Abstract
Objective
The development of effective cancer treatments depends on the availability of cell lines that faithfully recapitulate the cancer in question. This study definitively re-assigns the histologic identities of two ovarian cancer cell lines, COV434 (originally described as a granulosa cell tumour) and TOV-112D (originally described as grade 3 endometrioid carcinoma), both of which were recently suggested to represent small cell carcinoma of the ovary, hypercalcemic type (SCCOHT), based on their shared gene expression profiles and sensitivity to EZH2 inhibitors.
Methods
For COV434 and TOV-112D, we re-reviewed the original pathology slides and obtained clinical follow-up on the patients, when available, and performed immunohistochemistry for SMARCA4, SMARCA2 and additional diagnostic markers on the original formalin-fixed, paraffin-embedded (FFPE) clinical material, when available. For COV434, we further performed whole exome sequencing and validated SMARCA4 mutations by Sanger sequencing. We studied the growth of the cell lines at baseline and upon re-expression of SMARCA4 in vitro for both cell lines and evaluated the serum calcium levels in vivo upon injection into immunodeficient mice for COV434 cells.
Results
The available morphological, immunohistochemical, genetic, and clinical features indicate COV434 is derived from SCCOHT, and TOV-112D is a dedifferentiated carcinoma. Transplantation of COV434 into mice leads to increased serum calcium level. Re-expression of SMARCA4 in either COV434 and TOV-112D cells suppressed their growth dramatically.
Conclusions
COV434 represents a bona fide SCCOHT cell line. TOV-112D is a dedifferentiated ovarian carcinoma cell line.
Keywords: COV434, TOV-112D, SCCOHT, granulosa cell tumor, dedifferentiated carcinoma, SMARCA4
INTRODUCTION
The study of cancer in vitro requires validated cell line model systems. This is especially true for rare cancers for which few if any cell lines exist. We and others have validated several ovarian cancer cell lines as accurate model systems of ovarian cancer histotypes based on genetic, gene expression, and immunohistochemical similarities to human tumours.1,2 These analyses have also raised questions about several commonly used ovarian cancer cell lines because their molecular and immunohistochemical profiles do not correspond to known ovarian cancer histotypes.
Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) is a rare, deadly, and sometimes inherited ovarian cancer that afflicts young women and children.3–5 The cancer is typically diploid and characterized by near-universal mutations in SMARCA4, one of the two ATPase subunits of the SWI/SNF chromatin-remodelling complex.6–10 The resultant loss of SMARCA4, together with nonmutational loss of SMARCA2, the other SWI/SNF ATPase, is highly sensitive and specific for SCCOHT.8,9,11–16 Only two bona fide SCCOHT cell lines have been described: BIN67 and SCCOHT1;17,18 both show dual deficiency of SMARCA4/SMARCA2.16 In addition, dual loss of SMARCA4/SMARCA2 (or other combinations of SWI/SNF proteins) can also be observed in dedifferentiated carcinomas and undifferentiated carcinomas of the endometrium and ovary and other rare cancers including the recently described SMARCA4-deficient undifferentiated thoracic sarcoma.19–28 The latter has recently been re-classified to represent smoking-associated undifferentiated or dedifferentiated non-small cell lung carcinoma rather than primary thoracic sarcoma, named as SMARCA4-deficient sarcomatoid thoracic carcinoma.29
COV434 cells were derived from a 26 year-old woman who was diagnosed with a granulosa cell tumour.30 In the original report, the cells had a stable, hyperdiploid karyotype with gain of chromosome 5, with a subclone showing additional gain of chromosome 22q.30 Similar to normal granulosa cells, they can synthesize estradiol upon stimulation with follicle stimulating hormone in culture.31 Based on the young patient age and absence of the pathognomonic FOXL2 mutation of adult granulosa cell tumour (AGCT), the reported features of tumour are consistent with a juvenile granulosa cell tumour (JGCT).32 To our knowledge, no additional clinical information has been published about the patient at diagnosis or follow-up. TOV-112D cells were derived from a 42 year-old woman with stage IIIC grade 3 endometrioid carcinoma who died only three months after surgery.33 The cell line showed no microsatellite instability, displayed a complex karyotype, and harboured the same mutation in exon 5 of TP53 (p.R172H) in both the original tumour and cell line.
We set out to characterize and compare the pathology, immunohistochemistry, and genetics of the original tumors and cell lines with the goal of establishing definitive diagnoses for the original tumours in the context of our current understanding of ovarian cancer histotypes. Because the cell lines in question all show dual loss of SMARCA4 and SMARCA2, we sought to determine whether the original tumors potentially represent misdiagnosed cases of SCCOHT, which would provide new cell line models for this rare and deadly disease.
METHODS
Cell lines and tissues
This study was approved under the University of British Columbia Ethics Board protocol H02–61375. COV434, BIN67 and SVOG3e cells were grown in DMEM/F12 supplemented with 10% FBS. TOV-112D, NOY1 and AN3CA cells were grown in RPMI supplemented with 10% FBS. BIN67 and COV434 cells were provided by Drs. Barbara Vanderhyden and Mikko Anttonen, respectively. TOV-112D and AN3CA cells were obtained from ATCC. NOY1 cells were purchased from Kerafast (Boston MA). All cell lines were certified by STR analysis, tested regularly for Mycoplasma (Genetica DNA Laboratories, Burlington, NC) and used for the study within 6 months of thawing. The unique Cellosaurus identifiers for COV434 and TOV-112D are RRID:CVCL_2010 and RRID:CVCL_3612, respectively.
Exome sequencing, mapping and variant calling
Genomic DNA isolated from COV434 cells was fragmented to a target size of 150 to 200 bp on the Covaris E210 system and the whole-exome was sequenced as previously described.8 Fastq files were aligned with Burrows-Wheeler Aligner (BWA) 0.7.5a to hs37d5, and the SAM output file was converted into a sorted BAM file using SAMtools 0.1.19. BAM files underwent indel realignment, duplicate marking and recalibration steps in this order with the Genome Analysis Toolkit (GATK) 2.8–1, where dpsnp137 was used for known SNPs and Mills_and_1000G_gold_ standard.indels.b37.vcf was used for known indels. Variant calling was carried out in tumor-only mode with HaplotypeCaller, and output VCF files were recalibrated with VariantRecalibrator from GATK 2.8–1. SnpEff 3.2 and SnpSift 1.9c were then used to annotate these VCF files with database version GRCh37.70. Only variants with a minimum quality score of 20 were extracted. Thereafter, we excluded variants with a Global Minor Allele Frequency >= 0.01 and those somatic coding variants (SNVs) that either appeared in the 1000 Genomes Project database, the dbSNP database or the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project database, assuming that these SNVs might be of less importance for tumorigenesis.
Sanger sequencing
The identified SMARCA4 mutations, chr19:11145589: G:C in COV434 and chr19: 11145589: C del in TOV-112D, were validated by Sanger sequencing. M13 tagged PCR primer sets were designed to amplify regions of the putative SMARCA4 mutation sites: SMARCA4-Splice11145589-F (TGTAAAACGACGGCCAGTCCCTTCTGAACTCTCGGTGT) and SMARCA4-Splice11145589-R (CAGGAAACAGCTATGACTTCTCCTCCTCCTCCTCACA); SMARCA4-TOV112D-F (TGTAAAACGACGGCCAGTCCTCTGTAAGTGTTTGGTCTGG) and SMARCA4-TOV112D-R_(CAGGAAACAGCTATGACAACCTGCCGGGAATTCAAA). PCR products were purified using EXoSAP-IT (USB products Affymetrix). Sequencing reactions were done in both forward and reverse directions using M13 primers and the ABI BigDYe terminatore v3.1 cycle sequencing kit (Applied Biosystems). Amplified products were then sequenced using an ABI Prism 3130x Genetic Analyzer by Source Bioscience.
