Abstract
Penicillin-binding proteins (PBPs) make up an essential class of bacterial enzymes that carry out the final steps of peptidoglycan synthesis and regulate the recycling of this polymeric structure. PBPs are an excellent drug target and have been the most clinically relevant antibacterial target since the 1940s with the introduction of β-lactams. Despite this, a large gap in knowledge remains regarding the individual function and regulation of each PBP homologue in most bacteria. This can be attributed to a lack of chemical tools and methods that enable the study of individual PBPs in an activity-dependent manner and in their native environment. The development of such methods in Gram-negative bacteria has been particularly challenging due to the presence of an outer membrane and numerous resistance mechanisms. To address this, we have developed an optimized live-cell assay for screening inhibitors of the PBPs in Escherichia coli MG1655. We utilized EDTA to permeabilize Gram-negative cells, enabling increased penetration of our readout probe, Bocillin-FL, and subsequent analysis of PBP-inhibition profiles. To identify scaffolds for future development of PBP-selective activity-based probes, we screened ten β-lactams, one diazabicyclooctane, and one monobactam for their PBP-selectivity profiles in E. coli MG1655. These results demonstrate the utility of our assay for the screening of inhibitors in live, non-hypersusceptible Gram-negative organisms.
Keywords: penicillin-binding proteins, Gram-negative bacteria, Escherichia coli, IC50, outer-membrane, permeabilization
Graphical Abstract
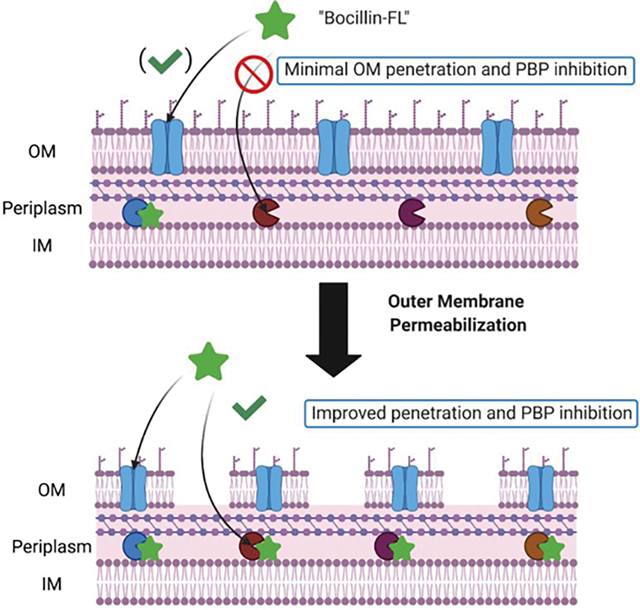
The cell envelope is an essential component for the survival of bacteria and has been exploited for decades in the development of antibiotics.1–4 In Gram-negative bacteria, the presence of an outer membrane in the cell envelope presents barriers to the effective penetration of antibiotics and other molecules. Additionally, Gram-negative species often harbor resistance mechanisms that prevent adequate intracellular accumulation of those antibiotics that transverse the cell envelope.5 For many species and strains of Gram-negative bacteria, the accumulation of β-lactam antibiotics is greatly impacted by the presence of β-lactamases, efflux pumps, and porins with reduced recognition of β-lactams as substrates.6–8 β-Lactams target an essential class of bacterial enzymes known as the penicillin-binding proteins (PBPs). PBPs function to catalyze the final two steps in the biosynthesis of the peptidoglycan layer—a polymeric mesh structure composed of repeating disaccharide units that are cross-linked via stem peptide chains—of the cell envelope: a process that is conserved among all eubacteria.9,10
Cross-linking of the stem peptide chains is carried out via a transpeptidation reaction that utilizes a ubiquitously conserved serine residue in the PBP active site.9,11 Reaction of this conserved residue with the β-lactams is crucial to the activity of these molecules, which have been exploited in both human and veterinary medicine for decades. β-Lactams with the lowest minimum-inhibitory concentrations (MIC) frequently target essential PBPs12,13 increasing their lethality but also resulting in rapid evolution of PBP mutations rendering these drugs the ineffective.14 Alternative strategies are to design PBP inhibitors that target multiple PBPs, including nonessential PBPs, and to explore combination therapies of molecules that exclusively target essential PBPs and those that also inhibit nonessential homologues.
Significant progress has been made in the fundamental understanding of the PBPs in various organisms.11,15–18 Different species of bacteria encode multiple PBP-isoforms, and yet the enzymatic functions and regulations of these proteins are often redundant or nonessential.9,19 This paradox lends to the following questions: What are the roles of individual PBP isoforms, and how does their coordination and regulation affect the cell division and cell wall biosynthesis processes? Importantly, aside from model Gram-positive and Gram-negative bacteria, there are large gaps in knowledge regarding individual PBP isoform functions in clinically relevant species. To address these questions, we have taken a chemical biology approach to develop PBP-isoform selective chemical probes that have enabled us to study the activity and spatial-temporal regulations of individual PBPs in different species.10,17,20,21
To identify PBP-selective molecules, we previously developed a live-cell screening assay for PBP inhibitors in Streptococcus pneumoniae,13 Bacillus subtilis,22 and hypersusceptible Escherichia coli.12 Historically, similar assays have been performed using the membrane fraction of bacterial cell lysates with either radiolabeled penicillins23–27 or Bocillin-FL (a fluorescent penicillin V analogue that labels most PBPs).28,29 While useful for mitigating cell penetration and accumulation issues, not all PBPs are labeled in lysates using Bocillin-FL. For example, PBP7 and PBP8 (a degradation product of PBP7) from E. coli are often not observed in PBP-labeling experiments carried out in lysates,28,30,31 while we were able to show consistent labeling of these two PBPs in our live-cell assay in E. coli DC2.12 It is also plausible that, in using lysates, native protein–protein interactions and activities are perturbed.32 While the use of our live-cell assay has enabled interrogation of entire PBP profiles in various species, its use has been limited to Gram-positive bacteria and E. coli DC2, a hypersusceptible mutant strain with defects in its outer membrane.33 There is one investigation in which live Acinetobacter baumannii was first treated with β-lactams, followed by cell lysis, and then treatment with Bocillin-FL on the membrane fraction to assess PBP inhibition.34 This assay is an improvement over those carried out strictly using cell lysates; however, use of Bocillin-FL and other small-molecule probes in vivo is ideal.
To investigate the utility of our assay in wild-type Gram-negative strains, we used E. coli MG1655. Use of our previously published protocol yielded either low or incomplete labeling of the PBPs likely due to the presence of a more complete outer membrane in E. coli MG1655 (lacks abs-2 mutation found in DC2 strain that leads to increased permeability) and the relatively large size of Bocillin-FL (>600 Da). It is known that large molecules (~700 Da or greater) often poorly penetrate the outer membrane of Gram-negative bacteria.35,36 Additionally, most β-lactams enter the periplasmic space through porins,37 and the addition of large fluorophore may prevent recognition. Thus, we investigated two methods to increase cell permeability to enable Bocillin-FL labeling of the entire PBP complement. We evaluated the membrane permeabilizing agent polymyxin B nonapeptide (PMBN)38–40 and the combination of tris(hydroxymethyl)aminomethane (Tris) and ethylenediaminetetraacetic acid (EDTA, Figure 1).41 Here, we describe the optimization of our gel-based assay and the first report of successful labeling of an entire PBP profile in live, non-hypersusceptible E. coli. We further show the PBP-selectivity profiles of 12 inhibitors in E. coli MG1655 providing a means to study the targets of the most clinically used class of antibiotics—the β-lactams—in organisms that represent some of the biggest threats to human health.
Figure 1.
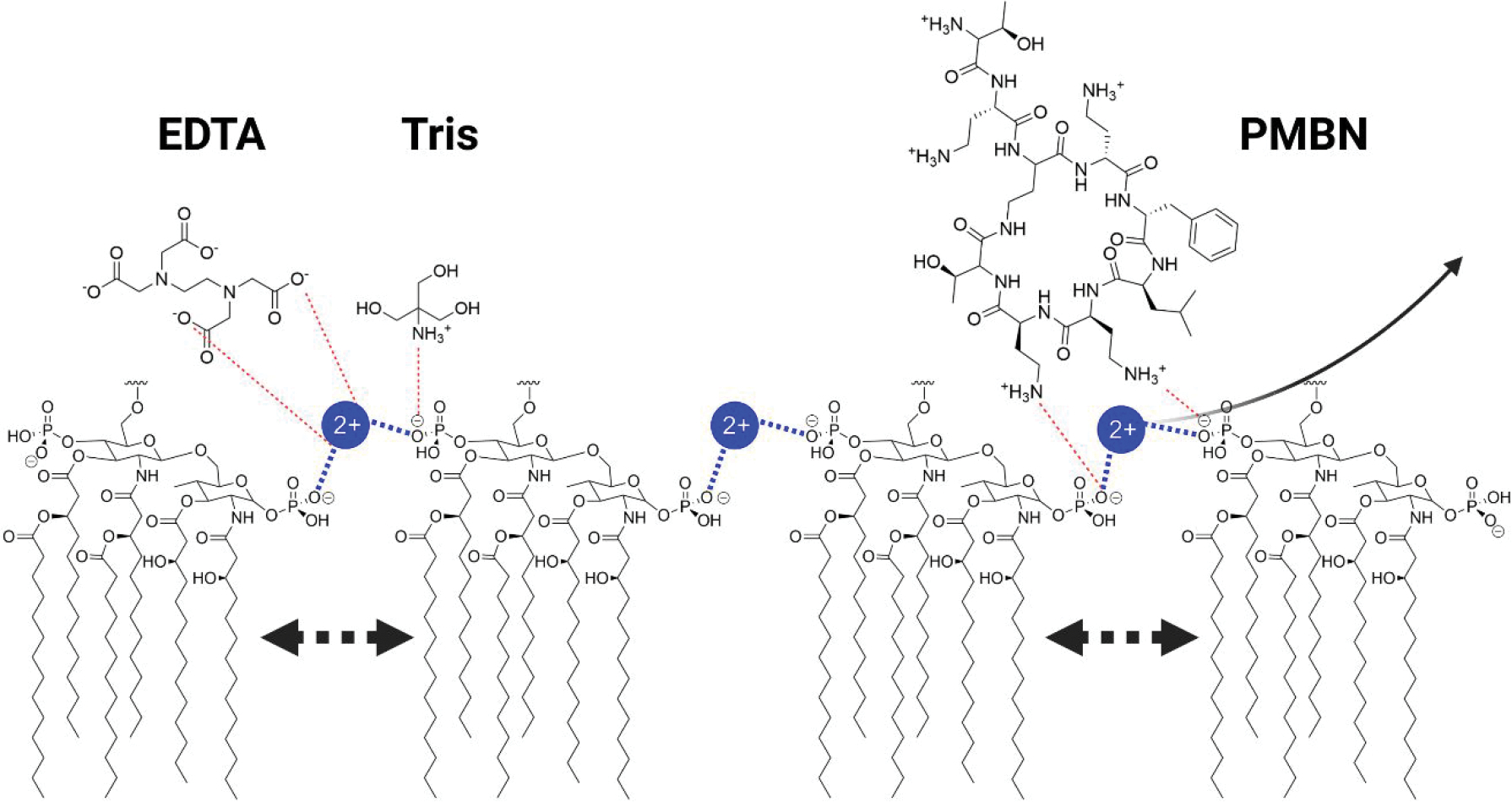
Permeabilization of the outer membrane via LPS disruption. EDTA and Tris work synergistically to displace divalent metals that bridge adjacent phosphate groups, while cationic groups on PMBN interact with adjacent LPS groups to displace divalent metals. Displacement of the metals results in destabilization of the LPS and pore formation.
