Summary
Alcohol-associated liver disease is accompanied by intestinal mycobiome dysbiosis yet the impacts on liver disease are unclear. We demonstrate that Candida albicans-specific Th17 cells are increased in the circulation and present in the liver of patients with alcohol-associated liver disease. Chronic ethanol administration to mice causes migration of C. albicans-reactive Th17 cells from the intestine to the liver. The antifungal agent nystatin decreased C. albicans-specific Th17 cells in the liver, and reduced ethanol-induced liver disease in mice. Transgenic mice expressing T-cell receptors (TCRs) reactive to Candida antigens developed more severe ethanol-induced liver disease than transgene-negative littermates. Adoptively transferring Candida-specific TCR transgenic T cells or polyclonal C. albicans-primed T cells exacerbated ethanol-induced liver disease in wild-type mice. IL17 receptor A signaling in Kupffer cells was required for the effects of polyclonal C. albicans-primed T cells. Our findings indicate that ethanol increases C. albicans-specific Th17 cells, which contribute to alcohol-associated liver disease.
Graphical Abstract
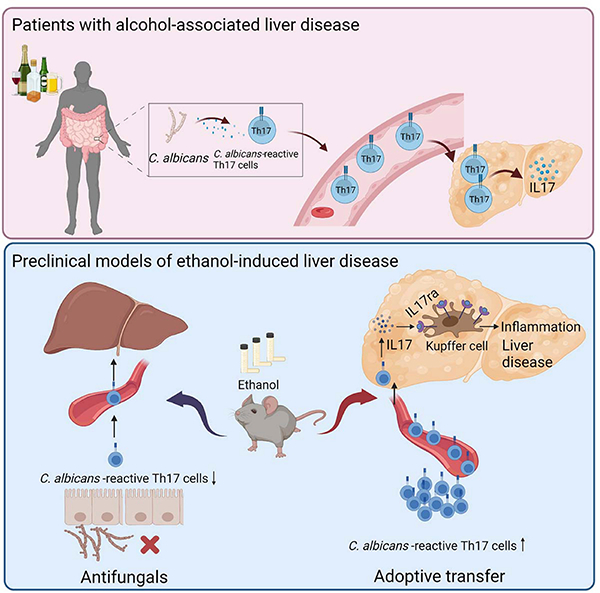
C. albicans-specific Th17 response contributes to alcohol-associated liver disease
Upper panel: Patients with alcohol user disorder and liver disease develop fungal dysbiosis with overgrowth of C. albicans. This results in the development of C. albicans-reactive Th17 cells, which migrate to the liver and secrete IL17. Lower panel: In mice, chronic ethanol feeding results in increased numbers of intestinal C. albicans-specific Th17 cells, which migrate to the liver. An antifungal agent prevents development of the C. albicans-specific Th17 cells and protects against ethanol-induced liver disease, whereas transfer of C. albicans-specific Th17 cells to wild-type mice increases ethanol-induced liver disease via IL17ra signaling in Kupffer cells. Created with BioRender.com.
eTOC Blurb
How fungal dysbiosis contributes to alcohol-associated liver disease is unclear. Zeng et al find that C. albicans-specific Th17 cells increase in the blood and liver of patients with alcohol-associated liver disease. In mice, C. albicans-specific Th17 cells migrate from the intestine to liver, where they contribute to ethanol-induced liver disease.
Introduction
Alcohol-associated liver disease is a spectrum of liver diseases in patients with alcohol use disorder. Almost 90% of patients with alcohol use disorder develop hepatic steatosis, which can be considered as the earliest stage of alcohol-associated liver disease. As disease progresses, more of the healthy liver undergoes inflammation, swelling and scarring, leading to alcohol-associated steatohepatitis, fibrosis, cirrhosis, and eventual liver failure.1,2 Alcohol-associated liver disease is now the leading indication for liver transplantation in the United States3 and is one of the main contributors to alcohol-attributable death and illness.4
The composition of the gut microbiota is an important factor in the development of alcohol-associated liver disease.5 Despite most research focusing on the bacterial component of the gut microbiota, the contribution of the fungal mycobiome to alcohol-associated liver disease is understudied and poorly defined.6,7 Chronic alcohol consumption leads to intestinal fungal dysbiosis and overgrowth. Patients with different stages of alcohol-associated liver disease had lower fungal stool diversity and increases in fecal C. albicans.8,9 Serum levels of specific anti-C. albicans immunoglobulin G (IgG) and IgM were significantly higher in patients with alcohol use disorder compared with non-alcoholic control participants,10 indicating an increase in systemic exposure and immune response to intestinal fungi.
Th17 cells mediate immune responses against extracellular bacteria and fungi at barrier sites, such as the intestinal tissues and the skin.11 In mice, Th17 induction depends on certain microbial species within the gastrointestinal tract.12–14 Th17 cells locally induced in the intestine are antigen-specific,12,14 but they can also migrate to peripheral organs.15 In humans, the microbial species that induce Th17 responses remain largely unknown, but C. albicans has been identified as a dominant driver of fungus-specific Th17 responses.16 Re-circulation of C. albicans-specific Th17 cells from the intestine and reactivation by local airborne fungi promotes pathogenic airway inflammation in susceptible patients via cross-reactive Th17 cells. This finding indicated that the CD4+ T cell-mediated Th17 response can become pathological if dysregulated or directed to inappropriate target tissues.16 Here, we investigated whether C. albicans-specific Th17 cells contribute to alcohol-associated liver disease.
Results
Patients with alcohol use disorder and liver disease have increased peripheral and hepatic C. albicans-specific Th17 responses
To determine whether patients with alcohol use disorder and liver disease have an increased C. albicans-reactive Th17 response, we analyzed peripheral CD4+ T cells specific for C. albicans in non-alcoholic subjects (controls, n=33) and patients with alcohol use disorder and liver disease (AUD, n=36) (Supplementary Table 1). We used the antigen-reactive T cell enrichment (ARTE) approach for highly sensitive detection and characterization of C. albicans-reactive CD4+ T cells.16–19 After ex vivo stimulation of peripheral blood mononuclear cells (PBMCs) with whole fungal lysates for 7 hrs, fungus-reactive T-helper cells were magnetically enriched using the T cell activation marker CD154 and analyzed for the production of effector cytokines. Specificity of the fungal lysate-activated T cells has been shown previously.16 Compared with controls, patients with alcohol use disorder and liver disease had higher frequencies of CD154+ memory CD4+ T cells after stimulation with C. albicans lysate (Fig. 1A). However, no statistically significant differences were observed between patients and controls in response to stimulation with Saccharomyces cerevisiae (S. cerevisiae) lysate, used as a control yeast (Fig. 1A). S. cerevisiae is a major component of the normal fungal mycobiota.20,21 Strikingly, C. albicans-reactive T cells from patients with alcohol use disorder and liver disease had increased production of interleukin 17A (IL17A) and IL22, compared with T cells from controls, whereas levels of IL21, interferon gamma (IFNG), IL2, IL10, GM-CSF, Ki67, and Integrin-β7 (IntB7) did not differ significantly (Fig. 1B–D, Supplementary Fig. 1A). In contrast, stimulation with S. cerevisiae did not induce different responses in T-helper cells from controls vs patients with alcohol use disorder and liver disease. Frequency of C. albicans-reactive Th17 cells correlated with liver stiffness in patients with alcohol use disorder and liver disease, a clinical indicator for progressive and fibrotic liver disease (Supplementary Fig. 1B). As body-mass index (BMI) was significantly higher in patients with alcohol use disorder and liver disease than control subjects (Supplementary Table 1) and may act as a possible confounder for higher frequency of C. albicans-reactive Th17 cells, we matched the patients with alcohol use disorder and liver disease, and control subjects (BMI median 22.8 (3.8) with n=14 and 22.6 (4.0) with n=14, P=0.84 per Mann-Whitney U test) and found patients with alcohol use disorder and liver disease still had significantly higher frequencies of IL17+CD154+CD4+ T cells after stimulation with C. albicans lysate compared with controls (3.53 × 10−4 (5.56 × 10−4) vs 7.16 × 10−5 (5.75 × 10−5), P=0.004 per Mann-Whitney U test), demonstrating that the difference in BMI did not skew our results. Together, these results indicate that C. albicans-reactive Th17 cells are specifically increased in patients with alcohol use disorder and liver disease.
Figure 1. Patients with alcohol use disorder and liver disease have increased C. albicans-reactive Th17 responses vs non-alcoholic controls.
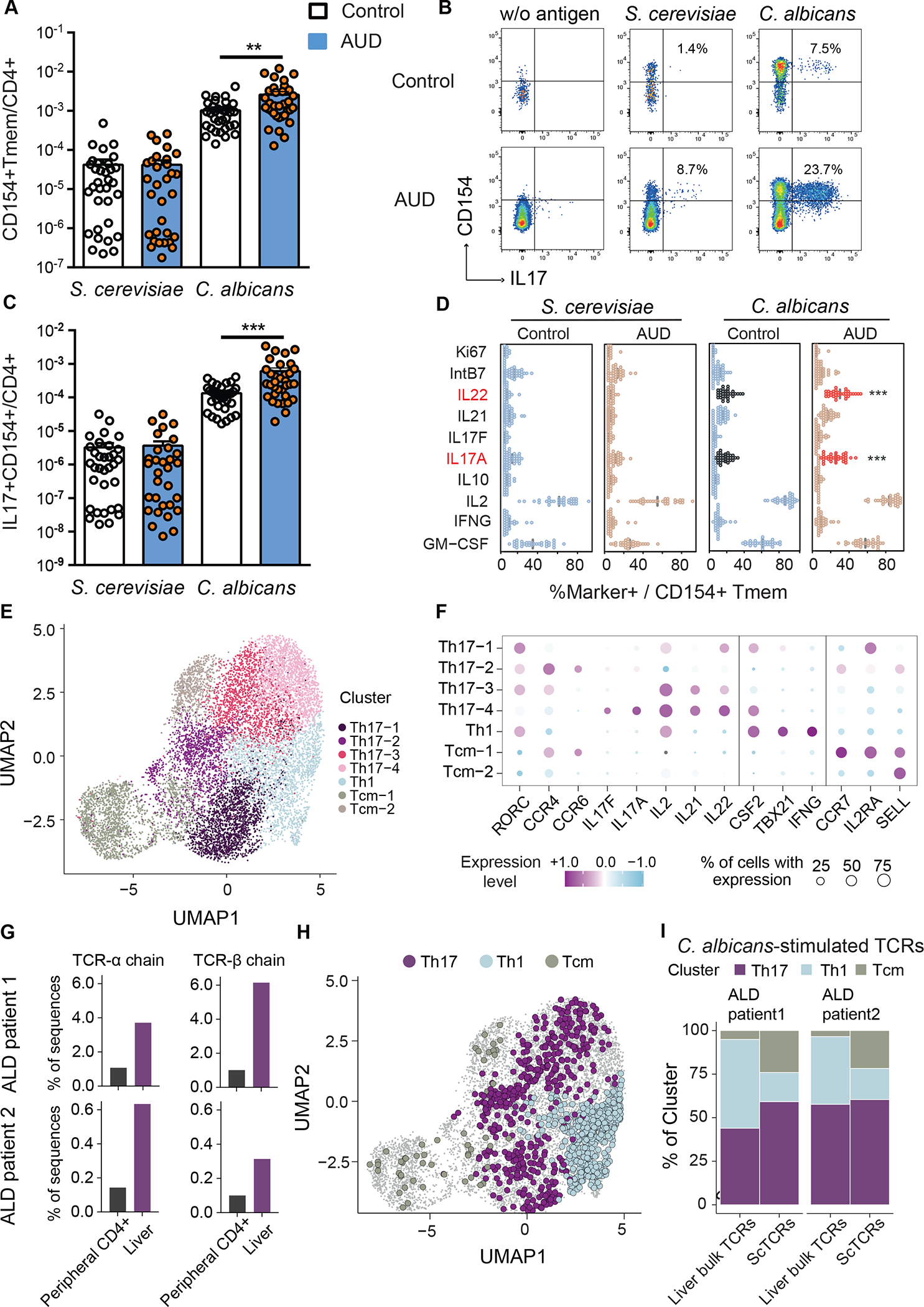
(A) Absolute frequencies of fungus-reactive CD154+CD45RA-memory CD4+ T cells (Tmem) in non-alcoholic controls (n=33) and patients with alcohol use disorder (AUD) and liver disease (n=36). (B) Flow cytometry plots of IL17A+CD154+ cells within CD4+ T cells. Numbers indicate percentage of IL17A+ cells within CD154+ memory T cells. (C) Absolute frequencies of fungus-reactive IL17A+ producers within total CD4+ T cells. (D) Cytokine production analyzed by ex-vivo fungus-reactive Tmem, from controls and patients with alcohol use disorder and liver disease, after stimulation with C. albicans or S. cerevisiae lysate. Relative frequencies of Ki67, IntB7, IL22, IL21, IL17F, IL17A, IL10, IL2, IFNG, GM-CSF within CD154+ Tmem are shown (%Marker+ / CD154+ Tmem). Significant differences are indicated by red text and asterisks. (E) C. albicans-stimulated CD154+ memory T cells from peripheral blood of patients with alcohol-associated liver disease (ALD) (n=2) were ex vivo FACS purified and analyzed by single-cell gene expression. UMAP visualization shows the subset composition of the C. albicans-stimulated cells colored by functional gene expression clusters. (F) Bubble plot visualization showing the gene expression of selected markers in each T cell cluster. Colors represent the Z-score normalized gene expression and the size of the bubbles indicates the proportion of cells expressing the respective genes. (G) Bulk TCR sequencing was performed from bulk liver biopsies and total CD4+ T cells from blood of the same individuals as the C. albicans scRNA seq dataset. Proportions of C. albicans-stimulated TCR-alpha and beta sequences within the liver and peripheral CD4+ T cells are shown for both donors. (H) C. albicans-stimulated TCR clonotypes showed identical TCR sequences with bulk TCR data from the liver samples are highlighted in the UMAP plot and color-coded according to the identified functional clusters. (I) Phenotype distribution of C. albicans-stimulated clonotypes showed identical TCR sequences with liver samples compared to total of C. albicans-stimulated clonotypes from PBMCs. Figure 1A–D was performed in 12 independent experiments with 2–5 patients and controls each time. Results are expressed as mean±SEM. P value determined by 1-way ANOVA with Holm-Sidak post-hoc test. **P<0.01, ***P<0.001. See also Figure S1.