Immunohistochemistry
Formalin fixed, paraffin embedded tissue blocks were sectioned at 4μm thickness onto Superfrost+ glass slides and were processed using the Ventana Discovery XT, and the Ventana Benchmark XT and Benchmark Ultra automated systems (Ventana Medical Systems, Tucson, AZ, USA). Antibodies used for immunohistochemistry are listed in Supplemental Table S1.
Western blotting
Whole-cell extracts were obtained for SDS-PAGE electrophoresis as previously described,34 and western blots were performed using the SMARCA4 and SMARCA2 antibodies described above. Vinculin (clone hVIN-1, V9131; Sigma) was used to confirm equal protein loading.
Plasmids and lentivirus packaging
The pLDpuro-SMARCA4 and pLDpuro-GFP plasmids were constructed by introducing the SMARCA4 and GFP cDNAs from entry vectors (Genecopeia) into the pLDpuro-EnVA destination vector (a gift of Dr. Jason Moffat) using Gateway reaction (Life Sciences). To produce lentivirus, pLDpuro-SMARCA4 and pLDpuro-GFP plasmid was co-transfected with packaging plasmids psPAX2 and pMD2.G into HEK293T cells. Supernatants were collected at 48 and 72 hours for lentivirus harvesting.34
Cell growth curves
Cells were infected with lentivirus expressing either GFP or SMARCA4, followed by selection with puromycin 24 hr post infection for 48 hours. Cells were then harvested for either Western blot analysis of SMARCA4 expression or reseeded at 5000/well in a 96-well plate and monitored by Incucyte live cell imaging monitor. Cell confluences were determined and plotted over time to determine the cell proliferation curve.
Mouse xenograft studies
All of the procedures related to animal handling, care and treatment in this study were performed according to the guidelines approved by the Animal Care Committee of the University of British Columbia. Briefly, COV434 cells, 4×106 cells per mice with a 1:1 mix of Matrigel (Corning, Corning, NY, USA) in a final volume of 200 μl, were injected subcutaneously into the back of NRG (NOD.Rag1KO.IL2RγcKO) mice (n=6). Tumor volumes and mouse weights were measured twice weekly. Tumor volume was calculated with the following formula: length × (width)2 × 0.52. Average tumor volumes were plotted over time. Serum were collected before inoculation and at the experimental endpoint. Serum calcium level was measured using a colorimetric Calcium Assay kit (Abcam, ab102505). For injection into the ovarian bursa, 1×105 COV434 cells in PBS in a final volume of 10 μL were inoculated into the left ovary of NRG mice. Mice were monitored for health until they reach humane endpoint for tumor isolation. Isolated tumors were fixed in 10% neutral buffered formalin and embedded for histological analysis by H&E staining.
Statistical analysis
The student’s t test was used to evaluate the significant difference between 2 groups of data in all in vitro experiments. P < 0.05 was considered significant.
RESULTS
Dual loss of SMARCA4 and SMARCA2 proteins in COV434 and TOV-112D cells
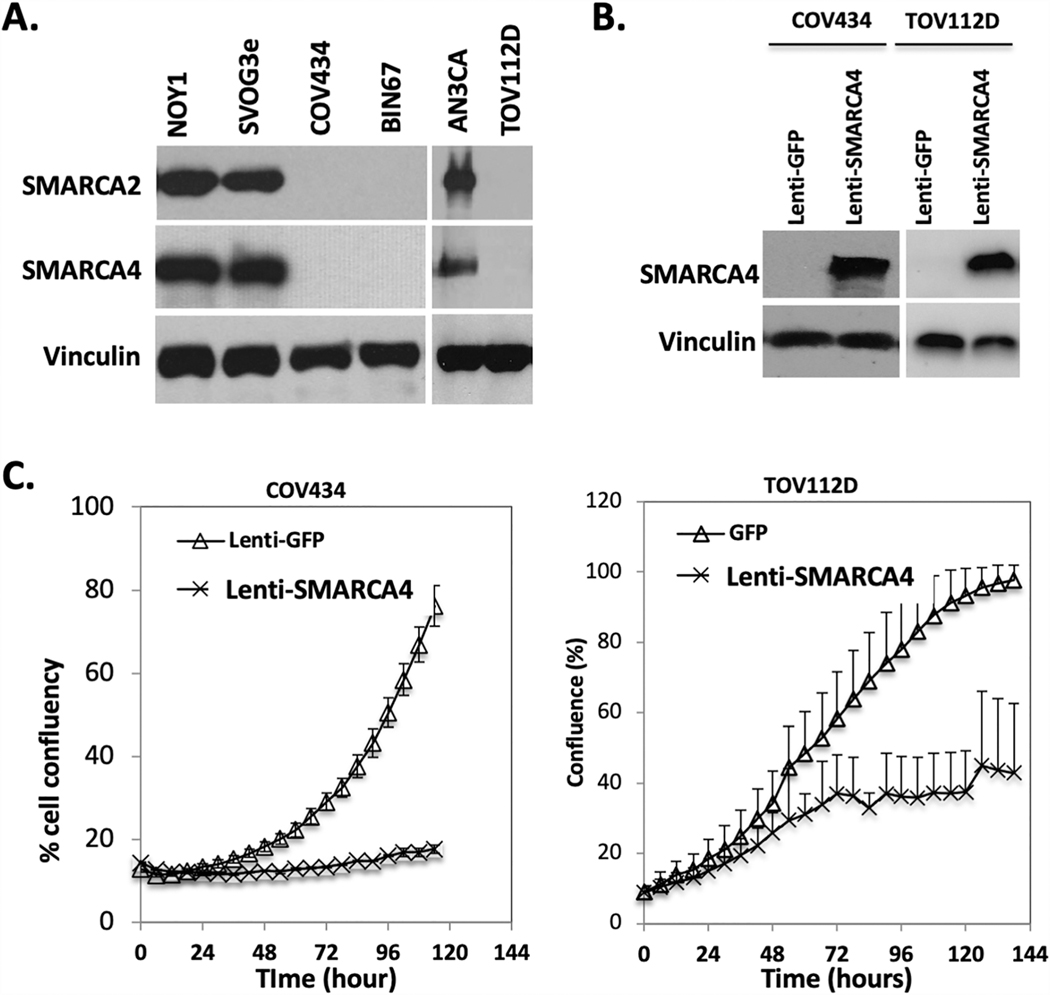
During our previous studies on BIN67 cells, a cell line derived from a patient with SCCOHT,17,35 we discovered that COV434 and TOV-112D cells also lack both SMARCA4 and SMARCA2 proteins, the two ATPase subunits of the SWI/SNF complex (Figure 1A). Lentivirus-mediated re-expression of SMARCA4 in both COV434 and TOV-112D cells strongly suppressed cell proliferation (Figure 1B and 1C), as we previously observed in BIN67 cells.16 Since dual loss of SWI/SNF ATPases is highly specific for SCCOHT,16 it raised the question whether these two cell lines were derived from primary ovarian tumors that were misclassified at diagnosis. To validate the diagnosis of the original tumors from which COV434 and TOV-112D cells were derived,30,33 we obtained representative slides of the original tumours for histopathological review. STR analysis confirmed that both COV434 and TOV-112D cells arose from the original tumour tissue we obtained (data not shown).
Figure 1.
Expression of SMARCA4 and SMARCA2 in COV434 and TOV-112D cell lines. A. Western blot analysis showing loss of SMARCA4 and SMARCA2 in COV434 and TOV-112D. B. Western blot analysis showing lentiviral-mediated re-expression of SMARCA4 in COV434 and TOV-112D. C. Growth curves demonstrate SMARCA4 re-expression dramatically inhibits the growth of COV434 and TOV-112D.