RESULTS
Permeabilization of E. coli MG1655 to Enable Bocillin-FL Labeling of the PBPs.
We previously described a gel-based assay for the determination of inhibitor IC50 values and corresponding selectivity profiles against the PBPs of E. coli DC2,12 in which live cells were treated with inhibitor followed by labeling of the uninhibited PBPs with Bocillin-FL (7.5 μM) in phosphate buffered saline (PBS). Attempts to directly translate this method into E. coli MG1655, a rough, non-hypersusceptible strain, yielded low or incomplete labeling of its eight PBPs (Figure 2). We hypothesized that because E. coli MG1655 is non-hypersusceptible and has a more complete cell envelope than DC2,42 Bocillin-FL had a reduced ability to penetrate the outer membrane. The literature supports this theory as poor penetration of fluorophore-conjugated molecules has previously been reported in Gram-negative bacteria.43–46 The inability to utilize a fluorescent readout probe for PBP activity in non-hypersusceptible strains poses a significant barrier to the assessment of inhibitor-PBP selectivity profiles.
Figure 2.
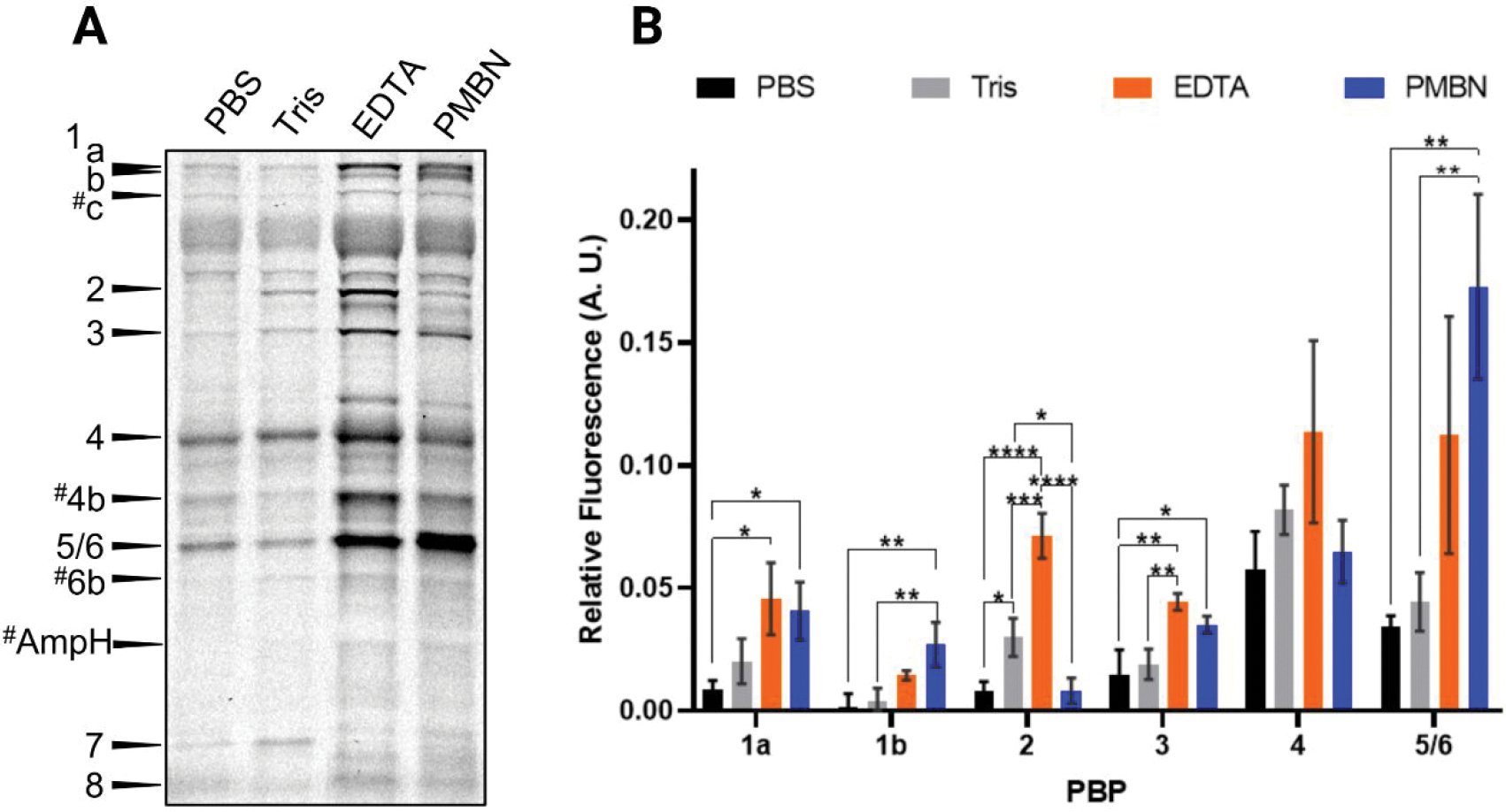
Comparison of Bocillin-FL labeling of PBPs using pretreatments with various permeabilization reagents. Live E. coli MG1655 were pretreated with 50 μL of PBS, Tris, Tris-EDTA (200 μM), or Tris-PMBN (200 μM) for 30 min, followed by incubation with 25 μM Bocillin-FL in either PBS (left lane) or Tris (remaining lanes) for 30 min. (A) An SDS-PAGE analysis showed the labeling of the entire PBP profile when cells were pretreated with Tris-EDTA or Tris-PMBN. The # indicates putative PBP assignments. One of three biological replicates is shown. (B) A comparison of the gel-band fluorescence showed significant increases in labeling for most PBPs when cells were pretreated with Tris-EDTA or Tris-PMBN. Mean fluorescence values that were corrected for protein loading differences via Coomassie stain from triplicate data sets for each PBP compared using one-way ANOVA multiple comparisons in GraphPad Prism [see the Methods section; P-values, 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****)].
To address this issue, we investigated two commonly reported methods to permeabilize the outer membrane of Gram-negative bacteria, Tris-EDTA or PMBN.38,41,46–49 Tris-EDTA causes a portion of the cell envelope lipopolysaccharides (LPS) to be released, enabling penetration of large molecules.41 PMBN is a polymyxin B derivative that lacks an acyl chain and has very poor antibacterial activity but is capable of permeabilizing the outer membrane.38 Cells pretreated with either Tris-EDTA or Tris-PMBN, followed by Bocillin-FL labeling, yielded vastly superior PBP labeling in E. coli MG1655 in comparison to either pretreatment with Tris alone or PBS (Figure 2). We ultimately selected Tris-EDTA as our condition of choice as this combination gave the most consistent results and is more cost-effective than PMBN. Intriguingly, not only were we able to visualize the eight PBPs previously seen in E. coli DC2, but we also saw additional Bocillin-FL-labeled proteins that may be PBP1c (pbpC), PBP4b (yfeW), PBP6b (dacD), and AmpH (ampH; intermittently detected) based on their apparent molecular weights (i.e., gel migration distances, Figure 2A). These PBPs are rarely reported in PBP-inhibitor studies12,30,50–52 but have been studied for their physiological roles.9,53,54
We next sought to determine if pretreatment of the cells was necessary to achieve adequate permeabilization or if these reagents could be used directly in the Bocillin-FL labeling step. A combined permeabilization/PBP labeling step would substantially decrease the amount of time that the cells were subjected to a low nutrient environment such as PBS or Tris (half the length of time—total of 30 min; Figures 3 and 4). We also sought to directly compare these results to our original protocol that was designed for E. coli DC2 (live cells incubated with 7.5 μM Bocillin-FL in PBS for 10 min, denoted as PBS†), including comparison of the higher concentration of Bocillin-FL utilized in these studies (25 μM; Figure 3, compare lanes 1 and 2). While slightly higher PBP labeling was observed when the Bocillin-FL concentration was increased, it was only statistically significant for PBP3 indicating that the simple use of more labeling agent was not sufficient. A similar result was obtained in Tris buffer, with only the labeling of PBP8 increasing substantially in comparison to the original DC2 protocol. Conversely, we saw dramatic differences in PBP labeling upon cotreatment of the cells with Tris-EDTA in comparison to Tris alone (1.2–5 fold increases, Figures 3 and 4), indicating that EDTA was an essential component. We also found that cotreatment of cells with Tris-EDTA and Bocillin-FL yielded similar results to pretreatment, revealing that a single incubation step could be utilized (Figures 2A and 3A). Finally, direct comparison of the EDTA-Tris conditions to the original DC2 method yielded fold increases ranging from 2 to 61, with statistically significant increases in Bocillin-FL labeling of all of the PBPs (Figure 4). These results demonstrate that our optimized conditions using Tris-EDTA to permeabilize the outer membrane of live E. coli MG1655 are both effective for obtaining a substantial fluorescent signal and superior to the original assay conditions that were developed in DC2.
Figure 3.
