Abstract
Multiple lines of evidence implicate dysregulated microglia-mediated synaptic pruning in the pathophysiology of schizophrenia. In vitro human cellular studies represent a promising means of pursuing this hypothesis, complementing efforts with animal models and postmortem human data while addressing their limitations. The challenges in culturing homogeneous populations of cells derived from postmortem or surgical biopsy brain material from patients, and their limited availability, has led to a focus on differentiation of induced pluripotent stem cells. These methods too have limitations, in that they disrupt the epigenome and can demonstrate line-to-line variability due in part to extended time in culture, partial reprogramming, and/or residual epigenetic memory from the cell source, yielding large technical artifacts. Yet another strategy uses direct transdifferentiation of peripheral mononuclear blood cells, or umbilical cord blood cells, to microglia-like cells. Any of these approaches can be paired with patient-derived synaptosomes from differentiated neurons as a simpler alternative to co-culture. Patient-derived microglia models may facilitate identification of novel modulators of synaptic pruning and identification of biomarkers that may allow more targeted early interventions.
Schizophrenia is a chronic, disabling, and strongly heritable illness. With a lifetime prevalence of 1% and an onset in late adolescence or early adulthood, schizophrenia makes a major contribution to disability worldwide. Existing treatments are only partially effective in restoring function and preventing relapse and are accompanied by adverse effects that are themselves disabling. Efforts to identify novel treatments have been hindered by a very limited understanding of disease pathogenesis, despite the identification of more than 250 regions of the genome associated with schizophrenia liability (1,2).
Normal postnatal brain development involves the generation of supernumerary neuronal synapses that are selectively eliminated during development through the activity-dependent process of synaptic pruning (3). These processes are necessary for proper brain development (4), while their disruption can lead to aberrations such as the reduced dendritic spine density implicated in schizophrenia (5). Although neuronal-specific factors contribute to synaptic refinement, numerous studies have also demonstrated the key role of microglia in this process. While the precise mechanisms by which microglia choose their synaptic targets is still unknown, studies confirm selective engulfment of synapses by microglia as critical for the development and maintenance of proper brain connectivity (6–9).
Feinberg first postulated in 1982 that defective synaptic pruning during adolescence contributes to the etiology of schizophrenia (10). Since then, convergent evidence implicates aberrant synaptic pruning, including postmortem studies showing changes in hippocampal and cortical spine density and live brain imaging studies demonstrating decreased cortical thickness and subcortical volumes in patients with schizophrenia (5,11–14). Extensive synaptic pruning in the human cerebral cortex normally occurs during late adolescence and early adulthood, notably coinciding with the typical age of clinical onset for schizophrenia.
Genomic association studies suggest a possible mechanism for the observed decrease in synaptic density in patients with schizophrenia: the locus most strongly linked to schizophrenia risk (1) associates with greater brain complement C4 expression, consistent with increased C4 expression in post-mortem samples (15). Thus, the complement pathway provides a probable mechanism to explain at least in part the structural changes observed in schizophrenia. Multiple lines of evidence suggest that this complement-associated synaptic pruning is mediated by microglia (5,10,13,16,17).
PATIENT-DERIVED IN VITRO MODELS OF MICROGLIA FUNCTION
Multiple functional mouse models of microglia-mediated synaptic pruning during neurodevelopment implicate the non-specialized mouse C4 gene (6,7). However, human models are required to test the hypothesis directly: the distinct and functionally specialized human C4 gene isotypes, linked to schizophrenia via C4A expression, do not exist in the rodent genome. In addition, the microglial transcriptome varies greatly across species (18), further supporting the need for human microglia for studies of human neurodevelopment and disease. However, sources of patient-derived in vitro microglia vary in their robustness and amenability to perform large-scale experiments.
Postmortem Isolation of In Vitro Microglia
Various methods have been developed to isolate viable microglia from primary human sources (19,20), using enzymatic or mechanical methods to dissociate brain tissue followed by further cell-specific purification (21–25). However, the robust in vitro culturing of primary microglia has been challenging, and microglial identity and transcriptomic signature have been shown to change rapidly in vitro (26). Furthermore, while these ex vivo microglia have provided insight into neurological disease, they are not optimal for adequately powered statistical comparisons of patients to healthy control individuals or for high-throughput drug screening. Variables such as antemortem neurological diagnosis and exposures, postmortem delay before harvest, high donor-to-donor variability in microglia yield (27), and rapid gene and marker expression changes upon in vitro culture of primary human microglia (26) preclude reproducible derivation of robust in vitro microglia cultures at scale.
Patient-Induced Pluripotent Stem Cell–Derived Models of In Vitro Microglia
Somatic cells can be reprogrammed toward induced pluripotent stem cells (iPSCs) through the exogenous introduction of transcription factors (28). Furthermore, the derivation of human iPSC-derived neuronal cells, for instance through SMAD inhibition (29), allows for the efficient derivation of patient-derived neuronal models. In addition, neuronal type–specific differentiation procedures from iPSCs have been developed for robust differentiation into relatively homogeneous neuronal subtypes of the neocortex such as dopaminergic (30–32), GABAergic (gamma-aminobutyric acidergic) (33,34), glutamatergic (35,36), serotonergic (37,38), and interneurons (39,40); one example is the efficient differentiation into glutamatergic cortical excitatory neurons by overexpression of the transcription factor NGN2 (35).
The derivation of microglia from iPSCs has been more challenging. Microglia are of mesodermal origin, originating from yolk sac Myb-independent erythromyeloid progenitors (EMPs) during early development before the emergence of Myb-dependent hematopoietic stem cells (41–43). Protocols have been developed to recapitulate the developmental ontogeny through stepwise formation of primitive mesoderm to hematopoietic precursor cells and erythromyeloid progenitors. Muffat et al. (44) demonstrated the differentiation of iPSC-derived microglia-like cells through intermediate erythromyeloid progenitor–like cells. These iPSC-derived microglia-like cells expressed canonical microglia markers, were migratory, and demonstrated phagocytic capacity using fluorescent polystyrene beads.
Since this initial study, several methodologies have been optimized for faster differentiation, greater yields, and/or a more mature microglial phenotype. Abud et al. (45) developed methods for iPSC-derived microglia-like cells through CD34+ myeloid precursors. Large-scale yield was further improved by deriving nonadherent embryonic macrophage progenitors that can be harvested over several weeks for microglial maturation either through co-culture with iPSC-derived neuronal cultures (46) or addition of neuronal factors (45). Furthermore, three-dimensional (3D) methods have been developed containing microglia with a more mature adult phenotype, albeit with significantly lower yields (47). Recently, Chen et al. (48) demonstrated that iPSCs can be converted to microglia via overexpression of transcription factors; these cells display microglial markers, a transcriptome resembling human fetal microglia, lipopolysaccharide/interferon-γ–induced inflammatory response, and phagocytic activity.
An advantage of using iPSC-derived microglia is the ability to perform transplantation studies with progenitors into mouse models to mimic endogenous microglia development and function in physiologically relevant environments (49–51) to model potential differences between patient- and healthy control subject–derived, or genetically modified, cells in an in vivo brain context.
