Abstract
A U-shaped contraction pattern was shown to be associated with a better Cardiac resynchronization therapy (CRT) response. The main goal of this study is to automatically recognize left ventricular contractile patterns using machine learning algorithms trained on conventional quantitative features (ConQuaFea) and radiomic features extracted from Gated single-photon emission computed tomography myocardial perfusion imaging (GSPECT MPI). Among 98 patients with standard resting GSPECT MPI included in this study, 29 received CRT therapy and 69 did not (also had CRT inclusion criteria but did not receive treatment yet at the time of data collection, or refused treatment). A total of 69 non-CRT patients were employed for training, and the 29 were employed for testing. The models were built utilizing features from three distinct feature sets (ConQuaFea, radiomics, and ConQuaFea + radiomics (combined)), which were chosen using Recursive feature elimination (RFE) feature selection (FS), and then trained using seven different machine learning (ML) classifiers. In addition, CRT outcome prediction was assessed by different treatment inclusion criteria as the study’s final phase. The MLP classifier had the highest performance among ConQuaFea models (AUC, SEN, SPE = 0.80, 0.85, 0.76). RF achieved the best performance in terms of AUC, SEN, and SPE with values of 0.65, 0.62, and 0.68, respectively, among radiomic models. GB and RF approaches achieved the best AUC, SEN, and SPE values of 0.78, 0.92, and 0.63 and 0.74, 0.93, and 0.56, respectively, among the combined models. A promising outcome was obtained when using radiomic and ConQuaFea from GSPECT MPI to detect left ventricular contractile patterns by machine learning.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-022-00705-9.
Keywords: Machine learning, CRT, GSPECT MPI, Radiomics, Quantitative features
Introduction
Heart failure (HF) is a relatively common cardiovascular disorder with prominent morbidity and mortality [1]. HF is closely related to left ventricular (LV) cardiac mechanical dyssynchrony, which reflects timing differences across various left ventricle regions [2]. Nonhomogeneous contraction patterns can be caused by the uncoordinated distribution of electrical activation in the heart pathways, known as cardiac dyssynchrony [3–5]. Thus, therapeutic measures are developed to resynchronize the left ventricular contraction in HF patients.
Cardiac resynchronization therapy (CRT) demonstrated significant success in treating patients with fatal HF [6, 7]. Patients undergoing CRT have substantiated the claims of most previous studies regarding the enhancements in several parameters, such as 6-min walking distance, New York Heart Association (NYHA) functional class, quality of life score, and peak O2 [6]. Nonetheless, estimations demonstrated that approximately one-third of chosen cases do not respond to this costly and invasive therapy [8–12] in spite of meeting current inclusion criteria for CRT therapy by the guidelines, which includes NYHA III or IV, left ventricular ejection fraction (LVEF) < 35%, and QRS duration ≥ 130 ms [13]. Consequently, the search for more specific criteria is still under investigation.
In line with finding new and more appropriate indicators for CRT patient selection, several studies have been conducted. In a study by Bax et al. [14], LV dyssynchrony was introduced as a desirable indicator. Furthermore, in another study conducted by Chen et al. [15], histogram bandwidth and phase standard deviation parameters extracted from phase analysis were suggested as important indicators. In addition, in examining the locations of CRT leads by Adelstein et al. [9], it was found that the best locations are far away from the scar tissue [9]. New LV mechanical dyssynchrony parameters were extracted from GSPECT MPI phase analysis with deep learning to aid CRT patient selection by He et al. [16].
Two types of left ventricular contraction patterns including U-shaped and non-U-shaped patterns have been recently proposed by different imaging modalities [17]. A U-shaped pattern is formed by a left ventricle–directed linear blockage which impedes contraction propagation [18, 19]. Conversely, a non-U-shaped pattern consists of two types, namely, “homogenous contraction” with apparent delay on all walls and “heterogeneous contraction” with multiple contraction delays in different sites [19]. An improved CRT response is discerned to be in association with a U-shaped contraction pattern [17–22].
Myocardial perfusion imaging (MPI) with gated single-photon emission computed tomography (GSPECT) is a practical technique to assess perfusion and function of left ventricle (LV). Evaluation of LV dyssynchrony and LV contraction patterns can also be ascertained by applying phase analysis to GSPECT MPI. The advantage of simultaneous assessment of perfusion abnormalities, such as the extent and severity of ischemia and scar, and functional parameters including LV dyssynchrony, LVEF, and LV volumes make this modality an eligible method for evaluation and diagnosis of patients with HF and LV dysfunction. Besides, the automated nature of phase analysis results in an acceptable repeatability and reproducibility of this technique. As a result, MPI GSPECT is a relevant modality for examining the left ventricular contractile pattern [19, 23].
Recent studies concerning quantitative radiomics analysis, through acting as biomarkers, have provided new insights into better handling of diseases, such as cancer [24] and coronary artery disease (CAD) to predict survival [25, 26], prognosis [27, 28], and therapeutic response [29, 30], different pathology classification [28, 31–33], and accumulating data for personalized medicine [34]. In fact, radiomics is an almost new science that has attracted many researchers’ attention and is therefore growing rapidly. However, it is not yet fully prepared to enter the clinical phase. In fact, radiomics analysis has been performed on magnetic resonance imaging (MRI), computed tomography (CT), and positron emission tomography (PET) images, but less on SPECT owing to its low spatial resolution and low sensitivity. It should be noted that in recent studies, radiomics has performed well in brain SPECT images as well as heart SPECT images [35–37]. Radiomic features are used to feed machine learning algorithms. Machine learning techniques, through the usage of computer algorithms and advanced statistical techniques, facilitate automatic extraction of prognostic knowledge or discriminatory patterns from data, often with the aim of making prediction on new data [38–40].
In this work, the purpose of automatic detection of left ventricular contractile patterns is fulfilled by radiomic and conventional quantitative features (ConQuaFea) using machine learning algorithms and MPI GSPECT images. This study aims to help clinicians to more confidently select patients who are eligible for CRT treatment and will also save them time and energy. In the last step, a comparison was made with the evaluation of response to CRT treatment using the left ventricular contraction pattern diagnosed by two experienced nuclear medicine physicians and the models presented in this study and standard criteria for prescribing CRT.
Materials and Methods
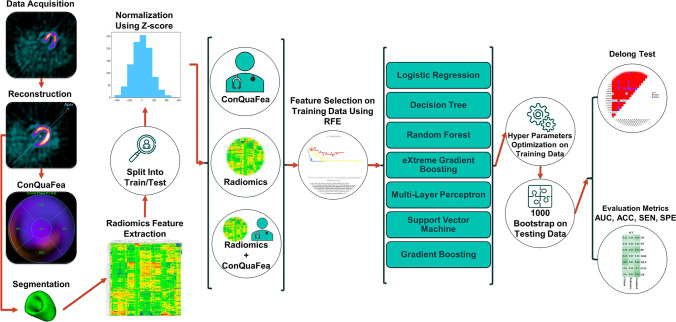
Figure 1 represents the graphical pipeline of the study. The whole workflow of the study, from data acquisition to evaluation of the proposed models, is elaborated in the following sub-sections as shown in Fig. 1.