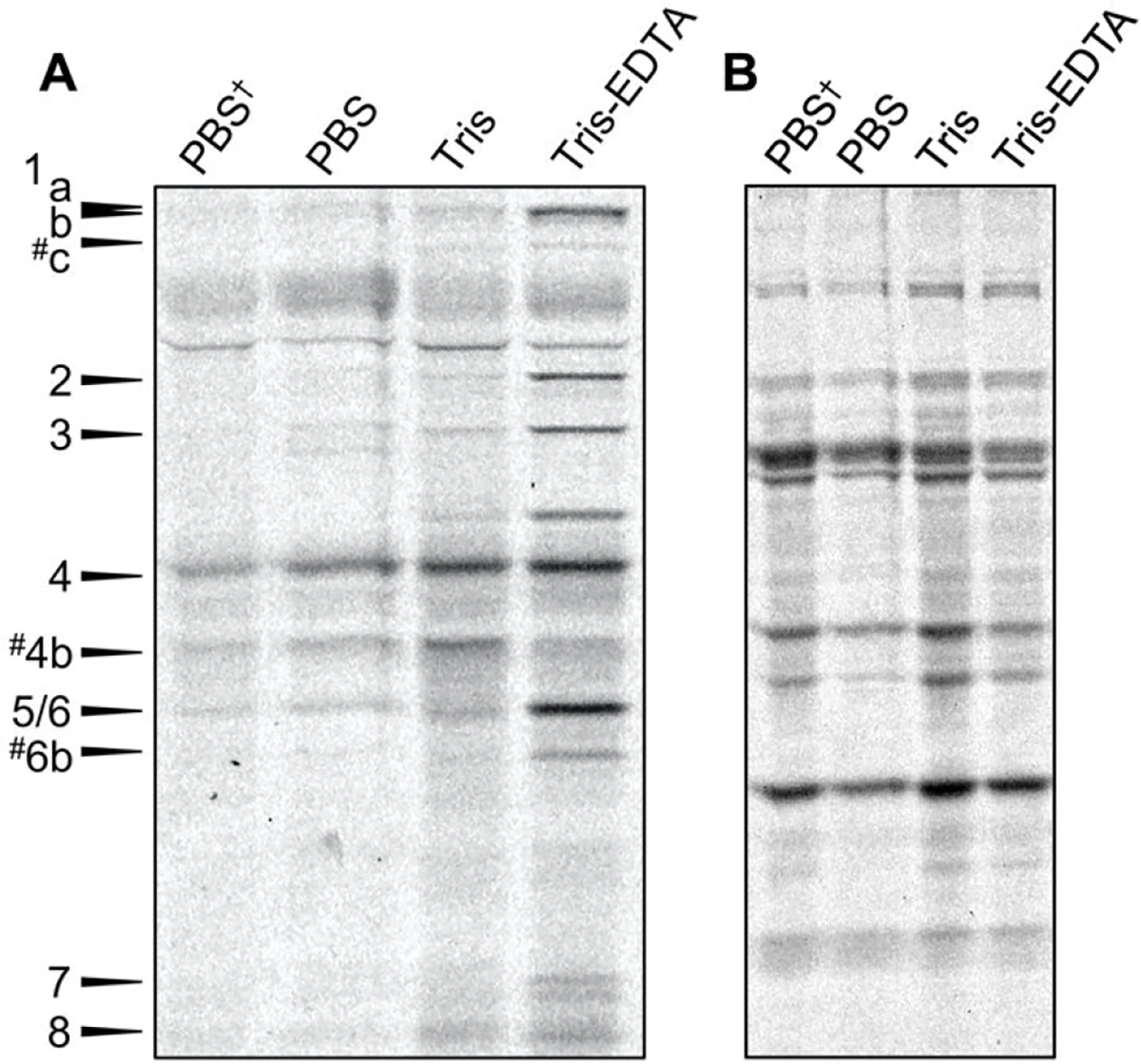
(A) Comparison of Bocillin-FL labeling using PBS, Tris, or Tris-EDTA. SDS-PAGE analysis revealed that treatment of E. coli using 25 μM Bocillin-FL in 50 mM Tris–200 μM EDTA for 30 min results in labeling of 11 or 12 PBPs in E. coli. (B). A Coomassie stain of the same gel indicates that the amount of proteome loaded into each lane is equivalent. PBS† indicates the previous DC2 procedure (7.5 μM Bocillin-FL in PBS, 10 min). # indicates putative PBP assignments. One of three biological replicates shown.
Figure 4.
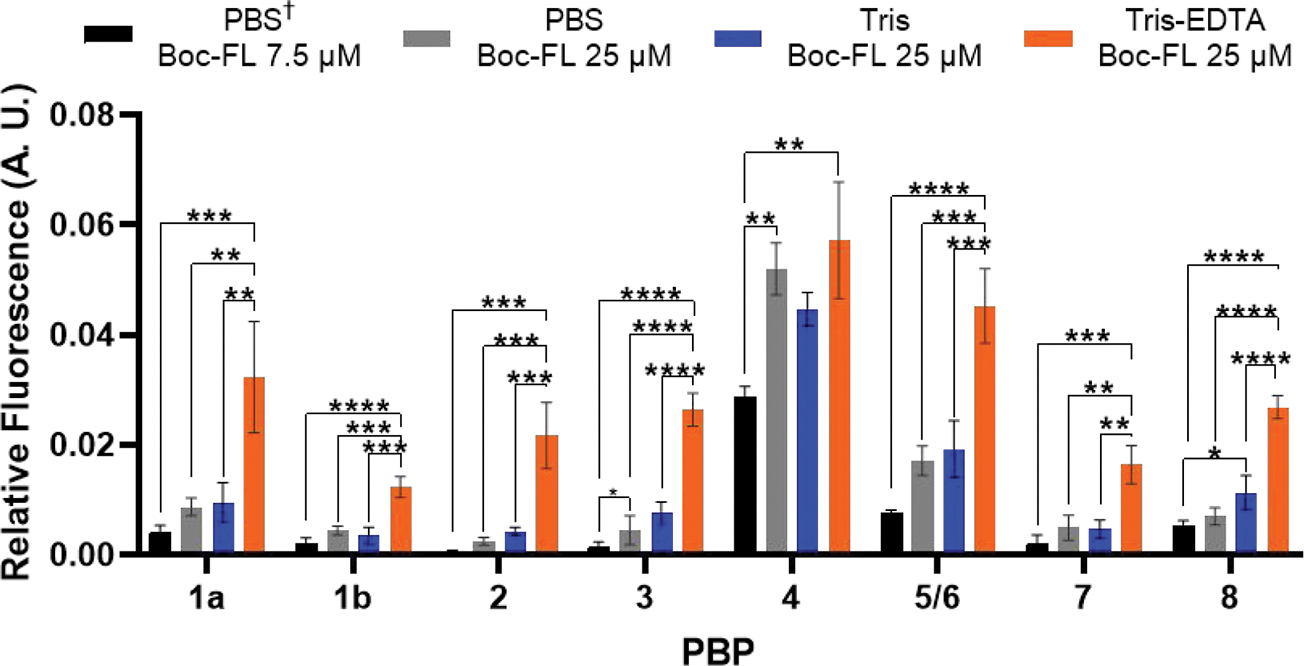
Comparison of PBP-labeling profiles when cells were treated with Bocillin-FL directly in permeabilization buffer conditions. Cells were treated with 50 μL of respective solutions and incubated for 30 min (PBS 7.5 μM Boc-FL for 10 min are original conditions published with DC212). Significantly higher fluorescence was observed for all PBPs when cells were treated with Bocillin-FL in Tris-EDTA. Mean fluorescence values that were corrected for protein loading differences via Coomassie stain from triplicate data sets for each PBP compared using one-way ANOVA multiple comparisons in GraphPad Prism [see the Methods section; P-values, 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****)].
Assessment of Membrane Damage and Cell Viability.
Ideally, inhibitor assessment would be performed in live cells to get the most realistic view of their effects on the PBPs. To evaluate the potential of permeabilization to cause a substantial loss of cellular viability, we investigated our optimized Tris-EDTA conditions and the next most effective method, treatment with Tris-PMBN, by growth curve analysis. Cells were treated with either of these conditions, Tris buffer (negative control) or colistin (positive control) for 30 min, the length of our labeling protocol. Next, we utilized a plate-based assay to measure A600 over 18 h (Figure 5). The growths of Tris-EDTA-, Tris-PMBN-, and Tris-treated cells were indistinguishable, indicating that neither Tris-EDTA nor Tris-PMBN affected cell viability. As expected, colistin treatment resulted in a substantial lag in growth (~10 h) indicating cell death.
Figure 5.
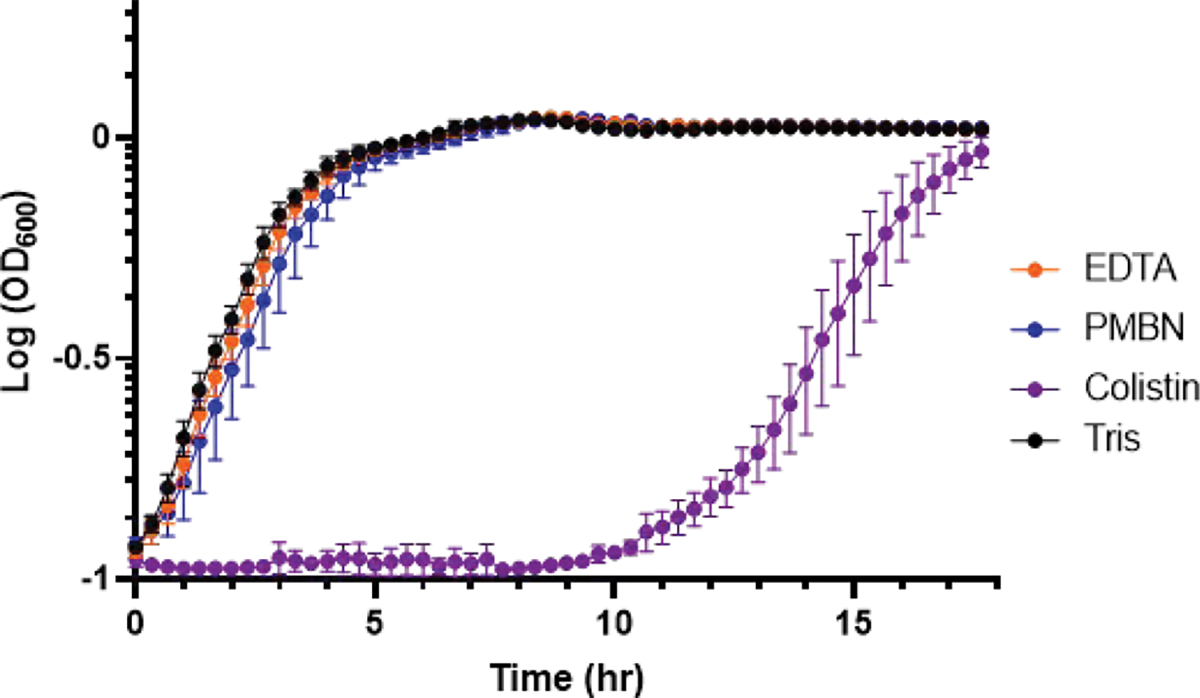
Viability of permeabilized E. coli MG1655. Cells were treated with Tris only or 200 μM EDTA, PMBN, or colistin in Tris for 30 min. Following a wash step, cells were resuspended in LB broth, and A600 was measured over 18 h at 37 °C, 220 rpm. Data points represent averages from triplicate data sets with corresponding standard deviations represented as error bars.
Next, we sought to determine the extent of damage to the membranes that was enabling PBP labeling. As stated, Tris-EDTA41 and PMBN38,39 damage the outer membrane. We utilized N-phenyl-1-naphthylamine (NPN), a hydrophobic molecule that weakly fluoresces in aqueous environments but exhibits increased fluorescence in hydrophobic environments,55 to determine the amount of outer membrane damage caused under our optimized conditions in a plate-based fluorescence assay. Permeabilization of the outer membrane will inherently enable increased interactions between the lipid tails of the phospholipid outer membrane and NPN, but the amount of NPN dye able to penetrate the outer membrane should be less than for those cells treated with a known lytic agent, such as colistin. Cells were treated with Tris only (“untreated”), Tris-EDTA, Tris-PMBN, or Tris-colistin (positive control) in the presence of NPN. The ratio of NPN uptake was determined by comparing the mean fluorescence values for Tris (“untreated”), Tris-EDTA, and Tris-PMBN exposed cells to the colistin positive control (Figure 6A). Both Tris-EDTA and Tris-PMBN caused significantly more NPN uptake than buffer alone, indicating substantial outer membrane damage. Intriguingly, Tris-PMBN yielded the most membrane permeabilization yet was less effective at enabling Bocillin-FL labeling overall.
Figure 6.