To analyze whether C. albicans-reactive T cells are present in livers of patients with alcohol-associated liver disease, we performed single-cell RNA sequencing (scRNAseq) and T cell receptor (TCR) sequencing of Fluorescence-activated cell sorting (FACS) - purified C. albicans-stimulated CD154+ memory T cells from PBMCs, as well as bulk TCR sequencing from liver biopsies and peripheral CD4+ T cells and compared their sequences of TCR clonotypes (Supplementary Fig. 1C; Supplementary Table 2). Uniform manifold approximation and projection (UMAP) cluster analysis of C. albicans-stimulated T cells revealed four Th17-associated clusters characterized by differential expression of Th17 cytokines (IL17A, IL17F, IL21, IL22), homing markers (CCR4, CCR6) and transcription factor RORC. In addition, one Th1 cluster (IFNG, TBX21) and two different central memory (Tcm) clusters, characterized by expressions of CCR7, SELL, IL2RA, were observed (Fig. 1E–F). Comparison of TCRs identified in paired liver and blood samples showed that identical C. albicans-stimulated TCR-alpha and beta sequences (clonotypes) recovered from circulating C. albicans-stimulated CD4+ T cell populations were enriched in liver samples compared with CD4+ T cells from PBMCs (Fig. 1G). We next analyzed the single-cell phenotype of C. albicans-stimulated TCRs that were also detected in the liver. To this end, the different Th17 and Tcm clusters were pooled. Several C. albicans-stimulated TCRs from liver had a Th17 phenotype, but also Th1 and Tcm cells were detected (Fig. 1H–I). Although the specificity of C. albicans-stimulated TCRs identified in the scRNAseq data set has not further been validated, e.g. by NFAT reporter assay-based MHC/peptide multimers, previous data demonstrated that fungal lysate-activated CD154+ T cells are highly specific.16 In summary, these data indicate that C. albicans-stimulated Th17 cells migrate to and are present in the liver of patients with alcohol use disorder and might contribute to liver disease.
Ethanol administration increases C. albicans-specific Th17 responses in mice
To investigate whether ethanol induces a C. albicans-specific Th17 cell-mediated response, wild-type mice were placed on a chronic plus binge ethanol diet or isocaloric (control) diet22 and fungus-activated Th17 cells were identified. Three different compartments were analyzed: mesenteric lymph nodes, where luminal antigens from the intestine are presented by antigen-producing cells to CD4+ T cells; portal vein blood, where cells travel from the intestine to the liver; and the liver itself. Fungus-activated Th17 cells are identified based on antigen-induced upregulation of CD154 and IL17A after 6 hrs in vitro stimulation with fungal lysates. In mice on the control diet, IL17A production by immune cells in response to C. albicans and S. cerevisiae lysate was low in mesenteric lymph nodes, compared with ethanol-fed mice, in which C. albicans induced a significantly higher proportion of Th17 cells (Fig. 2A, D; Supplementary Fig. 2A–B). Compared with S. cerevisiae, C. albicans also generated a robust Th17 response in immune cells from portal vein blood following ethanol feeding (Fig. 2B, D; Supplementary Fig. 2A, C). Furthermore, and similar to our findings in humans, the proportion of IL17+CD154+ CD4+ T cells in liver following ex vivo stimulation with C. albicans lysate was increased in mice on the ethanol diet (Fig. 2C–D; Supplementary Fig. 2A, D). Collectively, these data indicate that chronic ethanol administration increases C. albicans-activated Th17 cells in mesenteric lymph nodes, portal vein blood, and liver.
Figure 2. Ethanol administration increases C. albicans-specific Th17 cells in mice.
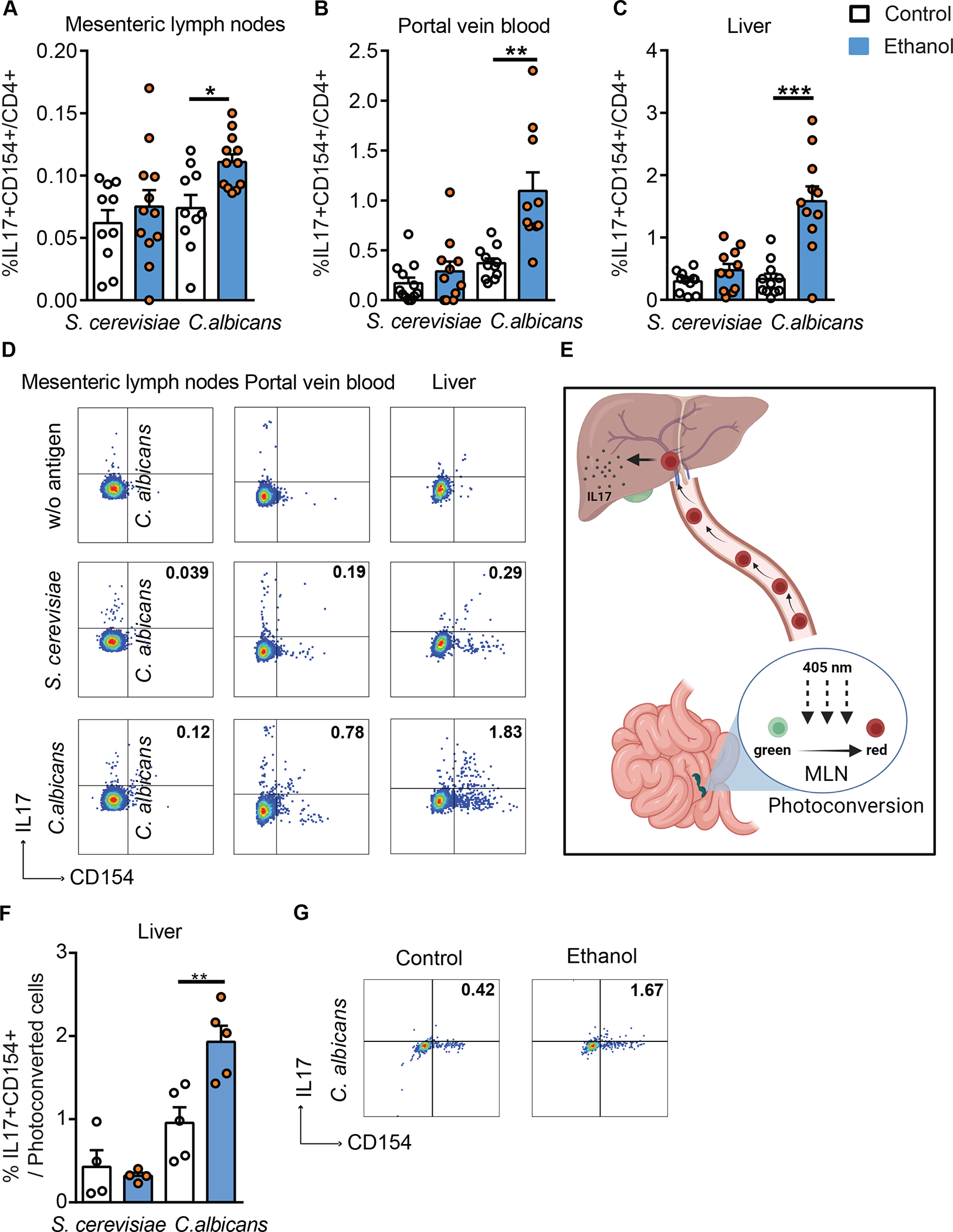
C57BL/6 mice were fed a chronic plus binge ethanol diet (ethanol, blue) or isocaloric diet (control, white). Fungus-activated Th17 cells in mesenteric lymph nodes (A), portal vein blood (B), and liver (C), detected after isolation of mononuclear cells following 6 hrs ex vivo stimulation with C. albicans or S. cerevisiae lysate. (D) Flow cytometry plots of IL17A+CD154+ cells among CD4+ T cells. Kaede mice were fed a chronic plus binge ethanol diet or isocaloric diet. (E) Migration of cells from mesenteric lymph nodes (MLN) to liver using Kaede mice. Photoconversion was performed in mesenteric lymph nodes, and livers were collected after 48 hrs. Created with BioRender.com. (F) Fungus-activated Th17 cells in liver, detected after isolation of mononuclear cells following 6 hrs ex vivo stimulation with C. albicans or S. cerevisiae lysate. (G) Flow cytometry plots of IL17A+CD154+ cells among migrated (photoconverted) cells after stimulation with C. albicans lysate. Figure 2A–C was performed in 3–4 independent experiments. Figure 2E–G was conducted in 2 independent experiments. Results are expressed as mean±SEM. P values among groups of mice fed with control diet or ethanol diet are determined by 1-way ANOVA with Tukeýs post-hoc test. *P<0.05, **P<0.01, ***P<0.001. See also Figure S2.
To directly monitor migration of C. albicans-specific Th17 cells from the intestine to the liver, we studied mice that express the Kaede transgene, and placed them on chronic ethanol or control diets. Kaede is a photoconvertible fluorescent protein that irreversibly changes from green to red upon exposure to violet light.23 Cells in mesenteric lymph nodes were photoconverted and their migration to liver was monitored (Fig. 2E). Hepatic mononuclear cells were isolated, stimulated for 6 hrs with C. albicans or S. cerevisiae lysates, and then analyzed for the presence of red fluorescence and the frequency of fungus-activated Th17 cells. Compared with sham-operated Kaede mice, hepatic mononuclear cells from photoconverted Kaede-transgenic mice were fluorescent red (Supplementary Fig. 2E–F). We found that the proportion of photoconverted (red) IL17+CD154+ CD4+T cells in the liver following stimulation with C. albicans, but not with S. cerevisiae lysate, was increased in mice on the chronic ethanol diet vs the control diet (Fig. 2F–G). This finding indicates that photoconverted C. albicans-specific Th17 cells migrate from mesenteric lymph nodes to the liver during ethanol feeding.
Reducing intestinal fungi decreases C. albicans-specific Th17 cells in the liver and attenuates ethanol-induced liver disease in mice
To study whether reducing intestinal fungi results in a decrease of C. albicans-specific Th17 cells in the liver, mice were placed on a chronic Lieber-DeCarli diet or isocaloric diet in the presence or absence of the poorly absorbable antifungal agent nystatin for the last 10 days. Nystatin decreased the proportion of C. albicans-specific IL17+CD154+ CD4+ T cells in the livers of ethanol-fed mice (Fig. 3A–B). Intestinal fungal overgrowth and Candida spp. were reduced in ethanol-fed mice following nystatin administration (Fig. 3C–E). Compared with mice given vehicle, mice given nystatin developed less-severe ethanol-induced liver injury, indicated by a significantly lower level of serum alanine aminotransferase (ALT) (Fig. 3F) and decreased hepatic steatosis, measured by hepatic triglycerides, and shown by oil red O-staining of liver sections (Fig. 3G–H). Nystatin also decreased hepatic mRNA expression of the macrophage marker F4/80 (Fig. 3I). Nystatin administration did not affect intestinal absorption or hepatic metabolism of ethanol, based on serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs, which encode enzymes that metabolize ethanol in the liver (Fig. 3J–L). Nystatin did not induce hepatic inflammation in control-fed mice (Supplementary Fig. 3A). Nystatin did not significantly affect the overall composition of the bacterial microbiota in control- or ethanol-fed mice (Supplementary Fig. 3B–D). Altogether, these findings indicate that reducing intestinal fungi protects against ethanol-induced liver disease, which is associated with a decrease in hepatic C. albicans-specific Th17 cells.
Figure 3. Antifungal agent reduces hepatic C. albicans-activated Th17 cells and attenuates ethanol-induced liver disease in mice.