While the ability to generate these cells from patient-derived iPSCs has allowed for in vitro characterization and manipulations from patient-derived sources, they still have limitations as model systems. The reprogramming of iPSCs from patient somatic cells results in epigenetic resetting to accomplish pluripotency (52), which risks eliminating potential epigenetic contributions (53) that have been implicated to be particularly relevant in complex psychiatric disorders such as schizophrenia [as reviewed in (54,55)]. Conversely, individual iPSC lines from the same patient can differ widely depending on partial reprogramming and/or residual epigenetic memory from the cell source; for example, while theoretical, it is possible that environmental exposures such as medications may affect patient-derived lines. Such line-to-line variability means that findings in case-control studies can be readily confounded by technical artifacts (56–59). Other factors, including the cellular age of the donor cells, are modified or reset during iPSC induction (60–64). Thus, age-related alterations in the bioenergetics and epigenome of the donor cells that may be important to cellular phenotypes may not be maintained (65).
Patient Peripheral Blood Mononuclear Cell–Derived Models of In Vitro Microglia
As an alternative to iPSC-derived differentiated human cellular models, transdifferentiation methodologies have been developed that maintain epigenetic and age-associated characteristics of patient donor cells (64). For example, transdifferentiation methods have been developed to derive induced neurons from fibroblasts using exogenous overexpression of transcription factors (66–68), microRNAs (69,70), or small molecules (71,72) without going through a pluripotent intermediate.
Peripheral blood mononuclear cells (PBMCs) are typically used as a DNA source for genomic studies (1,73), but they have also been applied to investigate transcriptomic differences between patients with schizophrenia and healthy control individuals (74,75). Several studies have suggested that PBMCs may be a useful proxy for aberrant brain function in patients with schizophrenia (76). PBMCs express more than 4100 brain transcripts and show similar alternate splicing patterns compared with brain samples (77); PBMCs from patients with schizophrenia demonstrate differences in gene expression (75,78,79), noncoding RNAs (80–84), epigenetics (85,86), bioenergetics (87,88), proteomics (89), and metabolomics (90). In one study, transcriptomic analysis between patient and control PBMCs and postmortem brain tissue shared similar differentially expressed genes implicated in common pathways (79). These observations further support the use of PBMCs as models, albeit indirectly, of potential central nervous system dysfunction.
A fraction of PBMCs (monocytes) possess high phenotypic plasticity and can adopt a microglial-like phenotype in vitro. In an early demonstration, rat monocytes cultured in astrocyte-conditioned media or in co-culture with astrocytes converted to a ramified morphology and expressed microglia-like membrane currents by electrophysiology (91,92). Leone et al. (93) showed that the culture of human PBMCs in astrocyte-conditioned media supplemented with granulocyte macrophage colony-stimulating factor (CSF) and macrophage CSF converted to ramified monocyte-derived microglia-like cells and expressed canonical microglia markers. Using a defined media cocktail including macrophage CSF, granulocyte macrophage CSF, nerve growth factor-β, and CCL2, Etemad et al. (94) demonstrated derivation of human monocyte-derived microglia that displayed microglial markers and chemokine receptors (e.g., CX3CR1) and were functional in phagocytosing labeled bacterial particles. Ohgidani et al. (95) further determined that a limited set of just two factors, interleukin 4 and granulocyte macrophage CSF, were sufficient to convert patient PBMCs to induced microglia-like cells (iMGs) to model microglial dysfunction in Nasu-Hakola disease.
The direct conversion of human monocytes to iMGs thus represents an emerging strategy for generating models from patient-derived blood (94–101). PBMCs can be isolated from small 5-mL blood draws up to whole units of blood and cryopreserved for large-scale biobanks of assay-ready cells. Using similar cytokine induction methodology, we further characterized PBMC-derived iMGs (99) with the addition of an extracellular matrix containing laminin, collagen, and the astrocyte-expressed extracellular component entactin (nidogen-1) (102) to derive iMGs from human PBMCs (hiMGs). Transcriptomic data demonstrated similarity to primary fetal microglia compared with monocyte-derived macrophages, immortalized primary microglia, and isolated adult postmortem microglia (103,104). Phagocytosis of iPSC-derived neural progenitors and isolated human synaptic vesicles (synaptosomes) from iPSC-derived differentiated neuronal cultures were used in conjunction with hiMGs in in vitro models of microglial engulfment. In particular, synaptosomes were readily engulfed by hiMGs, a process inhibited by pretreatment with an inactivating antibody to the αM subunit (CD11b) of C3R (105), demonstrating that phagocytosis was complement dependent. These procedures have been replicated in several studies, with transcriptomic data analysis validating that they closely resemble human primary microglia and iPSC-derived iMGs (96). Patient monocyte–derived microglia have recently been used to suggest novel microglial transcriptome and proteome contributions in schizophrenia (98). In addition to PBMCs from adult donors, we recently demonstrated that microglia-like cells can be derived from umbilical cord blood–derived mononuclear cells to model potential deleterious effects of maternal immune activation (106). In conjunction with longitudinal clinical outcomes, for example, derived from electronic health records or birth cohort studies, patient-specific models of microglia function at birth may represent a new strategy for stratifying risk for neurodevelopmental disorders early in postnatal development.
While direct conversion of human blood monocytes toward microglia-like in vitro models has several advantages compared with iPSC-derived systems, including rapid scalable generation and lack of a pluripotent intermediate, some limitations should be noted. Transdifferentiation methods are limited by low conversion efficiency without a proliferative stage, requiring a large amount of input cells to accommodate yields for large-scale study. In addition, direct conversion methods do not follow developmental ontogeny toward terminally differentiated end points thus are inadequate in understanding developmentally upstream factors.
PATIENT-DERIVED IN VITRO MODEL SYSTEMS OF MICROGLIAL SYNAPTIC PRUNING
Models of Schizophrenia in Patient-Derived In Vitro Neuronal/Microglial Co-culture
In vitro co-culture systems allow each cell type to be separately derived (Figure 1A), for example, from patients versus healthy control subjects or between different genetic backgrounds (Figure 1B), or differentially manipulated genetically through gene editing or biochemically with small molecules or biologics (Figure 1C) to elucidate their individual contributions to observed phenotypes. Patient subgroups can also be investigated similarly, for example, to understand sex-specific effects (107–109). Recently, a limited number of co-culture methods have been used to model human neuronal-microglial interactions using pluripotent-derived cells (45,46,110,111). Challenges remain in co-culturing human microglia in neuronal cultures; media used to grow microglia is often deficient in factors that neurons require for robust viability and activity, and typical additives often used for neuronal culture could be deleterious to microglial function, although recent advances allow improved co-culture maintenance (46). Microglia can be added transiently to neuronal cultures to model functional attributes such as synaptic pruning. We have used this technique to model dendritic spine density elimination in iPSC-derived cortical neuronal cultures (100), although quantification of spine density is low throughput and is highly dependent on optimized cell density for robust image-based analyses.
Figure 1.
