Abstract
The SARS-CoV-2 Africa dashboard is an interactive tool that enables visualization of SARS-CoV-2 genomic information in African countries. The customizable app allows users to visualize the number of sequences deposited in each country, and the variants circulating over time. Our dashboard enables near real-time exploration of public data that can inform policymakers, healthcare professionals and the public about the ongoing pandemic.
Coronavirus disease 2019 (COVID-19) is the clinical manifestation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection1. The COVID-19 pandemic has been ongoing for more than two years, with the first case of COVID-19 in Africa reported in Egypt in mid-February 20202. The first SARS-CoV-2 genome sequenced in Africa was reported in March 20203. Genomic sequencing and surveillance have had a crucial role in monitoring and mitigating the COVID-19 pandemic. There have been approximately 12 million cases and more than 256,000 deaths reported to date in Africa4, and African countries have contributed substantial amounts of genomic sequencing data to global agencies. For example, two of the five variants of concern (Beta and Omicron) were first identified in Africa through genomic surveillance systems and real-time sequencing and data release 5,6.
During the first year of the pandemic, SARS-CoV-2 genomes from Africa were mainly produced for a small number of countries with genomes available from 38 of the 54 African countries7. Subsequently, the Africa Centers for Disease Control (Africa CDC), and the World Health Organization Regional Office for Africa (WHO/AFRO) invested in capacity building and provided resources to equip more African countries to produce genomes locally8. For example, the African Union Commission and Africa CDC launched the Africa Pathogen Genomics Initiative (Africa PGI) with an initial investment of US$100 million. Currently, >100,000 genomes, originating from 51 African countries and four independent overseas territories, are publicly available at Global Initiative on Sharing Avian Influenza Data (GISAID)9.
Dashboards for live COVID-19 information
Online dashboards presenting global and regional COVID-19 data, including case numbers, reported deaths and vaccination rates, have proliferated since the onset of the pandemic 10–12. These dashboards have a vital role in guiding the public health response and decision-making by policymakers, public health officials and scientists13. Data visualization in dashboards also keeps the public abreast of the state of the pandemic. Examples of genomic dashboards include the Danish Covid-19 Genome Consortium dashboard (https://www.covid19genomics.dk/statistics), the Welcome Sanger Institute’s Covid-19 Genomic Surveillance dashboard (https://covid19.sanger.ac.uk/lineages/raw) and the COVID-19 Genomics UK Consortium (https://sars2.cvr.gla.ac.uk/cog-uk/). These dashboards include the number of genomes sequenced, the proportion of variants identified in the sequenced genomes as well as information on the mutations in the lineages of interest. Although these dashboards display important genomic information about Denmark and England, there was initially no genomics dashboard for the African continent, so we set out to devise one in order to provide real-time analytical tools to visualize a genomics-oriented understanding of the state of the pandemic on the African continent.
Data inputs for the SARS-CoV-2 Africa dashboard
The SARS-CoV-2 Africa dashboard is an open-source, web-based graphical user interface to present the data produced by genomic surveillance of COVID-19 on the continent and provide details of variants that are currently circulating. The dashboard is supported by the main web browsers commercially available, including Chrome, Firefox, Edge and Safari. The dashboard collates all sequencing data available in GISAID with metadata linking it to a country in Africa and uses these metadata to display temporal and spatial trends in SARS-CoV-2 evolution in Africa. Genomics data are incorporated into the dashboard by API (Application Programming Interface) via an agreement with GISAID. The web application processes it, and includes a data quality assessment that can eliminate poor quality registers, for example, sequences assigned to a variant that was submitted before the variant was identified (Figure 1A).
Figure 1. Incorporation of SARS-CoV-2 data into the SARS-CoV-2 Africa dashboard.