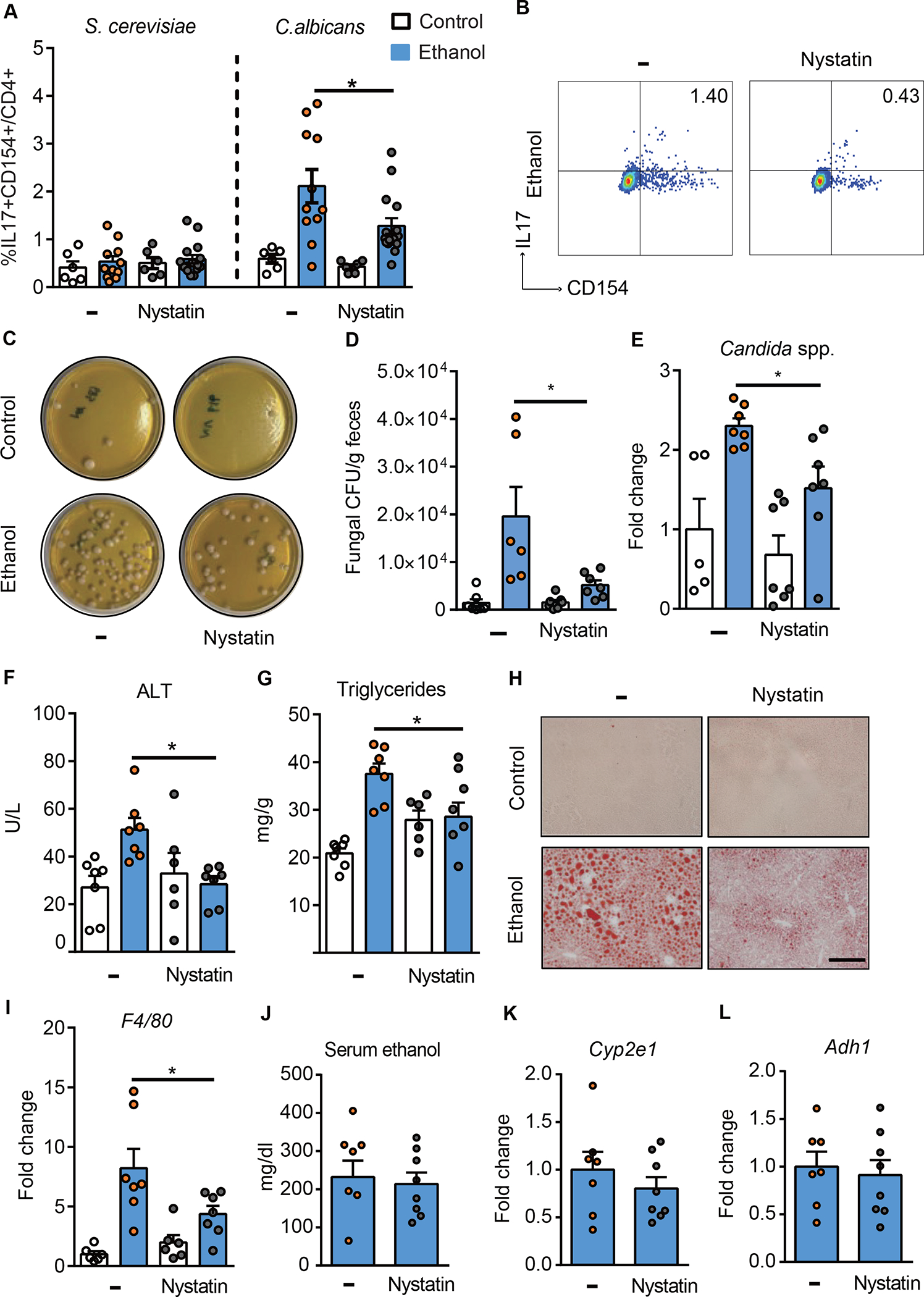
(A-B) C57BL/6 mice were fed a chronic plus-binge ethanol diet (NIAAA model) or an isocaloric (control) diet, with or without the anti-fungal drug nystatin. (A) Fungus-activated Th17 cells in liver detected after isolation of mononuclear cells, following 6 hrs ex vivo stimulation with C. albicans or S. cerevisiae lysate. (B) Flow cytometry plots of IL17A+CD154+ cells among CD4+ T cells. (C–L) C57BL/6 mice were placed on a chronic Lieber DeCarli diet or control diet for 8 weeks. Diets were supplemented with or without nystatin for the last 10 days. (C) Fecal samples from controls and ethanol-fed mice were cultured on YPD agar plates with antibiotics; representative agar plates are shown. (D) Colony forming units (CFUs) of fungi in fecal samples were counted. (E) Fecal fungal DNA was extracted, and the abundance of Candida spp. was detected by qPCR. Fold change was calculated relative to vehicle-treated mice on control diet. (F) Serum levels of ALT. (G) Hepatic triglyceride content. (H) Representative oil red O staining of liver sections (scale bar, 100 μm). (I) Hepatic levels of F4/80 mRNA. (J) Serum levels of ethanol. (K and L) Hepatic levels of Cyp2e1 and Adh1 mRNAs. Figure 3A–B was performed in 3 independent experiments. Figure 3C–L was conducted in 2 independent experiments. Results are expressed as mean±SEM. P values between groups of mice fed the ethanol diet, with vs without nystatin, were determined by 2-way ANOVA with Tukeýs post-hoc test (A, D, E, F, G, and I) or 2-sided Student t test (J-L). *P<0.05. See also Figure S3.
Candida-specific TCR transgenic T cells promote ethanol-induced liver disease.
To further define the importance of the C. albicans-specific Th17 cell-mediated response in the development of ethanol-induced liver disease, we placed Candida-specific TCR transgenic mice (Rag1−/−/CaTCRtg) and their transgene-negative littermates (Rag1−/−) on chronic plus binge ethanol diets. In these transgenic mice, peripheral CD4+ T cells express a transgene encoding TCRs that specifically recognize Candida-derived antigens.24 Compared with ethanol-fed transgene-negative littermates, Candida-specific TCR transgenic mice had higher frequencies of C. albicans-activated Th17 cells in the liver (Fig. 4A, B and Supplementary Fig. 4A–B) and developed more severe ethanol-induced liver injury (Fig. 4C), steatosis (Fig. 4D–E) and inflammation (Fig. 4F); intestinal absorption or hepatic metabolism of ethanol did not differ significantly between transgene-negative and positive mice (Fig. 4G–I).
Figure 4. Candida-specific TCR transgenic mice develop more severe ethanol-induced liver disease.
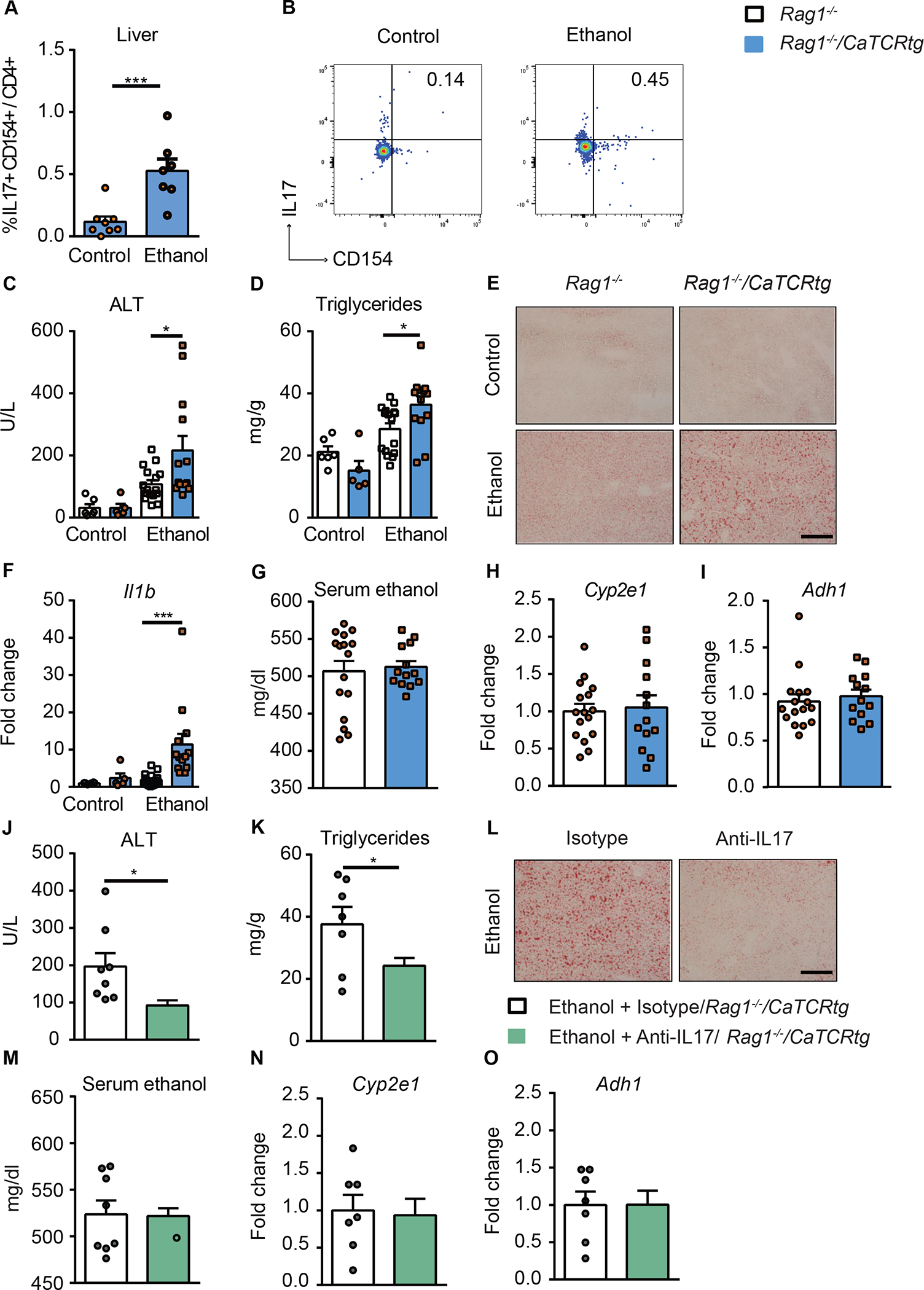
Candida-specific TCR transgenic mice (Rag1−/−/CaTCRtg) and their transgene-negative littermates (Rag1−/−) were fed a chronic plus binge ethanol diet or isocaloric diet. (A) Fungus-activated Th17 cells in liver were detected after isolation of hepatic mononuclear cells following 6 hrs ex vivo stimulation with C. albicans lysate. (B) Flow cytometry plots showing IL17A+ and CD154+ staining of CD4+ T cells. (C) Serum levels of ALT. (D) Hepatic triglyceride content. (E) Representative oil red O-stained liver sections (scale bar, 100 μm). (F) Hepatic levels of Il1b mRNA. (G) Serum levels of ethanol. (H and I) Hepatic levels of Cyp2e1 and Adh1 mRNAs. (J–O) Candida-specific TCR transgenic mice (Rag1−/−/CaTCR tg) were fed a chronic plus binge ethanol diet and given injections of an isotype antibody (control, white) or antibody against IL17A (green) 5 days before sacrifice. (J) Serum levels of ALT. (K) Hepatic triglyceride content. (L) Representative oil red O-stained liver sections (scale bar, 100 μm). (M) Serum levels of ethanol. (N and O) Hepatic levels of Cyp2e1 and Adh1 mRNAs. Figure 4A–B, Figure 4C–I and Figure 4J–O were conducted in 2 independent experiments. Results are expressed as mean±SEM. P values determined by 2-sided Student t test (A, G, H, I, J, K, M, N and O) and 2-way ANOVA with Tukeýs post-hoc test (C, D and F). *P<0.05, ***P<0.001. See also Figure S4.
To demonstrate that the Candida-induced Th17 response contributes to the development of ethanol-induced liver disease, we investigated the effects of an antibody against IL17 on liver damage in Candida-specific TCR transgenic mice. Administration of a neutralizing antibody against IL17 significantly decreased liver injury and steatosis in Rag1−/−/CaTCRtg mice (Fig. 4J–L) and did not affect serum ethanol levels or hepatic metabolism of ethanol (Fig. 4M–O). IL17 therefore mediates the disease-exacerbating effect of the Candida-specific TCR transgene.
Candida-specific T cells are a small subset of antigen-specific T cells within the entire polyclonal repertoire in a physiological context.24 To further study the disease promoting effect of Candida-specific TCR-transgenic T cells, we adoptively transferred C. albicans-primed T cells from Candida-specific TCR transgenic mice (Rag1−/−/CaTCRtg) or non-transgenic control wild-type mice into wild-type mice during chronic plus binge ethanol feeding. For the adoptive transfer of C. albicans-primed T cells, Rag1−/−/CaTCRtg and wild-type C57BL/6 mice were gavaged with C. albicans. CD4+ T cells from the spleen and lymph nodes were collected and cocultured with antigen presenting bone marrow-derived dendritic cells in the presence of C. albicans lysate. After 6 days of expansion, CD4+ T cells were transferred to wild-type recipient mice during chronic plus binge ethanol feeding (Fig. 5A, Supplementary Fig. 4C). C. albicans-primed Thy1.1+ CD4+ Vα2+ hector T cells from Rag1−/−/CaTCRtg mice were detected in livers of ethanol-fed wild-type recipient mice (Fig. 5B–C, Supplementary Fig. 4D), suggesting successful adoptive transfer and in vivo expansion. Compared with mice that received C. albicans-primed CD4+ T cells from wild-type mice, mice with adoptively transferred C. albicans-primed CD4+ T cells from Rag1−/−/CaTCRtg mice showed more severe ethanol-induced liver injury (Fig. 5D), steatosis (Fig. 5E–F), inflammation (Fig. 5G), but similar serum ethanol levels and hepatic metabolism of ethanol (Fig. 5H–J), indicating an exacerbating effect of C. albicans-primed hector Rag1−/−/CaTCRtg T cells for ethanol-induced liver disease.
Figure 5. Candida-specific TCR transgenic hector T cells promote ethanol-induced liver disease.