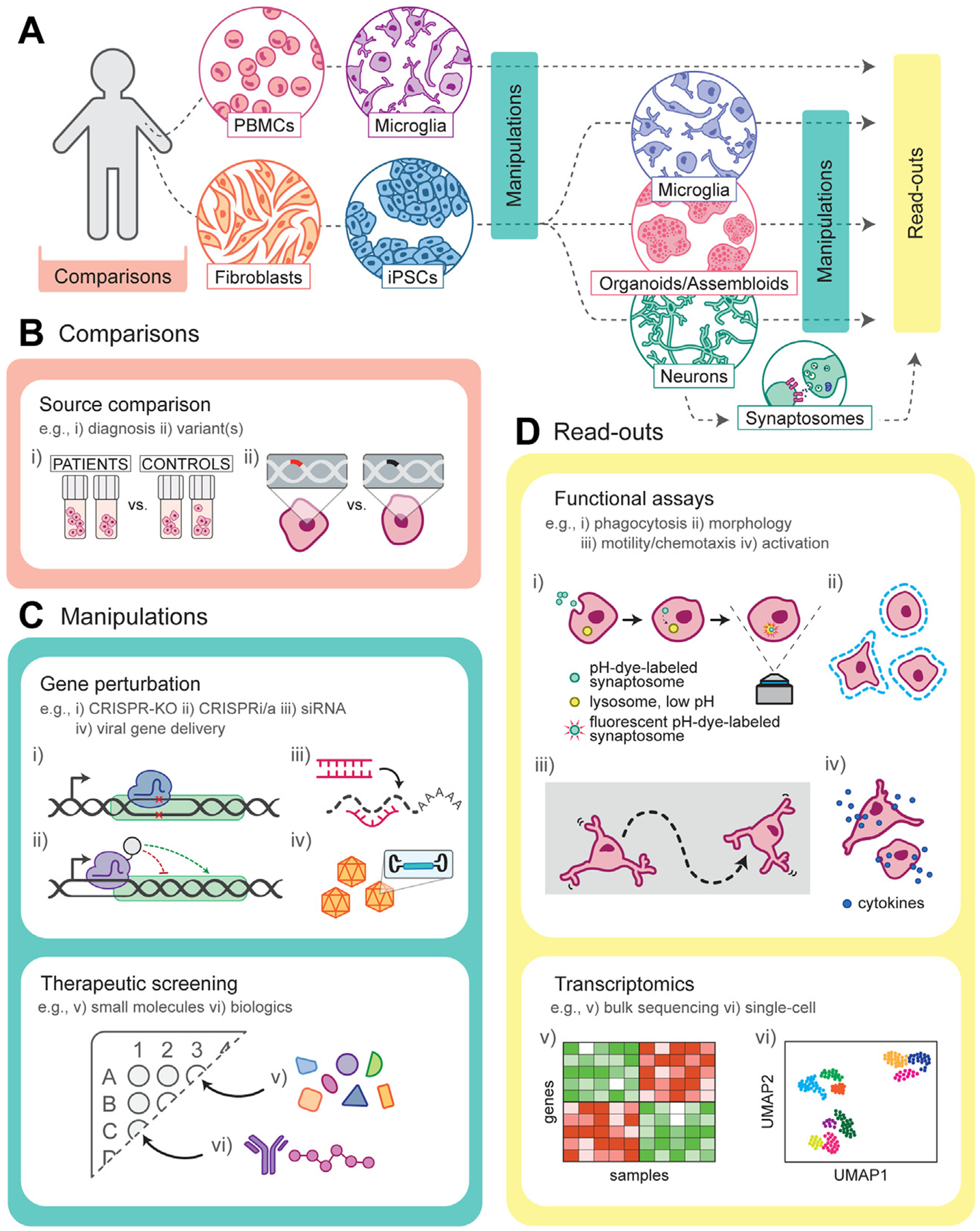
Patient-derived in vitro models of microglial function and synaptic engulfment in schizophrenia. (A) Overview of patient-derived microglial and neuronal in vitro cellular models to understand disease etiology and for therapeutic discovery. Dotted lines represent common experimental pathways. (B) Biobanks of patient cells offer a trove of information to compare by i) clinical diagnosis or by ii) specific genetic signatures such as risk variant(s). The ability to reprogram these cells is important to observe cell type–specific phenotypes and responses to perturbations. (C) Cell manipulations can be applied during different reprogramming stages and may be optimal in a cell type–specific manner. i) Targeted gene knockout, ii) gene regulation, iii) silencing, and iv) transgene expression can be used to manipulate individual genes implicated in schizophrenia. In addition, the effects of v) small molecules and vi) biologics can be screened for phenotypic reversal for therapeutic discovery and development. (D) Functional in vitro assays monitor cell-specific behavior, such as i) phagocytosis (e.g., synaptosomes), ii) alterations in morphology, iii) motility/chemotaxis, and iv) activation state. Furthermore, transcriptomic data, either in v) bulk or vi) single-cell resolution, allow for gene-and pathway-level information that can lead to novel targets for further study of potential therapeutic value. CRISPR, clustered regularly interspaced short palindromic repeats; i/a, inhibiting/activating; iPSCs, induced pluripotent stem cells; KO, knockout; PBMCs, peripheral blood mononuclear cells; siRNA, small interfering RNA.
Monolayer co-culture systems using microglia and/or neurons derived from patients can provide important insight into aberrant microglial-neuronal interactions. However, they poorly reflect the complexity found in the 3D brain, notably intrinsic and region-specific biochemical cellular cues. Newer methods of 3D culture may enable better representative models of development in utero, including structure, signaling, and multicellular systems. Cerebral organoids are iPSC-derived 3D cultures that to some extent can recapitulate the architecture of brain tissue in vitro. These models have been recently used to model neurodevelopmental (112,113) and neurodegenerative (114,115) disorders, but adaptation of the use of cerebral organoids for studying neuropsychiatric diseases such as schizophrenia has been limited. Ye et al. (116) showed disrupted mitosis and a reduction in the number of radial glia in forebrain cerebral organoids derived from patients with schizophrenia with mutations in the DISC1 gene. Stachowiak et al. (117) demonstrated abnormal distribution of proliferating neural progenitors in schizophrenia patient–derived cerebral organoids with reduced expression of nuclear FGFR1 in cortical neurons and abnormal cortical neurogenesis. These studies represent the utility of cerebral organoids to study dysregulated brain development but have been limited in their use to study more complex microglial-neuronal interactions due mainly to the limited contribution of microglia. Brain organoids are typically established through selective induction favoring neuroectoderm (118) while reducing the contribution of mesodermal derivatives, including microglia. However, small contributions of microglial cells have been demonstrated in organoids, likely as a result of incomplete neuroectodermal induction and/or mesodermal contribution present as already partially differentiated during their initial formation. These innate microglia were described by Ormel et al. (47) in a study that showed multilineage progenitors during the early stages of organoid development that contributed to the formation of microglia-like cells associated with neurons and engulfed synaptic markers suggesting pruning. Although promising, these innate microglia contribute a small fraction of the organoids that is highly variable between organoids, limiting their reproducibility. In addition, mix and match types of experiments, e.g., using microglia derived from patients in a healthy control neuronal background, are not feasible with these procedures because organoid self-assembly is derived from a single starting population of cells. Recent efforts have been initiated to control the microglial contribution through the co-culture of induced microglia with cerebral organoids showing some modest spontaneous microglial engraftment into the organoid (119) or through the production of assembloids in which cellular spheroids are preformed with multilineage progenitors (120) to control microglia/neuron ratio within each organoid (121). While cerebral organoid and assembloid techniques have improved recently and will allow further adaptation of patient-derived models of schizophrenia, currently the state of the art for modeling of synaptic pruning in these models remains challenging: it is difficult to develop robust assays given high heterogeneity among organoids (122), difficulty imaging more complex systems that entail approaches more similar to those in animal models, and in particular very low throughput.