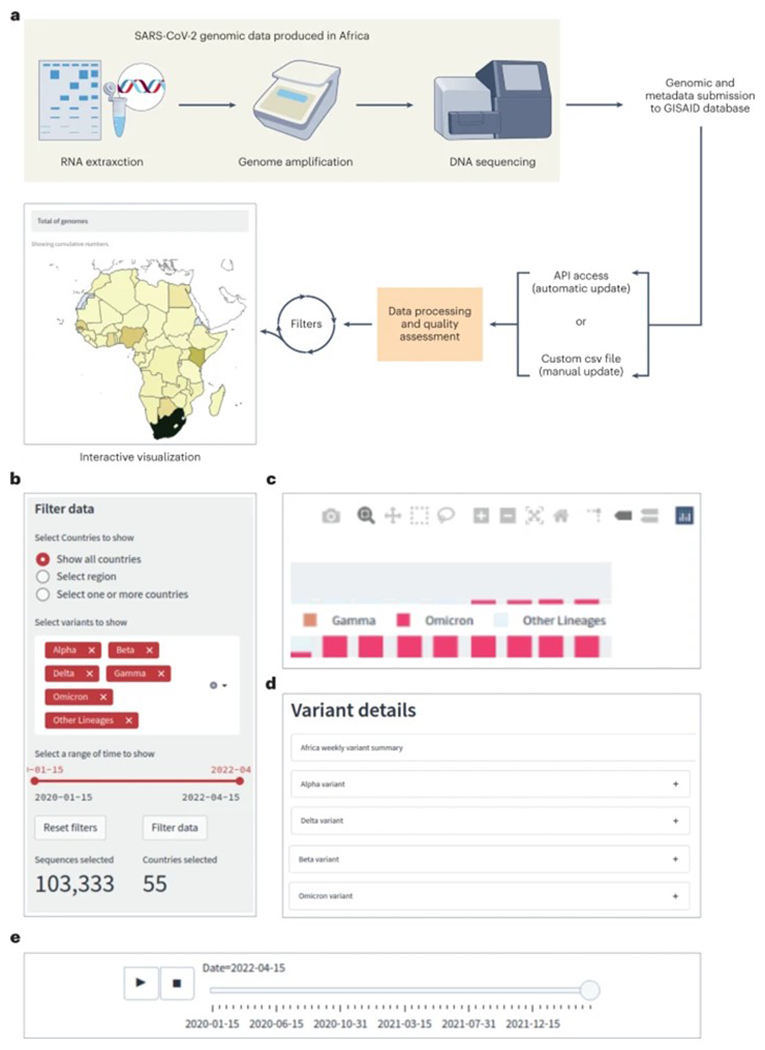
(A). The main features of the interface are: (B) General filters that allow users to select the data of interest, (C) Figure controls allow users to enable and disable legend elements, labels, select a part of the figure, zoom in, and out, and download the plot, (D) A tabulated description and mutation map is provided for variants that are of interest/importance at the time of access, and (E) Timeline player that displays the mapped progression of the pandemic over time, based on the filter selection.
Data processing in the SARS-CoV-2 Africa dashboard
SARS-CoV-2 genomes are accessioned on GISAID with contextual metadata (such as patient details, collection and sampling strategies, and sequencing and assembly methods) that is subjected to curation by GISAID prior to release. GISAID data can be freely accessed and downloaded by users after registration. The data acquisition and processing pipelines use Python 3.6 and the web interface is implemented using Streamlit (https://streamlit.io/), with charts created using Plotly14. The code can be locally installed for customization in a Conda environment15. Code and dependencies can be installed by cloning the Github repository available at https://github.com/CERI-KRISP/SARS-Cov-2-Africa-dashboard.
Performance of the SARS-CoV-2 Africa dashboard
In order to evaluate dashboard performance, we implemented an experiment using ApacheBench (https://httpd.apache.org/docs/2.4/programs/ab.html) version 2.3, varying for different levels of concurrency (10, 100, 500, and 1000 simultaneous access). For each level of concurrence, we performed 5,000 requisitions which showed that the dashboard performed well for simultaneous access. For example, in the last level (1000 requests), only 0.08% of the total requisitions were not answered. All requests for 0 to 1000 were completed within an average 1.862 to 25.841 milliseconds (Supplementary table 1). The dashboard provides interactive visualizations of the temporal and spatial distributions of SARS-CoV-2 variants and their prevalence across different African regions and countries. Several filters are provided to customize the visualizations according to user needs. The main features of the interface are four modules (Figure 1, panels B-E): General filters allow users to select the data of interest, figure controls allow users to customize the display and snapshot a desired plot, a tabulated weekly summary of variant details is provided, and drop-down mutation maps for variants that are of interest can be used. We also included a ‘timeline player’ that displays the progression of the pandemic over time, based on user-defined filter selections.