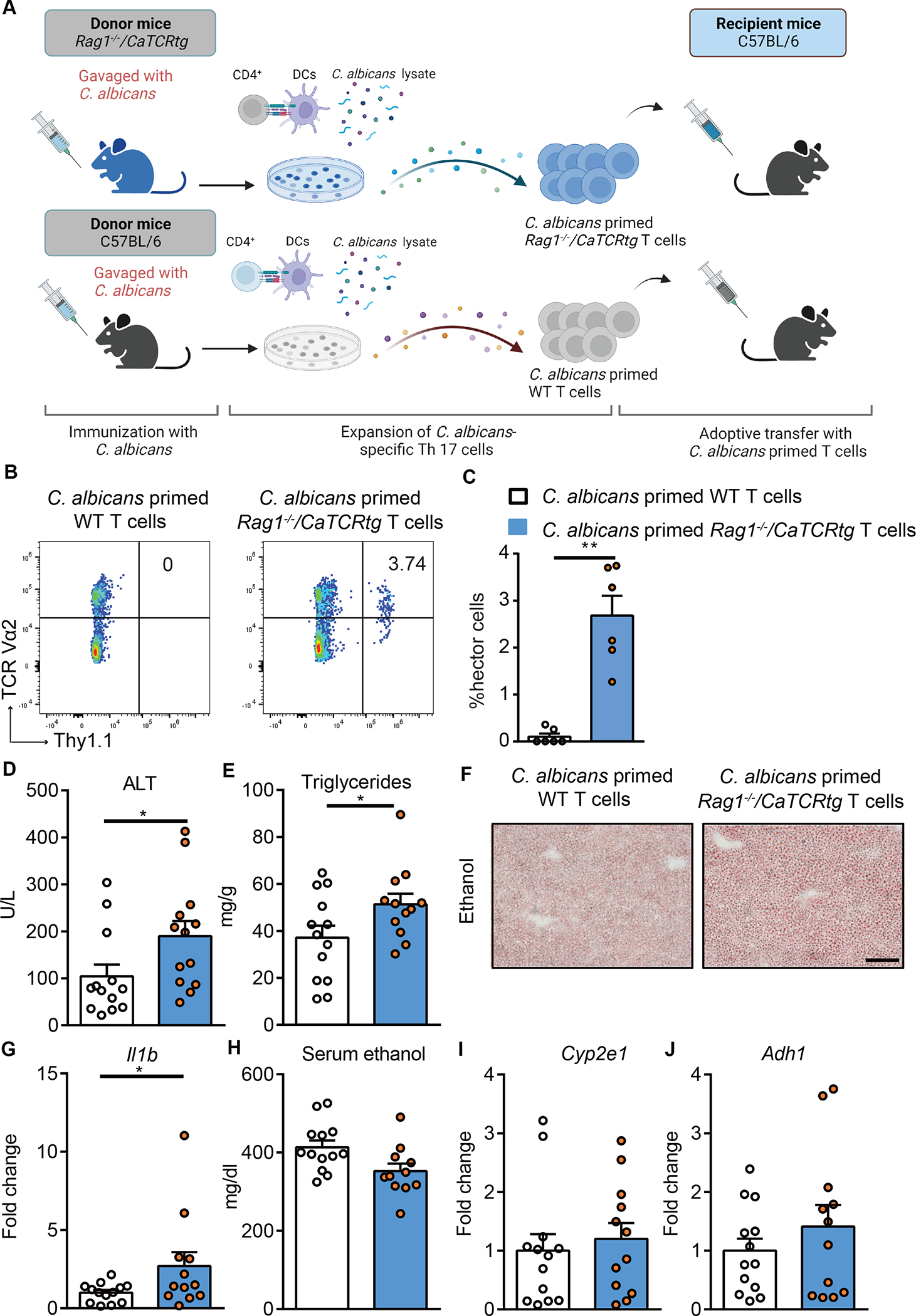
(A) Diagram of adoptive transfer of C. albicans-primed Rag1−/−/CaTCRtg and wild-type T cells to wild-type mice. On day 0, Candida-specific TCR transgenic (Rag1−/−/CaTCRtg) and C57BL/6 donor mice were gavaged with C. albicans. On day 10, CD4+ T cells from lymph nodes and spleen of donor mice were co-cultured with bone marrow-derived dendritic cells in the presence of C. albicans lysate for 6 days. On day 16, CD4+ T cells were injected intravenously to C57BL/6 mice (day 13 of chronic plus binge ethanol feeding). Created with BioRender.com. (B and C) Thy1.1+ CD4+ Vα2+ hector T cells were detected in livers of ethanol-fed recipient mice at the time of collection. (D) Serum levels of ALT. (E) Hepatic triglyceride content. (F) Representative oil red O-stained liver sections (scale bar, 100 μm). (G) Hepatic levels of Il1b mRNA. (H) Serum levels of ethanol. (I and J) Hepatic levels of Cyp2e1 and Adh1 mRNAs. Figure 5 was conducted in 2 independent experiments. Results are expressed as mean±SEM. Fold change was calculated relative to mice that were adoptively transferred with C. albicans-primed CD4+ T cells from wild-type C57BL/6 mice. P values determined by 2-sided Student t test. *P<0.05. See also Figure S4.
For the adoptive transfer of non-primed T cells from Candida-specific TCR transgenic mice (Rag1−/−/CaTCRtg) and wild-type C57BL/6 mice, CD4+ T cells from the spleen and lymph nodes were collected and transferred to wild-type recipient mice during chronic plus binge ethanol feeding (Supplementary Fig. 5A). Mice with adoptively transferred CD4+ T cells from Rag1−/−/CaTCRtg mice had more ethanol-induced liver disease as compared with mice that received CD4+ T cells from wild-type mice (Supplementary Fig. 5B–H), indicating that hector Rag1−/−/CaTCRtg T cells without C. albicans-priming also promote ethanol-induced liver disease.
Adoptively transferred C. albicans-primed polyclonal T cells exacerbate ethanol-induced liver disease in wild-type mice
To further investigate the role of C. albicans-primed polyclonal T cells in the development of ethanol-induced liver disease, we adoptively transferred C. albicans-primed polyclonal T cells into wild-type mice during chronic plus binge ethanol feeding. IL17A-eGFP reporter mice were gavaged with C. albicans or S. cerevisiae. CD4+ T cells from the spleen and lymph nodes were collected and cocultured with antigen presenting bone marrow-derived dendritic cells in the presence of C. albicans or S. cerevisiae lysate. After 6 days of expansion, polyclonal CD4+ T cells were assessed for IL17A-eGFP+ expression and then transferred to wild-type recipient mice during chronic plus binge ethanol feeding (Fig. 6A, Supplementary Fig. 6A). Polyclonal CD4+ T cells from C. albicans-gavaged mice that were stimulated with C. albicans lysate contained 7.04% IL17A-eGFP–producing T cells, whereas S. cerevisiae-primed T cells comprised 1.09% IL17A-eGFP–producing T cells (Fig. 6B, Supplementary Fig. 6B). A higher number of adoptively transferred, C. albicans-primed, IL17A-eGFP+ CD4+ T cells were found in livers of ethanol-fed mice than of S. cerevisiae-primed IL17A-eGFP+ CD4+ T cells (Fig. 6C–D, Supplementary Fig. 6C). Mice with adoptively transferred C. albicans-primed polyclonal CD4+ T cells had more ethanol-induced liver injury (Fig. 6E), steatosis (Fig. 6F–G), inflammation (Fig. 6H), but similar serum ethanol levels and hepatic metabolism of ethanol (Fig. 6I–K) compared with mice that received S. cerevisiae-primed CD4+ T cells, supporting the role of C. albicans-specific Th17 cells in the development of ethanol-induced liver disease.
Figure 6. C. albicans-primed polyclonal T cells exacerbate ethanol-induced liver disease.
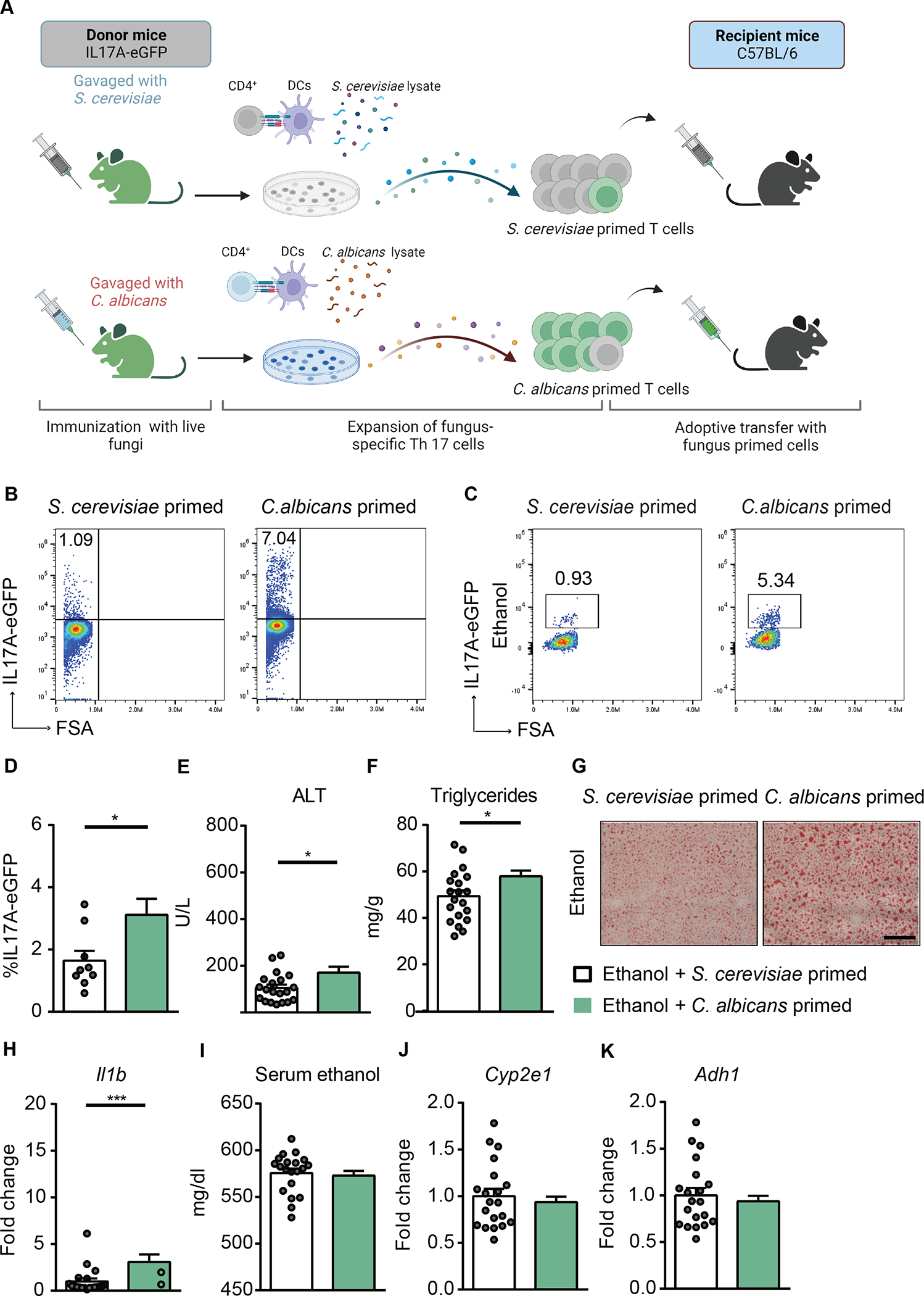
(A) Diagram of adoptive transfer of C. albicans-specific polyclonal T cells to mice. On day 0, IL17A-eGFP donor mice were gavaged with C. albicans or S. cerevisiae. On day 10, polyclonal CD4+ T cells from mesenteric lymph nodes and spleen of donor mice were co-cultured with bone marrow-derived dendritic cells in the presence of fungal lysates for 6 days. On day 16, polyclonal CD4+ T cells were injected intravenously to C57BL/6 mice (day 13 of chronic plus binge ethanol feeding). Created with BioRender.com. (B) The proportion of IL17A-eGFP+ cells among total CD4+ T cells after ex vivo stimulation for adoptive-transfer was assessed by flow cytometry. (C and D) IL17A-eGFP+ cells were detected in livers of ethanol-fed recipient mice at the time of collection. (E) Serum levels of ALT. (F) Hepatic triglyceride content. (G) Representative oil red O-stained liver sections (scale bar, 100 μm). (H) Hepatic levels of Il1b mRNA. (I) Serum levels of ethanol. (J and K) Hepatic levels of Cyp2e1 and Adh1 mRNAs. Figure 6 was conducted in 3 independent experiments. Results are expressed as mean±SEM. P values determined by 2-sided Student t test. *P<0.05, ***P<0.001. See also Figure S5.
Kupffer cells mediate the disease exacerbating effect of adoptively transferred C. albicans-primed polyclonal T cells
IL17 receptor A (IL17ra) is a common receptor for the IL17 family cytokines.25 To determine whether the disease promoting effect of C. albicans-primed T cells is mediated by IL17ra, we generated IL17ra bone marrow chimeric mice (IL17raΔBM) using a combination of clodronate-mediated Kupffer cell depletion, irradiation, and bone marrow transplantation.26 Wild-type recipient mice were transplanted with bone marrow cells from mice lacking IL17ra or wild-type mice. Chimeric mice were subjected to the chronic-plusbinge ethanol diet model and adoptively transferred C. albicans-primed polyclonal T cells (Supplementary Fig. 7A). Chimeric mice with bone marrow-derived cells lacking IL17ra and injected with C. albicans-primed polyclonal T cells showed less ethanol-induced liver injury, steatosis and inflammation compared with wild-type mice transplanted with wild-type bone marrow (Supplementary Fig. 7B–G). No significant difference was observed in serum ethanol level and hepatic expression of Adh1 and Cyp2e1 between groups (Supplementary Fig. 7H–J).
To further confirm the effector cells of hepatic Th17, we adoptively transferred IL17A-eGFP cells stimulated with C. albicans lysate into mice that lack IL17ra expression on Kupffer cells (IL17raΔKC) or hepatocytes (IL17raΔHep). Liver disease exacerbation of C. albicans-primed T cells was reduced in ethanol-fed IL17raΔKC as compared with their ethanol-fed IL17rafl/fl littermate controls (Fig. 7A–H). No differences in liver injury and steatosis were found between ethanol-fed IL17raΔHep mice and IL17rafl/fl controls following adoptive transfer of C. albicans-primed T cells (Supplementary Fig. 7K–Q). These results indicate that the disease promoting effect of adoptively transferred C. albicans-primed polyclonal T cells is mediated via IL17ra signaling on Kupffer cells following ethanol administration.