Review of COV434 original tumour pathology
The primary tumour of COV434 cells grows as diffuse sheets of cells with occasional pseudofollicles containing eosinophilic luminal material (Figure 2A, upper panels). Cytologically, the tumour is mostly composed of monotonous small cells with high nucleus to cytoplasm ratio, hyperchromatic nuclei, small nucleoli and minimal eosinophilic cytoplasm (upper panels). Scattered throughout the tumour are clusters of larger cells with vesicular chromatin and more abundant eosinophilic cytoplasm, some of which adopt a rhabdoid appearance (Figure 2A, lower panels). Mitotic activity is high. Sparse lymphocytes and little intervening stroma are present. The main entities in the histological differential diagnosis are JGCT and SCCOHT.
Figure 2.
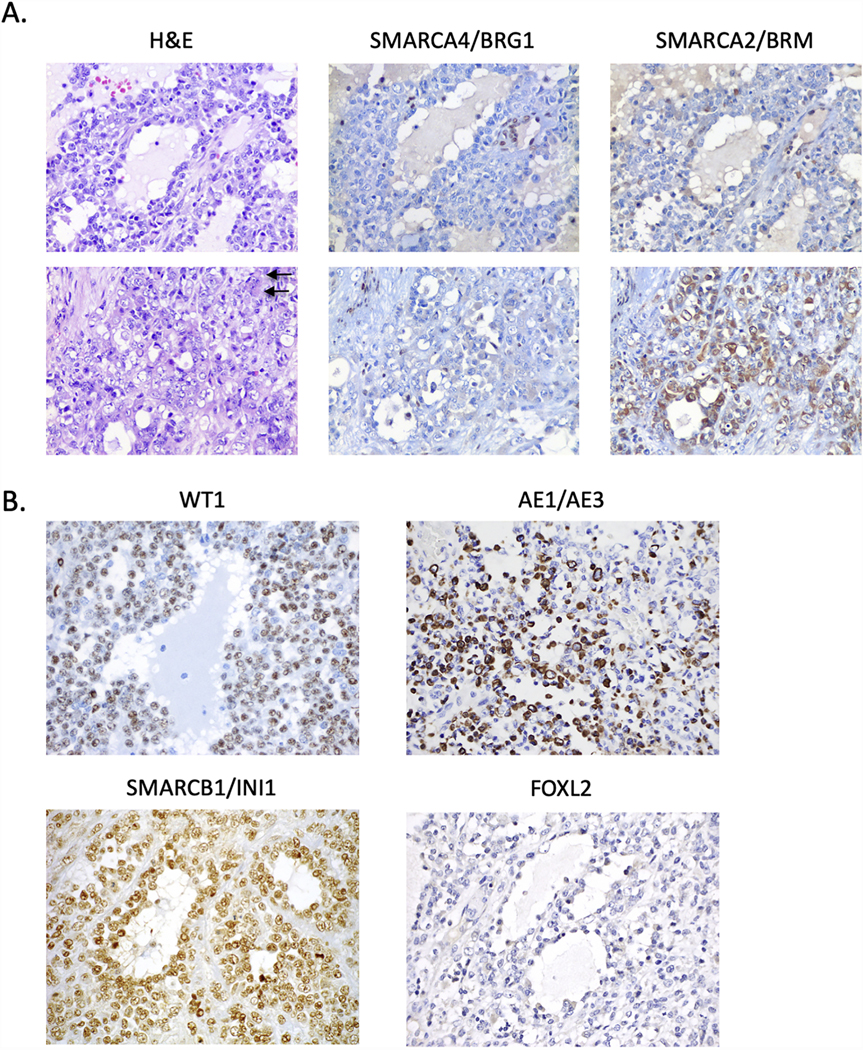
Histopathology and immunohistochemistry of COV434 original tumour. A. H&E-stained sections and immunohistochemistry for SMARCA4 and SMARCA2 of the original ovarian tumour. Two different regions of the tumour are depicted (upper and lower panels). Left images are H&E images which show the tumour grows as sheets of tumour cells with scattered follicle-like spaces. The tumour cells show complete loss of SMARCA4 (middle two panels) with foci of cytoplasmic expression of SMARCA2 (right two panels). B. Immunohistochemistry for WT1, keratin AE1/AE3, SMARCB1/INI1 and FOXL2. Tumour cells are positive for WT1 and keratin AE1/AE3 (particularly in regions with large cells, as depicted), negative for FOXL2, and have intact (normal) expression of SMARCB1/INI1. Arrow, rhabdoid cells. All images at 400x magnification.
Immunohistochemical staining demonstrates the tumour cells are negative for SMARCA4 and nuclear SMARCA2 (Figure 2A). Scattered foci of cells show cytoplasmic positivity for SMARCA2, in particular in the regions with larger cells (Figure 2A lower panels, as previously reported in SCCOHT16) and are positive for the core SWI/SNF protein SMARCB1/INI1 (Figure 2B). The tumour cells are diffusely positive for the WT1 transcription factor, focally positive for keratin AE1/AE3 in large cell regions (small cell regions showed scattered positive cells or are negative), and negative for the highly-sensitive sex cord-stromal tumour marker FOXL2 (Figure 2B). IHC staining results of these proteins and additional histologic markers are summarized in Table 1. The tumour cells are negative for CD10, the sex cord-stromal marker inhibin A, and the neuroendocrine tumour markers synaptophysin, chromogranin and CD56 (data not shown). p53 shows a wild type (variable) staining pattern, and the expression of mismatch repair proteins (MLH1, MSH2, MSH6 and PMS2) is normal/intact (data not shown). Taken together, the clinicopathological and immunohistochemical features are those of SCCOHT, not a granulosa cell tumour.
Table 1.
Summary of immunohistochemistry results in COV434 and TOV-112D original tumours.
| COV434 | TOV-112D | ||||
|---|---|---|---|---|---|
| Antibody | Staining | Slide | Well differentiated | Undifferentiated | Slide |
| SMARCA4 | Negative | Whole | Positive | Negative | Whole |
| SMARCA2 | Negative | Whole | Positive | Negative | Whole |
| SMARCB1 | Positive | Whole | Positive | Positive | Whole |
| WT1 | Positive | Whole | Negative | Negative | Whole |
| p53 | Wild type | Whole# | Missense mutant | Missense mutant | Whole |
| FOXL2 | Negative | Whole | Negative | Negative | TMA |
| Inhibin A | Negative | Whole | Negative | Negative | TMA |
| Calretinin | ND | Negative | Negative | TMA | |
| MMR proteins## | Intact | Whole | Intact | Intact | TMA |
| AE1/AE3 | ND | Positive | Negative | Whole | |
| MOC31 | ND | Positive | Negative | TMA | |
| Claudin-4 | ND | Positive | Negative | TMA | |
| EMA | ND | Positive, patchy | Negative | TMA | |
| PAX8 | ND | Negative | Negative | TMA | |
| p16 | ND | Negative | Diffuse strong | Whole | |
| ER | ND | Positive | Negative | Whole | |
| PR | ND | Positive | Negative | Whole | |
| Napsin A | ND | Negative | Negative | Whole | |
| HNF1 beta | ND | Negative | Negative | Whole | |
| ARID1A | ND | Positive | Positive | Whole | |
| PTEN | ND | Positive | Positive | Whole | |
| CD10 | Negative | Whole | ND | ND | |
| synaptophysin | Negative | Whole | ND | ND | |
| chromogranin | Negative | Whole | ND | ND | |
| CD56 | Negative | Whole | ND | ND | |
Notes:
, staining was done on whole slides of cell line xenografted tumour;
, mismatch repair (MMR) proteins include MLH1, MSH2, MSH6 and PMS2; ND, not determined.