Case study for Omicron spread
A hypothetical example of the application of this dashboard is shown in Figure 2. The scenario is that the Minister of Health in Namibia wants to understand the spread of the Omicron lineage in neighboring countries after reports of Omicron in South Africa and Botswana, to better understand how Namibia may be affected. They would use filters to display the number of Omicron lineage genomes in each neighboring country. In Figure 2, on the left-hand side panel of the dashboard, there are filters that allow for specific countries, specific regions, or for all countries’ data to be shown. In this example, Namibia and its neighboring countries (South Africa, Botswana, and Angola) have been selected. On the interactive map titled “Genomes per country”, the metric “Genomes by variant” has been selected and in this case, the Omicron variant was selected. As seen in Figure 2, if the cursor is hovering over a country on the map, the name of the country, the number of genomes produced by that country, and the date is displayed.
Figure 2.
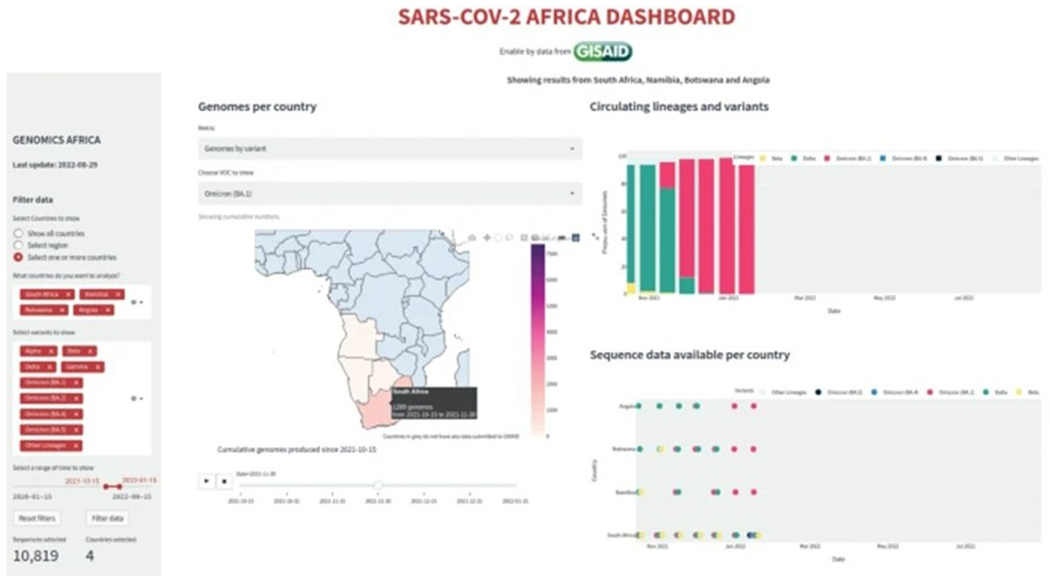
The case study scenario screenshot shows how to investigate Omicron spread in Namibia and neighbouring countries using the filters and charts provided by the SARS-CoV-2 Africa dashboard interface. Logo courtesy of the GISAID Initiative.
When studying the figures on the dashboard with these filters applied, one can see that the proportion of genomes deposited in GISAID at the end of October, 2021 is predominantly Delta lineage, with few remnant Beta genomes. Within the first two weeks of November, 2021 the Omicron BA.1 lineage rapidly increased to comprise 19% of all genomes. Watching the sliding scale animation for Omicron lineages on the map displays the early detection of the lineage in South Africa, with swift progression from a low (light pink) to high (dark purple) number of cumulative genomes. From these visualizations, the Namibian Minister of Health would be aware of the rapid spread of Omicron and its growth advantage over Delta, and would be able to see that, at that time, Omicron had the potential to be the dominant variant in southern Africa. The Minister would be empowered with the needed information to enable consultation with local researchers, public health officials and clinicians in order to provide local and regional public health responses needed to mitigate the effects of Omicron on the population.
Outlook
Numerous dashboards for global and regional COVID-19 data, such as case numbers, reported deaths, and vaccination rates, have proliferated since the onset of the COVID-19 pandemic. These dashboards have been vital in guiding the public health response and decision-making by policymakers, public health officials, and scientists13. Data visualizations produced by these dashboards have also been useful for keeping the public informed. Genomic surveillance of SARS-CoV-2 has been crucial in monitoring the progression of the pandemic, particularly in the low vaccination landscape of Africa, where globally important variants have emerged and are likely to continue to appear.