Fig. 1.
Flowchart describing the main steps involved in the presented study
Dataset and Image Acquisition
This is a retrospective study including 98 patients encompassing 29 patients who underwent CRT treatment and 69 patients who did not (not being treated yet at the time of data collection or refused treatment) but had the same inclusion criteria: (a) EF ≤ 35% based on echocardiography and (b) QRS duration ≥ 130 ms [13]. Eighty-three men and 15 women (mean age = 59.37) were selected as participants. Out of 98 patients, 48 had U-shaped and 50 had non-U-shaped contractile patterns (visually assessed by two experienced nuclear medicine physicians from the polar maps as described in detail in the following section). Overall, among the patients who underwent CRT, 13 had U-shaped and 16 had non-U-shaped patterns, and among those patients who did not undergo CRT, 35 had U-shaped and 34 had non-U-shaped patterns. All patients underwent MPI GSPECT with the same vendor and same acquisition parameters. Each patient registered, underwent conventional resting MPI GSPECT. At rest condition, 555–740 MBq of Tc-99 m sestamibi was intravenously administered, and the GSPECT scan started 45–90 min post-injection. The images were acquired on a dual-headed gamma camera (Symbia™ T2, Siemens Healthcare) with body auto-contour form 135° (RAO) to − 45° (LAO) in 180° orbit, 32 thirty-second steps and 16-bin gating, using a matrix size of 64 × 64 with an isotropic voxel size (6.591 × 6.591 × 6.591 mm3) in the reconstructed images. The photopeak was adjusted to 140 keV with 20% energy window. Image reconstruction was accomplished using filtered backprojection technique with a post-reconstruction Butterworth filter (order = 5, cut-off frequency of 0.45 cycles/mm).
Conventional Quantitative Features (ConQuaFea)
Phase analysis was also performed using quantitative gated SPECT (QGS) software (Table 1) to extract several quantitative image-based features. These are summarized in Table 1, “Features extracted from Quantitative Gated SPECT (QGS)” section. Furthermore, other categorical image-based features, such as perfusion, phase, and wall motion, were also collected from polar maps (Table 2). In addition, explanations related to the phase analysis and QGS features are provided in Supplementary Table A.1. The results report the mean and interquartile range (IQR). The p values were calculated for continuous features using t-test and for categorical features using chi-square. Moreover, the way of scoring different regions of the heart is available in Table 3 according to [41]. Quantitative image-based and phase analysis data were combined and are reported as conventional quantitative features (ConQuaFea) set in the rest of the manuscript.
Table 1.
First part of ConQuaFea including phase analysis and QGS features, separately for cohorts of patients with U-shaped and non-U-shaped contractile patterns. IQR interquartile range
| ConQuaFea first part |
Non-u-shaped (n = 50) Mean and IQR |
U-shaped (n = 48) Mean and IQR |
P value | ||
|---|---|---|---|---|---|
| Age (years) | 59.52 and 13.00 | 59.23 and 13.25 | 0.070 | ||
| Ejection fraction (Echo) (%) | 29.16 and 10.00 | 30.10 and 10.25 | 0.269 | ||
| Phase analysis indices | Apex | Bandwidth (ms) | 125.20 and 114.00 | 128.63 and 114.00 | 0.754 |
| Mean | 356.32 and 88.00 | 340.50 and 86.25 | 0.478 | ||
| Standard deviation | 35.70 and 35.00 | 37.96 and 35.00 | 0.947 | ||
| Entropy (%) | 39.11 and 33.90 | 39.51 and 34.03 | 0.535 | ||
| Lateral | Bandwidth (ms) | 123.70 and 104.00 | 164.73 and 112.50 | 0.018 | |
| Mean | 340.46 and 78.00 | 368.58 and 78.50 | 0.755 | ||
| Standard deviation | 35.30 and 27.00 | 46.96 and 30.25 | 0.032 | ||
| Entropy (%) | 45.87 and 18.80 | 48.06 and 19.43 | 0.134 | ||
| Inferior | Bandwidth (ms) | 121.88 and 93.00 | 143.92 and 92.25 | 0.523 | |
| Mean | 341.10 and 59.00 | 363.38 and 56.75 | 0.047 | ||
| Standard deviation | 34.24 and 27.00 | 41.46 and 27.50 | 0.395 | ||
| Entropy (%) | 46.97 and 17.40 | 49.02 and 17.15 | 0.723 | ||
| Septal | Bandwidth (ms) | 134.96 and 118.00 | 147.71 and 116.00 | 0.476 | |
| Mean | 344.86 and 78.00 | 350.96 and 78.75 | 0.138 | ||
| Standard deviation | 38.84 and 35.00 | 40.33 and 34.25 | 0.981 | ||
| Entropy (%) | 47.40 and 19.60 | 45.60 and 19.70 | 0.941 | ||
| Anterior | Bandwidth (ms) | 131.54 and 116.00 | 159.21 and 114.75 | 0.301 | |
| Mean | 333.36 and 57.00 | 350.90 and 57.00 | 0.288 | ||
| Standard deviation | 36.40 and 35.00 | 43.60 and 34.50 | 0.312 | ||
| Entropy (%) | 46.59 and 20.80 | 50.07 and 20.35 | 0.864 | ||
| Features extracted from quantitative gated SPECT (QGS) | Lung heart ratio | 0.36 and 0.11 | 0.40 and 0.11 | 0.155 | |
| Summed motion score | 27.00 and 20.00 | 25.90 and 19.25 | 0.000 | ||
| Summed thickening score | 18.26 and 16.00 | 18.10 and 14.50 | 0.004 | ||
| Summed motion (%) | 31.74 and 23.00 | 30.46 and 22.25 | 0.000 | ||
| Summed thickening (%) | 35.72 and 31.00 | 35.46 and 29.50 | 0.004 | ||
| End-diastolic volume (QGS) (ml) | 180.20 and 80.00 | 153.27 and 80.75 | 0.076 | ||
| End systolic volume (QGS) (ml) | 123.68 and 68.00 | 98.94 and 67.50 | 0.032 | ||
| Systolic volume (ml) | 56.80 and 26.00 | 54.29 and 26.00 | 0.732 | ||
| Ejection fraction (%) | 37.02 and 17.00 | 37.81 and 17.00 | 0.030 | ||
| Peak emptying rate (EDV/s) | − 2.07 and 1.04 | − 1.99 and 0.99 | 0.181 | ||
| Peak filling rate (EDV/s) | 1.44 and 0.85 | 1.72 and 0.85 | 0.594 | ||
| Peak filling rate2 (EDV/s) | 1.39 and 0.97 | 1.00 and 0.98 | 0.243 | ||
| Mean filling rate/3 (EDV/s) | 0.74 and 0.60 | 0.84 and 0.55 | 0.866 | ||
| Time to peak filling from ES (ms) | 138.25 and 80.00 | 159.56 and 79.50 | 0.914 | ||
| Beats per minute (beats/minute) | 773.20 and 224.00 | 856.54 and 235.50 | 0.788 | ||
Table 2.