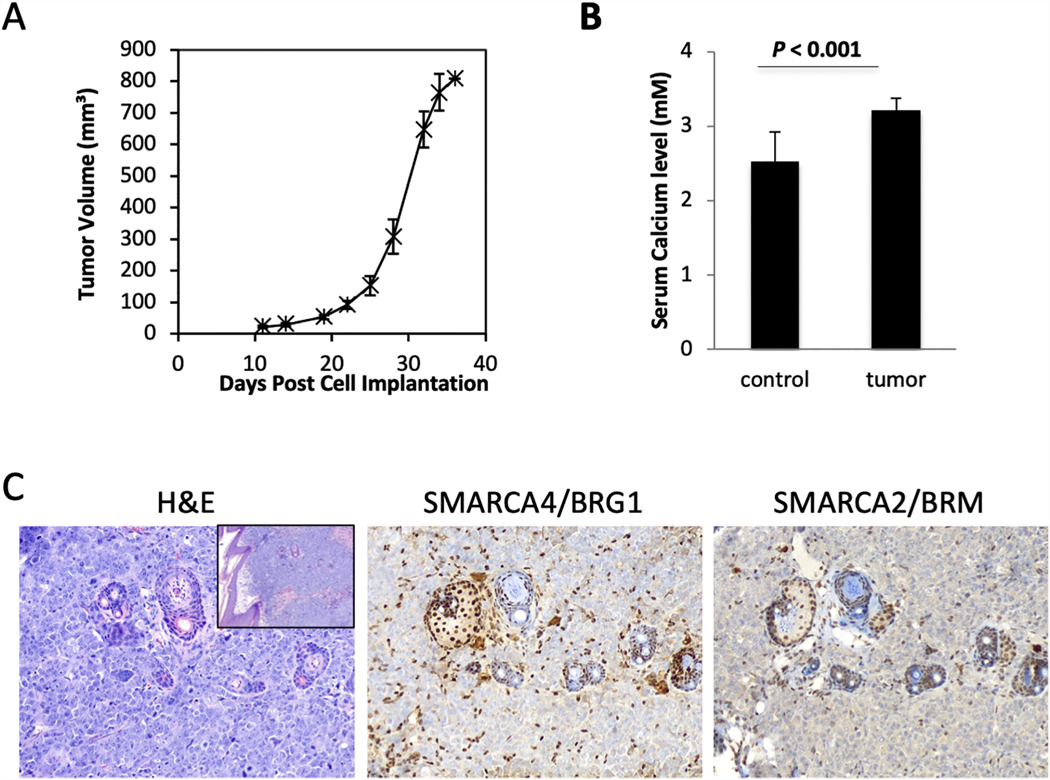
COV434 mouse xenografts
Hypercalcemia is seen in approximately two thirds of SCCOHT patients. To determine whether COV434 xenografts drive the development of hypercalcemia, we injected COV434 cells subcutaneously into immunodeficient NRG mice. This resulted in the rapid growth of tumours from COV434 cells (Figure 3A) with an increase in serum calcium levels from 2.42±0.11 mM to 2.87±0.05 mM (Figure 3B, P <0.001). Upon histologic examination, the COV434 xenografts are undifferentiated neoplasms composed of sheets of high-grade monomorphic cells with brisk mitotic activity lacking both SMARCA4 and SMARCA2 (Figure 3C).
Figure 3.
COV434 mouse xenografts. A. Growth curve upon subcutaneous injection of COV434 cells into immunodeficient NRG mice (n=6). B. Serum calcium measurements of control and tumour-bearing mice. C. H&E-stained sections and immunohistochemistry for SMARCA4 and SMARCA2 of tumor bearing xenografts. Cells with nuclear staining of both proteins represent internal positive controls: lymphocytes, stromal cells, and entrapped follicular structures. Main 3 images at 200x magnification. H&E inset (left panel) at 100x magnification.
DNA sequencing of COV434 and TOV-112D tumour cell lines and COV434 primary tumour
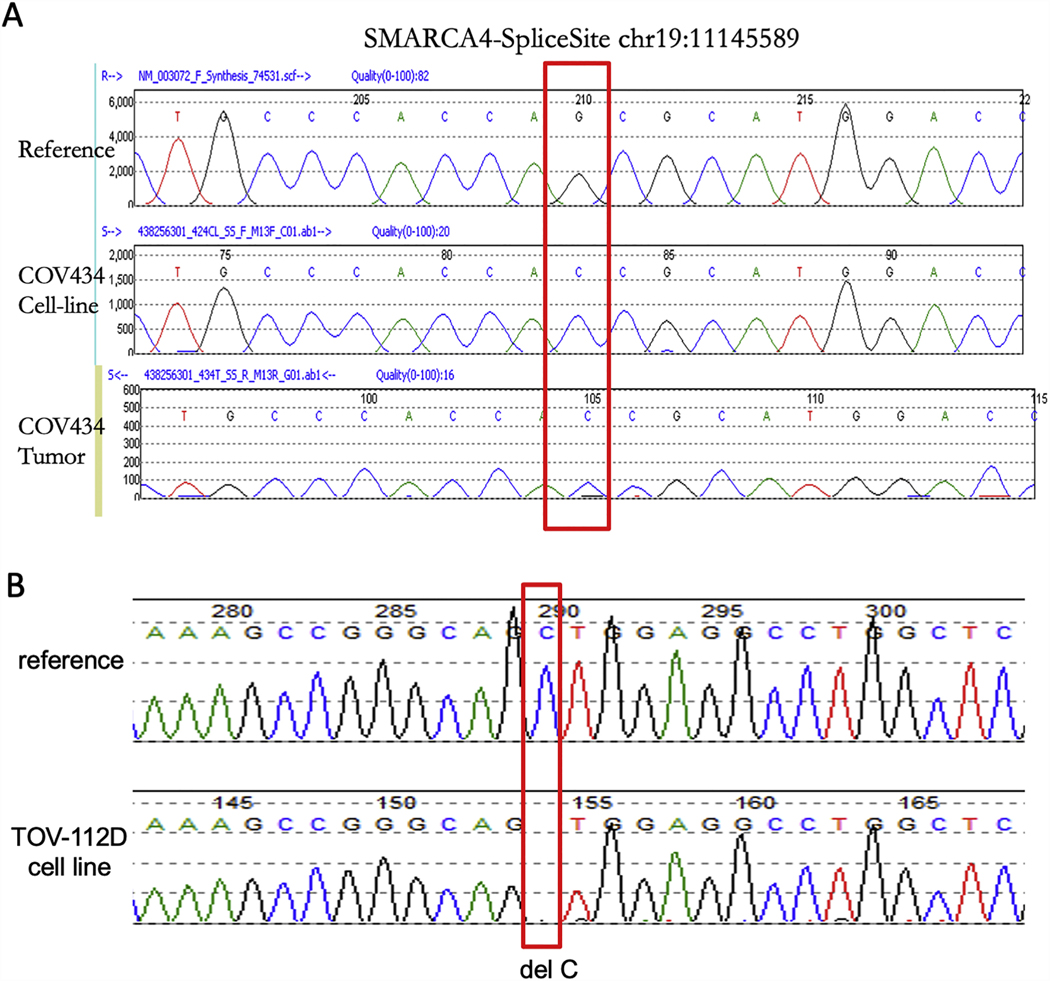
To determine how SMARCA4 and SMARCA2 are silenced in COV434 cells, we performed the whole exome sequencing and identified that SMARCA4 was the only known cancer driver gene that carries either missense hotspot or truncating mutations (Supplemental Table S2). A SMARCA4 splice-site mutation (chr19:11145589: G:C) was identified, which matches the exome sequencing data of COV434 cells in the Broad Institute Cancer Cell Line Encyclopedia (CCLE) database. Using Sanger sequencing, we further validated that both COV434 primary tumor and cell line harbor this homozygous splice-site mutation (Figure 4A). Similarly, the whole exome sequencing of TOV-112D cells by the CCLE demonstrated that there was a frame shift deletion in SMARCA4 gene (chr19: 11145589: C del, p.L639fs) (Supplemental Table S3); we confirmed the presence of this frameshift deletion mutation of SMARCA4, either hemizygous or homozygous, in TOV-112D cells using Sanger sequencing (Figure 4B). Furthermore, a TP53 missense hotspot mutation (p.R175H) was also identified in TOV-112D cells (Supplemental Table S3) as previously reported.33 While we do not have sufficient TOV-112D primary tumor sample for validation that the identical SMARCA4 and TP53 mutations are present in the original tumor, immunohistochemistry results for SMARCA4 and p53 in the original tumor are concordant with the sequencing results in the cell line (Figure 5B). No SMARCA2 mutations were identified in either cell line.