Figure 7. Kupffer cells mediate the disease exacerbating effect of adoptively transferred polyclonal C. albicans-primed T cells.
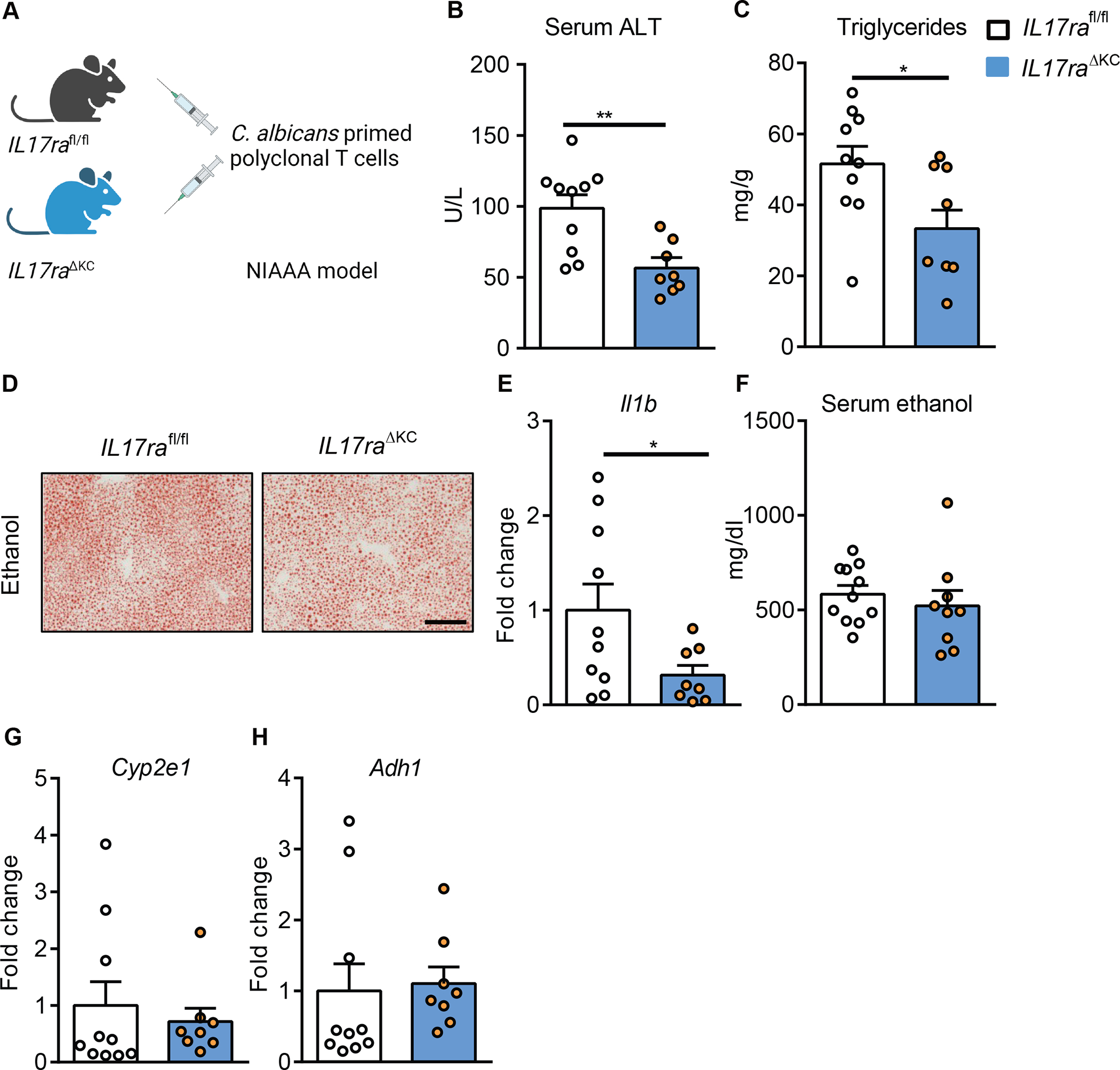
(A) Diagram of adoptive transfer of polyclonal C. albicans-primed T cells to mice lacking IL17ra on Kupffer cells (II17raΔKC) and their littermate II17rafl/fl mice. II17raΔKC and littermate II17rafl/fl mice were fed a chronic plus binge ethanol diet and injected C. albicans-primed polyclonal T cells intravenously 3 days before harvesting. (B) Serum levels of ALT. (C) Hepatic triglyceride content. (D) Representative oil red O-stained liver sections (scale bar, 100 μm). (E) Hepatic levels of Il1b mRNA. (F) Serum levels of ethanol. (G and H) Hepatic levels of Cyp2e1 and Adh1 mRNAs. Figure 7 was conducted in 2 independent experiments. Results are expressed as mean±SEM. P values determined by 2-sided Student t test. *P<0.05, **P<0.01. See also Figure S6 and Figure S7.
Discussion
Intestinal fungal dysbiosis is characterized by an increase in the fungal pathobiont C. albicans in patients with alcohol-associated liver disease.8–10 Th17 cells participate in immune surveillance in the barrier site to prevent dysbiosis, fungal overgrowth, and translocation across intestinal barriers. C. albicans is the strongest known inducer of the Th17 cell-mediated response in humans.16 In this study, Th17 cells directed against C. albicans increase in the blood and liver of patients with alcohol-associated liver disease. C. albicans-specific Th17 cells migrate from the intestine to the liver, where they contribute to ethanol-induced liver disease (Graphical Abstract). Our results provide evidence that the immune-protective Th17 cell-mediated response promotes liver disease via migration of these cells to the liver, continuous exposure of antigens to the liver via the portal vein, and secretion of IL17. Similar to this study, intestinal inflammation expanded C. albicans-specific and cross-reactive Th17 cells which can contribute to lung inflammation induced by airborne A. fumigatus.16 In that study and ours, intestine-primed C. albicans-specific CD4+ T cells migrated to other organs and induced a chronic inflammatory response upon exposure to fungal antigens, which are delivered via the airways to the lungs or the portal vein to the liver. Chronic alcohol use is associated with a dysfunctional gut barrier, which allows the portal vein to deliver microbial antigens from the intestine to the liver. The fungal proteins that prime C. albicans-specific Th17 cells remain largely unknown. Our previous work demonstrated that a fungal surface component, β-glucan, can translocate to liver and bind to C-type lectin domain family 7 member A (CLEC7A) on hepatic macrophages, which release IL1b and increase ethanol-induced liver disease in mice.26 Also, candidalysin, a toxic peptide secreted by C. albicans, can damage primary hepatocytes, and promote development of alcohol-associated liver disease.9 Deletion of ECE1 in C. albicans, which encodes candidalysin, reduced Th17 responses in mice after intestinal C. albicans colonization.27 Whether candidalysin itself is a target of C. albicans-specific Th17 cells is currently unknown. Studies showed that Als1 and Als3 (proteins encoded by the agglutinin-like sequence gene family), alcohol dehydrogenase (Adh1), cell wall-associated proteins including yeast wall protein1 (Ywp1), enolase (Eno1), fructose-bisphosphate aldolase 1 (Fba1), glyceraldehyde-3-phosphate dehydrogenase (GAP1), glyceraldehyde-3-phosphate dehydrogenase (G3pdh), hyphal wall protein-1 (Hwp1), methyltetrahydropteroyltriglutamate (Met6), and phosphoglycerate kinase (Pgk1) were involved in the Candida-induced Th17 differentiation in vitro.24 Studies are needed to identify the specific antigens of C. albicans that activate Th17 cells and promote development of alcohol-associated liver disease.
IL17 receptors have been detected on various liver cells, including hepatocytes, Kupffer cells, stellate cells, biliary epithelial cells, and sinusoidal endothelial cells to induce immune cell infiltration and liver damage.25 But the cell-specific role of IL17ra signaling can be context-specific in terms of the disease settings and source of IL17 producing cells. The role of cell type specific expression of IL17ra signaling was investigated in a hepatocellular carcinoma (HCC) model induced by DEN treatment together with high fat diet and ethanol feeding for 18 weeks.25 Mice lacking IL17ra expression on hepatocytes were protected from DEN-induced HCC and developed less steatosis and fibrosis. IL17 signaling in myeloid cells is also critical for hepatic tumorigenesis, steatosis, inflammation and fibrosis. In our study, we have shown the disease promoting effect of adoptively transferred C. albicans-primed Th17 cells is mediated via IL17ra signaling on Kupffer cells, not hepatocytes, using the chronic plus binge (NIAAA) model. In addition, our study investigated the specific role of C. albicans specific Th17 cells, not the total IL17 producing cells, for the development of ethanol-induced liver disease. Several other Th17 inducing bacterial and fungal microbes have been identified in mice including segmented filamentous bacteria (SFB),12,14 Citrobacter rodentium,28 Malassezia spp.,29 and Aspergillus fumigatus.16 We detected segmented filamentous bacteria (SFB) in fecal samples from C57BL/6 mice obtained from Charles River and used in our study (not shown). Th17 cells directed against and/ or induced by segmented filamentous bacteria or other microbial species could contribute to alcohol-associated liver disease, but their specific role for disease pathogenesis requires further studies.
Under homeostatic conditions, Th17 cells are largely confined to intestinal tissues. When dysregulated, Th17 cells can translocate to peripheral organs and induce inflammation.16 Circulating levels of IL17 and frequency of IL17 producing cells are elevated in patients with alcohol-associated liver disease.30,31 But little is known as to the cause-and-effect studies of IL17 in the development of alcohol-associated liver disease. IL17 recruits neutrophils to the site of inflammation through IL8/Cxcl8 chemokine production. The effects of IL17 can be amplified by synergistic interaction with various liver resident cells via inflammatory cytokines. For example, IL17 differentiation can be affected by Tumor necrosis factor (Tnf), IL6 and Transforming growth factor beta 1 (Tgfb1) release from Kupffer cells and hepatic stellate cells, resulting in Th17/Treg imbalance and uncontrolled inflammation.32 IL17 and Tnf act in synergy to induce IL6 and IL8 in hepatocytes, and activate hepatic stellate cells for collagen production and liver fibrosis.33 The uncontrolled and chronic IL17 production and its induction of inflammatory and fibrotic cytokines results in a vicious cycle that leads to chronic infiltration of immune cells and liver damage.
Th17 cells mainly produce IL17A and to a lesser extent IL22, IFNG, and IL21. IL22 has hepatoprotective and proliferative effects and reduces ethanol-induced liver injury.34,35 IL22 could counteract the injurious effects of IL17. Further studies are required to investigate the effects of C. albicans stimulated IL22 and reciprocal interactions with IL17 in the development of alcohol-associated liver disease.
In summary, our study demonstrates the important role of C. albicans-specific Th17 cells in the development of alcohol-associated liver disease. Chronic ethanol administration activates C. albicans-specific Th17 cell-mediated responses in patients and preclinical models. The increased C. albicans-specific Th17 cells in intestine could migrate to the liver, where they could produce IL17 that contributes to the development of alcohol-associated liver disease. Our findings provide insights into induction of intestinal Th17 cells and how they might contribute to development of liver disease. Antifungal drugs that target alcohol-associated overgrowth of C. albicans in the gut might reduce C. albicans-specific Th17 cell responses and be used in treatment of alcohol-related liver disease.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bernd Schnabl (beschnabl@ucsd.edu).
Materials availability
This study did not generate any unique reagents.
Date and code availability
Single cell sequencing data are currently deposited in a public database (GSE215931). 16S sequencing are registered at NCBI under BioProject PRJNA911776.
This study does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Actively drinking patients with a diagnosis of alcohol dependence were admitted for elective alcohol rehabilitation at St. Luc University Hospital in Brussels, Belgium. All patients were heavy drinkers, consuming more than 60 g of alcohol per day (self-reported consumption) for more than 1 year. On the day of admission, Fibroscan (Echosense, Paris, France) with controlled attenuation parameter (CAP) and liver stiffness in kPa was performed and blood samples were collected. Patients were excluded from the study if they used antibiotics, probiotics, or prebiotics during the 2 months preceding enrollment, were receiving immunosuppressive medications, or had diagnoses of diabetes, inflammatory bowel disease, liver disease of any other etiology, or clinically significant cardio-vascular, pulmonary, or renal co-morbidities. Patients with alcohol use disorder and liver disease were compared with non-alcoholic volunteers who drank less than 20 g of alcohol per day and were enrolled at St. Luc University Hospital in Brussels, Belgium and University of Kiel, Kiel, Germany. For the healthy controls, there were 15 male and 18 female subjects with median age of 34 years old (interquartile range 16). For the patients with alcohol use disorder and liver disease, there were 19 male and 17 female with median age of 50 years old (interquartile range 16.3) (Supplementary Table 1). Liver biopsies were collected from one male patient and one female patient with median age of 49 (interquartile range 4) when clinically indicated (Supplementary Table 2). Excess liver tissue was used for research. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the institution’s human research and ethical committee at the Université Catholique de Louvain, Brussels, Belgium (B403201422657) and at the University of Kiel, Germany. Written informed consent was obtained from all patients and non-alcoholic volunteers.