Second part of ConQuaFea including wall motion, phase, and perfusion features, separately for cohorts of patients with U-shaped and non-U-shaped contractile patterns
| ConQuaFea second part | Non-u-shaped | U-shaped | P value | ||
|---|---|---|---|---|---|
| Wall motion | Septal | # in classes (0, 1, 2, 3, 4, 5) | 2, 3, 8, 7, 10, 20 | 7, 4, 9, 5, 14, 9 | 0.150 |
| Anterior | 16, 7, 11, 5, 10, 1 | 17, 6, 11, 10, 3, 1 | 0.357 | ||
| Lateral | 19, 8, 9, 10, 3, 1 | 22, 7, 10, 6, 3, 0 | 0.806 | ||
| Inferior | 10, 8, 8, 9, 13, 2 | 14, 4, 9, 7, 13, 1 | 0.761 | ||
| Apex | 11, 6, 6, 7, 11, 9 | 10, 3, 7, 2, 18, 8 | 0.346 | ||
| Phase | Septal | # in classes (1, 2, 3, 4, 5) | 9, 11, 8, 9, 13 | 10, 7, 11, 8, 12 | 0.831 |
| Anterior | 12, 13, 10, 12, 3 | 11, 14, 11, 9, 3 | 0.972 | ||
| Lateral | 6, 12, 12, 8, 12 | 11, 10, 5, 9, 13 | 0.331 | ||
| Inferior | 9, 4, 14, 16, 7 | 5, 9, 8, 16, 10 | 0.268 | ||
| Apex | 17, 9, 6, 4, 14 | 10, 11, 14, 4, 9 | 0.180 | ||
| Perfusion | Septal | # in classes (0, 1, 2, 3, 4) | 28, 10, 4, 6, 2 | 31, 6, 4, 4, 3 | 0.788 |
| Anterior | 33, 3, 9, 3, 2 | 33, 6, 3, 3, 3 | 0.385 | ||
| Lateral | 38, 6, 2, 3, 1 | 38, 5, 4, 1, 0 | 0.606 | ||
| Inferior | 26, 8, 8, 6, 2 | 29, 11, 5, 3, 0 | 0.368 | ||
| Apex | 20, 7, 5, 8, 10 | 26, 4, 3, 5, 10 | 0.600 | ||
Table 3.
Scoring heart segments in terms of wall motion, perfusion, and phase
| Categorical features | Scores |
|---|---|
| Wall motion |
0 = normal 1 = mild hypokinesia 2 = moderate hypokinesia 3 = severe hypokinesia 4 = akinesia 5 = dyskinesia |
| Perfusion |
0 = normal 1 = mild hypoperfusion 2 = moderate hypoperfusion 3 = severe hypoperfusion 4 = absent perfusion |
| Phase |
1 = fastest contraction 5 = slowest contraction |
Investigation of Myocardial Contractile Patterns from Polar Maps
The myocardial uptake is divided into a limited number of raw perfusion samples after the left ventricle has been segmented, each represented by the mean or maximal wall photon counts at a specific place on the myocardial surface [42]. The objective is to parametrically represent myocardial perfusion to enable standard inter-subject comparability. A visual representation known as a “polar map” or “bull’s eye map” was created to make it easier to analyze the data obtained by polar sampling [43]. In clinical practice, polar maps are often employed because they provide a fast visual overview of myocardial perfusion data for the whole LV. Additionally, polar maps enable the myocardium to be divided into sections defined by vascular regions, LV wall, normalized 21-segment, 17-segment, or 5-segment status, which are useful for localizing the abnormalities [44, 45].
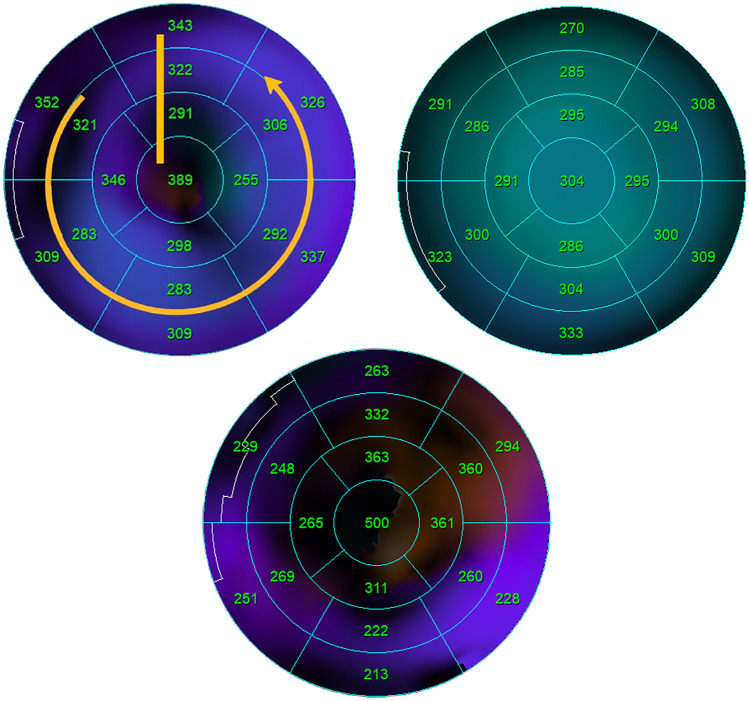
Mechanical left ventricular dyssynchrony can produce different patterns in polar map images as a result of two types of electrical activity propagation in the left ventricular myocardium, which include a U-shaped pattern and a non-U-shaped pattern. U-shaped pattern means that electrical activity propagation is blocked in a line while the other walls are almost contracted. For example, a contraction of an area starts from the anteroseptal wall and then goes to the apex, the lateral wall, and finally to the anterior wall. However, there are two types of non-U-shaped pattern. The first type is a homogeneous pattern; for example, contraction goes from the septal wall to the lateral wall. The second type is the heterogeneous pattern, in which there are multiple areas for myocardial contraction [19]. Different contraction patterns of the left ventricle can be seen in Fig. 2. In this study, the contraction pattern was assessed visually by two nuclear medicine physicians from the 17-segment mode polar maps polar maps.
Fig. 2.