Figure 4.
Validation of SMARCA4 mutation by Sanger sequencing in COV434 cell line and original tumor sample, and in TOV-112D cell line. DNA samples were extracted from both cell lines and the COV434 primary tumour, and Sanger Sequencing was performed. A. COV434 sequencing using primers that amplified the splice site (chr19:11145589) of SMARCA4 gene. The red rectangle denotes the identical mutation in COV434 cells and the original tumor sample compared to a reference sample. B. TOV-112D sequencing using primers that amplified regions around the chr19: 11145589 site of SMARCA4 gene.
Figure 5.
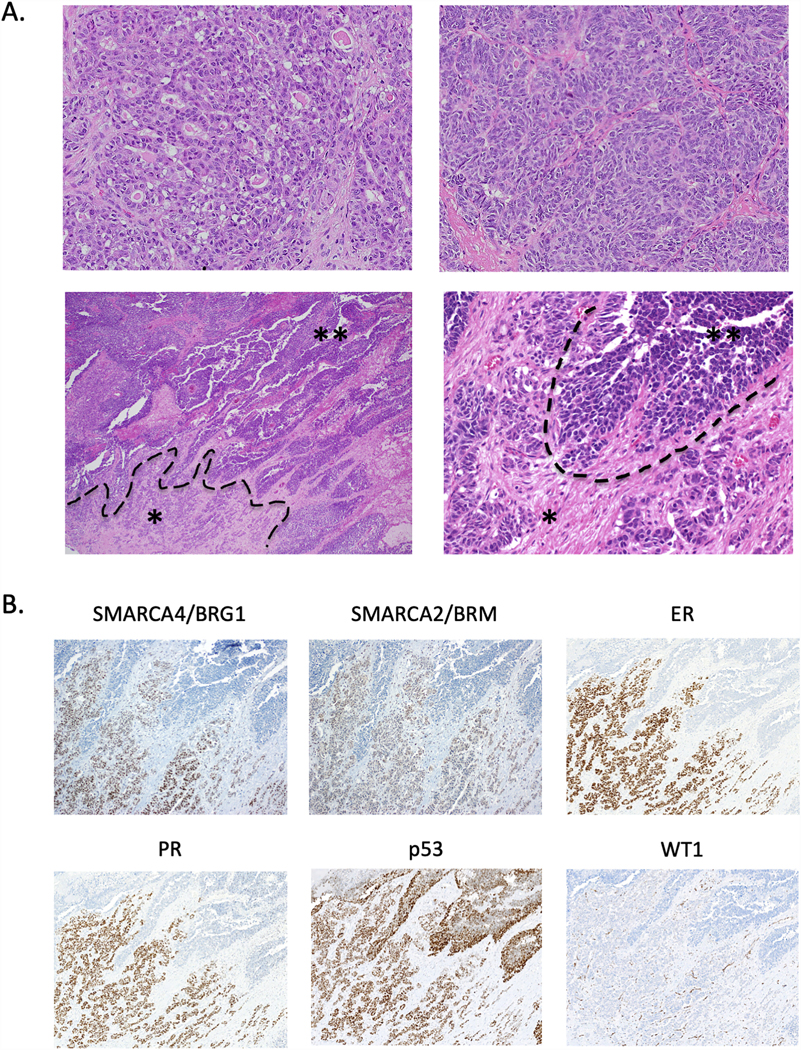
Histopathology and immunohistochemistry of TOV-112D original tumour. A. H&E-stained sections depicting multiple different regions of the original tumour. The tumour grows as glands (upper left) with focal sex cord differentiation (upper right). The tumor shows an abrupt transition from the glandular and corded region (asterisk *) to a broad undifferentiated region growing as solid sheets and nests (double asterisk **). B. Immunohistochemistry for SMARCA4/BRG1, SMARCA2/BRM, ER, PR, p53, and WT1 in the region showing loss of differentiation depicted in the lower panels in Part A. Images show loss of staining for SMARCA4/BRG1, SMARCA2/BRM, ER, and PR in the undifferentiated region and diffuse strong staining for p53 throughout the tumour, consistent with a missense mutation in the TP53 gene. No nuclear staining for WT1 is observed (only vascular staining, a positive internal control). H&E-stained images in Part A: upper panels at 100x magnification, lower left at 40x, lower right at 200x. Immunohistochemistry in Part B at 100x magnification.
Review of TOV-112D original tumour pathology
Representative slides from the original tumour of TOV-112D demonstrate that the tumour shows both well differentiated and undifferentiated regions. The tumour in the well differentiated region grows as glands/microcysts, anastomosing sex cord-like trabeculae, and closed sertoliform-like tubules, with foci of possible immature morules, consistent with the original reported diagnosis of endometrioid carcinoma (Figures 5A, upper panels, and Figure 6). The glands/microcysts contain eosinophilic luminal material and are lined by cells ranging from cuboidal to low columnar to columnar/pseudostratified. Cytologically, the cells have with moderate nuclear atypia (Figure 5A, upper panels).33 There is a relatively abrupt transition between this well differentiated component and the solid undifferentiated region of the tumour (Figure 5A, lower panels), which consists of anastomosing nests and sheets of high-grade, monomorphic tumour cells with vesicular chromatin, small nucleoli, and scant eosinophilic cytoplasm – morphologic features similar to those of SCCOHT.
Figure 6.
