. 2020;75:311–325. doi: 10.1111/all.13985. [DOI] [PubMed] [Google Scholar]
- 467.Peng T, Qi B, He J, Ke H, Shi J. Advances in the development of phosphodiesterase-4 inhibitors. J. Med. Chem. 2020;63:10594–10617. doi: 10.1021/acs.jmedchem.9b02170. [DOI] [PubMed] [Google Scholar]
- 468.Janosova V, et al. Phosphodiesterase 4 inhibitors in allergic rhinitis/rhinosinusitis. Front. Pharm. 2020;11:1135. doi: 10.3389/fphar.2020.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Vestergaard C, et al. European task force on atopic dermatitis position paper: treatment of parental atopic dermatitis during preconception, pregnancy and lactation period. J. Eur. Acad. Dermatol. Venereol. 2019;33:1644–1659. doi: 10.1111/jdv.15709. [DOI] [PubMed] [Google Scholar]
- 470.Drucker AM, et al. Systemic immunomodulatory treatments for patients with atopic dermatitis: a systematic review and network meta-analysis. JAMA Dermatol. 2020;156:659–667. doi: 10.1001/jamadermatol.2020.0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Arakawa H, et al. Japanese guidelines for childhood asthma 2020. Allergol. Int. 2020;69:314–330. doi: 10.1016/j.alit.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 472.Barnes PJ. New therapies for asthma: is there any progress? Trends Pharm. Sci. 2010;31:335–343. doi: 10.1016/j.tips.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 473.Li L, Liu R, Peng C, Chen X, Li J. Pharmacogenomics for the efficacy and side effects of antihistamines. Exp. Dermatol. 2022;31:993–1004. doi: 10.1111/exd.14602. [DOI] [PubMed] [Google Scholar]
- 474.Rollema C, et al. Pharmacology, particle deposition and drug administration techniques of intranasal corticosteroids for treating allergic rhinitis. Clin. Exp. Allergy. 2022;52:1247–1263. doi: 10.1111/cea.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Hatakeyama S, et al. The safety of pranlukast and montelukast during the first trimester of pregnancy: a prospective, two-centered cohort study in Japan. Congenit. Anom. 2022;62:161–168. doi: 10.1111/cga.12471. [DOI] [PubMed] [Google Scholar]
- 476.Pfaar O, Lou H, Zhang Y, Klimek L, Zhang L. Recent developments and highlights in allergen immunotherapy. Allergy. 2018;73:2274–2289. doi: 10.1111/all.13652. [DOI] [PubMed] [Google Scholar]
- 477.Hoffmann HJ, et al. Novel approaches and perspectives in allergen immunotherapy. Allergy. 2017;72:1022–1034. doi: 10.1111/all.13135. [DOI] [PubMed] [Google Scholar]
- 478.van Zelm MC, McKenzie CI, Varese N, Rolland JM, O’Hehir RE. Recent developments and highlights in immune monitoring of allergen immunotherapy. Allergy. 2019;74:2342–2354. doi: 10.1111/all.14078. [DOI] [PubMed] [Google Scholar]
- 479.Dorofeeva Y, et al. Past, present, and future of allergen immunotherapy vaccines. Allergy. 2021;76:131–149. doi: 10.1111/all.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Yao Y, et al. CD23 expression on switched memory B cells bridges T-B cell interaction in allergic rhinitis. Allergy. 2020;75:2599–2612. doi: 10.1111/all.14288. [DOI] [PubMed] [Google Scholar]
- 481.Bousquet J, et al. 2019 ARIA Care pathways for allergen immunotherapy. Allergy. 2019;74:2087–2102. doi: 10.1111/all.13805. [DOI] [PubMed] [Google Scholar]
- 482.Cevhertas L, et al. Advances and recent developments in asthma in 2020. Allergy. 2020;75:3124–3146. doi: 10.1111/all.14607. [DOI] [PubMed] [Google Scholar]
- 483.Jutel M, Bruggenjurgen B, Richter H, Vogelberg C. Real-world evidence of subcutaneous allergoid immunotherapy in house dust mite-induced allergic rhinitis and asthma. Allergy. 2020;75:2050–2058. doi: 10.1111/all.14240. [DOI] [PubMed] [Google Scholar]
- 484.Pfaar O, et al. Perspectives in allergen immunotherapy: 2019 and beyond. Allergy. 2019;74:3–25. doi: 10.1111/all.14077. [DOI] [PubMed] [Google Scholar]
- 485.Yao Y, et al. Allergen immunotherapy improves defective follicular regulatory T cells in patients with allergic rhinitis. J. Allergy Clin. Immunol. 2019;144:118–128. doi: 10.1016/j.jaci.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 486.Muraro A, et al. EAACI guidelines on allergen immunotherapy: executive statement. Allergy. 2018;73:739–743. doi: 10.1111/all.13420. [DOI] [PubMed] [Google Scholar]