Illustration of different contraction patterns of the left ventricle showing U-shaped (upper right), non-U-shaped (upper left) (homogeneous), and non-U-shaped (heterogeneous) (bottom)
Radiomics Feature Extraction
To extract MPI SPECT radiomic features, the left ventricle was segmented by an experienced nuclear medicine technologist and edited/verified by a nuclear medicine physician. Then, feature extraction was conducted utilizing pyradiomics, a library in python compliant with image biomarker standardization initiative (IBSI) [40, 46]. Re-sampling was performed for all images with order 3 interpolation using sitkBSpline to 6.591 × 6.591 × 6.591 mm3 voxels. Intensities within the volume of interest (VOI) were separated to 32 discrete gray levels with fixed bin number technique. A total of 107 radiomic features including shape, intensity, and second-/high-order texture features from GLDM, GLCM, GLRLM, GLSZM, and NGTDM families were extracted. The name and short description of all radiomic features are presented in Supplementary Table A.2.
Machine Learning Workflow
As can be seen in Fig. 1, after the feature extraction process, the data was split into train/test partitions in a way that 69 non-CRT patients were used for training and 29 CRT patients were used for testing. In addition, three feature sets were pursued for modeling. In the first case, only ConQuaFea was used for modeling. In the second case, only radiomic features and finally, in the third case, a combination of these two feature sets (combined) were used for modeling. In all models, the features extracted from the training dataset were normalized using Z-score method, and the calculated mean and standard deviation (SD) were applied on corresponding features extracted from the test dataset.
The feature selection method used in this study was recursive feature elimination (RFE). Modeling was performed using logistic regression (LR), decision tree (DT), random forest (RF), extreme gradient boosting (XGB), multi-layer perceptron (MLP), support vector machine (SVM), and gradient boosting (GB) algorithms. As a result, a total of 21 different models (3 kinds of feature-sets including ConQuaFea, radiomics, and combined × 1 feature selection method × 7 machine learning methods) were implemented. Hyper-parameters were optimized using GridSearch with 5-fold cross-validation in training data, and best values were selected to train model followed by applying the trained model on test data by 1000 bootstrap. The best hyper-parameters for each classifier are presented in Table A.3. Area under the ROC curve (AUC), accuracy (ACC), sensitivity (SEN), and specificity (SPE) metrics were used to evaluate the models. All analysis was performed on Python 3.8.8.
Delong Test
Using Delong test, comparisons between AUC of all models were performed, which was in turn followed by false discoveries rate (FDR) correction with Benjamini–Hochberg method applied on p values. Consequently, adjusted p values, also known as q values, were assessed. P values less than 0.05 were considered statistically meaningful.
Evaluation of CRT Response
In the final step of the study, the task of differentiating between patients responding to CRT or not was assessed by different inclusion criteria for the treatment, namely, (1) conventional criteria and (2) left ventricle contractile pattern status assessed visually by two nuclear medicine physicians or alternatively by machine learning and ConQuaFea/radiomic features. These patients considered as responders to CRT, experienced an improvement of at least 5% in LVEF during six months of follow-up [13].
Results
Selected Features
Based on RFE feature selection, 9 ConQuaFea, 18 combined, and 12 radiomic features were selected. Figure 3 illustrates the distribution of selected radiomic features over different feature families in the form of a pie chart. As it is shown in Fig. 3, GLCM and GLSZM features were selected the most, following them were GLDM and NGTDM which had the highest selection. This is while, first order in radiomic features and GLRLM in combined features were not selected by the feature selection methods.
Fig. 3.
Distribution of selected radiomic features by RFE feature selection method across different feature families. The left chart is related to the time when the features were selected from the radiomic data set, whereas the right chart is related to the time when the radiomic features were selected from combined features
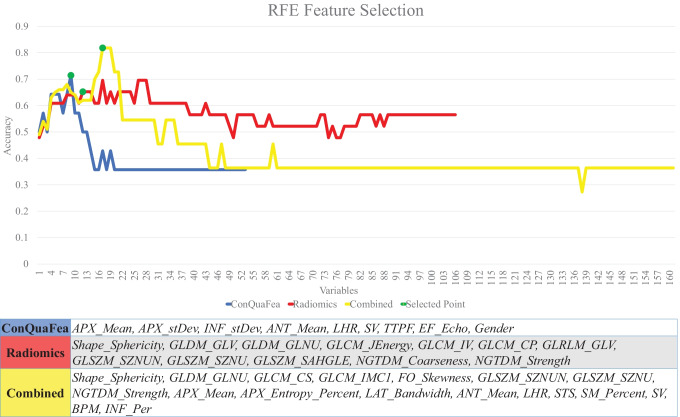
Figure 4 illustrates the selection process and lists the selected features by RFE feature selection for radiomics, ConQuaFea, and combined models.
Fig. 4.
The process of RFE feature selection and the resultant features. The green circles show the selected points (APX_Mean, apex mean; APX_stDev, apex standard deviation; INF_stDev, inferior standard deviation; ANT_Mean, anterior mean; LHR, lung heart ratio; SV, systolic volume; TTPF, time to peak filling from ES; EF_Echo, ejection fraction (Echo); APX_Entropy_Percent, apex entropy (%); LAT_Bandwidth, lateral bandwidth; STS, summed thickening score; SM_Percent, summed motion (%); BPM, beats per minute; INF_Per, inferior perfusion)
Models’ Performance
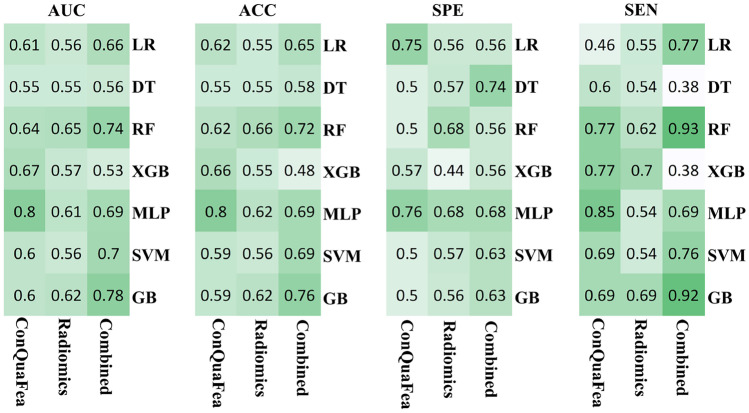
The models were developed using features from three different feature sets (ConQuaFea, radiomics, and combined), selected by RFE feature selection method, and trained with seven different machine learning methods (LR, DT, RF, XGB, MLP, SVM, and GB). Figure 5 illustrates performance metrics of all models. The performance metrics reported in Fig. 5 include AUC, accuracy, sensitivity, and specificity.
Fig. 5.
Performance metrics of ConQuaFea, radiomic, and combined features. The metrics include the area under the curve (AUC), accuracy (ACC), sensitivity (SEN), and specificity (SPE). Seven different classifiers with a feature selection method and 3 different feature sets are considered
The best performance among ConQuaFea models was achieved by MLP classifier (ACC, AUC, SEN, SPE = 0.80, 0.80, 0.85, 0.76, respectively). Among radiomics models the best models was RF model (ACC, AUC, SEN, SPE = 0.66, 0.65, 0.62, 0.68, respectively). Among the combined models, the best ones were GB and RF machines (ACC, AUC, SEN, SPE were 0.76, 0.78, 0.92, 0.63, and 0.72, 0.74, 0.93, 0.56, respectively).