MICROGLIA ENGULFMENT OF ISOLATED PATIENT-DERIVED SYNAPTOSOMES TO MODEL IN VITRO SYNAPTIC PRUNING
Building upon traditional approaches for studying synaptic function directly in cultured neurons, purified synaptic nerve terminals (synaptosomes) can facilitate large-scale functional (123) in vitro assays to model microglial processes such as synaptic pruning. Synaptosomes have been used to study synaptic function since first isolated in the 1950s (124). Following homogenization and fractionation (125), synaptosomal preparations are enriched in isolated pre- and post-synaptic components retaining their in vivo functions, including synaptic mitochondrial bioenergetics and respiration (126,127), neurotransmitter release (including glutamate), and endocytosis activities (123,128,129), and contain synaptic nucleotides including noncoding circular RNAs (130) and microRNAs (131). In addition, synaptosomal preparations can be cryopreserved while retaining their activity (132) to be banked as a ready-to-use reagent for downstream assays. Synaptosomes derived from postmortem patients have been used to model schizophrenia for the study of synaptic-specific protein levels and RNA expression specifically in synapses (131,133), aberrant synaptic protein content (134,135), synaptic proteomic studies (136,137), glutamate release (138,139), and complement deposition (140). More recently, synaptosomes isolated from iPSC-derived neuronal cultures have allowed for patient-specific in vitro models of synaptic dysfunction (141,142). The purity and ease of synaptosome preps allow for high-throughput quantitation and characterization of engulfment as well as increase of the signal to noise in comparison to using intact neural cultures. Until recently (99,100), they have not been used to generate models of schizophrenia patient–derived microglia-mediated synaptic pruning assays amenable to high throughput.
EVIDENCE FOR ABERRANT MICROGLIAL SYNAPTIC PRUNING IN FROM IN VITRO SCHIZOPHRENIA PATIENT–DERIVED MODELS
The application of these model systems facilitates study of microglial and neuronal contributions to aberrant synaptic pruning in a scalable manner. We recently developed and validated an in vitro model of microglia-mediated synapse engulfment using a large bank of patient-derived hiMGs in combination with correspondent iPSC-derived neuronal cultures and isolated synaptosomes to study aberrant synaptic engulfment in model systems derived from patients with schizophrenia (100).
To model synaptic engulfment at scale, iMG engulfment of purified synaptosomes derived from patient and control iPSC-derived neuronal cultures was measured using a pH-sensitive dye (pHrodo) indicating synaptosomal engulfment (Figure 1D) localized to acidic lysosomes, further confirmed by end point confocal microscopy of synaptic puncta inside cell bodies. To assess differences in synaptic vesicle engulfment between patient and control models, we compared pure disease models (both iMGs and synaptosomes derived from patients) to pure control models comprising both components derived from healthy control subjects. Measurement of phagocytic indices in these iMG-synaptosome models determined an increase in engulfment in the patient models compared with control subjects further confirmed in iMG-neuronal co-cultures. Furthermore, having distinct models from both patients and control subjects allowed for the generation of mixed models in which the synaptic structures from patients were added to iMGs derived from control subjects and vice versa to further demonstrate that the observed increased synapse engulfment in patient models can be attributed to both neural and microglial influences (143).
This study further evaluated the contributions of schizophrenia risk variants in the human complement C4 locus of the patient genotypes. Schizophrenia risk–associated variants within the C4 locus have been suggested to be associated with increased neuronal complement deposition and synapse up-take, in particular an increased copy number of the long-form C4A isotype (C4AL), showing a correlation with increased expression in human postmortem brain with schizophrenia risk (15). While excessive synapse elimination by microglia via a C4-dependent mechanism has been suggested in schizophrenia, this correlation had not been directly examined in vitro using patient-derived cellular models. We had previously shown that synaptosome engulfment by iMGs in vitro was complement dependent (99); therefore, this study further sought to determine if differences in C4 copy number (in particular the C4L isoform) resulted in higher C4 expression in iPSC-neuronal culture and whether this expression can influence uptake in isolated synaptic structures. Using patient iPSC–derived neuronal cultures, neuron-specific C4 expression, as measured using an indirect C3 complement deposition assay, was shown to strongly correlate with C4AL copy number. By stratifying the genotypes of the neuronal cultures used to isolate synaptosomes by C4AL copy number, a strong correlation was observed between C4AL copy number and iMG engulfment in this functional assay. These observations were exclusive to the neuronal component because no correlation was determined by adjusting for the C4AL copy number of the input iMGs.
Taken together, these observations support the use of in vitro model systems with patient-derived iMGs and neuronal models to develop robust and reproducible assays (Figure 1D) to further understand mechanisms of microglial-mediated synaptic pruning in genetically representative backgrounds. In addition, the scalability of these assays and the ability to easily manipulate them make them highly amenable to screening efforts for novel therapeutic modulators of synaptic pruning. Further adaptation of these in vitro cellular models of synaptic pruning (for example, through the use of genetically engineered iPSC-derived neurons and/or iMGs to manipulate individual genes implicated in schizophrenia) will enable further understanding of the potential role of genes involved in microglial-mediated synaptic refinement during normal and aberrant development implicated in schizophrenia.
MODEL FOR NOVEL DRUG/THERAPEUTIC DISCOVERY
If, as convergent lines of evidence suggest, schizophrenia is at least in part a disorder of pruning, it follows that modulators of pruning could arrest, or even reverse, disease progression. The goal of such a therapeutic strategy need not be complete blockade of pruning; indeed, a modest shift in trajectory would likely be sufficient to reduce the observed neuropathology. While beyond the scope of this review, the broader use of pruning modulators, and modulators of microglia activity more generally, for a host of neurodevelopmental and neurodegenerative disorders should be apparent.
The most obvious target to modulate pruning is the complement system, particularly C4A. This target has been celebrated as an illustration of the power of genome-wide association studies to identify treatment targets, but C4A may be the exception that proves the rule: more than a decade after the identification of the first risk loci for schizophrenia, there is not a single targeted therapeutic for schizophrenia in human studies based on genome-wide studies. Whether continued efforts to boil the genomic ocean will more clearly resolve candidate pathways beyond the synaptic biology already strongly implicated in schizophrenia and related disorders remains to be seen. Notably, cellular models may also facilitate efforts to understand the mechanisms by which these variants mediate disease risk, for example, by applying CRISPR (clustered regularly interspaced short palindromic repeats) to manipulate risk loci (144), laying the groundwork for pathway-focused screens.
An alternative to pathway-based screening is to identify modulators on the basis of functional assays such as those described, further followed by transcriptomics to discover key effected pathways upon modulation (Figure 1D). Key to their application is scalability, the ability to generate large numbers of reagents to enable screening in 96- or 384-well format. A further advantage is the simplicity of readout (e.g., imaging followed by simple quantification) and large signal-to-noise ratio that can be routinely performed in academic laboratories without robotics.
Demonstrating the potential use of these assays for screening, we previously demonstrated that the antibiotic minocycline is a potent inhibitor of phagocytosis in these in vitro microglia models (100). Minocycline was examined on the basis of its putative anti-inflammatory mechanism, as well as prior enthusiasm for its investigation in schizophrenia more broadly. To follow up these in vitro findings, we hypothesized that exposure to minocycline or doxycycline during adolescence, where they are commonly used for the treatment of acne vulgaris, might be associated with diminished risk of subsequent schizophrenia diagnosis. Using electronic health records, we found a significant reduction in risk (100). While preliminary, given the nonrandom design and substantial risk of confounding, this finding illustrates the power of in vitro investigations to nominate compounds for repositioning in schizophrenia. More generally, the minocycline example illustrates the importance of validating results of cellular models in other systems, showing that effects identified in vitro can be extrapolated to in vivo studies. Such assays could be implemented using standard cell lines, but the inclusion of patient-derived reagents allows for screens that investigate specificity of effect. For example, an investigator might elect to prioritize for follow-up compounds that show greater effects in schizophrenia-derived microglia or against schizophrenia-derived synaptosomes. Secondary screens could also consider patient-derived lines from treatment-resistant schizophrenia, aiming to identify compounds with greatest activity in particular patient subpopulations.