- 487.Malling H-J. Minimising the risks of allergen-specific injection immunotherapy. Drug Saf. 2000;23:323–332. doi: 10.2165/00002018-200023040-00005. [DOI] [PubMed] [Google Scholar]
- 488.Cox, L., Larenas-Linnemann, D., Lockey, R. F. & Passalacqua, G. Speaking the same language: the World Allergy Organization subcutaneous immunotherapy systemic reaction grading system. J. Allergy Clin. Immunol. 125, 569–574, 574 e561–574 e567 (2010). [DOI] [PubMed]
- 489.Bousquet J, et al. Highlights and recent developments in airway diseases in EAACI journals (2018) Allergy. 2019;74:2329–2341. doi: 10.1111/all.14068. [DOI] [PubMed] [Google Scholar]
- 490.Pfaar O, et al. Perspectives in allergen immunotherapy: 2017 and beyond. Allergy. 2018;73:5–23. doi: 10.1111/all.13355. [DOI] [PubMed] [Google Scholar]
- 491.Senti G, et al. Intralymphatic allergen administration renders specific immunotherapy faster and safer: a randomized controlled trial. Proc. Natl Acad. Sci. USA. 2008;105:17908–17912. doi: 10.1073/pnas.0803725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Komlosi ZI, et al. Highlights of novel vaccination strategies in allergen immunotherapy. Immunol. Allergy Clin. North Am. 2020;40:15–24. doi: 10.1016/j.iac.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 493.Fokkens WJ, et al. EUFOREA consensus on biologics for CRSwNP with or without asthma. Allergy. 2019;74:2312–2319. doi: 10.1111/all.13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Kraft S, Novak N. Fc receptors as determinants of allergic reactions. Trends Immunol. 2006;27:88–95. doi: 10.1016/j.it.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 495.Tsabouri S, Tseretopoulou X, Priftis K, Ntzani EE. Omalizumab for the treatment of inadequately controlled allergic rhinitis: a systematic review and meta-analysis of randomized clinical trials. J. Allergy Clin. Immunol. Pr. 2014;2:332–340.e331. doi: 10.1016/j.jaip.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 496.Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. 2021;184:1469–1485. doi: 10.1016/j.cell.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 497.Breiteneder H, et al. Future research trends in understanding the mechanisms underlying allergic diseases for improved patient care. Allergy. 2019;74:2293–2311. doi: 10.1111/all.13851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Eschenbacher W, Straesser M, Knoeddler A, Li RC, Borish L. Biologics for the treatment of allergic rhinitis, chronic rhinosinusitis, and nasal polyposis. Immunol. Allergy Clin. North Am. 2020;40:539–547. doi: 10.1016/j.iac.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Weinstein SF, et al. Efficacy and safety of dupilumab in perennial allergic rhinitis and comorbid asthma. J. Allergy Clin. Immunol. 2018;142:171–177.e171. doi: 10.1016/j.jaci.2017.11.051. [DOI] [PubMed] [Google Scholar]
- 500.Newsom M, Bashyam AM, Balogh EA, Feldman SR, Strowd LC. New and emerging systemic treatments for atopic dermatitis. Drugs. 2020;80:1041–1052. doi: 10.1007/s40265-020-01335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Misery L, Huet F, Gouin O, Stander S, Deleuran M. Current pharmaceutical developments in atopic dermatitis. Curr. Opin. Pharm. 2019;46:7–13. doi: 10.1016/j.coph.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 502.Meurs H, Zaagsma J, Maarsingh H, van Duin M. Recent Patents in Allergy/Immunology: use of arginase inhibitors in the treatment of asthma and allergic rhinitis. Allergy. 2019;74:1206–1208. doi: 10.1111/all.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Breiteneder H, et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy. 2020;75:3039–3068. doi: 10.1111/all.14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Oyama S, et al. Cynomolgus monkey model of interleukin-31-induced scratching depicts blockade of human interleukin-31 receptor A by a humanized monoclonal antibody. Exp. Dermatol. 2018;27:14–21. doi: 10.1111/exd.13236. [DOI] [PubMed] [Google Scholar]