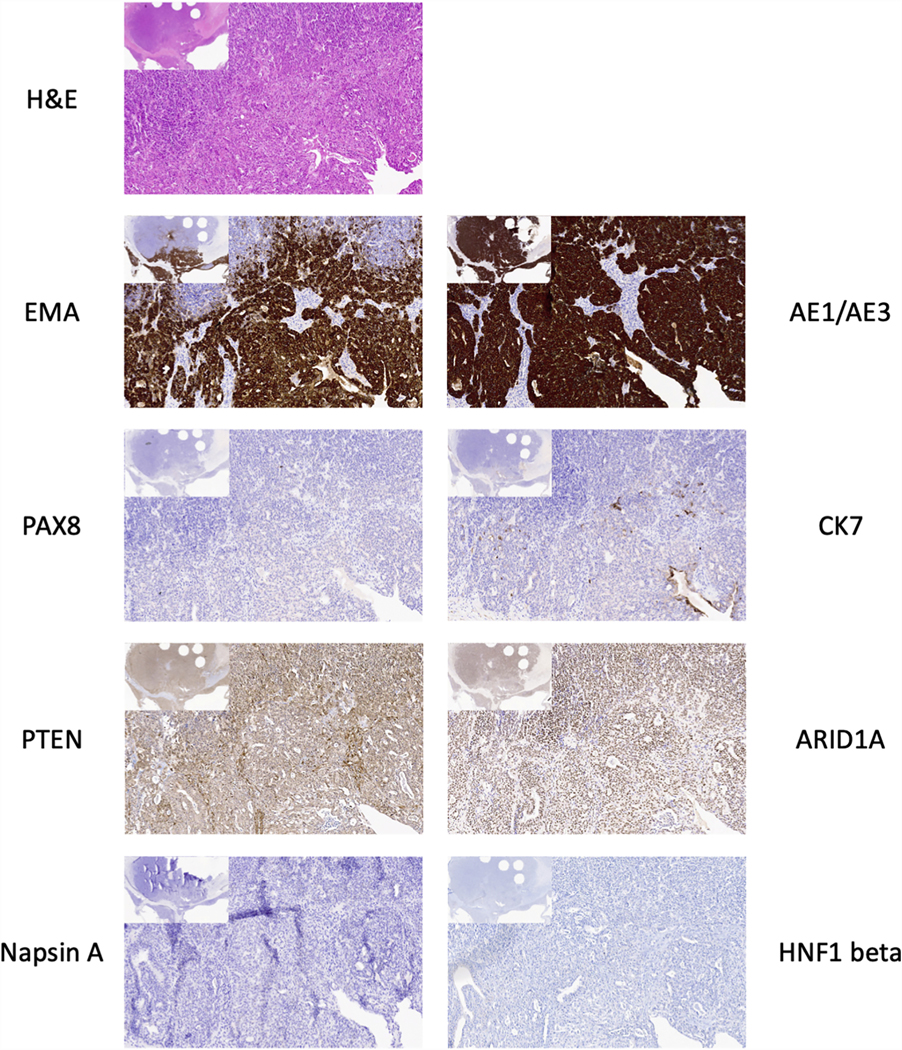
Additional immunohistochemistry on the well differentiated region of TOV-112D original tumour. H&E-stained slide and immunohistochemistry for AE1/AE3, EMA, cytokeratin 7 (CK7), PAX8, ARID1A, PTEN, HNF1 beta, and Napsin A. Larger images at 100x. Image insets at 20x magnification. The round holes in the tissue in the inset images are from tissue punches for TMA construction.
Immunohistochemical stains demonstrate the well differentiated portion of the tumour is positive for SMARCA4, SMARCA2, ER, PR, and several epithelial markers (AE1/AE3, MOC31, and EMA), whereas the undifferentiated portion is negative for these markers (Figure 5B and data not shown). Both regions show diffuse moderate to strong staining for p53, consistent with the reported missense mutation in the TP53 gene,33 and show intact mismatch repair proteins (MLH1, MSH2, MSH6 and PMS2). The well differentiated region is negative for PAX8, WT1, Napsin A, and HNF1 beta (Figure 6), as well as the sex cord-stromal tumour markers FOXL2, inhibin A, and calretinin (data not shown). ARID1A and PTEN are intact in both the well differentiated (Figure 6) and undifferentiated region (data not shown). IHC results are summarized in Table 1. Taken together, the clinicopathological and immunohistochemical features of this tumour are those of a p53-mutant dedifferentiated carcinoma, not SCCOHT.
DISCUSSION
Our analysis has clarified the diagnoses of the original tumours from which COV434 and TOV-112D cell lines were derived. COV434 was originally described as a granulosa cell tumour-derived cell line and was recently suggested instead to be from SCCOHT. The patient’s age, extremely aggressive clinical course, and characteristic histopathology indicate this tumour is indeed SCCOHT. This clinicopathological impression is supported by immunohistochemical, molecular, and cytogenetic data demonstrating loss of SMARCA4 and SMARCA2 expression by IHC, mutation of SMARCA4 in the original tumour, the diploid tumour status, and a lack of copy number alterations in the cell line. In addition, injection of the cell line into mice induced an increase in serum calcium, analogous to hypercalcemia observed in the majority of SCCOHT patients, which further supports the diagnosis.
TOV-112D was originally established from a tumour designated as a grade 3 endometrioid carcinoma based on tumour morphology and immunohistochemistry done at the time of diagnosis. While the loss of SMARCA4/SMARCA2 and the extremely aggressive clinical course are features of SCCOHT, the histopathological and immunohistochemical features and complex karyotype of the original tumour, and the molecular features of the cell line are incompatible with SCCOHT.
Our initial histopathological review of indicated the morphological features of the original TOV-112D tumour are those of a dedifferentiated neoplasm, which raised a broad differential diagnosis including a dedifferentiated carcinoma (DDC) of either endometrioid or non-endometrioid (i.e. high-grade serous carcinoma or clear cell carcinoma) type, a mesonephric-like adenocarcinoma, or even perhaps a heretofore undescribed type of dedifferentiated sex cord-stromal tumor. A sex cord-stromal tumor is excluded based on the expression of multiple epithelial markers (AE1/AE3, MOC31, claudin-4, EMA) and the negative sex cord-stromal markers (FOXL2, inhibin A, calretinin). SMARCA4 missense mutations have been reported in 2/17 mesonephric carcinomas 36, but the diffuse strong ER and PR, negative CD10, and dedifferentiated histology (which has not been reported in mesonephric carcinomas) are strong evidence against this entity. Similarly, the absence of a KRAS mutation 37, which is found in the majority of mesonephric carcinomas 36 is consistent with this notion. TP53 mutations are uncommon in mesonephric carcinoma; one published mesonephric carcinoma with a TP53 mutation lacked a KRAS mutation but harbored a mutation in BCOR 36. The IHC profile – strong diffuse ER/PR and negative Napsin/HNF1 beta – is the opposite of the typical immunoprofile of clear cell carcinomas, which makes this entity highly unlikely (only 3% of ovarian clear cell carcinomas in a large series showed a WT1 /ER +/ HNF1 beta - immunoprofile; 38). The relatively young patient age and mutant p53 suggest this could be a WT1-negative tubo-ovarian high-grade serous carcinoma. However, the morphological features are not typical of this entity, and the cell line shows no supportive evidence of mutations in BRCA1/2 to support the diagnosis 37. The morphologic features, the diffuse strong ER/PR in the well differentiated regions, and the presence of a CTNNB1 mutation in the cell line (Supplemental Table S3) – which occur in 31–60% of ovarian endometrioid carcinomas but extremely rarely (<1%) in HGSC 39–45-- are consistent with a dedifferentiated endometrioid carcinoma.
DDCs are aggressive tumours (similar to SCCOHT) that have been studied extensively in the endometrium where it is encountered more frequently; the ovarian counterpart has similar morphological and molecular features, which include the loss of SMARCA4, SMARCA2, and other epithelial markers in the undifferentiated component.46 The current thinking is that most dedifferentiated carcinomas contain a well differentiated portion that consists of a conventional low-grade (FIGO grade 1 or 2) endometrioid carcinoma with mismatch repair (MMR) proteins deficiency and wild type TP53. A recent study has highlighted the fact that in many dedifferentiated carcinomas, the differentiated portion is high-grade 47, as indeed we see in this case; most of these are endometrioid histotype, although occasionally the carcinoma has ambiguous features. In our view, the differentiated component of the TOV-112D tumor resembles an endometrioid carcinoma with sex cord-like differentiation, intact mismatch repair proteins, and mutations in TP53 and CTNNB1. The presence of a mutant p53 immunostaining pattern in SMARCA4-deficient DDC is uncommon, but it may be more common in DDC with intact SMARCA4 protein.23 In contrast to most DDC, microsatellite instability was not observed in TOV-112D cells, but the TP53 mutation may explain the complex karyotype observed in the cell line. PAX8 is positive in most but not all ovarian carcinomas, so the negative PAX8 does not preclude the diagnosis of an endometrioid (or any Mullerian) carcinoma. The negative cytokeratin 7 (CK7) admittedly is more unusual, but this has been reported in up to 3% of ovarian endometrioid carcinomas in one study,48 and we have also observed this in uterine endometrioid carcinomas. We speculate that the absence of PAX8 and CK7 throughout the tumor may reflect underlying genetic or epigenetic alterations that predisposed this tumor to dedifferentiation, and the undifferentiated component grew to become the TOV-112D cell line.