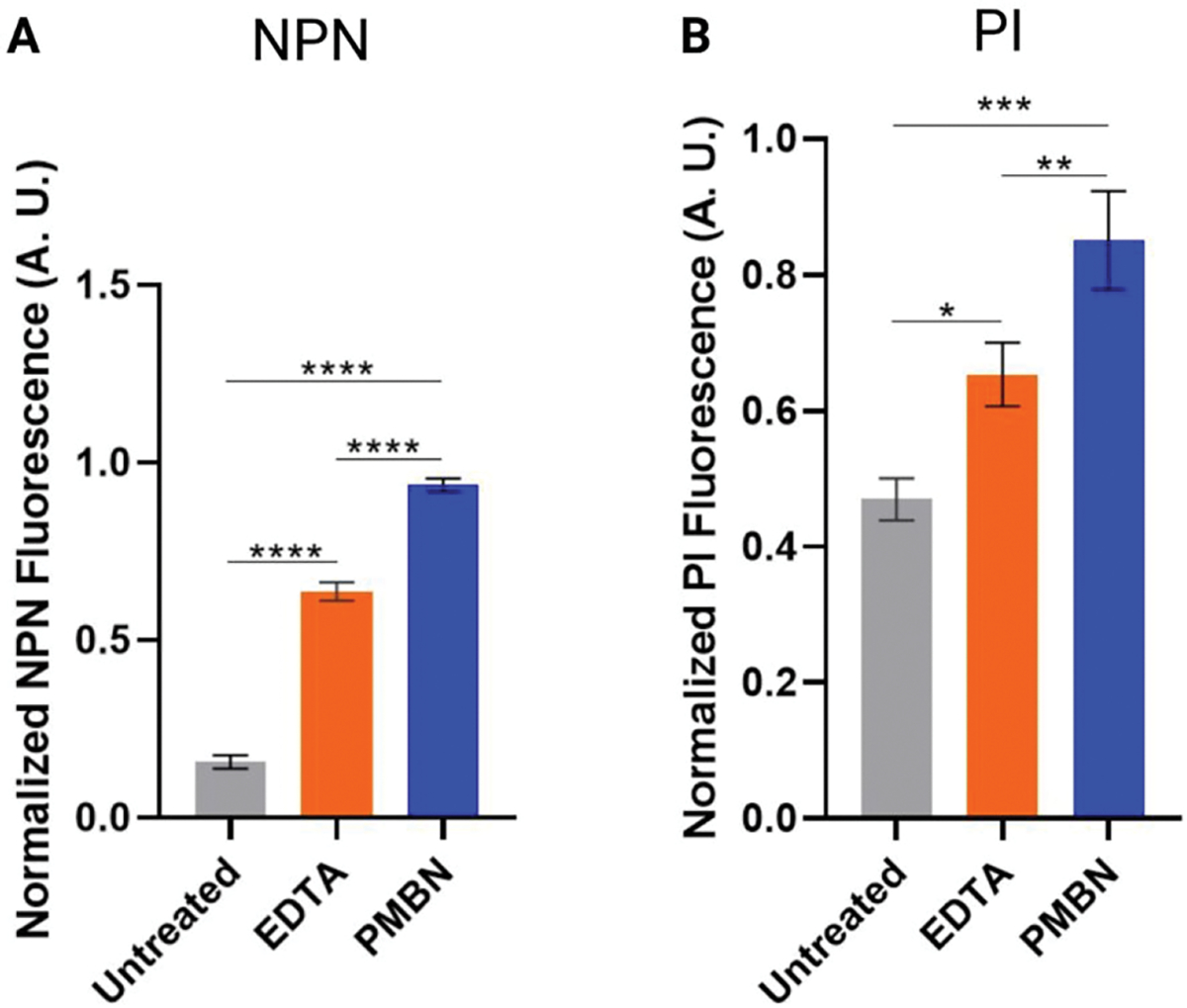
Membrane damage caused by Tris-EDTA and PMBN. E. coli MG1655 was treated with Tris only (untreated) or 200 μM EDTA, PMBN, or colistin (positive control) in Tris. Untreated cells were used as a negative control. (A) Outer membrane damage was assessed through the uptake of NPN. Fluorescence values for each condition were corrected to the colistin positive control (see the Methods section). (B) Inner membrane damage was determined by uptake of the membrane impermeable dye, propidium iodide (PI). Fluorescence values for each condition were corrected to the PI uptake of cells treated with colistin (see the Methods section). [One-way ANOVA multiple comparisons; P-values, 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****).]
Finally, to determine if Tris-EDTA or PMBN caused inner membrane damage, we measured the amount of membrane-impermeable propidium iodide (PI) that entered the cytosol and bound to DNA. Following a 30 min treatment of live cells with Tris (“untreated”), Tris-EDTA, Tris-PMBN, or Tris-colistin (positive control), DNA staining with PI was measured in a plate-based assay. While both Tris-EDTA and Tris-PMBN caused more PI labeling than buffer alone, neither caused as much damage as colistin (Figure 6B). While permeabilization of the inner membrane is often associated with loss of cell viability, our viability study indicates otherwise, which is consistent with the fact that others have shown that the use of PI as a measure of viability is not always accurate.56
Interestingly, phase-contrast imaging of cells treated with respective conditions revealed unexpected differences in cell morphologies (Figure S5). Treatment with PMBN yielded cell damage that was comparable to that observed in cells treated with colistin, as indicated by the numerous light-gray cells that likely represent empty sacculi. This aligns with our membrane damage studies that demonstrate that treating cells with PMBN perturbs both the inner and outer membrane nearly to the same level as an equivalent concentration of colistin. However, our growth curve analyses demonstrate no perturbations in cell growth of PMBN-treated cells relative to the buffer control. These results are unexpected, and future work will be required to correlate the growth curve findings with the extent of membrane damage that we observed. Expectedly, we observed little difference in the morphologies of cells treated with either Tris or EDTA. Collectively, the results from these studies demonstrate that treatment of cells with EDTA in the presence of Tris buffer is ideal for permeabilizing the outer membrane of E. coli MG1655 to enable the labeling of PBPs with Bocillin-FL without causing cell death. With this method in hand, we set out to assess the PBP-selectivity profiles of a series of known PBP-inhibitors in E. coli MG1655.
Titration of Inhibitors in E. coli MG1655 with Tris-EDTA Permeabilization.
To evaluate the PBP-inhibition profiles for a panel of compounds against E. coli MG1655, we used a fluorescence competition assay to determine the apparent IC50 values (IC50app) for a DBO β-lactamase inhibitor (avibactam), a monobactam (aztreonam), and ten molecules representing four classes of β-lactams (penicillins, carbapenems, penems, and cephalosporins; Figures S1 and S2). We utilized Tris-EDTA in the presence of increasing concentrations of these inhibitors to permeabilize the cells, enabling the use of Bocillin-FL as the readout probe in a gel-based assay. When literature data was available from live-cell or cell lysate experiments in any E. coli strain, we compared this with the IC50app and selectivity profiles generated in this study (Table 1). If a PBP-selectivity profile was not explicitly stated in literature reports, we inferred them based upon the specifications used in this study, where selectivity was achieved if the lowest IC50app was 4-fold lower than that of the next closest IC50app. Coselectivity was assigned if three or fewer PBPs had IC50app values within 4-fold of each other.
Table 1.
PBP Inhibitor IC50app Values in E. coli MG1655
| IC50 Values (μM) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | PBPla | PBP1b | PBP2 | PBP3 | PBP4 | PBP5/6 | PBP7 | PBP8 | PBP Selectivity | Reported PBP Selectivity |
| DBO a | ||||||||||
| avibactam | >104 | 310 ± 460 | <100g | >104 | >104 | 480 ± 430 | NDd | NDd | 2 | 2i |
| Monobactam | ||||||||||
| aztreonam | 970 ± 940 | 1.6 ± 0.9 | >104g | 0.09 ± 0.09 | >104g | >104g | >104g | >104g | 3 | 3h12, 3i |
| Penicillin | ||||||||||
| penicillin G | 170 ± 93 | 1.5 ± 0.7 | 65 ± 36 | 15 ± 9 | <1000e | >104 | <1000 | 0.8 ± 0.9 | 1b, 7f, 8 | 4, 7, 8h |
| piperacillin | 17 ± 15 | 12 ± 7.4 | 2.8 ± 2.8 | 0.001 ± 0.000 | 25 ± 24 | 10 ± 13 | 2.3 ± 2.5 | 1.2 ± 1.6 | 3 | 3h |
| methicillin | >104 | 180 ± 150 | 130 ± 200 | 23 ± 33b | 87 ± 51 | >104 | 58 ± 70 | 200 ± 370 | NSj | NSh |
| Penem | ||||||||||
| faropenem | 6.4 ± 3.6 | 0.4 ± 0.2 | 0.04 ± 0.04 | 12 ± 5.7 | 2.0 ± 1.6 | >104 | 1.2 ± 1.8 | 0.01 ± 0.01 | NSj | NSh |
| Carbapenem | ||||||||||
| meropenem | >104 | 110 ± 86 | 0.01 ± 0.00 | 0.2 ± 0.2 | >104 | 5.1 ± 2.7 | NDd | NDd | 2 | (2, 4)h, (2, 4, 6)i |
| Cephalosporin | ||||||||||
| cephalexin | >104 | >104 | 34 ± 12 | 210 ± 88 | 68 ± 79 | >104 | 200 ± 250 | 320 ± 230 | 2, 4 | 4h |
| cefaclor | >104 | >104 | 360 ± 210 | 15 ± 14 | >104 | >104g | 86 ± 66 | 68 ± 48 | 3, 7, 8 | |
| cefuroxime | 73 ± 42 | 1.6 ± 1.9 | 91 ± 65 | 0.02 ± 0.01 | 24 ± 22 | >104 | 25 ± 31 | 15 ± 13 | 3 | 3h |
| ceftriaxone | 4.9 ± 2.2b | 0.2 ± 0.2 | 1.5 ± 0.9 | 0.001 ± 0.003 | 3.0 ± 8.0 | 15 ± 17 | 9.5 ± 9.8 | 10 ± 7.9 | 3 | 3h, 3i |
| cefepime | 5.5 ± 2.6 | 2.6 ± 2.4 | 0.2 ± 0.1 | <10−4b,c | >104g | >104g | 0.2 ± 0.1 | 0.4 ± 0.2 | 3 | 3i |
Diazabicyclooctane.
Only two replicates were used due to one or more PBPs being missing from the control lane.
IC50 value could not be determined due to almost complete inhibition of Bocillin-FL labeling at the lowest concentration of inhibitor tested.
IC50 could not be determined due to low Bocillin-FL labeling in the control and treated lanes for all replicates.
IC50 was estimated based on visualization of the SDS-PAGE analysis due to ambiguous IC50 calculations in GraphPad.
Selectivity was based on visualization of the SDS-PAGE analysis. Calculated IC50 values did not have fully bound 95% CI.
IC50 values could not be calculated due to increased Bocillin-FL labeling at 10 mM concentrations of inhibitor.
Reported selectivity was determined in vivo.12
Not selective.
Of the 12 inhibitors tested, only avibactam did not inhibit PBP3, even at >10 mM (Table 1). The remaining molecules had IC50app values of 210 μM or less for PBP3, with five of these being selective for PBP3 with sub-micromolar IC50app values: aztreonam, piperacillin, cefuroxime, ceftriaxone, and cefepime. These results are consistent with both in vivo and in vitro data for inhibitors tested against various strains of E. coli (Table 1) and provided confidence that our optimized method is a reliable means to accurately evaluate the selectivity of inhibitors against PBPs in non-hypersusceptible strains of E. coli.