Africa has generated a wealth of genomic surveillance data, with more than 129,000 SARS-CoV-2 genomes currently available on GISAID. The SARS-CoV-2 Africa dashboard is the first detailed online, real-time, interactive tool produced for the Global South. It provides simple and clear graphics that are easy to interpret and equips developers to analyze and visualize the data themselves, by allowing manual input of data via custom csv files, formatted as per the provided template (Supplementary Information). Our dashboard makes often intimidating and complex genomic data accessible to all users and can be used to inform policy, and guide public health response in Africa and for Africa.
Supplementary Material
ACKNOWLEDGEMENTS
We thank GISAID for providing real-time access to metadata available on the database. SARS-CoV-2 sequencing at CERI is supported, in part, by grants from the South African Medical Research Council (SAMRC), World Health Organization, the Rockefeller Foundation, the Abbott Pandemic Defense Coalition (APDC), the National Institute of Health USA (U01 AI151698) for the United World Antivirus Research Network (UWARN) and the INFORM Africa project through IHVN (U54 TW012041), the South African Department of Science and Innovation (DSI) and the SAMRC under the BRICS JAF #2020/049. JSX was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (Capes) - Finance Code 001. DEVP received funding from an Oracle for Research Grant. All datasets used in our dashboard are in publicly accessible repositories. Genomic data are available from the GISAID database (https://www.gisaid.org/). The SARS-CoV-2 Africa dashboard is freely available at https://climade.health/dashboard/covid-africa/. Source code is available at https://github.com/CERI-KRISP/SARS-Cov-2-Africa-dashboard.
Footnotes
ETHICS & INCLUSION STATEMENT
Our research included local African researchers from the Centre for Epidemic Response and Innovation (CERI) and KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP). Local researchers participated in all steps from study design, to implementation and authorship (CB, EW, HT, JES, MM, NS, TdO and WAK). Participating authors from CERI and KRISP helped to guide the development of the dashboard so that it is locally relevant and useful in Africa. Roles and responsibilities were agreed among collaborators before the start of the project. Guidance on our dashboard has been disseminated to local African researchers via an instructional webinar, as part of the Africa COVID-19 Genomics Training Webinar Series, hosted by the African Union and the African CDC. Our research is not restricted to researchers Only, and does not pose any health, safety, security or other risks.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPLEMENTARY INFORMATION
Supplementary Methods are available at https://doi.org/10.25413/sun.19722025.
REFERENCES
- 1.Li Q et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med 382, 1199–1207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medhat MA & El Kassas M COVID-19 in Egypt: Uncovered figures or a different situation? J. Glob. Health 10, 010368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oluniyi P First African SARS-CoV-2 genome sequence from Nigerian COVID-19 case - Genome Reports - Virological. Virological https://virological.org/t/first-african-sars-cov-2-genome-sequence-from-nigerian-covid-19-case/421 (2020). [Google Scholar]
- 4.Ritchie H et al. Coronavirus Pandemic (COVID-19). Our World in Data https://ourworldindata.org/coronavirus (2020). [Google Scholar]
- 5.Viana R et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603, 679–686 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegally H et al. Rapid replacement of the Beta variant by the Delta variant in South Africa. medRxiv (2021) doi: 10.1101/2021.09.23.21264018. [DOI] [Google Scholar]
- 7.Wilkinson E et al. A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa. Science 374, 423–431 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tegally H et al. The evolving SARS-CoV-2 epidemic in Africa: Insights from rapidly expanding genomic surveillance. medRxiv (2022) doi: 10.1101/2022.04.17.22273906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khare S et al. Gisaid’s role in pandemic response. China CDC Wkly 3, 1049–1051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright DW et al. Tracking SARS-CoV-2 mutations and variants through the COG-UK-Mutation Explorer. Virus Evol. 8, veac023 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broad Covid Dashboard. https://covid-19-sequencing.broadinstitute.org/.
- 12.CDC COVID Data Tracker. CDC COVID Data Tracker https://covid.cdc.gov/covid-data-tracker.
- 13.Peeples L Lessons from the COVID data wizards. Nature 603, 564–567 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Plotly Technologies Inc. Collaborative data science. (Plotly Technologies Inc., 2015). [Google Scholar]
- 15.Anaconda Software Distribution. Conda. (Anaconda Software Distribution, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