The OVK18 cell line was also suggested to represent an SCCOHT cell line, based on the dual loss of SMARCA4 and SMARCA2.49 OVK18 cells were reported to be established from a 49 year-old woman with ovarian endometrioid carcinoma with squamous metaplasia involving both ovaries with spread to the myometrium and peritoneum.50 While we are cautious not to over interpret a representative histological image, Figure 1 in the original publication that described the cell line clearly depicts a villoglandular neoplasm that is compatible with the original diagnosis of endometrioid carcinoma,50 and the reported presence of squamous metaplasia supports the diagnosis of endometrioid carcinoma, not SCCOHT. Furthermore, mutational analysis of this cell line revealed mutations in KRAS, PTEN, ARID1A, and TP53,2 which are commonly found in endometrioid carcinoma and DDC51,52 but not SCCOHT.2 OVK18 shows a hypermutated profile (14.4 mutations/Mb),2 similar to tumours and cell lines (e.g. TOV21G) that harbour mutations in mismatch repair genes. Endometrioid carcinomas and DDC, but not SCCOHT, are known to harbour mismatch repair defect, so this further supports that OVK18 represents a hypermutated SMARCA4-deficient DDC cell line of the ovary. However, until the primary tumour is examined in more detail, it remains unclear whether SMARCA4 mutation and protein loss were present in the primary tumour or occurred during cell culture. Interestingly, the primary tumour of OVK18 cells was well differentiated, but xenograft transplantation of the cell line into nude mice grew an undifferentiated monomorphic tumour that was negative for estrogen and progesterone receptors and unresponsive to steroid hormones.50 Therefore, it is possible that the primary tumour of OVK18 contained a minor undifferentiated component that grew out during the establishment of the cell line (i.e. the original tumour was a DDC), or a dedifferentiated subclone emerged upon acquiring SMARCA4 mutation in culture.
Taken together, our current study provides evidence that COV434 is a bona fide SCCOHT cell line, whereas TOV-112D represents a non-conventional SMARCA4 and SMARCA2-deficient ovarian DDC cell line with TP53 and CTNNB1 mutations. In addition, despite our lack of access to the primary tumour tissue of OVK18, based on the histology provided in original publication and the published molecular characterization of this cell line, we suggest that OVK18 represents a hypermutated SMARCA4 and SMARCA2-deficient ovarian DDC cell line. The correct classification of these cell lines will guide future studies using these models to understand the biology of the diseases and to develop novel therapeutic strategies.
Supplementary Material
The original tumour of COV434 cells displayed morphological, immunohistochemical, genetic, and clinical features of SCCOHT;
The original tumour of TOV-112D cells was re-diagnosed as a dedifferentiated ovarian carcinoma;
Re-expression of SMARCA4 suppressed the growth of COV434 and TOV-112D cells dramatically.
Acknowledgements
We thank Drs. Barbara Vanderhyden, Mikko Anttonen, Vincent Smit and Jason Moffat for providing BIN67 cells, COV434 cells, COV434 primary tumor materials and the pLD-puro-EnVA vector, respectively. We thank Simon Cheung and Janine Senz for technical supports. We also thank Dr. Philip Clement for reviewing the slides. This work was supported by research funds from the Canadian Cancer Society Research Institute (#703458, to D.G.H.), the National Institute of Health (1R01CA195670-01, to D.G.H., J.T. and B.W.), the Terry Fox Research Institute Initiative New Frontiers Program in Cancer (D.G.H.). We thank Liliane Meunier of the molecular pathology core facility of the CRCHUM and funding for biobanking was provided by the Réseau de recherche sur le cancer of the Fonds de recherche Québec - Santé affiliated with the Canadian Tumor Repository Network.
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Anglesio MS, Wiegand KC, Melnyk N, et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS One. 2013;8(9):e72162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickersin GR, Kline IW, Scully RE. Small cell carcinoma of the ovary with hypercalcemia: a report of eleven cases. Cancer. 1982;49(1):188–197. [DOI] [PubMed] [Google Scholar]
- 4.Scully RE. Atlas of Tumor Pathology: Tumors of the Ovary and Maldeveloped Gonads. Vol 16. 2nd ed. Washington, DC: Armed Forces Institute of Pathology; 1979. [Google Scholar]
- 5.Young RH, Oliva E, Scully RE. Small cell carcinoma of the ovary, hypercalcemic type: a clinicopathological analysis of 150 cases. Am J of Surg Pathol. 1994;18(11):1102–1116. [DOI] [PubMed] [Google Scholar]
- 6.Jelinic P, Mueller JJ, Olvera N, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46(5):424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupryjanczyk J, Dansonka-Mieszkowska A, Moes-Sosnowska J, et al. Ovarian small cell carcinoma of hypercalcemic type - evidence of germline origin and SMARCA4 gene inactivation. a pilot study. Pol J Pathol. 2013;64(4):238–246. [DOI] [PubMed] [Google Scholar]
- 8.Ramos P, Karnezis AN, Craig DW, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46(5):427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witkowski L, Carrot-Zhang J, Albrecht S, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46(5):438–443. [DOI] [PubMed] [Google Scholar]
- 10.Ramos P, Karnezis AN, Hendricks WPD, et al. Loss of the tumor suppressor SMARCA4 in small cell carcinoma of the ovary, hypercalcemic type (SCCOHT). Rare Diseases. 2014;2(1):e967148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agaimy A, Thiel F, Hartmann A, Fukunaga M. SMARCA4-deficient undifferentiated carcinoma of the ovary (small cell carcinoma, hypercalcemic type): clinicopathologic and immunohistochemical study of 3 cases. Ann Diagn Pathol. 2015;19(5):283–287. [DOI] [PubMed] [Google Scholar]
- 12.Clarke BA, Witkowski L, Ton Nu TN, et al. Loss of SMARCA4 (BRG1) protein expression as determined by immunohistochemistry in small-cell carcinoma of the ovary, hypercalcaemic type distinguishes these tumours from their mimics. Histopathology. 2016;69(5):727–738. [DOI] [PubMed] [Google Scholar]
- 13.Conlon N, Silva A, Guerra E, et al. Loss of SMARCA4 Expression Is Both Sensitive and Specific for the Diagnosis of Small Cell Carcinoma of Ovary, Hypercalcemic Type. Am J Surg Pathol. 2016;40(3):395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jelinic P, Schlappe BA, Conlon N, et al. Concomitant loss of SMARCA2 and SMARCA4 expression in small cell carcinoma of the ovary, hypercalcemic type. Mod Pathol. 2016;29(1):60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karanian-Philippe M, Velasco V, Longy M, et al. SMARCA4 (BRG1) loss of expression is a useful marker for the diagnosis of ovarian small cell carcinoma of the hypercalcemic type (ovarian rhabdoid tumor): a comprehensive analysis of 116 rare gynecologic tumors, 9 soft tissue tumors, and 9 melanomas. Am J Surg Pathol. 2015;39(9):1197–1205. [DOI] [PubMed] [Google Scholar]
- 16.Karnezis AN, Wang Y, Ramos P, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2016;238(3):389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamwell LF, Gambaro K, Merziotis M, et al. Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics. Orphanet J Rare Dis. 2013;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otte A, Gohring G, Steinemann D, et al. A tumor-derived population (SCCOHT-1) as cellular model for a small cell ovarian carcinoma of the hypercalcemic type. Int J Oncol. 2012;41(2):765–775. [DOI] [PubMed] [Google Scholar]