Avibactam and meropenem were selective for PBP2, while cephalexin, a first-generation cephalosporin, was coselective for PBP2 and PBP4, which contradicts our previous findings in E. coli DC2, where it was selective for PBP4 alone and caused increased Bocillin-FL labeling of PBP2. Strikingly, our results show that all inhibitors except aztreonam were found to have IC50app values at concentrations <1 mM for PBP2. This is in stark contrast to other reports in the literature, in which predominately only carbapenems, third-generation cephalosporins, and select penicillins are capable of inhibiting PBP2 at concentrations <1 mM.12,30,52 This result is not completely unexpected, given that there are no literature reports of inhibitor IC50 values against the PBPs in E. coli MG1655, but it is worth noting as this is exceptionally important in the context of developing PBP-selective chemical probes for a given PBP within a species. Strain-to-strain variation in selectivity profiles could hinder the ability to broadly apply PBP-selective probes across multiple strains of a given species. For inhibitors that have been tested in other strains of E. coli, our results for PBP3 are congruent with the reported literature, indicating that the concern of strain-to-strain variability may not be an issue for all PBP isoforms, but researchers should be cognizant of this possibility.
Penicillin G was the only other inhibitor tested that showed distinct selectivity for PBPs other than PBP2 or PBP3. It was coselective for PBP1b, PBP7, and PBP8. The coselectivity of PBP7 was assigned based on inspection of the raw gel data from the SDS-PAGE analysis, as the calculated IC50app did not provide a fully bounded 95% CI preventing an accurate value from being reported. The coselectivity of PBP1b contrasts the results in E. coli DC2, in which PBP4 was instead coselective with PBP7 and PBP8. These variations provide further evidence that generalizations across related strains will not always provide an accurate picture.
A select set of inhibitors caused increased Bocillin-FL labeling for certain PBPs at the 10 mM inhibitor concentration in all three biological replicates. Aztreonam treatment resulted in increased Bocillin-FL labeling of PBP2, PBP4, PBP5/6, and PBP7. Increased Bocillin-FL labeling of PBP2 and PBP5/6 is consistent with our previous data generated in E. coli DC2, while increased labeling of PBP4 and PBP7 is not (Figure 7). Additionally, increased Bocillin-FL labeling was observed for PBP4 and PBP5/6 when cells were treated with 10 mM cefepime, and PBP5/6 labeling was increased when cells were treated with 10 mM cefaclor. These results were not due to a difference in the amount of proteome loading onto the SDS-PAGE gel as determined by analysis of the Coomassie stained gel (Figure S6). The exact cause of this is not clear, although it is plausible to suspect allosteric modulation of these PBPs by specific inhibitors, a phenomenon that has been demonstrated via protein X-ray crystallographic studies.57,58
Figure 7.
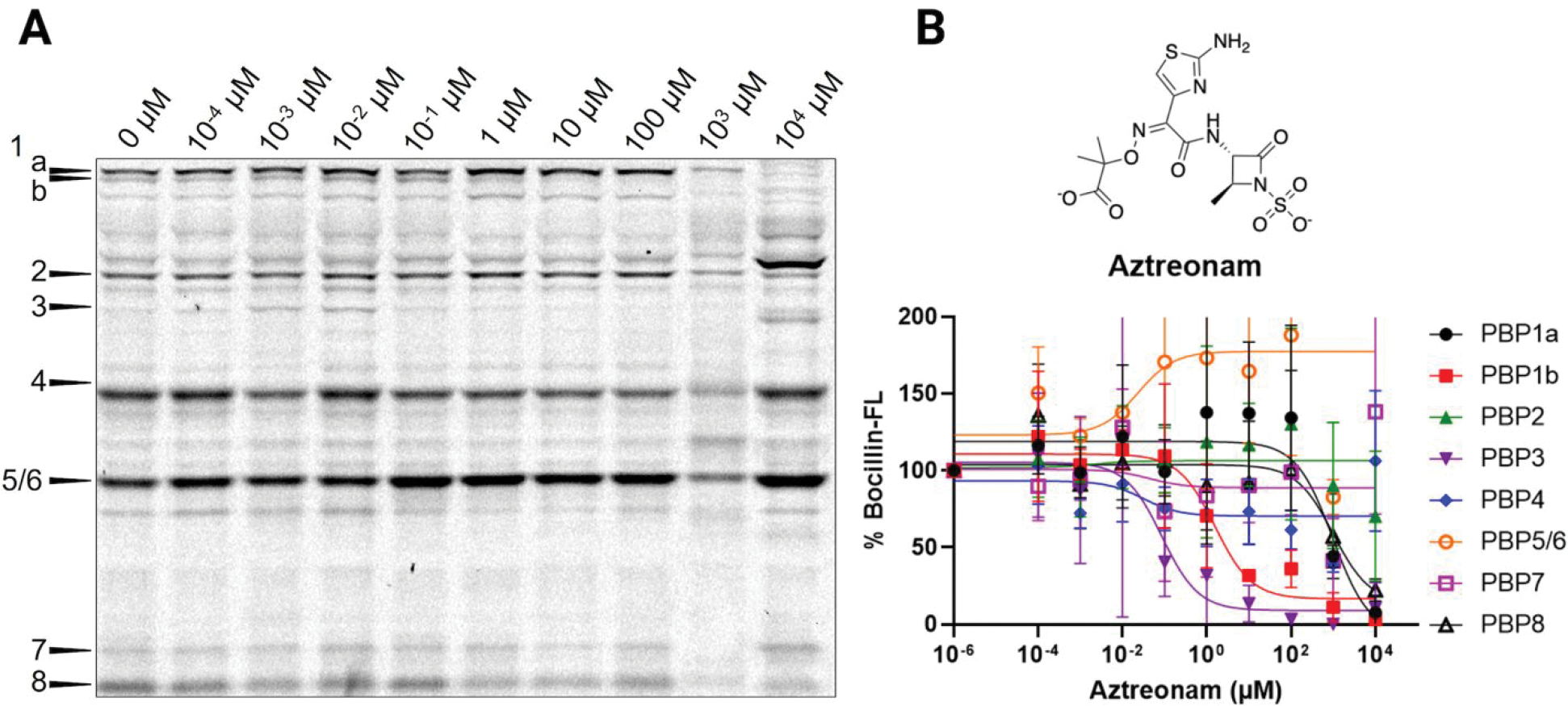
Aztreonam titration in E. coli MG1655. (A) Cells treated with increasing concentrations of aztreonam in Tris-EDTA, followed by labeling with Bocillin-FL. Selective inhibition of PBP3 was observed, along with increased Bocillin-FL labeling of PBP2, PBP4, PBP5/6, and PBP7 (10 mM). (B) Relative percent Bocillin-FL intensities plotted versus aztreonam concentration. Mean values from biological triplicates are plotted with error bars representing standard deviations.
As noted earlier, avibactam was found to be selective for PBP2, with an IC50app <100 μM, which is consistent with a previously reported value from an in vitro experiment.30 In this same study, the reported IC50 values for PBPs 1a, 1b, 3, 4, and 5/6 were >50 μM, as this was the highest concentration tested (PBP7 and PBP8 were not evaluated). We found that the IC50app values of avibactam against PBP1a and PBP4 were >10 mM and those of PBP1b and PBP5/6 were 300 and 480 μM, respectively. In some cases, we noted substantial errors associated with the IC50app determinations, likely due to the inherent variability of cell-based assays such as this. However, the obtained data still enabled us to determine the PBP selectivity for all inhibitors. Indeed, our results are highly informative given the lack of comprehensive PBP inhibition profiles of DBO molecules. A recent report by Durand-Reville et al.59 demonstrated a successful medicinal chemistry campaign to fine-tune the DBO class of molecules for increased PBP potency and enhanced in vivo efficacy against Gram-negative pathogens. Future work could employ these molecules as dual-functioning activity-based probes that target both β-lactamases and PBPs or to specifically target the PBPs in strains without β-lactamases (e.g., E. coli MG1655).
Most of our data confirm previous findings about the potency of various β-lactam antibiotics with the PBPs and the resulting isoform selectivity profiles. However, one result that strikingly contradicted other studies was that of meropenem, which has been reported as an inhibitor of PBP2, PBP4, and PBP6. While we also observed inhibition of PBP2 and PBP6, with the former having a much lower IC50app, we did not see significant PBP4 inhibition. We were unable to determine an IC50app for meropenem against PBP4 as 50% inhibition was not reached even at the highest concentration that we evaluated, 10 mM (Figure 8). In contrast, reported IC50 values range from 10−4 to 10−2 μM.12,51 This is an unexpected result, as MIC values for meropenem do not change this drastically between various strains of susceptible E. coli. While PBP2 is essential for cell growth, PBP4 is nonessential, but deletion of PBP4 results in decreased cross-linking of the peptidoglycan and causes an increase in sensitivity to bile salts.60 It was also recently found that PBP4 localization to the midcell during cell division is important for coordinating the assembly of the cell divisome.61 These results demonstrate that there is likely a synergistic effect of meropenem treatment by inhibition of PBP2 and PBP4, but inhibition of PBP2 is likely the major contributor to cell death, based on consistent MIC values between strains. To determine if the lack of PBP4 inhibition was potentially due to differences in the activity of PBP4 in native environments, we titrated meropenem against the PBPs in E. coli MG1655 lysate. This in vitro experiment also showed no PBP4 inhibition by meropenem, indicating that this difference is not due to protein environment or active site accessibility (Figure S3).
Figure 8.