THERAPEUTIC INTERVENTION
A key question in the application of pruning modulation is the optimal period for intervention. For neurodevelopmental disorders, it might be the case that modulation is required in early childhood or even in utero. The recognition that in utero environmental effects contribute to schizophrenia liability argues at minimum that early brain development contributes to risk (145). In contrast, as noted above, exposure to minocycline or doxycycline in adolescence was associated with diminished risk of schizophrenia or other psychotic disorders, suggesting the possibility of later intervention. Other lines of investigation also suggest that later interventions (146) can still diminish transition to psychosis. To date, the majority of clinical trials aimed at prevention of psychotic disorders enroll individuals who are already symptomatic, even if they have not developed a diagnosable disorder per se. This reflects in part a reluctance to expose presymptomatic individuals—and children more generally—to novel therapeutics. To justify exposing this population to risk, it is necessary to identify individuals who are at sufficiently high risk for developing disease to justify interventions that themselves carry risk.
Enrolling clinical high-risk participants—primarily children with first-degree relatives with schizophrenia—is one such strategy. An emerging alternative is the application of polygenic risk scores for schizophrenia, although these scores still capture only a fraction of disease liability (147,148). Patient-derived models of pruning may offer a third alternative as a functional biomarker of pruning integrity. That is, if peripheral blood can be applied to create a patient-specific pruning model, it might enable identification of individuals most likely to benefit from a modulator of pruning. Such biomarkers may also enable more powerful proof-of-concept studies, with parallel in vivo (in patient) and in vitro (in culture) characterization of response to a candidate therapeutic. Likewise, generation of microglia-like cells from cord blood at birth (106), enabling estimation of microglia activity, may facilitate risk stratification. If the association between activity in these cells and subsequent neurodevelopmental disease can be confirmed, it may be possible to intervene early on the basis of such a diagnostic tool or, at minimum, to identify high-risk individuals in whom novel therapeutics may have a greater ratio of benefit to risk.
ACKNOWLEDGMENTS AND DISCLOSURES
This work was supported by the National Institutes of Health (Grant No. R01-MH120227 [to RHP, principal investigator]).
RHP has received personal fees from Burrage Capital, RID Ventures, Genomind, Takeda, and Psy Therapeutics unrelated to the work described; he also holds equity in Psy Therapeutics. SDS has received personal fees from Thrive Bioscience, Inc. unrelated to the work described. JEH reports no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014): Biological insights from 108 schizophrenia-associated genetic loci. Nature 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Walters JTR, O’Donovan MC (2020): Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv. 10.1101/2020.09.12.20192922. [DOI] [Google Scholar]
- 3.Faust TE, Gunner G, Schafer DP (2021): Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat Rev Neurosci 22:657–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riccomagno MM, Kolodkin AL (2015): Sculpting neural circuits by axon and dendrite pruning. Annu Rev Cell Dev Biol 31:779–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konopaske GT, Lange N, Coyle JT, Benes FM (2014): Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry 71:1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, et al. (2007): The classical complement cascade mediates CNS synapse elimination. Cell 131:1164–1178. [DOI] [PubMed] [Google Scholar]
- 7.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. (2012): Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74:691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. (2011): Synaptic pruning by microglia is necessary for normal brain development. Science 333:1456–1458. [DOI] [PubMed] [Google Scholar]
- 9.Aguzzi A, Barres BA, Bennett ML (2013): Microglia: Scapegoat, saboteur, or something else? Science 339:156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg I (1982–1983): Schizophrenia: Caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res 17:319–334. [DOI] [PubMed] [Google Scholar]
- 11.Glausier JR, Lewis DA (2013): Dendritic spine pathology in schizophrenia. Neuroscience 251:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDonald ML, Alhassan J, Newman JT, Richard M, Gu H, Kelly RM, et al. (2017): Selective loss of smaller spines in schizophrenia. Am J Psychiatry 174:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannon TD, Chung Y, He G, Sun D, Jacobson A, van Erp TGM, et al. (2015): Progressive reduction in cortical thickness as psychosis develops: A multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol Psychiatry 77:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloomfield PS, Selvaraj S, Veronese M, Rizzo G, Bertoldo A, Owen DR, et al. (2016): Microglial activity in people at ultra high risk of psychosis and in schizophrenia: An [(11)C] PBR28 PET brain imaging study [published correction appears in Am J Psychiatry 2017; 174: 402]. Am J Psychiatry 173:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. (2016): Schizophrenia risk from complex variation of complement component 4 [published correction appears in Nature 2022; 601:E4–E5]. Nature 530:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM, et al. (1998): Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry 65:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glantz LA, Lewis DA (2000): Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry 57:65–73. [DOI] [PubMed] [Google Scholar]
- 18.Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, et al. (2019): Cross-species single-cell analysis reveals divergence of the primate microglia program [published correction appears in Cell 2020; 181:746]. Cell 179:1609–1622.e16. [DOI] [PubMed] [Google Scholar]
- 19.Hayes GM, Woodroofe MN, Cuzner ML (1988): Characterisation of microglia isolated from adult human and rat brain. J Neuroimmunol 19:177–189. [DOI] [PubMed] [Google Scholar]
- 20.Hassan NF, Campbell DE, Rifat S, Douglas SD (1991): Isolation and characterization of human fetal brain-derived microglia in in vitro culture. Neuroscience 41:149–158. [DOI] [PubMed] [Google Scholar]
- 21.Cardona AE, Huang D, Sasse ME, Ransohoff RM (2006): Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc 1:1947–1951. [DOI] [PubMed] [Google Scholar]
- 22.Pan J, Wan J (2020): Methodological comparison of FACS and MACS isolation of enriched microglia and astrocytes from mouse brain. J Immunol Methods 486:112834. [DOI] [PubMed] [Google Scholar]
- 23.Holt LM, Stoyanof ST, Olsen ML (2019): Magnetic cell sorting for in vivo and in vitro astrocyte, neuron, and microglia analysis. Curr Protoc Neurosci 88:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordt EA, Block CL, Petrozziello T, Sadri-Vakili G, Smith CJ, Edlow AG, Bilbo SD (2020): Isolation of microglia from mouse or human tissue. STAR Protoc 1:100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. (2016): New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113:E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. (2017): An environment-dependent transcriptional network specifies human microglia identity. Science 356:eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizee MR, Miedema SSM, van der Poel M, Adelia, Schuurman KG, van Strien ME, et al. (2017): Isolation of primary microglia from the human post-mortem brain: Effects of ante- and post-mortem variables. Acta Neuropathol Commun 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007): Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. [DOI] [PubMed] [Google Scholar]