Mice
C57BL/6 mice used in Figures 2, 3, 5, 6 and Supplementary Figures 2, 3, 4, 5, 6 were purchased from Charles River. Kaede transgenic mice on a C57BL/6 genetic background.15 Rag1−/−/CaTCRtg and Rag1−/− mice,24 and IL17A-eGFP mice (C57BL/6-Il17atm1Bcgen, Jackson Laboratory, strain #: 018472) have been described. Mice with a whole body IL17ra deficiency were generated by crossing IL17rafl/fl with E2a-Cre mice.36 Mice lacking IL17 on hepatocytes (IL17raΔHep) were generated by crossing IL17rafl/fl mice25 with albumin-Cre mice.37 Mice lacking IL17 on Kupffer cells (IL17raΔKC) were generated by crossing IL17rafl/fl mice with Kupffer cell specific Clec4f-Cre mice.38 All protocols were conducted in accordance with the guidelines in the Institutional Animal Care and Use Committee of the University of California, San Diego (La Jolla, CA).
Female mice (age, 9–10 weeks) were fed a chronic plus-binge ethanol diet (NIAAA model)22 or a chronic Lieber DeCarli diet model for 8 weeks.39 For the NIAAA model, mice received an isocaloric control liquid Lieber DeCarli diet from day 0 to day 5. Afterwards, mice were fed Lieber DeCarli diet containing 36% (vol/vol) ethanol for 10 days. On the last day, mice were gavaged with a single dose of ethanol in the night and sacrificed 9 hrs later (the next morning). Pair-fed control mice were placed on a control liquid diet during the 16 days and received isocaloric maltodextrin and euthanized 9 hrs later.
During chronic Lieber DeCarli feeding, the caloric intake from ethanol was 0 on day 1, 10% of total calories on days 2 and 3, 20% on days 4 and 5, 30% from day 6 until the end of 6 weeks, and 36% for the last 2 weeks. Control mice received an isocaloric amount of maltodextrin instead of ethanol.39
To study the effects of an antifungal drug on C. albicans-specific Th17 cells and ethanol-induced liver disease, mice received nystatin (40,000 unit/kg per day),40 which was dissolved in the liquid Lieber DeCarli ethanol-containing and control diets, for the last 10 days. Xanthan gum was added to liquid diet to increase viscosity of the liquid diet solutions and prevent gravitational separation of nystatin.
To block IL17 signaling in Rag1−/−/CaTCRtg mice, 500 μg of InVivoMab anti-mouse IL17A (clone 17F3) or isotype control antibodies (Purified Rat IgG2a) were intraperitoneally injected 5 days before sacrifice.
To generate bone marrow chimeric mice, C57BL/6 recipient female mice (age, 6 weeks) received 5 GY radiation twice using X-ray irradiation system. Bone marrow cells were collected from either female C57BL/6 donor mice or IL17rafl/fl mice crossed with E2a-Cre donor mice and injected to recipient mice via the tail vein. Two weeks later, mice were injected i.p. with 200 μL of clodronate liposomes (5 mg/mL; Vrije Universiteit, Amsterdam, Netherlands) to deplete radioresistant Kupffer cells.26 Mice were subjected to NIAAA model two weeks after bone marrow transplantation. Chimeric mice were injected with C. albicans-primed T cells three days before sacrificing the mice. To investigate the effector cells of hepatic Th17, Il17raΔKC mice, Il17raΔHepand their control Il17rafl/fl littermates (age, 9 weeks) were subjected to the NIAAA model and injected with C. albicans-primed polyclonal T cells three days before sacrificing the mice.
METHODS DETAILS
Antigen-reactive T cell enrichment
Antigen-reactive T cell enrichment (ARTE) was conducted as previously described.16 In brief, depending on the available cell number, 0.5–2×107 human peripheral blood mononuclear cells (PBMCs) were plated in RPMI-1640 medium (GIBCO), supplemented with 5% (v/v) human AB-serum (Sigma Aldrich, Schnelldorf, Germany) at a cell density of 1×107 PBMCs / 2 cm2 in cell culture plates and stimulated for 7 hrs with C. albicans (40μg/mL) and S. cerevisiae (40μg/mL) lysates in presence of 1 μg/mL CD40 and 1 μg/mL CD28 pure antibody (both Miltenyi Biotec, Bergisch Gladbach, Germany). 1 μg/mL Brefeldin A (Sigma Aldrich) was added for the last 2 hrs. Cells were labeled with CD154-Biotin followed by anti-Biotin MicroBeads (CD154 MicroBead Kit; Miltenyi Biotec) and magnetically enriched by two sequential MS columns (Miltenyi Biotec). Surface staining was performed on the first column, followed by fixation and intracellular staining on the second column. Frequencies of antigen-specific T cells were determined based on the total cell count of CD154+ cells after enrichment, normalized to the total number of CD4+ T cells applied on the column. For each stimulation, background cells enriched from the non-stimulated control were subtracted.
Single cell RNA sequencing and TCR sequencing
For single cell transcriptomics, CD154+ cells were isolated by ARTE and further purified by FACS sorting on a BD FACS ARIA Fusion (BD Bioscience) instrument based on dual expression of CD154 and CD69. Cells were sorted into pre-coated low-bind collection tubes, containing 500 μL sterile filtered 1xPBS + 0.2% AB serum. Cells were centrifuged for 10 min at 400xg. The supernatant was carefully removed leaving 20 μL for loading on a Chromium Chip K (10x Genomics) according to the manufacturer’s instructions for processing with the Chromium Next GEM Single Cell 5′Library and Gel Bead Kit v2. TCR single-cell libraries were subsequently prepared with the Chromium Single Cell V(D)J Enrichment Kit, Human T Cell. Libraries were sequenced on Illumina NovaSeq 6000 machine with 2×100 bp for gene expression, aiming for 50,000 reads per cell for gene expression and 5000 reads per cell for TCR libraries.
Single-cell transcriptome analysis
The preprocessing of the single cell RNA-data was performed with the 10x Genomics’ Cell Ranger software v6.0.0 using the reference GRCh38 v3.0.0 for the mappings. The resulting filtered feature-barcode matrix files were analyzed with the R package Seurat v.4.1.1.41 Thereby, all genes with a detected expression in less than 0.1% of the non-empty cells were excluded. Moreover, TCR genes were not considered for further analyses to avoid functional clustering of cells based on TCR information. To minimize the number of doublets, empty cells, and cells with a transcriptome in low quality, only cells harboring between 200 and 3500 RNA features and less than 5% mitochondrial RNA were selected for further processing. Afterwards, data were log-normalized and scaled based on all genes. After performing a PCA dimensionality reduction (20 dimensions) with the RunPCA function, the expression values were corrected for effects caused by different sample preparation time points using the R package Harmony v1.0.42 In the final steps, the Uniform Manifold Approximation and Projection (UMAP) dimensional reduction was performed with the RunUMAP function using 20 dimensions. A shared nearest neighbor graph was created with the FindNeighbors method, and the clusters identification was performed with a resolution of 0.8 using the FindClusters function. Clusters identified to be composed of contaminating cells (non-T cells) and cycling cells were excluded from further analyses. Finally, 11,068 cells were used for downstream analysis.
Single cell T cell receptor (TCR) sequence analysis
Single-cell T-cell receptor repertoire clonotype tables were generated using the VDJ command of the Cellranger software v6.0.0 from 10xGenomics and using the reference GRCh38 version 5.0.0. Clonotype tables were filtered to include cells that passed quality filtering in the gene expression analysis.
Bulk T cell receptor (TCR) sequencing
Liver biopsies were obtained in RNAlater and transferred into 2 mL tubes containing 600 μL RLT+ buffer + 1% β-Mercaptoethanol. Tissue was disrupted using the Qiagen tissue lyser. PBMCs were isolated from paired blood samples by density gradient centrifugation (Biocoll; Biochrom, Berlin, Germany) and CD4+ T cells were enriched using CD4 Microbeads (Miltenyi Biotec). RNA was isolated from liver samples and CD4+ T cells from PBMCs using the RNeasy Micro Kit (Qiagen). TCR libraries were prepared using human TCR RNA multiplex kit (MiLaboratories), according to the manufacturer’s protocol using 24 PCR cycles (liver) and 18 PCR cycles (blood) for the 1st PCR, and 14 PCR cycles (liver) and 16 PCR cycles (blood) for the 2nd PCR amplification. The libraries were sequenced on an Illumina NovaSeq6000 machine, SP v1.5 flow cells and 300 cycles. Data of both TCRα and β chains were obtained.
Bulk and single-cell TCR overlap analyses
PCR and sequencing error correction were performed through identification and selection of unique molecular identifiers using the software MiGEC, version 1.2.6.43 Filtered sequences were aligned on a TCR gene reference. Clonotypes were identified, grouped and CDR3 sequences were identified using the software MiXCR version 3.0.14.44 Clonotype tables containing clonotype counts, frequencies, CDR3 nucleotide and amino acid sequences and V(D)J genes were obtained and used for further analysis. TCR alpha and beta sequences were compared with the C. albicans-stimulated single-cell TCRs, thus identifying antigen-reactive TCRs present in the liver and in the total CD4+ T cells in the blood (bulk TCR samples). The percentage of C. albicans-stimulated TCR sequences was calculated as the sum of the counts of C. albicans-stimulated TCRs in the bulk TCR samples divided by the total count number of each sample and multiplied by one hundred.
Isolation of mononuclear cells
Isolated mouse mesenteric lymph nodes were placed in RPMI 1640 medium (Gibco) and squeezed through a 70-μm cell strainer. Cells were washed twice and resuspended in the T cell culture medium. Mouse portal vein blood (around 200 μL) was collected in heparin tubes, resuspended in 40% percoll, and overlayed onto 70% percoll, followed by density-gradient centrifugation (GE Healthcare, Sweden). Mononuclear cells from portal vein blood were collected from the percoll interface, washed twice, and resuspended in the T-cell culture medium. Mouse livers were homogenized and filtered through a 70 μm cell strainer. Mononuclear cells were isolated following the same density gradient centrifugation as portal vein blood. Red blood cell lysing buffer (Sigma, St. Louis, MO) was used to remove red blood cells from cell suspensions. Cells were washed twice and resuspended in the T cell culture medium.45
Fungus-activated T cell detection
1–2 ×106 mononuclear cells were resuspended in T cell culture medium and seeded into 96-well culture plates, followed by stimulation with whole fungal lysates (40 μg/mL) for 6 hrs in the presence of 1 mg/mL anti-CD40, 1 mg/mL anti-CD28 purified antibody (Biolegend), and BD GolgiStop™ (BD, Biosciences). Lyophilized extracts of Candida albicans (XPM15D3A5) and Saccharomyces cerevisiae (XPM67D3A2.5) were purchased from Greer Laboratories (Lenoir, NC, USA). Cells were stained with 7AAD, LIVE/DEAD dyes, CD4, CD154, IL17, and RORγt. Frequencies of fungus-specific T cells were determined based on counts of CD154+ IL17+ cells, normalized to the total number of CD4+ T cells.