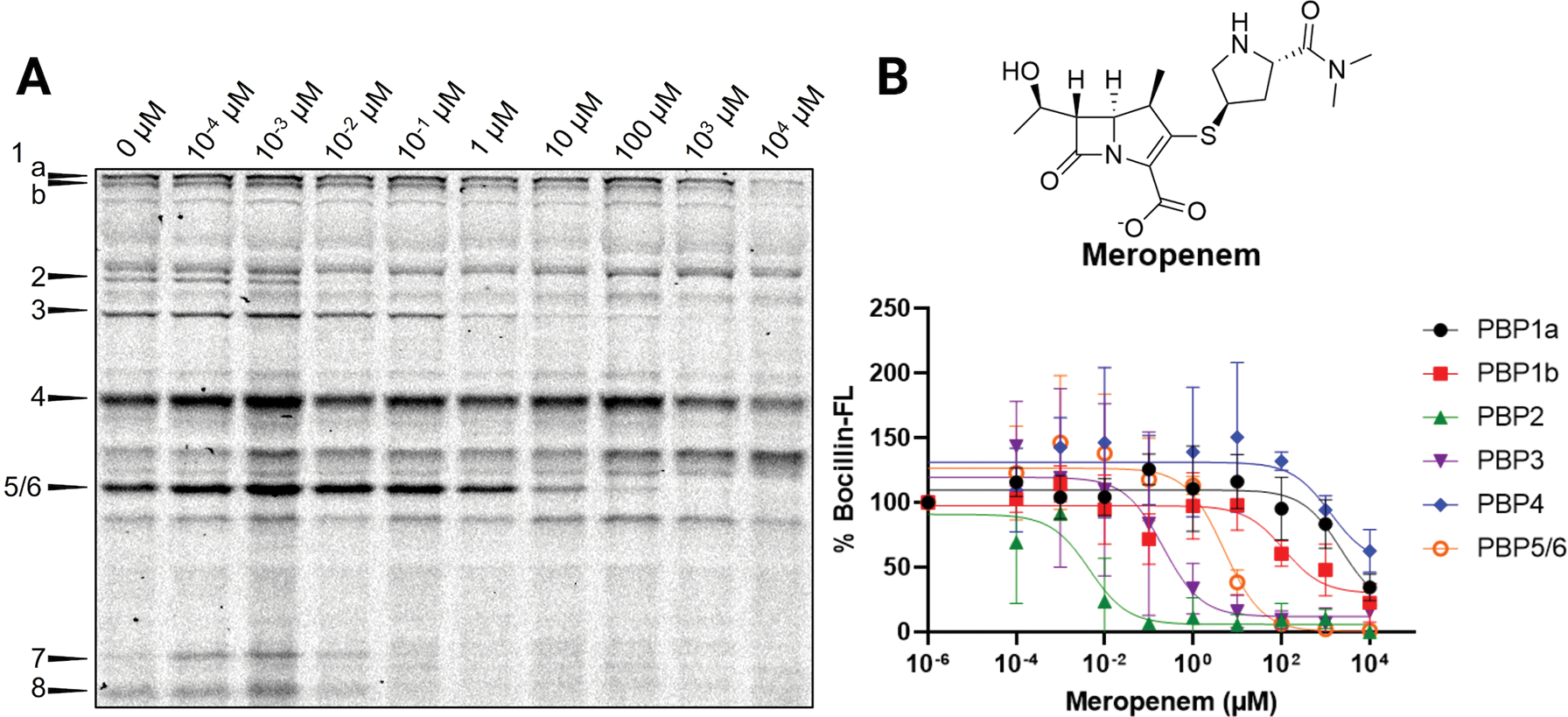
Meropenem inhibition profile in E. coli MG1655. (A) Cells treated with increasing concentrations of meropenem in Tris-EDTA, followed by Bocillin-FL labeling. PBP4 was minimally inhibited even at 10 mM meropenem. (B) Relative percent Bocillin-FL intensities plotted versus meropenem concentration. Mean values from biological triplicates are plotted with error bars representing standard deviations.
PBP4 was also poorly inhibited by aztreonam and cefaclor. Aztreonam has been reported to have an IC50 of 68 μM,12 and cefaclor has not been reported; however, cefaclor is a second-generation cephalosporin, and other inhibitors within the class have IC50 values of less than 100 μM.12 Aside from these three molecules and cefepime, which inhibited PBP4 but at 10 mM caused increased Bocillin-FL labeling and prevented the calculation of an IC50app, our data for the inhibition profiles of PBP4 match well with literature reported values.12,30,50–52 These results imply that there is a difference in the ability of select molecules to inhibit PBP4 in live E. coli MG1655 that has not been observed in other strains. Future work will be required to investigate if there is physiological significance to this difference.
DISCUSSION
In this study, we report an optimized gel-based assay for the evaluation of 12 inhibitors in live, non-hypersusceptible E. coli MG1655. Historically, such assays have been carried out in vitro, using either pure proteins or cell lysates.29,62–64 Results from these studies do not always translate to whole cells given the potential loss of native protein–protein interactions, as well as possible protein unfolding (i.e., the selectivity profile of a given inhibitor differs between live cells and lysates). Indeed, it has been demonstrated that Bocillin-FL labeling profiles in lysates are often missing some of the PBPs. Our work addresses this gap as we have devised a biochemical screening assay that is performed on live cells in the presence of a permeabilizing agent.
Outer membrane permeabilizing agents have been studied for decades, and two of the most frequently employed reagents are Tris-EDTA and PMBN. Though these two reagents work through different mechanisms, we found that both methods permeabilized the outer membrane sufficiently to yield increased Bocillin-FL labeling of the entire PBP complement in E. coli MG1655. Although both treatments resulted in varying degrees of inner membrane permeabilization, neither treatment caused as much damage as colistin nor did they kill a substantial number of cells, as demonstrated by growth curve analysis. Collectively, we have demonstrated that short treatments with either Tris-EDTA or PMBN are sufficient to permeabilize the outer membrane of E. coli MG1655 without the loss of cellular viability. We found that Tris-EDTA provided the most consistent improvements to PBP labeling.
Next, we used our Tris-EDTA conditions in titration experiments to investigate the PBP-inhibition profiles of 12 molecules using Bocillin-FL as our readout probe to determine IC50app and assign PBP-selectivity. For inhibitors with known IC50 values from in vivo or in vitro experiments, we compared our findings, resulting in both considerable similarities and differences. Selective inhibition of PBP3 by aztreonam, piperacillin, cefuroxime, ceftriaxone, and cefepime was consistent with the literature,12,50–52 and our reported IC50app values were largely in agreement with earlier reports. Our determination that avibactam was selective for PBP2 was also consistent with previously published data.30 Moreover, our titration results with avibactam yield a more comprehensive picture for inhibition against an entire PBP profile in vivo, compared to a previous report conducted in vitro using cell lysates.30
In stark contrast to the literature, we found that 11 out of 12 tested molecules inhibited PBP2 at concentrations of less than 500 μM. Aztreonam was the only inhibitor that was unable to prevent Bocillin-FL labeling, which contradicts literature reports12,50–52 wherein penicillins, third-generation cephalosporins, and carbapenems have predominately been found to inhibit PBP2. Conversely, we did not detect any appreciable inhibition of PBP4 by meropenem, while earlier reports indicated that both PBP2 and PBP4 are inhibited with similar IC50app values,12,51 possibly pointing to differences in the architecture, regulation, or function of this protein in strain MG1655.
The methodology described herein has a wide variety of potential applications, including the identification of scaffolds for the future development of PBP-selective activity-based probes (ABPs). ABPs are powerful chemical tools that will enable the further investigation of PBP activity in whole cells, including microscopy and proteomics applications. Indeed, previous work by us and others to identify PBP-selective inhibitors has yielded β-lactam-based ABPs.21,22,65,66 There is currently only one report in the literature describing the use of a similar assay in a non-hypersusceptible organism, the PA01 strain of Pseudomonas aeruginosa, that investigated the Bocillin-FL labeling profiles with dual-combination treatments.67 We have also observed Bocillin-FL labeling of the entire PBP complement in P. aeruginosa (Figure S4), and it is unclear why these results were obtained without the need of cell permeabilization. However, this is the first example of studies performed in non-hypersusceptible Gram-negative species that specifically investigated the PBP-selectivity profiles of a diverse group of molecules, facilitating a deeper understanding of the PBPs in future work for some of the most clinically important pathogens.
One of the major challenges in advancing to these complex experiments will be the further investigation of whether the devised permeabilization strategy, treatment with Tris-EDTA, will be needed with these ABPs and how it ultimately may affect cellular processes. Additionally, considerations for resistance mechanisms (porin mutations, efflux pumps, β-lactamases) present in strains under study must be made when applying these methods. This will be critically important for live-cell studies, interrogating PBP regulations and protein–protein interactions to ensure that the results will be translatable to clinically relevant organisms. The work presented here should not be perceived as a new method to quantitatively assess the potencies of time-dependent, covalent PBP inhibitors in Gram-negative species. It is important to note that we have determined apparent IC50 values that are relevant within the context of this assay, and the use of IC50app as a means of quantitative potency is inappropriate;68,69 instead kinact/KI values should be used as the gold-standard for time-dependent, irreversible PBP inhibitors. However, the use of IC50app values to establish PBP-isoform selectivity is appropriate and is highly useful in identifying scaffolds that would be ideal candidates for developing activity-based probes and for selective PBP inhibition in live cells.
Due to the clinical significance of Gram-negative bacteria and their impact in the global health threat of antibiotic resistance, it is imperative that we not only identify new drug targets but also deepen our understanding of current drug targets to enable more effective drug design. This assay will open new doors for understanding the roles of individual PBPs and the potential development of novel antibiotics that target the essential PBPs in various Gram-negative organisms. This includes the development of antibiotics that inhibit multiple PBPs and those that can disrupt or inhibit other proteins involved in the PBP-associated protein machinery (i.e., the divisome and elongasome). The screening strategy devised herein represents an improvement over current methods for the development of PBP isoform-selective inhibitors, which often rely on MIC determinations or in vitro experiments that cannot provide details about a specific PBP activity in a whole-cell context. The development of novel antibiotics is desperately needed as the antibiotic crisis continues to increase, and the work presented here provides a new means to address this threat in Gram-negative bacteria.
METHODS
Additional information regarding protocols and materials can be found in the Supporting Information.
Bacterial Strains and Culture Conditions.
E. coli MG1655 was purchased from ATCC (cat. 700926). Overnight cultures were grown by inoculating 6 mL of Lennox Luria–Bertani (Sigma-Aldrich, L3022) broth with a sterile, 200 μL tip-full of frozen E. coli MG1655 glycerol stock. Six serial dilutions were made by using 0.5 mL of the previous culture tube to inoculate 6 mL of LB broth. Cultures were grown for 14–16 h at 37 °C and shaken at 220 rpm. Fresh cultures were prepared by inoculating 9 mL of fresh LB broth in a culture tube with 1 mL of culture from the last tube in the serial dilution series. These cultures were grown at 37 °C with shaking at 220 rpm until an OD600 of ~0.4–0.5 was reached (~2 h).
Comparison of Bocillin-FL Labeling in Different Buffers.
Cultures of E. coli MG1655 were prepared as described above. Cells (1 mL) at OD600 = 0.4–0.5 were collected in respective Eppendorf tubes by centrifugation at 18 000g for 2 min at room temperature (RT). The supernatant was removed, and the cells were washed with 0.5 mL of 1× PBS pH 7.4 and harvested by centrifugation at 18 000g for 2 min at RT. After centrifugation, the supernatant was removed, and the cells were resuspended in 50 μL of the following respective solutions: (1) 1× PBS–7.5 μM Bocillin-FL, (2) 1× PBS–25 μM Bocillin-FL, (3) 50 mM Tris pH 7.8–25 μM Bocillin-FL, and (4) 50 mM Tris–200 mM EDTA–25 μM Bocillin-FL. Samples were incubated in the dark at RT for 30 min. After incubation, cells were harvested by centrifugation at 18 000g for 2 min at RT, washed with 0.5 mL of PBS, and centrifuged at 18 000g for 2 min at RT. The pellets were resuspended in 100 μL of 200 mM Tris pH 8–20 mM EDTA containing 10 mg/mL lysozyme. Samples were incubated statically at 37 °C for 20 min, followed by lysis using a Hielscher vial tweeter UP200 St (90% C, 95% A, 5% adjustment snap, 50 s on, 10 s off on ice for 10 min total run time). The membrane fraction was collected by centrifugation (21 000g for 15 min, 4 °C), and the supernatant was removed. The membrane pellets were resuspended in 50 mM Tris pH 7.4–0.5% SDS, vortexed briefly, and incubated at 4 °C for 10 min. It is noted that the incubation at 4 °C resulted in far less viscous solutions compared to RT or heated incubations. Protein concentrations were measured using a NanoPhotometer P330 instrument. Aliquots (30 μL) of each sample were taken into individual Eppi tubes, and 10 μL of 4× SDS Laemmli buffer were added to each sample. Proteins were denatured by boiling samples at 95 °C for 5 min. Following denaturation, 250 μg of each sample was loaded into respective wells of a 10% SDS-PAGE gel, and the protein bands were separated and analyzed as described in the Supporting Information. Protein labeling was found to be stable over several days if samples were frozen, as necessary.