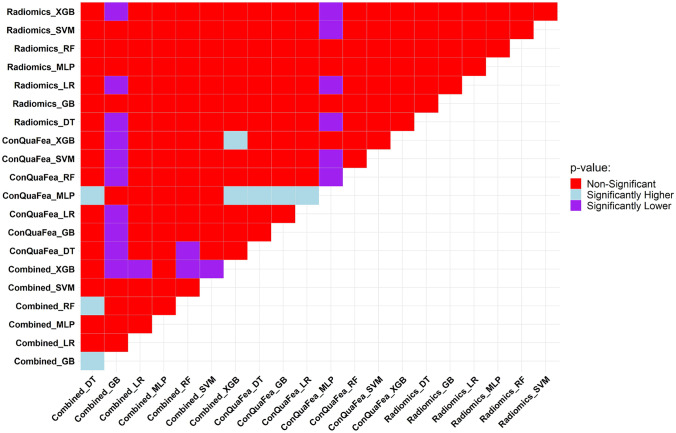
The results of the Delong test are illustrated in Fig. 6. The AUC of each model was compared with 20 other models. The results were classified as statistically significant (significantly lower or significantly higher) and non-significant. In Fig. 6, the MLP classifier on ConQuaFea had the best results with 11 significantly higher q values. Regarding radiomic features, the RF classifier did not have any significant q values. In terms of models with combined feature sets, GB and RF classifiers had 11 and 3 significantly higher q values, respectively.
Fig. 6.
Comparison of model’s performance through the Delong test being applied on the AUCs of models. We compare the pairwise model in this figure where the row models were tested against column models. Light blue, if the row model had significantly higher p value than the column model; purple, if the row model had significantly lower p value compared to the column model; red, if the comparison between the row model and column model had non-significant p value
CRT Response Prediction
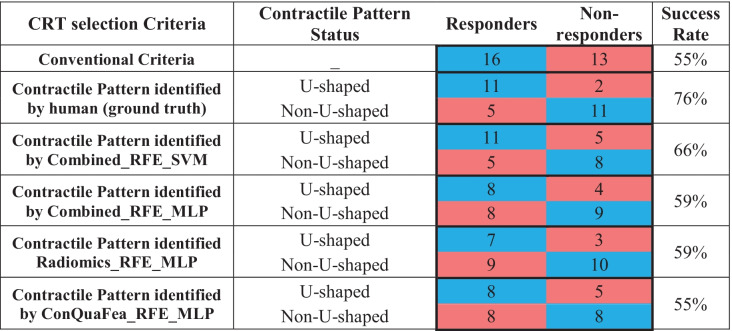
The primary aim of this study was to predict contractile patterns using GSPECT MPI, but the performance of the different models regarding the prediction of CRT response for 29 patients undergoing the treatment were also evaluated. Table 4 illustrates outcomes of 29 patients who underwent CRT with the confusion matrix regarding the prediction of models based on conventional criteria and myocardium contractile pattern (identified by two nuclear medicine physicians and different proposed machines) for prescription of the treatment. Table 4 is color coded as blue for correct decisions (true positive and negative) and red for wrong decisions (false positive and negative). The first row of Table 4 shows that based on the conventional criteria, from 29 patients prescribed with CRT, only 16 patients (55%) responded positively after the treatment whereas 13 other patients (45%) failed in treatment. The second row shows the scenario in which the treatment criteria was set as the contractile pattern statue (identified by humans). In this case, 13 patients were identified with U-shaped pattern, supposed to be sent for treatment, and if so, only 2 of them would have failed. In addition, from the 16 patients which were identified as non-U-shaped, they were not supposed to be sent for treatment, 5 of them would have responded positively to the treatment. The following rows (identified by machine) can be seen in Table 4.
Table 4.
Prediction of models based on conventional criteria and myocardium contractile pattern (identified by two nuclear medicine physicians and different proposed machines) for the prescription of treatment
Discussion
Echocardiography, non-contact mapping (NCM), cardiac magnetic resonance (CMR), and myocardial perfusion imaging (MPI) are discovering two types of contraction patterns in the left ventricle. NCM is considered the gold standard of assessing the patterns of LV electrical activation. Yet the hazardous and invasive nature of the procedure restricts its wide clinical application [18]. Accessibility, noninvasiveness and low cost are the advantages leading to wide usage of echocardiography. However, low repeatability of echo-based parameters, the state of being operator-dependent and suboptimal acoustic window for 20% of the patients have led to the approach being unpromising in selecting patients for CRT [52, 53]. CMR has emerged as a valuable tool to assess LV contraction patterns due to its high resolution and excellent tissue characterization which make it a promising approach for selecting responsive patients to CRT. Nevertheless, a high percentage of patients are disqualified on account of having pacemakers and implantable cardioverter defibrillators (ICDs) or as a result of being claustrophobic. In addition, the procedure is expensive, difficult to access, and extensively time-consuming [52–54]. GSPECT MPI transpires to be a practical technique to ascertain LV contraction patterns. Furthermore, GSPECT MPI is considerably being used for HF patients to identify LV dyssynchrony, LVEF, LV volumes, ischemia, viability, and scar tissue. Moreover, the advantages include the state of being ubiquitous and automated [19, 23, 49].
To the best of our knowledge, our study represents the first attempt to label left ventricular contractile patterns by ConQuaFea and radiomic features using machine learning and GSPECT MPI images. While this task has been previously studied in GSPECT MPI [19], it has not been investigated by machine learning approaches as well as radiomic features. In this study, we applied multiple machine learning and a feature selection method on feature sets, including radiomics, ConQuaFea/phase-analysis, and combination of both features to develop machines for the identification of LV contractile pattern from the MPI GSPECT images.
Toward the identification of most relative radiomic features, Fig. 3 illustrates the distribution of radiomic features selected by RFE feature selection algorithm over radiomic feature families. As it can be seen, GLCM and GLSZM features followed by GLDM and NGTDM were the most selected features, respectively, showing the highest correlation of the texture, hence the heterogeneity of the underlying biology of the myocardial tissue with the outcome of interest. First-order radiomic features and GLRLM in combined features were not selected by RFE. In addition, in both cases, only one feature in shape family was selected. Since the region of interest was set as the whole left ventricle, it was expected that morphological features do not show much correlation with the desired outcome.
Toward the identification of the optimum automated model, different combinations of RFE feature selection and machine learning algorithms were applied on the included feature sets (ConQuaFea, radiomics, and combined). ConQuaFea feature sets showed superiority over radiomic features. Nevertheless, radiomic features also showed an adequate performance. However, it was suggested that further studies should investigate the correlations between different feature sets in an independent analysis.