Flow cytometry
For human T cell analyses, cells were stained with fluorochrome-conjugated antibodies (see Key Resources Table). Viobility 405/520 Fixable Dye (Miltenyi Biotec) was used to exclude dead cells. For intracellular staining, cells were fixed and permeabilized with the Inside Stain Kit (Miltenyi Biotec). Data were acquired on a LSR Fortessa (BD Bioscience, San Jose, CA, USA). FlowJo (Treestar, Ashland, OR, USA) software was used for analysis.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC/Cyanine7 anti-mouse CD4 Antibody | Biolegend | Cat#100413; RRID: AB_312698 |
| FITC anti-mouse CD4 Antibody | Biolegend | Cat# 100509; RRID: AB_312712 |
| PE anti-mouse CD4 Antibody | Biolegend | Cat# 116005; RRID: AB_313690 |
| APC anti-mouse CD154 Antibody | biolegend | Cat# 106510; RRID: AB_2561561 |
| PE anti-mouse CD154 Antibody | Biolegend | Cat# 106505; RRID: AB_313270 |
| PE/Cy7 anti-mouse IL17A Antibody | Biolegend | Cat# 506922; RRID: AB_2125010 |
| BD Pharmingen™ 7-AAD | BD Biosciences | Cat# 559925; RRID: AB_2869266 |
| CD16/CD32 Monoclonal Antibody | Thermo Fisher | Cat# 14-0161-82; RRID: AB_467133 |
| Pacific Blue™ anti-rat CD90/mouse CD90.1 (Thy-1.1) Antibody | Biolegend | Cat# 202522; RRID: AB_1595477 |
| APC/Cyanine7 anti-mouse TCR Vα2 Antibody | Biolegend | Cat# 127818; RRID: AB_AB_10682897 |
| Purified anti-mouse CD28 Antibody | Biolegend | Cat# 102102; RRID: AB_312867 |
| Purified anti-mouse CD40 Antibody | Biolegend | Cat# 102802; RRID:AB_312935 |
| Anti-human CD4-APC-Vio770 (clone: M-T466) | Miltenyi Biotec | Cat#130-113-251, RRID:AB_2726053 |
| Anti-human CD8-VioGreen (clone: REA734) | Miltenyi Biotec | Cat#130-110-684, RRID:AB_2659241 |
| Anti-human CD14-VioGreen (clone: REA599) | Miltenyi Biotec | Cat#130-110-525, RRID:AB_2655057 |
| Anti-human CD20-VioGreen (clone: LT20) | Miltenyi Biotec | Cat#130-096-904, RRID:AB_2726147 |
| Anti-human CD45RA-PE-Cy5 (clone: HI100) | BioLegend | Cat#304110, RRID:AB_314414 |
| Anti-human CD154-FITC (clone: REA238) | Miltenyi Biotec | Cat#130-109-469, RRID:AB_2751146 |
| Anti-human IL-17A BV605 (clone: BL168) | BioLegend | Cat#512326, RRID: AB_2563887 |
| Anti-human IL22-PerCP-eFlour710 | Invitrogen | Cat# 46-7222-82, RRID: AB_2573839 |
| Anti-human IFNg-BV785 (clone: 4S.B3) | BioLegend | Cat#502542, RRID: AB_2563882 |
| Anti-human IL-17F BV650 (clone: O33-782) | BD Biosciences | Cat#564264, RRID: AB_2869555 |
| Anti-human IL-2-BV711 (clone: 5344.111) | BD Biosciences Miltenyi Biotec | Cat#563946, RRID:AB_2738501 |
| Anti-human GM-CSF APC (clone: REA1215) | Miltenyi Biotec | Cat#130-123-420, RRID: AB_2811516 |
| Anti-human IL-21-PE (clone: REA1039) | BioLegend | Cat#130-117-421, RRID:AB_2727941 |
| Anti-human IL-10-PE-Dazzle (clone: JES3-9D7) | BD Biosciences | Cat#501426, RRID:AB_2566744 |
| Anti-human Ki-67 Alexa Fluor 700 (clone: B56) | Miltenyi Biotec | Cat#561277, RRID:AB_10611571 |
| Anti-human IntB7 PE-Vio770 (clone: REA441) | Miltenyi Biotec | Cat# 130-106-442, AB_2652507 |
| CD154 MicroBead Kit, human | Miltenyi Biotec | Cat#130-092-658 |
| CD28 pure – functional grade, human (clone: 15E8) | Miltenyi Biotec | Cat#130-093-375, RRID: AB_1036134 |
| CD40 pure – functional grade, human (clone: HB14) | Miltenyi Biotec | Cat#130-050-201, RRID: AB_2665482 |
| Bacterial and virus strains | ||
| Candida albicans | M1477 | |
| Saccharomyces cerevisiae | ATCC | ATCC9763 |
| Lyophilized extracts of Saccharomyces cerevisiae | Greer Laboratories | Cat# XPM67D3A2.5 |
| Lyophilized extracts of Candida albicans | Greer Laboratories | Cat# XPM15D3A5 |
| Biological samples | ||
| Blood samples from human cohorts | Schnabl lab | This study |
| Liver samples from patients with alcohol related liver disease | Schnabl lab | This study |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI-1640 medium | Life technologies | Cat# 11875093 |
| FBS (heat inactivated) | CoreBio | Cat# FB-02 |
| Penicillin-Streptomycin | Thermo Fisher | Cat# 15140122 |
| Nystatin | Sigma | Cat# N1638-100ML |
| InVivo Mab anti-mouse IL17A (clone 17F3) | BioXcell /Fisher Sci | Cat# BE0173 |
| Recombinant Mouse GM-CSF Protein | R&D | Cat# 415-ML-010/CF |
| BD GolgiStop™ | BD | Cat# 554724 |
| Percoll | Fisher (Amersham) | Cat# 17-0891-01 |
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat# 00-5523-00 |
| EasySep™ Mouse CD11c Positive Selection Kit II | Stemcells | Cat# 18780 |
| EasySep™ Mouse CD4+ T Cell Isolation Kit | Stemcells | Cat# 19852 |
| DPBS, no calcium, no magnesium | Gibco™ | Cat# 14190144 |
| HBSS | Gibco/ Life Tech | Cat# 14175-095 |
| Critical commercial assays | ||
| TRIZOL | Life technologies/ Fisher scientific | Cat# 15596-018 |
| Infinity ALT (GPT) | Fisher | Cat# TR71121 |
| ALT (SGPT) Kinetic | TECO Diagnostics | Cat# A524-150 |
| Ethanol Colorimetric/Fluorometric Assay Kit | Abcam | Cat# ab65343 |
| Triglycerides Liquid, Reagent Set, 1x120 ml | Pointe Scientific Inc. | Cat# T7532-120 |
| iTaq UniverSYBR Green SMX 1000 | Core Bio service | Cat# 1725124 |
| DNeasy PowerSoil kit | Qiagen | Cat#12855-50 |
| High Capacity cDNA reverse Transcription Kit | Invitrogen (ABI) | Cat#4368814 |
| Deposited data | ||
| Raw single cell sequencing data | NCBI | GSE215931 |
| Raw 16S rRNA sequencing data | NCBI | PRJNA911776 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Charles River | NA |
| Mouse: Kaede mice | Mathis et al15 | NA |
| Mouse: Rag1−/−/CaTCRtg | LeibundGut-Landmann et al.24 | NA |
| Mouse: IL17A-eGFP mice | Jackson Laboratory | JAX: 018472 RRID: IMSR_JAX:018472 |
| Mouse: IL17rafl/fl | Kisseleva et al.37 | NA |
| Mouse: Il17rafl/fl E2a-Cre+ | Kolls et al.36 | NA |
| Mouse: Il17raΔBM | Schnabl lab | This study |
| Mouse: IL17raΔHep | Kisseleva et al.37 | NA |
| Mouse: Clec4f-cre-tdTomato | Glass et al.38 | NA |
| Mouse: IL17raΔKC | Schnabl lab | This study |
| Software and algorithms | ||
| FlowJo | Treestar | RRID:SCR_000410 |
| GraphPad Prism v9.3.1 | GraphPad Software, LLC | https://www.graphpad.com/scientific-software/prism/ |
| R Project for Statistical Computing | RRID:SCR_001905 | |
Mouse cells were incubated with Fc block (eBioscience) and stained in different combinations of fluorochrome-conjugated antibodies. For intracellular staining, cells were fixed and permeabilized with the eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set. Data were acquired by Cytek Aurora NL-3000 or NovoCyte flow cytometer (ACEA Biosciences).
Photoconversion of Kaede transgenic mice
For photoconversion of mesenteric lymph nodes, Kaede transgenic mice on a chronic plus binge ethanol diet (ethanol) or isocaloric diet were anesthetized, and laparotomy was performed on the 9th day of chronic ethanol or control diets. Mesenteric lymph nodes were exposed to violet light for 9 min (every lymph node was exposed for 3 min with pause in between) using a hand-held laser (Electra Pro Series Violet Handheld Laser, Laser glow Technologies, Toronto, ON, Canada, 405 nm) with surrounding tissue covered by aluminum foil. Intestines were rinsed with 0.9% normal saline and then incisions were closed with nylon sutures. Livers were collected from mice 48 hrs after the surgery and isolated cells were analyzed by flow cytometry as described.46
Real-time quantitative PCR
Mouse liver RNA was extracted using TRIzol reagent (Life Technologies) and converted to cDNA using High-Capacity cDNA reverse Transcription Kit (Invitrogen). Primer sets are listed in Supplementary Table 3. Gene expression was measured by ABI StepOnePlus real-time PCR system using SYBR Green (Bio-Rad Laboratories). Expression was normalized to 18S ribosomal RNA (rRNA).
Biochemical analysis and histology
Serum levels of ALT and hepatic triglyceride levels were determined using Infinity ALT kit (Thermo Scientific) or ALT (SGPT) Kinetic (Teco Diagnostics), and Triglyceride Liquid Reagents kit (Pointe Scientific), respectively. Serum levels of ethanol were measured using the Ethanol Colorimetric Assay kit (Abcam) according to the manufacturer’s instructions. To determine lipid accumulation, liver sections were analyzed by Oil Red O staining (Sigma-Aldrich). Representative pictures are shown in each group.
Fungi culture
C. albicans (M1477) and S. cerevisiae were cultured on YPD medium (1% yeast extract, 2% peptone, 2% D-glucose) with antibiotics (100 μg/mL gentamicin and 100 μg/mL chloramphenicol) at 30 °C for 18 hrs. Cultures were washed in sterile PBS and adjusted to the required cell density by measuring OD600 in a microplate reader (Molecular Devices).9
Count of Fungi CFU
Fungal CFUs in mouse feces were counted as previously reported.9 Briefly, 2 pellets of mouse feces (around 50 mg) were homogenized and resuspended in 500 μL PBS. Serial dilutions were made to adjust the solutions to proper concentration. 100 μL solution was added onto YPD agar plate with antibiotics and then incubated at 30 °C for 48 hrs. Colony numbers of each sample were counted. Fungal CFUs were normalized to weight of feces.
DNA extraction for fungal qPCR
DNA was extracted from cecum samples of mice using DNeasy PowerSoil kit (Qiagen) according to the manufacturer’s protocol. The abundance of Candida spp. was measured via qPCR using specific primers and normalized by total fungal 18S rRNA.47
16S rDNA sequencing
16S rDNA sequencing was performed as previously described.48 Briefly, bacterial DNA from mouse cecum was extracted using DNeasy PowerSoil kit (Qiagen). DNA (25 to 30 ng) was used to generate amplicons using Nextera DNA Library Prep XT kit (Illumina, Inc). V3-V4 hypervariable regions of prokaryotic 16S rDNA were selected for generating amplicons gene using forward primer 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3′ and reverse primer 5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3′. Sequencing was performed using paired-end configuration on the Illumina MiSeq platform (Illumina). Sequence data were analyzed using QIIME 2 pipeline and R. Sequence reads are available at NCBI under BioProject PRJNA911776.
Isolation of bone marrow-derived dendritic cells
Bone marrow cells were collected from femurs and tibias of mice and cultured in a Falcon 150 mm × 15 mm Not TC-treated Bacteriological Petri Dish in the presence of 10 ng/mL of recombinant mouse granulocyte-macrophage colony-stimulating factor (GM-CSF) (R&D, Minneapolis, MN, USA). On day 3, 25 mL fresh medium containing 10 ng/mL of GM-CSF was added to the plate. On day 6, 10 mL medium was removed and centrifuged. The pellets were resuspended in 10 mL of fresh media with 10 ng/mL GM-CSF and added back into the original culture media. On day 7, the cells were harvested and purified using EasySep™ Mouse CD11c Positive Selection Kit (STEMCELL).49
Adoptive transfer of T cells
For adoptive transfer of ex vivo C. albicans-primed hector T cells from Candida-specific TCR transgenic mice, on day 0 and day 9, Candida-specific TCR transgenic (Rag1−/−/CaTCRtg) and C57BL/6 donor mice were gavaged with 1× 108 CFU live C. albicans (M1477) per mouse. On day 10, CD4+ T cells from spleen and lymph nodes of the gavaged mice were magnetically sorted, using EasySep Mouse CD4+ T-Cell Isolation Kit. CD4+ T cells (1 × 106 well−1) were co-cultured in a 24-well plate with bone marrow-derived dendritic cells (0.5 × 106 well−1) in the presence of C. albicans lysate (60 μg/mL). Six days later, C. albicans-primed CD4+ T cells from Candida-specific TCR transgenic (Rag1−/−/CaTCRtg) and C57BL/6 mice were intravenously injected into ethanol-fed mice (NIAAA model), which were sacrificed 3 days later.50
For adoptive transfer of hector T cells from Candida-specific TCR transgenic mice, CD4+ T cells from spleen and lymph nodes of Candida-specific TCR transgenic (Rag1−/−/CaTCRtg) and C57BL/6 donor mice were magnetically sorted, using EasySep Mouse CD4+ T-Cell Isolation Kit and intravenously injected into ethanol-fed mice (NIAAA model), which were sacrificed 3 days later.50
For adoptive transfer of ex vivo fungi-primed polyclonal CD4+ T cells, on day 0 and day 9, IL17A-eGFP mice were gavaged with 1×108 CFU live C. albicans (M1477) or S. cerevisiae (ATCC 9763) per mouse. On day 10, polyclonal CD4+ T cells from spleen and lymph nodes of the gavaged IL17A-eGFP mice were magnetically sorted, using EasySep Mouse CD4+ T-Cell Isolation Kit. CD4+ T cells (1×106 well−1) were co-cultured in a 24-well plate with bone marrow-derived dendritic cells (0.5 × 106 well−1) in the presence of C. albicans or S. cerevisiae lysate (60 μg/mL). Six days later, fungus-primed CD4+ T cells were assessed for IL17A-eGFP+ expression; 1×106 fungi-primed polyclonal CD4+ T cells were intravenously injected into ethanol-fed mice (NIAAA model), which were sacrificed 3 days later.50
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analyses were conducted with GraphPad PRISM software 7.0 (GraphPad Software, La Jolla, CA, USA) and R statistical software (R version 2022.07.1 for Mac). The R Foundation for Statistical Computing is based on appropriate assumptions according to data distribution and variance characteristics. Results are presented as mean±SEM (except when stated otherwise). Significance of differences between groups was evaluated using 2-sided unpaired Student t test. Significance of differences among multiple groups was evaluated using 1-way analysis of variance (ANOVA) with Tukey’s or Holm-Sidak post-hoc test. To correlate clinical and laboratory variables, Pearson correlation was conducted. Matching of patients with alcohol use disorder and liver disease, and non-alcoholic controls according to their BMI was performed employing the MatchIt function in R using the nearest neighbor matching and glm method of estimating propensity scores. P<0.05 was considered to be statistically significant (adjusted for multiple comparison when performing multiple tests).
Supplementary Material
Highlights.