Inhibitor Titration Experiments in E. coli MG1655.
Cultures were grown as described above. Cells (1 mL) at OD600 = 0.4–0.5 were collected in respective Eppendorf tubes by centrifugation at 18 000g for 2 min at RT. The supernatant was removed, and the cells were washed with 0.5 mL of 1× PBS pH 7.4 and harvested by centrifugation at 18 000g for 2 min at RT. The supernatant was removed, and the cells were resuspended in 50 μL of the following: Tris pH 7.8–200 μM EDTA only (negative control; 0 μM), and 50 mM Tris pH 7.8–200 μM EDTA with increasing concentrations of inhibitor from 10−4 to 104 μM. Samples were incubated at RT for 30 min. Cells were harvested by centrifugation at 18 000g for 2 min at RT, followed by washing with 0.5 mL of 1× PBS and centrifugation at 18 000g for 2 min at RT. Following the wash step, cells were resuspended in 50 μL of 50 mM Tris pH 7.8 containing 25 μM Bocillin-FL (final concentration), and samples were incubated in the dark at RT for 30 min. After incubation, cells were harvested by centrifugation at 18 000g for 2 min at RT, washed with 0.5 mL of PBS, and harvested by centrifugation at 18 000g for 2 min at RT. Cells were lysed, membrane fractions collected, protein measurements taken, and samples ran on SDS-PAGE gels as described above. Protein labeling was found to be stable over several days if samples were frozen, as necessary.
NPN Outer Membrane Permeabilization.
E. coli MG1655 was cultured as described above. Fresh cultures were grown to an OD600 of 0.5, and 2 mL of cells were harvested for each replicate. Cell pellets were washed with 1 mL of PBS and centrifuged at 18 000g for 2 min at RT. The supernatant was removed, and the cells were resuspended in 1 mL of 50 mM Tris pH 7.8. To respective wells in a clear, flat-bottom 96-well plate (Corning, Costar 3596) was added 100 μL of the resuspended cells. For the buffer-only control wells, 100–150 μL of Tris buffer was added. 50 μL of 200 μM EDTA, PMBN, or colistin in Tris containing NPN at a final concentration of 10 μM was added to respective wells. An untreated control was included in which 50 μL of Tris containing 10 μM NPN was added to the well. A Tecan Spark plate reader was used to measure fluorescence using a monochromator (ex, 355 ± 20 nm; em, 405 ± 20 nm) with the gain set to optimal and fluorescence reading from the top. A measurement was taken every minute over the course of 1 h. The measurements taken at 30 min were used, and the ratio of NPN uptake was calculated using the following equation:
where fluorescencetreated was the relative fluorescence measured for Tris-, Tris-EDTA-, or Tris-PMBN-treated samples; fluorescenceNPN was the fluorescence of NPN in Tris; and fluorescencecolistin was the fluorescence measured in the Tris-colistin treated wells. The experiment was carried out in biological triplicate, and the relative NPN uptake ratios are reported as averages with standard deviations represented as error bars.
Propidium Iodide Inner Membrane Permeabilization.
E. coli MG1655 was cultured as described above. Fresh cultures were grown to an OD600 of 0.5, and 2 mL of cells were harvested for each replicate. Cell pellets were washed with 1 mL of PBS and centrifuged at 18 000g for 2 min at RT. The supernatant was removed, and the cells were resuspended in 1 mL of 50 mM Tris pH 7.8. To respective wells in a clear, flat-bottom 96-well plate (Corning, Costar 3596) was added 100 μL of the resuspended cells. For the buffer-only control wells, 100–150 μL of Tris buffer was added. 50 μL of 200 μM EDTA, PMBN, or colistin in Tris containing propidium iodide at a final concentration of 5 μM was added to respective wells. An untreated control was included in which 50 μL of Tris containing 5 μM NPN was added to the well. A Tecan Spark plate reader was used to measure fluorescence using a monochromator (ex, 535 ± 20 nm; em, 617 ± 20 nm) with the gain set to optimal and fluorescence reading from the top. A measurement was taken every minute over the course of 1 h. The measurements taken at 30 min were used, and the ratio of NPN uptake was calculated using the following equation:
where fluorescencetreated was the relative fluorescence measured for Tris-, Tris-EDTA-, or Tris-PMBN-treated samples; fluorescencePI was the fluorescence of PI in Tris; and Fluorescencecolistin was the fluorescence measured in the Tris-colistin treated wells. The experiment was carried out in biological triplicate, and the relative PI uptake ratios are reported as averages with standard deviations represented as error bars.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R01 GM128439 A1, E.E.C.) and the University of Minnesota, Department of Chemistry. J.D.S. was supported by the National Institutes of Health’s National Center for Advancing Translational Sciences, grants TL1R002493 and UL1TR002494. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translation Sciences. Graphical figures created with GraphPad Prism and BioRender.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.2c00004.
Additional experimental details, materials, and methods including representative SDS-PAGE gels for inhibitor titrations with corresponding IC50 plots; representative Coomassie stained gel demonstrating how protein bands were measured and used to correct the fluorescence intensities measured with Bocillin-FL; meropenem titration in cell lysates; SDS-PAGE analysis demonstrating labeling of PBPs in P. aeruginosa PA14 and PA01 without permeabilizing reagents; phase-contrast microscopy of E. coli MG1655 with and without permeabilizing reagent treatment; and Coomassie stain of the aztreonam titration gel shown in Figure 7 (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.2c00004
Contributor Information
Joshua D. Shirley, Department of Medicinal Chemistry, University of Minnesota, Minneapolis, Minnesota 55454, United States
Kelsie M. Nauta, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States
Erin E. Carlson, Department of Medicinal Chemistry, University of Minnesota, Minneapolis, Minnesota 55454, United States; Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States; Department of Biochemistry, Molecular Biology, and Biophysics and Department of Pharmacology, University of Minnesota, Minneapolis, Minnesota 55454, United States
REFERENCES
- (1).Silhavy TJ; Kahne D; Walker S The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010, 2 (5), a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kuhn A The Bacterial Cell Wall and Membrane-A Treasure Chest for Antibiotic Targets. Subcell Biochem 2019, 92, 1–5. [DOI] [PubMed] [Google Scholar]
- (3).Rajagopal M; Walker S Envelope Structures of Gram-Positive Bacteria. Curr. Top Microbiol Immunol 2015, 404, 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dorr T; Moynihan PJ; Mayer C Editorial: Bacterial Cell Wall Structure and Dynamics. Front Microbiol 2019, 10, 2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Eichenberger EM; Thaden JT Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics (Basel) 2019, 8 (2), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bush K Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62 (10), e01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bush K; Bradford PA Interplay between β-lactamases and new β-lactamase inhibitors. Nat. Rev. Microbiol 2019, 17 (5), 295–306. [DOI] [PubMed] [Google Scholar]
- (8).Bush K Overcoming β-lactam resistance in Gram-negative pathogens. Future Med. Chem. 2016, 8 (9), 921–4. [DOI] [PubMed] [Google Scholar]
- (9).Sauvage E; Kerff F; Terrak M; Ayala JA; Charlier P The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008, 32 (2), 234–58. [DOI] [PubMed] [Google Scholar]
- (10).Kocaoglu O; Carlson EE Penicillin-binding protein imaging probes. Curr. Protoc Chem. Biol. 2013, 5 (4), 239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Fisher JF; Mobashery S Constructing and deconstructing the bacterial cell wall. Protein Sci. 2020, 29 (3), 629–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kocaoglu O; Carlson EE Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob. Agents Chemother. 2015, 59 (5), 2785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kocaoglu O; Tsui HC; Winkler ME; Carlson EE Profiling of β-lactam selectivity for penicillin-binding proteins in Streptococcus pneumoniae D39. Antimicrob. Agents Chemother. 2015, 59 (6), 3548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zapun A; Contreras-Martel C; Vernet T Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol Rev. 2008, 32 (2), 361–85. [DOI] [PubMed] [Google Scholar]
- (15).Vollmer W; Massidda O; Tomasz A The Cell Wall of Streptococcus pneumoniae. Microbiol Spectr 2019, 7 (3), 7.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Miyachiro MM; Contreras-Martel C; Dessen A Penicillin-Binding Proteins (PBPs) and Bacterial Cell Wall Elongation Complexes. Subcell Biochem 2019, 93, 273–289. [DOI] [PubMed] [Google Scholar]
- (17).Sharifzadeh S; Boersma MJ; Kocaoglu O; Shokri A; Brown CL; Shirley JD; Winkler ME; Carlson EE Novel Electrophilic Scaffold for Imaging of Essential Penicillin-Binding Proteins in Streptococcus pneumoniae. ACS Chem. Biol. 2017, 12 (11), 2849–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Perez AJ; Boersma MJ; Bruce KE; Lamanna MM; Shaw SL; Tsui HT; Taguchi A; Carlson EE; VanNieuwenhze MS; Winkler ME Organization of peptidoglycan synthesis in nodes and separate rings at different stages of cell division of Streptococcus pneumoniae. Mol. Microbiol. 2021, 115 (6), 1152–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sassine J; Xu M; Sidiq KR; Emmins R; Errington J; Daniel RA Functional redundancy of division specific penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 2017, 106 (2), 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sharifzadeh S; Shirley JD; Carlson EE Activity-Based Protein Profiling Methods to Study Bacteria: The Power of Small-Molecule Electrophiles. Curr. Top Microbiol Immunol 2018, 420, 23–48. [DOI] [PubMed] [Google Scholar]
- (21).Kocaoglu O; Calvo RA; Sham LT; Cozy LM; Lanning BR; Francis S; Winkler ME; Kearns DB; Carlson EE Selective Penicillin-Binding Protein Imaging Probes Reveal Substructure in Bacterial Cell Division. ACS Chem. Biol. 2012, 7 (10), 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sharifzadeh S; Dempwolff F; Kearns DB; Carlson EE Harnessing β-Lactam Antibiotics for Illumination of the Activity of Penicillin-Binding Proteins in Bacillus subtilis. ACS Chem. Biol. 2020, 15 (5), 1242–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Masson JM; Labia R Synthesis of a 125I-radiolabeled penicillin for penicillin-binding proteins studies. Anal. Biochem. 1983, 128 (1), 164–8. [DOI] [PubMed] [Google Scholar]
- (24).Preston DA; Wu CY; Blaszczak LC; Seitz DE; Halligan NG Biological characterization of a new radioactive labeling reagent for bacterial penicillin-binding proteins. Antimicrob. Agents Chemother. 1990, 34 (5), 718–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Spratt BG Properties of the penicillin-binding proteins of Escherichia coli K12. Eur. J. Biochem. 1977, 72 (2), 341–52. [DOI] [PubMed] [Google Scholar]