In summary, our study highlighted the potential of MPI SPECT ConQuaFea. Radiomic features for the identification of left ventricular contractile patterns also showed an acceptable performance. Physicians may benefit from evaluating each patient’s specific condition and determine if CRT is needed. Moreover, it can reduce the workload and time spent by physicians, since the detection of U-shaped contractile patterns takes an average of 5 min per patient. In addition, excessive workloads, long working hours, sleep deprivation, expertise of the physician, and other factors can lead to misdiagnosis. Hence, this study attempts to help physicians making better and more precise decisions in short time.
As mentioned earlier, despite the effectiveness and importance of CRT [6, 7], at least 30% of patients selected for this treatment do not respond well to this costly and invasive treatment [8–12], showing insufficiency of current inclusion criteria for this treatment. Moreover, from the 29 patients treated with CRT, 13 patients (45%) did not respond to the treatment (Table 4). A number of studies attempted to introduce new criteria to improve the response rate to treatment. In Bax et al. [14, 47], the presence of left ventricular mechanical dyssynchrony was more common in patients responding to CRT treatment, and therefore, it was suggested as a criterion to improve response to CRT treatment. A study by Adelstein et al. [9] also suggested that CRT leads should not be placed in areas of scar tissue diagnosed by MPI prior to implantation, as otherwise the response to treatment will be reduced. In the study of Chen et al. [15], the authors realized that the only characteristics which were different between the CRT patients who were responders and those who were not, were the histogram bandwidth and phase standard deviation (extracted from phase analysis using Emory Cardiac Toolbox at the baseline (before CRT)), which is related to left ventricular mechanical dyssynchrony. He et al. [16] conducted research with the goal of using deep learning to extract new LVMD features using GSPECT MPI phase analysis to help selecting CRT patients. New LVMD parameters retrieved by automated Autoencoder (AE) from GSPECT MPI have the potential to enhance response prediction prior to CRT. The study by Rastgou et al. [48] stated that the phase analysis parameters of Emory Cardiac Toolbox and QGS program are well correlated, but these parameters should not be used interchangeably. Furthermore, in relation to entropy (a parameter in phase analysis), the lower value of this parameter indicates synchronized heart contraction whereas the higher value indicates desynchronize heart contraction [49].
In our study, the QGS program was used to extract phase analysis indices, and when the features were selected by RFE, the apex standard deviation and inferior standard deviation from ConQuaFea data set and apex entropy and lateral bandwidth from combined features were selected. Since these features are related to the left ventricular contractile pattern and the left ventricular contractile pattern to the type of CRT response, the phase analysis parameters, as mentioned in the above studies, are related to the type of CRT response.
In a study by Feeny et al. [50], a machine learning model was developed for CRT outcome prediction by enrolling 925 patients, with 9 clinical features (QRS morphology, QRS duration, New York Heart Association classification, left ventricular ejection fraction and end-diastolic diameter, sex, ischemic cardiomyopathy, atrial fibrillation, and epicardial left ventricular lead) used as input for the Naïve Bayes classifier. Their machine learning model outperformed the conventional guideline with an increased AUC, 0.70 vs. 0.65 (p value < 0.02), and an increased event-free survival with concordance index = 0.61 vs. 0.56 (p value < 0.001).
In a study by Tao et al. [19], contractile patterns were used to analyze CRT responses. Their results showed that 89% of U-shaped group were responders to CRT and 11% were non-responders. In our study, of the 29 patients who performed CRT, 16 patients had U-shaped contractile patterns, from which 11 responded positively, and 13 had non-U-shaped contractile patterns, among which 11 did not respond to the treatment (Table 4). It led to a success rate of 76% compared to traditional patient selection (16 responders out of 29 (55% responders)) in predicting treatment outcome based on contractile patterns. We also analyzed the treatment response, in case CRT was prescribed based on the left ventricle contractile pattern detected by our proposed models (Table 4). The results were slightly lower than the model based on the ground truth status of the left ventricle contractile pattern identified by the two nuclear medicine physicians which was expected since the error of pattern identification is also introduced to the final results. Moreover, it should be noted that according to the study conducted by Hartlage et al. [17], LV lead concordant to the latest contracting site would be more likely to produce a superior CRT response beside the left ventricular contractile pattern. In fact, in their study, patients with a U-shaped contraction pattern and the LV lead concordant to the last contraction site were 92% respondent, which indicates the importance of examining the pattern as well as the correct location of the lead in CRT patients. In our study, the main purpose was to detect the left ventricular contractile pattern, and therefore, the correct location of the leads was not investigated.
This study inherently bears a few limitations. Regarding the task of CRT outcome prediction, we set the criteria on the basis of the left ventricular contractile pattern and did not consider matters, such as the location of the CRT leads. Future studies might consider both to develop more comprehensive automated models for this important task. In addition, our dataset was obtained from one institute which undermines the robustness of the findings. Further studies might gather larger and diverse dataset obtained from multiple institutes with different image acquisition parameters and patients’ ethnicity to improve the reproducibility of the models [51]. However, for proof-of-concept, this study contained enough patients.
Conclusion
In this study, machine learning models were developed to predict left ventricular contractile patterns with ConQuaFea and radiomic features from GSPECT MPI using different machine learning approaches with acceptable and promising results. ConQuaFea performed better than radiomic features in recognizing left ventricular contractile pattern. In addition, by diagnosing the patients’ left ventricular contraction pattern, it is possible to improve the patient’s selection for CRT treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- ConQuaFea
Conventional quantitative features
- HF
Heart failure
- CRT
Cardiac resynchronization therapy
- NYHA
New York Heart Association
- LVEF
Left ventricular ejection fraction
- LV
Left ventricle
- CAD
Coronary artery disease
- GSPECT MPI
Gated single-photon emission computed tomography myocardial perfusion imaging
- NCM
Non-contact mapping
- CMR
Cardiac magnetic resonance
- MRI
Magnetic resonance imaging
- CT
Computed tomography
- PET
Positron emission tomography
- QGS
Quantitative gated SPECT
- IBSI
Image Biomarker Standardization Initiative
- VOI
Volume of interest
- SD
Standard deviation
- RFE
Recursive feature elimination
- LR
Logistic regression
- DT
Decision tree
- RF
Random forest
- XGB
Extreme gradient boosting
- MLP
Multi-layer perceptron
- SVM
Support vector machine
- GB
Gradient boosting
- AUC
Area under the ROC curve
- ACC
Accuracy
- SEN
Sensitivity
- SPE
Specificity
- FDR
False discoveries rate
Funding
Open access funding provided by University of Geneva. This work was supported by Iran University of Medical Sciences and Rajaie Cardiovascular Medical and Research Center under grant number IR.IUMS.FMD.REC.1400.087 and the Swiss National Science Foundation under Grant SNRF 320030_176052.
Availability of Data and Material
Not applicable.
Code Availability
Open-source library including 3D-Slicer and Python library used in this study.