Alcohol increases intestinal C. albicans-specific Th17 cells that migrate to the liver
An antifungal agent reduces C. albicans-specific Th17 cells in the liver
Adoptive transfer of C. albicans-specific Th17 cells promotes liver disease
C. albicans-specific Th17 cells promote liver disease via IL17ra on Kupffer cells
Acknowledgements
This study was supported in part by National Institutes of Health (NIH) grant K12 HD85036, University of California San Diego Altman Clinical and Translational Research Institute (ACTRI)/NIH grant KL2TR001444, Pinnacle Research Award in Liver Diseases Grant #PNC22-159963 from the American Association for the Study of Liver Diseases Foundation (to P.H.), NIH grants R01 AA24726, R37 AA020703, U01 AA026939, U01 AA026939-04S1, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and a Harrington Discovery Institute Foundation Grant (to B.S.), R35 HL139930 and the Louisiana Board of Regents Endowed Chairs for Eminent Scholars program (to J.K.K), services provided by NIH centers P30 DK120515 and P50 AA011999, by the German Research Foundation (DFG) under Germany’s Excellence Strategy - EXC 2167-390884018 “Precision Medicine in Chronic Inflammation” (to P.B.), NIH grants DK099205, AA028550, DK101737, AA011999, DK120515, AA029019, DK091183 (T.K.), P42ES010337 and R44DK115242 (D.B.). S.M. and W.J.M.H. are funded by the NIH (R01 GM124494 to WJM Huang). P. S. received grant support from Fond National de Recherche Scientifique Belgium (J.0146.17 and T.0217.18) and Action de Recherche Concertée (ARC), Université Catholique de Louvain, Belgium.
Abbreviations:
- Th17
T-helper 17 cells
- IL17
interleukin 17
- C. albicans
Candida albicans
- TCR
T-cell receptor
- IL17ra
IL17 receptor A
- ARTE
antigen-reactive T cell enrichment
- PBMCs
peripheral blood mononuclear cells
- Tmem
memory T cells
- Tcm
central memory T cells
- Tnf
Tumor necrosis factor
- S. cerevisiae
Saccharomyces cerevisiae
- UMAP
uniform manifold approximation and projection
- ALT
alanine amino transferase
- ANOVA
analysis of variance
- CFUs
colony forming units
- qPCR
quantitative PCR
- FACS
Fluorescence-activated cell sorting
Footnotes
Declaration of interests
B.S. has been consulting for Ambys Medicines, Ferring Research Institute, Gelesis, HOST Therabiomics, Intercept Pharmaceuticals, Mabwell Therapeutics, Patara Pharmaceuticals and Takeda. B.S.’s institution UC San Diego has received research support from Artizan Biosciences, Axial Biotherapeutics, BiomX, CymaBay Therapeutics, NGM Biopharmaceuticals, Prodigy Biotech and Synlogic Operating Company. B.S. is founder of Nterica Bio. UC San Diego has filed several patents with B.S. and Y.D. as inventors related to this work. P.S. received grant support from Gilead Sciences Belgium.
SUPPLEMENTARY INFORMATION
Supplemental Information includes figure legends, 3 tables and 7 figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lackner C, and Tiniakos D (2019). Fibrosis and alcohol-related liver disease. J. Hepatol. 70, 294–304. 10.1016/j.jhep.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Gustot T, and Jalan R (2019). Acute-on-chronic liver failure in patients with alcohol-related liver disease. J. Hepatol. 70, 319–327. 10.1016/j.jhep.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Lee BP, Vittinghoff E, Dodge JL, Cullaro G, and Terrault NA (2019). National Trends and Long-term Outcomes of Liver Transplant for Alcohol-Associated Liver Disease in the United States. JAMA Intern. Med. 179, 340–348. 10.1001/jamainternmed.2018.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shield K, and Rehm J (2020). The importance of preventing alcohol-related liver disease in the USA. The Lancet. Public health 5, e298–e299. 10.1016/s2468-2667(20)30118-3. [DOI] [PubMed] [Google Scholar]
- 5.Sarin SK, Pande A, and Schnabl B (2019). Microbiome as a therapeutic target in alcohol-related liver disease. J. Hepatol. 70, 260–272. 10.1016/j.jhep.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L, Stärkel P, Fan JG, Fouts DE, Bacher P, and Schnabl B (2021). The gut mycobiome: a novel player in chronic liver diseases. J. Gastroenterol. 56, 1–11. 10.1007/s00535-020-01740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng S, and Schnabl B (2022). Roles for the Mycobiome in Liver Disease. Liver Int. 42, 729–741. 10.1111/liv.15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang S, Duan Y, Liu J, Torralba MG, Kuelbs C, Ventura-Cots M, Abraldes JG, Bosques-Padilla F, Verna EC, Brown RS Jr., et al. (2020). Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatol. 71, 522–538. 10.1002/hep.30832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu H, Duan Y, Lang S, Jiang L, Wang Y, Llorente C, Liu J, Mogavero S, Bosques-Padilla F, Abraldes JG, et al. (2020). The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J. Hepatol. 72, 391–400. 10.1016/j.jhep.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann P, Lang S, Zeng S, Duan Y, Zhang X, Wang Y, Bondareva M, Kruglov A, Fouts DE, Stärkel P, and Schnabl B (2021). Dynamic Changes of the Fungal Microbiome in Alcohol Use Disorder. Front. Physiol 12, 699253. 10.3389/fphys.2021.699253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, and Mathis D (2014). Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc. Natl. Acad. Sci. U.S.A. 111, 6696–6701. 10.1073/pnas.1405634111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, Röhmel J, Eschenhagen P, Grehn C, Seidel K, et al. (2019). Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355.e1315. 10.1016/j.cell.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 17.Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, Iwert C, Vogt K, Kollet J, Babel N, Sawitzki B, et al. (2016). Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell 167, 1067–1078.e1016. 10.1016/j.cell.2016.09.050. [DOI] [PubMed] [Google Scholar]
- 18.Bacher P, Steinbach A, Kniemeyer O, Hamprecht A, Assenmacher M, Vehreschild MJ, Vehreschild JJ, Brakhage AA, Cornely OA, and Scheffold A (2015). Fungus-specific CD4(+) T cells for rapid identification of invasive pulmonary mold infection. Am. J. Respir. Crit. 191, 348–352. 10.1164/rccm.201407-1235LE. [DOI] [PubMed] [Google Scholar]
- 19.Bacher P, Kniemeyer O, Teutschbein J, Thön M, Vödisch M, Wartenberg D, Scharf DH, Koester-Eiserfunke N, Schütte M, Dübel S, et al. (2014). Identification of immunogenic antigens from Aspergillus fumigatus by direct multiparameter characterization of specific conventional and regulatory CD4+ T cells. J. Immunol. 193, 3332–3343. 10.4049/jimmunol.1400776. [DOI] [PubMed] [Google Scholar]
- 20.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. (2019). Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 25, 377–388.e376. 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang TT, Shao T-Y, Ang WXG, Kinder JM, Turner LH, Pham G, Whitt J, Alenghat T, and Way SS (2017). Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host Microbe 22, 809–816.e804. 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertola A, Mathews S, Ki SH, Wang H, and Gao B (2013). Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc. 8, 627–637. 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomura M, Yoshida N, Tanaka J, Karasawa S, Miwa Y, Miyawaki A, and Kanagawa O (2008). Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 105, 10871–10876. 10.1073/pnas.0802278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trautwein-Weidner K, Gladiator A, Kirchner FR, Becattini S, Rülicke T, Sallusto F, and LeibundGut-Landmann S (2015). Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis. PLoS Pathog. 11, e1005164–e1005164. 10.1371/journal.ppat.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma HY, Yamamoto G, Xu J, Liu X, Karin D, Kim JY, Alexandrov LB, Koyama Y, Nishio T, Benner C, et al. (2020). IL-17 signaling in steatotic hepatocytes and macrophages promotes hepatocellular carcinoma in alcohol-related liver disease. J. Hepatol. 72, 946–959. 10.1016/j.jhep.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang A-M, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, et al. (2017). Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest. 127, 2829–2841. 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XV, Leonardi I, Putzel GG, Semon A, Fiers WD, Kusakabe T, Lin W-Y, Gao IH, Doron I, Gutierrez-Guerrero A, et al. (2022). Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 603, 672–678. 10.1038/s41586-022-04502-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Friedrich C, Hagemann SC, Korte WH, Goharani N, Cording S, Eberl G, Sparwasser T, and Lochner M (2014). Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium. Mucosal Immunol. 7, 1290–1301. 10.1038/mi.2014.17. [DOI] [PubMed] [Google Scholar]
- 29.Sparber F, De Gregorio C, Steckholzer S, Ferreira FM, Dolowschiak T, Ruchti F, Kirchner FR, Mertens S, Prinz I, Joller N, et al. (2019). The Skin Commensal Yeast Malassezia Triggers a Type 17 Response that Coordinates Anti-fungal Immunity and Exacerbates Skin Inflammation. Cell Host Microbe 25, 389–403.e386. 10.1016/j.chom.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Lemmers A, Moreno C, Gustot T, Maréchal R, Degré D, Demetter P, de Nadai P, Geerts A, Quertinmont E, Vercruysse V, et al. (2009). The interleukin-17 pathway is involved in human alcoholic liver disease. Hepatol. 49, 646–657. 10.1002/hep.22680. [DOI] [PubMed] [Google Scholar]
- 31.Kasztelan-Szczerbińska B, Surdacka A, Celiński K, Roliński J, Zwolak A, Miącz S, and Szczerbiński M (2015). Prognostic Significance of the Systemic Inflammatory and Immune Balance in Alcoholic Liver Disease with a Focus on Gender-Related Differences. PloS One 10, e0128347. 10.1371/journal.pone.0128347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beringer A, and Miossec P (2018). IL-17 and IL-17-producing cells and liver diseases, with focus on autoimmune liver diseases. Autoimmun. Rev 17, 1176–1185. 10.1016/j.autrev.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Beringer A, Thiam N, Molle J, Bartosch B, and Miossec P (2018). Synergistic effect of interleukin-17 and tumour necrosis factor-α on inflammatory response in hepatocytes through interleukin-6-dependent and independent pathways. Clin. Exp. Immunol 193, 221–233. 10.1111/cei.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ki SH, Park O, Zheng M, Morales-Ibanez O, Kolls JK, Bataller R, and Gao B (2010). Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatol. 52, 1291–1300. 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, Stärkel P, Ho SB, Gao B, Fiehn O, et al. (2019). Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515. 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwanaga N, Chen K, Yang H, Lu S, Hoffmann JP, Wanek A, McCombs JE, Song K, Rangel-Moreno J, Norton EB, and Kolls JK (2021). Vaccine-driven lung TRM cells provide immunity against Klebsiella via fibroblast IL-17R signaling. Sci. Immunol 6, eabf1198. 10.1126/sciimmunol.abf1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bluemel S, Wang Y, Lee S, and Schnabl B (2020). Tumor necrosis factor alpha receptor 1 deficiency in hepatocytes does not protect from non-alcoholic steatohepatitis, but attenuates insulin resistance in mice. World J. Gastroenterol. 26, 4933–4944. 10.3748/wjg.v26.i33.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, Ego KM, Bruni CM, Deng Z, Schlachetzki JCM, et al. (2019). Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 51, 655–670.e658. 10.1016/j.immuni.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan Y, Chu H, Brandl K, Jiang L, Zeng S, Meshgin N, Papachristoforou E, Argemi J, Mendes BG, Wang Y, et al. (2021). CRIg on liver macrophages clears pathobionts and protects against alcoholic liver disease. Nat. Commun. 12, 7172. 10.1038/s41467-021-27385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsubara V, Silva E, Paula C, Ishikawa K, and Nakamae A (2012). Treatment with probiotics in experimental oral colonization by Candida albicans in murine model (DBA/2). Oral Dis. 18, 260–264. 10.1111/j.1601-0825.2011.01868.x. [DOI] [PubMed] [Google Scholar]
- 41.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shugay M, Britanova OV, Merzlyak EM, Turchaninova MA, Mamedov IZ, Tuganbaev TR, Bolotin DA, Staroverov DB, Putintseva EV, Plevova K, et al. (2014). Towards error-free profiling of immune repertoires. Nat. Methods 11, 653–655. 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 44.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381. 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Dong Z, Zhou R, Luo D, Wei H, and Tian Z (2005). Isolation of lymphocytes and their innate immune characterizations from liver, intestine, lung and uterus. Cell. Mol. Immunol. 2, 271–280. [PubMed] [Google Scholar]
- 46.Lee K-C, Chen P, Maricic I, Inamine T, Hu J, Gong S, Sun JC, Dasgupta S, Lin H-C, Lin Y-T, et al. (2019). Intestinal iNKT cells migrate to liver and contribute to hepatocyte apoptosis during alcoholic liver disease. Am. J. Physiol. Gastrointest. Liver. Physiol 316, G585–G597. 10.1152/ajpgi.00269.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Hung GC, Nagamine K, Li B, Tsai S, and Lo SC (2016). Development of Candida-Specific Real-Time PCR Assays for the Detection and Identification of Eight Medically Important Candida Species. Microbiol. Insights 9, 21–28. 10.4137/mbi.S38517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu H, Jiang L, Gao B, Gautam N, Alamoudi JA, Lang S, Wang Y, Duan Y, Alnouti Y, Cable EE, and Schnabl B (2021). The selective PPAR-delta agonist seladelpar reduces ethanol-induced liver disease by restoring gut barrier function and bile acid homeostasis in mice. Transl Res. 227, 1–14. 10.1016/j.trsl.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Zhang J, Chung Y-J, Kim JH, Kook CM, González-Navajas JM, Herdman DS, Nürnberg B, Insel PA, Corr M, et al. (2020). Inhibition of IRF4 in dendritic cells by PRR-independent and -dependent signals inhibit Th2 and promote Th17 responses. Elife 9, e49416. 10.7554/eLife.49416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tasaki S, Cho T, Nagao J-I, Ikezaki S, Narita Y, Arita-Morioka K-I, Yasumatsu K, Toyoda K, Kojima H, and Tanaka Y (2018). Th17 cells differentiated with mycelial membranes of Candida albicans prevent oral candidiasis. FEMS Yeast Res. 18, foy018. 10.1093/femsyr/foy018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