- 29.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L (2009): Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling [published correction appears in Nat Biotechnol 2009; 27:485]. Nat Biotechnol 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theka I, Caiazzo M, Dvoretskova E, Leo D, Ungaro F, Curreli S, et al. (2013): Rapid generation of functional dopaminergic neurons from human induced pluripotent stem cells through a single-step procedure using cell lineage transcription factors. Stem Cells Transl Med 2:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng YH, Chanda S, Janas JA, Yang N, Kokubu Y, Südhof TC, Wernig M (2021): Efficient generation of dopaminergic induced neuronal cells with midbrain characteristics. Stem Cell Reports 16:1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundberg M, Pinson H, Smith RS, Winden KD, Venugopal P, Tai DJC, et al. (2021): 16p11.2 deletion is associated with hyperactivation of human iPSC-derived dopaminergic neuron networks and is rescued by RHOA inhibition in vitro. Nat Commun 12:2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, et al. (2017): Generation of pure GABAergic neurons by transcription factor programming. Nat Methods 14:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun AX, Yuan Q, Tan S, Xiao Y, Wang D, Khoo ATT, et al. (2016): Direct induction and functional maturation of forebrain GABAergic neurons from human pluripotent stem cells. Cell Rep 16:1942–1953. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. (2013): Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao SY, Hu Y, Chen C, Yuan F, Xu M, Li Q, et al. (2017): Enhanced derivation of human pluripotent stem cell-derived cortical glutamatergic neurons by a small molecule. Sci Rep 7:3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J, Zhong X, Liu H, Hao L, Huang CT-L, Sherafat MA, et al. (2016): Generation of serotonin neurons from human pluripotent stem cells. Nat Biotechnol 34:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vadodaria KC, Stern S, Marchetto MC, Gage FH (2018): Serotonin in psychiatry: In vitro disease modeling using patient-derived neurons. Cell Tissue Res 371:161–170. [DOI] [PubMed] [Google Scholar]
- 39.Ni P, Noh H, Park GH, Shao Z, Guan Y, Park JM, et al. (2020): iPSC-derived homogeneous populations of developing schizophrenia cortical interneurons have compromised mitochondrial function [published correction appears in Mol Psychiatry 2020; 25: 3103–3104]. Mol Psychiatry 25:2873–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allison T, Langerman J, Sabri S, Otero-Garcia M, Lund A, Huang J, et al. (2021): Defining the nature of human pluripotent stem cell-derived interneurons via single-cell analysis. Stem Cell Reports 16:2548–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. (2010): Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. (2012): A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336:86–90. [DOI] [PubMed] [Google Scholar]
- 43.Hoeffel G, Ginhoux F (2018): Fetal monocytes and the origins of tissue-resident macrophages. Cell Immunol 330:5–15. [DOI] [PubMed] [Google Scholar]
- 44.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. (2016): Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22:1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. (2017): iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94:278–293.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. (2017): A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports 8:1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. (2018): Microglia innately develop within cerebral organoids. Nat Commun 9:4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen SW, Hung YS, Fuh JL, Chen NJ, Chu YS, Chen SC, et al. (2021): Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors. Stem Cell Reports 16:1363–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svoboda DS, Barrasa MI, Shu J, Rietjens R, Zhang S, Mitalipova M, et al. (2019): Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc Natl Acad Sci U S A 116:25293–25303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu R, Li X, Boreland AJ, Posyton A, Kwan K, Hart RP, Jiang P (2020): Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat Commun 11:1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fattorelli N, Martinez-Muriana A, Wolfs L, Geric I, De Strooper B, Mancuso R (2021): Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat Protoc 16:1013–1033. [DOI] [PubMed] [Google Scholar]
- 52.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, et al. (2007): Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1:55–70. [DOI] [PubMed] [Google Scholar]
- 53.Hewitt KJ, Garlick JA (2013): Cellular reprogramming to reset epigenetic signatures. Mol Aspects Med 34:841–848. [DOI] [PubMed] [Google Scholar]
- 54.Smigielski L, Jagannath V, Rössler W, Walitza S, Grünblatt E (2020): Epigenetic mechanisms in schizophrenia and other psychotic disorders: A systematic review of empirical human findings. Mol Psychiatry 25:1718–1748. [DOI] [PubMed] [Google Scholar]
- 55.Richetto J, Meyer U (2021): Epigenetic modifications in schizophrenia and related disorders: Molecular scars of environmental exposures and source of phenotypic variability. Biol Psychiatry 89:215–226. [DOI] [PubMed] [Google Scholar]
- 56.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. (2010): Epigenetic memory in induced pluripotent stem cells. Nature 467:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, et al. (2010): Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28:848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. (2011): Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells [published correction appears in Nature 2014; 514:126]. Nature 471:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, et al. (2011): Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13:541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suhr ST, Chang EA, Tjong J, Alcasid N, Perkins GA, Goissis MD, et al. (2010): Mitochondrial rejuvenation after induced pluripotency. PLoS One 5:e14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J (2010): The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28:721–733. [DOI] [PubMed] [Google Scholar]
- 62.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, et al. (2009): Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4:141–154. [DOI] [PubMed] [Google Scholar]
- 63.Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, et al. (2009): Telomere dynamics in human cells reprogrammed to pluripotency [published correction appears in PLoS One 2010; 5]. PLoS One 4:e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mertens J, Reid D, Lau S, Kim Y, Gage FH (2018): Aging in a dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu Rev Genet 52:271–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahmoudi S, Brunet A (2012): Aging and reprogramming: A two-way street. Curr Opin Cell Biol 24:744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Z, Jiang H, Zhong P, Yan Z, Chen S, Feng J (2016): Direct conversion of human fibroblasts to induced serotonergic neurons. Mol Psychiatry 21:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. (2011): Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A 108:10343–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. (2011): Induction of human neuronal cells by defined transcription factors. Nature 476:220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cates K, McCoy MJ, Kwon JS, Liu Y, Abernathy DG, Zhang B, et al. (2021): Deconstructing stepwise fate conversion of human fibroblasts to neurons by microRNAs. Cell Stem Cell 28:127–140.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richner M, Victor MB, Liu Y, Abernathy D, Yoo AS (2015): MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nat Protoc 10:1543–1555. [DOI] [PubMed] [Google Scholar]
- 71.Hu W, Qiu B, Guan W, Wang Q, Wang M, Li W, et al. (2015): Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17:204–212. [DOI] [PubMed] [Google Scholar]