- (26).Spratt BG; Pardee AB Penicillin-binding proteins and cell shape in E. coli. Nature 1975, 254 (5500), 516–7. [DOI] [PubMed] [Google Scholar]
- (27).Spratt BG Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U. S. A. 1975, 72 (8), 2999–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhao G; Meier TI; Kahl SD; Gee KR; Blaszczak LC BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 1999, 43 (5), 1124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sutaria DS; Moya B; Green KB; Kim TH; Tao X; Jiao Y; Louie A; Drusano GL; Bulitta JB First Penicillin-Binding Protein Occupancy Patterns of β-Lactams and β-Lactamase Inhibitors in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2018, 62 (6), e00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Asli A; Brouillette E; Krause KM; Nichols WW; Malouin F Distinctive Binding of Avibactam to Penicillin-Binding Proteins of Gram-Negative and Gram-Positive Bacteria. Antimicrob. Agents Chemother. 2016, 60 (2), 752–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lacasse E; Brouillette E; Larose A; Parr TR Jr.; Rubio A; Malouin F In Vitro Activity of Tebipenem (SPR859) against Penicillin-Binding Proteins of Gram-Negative and Gram-Positive Bacteria. Antimicrob. Agents Chemother. 2019, 63 (4), e02181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Dougherty TJ; Kennedy K; Kessler RE; Pucci MJ Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J. Bacteriol. 1996, 178 (21), 6110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Clark D Novel antibiotic hypersensitive mutants of Esherichia coli genetic mapping and chemical characterization. FEMS Microbiology Letters 1984, 21 (2), 189–195. [Google Scholar]
- (34).Lang Y; Shah NR; Tao X; Reeve SM; Zhou J; Moya B; Sayed ARM; Dharuman S; Oyer JL; Copik AJ; Fleischer BA; Shin E; Werkman C; Basso KB; Deveson Lucas D; Sutaria DS; Megroz M; Kim TH; Loudon-Hossler V; Wright A; Jimenez-Nieves RH; Wallace MJ; Cadet KC; Jiao Y; Boyce JD; LoVullo ED; Schweizer HP; Bonomo RA; Bharatham N; Tsuji BT; Landersdorfer CB; Norris MH; Soo Shin B; Louie A; Balasubramanian V; Lee RE; Drusano GL; Bulitta JB Combating Multidrug-Resistant Bacteria by Integrating a Novel Target Site Penetration and Receptor Binding Assay Platform Into Translational Modeling. Clin Pharmacol Ther 2021, 109 (4), 1000–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhao S; Adamiak JW; Bonifay V; Mehla J; Zgurskaya HI; Tan DS Defining new chemical space for drug penetration into Gram-negative bacteria. Nat. Chem. Biol. 2020, 16 (12), 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zgurskaya HI; Lopez CA; Gnanakaran S Permeability Barrier of Gram-Negative Cell Envelopes and Approaches To Bypass It. ACS Infect Dis 2015, 1 (11), 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).James CE; Mahendran KR; Molitor A; Bolla JM; Bessonov AN; Winterhalter M; Pages JM How β-lactam antibiotics enter bacteria: a dialogue with the porins. PLoS One 2009, 4 (5), e5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Vaara M The outer membrane permeability-increasing action of linear analogues of polymyxin B nonapeptide. Drugs Exp Clin Res. 1991, 17 (9), 437–443. [PubMed] [Google Scholar]
- (39).Kimura Y; Matsunaga H; Vaara M Polymyxin B octapeptide and polymyxin B heptapeptide are potent outer membrane permeability-increasing agents. J. Antibiot (Tokyo) 1992, 45 (5), 742–9. [DOI] [PubMed] [Google Scholar]
- (40).Vaara M; Viljanen P Binding of polymyxin B nonapeptide to gram-negative bacteria. Antimicrob. Agents Chemother. 1985, 27 (4), 548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Vaara M Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992, 56 (3), 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Curtis NA; Brown C; Boxall M; Boulton MG Inhibition of Escherichia coli K-12 by beta-lactam antibiotics with poor antibacterial activity: interaction of permeability and intrinsic activity against penicillin-binding proteins. Antimicrob. Agents Chemother. 1979, 15 (3), 332–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Stone MRL; Butler MS; Phetsang W; Cooper MA; Blaskovich MAT Fluorescent Antibiotics: New Research Tools to Fight Antibiotic Resistance. Trends Biotechnol 2018, 36 (5), 523–536. [DOI] [PubMed] [Google Scholar]
- (44).Kocaoglu O; Carlson EE Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol. 2016, 12 (7), 472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Hira J; Uddin MJ; Haugland MM; Lentz CS From Differential Stains to Next Generation Physiology: Chemical Probes to Visualize Bacterial Cell Structure and Physiology. Molecules 2020, 25 (21), 4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Allam A; Maigre L; Vergalli J; Dumont E; Cinquin B; Alves de Sousa R; Pajovic J; Pinet E; Smith N; Herbeuval JP; Refregiers M; Artaud I; Pages JM Microspectrofluorimetry to dissect the permeation of ceftazidime in Gram-negative bacteria. Sci. Rep 2017, 7 (1), 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Alakomi HL; Paananen A; Suihko ML; Helander IM; Saarela M Weakening effect of cell permeabilizers on gram-negative bacteria causing biodeterioration. Appl. Environ. Microbiol. 2006, 72 (7), 4695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Clifton LA; Skoda MW; Le Brun AP; Ciesielski F; Kuzmenko I; Holt SA; Lakey JH Effect of divalent cation removal on the structure of gram-negative bacterial outer membrane models. Langmuir 2015, 31 (1), 404–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Amro NA; Kotra LP; Wadu-Mesthridge K; Bulychev A; Mobashery S; Liu G.-y. High-Resolution Atomic Force Microscopy Studies of the Escherichia coli Outer Membrane: Structural Basis for Permeability. Langmuir 2000, 16, 2789–2796. [Google Scholar]
- (50).Pucci MJ; Boice-Sowek J; Kessler RE; Dougherty TJ Comparison of cefepime, cefpirome, and cefaclidine binding affinities for penicillin-binding proteins in Escherichia coli K-12 and Pseudomonas aeruginosa SC8329. Antimicrob. Agents Chemother. 1991, 35 (11), 2312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Davies TA; Shang W; Bush K; Flamm RK Affinity of doripenem and comparators to penicillin-binding proteins in Escherichia coli and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52 (4), 1510–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Davies TA; Page MG; Shang W; Andrew T; Kania M; Bush K Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2007, 51 (7), 2621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Peters K; Kannan S; Rao VA; Biboy J; Vollmer D; Erickson SW; Lewis RJ; Young KD; Vollmer W The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia coli. mBio 2016, 7 (3), e00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Chowdhury C; Kar D; Dutta M; Kumar A; Ghosh AS Moderate deacylation efficiency of DacD explains its ability to partially restore β-lactam resistance in Escherichia coli PBP5 mutant. FEMS Microbiol Lett. 2012, 337 (1), 73–80. [DOI] [PubMed] [Google Scholar]
- (55).Sabnis A; Hagart KL; Klockner A; Becce M; Evans LE; Furniss RCD; Mavridou DA; Murphy R; Stevens MM; Davies JC; Larrouy-Maumus GJ; Clarke TB; Edwards AM Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 2021, 10, e65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Rosenberg M; Azevedo NF; Ivask A Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep 2019, 9 (1), 6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Bernardo-Garcia N; Mahasenan KV; Batuecas MT; Lee M; Hesek D; Petrackova D; Doubravova L; Branny P; Mobashery S; Hermoso JA Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae. ACS Chem. Biol. 2018, 13 (3), 694–702. [DOI] [PubMed] [Google Scholar]
- (58).Otero LH; Rojas-Altuve A; Llarrull LI; Carrasco-Lopez C; Kumarasiri M; Lastochkin E; Fishovitz J; Dawley M; Hesek D; Lee M; Johnson JW; Fisher JF; Chang M; Mobashery S; Hermoso JA How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (42), 16808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Durand-Reville TF; Miller AA; O’Donnell JP; Wu X; Sylvester MA; Guler S; Iyer R; Shapiro AB; Carter NM; Velez-Vega C; Moussa SH; McLeod SM; Chen A; Tanudra AM; Zhang J; Comita-Prevoir J; Romero JA; Huynh H; Ferguson AD; Horanyi PS; Mayclin SJ; Heine HS; Drusano GL; Cummings JE; Slayden RA; Tommasi RA Rational design of a new antibiotic class for drug-resistant infections. Nature 2021, 597 (7878), 698–702. [DOI] [PubMed] [Google Scholar]
- (60).Iwaya M; Strominger JL Simultaneous deletion of D-alanine carboxypeptidase IB-C and penicillin-binding component IV in a mutant of Escherichia coli K12. Proc. Natl. Acad. Sci. U. S. A. 1977, 74 (7), 2980–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Verheul J; Lodge A; Yau HCL; Liu X; Liu X; Solovyova AS; Typas A; Banzhaf M; Vollmer W; Blaauwen T. d. Midcell localization of PBP4 of Escherichia coli modulates the timing of divisome assembly. bioRxiv, 2021. DOI: 10.1101/2020.07.30.230052. [DOI] [Google Scholar]
- (62).Curtis NA; Orr D; Ross GW; Boulton MG Competition of β-lactam antibiotics for the penicillin-binding proteins of Pseudomonas aeruginosa, Enterobacter cloacae, Klebsiella aerogenes, Proteus rettgeri, and Escherichia coli: comparison with antibacterial activity and effects upon bacterial morphology. Antimicrob. Agents Chemother. 1979, 16 (3), 325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Shapiro AB; Comita-Prevoir J; Sylvester M 5-Carboxytetramethylrhodamine-Ampicillin Fluorescence Anisotropy-Based Assay of Escherichia coli Penicillin-Binding Protein 2 Transpeptidase Inhibition. ACS Infect Dis 2019, 5 (6), 863–872. [DOI] [PubMed] [Google Scholar]
- (64).Shapiro AB; Gu RF; Gao N; Livchak S; Thresher J Continuous fluorescence anisotropy-based assay of BOCILLIN FL penicillin reaction with penicillin binding protein 3. Anal. Biochem. 2013, 439 (1), 37–43. [DOI] [PubMed] [Google Scholar]
- (65).Staub I; Sieber SA β-lactam probes as selective chemical-proteomic tools for the identification and functional characterization of resistance associated enzymes in MRSA. J. Am. Chem. Soc. 2009, 131 (17), 6271–6. [DOI] [PubMed] [Google Scholar]
- (66).Levine SR; Beatty KE Investigating β-Lactam Drug Targets in Mycobacterium tuberculosis Using Chemical Probes. ACS Infect Dis 2021, 7 (2), 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Siriyong T; Murray RM; Bidgood LE; Young SA; Wright F; Parcell BJ; Voravuthikunchai SP; Coote PJ Dual β-lactam combination therapy for multi-drug resistant Pseudomonas aeruginosa infection: enhanced efficacy in vivo and comparison with monotherapies of penicillin-binding protein inhibition. Sci. Rep 2019, 9 (1), 9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Miyahisa I; Sameshima T; Hixon MS Rapid Determination of the Specificity Constant of Irreversible Inhibitors (kinact/KI) by Means of an Endpoint Competition Assay. Angew. Chem., Int. Ed. Engl. 2015, 54 (47), 14099–102. [DOI] [PubMed] [Google Scholar]
- (69).Strelow JM A Perspective on the Kinetics of Covalent and Irreversible Inhibition. SLAS Discov 2017, 22 (1), 3–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