Declarations
Ethics Approval
This retrospective study was approved by the ethics committee of Iran University of Medical Sciences (IR.IUMS.FMD.REC.1400.087).
Consent to Participate
Informed consent was waived by ethics groups.
Consent for Publication
Informed consent was waived by ethics groups.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ahmad Bitarafan Rajabi, Email: bitarafan@hotmail.com.
Isaac Shiri, Email: isaac.shirilord@unige.ch.
Habib Zaidi, Email: habib.zaidi@hcuge.ch.
References
- 1.Groenewegen A, Rutten FH, Mosterd A, Hoes AW. Epidemiology of heart failure. Eur J Heart Fail. 2020;22:1342–1356. doi: 10.1002/ejhf.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagueh SF. Mechanical dyssynchrony in congestive heart failure: diagnostic and therapeutic implications. J Am Coll Cardiol. 2008;51:18–22. doi: 10.1016/j.jacc.2007.08.052. [DOI] [PubMed] [Google Scholar]
- 3.Levy WC, et al. The Seattle Heart Failure Model: prediction of survival in heart failure. Circulation. 2006;113:1424–1433. doi: 10.1161/CIRCULATIONAHA.105.584102. [DOI] [PubMed] [Google Scholar]
- 4.Marín-Oyaga V, Gutiérrez-Villamil C, Dueñas-Criado K, Arévalo-Leal S: Análisis de fase para la valoración del sincronismo mecánico del ventrículo izquierdo mediante Gated-SPECT de perfusión miocárdica. Importancia de los parámetros clínicos y técnicos. Rev Fac Med 65:453–459, 2017
- 5.Yaghoobi N, Malek H: The age of reason for gated SPECT MPI to deal with cardiac dyssynchrony. Res Cardiovasc Med 4, 2015 [DOI] [PMC free article] [PubMed]
- 6.Bax JJ, et al. Cardiac resynchronization therapy: part 1—issues before device implantation. J Am Coll Cardiol. 2005;46:2153–2167. doi: 10.1016/j.jacc.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien T, Park M-S, Youn J-C, Chung ES. The past, present and future of cardiac resynchronization therapy. Korean Circ J. 2019;49:384–399. doi: 10.4070/kcj.2019.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham WT, Hayes DL. Cardiac resynchronization therapy for heart failure. Circulation. 2003;108:2596–2603. doi: 10.1161/01.CIR.0000096580.26969.9A. [DOI] [PubMed] [Google Scholar]
- 9.Adelstein EC, Saba S. Scar burden by myocardial perfusion imaging predicts echocardiographic response to cardiac resynchronization therapy in ischemic cardiomyopathy. Am heart J. 2007;153:105–112. doi: 10.1016/j.ahj.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Auricchio A, et al. Effect of pacing chamber and atrioventricular delay on acute systolic function of paced patients with congestive heart failure. Circulation. 1999;99:2993–3001. doi: 10.1161/01.CIR.99.23.2993. [DOI] [PubMed] [Google Scholar]
- 11.Leclercq C, Hare JM. Ventricular resynchronization: current state of the art. Circulation. 2004;109:296–299. doi: 10.1161/01.CIR.0000113458.76455.03. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq C, Kass DA. Retiming the failing heart: principles and current clinical status of cardiac resynchronization. J Am Coll Cardiol. 2002;39:194–201. doi: 10.1016/S0735-1097(01)01747-8. [DOI] [PubMed] [Google Scholar]
- 13.Glikson M, et al. 2021 ESC Guidelines on cardiac pacing and cardiac resynchronization therapy: Developed by the Task Force on cardiac pacing and cardiac resynchronization therapy of the European Society of Cardiology (ESC) With the special contribution of the European Heart Rhythm Association (EHRA) Eur Heart J. 2021;42:3427–3520. doi: 10.1093/eurheartj/ehab364. [DOI] [PubMed] [Google Scholar]
- 14.Bax JJ, et al. Left ventricular dyssynchrony predicts response and prognosis after cardiac resynchronization therapy. J Am Coll Cardiol. 2004;44:1834–1840. doi: 10.1016/j.jacc.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, et al. Assessment of left ventricular mechanical dyssynchrony by phase analysis of ECG-gated SPECT myocardial perfusion imaging. J Nucl Cardiol. 2008;15:127–136. doi: 10.1016/j.nuclcard.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 16.He Z, et al.: A method using deep learning to discover new predictors of CRT response from mechanical dyssynchrony on gated SPECT MPI. J Nucl Cardiol :210601355, 2021
- 17.Hartlage GR, et al. Prediction of response to cardiac resynchronization therapy using left ventricular pacing lead position and cardiovascular magnetic resonance derived wall motion patterns: a prospective cohort study. J Cardiovasc Magn Reson. 2015;17:1–10. doi: 10.1186/s12968-015-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohal M, et al. Noninvasive assessment of LV contraction patterns using CMR to identify responders to CRT. JACC Cardiovasc Imaging. 2013;6:864–873. doi: 10.1016/j.jcmg.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Tao N, et al. Assessment of left ventricular contraction patterns using gated SPECT MPI to predict cardiac resynchronization therapy response. J Nucl Cardiol. 2018;25:2029–2038. doi: 10.1007/s12350-017-0949-1. [DOI] [PubMed] [Google Scholar]
- 20.Duckett SG, et al. Relationship between endocardial activation sequences defined by high-density mapping to early septal contraction (septal flash) in patients with left bundle branch block undergoing cardiac resynchronization therapy. Europace. 2012;14:99–106. doi: 10.1093/europace/eur235. [DOI] [PubMed] [Google Scholar]
- 21.Fung JW, et al. Effect of left ventricular endocardial activation pattern on echocardiographic and clinical response to cardiac resynchronization therapy. Heart. 2007;93:432–437. doi: 10.1136/hrt.2007.115295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson T, et al. A U-shaped type II contraction pattern in patients with strict left bundle branch block predicts super-response to cardiac resynchronization therapy. Heart rhythm. 2014;11:1790–1797. doi: 10.1016/j.hrthm.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Boogers MM, Chen J, Bax JJ. Role of nuclear imaging in cardiac resynchronization therapy. Expert Rev Cardiovasc Ther. 2009;7:65–72. doi: 10.1586/14779072.7.1.65. [DOI] [PubMed] [Google Scholar]
- 24.Manafi-Farid R, et al.: [(18)F]FDG-PET/CT radiomics and artificial intelligence in lung cancer: Technical aspects and potential clinical applications. Semin Nucl Med, 2022 [DOI] [PubMed]