- 72.Yang Y, Chen R, Wu X, Zhao Y, Fan Y, Xiao Z, et al. (2019): Rapid and efficient conversion of human fibroblasts into functional neurons by small molecules. Stem Cell Reports 13:862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium (2011): Genome-wide association study identifies five new schizophrenia loci. Nat Genet 43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gardiner EJ, Cairns MJ, Liu B, Beveridge NJ, Carr V, Kelly B, et al. (2013): Gene expression analysis reveals schizophrenia-associated dysregulation of immune pathways in peripheral blood mononuclear cells. J Psychiatr Res 47:425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petralia MC, Ciurleo R, Saraceno A, Pennisi M, Basile MS, Fagone P, et al. (2020): Meta-analysis of transcriptomic data of dorsolateral prefrontal cortex and of peripheral blood mononuclear cells identifies altered pathways in schizophrenia. Genes (Basel) 11:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goossens J, Morrens M, Coppens V (2021): The potential use of peripheral blood mononuclear cells as biomarkers for treatment response and outcome prediction in psychiatry: A systematic review. Mol Diagn Ther 25:283–299. [DOI] [PubMed] [Google Scholar]
- 77.Rollins B, Martin MV, Morgan L, Vawter MP (2010): Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet 153B:919–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leirer DJ, Iyegbe CO, Di Forti M, Patel H, Carra E, Fraietta S, et al. (2019): Differential gene expression analysis in blood of first episode psychosis patients. Schizophr Res 209:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song X, Liu Y, Pu J, Gui S, Zhong X, Chen X, et al. (2021): Transcriptomics analysis reveals shared pathways in peripheral blood mononuclear cells and brain tissues of patients with schizophrenia. Front Psychiatry 12:716722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun XY, Lu J, Zhang L, Song HT, Zhao L, Fan HM, et al. (2015): Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J Clin Neurosci 22:570–574. [DOI] [PubMed] [Google Scholar]
- 81.Beveridge NJ, Cairns MJ (2012): MicroRNA dysregulation in schizophrenia. Neurobiol Dis 46:263–271. [DOI] [PubMed] [Google Scholar]
- 82.Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ (2012): Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry 17:827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. (2011): MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One 6:e21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ghafouri-Fard S, Eghtedarian R, Taheri M, Brühl AB, Sadeghi-Bahmani D, Brand S (2021): A review on the expression of noncoding RNAs in patients with schizophrenia: With a special focus on peripheral blood as a source of expression analysis. Front Psychiatry 12:640463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gavin DP, Sharma RP (2010): Histone modifications, DNA methylation, and schizophrenia. Neurosci Biobehav Rev 34:882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gavin DP, Sharma RP (2009): Chromatin from peripheral blood mononuclear cells as biomarkers for epigenetic abnormalities in schizophrenia. Cardiovasc Psychiatry Neurol 2009:409562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu ML, Zhang XT, Du XY, Fang Z, Liu Z, Xu Y, et al. (2015): Severe disturbance of glucose metabolism in peripheral blood mononuclear cells of schizophrenia patients: A targeted metabolomic study. J Transl Med 13:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gubert C, Stertz L, Pfaffenseller B, Panizzutti BS, Rezin GT, Massuda R, et al. (2013): Mitochondrial activity and oxidative stress markers in peripheral blood mononuclear cells of patients with bipolar disorder, schizophrenia, and healthy subjects. J Psychiatr Res 47:1396–1402. [DOI] [PubMed] [Google Scholar]
- 89.Herberth M, Koethe D, Cheng TMK, Krzyszton ND, Schoeffmann S, Guest PC, et al. (2011): Impaired glycolytic response in peripheral blood mononuclear cells of first-onset antipsychotic-naive schizophrenia patients. Mol Psychiatry 16:848–859. [DOI] [PubMed] [Google Scholar]
- 90.Liu ML, Zheng P, Liu Z, Xu Y, Mu J, Guo J, et al. (2014): GC-MS based metabolomics identification of possible novel biomarkers for schizophrenia in peripheral blood mononuclear cells. Mol Biosyst 10:2398–2406. [DOI] [PubMed] [Google Scholar]
- 91.Schmidtmayer J, Jacobsen C, Miksch G, Sievers J (1994): Blood monocytes and spleen macrophages differentiate into microglia-like cells on monolayers of astrocytes: Membrane currents. Glia 12:259–267. [DOI] [PubMed] [Google Scholar]
- 92.Sievers J, Parwaresch R, Wottge HU (1994): Blood monocytes and spleen macrophages differentiate into microglia-like cells on monolayers of astrocytes: Morphology. Glia 12:245–258. [DOI] [PubMed] [Google Scholar]
- 93.Leone C, Le Pavec G, Même W, Porcheray F, Samah B, Dormont D, Gras G (2006): Characterization of human monocyte-derived microglia-like cells. Glia 54:183–192. [DOI] [PubMed] [Google Scholar]
- 94.Etemad S, Zamin RM, Ruitenberg MJ, Filgueira L (2012): A novel in vitro human microglia model: Characterization of human monocyte-derived microglia. J Neurosci Methods 209:79–89. [DOI] [PubMed] [Google Scholar]
- 95.Ohgidani M, Kato TA, Setoyama D, Sagata N, Hashimoto R, Shigenobu K, et al. (2014): Direct induction of ramified microglia-like cells from human monocytes: Dynamic microglial dysfunction in Nasu-Hakola disease. Sci Rep 4:4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Banerjee A, Lu Y, Do K, Mize T, Wu X, Chen X, Chen J (2021): Validation of induced microglia-like cells (iMG cells) for future studies of brain diseases. Front Cell Neurosci 15:629279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Noto D, Sakuma H, Takahashi K, Saika R, Saga R, Yamada M, et al. (2014): Development of a culture system to induce microglia-like cells from haematopoietic cells. Neuropathol Appl Neurobiol 40:697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ormel PR, Böttcher C, Gigase FAJ, Missall RD, van Zuiden W, Fernández Zapata MC, et al. (2020): A characterization of the molecular phenotype and inflammatory response of schizophrenia patient-derived microglia-like cells. Brain Behav Immun 90:196–207. [DOI] [PubMed] [Google Scholar]
- 99.Sellgren CM, Sheridan SD, Gracias J, Xuan D, Fu T, Perlis RH (2017): Patient-specific models of microglia-mediated engulfment of synapses and neural progenitors. Mol Psychiatry 22:170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, et al. (2019): Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci 22:374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ryan KJ, White CC, Patel K, Xu J, Olah M, Replogle JM, et al. (2017): A human microglia-like cellular model for assessing the effects of neurodegenerative disease gene variants. Sci Transl Med 9:eaai7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grimpe B, Probst JC, Hager G (1999): Suppression of nidogen-1 translation by antisense targeting affects the adhesive properties of cultured astrocytes. Glia 28:138–149. [DOI] [PubMed] [Google Scholar]
- 103.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. (2015): A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA 112:7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. (2014): Identification of a unique TGF-b-dependent molecular and functional signature in microglia [published correction appears in Nat Neurosci 2014; 17:1286]. Nat Neurosci 17:131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reichert F, Rotshenker S (2003): Complement-receptor-3 and scavenger-receptor-AI/II mediated myelin phagocytosis in microglia and macrophages. Neurobiol Dis 12:65–72. [DOI] [PubMed] [Google Scholar]