- 505.Kruse RL, Vanijcharoenkarn K. Drug repurposing to treat asthma and allergic disorders: progress and prospects. Allergy. 2018;73:313–322. doi: 10.1111/all.13305. [DOI] [PubMed] [Google Scholar]
- 506.Schuliga M. NF-kappaB signaling in chronic inflammatory airway disease. Biomolecules. 2015;5:1266–1283. doi: 10.3390/biom5031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Sherman, D. J. & Li, J. Proteasome inhibitors: harnessing proteostasis to combat disease. Molecules25 (2020). [DOI] [PMC free article] [PubMed]
- 508.Ramadass, V., Vaiyapuri, T. & Tergaonkar, V. Small molecule NF-kappaB pathway inhibitors in clinic. Int. J. Mol. Sci.21 (2020). [DOI] [PMC free article] [PubMed]
- 509.Hua W, et al. Rapamycin inhibition of eosinophil differentiation attenuates allergic airway inflammation in mice. Respirology. 2015;20:1055–1065. doi: 10.1111/resp.12554. [DOI] [PubMed] [Google Scholar]
- 510.KleinJan A, et al. The Notch pathway inhibitor stapled alpha-helical peptide derived from mastermind-like 1 (SAHM1) abrogates the hallmarks of allergic asthma. J. Allergy Clin. Immunol. 2018;142:76–85.e78. doi: 10.1016/j.jaci.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Zhou Z, et al. Avasimibe alleviates disruption of the airway epithelial barrier by suppressing the Wnt/beta-Catenin signaling pathway. Front Pharm. 2022;13:795934. doi: 10.3389/fphar.2022.795934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Jha A, et al. Increased nasal mucosal interferon and CCL13 response to a TLR7/8 agonist in asthma and allergic rhinitis. J. Allergy Clin. Immunol. 2021;147:694–703.e612. doi: 10.1016/j.jaci.2020.07.012. [DOI] [PubMed] [Google Scholar]
- 513.Siddall H, et al. Intranasal GSK2245035, a Toll-like receptor 7 agonist, does not attenuate the allergen-induced asthmatic response in a randomized, double-blind, placebo-controlled experimental medicine study. PLoS ONE. 2020;15:e0240964. doi: 10.1371/journal.pone.0240964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Menzies-Gow A, et al. Tezepelumab in adults and adolescents with severe, uncontrolled asthma. N. Engl. J. Med. 2021;384:1800–1809. doi: 10.1056/NEJMoa2034975. [DOI] [PubMed] [Google Scholar]
- 515.Chinthrajah, S. et al. Phase 2a randomized, placebo-controlled study of anti-IL-33 in peanut allergy. JCI Insight4 (2019). [DOI] [PMC free article] [PubMed]
- 516.Angkasekwinai P, Dong C. IL-9-producing T cells: potential players in allergy and cancer. Nat. Rev. Immunol. 2021;21:37–48. doi: 10.1038/s41577-020-0396-0. [DOI] [PubMed] [Google Scholar]
- 517.Kupczyk M, Kuna P. Targeting the PGD2/CRTH2/DP1 signaling pathway in asthma and allergic disease: current status and future perspectives. Drugs. 2017;77:1281–1294. doi: 10.1007/s40265-017-0777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Corren J. New targeted therapies for uncontrolled asthma. J. Allergy Clin. Immunol. Pr. 2019;7:1394–1403. doi: 10.1016/j.jaip.2019.03.022. [DOI] [PubMed] [Google Scholar]
- 519.Colagiovanni DB, et al. A nonclinical safety and pharmacokinetic evaluation of N6022: a first-in-class S-nitrosoglutathione reductase inhibitor for the treatment of asthma. Regul. Toxicol. Pharm. 2012;62:115–124. doi: 10.1016/j.yrtph.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 520.Sun X, et al. Discovery of s-nitrosoglutathione reductase inhibitors: potential agents for the treatment of asthma and other inflammatory diseases. ACS Med. Chem. Lett. 2011;2:402–406. doi: 10.1021/ml200045s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Que LG, et al. Effect of the S-nitrosoglutathione reductase inhibitor N6022 on bronchial hyperreactivity in asthma. Immun. Inflamm. Dis. 2018;6:322–331. doi: 10.1002/iid3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Zhang Y, Lan F, Zhang L. Update on pathomechanisms and treatments in allergic rhinitis. Allergy. 2022;77:3309–3319. doi: 10.1111/all.15454. [DOI] [PubMed] [Google Scholar]
- 523.Licari A, et al. Biologics in children with allergic diseases. Curr. Pediatr. Rev. 2020;16:140–147. doi: 10.2174/1573396315666191029123822. [DOI] [PubMed] [Google Scholar]