- 25.Khodabakhshi Z, et al. Overall Survival Prediction in Renal Cell Carcinoma Patients Using Computed Tomography Radiomic and Clinical Information. J Digit Imaging. 2021;34:1086–1098. doi: 10.1007/s10278-021-00500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amini M, et al. Overall Survival Prognostic Modelling of Non-small Cell Lung Cancer Patients Using Positron Emission Tomography/Computed Tomography Harmonised Radiomics Features: The Quest for the Optimal Machine Learning Algorithm. Clin Oncol (R Coll Radiol) 2022;34:114–127. doi: 10.1016/j.clon.2021.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Amini M, et al.: Multi-level multi-modality (PET and CT) fusion radiomics: prognostic modeling for non-small cell lung carcinoma. Phys Med Biol 66, 2021 [DOI] [PubMed]
- 28.Khodabakhshi Z, Mostafaei S, Arabi H, Oveisi M, Shiri I, Zaidi H. Non-small cell lung carcinoma histopathological subtype phenotyping using high-dimensional multinomial multiclass CT radiomics signature. Comput Biol Med. 2021;136:104752. doi: 10.1016/j.compbiomed.2021.104752. [DOI] [PubMed] [Google Scholar]
- 29.Arian F, et al.: Myocardial Function Prediction After Coronary Artery Bypass Grafting Using MRI Radiomics Features and Machine Learning Algorithms. J Digit Imaging, 2022 [DOI] [PMC free article] [PubMed]
- 30.Shayesteh S, et al. Treatment response prediction using MRI-based pre-, post-, and delta-radiomic features and machine learning algorithms in colorectal cancer. Med Phys. 2021;48:3691–3701. doi: 10.1002/mp.14896. [DOI] [PubMed] [Google Scholar]
- 31.Avard E, et al. Non-contrast Cine Cardiac Magnetic Resonance image radiomics features and machine learning algorithms for myocardial infarction detection. Comput Biol Med. 2022;141:105145. doi: 10.1016/j.compbiomed.2021.105145. [DOI] [PubMed] [Google Scholar]
- 32.Shiri I, et al.: Diagnosis of COVID-19 using CT image radiomics features: a comprehensive machine learning study involving 26,307 patients. medRxiv, 2021
- 33.Shiri I, et al. High-Dimensional Multinomial Multiclass Severity Scoring of COVID-19 Pneumonia Using CT Radiomics Features and Machine Learning Algorithms. Sci Rep: In press; 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiri I, et al. Impact of feature harmonization on radiogenomics analysis: Prediction of EGFR and KRAS mutations from non-small cell lung cancer PET/CT images. Comput Biol Med. 2022;142:105230. doi: 10.1016/j.compbiomed.2022.105230. [DOI] [PubMed] [Google Scholar]
- 35.Ashrafinia S, Dalaie P, Sadaghiani MS, Schindler T, Pomper MG, Rahmim A: Radiomics analysis of clinical myocardial perfusion stress SPECT images to identify coronary artery calcification. medRxiv, 2021 [DOI] [PMC free article] [PubMed]
- 36.Rahmim A, et al.: Improved prediction of outcome in Parkinson’s disease using radiomics analysis of longitudinal DAT SPECT images. NeuroImage: Clinical 16:539–544, 2017 [DOI] [PMC free article] [PubMed]
- 37.Edalat-Javid M, et al.: Cardiac SPECT radiomic features repeatability and reproducibility: A multi-scanner phantom study. J Nucl Cardiol :1–15, 2020 [DOI] [PubMed]
- 38.Bi Q, Goodman KE, Kaminsky J, Lessler J, et al. What is machine learning? A primer for the epidemiologist. Am. J. Epidemiol. 2019;188:2222–2239. doi: 10.1093/aje/kwz189. [DOI] [PubMed] [Google Scholar]
- 39.Martin-Isla C, et al. Image-based cardiac diagnosis with machine learning: a review. Front Cardiovasc Med. 2020;7:1. doi: 10.3389/fcvm.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwanenburg A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaret BL, Beller GA: Clinical Nuclear Cardiology: State of the Art and Future Directions E-Book, 2010
- 42.Garcia EV, et al. Quantification of rotational thallium-201 myocardial tomography. J. Nucl. Med. 1985;26:17–26. [PubMed] [Google Scholar]
- 43.Caldwell JH, Williams DL, Harp GD, Stratton J, Ritchie J. Quantitation of size of relative myocardial perfusion defect by single-photon emission computed tomography. Circulation. 1984;70:1048–1056. doi: 10.1161/01.CIR.70.6.1048. [DOI] [PubMed] [Google Scholar]
- 44.D. Cerqueira NJW, Vasken Dilsizian, Alice K. Jacobs, Sanjiv Kaul, Warren K. Laskey: Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation:539–542, 2002 [DOI] [PubMed]
- 45.Rubeaux M, Xu Y, Germano G, Berman DS, Slomka PJ. Normal databases for the relative quantification of myocardial perfusion. Curr. Cardiovasc. Imaging Rep. 2016;9:1–11. doi: 10.1007/s12410-016-9385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Griethuysen JJ, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bax JJ, et al. Left ventricular dyssynchrony predicts benefit of cardiac resynchronization therapy in patients with end-stage heart failure before pacemaker implantation. Am. J. Cardiol. 2003;92:1238–1240. doi: 10.1016/j.amjcard.2003.06.016. [DOI] [PubMed] [Google Scholar]
- 48.Rastgou F, et al. Assessment of left ventricular mechanical dyssynchrony by phase analysis of gated-SPECT myocardial perfusion imaging and tissue Doppler imaging: comparison between QGS and ECTb software packages. J. Nucl. Cardiol. 2014;21:1062–1071. doi: 10.1007/s12350-014-9941-1. [DOI] [PubMed] [Google Scholar]
- 49.Baughan N, Poitrasson-Rivière A, Moody JB, Lee BC, Ficaro EP. Quantification of Cardiac Dyssynchrony Entropy Implemented in a Commercial Nuclear Medicine Software Application. Nucl. 2020;206:977–983. [Google Scholar]
- 50.Feeny AK, et al. Machine learning prediction of response to cardiac resynchronization therapy: improvement versus current guidelines. Circ Arrhythm Electrophysiol. 2019;12:e007316. doi: 10.1161/CIRCEP.119.007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiri I, et al. Decentralized Distributed Multi-institutional PET Image Segmentation Using a Federated Deep Learning Framework. Clin Nucl Med. 2022;47:606–617. doi: 10.1097/RLU.0000000000004194. [DOI] [PubMed] [Google Scholar]
- 52.M. M. Henneman et al.: Can LV dyssynchrony as assessed with phase analysis on gated myocardial perfusion SPECT predict response to CRT?, J. Nucl. Med., 48:1104–1111, 2007 [DOI] [PubMed]
- 53.H. Abu Daya, S. Malhotra, and P. Soman: Radionuclide Assessment of Left Ventricular Dyssynchrony, Cardiol. Clin., 34:101–118, 2016 [DOI] [PubMed]
- 54.STELLBRINK C, et al. How to discriminate responders from non-responders to cardiac resynchronisation therapy. European Heart Journal Supplements. 2004;6:D101–D105. doi: 10.1016/j.ehjsup.2004.05.008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.
Open-source library including 3D-Slicer and Python library used in this study.