- 19.Altrabulsi B, Malpica A, Deavers MT, Bodurka DC, Broaddus R, Silva EG. Undifferentiated carcinoma of the endometrium. Am J Surg Pathol. 2005;29(10):1316–1321. [DOI] [PubMed] [Google Scholar]
- 20.Silva EG, Deavers MT, Bodurka DC, Malpica A. Association of low-grade endometrioid carcinoma of the uterus and ovary with undifferentiated carcinoma: a new type of dedifferentiated carcinoma? Int J Gynecol Pathol. 2006;25(1):52–58. [DOI] [PubMed] [Google Scholar]
- 21.Silva EG, Tornos C, Bailey MA, Morris M. Undifferentiated carcinoma of the ovary. Arch Pathol Lab Med. 1991;115(4):377–381. [PubMed] [Google Scholar]
- 22.Coatham M, Li X, Karnezis AN, et al. Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian dedifferentiated carcinomas. Mod Pathol. 2016;29(12):1586–1593. [DOI] [PubMed] [Google Scholar]
- 23.Hoang LN, Lee YS, Karnezis AN, et al. Immunophenotypic features of dedifferentiated endometrial carcinoma - insights from BRG1/INI1-deficient tumours. Histopathology. 2016;69(4):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karnezis AN, Hoang LN, Coatham M, et al. Loss of switch/sucrose non-fermenting complex protein expression is associated with dedifferentiation in endometrial carcinomas. Mod Pathol. 2016;29(3):302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobel M, Hoang LN, Tessier-Cloutier B, et al. Undifferentiated Endometrial Carcinomas Show Frequent Loss of Core Switch/Sucrose Nonfermentable Complex Proteins. Am J Surg Pathol. 2018;42(1):76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart CJ, Crook ML. SWI/SNF complex deficiency and mismatch repair protein expression in undifferentiated and dedifferentiated endometrial carcinoma. Pathology. 2015;47(5):439–445. [DOI] [PubMed] [Google Scholar]
- 27.Strehl JD, Wachter DL, Fiedler J, et al. Pattern of SMARCB1 (INI1) and SMARCA4 (BRG1) in poorly differentiated endometrioid adenocarcinoma of the uterus: analysis of a series with emphasis on a novel SMARCA4-deficient dedifferentiated rhabdoid variant. Ann Diagn Pathol. 2015;19(4):198–202. [DOI] [PubMed] [Google Scholar]
- 28.Le Loarer F, Watson S, Pierron G, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47(10):1200–1205. [DOI] [PubMed] [Google Scholar]
- 29.Rekhtman N, Montecalvo J, Chang JC, et al. SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J Thorac Oncol. 2020;15(2):231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg-Bakker CA, Hagemeijer A, Franken-Postma EM, et al. Establishment and characterization of 7 ovarian carcinoma cell lines and one granulosa tumor cell line: growth features and cytogenetics. Int J Cancer. 1993;53(4):613–620. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Vollmer M, De Geyter M, et al. Characterization of an immortalized human granulosa cell line (COV434). Mol Hum Reprod. 2000;6(2):146–153. [DOI] [PubMed] [Google Scholar]
- 32.Jamieson S, Butzow R, Andersson N, et al. The FOXL2 C134W mutation is characteristic of adult granulosa cell tumors of the ovary. Mod Pathol. 2010;23(11):1477–1485. [DOI] [PubMed] [Google Scholar]
- 33.Provencher DM, Lounis H, Champoux L, et al. Characterization of four novel epithelial ovarian cancer cell lines. In Vitro Cell Dev Biol Anim. 2000;36(6):357–361. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Chen SY, Karnezis AN, et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. J Pathol. 2017;242(3):371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upchurch KS, Parker LM, Scully RE, Krane SM. Differential cyclic AMP responses to calcitonin among human ovarian carcinoma cell lines: a calcitonin-responsive line derived from a rare tumor type. J Bone Miner Res. 1986;1(3):299–304. [DOI] [PubMed] [Google Scholar]
- 36.Mirkovic J, Sholl LM, Garcia E, et al. Targeted genomic profiling reveals recurrent KRAS mutations and gain of chromosome 1q in mesonephric carcinomas of the female genital tract. Mod Pathol. 2015;28(11):1504–1514. [DOI] [PubMed] [Google Scholar]
- 37.Ouellet V, Zietarska M, Portelance L, et al. Characterization of three new serous epithelial ovarian cancer cell lines. BMC Cancer. 2008;8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobel M, Kalloger SE, Carrick J, et al. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33(1):14–21. [DOI] [PubMed] [Google Scholar]
- 39.Cybulska P, Paula ADC, Tseng J, et al. Molecular profiling and molecular classification of endometrioid ovarian carcinomas. Gynecol Oncol. 2019;154(3):516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McConechy MK, Ding J, Senz J, et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 2014;27(1):128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollis RL, Thomson JP, Stanley B, et al. Molecular stratification of endometrioid ovarian carcinoma predicts clinical outcome. Nat Commun. 2020;11(1):4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto T, Yamazaki M, Takahashi H, et al. Distinct beta-catenin and PIK3CA mutation profiles in endometriosis-associated ovarian endometrioid and clear cell carcinomas. Am J Clin Pathol. 2015;144(3):452–463. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Rambau PF, Kelemen LE, et al. Nuclear beta-catenin and CDX2 expression in ovarian endometrioid carcinoma identify patients with favourable outcome. Histopathology. 2019;74(3):452–462. [DOI] [PubMed] [Google Scholar]
- 45.Pierson WE, Peters PN, Chang MT, et al. An integrated molecular profile of endometrioid ovarian cancer. Gynecol Oncol. 2020;157(1):55–61. [DOI] [PubMed] [Google Scholar]
- 46.Yemin Wang LH, Jennifer Ji X, Huntsman David G. SWI/SNF Complex Mutations in Gynecologic Cancers: Molecular Mechanisms and Models. Annual review of Pathology-Mechanisms of disease 2020;15:467–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busca A, Parra-Herran C, Nofech-Mozes S, et al. Undifferentiated endometrial carcinoma arising in the background of high-grade endometrial carcinoma - Expanding the definition of dedifferentiated endometrial carcinoma. Histopathology. 2020;77(5):769–780. [DOI] [PubMed] [Google Scholar]
- 48.Zhao C, Bratthauer GL, Barner R, Vang R. Comparative analysis of alternative and traditional immunohistochemical markers for the distinction of ovarian sertoli cell tumor from endometrioid tumors and carcinoid tumor: A study of 160 cases. Am J Surg Pathol. 2007;31(2):255–266. [DOI] [PubMed] [Google Scholar]
- 49.Chan-Penebre E, Armstrong K, Drew A, et al. Selective Killing of SMARCA2- and SMARCA4-deficient Small Cell Carcinoma of the Ovary, Hypercalcemic Type Cells by Inhibition of EZH2: In Vitro and In Vivo Preclinical Models. Mol Cancer Ther. 2017;16(5):850–860. [DOI] [PubMed] [Google Scholar]
- 50.Uehara S, Abe H, Hoshiai H, Yajima A, Suzuki M. Establishment and characterization of ovarian endometrioid carcinoma cell line. Gynecol Oncol. 1984;17(3):314–325. [DOI] [PubMed] [Google Scholar]
- 51.DeLair DF, Burke KA, Selenica P, et al. The genetic landscape of endometrial clear cell carcinomas. J Pathol. 2017;243(2):230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuhn E, Ayhan A, Bahadirli-Talbott A, Zhao C, Shih Ie M. Molecular characterization of undifferentiated carcinoma associated with endometrioid carcinoma. Am J Surg Pathol. 2014;38(5):660–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.