- 106.Sheridan SD, Thanos JM, De Guzman RM, McCrea LT, Horng JE, Fu T, et al. (2021): Umbilical cord blood-derived microglia-like cells to model COVID-19 exposure. Transl Psychiatry 11:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tiihonen J, Koskuvi M, Storvik M, Hyötyläinen I, Gao Y, Puttonen KA, et al. (2019): Sex-specific transcriptional and proteomic signatures in schizophrenia. Nat Commun 10:3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han J, Fan Y, Zhou K, Blomgren K, Harris RA (2021): Uncovering sex differences of rodent microglia. J Neuroinflammation 18:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hui CW, Vecchiarelli HA, Gervais É, Luo X, Michaud F, Scheefhals L, et al. (2020): Sex differences of microglia and synapses in the hippocampal dentate gyrus of adult mouse offspring exposed to maternal immune activation. Front Cell Neurosci 14:558181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grubman A, Vandekolk TH, Schröder J, Sun G, Hatwell-Humble J, Chan J, et al. (2020): A CX3CR1 reporter hESC line facilitates integrative analysis of in-vitro-derived microglia and improved microglia identity upon neuron-glia co-culture. Stem Cell Reports 14:1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guttikonda SR, Sikkema L, Tchieu J, Saurat N, Walsh RM, Harschnitz O, et al. (2021): Fully defined human pluripotent stem cell-derived microglia and tri-culture system model C3 production in Alzheimer’s disease. Nat Neurosci 24:343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mellios N, Feldman DA, Sheridan SD, Ip JPK, Kwok S, Amoah SK, et al. (2018): Mop-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry 23:1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baldassari S, Musante I, Iacomino M, Zara F, Salpietro V, Scudieri P (2020): Brain organoids as model systems for genetic neurodevelopmental disorders. Front Cell Dev Biol 8:590119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Venkataraman L, Fair SR, McElroy CA, Hester ME, Fu H (2022): Modeling neurodegenerative diseases with cerebral organoids and other three-dimensional culture systems: Focus on Alzheimer’s disease. Stem Cell Rev Rep 18:696–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pellegrini L, Lancaster MA (2021): Modeling neurodegeneration with mutant-tau organoids. Cell 184:4377–4379. [DOI] [PubMed] [Google Scholar]
- 116.Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, et al. (2017): DISC1 regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during mitosis [published correction appears in Neuron 2017; 96:1204]. Neuron 96:1041–1054.e5.29103808 [Google Scholar]
- 117.Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, et al. (2017): Cerebral organoids reveal early cortical maldevelopment in schizophrenia—Computational anatomy and genomics, role of FGFR1. Transl Psychiatry 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. (2013): Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song L, Yuan X, Jones Z, Vied C, Miao Y, Marzano M, et al. (2019): Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci Rep 9:11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Paşca SP (2019): Assembling human brain organoids. Science 363:126–127. [DOI] [PubMed] [Google Scholar]
- 121.Xu R, Boreland AJ, Li X, Erickson C, Jin M, Atkins C, et al. (2021): Developing human pluripotent stem cell-based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. Stem Cell Reports 16:1923–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hernández D, Rooney LA, Daniszewski M, Gulluyan L, Liang HH, Cook AL, et al. (2022): Culture variabilities of human iPSC-derived cerebral organoids are a major issue for the modelling of phenotypes observed in Alzheimer’s disease. Stem Cell Rev Rep 18:718–731. [DOI] [PubMed] [Google Scholar]
- 123.Daniel JA, Malladi CS, Kettle E, McCluskey A, Robinson PJ (2012): Analysis of synaptic vesicle endocytosis in synaptosomes by high-content screening. Nat Protoc 7:1439–1455. [DOI] [PubMed] [Google Scholar]
- 124.Whittaker VP, Michaelson IA, Kirkland RJ (1964): The separation of synaptic vesicles from nerve-ending particles (‘synaptosomes’). Biochem J 90:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tenreiro P, Rebelo S, Martins F, Santos M, Coelho ED, Almeida M, et al. (2017): Comparison of simple sucrose and Percoll based methodologies for synaptosome enrichment. Anal Biochem 517:1–8. [DOI] [PubMed] [Google Scholar]
- 126.Khattar NK, Yablonska S, Baranov SV, Baranova OV, Kretz ES, Larkin TM, et al. (2016): Isolation of functionally active and highly purified neuronal mitochondria from human cortex. J Neurosci Methods 263:1–6. [DOI] [PubMed] [Google Scholar]
- 127.Lores-Arnaiz S, Rodríguez de Lores Arnaiz G, Karadayian AG, Bustamante J (2018): Synaptosome bioenergetics and calcium handling: Aging response. In: Murphy KM, editor. Synaptosomes. New York: Humana Press, 131–151. [Google Scholar]
- 128.Nicholls DG (2010): Stochastic aspects of transmitter release and bioenergetic dysfunction in isolated nerve terminals. Biochem Soc Trans 38:457–459. [DOI] [PubMed] [Google Scholar]
- 129.Johansen L, Roberg B, Kvamme E (1987): Uptake and release for glutamine and glutamate in a crude synaptosomal fraction from rat brain. Neurochem Res 12:135–140. [DOI] [PubMed] [Google Scholar]
- 130.Curry-Hyde A, Ueberham U, Chen BJ, Zipfel I, Mills JD, Bochmann J, et al. (2020): Analysis of the circular transcriptome in the synaptosomes of aged mice. Neuroscience 449:202–213. [DOI] [PubMed] [Google Scholar]
- 131.Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, Dwivedi Y (2014): Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS One 9:e86469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gleitz J, Beile A, Wilffert B, Tegtmeier F (1993): Cryopreservation of freshly isolated synaptosomes prepared from the cerebral cortex of rats. J Neurosci Methods 47:191–197. [DOI] [PubMed] [Google Scholar]
- 133.Talbot K, Louneva N, Cohen JW, Kazi H, Blake DJ, Arnold SE (2011): Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS One 6:e16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thompson PM, Sower AC, Perrone-Bizzozero NI (1998): Altered levels of the synaptosomal associated protein SNAP-25 in schizophrenia. Biol Psychiatry 43:239–243. [DOI] [PubMed] [Google Scholar]
- 135.Fatemi SH, Earle JA, Stary JM, Lee S, Sedgewick J (2001): Altered levels of the synaptosomal associated protein SNAP-25 in hippo-campus of subjects with mood disorders and schizophrenia. Neuroreport 12:3257–3262. [DOI] [PubMed] [Google Scholar]
- 136.MacDonald ML, Garver M, Newman J, Sun Z, Kannarkat J, Salisbury R, et al. (2020): Synaptic proteome alterations in the primary auditory cortex of individuals with schizophrenia. JAMA Psychiatry 77:86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Velásquez E, Nogueira FCS, Velásquez I, Schmitt A, Falkai P, Domont GB, Martins-de-Souza D (2017): Synaptosomal proteome of the orbitofrontal cortex from schizophrenia patients using quantitative label-free and iTRAQ-based shotgun proteomics. J Proteome Res 16:4481–4494. [DOI] [PubMed] [Google Scholar]
- 138.Sherman AD, Hegwood TS, Baruah S, Waziri R (1991): Deficient NMDA-mediated glutamate release from synaptosomes of schizophrenics. Biol Psychiatry 30:1191–1198. [DOI] [PubMed] [Google Scholar]
- 139.Sherman AD, Davidson AT, Baruah S, Hegwood TS, Waziri R (1991): Evidence of glutamatergic deficiency in schizophrenia. Neurosci Lett 121:77–80. [DOI] [PubMed] [Google Scholar]
- 140.Györffy BA, Kun J, Török G, Bulyáki É, Borhegyi Z, Gulyássy P, et al. (2018): Local apoptotic-like mechanisms underlie complement-mediated synaptic pruning. Proc Natl Acad Sci U S A 115:6303–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Odaka H, Numakawa T, Soga M, Kido J, Matsumoto S, Kajihara R, et al. (2021): An iPSC-based neural model of sialidosis uncovers glycolytic impairment-causing presynaptic dysfunction and deregulation of Ca2+ dynamics. Neurobiol Dis 152:105279. [DOI] [PubMed] [Google Scholar]
- 142.Prots I, Grosch J, Brazdis RM, Simmnacher K, Veber V, Havlicek S, et al. (2018):α-Synuclein oligomers induce early axonal dysfunction in human iPSC-based models of synucleinopathies. Proc Natl Acad Sci U S A 115:7813–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang M, Zhang L, Gage FH (2019): Microglia, complement and schizophrenia. Nat Neurosci 22:333–334. [DOI] [PubMed] [Google Scholar]
- 144.Deans PJM, Brennand KJ (2021): Applying stem cells and CRISPR engineering to uncover the etiology of schizophrenia. Curr Opin Neurobiol 69:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Al-Haddad BJS, Jacobsson B, Chabra S, Modzelewska D, Olson EM, Bernier R, et al. (2019): Long-term risk of neuropsychiatric disease after exposure to infection in utero. JAMA Psychiatry 76:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bosnjak Kuharic D, Kekin I, Hew J, Rojnic Kuzman M, Puljak L (2019): Interventions for prodromal stage of psychosis. Cochrane Database Syst Rev 2019:CD012236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Visscher PM, Yengo L, Cox NJ, Wray NR (2021): Discovery and implications of polygenicity of common diseases. Science 373:1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Murray GK, Lin T, Austin J, McGrath JJ, Hickie IB, Wray NR (2021): Could polygenic risk scores be useful in psychiatry?: A review. JAMA Psychiatry 78:210–219. [DOI] [PubMed] [Google Scholar]