- 524.Cox L. Biologics and allergy immunotherapy in the treatment of allergic diseases. Immunol. Allergy Clin. North Am. 2020;40:687–700. doi: 10.1016/j.iac.2020.06.008. [DOI] [PubMed] [Google Scholar]
- 525.Jung, H. J., Chung, Y. J., Choi, Y .S., Chung, P. S. & Mo, J. H. Clinical efficacy and safety of low-level laser therapy in patients with perennial allergic rhinitis: a randomized, double-blind, Placebo-controlled trial. J. Clin. Med. 10 (2021). [DOI] [PMC free article] [PubMed]
- 526.Costa TMR, et al. Rhinophototherapy, an alternative treatment of allergic rhinitis: Systematic review and meta-analysis. Braz. J. Otorhinolaryngol. 2021;87:742–752. doi: 10.1016/j.bjorl.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Natsume O, et al. Two-step egg introduction for prevention of egg allergy in high-risk infants with eczema (PETIT): a randomised, double-blind, placebo-controlled trial. Lancet. 2017;389:276–286. doi: 10.1016/S0140-6736(16)31418-0. [DOI] [PubMed] [Google Scholar]
- 528.Nakao A. Circadian regulation of the biology of allergic disease: clock disruption can promote allergy. Front. Immunol. 2020;11:1237. doi: 10.3389/fimmu.2020.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Hong X, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat. Commun. 2015;6:6304. doi: 10.1038/ncomms7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Andiappan AK, et al. Genome-wide association study for atopy and allergic rhinitis in a Singapore Chinese population. PLoS ONE. 2011;6:e19719. doi: 10.1371/journal.pone.0019719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Ramasamy A, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J. Allergy Clin. Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 532.Shirkani A, et al. The Role of interleukin-4 and 13 gene polymorphisms in allergic rhinitis: a case control study. Rep. Biochem. Mol. Biol. 2019;8:111–118. [PMC free article] [PubMed] [Google Scholar]
- 533.Amo G, et al. A Nonsynonymous FCER1B SNP is associated with risk of developing allergic rhinitis and with IgE levels. Sci. Rep. 2016;6:19724. doi: 10.1038/srep19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Zhao CN, et al. The association of GSDMB and ORMDL3 gene polymorphisms with asthma: a meta-analysis. Allergy Asthma Immunol. Res. 2015;7:175–185. doi: 10.4168/aair.2015.7.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Nieto-Fontarigo JJ, et al. The CD14 (-159 C/T) SNP is associated with sCD14 levels and allergic asthma, but not with CD14 expression on monocytes. Sci. Rep. 2018;8:4147. doi: 10.1038/s41598-018-20483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Nowakowska J, et al. Nitric oxide synthase 2 promoter polymorphism is a risk factor for allergic asthma in children. Med. (Kaunas.) 2021;57:1341. doi: 10.3390/medicina57121341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Beigh AH, et al. Influence of single gene variants of FOXP3 on allergic asthma predisposition. Gene. 2020;763:145073. doi: 10.1016/j.gene.2020.145073. [DOI] [PubMed] [Google Scholar]
- 538.Stevens ML, et al. Disease-associated KIF3A variants alter gene methylation and expression impacting skin barrier and atopic dermatitis risk. Nat. Commun. 2020;11:4092. doi: 10.1038/s41467-020-17895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Li Y, et al. Association of UBASH3A gene polymorphism and atopic dermatitis in the Chinese Han population. Genes Immun. 2017;18:158–162. doi: 10.1038/gene.2017.15. [DOI] [PubMed] [Google Scholar]
- 540.Dytiatkovskyi V, Drevytska T, Lapikova-Bryhinska T, Dosenko V, Abaturov O. Genotype associations with the different phenotypes of atopic dermatitis in children. Acta Med. 2021;64:96–100. doi: 10.14712/18059694.2021.17. [DOI] [PubMed] [Google Scholar]
- 541.Hirota T, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat. Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 542.Ramadass, V., Vaiyapuri, T. & Tergaonkar, V. Small molecule NF-κB pathway inhibitors in clinic. Int. J. Mol. Sci.21 (2020). [DOI] [PMC free article] [PubMed]